Abstract
Liquid biopsies have been heralded as a game changer in cancer management. Blood tests offer a minimally invasive, safe and sensitive complementary (or even alternative) approach for tissue biopsies. With lung cancer being the second most commonly diagnosed cancer and the leading cause of cancer deaths worldwide, due to the limitations of tissue sampling, liquid biopsies must urgently materialize in the clinic. In this short review, we will present the current applications of cell-free DNA (cfDNA) in lung cancer management, emphasizing on our own experience and previous work. We will also shortly comment on the challenges and need for a coordinated collaboration combining disciplines and sectors (from academia to health economies) in order to accelerate liquid biopsy development in lung cancer and other cancers.
Keywords: Lung cancer, cell-free DNA (cfDNA), epidermal growth factor receptor (EGFR), liquid biopsies
Introduction
Many targeted therapies that inhibit genetic alterations, required to stop tumor growth and metastasis, have demonstrated proven activity in patients with lung adenocarcinoma (1,2). The use of matched therapy is strongly related to the level of evidence that the identified mutation can really predict response to the treatment. For instance, epidermal growth factor receptor (EGFR) mutations, anaplastic lymphoma kinase (ALK), ROS proto-oncogene 1, receptor tyrosine kinase (ROS1) fusions (level 1) and rearranged during transfection (RET) fusion, BRAFV600E mutations and hepatocyte growth factor receptor (MET) mutations or amplification (level 2) are the ones that are almost universally recognized to predict response to a specific drug (2). Immune checkpoint blockade has now emerged as a promising therapeutic approach for cancer patients including those with lung cancer (3). Still, durable tumor responses and prolongation of survival with immunotherapy occur in a minority of patients and we lack efficient tumor biomarker assays predictive of response to these novel therapies.
Current clinical oncology practice relies on the removal of tumor tissue through biopsies for analysis of tumor-linked genetic alterations. Although tumor tissue biopsy is the current gold standard for cancer diagnosis, it is an invasive approach, which poses a limitation for repeated sampling that is commonly needed for monitoring treatment response and resistance to targeted therapies. Furthermore, it has become clear that the information acquired from a single biopsy provides a spatially and temporarily limited snapshot of a tumor and often fails to reflect the heterogeneity of the disease (4,5). Liquid biopsies could provide a potential revolution in cancer diagnostics as a minimally invasive method for detecting and monitoring diseases, complementary to current tissue biopsy approaches (6). Indeed, there are several blood sources from which tumor DNA or RNA can be obtained and used for the detection of specific mutations. Current blood-based biopsy assays focus on four biosources, which are, cell-free DNA (cfDNA), circulating tumor cells (CTCs), extracellular vesicles (EVs, exosomes, oncosomes) and tumor-educated platelets (TEPs) (7).
The presence of cfDNA in plasma was for the first time reported in 1948 by Mandel and Metais (8). This cfDNA is double-stranded and highly fragmented with an approximate length of 150 bp. Many years later, it was discovered that cancer patients have higher levels of cfDNA in their blood compared to healthy individuals (9). In addition, patients with metastatic cancer have higher levels of cfDNA in their blood compared to non-metastatic cancer patients (9). In 1989, it was proven that cfDNA in cancer patients contains tumor DNA (10). This circulating tumor DNA (ctDNA) is so much more diluted in normal DNA, that only mutation specific polymerase-chain-reaction (PCR) assays were initially used to noninvasively analyze tumor genomes with sufficient sensitivity and specificity. However, the fact that most tumors consist of subclonal populations and only some of the somatic mutations are carried by all cells, poses difficulties about the use of mutation specific PCR assays. The recent use of next generation sequencing (NGS) based options has partially overcome this problem. The current methods for the detection of specific DNA aberrations are PCR-based techniques like our protein nucleic acid (PNA) clamp-based PCR assay (TaqMan assay) (11-13), digital PCR, droplet digital PCR (ddPCR) (14), BEAMing (for Beads, Emulsions, Amplification, and Magnetics) technology (15) or amplification refractory mutation system (ARMS) PCR (16). Multiplex PCR techniques are also lately used, such as tagged amplicon sequencing (17), as well as ligation and hybridization-capture methods (18), with the best example of this strategy being found in the study of Newman and colleagues (19). Needless to say, apart from blood, cfDNA can also be extracted from other body fluids, like urine. In a recent study, KRAS mutations were detected with mutation enrichment NGS in urine cfDNA of cancer patients and they provided good concordance with archival tumor tissue (20).
In our group we have worked since 1998 with cfDNA in non-small cell lung cancer (NSCLC) patients (21-25). Herein, we will review the use of cfDNA in lung cancer diagnostics and disease monitoring, emphasizing our own experience, as well as the current limitations for the development and commercialization of a liquid biopsy test.
