Abstract
The eukaryotic 26S proteasome is a large multi-subunit complex that degrades the majority of proteins in the cell under normal conditions. The 26S proteasome can be divided into two subcomplexes: the 19S regulatory particle (RP) and the 20S core particle (CP). Most substrates are first covalently modified by ubiquitin, which then directs them to the proteasome. The function of the RP is to recognize, unfold, deubiquitylate and translocate substrates into the CP, which contains the proteolytic sites of the proteasome. Given the abundance and subunit complexity of the proteasome, the assembly of this ~2.5 MDa complex must be carefully orchestrated to ensure its correct formation. In recent years, significant advances have been made in the understanding of proteasome assembly, structure and function. Technical advances in cryo-electron microscopy have resulted in a series of atomic cryo-EM structures of both human and yeast 26S proteasomes. These structures have illuminated new intricacies and dynamics of the proteasome. In this review, we focus on the mechanisms of proteasome assembly, particularly in light of recent structural information.
Graphical abstract
Introduction
Protein degradation is an essential facet of cellular function. In eukaryotes, most regulated protein degradation is performed by the ubiquitin-proteasome system[1–4]. The concerted action of a series of enzymes activate and affix one or more ubiquitin moieties to a target protein[1,2]. Attachment of ubiquitin or ubiquitin polymers to proteins marks them as substrates of the proteasome[1,5–7]. The proteasome is a large, multimeric protease complex that binds, deubiquitylates, and unfolds its substrates prior to completing their degradation[3].
Proteasomes are found in eukaryotes, archaea and some bacteria. For full activity, proteasomal activators associate with one or both ends of the proteasome core particle (CP)[3]. Known activators include the 19S regulatory particle (RP), proteasome activator PA28, and PA200/Blm10; hybrid proteasomes with different activators on either end of the CP cylinder have also been observed[8]. In this review, we will focus on the eukaryotic RP-activated proteasome, i.e., the 26S proteasome (Figure 1). Under certain conditions, the RP can be split into two large subcomplexes, called the base and lid[9]. The base and lid are major assembly intermediates that form en route to full proteasome formation[3]. Functionally, the major role of the lid is to coordinate substrate deubiquitylation with the unfolding and translocation of polypeptide substrates by the base. The base includes a heterohexameric ring of AAA ATPases (Rpt subunits) that is responsible for these latter activities.
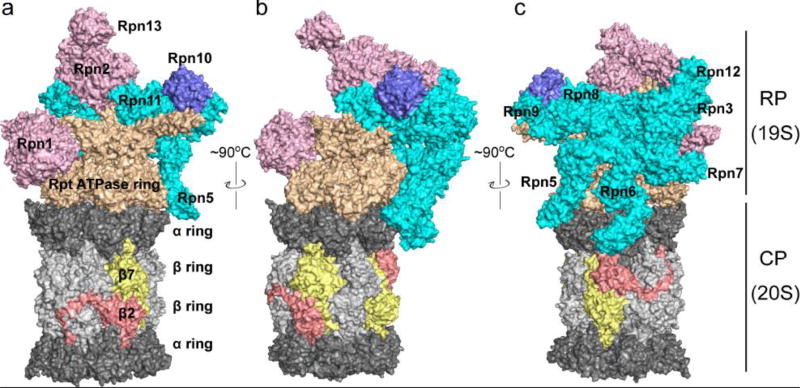
Figure 1.
The structure of the 26S proteasome. The CP is in gray with α rings in dark gray and β rings in light gray. The Rpt ATPase ring subunits of the RP base are colored light brown, while the Rpn1, Rpn2 and Rpn13 base subunits are pink. Individual lid subunits labeled and depicted in cyan, and Rpn10 is blue. The CP subunits β2 and β7 are depicted in copper and yellow, respectively, to highlight their C-terminal tails that extend to neighboring subunits. The positions of individual CP and base subunits are indicated in Figure 2 and Figure 4 due to space limitations in Figure 1. Figures were generated from PDB 3JCP.
Interestingly, both the CP and base require exogenous assembly factors or chaperones for their efficient assembly, but the lid apparently does not[3,10,11]. Five assembly chaperones have been identified for base formation, while three factors are well established as important CP assembly chaperones. In the following sections, we will discuss recent advances in our understanding of the structure of each of these proteasomal complexes, and we will focus principally on their assembly mechanisms.
The 20S Core Particle (CP)
The dyad-symmetric CP is a highly conserved, barrel-shaped complex composed of four stacked heptameric rings[8]. Narrow substrate entry channels are created by the two outer rings, which are each formed by seven α subunits. The two inner rings create an internal chamber that houses the proteolytic active sites responsible for protein cleavage; these rings are each formed by seven β subunits. CP α subunits include highly conserved N-terminal extensions that are absent from β subunits. These N-termini form a gate that controls substrate passage through the central α-ring channel[12]. Archaeal and bacterial CPs usually have a single type of α subunit and β subunit, each present in 14 copies in each particle. Thus, these proteasomes have 14 active sites arrayed within their central chambers.
In eukaryotes, seven distinct α-subunit paralogs form each heptameric outer ring and seven distinct β-subunit paralogs form each inner ring. The N-terminal peptide extension of the eukaryotic α3 subunit is the key contributor to the α-ring gate[12]. This gate is most often closed in the absence of activators[13]. Only three of the seven eukaryotic β subunits (β1, β2 and β5) retain an intact active site, so each eukaryotic CP has six proteolytic active sites. The proteasome is a threonine protease in which the active-site Thr residue is at the N-terminus of the β subunit. These subunits are activated following autocatalytic processing of an N-terminal propeptide[14,15]. Recent biochemical and structural analyses indicate that propeptide autocleavage and substrate proteolysis utilize closely related mechanisms[16]. The Thr residue is part of a conserved Thr-Lys-Asp catalytic triad, which functions similarly in both processes[16].
Overview of CP Assembly
CP biogenesis in eukaryotes is assisted by many factors to help ensure proper assembly. These factors include the intrinsic self-assembly properties of subunits, the N- and C-terminal extensions of specific subunits, and dedicated assembly chaperones[8]. Assembly is less complex for the simpler prokaryotic proteasomes, and only some assembly features are shared among bacterial, archaeal and eukaryotic CPs. We will discuss these in turn.
Bacterial CP Assembly
Proteasomes are found in the bacterial orders Actinomycetales and Nitrospirales[17]. However, a very recent bioinformatic analysis has uncovered proteasome-related genes in additional bacteria as well[18]. Proteasome assembly has been examined only in Actinobacteria to date. Based on in vitro analysis, actinobacterial CP assembly does not require prior formation of an α ring (Figure 2a(i)). Instead, individual α and β subunits form α-β heterodimers, which oligomerize to form a double-ring half-proteasome, followed by dimerization of half-mers[19–21]. Upon dimerization, the N-terminal propeptides of the β-subunits are autocatalytically removed and the N-terminal catalytic threonine nucleophiles are activated. Bacterial α or β subunits alone cannot self-assemble into homoheptameric ring structures due to the limited α-α and β-β contact surfaces between subunits[21,22]. Instead, the N-terminal propeptides of β subunits make significant contact with α subunits and augment α-α contacts, facilitating α-β heterodimer multimerization during half-proteasome formation[19–21]. In some species, such as Mycobacterium tuberculosis, the β propeptides protrude from the β face of the half-proteasome and inhibit the completion of assembly[19]. To date, no proteasome assembly chaperones have been identified in bacteria.
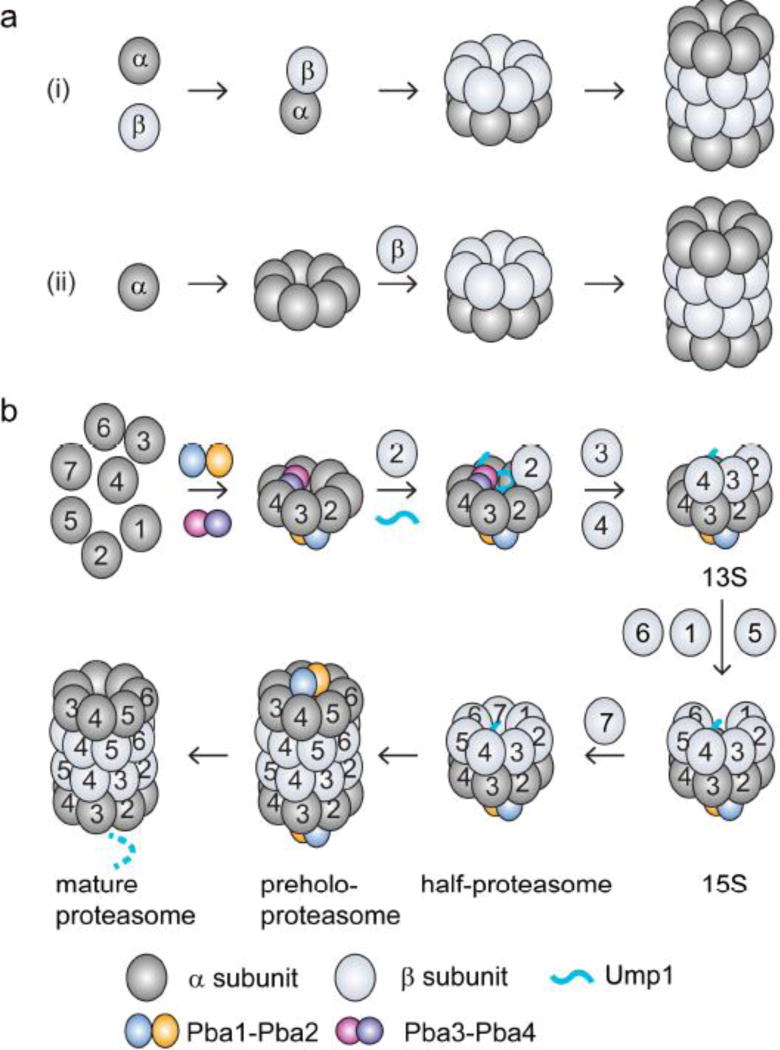
Figure 2.
Models for CP assembly pathways. (a) Two proposed assembly pathways are the (i) α-ring-independent (or α-β heterodimer-initiated) pathway and (ii) α-ring-dependent pathway. (b) Assembly pathway of the constitutive CP in eukaryotes. The α-ring is assembled with the help of two pairs of chaperone proteins: Pba3–Pba4 and Pba1–Pba2. β2 and Ump1 then associate with the α-ring. The association of β3 and β4 is accompanied by the dissociation of Pba3–Pba4. Then β5, β6 and β1 are incorporated sequentially, followed by β7. The insertion of β7 leads to dimerization of two half-proteasomes to form the preholoproteasome. In this complex, Pba1–Pba2 and Ump1 are still associated. In the last step, Pba1–Pba2 dissociates from the CP, the propeptides of the β subunits undergo autoprocessing to activate the CP, and Ump1 is degraded, forming the mature CP.
Archaeal CP Assembly
Archaeal proteasomes are similar to bacterial proteasomes in that they often contain only one type each of α and β subunits, although some have highly similar variants of the subunit, which likely reflect recent gene duplications. It has been proposed that the archaeal CP assembles in an α ring-dependent manner (Figure 2a(ii)), based on detailed in vitro studies of the assembly of the Thermoplasma acidophilum CP[23]. When T. acidophilum α and β subunits are coexpressed in E. coli, the α-ring appears to form first, serving as a template for the attachment of β subunits, leading to formation of half-proteasomes that dimerize to form an active CP. The short N-terminal propeptides of these archaeal β subunits have no obvious effect on CP assembly in vitro[23]. It is possible that in archaeal cells, the β propeptides prevent N-terminal acetylation and the consequent irreversible inactivation of the catalytic Thr1 residues prior to assembly, as is true for eukaryotic active site-bearing β subunits[24].
The conserved N-terminal helices in archaeal α subunits are required for formation of the α-ring gate[14]. They are also essential for α-ring self-assembly, at least in vitro[14]. Given the compositional simplicity of archaeal proteasomes, it was not anticipated that trans-acting factors would be required for assembly. Nevertheless, proteins related to the Pba1–Pba2 (PAC1–PAC2) proteasome assembly chaperones of eukaryotes (see below) have been found in diverse archaeal species, and evidence that they promote CP assembly has been found from in vitro studies[25].
Both archaeal and bacterial proteasomes usually contain only one type of α and one type of β subunit, and both archaeal and bacterial CPs have similar structures[8]. A lingering question has been why the assembly of archaeal proteasomes seems to depend on initial formation of a full α-ring heptamer, while bacterial proteasome assembly is α ring-independent. This puzzle may have been resolved based on the recent report of a second potential pathway of archaeal proteasome assembly[26]. This pathway is α ring-independent, similar to that of actinobacteria (Figure 2a(i)). For the archaeaon Methanococcus maripaludis, the formation of an α ring is not required for efficient assembly of the CP when both M. maripaludis α and β subunits are coexpressed in E. coli. This conclusion is based on an M. maripaludis α-subunit mutant that cannot form a free α ring but is still able to form heterodimers with coexpressed wild-type M. maripaludis β subunits, leading to formation of functional archaeal CPs[26]. The existence of two archaeal CP assembly pathways suggests that proteasome-containing bacterial species, which are relatively rare, may have acquired proteasome genes from ancient archaea but retained only the α ring-independent assembly pathway[26]. Since proteasome architecture is conserved in all three domains of life, it is possible that CP assembly without prior α-ring formation also occurs in eukaryotes, but this has not yet been documented.
Eukaryotic CP Assembly
Overview
Not surprisingly, eukaryotic CP assembly is more complicated than that in bacteria or archaea since eukaryotes express at least fourteen different paralogous CP subunits[8]. Most evidence to date strongly supports a chaperone-assisted, α ring-dependent assembly mechanism (Figure 2b), similar to the first CP assembly pathway described for archaea (Figure 2a(ii)). Two pairs of heterodimeric chaperone proteins, yeast Pba1–Pba2 (PAC1–PAC2 in humans) and Pba3–Pba4 (human PAC3–PAC4), are thought to promote α-ring assembly, while an additional chaperone protein, Ump1 (hUMP1, POMP or proteassemblin in humans), functions in proper incorporation of β subunits and half-mer dimerization. The order of β-subunit incorporation varies somewhat for different proteasomal isoforms and possibly different species. β7 is always the last subunit to incorporate; its insertion into the β ring completes half-mer formation, which is closely coupled to dimerization of two half-mers. The β subunit precursors in the resulting “preholoproteasome” are then autoprocessed, with the released propeptides being degraded along with Ump1 to yield a mature, active 20S proteasome (CP).
Assembly of the α ring
In eukaryotes, the α ring has been proposed to act as an assembly template based on the observation that both Trypanosoma brucei α5[27] and human α7[28] can self-assemble into homoheptameric rings. Whether either heteroheptameric or simpler α rings are common assembly intermediates in vivo is still not clear. Isolated human α7 subunits can form double heptameric ring structures[28,29], similar to the half-proteasome structure. These would be expected to be dead-end complexes, but the stacked α7 double-ring readily disassembles upon introduction of α6 subunits[29]. This suggests that proteasome assembly may involve disassembly of non-native oligomers. Additionally, the S. cerevisiae α4 subunit can form high molecular weight oligomers in vivo; when examined using site-specific disulfide crosslinking and SDS gel analysis, an α4 laddering pattern is revealed, suggesting that these structures are α4 rings[30]. The same α4 laddering pattern was observed in mammalian cells[31]. Despite the apparent existence of α4 rings in both yeast and mammals, their physiological contribution to proteasome assembly remains unclear.
As noted above, eukaryotic CP assembly is generally thought to be α ring-dependent. Support for this comes from the detection of an α-ring complex in mammalian cell lysates[32]. All seven different α-subunits and the dimeric CP assembly chaperone PAC1–PAC2 are present in this complex. The association of PAC1–PAC2 suggests it may function in assisting α-ring assembly[32–34]. PAC3–PAC4 (Pba3–Pba4) also appears to aid assembly in the early stages of CP formation[31,32,34–37]. The exact sequence of α-subunit additions during formation of an α ring is still unknown.
