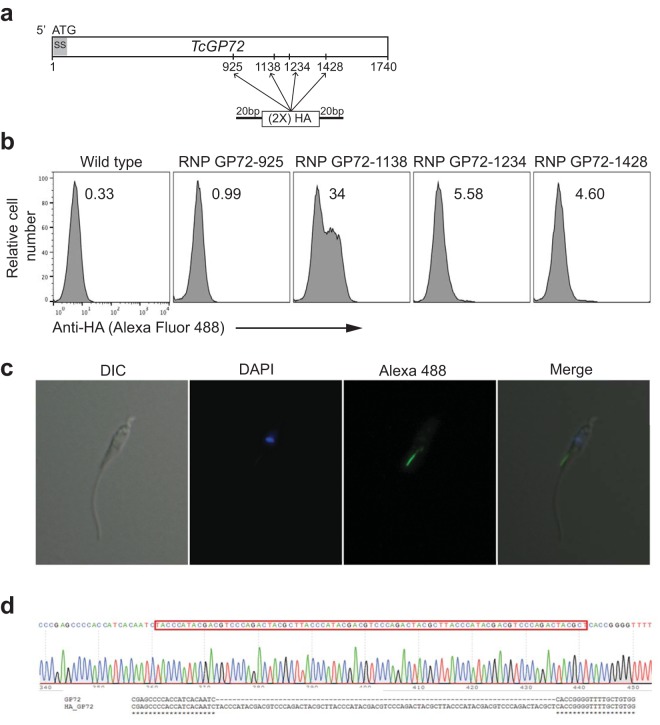
FIG 5 .
Endogenous gene tagging in T. cruzi using SaCas9 RNP complexes. (a) Four guide RNAs named with respect to the target nucleotide of the Cas9 cut site (sgRNA GP72-925, sgRNA GP72-1138, sgRNA GP72-1234, and sgRNA GP72-1428) were selected to cut the T. cruzi GP72 (TcGP72) gene at the indicated positions, avoiding the N- and C-terminal regions. ss, predicted signal sequence. For each guide, an in-frame DNA template was designed, containing 20-bp homology arms (each side) and two tandem sequences of the HA epitope. (b) Three days after electroporation, parasites were harvested by centrifugation and stained for detection of surface HA. Parasites receiving RNP complexes containing sgRNA GP72-1138 showed a shift in fluorescence for approximately 35% of the population, indicating incorporation of the HA tag. (c) The same parasites used in the experiment whose results are shown in panel b were adhered to poly l-lysine-coated coverslips, mounted in microscope slides, and observed via fluorescence microscopy. The HA tag fluorescence was observed in the flagellar attachment zone, as expected for GP72. DIC, differential inference contrast; DAPI, 4′,6-diamidino-2-phenylindole. (d) Sequencing of the GP72 gene in Alexa Fluor 488-positive parasites obtained from the experiment whose results are shown in panel b showed the correct insertion of the HA tag (in frame and in the expected position) in representative clones.