Detection of somatic mutations in cfDNA–the reality until now for lung cancer patients
From 2005 until 2008, the Spanish Lung Cancer Group performed a large scale screening study for EGFR mutations in patients with metastatic NSCLC (26). EGFR mutations were found in 350 out of 2,105 patients (16.6%) (27). Baseline blood samples were available from 164 patients, and the EGFR mutation could be detected in 97 of them (59.1%). Deletions in exon 19 were determined by length analysis after PCR amplification and exon 21 L858R mutations were detected with a 5’nuclease PCR assay (TaqMan assay). Both reactions were performed in the presence of a PNA clamp, designed to inhibit the amplification of the wild-type allele. The adjusted hazard ratio for the duration of progression-free survival (PFS) was 1.68 for the presence of the L858R mutation in paired serum DNA, as compared with the absence of the mutation (P=0.02) (Table 1). In a prespecified analysis of a secondary objective of the European Tarceva versus Chemotherapy (EURTAC) study, the rate of EGFR mutations in cfDNA was examined with the same methodology (26) and with a 53.2% sensitivity (27,39). The presence of EGFR mutations in cfDNA was associated with shorter PFS (HR 0.43; 95% CI: 0.26–0.73; P=0.002). However, when the blood samples were reanalyzed with an optimized rapid and sensitive PNA-based PCR assay (TaqMan assay), EGFR mutations (both deletions 19 and L858R mutations) could be detected in 78% of patients with usable blood samples (11). With the same optimized technique, we have been able to detect and quantify BRAFV600E in mixed archival serum and plasma samples of melanoma, colon and NSCLC patients with a specificity of 100% and a sensitivity of 57.7% (12). We have also demonstrated the negative prognostic significance of the EGFR L858R in cfDNA (10). On average, the median overall survival (OS) for erlotinib-treated NSCLC patients with the EGFR L858R mutation detected in their tissue was 17.7 (95% CI: 10.0–23.5) months. Among them, median OS was 13.7 (95% CI: 7.1–17.7) months for those with the L858R mutation also detected in their blood compared to 27.7 (95% CI: 16.1–46.2) months for patients without the mutation detected in their blood (P=0.02) (11). We have now performed a large-scale prospective screening of EGFR mutations in the blood of unselected advanced NSCLC patients with our PNA-based PCR assay. Among 1,026 assessable blood samples of advanced NSCLC patients with no biopsy or insufficient tumor tissue, sensitizing EGFR mutations were detected in 113 of them. An objective response rate of 72% was observed among 18 patients, treated with an EGFR tyrosine kinase inhibitor (TKI), based exclusively on the results of the EGFR testing in the blood (28).
Table 1. Clinical studies examining EGFR mutations in cfDNA.
| First author | Name of the study | Population | No | Technique used | Results | Reference |
|---|---|---|---|---|---|---|
| Rosell | NEJM | EGFR mutant | 164 | Del 19: length analysis with PNA; L858R: PCR TaqMan assay with PNA | (I) EGFR mutations detected in 59.1% of samples at baseline; (II) L858R mutation in cfDNA related with shorter PFS |
(26) |
| Rosell | EURTAC | EGFR mutant | 109 | Del 19: length analysis with PNA; L858R: PCR TaqMan assay with PNA | (I) EGFR mutations detected in 53.2% of samples at baseline; (II) EGFR mutations in cfDNA related with shorter PFS |
(27) |
| Karachaliou | EURTAC | EGFR mutant | 97 | PCR TaqMan assay with PNA | EGFR mutations detected in 78% of samples at baseline | (11) |
| Mayo-de-las-Casas | Prospective screening study | Advanced NSCLC | 1,033 | PCR TaqMan assay with PNA | EGFR mutations detected in 11% of samples at presentation | (28) |
| Douillard | IFUM | Advanced NSCLC | 652 | TheraScreen EGFR RGQ PCR Kit | EGFR mutations detected in 65.7% of samples at presentation (99.8% specificity) | (29) |
| Wu | LUX-Lung 3/6 | EGFR mutant | 661 (LUX-Lung 3, 287; LUX-Lung 6, 334) | TheraScreen EGFR RGQ PCR Kit | Overall concordance: 28.6% (LUX-Lung 3) and 60.5% (LUX-Lung 6) | (30) |
| Mok | FASTACT-2 | Advanced NSCLC | 241 | Cobas test | (I) 75% sensitivity, 95% specificity and 88% concordance; (II) predictive role of EGFR mutations dynamic changes in cfDNA |
(31) |
| Wu | ENSURE | Advanced NSCLC | 517 | cobas EGFR Mutation Test v2 | (I) 76.7% sensitivity and 98.2% specificity; (II) FDA approval of the test |
(32,33) |
| Mok | AURA3 | EGFR T790M-positive NSCLC | 399 | cobas EGFR Mutation Test v2 | (I)4 6.57% sensitivity and 71.83% specificity; (II) PFS with osimertinib was similar regardless of selection by tissue or plasma T790M-positive status |
(34,35) |
| Oxnard | AURA | EGFR mutant progressing to EGFR TKI | 216 | Beaming | EGFR T790M: 70.3% sensitivity, 69% specificity | (36) |
