Abstract
Rare patients who spontaneously control HIV replication provide a useful model to inform HIV vaccine development. HIV controllers develop particularly efficient antiviral CD4+ T cell responses mediated by shared high-affinity TCRs. To determine whether the candidate DNA vaccine ADVAX could induce similar responses, we analyzed Gag-specific primary CD4+ T cells from healthy volunteers who received ADVAX DNA by electroporation. Vaccinated volunteers had an immunodominant response to the Gag293 epitope with a functional avidity intermediate between that of controllers and treated patients. The TCR repertoire of Gag293-specific CD4+ T cells proved highly biased, with a predominant usage of the TRBV2 gene in vaccinees as well as controllers. TRAV gene usage was more diverse, with the dominance of TRAV29 over TRAV24 genes in vaccinees, while TRAV24 predominated in controllers. Sequence analysis revealed an unexpected degree of overlap between the specific repertoires of vaccinees and controllers, with the sharing of TRAV24 and TRBV2 public motifs (>30%) and of public clonotypes characteristic of high-affinity TCRs. MHC-II tetramer binding revealed a broad HLA-DR cross-restriction, explaining how Gag293-specific public clonotypes could be selected in individuals with diverse genetic backgrounds. TRAV29 clonotypes also proved cross-restricted, but conferred responses of lower functional avidity upon TCR transfer. In conclusion, DNA vaccination by electroporation primed for TCR clonotypes that were associated with HIV control, highlighting the potential of this vaccine delivery method. This study provides the first proof-of-concept that clonotypic analysis may be used as a tool to monitor the quality of vaccine-induced responses and modulate these towards “controller-like” responses.
INTRODUCTION
With close to 37 millions of persons living with HIV-1 worldwide, the development of an effective HIV vaccine is viewed as essential to end the HIV-1 pandemic (1). However, HIV biology poses considerable challenges to vaccine development, due to its capacity to evade immune responses and persist in latently infected cells for extended periods of time (2). So far, a single vaccine efficacy trial conducted in Thailand has demonstrated a degree of protection against HIV infection, with an estimated 31% efficacy (3)(4). In this trial, HLA class II alleles modulated the quality of vaccine-induced antibodies and had an impact on protective efficacy, suggesting that HLA-II restricted CD4+ T cells influenced protective vaccine responses through the control of antibody maturation (5). T cell responses may also directly contribute to vaccine-induced protection, as suggested in a CMV vector-based vaccination model that lead to simian immunodeficiency virus control (6, 7). Considering that complete sterilizing immunity is rarely achieved with anti-viral vaccines, it is critical that a candidate HIV vaccine should also control HIV replication at its entry site.
Rare cases of spontaneous control of HIV infection reveal that the human immune system has the capacity to mount an efficient antiviral response against HIV. Patients who contain HIV replication in the absence of antiretroviral therapy, called HIV controllers, or elite controllers, show signs of particularly efficient T cell responses, and maintain full CD4 helper function in the long term (8, 9). Converging evidence indicate that HIV controllers develop both CD4+ and CD8+ T cell responses with a high sensitivity to HIV Gag antigens (10, 11). As a consequence, controller T cells are particularly efficient at sensing low amounts of virus and at eliminating infected cells. (12–14). Controller CD4+ T cells show preserved central memory responses (15, 16),, but also maintain a highly differentiated Th1 effector compartment,, while such effector cells are progressively lost in patients who receive antiretroviral therapy (17, 18). Thus, spontaneous HIV control emerges as an active process enforced by T cells that retain effector function in the long term, in spite of the very low antigenemia available to stimulate such responses.
Emerging evidence points to the role of particular T cell receptor (TCR) clonotypes in conferring the efficient T cell responses characteristic of HIV controllers (19). TCR clonotypes expressed by controller CD8+ T cells are responsible for their efficient cytotoxic responses, while HLA-matched non-controller patients show clonotypes of lower efficacy (20, 21). The nature of TCR clonotypes expressed by Controller CD4+ T cells had until recently remained unexplored. Analysis of the response to the most immunodominant HIV-1 CD4 epitope, termed Gag293, had pointed to the presence of a CD4+ T cell population with high TCR affinity in HIV controllers, while this population was absent in treated patients (17). The expression of high affinity TCRs helped explain how controllers maintained CD4+ T cell effector functions, as minimal amounts of viral antigens were sufficient to trigger full effector differentiation. We recently characterized the set of TCRs directed at Gag293, and uncovered a highly biased repertoire characterized by the preferential expression of the TCR variable genes TRAV24 and TRBV2, with the bias being more marked in controllers than in treated patients (22). Of note, the degree of TCR clonotype sharing was unusually high in the HIV controller group, with close to half of TCR sequences sharing common CDR3 motifs. Furthermore, between 5 and 10% of TCR sequences from the controller group coded for public clonotypes, which are defined as identical CDR3 sequences found in at least two individuals. Functional analysis of the most prevalent public clonotypes by TCR transfer showed they conferred highly sensitive, polyfunctional Gag-specific CD4+ T cell responses to healthy donor cells. Thus, the transfer of public TCR clonotypes was sufficient to recapitulate CD4+ T cell properties associated with HIV control.
Inducing T cell responses as efficient as those achieved in controlled HIV infection represents a key objective in vaccine development. Whether candidate HIV vaccines can induce the TCR clonotypes associated with HIV control remains to be explored. Importantly, the high degree of clonotype sharing observed in controller CD4+ T cells opens the possibility to search for these clonotypes in a diverse population of vaccinated volunteers. We focused the clonotypic analysis on Gag293 specific cells, as this epitope is remarkably immunodominant, independently of the MHC II background (11, 23, 24). In addition, the Gag293 epitope is located in the most conserved region of HIV-1 capsid, called the major homology region, and is thus rarely mutated, with two cases of mutational escape in minor viral variants reported so far (25, 26). Given these advantageous properties, we set to evaluate the degree of overlap in the Gag293-specific clonotypic repertoire induced by vaccination and controlled HIV infection.
As a proof of concept, we analyzed Gag293-specific responses in healthy volunteers who had received the ADVAX DNA vaccine, in the frame of a phase I trial conducted at the Rockefeller University (27). ADVAX was the first candidate HIV vaccine to be administered by intramuscular electroporation (EP), a delivery modality that increased T cell responses up to 70 fold as compared to standard intramuscular (IM) syringe injection. ADVAX consisted in a set of two plasmids containing mutated versions of HIV-1 env/gag and pol/nef/tat genes, respectively, with viral sequences derived from a clade B′/C HIV-1 strain predominant in the province of Yunnan, China (28, 29). ADVAX DNA electroporation was proven to be safe, tolerable, and effective in improving the magnitude and durability of T cell responses (27). Of note, the breadth of T cell responses induced by this DNA vaccine was comparable to that induced by more complex viral vectored vaccine regimens (30), making ADVAX a good candidate for TCR clonotypic analysis. In the present study, we report that ADVAX DNA vaccination by electroporation induced a Gag293-specific repertoire that included multiple public motifs and public clonotypes shared with HIV controllers, while repertoire overlap with that of treated patients was limited. Thus, DNA vaccination by electroporation not only improves the magnitude of T cell responses, but also primes for TCR clonotypes that are associated with HIV control, suggesting the induction of a high quality clonotypic repertoire. Based on the extent of clonotype sharing observed in the present study, we propose that TCR clonotypic analysis could be used more broadly as a novel approach to guide vaccine development.
MATERIALS AND METHODS
Clinical samples
The study was carried out on cryopreserved PBMC samples collected from participants in the ADVAX-EP vaccination trial. This clinical trial aimed at evaluating the safety and immunogenicity of an intramuscular prime and boost injection of the ADVAX DNA-based HIV vaccine via electroporation compared to standard intramuscular injection or placebo (registration NCT00545987). Among participants in the EP groups (n=24), those in the EP high dose group (n=8) received 4 mg of ADVAX DNA administered at weeks 0, 8, and 36 via the TriGrid device (Ichor Medical System). Participants in the IM group (n=8) received 4 mg of ADVAX DNA at weeks 0 and 8 via syringe intramuscular injection. Participants in the Placebo group received saline via electroporation at weeks 0 and 8 (n=5) or weeks 0, 8, and 36 (n=3). The clinical trial was completed in 2011, and showed that EP DNA delivery was safe, well tolerated, and that it enhanced vaccine immunogenicity (27).
HIV controllers (HIC group; n=14) were recruited through the CO21 CODEX cohort set up by Agence Nationale de Recherche sur le SIDA et les hépatites virales (ANRS). HIV controllers were defined as HIV-1 infected patients who had been seropositive for >5 years, had received no antiretroviral treatment, and for whom >90% of plasma viral load measurements were undetectable by standard assays. All HIV controllers included in the study had current viral loads <50 copies/mL. These patients had a median duration of viral control of 19.6 years (IQR: 9.8–26.0) and a median CD4+ T cell count of 875 (IQR: 648–1,400). The treated patients (HAART group; n=14) had received antiretroviral therapy for a minimum of 5 years and showed efficient HIV-1 suppression with viral loads <50 copies/mL. The treated patients had a median CD4+ T cell count of 570 (IQR: 270–1,534). The Gag293-specific repertoire of patients from the HIC and HAART groups was previously reported in (22).
Ethical statement
The study was approved by the Institutional Review Board of the Rockefeller University Hospital (reference DHO-0614). All participants in this study provided written informed consent after appropriate review, discussion and counseling by the clinical study team.
Antibodies
The following antibodies were used for cell surface staining: CD3-eFluor 780-Allophycocyanin (eF780-APC, clone UCHT1), TCR-Allophycocyanin (APC, clone IP26) (all from eBioscience); CD4 BD Horizon R-phycoerythrin-CF594 (PE-CF594, clone RPA-T4), CD4-R-phycoerythrin-cyanine 7 (PE-Cy7, clone SK3), CD69-R-phycoerythrin (PE, clone FN50), (all from BD Biosciences); CD14-Viogreen (clone TÜK4), and CD19-VioGreen (clone LT19) (from Miltenyi Biotec); TRBV2-PE (clone IMMU 546, Beckman Coulter); CD8-Brilliant Violet 785 (BV785, clone RPA-T8), HLA-DR-PE-Cy7 (clone LN3), CD45RA-Brilliant Violet 421 (BV421, clone HI100), CCR7-PE-Cy7 (clone G043H7) (all from Biolegend). The fixable viability dye eFluor 506 (eF506, eBioscience) was added to gate out dead cells.
Generation of primary CD4+ T cell lines
Cryopreserved PBMCs from vaccinated volunteers were thawed, stimulated with immunodominant Gag peptides, and propagated as short term primary CD4+ T cell lines. PBMC were plated at 2×106 cells in 2 mL in 24-well plates in complete RPMI, consisting in RPMI 1640 medium with 2mM L-glutamine, 10mM HEPES, 100 μg/mL penicillin/streptomycin, and were supplemented with 10% human AB serum (Pan Biotech). Each PBMC sample (>107 cells) was subdivided and stimulated with peptides Gag293 (FRD YVD RFY KTL RAE QAS QE), Gag263 (KRW IIL GLN KIV RMY SPT SI), Gag161 (EKA FSP EVI PMF SAL SEG AT), or with control peptide buffer. The peptides used for stimulation were 20-mers with >95% purity (Proteogenix). Cell were stimulated with 10−5 M Gag peptide in the presence of 5 ng/mL recombinant IL-7 (Cytheris) and cultured for 2 weeks prior to analysis. Recombinant IL-2 (Chiron, Novartis) was added to a final concentration of 100 U/ml starting from day 2 of culture, and every 2 days afterwards
ELISpot assay
ELISpot assays were performed to quantify IFN-γ secretion by Gag-specific CD4+ T cell lines. 96-well nitrocellulose plates (Millipore Multiscreen HTS plate, MSHAS410) were coated with 1 μg/mL human IFN-γ capture monoclonal antibody (Clone 1-D1K, Mabtech) and incubated overnight. On the day of the ELISpot, the plates were blocked with 100 μl/well RPMI supplemented with 5% FBS for 2h at 37°C. Cell lines deprived of IL-2 for 16h before analysis were plated in duplicate at 30,000 cells/well in coated ELISpot plates, and incubated with serial dilutions of corresponding peptides for 16h at 37°C. Wells were then washed 5 times with PBS+0.1% Tween-20. The plate was then incubated with 1 μg/mL of biotinylated IFN-γ detection antibody (Clone 7B6-1, Mabtech) for 90 min at 37°C, followed by 4 washes with PBS+0.1% Tween-20. Next, wells were treated with 100 μl/well alkaline phosphatase-labeled streptavidin (Sigma-Aldrich Extravidin Alkaline Phosophatase) for 45 min at 37°C and washed 3x with PBS 0.1% Tween-20. The ELISpot were developed with the phosphatase-based chromogenic substrate BCIP/NBT (KPL, Eurobio) for 30 min at 37°C. The plate was then washed extensively with water and dried. IFN-γ spot-forming cells (SFC) were automatically counted with a Bioreader 4000 system (Bio-Sys). The ELISpot response was expressed as SFC/106 cells after subtracting background. Wells were counted as positive if the number of SFC was at least two times above background level. ELISpot responses were measured in response to serial peptide dilutions from 4×10−6 to 10−11 M, and the last dilution that gave a number of SFC at least two times above background was used to measure antigen sensitivity.
MHC class II tetramer labeling
CD4+ T cell lines displaying a positive IFN-γ ELISpot response to Gag293 (spanning amino acids 293–312 in HIV-1 p24-Gag) were labeled with HLA-DR-matched Gag293-loaded tetramer and FACS sorted. Vaccinees were first genotyped for the HLA-DRB1 gene using the INNO-LiPA HLA-DRB1 Plus kit as per the manufacturer’s protocol (Fujirebio). APC-labeled MHC II tetramers for the HLA-DRB1*0101 (DR1), HLA-DRB1*0701 (DR7), HLA-DRB1*1502 (DR15), and HLA-DRB5*0101 (DRB5) alleles were obtained through the NIH Tetramer Core Facility at Emory University, USA. HLA-DRB1*1101 (DR11) biotinylated monomers were obtained through the Tetramer Core Laboratory of the Benaroya Research Institute (Seattle, USA). Monomers were loaded with 0.2 mg/mL peptide by incubation at 37°C for 72h in the presence of 2.5 mg/ml n-octyl-β-D-glucopyranoside and protease inhibitors in sodium phosphate loading buffer. Peptide-loaded monomers were tetramerized using 5.9 μg APC-conjugated streptavidin (eBioscience) for every 10 μg monomer. For each tetramer loaded with the Gag293 peptide, a corresponding control tetramer was loaded with the control CLIP peptide (PVS KMR MAT PLL MQA).
