Abstract
Three deubiquitinating enzymes–Rpn11, Usp14, and Uch37–are associated with the proteasome regulatory particle. These enzymes allow proteasomes to remove ubiquitin from substrates before they are translocated into the core particle to be degraded. Although the translocation channel is too narrow for folded proteins, the force of translocation unfolds them mechanically. As translocation proceeds, ubiquitin chains bound to substrate are drawn to the channel’s entry port, where they can impede further translocation. Rpn11, situated over the port, can remove these chains without compromising degradation because substrates must be irreversibly committed to degradation before Rpn11 acts. This coupling between deubiquitination and substrate degradation is ensured by the Ins-1 loop of Rpn11, which controls ubiquitin access to its catalytic site. In contrast to Rpn11, Usp14 and Uch37 can rescue substrates from degradation by promoting substrate dissociation from the proteasome prior to the commitment step. Uch37 is unique in being a component of both the proteasome and a second multisubunit assembly, the INO80 complex. However, only recruitment into the proteasome activates Uch37. Recruitment to the proteasome likewise activates Usp14. However, the influence of Usp14 on the proteasome depends on the substrate, due to its marked preference for proteins that carry multiple ubiquitin chains. Usp14 exerts complex control over the proteasome, suppressing proteasome activity even when inactive in deubiquitination. A major challenge for the field will be to elucidate the specificities of Rpn11, Usp14, and Uch37 in greater depth, employing not only model in vitro substrates, but their endogenous targets.
Graphical Abstract
The Ubiquitin-Proteasome System
The proteasome controls the levels of myriad regulatory proteins and maintains cellular homeostasis by removing misfolded and potentially toxic proteins (reviewed in this issue by Hochstrasser; see also [1–3]). Proteins are marked for proteasomal degradation with ubiquitin, a highly conserved, 76-residue protein. Ubiquitination of substrate proteins is achieved via the sequential action of E1 (ubiquitin-activating), E2 (ubiquitin-conjugating) and E3 (ubiquitin-ligating) enzymes, resulting in the formation of a covalent isopeptide bond between the C-terminus of ubiquitin and a nucleophilic group on the substrate, typically on the ε-amino group of a lysine residue. Such isopeptide bonds can be cleaved by deubiquitinating enzymes (DUBs) to regenerate free ubiquitin and, in the case of many DUBs, to antagonize substrate degradation. In mammals, there are nearly 100 DUBs, pointing to the intricacy and importance of these reactions [4,5]. Ubiquitin itself contains seven lysines (K6, K11, K27, K29, K33, K48 and K63) and a free N-terminal amino group, all of which can be modified by another ubiquitin molecule, allowing the formation of diverse chain structures [6–8]. Therefore, DUBs select substrates from a remarkably complex pool of conjugated ubiquitin, which is found not only on hundreds of different target proteins, but within ubiquitin chains of varied length and topology.
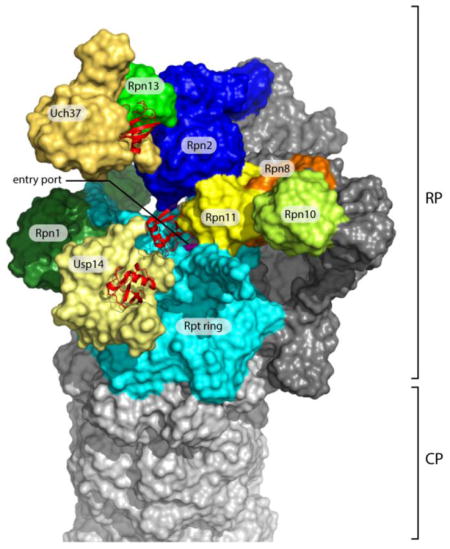
The 26S proteasome (Fig. 1) consists of a cylindrical 20S core particle (CP), capped at one or both ends by 19S regulatory particles (RPs). The CP consists of four stacked heteroheptameric ring complexes that form an internal chamber containing the proteolytic sites of the proteasome. The RP can be divided into the 9-subunit base, which binds the cylinder end of the CP, and the 9-subunit lid [9]. The RP tightly regulates entry into the CP in order to prevent adventitious degradation of non-targeted proteins [10,11].
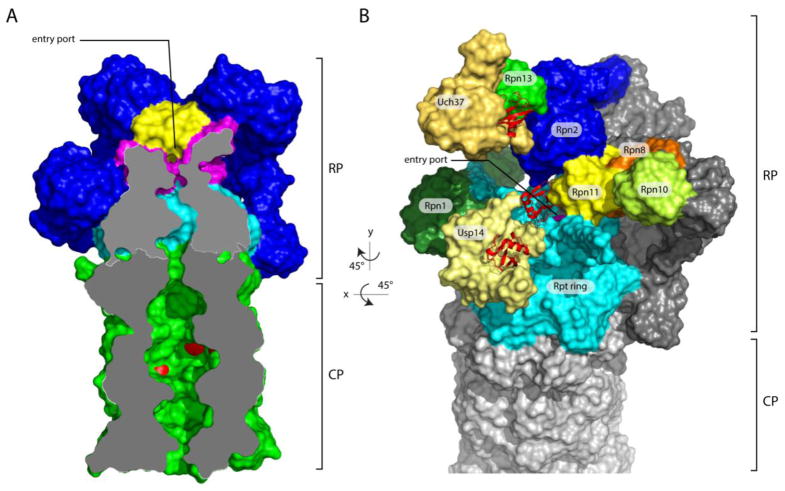
Figure 1. Overview of the axial channel and the position of the three proteasomal-associated DUBs on the human proteasome.
(A) The axial channels of the RP and CP of the human proteasome in a substrate-translocating, s3-like state. This representation was generated using structural data from PDB: 5T0J [26]. The grey surface represents a medial cut-away plane, which is only applied to the CP and the Rpt ring. The substrate entry port is formed by the OB domains, also known as N-domains. Rpn11 is painted yellow, the OB domains of the Rpt ring are in magenta, the ATPase domains are in cyan, and the CP is in green. The proteolytic active sites β1, β2 and β5’ are highlighted in red. To allow visualization of the internal space of the proteasome, certain subunits were removed. (B) Representation of the structure of the human s1 proteasome as determined by Huang et al. (PDB: 5GJQ) [54]. The three DUBs are shown in shades of yellow, whereas the three Ub/Ubl receptors are colored in various shades of green. Ubiquitin bound to the DUBs is shown in red ribbon representations. Rpn8, part of the MPN heterodimer with Rpn11, is shown in orange. Rpn13, Uch37, and ubiquitin were modeled onto the RP based on the position of Rpn13 on the yeast proteasome [53] (PDB: 5A5B) and the co-crystal structure of Uch37-Rpn13 (PDB: 4UEL, PDB: 4WLQ) [60,61]. The substrate entry port was built digitally by filling the hole with H2O molecules using HOLLOW [157].
Given that the proteasome typically cannot recognize its substrates unless they are modified by ubiquitin, it is remarkable that the proteasome has three distinct components–Rpn11, Usp14, and Uch37–that function to cleave ubiquitin from substrates. Indeed, deubiquitination events at the proteasome have the potential to derail substrate degradation by promoting the dissociation of substrate from the proteasome before degradation is initiated. These considerations imply that deubiquitination at the proteasome should be held under strict control to minimize futile cycles of ubiquitination and deubiquitination. In fact, deubiquitination events at the proteasome are closely coordinated with distinct steps in the pathway of substrate degradation. In this review, we first outline how the proteasome encounters, translocates, unfolds, and degrades a substrate. We will then discuss the proteasomal DUBs, focusing on their mechanisms of action, functions, and substrate specificity. For more general reviews of DUBs, see [5,12].
Substrate recognition and translocation
Ubiquitinated proteins associate with the proteasome through a set of integral RP subunits that serve as substrate receptors: Rpn10, Rpn13, and Rpn1 [1,13–17] (Fig. 1). Each of these receptors is capable of recognizing ubiquitin directly as well as indirectly; in ubiquitin recognition, Rpn10 is the strongest. Indirect recognition is mediated by a family of proteins often referred to as shuttling ubiquitin receptors. These shuttle proteins have an N-terminal ubiquitin-like (UBL) domain [18], which targets them to Rpn10, Rpn13, or Rpn1 [16,17,19–21], and a C-terminal ubiquitin-associated (UBA) domain, which binds ubiquitin conjugates [22]. Thus, before proteins are deubiquitinated at the proteasome, a prior step of ubiquitin binding may occur where many distinct interactions between ubiquitin and the proteasome-associated receptors are possible. Given the multiplicity of ubiquitin receptors, the frequent multiplicity of ubiquitin chains on a single substrate, and the varied architectures of these chains, it can be expected that different substrates will be processed by proteasomal DUBs in distinct ways. We will return to this point below.
Substrate binding will lead to degradation only if the substrate contains a constitutively or transiently unfolded segment–an initiation region–that can thread itself into the proteasome’s substrate entry port, which is formed by a heterohexameric ATPase ring (also known as the Rpt ring) within the RP [2,23]. The port opens to a narrow channel at the center of the ATPase ring (Fig. 1A), where loops from the ATPases can contact the substrate and exert a pulling force towards the CP. The initiation of substrate translocation by the ATPases is known as substrate engagement. Using the energy of ATP hydrolysis, the ATPases can progressively translocate the entire substrate into the CP, even unfolding globular domains to allow passage through the narrow RP channel. The CP contains an axial channel as well, which is aligned with that of the RP upon substrate engagement (Fig. 1A). Once these two channels are traversed, the substrate reaches the central cavity of the CP, which houses multiple proteolytic sites. A fundamental property of the proteasome is that it degrades most proteins processively, and this feature reflects the ability of the proteasome to complete translocation of an engaged substrate.
To date, cryo-electron microscopy (cryo-EM) studies have resolved four distinct conformational states of the proteasome, termed s1-s4 [24–29]. Idle proteasomes exist predominantly in the s1 state, which is thought to represent the substrate-accepting conformation. Substrate engagement induces a broad reconfiguration of the proteasome, particularly of the RP. In these high-energy states (s2–s4), many of the functionalities of the proteasome are activated, including deubiquitinating activities. Proteasomes can be driven into the s3 state by artificially blocking the translocation of an engaged substrate or, in the absence of added substrate, using the poorly hydrolyzed ATPγS in place of ATP. The s3 state is the most heavily populated of the substrate-engaged states, and the functional significance of the minor states s2 and s4 is not yet well understood. Therefore, the terms s3 state proteasomes and substrate-engaged proteasomes will be used essentially interchangeably below.
Although ubiquitin targets proteins to the proteasome, it can also hinder subsequent processing of the substrate. Ubiquitin has evolved to be exceptionally stable physically [30], so that it is refractory, though not absolutely, to unfolding and translocation by the ATPases of the RP. It seems likely that ubiquitin’s resistance to unfolding evolved as a strategy to limit turnover of substrate-bound ubiquitin [31–34], and a resulting depletion of ubiquitin, particularly under conditions of high substrate load or poor nutrient supply. However, this evolutionary strategy could only be viable if a mechanism for timely substrate deubiquitination evolved in tandem–otherwise the recalcitrance of ubiquitin to unfolding and translocation would also prevent the substrate from being fully translocated and hence processively degraded.
Although the scenario described above points to the facilitation of substrate degradation by deubiquitination, DUB activities can also compete with degradation. A model by which to understand this distinction is the commitment step in degradation. Although the commitment step has not been precisely defined experimentally, it is presumed to occur when the ATPases establish a firm grip on the initiation region. This model implies that deubiquitination reactions taking place after commitment cannot abort degradation, but only promote it by preventing ubiquitin from blocking the channel. In contrast, when deubiquitination at the proteasome precedes commitment, it can reduce the probability of commitment by facilitating premature substrate dissociation [34,35]. Whether deubiquitination at the proteasome functions to promote or inhibit degradation depends on the particular DUB, as discussed below.
Proteasome-associated DUBs
There are three DUBs intrinsic to or associated with mammalian proteasomes: proteasome subunit Rpn11/Poh1/PSMD14, a member of the JAB1/MPN/Mov34 (JAMM) family of DUBs; Usp14 (and its S. cerevisiae ortholog Ubp6), of the ubiquitin specific protease (USP/UBP) family; and Uch37/UCHL5, of the ubiquitin C-terminal hydrolase (UCH) family. Because Rpn11, Usp14, and Uch37 belong to different families of enzymes, their association with the proteasome must have arisen independently in evolution. To what extent these enzymes function redundantly is debated and in our view still unresolved, but overall these enzymes show dramatic functional differences and may have limited overlap in specificity.
The functions of Usp14 and Uch37 in the proteasome have been investigated for decades through the use of active site directed probes such as ubiquitin-aldehyde (UbAl) [36] and ubiquitin-vinylsulfone (UbVS) [37]. Such reagents form a covalent bond with the active-site cysteine of thiol proteases. In contrast to Usp14 and Uch37, Rpn11 is a metalloprotease. In the JAMM motif of Rpn11, EXnHXHX10D [38], the histidines and aspartate coordinate a Zn2+ ion, whereas the glutamate residue is expected to function as a proton acceptor to generate a hydroxide ion, which serves as the nucleophile [39]. Therefore probes such as UbVS do not label or inhibit Rpn11.
Rpn11
Rpn11 functions to promote protein degradation, presumably because Rpn11-mediated deubiquitination events take place characteristically after the commitment step. Rpn11 is positioned directly above the substrate entry port of the RP (Figs. 1A, 1B and 2) [24,26,27]. Therefore, as substrates are translocated through the RP channel, their attached ubiquitin chains are drawn to the entry port and to the active site of Rpn11. At this point, translocation may potentially pause, as the resistance of ubiquitin to unfolding hinders its passage through the entry port. Indeed, ubiquitin should in most cases not unfold at all, as chain removal appears kinetically favored over unfolding [34].