| Sacher | NCT02279004 | Advanced NSCLC | 180 | ddPCR | (I) EGFR exon 19 deletions: 82% sensitivity, 100% specificity; (II) EGFR L858R: 74% sensitivity, 100% specificity; (III) EGFR T790M: 77% sensitivity, 79% specificity; (IV) KRAS: 64% sensitivity, 100% specificity |
(37) |
| Reck | ASSESS | Advanced NSCLC | 1,311 (1,288 eligible) | Therascreen EGFR RGQ PCR kit, Roche cobas EGFR Mutation test, PNA-LNA PCR Clamp, Cycleave, others | 89% concordance, 46% sensitivity and 97% specificity | (38) |
RGQ, rotor-gene Q; PNA-LNA, peptide nucleic acid-locked nucleic acid; EGFR, epidermal growth factor receptor; cfDNA, cell-free DNA; PCR, polymerase-chain-reaction; NSCLC, non-small cell lung cancer; TKI, tyrosine kinase inhibitor; EURTAC, the European Tarceva versus Chemotherapy; PFS, progression-free survival;
Other investigators have explored the utility of the therascreen EGFR rotor-gene Q (RGQ) PCR Kit, the cobas EGFR Mutation Test or other methodologies for the detection of EGFR mutations in the blood (Table 1). The IFUM study was a phase IV single arm study to assess the efficacy and safety of gefitinib as first line therapy in EGFR mutant Caucasian NSCLC patients (29). In an exploratory biomarker analysis, the Therascreen EGFR RGQ PCR kit was used for the detection of EGFR mutations in cfDNA, with high concordance, specificity and sensitivity (29). The same test was used in the phase III LUX-Lung 3 and 6 trials (30) (Table 1). In the FASTACT-2 study in which chemotherapy with sequential erlotinib was compared to chemotherapy alone in NSCLC patients, EGFR mutations were assessed in the blood with the Cobas test, with a sensitivity and specificity of 75% and 95%, respectively, and a concordance between tissue and blood tests of 88%. Furthermore, the dynamic changes of EGFR mutations in cfDNA were able to predict treatment outcome (31). The cobas EGFR Mutation Test v2 was used for the detection of EGFR mutations in the plasma samples of the patients enrolled in the ENSURE trial (32). The test demonstrated a sensitivity of 76.7% and a specificity of 98.2% (33). The same test was used in the AURA3 study for the detection of the EGFR resistant mutation, T790M, with a sensitivity of 46.57% and a specificity of 71.83% (34,35). In a retrospective analysis of the AURA study, the group of Geoffrey Oxnard demonstrated that Beads, Emulsion, Amplification and Magnetics type digital polymerase chain reaction (BEAMing dPCR) could detect the T790M mutation in cfDNA at the time of acquired resistance to EGFR TKI with a sensitivity of 70% (36,40). The results of this study indicated that blood-based testing can identify patients who have progressed on an EGFR TKI and have developed the T790M mutation, but for the 30% of patients with false-negative plasma genotyping, a tumor biopsy is still required to determine the presence, or absence, of the resistant mutation (36,40). The same group had previously performed a prospective study to evaluate the performance of the ddPCR for the detection of EGFR (including T790M) and KRAS mutations in the blood of advanced NSCLC patients (37,41). Plasma ddPCR could detect with a median turnaround time of 3 days EGFR exon 19 deletions with a sensitivity of 82% and a specificity of 100%, EGFR L858R with a sensitivity of 74% and a specificity of 100%, EGFR T790M with a sensitivity of 77% and a specificity of 79% and KRAS mutations with a sensitivity of 64% and a specificity of 100% (37). Finally, the ASSESS study was performed in an effort to evaluate the diagnostic performance of various assays for the detection of EGFR mutations in cfDNA (38). The study enrolled 1,311 patients from 56 centers in Europe and Japan and the mutation testing methods in the plasma were mainly the Therascreen EGFR RGQ PCR kit, the Roche cobas EGFR mutation test, peptide nucleic acid-locked nucleic acid (PNA-LNA) PCR Clamp and Cycleave. Overall, the concordance of EGFR mutation status in tissue and plasma samples was 89%, with 97% specificity and 46% sensitivity. The Qiagen therascreen EGFR RGQ PCR kit and the Roche cobas EGFR mutation test surfaced as the most sensitive (73% and 75% respectively) among the plasma assays evaluated (38).
Aside from EGFR mutations, we have less experience from clinical trials and liquid biopsy assays for the detection of other alterations, like ALK fusions or KRAS mutations, in cfDNA. Capture-based NGS has been used for the detection of ALK fusions in cfDNA with an overall sensitivity, specificity and accuracy of 54%, 100% and 72%, respectively (42). Very interestingly Bordi and colleagues have shown that KRAS mutations can emerge as a mechanism of resistance to crizotinib in ALK positive NSCLC patients (43). They analyzed using ddPCR, cfDNA of 20 ALK positive NSCLC patients progressing to crizotinib. Three of them patients were carriers of both ALK resistant mutations and the KRAS point mutation p.G12D, while in seven patients only a KRAS point mutation in codon 12 was detected at the time of progression to crizotinib (43).