MHC II tetramer labeling was performed as recently described (31). Briefly, primary CD4+ T cells lines were incubated with 1μg MHC II tetramer/106 cells in complete RPMI supplemented with 15% human AB serum for 60 min at 4°C. The following combination of cell surface antibodies was added for an additional 30 min: CD3-eF780-APC, CD4-PE-CF594, CD8-BV785, CD45RA-BV421, CCR7-PE-Cy7, CD14-VioGreen, and CD19-VioGreen. Gag293-specific tetramer-labeled (Tet+) cells were visualized in the singlet, viable, CD3+, CD4+, CD14−, CD19−, CD8− lymphocyte gate, and were sorted using a FACSAria II cell sorter (BD Biosciences) installed in a microbiological safety cabinet. Each Gag293-tetramer labeled sample was matched with a control-tetramer labeled sample processed in identical conditions. Sorted Tet+ cells were re-suspended in RLT buffer (Qiagen) and kept frozen at −80°C until RNA extraction.
TCR CDR3 sequence analysis
The expression of 35 TCRα variable gene (TRAV) families and of 24 TCRβ variable gene (TRBV) families was measured by quantitative RT-PCR. Briefly, total RNA was extracted from Tet+ cells using the RNeasy mini or micro kits (Qiagen) according to the manufacturer’s protocol. Next, cDNA was obtained by reverse transcription with 500 μg/mL oligo (dT)17 and 200 U of Superscript II reverse transcriptase (Life technologies). A cDNA aliquot was amplified with each of 35 TRAV and 24 TRBV family-specific primers, in combination with a constant region TRAC or a TRBC primer, respectively, as described by A. Lim and coll. (32). Amplification was performed in the presence of a TRAV- or TRBV-specific TaqMan probe on an ABI 7300 real time PCR device (Applied Biosystems).
PCR products corresponding to the three amplified families (TRAV24, TRAV29 and TRBV2), were cloned and sequenced. The primers used to amplify the CDR3 junctions were: TRAV24 forward primer: 5′-CCG AGG CCT TGT TTG TAA TG-3′; TRAV29 forward primer: 5′-ACC CTG CTG AAG GTC CTA CAT TCC-3′; TRAC reverse primer: 5′GTG AAT AGG CAG ACA GAC TTG T-3′; TRBV2 forward primer: 5′-GGT CCG GAA TGG ATA CCT GGC TCG TAT GCT GGG C-3′; TRBC reverse primer: 5′-CCG GTC GAC CTA GCC TCT GGA ATC CTT TCT CTT GAC C-3′. PCR products were cloned into the pCR-Blunt-II-TOPO vector (Life Technologies), transformed into competent E. coli, and analyzed by DNA sequencing (Eurofins Genomics, Germany).
Bioinformatic analysis of the HIV-specific TCR repertoire
TRA and TRB sequences were analyzed with the software suite from the International ImMunoGeneTics (IMGT) Information System (33). The V(D)J gene nomenclature used is that of the IMGT database (www.imgt.org). Clonotypic diversity of the TRAV24, TRAV29 and TRBV2 repertoires was evaluated (i) by counting the number of unique amino acid clonotypes (clonotypes AA) per 100 CDR3 nucleotide sequences (ii) by computing Simpson’s diversity index. Simpson’s diversity index takes into account both the number of clonotypes and the frequency of each clonotype in the dataset, and is maximal when all clonotypes have an equal representation. The number of N and P mutations introduced during the V(D)J recombination process was determined by comparing the observed CDR3 sequences to their germline counterparts, using the Junction analysis module of the IMGT/HighV-QUEST software (33). The distribution of TRAV, TRAJ, TRBV, TRBJ, and TRBD genes in the sequence set was computed with the statistics module of IMGT/HighV-QUEST. The CDR3 lengths corresponded to the number of amino acids between, but not including, the two conserved residues C104 and F/W118, as defined by the IMGT-ONTOLOGY unique numbering system. In contrast, the “CDR3 junctions” included the conserved C104 and F/W118 residues. Kurtosis, which measures the “peakedness” of a distribution, was used to evaluate biases in CDR3 lengths. Kurtosis was measured in the Prism v6.0 software (GraphPad). A Gaussian distribution has a kurtosis of zero, while a flatter distribution has negative kurtosis, and a more “peaked” distribution has positive kurtosis.
Motifs enriched in the TRAV24, TRAV29 and TRBV2 CDR3 sequence sets were first identified with the MEME motif discovery software version 4.10.0 (34) available at http://meme-suite.org. The MEME software chooses the width and number of occurrence of each motif automatically in order to minimize the “E-value” of the motif, i.e. the probability of finding an equally well-conserved pattern in random sequences. Motifs were represented as sequence logos, where the relative sizes of the letters indicate their frequencies in the sequence set, and the total height of the letters represents the information content of the position, in bits. Based on the initial motif analysis, simpler public motifs included within the MEME motifs were identified and counted using the “Protein Pattern Find” module of the Sequence Manipulation Suite (35).
TCR lentivector construction and pseudo-virus production
For functional studies, full-length TCRα and TCRβ chains amplified from Gag293-specific Tet+ cells were cloned into lentiviral expression vectors. Full-length TRAV24 chains were amplified with a forward primer containing the TRAV24 leader sequence with an NheI restriction site and a Kozak sequence added in 5′ (5′-CGG CTA GCC GCC ACC ATG GAG AAG AAT CCT TTG GCA GCC-3′) and a reverse primer containing the 3′ region of TRAC and a NotI site (5′-TTA GCG GCC GCG CTG GAC CAC AGC CGC AGC G-3′). Full-length TRAV29 chains were amplified similarly a forward primer containing the TRAV29 leader sequence (5′-CGG CTA GCC GCC ACC ATG GCC ATG CTC CTG GGG G-3′). Full-length TRBV2 chains were amplified with a forward primer containing the TRBV2 leader sequence and a BspEI site in 5′ (5′-GGT CCG GAA TGG ATA CCT GGC TCG TAT GCT GGG C-3′) and a reverse primer containing the 3′ of TRBC and a SalI site (5′-CCG GTC GAC CTA GCC TCT GGA ATC CTT TCT CTT GAC C-3′). The TCRα and TCRβ chains were first cloned separately in the pCR-Blunt-II-TOPO vector, and then combined into the pCDH-EF1-MCS-T2A vector (SBI System Bioscience), with a self-cleaving T2A sequence inserted in between, ensuring an equimolar expression of the two chains from the same transcript. All constructs were verified by sequencing (Eurofins Genomics).
Lentiviral particles encoding TCRs were prepared by transient transfection of HEK 293Tn cells (SBI System Bioscience) using Lipofectamine 2000 reagent (Life Technologies) according to the manufacturer’s protocol. The lentiviral vector pCDH-EF1-MCS-T2A containing the TCR inserts, the packaging plasmid psPAXII (Addgene), the pCMV-VSV-G envelope plasmid, and the pRev plasmid (a kind gift from P. Charneau) were mixed at a 3:2:1:1 ratio and transfected at 65 μg per 162 cm2 cell flask. 48h after transfection, supernatants were collected, passed on a 0.45 μm filter, and concentrated by ultracentrifugation at 23,000 g for 2h at 4°C on a 20% sucrose cushion. Viral particles were re-suspended in PBS and frozen in aliquots at −80°C. Gag p24 antigen concentration was measured with the Alliance HIV-1 p24 Antigen ELISA kit (Perkin Elmer).
TCR functional analysis
Gag293-specific TCRs amplified in ADVAX recipients were transduced in the J76 cell line, which is a Jurkat derivative lacking endogenous TCR expression (36). J76 cells were kindly provided by Dr Mirjam Heemskerk (Leids Universitair Medisch Centrum, The Netherlands). Function was evaluated by the induction of the early activation marker CD69 on TCR-transduced J76 cells stimulated with decreasing doses of Gag293 peptide.
TCR transduction in J76 cells
J76 cells were maintained in complete RPMI in the presence of 10% fetal bovine serum (FBS). For TCR transfer, 0.5 x 106 J76 cells were resuspended in 0.5 mL complete RPMI medium supplemented with 10% FBS, and plated in a 24-well plate. TCR lentiviral particles corresponding to 200 ng of p24 antigen were added to each well and thoroughly resuspended by pipetting. After 3 hours, 0.5 mL fresh medium was added to each well. Three days later, J76 cells were labeled with CD4-PE-CF594, TCR-APC, CD3-eF780-APC antibodies and the eF506-conjugated viability dye, and analyzed by flow cytometry. Transduction efficiency was determined by the percentage of upregulation of the CD3 and TCR molecules at the cell surface.
J76 cell activation assay
Murine fibroblasts (L cells) stably transfected to express a single human HLA-DR allele (DR1, DR11, DR15, or DRB5) were used as antigen presenting cells (37). L cells were kindly provided by Dr. Bernard Maillère (Saclay Institute of Biology and Technologies, France). L cells were maintained in complete RPMI supplemented with 10% FBS and 1% Non-Essential Amino Acids (Life Technologies). L-cells were loaded with serial Gag293 peptide dilutions ranging from 2 x 10−5 to 10−11 M. Aliquot of 5 x 104 peptide-loaded L-cells were co-cultured with 5 x 104 TCR-transduced J76 cells in a 96-well plate for 16h at 37°C. The next day, cells were washed and labeled with CD69-PE, CD4-PE-Cy7, TCR-APC, CD3-eF780-APC antibodies and the eF506-conjugated viability dye. Events were acquired using a FACSCanto II flow cytometer (BD Biosciences) and analyzed to determine the cell surface expression of CD69 as a measure of J76 cell activation upon Gag293 recognition.
Statistical analyses
Statistics were computed with the Prism v7.0 software (GraphPad). P values <0.05 were considered statistically significant. Differences between groups were analyzed with the non-parametric Mann-Whitney U test. Differences in motif overlap frequencies were analyzed in contingency tables with Fisher’s exact test. Half maximal effective concentrations (EC50) were obtained after non-linear curve fit using a sigmoidal dose response model in Prism. All significant differences between groups (P < 0.05) are reported in data plots.
RESULTS
Increased Gag293-specific CD4 responses upon EP DNA vaccination
The ADVAX DNA vaccine was administered to healthy volunteers as a series of 3 electroporations of 4 mg DNA at weeks 0, 8, and 36 in the EP high dose group (n=8), and as two intramuscular injections of 4 mg DNA at weeks 0 and 8 in the IM group (n=8). Ex vivo analyses of PBMC in the ADVAX trial had shown detectable Gag-specific ELISpot responses in vaccinated volunteers of the EP high-dose group but not in the IM group (27). To increase the sensitivity of Gag-specific CD4+ T cell detection, we generated short-term primary CD4+ T cell lines primed with one of three immunodominant Gag peptides, and analyzed IFN-γ ELISpot responses after peptide restimulation. The three 20-mer peptides Gag293, Gag263, and Gag161 were chosen as together these induce a CD4 response in a majority of HIV-1 infected patients, and in over 80% of HIV controllers tested (11, 17). Primary CD4+ T cell lines were obtained from PBMC collected 2 weeks after the second vaccination (wk10) for both the EP and IM groups (n=7 and n=6, respectively; see Supplemental Fig. 1A for a list of samples tested). A second sample obtained 2 weeks after the third vaccination (wk38) was also tested for a subset of volunteers of the EP group (n=5). Controls included volunteers of the Placebo group (n=4), who had received saline instead of ADVAX DNA during the electroporation procedure.
Gag293-specific CD4 responses proved significantly higher in the EP group than in the IM group (P<0.05), confirming the enhanced immunogenicity conferred by DNA electroporation (Fig 1A). Responses to the other two Gag peptides, Gag263 and Gag161, were less frequent and did not significantly differ between the EP and IM group (Fig. 1B and C). One member of the Placebo group showed a positive response to Gag263, which may have resulted from in vitro priming of naive CD4+ T cell precursors, as previously reported (38, 39).
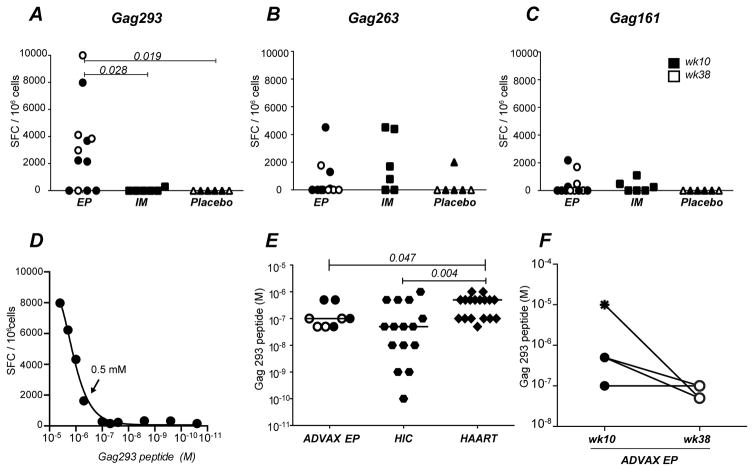
Figure 1. EP DNA vaccination induces high sensitivity Gag293 specific responses.
(A, B, C) IFNγ ELISpot responses in ADVAX vaccine recipients: production of IFNγ by CD4+ T cell lines specific for Gag293 (A), Gag263 (B), and Gag161 (C) were compared between healthy volunteers who received the ADVAX vaccine via electroporation (EP; circles), by intramuscular injection (IM; squares; n=6), or who received a saline solution by electroporation (Placebo; triangles; week 10, n=4; week 38, n=2). In the EP group, samples obtained at week 10 (solid symbol, n=7) or week 38 (open symbol, n=5) are distinguished. IFNγ production was measured by ELISpot assay on CD4+ T cell lines restimulated with 10−5 M peptide, and was expressed by the number of spot forming cells (SFC) per million cells. Statistically significant differences (P < 0.05) computed using the Mann Whitney U test are reported.
(D) Representative example of antigen sensitivity measurement in a Gag293-specific cell line from a volunteer in the ADVAX EP group (1 out of 24 experiments is shown). The IFNγ ELISpot response was measured in function of decreasing doses of Gag293 peptide, and antigen sensitivity was determined as the last positive dilution of peptide which elicited a positive IFNγ response 2-fold over background. Arrow indicates the last positive dilution in this example (0.5μM).
(E) Comparison of antigen sensitivity in Gag293-specific CD4+ T cell lines derived from HIV Controllers (HIC, hexagonal symbols, n=15), treated patients (HAART, diamonds, n=17), and vaccinees (ADVAX EP, circles) analyzed at week 10 (solid symbols, n=4) and week 38 (open symbols n=4). ADVAX samples with undetectable Gag293 responses are not shown. Significant differences (P < 0.05) obtained by the Mann-Whitney U test are reported.