Assembly chaperone Pba1–Pba2 (PAC1–PAC2)
Human PAC1 and PAC2 form a heterodimer and stabilize each other in vivo[32]. In mammalian cells, knockdown of PAC1, PAC2 or both using siRNA leads to slow cell growth, reduced levels of α rings and increased levels of misassembled α-ring dimers. This suggests that PAC1 and PAC2 help assemble the α ring at least in part by preventing α rings from dimerizing, maintaining the α ring in a state capable of β-subunit incorporation[32]. Like PAC1 and PAC2, yeast Pba1 and Pba2 function together as a heterodimer[33,34]. In contrast to PAC1–PAC2, Pba1–Pba2 is metabolically stable[32]. Deletion of PBA1 and/or PBA2 results in no obvious growth defect; however, either deletion can exacerbate the growth defect observed in certain proteasome mutants[33,34]. Additionally, Pba1–Pba2 was found to be exclusively associated with CP assembly intermediates in vivo, consistent with a role of Pba1–Pba2 in proteasome assembly[33].
Structures of three specific yeast CP assembly intermediates bearing Pba1–Pba2 have been characterized: the “15S intermediate” (Ump1, Pba1–Pba2, all α and all β subunits except β7), the “preholoproteasome” (immature full CP plus Ump1 and Pba1–Pba2)[38], and a reconstituted Pba1–Pba2-CP complex[39] (Figure 3b). The first two structures were derived from negative-stain EM while the third was determined by X-ray crystallography. In the 15S intermediate (Figure 3b top left), Pba1–Pba2 is partially embedded in the central cavity of the α ring, where the diameter of the pore is larger than that of a mature CP. Pba1–Pba2 can be seen interacting with all alpha subunits except α1 and α5 and helping to maintain the integrity of the α ring[38]. The second state of Pba1–Pba2 is shown in the negative-stain EM structure of the preholoproteasome (Figure 3b top right), where β7 has inserted and the two CP half-mers have come together. Pba1–Pba2 is shifted out of the central cavity, and located away from the center towards α5. Compared to the 15S structure, the Pba2-α7 interaction disappears in the preholoproteasome, while Pba1 interacts with the preholoproteasome at the α5/α6 interface. The disappearance and appearance of these interactions suggest that Pba1–Pba2 has distinct roles during different stages of CP assembly.
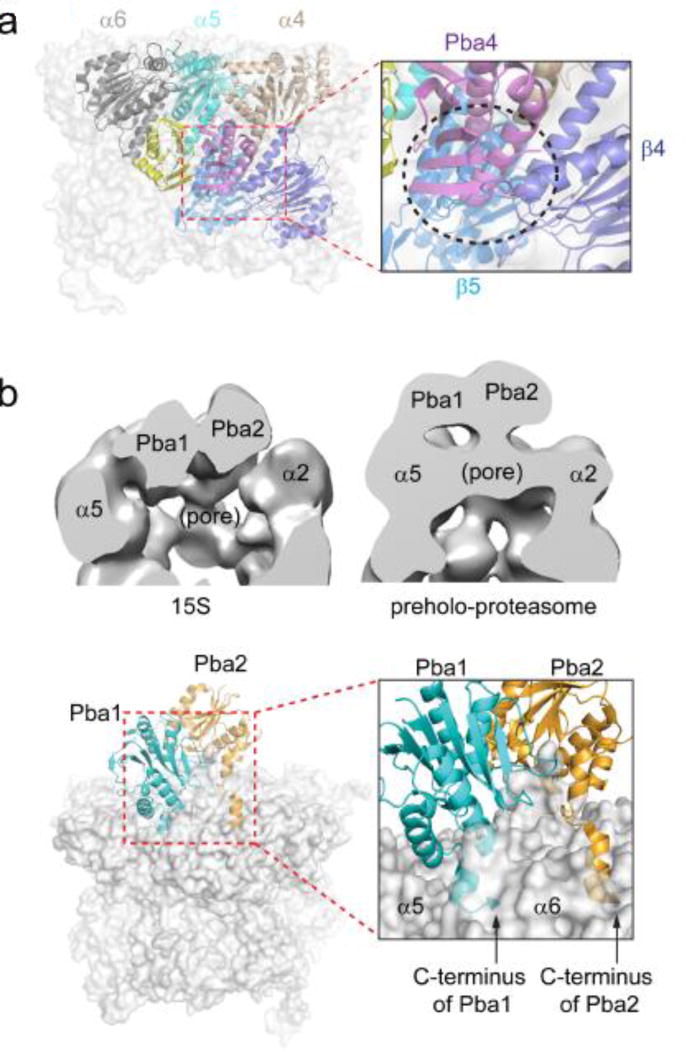
Figure 3.
Two pairs of CP assembly chaperone proteins. (a) Pba3–Pba4 associates with α5 and makes contacts with α4 and α6. The steric clash of Pba3–Pba4 with β4 and β5 is shown. Yeast Pba3–Pba4-α5 X-ray structure was modeled into the full CP structure by superimposing α5 from the two structures. The view is from the axial channel of the CP. α4: light brown; α5: cyan; α6: in dark gray; β4: purple; β5: light blue; Pba3: yellow; Pba4: magenta. The black circle in the inset highlights the steric conflict of Pba3–Pba4 with β4 and β5. Figures were generated from Pba3–Pba4-α5 (PDB ID: 2Z5C) and yeast CP (PDB ID: 5CZ4) structures. (b) Different positions of Pba1–Pba2 during CP assembly. Top left: Negative-stain EM structure of the 15S intermediate. Pba1–Pba2 locates in the widened pore of the α-ring. The figure was generated from EMD-2656 using Chimera. Top right: Negative-stain EM structure of the preholoproteasome. Pba1–Pba2 sits on top of the α-ring. The figure was generated using EMD-2658. Bottom left: X-ray crystal structure of a reconstituted Pba1–Pba2-20S complex. Bottom right: highlight of the insertion of the HbYX motifs of Pba1 and Pba2 into CP α-ring pockets. The C-termini of Pba1 and Pba2 are marked with arrows (PDB ID: 4G4S).
The third complex containing Pba1–Pba2 that was captured was a crystal structure of a reconstituted Pba1–Pba2-CP complex (Figure 3b bottom). It represents a snapshot of the interaction between Pba1–Pba2 and mature CP, presumably when Pba1–Pba2 is poised for release. Pba1–Pba2 has been shown to dissociate easily from mature CP at physiological salt concentrations[39]. Both Pba1 and Pba2 have a C-terminal HbYX (hydrophobic-tyrosine-any amino acid) motif[25], which is found in several proteasome activators and is required for the activators to interact with the α pocket formed by two adjacent α subunits. In this crystal structure, the C-terminus of Pba1 binds to the α5/α6 pocket and the C-terminus of Pba2 to the α6/α7 pocket. Similar to the preholoproteasome, the gate of the α ring is closed in the Pba1–Pba2-CP complex[39].
Pba1–Pba2 binds to the immature CP three orders magnitude more tightly than it does to mature CP[40]. This affinity switch is likely influenced by both the HbYX motifs of Pba1 and Pba2 as well as allosteric communication from β-subunit active sites to the outer surface of the α ring[25,40]. Tight binding of Pba1–Pba2 to CP precursors prevents their association with the RP, potentially ensuring RP-CP interaction only when the CP is properly processed to its mature form.
PbaA and PbaB were identified as archaeal relatives of Pba1 and Pba2, respectively[25]. Since archaeal proteasome subunits can form CPs spontaneously without dedicated chaperones, the physiological roles of these proteins remain to be determined. Studies have shown that PbaA and PbaB have distinctive functions in different species. In certain species, PbaA, but not PbaB, interacts with immature archaeal proteasomes at α pockets using a C-terminal HbYX motif[25]. In other species, PbaB forms a homotetramer and acts as an ATP-independent proteasome activator[41], while PbaA forms a homopentamer that is incapable of binding to the mature CP[42].
Assembly chaperone Pba3–Pba4 (PAC3–PAC4)
Similar to PAC1 and PAC2, human PAC3 and PAC4 form a heterodimer[34]. Knockdown of PAC3 with siRNA in mammalian cells leads to free α-subunit accumulation and proteasome assembly defects, supporting the hypothesis that PAC3 is involved in α-ring assembly[43]. In addition, knockdown of PAC3 leads to accumulation of proteasomal subparticles that lack the α4 subunit; these complexes appear to be aberrant dead-end assembly species[31].
Yeast Pba3 and Pba4 also form a heterodimer and interact most strongly with α5 subunits early in proteasome assembly[35,36]. Deletion of PBA3 and/or PBA4 results in accumulation of polyubiquitinated proteins, decreased proteasome activity, and accumulation of CP assembly intermediates, supporting the role of Pba3–Pba4 as a proteasome assembly chaperone.
In budding yeast, α3 is the only nonessential CP subunit, and lack of α3 leads to formation of an alternative proteasome isoform where a second α4 subunit incorporates at the α3 position. These particles, which are functional, have been called ‘α4-α4’ proteasomes[44]. The ability to form α4-α4 proteasomes appears to be conserved from yeast to humans[31,44]. Overexpression of PAC3 reduces the formation of this isoform, and conversely, PAC3 knockdown enhances its levels[31]. This suggests that PAC3 plays an important role in regulating the assembly of the α4-α4 proteasome. Similarly, in yeast pba3 or pba4 null mutants, ~20 – 50% of CP particles are of the α4-α4 type[35]. In yeast, the absence of the Pba3–Pba4 assembly chaperone also results in the accumulation of a dead-end complex that is composed of seven α subunits plus β2, β3, and β4, but two copies of α2 are present and no α4 is incorporated[36,37]. Thus, Pba3–Pba4 and PAC3–PAC4 likely guide correct positioning of α2, α3 and α4 subunits[35] and prevent off-pathway complex formation[36,37].
Crystal structures of Pba3–Pba4 and a Pba3–Pba4- α5 complex reveal that the chaperone heterodimer structurally resembles individual α and β subunits of the CP[36]. However, the interaction of α5 with Pba3–Pba4 in the ternary complex is different from how α5 binds its CP neighbors α4, α6 and β5. When modeled into a full 20S proteasome structure by aligning α5 in the two complexes, Pba3–Pba4 binds α5 at a position near the axial channel of the α ring (Figure 3a). Pba4 interacts with α4 to promote incorporation of the latter next to α5, explaining why α4 is missing in the dead-end complex in pba4 knockouts[37]. Furthermore, the location of Pba3–Pba4 with respect to α5 would clash sterically with the β4 subunit in the mature CP; therefore, Pba3–Pba4 must dissociate from α5 prior to β4 incorporation during proteasome assembly[37]. This accounts for the absence of Pba3–Pba4 in the “13S intermediate” (all α subunits, β2, β3, β4, Pba1–Pba2 and Ump1)[33], the earliest CP intermediate detected in yeast so far, indicating that Pba3–Pba4 contributes to proteasome assembly before this intermediate forms.
Incorporation of β subunits
Based on single gene knockdowns in cultured mammalian cells, each β subunit appears to be added sequentially to a preformed α ring to form a half-proteasome[45]. Among the seven β-subunits, β2 is the first to be added to the α-ring, followed by β3, β4, β5, β6, β1 and finally β7[45]. N-terminal propeptides and C-terminal tails of several β subunits act as ‘intramolecular chaperones’ to promote the process[45]. A dedicated extrinsic chaperone, Ump1, assists in assembly as well. The contributions of these factors are described below.
As the first subunit to join the α ring, β2 uses its unusually long C-terminal tail to help dictate the directionality and specificity of proteasome assembly. The β2 C-terminal tail, which is essential for viability, wraps around β3 and makes contact with β4[24,45,46] in the mature CP (Figure 1). β3 and β4 are thought to incorporate sequentially after β2. It should be noted, however, that the experiments performed could not distinguish whether single subunits are added or small oligomers; the β2 and β3 subunits, for example, could potentially dimerize prior to associating with the α ring.
The three active site-bearing subunits (β1, β2, and β5) all have N-terminal propeptides that are autocatalytically removed near the end of CP assembly. These propeptides all play some role in subunit processing and assembly of the CP[24]. The most prominent CP intramolecular chaperone is the N-terminal propeptide of β5 (β5pro), which is 75 residues in length in yeast[15] and is essential for proteasome biogenesis in both yeast and humans[15,45]. β5pro appears to function with Ump1 to promote propeptide autocleavage during proteasome maturation, and it facilitates dimerization of two half-proteasomes, similar to the dimerization function of the C-terminal tail of β7[15,47]. A detailed structure-function analysis of β5pro further clarified its contribution to CP assembly[47]. The N-terminal half of β5pro is poorly conserved and is not essential for viability, although substantial defects in proteasome maturation are observed when this half of β5pro is absent. Sequences closer to the cleavage site are more conserved and contribute to proteasome assembly downstream of 13S intermediate formation. Sequences immediately adjacent to the autoprocessing site are not critical for assembly but contribute strongly to autocleavage[47]. In mammalian cells, β5pro is required for β6 incorporation but is not necessary for its own correct placement[45,48]. The N-terminal propeptides of β6, β1, β2 and β7 are not essential for proteasome assembly in yeast[24,33,49]; the same is true for mammalian cells except for the β2 propeptide, which is essential for β3 incorporation[45].
Dimerization of half-proteasomes
Because β7 incorporation and half-mer dimerization are very tightly coupled temporally, it had been unclear which occurred first. The entry point of β7 was clarified by a recent study in yeast[47]. In a strain that retards CP assembly, an intermediate of a size consistent with a half-proteasome was identified and found to contain the unprocessed precursor β7 subunit. The results are most consistent with β7 incorporating to form half proteasomes prior to dimerization rather than dimerization preceding β7 insertion. β7 possesses a unique long C-terminal tail that promotes half-mer dimerization[33,46,47,50]. This tail extends into the interface between β1 and β2 on the opposite β ring and clamps the two half-proteasomes together (Figure 1). Deletion of this region of β7 causes accumulation of proteasome assembly intermediates and active-site autoprecessing defects[33,46]. Based on negative-stain EM structures of 15S and preholoproteasome complexes, the incorporation of β7 appears to cause movements of β subunits into their ultimate positions, which leads to changes in the positioning of α subunits as well[38]. Together, these movements result in the final formation of the gate of the α ring and a shift upwards in the location of Pba1–Pba2[38].
Assembly chaperone Ump1 (hUmp1)
In addition to the aforementioned intra- and intermolecular chaperones, the dedicated CP chaperone Ump1 promotes the precise assembly of β subunits[51]. Ump1 is an intrinsically disordered protein and was the first proteasome chaperone protein discovered[38,51–53]. In yeast, Ump1 acts early in the incorporation of β subunits into the CP. It associates with the CP intermediates along with or shortly after the first β subunits (β2, β3 and β4) are added to the α-ring[33]. In human cells, hUMP1 is essential for the recruitment of β2 to the α ring, a role that is not shared with yeast Ump1[45]. A negative-stain EM structure of the yeast 15S CP assembly intermediate purified via Ump1, together with crosslinking data, give some hints regarding the interactions between Ump1 and the 15S complex[38]. Ump1 loops around the inner chamber of the complex, making contacts with α1, α4, β4 and β6. The N-terminus of Ump1 is located near the interface between β6 and the incoming β7, possibly serving as a detector to sense the arrival of β7. The N-terminal region of Ump1 is expected to be in the vicinity of β5pro in the 15S complex, potentially permitting a direct interaction between these proteins. However, the precise nature of how Ump1 and β5pro interact with one another remains unclear[38].
Alternative CP Isoforms
In mammalian cells, several CP variants have been described in which the active subunits of the “constitutive” 20S proteasome have been replaced by inducible or tissue-specific paralogs. The “immunoproteasome” is the best-studied of these variants[54]. It has three β-subunit substitutions; β1i, β2i and β5i replace β1, β2 and β5, respectively. These alternative subunits are ~60% identical to the subunits they replace. The immunoproteasome alters the population of peptides generated for MHC class I antigen presentation.
Notably, the assembly pathway of such alternative proteasomes differs from that of constitutive CPs. Both β1i and β2i are assembled onto the α ring ahead of other β subunits, initiating β-ring assembly[54]. After incorporation of β3, β5i is incorporated prior to β4, unlike the order of β-subunit addition in constitutive proteasome assembly. The propeptides of the individual immunosubunits help drive their selective incorporation specifically into immunoproteasomes [55,56]. Most immunoproteasomes bear all three βi subunits in each β ring, but some are assembled as mixed proteasomes containing both constitutive β and βi subunits[57]. This implies some flexibility in the order of subunit addition during proteasome assembly. It is also expected to further enhance peptide diversity for antigen presentation.