KRAS mutations are the most common oncogenic alterations detected in approximately 20–30% of NSCLC patients (44). The presence of KRAS mutations is usually associated with the use of tobacco (1,45,46). Co-occurring genetic alterations in STK11 (LKB1), TP53 and CDKN2A/B can define three major subgroups of KRAS mutant lung adenocarcinoma with distinct response to immunotherapy. Specifically tumors that carry both KRAS and TP53 mutations have higher levels of somatic mutations and inflammatory markers, indicating a higher probability of response to immune checkpoint blockade. This is not the case for triple mutant tumors (KRAS, TP53 and STK11) that are lacking an immune system engagement and the probabilities of response to immune checkpoint blockade are lower in comparison with the double mutant tumors (47). KRAS mutations indicate a poor outcome of the patients and currently there is not any efficient targeted therapy for this disease (48).
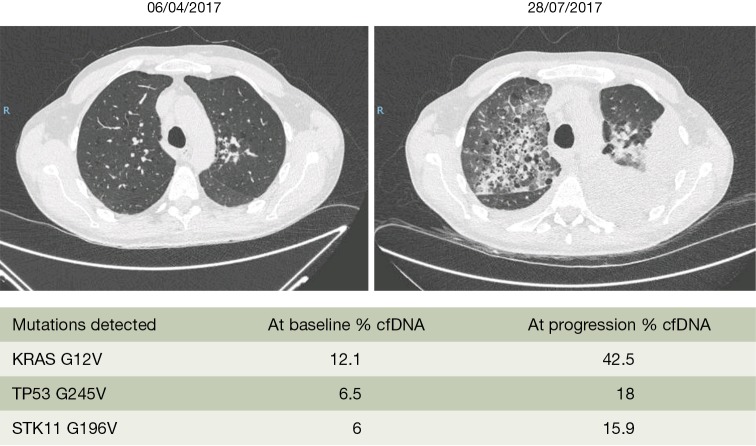
The data until now from several studies as well as a meta-analysis indicate that KRAS mutations in cfDNA may not be useful to predict response to chemotherapy (49-51). However, our everyday clinical experience allows us to say that in KRAS mutant NSCLC patients, when the mutation is also detected in the cfDNA the prognosis of the patients is detrimental. We present the example of a 48-year-old man who was diagnosed with a KRAS pG12V lung adenocarcinoma in April 2017 (Figure 1). The patient had brain and bone metastasis at presentation. Before starting first line chemotherapy we extracted blood, which revealed the KRAS G12V mutation in cfDNA with a 12.1% mutant allele fraction, together with a TP53 G245V (6.5% mutant allele fraction) and a STK11 G196V (6% mutant allele fraction) mutation. After three cycles of chemotherapy with cisplatin, pemetrexed and bevacizumab, computed tomography (CT) scan imaging demonstrated massive progression in all sites of the disease. The Guardant360 assay at the time of disease progression revealed a significant increase of the KRAS mutant allele fraction (Figure 1). We managed to include the patient in a phase 2, open-label, randomized study of the anti-CD38 monoclonal antibody daratumumab administered in combination with atezolizumab compared with atezolizumab alone. The patient was randomized to the single arm therapy but unfortunately passed away after the first administration of atezolizumab.
Figure 1.
The clinical case of a 48-year-old man, diagnosed with a stage IV lung adenocarcinoma KRAS mutant. The patient was included in the SLIPP study. The allelic fraction of the KRAS mutant allele was significantly increased at the time of progression to first line chemotherapy. cfDNA, cell-free DNA; SLIPP, the Spanish Lung Liquid versus Invasive Biopsy Program.
Finally apart from detecting mutations and resistant mutations to targeted therapies in cfDNA, massive parallel sequencing has been used to predict response to chemotherapy or immunotherapy in NSCLC through the detection of gains and losses of chromosomal regions in cfDNA and defining genomic copy number instability (CNI) (52,53).
Commercially developed liquid biopsy kits
In September 2014, the Committee for Medicinal Products for Human Use of the European Medicines Agency (EMA) adopted an amendment to the license of gefitinib stating that cfDNA may be used for the detection of EGFR mutations if a tumor sample is not available, based on the data for the therascreen EGFR RGQ PCR Kit (29). The cobas EGFR Mutation Test v2 was the first liquid biopsy test to be approved by the United States Food and Drug Administration (US FDA), in June 2016, for the detection of exon 19 deletions and exon 21 L858R mutations and the use of erlotinib (32,54-56). Three months later (September 2016), the same test received FDA and EMA approval for the detection of the T790M and the use of osimertinib (57).
There are currently at least seven commercially available kits for the detection of genetic alterations in cfDNA of cancer, including NSCLC, patients (Table 2). As previously commented, the therascreen EGFR RGQ PCR Kit is EMA approved, while the cobas EGFR Mutation Test v2 is the first and only liquid biopsy test approved by the FDA. Targeted NGS is recently used for the non-invasive detection of genetic alterations in lung cancer patients (Table 2). There are three tests (Guardant360, FoundationACT and OncotypeSEQ) that, though not still FDA approved, have demonstrated good accuracy when validated with clinical samples (59). Among them, Guardant360 is the one most commonly used by the oncology community since its commercial introduction in 2014. It is a cfDNA targeted NGS panel that has been studied in more than 20,000 clinical specimens with an almost 100% analytic sensitivity and a comparable clinical sensitivity with tissue derived NGS (60). To prospectively validate Guardant360 for the rapid detection of genetic alterations in lung adenocarcinoma, we have conducted the Spanish Lung Liquid versus Invasive Biopsy Program (SLIPP) study. The SLIPP is a multicenter non-inferiority observational study, in which blood is collected prior to first line chemotherapy and upon progression from patients with metastatic non-squamous NSCLC (the case report mentioned above has been included in the SLIPP study). The results of the blood assay will be compared with tissue testing performed as standard of care in Spain, using a variety of “real life” techniques, including, but not limited to, USA FDA-approved companion diagnostics (EGFR PCR tests and ALK FISH testing). The accrual of 182 patients has been completed and the results are awaited.
