Summary
MicroRNAs (miRNAs) are small RNA molecules that regulate gene expression associated with many complex biological processes. By comparing miRNA expression between long‐lived cohorts of Drosophila melanogaster that were fed a low‐nutrient diet with normal‐lived control animals fed a high‐nutrient diet, we identified miR‐184, let‐7, miR‐125, and miR‐100 as candidate miRNAs involved in modulating aging. We found that ubiquitous, adult‐specific overexpression of these individual miRNAs led to significant changes in fat metabolism and/or lifespan. Most impressively, adult‐specific overexpression of let‐7 in female nervous tissue increased median fly lifespan by ~22%. We provide evidence that this lifespan extension is not due to alterations in nutrient intake or to decreased insulin signaling.
Keywords: aging, diet restriction, let‐7, miR‐100, miR‐125, miR‐184
Introduction, results, and discussion
Aging is a complex, dynamic process in which healthy individuals deteriorate. The rate of this decline is influenced by both genetic and environmental factors, many of which are known to slow its progression and increase lifespan. These effects often manifest quite rapidly. Changes in diet or social conditions, for example, altered mortality rates in fruit flies (Drosophila melanogaster) and nematode worms (Caenorhabditis elegans) in as little as 12 h and were accompanied by concurrent alterations in physiology and behavior (Mair et al., 2003; Smith et al., 2008; Gendron et al., 2014).
MicroRNA (miRNA) molecules play an important role in the dynamic regulation of a wide variety of complex physiological and pathophysiological processes, and there is growing evidence of their importance in metabolism and aging (Boehm & Slack, 2006; Inukai & Slack, 2013). Overexpression of miRNA lin‐4 in C. elegans significantly increased worm lifespan in a manner that was dependent on both the insulin signaling transcription factor daf‐16/FOXO (a known metabolic regulator) and on the heat shock transcription factor hsf‐1 (Boehm & Slack, 2005). miR‐34 (Yang et al., 2013), miR‐71 (de Lencastre et al., 2010; Boulias & Horvitz, 2012), miR‐80 (Vora et al., 2013), miR‐238, miR‐239, and miR‐246 (de Lencastre et al., 2010) have also been shown to influence nematode lifespan, with miR‐71, miR‐80, and miR‐239 showing dependence on insulin signaling to modulate lifespan. In flies, miR‐34 (Liu et al., 2012) and miR‐277 (Esslinger et al., 2013) have been shown to affect lifespan; miR‐34 overexpression increased lifespan whereas miR‐277 overexpression shortened it. While the ability of miR‐34 to influence metabolism is yet to be investigated, miR‐277 has been shown to modulate target of rapamycin (TOR) signaling.
To identify new miRNAs that mediate aging in Drosophila, we used dietary ‘switch’ experiments. In these experiments, animals switched from a standard to a low‐nutrient environment experience a rapid drop in their age‐specific mortality rate, whereas those moved to a high‐nutrient diet experience mortality increases (Mair et al., 2003). We isolated small RNAs from female Drosophila melanogaster 3 days after the diet switch. Deep sequencing identified let‐7, miR‐184, miR‐34, and miR‐8 as differentially expressed between the two diets (Fig. S1A). miR‐8 abundance appeared to be increased in fully fed conditions while the other three were more abundant in conditions of diet restriction, suggesting that their expression may suppress aging.
Given that miR‐34 is known to influence lifespan and neurodegeneration (Liu et al., 2012), we focused our investigation on miR‐184 and let‐7. To determine whether increased miR‐184 expression promotes lifespan, we used the Gene Switch (GS) system to induce miR‐184 expression broadly in the adult animal (GS‐tubulin‐GAL4 > UAS‐miR‐184). Flies fed the transcriptional activator RU‐486 for 3 days showed a fourfold increase in miR‐184 expression compared to isogenic flies fed vehicle (Fig. S1B). In male and female flies, ubiquitous miR‐184 overexpression severely reduced lifespan, independent of diet, and failed to inhibit lifespan extension through dietary restriction (Fig. 1A and B). These data suggest that adult‐specific miR‐184 overexpression is deleterious regardless of diet.
Figure 1.