The thymus-specific proteasome (thymoproteasome) is another alternative CP isoform, assembled exclusively with β1i, β2i and β5t at the expected positions in the β ring even in the presence of β5i[54]. Similar to β5i, β5t is incorporated after β3 incorporation but independently of β4 insertion. Interestingly, the propeptide of β5t is what drives its early incorporation, whereas the mature domain of β5i is responsible for its ability to insert prior to β4 addition[54]. The thymoproteasome plays an essential role in positive selection of immune CD8+ T cells[58].
In addition to the β subunit-substituted proteasomes, there are CPs that contain alternative α subunits. The assembly process of α subunit-substituted proteasomes is not as clear as that of the β subunit-substituted ones. As introduced above, the α4-α4 proteasome is formed under conditions that include deletion of α3, overexpression of α4, deletion of PBA3 (PAC3) or PBA4 (PAC4), or environmental stimuli such as oxidative stress[30,31,35]. The exact assembly pathway of the α4-α4 proteasomes is unclear. One clear difference from constitutive proteasome assembly is that the α4-α4 CP does not require the assembly chaperone Pba3–Pba4 (PAC3–PAC4).
Several other α subunit-substituted proteasomes have been documented. These include spermatoproteasomes, which contain an alternative α4 subunit, α4s, which is synthesized exclusively in male germ cells after their differentiation into spermatocytes[59,60], and testis-specific proteasomes, containing an alternative α6 subunit (α6T), which is essential for fertility in Drosophila[61,62]. Selective assembly of α subunit-substituted alternative CPs must distinguish alternative α subunits from their constitutive counterparts. The mechanisms by which this occurs are still unknown.
Other Factors Assisting CP Assembly
Many other factors may have an impact on CP assembly. Comprehensive proteomic analyses of co- and post-translational modifications of proteasome subunits have revealed a plethora of modifications, both constitutive and transient[63]. At least some of these modifications may modulate proteasome assembly. The only example of this so far is from the archaeon Haloferax volcanii. Specifically, changes in Nα-acetylation of the H. volcanii α1 subunit alter the rate or the level of α-ring assembly[64]. Additional trans-acting factors can also contribute. Proteins in the TRC/GET (transmembrane recognition complex in mammals/guided entry of tail-anchored proteins in yeast) pathway[65] and a zygote-specific proteasome assembly chaperone (ZPAC) in mouse[66] were shown to participate in eukaryotic CP assembly.
Yeast Blm10 (human PA200), a large dome-shape HEAT-repeat protein, has been reported to be associated with CP assembly intermediates[33,50,67,68], suggesting a potential role of Blm10 in modulating CP assembly. However, Blm10 also associates with mature proteasomes and has apparent functions beyond CP assembly[69–73]. Yeast Fub1 (human PI31), a proline-rich protein[74], has been reported to be associated with several CP subunits; loss of Fub1 exacerbates defects caused by mutations in proteasome subunits or assembly chaperones[75,76]. These data suggest a possible role of Fub1 in modulating proteasome assembly. However, human PI31 has no effect on proteasome content and function[77]. A study on the Drosophila melanogaster PI31 did implicate DmPI31 in promoting RP assembly[78]. More studies will be required to establish contributions of Blm10/PA200 and Fub1/PI31 to proteasome assembly. Given the various types of CPs, more proteins that modulate proteasome assembly are likely to exist.
The 19S Regulatory Particle (RP)
The RP appears to be unique to eukaryotes and highly conserved among them; only a modest degree of subunit variability has been documented. Nineteen subunits comprise the basic RP core in yeast, but a wide range of proteasome-interacting proteins (PIPs) are also known[79]. Its two main components, the 9-subunit base and 9-subunit lid, form a conformationally dynamic complex that binds an additional subunit, Rpn10. Rpn10 is one of several ubiquitin receptors in the RP, and it also stabilizes the interaction between lid and base. In the following sections, we review structural aspects of the RP and its assembly.
The RP Base
Base Composition, Structure and Function
The base is composed of six paralogous AAA-ATPases termed Rpt1–Rpt6 and three non-ATPases, Rpn1, Rpn2, and Rpn13. Every Rpt subunit has an N-terminal α helix followed by an oligonucleotide/oligosaccharide-binding (OB) fold, a large AAA-ATPase domain and a C-terminal helical domain[80,81]. The six Rpt subunits associate in three pairs (Rpt1–Rpt2, Rpt3–Rpt6, Rpt4–Rpt5), which arrange into a ring to form a trimer of dimers in the order Rpt1-Rpt2-Rpt6-Rpt3-Rpt4-Rpt5 (Figure 4a). This order was initially determined via disulfide crosslinking[82] and was validated by numerous cryo-EM structures of the 26S proteasome. Each Rpt subunit makes extensive interactions with its Rpt heterodimer partner, but the specificity of these pairwise interactions is believed to be largely dictated by the coiled coils (CCs) formed by their N-terminal helices[80,81].
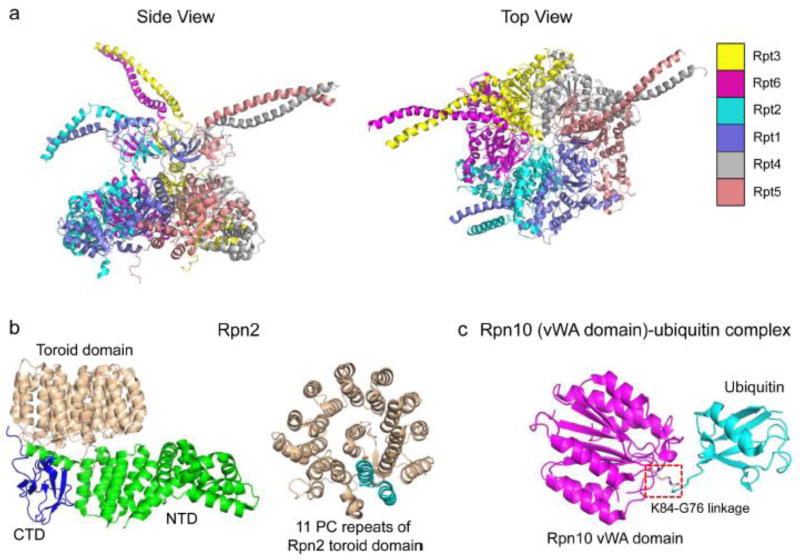
Figure 4.
Structures of the Rpt heterohexameric ring, Rpn2 and an Rpn10-ubiquitin conjugate. (a) Top and side views of the S. cerevisiae Rpt hexameric ring. The side view reveals that the ring arranges into two roughly co-axial rings or spirals. The upper ring of OB-folds follows the coiled-coils, extending upward, that join pairs of ATPases. The lower tier consists of the AAA-ATPase and C-terminal domains. The top view shows the narrow central pore of the Rpt ring through which substrates are threaded (PDB ID: 5MP9). (b) Crystal structure of yeast Rpn2. The structure on the left illustrates the N-terminal, C-terminal and toroidal domains of Rpn2. The structure on the right highlights a single repeat (in cyan) of the eleven PC repeats of the toroidal domain (PDB ID: 4ADY). (c) Crystal structure of yeast Rpn10 vWA domain in complex with ubiquitin. The isopeptide bond formed between Ubiquitin-G76 and Rpn10-K84 residues is boxed (PDB ID: 5LN1).
AAA-ATPase structure and conformational changes
Cryo-EM structures revealed that the Rpt subunit domains arrange in a two-tiered ring configuration (Figure 4a)[83,84]. The upper ring is made from the OB-folds of the Rpt subunits, from which the N-terminal CC domains radiate upward and outward[83,84]. The lower, larger ring is composed of the large ATPase and C-terminal helical domains of these subunits. The CC of Rpt1–Rpt2 interacts with Rpn1 while the CC of Rpt3–Rpt6 contacts Rpn2[83,84]. By contrast, the Rpt4–Rpt5 CC extends significantly from the body of the base without interaction with other proteasome subunits in the basal s1 state (see below)[83]. The three CCs also differ in length, with the Rpt3–Rpt6 CC being the longest, followed by Rpt4–Rpt5 and Rpt1–Rpt2[83,84]. These data all suggest that the CCs have distinct functions within the RP.
The unstructured C-terminal tails of specific Rpt subunits bind to pockets between adjacent α subunits of the CP, and one or more of these interactions help trigger gate opening of the CP, as described earlier. Gate opening allows translocation of unfolded substrates into the proteolytic chamber for degradation[85]. The C-terminal tails of Rpt2, Rpt3, and Rpt5 contain the conserved HbYX motif thought to be important for CP gate opening, although recent cryo-EM data on the 26S proteasome indicate the binding of these three tails is not sufficient for opening[86,87].
Initial sub-nanometer-resolution cryo-EM structures of the proteasome revealed that the Rpt ATPase ring is tilted at an angle of ~10° relative to the CP α-ring surface and toward the lid[88]. Additionally, the ATPase domains of the Rpt subunits in the ring adopt a spiral staircase-like configuration in the order of Rpt3-Rpt4-Rpt5-Rpt1-Rpt2 from top to bottom relative to the CP, with Rpt6 at about the same height as Rpt5 and bridging the vertical displacement between the highest (Rpt3) and lowest (Rpt2) subunits[84,88,89]. When the ring binds to a substrate, the conformation of the ATPase ring transitions to a more planar configuration with alignment of the RP ATPase and CP central channels[90].
The ability to reconstitute the full base by co-expression of yeast subunits in E. coli has enabled the determination of the distinct roles of each Rpt subunit[90]. Mutational analyses of the ATP-hydrolyzing activity of the Rpt subunits has indicated that subunits located at the top of the ring, such as Rpt3 and Rpt4, contribute more to binding and translocation of substrates compared to subunits positioned lower in the ring, such as Rpt1 and Rpt2[90].
Published cryoEM structures of the yeast proteasome also indicate that the base is dynamic and exists in multiple conformational states (see Figure 9 for similar models of the human proteasome). Several of these conformations have been captured using large samples of proteasomes incubated with various nucleotides and nucleotide analogs; the imaged particles were subsequently subjected to deep classification to tease out different conformations. Four distinct conformations have been elucidated thus far: the substrate-accepting (s1), substrate-commitment (s2), substrate-processing (s3), and the recently identified gate-opening of CP (s4) states[87,91]. A major feature of the s1 state is the structural heterogeneity of Rpn1; this is likely related to Rpn1’s role in recruiting various extrinsic ubiquitin receptors or ubiquitinated substrates[91]. Rpn1 in this state is also bound to the CC of Rpt1–Rpt2[91]. The base conformations in s1 and s2 show several subtle differences. In the s2 conformation, the ATPase ring is less tilted than s1 relative to the CP rings[91]. The N-terminal CCs of the Rpt subunits show the most changes between s1 and s2: the Rpt4–Rpt5 CC engages the ubiquitin receptor Rpn10, the Rpt3–Rpt6 CC interacts with Rpn2 and the lid, and according to a recent report, the Rpt1–Rpt2 CC develops a kink[87,91].
Figure 9.
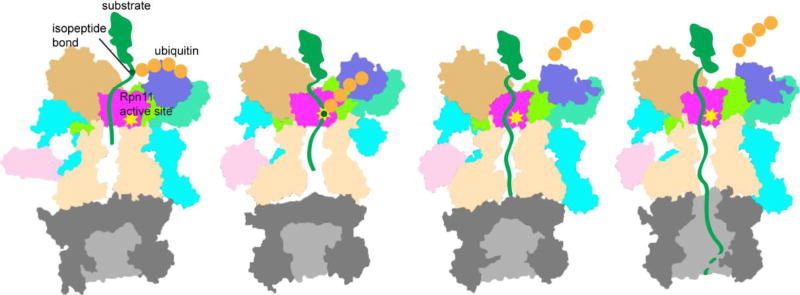
Model of substrate degradation by the proteasome based on cryo-EM structures of the human proteasome[167]. Four conformations of the proteasome, named SA – SD, are depicted. Figures were made based on PDB entries 5T0G, 5T0H, 5T0I and 5T0J using Pymol to show the cutaway surface of each state.
In the s3 conformation, the ATPase ring is further flattened relative to s2 and is virtually planar relative to the CP[91]. This change allows the pore of the ATPase ring to align with the (still occluded) substrate channel of the CP[87,91]. The HbYX motifs of Rpt2, Rpt3, and Rpt5 are bound to α-subunit pockets on the CP in all four conformations, indicating the binding of these HbYX motifs is not sufficient to open the CP gate since gate opening is not observed in s1–s3[87]. In s4, the density of the C-terminal tail of Rpt6 is detected in the pocket between the α3 and α4 subunits, suggesting that Rpt6 tail binding may be necessary for opening of the CP gate, although further investigation is required to confirm this[87].
Non-ATPase subunits of the base and ubiquitin receptors
The base Rpn1 and Rpn2 subunits are the two largest subunits of the proteasome. The central regions of both proteins contain eleven proteasome/cyclosome (PC) repeats[92]. Each repeat is roughly 35–40 residues in length and consists of a pair of antiparallel α helices[92]. A crystal structure of Rpn2 revealed a central domain in which the PC repeats form a toroidal structure (Figure 4b)[92]. This central region is flanked by an N-terminal domain consisting of stacked α helices forming a rod-like structure, and a globular β-sandwich C-terminal domain[92]. Similarly, the PC repeats of Rpn1 have also been predicted to form a toroid-shaped structure[92]. The PC repeats of both Rpn1 and Rpn2 serve as scaffolds, potentially for the assembly of regulatory particle subunits and also to allow docking and facilitate delivery of substrates to the Rpt ring for unfolding and translocation[92].
Both Rpn1 and Rpn2 also recognize ubiquitin-like domains (UBLs) in proteins such as Rad23, Dsk2, and Rpn13[93,94]. Rad23 and Dsk2, which also contain ubiquitin-binding UBA domains, are not constitutive elements of the proteasome, but rather function as extrinsic ubiquitin receptors that bind ubiquitinated substrates and deliver them to the proteasome for degradation via binding to Rpn1[93,94]. Another UBL-UBA protein, Ddi1, has also been identified as an extrinsic ubiquitin receptor[94]. Interestingly, a recent report suggests that Rpn1 itself functions as a ubiquitin receptor[95]. A specific site on the Rpn1 toroid was found to bind both ubiquitin and UBL proteins, specifically Rad23, whereas a different site on the toroid binds to the UBL domain of the deubiquitinase (DUB) Ubp6[95]. Thus, the predicted Rpn1 toroid acts as a multipurpose scaffold that binds not only to substrates directly via ubiquitin binding or indirectly via extrinsic ubiquitin receptors, but also to the DUB Ubp6[95,96].
In addition to binding Rpn1, Ubp6 also interacts with the Rpt1 ATPase subunit, and these interactions stabilize the substrate-committed (s2) conformation of the proteasome. This may enable the engaged substrate to be translocated into the Rpt ring for unfolding [97,98]. These interactions also place Ubp6 in close proximity to the intrinsic proteasomal DUB Rpn11, likely allowing these two DUBs to coordinate substrate deubiquitination [97,98]. The binding of ubiquitin-bound Ubp6 is also thought to prevent interference from additional substrates by ensuring that the proteasome remains in the s2 conformation until the engaged substrate is completely translocated and has its polyubiquitin chains removed[97].
The base subunit Rpn13 is a ubiquitin receptor that binds to substrates via a conserved pleckstrin-like receptor for ubiquitin (PRU) domain in its N-terminal region[99,100]. This domain consists of four antiparallel β strands packed against two antiparallel β strands[99,100]. The structure of human Rpn13 in complex with ubiquitin revealed that Rpn13 binds ubiquitin via residues on loops between the three β strands of the PRU domain[99]. The PRU domain is also responsible for binding Rpn2, whereas a distinct C-terminal domain, not present in yeast Rpn13, binds to the DUB Uch37[100,101]. Rpn13 docks onto the proteasome by interaction with the N-terminal region of Rpn2[101]. When human Rpn13 is not bound to the proteasome, its Uch37-binding domain and ubiquitin-binding domain (UBD) bind one another, rendering the UBD inaccessible[101]. Rpn13 binding to Rpn2 releases this interaction and activates Rpn13, allowing Rpn13 to bind ubiquitinated substrates[101].