Table 2. Commercially developed liquid biopsy kits.
| Test | Company | Methodology | Mutations detected | Status | Reference |
|---|---|---|---|---|---|
| Cobas EGFR Mutation test v2 | Roche Molecular diagnostics | Real-time PCR amplification test | Exon 18 (G719X) substitutions, exon 19 deletions, exon 20 insertions and substitutions (T790M, S768I), exon 21 substitutions (L858R, L861Q) |
FDA approved [2016] for erlotinib and osimertinib | – |
| TheraScreen EGFR RGQ PCR Kit | Qiagen | PCR amplification test using ARMS and Scorpions Technologies | Exon 18 (G719X) substitutions, exon 19 deletions, exon 20 insertions and substitutions (T790M, S768I), exon 21 substitutions (L858R, L861Q) |
EMA approved [2014] for gefitinib | (38) |
| Target-Selector | Biocept | Real-time PCR amplification test | EGFR mutations | – | – |
| Trovera | Trovagene | PCR in blood and urine cfDNA | EGFR, KRAS and BRAF mutations | – | – |
| Guardant 360 | Guardant Health | Digital sequencing | Single base mutations in 73 genes, amplifications in 18 genes, fusions in 6 genes, indels in 23 genes | CLIA certified laboratory developed test since February 2014 | – |
| FoundationAct | Foundation Medicine | High depth sequencing (Illumina HiSeq) | 62 genes and 6 gene fusions | CLIA certified laboratory developed test since May 2016 | (58) |
| OncotypeSEQ | Genomic Health | NGS | 17 genes | CLIA certified laboratory developed test since June 2016 | – |
RGQ, rotor-gene Q; EGFR, epidermal growth factor receptor; PCR, polymerase-chain-reaction; FDA, Food and Drug Administration; EMA, European Medicines Agency; cfDNA, cell-free DNA; NGS, next generation sequencing.
Conclusions
There is no doubt that blood represents a rich source of information through which solid cancers can be detected, identified and classified, and matched to a specific therapy. Although cfDNA can be an efficient biosource for the detection of somatic mutations in lung cancer patients, as well as for monitoring treatment, we consider that other alterations like ALK rearrangements or MET exon 14 skipping mutations, that result in defecting messenger RNA splicing, can be more accurately detected using RNA based assays (61). For instance, platelets can sequester tumor RNA through a microvesicle driven mechanism and can be transformed to TEPs. We have worked extensively in order to demonstrate that genetic alterations such as EML4-ALK fusion transcripts can be detected in RNA isolated from platelets (62,63). Furthermore, platelet RNA biomarker signatures can be altered in the presence of cancer and can be used for cancer detection (64,65). Other investigators have also shown that detection of cancer at early stages is feasible using cfDNA (66).
Several research and development efforts regarding liquid biopsy diagnostics exist throughout Europe and the US, but they lack an integration of concepts, including, discovery, development, standardization, clinical validation and implementation. To this scope, we have developed a consortium funded by the European Union (European Liquid Biopsy Academy; ELBA), in which we will combine the experience of academic partners and small-medium enterprises (SMEs) to create a large integrated project that will cover the development and implementation of innovative diagnostics and comparison of the four blood-based biosources (cfDNA, CTCs, EVs, and TEPs) and technologies aimed at NSCLC.
Acknowledgements
Funding: This work was funded by La Caixa Foundation and Red Tematica de Investigacion Cooperativa en Cancer (RTICC; grant RD12/0036/ 0072).
