Abstract
Precision medicine approaches in oncology are reliant on the accurate genomic characterization of tumors. While tissue remains the mainstay specimen for molecular testing, tumor biopsies are riddled with challenges and limitations due to their invasive and site-specific nature. Tumor inaccessibility and intratumoral heterogeneity, in particular, represent significant obstacles to the identification of actionable genetic alterations and hence effective mono- and combination therapy strategies. Proof-of-concept studies indicate that circulating tumor DNA (ctDNA) released from multiple tumor regions and anatomical locations is more reflective of intra- and intertumoral heterogeneity. Non-invasive liquid biopsy approaches that allow for the analysis of ctDNA are thus being increasingly implemented in routine patient care for the detection and monitoring of cancer-associated mutations. Indeed, the use of plasma testing to screen for epidermal growth factor receptor (EGFR) T790M mutant positive non-small cell lung cancer (NSCLC) patients eligible for treatment with third-generation EGFR inhibitors was recently approved by the U.S. Food and Drug Administration and is incorporated into the most recent version of the National Comprehensive Cancer Center guidelines as an alternative to tissue biopsy. Urine represents another liquid biopsy specimen that is distinguished by its ease of collection, option for home collection, and lack of temporal and volumetric collection restrictions. Importantly, there is an accumulating body of evidence supporting the clinical validity of urinary EGFR mutant testing for the identification and stratification of patients likely to benefit from EGFR-directed therapies and as a means to assess patient response, the presence of residual disease, and emergence of resistant tumor cell populations.
Keywords: Circulating tumor DNA (ctDNA), urine test, liquid biopsy, EGFR mutations, T790M
Introduction
The identification of activating and tyrosine kinase inhibitor (TKI)-sensitizing epidermal growth factor receptor (EGFR) mutations just over a decade ago transformed the clinical management of patients with non-small cell lung cancer (NSCLC) (1,2). The presence of EGFR L858R and exon 19 deletion mutants, among others, is observed in approximately a third of patients with NSCLC and is associated with a profound response to first-generation EGFR inhibitors such as erlotinib, gefitinib, and afatinib (3,4). For instance, erlotinib was approved for the first-line treatment of advanced NSCLC patients harboring EGFR activating mutations based on results from a randomized, multicenter, open-label trial (n=174) indicating a progression-free survival (PFS) benefit of 10.4 versus 5.2 months and an objective response rate (ORR) of 65% versus 16% with erlotinib compared to platinum-based doublet chemotherapy (5,6). As such, current clinical practice guidelines recommend the upfront testing for EGFR and other actionable oncogenic gene alterations, such as ALK and ROS1 rearrangements and BRAF mutations, in patients with NSCLC presenting with adenocarcinomas.
These clinical responses, however, are inevitably short-lived with acquired resistance to this class of inhibitors developing within 10–16 months of treatment initiation (6-9). While several mechanisms of resistance have been described, emergence of the EGFR T790M gatekeeper mutation is attributed to resistance in over half of these cases (10-12). To circumvent drug resistance in the latter patient population, third-generation, mutant-selective inhibitors have been developed to target EGFR activating and T790M resistance mutations (13-17). The clinical activity of third-generation inhibitors in patients with EGFR T790M resistance mutation-positive tumors has proven to be robust across several trials with ORRs and PFS times in the order of those reported for first-generation inhibitors in patients exhibiting activating EGFR mutations. In a randomized, international, open-label phase III trial of osimertinib in patients with EGFR T790M-positive tumors who had progressed on first-line EGFR-TKI therapy (n=419), a median PFS of 10.1 months and ORR of 71% were achieved (versus 4.4 months and 31% in the control arm, respectively) (18). These encouraging results were underscored by the approval of osimertinib in the treatment of EGFR T790M-positive patients that are refractory to other EGFR-TKIs.
In this era of precision medicine, the ability to detect and monitor actionable activating and resistance mutations with high sensitivity and specificity is thus central in improving patient outcomes. Tumor tissue genotyping is the current standard-of-care practice but is associated with many limitations including tumor inaccessibility, intratumoral and intertumoral heterogeneity, and biopsy-related adverse events (19-22). These challenges and risks are further pronounced in patients with NSCLC who have developed TKI resistance and require a second biopsy. Approximately 25% of patients are ineligible for repeat biopsy due to the presence of metastatic disease or compromised health status (19). Of those eligible for re-biopsy up to 20% are uninformative due to insufficient genetic material or absence of tumor component in samples. The non-invasive genotyping of circulating tumor DNA (ctDNA) in plasma, and more recently urine, has emerged as a viable alternative that avoids many of the pitfalls of tissue biopsies (23,24). Here, we discuss the clinical utility of urine testing for the detection and longitudinal monitoring of oncogenic driver and resistance EGFR mutations in NSCLC.