(F) Comparison of the antigen sensitivity of Gag293-specific CD4+ T cell lines from ADVAX EP samples (n=4) obtained at week 10 (solid circle), two weeks after 2nd EP vaccination, or at week 38 (open circle), two weeks after the third EP vaccination. The data point with an undetectable IFN-γ ELISpot response has been arbitrarily assigned an antigen sensitivity of 10−5 M (star symbol), corresponding to the limit of detection of the assay.
EP DNA vaccination induces Gag293-specific responses of higher sensitivity than progressive HIV infection
We next analyzed the antigen sensitivity of the immunodominant Gag293-specific response, by restimulating the primary CD4+ T cell lines with decreasing doses of Gag293 peptide. The antigen sensitivity was defined as the last peptide dilution that gave a positive IFN-γ ELISpot response 2x higher than background, with a higher antigen sensitivity corresponding to a lower peptide dose (Fig. 1D). We reported previously that this measurement clearly distinguished HIV controllers (HIC) from patients who were treated in the chronic stage of the infection (HAART), with a higher antigen sensitivity of the Gag293-specific response in the HIC group (11). Vaccinated volunteers from the ADVAX EP group showed a median antigen sensitivity (10−7M) that was close to that found in HIV controllers (5 x 10−8M) and significantly higher than that found in treated patients (5x10−7M; p<0.05) (Fig. 1E). However, antigen sensitivity in vaccinees did not reach the very high levels seen in a subset of the HIV controllers (up to 10−10M).
Comparison of antigen sensitivities of the Gag293-specific responses at week 10, after the second DNA EP vaccination, and at week 38, after the third DNA EP vaccination, showed a non-significative trend towards higher antigen sensitivities (Fig. 1F), suggesting a possible maturation of the CD4 response upon vaccine boosting. Taken together, these findings indicated that EP DNA vaccination induced Gag293-specific responses of higher antigen sensitivity than treated chronic HIV infection, even though the levels and duration of antigenic stimulation were much lower in the case of vaccination.
Biased TCR variable gene usage in Gag293-specific CD4+ T cells amplified by EP DNA vaccination
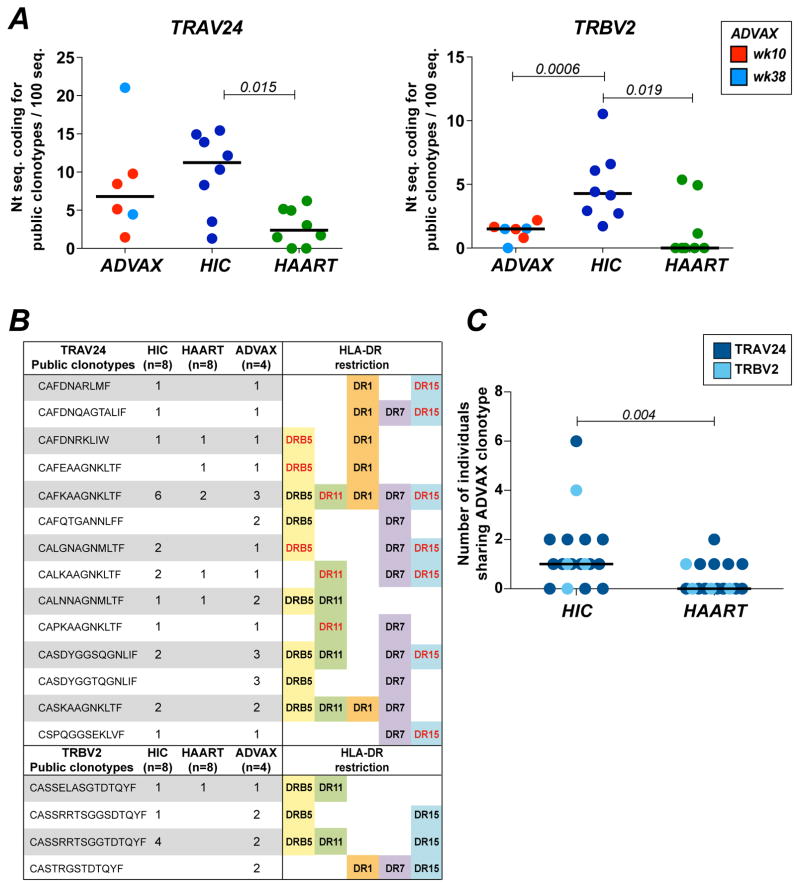
To characterize the Gag293-specific TCR clonotypic repertoire, vaccinees were first typed for HLA DRB1. Primary CD4+ T cell lines were then evaluated for the presence of Gag293-specific cells by labeling with matched HLA-DR tetramers loaded with the Gag293 peptide. Five tetramers were used for this study, based on HLA alleles DRB1*0101 (DR1), DRB1*0701 (DR7), DRB1*1101 (DR11), DRB1*1502 (DR15), and DRB5*0101 (DRB5). Matched tetramers loaded with an irrelevant CLIP peptide were used for negative controls, as shown for representative examples (Fig. 2A and Supplemental Fig. 1B). Additional negative controls consisted in healthy donor PBMC (Supplemental Fig. 1C). Five vaccinees from the EP group (hereafter referred to as the ADVAX group) showed detectable tetramer-positive (Tet+) cell populations, which were sorted for TCR clonotypic analysis. When feasible, samples were sorted with two HLA-DR tetramers, and at two available time point. The frequency and mean fluorescence intensity (MFI) of the Tet+ populations are reported in Supplemental Fig. 1A. Tet+ cell samples were analyzed by a series of 35 TRAV-specific and 24 TRBV-specific real time RT-qPCR reactions to quantify the distribution of the variable TCR gene families in the Gag293-specific CD4+ T cell population (representative example shown in Fig. 2B). The TRAV and TRBV qPCR results obtained for the ADVAX group (Fig. 2C, top panels) were compared to those previously reported for the HIC and HAART groups (Fig. 2C, middle and bottom panels) (22).
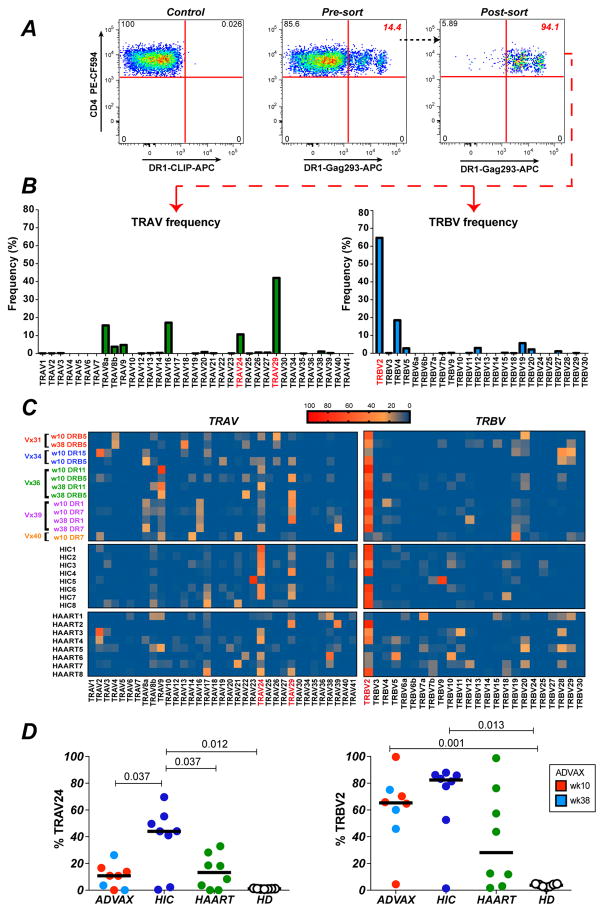
Figure 2. Biased TCR variable gene usage in Gag293-specific CD4+ T cells amplified by EP DNA vaccination.
(A) Example of Gag293-specific CD4+ T cells sorting after HLA-DR tetramer labeling. A primary CD4+ T cell line derived from an ADVAX recipient was labelled with an HLA-DRB1*0101 (DR1) tetramer loaded with Gag293. The percentage of tetramer+ (Tet+) cells in the total CD4+ T cell population (middle plot) and in the sorted Tet+ population (right plot) is reported in red. A sample labeled with the DR1 tetramer loaded with an irrelevant peptide (CLIP) was used as a negative control (left plot).
(B) Representative example of qPCR analysis (1 out of 8 experiments) to quantify the relative expression of the TRAV and TRBV variable genes in sorted Gag293-specific CD4+ T cells from an ADVAX recipient.
(C) Heatmaps depicting the TRAV and TRBV gene usage in Gag293-specific CD4+ T cells from vaccinee samples (n=5) sorted with different HLA-DR tetramers. The TRAV and TRBV gene usage in Gag293-specific CD4+ T cells from HIV controllers (HIC) and treated patients (HAART) are reported below. The most prevalent TRAV and TRBV genes are marked in red.
(D) Comparison of the expression of TRAV24 and TRBV2 in Gag93-specific cells from vaccinees (week 10, red, n=5; week 38, light blue, n=3), HIV controllers (HIC, dark blue, n=8) and treated patients (HAART, green, n=8). The frequency of TRAV24 and TRBV2 in the total TCR repertoire from healthy donors (HD, open circles, n=7) is reported for comparison. Significant differences (P < 0.05) obtained with the Mann-Whitney U test are reported.
TRAV gene distribution proved relatively diverse in the ADVAX group, with a predominance of the TRAV29 family, followed by TRAV24 and TRAV 9 (Fig. 2C, left). Comparison with HIV infected patients showed an overall similar pattern, though the clonotypic repertoire was more focused on TRAV24 in HIV Controllers compared to both vaccinees and treated patients. Quantitation of the percentage of TRAV24 expression confirmed the significantly higher expression of TRAV24 in the HIC group, as compared to the ADVAX and HAART groups (P<0.05 in both cases; Fig. 2D, left). Of note, Gag293-specific cells from the 3 groups showed a bias in TRAV24 expression compared to healthy donor T cells (HD), where TRAV24 was only expressed at low levels, with a median value of 1%.
TRBV gene distribution was highly biased in the ADVAX group, with a marked dominance of the TRBV2 family (Fig. 2C, right). The TRBV2 bias in vaccinees appeared as marked as that seen in HIV controllers, and more prevalent than that seen in treated patients. Quantitation of TRBV2 expression showed high median values in the 3 groups (82% in HIC; 65% in ADVAX; 28% in HAART) compared to those observed in healthy donor T cells (4% in HD; Fig. 2D, right). Taken together, this analysis revealed that EP DNA vaccination induced a strongly biased Gag293-specific TCR repertoire, with preferential expression of TCR variable family genes that were also amplified in HIV infection. Interestingly, vaccinees shared a marked TRBV2 bias with HIV controllers, raising the possibility of a shared clonotypic repertoire between the two groups.
Intermediate levels of clonotypic diversity in vaccinated volunteers
Clonotypic repertoire analysis of ADVAX samples was first carried out for the two variable gene families preferentially shared with HIV controllers, TRAV24 and TRBV2, to allow comparisons between groups. Positive PCR products (>5%) corresponding to these families were cloned, sequenced, and analyzed with the software tools provided by the ImmunoGeneTics information system (IMGT) (33). A minimum of 50 productive CDR3 sequences were analyzed per ADVAX sample, with the full list of sequences provided in Supplemental Table 1. This dataset, which consisted in 473 TRAV24 sequences and 723 TRBV2 sequences, was compared to those previously reported for 8 HIV controllers (HIC: 584 TRAV24 and 716 TRBV2 sequences) and 8 efficiently treated patients (HAART: 496 TRAV24 and 566 TRBV2 sequences) (22).
Productive TRAV24 and TRBV2 sequences were evaluated for diversity by counting the number of distinct clonotypes, i.e. the number of unique CDR3 amino acid (a.a.) sequences, normalized to a total of 100 sequences. The diversity of TRAV24 clonotypes proved variable among vaccinees, with a few samples that could be as diverse as those of controllers (Fig. 3A, left). The diversity of TRBV2 clonotypes was significantly lower in vaccinees than in controllers (P<0.05), and comparable to that observed in treated patients (Fig. 3A, right). Further analyses based on the Simpson’s diversity index, which takes into account both the number of clonotypes and the frequency of each clonotype in the dataset, supported the notion that clonotypic diversity in the ADVAX group was intermediate between those observed for controllers and treated patients (Supplemental Fig. 2A). Thus, vaccination with DNA alone was sufficient to induce a relatively diverse Gag293-specific clonotypic repertoire.
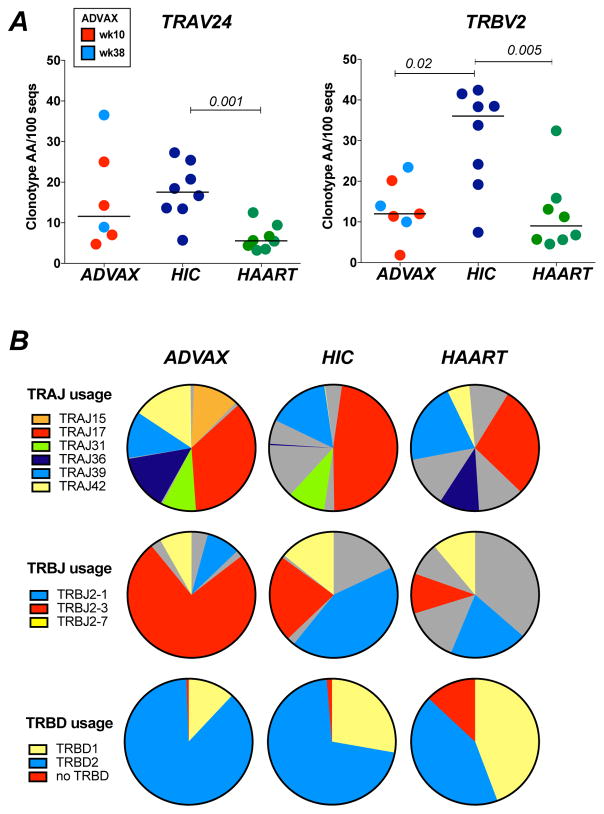
Figure 3. Analysis of clonotypic diversity in the Gag293-specific repertoire.
(A) Diversity of the TRAV24 (left) and TRBV2 (right) clonotype datasets was evaluated by the number of unique CDR3 amino acid sequences (clonotype AA) obtained per 100 cloned nucleotide sequences. Clonotypic diversity was compared between ADVAX recipients (week 10, red, TRAV24 n=4, TRBV2 n=4; week 38, light blue, TRAV24 n=2, TRBV2 n=3), HIV Controllers (HIC, dark blue, n=8), and treated patients (HAART, green, n=8). Significant differences (P < 0.05) obtained with the Mann-Whitney U test are reported.