Rpn10 was the first identified intrinsic ubiquitin receptor of the proteasome[102,103] It bears an N-terminal von Willebrand type A factor (vWA) domain, which interacts with proteasome subunits, and a C-terminal ubiquitin-interacting motif (UIM) domain[104]. Mutation of the vWA domain promotes dissociation of the lid and base subcomplexes, suggesting a role for Rpn10 is stabilizing their interaction. Interestingly, a hydrophobic patch on the vWA domain has been recently shown also to bind ubiquitin, revealing a second ubiquitin-binding domain on Rpn10, which promotes ubiquitination of the subunit at a specific lysine, Lys84 (Figure 4c)[104]. Monoubiquitination of Rpn10 inhibits its ability to recruit substrates for degradation, therefore decreasing proteasome activity[105]. The modification of Lys84 of the subunit was shown to promote its release from the proteasome[104]. Monoubiquitination of Rpn10 decreases under stress conditions such as heat or cold shock, suggesting that this modification is part of a physiological mechanism to regulate proteasome activity[105].
Overview of Base Assembly
The six eukaryotic Rpt subunits have high sequence and structural homology to one another (Figure 4a). Ordered assembly of the base complex is ensured by a set of dedicated assembly chaperones[11,106–110] (Figure 5). The Nas2, Nas6, Rpn14, and Hsm3 chaperones all interact with the C-terminal helical domains of their respective Rpt subunits (Nas2-Rpt5, Nas6-Rpt3, Rpn14-Rpt6, and Hsm3-Rpt1)[11,106–109]. By contrast, the Adc17 chaperone binds to the N-terminal CC-OB domain of Rpt6[110]. None of these assembly factors is essential for viability, but loss of specific combinations of them is lethal under proteotoxic stress conditions. The details of base assembly are still poorly understood. Assembly may follow multiple routes, which could also vary among species. We will discuss base assembly primarily from analyses done with the yeast model system and compare these results with base assembly in mammalian cells (Figure 6).
Figure 5.
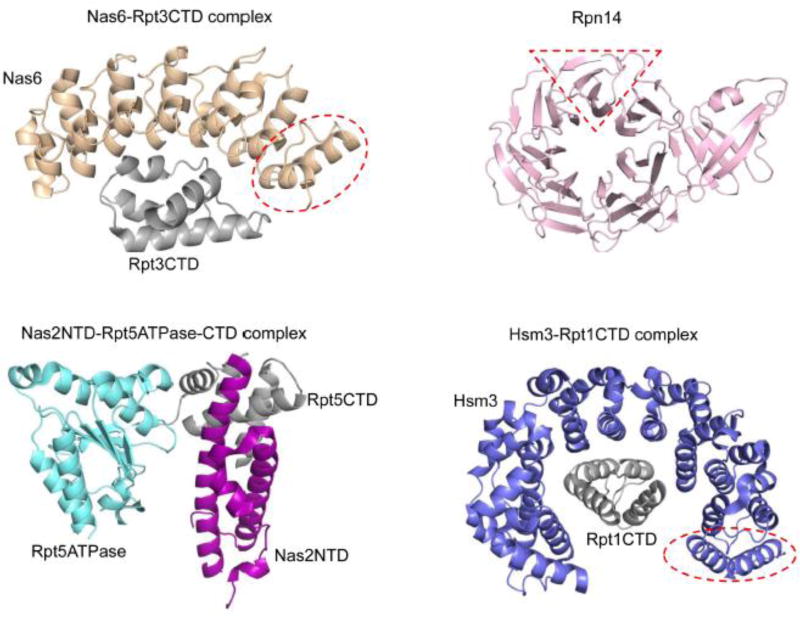
Crystal structures of base assembly chaperones alone or in complex with their respective Rpt partners. All chaperones and Rpt subunits depicted are from S. cerevisiae with the exception of the Rpt5ATPase-CTD. The Rpt5ATPase-CTD construct consists of the ATPase domain from Pyrococcus furiosus PAN (an archaeal homolog of Rpt subunits) followed by the C-terminal peptide from S. cerevisiae Rpt5. Regions highlighted in red on Nas6, Rpn14, and Hsm3 represent a single structural repeat of the chaperones. PDB IDs: 2DZN (Nas6-Rpt3CTD), 3WHL (Nas2NTD-Rpt5ATPase-CTD), 3ACP (Rpn14) and 4A3V (Hsm3-Rpt1CTD).
Figure 6.
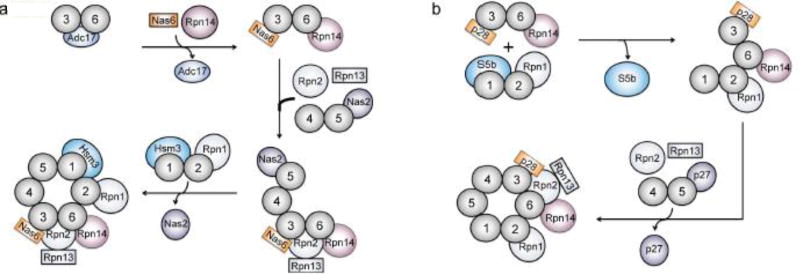
Models of proteasome base assembly in (a) yeast and (b) human. The models are based primarily on characterization of complexes isolated from cells and analyzed by mass spectrometry. The ordering of events is therefore speculative and distinct assembly pathways remain possible. Rpt1–Rpt6 subunits are numbered 1–6 in the figure. Potential differences can be seen in the association order of Rpt dimer complexes in S. cerevisiae and human, but both pathways may occur in both species.
One of the earliest base assembly events is thought to be the formation of Rpt dimer pairs: Rpt1–Rpt2, Rpt3–Rpt6, and Rpt4–Rpt5. In yeast, two assembly chaperones, Adc17 and Hsm3, have been shown to facilitate Rpt dimerization. Adc17 functions to bring together Rpt3 and Rpt6 and likely dissociates early during base assembly since it has not been found in larger subcomplexes[110]. In contrast, Hsm3 provides a scaffold to assist in the formation of the Rpt1–2 heterodimer (Rpn1 is also present in the complex and contacts Hsm3) and only dissociates after formation of the full RP complex but before incorporation into the 26S proteasome[107,111].
It is predicted that the Rpt subunits form subassemblies consisting of the heterodimer ATPase pairs and their respective chaperones en route to formation of the mature base: Hsm3-Rpt1-Rpt2-Rpn1 (Hsm3 module), Nas2-Rpt5-Rpt4 (Nas2 module), and Nas6-Rpt3-Rpt6-Rpn14 (Nas6-Rpn14 module)[11,82,106,107]. The Hsm3 module and Nas2 modules have been detected in yeast, whereas the Nas6-Rpn14 module has only been detected in mammalian cells to date[11,82,106,107]. In yeast, the Nas2 and (presumptive) Nas6-Rpn14 modules are able to associate, forming an intermediate-sized complex with all components of these modules as well as Rpn2 and Rpn13[82,112]. Nas2 must dissociate before or during binding of the Hsm3 module to this intermediate as a complex that contains both Hsm3 and Nas2 has never been observed[82]. Hsm3 has been detected in full base complexes whereas Nas2 has not[82].
Mammalian base assembly might proceed slightly differently from that in yeast. In particular, in mammalian cells, the Hsm3 and Nas6-Rpn14 modules have been observed to form a complex prior to binding of the Nas2 module[11]. Rpn2 and Rpn13 are also thought to bind to the complex containing the Hsm3 and Nas6-Rpn14 modules in mammals. It is still unclear whether Rpn2/Rpn13 or the Nas2 module bind first to this complex[11,106,112]. For both yeast and mammalian cells, most –but not all– data suggest that Rpn10 and the lid associate only with fully formed base complexes, completing assembly of the regulatory particle[11,82,106,107,113].
Potential role of the CP as a template for base assembly
An interesting question that remains in the understanding of base assembly is whether the CP can serve as a ‘template’ for the assembly of base subcomplexes. The CP templating hypothesis was first put forth when it was discovered that base intermediates accumulate if CP assembly is compromised; this was seen in mutants lacking CP assembly chaperones or the α3 CP subunit[35]. In a subsequent study, accumulation of base intermediates, instead of free, fully assembled base or RP, was detected in yeast strains containing mutations in the C-termini of Rpt subunits that specifically bind to the α-ring pockets[114]. These observations suggested that the anchoring of the Rpt tails into the CP α-ring pockets could promote the correct placement of base subunits during RP assembly. Consistent with the CP templating hypothesis, a report showed that Rpt1-α4 and Rpt2-α4 fusions in yeast do not disrupt the formation of full 26S proteasomes[115]. While this suggests the base could potentially assemble on the α-ring surface of the CP, assembly without the subunit fusions might not normally favor this route. To date, a complex of the CP with a partially assembled base complex has not been reported. Such intermediates would be expected if the CP truly acts as a base assembly template. It has been suggested that intermediates of this kind might be difficult to capture due to their unstable nature and rapid association of base modules with the CP template [116].
A CP-independent base-assembly pathway has been inferred primarily from the ability to reconstitute the full yeast base complex in the absence of CP in E. coli[89]. This functional base complex was reconstituted by co-expressing all six Rpt subunits, Rpn1, Rpn2, Rpn13 and four base assembly chaperones Hsm3, Nas2, Nas6 and Rpn14[89]. Although full base complex can be assembled independently of CP in vitro, it does not necessarily rule out a CP-templating mechanism for enhancing base assembly. CP–dependent and –independent assembly pathways are not mutually exclusive. However, further experiments will be required to test the CP template model.
Base Assembly Chaperones
Nas2 (p27 in human)
Nas2 contains a C-terminal PDZ domain that binds to the HbYX motif of Rpt5[106,107,117]. This interaction is thought to prevent premature proteasome association and activation since the Rpt5 HbYX motif contributes to CP association and opening of the CP gate, as discussed earlier[85]. A crystal structure of the N-terminal domain of Nas2 in complex with the C-terminal helical domain of Rpt5 revealed a second function for the chaperone (Figure 5). This region of Rpt5 normally binds Rpt1, its neighbor in the ATPase ring[118]. Nas2 binding would therefore block incorporation of Rpt1 (as part of the Hsm3 module) or any incorrect modules. This might limit assembly of nonnative ATPase hexamers, while Nas2-PDZ binding of the Rpt5 HbYX peptide would block CP association with incorrect base assemblies[118]. These results are fully consistent with earlier biochemical data, which predicted that Nas2 dissociates immediately before or during the binding of the Hsm3 module[118]. It is likely that the correct positioning of Rpt1 next to Rpt5 results in the release of Nas2.
In summary, Nas2 likely has two roles: preventing premature association of base intermediates with the CP and ensuring the proper joining of base assembly modules to form the correctly arranged base subcomplex.
Hsm3 (S5b in human)
Crystal structures of Hsm3 revealed a curved structure that consists of 11 HEAT repeats, where each repeat consists of two anti-parallel α helices (Figure 5)[111,119,120]. The central region of the concave surface of Hsm3 binds to the C-terminal helical domain of Rpt1[111,119,120]. Mutations of hydrophobic residues at this interface are strongly detrimental to binding and cause proteasome assembly defects in vivo[119]. The C-terminal domain of Hsm3 also interacts with the AAA-ATPase domain of Rpt2[111,119]. Hence, Hsm3 is thought to act as a scaffold to bridge Rpt1 and Rpt2[111,119]. Additionally, yeast two-hybrid and in vitro binding analyses showed that Hsm3 weakly interacts with Rpn1, further highlighting the importance of Hsm3 in scaffolding assembly of the Hsm3-Rpt1-Rpt2-Rpn1 module[111].
Nas6 (p28/gankyrin in human)
A crystal structure of Nas6 bound to the C-terminal domain of Rpt3 revealed that Nas6 consists of seven ankyrin repeats; each repeat is formed by ~30 residues arranged in two antiparallel helices followed by a β-hairpin loop (Figure 5)[121]. Similarly to the Hsm3 HEAT repeats, the Nas6 ankyrin repeats create a curved structure whose concave surface binds to the C-terminal α-helical bundle of Rpt3 via extensive van der Waals interactions and complementary charged patches[121].
Interestingly, when Nas6 is bound to Rpt3, it is predicted to clash sterically with the CP and block interaction of the base and CP. This could prevent premature or potentially incorrect docking of the RP complex to the CP[108]. When the free base docks onto the CP, the C-terminal tail of Rpt6 appears to bind to the pocket between the CP α2 and α3 subunits, while Nas6 remains bound to Rpt3. This Rpt6 binding to the CP may then promote the release of Nas6[122]. Specifically, Rpt6 tail-CP binding is thought to promote Rpt3 tail insertion into a CP α-ring pocket, causing a steric clash between Nas6 and the CP and Nas6 dissociation[122]. A recent report showed that Nas6 also prevents association of the base and lid, but specifically when ATP hydrolysis is inhibited, suggesting a switch in Nas6 positioning that may coordinate proper assembly of the full 26S proteasome[116]. A role for Nas6 in lid-base joining had been predicted from in vivo studies[107].
Rpn14 (PAAF1 in human)
A high-resolution crystal structure of Rpn14 confirmed that this protein contains a C-terminal WD40-repeat domain with a seven-bladed β-propeller fold in which each blade consists of four anti-parallel β-strands (Figure 5)[123]. NMR analyses of Rpn14 and Rpt6 revealed that Rpn14 binds with high affinity to a four-helix bundle in the C-terminal domain of Rpt6[124]. NMR analyses of the C-terminal domain of Rpt6 revealed that it assumes two dynamic conformational states: a partially unfolded state and an ordered four-helix bundle[124]. The switch between these states is thought to allow Rpn14 binding and release from the C-terminal domain of Rpt6[124]. Despite the available structural and biochemical data on Rpn14, its exact role in base assembly remains unclear.
Adc17
Adc17 is a recently discovered base assembly chaperone that was identified via a suppressor screen with a yeast rpt6 temperature-sensitive mutant[110]. No sequence homolog of Adc17 in vertebrates has been identified. Notably, Adc17 is the only base proteasome assembly chaperone identified to date that interacts with the N-terminal region of an Rpt subunit[110]. The predicted role of Adc17 in Rpt3–Rpt6 heterodimer formation is supported by in vitro binding assays which revealed that Adc17 binds both Rpt6 alone and in combination with Rpt3, in the latter case forming a ternary protein complex, but it cannot interact with Rpt3 alone[110]. Additionally, mutation of a conserved residue in a predicted Adc17 CC abolished its ability to bind Rpt6[110]. These results suggest that Adc17 first binds Rpt6 and subsequently assists dimerization of Rpt6 with Rpt3[110], potentially facilitating formation of the CC between the ATPase N-terminal helices. Interestingly, Adc17 does not appear to interact with the full Nas6-Rpt3-Rpt6-Rpn14 module; therefore, it probably dissociates shortly after Rpt3–Rpt6 dimerization[110].
The RP Lid
Lid composition
The lid has nine subunits: Rpn3, 5, 6, 7, 8, 9, 11, 12, and 15 (Sem1/DSS1) (Figure 7) and shows significant sequence and structural similarity to both the COP9 signalosome and eIF3[9,125–127]. Each of these complexes contains multiple subunits bearing a PCI (Proteasome, COP9 signalosome, eIF3) fold. In the 26S proteasome, the deubiquitinase activity of the lid Rpn11 subunit cleaves ubiquitin chains from proteasomal substrates. Similarly, the COP9 signalosome removes the UBL Nedd8 from substrate proteins, principally cullins. The eIF3 complex is involved in the initiation of protein translation (a UBL-cleaving activity has not been found). These complexes can even share subunits. In yeast, Rpn5 is both a proteasome lid subunit and a component of the COP9 signalosome[128,129].
Figure 7.