Urine as a specimen type
The presence of cell-free DNA (cfDNA) in the bloodstream has been recognized for many years. Genetic material is released into circulation via several mechanisms including cell apoptosis, necrosis, and exocytosis (i.e., active secretion) (25). Various physiological and clinical conditions, ranging from exercise to trauma or infection, are known to result in increased cfDNA concentrations (23). One of the most widely adopted applications of cfDNA analysis is for noninvasive prenatal testing of fetal cfDNA in maternal plasma (26-28). The discoveries that cancer patients have elevated levels of cfDNA in general and more importantly that tumor-specific TP53 or KRAS ctDNA mutations could be detected in blood, and other bodily fluids such as urine, marked the beginning of the use of liquid biopsies in the detection and monitoring of cancer biomarkers (29-31). A particularly valuable feature of ctDNA analysis is its potential to more thoroughly characterize the genetic landscape of a tumor since it, by definition, entails the simultaneous sampling from multiple primary and metastatic disease sites. Monitoring ctDNA dynamics can thus, in principal, capture tumor heterogeneity and evolution during the course of disease progression or in response to therapy. Urine is a completely non-invasive specimen type amenable to home collection unlike blood which entails a medical procedure requiring clinical and logistical coordination. Moreover, urine testing allows for serial sample collection for monitoring purposes and is not limited by a patient’s performance status or other physical restrictions (e.g., maximum allowable blood draw volumes).
Two fractions of urinary cfDNA have been described: high molecular weight nucleic acids originating from urinary tract and endothelial cells and low molecular weight [50–250 base pairs (bp)] circulating DNA fragments excreted into urine following glomerular filtration by the kidneys (32-35). Unlike plasma cfDNA which is thought to be protected from degradation by nucleoprotein complexes or extracellular vesicles, transrenal cfDNA undergoes additional cleavage by urine nucleases resulting in even shorter fragments in the range of 50–100 bp (36). The highly-fragmented nature of urinary ctDNA along with its low abundance (<0.01%) in a pool of wild-type cfDNA presents a major technical challenge in the extraction and detection of mutant alleles. The sensitive and quantitative detection of EGFR, BRAF, and KRAS mutations in urine has been achieved using a short-footprint mutation enrichment polymerase chain reaction (PCR) coupled with next-generation sequencing (NGS) approach (37-41). Reported limits of detection (LOD) for the EGFR L858R, exon 19 deletion, and T790M mutations in urine, or plasma, using this platform is 0.006%, 0.006%, and 0.01%, respectively, comparing favorably with the FDA-approved cobas® EGFR Mutation Test (LOD ≥0.2%) (37,42,43).
Detection of EGFR oncogenic driver and resistance mutations
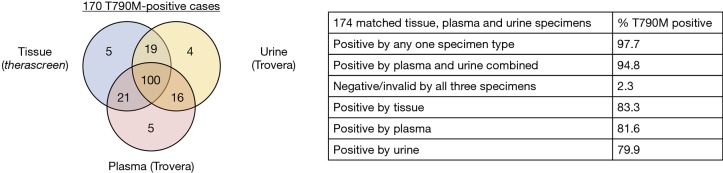
The clinical viability of assessing EGFR mutation status from urine was first interrogated in a retrospective study of samples collected from 63 patients with advanced NSCLC enrolled in the TIGER-X, phase I/II study of the third-generation EGFR-TKI, rociletinib (37). Quantitative analysis of the most common, recurrent EGFR hotspot mutations was performed using the aforementioned mutation enrichment PCR/NGS platform. With tissue as the reference, the sensitivity of EGFR mutation detection in urine ranged from 67–75% (Table 1) but was as high as 80–93% in samples that met recommended urine volumes (90–100 mL, a third of a normal urinary void) (37). Potential predictors for the successful detection of urine ctDNA including factors such as sample volume, DNA concentration, degree of hydration, and timing of voids are still being interrogated. Importantly, a high specificity of 94–100% was attained by the EGFR urine testing assay, as determined in samples obtained from healthy volunteers (Table 1), easing the concern of and potential for repercussions of false-positive calls. Further evaluation of the concordance between urine and tissue EGFR T790M results in an expanded dataset (n=213) yielded a somewhat improved positive percent agreement (PPA) of 81% and equivalence in rate of detection as compared to plasma (Table 1). The negative percent agreement (NPA) was 31%, highlighting the degree of discrepancies between tissue and liquid biopsy results likely attributable to tumor sampling errors and intratumoral heterogeneity. An in-depth comparison of matched, pretreatment EGFR T790M results in tissue, plasma, and urine revealed that the different specimen types offered complementary information with each identifying unique mutation-positive patients (Figure 1) (44). This accounted for approximately 8% (14 of 170) of a subset of patients that underwent tissue, plasma, and urine testing in the TIGER-X study. Importantly, the ORR to rociletinib in the expanded dataset was similar regardless of whether a tissue, plasma, or urine test was utilized to detect the EGFR T790M mutation [34% (95% CI, 30–39%), 32% (95% CI, 27–37%), and 37% (95% CI, 29–44%), respectively] (45).