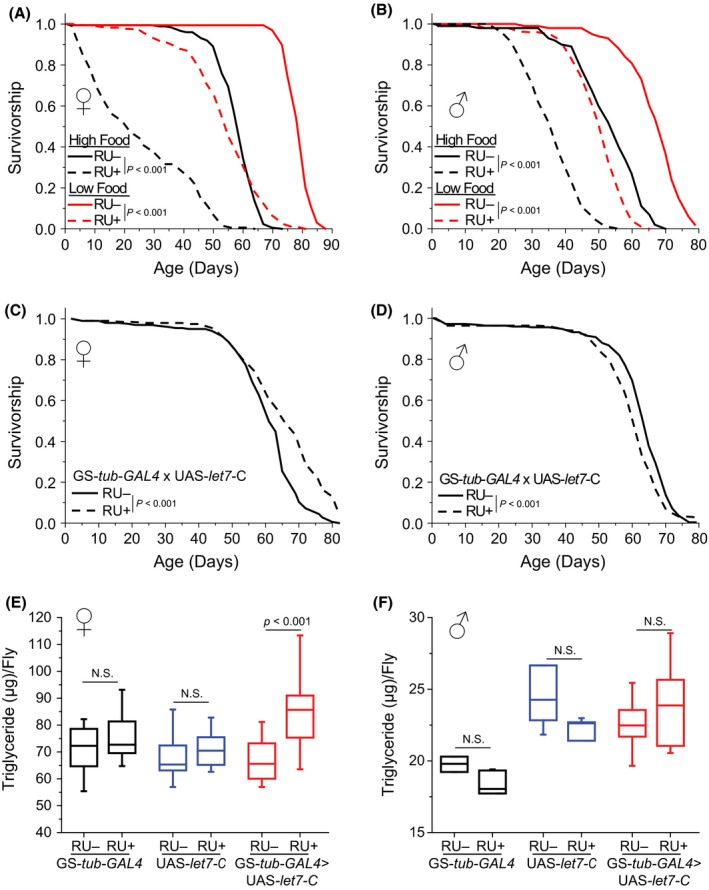
Adult‐specific, ubiquitous overexpression of miR‐184 and the let‐7‐C alters fly lifespan and metabolism. (A) Ubiquitous miR‐184 overexpression in females drastically shortened lifespan, regardless of food type (N = 174 flies for high food RU−, 171 flies for high food RU+, 167 flies for low food RU−, and 170 flies for low food RU+). (B) Ubiquitous miR‐184 overexpression in males drastically shortened lifespan, regardless of food type (N = 192 flies for both high food RU− and RU+, 191 flies for low food RU−, and 194 flies for low food RU+). (C) Ubiquitous overexpression of let‐7‐C significantly increased both the median (from 61 to 66 days) and maximal (from 75 to 83 days in the 10% longest lived) lifespan of female flies kept on a high‐nutrient diet (N = 201 flies for RU− food and 197 flies for RU+ food). (D) Ubiquitous overexpression of let‐7‐C significantly decreased male lifespan in flies kept on a high‐nutrient diet (N = 249 flies for RU− food and 244 flies for RU+ food). (E) Ubiquitous overexpression of let‐7‐C significantly increased triglycerides in female flies (N = 50 flies per genotype/food treatment). (F) Ubiquitous overexpression of let‐7‐C had no effect on male triglyceride amounts (N = 25 flies for each GS‐tub‐GAL4 and UAS‐let7‐C food treatment, N = 50 flies for each GS‐tub‐GAL4 > UAS‐let7‐C food treatment).
Aging and metabolic homeostasis are often linked (Finkel, 2015). To exemplify, dietary restriction not only increases lifespan, but also increases fat (Kapahi et al., 2016). Metabolic state is often indicated by triglyceride (TAG) abundance, the primary storage lipid in the fly. We therefore examined the effect of miR‐184 overexpression on the abundance of triglyceride (TAG). We observed no effect of miR‐184 overexpression on TAG abundances after 3 days of RU‐486 feeding, when >92% of the flies are still alive (Fig. S1C and D).