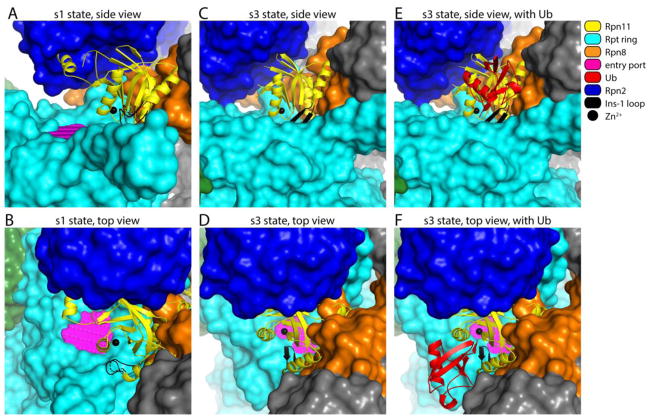
Figure 2. Overview of Rpn11 in its free and ubiquitin-bound forms, in the context of proteasomes in the s1 and s3 states.
Structural representation of yeast proteasome in s1 (A–B) or s3 (C–F) state, based on PDB: 4CR2 and PDB: 4CR4 [24]. The substrate entry port was built as described in Fig. 1B. (A–B) The alternative configuration of the Ins-1 loop (dashed line), as well as the position of the catalytic Zn2+ ion (black sphere), are based on PDB: 4O8X [64]. (C–F) The configuration of Rpn11 as well as the position of the bound distal ubiquitin in panels E and F were based on the crystal structure of ubiquitin-Rpn11-Rpn8 (PDB: 5U4P [34]). Color coding is given in the key.
A key feature of Rpn11, distinguishing it from Usp14 and Uch37, is the ATP-dependence of its deubiquitinating activity in the context of the proteasome [38,40]. Substrate translocation from the RP to the CP is also ATP-dependent, and this shared feature suggests a fundamental coupling of Rpn11 activity to translocation. The ATP-dependence of Rpn11 activity may reflect not only the energy requirement for translocation, but also that the conversion of the proteasome from its basal s1 state to the substrate-engaged s3 state requires ATP. Thus, when substrate translocation is initiated, Rpn11 is shifted towards the entry port by approximately 10 Å, which may increase the efficiency by which it cleaves chains (Fig. 2) [24,27].
Usp14
Unlike Rpn11, Usp14 and Ubp6 are not integral subunits or stoichiometric components of the proteasome [31]. Despite having a high affinity for the proteasome (in the low nanomolar range [41]), Usp14 is readily lost from proteasomes during conventional purification and was consequently long overlooked as a proteasome component. Interestingly, the fraction of proteasomes loaded with Usp14 is elevated when cells are treated with either proteasome inhibitors or Usp14 inhibitors [37,42].
Usp14 and Ubp6 contain an N-terminal ubiquitin-like (UBL) domain and a C-terminal catalytic domain. The only known role of the Ubp6UBL domain is to tether Ubp6 to the proteasome. The proteasomal receptor for Ubp6UBL is subunit Rpn1 (Fig. 1B), in particular the same toroidal repeat domain that also binds the UBL domains of shuttling ubiquitin receptors as well as ubiquitin itself [17,31]. However, the Ubp6UBL binding site in Rpn1 (T2) is distinct from the site that binds ubiquitin and the shuttling ubiquitin receptors (T1) [17]. The two sites are adjacent but there is little or no competition between Ubp6 and the shuttling ubiquitin receptors [17,43,44].
An interesting feature of Ubp6 is its “noncatalytic effect,” through which it inhibits protein degradation by the proteasome. This effect was discovered using an active site substitution mutant, C118A [45]. The addition of this mutant form of Ubp6 to proteasomes lacking Ubp6 strongly attenuates degradation of the model substrate cyclin B. The effect is not peculiar to cyclin B, as it was more recently observed by Bashore et al. using a titin-GFP model substrate [46]. Expression of the C118A mutant in yeast leads to the induction of proteasome synthesis [47]. If this adaptive proteasome stress response is impaired, the C118A mutation produces a severe growth defect. Noncatalytic effects have been reported for Usp14 as well [41,48,49], and were recently characterized in some detail using murine proteasomes [50].
Ubiquitin promotes the Ubp6/Usp14 noncatalytic effect [46,50–52]. This regulatory effect can also be observed with UbAl and UbVS, indicating that inhibition of the proteasome results from ubiquitin docking into the active site of Ubp6. Although the noncatalytic effect is nicely visualized using such chemically modified forms of ubiquitin, it is most likely ubiquitin conjugated to substrate that exerts these effects in a physiological setting. If so, the capacity of a ubiquitin-protein conjugate to induce the noncatalytic effect may depend on the arrangement of ubiquitin groups on the substrate.
Structural studies have provided important insights into the noncatalytic effect. In the complex between Ubp6 and the proteasome, the UBL domain can be visualized by cryo-EM at the T2 site of Rpn1, but the catalytic domain of Ubp6 is not observed, suggesting that it is unconstrained in the complex [46,53]. However, when Ubp6 charged with UbVS or UbAl is complexed with the proteasome, the catalytic domain is bound to the ATPase ring, most closely to Rpt1 and Rpt2 [46,53,54]. Thus, different proteasome subunits serve as receptors for the UBL and catalytic domains of Ubp6. Since Ubp6-UbVS and Ubp6-UbAl induce the noncatalytic effect, the structural data suggest that Ubp6 inhibits protein degradation by the proteasome when its ubiquitin-bound catalytic domain interacts with the ATPase ring.
Why does interaction between the Ubp6 catalytic domain and the ATPase ring inhibit proteasome activity? Usp14/Ubp6 can enhance the ATPase activity of the proteasome and partially open the CP gate [46,50–52]. However, as these are positive effects, there must also be negative effects underlying inhibition of substrate turnover. Interestingly, cryo-EM data suggest that the Ubp6-ubiquitin complex strongly prefers a substrate-engaged conformational state of the proteasome [46,53]. As a result of this preference, Ubp6 biases the fraction of proteasomes in a given state [53]. Perhaps the effectiveness of this inhibitory strategy does not depend on the state in which the proteasome is trapped but in the suppression of state transitions, which could be required to complete a round of degradation.
Uch37
Uch37 binds proteasomes through its C-terminal Uch37-like domain (ULD), which is recognized by proteasome subunit Rpn13 (Fig. 1B). The C-terminal DEUBiquitinase ADaptor (DEUBAD) [55] domain of Rpn13 (termed Rpn13DEU) mediates this interaction [56–61]. The DEUBAD domain of Rpn13 is present in S. pombe and in metazoan species, all of which express Uch37, but missing in S. cerevisiae. Accordingly, S. cerevisiae lack Uch37 as well.
Perhaps the most striking feature of Uch37 is that it is not only present on the proteasome, but also associates with the chromatin-remodeling complex INO80. The Uch37 receptor within INO80 is subunit NFRKB [62]. Interestingly, NFRKB expression is restricted to metazoans, implying that Uch37 cannot be recruited to the INO80 complex in Uch37-expressing protozoans such as S. pombe. Like Rpn13, NFRKB employs a DEUBAD domain (NFRKBDEU) to interact with the ULD domain of Uch37 [60,61]. Thus, Uch37 does not bridge these complexes but rather its recruitment into them is competitive. Interestingly, however, Rpn13 binding promotes the catalytic activity of Uch37, whereas NFRKB binding suppresses it [60–62]. The basis of this distinction has been elegantly elucidated through structural analysis of Rpn13DEU and NFRKBDEU, as discussed below. Although the catalytic activity of Uch37 is suppressed within the INO80 complex, it is possible that certain conditions or substrates could relieve or overcome this suppression. Considering that INO80 functions in transcription and DNA repair through chromatin remodeling [63], histones and transcription factors are among likely candidates for substrates of INO80-bound Uch37. The placement of Uch37 within the proteasome and INO80 points to a potential functional relationship between these complexes, but the nature of this connection is unclear.
Enzymatic activity and its regulation
All three proteasomal DUBs are highly regulated. They are active preferentially in the context of the proteasome, with a degree of activation as high as 1000-fold in the case of Usp14 [41]. However, even when assembled into the proteasome, Usp14 and Rpn11 are not constitutively in an active state. A common theme among these enzymes is the existence of loop segments that restrict the access of ubiquitin to the catalytic site. These loops are displaced to activate the enzyme. The structural details of this event are remarkably different from enzyme to enzyme.
Rpn11
Rpn11 must quickly deubiquitinate a translocating proteasome substrate to avoid slowing substrate degradation. However, its activity on translocating substrates must be held in check before the commitment step. The restriction of Rpn11 to committed substrates may be accounted for in part by substrate-bound ubiquitin chains being drawn to Rpn11’s active site by translocation. In addition, a second important mechanism enforces deubiquitination-degradation coupling [34]. This involves a conformational switch in a loop segment within the active site of Rpn11.
Rpn11 contains two loop regions that extend from its folded globular MPN domain, known as Ins-1 and Ins-2 [64,65]. The positioning of Ins-1 and Ins-2 is similar to that previously described for the non-proteasomal mammalian DUBs AMSH and AMSH-LP [66], which regulate endosomal trafficking, as well as the AMSH ortholog from S. pombe, Sst2 [67]. However, both loops show interesting differences between AMSH-LP and Rpn11. Ins-1 is of particular importance for Rpn11 as it undergoes conformational transitions between inactive and active states [34]. These states have been defined by crystallographic analysis of dimeric Rpn11-Rpn8 complexes. Rpn8 is an MPN domain protein, like Rpn11, but its MPN domain does not bind Zn2+ and is catalytically inactive. The Ins-1 loop of Rpn11 is found in the inactive state in Rpn11-Rpn8 complexes [64,65] and in the active state in Rpn11-Rpn8-ubiquitin ternary complexes [34]. Although the interaction between ubiquitin and Rpn11 is notably weak (Ki ~2.3 mM), it can nonetheless drive the conformational transition of Ins-1 [34].
In the closed, or low-activity state of Ins-1, this loop, though somewhat flexible, blocks access of the C-terminus of ubiquitin to the active site (Fig. 3A). In the active state, Ins-1 is not simply displaced but undergoes a conformational transition to a β-hairpin (Fig. 3B) [34]. This hairpin forms one side of the catalytic groove in the active conformation of Rpn11, thus assisting in the proper positioning of the C-terminal ubiquitin tail for catalysis. The active β-hairpin state of Ins-1 is comparable to the constitutive state of Ins-1 in AMSH-LP and Sst2. In all cases, a short segment within ubiquitin’s tail, as it reaches to the catalytic center of the enzyme, adopts a β-strand structure to form a β-sheet with Ins-1. The Ins-1 loop of Rpn11 is unique however in its ability to assume active and inactive states. This is evidently an adaptation that refines deubiquitination-degradation coupling [34].
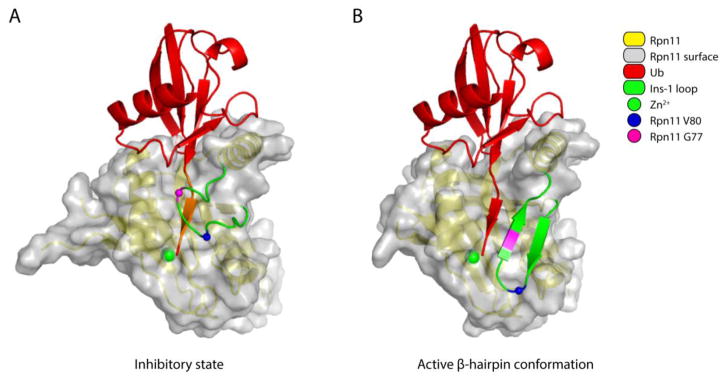
Figure 3. Conformational changes in the Ins-1 loop activate Rpn11.
(A) The inactive form of Rpn11 is based on the free Rpn11-Rpn8 model (PDB: 4O8X [64]). Ubiquitin was positioned by superimposing the ubiquitin-bound Rpn11 (PDB: 5U4P [34]) onto the free Rpn11 model. The residues in the C-terminal tail of ubiquitin that are within 1.4 Å of the Ins-1 loop are painted orange to indicate steric hindrance. (B) The active conformation of Rpn11 is based on the structure of Ub-Rpn11-Rpn8 (PDB: 5U4P [34]). In both A and B, the surface of the Ins-1 loop was removed to allow better visualization of the beta-sheet formed between the C-terminal tail of ubiquitin and this loop. Other color coding is given in the key.
The model of Ins-1 regulation described above has been supported through biochemical analysis of substitution mutations within this element [34]. In the hairpin state, residue V80 of Ins-1 inserts into a hydrophobic pocket within the body of Rpn11, and thus plays a critical role in stabilizing this state. Using a synthetic fluorescent substrate, Ub-GC-TAMRA, a V80A mutant was found to reduce the kcat of a Rpn11-Rpn8 dimer by over 100-fold [34], indicating that β-hairpin formation activates Rpn11. Residue G77 is found in the β-turn of the Ins-1 hairpin, in a position that can readily tolerate a proline residue and which is indeed occupied by proline in AMSH-LP and Sst2. However, a G77P substitution destabilizes the inactive state of Ins-1. While not inducing constitutive hairpin formation, G77P apparently increases the propensity of Rpn11 for this transition, with kcat for Ub-GC- TAMRA elevated substantially for proteasomal Rpn11 (5.4-fold) [34].
How do these mutations affect the processing of ubiquitin-protein conjugates? The rate of degradation of a model conjugate by the V80A mutant is reduced by ~4-fold, with some degree of deubiquitination being evident [34]. Thus, the effect with proteasomes is similar in nature but weaker than that seen with the synthetic substrate and the Rpn11-Rpn8 complex. Perhaps against expectation, G77P also slows substrate degradation, but in this case the rate effect is apparently the result of premature deubiquitination, which diverts conjugates from the pathway of substrate degradation. The premature deubiquitination seen with G77P is of special interest as it clearly implicates Ins-1 in the repression of pre-commitment deubiquitination. A different anomaly of deubiquitination is seen with the V80A mutant. When assayed for degradation in the presence of inhibitors of the proteolytic activity of the proteasome, V80A does not produce homogeneously deubiquitinated (Ub0) substrate, as does wild-type, but a mixture of Ub0 and Ub1 forms [34]. While the Ub0 form simply reflects the canonical en bloc cleavage mode of Rpn11, the Ub1 form reflects cleavage between the first and second ubiquitin groups in the chain. It may be assumed that this “endo” cleavage requires unfolding of the first (proximal) ubiquitin (see below). Thus, when deubiquitination is sufficiently slowed, a direct competition between deubiquitination and proximal ubiquitin unfolding becomes evident. However, ubiquitin unfolding is slow and should occur in only a minor fraction of degradation events with wild-type proteasomes.
