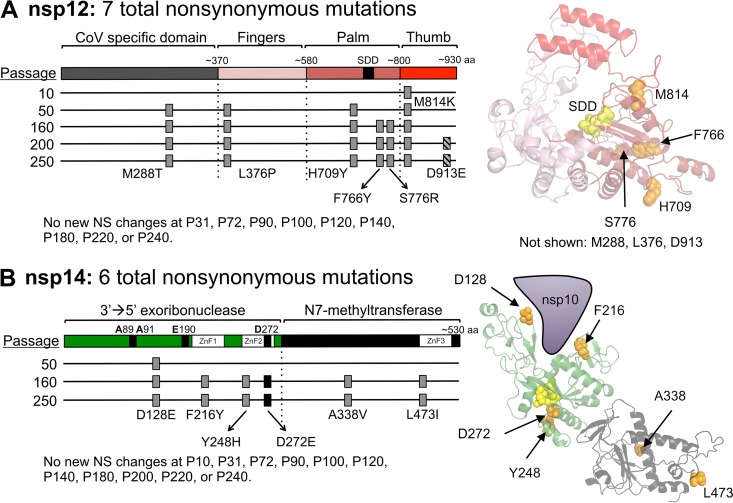
FIG 6 .
The timing of fixation of mutations in nsp12-RdRp and nsp14-ExoN within MHV-ExoN(-). (A) A schematic of nsp12-RdRp with the CoV-specific region and the canonical finger, palm, and thumb domains of RdRps is shown. The nsp12-RdRp coding region was sequenced at the indicated passage, and the nonsynonymous changes are plotted; gray boxes indicate consensus changes, and hatched boxes indicate variants shown to be present in <50% of the population by di-deoxy sequencing. At right, mutations are marked in orange on a Phyre2-modeled structure of MHV-nsp12, with the active site residues marked in yellow (17). RdRp domains are colored according to the linear schematic. M288T, L376P, and D913E lie outside the modeled region and thus are not marked. (B) A schematic of nsp14 with the ExoN and N7-methyltransferase domains is shown, with mutation plotting depicted as described for panel A. The black box denotes a mutation to ExoN motif III. At right, mutations are marked in orange on a Phyre2-modeled structure of MHV-nsp14. Domains are colored according to the linear schematic.