We next asked whether let‐7 influences aging. In flies, let‐7 is co‐transcribed as part of the let‐7‐complex (let‐7‐C), which is a single RNA transcript comprised of miR‐100, miR‐125, and let‐7 (Fig. S2A) (Pasquinelli et al., 2000). We found that broad let‐7‐C overexpression significantly increased both the median and maximal lifespan of females kept on a high‐nutrient diet (Fig. 1C), as well as the median lifespan of female flies kept in low‐nutrient conditions (Fig. S2B). Female lifespan was also extended using a second putatively ubiquitous GS driver (Fig. S2C) but not in control crosses (Fig. S2D,E, and F). Male flies overexpressing let‐7‐C were modestly, but significantly, shorter‐lived (Fig. 1D), revealing a sexually dimorphic effect. Let‐7‐C overexpression also led to increased TAG stores in female flies but not males (Fig. 1E and F). Flies lacking let‐7‐C are known to experience severe developmental lethality with a small percentage of escapers that exhibit a shortened lifespan (Caygill & Johnston, 2008). We found that surviving female adults had less TAG abundance, which is consistent with the notion that let‐7‐C expression promotes TAG storage (Fig. S3A). Of note, the fecundity of let‐7‐C overexpression flies was similar to control flies, establishing that the increased lifespan seen in females does not require changes in reproduction (Fig. S3B).
Attempts to identify a single tissue in which adult‐specific overexpression of the let‐7‐C is sufficient to extend lifespan were not successful. Overexpression in the fat body (using the GS‐S1106‐GAL4 driver line), nervous system (using the GS‐elav‐GAL4 driver line), or gut (using the GS‐TIGS2‐GAL4 driver line) had no effect on female lifespan (Fig. S4A,B, and C). These data suggest that let‐7‐C overexpression is required in a currently untested tissue type and/or a combination of tissues to promote lifespan extension.
To identify which components of the let‐7‐C are responsible for extended female lifespan and increased TAG, we ubiquitously overexpressed individual miRNAs. The specificity of our transgenic constructs was confirmed by qPCR (Fig. S5A). Surprisingly, broad overexpression of individual let‐7‐C members either significantly decreased lifespan (let‐7 and miR‐125; Fig. 2A and Fig. S5B) or had no effect (miR‐100; Fig. S5C). The negative effect of miR‐125 overexpression on lifespan was unexpected given that Drosophila miR‐125 is the homologue of C. elegans lin‐4, a miRNA that was previously demonstrated to increase worm lifespan when overexpressed (Boehm & Slack, 2005). These data indicate that miR‐125 and lin‐4 have different functions between species. Ubiquitous overexpression of let‐7 significantly increased TAG levels (Fig. S5D), whereas ubiquitous overexpression of miR‐125 or miR‐100 did not (Fig. S5E and F), suggesting that the increased TAG seen with ubiquitous let‐7‐C overexpression is due to let‐7 itself.
Figure 2.
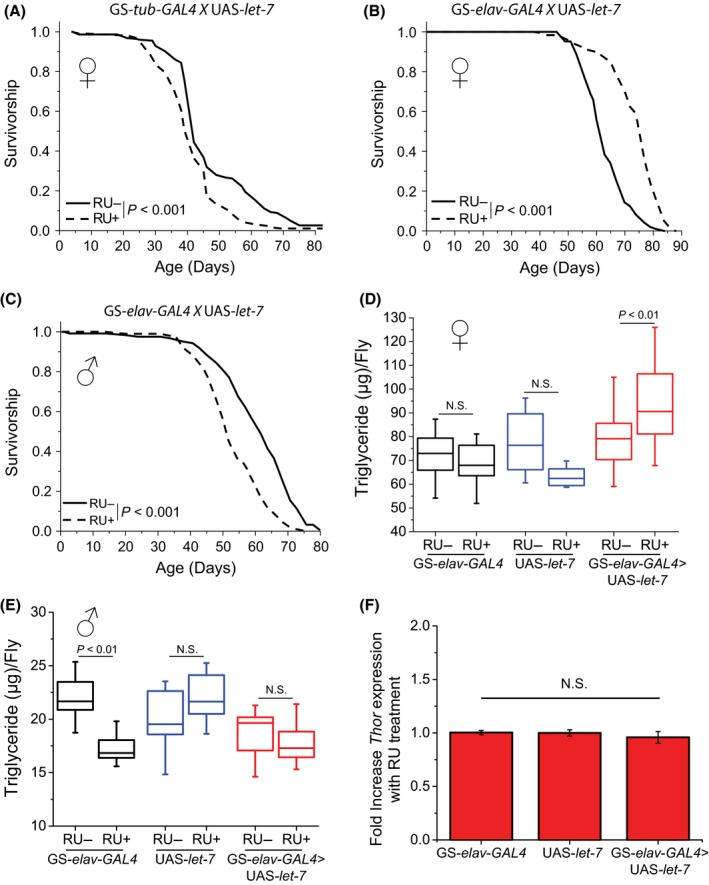