Rpn11-mediated deubiquitination is clearly faster for translocating ubiquitin-protein conjugates than for Ub-GC-TAMRA. An interesting interpretation of this difference, proposed by Worden et al. [34], is that the mechanical force exerted on the substrate by ATPases of the proteasome is used to accelerate the transition of Ins-1 from the closed to the hairpin state. This model postulates an initial interaction of the C-terminal tail of ubiquitin with Ins-1 in its closed form. It will be interesting to test this model for the control of Ins-1 conformational states.
Rpn11 activity is also regulated in the course of proteasome assembly. In the lid, Rpn11 activity is inhibited by interactions of the Rpn8/Rpn11 heterodimer with Rpn5 and Rpn9 [68]. Alpha-helix 13 of Rpn5 locks the Ins-1 loop of Rpn11 into the ‘closed’ state while interacting with other loops surrounding the Rpn11 active site to obstruct the catalytic groove. Furthermore, Asn275 of Rpn5 mediates stable tetrahedral coordination of the Rpn11 catalytic Zn2+ ion, most likely via a bridging water molecule, thus inhibiting Rpn11 isopeptidase activity [68]. Rpn9 also interacts with Rpn8 to stabilize the inhibited conformation of the Rpn8/Rpn11 heterodimer [68]. Point mutations of the responsible residues in Rpn5 and Rpn9 partially relieve Rpn11 inhibition in the isolated lid [68].
When the lid is incorporated into the 26S proteasome [69], the Rpn8/Rpn11 MPN heterodimer rotates 90°, resulting in disruption of the Rpn11-Rpn5 and Rpn8-Rpn9 contact sites, and positioning of Rpn11 adjacent to the substrate entry port [65,68]. Interestingly, Rpn11 is significantly more active in the proteasome holoenzyme than in isolated MPN heterodimers [68]. This may be caused by interactions between Rpn2 and the Rpn11 Ins-2 loop [65], which are suggested to stabilize the extended active conformation of the MPN Rpn8/Rpn11 heterodimer [68].
Usp14
The catalytic domains of Usp14 and Ubp6 adopt a typical USP architecture, likened to an extended right hand consisting of Fingers, Palm and Thumb domains [70] (Fig. 4). Usp14 binds ubiquitin using its Fingers domain and a binding groove between the Palm and Thumb domains. Related USP-type DUBs such as HAUSP are inhibited prior to substrate binding via a misalignment of the catalytic triad residues [71], but this is not the case for the active site of Usp14 [70]. Yet the DUB activities of Usp14 and its yeast ortholog Ubp6 are activated 300–1000 fold upon association with the proteasome [31,41,72]. The crystal structure of a Usp14-UbAl adduct provides a model of the activated state [70] (Fig. 4). UbAl adduct formation is accompanied by substantial movements of two loop segments known as blocking loops BL1 and BL2, which emanate from the Palm domain. In the free form of the enzyme, BL1 and BL2 extend into the ubiquitin binding groove to occlude substrate access. In the UbAl-bound form of Usp14, BL1 and BL2 are displaced, allowing the ubiquitin C-terminus access to the binding groove [70]. Most likely the proteasome promotes BL1 and BL2 displacement to activate Usp14 [46,53].
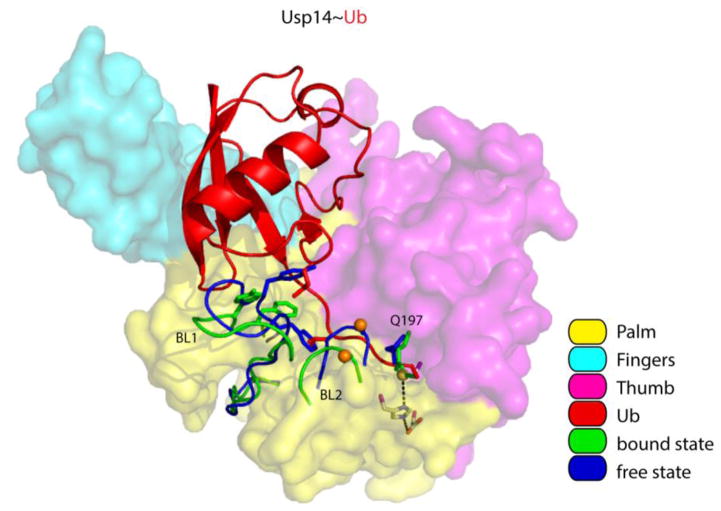
Figure 4. Displacement of the BL1 and BL2 loops activates Usp14.
Overview of Usp14 and UbAl, based on PDB: 2AYN and PDB: 2AYO [70]. The surfaces of the Palm, Thumb and Fingers domains were made semi-transparent to show the side chains of the catalytic triad. Usp14 regions that reorient upon binding (including the BL1 and BL2 loop) are displayed in blue for the free state and in green for the bound state. We note that the side chain of Q197 in the Thumb domain of Usp14 flips when Usp14 binds ubiquitin. The thiol group of the catalytic cysteine is shown as a yellow sphere, and phosphorylation site Ser432 on BL2 as an orange sphere. The isopeptide bond between UbAl and the catalytic cysteine is in green. Several side chains on the BL1 loop that interfere with ubiquitin binding are shown as stick models in addition to the cartoon backbone in both blue (free) and green (bound). Other color coding is explained in the key.
The relief of Usp14 autoinhibition can be achieved not only by proteasomes, but also by phosphorylation. The highly conserved Ser432 residue of Usp14 was recently found to be phosphorylated by AKT, a key component of growth signaling pathways [73]. Ser432 is located within the BL2 loop of Usp14, in proximity to a negatively charged patch on the Thumb domain. It has been proposed that the negative charge of the phosphate group on Ser432 is repelled by the negatively charged patch on the Thumb domain, resulting in BL2 displacement and the concomitant relief of the BL2 inhibitory effect. Indeed, phosphorylation of Usp14 (or the use of a Usp14 phosphomimetic) results in increased activity in assays with the fluorogenic substrate ubiquitin-7-amino-4-methylcoumarin (Ub-AMC), for both Usp14 in isolation and Usp14 in the proteasome, suggesting that phosphorylation and proteasome binding activate Usp14 via unrelated mechanisms [73]. Usp14 activity is elevated in LPS-treated-cells, in parallel with its phosphorylation, possibly by AKT [74,75]. These and other observations suggest an anti-inflammatory effect of Usp14 inhibition [74–78].
Phosphorylated Ser432 Usp14 was detected in proteasome-free fractions using glycerol gradient centrifugation, whereas unphosphorylated Usp14 was mostly found in the proteasome fractions [73]. These observations may suggest that phosphorylated Usp14 could “moonlight” as an active enzyme outside of the proteasome. However, the proteasome-free form of Usp14 phosphomimetic mutant, though quite active on the standard DUB tracer Ub-AMC, showed little or no activity against ubiquitin-protein conjugates [35]. Therefore it remains unclear whether Ser432 phosphorylation generates a proteasome-independent active form of Usp14.
The activation of Usp14 by AKT may provide a mechanism whereby growth signals can suppress catabolism through targeting proteasome activity. However, a variety of signals may converge on Usp14 and Ubp6. In yeast, Ubp6 is prominently regulated at the transcriptional level by signaling pathways that appear to monitor the state of the ubiquitin-proteasome pathway, and particularly the availability of free ubiquitin [47]. Ubp6 induction upon a decline in ubiquitin levels leads to higher Ubp6 occupancy on the proteasome. This helps to preserve the ubiquitin pool, through Ubp6’s DUB activity, which can enhance the efficiency of ubiquitin recycling at the proteasome, and perhaps as well by its noncatalytic activity, which can reduce substrate flux through the proteasome [47].
Uch37
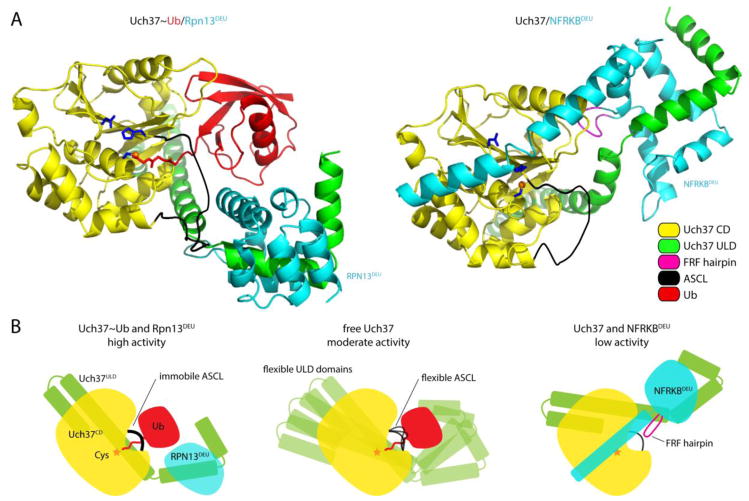
As noted above, Uch37 activity is controlled by its association with DEUBAD domains. In the case of the proteasomal complex, Rpn13DEU relieves autoinhibition of Uch37, which arises from two sources. First, the ULD domain of Uch37 is inhibitory to Uch37 activity [56,79,80], apparently because it can adopt conformations which sterically impede ubiquitin binding [60,61]. When bound to Rpn13DEU, the ULD domain becomes fixed in an orientation that is permissive for substrate binding (Fig. 5A and 5B). Second, Uch37 activity is suppressed by a loop segment that reaches over the active site. This is known as the active site crossover loop, or ASCL–a feature shared by all UCH family members. As with the ULD domain, the ASCL is displaced from its inhibitory position near the active site upon binding of Rpn13DEU. Redirection of the ASCL loop is mediated by a direct interaction between Rpn13DEU and ASCL residues Met148 and Phe149. Taken together, Rpn13DEU-induced conformational changes significantly improve Uch37’s affinity for substrates (i.e., KM is reduced), and thereby increase its catalytic efficiency [60,61].
Figure 5. Regulation of Uch37 by interactions with Rpn13DEU and NFRKBDEU.
(A) Overview of Uch37 in complex with Rpn13DEU and Ub, or NFRKBDEU, based on their crystal structures (PDB: 4UEL and PDB: 4UF5) [60,61]. Side chains of the Uch37 catalytic triad in the Uch37 catalytic domain (CD) are shown in blue. The ASCL has not been resolved over its entire length, but is shown as a thick black line to indicate how the C-terminus of ubiquitin passes underneath the loop. Color coding is explained in the key. (B) Schematic representation of free Uch37 (center), and Uch37 bound to Rpn13DEU (left) and NFRKBDEU (right). Mobility of the flexible ASCL loop in free Uch37 is indicated by multiple black lines.
Why does NFRKBDEU fail to activate Uch37? Unlike the Rpn13DEU domain, NFRKBDEU contains a conserved, six-residue element referred to as the FRF hairpin [61] or the F100 loop [60], as well as a long, helical structure at its C-terminus [60,61]. The FRF hairpin inserts into the pocket of Uch37 that normally binds ubiquitin, and is clasped in place by binding of the long, C-terminal helix of NFRKBDEU to the catalytic domain of Uch37, which forces the second helix in the Uch37 ULD domain to bend [60,61,81] (Fig. 5A and 5B). These interactions lower the substrate affinity of Uch37. Furthermore, interactions between NFRKBDEU residue Tyr135 and the catalytic His164 of Uch37 lead to disruption of Uch37’s active site geometry [60]. Together, the effects of NFRKBDEU on Uch37 substrate affinity and active site geometry result in dramatically reduced deubiquitinating activity.
Substrate specificity
The canonical substrate of the proteasome is traditionally considered to be modified by a single polyubiquitin chain containing four or more ubiquitin groups linked via residue K48 of ubiquitin. However, the architecture of ubiquitin modification is now understood to be highly diverse. Each of the seven lysines within ubiquitin can be used for chain formation as well as the N-terminal amino group. Using these target sites, mixed-linkage chains as well as branched chains of various lengths may be formed [6]. On the other hand, the proteasome does not require chain formation per se; at least some substrates modified on multiple sites by single ubiquitin groups can be efficiently degraded as well [35,82–84].
Proteasomal DUBs are thus presented with a tremendously varied array of potential substrates. Indeed, the potential substrates are so varied that it is a major technical problem to define the substrate specificity of these enzymes, and our current understanding is far from complete. Another aspect of this problem, highlighting how the proteasomal DUBs are exceptional within the DUB family, is that substrate selection by these DUBs may be conditioned by the selectivity of other elements of the proteasome such as ubiquitin receptors and the ATPases that move the substrate through the proteasome.
In vitro DUB specificity is typically characterized with respect to the following properties: chain linkage preference, chain length preference, and preference for the leaving group (which is often, as in the case of true conjugates, the protein modified by ubiquitin [5,85,86]). DUBs frequently exhibit marked specificity for where they cleave within a chain. Distal cleavage removes a single ubiquitin from the end of the chain that is furthest from the modified protein. Proximal or en bloc cleavage separates the entire chain from the modified protein. Finally, cleavages that remove multiple ubiquitin groups at once, but not the entire chain, are known as endo cleavages. These specificity features are critical aspects of how Rpn11, Usp14, and Uch37 control proteasome activity.
Rpn11
Purified proteasomes lacking in Rpn11 activity are highly deficient in substrate degradation [38,40]. It is generally assumed that this reflects a failure to translocate substrate into the CP, and therefore that the ubiquitin chain, if not removed, provides a barrier to translocation. Thus, the principal barrier to translocation could be ubiquitin unfolding [34,38,40]. If ubiquitin were unfolded, translocation could still be hindered in the absence of Rpn11 activity by the requirement to translocate more than one polypeptide chain through the pore at once. However, the proteasome has been shown to be capable of translocating multiple chains concurrently [34,87], so that models in which the barrier to translocation is the folded structure of ubiquitin are most consistent with existing data.