(B) J and D gene usage in Gag293-specific clonotypes: Frequencies of TRAJ genes (top row), TRBJ genes (middle row), and TRBD genes (bottom row) in Gag293-specific TRAV24 or TRBV2 sequences were analyzed in the ADVAX, HIC, and HAART groups. TRAJ, TRBJ and TRBD gene families that predominate in in vaccinees (>8%) are depicted in colours, while all other genes families are shown in grey. Number of sequences analyzed: ADVAX TRA: 473; HIC TRA: 584; HAART TRA: 496; ADVAX TRB: 723; HIC TRB: 716; HAART TRB: 566.
Biased J and D gene usage in Gag293-specific CD4+ T cells amplified by DNA vaccination
Analysis of the distribution of junction (J) and diversity (D) TCR gene segments highlighted a restricted usage of these genes in the ADVAX sequence dataset (Fig. 3B). Six TRAJ genes, out of a total of 61 TRAJ genes reported in the IMGT database, were dominant in the ADVAX dataset, representing a total 98% of ADVAX TRAV24 sequences (Fig. 3B, top row). These 6 TRAJ genes were shared in 74% of controller sequences and 70% of treated patient sequences, suggesting a common TRAJ bias. TRAJ17 was dominant among the TRAV24 clonotypes from the three groups (36% in ADVAX, 48% in HIC, 27% in HAART), supporting the notion of a common TCR bias.
In the TRBV2 sequence dataset, a strong bias for TRBJ2–3 was apparent in the ADVAX group (75% of sequences), while this bias was less marked in the controller group (22%) and even less so in the HAART group (10%) (Fig. 3B, middle row). The 3 most prevalent TRBJ genes in ADVAX dataset, which together represented 91% sequences, were shared in 79% of controller sequences and 41% of treated patient sequences, suggesting a higher degree of similarity between vaccine and controller TRBV2 sequences. This notion was confirmed by the analysis of D gene distribution, as vaccinee and controller TRBV2 sequences preferentially used TRBD2, while treated patient sequences used TRBD1 as frequently as TRBD2 (Fig. 3B, bottom row).
Analysis of CDR3 length distribution in TRAV24 sequences showed a peak at 10 aa in the 3 groups, using the IMGT definition for CDR3 length, which excludes the conserved C104 and F/W118 terminal residues (Supplemental Fig. 2B, left). The kurtosis parameter measures the peakedness of a distribution, and takes negative values for distributions “flatter” or with more outliers than the Gaussian distribution. Kurtosis was higher in the ADVAX and HIC groups than in the HAART group, indicating a distribution of TRAV24 CDR3 lengths more focused around the 10 aa peak in vaccinees and controllers compared to treated patients (Supplemental Fig. 2C, left). In the TRBV2 sequence dataset, CDR3 lengths peaked at 13 aa in the ADVAX and HIC groups, while CDR3 lengths were more diverse in the HAART group (Supplemental Fig. 2B, right). This notion was supported by positive kurtosis values in the ADVAX and HIC groups, and a negative value in the HAART group (Supplemental Fig. 2C, right). Taken together, both J/D gene usage and CDR3 length analyses suggested that vaccinee CDR3 sequences shared more features with those of controllers than those of treated patients.
High prevalence of public motifs shared between vaccinated volunteers and HIV controller CDR3 sequences
We next evaluated whether the Gag293-specific CDR3 sequences from vaccinated volunteers and HIV controllers shared common amino acid motifs. A first analysis using the MEME motif discovery software (34) showed similarities in the dominant motifs derived from the ADVAX and HIC TRAV24 datasets, with these similarities being particularly marked in the C-terminal half of the CDR3 region, consistent with shared TRAJ usage (Supplemental Fig. 3A, left). Analysis of the TRBV2 datasets revealed more complex motifs, with conserved residues apparent in the N-terminus, the central portion (positions 8–9), and the C-terminus of the CDR3 region (Supplemental Fig. 3A, right).
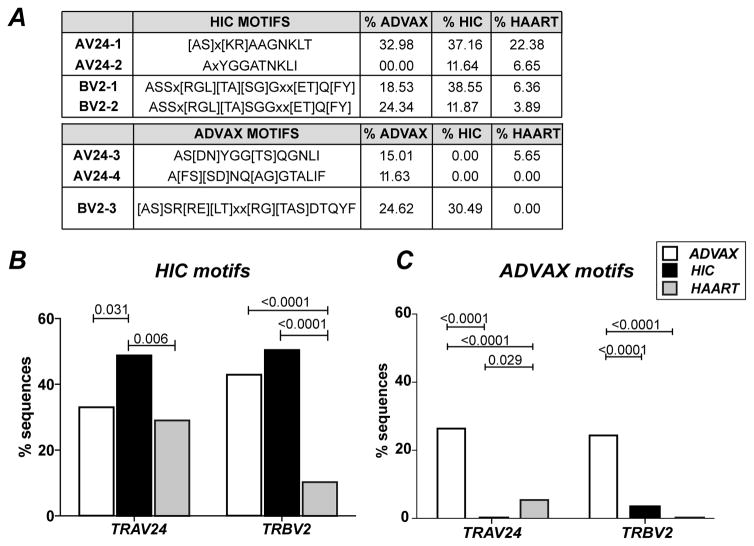
To further evaluate the degree of CDR3 motif sharing, we quantified the frequency of simpler motifs that were previously found to be highly prevalent in HIV controller Gag293-specific CDR3 sequences (22). The first TRAV24 motif, [AS]x[KR]AAGNKLT, was found in 33% of ADVAX TRAV24 sequences, compared to 37% of HIC and 22% of HAART sequences (Fig. 4A, top). The second TRAV24 motif AxYGGATNKLI defined in controllers was not found in the ADVAX sequence dataset. The sum of frequencies for these two motifs remained significantly higher in HIV controllers than in the other two groups as measured by Fisher exact test (Fig. 4B, left). In contrast, the two TRBV2 motifs, ASSx[RGL][TA][SG]Gxx[ET]Q[FY] and ASSx[RGL][TA][SGGxx[ET]Q[FY], were found to be almost as prevalent in the TRBV2 repertoire of vaccinees (42.87%) as in that of HIV controllers (50.42%), while these motifs were infrequent in treated patients (10.25%; ADVAX vs HAART: P<0.0001; Fig. 4B, right). Thus, one third or more of Gag293-specific clonotypes induced by DNA vaccination shared CDR3 motifs with those of HIV controllers, suggesting a significant degree of TCR repertoire overlap in spite of very different modes of antigenic priming.
Figure 4. Frequent sharing of public CDR3 motifs between HIV controllers and ADVAX recipients.
(A) The percentage of motif occurrence in the CDR3 sequence datasets from vaccinees (ADVAX), HIV controllers (HIC), and treated patients (HAART) was determined by using the sequence manipulation suite (http://www.bioinformatics.org/sms2/). Motifs are reported in single letter a.a. code; X stands for “any a.a.”; when several a.a. are possible at a given position in the motif, they are shown in brackets.
(B) The sum of AV24-1 and AV24-2 motif frequencies (left) and of BV2-1 an BV-2 motif frequencies (right) are plotted. (C) The sum of AV24-3 and AV24-4 frequencies (left) and the BV2-3 motif frequency (right) are plotted. (B, C) Significant differences (P < 0.05) between groups obtained with Fischer’s exact test are reported. Number of sequences analyzed: ADVAX TRA: 473; HIC TRA: 584; HAART TRA: 496; ADVAX TRB: 723; HIC TRB: 716; HAART TRB: 566.
We next sought to better characterize the part of the vaccine-induced clonotypic repertoire that wasn’t shared with HIV controllers. To do so, we searched for a.a. motifs that were prevalent in the ADVAX sequence dataset but did not overlap with the controller-derived CDR3 motifs. Using the Protein Pattern Find module from the Sequence Manipulation Suite (35), we identified two TRAV24 motifs and one TRBV2 motif that fit this definition (Fig. 4A, bottom). The two TRAV24 motifs, AS[DN]YGG[TS]QGNLI and A[FS][SD]NQ[AG]GTALIF, were present in a total of 26% of ADVAX sequences, but were absent in the controller sequence set and made only 5.6% of the treated patient sequences (p <0.0001 in both cases; Fig. 4C, left). Similarly, the BV2 motif [A/S]SR[R/E][L/T]xx[R/G][T/A/S]DTQYF was frequent in the ADVAX repertoire (24.6%) and rare or absent in patient groups (p <0.0001 in both cases; Fig. 4C, right). Thus, the Gag293-specific repertoire induced by DNA vaccination comprised a set of clonotypes characteristic of vaccinees, in addition to a set of clonotypes similar to those of HIV controllers.
DNA vaccination induces HLA-DR cross-restricted public clonotypes shared with HIV controllers
We next evaluated the frequency of Gag293-specific public clonotypes induced by DNA vaccination. A public clonotype was defined as a CDR3 a.a. sequence shared by at least two individuals belonging to any of the three groups studied, based on the ADVAX sequence dataset reported in the present study (Supplemental Table 1) and on the previously reported HIC and HAART total sequence datasets (22). This definition of public clonotypes implied the detection of identical CDR3 a.a. sequences, without any mismatch tolerated. A total of 14 TRAV24 and 4 TRBV2 public clonotypes were detected in the Gag293-specific sequences obtained from vaccinees, which represented 31.1% and 5.7% of the total number of TRAV24 and TRBV2 a.a. clonotypes, respectively (Fig. 5B). Thus, clonotype sharing appeared frequent, even though the subject studied had diverse HLA-DR genotypes. To better understand this phenomenon, we reported for each public clonotype the HLA-DR alleles corresponding to the MHC class II tetramers used for sorting, in the present study (black type) or in the previous one (red type) (22). This analysis highlighted that all public clonotypes showed a degree of HLA-DR cross-restriction, as indicated by their reactivity with at least two distinct HLA-DR tetramers. HLA-DR cross-restriction was particularly broad for the set of TRAV24 public clonotypes, with up to 5 distinct HLA-DR alleles recognized. These findings helped explain the extent of Gag293-specific clonotypes sharing in individuals of diverse HLA background.
Figure 5. DNA vaccination induces Gag293-specific public clonotypes shared with HIV controllers.
(A) Frequency of nucleotide sequences coding for TRAV24 (left) and TRBV2 (right) public clonotypes, normalized to 100 sequences. Public clonotypes are defined as identical CDR3 a.a. sequences shared by at least 2 individuals. Public clonotype frequencies were compared between ADVAX recipients (week 10, red, TRAV24 n=4, TRBV2 n=4; week 38, light blue, TRAV24 n=2, TRBV2 n=3), HIV Controllers (HIC, dark blue, n=8), and treated patients (HAART, green, n=8).. Significant differences (P < 0.05) obtained with the Mann-Whitney U test are reported.
(B) List public clonotypes found in the TRAV24 and TRBV2 repertoire of Gag293-specific CD4+ T cells from ADVAX recipients. HLA-DR cross-restriction is evidenced by the reporting the diverse MHC II tetramers that could be used to sort a given clonotype, either in the present vaccine study (black type) or in the previous HIV controller study (red type) (22).
(C) Clonotypic repertoire overlap between groups was evaluated by counting for each public clonotype found in vaccinees the number of patients in the HIC (left) and HAART (right) groups who shared this clonotype. TRAV24 (n=14) and TRBV2 (n=4) public clonotypes are represented by dark blue and light blue circles, respectively. The significant difference (P < 0.05) obtained with the Mann-Whitney U test is reported.
To compare public clonotype frequencies between groups, we computed the number of nucleotide sequences coding for public clonotypes, normalized to 100 sequences, using the datasets generated from Gag293-specific primary T cell lines (Fig. 5A). Public clonotype frequencies in the ADVAX group appeared intermediate between those obtained for HIV controllers and treated patients. Specifically, the TRAV24 public clonotype frequency in the HIC group was higher than in the HAART group (P=0.015) but did not differ significantly from the frequency measured in the ADVAX group. For the TRBV2 dataset, public clonotype frequencies were higher in the HIC group than in both the ADVAX and HAART groups (P=0.0006 and P=0.019, respectively).
It was noteworthy that the clonotypic repertoire of vaccinees contained the most prevalent public clonotypes previously reported in HIV controllers (Fig. 5B). Indeed, the most prevalent TRAV24 public clonotype (CAFKAAGNKLTF) found in 6/8 HIC and 2/8 HAART was also detected in 3/4 of the vaccinees. Similarly, the most prevalent TRBV2 public clonotype (CASSRRTSGGTDTQYF) found in 4/8 HIC was also detected in 2/4 of the vaccinees. To better evaluate clonotypic repertoire overlap between groups, we counted for each public clonotype found in vaccinees the number of patients in the HIC and HAART groups who shared this clonotype (Fig. 5C). This analysis showed that the Gag293-specific clonotypes induced by vaccination were more frequently shared with HIV controllers than with treated patients (P=0.004). Taken together, these findings highlighted the capacity of DNA vaccination to induce a set of broadly cross-restricted TCR clonotypes preferentially expressed in controlled HIV infection.
Shared CDR3 features between TRAV29 and TRAV24 Gag293-specific clonotypes
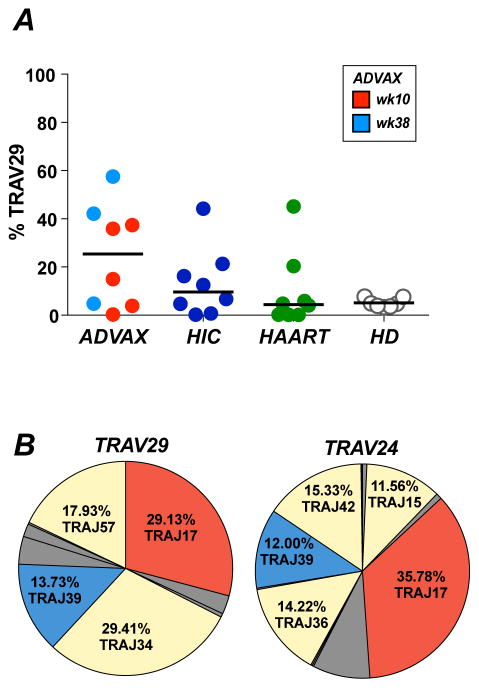
While TRBV2 was highly dominant in the Gag293-specific repertoire of vaccinees, TRAV usage was more diverse, with TRAV29 often predominating over TRAV24 (Fig. 2C). Quantitation of TRAV29 usage among Gag293-specific CD4+ T cells showed a trend for a higher frequency of TRAV29 expression in vaccinees than in HIV controllers and treated patients (medians: ADVAX 25.4%; HIC 9.6%; HAART 4.4%), though the differences did not reach significance (Fig. 6A). To compare the properties of TRAV29 and TRAV24 clonotypes, we sequenced the TRAV29-positive Gag293-specific clonotypic repertoire in vaccinees (Supplemental Table 1). Analysis of 357 sequences obtained from 3 vaccinees with high TRAV29 expression revealed a clonotypic repertoire that was less diverse than the one obtained for TRAV24 (median number of a.a. clonotypes/100 sequences: 8.2 for TRAV29 vs. 11.6 for TRAV24; Simpson diversity index: 0.46 for TRAV29 vs. 0.69 for TRAV24; not shown). Shared clonotypes were not detected between the 3 vaccinated volunteers analyzed, suggesting a low frequency of TRAV29 public clonotypes.