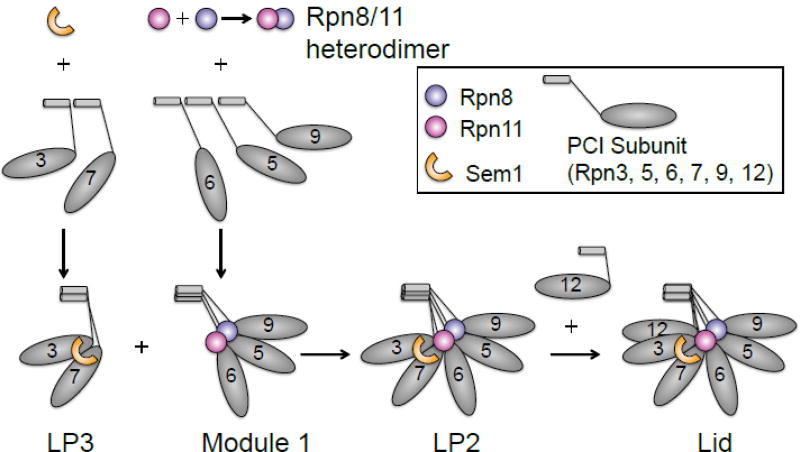
Hierarchical assembly pathway model for the RP lid. In the model, the lid assembles along a two-pronged pathway in which the subcomplexes LP3 and Module 1 are formed first, and then associate to form LP2. The final step in assembly occurs when Rpn12 joins LP2 to form the mature lid. The full lid, but not LP2, is able to bind the free base. PCI subunits (gray) are labeled with the yeast Rpn subunit numbers.
Two of the lid proteins, Rpn11 and Rpn8, adopt an MPN (Mpr1/Pad1 N-terminal) fold. Rpn11 is a JAMM metalloprotease; Rpn8 is inactive but heterodimerizes with Rpn11, yielding a complex with modest DUB activity[130,131]. Sem1 (Rpn15) is a small intrinsically disordered protein that is also part of several other nonproteasomal protein complexes [132]. In the proteasome lid, Sem1 binds both Rpn3 and Rpn7[133–136]. The remaining six lid subunits have the PCI fold, which consists of an extended helical domain followed by a conserved winged-helix motif and C-terminal α helix[137]. The winged-helix domains associate into a horseshoe shape with the N-terminal helical domains extending like fingers from the horseshoe. The C-terminal helices of the PCI subunits and the two MPN subunits associate into a complex helical bundle[130].
Lid Assembly
There have been significant advances in recent years toward a better understanding of lid assembly. Importantly, both the lid[88,133,138,139] and the entire RP[140] can be reconstituted by recombinant yeast protein expression in E. coli. Additionally, the majority of lid subunits can be tagged with GFP or other epitopes and show no apparent phenotypic defects[139], although some subunits are sensitive to tag location[140]. These tools have greatly expanded our ability to study the assembly and structure of the complex.
Lid assembly appears to proceed through an ordered series of subcomplexes that ultimately yield the mature lid (Figure 7)[138]. Lid formation does not depend on the presence of a wild-type RP base or CP. Many of the early assembly steps are still undefined, but the formation of the C-terminal helical bundle from the MPN and PCI lid subunits seems to dictate the order of later steps in assembly[138,141]. Rpn8 and Rpn11, as noted, can form a heterodimer. Rpn5, Rpn6, and Rpn9 join the dimer to form ‘Module 1’. In a second branch of the assembly pathway, Sem1 brings Rpn3 and Rpn7 together to form lid particle 3 (LP3). Module 1 then joins with LP3 to form LP2, which contains all lid subunits except Rpn12[138,141,142]. Rpn12 comes in at the end of the pathway, inserting its C-terminal helix into an extended groove formed by the nearly completed helical bundle. Incorporation of the terminal helix of Rpn12 into the bundle triggers extensive conformational changes that facilitate association of the lid with the base to complete RP formation; LP2, in contrast, has little apparent affinity for the base[140].
There is evidence, however, that certain lid subassemblies can associate with base-CP complexes. In yeast strains expressing a mutant Rpn11 subunit with a truncated C-terminus, Module 1 accumulates, and incomplete proteasomes can be isolated with this subcomplex bound to base-CP complexes[142]. Addition of the missing C-terminal Rpn11 fragment, either in cells or to extracts, allows restoration of full proteasome formation. The extent to which wild-type proteasomes follow such a pathway remains to be determined.
Structure and Conformational Changes in the Lid
Many of the advances in our understanding of the lid and its assembly have been aided by recent higher resolution structural studies. Cryo-EM reconstructions of the recombinant lid as well as the full 26S proteasome reveal large-scale and local conformational shifts during assembly and substrate processing[125]. Substantial conformational changes have also been documented by a combination of negative-stain EM and chemical crosslinking-mass spectrometry[140]. In addition, structures of subunits or subcomplexes of the lid have also been derived using X-ray crystallography[3]. Here we discuss aspects of lid assembly and function in the context of these new structural data.
The Rpn8/Rpn11 Heterodimer
The Rpn8/Rpn11 heterodimer, which bears the major DUB activity of the proteasome, is thought to be an important early intermediate in the assembly of the lid[141]. The C-terminal helices of Rpn8 are of particular importance due to their central location in the lid helical bundle; without them, lid assembly is severely impaired both in vivo and when the recombinant lid subunits are co-expressed in E. coli[138,141]. The globular cores of Rpn8 and Rpn11 interact through two regions of primarily α-helical structure[130,131]. One of these interfaces consists of a four-helix bundle, in which one helix from each subunit extends to interact with the MPN domain of the other protein, anchoring the heterodimer[130,131]. Later in lid assembly, there is a rigid-body rotation of the Rpn8/Rpn11 heterodimer during the transition from LP2 to lid; this is followed by additional large conformational changes when the lid is incorporated into the full proteasome [125,140].
Structural Insight into Activation of the Rpn11 DUB
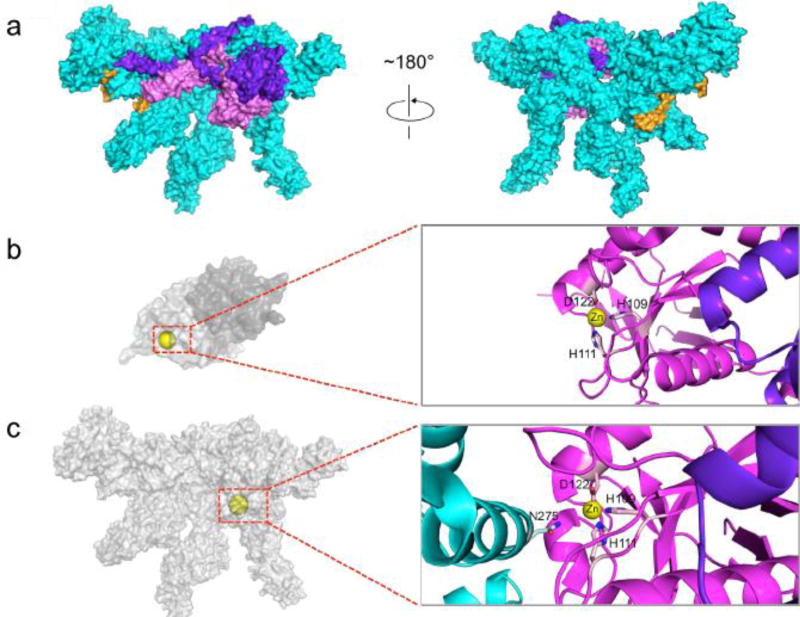
Rpn11 is at a position in the RP where it can recognize and deubiquitinate polyubiquitinated substrates as they are channeled into the central pore of the ATPase ring[83]. An insertion sequence in Rpn11, Ins-2, helps ensure the proper positioning of Rpn11 over the ATPase channel via interactions with the base subunit Rpn2[131]. In other DUBs, the Ins-2 segment is important for maintaining enzyme structural integrity [143]. High-resolution structural analyses have clarified additional details regarding the mechanism of Rpn11 action and its inhibition when not incorporated into proteasomes[125,131]. The active-site zinc is tetrahedrally coordinated by His109, His111, Asp122 and a water molecule; the latter interacts with nearby Asn275 of Rpn5[125,130] (Figure 8b,c). The Rpn5-Asn275 sidechain stabilizes the tetrahedral coordination of the Zn2+ ion, inhibiting Rpn11 catalytic activity. Additional interactions between the insertion motif Ins-1 of Rpn11 and Rpn5 and between Rpn8 and Rpn9 further stabilize the inhibited state in the isolated lid. When the lid incorporates into the proteasome, the Rpn8–Rpn11 heterodimer rotates ~90 degrees away from Rpn5, relieving these inhibitory effects as well as steric interference caused by Rpn5 helix-13[125,131].
Figure 8.
Structure of the recombinant RP lid. (a) The high-resolution cryoEM structure of free lid (PDB ID 3JCK). As in Fig. 7, Rpn8 is shown in purple, Rpn11 in pink, and Sem1 in gold. The six PCI proteins are shown in cyan. (b) The Rpn8–Rpn11 heterodimer (PDB ID 4O8X). Rpn11, light grey, which harbors the DUB activity of the lid, is shown in the context of the Rpn8–11 heterodimer. Active site residues are shown in light pink, and the Zn2+ in yellow. The zinc atom is not depicted to scale for clarity. The Rpn11 active site is easily accessible. (c) The active site of Rpn11 is depicted in the context of the full lid. The inset shows the interactions among Rpn11 (pink), Rpn8 (purple), and Rpn5 (cyan), blocking the Rpn11 active site. Again, the zinc atom is not depicted to scale for clarity.
These results are in good accord with available biochemical data: free lid exhibits fivefold less DUB activity than the purified Rpn8/Rpn11 heterodimer, and Rpn5 and Rpn8 mutations in regions that normally limit access to or full formation of the Rpn11 active site, exhibit increased DUB activity in free lid particles. The majority of the data suggests that Rpn11 DUB activity decreases as the lid proceeds along its assembly pathway. One notable exception seems to be when assembly halts at Module 1 in certain lid assembly mutants; here, Module 1 shows increased DUB activity in comparison to free Rpn11[142]. As noted above, this complex has also been reported to interact with the base, unlike other lid assembly precursors[142]; whether DUB activity in this context is relevant in vivo is uncertain. In addition to other inhibitory mechanisms, helix-13 of Rpn5 has been shown to interact with the catalytic groove of Rpn11, contributing to the steric inhibition of Rpn11 DUB activity in the free lid (Figure 8c)[125].
The structural data also suggest an explanation for why Rpn11 can cleave polyubiquitin chains linked to virtually any substrate: the DUB interacts with ubiquitinated substrates only through contacts to the ubiquitin molecule that is distal to the cleaved isopeptide bond. Comparisons of Rpn11 active-site accessibility in the proteasome between different conformational states also revealed that the active site is occluded in the resting state of the proteasome but moves to a position over the ATPase outer-ring channel upon proteasome engagement with substrate[4,91,144]. This allows placement of the bond between the substrate and ubiquitin chain into the unobstructed Rpn11 active site as the substrate is threading through the ATPase pore, leading to amputation of the ubiquitin chain and continued substrate translocation.
Sem1 (DSS1/Rpn15)
Sem1 functions as a molecular tether that facilitates formation of the lid assembly intermediate known as LP3[133,145]. Sem1, together with Rpn3 and Rpn7, comprise LP3. Two short segments in Sem1 connected by a disordered linker hold the other two subunits together. Later in assembly when LP3 and Module 1 have joined, this tethering function of Sem1 is no longer required[133], although the short polypeptide has additional, post-assembly functions in the proteasome as well[146]. Because most of Sem1 is intrinsically disordered, only a few short segments have been visualized in any of the 26S proteasome cryo-EM structures. Interestingly, Sem1 has also been reported to bind to ubiquitin[134], but whether it has this function in the context of the proteasome is unclear because the ubiquitin and RP subunit-binding sites in Sem1 overlap.
PCI Subunits and the Role of Rpn12 in RP Assembly
In the lid, the horseshoe-like structure formed by the winged-helix domains of the six PCI proteins cradles the MPN core of Rpn8/Rpn11 (Figures 7, 8a) [4,88,91,133,144]. The lid undergoes substantial conformational changes upon formation of the 26S proteasome, where it is splayed onto side of the Rpt3/Rpt6 base sector and makes contact with the CP as well[125]. The lid also rotates and translates its position on the proteasome during the proteolytic cycle[83]. These movements may coordinate displacement of substrates from their original binding sites in the RP to positions allowing their unfolding by the base ATPases and deubiquitination by Rpn11.
In what is likely the most common pathway of lid assembly, subunits assemble first into the 8-subunit LP2 intermediate and then bind the remaining PCI subunit, Rpn12. This completes lid formation and establishes competence for base binding[138,140]. LP2 is unable to bind to the base. Large-scale and more local conformational changes are triggered by LP2 binding to Rpn12[140]. The key to these changes is the integration of the C-terminal helix of Rpn12 into the nascent lid helical bundle. This is thought to create an ‘open’ state of the lid that can associate stably with the base. These conformational changes are necessary to relieve steric clashes that prevent LP2 from interacting with the base and forming the RP[125,140]. When the lid integrates into the RP and 26S proteasome, it undergoes significant conformational changes again. The entire PCI horseshoe compresses and Rpn3, Rpn7 and Rpn12 rotate toward the base[125].
Regulation of proteasome expression
Proteasome homeostasis is essential to ensuring normal cell growth and viability. Specific cell signaling pathways are responsible for regulating expression of proteasome subunits and assembly chaperones. The best characterized homeostatic mechanism described to date centers on yeast Rpn4, a zinc-finger transcription factor, which is part of a proteasome-dependent negative feedback loop[147,148]. The first indication that such a mechanism might exist came from studies of CP mutants with severely impaired activity; massive accumulation of proteasome subunits was observed[15]. Notably, the proteasome degrades Rpn4 with a half-life of ~two minutes[147]. Thus, cellular Rpn4 levels are inversely related to proteasome activity.
The transcriptional targets of Rpn4 are genes with a proteasome-associated control element (PACE) in their promoters; this 9-nucleotide sequence or slightly different variants are found upstream of almost all proteasomal subunit genes[149]. When proteasome activity is compromised, such as during proteotoxic stress, Rpn4 becomes more stable and accumulates[148]. This increase in Rpn4 levels leads in turn to increased rates of proteasomal gene transcription and formation of more proteasomes[147–149]. Restoration of proteolytic capacity then leads to enhanced Rpn4 degradation, returning proteasome gene transcription to its basal rate.
PACE sequences are not found in the promoter regions of most yeast proteasome assembly chaperones, although several have PACE-like sequences[150]. It was speculated that some assembly chaperones need not be expressed at high levels, particularly as they are utilized repeatedly[150]. Alternatively, it is possible that a different pathway regulates the levels of assembly chaperones. This turns out to be the case, as it was recently discovered that the evolutionarily conserved TORC1 pathway regulates levels of the RP assembly chaperones (RACs) in both human and yeast[151].
TORC1 is a highly conserved protein kinase involved in many signaling pathways and is a major regulator of cell growth[152]. A mitogen-activated protein kinase Mpk1, found downstream in one of the TORC1 pathways, induces expression of RACs under specific stress conditions[151]. TORC1 is an inhibitor of Mpk1[151]. Upon stress, TORC1 is inhibited, allowing Mpk1 to be activated and subsequently increasing the expression of RACs[151] The exact mechanism of RAC upregulation remains to be determined, but interestingly, the data are most consistent with translational control. The TORC1 pathway does not control the levels of all proteasome assembly chaperones[151]. Levels of the core particle assembly chaperones Pba1 and Pba2 were not altered by MPK1 deletion, indicating that they are likely regulated in a different manner[151], possibly via the Rpn4 feedback loop[150]. Further work is required to clarify the links between Rpn4- and TORC1-regulation of proteasome assembly.
In humans, the functional equivalents of Rpn4 are the Nrf1[153–155] and Nrf2[156–159] transcription factors. Nrf1 regulates basal levels of proteasome subunits and mediates proteasome gene up-regulation in response to short-term, low-dose proteasome inhibition, whereas Nrf2 induces proteasome gene transcription during oxidative stress, but not during proteasome inhibition. Nrf1 and Nrf2 both contain a unique leucine-zipper domain and are members of the Cap’n’Collar (CNC) transcription factor family. Like Rpn4, both Nrf1 and Nrf2 are rapidly degraded by the proteasome with half-lives of 12–13 minutes[154,160]. Under normal conditions, Nrf1 is associated with the endoplasmic reticulum (ER)[161], whereas Nrf2 localizes in the cytoplasm and at mitochondria[162].