The removal of ubiquitin groups that would otherwise impair substrate translocation from the RP into the CP calls for an enzyme that acts quickly and with relaxed specificity. In the purified Rpn8/Rpn11 heterodimer, Rpn11 is a highly promiscuous DUB that efficiently cleaves within various types of polyubiquitin chains [64]. However, when incorporated into the proteasome, Rpn11 does not cleave efficiently within chains–a key feature of its specificity [40]. For any endo cleavage, the distal ubiquitin is accommodated by the active site of the enzyme, whereas the proximal ubiquitin is the leaving group. Because the active site of Rpn11 is so closely apposed to the substrate entry port of the proteasome, there is no space to fit a proximal ubiquitin group, preventing any endo or distal cleavages [64] (Fig. 6A). This is presumably by design, as only en bloc cleavages remove the impediment to translocation.
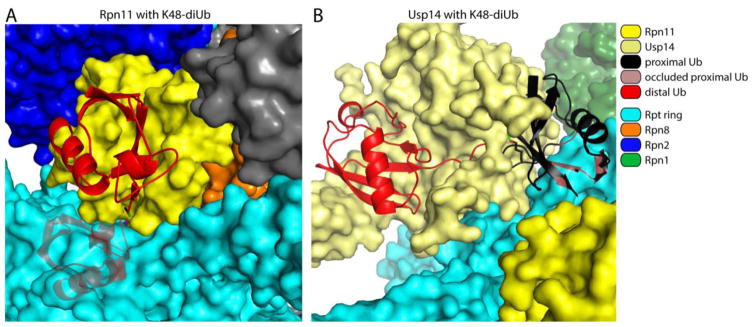
Figure 6. Proteasome-associated Rpn11 and Usp14 cannot accommodate folded proximal ubiquitin molecules.
Molecular modeling was used to show potential steric exclusion for the proximal ubiquitin within a di-ubiquitin element docked at either Rpn11 (A) or Usp14 (B, modified from [35]) within the proteasome. In short, isopeptide from di-ubiquitin (PDB: 1TBE [158]) was fit to the C-terminus of the bound distal ubiquitin on Rpn11 (PDB: 5U4P [34]) and Usp14 (PDB: 5GJQ [54]). The position of the proximal ubiquitin was then varied through all possible configurations of the Lys side chain for all the lysines in ubiquitin, from which no clash-free position could be found. The configurations for the proximal ubiquitins with the least steric conflict are shown, using a K48-linked ubiquitin-dimer as an example. The proximal ubiquitin is painted black while the portion that clashes with the Rpt subunits is shaded grey with red highlights. The isopeptide bond between the two ubiquitin molecules is in green.
As noted above, the active site of Rpn11 is similar to that of AMSH-LP. The latter shows K63 linkage specificity, due to a binding site for proximal ubiquitin, which is oriented so as to present K63 to the active site cysteine [39]. Many AMSH-LP residues that mediate binding of distal ubiquitin are conserved in Rpn11, but those which mediate proximal ubiquitin binding are not [64]. In fact, whereas the Ins-2 loop of AMSH-LP contacts the proximal ubiquitin, restricting linkage specificity [39], in Rpn11 it anchors the enzyme to the proteasome [64,65].
The constraints on proximal ubiquitin apply equally to the protein modified by ubiquitin: it must fit into the tight space between Rpn11 and the proteasome. Proximal ubiquitin is excluded from this space because it is globular. Accordingly, when a protein that is ubiquitinated cannot be unfolded, it is expected to be a poor substrate for Rpn11 in the proteasome [65]. However, translocation also drives unfolding of the substrate, converting what might be a poor substrate into a favorable one [2].
Usp14
Usp14 and Ubp6 are routinely assayed using artificial substrates such as Ub-AMC. However, with bona fide ubiquitin-protein conjugates as substrates, it has often been difficult to convincingly observe Usp14 and Ubp6 activity [88]. This has been the case even when Usp14/Ubp6 is associated with the proteasome, and thus in an activated state, and even when using established proteasome substrates. As free Usp14 and Ubp6 can hydrolyze free ubiquitin chains, these have been widely used as a measure of activity, although such reactions proceed at unexpectedly slow rates. Using free chains as substrates, chain linkage preferences have been noted [89], as well as a distal cleavage preference [70]. Distal cleavages should result in progressive chain shortening, sometimes referred to as chain “nibbling”. However, the prevailing notion that Usp14 and Ubp6 are sluggish enzymes has been difficult to reconcile with the phenotype of ubiquitin depletion seen in ubp6 null mutants in yeast [31–33,45,90] and Usp14 mutations in the mouse [91–93]. The dynamic balance of the ubiquitin pool as a whole can presumably depend only on highly active DUBs. The resolution of this conundrum required the identification of a preferred substrate of Usp14 and of the properties that govern substrate preference.
Cyclin B, ubiquitinated by its physiological ligase, the APC, has been used as a model substrate for deubiquitination by proteasome-activated Usp14 and for degradation by the proteasome [35,41,82,84]. Both reactions are extremely fast with this substrate; using quench flow methods, products of Usp14-mediated deubiquitination from cyclin B can be followed on a time scale of milliseconds to seconds [35]. In contrast, free chain cleavage is minimal, even after hours of incubation. The architecture of ubiquitin chains formed by the APC depends on the E2/UBC enzyme with which it cooperates; when used with either Ubc4 or UbcH10, the APC produces multiple short chains on the N-terminus of cyclin B, which is rich with lysine residues [82,94]. Using mutants in which all lysines but one have been substituted with arginine, one can test the effect of this multichain architecture on substrate preference. Surprisingly, cyclin B mutants modified by a single ubiquitin chain were not detectably cleaved by Usp14, or by Ubp6, despite carrying the same number of ubiquitin groups as the wild-type form [35]. This does not depend on the nature of the chain, since the same result was obtained using different types of chain linkage. Thus, Usp14 and Ubp6 show a dramatic preference in vitro for substrates that carry multiple ubiquitin chains.
Usp14 and Ubp6 rarely deubiquitinate a protein to completion in standard in vitro assays. Instead, a strong stop to deubiquitination is seen through the accumulation of oligo-ubiquitinated species [35,41]. An obvious scenario that could explain this phenomenon is that progressive shortening of a chain stops when the substrate is left with too few ubiquitin groups to associate with the proteasome. However, this is not the case, as shown by modifying cyclin B with multiple tetraubiquitin chains rather than ubiquitin. Deubiquitination of this multi-tetraubiquitinated cyclin B yields homogeneous products of free tetraubiquitin chains as well as tetraubiquitinated cyclin B, indicating that Usp14, in this fast mode, evidently removes chains en bloc rather than by distal nibbling. It is well established that tetraubiquitin chains have a significant affinity for the proteasome [95], so Usp14 comes to a stop at tetraubiquitinated cyclin B not because the substrate is displaced from the proteasome, but because there is only a single chain on this species.
Why do Usp14 and Ubp6 remove chains en bloc? Based on cryo-EM data, it was proposed that, in the enzymatically active form of Usp14, its catalytic domain is positioned so closely to the proteasome that a proximal ubiquitin cannot be accommodated [35] (Fig. 6B). As described above for the case of Rpn11, this simple constraint may be sufficient to enforce an en bloc mode of cleavage. Furthermore, if the proposal is valid, Usp14 should prefer substrates in which ubiquitin chains modify lysine residues within flexible or unfolded segments of the target protein. This possibility remains to be tested, but the proposed specificity of Usp14 is strikingly similar to that of Rpn11. An interesting distinction between the two enzymes is that the unfolded segments presented to Rpn11 may be in many cases natively folded, but converted to an unfolded state as a consequence of substrate translocation. In summary, Usp14 acts highly preferentially on substrates with multiple ubiquitin modifications, and substrate flexibility might be an important specificity rule for Usp14.
The deep similarities between Usp14 and Rpn11 are unlikely to be coincidental. They suggest that these enzymes cooperate in removing ubiquitin chains to prevent chains from stalling translocation. While Rpn11 is thought to act after the commitment step, the activity of Usp14 does not require ATP, and it can act prior to commitment. Thus, Usp14 and Rpn11 may function in a temporal progression; a multichain substrate might be subject to initial Usp14-dependent en bloc cuts, leaving in the simplest case a single chain intact, which will, after commitment, be removed by Rpn11. Chains on globular domains would preferentially be spared for Rpn11, since these domains are presumably unfolded prior to their presentation to Rpn11, as discussed above. In the limiting version of this model, Ubp6 mutants would have a rather weak phenotype, because the chains that it fails to remove would simply be redirected to Rpn11. The strong phenotype of Ubp6 mutants, and particularly their ubiquitin depletion phenotype, suggests that this is not entirely the case. It is expected that some feature of chain removal by Ubp6, most likely its specificity, underlies the functional differentiation of these enzymes. It will be important to resolve this problem in future studies.
Uch37
While considerable progress has been achieved in understanding the substrate specificities of Rpn11 and Usp14, that of Uch37 remains mysterious and confounding. RP-associated Uch37 was found to progressively trim ubiquitin chains from the distal end, removing one ubiquitin moiety at a time [96,97]. Both proteasome-associated Uch37 and the isolated Uch37 catalytic domain have been shown to act on chains of varied linkage types [88,98,99], and RP-associated Uch37 has been reported to remove ubiquitin moieties from α-globin [96]. The efficiency of Uch37-mediated hydrolysis, however, seems strikingly low: whereas Usp14 acts on its preferred substrates within seconds [35], Uch37-mediated cleavage of Ub-chains takes much longer, with incubations of 30 minutes and even 8 hours [88,96,98,99]. It may be suggested that Uch37 acts slowly in order to provide the proteasome with ample time to degrade bona fide substrates [60], slowly deubiquitinating substrates that stall the proteasome until they are released. Perhaps it is too slow even for this model to apply. Instead, Uch37 may have preferred substrates that have not yet been identified.
When Ub-AMC is used as substrate, Uch37 is by far the most active of the three DUBs on the proteasome [41,57,98], but when ubiquitin-protein conjugates are used as substrates [35,84], the situation is dramatically reversed. The specificity properties of Uch37 are for the most part generic to the UCH family of DUBs. These enzymes show striking preference for ubiquitin adducts in which the leaving group is small, as is the case for ubiquitin-AMC [100,101]. This preference seems to be imposed by the inhibitory active-site crossover loop (ASCL) described above [100]. The key feature of this loop is that it paradoxically forms a bridge over the active site. In contrast, the BL1 and BL2 loops of Usp14 encroach on the ubiquitin binding groove from a lateral position. Retraction of BL1 and BL2 thus leaves the Usp14 active site fully unhindered. However, because the ASCL is anchored on both sides of the active site, there seems to be no position available to this flexible segment that does not compromise substrate access topologically. This may explain why Uch37 tends to act very slowly on tested ubiquitin-protein conjugates. Likewise, a proximal ubiquitin group may be hindered by the ASCL. On the other hand, ubiquitin adducts with small leaving groups, such as Ub-AMC, should be able to slip under the ASCL with little difficulty.
How then does Uch37 act on ubiquitin-protein conjugates at all? One resolution, which we do not favor, is that ubiquitin-protein conjugates are not the physiological substrates of the enzyme. The most interesting solution to the problem, proposed by VanderLinden et al. [60], is that, for ubiquitin-protein conjugates, the body of the target protein must be positioned on the same side of the ASCL as ubiquitin itself. Thus, two segments from the conjugate would pass under the loop, one from the substrate and one from the C-terminus of ubiquitin. Fully extended, the loop can span 20 Å, easily adequate for the passage of such a hairpin. It will be interesting to see tests of this model. The model implies that two prominent features proposed to be shared by Rpn11 and Usp14–en bloc chain removal and preference for ubiquitin groups situated on flexible regions of the substrate–may well extend to Uch37. If this scenario holds, it will be remarkable how the three enzymes use very different mechanisms to achieve specificity features that are broadly comparable. In the case of Uch37, global proteomic analysis will likely be required to determine its true substrate specificity.
Effects of deubiquitination on substrate degradation
As discussed above, deubiquitination by Rpn11 stimulates the degradation of substrates that are already committed to degradation by removing bulky ubiquitin chains that otherwise might stall the proteasome. In contrast, deubiquitination by the ATP-independent DUBs Usp14/Ubp6 and Uch37 has been proposed to suppress degradation through promoting premature substrate dissociation [35,41,45,96]. Most of the work on this problem has concerned Usp14/Ubp6, so we will focus on this enzyme.
Loss of Ubp6 in yeast destabilizes several model and physiological proteasome substrates in vivo [45], and inhibition of the DUB activity of Usp14 in mammalian cells stimulates the degradation of specific proteasome substrates [41,102–108]. Accordingly, when Usp14 is activated by AKT, the degradation of multiple proteins appears to be suppressed [73]. The suppression of proteasome activity by Ubp6 and Usp14 is a two-fold effect in which substrate deubiquitination and the aforementioned noncatalytic effect can both take part. The confluence of these two mechanistically distinct effects is an interpretive challenge that runs through much of the Usp14/Ubp6 literature. Kim and Goldberg recently reported that the overall rate of protein degradation is accelerated by an impressive 45% in murine embryonic fibroblasts (MEFs) in which the Usp14 gene has been deleted, and enhanced degradation was seen for both short-lived and long-lived proteins [50]. The fraction of this effect contributed by deubiquitination was not determined, but interestingly, when long-lived proteins were examined, a stimulatory effect of the mutation was seen not only in the proteasome pathway, but also on autophagy [50]. Thus, Usp14/Ubp6 appears to suppress protein degradation through three mechanisms: deubiquitination of proteasome substrates, the “noncatalytic effect,” which operates directly on the proteasome, and through regulating autophagy. The mechanism of the autophagy effect, which was originally reported by Xu et al., remains to be determined, but it apparently involves the deubiquitinating activity of Usp14 [109].
A small-molecule inhibitor of Usp14, known as IU1, was found to accelerate the degradation of several proteins [41]. These effects were not seen in Usp14 null MEFs, providing evidence for the on-target specificity of IU1 in these experiments. IU1 does not appear to induce the noncatalytic effect [41]; therefore this compound provides one method to discriminate between catalytic and noncatalytic suppression of proteasome activity. Recently a 10-fold more potent variant of IU1, known as IU1-47, has been reported ([110]).