Table 1. Concordance in EGFR mutation detection using tissue as the reference.
| Study | Patient population | # of patients (Urine Submitted) | EGFR mutation | Urine | Plasma | Reference | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PPA (%) | NPA (%) | Specificity (%) | PPA (%) | NPA (%) | Specificity (%) | ||||||
| TIGER-X | NSCLC (Stage IIIB to IV) | 63 | L858R | 75 (12/16) | 100 (50/50) | 100 (17/17) | 100 (48/48) | Reckamp (2016), J Thorac Oncol | |||
| D Exon 19 | 67 (28/42) | 94 (47/50) | 87 (34/39) | 96 (47/49) | |||||||
| T790M | 72 (34/47) | 96 (54/56) | 93 (38/41) | 94 (60/64) | |||||||
| TIGER-X | NSCLC (Stage IIIB to IV) | 213 | T790M | 81 (142/175) | 31 (5/16) | 81 (313/387) | 43 (17/40) | Wakelee (2016), J Clin Oncol | |||
EGFR, epidermal growth factor receptor; NSCLC, non-small cell lung cancer; PPA, positive percent agreement; NPA, negative percent agreement. Assay specificity determined in samples from healthy volunteers.
Figure 1.
EGFR T790M detection rate in matched tissue, plasma, and urine from a subset of 174 patients enrolled in the TIGER-X trial. (A) Venn diagram of 170 samples where at least one specimen type had a positive test result; (B) percentage of EGFR T790M-positive patients identified by various sample types. EGFR, epidermal growth factor receptor. Adapted from: Wakelee 2017, J Thorac Oncol.
A recently published case study documented the clinical course of a patient with an EGFR L858R mutation positive tumor who progressed on erlotinib subsequent to two uninformative repeat tissue biopsies (46). Nine months post radiological progression and two failed therapeutic regimens (chemotherapy and afatinib), urine ctDNA analysis identified the presence of EGFR T790M and the patient was started on osimertinib which led to a symptomatic and imaging response. This example once again illustrates the utility of independent specimen types in identifying actionable mutations and effective therapies.
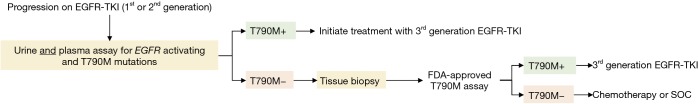
Multiple groups have reported that response rates to the third-generation EGFR-TKIs, osimertinib and rociletinib, are equivalent regardless of the specimen type used to identify EGFR T790M status (47,48). In a retrospective analysis of 216 patients enrolled in the phase I AURA trial of osimertinib, patients positive for EGFR T790M by plasma testing (ORR, 63%; PFS 9.7 months) had similar outcomes as those positive by tissue testing (ORR, 92%, PFS, 9.7 months) (48). The investigators subsequently proposed a patient management paradigm where plasma genotyping serves as the reflex test upon progression on first-generation EGFR-TKIs to circumvent the challenges associated with repeat tissue biopsies. Since a risk of false-negative results exists, a negative ctDNA result should prompt tissue genotyping, when available, to increase confidence in the mutational status. Recent data on the validity of urine testing endorses its inclusion in the above-mentioned model, whereby urine- and plasma-based testing precede tissue biopsies (Figure 2). Wakelee et al. presented that combined urine and plasma testing identified more EGFR T790M-positive cases (95%) compared to tissue biopsy alone (83%) (Figure 1) (44). Moreover, combined urine and plasma testing allowed for the sensitive detection of EGFR T790M in the intrathoracic (M1a/M0) disease setting (91% detection rate versus 96% in patients with distal metastases) further supporting an upfront liquid biopsy-based mutation screening strategy. One can envision the implementation of a similar schema in the identification of cancer-associated mutations in patients newly-diagnosed with NSCLC as a means to more comprehensively genetically profile the tumor.
Figure 2.
Proposed patient management paradigm upon progression on first- or second-generation EGFR-TKIs. EGFR, epidermal growth factor receptor; TKI, tyrosine kinase inhibitor.