When proteasomes are inhibited, the N-terminal transmembrane region of Nrf1 is proteolytically processed, and active Nrf1 is released from the ER and translocated into the nucleus to regulate gene expression[155,163]. Similarly, in response to oxidative stress, Nrf2 translocates from the cytoplasm to the nucleus where it activates its target genes[159]. Both Nrf1 and Nrf2 recognize the antioxidant response element (ARE) in the regulatory regions of their target genes[164]. It is speculated that Nrf1 and Nrf2 up-regulate proteasome genes through recognition of upstream ARE sequences. Ubiquitin-specific protease 15 (USP15) has been suggested to activate Nrf1 function in the nucleus by stabilizing Nrf1 through its deubiquitination[165]. Furthermore, activation of mTORC1 signaling stimulates Nrf1 activity, leading to increased levels of proteasomes[166]. However, the connection between Nrf1/Nrf2 and TORC1-mediated regulation of proteasome assembly chaperones remains unknown.
Perspectives
Over the past few years, significant progress has been made towards understanding the assembly mechanisms of the proteasome. Despite this, there are still many questions that remain. Regarding the CP, we still do not know how each α subunit incorporates at the correct position and whether there is an α ring-independent assembly pathway in eukaryotes. We also do not yet know the physiological roles of the alternative proteasomes containing α-subunit substitutions.
It remains possible that there are more assembly chaperones left to discover for both the CP and the RP base and potentially even the lid. In the future, it will be important to document proteasomal conformational changes upon binding and hydrolysis of ATP by specific Rpt subunits not only to link these changes with substrate unfolding, deubiquitination, and degradation (Figure 9), but also to determine how the ATPase cycle might be linked to proteasome assembly. The differences between base assembly models for mammalian and yeast proteasomes remain to be further explored, and the question of whether the CP can function as a template for RP base ATPase arrangement in vivo is still unanswered. With respect to the lid, the structures of most of the known assembly intermediates are not known. In the case of LP2, high-resolution structures may pinpoint the exact reasons why it cannot bind the base and may give hints to how binding of the Rpn12 C-terminal peptide triggers the conformational changes that accompany completion of lid assembly and enable base association. Given the tools now available, we can expect rapid advances in addressing these issues.
Highlights.
There has been significant advancement in the understanding of proteasome structure and assembly in recent years
An overview is provided of the high resolution 26S proteasome structure and its functionally distinct conformations
Current understanding of assembly mechanisms of proteasomal subcomplexes is reviewed
Acknowledgments
We thank Judy Ronau for her critical reading of the manuscript and many helpful suggestions for the figures. Work on the proteasome in our group has been supported by NIH grants GM046904 and GM083050 to M.H. L.B. was funded in part by an award from the NSF Graduate Research Fellowships Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Saeki Y, Tanaka K. Assembly and Function of the Proteasome. Ubiquitin Family Modifiers and the Proteasome. 2012;832:315–337. doi: 10.1007/978-1-61779-474-2_22. [DOI] [PubMed] [Google Scholar]
- 2.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 3.Tomko RJ, Jr, Hochstrasser M. Molecular Architecture and Assembly of the Eukaryotic Proteasome. Annu. Rev. Biochem. 2013;82:415–445. doi: 10.1146/annurev-biochem-060410-150257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wehmer M, Sakata E. Recent advances in the structural biology of the 26S proteasome. Int. J. Biochem. Cell Biol. 2016;79:437–442. doi: 10.1016/j.biocel.2016.08.008. [DOI] [PubMed] [Google Scholar]
- 5.Lu Y, Lee B-H, King RW, Finley D, Kirschner MW. Substrate degradation by the proteasome: A single-molecule kinetic analysis. Science. 2015;348:1250834–1250834. doi: 10.1126/science.1250834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vittal V, Stewart MD, Brzovic PS, Klevit RE. Regulating the Regulators: Recent Revelations in the Control of E3 Ubiquitin Ligases. Journal of Biological Chemistry. 2015;290:21244–21251. doi: 10.1074/jbc.R115.675165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saeki Y. Ubiquitin recognition by the proteasome. J. Biochem. 2017 doi: 10.1093/jb/mvw091. mvw091. [DOI] [PubMed] [Google Scholar]
- 8.Kunjappu MJ, Hochstrasser M. Assembly of the 20S proteasome. Biochim. Biophys. Acta. 2014;1843:2–12. doi: 10.1016/j.bbamcr.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glickman MH, Rubin DM, Fried VA, Finley D. The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol. Cell. Biol. 1998;18:3149–3162. doi: 10.1128/mcb.18.6.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Isono E, Nishihara K, Saeki Y, Yashiroda H, Kamata N, Ge L, et al. The assembly pathway of the 19S regulatory particle of the yeast 26S proteasome. Mol. Biol. Cell. 2007;18:569–580. doi: 10.1091/mbc.E06-07-0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaneko T, Hamazaki J, Iemura S-I, Sasaki K, Furuyama K, Natsume T, et al. Assembly pathway of the Mammalian proteasome base subcomplex is mediated by multiple specific chaperones. Cell. 2009;137:914–925. doi: 10.1016/j.cell.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 12.Groll M, Bajorek M, Köhler A, Moroder L, Rubin DM, Huber R, et al. A gated channel into the proteasome core particle. Nat. Struct. Mol. Biol. 2000;7:1062–1067. doi: 10.1038/80992. [DOI] [PubMed] [Google Scholar]
- 13.Osmulski PA, Hochstrasser M, Gaczynska M. A tetrahedral transition state at the active sites of the 20S proteasome is coupled to opening of the alpha-ring channel. Structure. 2009;17:1137–1147. doi: 10.1016/j.str.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lowe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 15.Chen P, Hochstrasser M. Autocatalytic subunit processing couples active site formation in the 20S proteasome to completion of assembly. Cell. 1996;86:961–972. doi: 10.1016/s0092-8674(00)80171-3. [DOI] [PubMed] [Google Scholar]
- 16.Huber EM, Heinemeyer W, Li X, Arendt CS, Hochstrasser M, Groll M. A unified mechanism for proteolysis and autocatalytic activation in the 20S proteasome. Nat Commun. 2016;7:10900. doi: 10.1038/ncomms10900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Becker SH, Darwin KH. Bacterial Proteasomes: Mechanistic and Functional Insights. Microbiol. Mol. Biol. Rev. 2017;81:e00036–16. doi: 10.1128/MMBR.00036-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lehmann G, Udasin RG, Livneh I, Ciechanover A. Identification of UBact, a ubiquitin-like protein, along with other homologous components of a conjugation system and the proteasome in different gram-negative bacteria. Biochem. Biophys. Res. Commun. 2017;483:946–950. doi: 10.1016/j.bbrc.2017.01.037. [DOI] [PubMed] [Google Scholar]
- 19.Li D, Li H, Wang T, Pan H, Lin G, Li H. Structural basis for the assembly and gate closure mechanisms of the Mycobacterium tuberculosis 20S proteasome. The EMBO Journal. 2010;29:2037–2047. doi: 10.1038/emboj.2010.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin G, Hu G, Tsu C, Kunes YZ, Li H, Dick L, et al. Mycobacterium tuberculosis prcBA genes encode a gated proteasome with broad oligopeptide specificity. Mol. Microbiol. 2006;59:1405–1416. doi: 10.1111/j.1365-2958.2005.05035.x. [DOI] [PubMed] [Google Scholar]
- 21.Zühl F, Seemüller E, Golbik R, Baumeister W. Dissecting the assembly pathway of the 20S proteasome. FEBS Lett. 1997;418:189–194. doi: 10.1016/S0014-5793(97)01370-7. [DOI] [PubMed] [Google Scholar]
- 22.Kwon YD, Nagy I, Adams PD, Baumeister W, Jap BK. Crystal structures of the Rhodococcus proteasome with and without its pro-peptides: implications for the role of the pro-peptide in proteasome assembly. Journal of Molecular Biology. 2004;335:233–245. doi: 10.1016/j.jmb.2003.08.029. [DOI] [PubMed] [Google Scholar]
- 23.Zwickl P, Kleinz J, Baumeister W. Critical elements in proteasome assembly. Nat. Struct. Mol. Biol. 1994;1:765–770. doi: 10.1038/nsb1194-765. [DOI] [PubMed] [Google Scholar]
- 24.Arendt CS, Hochstrasser M. Eukaryotic 20S proteasome catalytic subunit propeptides prevent active site inactivation by N-terminal acetylation and promote particle assembly. The EMBO Journal. 1999;18:3575–3585. doi: 10.1093/emboj/18.13.3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kusmierczyk AR, Kunjappu MJ, Kim RY, Hochstrasser M. A conserved 20S proteasome assembly factor requires a C-terminal HbYX motif for proteasomal precursor binding. Nat. Struct. Mol. Biol. 2011;18:622–629. doi: 10.1038/nsmb.2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Panfair D, Ramamurthy A, Kusmierczyk AR. Alpha-ring Independent Assembly of the 20S Proteasome. Sci Rep. 2015;5:13130. doi: 10.1038/srep13130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yao Y, Toth CR, Huang L, Wong ML, Dias P, Burlingame AL, et al. alpha5 subunit in Trypanosoma brucei proteasome can self-assemble to form a cylinder of four stacked heptamer rings. Biochemical Journal. 1999;344(Pt 2):349–358. doi: 10.1042/bj3440349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gerards WL, Enzlin J, Häner M, Hendriks IL, Aebi U, Bloemendal H, et al. The human alpha-type proteasomal subunit HsC8 forms a double ringlike structure, but does not assemble into proteasome-like particles with the beta-type subunits HsDelta or HsBPROS26. J. Biol. Chem. 1997;272:10080–10086. doi: 10.1074/jbc.272.15.10080. [DOI] [PubMed] [Google Scholar]
- 29.Ishii K, Noda M, Yagi H, Thammaporn R, Seetaha S, Satoh T, et al. Disassembly of the self-assembled, double-ring structure of proteasome α7 homo-tetradecamer by α6. Sci Rep. 2015;5:18167. doi: 10.1038/srep18167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hammack LJ, Kusmierczyk AR. Assembly of proteasome subunits into non-canonical complexes in vivo. Biochem. Biophys. Res. Commun. 2016 doi: 10.1016/j.bbrc.2016.11.024. [DOI] [PubMed] [Google Scholar]
- 31.Padmanabhan A, Vuong SA-T, Hochstrasser M. Assembly of an Evolutionarily Conserved Alternative Proteasome Isoform in Human Cells. Cell Reports. 2016;14:2962–2974. doi: 10.1016/j.celrep.2016.02.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirano Y, Hendil KB, Yashiroda H, Iemura S-I, Nagane R, Hioki Y, et al. A heterodimeric complex that promotes the assembly of mammalian 20S proteasomes. Nature. 2005;437:1381–1385. doi: 10.1038/nature04106. [DOI] [PubMed] [Google Scholar]
- 33.Li X, Kusmierczyk AR, Wong P, Emili A, Hochstrasser M. beta-Subunit appendages promote 20S proteasome assembly by overcoming an Ump1-dependent checkpoint. The EMBO Journal. 2007;26:2339–2349. doi: 10.1038/sj.emboj.7601681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le Tallec B, Barrault M-B, Courbeyrette R, Guérois R, Marsolier-Kergoat M-C, Peyroche A. 20S proteasome assembly is orchestrated by two distinct pairs of chaperones in yeast and in mammals. Molecular Cell. 2007;27:660–674. doi: 10.1016/j.molcel.2007.06.025. [DOI] [PubMed] [Google Scholar]
- 35.Kusmierczyk AR, Kunjappu MJ, Funakoshi M, Hochstrasser M. A multimeric assembly factor controls the formation of alternative 20S proteasomes. Nat. Struct. Mol. Biol. 2008;15:237–244. doi: 10.1038/nsmb.1389. [DOI] [PubMed] [Google Scholar]
- 36.Yashiroda H, Mizushima T, Okamoto K, Kameyama T, Hayashi H, Kishimoto T, et al. Crystal structure of a chaperone complex that contributes to the assembly of yeast 20S proteasomes. Nat. Struct. Mol. Biol. 2008;15:228–236. doi: 10.1038/nsmb.1386. [DOI] [PubMed] [Google Scholar]
- 37.Takagi K, Saeki Y, Yashiroda H, Yagi H, Kaiho A, Murata S, et al. Pba3–Pba4 heterodimer acts as a molecular matchmaker in proteasome α-ring formation. Biochem. Biophys. Res. Commun. 2014;450:1110–1114. doi: 10.1016/j.bbrc.2014.06.119. [DOI] [PubMed] [Google Scholar]
- 38.Kock M, Nunes MM, Hemann M, Kube S, Jürgen Dohmen R, Herzog F, et al. Proteasome assembly from 15S precursors involves major conformational changes and recycling of the Pba1–Pba2 chaperone. Nat Commun. 2015;6:6123. doi: 10.1038/ncomms7123. [DOI] [PubMed] [Google Scholar]
- 39.Stadtmueller BM, Kish-Trier E, Ferrell K, Petersen CN, Robinson H, Myszka DG, et al. Structure of a proteasome Pba1–Pba2 complex: implications for proteasome assembly, activation, and biological function. Journal of Biological Chemistry. 2012;287:37371–37382. doi: 10.1074/jbc.M112.367003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wani PS, Rowland MA, Ondracek A, Deeds EJ, Roelofs J. Maturation of the proteasome core particle induces an affinity switch that controls regulatory particle association. Nat Commun. 2015;6:6384. doi: 10.1038/ncomms7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumoi K, Satoh T, Murata K, Hiromoto T, Mizushima T, Kamiya Y, et al. An Archaeal Homolog of Proteasome Assembly Factor Functions as a Proteasome Activator. PLoS ONE. 2013;8:e60294. doi: 10.1371/journal.pone.0060294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sikdar A, Satoh T, Kawasaki M, Kato K. Crystal structure of archaeal homolog of proteasome-assembly chaperone PbaA. Biochem. Biophys. Res. Commun. 2014;453:493–497. doi: 10.1016/j.bbrc.2014.09.114. [DOI] [PubMed] [Google Scholar]
- 43.Hirano Y, Hayashi H, Iemura S-I, Hendil KB, Niwa S-I, Kishimoto T, et al. Cooperation of multiple chaperones required for the assembly of mammalian 20S proteasomes. Molecular Cell. 2006;24:977–984. doi: 10.1016/j.molcel.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 44.Velichutina I, Connerly PL, Arendt CS, Li X, Hochstrasser M. Plasticity in eucaryotic 20S proteasome ring assembly revealed by a subunit deletion in yeast. The EMBO Journal. 2004;23:500–510. doi: 10.1038/sj.emboj.7600059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hirano Y, Kaneko T, Okamoto K, Bai M, Yashiroda H, Furuyama K, et al. Dissecting β-ring assembly pathway of the mammalian 20S proteasome. The EMBO Journal. 2008;27:2204–2213. doi: 10.1038/emboj.2008.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramos PC, Marques AJ, London MK, Dohmen RJ. Role of C-terminal extensions of subunits beta2 and beta7 in assembly and activity of eukaryotic proteasomes. J. Biol. Chem. 2004;279:14323–14330. doi: 10.1074/jbc.M308757200. [DOI] [PubMed] [Google Scholar]
- 47.Li X, Li Y, Arendt CS, Hochstrasser M. Distinct Elements in the Proteasomal β5 Subunit Propeptide Required for Autocatalytic Processing and Proteasome Assembly. Journal of Biological Chemistry. 2016;291:1991–2003. doi: 10.1074/jbc.M115.677047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Witt E, Zantopf D, Schmidt M, Kraft R, Kloetzel PM, Krüger E. Characterisation of the newly identified human Ump1 homologue POMP and analysis of LMP7(beta 5i) incorporation into 20 S proteasomes. Journal of Molecular Biology. 2000;301:1–9. doi: 10.1006/jmbi.2000.3959. [DOI] [PubMed] [Google Scholar]
- 49.Jäger S, Groll M, Huber R, Wolf DH, Heinemeyer W. Proteasome beta-type subunits: unequal roles of propeptides in core particle maturation and a hierarchy of active site function. Journal of Molecular Biology. 1999;291:997–1013. doi: 10.1006/jmbi.1999.2995. [DOI] [PubMed] [Google Scholar]