Since the original study on IU1, several groups have reported proteins that show accelerated degradation under IU1 treatment, including the Prion protein PrP [102,103], cGAS [108], phosphorylated tyrosine hydroxylase [104], the androgen receptor [105], vimentin [106], aurora kinase [111], CREB-binding protein [75], YFP-tagged CD3δ [107], and the model UPS substrate GFPu [107]. In many of these studies, the on-target nature of the IU1 effect is supported by genetic confirmation [102,105–108]. For example, PrP is degraded more rapidly in the presence of IU1 or when Usp14 expression is reduced, and more slowly when Usp14 is overexpressed [102]. Like PrP, cGAS, a key regulator of innate immunity, is an intriguing target [108]. In the absence of Usp14 activity, the ubiquitination of cGAS is elevated, which promotes its degradation not through the proteasome but through autophagy. Further studies on the mechanism by which Usp14 inhibition alters ubiquitination and degradation of cGAS may help elucidate the mechanism by which Usp14 modulates autophagy.
A corollary to the hypothesis that Usp14 suppresses proteasome activity through deubiquitination is that Usp14 must act quickly with respect to the proteasome, since deubiquitination would essentially be in kinetic competition with the commitment step. This scenario has been assessed in vitro using quench-flow and single-molecule-based methods [35]. Although cyclin B is an exceptionally favorable substrate of the proteasome, its turnover in a single encounter with the proteasome is considerably slower than the rate of deubiquitination. Single-molecule analysis allows for the dwell time of the substrate on the proteasome to be measured, and this is indeed controlled by Usp14; the substrate dissociates from the proteasome prematurely as a consequence of deubiquitination. Thus, for cyclin B, a kinetic competition model is in agreement with the data. It would be interesting to extend this approach to other targets of Usp14.
Although deubiquitination by Usp14/Ubp6 is genetically separable from the noncatalytic effect, they are undoubtedly linked in the wild-type enzyme. It was initially proposed that the noncatalytic effect may buy time for the deubiquitination to be completed [45]. Recent data agree with this interpretation [35]. Although the noncatalytic effect is strongly stimulated by ubiquitin engagement, catalytically inactive Usp14 can also suppress the in vitro degradation of a non-ubiquitinated proteasome substrate [46,50] in what may be a basal suppressive influence. It has therefore been suggested that Usp14’s inhibitory effect could serve as an additional function in biasing the proteasome against nonubiquitinated substrates [50].
Physiological function
While there has been strong progress in the biochemistry and structural biology of the proteasomal DUBs, our understanding of their physiological roles in mammals is still at an early stage. All three of these enzymes are essential in the mouse [112–114], suggestive of distinct physiological functions. However, it is not clear for any of these DUBs whether the functional requirement applies to their DUB activity per se. No conditional mutants exist. In cultured cells there does not seem to be a general requirement for either Usp14 or Uch37 (unpubl. data), so the lethal phenotypes of the mouse null mutants are presumably rooted in transient processes associated with early development.
Rpn11
Rpn11 is an essential protein in yeast, D. melanogaster, and human cells [38,40,115,116]. In yeast, Rpn11 active site mutants appear to be viable [117,118], implying that the lethal phenotype of the null reflects an indispensable role of Rpn11 in proteasome assembly. Loss of Rpn11 indeed disrupts 26S proteasome assembly [38,72]. Catalytic site mutants are more useful, as they assemble normally but fail to deubiquitinate or degrade model substrates in vitro [38,40]. In addition, active-site mutants in yeast show an accumulation of high molecular weight ubiquitin conjugates and stabilization of model proteasome substrates [117]. This mutation is synthetically lethal with a UBP6 deletion, which could possibly indicate a shared function or substrate [118]. Numerous defects have been reported in Rpn11 knockdown cell lines [72,115,116,119–124]. It is unclear whether these defects result from effects on deubiquitination or proteasome assembly. A specific small-molecule inhibitor of Rpn11, capzimin, has recently been described, and is under consideration as an anti-cancer compound [125,126]. Treating cells with capzimin may provide a simple way to test for acute effects of loss of Rpn11 deubiquitinating activity.
Usp14
Experiments in various model systems have revealed a multitude of processes in which Usp14 and Ubp6 may function [31–33,45,47,48,50,76–78,90,91,93,109,113,127–138]. An interesting phenotype of ubp6 null mutants in yeast is their exceptional and perhaps unique ability to tolerate aneuploid chromosomes [135–137]. Aneuploidy gives rise to substantial proteotoxic stress, as evidenced by the accumulation of aberrant protein aggregates in aneuploid strains. Enhanced protein degradation in ubp6 mutants may underlie their accommodation of the aneuploid state, though the exact mechanism has not been identified.
For Usp14, the best mutation to date is a hypomorphic allele found in the axj mouse [139]. The reduced Usp14 expression in this mutant does not appear to confer a pleiotropic phenotype, but it produces defects in the structure and function of the neuromuscular junction (NMJ) [91,139,140]. Thus, Usp14 seems especially critical in motor neurons, or in their development, although the mechanistic basis of this phenotype is not known. The axj phenotype can be ameliorated by overexpression of ubiquitin, in accord with a reduction of ubiquitin levels in neurons of the axj mutant [48,130]. Thus, perturbation of ubiquitin pools may be required for the full effect of the axj mutation, though it remains possible that ubiquitin overexpression can suppress a defect that is independent of ubiquitin pools. More interestingly, overexpression of a catalytically dead C114A mutant of Usp14 restores a subset of axj phenotypes [49,131], suggesting that some of the mutant defects might result from a deficiency of the noncatalytic effect, rather than deubiquitination. Expression of Usp14-C114A does not rescue the ubiquitin deficiency phenotype [92], underscoring the dependence of ubiquitin recycling on Usp14 catalytic activity. A final genetic insight is that the severity of the axj phenotype is highly dependent on genetic background [93]. Identification of the genetic modifier that determines phenotypic severity in inbred mouse lines might provide insight into the NMJ defect.
Uch37
Uch37 has been implicated in DNA replication and cell cycle [141], TGF-β signaling [142–145], WNT signaling via Tcf7 [146], longevity [147], cell migration and invasion [148], and the reversal of ubiquitination of proteasome subunits [138]. It is difficult to resolve common themes from the physiological studies reported to date. DNA double strand break repair is also regulated by Uch37, though presumably by the INO80-associated species, since Uch37-mediated stabilization of INO80 subunit NFRKB is required for INO80-mediated recruitment of the resection factor EXO1 [149]. Elevated levels of Uch37 have been associated with several cancers, and Uch37 overexpression is correlated with outcome and recurrence in several cancers [148,150–153]. Furthermore, the role of Uch37 in DNA double strand break repair [149] suggests that Uch37 inhibition might also be effective in cancers with defective DNA repair. Taken together, these findings, reviewed by Chen et al. [154], make Uch37 a potential target for anti-cancer therapy.
Challenges for the future
Over the last two decades, major progress has been made in our understanding of the proteasome-associated DUBs. However, the picture of proteasomal deubiquitination that emerges from this work remains poorly defined in many respects. Most notably, to date we have only a preliminary account of the endogenous substrates of Usp14 and Uch37. Solving this problem will require application of proteomic methods in conjunction with careful in vitro biochemistry to show that effects observed in vivo are direct. In vivo testing of the in vitro multichain specificity of Usp14 may require innovative proteomic approaches, as most current methods rely on proteolytic digestion, which prevents one from determining whether multiple lysines are simultaneously ubiquitinated on a single protein molecule. For Uch37, favorable in vitro substrates that are true ubiquitin-protein conjugates have yet to be identified, and we can only guess at the principles underlying its seemingly restrictive specificity. It is generally assumed that Rpn11 simply removes chains that are translocated into the proximity of its active site, but it is well-positioned–near the substrate entry port–to execute a checkpoint or surveillance function, and the possibility that significant constraints on its specificity may exist has not been tested to our knowledge.
The substrates of the proteasomal DUBs must also be related to the phenotypes of the corresponding mutants. In the case of Usp14 and Uch37, it is oddly difficult at this time to relate the known phenotypes to defects in proteasome function. Indeed, it is possible that key substrates of these enzymes, even as they function on the proteasome, are not substrates of the proteasome itself. However, before exploring this line of thought, we will need a more precise and complete description of the phenotypes of Usp14 and particularly Uch37 mutants. Furthermore, for Usp14, the distinction between catalytic and noncatalytic phenotypes needs to be established more clearly, and, likewise, for Uch37, its proteasome-related and INO80-related phenotypes must be systematically distinguished from one another. In principle, small-molecule inhibitors of the proteasomal DUBs have great potential to support such functional studies, but their utility for research will be limited until inhibitors of greater specificity and potency are available.
A final question concerns the potential role of ubiquitin receptors in regulating the deubiquitinating activity of the proteasome. It is intriguing that the binding partners of Usp14 and Uch37 on the proteasome—Rpn1 and Rpn13—both function as intrinsic receptors for ubiquitin, and for ubiquitin-like proteins that themselves bind ubiquitin. In addition, Rpn11 sits above the substrate entry port, close to the ubiquitin receptor Rpn10 [69,155,156]. These observations suggest that ubiquitin receptors on the proteasome might deliver, stabilize, or orient substrates for deubiquitination. The importance of such positioning for DUB specificity remains to be determined but could be readily addressed for Ubp6 and Rpn11 through the use of existing yeast mutants in these receptor sites [17].
Research Highlights.
The three proteasomal DUBs differ strongly in mechanism, specificity, and function
Rpn11 removes ubiquitin chains from substrates that are committed to degradation
Usp14 and Uch37 can work before commitment to rescue substrates from degradation
Usp14 deubiquitinates substrates with multiple Ub modifications
Uch37 is found in both the proteasome and the INO80 chromatin remodeling complex
Acknowledgments
We thank members of the Finley lab, particularly S. Elsasser, for critical reading of the manuscript. Funding was provided by grants from the National Institutes of Health to D.F. (R01GM043601) and the Dutch Cancer Foundation to S.d.P. (BUIT 2015-7517).
Footnotes
Disclosure of competing financial interests
The Usp14 inhibitor IU1 is under patent, which is held by D.F. and others.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Saeki Y. Ubiquitin recognition by the proteasome. J Biochem. 2017;161:113–124. doi: 10.1093/jb/mvw091. [DOI] [PubMed] [Google Scholar]
- 2.Yu H, Matouschek A. Recognition of Client Proteins by the Proteasome. Annu Rev Biophys. 2017;46:149–173. doi: 10.1146/annurev-biophys-070816-033719. [DOI] [PubMed] [Google Scholar]
- 3.Collins GA, Goldberg AL. The Logic of the 26S Proteasome. Cell. 2017;169:792–806. doi: 10.1016/j.cell.2017.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fraile JM, Quesada V, Rodríguez D, Freije JMP, López-Otín C. Deubiquitinases in cancer: new functions and therapeutic options. Oncogene. 2012;31:2373–2388. doi: 10.1038/onc.2011.443. [DOI] [PubMed] [Google Scholar]
- 5.Mevissen TET, Komander D. Mechanisms of Deubiquitinase Specificity and Regulation. Annu Rev Biochem. 2017;86:159–192. doi: 10.1146/annurev-biochem-061516-044916. [DOI] [PubMed] [Google Scholar]
- 6.Yau R, Rape M. The increasing complexity of the ubiquitin code. Nat Cell Biol. 2016;18:579–586. doi: 10.1038/ncb3358. [DOI] [PubMed] [Google Scholar]
- 7.Komander D, Rape M. The Ubiquitin Code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 8.Behrends C, Harper JW. Constructing and decoding unconventional ubiquitin chains. Nat Struct Mol Biol. 2011;18:520–8. doi: 10.1038/nsmb.2066. [DOI] [PubMed] [Google Scholar]
- 9.Glickman MH, Rubin DM, Coux O, Wefes I, Pfeifer G, Cjeka Z, Baumeister W, Fried VA, Finley D. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell. 1998;94:615–23. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- 10.Groll M, Bajorek M, Köhler A, Moroder L, Rubin DM, Huber R, Glickman MH, Finley D. A gated channel into the proteasome core particle. Nat Struct Biol. 2000;7:1062–7. doi: 10.1038/80992. [DOI] [PubMed] [Google Scholar]