In addition to potentially unveiling the presence of mutations missed in other specimen types, liquid biopsy-based tumor DNA analysis has a rapid result turnaround time of 1–2 weeks (49,50). This is markedly different from the 3–6 weeks typically required from the time a tissue biopsy is ordered by the treating physician to the reporting of results (51,52). A recent pharmacoeconomic analysis of the total cost of care of a urine- versus tissue-testing strategy for EGFR T790M detection revealed a savings of $1,243–1,680 per patient due to avoidance of biopsy procedures and associated complications as well as tissue-based molecular testing in 56% of patients (53). The authors further reported that urine testing resulted in prolonged PFS and OS compared with tissue testing due to a 7% increase in EGFR T790M detection and timely initiation of treatment with a third-generation EGFR-TKI. Hence, urine and/or blood genotyping can expedite the identification of actionable mutations and treatment initiation while avoiding invasive tissue biopsies and their associated costs.
Longitudinal monitoring of response to targeted therapies
Liquid biopsies offer the significant advantage of enabling the monitoring of dynamic changes in tumor cell populations over time (54-59). More specifically, the longitudinal assessment of EGFR mutant allele burden represents a non-invasive and cost-effective mechanism to evaluate patient response to EGFR-TKIs compared to repeat tissue biopsies or imaging. Several proof-of-concept studies show a clear correlation between urinary EGFR mutant levels and response to EGFR-TKIs, whereby a near-complete and sustained decline from baseline of ctDNA levels is associated with radiographic response (37,42). A case series in which urinary EGFR mutational burden was assessed during the course of treatment with first-line and approved or experimental third-generation EGFR-TKIs clearly depicted the utility of urine testing in the detection of response and resistance (60). A reduction in EGFR mutant allele burden was observed four weeks after initiation of EGFR-TKI therapy and subsequently corroborated by evidence of radiological improvement in three of the described cases. Conversely, an increase in urinary EGFR L858R levels and the emergence of the T790M mutation, respectively, preceded the definitive confirmation of progressive disease in two patients treated with erlotinib who had several inconclusive imaging results. While both EGFR activating and T790M mutant ctDNA load often track in response to therapies, monitoring the original oncogenic driver mutation in conjunction with the resistance, or other founder, mutations provides a means to more comprehensively characterize the tumor cell populations affected by a given therapy. For instance, ctDNA originating from non-T790M resistant (e.g., MET amplified) cells in a heterogeneous tumor population harboring EGFR activating mutations may persist in response to a third-generation EGFR-TKI while T790M levels decrease as expected. Such a scenario could be indicative of incomplete response due to the outgrowth of tumor cells exploiting alternative bypass signaling pathways.
A recent evaluation of the day-to-day kinetics of urinary EGFR mutant load also unveiled the potential clinical utility of urine testing as an early indicator of response to targeted therapies (42). Daily monitoring of EGFR mutations in urine of patients with NSCLC receiving EGFR-TKIs revealed a pattern of intermittent spikes throughout the first week of treatment. These surges in levels post-treatment are likely reflective of tumor cell apoptosis and the concomitant release of mutant ctDNA. As observed with the long-term monitoring of urine EGFR mutant levels, an overall decrease in mutant allele fractions from baseline to day 7 preceded radiographic response as assessed at 6–12 weeks. These data argue that urinary EGFR mutant allele fraction can essentially serve as real-time biomarker of response and should be further assessed in larger patient cohorts.
Future directions and conclusions
While the utility of urine EGFR mutant detection and monitoring requires further clinical evaluation in prospective studies allowing for clinical intervention based on ctDNA analysis results, the above-discussed findings imply several other clinical applications for urine genotyping. A clear extension would be in the context of early disease detection. Early detection and diagnosis has been shown to improve overall patient survival in several cancer types. To date, the clinical utility of urine-based testing for the identification of EGFR mutations has been exclusively studied in patients with advanced NSCLC. It has been demonstrated across several tumor types that the amount of ctDNA in plasma is a function of disease stage with detection rates dropping from 82% to less than 50% for patients with stage IV versus stage I disease, respectively (61). It is hypothesized that the amount of ctDNA is a function of overall tumor burden, localization, and access to vascularization. To this end it has been proposed that specimens most proximal to the anatomical location of the tumor may have the highest ctDNA content (urine for genitourinary cancer, bronchial brushings for lung cancer, cerebrospinal fluid for brain cancer, etc.) (23). As such future studies should be focused on comparing the ability of urine testing to detect ctDNA in not only early stage NSCLC but in other indications as well.