Figure 6. Shared CDR3 features between TRAV29 and TRAV24 Gag293-specific clonotypes.
(A) Frequency of TRAV29 expression in Gag293-specific clonotypes from vaccinees (week 10, red, n=5; week 38, light blue, n=3), HIV controllers (HIC, dark blue, n=8) and treated patients (HAART, green, n=8). The frequency of TRAV29 in the total clonotypic repertoire of healthy donors (HD, clear circles, n=7) is shown for comparison.
(B) Pie charts depicting the distribution of TRAJ gene usage in Gag293-specific TRAV24 (top) or TRAV29 (right) sequences from ADVAX recipients. The dominant J genes shared between the TRAV24 and TRAV29 repertoires are colored in red and blue, while other amplified J genes (>10%) are shown in yellow. Number of sequences analyzed: ADVAX TRAV29: 359; ADVAX TRAV24: 473.
Interestingly, analysis of TRAJ distribution showed that TRAJ17, which was the most prevalent J gene in the TRAV24 dataset (35.78%), was also dominant in the TRAV29 dataset (29.13%) (Fig. 6B). TRAJ39 was also used at 12 to 13% in both datasets, resulting in over 40% of common J gene usage. Analysis of a.a. motifs with the MEME software revealed similarities in the C-terminal part of the dominant motifs found in TRAV29 and TRAV24 sequences, consistent with J gene sharing (Supplemental Fig. 3B). Specifically, an AGNKLTF motif, which was encoded by TRAJ17, predominated in the C-terminal region of the CDR3 for both datasets. Thus, comparison of the TRAV24 and TRAV29 clonotypes in vaccinees helped identify a set of conserved CDR3 residues that were likely to be involved in Gag293 recognition.
While the TRAV24 and TRAV29 clonotypes from vaccinees shared some conserved features, they differed in CDR3 length, which was in median 2 a.a. longer for TRAV29 clonotypes (median length: 12 versus 10 a.a., not shown). We analyzed the numbers of mutations introduced during the V(D)J recombination process, to determine whether a higher number of insertions or a lower number of deletions could account for the increased length of the TRAV29 clonotypes (Supplemental Fig. 3C). This analysis showed that compared to germline sequences, the TRAV29 clonotypes had a number of P+N mutations that was only slightly higher than that of TRAV24 private clonotypes (TRAV29 median=5; TRAV24 median=4; P=0.04). In contrast, TRAV29 clonotypes showed a low number of nucleotides trimmed from germline sequences (median=5) as compared to private TRAV24 clonotypes (median=10; P<0.0001). Thus, TRAV29 clonotypes had undergone only limited trimming upon V(D)J recombination, which could account for their longer length. Limited trimming may also help explain the frequent amplification of TRAV29 clonotypes during vaccination, as fewer steps are needed to generate such clonotypes during the V(D)J recombination process.
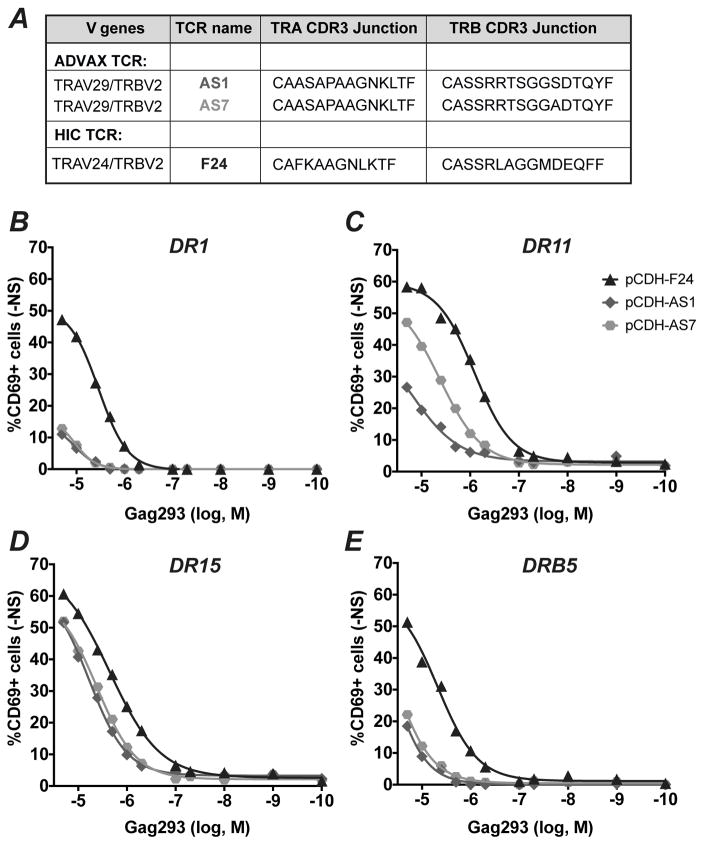
A dominant TRAV29 clonotype generates HLA-DR cross-restricted TCRs
We then set to determine whether a TRAV29 clonotype could confer functional properties associated with TRAV24 public clonotypes, such as HLA cross-restriction and high antigen sensitivity. To do so, we selected the most dominant TRAV29 clonotype (CAASAPAAGNKLTF), which represented 100% of TRAV29 sequences in sample Vx36 wk38 (Supplemental Table 1), and for which we could predict pairing with two highly expressed TRBV2 clonotypes (CASSRRTSGGADTQYF and CASSRRTSGGSDTQYF). Of note, the chosen TRAV29 clonotype comprised TRAJ17 and hence the dominant AGNKLT C-terminal motif identified in the MEME analysis. The associated TRBV2 clonotypes each differed by a single residue from the most prevalent TRBV2 public clonotype found in controllers and vaccinees (CASSRRTSGGTDTQYF). The TRAV29 and TRBV2 sequences were paired by insertion in a T2A lentivector, to generate the AS1 and AS7 TCRs (Fig. 7A). The TCR lentivectors were transduced in the TCR-negative J76 CD4+ T cell line (36), and tested for function by measuring the induction of the CD69 activation marker in the presence of APC pulsed with the decreasing doses Gag293 peptide. L cells stably expressing a single HLA-DR allele were used as APC, allowing for a precise control of the restricting HLA-DR molecule. The F24 TCR, a high avidity public TCR derived from HIV controllers (22), was used as a positive control. The AS1 and AS7 TCRs proved functional in 4 of the 7 HLA-DR contexts tested, as they yielded a detectable response in the presence of DR1, DR11, DR15, and DRB5 APC (Fig. 7B to D), but did not respond in the presence of DR3, DR4, and DR7 APC (not shown). However, the AS1 and AS7 TCRs showed lower antigen sensitivities than the controller-derived TCR F24: specifically, the AS1 and AS7 TCR responded relatively efficiently when restricted by DR11 and DR15, with antigen sensitivities measured by EC50 values in the 10−5 to 10−6 M range (AS1/DR11: 1.54x10−5; AS7/DR11: 4.14x10−6; AS1/DR15: 5.95x10−6; AS7/DR15: 4.15x10−6), while responses were low when restricted by DR1 and DRB5. The two TCRs showed comparable responses except in the context of DR11 restriction, where AS7 proved more sensitive to Gag293 than AS1. These two TCRs differed by only one residue at position 11 in the CDR3b region, indicating that an A rather than an S at this position promoted a more efficient detection of the Gag293/HLA-DR11 complex. Thus, a single residue within the CDR3b could modulate the extent of HLA-DR cross-restriction.
Figure 7. Analysis of TRAV29-mediated functional response to Gag293 stimulation.
(A) A highly prevalent TRAV29 clonotype was paired with two TRBV2 clonotypes derived from the ADVAX-induced Gag293-specific repertoire, to generate the AS1 and AS7 TCRs. F24, a high-avidity TCR isolated from HIV controllers was used as a positive control. The F24 TCR comprised the most prevalent TRAV24 public clonotype paired to one TRBV2 public clonotype.
(B to D) The AS1 and AS7 TCRs were expressed in Jurkat J76 cells, which lack an endogenous TCR complex. Transduced J76 cell were stimulated with APCs expressing human DR1 (B), DR11 (C), DR15 (D), or DRB5 (E) pulsed with decreasing doses of Gag293 peptide. T cell activation was measured by the expression of CD69 following overnight stimulation. One representative experiment out of three is depicted.
Taken together, the analysis of TRAV29 clonotypes induced by DNA vaccination showed conserved features with TRAV24 clonotypes, including shared TRAJ gene usage and C-terminal motifs, as well as the capacity to generate HLA-cross-restricted TCRs specific for Gag293. However, functional analysis of two highly expressed TRAV29-containing TCRs pointed to intermediate responses, suggesting that TRAV24 rather than TRAV29 clonotypes were responsible for the high avidity component of the Gag293-specific response.
High precursor frequencies contribute to the frequent sharing of TRAV24 but not TRBV2 clonotypes
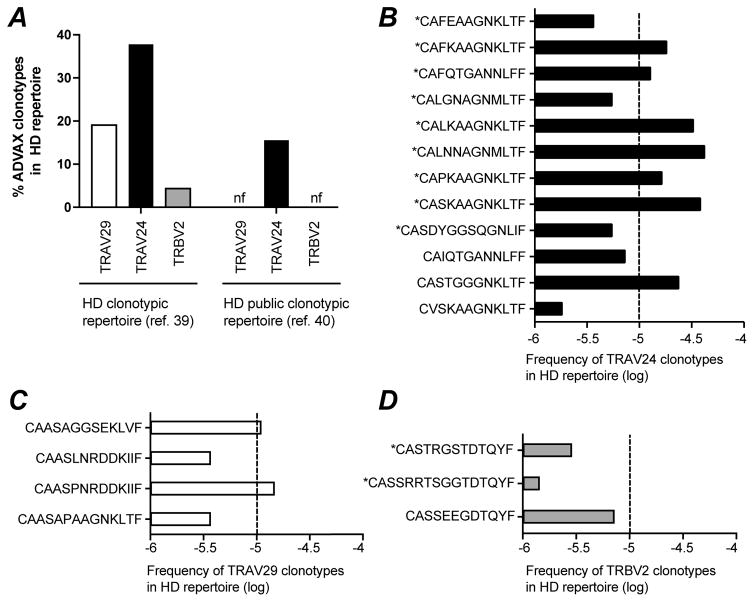
To better define factors that shaped the clonotypic repertoire induced by DNA vaccination, we set to estimate the precursor frequency of the Gag293-specific clonotypes identified in ADVAX recipients. To do so, we counted the occurrences of each vaccinee clonotype in a large dataset of total T cell clonotypes obtained from healthy donors. For this analysis, we used the clonotypic dataset recently published by J. Heather and colleagues (40), which consisted in 541,401 productive TRA sequences and 693,571 productive TRB sequences collected from 11 healthy donors (HD). The percentage of ADVAX clonotypes found in the HD repertoire was found to be high for TRAV24 clonotypes (37.8%), intermediate for TRAV29 clonotypes (19%), and low for TRBV2 clonotypes (4.3%) (Fig. 8A, left). The presence of ADVAX clonotypes was then explored in a second HD dataset reported by S. Mangul and colleagues, and consisting exclusively of public TCR clonotypes (41). The public clonotypes were extracted from RNA-Seq data generated by the Genotype-Tissue Expression Consortium, using samples from 544 individuals, and based on the reconstruction of approximately 200,000 TCR CDR3 sequences. 15.6% of the ADVAX TRAV24 clonotypes, but none of the TRAV29 clonotypes could be detected among the 4,406 TRA public clonotypes reported. Similarly, none of the ADVAX TRBV2 clonotypes could be detected in the set of 3,101 TRB public clonotypes (Fig. 8A, right). Thus, analysis of two independent datasets showed that Gag293-specific TRAV24 clonotypes were frequently detected in the clonotypic repertoire of individuals who were not infected by HIV.
Figure 8. Precursor frequencies of Gag293-specific clonotypes.
(A) The percentage of ADVAX-induced Gag293-specific clonotypes was computed in healthy donor clonotypic repertoires obtained from total T cells. Left: % found in a dataset of 541,401 TRA clonotypes and 693,571 TRB clonotypes from healthy donors (HD) (40); Right: % found in a dataset of 4,406 TRA public clonotypes and 3,101 TRB public clonotypes from healthy donors (HD) (41); The percentage of TRAV29 (white bar), TRAV24 (black bar), and TRBV2 (grey bar) Gag293-specific clonotypes found at least once in the HD repertoires is reported.
(B to D) Frequency of the TRAV24 (B), TRAV29 (C), and TRBV2 (D) Gag293-specific clonotypes found within the dataset of 541,401 TRA clonotypes and 693,571 TRB clonotypes from healthy donors (HD) reported by J. heather et al (40). Dotted lines mark the 10−5 frequency threshold. Public clonotypes are marked with an asterisk (*).
We then computed the frequency of each ADVAX clonotype found in the HD repertoire reported by Heather et al., which provided an estimate of the clonotype precursor frequency in the naive T cell repertoire, prior to Gag-dependent clonal selection (Fig. 8B to D). Multiple TRAV24 ADVAX clonotypes showed remarkably high precursor frequency, above the 10−5 threshold (Fig. 8B). Among these high frequency clonotypes, the majority (6/7) were public in the ADVAX/HIC/HAART repertoires, suggesting a link between high precursor frequency and publicity. TRAV29 clonotypes showed intermediate precursor frequencies, while TRBV2 clonotypes showed low precursor frequencies that remained below the 10−5 threshold (Fig. 8C and D). Thus, high precursor frequency likely contributed to the frequent sharing of TRAV24 clonotypes among vaccinees and HIV Controllers. In contrast, the sharing of TRBV2 clonotypes could not be accounted for by high precursor frequency, and must therefore have resulted from selection of clonotypes that were highly efficient at Gag293 recognition.
DISCUSSION
Analyses of the TCR clonotypic repertoire are emerging as a new approach to evaluate the quality of T cell responses at the molecular level. Our study of the Gag293-specific clonotypic repertoire induced by EP DNA vaccination revealed an unexpected degree of motif and clonotype sharing with HIV controllers, while sharing was more restricted with treated patients. These findings were consistent with the functional analysis, as Gag293-specific CD4+ T cell responses in ADVAX recipients showed an antigen sensitivity that was close to that measured in HIV controllers, and higher than that measured in treated patients. It was noteworthy that the most frequent TRAV24 and TRBV2 public clonotypes found in HIV controllers, which can generate high affinity TCRs, were also induced by EP DNA vaccination. Intramuscular injection of the same DNA vaccine did not yield detectable Gag293-specific responses, pointing to the benefit of DNA delivery by electroporation, which in the ADVAX trial increased immunogenicity by up to 70x fold (27). The present study demonstrates that EP DNA delivery not only increases T cell responses quantitatively, but also primes for TCR clonotypes that are associated with viral control.