Adult‐specific, neuronal let‐7 overexpression increases female lifespan and TAG. (A) Ubiquitous let‐7 overexpression in females significantly decreased female lifespan (N = 225 flies for both food treatments). (B) Neuronal let‐7 overexpression significantly increased female lifespan (N = 167 flies for RU− food and 61 for RU+ food), while having the opposite effect in males (C; N = 101 flies for RU− food and 123 flies for RU+ food). (D) Neuronal let‐7 overexpression significantly increased female TAG levels (N = 50 flies per genotype/food treatment). (E) Neuronal let‐7 overexpression had no significant effect on male TAG levels (N = 50 flies per genotype/food treatment). (F) 4E‐BP mRNA levels are unaltered in females with neuronal let‐7 overexpression (N = 50 flies per genotype/food treatment).
Although overexpression of let‐7‐C in specific tissues was unable to increase lifespan, it is possible that the tissue‐specific effects of miRNAs of opposite valence combined to zero effect. let‐7 itself has been implicated in both neuronal proliferation and differentiation (Meza‐Sosa et al., 2012). We therefore asked whether neuronal overexpression of let‐7, or other members of the let‐7‐C, would affect longevity and/or metabolism. We found that let‐7 overexpression in neurons caused a significant increase in female median (22%) and maximum (14%) lifespan (Fig. 2B). In contrast, overexpression of miR‐125 in neurons reduced lifespan, while miR‐100 overexpression had no effect (Fig. S6A and B). Mirroring the sex‐specific effects of let‐7‐C overexpression, we found that let‐7 overexpression in male neurons significantly decreased lifespan (Fig. 2C). Female TAG levels were significantly elevated when let‐7 alone was overexpressed in neurons only (Fig. 2D). Male TAG levels may also be increased given that the significant TAG decrease seen in the driver‐only control disappears with let‐7 overexpression (Fig. 2E).
Next, we investigated whether lifespan extension from neuronal let‐7 overexpression was caused by either self‐imposed diet restriction or decreased insulin signaling. We used the FLIC (Fly Liquid Interaction Counter; Ro et al., 2014) to measure feeding behavior and found that neuronal let‐7 overexpression did not alter total feeding interactions, implying that both transgenic and control flies taste and eat food with similar duration and frequency (Fig. S7). QPCR data examining the effect of neuronal let‐7 overexpression on systemic Thor (4E‐BP) mRNA levels revealed no significant changes (Fig. 2F), suggesting that neuronal let‐7 overexpression increases lifespan in a manner that is independent of systemic insulin signaling.
The mechanism through which neuronal let‐7 expression influences metabolism and lifespan remains to be determined. let‐7 is expressed in different brain neuropil regions, including the optic lobes, the antennal lobes (homologous to the mammalian olfactory bulb), the central complex (involved in locomotor and visual behavior), and the mushroom body (homologous to the mammalian hypothalamus) (Kucherenko et al., 2012). Furthermore, let‐7 is predicted to regulate ~48 different mRNA molecules [PICTAR‐FLY; (Grun et al., 2005)], many of which are neuronally expressed (76% of profiled mRNA targets). None of these targets are currently known to be involved in fat accumulation and/or turnover. A candidate lifespan screen overexpressing let‐7 in specific sets of neurons that express predicted let‐7 targets (such as Dh44‐ and ETHR‐expressing neurons) failed to implicate specific targets (Fig. S8).