The detection of minimal residual disease (MRD) or disease recurrence following surgical resection is another area where urine ctDNA analysis holds great promise. The detection of enduring ctDNA may identify patients at high-risk of recurrence and could warrant more aggressive adjuvant therapies (62,63). On the other hand, the absence of ctDNA may be a favorable prognostic factor used to spare patients from drug-related toxicities and improve their quality of life. The post-operative reappearance of urine ctDNA could also serve as an indicator of clinical relapse which would enable earlier therapeutic intervention prior to symptomatic or imaging recurrence. While monitoring for known mutations identified upon tissue biopsy following initial surgical resection may be sufficient to monitor MRD, a full characterization via liquid biopsy specimens may once again provide a more complete representation of genetically variable tumor cell populations. Additionally, liquid biopsies provide an avenue for monitoring tumor cell evolution during disease progression or in response to therapy (55,64). For this purpose, targeted gene panels may need to be developed that cover a broader range of cancer-associated mutations while maintaining the high sensitivity required for detection of low abundance gene mutations in urine and other specimen types alike.
In conclusion, urine-based liquid biopsies have demonstrated high concordance with tissue EGFR mutation test results and in some cases captured mutations missed by tumor biopsy. In stark opposition to tissue biopsies, urine can be collected with ease, does not require medical procedures or supervision, and is not hindered by a patient’s health status or physical limitations to sample collection. Given that clinical responses to third-generation EGFR-TKIs appear to be independent of the biopsy specimen-type used to identify the EGFR T790M mutation, we propose a framework whereby urine and plasma testing is performed immediately upon progression on a first-generation inhibitor. A positive result would not only avoid a delay in onset of treatment due to the quick turnaround time relative to tissue biopsies but would spare patients from invasive tissue biopsies and eliminate procedure-related risks (Figure 2). Future studies should be geared at the prospective evaluation of this patient stratification paradigm in larger cohorts to formally demonstrate the equivalence, and perhaps superiority, of liquid biopsy-based genotyping over tissue testing.
Acknowledgements
None.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004;304:1497-500. 10.1126/science.1099314 [DOI] [PubMed] [Google Scholar]
- 2.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129-39. 10.1056/NEJMoa040938 [DOI] [PubMed] [Google Scholar]
- 3.Linardou H, Dahabreh IJ, Bafaloukos D, et al. Somatic EGFR mutations and efficacy of tyrosine kinase inhibitors in NSCLC. Nat Rev Clin Oncol 2009;6:352-66. 10.1038/nrclinonc.2009.62 [DOI] [PubMed] [Google Scholar]
- 4.Chong CR, Jänne PA. The quest to overcome resistance to EGFR-targeted therapies in cancer. Nat Med 2013;19:1389-400. 10.1038/nm.3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khozin S, Blumenthal GM, Jiang X, et al. U.S. Food and Drug Administration Approval Summary: Erlotinib for the First-Line Treatment of Metastatic Non-Small Cell Lung Cancer With Epidermal Growth Factor Receptor Exon 19 Deletions or Exon 21 (L858R) Substitution Mutations. The Oncologist 2014;19:774-9. 10.1634/theoncologist.2014-0089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-46. 10.1016/S1470-2045(11)70393-X [DOI] [PubMed] [Google Scholar]
- 7.Rosell R, Moran T, Queralt C, et al. Screening for Epidermal Growth Factor Receptor Mutations in Lung Cancer. N Engl J Med 2009;361:958-67. 10.1056/NEJMoa0904554 [DOI] [PubMed] [Google Scholar]
- 8.Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or Chemotherapy for Non-Small-Cell Lung Cancer with Mutated EGFR. N Engl J Med 2010;362:2380-8. 10.1056/NEJMoa0909530 [DOI] [PubMed] [Google Scholar]
- 9.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or Carboplatin-Paclitaxel in Pulmonary Adenocarcinoma. N Engl J Med 2009;361:947-57. 10.1056/NEJMoa0810699 [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR Mutation and Resistance of Non-Small-Cell Lung Cancer to Gefitinib. N Engl J Med 2005;352:786-92. 10.1056/NEJMoa044238 [DOI] [PubMed] [Google Scholar]