- 50.Marques AJ, Glanemann C, Ramos PC, Dohmen RJ. The C-terminal extension of the beta7 subunit and activator complexes stabilize nascent 20 S proteasomes and promote their maturation. J. Biol. Chem. 2007;282:34869–34876. doi: 10.1074/jbc.M705836200. [DOI] [PubMed] [Google Scholar]
- 51.Ramos PC, Höckendorff J, Johnson ES, Varshavsky A, Dohmen RJ. Ump1p is required for proper maturation of the 20S proteasome and becomes its substrate upon completion of the assembly. Cell. 1998;92:489–499. doi: 10.1016/s0092-8674(00)80942-3. [DOI] [PubMed] [Google Scholar]
- 52.Sá-Moura B, Simões AM, Fraga J, Fernandes H, Abreu IA, Botelho HM, et al. Biochemical and biophysical characterization of recombinant yeast proteasome maturation factor ump1. Comput Struct Biotechnol J. 2013;7:e201304006–9. doi: 10.5936/csbj.201304006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Uekusa Y, Okawa K, Yagi-Utsumi M, Serve O, Nakagawa Y, Mizushima T, et al. Backbone 1H, 13C and 15N assignments of yeast Ump1, an intrinsically disordered protein that functions as a proteasome assembly chaperone. Biomol NMR Assign. 2014;8:383–386. doi: 10.1007/s12104-013-9523-1. [DOI] [PubMed] [Google Scholar]
- 54.Bai M, Zhao X, Sahara K, Ohte Y, Hirano Y, Kaneko T, et al. Assembly mechanisms of specialized core particles of the proteasome. Biomolecules. 2014;4:662–677. doi: 10.3390/biom4030662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.De M, Jayarapu K, Elenich L, Monaco JJ, Colbert RA, Griffin TA. Beta 2 subunit propeptides influence cooperative proteasome assembly. J. Biol. Chem. 2003;278:6153–6159. doi: 10.1074/jbc.M209292200. [DOI] [PubMed] [Google Scholar]
- 56.Kingsbury DJ, Griffin TA, Colbert RA. Novel propeptide function in 20 S proteasome assembly influences beta subunit composition. J. Biol. Chem. 2000;275:24156–24162. doi: 10.1074/jbc.M001742200. [DOI] [PubMed] [Google Scholar]
- 57.Kammerl IE, Meiners S. Proteasome function shapes innate and adaptive immune responses. Am. J. Physiol. Lung Cell Mol. Physiol. 2016;311:L328–36. doi: 10.1152/ajplung.00156.2016. [DOI] [PubMed] [Google Scholar]
- 58.Murata S, Takahama Y, Tanaka K. Thymoproteasome: probable role in generating positively selecting peptides. Curr. Opin. Immunol. 2008;20:192–196. doi: 10.1016/j.coi.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 59.Qian M-X, Pang Y, Liu CH, Haratake K, Du B-Y, Ji D-Y, et al. Acetylation-mediated proteasomal degradation of core histones during DNA repair and spermatogenesis. Cell. 2013;153:1012–1024. doi: 10.1016/j.cell.2013.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Uechi H, Hamazaki J, Murata S. Characterization of the testis-specific proteasome subunit α4s in mammals. Journal of Biological Chemistry. 2014;289:12365–12374. doi: 10.1074/jbc.M114.558866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhong L, Belote JM. The testis-specific proteasome subunit Prosalpha6T of D. melanogaster is required for individualization and nuclear maturation during spermatogenesis. Development. 2007;134:3517–3525. doi: 10.1242/dev.004770. [DOI] [PubMed] [Google Scholar]
- 62.Belote JM, Miller M, Smyth KA. Evolutionary conservation of a testes-specific proteasome subunit gene in Drosophila. Gene. 1998;215:93–100. doi: 10.1016/s0378-1119(98)00256-x. [DOI] [PubMed] [Google Scholar]
- 63.Kikuchi J, Iwafune Y, Akiyama T, Okayama A, Nakamura H, Arakawa N, et al. Co- and post-translational modifications of the 26S proteasome in yeast. Proteomics. 2010;10:2769–2779. doi: 10.1002/pmic.200900283. [DOI] [PubMed] [Google Scholar]
- 64.Humbard MA, Zhou G, Maupin-Furlow JA. The N-terminal penultimate residue of 20S proteasome alpha1 influences its N(alpha) acetylation and protein levels as well as growth rate and stress responses of Haloferax volcanii. J. Bacteriol. 2009;191:3794–3803. doi: 10.1128/JB.00090-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Akahane T, Sahara K, Yashiroda H, Tanaka K, Murata S. Involvement of Bag6 and the TRC pathway in proteasome assembly. Nat Commun. 2013;4:2234. doi: 10.1038/ncomms3234. [DOI] [PubMed] [Google Scholar]
- 66.Shin S-W, Shimizu N, Tokoro M, Nishikawa S, Hatanaka Y, Anzai M, et al. Mouse zygote-specific proteasome assembly chaperone important for maternal-to-zygotic transition. Biol Open. 2013;2:170–182. doi: 10.1242/bio.20123020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Savulescu AF, Glickman MH. Proteasome Activator 200: The HEAT is on…. Mol. Cell Proteomics. 2011;10:R110.006890–R110.006890. doi: 10.1074/mcp.R110.006890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fehlker M, Wendler P, Lehmann A, Enenkel C. Blm3 is part of nascent proteasomes and is involved in a late stage of nuclear proteasome assembly. EMBO Reports. 2003;4:959–963. doi: 10.1038/sj.embor.embor938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dange T, Smith D, Noy T, Rommel PC, Jurzitza L, Cordero RJB, et al. Blm10 protein promotes proteasomal substrate turnover by an active gating mechanism. Journal of Biological Chemistry. 2011;286:42830–42839. doi: 10.1074/jbc.M111.300178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weberruss MH, Savulescu AF, Jando J, Bissinger T, Harel A, Glickman MH, et al. Blm10 facilitates nuclear import of proteasome core particles. The EMBO Journal. 2013;32:2697–2707. doi: 10.1038/emboj.2013.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lehmann A, Jechow K, Enenkel C. Blm10 binds to pre-activated proteasome core particles with open gate conformation. EMBO Reports. 2008;9:1237–1243. doi: 10.1038/embor.2008.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schmidt M, Haas W, Crosas B, Santamaria PG, Gygi SP, Walz T, et al. The HEAT repeat protein Blm10 regulates the yeast proteasome by capping the core particle. Nat. Struct. Mol. Biol. 2005;12:294–303. doi: 10.1038/nsmb914. [DOI] [PubMed] [Google Scholar]
- 73.Sadre-Bazzaz K, Whitby FG, Robinson H, Formosa T, Hill CP. Structure of a Blm10 complex reveals common mechanisms for proteasome binding and gate opening. Molecular Cell. 2010;37:728–735. doi: 10.1016/j.molcel.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McCutchen-Maloney SL, Matsuda K, Shimbara N, Binns DD, Tanaka K, Slaughter CA, et al. cDNA cloning, expression, and functional characterization of PI31, a proline-rich inhibitor of the proteasome. J. Biol. Chem. 2000;275:18557–18565. doi: 10.1074/jbc.M001697200. [DOI] [PubMed] [Google Scholar]
- 75.Yashiroda H, Toda Y, Otsu S, Takagi K, Mizushima T, Murata S. N-terminal α7 deletion of the proteasome 20S core particle substitutes for yeast PI31 function. Mol. Cell. Biol. 2014 doi: 10.1128/MCB.00582-14. MCB.00582–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hatanaka A, Chen B, Sun J-Q, Mano Y, Funakoshi M, Kobayashi H, et al. Fub1p, a novel protein isolated by boundary screening, binds the proteasome complex. Genes Genet. Syst. 2011;86:305–314. doi: 10.1266/ggs.86.305. [DOI] [PubMed] [Google Scholar]
- 77.Li X, Thompson D, Kumar B, DeMartino GN. Molecular and cellular roles of PI31 (PSMF1) protein in regulation of proteasome function. Journal of Biological Chemistry. 2014;289:17392–17405. doi: 10.1074/jbc.M114.561183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bader M, Benjamin S, Wapinski OL, Smith DM, Goldberg AL, Steller H. A Conserved F Box Regulatory Complex Controls Proteasome Activity in Drosophila. Cell. 2011;145:371–382. doi: 10.1016/j.cell.2011.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Guerrero C, Tagwerker C, Kaiser P, HUANG L. An integrated mass spectrometry-based proteomic approach: quantitative analysis of tandem affinity-purified in vivo cross-linked protein complexes (QTAX) to decipher the 26 S proteasome-interacting network. Mol. Cell Proteomics. 2006;5:366–378. doi: 10.1074/mcp.M500303-MCP200. [DOI] [PubMed] [Google Scholar]
- 80.Djuranovic S, Hartmann MD, Habeck M, Ursinus A, Zwickl P, Martin J, et al. Structure and activity of the N-terminal substrate recognition domains in proteasomal ATPases. Molecular Cell. 2009;34:580–590. doi: 10.1016/j.molcel.2009.04.030. [DOI] [PubMed] [Google Scholar]
- 81.Zhang F, Hu M, Tian G, Zhang P, Finley D, Jeffrey PD, et al. Structural Insights into the Regulatory Particle of the Proteasome from Methanocaldococcus jannaschii. Molecular Cell. 2009;34:473–484. doi: 10.1016/j.molcel.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tomko RJ, Funakoshi M, Schneider K, Wang J, Hochstrasser M. Heterohexameric ring arrangement of the eukaryotic proteasomal ATPases: implications for proteasome structure and assembly. Molecular Cell. 2010;38:393–403. doi: 10.1016/j.molcel.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Luan B, Huang X, Wu J, Mei Z, Wang Y, Xue X, et al. Structure of an endogenous yeast 26S proteasome reveals two major conformational states. Proceedings of the National Academy of Sciences. 2016:201601561. doi: 10.1073/pnas.1601561113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Beck F, Unverdorben P, Bohn S, Schweitzer A, Pfeifer G, Sakata E, et al. Near-atomic resolution structural model of the yeast 26S proteasome. Proceedings of the National Academy of Sciences. 2012;109:14870–14875. doi: 10.1073/pnas.1213333109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Smith DM, Chang S-C, Park S, Finley D, Cheng Y, Goldberg AL. Docking of the proteasomal ATPases“ carboxyl termini in the 20S proteasome”s alpha ring opens the gate for substrate entry. Molecular Cell. 2007;27:731–744. doi: 10.1016/j.molcel.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ding Z, Fu Z, Xu C, Wang Y, Wang Y, Wang Y, et al. High-resolution cryo-EM structure of the proteasome in complex with ADP-AlFx. Cell Research. 2017;27:373–385. doi: 10.1038/cr.2017.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wehmer M, Rudack T, Beck F, Aufderheide A, Pfeifer G, Plitzko JM, et al. Structural insights into the functional cycle of the ATPase module of the 26S proteasome. Proceedings of the National Academy of Sciences. 2017:201621129. doi: 10.1073/pnas.1621129114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lander GC, Estrin E, Matyskiela ME, Bashore C, Nogales E, Martin A. Complete subunit architecture of the proteasome regulatory particle. Nature. 2012;482:186–191. doi: 10.1038/nature10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Beckwith R, Worden E, Estrin E, Martin A. Reconstitution of the 26S Proteasome Reveals Functional Asymmetries in its Heterohexameric AAA+ Unfoldase. Biophysical Journal. 2014;106:468a. doi: 10.1016/j.bpj.2013.11.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Beckwith R, Estrin E, Worden EJ, Martin A. Reconstitution of the 26S proteasome reveals functional asymmetries in its AAA+ unfoldase. Nat. Struct. Mol. Biol. 2013;20:1164–1172. doi: 10.1038/nsmb.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Unverdorben P, Beck F, Śledź P, Schweitzer A, Pfeifer G, Plitzko JM, et al. Deep classification of a large cryo-EM dataset defines the conformational landscape of the 26S proteasome. Proceedings of the National Academy of Sciences. 2014;111:5544–5549. doi: 10.1073/pnas.1403409111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.He J, Kulkarni K, da Fonseca PCA, Krutauz D, Glickman MH, Barford D, et al. The structure of the 26S proteasome subunit Rpn2 reveals its PC repeat domain as a closed toroid of two concentric α-helical rings. Structure. 2012;20:513–521. doi: 10.1016/j.str.2011.12.015. [DOI] [PubMed] [Google Scholar]
- 93.Elsasser S, Gali RR, Schwickart M, Larsen CN, Leggett DS, Müller B, et al. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat. Cell Biol. 2002;4:725–730. doi: 10.1038/ncb845. [DOI] [PubMed] [Google Scholar]
- 94.Saeki Y, Saitoh A, Toh-e A, Yokosawa H. Ubiquitin-like proteins and Rpn10 play cooperative roles in ubiquitin-dependent proteolysis. Biochem. Biophys. Res. Commun. 2002;293:986–992. doi: 10.1016/S0006-291X(02)00340-6. [DOI] [PubMed] [Google Scholar]
- 95.Shi Y, Chen X, Elsasser S, Stocks BB, Tian G, Lee B-H, et al. Rpn1 provides adjacent receptor sites for substrate binding and deubiquitination by the proteasome. Science. 2016;351:aad9421–aad9421. doi: 10.1126/science.aad9421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, Baker RT, et al. Multiple associated proteins regulate proteasome structure and function. Molecular Cell. 2002;10:495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- 97.Bashore C, Dambacher CM, Goodall EA, Matyskiela ME, Lander GC, Martin A. Ubp6 deubiquitinase controls conformational dynamics and substrate degradation of the 26S proteasome. Nat. Struct. Mol. Biol. 2015;22:712–719. doi: 10.1038/nsmb.3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Aufderheide A, Beck F, Stengel F, Hartwig M, Schweitzer A, Pfeifer G, et al. Structural characterization of the interaction of Ubp6 with the 26S proteasome. Proceedings of the National Academy of Sciences. 2015:201510449. doi: 10.1073/pnas.1510449112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schreiner P, Chen X, Husnjak K, Randles L, Zhang N, Elsasser S, et al. Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature. 2008;453:548–552. doi: 10.1038/nature06924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, et al. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453:481–488. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chen X, Lee B-H, Finley D, Walters KJ. Structure of proteasome ubiquitin receptor hRpn13 and its activation by the scaffolding protein hRpn2. Molecular Cell. 2010;38:404–415. doi: 10.1016/j.molcel.2010.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Elsasser S, Chandler-Militello D, Müller B, Hanna J, Finley D. Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. J. Biol. Chem. 2004;279:26817–26822. doi: 10.1074/jbc.M404020200. [DOI] [PubMed] [Google Scholar]
- 103.Deveraux Q, Ustrell V, Pickart C, Rechsteiner M. A 26 S protease subunit that binds ubiquitin conjugates. J. Biol. Chem. 1994;269:7059–7061. [PubMed] [Google Scholar]