Herein, we have uncovered a new role for the conserved miRNA let‐7 in aging. While previous work has suggested that adult maintenance of let‐7‐C expression is required for healthy male lifespan (Chawla et al., 2016), this is the first work demonstrating that let‐7‐C overexpression is sufficient to increase normal lifespan. Furthermore, the ability of let‐7‐C and neuronal let‐7 to increase lifespan and alter metabolism is sexually dimorphic, showing significant increases in female lifespan and TAG while having little or even the opposite effect in males. This result may not be surprising given the role of let‐7 in male germline stem cell behavior (Toledano et al., 2012) and in cell‐specific sexual identity (Fagegaltier et al., 2014).
Funding
This work was supported by the following sources: the National Institute of Health grants R01AG030593, R01GM102279, and RO1AG023166 from the National Institute on Aging (NIA).
Conflict of interest
The authors declare that they do not have any conflict of interests.
Author contributions
C. Gendron designed the experiments, performed the experiments, and wrote the article. S. Pletcher designed the experiments and wrote the article.
Supporting information
Appendix S1. Methods.
Fig. S1 Identification of miRNA that are altered through diet, and analysis of miR‐184 overexpression flies. (A) Several miRNA are altered in flies when given either a high‐nutrient diet compared to those a low‐nutrient diet. Here, we highlight 4 miRNA that appeared to show some diet dependency: let‐7, miR‐8, miR‐34, and miR‐184. (B) qPCR of GS‐tubulin‐GAL4 > UAS‐miR‐184 flies show that feeding RU‐486 induces a 4‐fold increase in miR‐184 levels (N = 10 flies per food type). Ubiquitous overexpression of miR‐184 has no effect on TAG levels in females (C) or in males (D). In panel (C), N = 50 female flies for genotype/food treatment; in panel (D) N = 30 male flies for genotype/food treatment.
Fig. S2 Adult‐specific let‐7‐C overexpression increases female lifespan, regardless of diet or type of ubiquitous driver. (A) Cartoon of the let‐7‐complex. All 3 miRNA of the let‐7‐C (miR‐100, let‐7, and miR‐125) are transcribed as a polycistronic mRNA molecule. (B) Adult‐specific let‐7‐C overexpression using the GS‐tubulin‐GAL4 driver significantly increases female fly lifespan, regardless of diet (N = 201 flies for high food RU‐, 197 flies for high food RU+, 198 flies for low food RU‐, and 199 flies for low food RU+). (C) Adult‐specific let‐7‐C overexpression using the GS‐daughterless‐GAL4 driver also significantly increases female fly lifespan (N = 170 flies for RU‐ food and 173 flies for RU+ food). Control crosses consisting of the GS‐tubulin‐GAL4 driver (D; N = 200 flies for RU‐ food and 196 flies for RU+ food), the GS‐daughterless‐GAL4 driver (E; N = 169 flies for both food types), or UAS‐let7‐C (F; N = 195 flies for RU‐ food and 185 flies for RU+ food), with or without RU‐486 feeding, has no significant effect on lifespan.
Fig. S3 TAG is significantly decreased in let‐7‐C mutant female flies (A) and let‐7‐C overexpression has no effect on fecundity (B). In (A), N = 50 flies for both genotypes. (B) The number of eggs laid from female GS‐tubulin‐GAL4 > UAS‐let7‐C were counted every day for 7 days (N = 15 flies per food treatment).
Fig. S4 Adult‐specific overexpression of the let‐7‐C in the fat body (A), the nervous system (B), or the gut (C) has no significant effect on female lifespan. For GS‐S 1 106‐GAL4 X UAS‐let7‐C, N = 173 flies for RU‐ food and 174 flies for RU+ food. For GS‐elav‐GAL4 x UAS‐let7‐C, N = 171 flies for RU‐ food and 174 flies for RU+ food. For GS‐TIGS2‐GAL4 x UAS‐let7‐C, N = 175 flies for RU‐ food and 173 flies for RU+ food.