A high percentage of the clonotypes identified in ADVAX recipients were public (31.1% of TRAV24 and 5.7% of TRBV2 clonotypes), even though the study participants expressed diverse HLA-DR genotypes. The extent of clonotypic sharing could be explained in part by HLA cross-restriction, as all of the public clonotypes identified in vaccinees could be isolated with at least two distinct HLA-DR tetramers. CD4+ T cell responses have been reported to be more frequently cross-restricted than CD8+ T cell responses, one reason being that CD4 epitopes can bind a greater diversity of MHC alleles than their CD8 counterparts (42). This greater flexibility may be accounted by structural features of the peptide / MHC II interaction, such as the possibility for the peptide to bind into different registers within the open MHC II grove. The Gag293 peptide and its shorter variants are able to bind efficiently to an unusually large array of HLA-DR molecules, with up to 13 alleles recognized (43, 44). This promiscuous HLA II binding likely contributes the frequent detection of the Gag293-specific response, which is the most immunodominant among CD4 responses directed at the HIV proteome (11, 24, 43, 44). HLA cross-restriction also depends on TCR clonotype properties, as by definition an HLA-cross-restricted TCR can recognize multiple peptide/MHC complexes. Of note, we previously reported a correlation between the functional avidity of Gag293-specific TCRs from controllers and their degree of HLA-DR cross-restriction (22). This relationship likely reflects the capacity of high avidity TCRs to better tolerate suboptimal variants of HLA contact residues in the context of a strong TCR/pMHC interaction. A similar phenomenon may occur in EP DNA vaccination, with the amplification of clonotypes involved in high avidity TCR interactions, and thus better able to tolerate multiple MHC II restricting alleles. Therefore, the extent of HLA cross-restriction may reflect the capacity EP DNA vaccination to induce a high avidity Gag293-specific response.
Analysis of clonotypic repertoires from healthy individuals suggests that a high precursor frequency also contributes to the amplification of TRAV24 public clonotypes in ADVAX recipients. More than one third of the Gag293-specific TRAV24 clonotypes from vaccinees were present in the dataset reported by Heather et al. (40), and half of those were also present in an independently obtained public clonotype dataset reported by Mangul. et al (41). Multiple Gag293-specific TRAV24 clonotypes could be detected in the healthy repertoire at frequencies above 10−5, which is in the high range of T cell precursor frequencies estimated for HIV-derived CD4 epitopes (39, 45, 46). Of note, HIV-specific CD4+ T cells have been detected not only in the naive but also in the memory CD4+ T cell compartment of healthy uninfected individuals, suggesting that cross-reactivity with environmental (possibly microbial) antigens may increase the preexisting frequencies of some HIV-specific clonotypes (39, 46). Another reason for the high precursor frequency of Gag293-specific TRAV24 clonotypes likely results from the ease by which they can be generated during V(D)J recombination. Consistent with the notion of convergent recombination (47), TRAV24 public clonotypes had a low number of non-templated CDR3 mutations and required little trimming from germline sequences, which increases the probability of generating these clonotypes during the random V(D)J recombination process. Large scale repertoire analyses have confirmed that public clonotypes tend to harbor fewer mutations and are detected at higher precursor frequencies than private clonotypes (48, 49). Importantly, public clonotypes preexisting at high precursor frequencies may provide a critical advantage upon encounter with a rapidly replicating pathogen such as HIV. Indeed, such public clonotypes could be sufficiently abundant to trigger a rapid T cell response prior to systemic virus dissemination, thus limiting T cell exhaustion and depletion. It is relevant that a high frequency of Gag-specific CD8 public clonotypes has been associated with a favorable outcome in the SIV simian model of AIDS, both in primary infection and in a vaccination setting (50). It is also becoming apparent that particular Gag-specific CD8 clonotypes are associated with HIV control (19–21), though not all of these clonotypes are generated at high precursor frequencies (51). There are still very few studies of the clonotypic repertoire of HIV-specific CD4+ T cells, due to their scarcity and their preferential depletion in the setting of progressive HIV infection (52, 53). However, the fact that Gag293-specific public clonotypes predominate in HIV controllers as compared to patients who had progressive infection also argues for a protective role of CD4 public clonotypes. Therefore, there is a strong rationale to devise vaccination strategies that increase the frequency of both CD4 and CD8 public clonotypes that predominate in controlled HIV infection.
While TRBV2 public clonotypes were detected in ADVAX recipients, they were less common than TRAV24 public clonotypes, which may have resulted from the lower precursor frequency of individual TRBV2 clonotypes. However, the Gag293-specific repertoire as a whole was extremely biased towards TRBV2 usage, in vaccinees as well as infected patients, suggesting that conserved structural determinants of the TRBV2 chain make an essential contribution to the recognition of the Gag293 peptide. Of note, public motifs identified in controllers were present in over 40% of TRBV2 clonotypes from vaccinees, well above the frequency of such motifs in the treated patient group (10%). This finding further highlights the capacity of EP DNA vaccination to induce a set of clonotypes associated with HIV control. In contrast, patients who had progressive HIV infection may have lost a significant portion of their HIV-specific TCR repertoire, as suggested by an overall decrease of TCR clonotypic diversity in such patients (40, 54). It is also possible that T cell priming in the context of productive HIV infection prevented the selection or amplification of efficient clonotypes at an early stage. Further research into factors that shape the composition of the HIV-specific T cell repertoire during primary HIV infection may help distinguish between these possibilities, and may shed light on the mechanisms underlying the early loss of CD4 helper function.
While the Gag293-specific clonotypic repertoire of ADVAX recipients contained “controller-like” clonotypes, it also comprised an additional population of clonotypes that appeared uniquely induced by vaccination. This was apparent from the TRAV24 and TRBV2 public motif analyses, which identified a set of motifs shared with controllers, and another set of motifs almost exclusively used in clonotypes from vaccinees. This was also apparent in the distribution of V gene usage, as the TRAV24 bias characteristic of controllers was less marked in vaccinees, who showed a more frequent use of the TRAV29 chain. CDR3 sequences from TRAV24 and TRAV29 clonotypes did show common features, such as a shared GNKLTF C-terminal motif, but also differed significantly in size and diversity, with the TRAV29 clonotypes being in mean longer by 2 residues and less diverse. Functional analysis of two highly-expressed TRAV29+ TCRs suggested that these TCRs could share some of properties of TRAV24+ TCRs from controllers, such as broad HLA-DR cross-restriction, but did not achieve high antigen sensitivity in all the HLA-DR contexts tested. These findings, associated to the predominance of TRAV24 over TRAV29 clonotypes in HIV controllers, suggest that TRAV24 rather than TRAV29 clonotypes make the dominant contribution to the high-avidity Gag293-specific response.
The factors involved in skewing the vaccine response towards the induction of a controller-like clonotypic repertoire remain to be elucidated. It is clear that controllers are subjected to recurrent stimulation with HIV antigens over periods of years, as indicated by the persistent detection of low levels of viral replication, as well as by signs of low grade chronic immune activation (16, 55, 56). Chronic antigenic stimulation is likely to stimulate the amplification of high avidity TCR clonotypes, as such clonotypes confer better T cell proliferative capacity (57) and are preferentially amplified upon recall responses (58). Analyses of persistent CMV and EBV infections support the notion public clonotypes are selected and amplified due to high affinity interactions with antigen-MHC complexes (59–61). Repeated vaccine boosting may in part mimic chronic antigenic stimulation and promote the amplification of high avidity TCR clonotypes. Of note, the T cell responses induced by ADVAX DNA electroporation were diverse and of relatively high avidity, but were not of high magnitude (27). It is likely that combining EP DNA vaccination with a series of protein-based HIV antigen boosts could further expand the subset of high-avidity Gag-specific clonotypes. Indeed, combining DNA priming with protein boosting has shown superior immunogenicity compared to DNA vaccination alone in multiple models (62, 63), and may be a strategy to further bias the clonotypic repertoire towards controller-like signatures. On the other hand, immune mechanisms that naturally maintain the diversity of the clonotypic repertoire may have to be countered to allow the emergence of high avidity public clonotypes. For instance, specific B cells that emerge in the course of infection or vaccination have the capacity to prime low avidity CD4+ T cells, due to their very efficient capture of antigen through Ig receptors (64) Thus, the focusing of the specific TCR repertoire on high avidity clonotypes may have to be balanced with the development of the specific B cell response.
Adjuvants also impact the quality of the vaccine-induced-clonotypic repertoire, as shown for instance by the capacity of a combination of TLR-4 and TLR-9 ligands to induce the selection high-avidity CD4 clonotypes (65). Of note, electroporation per se was shown to have an adjuvant effect by causing a transient inflammation that recruits antigen presenting cells to the site of DNA injection (66, 67). This phenomenon likely contributes to the notable increase in immunogenicity of the ADVAX vaccine administered by electroporation as compared to simple intramuscular injection. It will be of interest to test additional vaccination modalities, such as viral vector delivery (68), and to determine which is best able to mimic the clonotypic repertoire typical of HIV controllers. Parameters such as the TRAV24/TRAV29 ratio, the frequency of public motifs, and the frequency of public clonotypes shared with the controller dataset could be analyzed. Monitoring the emergence of high avidity public clonotypes known to confer efficient T cells responses upon TCR transfer will be of particular interest, as this approach will directly inform on specific T cell function.
The present study shows that comparing the specific clonotypic repertoire of vaccinees and HIV controllers is feasible even while studying individuals who were not selected for a particular HLA genotype. The study of a relatively limited number of individuals was sufficient to evidence a high degree of clonotypic sharing, which opens the possibility of applying this approach to multiple vaccine candidates, especially if they induce HLA cross-restricted CD4 responses. The concept is applicable to a variety of infection and vaccination settings where protective TCR clonotypes have been identified or are thought to occur (69, 70). It is for instance relevant that high avidity CD8 TCR clonotypes have been associated to the spontaneous control of HCV infection in several studies (71, 72). Recent studies have started exploring the dynamics of the specific clonotypic repertoire upon vaccination against diverse viral pathogens, and have for instance revealed the recurrent expansion of influenza virus specific clonotypes upon annual flu vaccination (73, 74). Clonotypic analyses also open the possibility to rationally optimize vaccination protocols, by monitoring at an early stage the effects of parameters such as antigen dose and vectorization on the quality of the vaccine-induced T cell response (75, 76). As such, clonotypic analyses can complement and integrate usefully with the developing field of systems vaccinology, which aims at deconvoluting the complex network of immune genes that determine vaccination outcome (77, 78). Implementing these system-based approaches should help identify predictors of efficacy at an early stage in vaccination trials. This could in turn inform vaccine design, and ultimately accelerate vaccine development.
In conclusion, we demonstrate that DNA vaccination administered by electroporation induces a Gag-specific clonotypic repertoire with significant overlap with that of HIV Controllers. The extent of clonotypic sharing could be attributed in part to broad HLA cross-restriction and high clonotype precursor frequencies, in addition to selection for high avidity antigen recognition. The study provides a proof of concept for TCR clonotypic analyses in the HIV vaccine field, and highlights the interest of comparing vaccine candidates based on the induction of clonotypic signatures associated with HIV control.
Supplementary Material
Acknowledgments
FUNDING SOURCES
This work was supported by Sidaction, Agence Nationale de Recherche sur le SIDA et les hépatites virales (ANRS), and the Pasteur Institute, Paris, France.
The ADVAX vaccine study was funded by the International AIDS Vaccine Initiative and its donors, including the generous support of the American people through the United States Agency for International Development (USAID; USAID Cooperative Agreement GPO-A-00-06-00006-00). The full list of IAVI donors is available at http://www.iavi.org. The contents of this manuscript are the responsibility of IAVI and do not necessarily reflect the views of USAID or the US Government.
L.A.C. is supported by grants from Sidaction (AI25-1-02343/2344/2345) and ANRS (ECTZ3003). M.M. is a scholar in the Pasteur - Paris University (PPU) International doctoral program with the Paris 7 University Doctoral School Bio Sorbonne Paris Cité, and received a stipend from Pasteur Carnot Maladies Infectieuses, followed by a fellowship from the ANRS. M.G. is supported by a stipend from Agence Nationale de la Recherche (ANR). M.P. is the recipient of a postdoctoral fellowship from ANRS.
The views expressed are those of the authors and should not be construed as official or representing the positions of the U.S. Departments of the Army or Defense.
We thank Patricia Fast from the International AIDS Vaccine Initiative (IAVI) for review of the manuscript and support of the study; Annick Lim for help with TCR variable gene quantitation; James Heather and Serghei Mangul for the sharing of TCR clonotype datasets; Pierre Charneau for the gift of plasmids; Mirjam Heemskerk and Bernard Maillère for the gift of cell lines; Anaïs Perilhou for help with regulatory matters; the teams of the Center of Human Immunology and of the Flow Cytometry Platform (PFC) of the Pasteur Institute for help with cell sorting and flow cytometry. MHC class II tetramer reagents were provided by the NIH Tetramer Core facility at Emory University.