- 11.Smith DM, Chang SC, Park S, Finley D, Cheng Y, Goldberg AL. Docking of the proteasomal ATPases’ carboxyl termini in the 20S proteasome’s alpha ring opens the gate for substrate entry. Mol Cell. 2007;27:731–44. doi: 10.1016/j.molcel.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clague MJ, Barsukov I, Coulson JM, Liu H, Rigden DJ, Urbe S. Deubiquitylases From Genes to Organism. Physiol Rev. 2013;93:1289–1315. doi: 10.1152/physrev.00002.2013. [DOI] [PubMed] [Google Scholar]
- 13.Deveraux Q, Ustrell V, Pickart C, Rechsteiner M. A 26 S protease subunit that binds ubiquitin conjugates. J Biol Chem. 1994;269:7059–61. [PubMed] [Google Scholar]
- 14.van Nocker S, Sadis S, Rubin DM, Glickman M, Fu H, Coux O, Wefes I, Finley D, Vierstra RD. The multiubiquitin-chain-binding protein Mcb1 is a component of the 26S proteasome in Saccharomyces cerevisiae and plays a nonessential, substrate-specific role in protein turnover. Mol Cell Biol. 1996;16:6020–8. doi: 10.1128/mcb.16.11.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schreiner P, Chen X, Husnjak K, Randles L, Zhang N, Elsasser S, Finley D, Dikic I, Walters KJ, Groll M. Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature. 2008;453:548–52. doi: 10.1038/nature06924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D, Dikic I. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453:481–488. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi Y, Chen X, Elsasser S, Stocks BB, Tian G, Lee BH, Shi Y, Zhang N, de Poot SAH, Tuebing F, Sun S, Vannoy J, Tarasov SG, Engen JR, Finley D, Walters KJ. Rpn1 provides adjacent receptor sites for substrate binding and deubiquitination by the proteasome. Science. 2016;351 doi: 10.1126/science.aad9421. pii: aad9421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elsasser S, Finley D. Delivery of ubiquitinated substrates to protein-unfolding machines. Nat Cell Biol. 2005;7:742–749. doi: 10.1038/ncb0805-742. [DOI] [PubMed] [Google Scholar]
- 19.Chen X, Randles L, Shi K, Tarasov SG, Aihara H, Walters KJ. Structures of Rpn1 T1:Rad23 and hRpn13:hPLIC2 Reveal Distinct Binding Mechanisms between Substrate Receptors and Shuttle Factors of the Proteasome. Structure. 2016;24:1257–70. doi: 10.1016/j.str.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hiyama H, Yokoi M, Masutani C, Sugasawa K, Maekawa T, Tanaka K, Hoeijmakers JH, Hanaoka F. Interaction of hHR23 with S5a. The ubiquitin-like domain of hHR23 mediates interaction with S5a subunit of 26 S proteasome. J Biol Chem. 1999;274:28019–25. doi: 10.1074/jbc.274.39.28019. [DOI] [PubMed] [Google Scholar]
- 21.Fujiwara K, Tenno T, Sugasawa K, Jee JG, Ohki I, Kojima C, Tochio H, Hiroaki H, Hanaoka F, Shirakawa M. Structure of the ubiquitin-interacting motif of S5a bound to the ubiquitin-like domain of HR23B. J Biol Chem. 2004;279:4760–7. doi: 10.1074/jbc.M309448200. [DOI] [PubMed] [Google Scholar]
- 22.Wilkinson CRM, Seeger M, Hartmann-Petersen R, Stone M, Wallace M, Semple C, Gordon C. Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nat Cell Biol. 2001;3:939–943. doi: 10.1038/ncb1001-939. [DOI] [PubMed] [Google Scholar]
- 23.Prakash S, Tian L, Ratliff KS, Lehotzky RE, Matouschek A. An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat Struct Mol Biol. 2004;11:830–837. doi: 10.1038/nsmb814. [DOI] [PubMed] [Google Scholar]
- 24.Unverdorben P, Beck F, ŒledŸ P, Schweitzer A, Pfeifer G, Plitzko JM, Baumeister W, Förster F. Deep classification of a large cryo-EM dataset defines the conformational landscape of the 26S proteasome. Proc Natl Acad Sci. 2014;111:5544–9. doi: 10.1073/pnas.1403409111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luan B, Huang X, Wu J, Mei Z, Wang Y, Xue X, Yan C, Wang J, Finley DJ, Shi Y, Wang F. Structure of an endogenous yeast 26S proteasome reveals two major conformational states. Proc Natl Acad Sci. 2016;113:2642–7. doi: 10.1073/pnas.1601561113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen S, Wu J, Lu Y, Ma YB, Lee BH, Yu Z, Ouyang Q, Finley DJ, Kirschner MW, Mao Y. Structural basis for dynamic regulation of the human 26S proteasome. Proc Natl Acad Sci. 2016;113:12991–12996. doi: 10.1073/pnas.1614614113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matyskiela ME, Lander GC, Martin A. Conformational switching of the 26S proteasome enables substrate degradation. Nat Struct Mol Biol. 2013;20:781–8. doi: 10.1038/nsmb.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.ŒledŸ P, Unverdorben P, Beck F, Pfeifer G, Schweitzer A, Förster F, Baumeister W. Structure of the 26S proteasome with ATP-γS bound provides insights into the mechanism of nucleotide-dependent substrate translocation. Proc Natl Acad Sci. 2013;110:7264–9. doi: 10.1073/pnas.1305782110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wehmer M, Rudack T, Beck F, Aufderheide A, Pfeifer G, Plitzko JM, Förster F, Schulten K, Baumeister W, Sakata E. Structural insights into the functional cycle of the ATPase module of the 26S proteasome. Proc Natl Acad Sci. 2017;114:1305–1310. doi: 10.1073/pnas.1621129114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lenkinski RE, Chen DM, Glickson JD, Goldstein G. Nuclear magnetic resonance studies of the denaturation of ubiquitin. Biochim Biophys Acta. 1977;494:126–30. doi: 10.1016/0005-2795(77)90140-4. [DOI] [PubMed] [Google Scholar]
- 31.Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, Baker RT, Walz T, Ploegh H, Finley D. Multiple associated proteins regulate proteasome structure and function. Mol Cell. 2002;10:495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- 32.Hanna J, Leggett DS, Finley D. Ubiquitin depletion as a key mediator of toxicity by translational inhibitors. Mol Cell Biol. 2003;23:9251–61. doi: 10.1128/MCB.23.24.9251-9261.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chernova TA, Allen KD, Wesoloski LM, Shanks JR, Chernoff YO, Wilkinson KD. Pleiotropic effects of Ubp6 loss on drug sensitivities and yeast prion are due to depletion of the free ubiquitin pool. J Biol Chem. 2003;278:52102–15. doi: 10.1074/jbc.M310283200. [DOI] [PubMed] [Google Scholar]
- 34.Worden EJ, Dong KC, Martin A. An AAA Motor-Driven Mechanical Switch in Rpn11 Controls Deubiquitination at the 26S Proteasome. Mol Cell. 2017;67:799–811.e8. doi: 10.1016/j.molcel.2017.07.023. [DOI] [PubMed] [Google Scholar]
- 35.Lee BH, Lu Y, Prado MA, Shi Y, Tian G, Sun S, Elsasser S, Gygi SP, King RW, Finley D. USP14 deubiquitinates proteasome-bound substrates that are ubiquitinated at multiple sites. Nature. 2016;532:398–401. doi: 10.1038/nature17433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hershko A, Rose IA. Ubiquitin-aldehyde: a general inhibitor of ubiquitin-recycling processes. Proc Natl Acad Sci. 1987;84:1829–33. doi: 10.1073/pnas.84.7.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borodovsky A, Kessler BM, Casagrande R, Overkleeft HS, Wilkinson KD, Ploegh HL. A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. EMBO J. 2001;20:5187–96. doi: 10.1093/emboj/20.18.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verma R, Aravind L, Oania R, McDonald WH, Yates JR, Koonin EV, Deshaies RJ. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–5. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- 39.Sato Y, Yoshikawa A, Yamagata A, Mimura H, Yamashita M, Ookata K, Nureki O, Iwai K, Komada M, Fukai S. Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature. 2008;455:358–62. doi: 10.1038/nature07254. [DOI] [PubMed] [Google Scholar]
- 40.Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- 41.Lee BH, Lee MJ, Park S, Oh DC, Elsasser S, Chen PC, Gartner C, Dimova N, Hanna J, Gygi SP, Wilson SM, King RW, Finley D. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467:179–184. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuo CL, Goldberg AL. Ubiquitinated proteins promote the association of proteasomes with the deubiquitinating enzyme Usp14 and the ubiquitin ligase Ube3c. Proc Natl Acad Sci. 2017;114:E3404–E3413. doi: 10.1073/pnas.1701734114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elsasser S, Chandler-Militello D, Muller B, Hanna J, Finley D. Rad23 and Rpn10 Serve as Alternative Ubiquitin Receptors for the Proteasome. J Biol Chem. 2004;279:26817–26822. doi: 10.1074/jbc.M404020200. [DOI] [PubMed] [Google Scholar]
- 44.Yu H, Kago G, Yellman CM, Matouschek A. Ubiquitin-like domains can target to the proteasome but proteolysis requires a disordered region. EMBO J. 2016;35:1522–36. doi: 10.15252/embj.201593147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hanna J, Hathaway NA, Tone Y, Crosas B, Elsasser S, Kirkpatrick DS, Leggett DS, Gygi SP, King RW, Finley D. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell. 2006;127:99–111. doi: 10.1016/j.cell.2006.07.038. [DOI] [PubMed] [Google Scholar]
- 46.Bashore C, Dambacher CM, Goodall EA, Matyskiela ME, Lander GC, Martin A. Ubp6 deubiquitinase controls conformational dynamics and substrate degradation of the 26S proteasome. Nat Struct Mol Biol. 2015;22:712–9. doi: 10.1038/nsmb.3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hanna J, Meides A, Zhang DP, Finley D. A Ubiquitin Stress Response Induces Altered Proteasome Composition. Cell. 2007;129:747–759. doi: 10.1016/j.cell.2007.03.042. [DOI] [PubMed] [Google Scholar]
- 48.Vaden JH, Watson JA, Howard AD, Chen PC, Wilson JA, Wilson SM. Distinct effects of ubiquitin overexpression on NMJ structure and motor performance in mice expressing catalytically inactive USP14. Front Mol Neurosci. 2015;8:11. doi: 10.3389/fnmol.2015.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walters BJ, Hallengren JJ, Theile CS, Ploegh HL, Wilson SM, Dobrunz LE. A catalytic independent function of the deubiquitinating enzyme USP14 regulates hippocampal synaptic short-term plasticity and vesicle number. J Physiol. 2014;592:571–86. doi: 10.1113/jphysiol.2013.266015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim HT, Goldberg AL. The Deubiquitinating Enzyme Usp14 Allosterically Inhibits Multiple Proteasomal Activities and Ubiquitin-Independent Proteolysis. J Biol Chem. 2017;292:9830–9839. doi: 10.1074/jbc.M116.763128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peth A, Besche HC, Goldberg AL. Ubiquitinated proteins activate the proteasome by binding to Usp14/Ubp6, which causes 20S gate opening. Mol Cell. 2009;36:794–804. doi: 10.1016/j.molcel.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peth A, Kukushkin N, Bossé M, Goldberg AL. Ubiquitinated proteins activate the proteasomal ATPases by binding to Usp14 or Uch37 homologs. J Biol Chem. 2013;288:7781–7790. doi: 10.1074/jbc.M112.441907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aufderheide A, Beck F, Stengel F, Hartwig M, Schweitzer A, Pfeifer G, Goldberg AL, Sakata E, Baumeister W, Förster F. Structural characterization of the interaction of Ubp6 with the 26S proteasome. Proc Natl Acad Sci. 2015;112:8626–31. doi: 10.1073/pnas.1510449112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang X, Luan B, Wu J, Shi Y. An atomic structure of the human 26S proteasome. Nat Struct Mol Biol. 2016;23:778–785. doi: 10.1038/nsmb.3273. [DOI] [PubMed] [Google Scholar]
- 55.Sanchez-Pulido L, Kong L, Ponting CP. A common ancestry for BAP1 and Uch37 regulators. Bioinformatics. 2012;28:1953–6. doi: 10.1093/bioinformatics/bts319. [DOI] [PubMed] [Google Scholar]
- 56.Yao T, Song L, Xu W, DeMartino GN, Florens L, Swanson SK, Washburn MP, Conaway RC, Conaway JW, Cohen RE. Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nat Cell Biol. 2006;8:994–1002. doi: 10.1038/ncb1460. [DOI] [PubMed] [Google Scholar]
- 57.Hamazaki J, Iemura SI, Natsume T, Yashiroda H, Tanaka K, Murata S. A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes. EMBO J. 2006;25:4524–36. doi: 10.1038/sj.emboj.7601338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qiu XB, Ouyang SY, Li CJ, Miao S, Wang L, Goldberg AL. hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37. EMBO J. 2006;25:5742–53. doi: 10.1038/sj.emboj.7601450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen X, Lee BH, Finley D, Walters KJ. Structure of Proteasome Ubiquitin Receptor hRpn13 and Its Activation by the Scaffolding Protein hRpn2. Mol Cell. 2010;38:404–415. doi: 10.1016/j.molcel.2010.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.VanderLinden RT, Hemmis CW, Schmitt B, Ndoja A, Whitby FG, Robinson H, Cohen RE, Yao T, Hill CP. Structural Basis for the Activation and Inhibition of the UCH37 Deubiquitylase. Mol Cell. 2015;57:901–911. doi: 10.1016/j.molcel.2015.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sahtoe DD, van Dijk WJ, El Oualid F, Ekkebus R, Ovaa H, Sixma TK. Mechanism of UCH-L5 activation and inhibition by DEUBAD domains in RPN13 and INO80G. Mol Cell. 2015;57:887–900. doi: 10.1016/j.molcel.2014.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yao T, Song L, Jin J, Cai Y, Takahashi H, Swanson SK, Washburn MP, Florens L, Conaway RC, Cohen RE, Conaway JW. Distinct modes of regulation of the Uch37 deubiquitinating enzyme in the proteasome and in the Ino80 chromatin-remodeling complex. Mol Cell. 2008;31:909–17. doi: 10.1016/j.molcel.2008.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Conaway RC, Conaway JW. The INO80 chromatin remodeling complex in transcription, replication and repair. Trends Biochem Sci. 2009;34:71–77. doi: 10.1016/j.tibs.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 64.Worden EJ, Padovani C, Martin A. Structure of the Rpn11-Rpn8 dimer reveals mechanisms of substrate deubiquitination during proteasomal degradation. Nat Struct Mol Biol. 2014;21:220–7. doi: 10.1038/nsmb.2771. [DOI] [PubMed] [Google Scholar]