Fig. S5 Characterizing the overexpression of individual let‐7‐C members on lifespan and TAG. (A) QPCR of each fly genotype used to overexpress miR‐100, let‐7, or miR‐125 (N = 20 flies each per genotype/food treatment). (B) Ubiquitous, adult‐specific overexpression of miR‐125 significantly decreases female lifespan (N = 251 flies for RU‐ food and 250 flies for RU+ food). (C) Ubiquitous, adult‐specific overexpression of miR‐100 has no effect on female lifespan (N = 222 flies for RU‐ food and 218 for RU+ food). (D) Ubiquitous, adult‐specific overexpression of let‐7 significantly increases TAG levels (N = 50 flies per genotype/treatment with the exception of GS‐tub‐GAL4 > UAS‐let‐7 on RU‐ food, where N = 40 flies). There is no significant effect of miR‐125 (E; N = 150 flies per genotype/treatment with the exception of UAS‐miR‐125 where N = 50 flies/treatment) or miR‐100 (F; N = 50 flies per genotype/treatment) overexpression on TAG.
Fig. S6 Neuronal overexpression of miR‐125 or miR‐100 does not increase fly lifespan. (A) Neuronal overexpression of miR‐125 significantly decreases female lifespan (N = 172 flies for RU‐ food and 171 flies for RU+ food). (B) Neuronal overexpression of miR‐100 has no effect on female lifespan (N = 173 flies for RU‐ food and 172 flies for RU+ food).
Fig.S7 Overexpressing let‐7 in neurons does not affect female feeding. N = 12 female flies per genotype/treatment.
Fig. S8 Let‐7 overexpression in specific neuronal subpopulations, Dh44‐expressing (A) or ETHR‐expressing (B) neurons, does not significantly increase female lifespan. In panel (A), N = 197 flies for Dh44‐GAL4 > UAS‐let‐7 and yw x UAS‐let‐7; N = 198 flies for Dh44‐GAL4 x yw. In panel (B), N = 199 flies for ETHR‐GAL4 > UAS‐let‐7 and w 1118 x UAS‐let‐7; N = 200 flies for ETHR‐GAL4 x yw.
Acknowledgments
We would like to acknowledge V. Ambrose, L. Johnston, L. Partridge, and U. Gaul for providing vital Drosophila stocks for our experiments.
References
- Boehm M, Slack F (2005) A developmental timing microRNA and its target regulate life span in C. elegans . Science 310, 1954–1957. [DOI] [PubMed] [Google Scholar]
- Boehm M, Slack FJ (2006) MicroRNA control of lifespan and metabolism. Cell Cycle 5, 837–840. [DOI] [PubMed] [Google Scholar]
- Boulias K, Horvitz HR (2012) The C. elegans microRNA mir‐71 acts in neurons to promote germline‐mediated longevity through regulation of DAF‐16/FOXO. Cell Metab. 15, 439–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caygill EE, Johnston LA (2008) Temporal regulation of metamorphic processes in Drosophila by the let‐7 and miR‐125 heterochronic microRNAs. Curr. Biol. 18, 943–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla G, Deosthale P, Childress S, Wu YC, Sokol NS (2016) A let‐7‐to‐miR‐125 microRNA switch regulates neuronal integrity and lifespan in Drosophila . PLoS Genet. 12, e1006247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esslinger SM, Schwalb B, Helfer S, Michalik KM, Witte H, Maier KC, Martin D, Michalke B, Tresch A, Cramer P, Forstemann K (2013) Drosophila miR‐277 controls branched‐chain amino acid catabolism and affects lifespan. RNA Biol. 10, 1042–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagegaltier D, Konig A, Gordon A, Lai EC, Gingeras TR, Hannon GJ, Shcherbata HR (2014) A genome‐wide survey of sexually dimorphic expression of Drosophila miRNAs identifies the steroid hormone‐induced miRNA let‐7 as a regulator of sexual identity. G3: Genes ‐ Genomes ‐ Genetics, 198, 647–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T (2015) The metabolic regulation of aging. Nat. Med. 21, 1416–1423. [DOI] [PubMed] [Google Scholar]