References
- 1.Medlock J, Pandey A, Parpia AS, Tang A, Skrip LA, Galvani AP. Effectiveness of UNAIDS targets and HIV vaccination across 127 countries. Proc Natl Acad Sci U S A. 2017;114:4017–4022. doi: 10.1073/pnas.1620788114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haynes BF, Shaw GM, Korber B, Kelsoe G, Sodroski J, Hahn BH, Borrow P, McMichael AJ. HIV-Host Interactions: Implications for Vaccine Design. Cell Host Microbe. 2016;19:292–303. doi: 10.1016/j.chom.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, Benenson M, Gurunathan S, Tartaglia J, McNeil JG, Francis DP, Stablein D, Birx DL, Chunsuttiwat S, Khamboonruang C, Thongcharoen P, Robb ML, Michael NL, Kunasol P, Kim JH Investigators MT. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med. 2009;361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 4.Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, Liao HX, DeVico AL, Lewis GK, Williams C, Pinter A, Fong Y, Janes H, DeCamp A, Huang Y, Rao M, Billings E, Karasavvas N, Robb ML, Ngauy V, de Souza MS, Paris R, Ferrari G, Bailer RT, Soderberg KA, Andrews C, Berman PW, Frahm N, De Rosa SC, Alpert MD, Yates NL, Shen X, Koup RA, Pitisuttithum P, Kaewkungwal J, Nitayaphan S, Rerks-Ngarm S, Michael NL, Kim JH. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med. 2012;366:1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prentice HA, Tomaras GD, Geraghty DE, Apps R, Fong Y, Ehrenberg PK, Rolland M, Kijak GH, Krebs SJ, Nelson W, DeCamp A, Shen X, Yates NL, Zolla-Pazner S, Nitayaphan S, Rerks-Ngarm S, Kaewkungwal J, Pitisuttithum P, Ferrari G, McElrath MJ, Montefiori DC, Bailer RT, Koup RA, O’Connell RJ, Robb ML, Michael NL, Gilbert PB, Kim JH, Thomas R. HLA class II genes modulate vaccine-induced antibody responses to affect HIV-1 acquisition. Sci Transl Med. 2015;7:296ra112. doi: 10.1126/scitranslmed.aab4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hansen SG, Piatak M, Jr, Ventura AB, Hughes CM, Gilbride RM, Ford JC, Oswald K, Shoemaker R, Li Y, Lewis MS, Gilliam AN, Xu G, Whizin N, Burwitz BJ, Planer SL, Turner JM, Legasse AW, Axthelm MK, Nelson JA, Fruh K, Sacha JB, Estes JD, Keele BF, Edlefsen PT, Lifson JD, Picker LJ. Immune clearance of highly pathogenic SIV infection. Nature. 2013;502:100–104. doi: 10.1038/nature12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hansen SG, Wu HL, Burwitz BJ, Hughes CM, Hammond KB, Ventura AB, Reed JS, Gilbride RM, Ainslie E, Morrow DW, Ford JC, Selseth AN, Pathak R, Malouli D, Legasse AW, Axthelm MK, Nelson JA, Gillespie GM, Walters LC, Brackenridge S, Sharpe HR, Lopez CA, Fruh K, Korber BT, McMichael AJ, Gnanakaran S, Sacha JB, Picker LJ. Broadly targeted CD8(+) T cell responses restricted by major histocompatibility complex E. Science. 2016;351:714–720. doi: 10.1126/science.aac9475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chakrabarti LA, Simon V. Immune mechanisms of HIV control. Curr Opin Immunol. 2010;22:488–496. doi: 10.1016/j.coi.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walker BD, Yu XG. Unravelling the mechanisms of durable control of HIV-1. Nat Rev Immunol. 2013;13:487–498. doi: 10.1038/nri3478. [DOI] [PubMed] [Google Scholar]
- 10.Almeida JR, Price DA, Papagno L, Arkoub ZA, Sauce D, Bornstein E, Asher TE, Samri A, Schnuriger A, Theodorou I, Costagliola D, Rouzioux C, Agut H, Marcelin AG, Douek D, Autran B, Appay V. Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. J Exp Med. 2007;204:2473–2485. doi: 10.1084/jem.20070784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vingert B, Perez-Patrigeon S, Jeannin P, Lambotte O, Boufassa F, Lemaitre F, Kwok WW, Theodorou I, Delfraissy JF, Theze J, Chakrabarti LA AEHCS Group. HIV controller CD4+ T cells respond to minimal amounts of Gag antigen due to high TCR avidity. PLoS Pathog. 2010;6:e1000780. doi: 10.1371/journal.ppat.1000780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saez-Cirion A, Lacabaratz C, Lambotte O, Vermisse P, Urrutia A, Boufassa F, Barre-Sinoussi F, Delfraissy J, Sinet M, Pancino G, Venet A ANRS EP36 HIV CONTROLLERS study group. HIV controllers exhibit potent CD8 T cell capacity to suppress HIV infection ex vivo and peculiar CTL activation phenotype. Proc Natl Acad Sci U S A. 2007;104:6776–6781. doi: 10.1073/pnas.0611244104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Almeida JR, Sauce D, Price DA, Papagno L, Shin SY, Moris A, Larsen M, Pancino G, Douek DC, Autran B, Saez-Cirion A, Appay V. Antigen sensitivity is a major determinant of CD8+ T-cell polyfunctionality and HIV-suppressive activity. Blood. 2009;113:6351–6360. doi: 10.1182/blood-2009-02-206557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Migueles SA, Osborne CM, Royce C, Compton AA, Joshi RP, Weeks KA, Rood JE, Berkley AM, Sacha JB, Cogliano-Shutta NA, Lloyd M, Roby G, Kwan R, McLaughlin M, Stallings S, Rehm C, O’Shea MA, Mican J, Packard BZ, Komoriya A, Palmer S, Wiegand AP, Maldarelli F, Coffin JM, Mellors JW, Hallahan CW, Follman DA, Connors M. Lytic granule loading of CD8+ T cells is required for HIV-infected cell elimination associated with immune control. Immunity. 2008;29:1009–1021. doi: 10.1016/j.immuni.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Younes SA, Yassine-Diab B, Dumont AR, Boulassel MR, Grossman Z, Routy JP, Sekaly RP. HIV-1 viremia prevents the establishment of interleukin 2-producing HIV-specific memory CD4+ T cells endowed with proliferative capacity. J Exp Med. 2003;198:1909–1922. doi: 10.1084/jem.20031598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Potter SJ, Lacabaratz C, Lambotte O, Perez-Patrigeon S, Vingert B, Sinet M, Colle JH, Urrutia A, Scott-Algara D, Boufassa F, Delfraissy JF, Theze J, Venet A, Chakrabarti LA. Preserved central memory and activated effector memory CD4+ T-cell subsets in human immunodeficiency virus controllers: an ANRS EP36 study. J Virol. 2007;81:13904–13915. doi: 10.1128/JVI.01401-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vingert B, Benati D, Lambotte O, de Truchis P, Slama L, Jeannin P, Galperin M, Perez-Patrigeon S, Boufassa F, Kwok WW, Lemaitre F, Delfraissy JF, Theze J, Chakrabarti LA. HIV controllers maintain a population of highly efficient Th1 effector cells in contrast to patients treated in the long term. J Virol. 2012;86:10661–10674. doi: 10.1128/JVI.00056-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferre AL, Hunt PW, McConnell DH, Morris MM, Garcia JC, Pollard RB, Yee HF, Jr, Martin JN, Deeks SG, Shacklett BL. HIV controllers with HLA-DRB1*13 and HLA-DQB1*06 alleles have strong, polyfunctional mucosal CD4+ T-cell responses. J Virol. 2010;84:11020–11029. doi: 10.1128/JVI.00980-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lissina A, Chakrabarti LA, Takiguchi M, Appay V. TCR clonotypes: molecular determinants of T-cell efficacy against HIV. Curr Opin Virol. 2016;16:77–85. doi: 10.1016/j.coviro.2016.01.017. [DOI] [PubMed] [Google Scholar]
- 20.Ladell K, Hashimoto M, Iglesias MC, Wilmann PG, McLaren JE, Gras S, Chikata T, Kuse N, Fastenackels S, Gostick E, Bridgeman JS, Venturi V, Arkoub ZA, Agut H, van Bockel DJ, Almeida JR, Douek DC, Meyer L, Venet A, Takiguchi M, Rossjohn J, Price DA, Appay V. A molecular basis for the control of preimmune escape variants by HIV-specific CD8+ T cells. Immunity. 2013;38:425–436. doi: 10.1016/j.immuni.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 21.Chen H, Ndhlovu ZM, Liu D, Porter LC, Fang JW, Darko S, Brockman MA, Miura T, Brumme ZL, Schneidewind A, Piechocka-Trocha A, Cesa KT, Sela J, Cung TD, Toth I, Pereyra F, Yu XG, Douek DC, Kaufmann DE, Allen TM, Walker BD. TCR clonotypes modulate the protective effect of HLA class I molecules in HIV-1 infection. Nat Immunol. 2012;13:691–700. doi: 10.1038/ni.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benati D, Galperin M, Lambotte O, Gras S, Lim A, Mukhopadhyay M, Nouel A, Campbell KA, Lemercier B, Claireaux M, Hendou S, Lechat P, de Truchis P, Boufassa F, Rossjohn J, Delfraissy JF, Arenzana-Seisdedos F, Chakrabarti LA. Public T cell receptors confer high-avidity CD4 responses to HIV controllers. J Clin Invest. 2016;126:2093–2108. doi: 10.1172/JCI83792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ranasinghe S, Flanders M, Cutler S, Soghoian DZ, Ghebremichael M, Davis I, Lindqvist M, Pereyra F, Walker BD, Heckerman D, Streeck H. HIV-Specific CD4 T Cell Responses to Different Viral Proteins Have Discordant Associations with Viral Load and Clinical Outcome. J Virol. 2012;86:277–283. doi: 10.1128/JVI.05577-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laher F, Ranasinghe S, Porichis F, Mewalal N, Pretorius K, Ismail N, Buus S, Stryhn A, Carrington M, Walker BD, Ndung’u T, Ndhlovu ZM. HIV Controllers Exhibit Enhanced Frequencies of Major Histocompatibility Complex Class II Tetramer+ Gag-Specific CD4+ T Cells in Chronic Clade C HIV-1 Infection. J Virol. 2017:91. doi: 10.1128/JVI.02477-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones RB, Yue FY, Gu XX, Hunter DV, Mujib S, Gyenes G, Mason RD, Mohamed R, MacDonald KS, Kovacs C, Ostrowski MA. Human immunodeficiency virus type 1 escapes from interleukin-2-producing CD4+ T-cell responses without high-frequency fixation of mutations. J Virol. 2009;83:8722–8732. doi: 10.1128/JVI.00433-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ranasinghe S, Lamothe PA, Soghoian DZ, Kazer SW, Cole MB, Shalek AK, Yosef N, Jones RB, Donaghey F, Nwonu C, Jani P, Clayton GM, Crawford F, White J, Montoya A, Power K, Allen TM, Streeck H, Kaufmann DE, Picker LJ, Kappler JW, Walker BD. Antiviral CD8+ T Cells Restricted by Human Leukocyte Antigen Class II Exist during Natural HIV Infection and Exhibit Clonal Expansion. Immunity. 2016;45:917–930. doi: 10.1016/j.immuni.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vasan S, Hurley A, Schlesinger SJ, Hannaman D, Gardiner DF, Dugin DP, Boente-Carrera M, Vittorino R, Caskey M, Andersen J, Huang Y, Cox JH, Tarragona-Fiol T, Gill DK, Cheeseman H, Clark L, Dally L, Smith C, Schmidt C, Park HH, Kopycinski JT, Gilmour J, Fast P, Bernard R, Ho DD. In vivo electroporation enhances the immunogenicity of an HIV-1 DNA vaccine candidate in healthy volunteers. PLoS One. 2011;6:e19252. doi: 10.1371/journal.pone.0019252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang Y, Chen Z, Zhang W, Gurner D, Song Y, Gardiner DF, Ho DD. Design, construction, and characterization of a dual-promoter multigenic DNA vaccine directed against an HIV-1 subtype C/B′ recombinant. J Acquir Immune Defic Syndr. 2008;47:403–411. doi: 10.1097/QAI.0b013e3181651b9d. [DOI] [PubMed] [Google Scholar]
- 29.Vasan S, Schlesinger SJ, Huang Y, Hurley A, Lombardo A, Chen Z, Than S, Adesanya P, Bunce C, Boaz M, Boyle R, Sayeed E, Clark L, Dugin D, Schmidt C, Song Y, Seamons L, Dally L, Ho M, Smith C, Markowitz M, Cox J, Gill DK, Gilmour J, Keefer MC, Fast P, Ho DD. Phase 1 safety and immunogenicity evaluation of ADVAX, a multigenic, DNA-based clade C/B′ HIV-1 candidate vaccine. PLoS One. 2010;5:e8617. doi: 10.1371/journal.pone.0008617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kopycinski J, Cheeseman H, Ashraf A, Gill D, Hayes P, Hannaman D, Gilmour J, Cox JH, Vasan S. A DNA-based candidate HIV vaccine delivered via in vivo electroporation induces CD4 responses toward the alpha4beta7-binding V2 loop of HIV gp120 in healthy volunteers. Clin Vaccine Immunol. 2012;19:1557–1559. doi: 10.1128/CVI.00327-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Galperin M, Benati D, Claireaux M, Mukhopadhyay M, Chakrabarti LA. MHC Class II Tetramer Labeling of Human Primary CD4+ T Cells from HIV Infected Patients. Bio-protocol. 2017;7:e2187. doi: 10.21769/BioProtoc.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim A, Trautmann L, Peyrat MA, Couedel C, Davodeau F, Romagne F, Kourilsky P, Bonneville M. Frequent contribution of T cell clonotypes with public TCR features to the chronic response against a dominant EBV-derived epitope: application to direct detection of their molecular imprint on the human peripheral T cell repertoire. J Immunol. 2000;165:2001–2011. doi: 10.4049/jimmunol.165.4.2001. [DOI] [PubMed] [Google Scholar]
- 33.Li S, Lefranc MP, Miles JJ, Alamyar E, Giudicelli V, Duroux P, Freeman JD, Corbin VD, Scheerlinck JP, Frohman MA, Cameron PU, Plebanski M, Loveland B, Burrows SR, Papenfuss AT, Gowans EJ. IMGT/HighV QUEST paradigm for T cell receptor IMGT clonotype diversity and next generation repertoire immunoprofiling. Nat Commun. 2013;4:2333. doi: 10.1038/ncomms3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bailey TL, Johnson J, Grant CE, Noble WS. The MEME Suite. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stothard P. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques. 2000;28:1102, 1104. doi: 10.2144/00286ir01. [DOI] [PubMed] [Google Scholar]
- 36.Heemskerk MH, Hoogeboom M, de Paus RA, Kester MG, van der Hoorn MA, Goulmy E, Willemze R, Falkenburg JH. Redirection of antileukemic reactivity of peripheral T lymphocytes using gene transfer of minor histocompatibility antigen HA-2-specific T-cell receptor complexes expressing a conserved alpha joining region. Blood. 2003;102:3530–3540. doi: 10.1182/blood-2003-05-1524. [DOI] [PubMed] [Google Scholar]
- 37.Klohe EP, Watts R, Bahl M, Alber C, Yu WY, Anderson R, Silver J, Gregersen PK, Karr RW. Analysis of the molecular specificities of anti-class II monoclonal antibodies by using L cell transfectants expressing HLA class II molecules. J Immunol. 1988;141:2158–2164. [PubMed] [Google Scholar]
- 38.Lubong Sabado R, Kavanagh DG, Kaufmann DE, Fru K, Babcock E, Rosenberg E, Walker B, Lifson J, Bhardwaj N, Larsson M. In vitro priming recapitulates in vivo HIV-1 specific T cell responses, revealing rapid loss of virus reactive CD4 T cells in acute HIV-1 infection. PLoS One. 2009;4:e4256. doi: 10.1371/journal.pone.0004256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Campion SL, Brodie TM, Fischer W, Korber BT, Rossetti A, Goonetilleke N, McMichael AJ, Sallusto F. Proteome-wide analysis of HIV-specific naive and memory CD4(+) T cells in unexposed blood donors. J Exp Med. 2014;211:1273–1280. doi: 10.1084/jem.20130555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heather JM, Best K, Oakes T, Gray ER, Roe JK, Thomas N, Friedman N, Noursadeghi M, Chain B. Dynamic Perturbations of the T-Cell Receptor Repertoire in Chronic HIV Infection and following Antiretroviral Therapy. Front Immunol. 2015;6:644. doi: 10.3389/fimmu.2015.00644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mangul S, Mandric I, Yang HT, Strauli N, Montoya D, Rotman J, Van Der Wey W, Ronas JR, Statz B, Zelikovsly A, Sreafico R, Shifman S, Zaitlrn N, Rossetti M, Ansel KM, Eskin E. Profiling adaptive immune repertoires across multiple human tissues by RNA Sequencing. 2017 bioRxiv http://biorxiv.org/content/early/2017/03/25/089235.