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Rosell R, Karachaliou N. Large-scale screening for somatic mutations in lung cancer. Lancet 2016;387:1354-6. 10.1016/S0140-6736(15)01125-3 [DOI] [PubMed] [Google Scholar]
- 2.Jordan EJ, Kim HR, Arcila ME, et al. Prospective Comprehensive Molecular Characterization of Lung Adenocarcinomas for Efficient Patient Matching to Approved and Emerging Therapies. Cancer Discov 2017;7:596-609. 10.1158/2159-8290.CD-16-1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pardoll DM. Immunology beats cancer: a blueprint for successful translation. Nat Immunol 2012;13:1129-32. 10.1038/ni.2392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Bruin EC, McGranahan N, Mitter R, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science 2014;346:251-6. 10.1126/science.1253462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bedard PL, Hansen AR, Ratain MJ, et al. Tumour heterogeneity in the clinic. Nature 2013;501:355-64. 10.1038/nature12627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diaz LA, Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 2014;32:579-86. 10.1200/JCO.2012.45.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Di Meo A, Bartlett J, Cheng Y, et al. Liquid biopsy: a step forward towards precision medicine in urologic malignancies. Mol Cancer 2017;16:80. 10.1186/s12943-017-0644-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mandel P, Metais P. Les acides nucléiques du plasma sanguin chez l'homme. C R Seances Soc Biol Fil 1948;142:241-3. [PubMed] [Google Scholar]
- 9.Leon SA, Shapiro B, Sklaroff DM, et al. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res 1977;37:646-50. [PubMed] [Google Scholar]
- 10.Stroun M, Anker P, Maurice P, et al. Neoplastic characteristics of the DNA found in the plasma of cancer patients. Oncology 1989;46:318-22. 10.1159/000226740 [DOI] [PubMed] [Google Scholar]
- 11.Karachaliou N, Mayo-de las Casas C, Queralt C, et al. Association of EGFR L858R Mutation in Circulating Free DNA With Survival in the EURTAC Trial. JAMA Oncol 2015;1:149-57. 10.1001/jamaoncol.2014.257 [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez-Cao M, Mayo-de-Las-Casas C, Molina-Vila MA, et al. BRAF mutation analysis in circulating free tumor DNA of melanoma patients treated with BRAF inhibitors. Melanoma Res 2015;25:486-95. 10.1097/CMR.0000000000000187 [DOI] [PubMed] [Google Scholar]
- 13.Vogelstein B, Kinzler KW. Digital PCR. Proc Natl Acad Sci U S A 1999;96:9236-41. 10.1073/pnas.96.16.9236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hindson BJ, Ness KD, Masquelier DA, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem 2011;83:8604-10. 10.1021/ac202028g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diehl F, Li M, Dressman D, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci U S A 2005;102:16368-73. 10.1073/pnas.0507904102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whitcombe D, Theaker J, Guy SP, et al. Detection of PCR products using self-probing amplicons and fluorescence. Nat Biotechnol 1999;17:804-7. 10.1038/11751 [DOI] [PubMed] [Google Scholar]
- 17.Forshew T, Murtaza M, Parkinson C, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med 2012;4:136ra68. 10.1126/scitranslmed.3003726 [DOI] [PubMed] [Google Scholar]
- 18.Ng SB, Turner EH, Robertson PD, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature 2009;461:272-6. 10.1038/nature08250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548-54. 10.1038/nm.3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujii T, Barzi A, Sartore-Bianchi A, et al. Mutation-Enrichment Next-Generation Sequencing for Quantitative Detection of KRAS Mutations in Urine Cell-Free DNA from Patients with Advanced Cancers. Clin Cancer Res 2017;23:3657-66. 10.1158/1078-0432.CCR-16-2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sánchez-Céspedes M, Monzó M, Rosell R, et al. Detection of chromosome 3p alterations in serum DNA of non-small-cell lung cancer patients. Ann Oncol 1998;9:113-6. 10.1023/A:1008230331221 [DOI] [PubMed] [Google Scholar]
- 22.Esteller M, Sanchez-Cespedes M, Rosell R, et al. Detection of aberrant promoter hypermethylation of tumor suppressor genes in serum DNA from non-small cell lung cancer patients. Cancer Res 1999;59:67-70. [PubMed] [Google Scholar]
- 23.Balaña C, Ramirez JL, Taron M, et al. O6-methyl-guanine-DNA methyltransferase methylation in serum and tumor DNA predicts response to 1,3-bis(2-chloroethyl)-1-nitrosourea but not to temozolamide plus cisplatin in glioblastoma multiforme. Clin Cancer Res 2003;9:1461-8. [PubMed] [Google Scholar]
- 24.Ramirez JL, Sarries C, de Castro PL, et al. Methylation patterns and K-ras mutations in tumor and paired serum of resected non-small-cell lung cancer patients. Cancer Lett 2003;193:207-16. 10.1016/S0304-3835(02)00740-1 [DOI] [PubMed] [Google Scholar]