- Gendron CM, Kuo TH, Harvanek ZM, Chung BY, Yew JY, Dierick HA, Pletcher SD (2014) Drosophila life span and physiology are modulated by sexual perception and reward. Science 343, 544–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grun D, Wang YL, Langenberger D, Gunsalus KC, Rajewsky N (2005) microRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLoS Comput. Biol. 1, e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inukai S, Slack F (2013) MicroRNAs and the genetic network in aging. J. Mol. Biol. 425, 3601–3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapahi P, Kaeberlein M, Hansen M (2016) Dietary restriction and lifespan: lessons from invertebrate models. Ageing Res. Rev. 39, 3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucherenko MM, Barth J, Fiala A, Shcherbata HR (2012) Steroid‐induced microRNA let‐7 acts as a spatio‐temporal code for neuronal cell fate in the developing Drosophila brain. EMBO J. 31, 4511–4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lencastre A, Pincus Z, Zhou K, Kato M, Lee SS, Slack FJ (2010) MicroRNAs both promote and antagonize longevity in C. elegans . Curr. Biol. 20, 2159–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Landreh M, Cao K, Abe M, Hendriks GJ, Kennerdell JR, Zhu Y, Wang LS, Bonini NM (2012) The microRNA miR‐34 modulates ageing and neurodegeneration in Drosophila . Nature 482, 519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair W, Goymer P, Pletcher SD, Partridge L (2003) Demography of dietary restriction and death in Drosophila . Science 301, 1731–1733. [DOI] [PubMed] [Google Scholar]
- Meza‐Sosa KF, Valle‐Garcia D, Pedraza‐Alva G, Perez‐Martinez L (2012) Role of microRNAs in central nervous system development and pathology. J. Neurosci. Res. 90, 1–12. [DOI] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, Spring J, Srinivasan A, Fishman M, Finnerty J, Corbo J, Levine M, Leahy P, Davidson E, Ruvkun G (2000) Conservation of the sequence and temporal expression of let‐7 heterochronic regulatory RNA. Nature 408, 86–89. [DOI] [PubMed] [Google Scholar]
- Ro J, Harvanek ZM, Pletcher SD (2014) FLIC: high‐throughput, continuous analysis of feeding behaviors in Drosophila . PLoS ONE 9, e101107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ED, Kaeberlein TL, Lydum BT, Sager J, Welton KL, Kennedy BK, Kaeberlein M (2008) Age‐ and calorie‐independent life span extension from dietary restriction by bacterial deprivation in Caenorhabditis elegans . BMC Dev. Biol. 8, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledano H, D'Alterio C, Czech B, Levine E, Jones DL (2012) The let‐7‐Imp axis regulates ageing of the Drosophila testis stem‐cell niche. Nature 485, 605–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vora M, Shah M, Ostafi S, Onken B, Xue J, Ni JZ, Gu S, Driscoll M (2013) Deletion of microRNA‐80 activates dietary restriction to extend C. elegans healthspan and lifespan. PLoS Genet. 9, e1003737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Chen D, He Y, Melendez A, Feng Z, Hong Q, Bai X, Li Q, Cai G, Wang J, Chen X (2013) MiR‐34 modulates Caenorhabditis elegans lifespan via repressing the autophagy gene atg9. Age (Dordr) 35, 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1. Methods.
Fig. S1 Identification of miRNA that are altered through diet, and analysis of miR‐184 overexpression flies. (A) Several miRNA are altered in flies when given either a high‐nutrient diet compared to those a low‐nutrient diet. Here, we highlight 4 miRNA that appeared to show some diet dependency: let‐7, miR‐8, miR‐34, and miR‐184. (B) qPCR of GS‐tubulin‐GAL4 > UAS‐miR‐184 flies show that feeding RU‐486 induces a 4‐fold increase in miR‐184 levels (N = 10 flies per food type). Ubiquitous overexpression of miR‐184 has no effect on TAG levels in females (C) or in males (D). In panel (C), N = 50 female flies for genotype/food treatment; in panel (D) N = 30 male flies for genotype/food treatment.