- 11.Pao W, Miller VA, Politi KA, et al. Acquired Resistance of Lung Adenocarcinomas to Gefitinib or Erlotinib Is Associated with a Second Mutation in the EGFR Kinase Domain. PLoS Med 2005;2:e73. 10.1371/journal.pmed.0020073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu HA, Arcila ME, Rekhtman N, et al. Analysis of Tumor Specimens at the Time of Acquired Resistance to EGFR-TKI Therapy in 155 Patients with EGFR-Mutant Lung Cancers. Clin Cancer Res 2013;19:2240-7. 10.1158/1078-0432.CCR-12-2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sequist LV, Soria JC, Goldman JW, et al. Rociletinib in EGFR -Mutated Non-Small-Cell Lung Cancer. N Engl J Med 2015;372:1700-9. 10.1056/NEJMoa1413654 [DOI] [PubMed] [Google Scholar]
- 14.Jänne PA, Yang JC, Kim DW, et al. AZD9291 in EGFR Inhibitor-Resistant Non-Small-Cell Lung Cancer. N Engl J Med 2015;372:1689-99. 10.1056/NEJMoa1411817 [DOI] [PubMed] [Google Scholar]
- 15.Lelais G, Epple R, Marsilje TH, et al. Discovery of (R, E)- N -(7-Chloro-1-(1-[4-(dimethylamino)but-2-enoyl]azepan-3-yl)-1 H -benzo[ d ]imidazol-2-yl)-2-methylisonicotinamide (EGF816), a Novel, Potent, and WT Sparing Covalent Inhibitor of Oncogenic (L858R, ex19del) and Resistant (T790M) EGFR Mutants for the Treatment of EGFR Mutant Non-Small-Cell Lung Cancers. J Med Chem 2016;59:6671-89. 10.1021/acs.jmedchem.5b01985 [DOI] [PubMed] [Google Scholar]
- 16.Planken S, Behenna DC, Nair SK, et al. Discovery of N -((3 R, 4 R)-4-Fluoro-1-(6-((3-methoxy-1-methyl-1 H -pyrazol-4-yl)amino)-9-methyl-9 H -purin-2-yl)pyrrolidine-3-yl)acrylamide (PF-06747775) through Structure-Based Drug Design: A High Affinity Irreversible Inhibitor Targeting Oncogenic EGFR Mutants with Selectivity over Wild-Type EGFR. J Med Chem 2017;60:3002-19. 10.1021/acs.jmedchem.6b01894 [DOI] [PubMed] [Google Scholar]
- 17.Park K, Lee JS, Han JY, et al. 1300: Efficacy and safety of BI 1482694 (HM61713), an EGFR mutant-specific inhibitor, in T790M-positive NSCLC at the recommended phase II dose. J Thorac Oncol 2016;11:S113. 10.1016/S1556-0864(16)30243-X27198272 [DOI] [Google Scholar]
- 18.Mok TS, Wu YL, Ahn MJ, et al. Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N Engl J Med 2017;376:629-40. 10.1056/NEJMoa1612674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoon HJ, Lee HY, Lee KS, et al. Repeat Biopsy for Mutational Analysis of Non-Small Cell Lung Cancers Resistant to Previous Chemotherapy: Adequacy and Complications. Radiology 2012;265:939-48. 10.1148/radiol.12112613 [DOI] [PubMed] [Google Scholar]
- 20.Douillard JY, Ostoros G, Cobo M, et al. Gefitinib treatment in EGFR mutated caucasian NSCLC: circulating-free tumor DNA as a surrogate for determination of EGFR status. J Thorac Oncol 2014;9:1345-53. 10.1097/JTO.0000000000000263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawamura T, Kenmotsu H, Taira T, et al. Rebiopsy for patients with non-small-cell lung cancer after epidermal growth factor receptor-tyrosine kinase inhibitor failure. Cancer Sci 2016;107:1001-5. 10.1111/cas.12963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piotrowska Z, Niederst MJ, Karlovich CA, et al. Heterogeneity Underlies the Emergence of EGFRT790 Wild-Type Clones Following Treatment of T790M-Positive Cancers with a Third-Generation EGFR Inhibitor. Cancer Discov 2015;5:713-22. 10.1158/2159-8290.CD-15-0399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wan JC, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 2017;17:223-38. 10.1038/nrc.2017.7 [DOI] [PubMed] [Google Scholar]
- 24.Bardelli A, Pantel K. Liquid Biopsies, What We Do Not Know (Yet). Cancer Cell 2017;31:172-9. 10.1016/j.ccell.2017.01.002 [DOI] [PubMed] [Google Scholar]
- 25.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 2011;11:426-37. 10.1038/nrc3066 [DOI] [PubMed] [Google Scholar]
- 26.Lo YM. Molecular testing of urine: catching DNA on the way out. Clin Chem 2000;46:1039-40. [PubMed] [Google Scholar]
- 27.Tsui NB, Jiang P, Chow KC, et al. High Resolution Size Analysis of Fetal DNA in the Urine of Pregnant Women by Paired-End Massively Parallel Sequencing. Oudejans C, editor. PLoS One 2012;7:e48319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu SC, Lee SW, Jiang P, et al. High-Resolution Profiling of Fetal DNA Clearance from Maternal Plasma by Massively Parallel Sequencing. Clin Chem 2013;59:1228-37. 10.1373/clinchem.2013.203679 [DOI] [PubMed] [Google Scholar]