- 65.Pathare GR, Nagy I, ŒledŸ P, Anderson DJ, Zhou HJ, Pardon E, Steyaert J, Förster F, Bracher A, Baumeister W. Crystal structure of the proteasomal deubiquitylation module Rpn8-Rpn11. Proc Natl Acad Sci. 2014;111:2984–9. doi: 10.1073/pnas.1400546111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Davies CW, Paul LN, Kim MI, Das C. Structural and thermodynamic comparison of the catalytic domain of AMSH and AMSH-LP: nearly identical fold but different stability. J Mol Biol. 2011;413:416–29. doi: 10.1016/j.jmb.2011.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shrestha RK, Ronau JA, Davies CW, Guenette RG, Strieter ER, Paul LN, Das C. Insights into the mechanism of deubiquitination by JAMM deubiquitinases from cocrystal structures of the enzyme with the substrate and product. Biochemistry. 2014;53:3199–217. doi: 10.1021/bi5003162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dambacher CM, Worden EJ, Herzik MA, Martin A, Lander GC. Atomic structure of the 26S proteasome lid reveals the mechanism of deubiquitinase inhibition. Elife. 2016;5:e13027. doi: 10.7554/eLife.13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lander GC, Estrin E, Matyskiela ME, Bashore C, Nogales E, Martin A. Complete subunit architecture of the proteasome regulatory particle. Nature. 2012;482:186–91. doi: 10.1038/nature10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hu M, Li P, Song L, Jeffrey PD, Chenova TA, Wilkinson KD, Cohen RE, Shi Y. Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. EMBO J. 2005;24:3747–3756. doi: 10.1038/sj.emboj.7600832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hu M, Li P, Li M, Li W, Yao T, Wu JW, Gu W, Cohen RE, Shi Y. Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell. 2002;111:1041–54. doi: 10.1016/s0092-8674(02)01199-6. [DOI] [PubMed] [Google Scholar]
- 72.Koulich E, Li X, DeMartino GN. Relative structural and functional roles of multiple deubiquitylating proteins associated with mammalian 26S proteasome. Mol Biol Cell. 2008;19:1072–82. doi: 10.1091/mbc.E07-10-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xu D, Shan B, Lee BH, Zhu K, Zhang T, Sun H, Liu M, Shi L, Liang W, Qian L, Xiao J, Wang L, Pan L, Finley D, Yuan J. Phosphorylation and activation of ubiquitin-specific protease-14 by Akt regulates the ubiquitin-proteasome system. Elife. 2015;4:e10510. doi: 10.7554/eLife.10510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mialki RK, Zhao J, Wei J, Mallampalli DF, Zhao Y. Overexpression of USP14 protease reduces I-κB protein levels and increases cytokine release in lung epithelial cells. J Biol Chem. 2013;288:15437–41. doi: 10.1074/jbc.C112.446682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wei J, Dong S, Bowser RK, Khoo A, Zhang L, Jacko AM, Zhao Y, Zhao J. Regulation of the ubiquitylation and deubiquitylation of CREB-binding protein modulates histone acetylation and lung inflammation. Sci Signal. 2017;10 doi: 10.1126/scisignal.aak9660. pii: eaak9660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sun Y, Qin Z, Li Q, Wan JJ, Cheng MH, Wang PY, Su DF, Yu JG, Liu X. MicroRNA-124 negatively regulates LPS-induced TNF-α production in mouse macrophages by decreasing protein stability. Acta Pharmacol Sin. 2016;37:889–97. doi: 10.1038/aps.2016.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xu YH, Guo NL. USP14 inhibitor IU1 prevents ventilator-induced lung injury in rats. Cell Mol Biol (Noisy-Le-Grand) 2014;60:50–4. [PubMed] [Google Scholar]
- 78.Liu N, Kong T, Chen X, Hu H, Gu H, Liu S, Chen X, Yang Q, Li A, Xiong X, Zhang Z. Ubiquitin-specific protease 14 regulates LPS-induced inflammation by increasing ERK1/2 phosphorylation and NF-κB activation. Mol Cell Biochem. 2017;431:87–96. doi: 10.1007/s11010-017-2978-0. [DOI] [PubMed] [Google Scholar]
- 79.Burgie SE, Bingman CA, Soni AB, Phillips GN. Structural characterization of human Uch37. Proteins Struct Funct Bioinforma. 2012;80:649–654. doi: 10.1002/prot.23147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jiao L, Ouyang S, Shaw N, Song G, Feng Y, Niu F, Qiu W, Zhu H, Hung L-W, Zuo X, Eleonora Shtykova V, Zhu P, Dong Y-H, Xu R, Liu Z-J. Mechanism of the Rpn13-induced activation of Uch37. Protein Cell. 2014;5:616–30. doi: 10.1007/s13238-014-0046-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen X, Walters KJ. Structural Plasticity Allows UCH37 to Be Primed by RPN13 or Locked Down by INO80G. Mol Cell. 2015;57:767–768. doi: 10.1016/j.molcel.2015.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dimova NV, Hathaway NA, Lee BH, Kirkpatrick DS, Berkowitz ML, Gygi SP, Finley D, King RW. APC/C-mediated multiple monoubiquitylation provides an alternative degradation signal for cyclin B1. Nat Cell Biol. 2012;14:168–76. doi: 10.1038/ncb2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Braten O, Livneh I, Ziv T, Admon A, Kehat I, Caspi LH, Gonen H, Bercovich B, Godzik A, Jahandideh S, Jaroszewski L, Sommer T, Kwon YT, Guharoy M, Tompa P, Ciechanover A. Numerous proteins with unique characteristics are degraded by the 26S proteasome following monoubiquitination. Proc Natl Acad Sci. 2016;113:E4639–E4647. doi: 10.1073/pnas.1608644113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lu Y, Lee B, King RW, Finley D, Kirschner MW. Substrate degradation by the proteasome: a single-molecule kinetic analysis. Science. 2015;348:1250834. doi: 10.1126/science.1250834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schaefer JB, Morgan DO. Protein-linked ubiquitin chain structure restricts activity of deubiquitinating enzymes. J Biol Chem. 2011;286:45186–96. doi: 10.1074/jbc.M111.310094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wilkinson KD, Tashayev VL, O’Connor LB, Larsen CN, Kasperek E, Pickart CM. Metabolism of the polyubiquitin degradation signal: structure, mechanism, and role of isopeptidase T. Biochemistry. 1995;34:14535–46. doi: 10.1021/bi00044a032. [DOI] [PubMed] [Google Scholar]
- 87.Lee C, Prakash S, Matouschek A. Concurrent translocation of multiple polypeptide chains through the proteasomal degradation channel. J Biol Chem. 2002;277:34760–5. doi: 10.1074/jbc.M204750200. [DOI] [PubMed] [Google Scholar]
- 88.Jacobson AD, Zhang NY, Xu P, Han KJ, Noone S, Peng J, Liu CW. The Lysine 48 and Lysine 63 Ubiquitin Conjugates Are Processed Differently by the 26 S Proteasome. J Biol Chem. 2009;284:35485–35494. doi: 10.1074/jbc.M109.052928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mansour W, Nakasone MA, von Delbrück M, Yu Z, Krutauz D, Reis N, Kleifeld O, Sommer T, Fushman D, Glickman MH. Disassembly of Lys 11 and Mixed Linkage Polyubiquitin Conjugates Provides Insights into Function of Proteasomal Deubiquitinases Rpn11 and Ubp6. J Biol Chem. 2015;290:4688–4704. doi: 10.1074/jbc.M114.568295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Amerik AY, Li SJ, Hochstrasser M. Analysis of the Deubiquitinating Enzymes of the Yeast Saccharomyces cerevisiae. Biol Chem. 2000;381:981–92. doi: 10.1515/BC.2000.121. [DOI] [PubMed] [Google Scholar]
- 91.Anderson C, Crimmins S, Wilson JA, Korbel GA, Ploegh HL, Wilson SM. Loss of Usp14 results in reduced levels of ubiquitin in ataxia mice. J Neurochem. 2005;95:724–31. doi: 10.1111/j.1471-4159.2005.03409.x. [DOI] [PubMed] [Google Scholar]
- 92.Crimmins S, Jin Y, Wheeler C, Huffman AK, Chapman C, Dobrunz LE, Levey A, Roth KA, Wilson JA, Wilson SM. Transgenic rescue of ataxia mice with neuronal-specific expression of ubiquitin-specific protease 14. J Neurosci. 2006;26:11423–31. doi: 10.1523/JNEUROSCI.3600-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Marshall AG, Watson JA, Hallengren JJ, Walters BJ, Dobrunz LE, Francillon L, Wilson JA, Phillips SE, Wilson SM. Genetic background alters the severity and onset of neuromuscular disease caused by the loss of ubiquitin-specific protease 14 (usp14) PLoS One. 2013;8:e84042. doi: 10.1371/journal.pone.0084042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kirkpatrick DS, Hathaway NA, Hanna J, Elsasser S, Rush J, Finley D, King RW, Gygi SP. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol. 2006;8:700–710. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- 95.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lam YA, Xu W, DeMartino GN, Cohen RE. Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature. 1997;385:737–740. doi: 10.1038/385737a0. [DOI] [PubMed] [Google Scholar]
- 97.Lam YA, DeMartino GN, Pickart CM, Cohen RE. Specificity of the ubiquitin isopeptidase in the PA700 regulatory complex of 26 S proteasomes. J Biol Chem. 1997;272:28438–46. doi: 10.1074/jbc.272.45.28438. [DOI] [PubMed] [Google Scholar]
- 98.Stone M, Hartmann-Petersen R, Seeger M, Bech-Otschir D, Wallace M, Gordon C. Uch2/Uch37 is the major deubiquitinating enzyme associated with the 26S proteasome in fission yeast. J Mol Biol. 2004;344:697–706. doi: 10.1016/j.jmb.2004.09.057. [DOI] [PubMed] [Google Scholar]
- 99.Lu X, Nowicka U, Sridharan V, Liu F, Randles L, Hymel D, Dyba M, Tarasov SG, Tarasova NI, Zhao XZ, Hamazaki J, Murata S, Burke TR, Walters KJ. Structure of the Rpn13-Rpn2 complex provides insights for Rpn13 and Uch37 as anticancer targets. Nat Commun. 2017;8:15540. doi: 10.1038/ncomms15540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhou ZR, Zhang YH, Liu S, Song AX, Hu HY. Length of the active-site crossover loop defines the substrate specificity of ubiquitin C-terminal hydrolases for ubiquitin chains. Biochem J. 2012;441:143–9. doi: 10.1042/BJ20110699. [DOI] [PubMed] [Google Scholar]
- 101.Larsen CN, Krantz BA, Wilkinson KD. Substrate Specificity of Deubiquitinating Enzymes: Ubiquitin C-Terminal Hydrolases. Biochemistry. 1998;37:3358–3368. doi: 10.1021/bi972274d. [DOI] [PubMed] [Google Scholar]
- 102.Homma T, Ishibashi D, Nakagaki T, Fuse T, Mori T, Satoh K, Atarashi R, Nishida N. Ubiquitin-specific protease 14 modulates degradation of cellular prion protein. Sci Rep. 2015;5:11028. doi: 10.1038/srep11028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.McKinnon C, Goold R, Andre R, Devoy A, Ortega Z, Moonga J, Linehan JM, Brandner S, Lucas JJ, Collinge J, Tabrizi SJ. Prion-mediated neurodegeneration is associated with early impairment of the ubiquitin-proteasome system. Acta Neuropathol. 2016;131:411–25. doi: 10.1007/s00401-015-1508-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nakashima A, Ohnuma S, Kodani Y, Kaneko YS, Nagasaki H, Nagatsu T, Ota A. Inhibition of deubiquitinating activity of USP14 decreases tyrosine hydroxylase phosphorylated at Ser19 in PC12D cells. Biochem Biophys Res Commun. 2016;472:598–602. doi: 10.1016/j.bbrc.2016.03.022. [DOI] [PubMed] [Google Scholar]
- 105.Liao Y, Liu N, Hua X, Cai J, Xia X, Wang X, Huang H, Liu J. Proteasome-associated deubiquitinase ubiquitin-specific protease 14 regulates prostate cancer proliferation by deubiquitinating and stabilizing androgen receptor. Cell Death Dis. 2017;8:e2585. doi: 10.1038/cddis.2016.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhu Y, Zhang Y, Sui Z, Zhang Y, Liu M, Tang H. USP14 de-ubiquitinates vimentin and miR-320a modulates USP14 and vimentin to contribute to malignancy in gastric cancer cells. Oncotarget. 2016;8:48725–48736. doi: 10.18632/oncotarget.10706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sareen-Khanna K, Papillon J, Wing SS, Cybulsky AV. Role of the deubiquitinating enzyme ubiquitin-specific protease-14 in proteostasis in renal cells. Am J Physiol Renal Physiol. 2016;311:F1035–F1046. doi: 10.1152/ajprenal.00252.2016. [DOI] [PubMed] [Google Scholar]
- 108.Chen M, Meng Q, Qin Y, Liang P, Tan P, He L, Zhou Y, Chen Y, Huang J, Wang RF, Cui J. TRIM14 Inhibits cGAS Degradation Mediated by Selective Autophagy Receptor p62 to Promote Innate Immune Responses. Mol Cell. 2016;64:105–119. doi: 10.1016/j.molcel.2016.08.025. [DOI] [PubMed] [Google Scholar]
- 109.Xu D, Shan B, Sun H, Xiao J, Zhu K, Xie X, Li X, Liang W, Lu X, Qian L, Yuan J. USP14 regulates autophagy by suppressing K63 ubiquitination of Beclin 1. Genes Dev. 2016;30:1718–30. doi: 10.1101/gad.285122.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Boselli M, Lee B-H, Robert J, Prado MA, Min S, Cheng C, Silva MC, Seong C, Elsasser S, Hatle KM, Gahman TC, Gygi SP, Haggerty SJ, Gan L, King RW, Finley D. An inhibitor of the proteasomal deubiquitinating enzyme USP14 induces elimination of phosphorylated tau in cultured neurons. J Biol Chem. 2017 doi: 10.1074/jbc.M117.815126. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Song C, Ma R, Yang X, Pang S. The Deubiquitinating Enzyme USP14 Regulates Leukemic Chemotherapy Drugs-Induced Cell Apoptosis by Suppressing Ubiquitination of Aurora Kinase B. Cell Physiol Biochem. 2017;42:965–973. doi: 10.1159/000478679. [DOI] [PubMed] [Google Scholar]
- 112.Al-Shami A, Jhaver KG, Vogel P, Wilkins C, Humphries J, Davis JJ, Xu N, Potter DG, Gerhardt B, Mullinax R, Shirley CR, Anderson SJ, Oravecz T. Regulators of the proteasome pathway, Uch37 and Rpn13, play distinct roles in mouse development. PLoS One. 2010;5:e13654. doi: 10.1371/journal.pone.0013654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Crimmins S, Sutovsky M, Chen PC, Huffman A, Wheeler C, Swing DA, Roth K, Wilson J, Sutovsky P, Wilson S. Transgenic rescue of ataxia mice reveals a male-specific sterility defect. Dev Biol. 2009;325:33–42. doi: 10.1016/j.ydbio.2008.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.http://www.mousephenotype.org/data/genes/MGI:1913284, (n.d.).