Fig. S2 Adult‐specific let‐7‐C overexpression increases female lifespan, regardless of diet or type of ubiquitous driver. (A) Cartoon of the let‐7‐complex. All 3 miRNA of the let‐7‐C (miR‐100, let‐7, and miR‐125) are transcribed as a polycistronic mRNA molecule. (B) Adult‐specific let‐7‐C overexpression using the GS‐tubulin‐GAL4 driver significantly increases female fly lifespan, regardless of diet (N = 201 flies for high food RU‐, 197 flies for high food RU+, 198 flies for low food RU‐, and 199 flies for low food RU+). (C) Adult‐specific let‐7‐C overexpression using the GS‐daughterless‐GAL4 driver also significantly increases female fly lifespan (N = 170 flies for RU‐ food and 173 flies for RU+ food). Control crosses consisting of the GS‐tubulin‐GAL4 driver (D; N = 200 flies for RU‐ food and 196 flies for RU+ food), the GS‐daughterless‐GAL4 driver (E; N = 169 flies for both food types), or UAS‐let7‐C (F; N = 195 flies for RU‐ food and 185 flies for RU+ food), with or without RU‐486 feeding, has no significant effect on lifespan.
Fig. S3 TAG is significantly decreased in let‐7‐C mutant female flies (A) and let‐7‐C overexpression has no effect on fecundity (B). In (A), N = 50 flies for both genotypes. (B) The number of eggs laid from female GS‐tubulin‐GAL4 > UAS‐let7‐C were counted every day for 7 days (N = 15 flies per food treatment).
Fig. S4 Adult‐specific overexpression of the let‐7‐C in the fat body (A), the nervous system (B), or the gut (C) has no significant effect on female lifespan. For GS‐S 1 106‐GAL4 X UAS‐let7‐C, N = 173 flies for RU‐ food and 174 flies for RU+ food. For GS‐elav‐GAL4 x UAS‐let7‐C, N = 171 flies for RU‐ food and 174 flies for RU+ food. For GS‐TIGS2‐GAL4 x UAS‐let7‐C, N = 175 flies for RU‐ food and 173 flies for RU+ food.
Fig. S5 Characterizing the overexpression of individual let‐7‐C members on lifespan and TAG. (A) QPCR of each fly genotype used to overexpress miR‐100, let‐7, or miR‐125 (N = 20 flies each per genotype/food treatment). (B) Ubiquitous, adult‐specific overexpression of miR‐125 significantly decreases female lifespan (N = 251 flies for RU‐ food and 250 flies for RU+ food). (C) Ubiquitous, adult‐specific overexpression of miR‐100 has no effect on female lifespan (N = 222 flies for RU‐ food and 218 for RU+ food). (D) Ubiquitous, adult‐specific overexpression of let‐7 significantly increases TAG levels (N = 50 flies per genotype/treatment with the exception of GS‐tub‐GAL4 > UAS‐let‐7 on RU‐ food, where N = 40 flies). There is no significant effect of miR‐125 (E; N = 150 flies per genotype/treatment with the exception of UAS‐miR‐125 where N = 50 flies/treatment) or miR‐100 (F; N = 50 flies per genotype/treatment) overexpression on TAG.
Fig. S6 Neuronal overexpression of miR‐125 or miR‐100 does not increase fly lifespan. (A) Neuronal overexpression of miR‐125 significantly decreases female lifespan (N = 172 flies for RU‐ food and 171 flies for RU+ food). (B) Neuronal overexpression of miR‐100 has no effect on female lifespan (N = 173 flies for RU‐ food and 172 flies for RU+ food).
Fig.S7 Overexpressing let‐7 in neurons does not affect female feeding. N = 12 female flies per genotype/treatment.
Fig. S8 Let‐7 overexpression in specific neuronal subpopulations, Dh44‐expressing (A) or ETHR‐expressing (B) neurons, does not significantly increase female lifespan. In panel (A), N = 197 flies for Dh44‐GAL4 > UAS‐let‐7 and yw x UAS‐let‐7; N = 198 flies for Dh44‐GAL4 x yw. In panel (B), N = 199 flies for ETHR‐GAL4 > UAS‐let‐7 and w 1118 x UAS‐let‐7; N = 200 flies for ETHR‐GAL4 x yw.