- 29.Leon SA, Shapiro B, Sklaroff DM, et al. Free DNA in the Serum of Cancer Patients and the Effect of Therapy. Cancer Res 1977;37:646-50. [PubMed] [Google Scholar]
- 30.Sidransky D, Von Eschenbach A, Tsai YC, et al. Identification of p53 gene mutations in bladder cancers and urine samples. Science 1991;252:706-9. 10.1126/science.2024123 [DOI] [PubMed] [Google Scholar]
- 31.Sorenson GD, Pribish DM, Valone FH, et al. Soluble normal and mutated DNA sequences from single-copy genes in human blood. Cancer Epidemiol Biomarkers Prev 1994;3:67-71. [PubMed] [Google Scholar]
- 32.Su YH, Wang M, Brenner DE, et al. Human Urine Contains Small, 150 to 250 Nucleotide-Sized, Soluble DNA Derived from the Circulation and May Be Useful in the Detection of Colorectal Cancer. J Mol Diagn 2004;6:101-7. 10.1016/S1525-1578(10)60497-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Botezatu I, Serdyuk O, Potapova G, et al. Genetic analysis of DNA excreted in urine: a new approach for detecting specific genomic DNA sequences from cells dying in an organism. Clin Chem 2000;46:1078-84. [PubMed] [Google Scholar]
- 34.Bryzgunova OE, Skvortsova TE, Kolesnikova EV, et al. Isolation and Comparative Study of Cell-Free Nucleic Acids from Human Urine. Ann N Y Acad Sci 2006;1075:334-40. 10.1196/annals.1368.045 [DOI] [PubMed] [Google Scholar]
- 35.Su YH, Wang M, Aiamkitsumrit B, et al. Detection of a K-ras mutation in urine of patients with colorectal cancer. Cancer Biomark 2005;1:177-82. 10.3233/CBM-2005-12-305 [DOI] [PubMed] [Google Scholar]
- 36.Melkonyan HS, Feaver WJ, Meyer E, et al. Transrenal Nucleic Acids: From Proof of Principle to Clinical Tests. Ann N Y Acad Sci 2008;1137:73-81. 10.1196/annals.1448.015 [DOI] [PubMed] [Google Scholar]
- 37.Reckamp KL, Melnikova VO, Karlovich C, et al. A Highly Sensitive and Quantitative Test Platform for Detection of NSCLC EGFR Mutations in Urine and Plasma. J Thorac Oncol 2016;11:1690-700. 10.1016/j.jtho.2016.05.035 [DOI] [PubMed] [Google Scholar]
- 38.Hyman DM, Diamond EL, Vibat CR, et al. Prospective Blinded Study of BRAFV600E Mutation Detection in Cell-Free DNA of Patients with Systemic Histiocytic Disorders. Cancer Discov 2015;5:64-71. 10.1158/2159-8290.CD-14-0742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Janku F, Vibat CR, Kosco K, et al. BRAF V600E mutations in urine and plasma cell-free DNA from patients with Erdheim-Chester disease. Oncotarget 2014;5:3607. 10.18632/oncotarget.1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klempner SJ, Gershenhorn B, Tran P, et al. BRAFV600E Mutations in High-Grade Colorectal Neuroendocrine Tumors May Predict Responsiveness to BRAF-MEK Combination Therapy. Cancer Discov 2016;6:594-600. 10.1158/2159-8290.CD-15-1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fujii T, Barzi A, Sartore-Bianchi A, et al. Mutation-Enrichment Next-Generation Sequencing for Quantitative Detection of KRAS Mutations in Urine Cell-Free DNA from Patients with Advanced Cancers. Clin Cancer Res 2017;23:3657-66. 10.1158/1078-0432.CCR-16-2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Husain H, Melnikova VO, Kosco K, et al. Monitoring Daily Dynamics of Early Tumor Response to Targeted Therapy by Detecting Circulating Tumor DNA in Urine. Clin Cancer Res 2017. [Epub ahead of print]. 10.1158/1078-0432.CCR-17-0454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thress KS, Brant R, Carr TH, et al. EGFR mutation detection in ctDNA from NSCLC patient plasma: A cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer 2015;90:509-15. 10.1016/j.lungcan.2015.10.004 [DOI] [PubMed] [Google Scholar]
- 44.Wakelee H, Melnikova V, Karlovich C, et al. MA08.01 A Highly Sensitive Next-Generation Sequencing Platform for Detection of NSCLC EGFR T790M Mutation in Urine and Plasma. J Thorac Oncol 2017;12:S384-5. 10.1016/j.jtho.2016.11.434 [DOI] [PubMed] [Google Scholar]
- 45.Wakelee HA, Gadgeel SM, Goldman JW, et al. Epidermal growth factor receptor (EGFR) genotyping of matched urine, plasma and tumor tissue from non-small cell lung cancer (NSCLC) patients (pts) treated with rociletinib. J Clin Oncol 2016;34:abstr 9001.