- 25.Ramirez JL, Rosell R, Taron M, et al. 14-3-3sigma methylation in pretreatment serum circulating DNA of cisplatin-plus-gemcitabine-treated advanced non-small-cell lung cancer patients predicts survival: The Spanish Lung Cancer Group. J Clin Oncol 2005;23:9105-12. 10.1200/JCO.2005.02.2905 [DOI] [PubMed] [Google Scholar]
- 26.Rosell R, Moran T, Queralt C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med 2009;361:958-67. 10.1056/NEJMoa0904554 [DOI] [PubMed] [Google Scholar]
- 27.Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-46. 10.1016/S1470-2045(11)70393-X [DOI] [PubMed] [Google Scholar]
- 28.Mayo-de-las-Casas C, Jordana-Ariza N, Garzon-Ibanez M, et al. Large scale, prospective screening of EGFR mutations in the blood of advanced NSCLC patients to guide treatment decisions. Ann Oncol 2017;28:2248-55. 10.1093/annonc/mdx288 [DOI] [PubMed] [Google Scholar]
- 29.Douillard JY, Ostoros G, Cobo M, et al. Gefitinib treatment in EGFR mutated caucasian NSCLC: circulating-free tumor DNA as a surrogate for determination of EGFR status. J Thorac Oncol 2014;9:1345-53. 10.1097/JTO.0000000000000263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu YL, Sequist LV, Hu CP, et al. EGFR mutation detection in circulating cell-free DNA of lung adenocarcinoma patients: analysis of LUX-Lung 3 and 6. Br J Cancer 2017;116:175-85. 10.1038/bjc.2016.420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mok T, Wu YL, Lee JS, et al. Detection and Dynamic Changes of EGFR Mutations from Circulating Tumor DNA as a Predictor of Survival Outcomes in NSCLC Patients Treated with First-line Intercalated Erlotinib and Chemotherapy. Clin Cancer Res 2015;21:3196-203. 10.1158/1078-0432.CCR-14-2594 [DOI] [PubMed] [Google Scholar]
- 32.Wu YL, Zhou C, Liam CK, et al. First-line erlotinib versus gemcitabine/cisplatin in patients with advanced EGFR mutation-positive non-small-cell lung cancer: analyses from the phase III, randomized, open-label, ENSURE study. Ann Oncol 2015;26:1883-9. 10.1093/annonc/mdv270 [DOI] [PubMed] [Google Scholar]
- 33.cobas EGFR Mutation Test v2. 2016. Available online: http://www.fda.gov/Drugs/InformationOnDrugs/ApprovedDrugs/ucm504540.htm
- 34.Wu YL, Jenkins S, Ramalingam S, et al. MA08.03 Osimertinib vs Platinum-Pemetrexed for T790M-Mutation Positive Advanced NSCLC (AURA3): Plasma ctDNA Analysis. J Thorac Oncol 2017;12:S386. [Google Scholar]
- 35.Mok TS, Wu YL, Ahn MJ, et al. Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N Engl J Med 2017;376:629-40. 10.1056/NEJMoa1612674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oxnard GR, Thress KS, Alden RS, et al. Association Between Plasma Genotyping and Outcomes of Treatment With Osimertinib (AZD9291) in Advanced Non-Small-Cell Lung Cancer. J Clin Oncol 2016;34:3375-82. 10.1200/JCO.2016.66.7162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sacher AG, Paweletz C, Dahlberg SE, et al. Prospective Validation of Rapid Plasma Genotyping for the Detection of EGFR and KRAS Mutations in Advanced Lung Cancer. JAMA Oncol 2016;2:1014-22. 10.1001/jamaoncol.2016.0173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reck M, Hagiwara K, Han B, et al. ctDNA Determination of EGFR Mutation Status in European and Japanese Patients with Advanced NSCLC: The ASSESS Study. J Thorac Oncol 2016;11:1682-9. 10.1016/j.jtho.2016.05.036 [DOI] [PubMed] [Google Scholar]
- 39.Rosell R, Molina MA, Serrano MJ. EGFR mutations in circulating tumour DNA. Lancet Oncol 2012;13:971-3. 10.1016/S1470-2045(12)70369-8 [DOI] [PubMed] [Google Scholar]
- 40.Rosell R, Karachaliou N. Implications of Blood-Based T790M Genotyping and Beyond in Epidermal Growth Factor Receptor-Mutant Non-Small-Cell Lung Cancer. J Clin Oncol 2016;34:3361-2. 10.1200/JCO.2016.68.3458 [DOI] [PubMed] [Google Scholar]
- 41.Rosell R, Karachaliou N. Lung cancer: Using ctDNA to track EGFR and KRAS mutations in advanced-stage disease. Nat Rev Clin Oncol 2016;13:401-2. 10.1038/nrclinonc.2016.83 [DOI] [PubMed] [Google Scholar]
- 42.Cui S, Zhang W, Xiong L, et al. Use of capture-based next-generation sequencing to detect ALK fusion in plasma cell-free DNA of patients with non-small-cell lung cancer. Oncotarget 2017;8:2771-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bordi P, Tiseo M, Rofi E, et al. Detection of ALK and KRAS Mutations in Circulating Tumor DNA of Patients With Advanced ALK-Positive NSCLC With Disease Progression During Crizotinib Treatment. Clin Lung Cancer 2017. [Epub ahead of print]. 10.1016/j.cllc.2017.04.013 [DOI] [PubMed] [Google Scholar]
- 44.Karachaliou N, Mayo C, Costa C, et al. KRAS mutations in lung cancer. Clin Lung Cancer 2013;14:205-14. 10.1016/j.cllc.2012.09.007 [DOI] [PubMed] [Google Scholar]
- 45.Barlesi F, Mazieres J, Merlio JP, et al. Routine molecular profiling of patients with advanced non-small-cell lung cancer: results of a 1-year nationwide programme of the French Cooperative Thoracic Intergroup (IFCT). Lancet 2016;387:1415-26. 10.1016/S0140-6736(16)00004-0 [DOI] [PubMed] [Google Scholar]