- 42.Greenbaum J, Sidney J, Chung J, Brander C, Peters B, Sette A. Functional classification of class II human leukocyte antigen (HLA) molecules reveals seven different supertypes and a surprising degree of repertoire sharing across supertypes. Immunogenetics. 2011;63:325–335. doi: 10.1007/s00251-011-0513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaufmann DE, Bailey PM, Sidney J, Wagner B, Norris PJ, Johnston MN, Cosimi LA, Addo MM, Lichterfeld M, Altfeld M, Frahm N, Brander C, Sette A, Walker BD, Rosenberg ES. Comprehensive analysis of human immunodeficiency virus type 1-specific CD4 responses reveals marked immunodominance of gag and nef and the presence of broadly recognized peptides. J Virol. 2004;78:4463–4477. doi: 10.1128/JVI.78.9.4463-4477.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ranasinghe S, Cutler S, Davis I, Lu R, Soghoian DZ, Qi Y, Sidney J, Kranias G, Flanders MD, Lindqvist M, Kuhl B, Alter G, Deeks SG, Walker BD, Gao X, Sette A, Carrington M, Streeck H. Association of HLA-DRB1-restricted CD4 T cell responses with HIV immune control. Nature Medicine. 2013;19:930–933. doi: 10.1038/nm.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Castelli FA, Szely N, Olivain A, Casartelli N, Grygar C, Schneider A, Besse A, Levy Y, Schwartz O, Maillere B. Hierarchy of CD4 T cell epitopes of the ANRS Lipo5 synthetic vaccine relies on the frequencies of pre-existing peptide-specific T cells in healthy donors. J Immunol. 2013;190:5757–5763. doi: 10.4049/jimmunol.1300145. [DOI] [PubMed] [Google Scholar]
- 46.Su LF, Kidd BA, Han A, Kotzin JJ, Davis MM. Virus-specific CD4(+) memory-phenotype T cells are abundant in unexposed adults. Immunity. 2013;38:373–383. doi: 10.1016/j.immuni.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Venturi V, Quigley MF, Greenaway HY, Ng PC, Ende ZS, McIntosh T, Asher TE, Almeida JR, Levy S, Price DA, Davenport MP, Douek DC. A mechanism for TCR sharing between T cell subsets and individuals revealed by pyrosequencing. J Immunol. 2011;186:4285–4294. doi: 10.4049/jimmunol.1003898. [DOI] [PubMed] [Google Scholar]
- 48.Robins HS, Srivastava SK, Campregher PV, Turtle CJ, Andriesen J, Riddell SR, Carlson CS, Warren EH. Overlap and effective size of the human CD8+ T cell receptor repertoire. Sci Transl Med. 2010;2:47ra64. doi: 10.1126/scitranslmed.3001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Venturi V, Kedzierska K, Price DA, Doherty PC, Douek DC, Turner SJ, Davenport MP. Sharing of T cell receptors in antigen-specific responses is driven by convergent recombination. Proc Natl Acad Sci U S A. 2006;103:18691–18696. doi: 10.1073/pnas.0608907103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Price DA, Asher TE, Wilson NA, Nason MC, Brenchley JM, Metzler IS, Venturi V, Gostick E, Chattopadhyay PK, Roederer M, Davenport MP, Watkins DI, Douek DC. Public clonotype usage identifies protective Gag-specific CD8+ T cell responses in SIV infection. J Exp Med. 2009;206:923–936. doi: 10.1084/jem.20081127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iglesias MC, Briceno O, Gostick E, Moris A, Meaudre C, Price DA, Ungeheuer MN, Saez-Cirion A, Mallone R, Appay V. Immunodominance of HLA-B27-restricted HIV KK10-specific CD8(+) T-cells is not related to naive precursor frequency. Immunol Lett. 2013;149:119–122. doi: 10.1016/j.imlet.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 52.Douek DC, Brenchley JM, Betts MR, Ambrozak DR, Hill BJ, Okamoto Y, Casazza JP, Kuruppu J, Kunstman K, Wolinsky S, Grossman Z, Dybul M, Oxenius A, Price DA, Connors M, Koup RA. HIV preferentially infects HIV-specific CD4+ T cells. Nature. 2002;417:95–98. doi: 10.1038/417095a. [DOI] [PubMed] [Google Scholar]
- 53.Seth N, Kaufmann D, Lahey T, Rosenberg ES, Wucherpfennig KW. Expansion and contraction of HIV-specific CD4 T cells with short bursts of viremia, but physical loss of the majority of these cells with sustained viral replication. J Immunol. 2005;175:6948–6958. doi: 10.4049/jimmunol.175.10.6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gorochov G, Neumann AU, Kereveur A, Parizot C, Li T, Katlama C, Karmochkine M, Raguin G, Autran B, Debre P. Perturbation of CD4+ and CD8+ T-cell repertoires during progression to AIDS and regulation of the CD4+ repertoire during antiviral therapy. Nat Med. 1998;4:215–221. doi: 10.1038/nm0298-215. [DOI] [PubMed] [Google Scholar]
- 55.Pereyra F, Palmer S, Miura T, Block BL, Wiegand A, Rothchild AC, Baker B, Rosenberg R, Cutrell E, Seaman MS, Coffin JM, Walker BD. Persistent low-level viremia in HIV-1 elite controllers and relationship to immunologic parameters. J Infect Dis. 2009;200:984–990. doi: 10.1086/605446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hunt PW, Brenchley J, Sinclair E, McCune JM, Roland M, Page-Shafer K, Hsue P, Emu B, Krone M, Lampiris H, Douek D, Martin JN, Deeks SG. Relationship between T cell activation and CD4+ T cell count in HIV-seropositive individuals with undetectable plasma HIV RNA levels in the absence of therapy. J Infect Dis. 2008;197:126–133. doi: 10.1086/524143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zehn D, Lee SY, Bevan MJ. Complete but curtailed T-cell response to very low-affinity antigen. Nature. 2009;458:211–214. doi: 10.1038/nature07657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cukalac T, Chadderton J, Handel A, Doherty PC, Turner SJ, Thomas PG, La Gruta NL. Reproducible selection of high avidity CD8+ T-cell clones following secondary acute virus infection. Proc Natl Acad Sci U S A. 2014;111:1485–1490. doi: 10.1073/pnas.1323736111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Price DA, Brenchley JM, Ruff LE, Betts MR, Hill BJ, Roederer M, Koup RA, Migueles SA, Gostick E, Wooldridge L, Sewell AK, Connors M, Douek DC. Avidity for antigen shapes clonal dominance in CD8+ T cell populations specific for persistent DNA viruses. J Exp Med. 2005;202:1349–1361. doi: 10.1084/jem.20051357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trautmann L, Rimbert M, Echasserieau K, Saulquin X, Neveu B, Dechanet J, Cerundolo V, Bonneville M. Selection of T cell clones expressing high-affinity public TCRs within Human cytomegalovirus-specific CD8 T cell responses. J Immunol. 2005;175:6123–6132. doi: 10.4049/jimmunol.175.9.6123. [DOI] [PubMed] [Google Scholar]
- 61.Gras S, Kjer-Nielsen L, Burrows SR, McCluskey J, Rossjohn J. T-cell receptor bias and immunity. Curr Opin Immunol. 2008;20:119–125. doi: 10.1016/j.coi.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 62.Hutnick NA, Myles DJ, Bian CB, Muthumani K, Weiner DB. Selected approaches for increasing HIV DNA vaccine immunogenicity in vivo. Curr Opin Virol. 2011;1:233–240. doi: 10.1016/j.coviro.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xiang B, Baybutt TR, Berman-Booty L, Magee MS, Waldman SA, Alexeev VY, Snook AE. Prime-Boost Immunization Eliminates Metastatic Colorectal Cancer by Producing High-Avidity Effector CD8+ T Cells. J Immunol. 2017 doi: 10.4049/jimmunol.1502672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Merkenschlager J, Ploquin MJ, Eksmond U, Andargachew R, Thorborn G, Filby A, Pepper M, Evavold B, Kassiotis G. Stepwise B-cell-dependent expansion of T helper clonotypes diversifies the T-cell response. Nat Commun. 2016;7:10281. doi: 10.1038/ncomms10281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Malherbe L, Mark L, Fazilleau N, McHeyzer-Williams LJ, McHeyzer-Williams MG. Vaccine adjuvants alter TCR-based selection thresholds. Immunity. 2008;28:698–709. doi: 10.1016/j.immuni.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Babiuk S, Baca-Estrada ME, Foldvari M, Middleton DM, Rabussay D, Widera G, Babiuk LA. Increased gene expression and inflammatory cell infiltration caused by electroporation are both important for improving the efficacy of DNA vaccines. J Biotechnol. 2004;110:1–10. doi: 10.1016/j.jbiotec.2004.01.015. [DOI] [PubMed] [Google Scholar]
- 67.Martinon F, Kaldma K, Sikut R, Culina S, Romain G, Tuomela M, Adojaan M, Mannik A, Toots U, Kivisild T, Morin J, Brochard P, Delache B, Tripiciano A, Ensoli F, Stanescu I, Le Grand R, Ustav M. Persistent immune responses induced by a human immunodeficiency virus DNA vaccine delivered in association with electroporation in the skin of nonhuman primates. Hum Gene Ther. 2009;20:1291–1307. doi: 10.1089/hum.2009.044. [DOI] [PubMed] [Google Scholar]
- 68.Vasan S, Schlesinger SJ, Chen Z, Hurley A, Lombardo A, Than S, Adesanya P, Bunce C, Boaz M, Boyle R, Sayeed E, Clark L, Dugin D, Boente-Carrera M, Schmidt C, Fang Q, Lei Ba, Huang Y, Zaharatos GJ, Gardiner DF, Caskey M, Seamons L, Ho M, Dally L, Smith C, Cox J, Gill D, Gilmour J, Keefer MC, Fast P, Ho DD. Phase 1 safety and immunogenicity evaluation of ADMVA, a multigenic, modified vaccinia Ankara-HIV-1 B′/C candidate vaccine. PLoS One. 2010;5:e8816. doi: 10.1371/journal.pone.0008816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Miles JJ, Douek DC, Price DA. Bias in the alphabeta T-cell repertoire: implications for disease pathogenesis and vaccination. Immunol Cell Biol. 2011;89:375–387. doi: 10.1038/icb.2010.139. [DOI] [PubMed] [Google Scholar]
- 70.Thorborn G, Young GR, Kassiotis G. Effective T helper cell responses against retroviruses: are all clonotypes equal? J Leukoc Biol. 2014;96:27–37. doi: 10.1189/jlb.2RI0613-347R. [DOI] [PubMed] [Google Scholar]
- 71.Neveu B, Debeaupuis E, Echasserieau K, le Moullac-Vaidye B, Gassin M, Jegou L, Decalf J, Albert M, Ferry N, Gournay J, Houssaint E, Bonneville M, Saulquin X. Selection of high-avidity CD8 T cells correlates with control of hepatitis C virus infection. Hepatology. 2008;48:713–722. doi: 10.1002/hep.22379. [DOI] [PubMed] [Google Scholar]
- 72.Abdel-Hakeem MS, Boisvert M, Bruneau J, Soudeyns H, Shoukry NH. Selective expansion of high functional avidity memory CD8 T cell clonotypes during hepatitis C virus reinfection and clearance. PLoS Pathog. 2017;13:e1006191. doi: 10.1371/journal.ppat.1006191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Herati SR, Muselman A, Vella L, Bengsch B, Parkhouse K, Del Alcazar D, Kotzin J, Doyle SA, Tebas P, Hensley SE, Su LF, Schmader KE, Wherry JE. Successive annual influenza vaccination induces a recurrent oligoclonotypic memory response in circulating T follicular helper cells. Science Immunology. 2017;2:1–14. doi: 10.1126/sciimmunol.aag2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Qi Q, Cavanagh MM, Le Saux S, NamKoong H, Kim C, Turgano E, Liu Y, Wang C, Mackey S, Swan GE, Dekker CL, Olshen RA, Boyd SD, Weyand CM, Tian L, Goronzy JJ. Diversification of the antigen-specific T cell receptor repertoire after varicella zoster vaccination. Sci Transl Med. 2016;8:332ra346. doi: 10.1126/scitranslmed.aaf1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hill BJ, Darrah PA, Ende Z, Ambrozak DR, Quinn KM, Darko S, Gostick E, Wooldridge L, van den Berg HA, Venturi V, Larsen M, Davenport MP, Seder RA, Price DA, Douek DC. Epitope specificity delimits the functional capabilities of vaccine-induced CD8 T cell populations. J Immunol. 2014;193:5626–5636. doi: 10.4049/jimmunol.1401017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Thorborn G, Ploquin MJ, Eksmond U, Pike R, Bayer W, Dittmer U, Hasenkrug KJ, Pepper M, Kassiotis G. Clonotypic composition of the CD4+ T cell response to a vectored retroviral antigen is determined by its speed. J Immunol. 2014;193:1567–1577. doi: 10.4049/jimmunol.1400667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pulendran B. Systems vaccinology: probing humanity’s diverse immune systems with vaccines. Proc Natl Acad Sci U S A. 2014;111:12300–12306. doi: 10.1073/pnas.1400476111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gaucher D, Therrien R, Kettaf N, Angermann BR, Boucher G, Filali-Mouhim A, Moser JM, Mehta RS, Drake DR, 3rd, Castro E, Akondy R, Rinfret A, Yassine-Diab B, Said EA, Chouikh Y, Cameron MJ, Clum R, Kelvin D, Somogyi R, Greller LD, Balderas RS, Wilkinson P, Pantaleo G, Tartaglia J, Haddad EK, Sekaly RP. Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. J Exp Med. 2008;205:3119–3131. doi: 10.1084/jem.20082292. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.