- 104.Keren-Kaplan T, Zeev Peters L, Levin-Kravets O, Attali I, Kleifeld O, Shohat N, et al. Structure of ubiquitylated-Rpn10 provides insight into its autoregulation mechanism. Nat Commun. 2016;7:12960. doi: 10.1038/ncomms12960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Isasa M, Katz EJ, Kim W, Yugo V, González S, Kirkpatrick DS, et al. Monoubiquitination of RPN10 regulates substrate recruitment to the proteasome. Molecular Cell. 2010;38:733–745. doi: 10.1016/j.molcel.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Saeki Y, Toh-e A, Kudo T, Kawamura H, Tanaka K. Multiple Proteasome-Interacting Proteins Assist the Assembly of the Yeast 19S Regulatory Particle. Cell. 2009;137:900–913. doi: 10.1016/j.cell.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 107.Funakoshi M, Tomko RJ, Kobayashi H, Hochstrasser M. Multiple assembly chaperones govern biogenesis of the proteasome regulatory particle base. Cell. 2009;137:887–899. doi: 10.1016/j.cell.2009.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Roelofs J, Park S, Haas W, Tian G, McAllister FE, Huo Y, et al. Chaperone-mediated pathway of proteasome regulatory particle assembly. Nature. 2009;459:861–865. doi: 10.1038/nature08063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Le Tallec B, Barrault M-B, Guérois R, Carré T, Peyroche A. Hsm3/S5b Participates in the Assembly Pathway of the 19S Regulatory Particle of the Proteasome. Molecular Cell. 2009;33:389–399. doi: 10.1016/j.molcel.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 110.Hanssum A, Zhong Z, Rousseau A, Krzyzosiak A, Sigurdardottir A, Bertolotti A. An inducible chaperone adapts proteasome assembly to stress. Molecular Cell. 2014;55:566–577. doi: 10.1016/j.molcel.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Barrault M-B, Richet N, Godard C, Murciano B, Le Tallec B, Rousseau E, et al. Dual functions of the Hsm3 protein in chaperoning and scaffolding regulatory particle subunits during the proteasome assembly. Proceedings of the National Academy of Sciences. 2012;109:E1001–10. doi: 10.1073/pnas.1116538109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tomko RJ, Hochstrasser M. Order of the proteasomal ATPases and eukaryotic proteasome assembly. Cell Biochem. Biophys. 2011;60:13–20. doi: 10.1007/s12013-011-9178-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Thompson D, Hakala K, DeMartino GN. Subcomplexes of PA700, the 19 S regulator of the 26 S proteasome, reveal relative roles of AAA subunits in 26 S proteasome assembly and activation and ATPase activity. Journal of Biological Chemistry. 2009;284:24891–24903. doi: 10.1074/jbc.M109.023218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Park S, Roelofs J, Kim W, Robert J, Schmidt M, Gygi SP, et al. Hexameric assembly of the proteasomal ATPases is templated through their C termini. Nature. 2009;459:866–870. doi: 10.1038/nature08065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pack C-G, Yukii H, Toh-e A, Kudo T, Tsuchiya H, Kaiho A, et al. Quantitative live-cell imaging reveals spatio-temporal dynamics and cytoplasmic assembly of the 26S proteasome. Nat Commun. 2014;5:3396. doi: 10.1038/ncomms4396. [DOI] [PubMed] [Google Scholar]
- 116.Li F, Tian G, Langager D, Sokolova V, Finley D, Park S. Nucleotide-dependent switch in proteasome assembly mediated by the Nas6 chaperone. Proceedings of the National Academy of Sciences. 2017;114:1548–1553. doi: 10.1073/pnas.1612922114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lee SY-C, De la Mota-Peynado A, Roelofs J. Loss of Rpt5 protein interactions with the core particle and Nas2 protein causes the formation of faulty proteasomes that are inhibited by Ecm29 protein. Journal of Biological Chemistry. 2011;286:36641–36651. doi: 10.1074/jbc.M111.280875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Satoh T, Saeki Y, Hiromoto T, Wang Y-H, Uekusa Y, Yagi H, et al. Structural basis for proteasome formation controlled by an assembly chaperone nas2. Structure. 2014;22:731–743. doi: 10.1016/j.str.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 119.Takagi K, Kim S, Yukii H, Ueno M, Morishita R, Endo Y, et al. Structural basis for specific recognition of Rpt1p, an ATPase subunit of 26 S proteasome, by proteasome-dedicated chaperone Hsm3p. Journal of Biological Chemistry. 2012;287:12172–12182. doi: 10.1074/jbc.M112.345876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Park S, Li X, Kim HM, Singh CR, Tian G, Hoyt MA, et al. Reconfiguration of the proteasome during chaperone-mediated assembly. Nature. 2013;497:512–516. doi: 10.1038/nature12123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Nakamura Y, Umehara T, Tanaka A, Horikoshi M, Padmanabhan B, Yokoyama S. Structural basis for the recognition between the regulatory particles Nas6 and Rpt3 of the yeast 26S proteasome. Biochem. Biophys. Res. Commun. 2007;359:503–509. doi: 10.1016/j.bbrc.2007.05.138. [DOI] [PubMed] [Google Scholar]
- 122.Sokolova V, Li F, Polovin G, Park S. Proteasome Activation is Mediated via a Functional Switch of the Rpt6 C-terminal Tail Following Chaperone-dependent Assembly. Sci Rep. 2015;5:14909. doi: 10.1038/srep14909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kim S, Saeki Y, Fukunaga K, Suzuki A, Takagi K, Yamane T, et al. Crystal structure of yeast rpn14, a chaperone of the 19 S regulatory particle of the proteasome. Journal of Biological Chemistry. 2010;285:15159–15166. doi: 10.1074/jbc.M110.104042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ehlinger A, Park S, Fahmy A, Lary JW, Cole JL, Finley D, et al. Conformational dynamics of the Rpt6 ATPase in proteasome assembly and Rpn14 binding. Structure. 2013;21:753–765. doi: 10.1016/j.str.2013.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dambacher CM, Worden EJ, Herzik MA, Martin A, Lander GC. Atomic structure of the 26S proteasome lid reveals the mechanism of deubiquitinase inhibition. eLife Sciences. 2016;5:e13027. doi: 10.7554/eLife.13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Erzberger JP, Stengel F, Pellarin R, Zhang S, Schaefer T, Aylett CHS, et al. Molecular architecture of the 40S·eIF1·eIF3 translation initiation complex. Cell. 2014;158:1123–1135. doi: 10.1016/j.cell.2014.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lingaraju GM, Bunker RD, Cavadini S, Hess D, Hassiepen U, Renatus M, et al. Crystal structure of the human COP9 signalosome. Nature. 2014;512:161–165. doi: 10.1038/nature13566. [DOI] [PubMed] [Google Scholar]
- 128.Yu Z, Kleifeld O, Lande-Atir A, Bsoul M, Kleiman M, Krutauz D, et al. Dual function of Rpn5 in two PCI complexes, the 26S proteasome and COP9 signalosome. Mol. Biol. Cell. 2011;22:911–920. doi: 10.1091/mbc.E10-08-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Meister C, Gulko MK, Köhler AM, Braus GH. The devil is in the details: comparison between COP9 signalosome (CSN) and the LID of the 26S proteasome. Curr. Genet. 2016;62:129–136. doi: 10.1007/s00294-015-0525-7. [DOI] [PubMed] [Google Scholar]
- 130.Pathare GR, Nagy I, Śledź P, Anderson DJ, Zhou H-J, Pardon E, et al. Crystal structure of the proteasomal deubiquitylation module Rpn8–Rpn11. Proceedings of the National Academy of Sciences. 2014;111:2984–2989. doi: 10.1073/pnas.1400546111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Worden EJ, Padovani C, Martin A. Structure of the Rpn11−Rpn8 dimer reveals mechanisms of substrate deubiquitination during proteasomal degradation. Nat. Struct. Mol. Biol. 2014;21:220–227. doi: 10.1038/nsmb.2771. [DOI] [PubMed] [Google Scholar]
- 132.Kragelund BB, Schenstrøm SM, Rebula CA, Panse VG, Hartmann-Petersen R. DSS1/Sem1, A Multifunctional and Intrinsically Disordered Protein. Trends in Biochemical Sciences. 2016 doi: 10.1016/j.tibs.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 133.Tomko RJ, Hochstrasser M. The intrinsically disordered Sem1 protein functions as a molecular tether during proteasome lid biogenesis. Molecular Cell. 2014;53:433–443. doi: 10.1016/j.molcel.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Paraskevopoulos K, Kriegenburg F, Tatham MH, Rösner HI, Medina B, Larsen IB, et al. Dss1 Is a 26S Proteasome Ubiquitin Receptor. Molecular Cell. 2014 doi: 10.1016/j.molcel.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gudmundsdottir K, Lord CJ, Ashworth A. The proteasome is involved in determining differential utilization of double-strand break repair pathways. Oncogene. 2007;26:7601–7606. doi: 10.1038/sj.onc.1210579. [DOI] [PubMed] [Google Scholar]
- 136.Wei S-J, Williams JG, Dang H, Darden TA, Betz BL, Humble MM, et al. Identification of a specific motif of the DSS1 protein required for proteasome interaction and p53 protein degradation. Journal of Molecular Biology. 2008;383:693–712. doi: 10.1016/j.jmb.2008.08.044. [DOI] [PubMed] [Google Scholar]
- 137.Ellisdon AM, Stewart M. Structural biology of the PCI-protein fold. Bioarchitecture. 2012;2:118–123. doi: 10.4161/bioa.21131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Tomko RJ, Hochstrasser M. Incorporation of the Rpn12 subunit couples completion of proteasome regulatory particle lid assembly to lid-base joining. Molecular Cell. 2011;44:907–917. doi: 10.1016/j.molcel.2011.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Fukunaga K, Kudo T, Toh-e A, Tanaka K, Saeki Y. Dissection of the assembly pathway of the proteasome lid in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 2010;396:1048–1053. doi: 10.1016/j.bbrc.2010.05.061. [DOI] [PubMed] [Google Scholar]
- 140.Tomko RJ, Jr, Taylor DW, Chen ZA, Wang H-W, Rappsilber J, Hochstrasser M. A Single α Helix Drives Extensive Remodeling of the Proteasome Lid and Completion of Regulatory Particle Assembly. 2015;163:432–444. doi: 10.1016/j.cell.2015.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Estrin E, Lopez-Blanco JR, Chacón P, Martin A. Formation of an intricate helical bundle dictates the assembly of the 26S proteasome lid. Structure. 2013;21:1624–1635. doi: 10.1016/j.str.2013.06.023. [DOI] [PubMed] [Google Scholar]
- 142.Yu Z, Livnat-Levanon N, Kleifeld O, Mansour W, Nakasone MA, Castaneda CA, et al. Base-CP proteasome can serve as a platform for stepwise lid formation. Biosci. Rep. 2015;35:1–14. doi: 10.1042/BSR20140173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ronau JA, Beckmann JF, Hochstrasser M. Substrate specificity of the ubiquitin and Ubl proteases. Cell Research. 2016;26:441–456. doi: 10.1038/cr.2016.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Schweitzer A, Aufderheide A, Rudack T, Beck F, Pfeifer G, Plitzko JM, et al. Structure of the human 26S proteasome at a resolution of 3.9 Å. Proceedings of the National Academy of Sciences. 2016;113:7816–7821. doi: 10.1073/pnas.1608050113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bohn S, Sakata E, Beck F, Pathare GR, Schnitger J, Nagy I, et al. Localization of the regulatory particle subunit Sem1 in the 26S proteasome. Biochem. Biophys. Res. Commun. 2013;435:250–254. doi: 10.1016/j.bbrc.2013.04.069. [DOI] [PubMed] [Google Scholar]
- 146.Sone T, Saeki Y, Toh-e A, Yokosawa H. Sem1p is a novel subunit of the 26 S proteasome from Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:28807–28816. doi: 10.1074/jbc.M403165200. [DOI] [PubMed] [Google Scholar]
- 147.Xie Y, Varshavsky A. RPN4 is a ligand, substrate, and transcriptional regulator of the 26S proteasome: a negative feedback circuit. Proc. Natl. Acad. Sci. U.S.a. 2001;98:3056–3061. doi: 10.1073/pnas.071022298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ju D, Wang L, Mao X, Xie Y. Homeostatic regulation of the proteasome via an Rpn4-dependent feedback circuit. Biochem. Biophys. Res. Commun. 2004;321:51–57. doi: 10.1016/j.bbrc.2004.06.105. [DOI] [PubMed] [Google Scholar]
- 149.Mannhaupt G, Schnall R, Karpov V, Vetter I, Feldmann H. Rpn4p acts as a transcription factor by binding to PACE, a nonamer box found upstream of 26S proteasomal and other genes in yeast. FEBS Lett. 1999;450:27–34. doi: 10.1016/s0014-5793(99)00467-6. [DOI] [PubMed] [Google Scholar]
- 150.Shirozu R, Yashiroda H, Murata S. Identification of minimum Rpn4-responsive elements in genes related to proteasome functions. FEBS Lett. 2015;589:933–940. doi: 10.1016/j.febslet.2015.02.025. [DOI] [PubMed] [Google Scholar]
- 151.Rousseau A, Bertolotti A. An evolutionarily conserved pathway controls proteasome homeostasis. Nature. 2016;536:184–189. doi: 10.1038/nature18943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Laplante M, Sabatini DM. mTOR Signaling in Growth Control and Disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Radhakrishnan SK, Lee CS, Young P, Beskow A, Chan JY, Deshaies RJ. Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Molecular Cell. 2010;38:17–28. doi: 10.1016/j.molcel.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Steffen J, Seeger M, Koch A, Krüger E. Proteasomal degradation is transcriptionally controlled by TCF11 via an ERAD-dependent feedback loop. Molecular Cell. 2010;40:147–158. doi: 10.1016/j.molcel.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 155.Sha Z, Goldberg AL. Proteasome-mediated processing of Nrf1 is essential for coordinate induction of all proteasome subunits and p97. Curr. Biol. 2014;24:1573–1583. doi: 10.1016/j.cub.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kraft DC, Deocaris CC, Wadhwa R, Rattan SIS. Preincubation with the Proteasome Inhibitor MG-132 Enhances Proteasome Activity via the Nrf2 Transcription Factor in Aging Human Skin Fibroblasts. Annals of the New York Academy of Sciences. 2006;1067:420–424. doi: 10.1196/annals.1354.060. [DOI] [PubMed] [Google Scholar]
- 157.Pickering AM, Linder RA, Zhang H, Forman HJ, Davies KJA. Nrf2-dependent induction of proteasome and Pa28αβ regulator are required for adaptation to oxidative stress. Journal of Biological Chemistry. 2012;287:10021–10031. doi: 10.1074/jbc.M111.277145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kapeta S, Chondrogianni N, Gonos ES. Nuclear erythroid factor 2-mediated proteasome activation delays senescence in human fibroblasts. Journal of Biological Chemistry. 2010;285:8171–8184. doi: 10.1074/jbc.M109.031575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kwak M-K, Wakabayashi N, Greenlaw JL, Yamamoto M, Kensler TW. Antioxidants enhance mammalian proteasome expression through the Keap1-Nrf2 signaling pathway. Mol. Cell. Biol. 2003;23:8786–8794. doi: 10.1128/MCB.23.23.8786-8794.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Stewart D, Killeen E, Naquin R, Alam S, Alam J. Degradation of transcription factor Nrf2 via the ubiquitin-proteasome pathway and stabilization by cadmium. J. Biol. Chem. 2003;278:2396–2402. doi: 10.1074/jbc.M209195200. [DOI] [PubMed] [Google Scholar]
- 161.Wang W, Chan JY. Nrf1 is targeted to the endoplasmic reticulum membrane by an N-terminal transmembrane domain. Inhibition of nuclear translocation and transacting function. J. Biol. Chem. 2006;281:19676–19687. doi: 10.1074/jbc.M602802200. [DOI] [PubMed] [Google Scholar]
- 162.Lo S-C, Hannink M. PGAM5 tethers a ternary complex containing Keap1 and Nrf2 to mitochondria. Experimental Cell Research. 2008;314:1789–1803. doi: 10.1016/j.yexcr.2008.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Radhakrishnan SK, den Besten W, Deshaies RJ. p97-dependent retrotranslocation and proteolytic processing govern formation of active Nrf1 upon proteasome inhibition. eLife Sciences. 2014;3:e01856. doi: 10.7554/eLife.01856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ohtsuji M, Katsuoka F, Kobayashi A, Aburatani H, Hayes JD, Yamamoto M. Nrf1 and Nrf2 play distinct roles in activation of antioxidant response element-dependent genes. J. Biol. Chem. 2008;283:33554–33562. doi: 10.1074/jbc.M804597200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Fukagai K, Waku T, Chowdhury AMMA, Kubo K, Matsumoto M, Kato H, et al. USP15 stabilizes the transcription factor Nrf1 in the nucleus, promoting the proteasome gene expression. Biochem. Biophys. Res. Commun. 2016;478:363–370. doi: 10.1016/j.bbrc.2016.07.045. [DOI] [PubMed] [Google Scholar]
- 166.Zhang Y, Manning BD. mTORC1 signaling activates NRF1 to increase cellular proteasome levels. Cell Cycle. 2015;14:2011–2017. doi: 10.1080/15384101.2015.1044188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Chen S, Wu J, Lu Y, Ma Y-B, Lee B-H, Yu Z, et al. Structural basis for dynamic regulation of the human 26S proteasome. Proceedings of the National Academy of Sciences. 2016;113:12991–12996. doi: 10.1073/pnas.1614614113. [DOI] [PMC free article] [PubMed] [Google Scholar]