- 46.Kris MG, Johnson BE, Berry LD, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA 2014;311:1998-2006. 10.1001/jama.2014.3741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skoulidis F, Byers LA, Diao L, et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov 2015;5:860-77. 10.1158/2159-8290.CD-14-1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lohinai Z, Klikovits T, Moldvay J, et al. KRAS-mutation incidence and prognostic value are metastatic site-specific in lung adenocarcinoma: poor prognosis in patients with KRAS mutation and bone metastasis. Sci Rep 2017;7:39721. 10.1038/srep39721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ai B, Liu H, Huang Y, et al. Circulating cell-free DNA as a prognostic and predictive biomarker in non-small cell lung cancer. Oncotarget 2016;7:44583-95. 10.18632/oncotarget.10069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garzón M, Villatoro S, Teixidó C, et al. KRAS mutations in the circulating free DNA (cfDNA) of non-small cell lung cancer (NSCLC) patients. Transl Lung Cancer Res 2016;5:511-6. 10.21037/tlcr.2016.10.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Camps C, Sirera R, Bremnes R, et al. Is there a prognostic role of K-ras point mutations in the serum of patients with advanced non-small cell lung cancer? Lung Cancer 2005;50:339-46. 10.1016/j.lungcan.2005.06.007 [DOI] [PubMed] [Google Scholar]
- 52.Weiss GJ, Beck J, Braun DP, et al. Tumor Cell-Free DNA Copy Number Instability Predicts Therapeutic Response to Immunotherapy. Clin Cancer Res 2017;23:5074-81. 10.1158/1078-0432.CCR-17-0231 [DOI] [PubMed] [Google Scholar]
- 53.Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009;45:228-47. 10.1016/j.ejca.2008.10.026 [DOI] [PubMed] [Google Scholar]
- 54.Park K, Yu CJ, Kim SW, et al. First-Line Erlotinib Therapy Until and Beyond Response Evaluation Criteria in Solid Tumors Progression in Asian Patients With Epidermal Growth Factor Receptor Mutation-Positive Non-Small-Cell Lung Cancer: The ASPIRATION Study. JAMA Oncol 2016;2:305-12. 10.1001/jamaoncol.2015.4921 [DOI] [PubMed] [Google Scholar]
- 55.Goss G, Tsai CM, Shepherd FA, et al. Osimertinib for pretreated EGFR Thr790Met-positive advanced non-small-cell lung cancer (AURA2): a multicentre, open-label, single-arm, phase 2 study. Lancet Oncol 2016;17:1643-52. 10.1016/S1470-2045(16)30508-3 [DOI] [PubMed] [Google Scholar]
- 56.Mok T, Ladrera G, Srimuninnimit V, et al. Tumor marker analyses from the phase III, placebo-controlled, FASTACT-2 study of intercalated erlotinib with gemcitabine/platinum in the first-line treatment of advanced non-small-cell lung cancer. Lung Cancer 2016;98:1-8. 10.1016/j.lungcan.2016.04.023 [DOI] [PubMed] [Google Scholar]
- 57.cobas® EGFR Mutation Test v2 - P150047. Available online: http://www.fda.gov/medicaldevices/productsandmedicalprocedures/deviceapprovalsandclearances/recently-approveddevices/ucm519922.htm
- 58.Boyle SE, Fedele CG, Corbin V, et al. CD271 Expression on Patient Melanoma Cells Is Unstable and Unlinked to Tumorigenicity. Cancer Research 2016;76:3965-77. 10.1158/0008-5472.CAN-15-2377 [DOI] [PubMed] [Google Scholar]
- 59.Pan Y, Ji JS, Jin JG, et al. Cancer Liquid Biopsy: Is It Ready for Clinic? IEEE Pulse 2017;8:23-7. 10.1109/MPUL.2016.2630838 [DOI] [PubMed] [Google Scholar]
- 60.Lanman RB, Mortimer SA, Zill OA, et al. Analytical and Clinical Validation of a Digital Sequencing Panel for Quantitative, Highly Accurate Evaluation of Cell-Free Circulating Tumor DNA. PLoS One 2015;10:e0140712. 10.1371/journal.pone.0140712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reungwetwattana T, Ou SH. MET exon 14 deletion (METex14): finally, a frequent-enough actionable oncogenic driver mutation in non-small cell lung cancer to lead MET inhibitors out of "40 years of wilderness" and into a clear path of regulatory approval. Transl Lung Cancer Res 2015;4:820-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aguado C, Gimenez-Capitan A, Karachaliou N, et al. Fusion gene and splice variant analyses in liquid biopsies of lung cancer patients. Transl Lung Cancer Res 2016;5:525-31. 10.21037/tlcr.2016.09.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nilsson RJ, Karachaliou N, Berenguer J, et al. Rearranged EML4-ALK fusion transcripts sequester in circulating blood platelets and enable blood-based crizotinib response monitoring in non-small-cell lung cancer. Oncotarget 2016;7:1066-75. 10.18632/oncotarget.6279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Best MG, Sol N, Kooi I, et al. RNA-Seq of Tumor-Educated Platelets Enables Blood-Based Pan-Cancer, Multiclass, and Molecular Pathway Cancer Diagnostics. Cancer Cell 2015;28:666-76. 10.1016/j.ccell.2015.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Best MG, Sol N, In't Veld SGJG, et al. Swarm Intelligence-Enhanced Detection of Non-Small-Cell Lung Cancer Using Tumor-Educated Platelets. Cancer Cell 2017;32:238-252.e9. 10.1016/j.ccell.2017.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Phallen J, Sausen M, Adleff V, et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med 2017;9:eaan2415. [DOI] [PMC free article] [PubMed]