- 46.Berz D, Raymond VM, Garst JH, et al. Non-invasive urine testing of EGFR activating mutation and T790M resistance mutation in non-small cell lung cancer. Exp Hematol Oncol 2016;5:24. 10.1186/s40164-016-0052-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karlovich C, Goldman JW, Sun JM, et al. Assessment of EGFR Mutation Status in Matched Plasma and Tumor Tissue of NSCLC Patients from a Phase I Study of Rociletinib (CO-1686). Clin Cancer Res 2016;22:2386-95. 10.1158/1078-0432.CCR-15-1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oxnard GR, Thress KS, Alden RS, et al. Association Between Plasma Genotyping and Outcomes of Treatment With Osimertinib (AZD9291) in Advanced Non-Small-Cell Lung Cancer. J Clin Oncol 2016;34:3375-82. 10.1200/JCO.2016.66.7162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sacher AG, Dahlberg SE, Heng J, et al. Association Between Younger Age and Targetable Genomic Alterations and Prognosis in Non-Small-Cell Lung Cancer. JAMA Oncol 2016;2:313. 10.1001/jamaoncol.2015.4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwaederle M, Husain H, Fanta PT, et al. Use of Liquid Biopsies in Clinical Oncology: Pilot Experience in 168 Patients. Clin Cancer Res 2016;22:5497-505. 10.1158/1078-0432.CCR-16-0318 [DOI] [PubMed] [Google Scholar]
- 51.Schwaederle M, Parker BA, Schwab RB, et al. Molecular Tumor Board: The University of California San Diego Moores Cancer Center Experience. The Oncologist 2014;19:631-6. 10.1634/theoncologist.2013-0405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sacher AG, Paweletz C, Dahlberg SE, et al. Prospective Validation of Rapid Plasma Genotyping for the Detection of EGFR and KRAS Mutations in Advanced Lung Cancer. JAMA Oncol 2016;2:1014-22. 10.1001/jamaoncol.2016.0173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sands J, Hornberger J. P3.02b-010 Urine Detection of EGFR T790M Mutation in Non-Small-Cell Lung Cancer: An Outcomes and Total Cost of Care Analysis. J Thorac Oncol 2017;12:S1191 10.1016/j.jtho.2016.11.1677 [DOI] [PubMed] [Google Scholar]
- 54.Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548-54. 10.1038/nm.3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Siravegna G, Mussolin B, Buscarino M, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med 2015;21:795-801. 10.1038/nm.3870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thress KS, Paweletz CP, Felip E, et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med 2015;21:560-2. 10.1038/nm.3854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med 2008;14:985-90. 10.1038/nm.1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Forshew T, Murtaza M, Parkinson C, et al. Noninvasive Identification and Monitoring of Cancer Mutations by Targeted Deep Sequencing of Plasma DNA. Sci Transl Med 2012;4:136ra68. 10.1126/scitranslmed.3003726 [DOI] [PubMed] [Google Scholar]
- 59.Dawson SJ, Tsui DW, Murtaza M, et al. Analysis of Circulating Tumor DNA to Monitor Metastatic Breast Cancer. N Engl J Med 2013;368:1199-209. 10.1056/NEJMoa1213261 [DOI] [PubMed] [Google Scholar]
- 60.Tchekmedyian N, Mudad R, Blanco FF, et al. Longitudinal monitoring of ctDNA EGFR mutation burden from urine correlates with patient response to EGFR TKIs: A case series. Lung Cancer 2017;108:22-8. 10.1016/j.lungcan.2017.02.010 [DOI] [PubMed] [Google Scholar]
- 61.Bettegowda C, Sausen M, Leary RJ, et al. Detection of Circulating Tumor DNA in Early- and Late-Stage Human Malignancies. Sci Transl Med 2014;6:224ra24. 10.1126/scitranslmed.3007094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tie J, Wang Y, Tomasetti C, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med 2016;8:346ra92. 10.1126/scitranslmed.aaf6219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Garcia-Murillas I, Schiavon G, Weigelt B, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med 2015;7:302ra133. 10.1126/scitranslmed.aab0021 [DOI] [PubMed] [Google Scholar]
- 64.Murtaza M, Dawson SJ, Pogrebniak K, et al. Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer. Nat Commun 2015;6:8760. 10.1038/ncomms9760 [DOI] [PMC free article] [PubMed] [Google Scholar]