- 115.Gallery M, Blank JL, Lin Y, Gutierrez JA, Pulido JC, Rappoli D, Badola S, Rolfe M, Macbeth KJ. The JAMM motif of human deubiquitinase Poh1 is essential for cell viability. Mol Cancer Ther. 2007;6:262–8. doi: 10.1158/1535-7163.MCT-06-0542. [DOI] [PubMed] [Google Scholar]
- 116.Lundgren J, Masson P, Realini CA, Young P. Use of RNA interference and complementation to study the function of the Drosophila and human 26S proteasome subunit S13. Mol Cell Biol. 2003;23:5320–30. doi: 10.1128/MCB.23.15.5320-5330.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Maytal-Kivity V, Reis N, Hofmann K, Glickman MH. MPN+, a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function. BMC Biochem. 2002;3:28. doi: 10.1186/1471-2091-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Guterman A, Glickman MH. Complementary roles for Rpn11 and Ubp6 in deubiquitination and proteolysis by the proteasome. J Biol Chem. 2004;279:1729–38. doi: 10.1074/jbc.M307050200. [DOI] [PubMed] [Google Scholar]
- 119.Byrne A, McLaren RP, Mason P, Chai L, Dufault MR, Huang Y, Liang B, Gans JD, Zhang M, Carter K, Gladysheva TB, Teicher BA, Biemann HPN, Booker M, Goldberg MA, Klinger KW, Lillie J, Madden SL, Jiang Y. Knockdown of human deubiquitinase PSMD14 induces cell cycle arrest and senescence. Exp Cell Res. 2010;316:258–71. doi: 10.1016/j.yexcr.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 120.Butler LR, Densham RM, Jia J, Garvin AJ, Stone HR, Shah V, Weekes D, Festy F, Beesley J, Morris JR. The proteasomal de-ubiquitinating enzyme POH1 promotes the double-strand DNA break response. EMBO J. 2012;31:3918–34. doi: 10.1038/emboj.2012.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang B, Ma A, Zhang L, Jin WL, Qian Y, Xu G, Qiu B, Yang Z, Liu Y, Xia Q, Liu Y. POH1 deubiquitylates and stabilizes E2F1 to promote tumour formation. Nat Commun. 2015;6:8704. doi: 10.1038/ncomms9704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nabhan JF, Ribeiro P. The 19 S Proteasomal Subunit POH1 Contributes to the Regulation of c-Jun Ubiquitination, Stability, and Subcellular Localization. J Biol Chem. 2006;281:16099–16107. doi: 10.1074/jbc.M512086200. [DOI] [PubMed] [Google Scholar]
- 123.Liu H, Buus R, Clague MJ, Urbé S. Regulation of ErbB2 receptor status by the proteasomal DUB POH1. PLoS One. 2009;4:e5544. doi: 10.1371/journal.pone.0005544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Buckley SM, Aranda-Orgilles B, Strikoudis A, Apostolou E, Loizou E, Moran-Crusio K, Farnsworth CL, Koller AA, Dasgupta R, Silva JC, Stadtfeld M, Hochedlinger K, Chen EI, Aifantis I. Regulation of Pluripotency and Cellular Reprogramming by the Ubiquitin-Proteasome System. Cell Stem Cell. 2012;11:783–798. doi: 10.1016/j.stem.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Li J, Yakushi T, Parlati F, Mackinnon AL, Perez C, Ma Y, Carter KP, Colayco S, Magnuson G, Brown B, Nguyen K, Vasile S, Suyama E, Smith LH, Sergienko E, Pinkerton AB, Chung TDY, Palmer AE, Pass I, Hess S, Cohen SM, Deshaies RJ. Capzimin is a potent and specific inhibitor of proteasome isopeptidase Rpn11. Nat Chem Biol. 2017;13:486–493. doi: 10.1038/nchembio.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Perez C, Li J, Parlati F, Rouffet M, Ma Y, Mackinnon AL, Chou TF, Deshaies RJ, Cohen SM. Discovery of an Inhibitor of the Proteasome Subunit Rpn11. J Med Chem. 2017;60:1343–1361. doi: 10.1021/acs.jmedchem.6b01379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Walters BJ, Campbell SL, Chen PC, Taylor AP, Schroeder DG, Dobrunz LE, Artavanis-Tsakonas K, Ploegh HL, Wilson JA, Cox GA, Wilson SM. Differential effects of Usp14 and Uch-L1 on the ubiquitin proteasome system and synaptic activity. Mol Cell Neurosci. 2008;39:539–548. doi: 10.1016/j.mcn.2008.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sakata E, Stengel F, Fukunaga K, Zhou M, Saeki Y, Förster F, Baumeister W, Tanaka K, Robinson CV. The catalytic activity of Ubp6 enhances maturation of the proteasomal regulatory particle. Mol Cell. 2011;42:637–49. doi: 10.1016/j.molcel.2011.04.021. [DOI] [PubMed] [Google Scholar]
- 129.Bhattacharyya BJ, Wilson SM, Jung H, Miller RJ. Altered neurotransmitter release machinery in mice deficient for the deubiquitinating enzyme Usp14. Am J Physiol Cell Physiol. 2012;302:C698–708. doi: 10.1152/ajpcell.00326.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chen PC, Bhattacharyya BJ, Hanna J, Minkel H, Wilson JA, Finley D, Miller RJ, Wilson SM. Ubiquitin Homeostasis Is Critical for Synaptic Development and Function. J Neurosci. 2011;31:17505–17513. doi: 10.1523/JNEUROSCI.2922-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Vaden JH, Bhattacharyya BJ, Chen PC, Watson JA, Marshall AG, Phillips SE, Wilson JA, King GD, Miller RJ, Wilson SM. Ubiquitin-specific protease 14 regulates c-Jun N-terminal kinase signaling at the neuromuscular junction. Mol Neurodegener. 2015;10:3. doi: 10.1186/1750-1326-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Jung H, Kim BG, Han WH, Lee JH, Cho JY, Park WS, Maurice MM, Han JK, Lee MJ, Finley D, Jho EH. Deubiquitination of Dishevelled by Usp14 is required for Wnt signaling. Oncogenesis. 2013;2:e64. doi: 10.1038/oncsis.2013.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mines MA, Goodwin JS, Limbird LE, Cui FF, Fan GH. Deubiquitination of CXCR4 by USP14 is critical for both CXCL12-induced CXCR4 degradation and chemotaxis but not ERK ativation. J Biol Chem. 2009;284:5742–52. doi: 10.1074/jbc.M808507200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ponnappan S, Palmieri M, Sullivan DH, Ponnappan U. Compensatory increase in USP14 activity accompanies impaired proteasomal proteolysis during aging. Mech Ageing Dev. 2013;134:53–59. doi: 10.1016/j.mad.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Torres EM, Dephoure N, Panneerselvam A, Tucker CM, Whittaker CA, Gygi SP, Dunham MJ, Amon A. Identification of aneuploidy-tolerating mutations. Cell. 2010;143:71–83. doi: 10.1016/j.cell.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Dephoure N, Hwang S, O’Sullivan C, Dodgson SE, Gygi SP, Amon A, Torres EM. Quantitative proteomic analysis reveals posttranslational responses to aneuploidy in yeast. Elife. 2014;3:e03023. doi: 10.7554/eLife.03023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Oromendia AB, Dodgson SE, Amon A. Aneuploidy causes proteotoxic stress in yeast. Genes Dev. 2012;26:2696–708. doi: 10.1101/gad.207407.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Jacobson AD, MacFadden A, Wu Z, Peng J, Liu CW. Autoregulation of the 26S proteasome by in situ ubiquitination. Mol Biol Cell. 2014;25:1824–1835. doi: 10.1091/mbc.E13-10-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wilson SM, Bhattacharyya B, Rachel RA, Coppola V, Tessarollo L, Householder DB, Fletcher CF, Miller RJ, Copeland NG, Jenkins NA. Synaptic defects in ataxia mice result from a mutation in Usp14, encoding a ubiquitin-specific protease. Nat Genet. 2002;32:420–5. doi: 10.1038/ng1006. [DOI] [PubMed] [Google Scholar]
- 140.Chen PC, Qin LN, Li XM, Walters BJ, Wilson JA, Mei L, Wilson SM. The proteasome-associated deubiquitinating enzyme Usp14 is essential for the maintenance of synaptic ubiquitin levels and the development of neuromuscular junctions. J Neurosci. 2009;29:10909–19. doi: 10.1523/JNEUROSCI.2635-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Randles L, Anchoori RK, Roden RBS, Walters KJ. The Proteasome Ubiquitin Receptor hRpn13 and Its Interacting Deubiquitinating Enzyme Uch37 Are Required for Proper Cell Cycle Progression. J Biol Chem. 2016;291:8773–83. doi: 10.1074/jbc.M115.694588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wicks SJ, Grocott T, Haros K, Maillard M, ten Dijke P, Chantry A. Reversible ubiquitination regulates the Smad/TGF-beta signalling pathway. Biochem Soc Trans. 2006;34:761–3. doi: 10.1042/BST0340761. [DOI] [PubMed] [Google Scholar]
- 143.Wicks SJ, Haros K, Maillard M, Song L, Cohen RE, Ten Dijke P, Chantry A. The deubiquitinating enzyme UCH37 interacts with Smads and regulates TGF-beta signalling. Oncogene. 2005;24:8080–4. doi: 10.1038/sj.onc.1208944. [DOI] [PubMed] [Google Scholar]
- 144.Cutts AJ, Soond SM, Powell S, Chantry A. Early phase TGFβ receptor signalling dynamics stabilised by the deubiquitinase UCH37 promotes cell migratory responses. Int J Biochem Cell Biol. 2011;43:604–612. doi: 10.1016/j.biocel.2010.12.018. [DOI] [PubMed] [Google Scholar]
- 145.Nan L, Jacko AM, Tan J, Wang D, Zhao J, Kass DJ, Ma H, Zhao Y. Ubiquitin carboxyl-terminal hydrolase-L5 promotes TGFβ-1 signaling by de-ubiquitinating and stabilizing Smad2/Smad3 in pulmonary fibrosis. Sci Rep. 2016;6:33116. doi: 10.1038/srep33116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Han W, Lee H, Han JK. Ubiquitin C-terminal hydrolase37 regulates Tcf7 DNA binding for the activation of Wnt signalling. Sci Rep. 2017;7:42590. doi: 10.1038/srep42590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Matilainen O, Arpalahti L, Rantanen V, Hautaniemi S, Holmberg CI. Insulin/IGF-1 signaling regulates proteasome activity through the deubiquitinating enzyme UBH-4. Cell Rep. 2013;3:1980–95. doi: 10.1016/j.celrep.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 148.Fang Y, Fu D, Tang W, Cai Y, Ma D, Wang H, Xue R, Liu T, Huang X, Dong L, Wu H, Shen X. Ubiquitin C-terminal Hydrolase 37, a novel predictor for hepatocellular carcinoma recurrence, promotes cell migration and invasion via interacting and deubiquitinating PRP19. Biochim Biophys Acta. 2013;1833:559–72. doi: 10.1016/j.bbamcr.2012.11.020. [DOI] [PubMed] [Google Scholar]
- 149.Nishi R, Wijnhoven P, le Sage C, Tjeertes J, Galanty Y, Forment JV, Clague MJ, Urbé S, Jackson SP. Systematic characterization of deubiquitylating enzymes for roles in maintaining genome integrity. Nat Cell Biol. 2014;16:1016–1026. doi: 10.1038/ncb3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Chen Y, Fu D, Xi J, Ji Z, Liu T, Ma Y, Zhao Y, Dong L, Wang Q, Shen X. Expression and clinical significance of UCH37 in human esophageal squamous cell carcinoma. Dig Dis Sci. 2012;57:2310–7. doi: 10.1007/s10620-012-2181-9. [DOI] [PubMed] [Google Scholar]
- 151.Wang L, Chen YJ, Xu K, Wang YY, Shen XZ, Tu RQ. High expression of UCH37 is significantly associated with poor prognosis in human epithelial ovarian cancer. Tumour Biol. 2014;35:11427–33. doi: 10.1007/s13277-014-2446-3. [DOI] [PubMed] [Google Scholar]
- 152.Tian Z, D’Arcy P, Wang X, Ray A, Tai YT, Hu Y, Carrasco RD, Richardson P, Linder S, Chauhan D, Anderson KC. A novel small molecule inhibitor of deubiquitylating enzyme USP14 and UCHL5 induces apoptosis in multiple myeloma and overcomes bortezomib resistance. Blood. 2014;123:706–16. doi: 10.1182/blood-2013-05-500033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Rolén U, Kobzeva V, Gasparjan N, Ovaa H, Winberg G, Kisseljov F, Masucci MG. Activity profiling of deubiquitinating enzymes in cervical carcinoma biopsies and cell lines. Mol Carcinog. 2006;45:260–269. doi: 10.1002/mc.20177. [DOI] [PubMed] [Google Scholar]
- 154.Chen YJ, Ma YS, Fang Y, Wang Y, Fu D, Shen XZ. Power and promise of ubiquitin carboxyl-terminal hydrolase 37 as a target of cancer therapy. Asian Pac J Cancer Prev. 2013;14:2173–9. doi: 10.7314/apjcp.2013.14.4.2173. [DOI] [PubMed] [Google Scholar]
- 155.Beck F, Unverdorben P, Bohn S, Schweitzer A, Pfeifer G, Sakata E, Nickell S, Plitzko JM, Villa E, Baumeister W, Forster F. Near-atomic resolution structural model of the yeast 26S proteasome. Proc Natl Acad Sci. 2012;109:14870–14875. doi: 10.1073/pnas.1213333109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Sakata E, Bohn S, Mihalache O, Kiss P, Beck F, Nagy I, Nickell S, Tanaka K, Saeki Y, Förster F, Baumeister W. Localization of the proteasomal ubiquitin receptors Rpn10 and Rpn13 by electron cryomicroscopy. Proc Natl Acad Sci U S A. 2012;109:1479–84. doi: 10.1073/pnas.1119394109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ho BK, Gruswitz F. HOLLOW: Generating Accurate Representations of Channel and Interior Surfaces in Molecular Structures. BMC Struct Biol. 2008;8:49. doi: 10.1186/1472-6807-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Cook WJ, Jeffrey LC, Kasperek E, Pickart CM. Structure of tetraubiquitin shows how multiubiquitin chains can be formed. J Mol Biol. 1994;236:601–9. doi: 10.1006/jmbi.1994.1169. [DOI] [PubMed] [Google Scholar]