Abstract
One of the most obvious manifestations of polarity in epithelia is the subdivision of the cell surface by cell junctions into apical and basolateral domains. crumbs genes are among key regulators of this form of polarity. Loss of crumbs function disrupts the apical cell junction belt and crumbs overexpression expands the apical membrane size. Crumbs proteins contain a single transmembrane domain and localize to cell junction area at the apical surface of epithelia. In some tissues, they are also found in cilia. To test their role in ciliogenesis, we investigated mutant phenotypes of zebrafish crumbs genes. In zebrafish, mutations of three crumbs genes, oko meduzy/crb2a, crb3a, and crb2b, affect cilia length in a subset of tissues. In oko meduzy (ome), this is accompanied by accumulation of other Crumbs proteins in the ciliary compartment. Moreover, intraflagellar transport (IFT) particle components accumulate in the ciliary shaft of ome;crb3a double mutants. Consistent with the above, Crb3 knockdown in mammalian cells affects the dynamics of IFT particle movement. These findings reveal crumbs-dependent mechanisms that regulate the localization of ciliary proteins, including Crumbs proteins themselves, and show that crumbs genes modulate intraflagellar transport and cilia elongation.
Keywords: cilia, Crumbs, cristae, IFT, apico-basal polarity
Cilia are finger-like cell surface protrusions that house components of many signal transduction cascades (Schou et al. 2015; Mourão et al. 2016; Malicki and Johnson 2017). The detection of photons by photoreceptors and chemicals by olfactory sensory neurons is mediated by signal transduction mechanisms inside the ciliary shaft (Jenkins et al. 2009; Kennedy and Malicki 2009). Vertebrate hedgehog signaling requires cilia and wnt, the platelet-derived growth factor and mTOR pathways are modulated by them [reviewed in Schou et al. (2015), Mourão et al. (2016), Malicki and Johnson (2017)]. In addition to signaling functions, cilia have a hydrodynamic role: their movement drives the flow of fluid in ducts and vesicles, such as the pronephric duct in zebrafish or the embryonic node in the mouse (Kramer-Zucker et al. 2005; Hirokawa et al. 2012). They also propel cells, such as sperm cells.
In cells that display apico-basal polarity, almost without exception, cilia form at the apical surface. Consequently, the ciliary membrane is an apical surface subcompartment, characterized by a unique protein and lipid content (Craige et al. 2010; Hu et al. 2010; Mukhopadhyay et al. 2010; Chih et al. 2011). Ciliated cells of epithelial sheets thus feature two cell membrane subdivisions: the one that separates the apical and basolateral domains and another one that separates the ciliary membrane from the rest of the apical surface. crumbs genes were initially discovered as essential regulators of the apico-basal cell membrane subdivision in fly embryonic epithelia (Jurgens et al. 1984; Tepass et al. 1990; Wodarz et al. 1995). They encode transmembrane (TM) proteins that localize to the vicinity of epithelial cell junctions, feature a short cytoplasmic tail and an extracellular domain of varying size (Tepass et al. 1990; van den Hurk et al. 2005; Omori and Malicki 2006). Loss of crumbs function in the fly disrupts the cell junction belt at the boundary of the apical and the basolateral surface, and crumbs overexpression expands apical membrane size (Wodarz et al. 1995; Grawe et al. 1996). A similar function of crumbs genes has been observed in vertebrates; mutations in one of the zebrafish crumbs loci, oko meduzy, and the locus encoding a related apico-basal polarity determinant, nagie oko, a fly stardust homolog, cause loss of apical–basal polarity in the eye neuroepithelium and a severe neuronal patterning defect in the retina (Malicki et al. 1996; Malicki and Driever 1999; Wei and Malicki 2002; Omori and Malicki 2006). A related crumbs function in apico-basal polarity is also evident in fly and zebrafish photoreceptor cells (Pellikka et al. 2002; Hsu et al. 2006; Omori and Malicki 2006). Finally, while the apico-basal polarity function is mostly mediated by its intracellular tail (Wodarz et al. 1995), Crumbs extracellular domains mediate cell adhesion in the zebrafish photoreceptor cell layer (Zou et al. 2012) and human CRB1 mutations cause severe, early-onset retinal degeneration (den Hollander et al. 1999).
Vertebrate Crumbs and related apico-basal polarity determinants also affect cilia formation. While one crumbs gene exists in the fly, the human and zebrafish genomes contains three and five crumbs genes, respectively (van den Hurk et al. 2005; Omori and Malicki 2006; Gosens et al. 2008). Zebrafish crumbs genes display distinct expression patterns. crb2b, for example, is highly enriched in the pronephros and in photoreceptor cells (Hsu et al. 2006; Omori and Malicki 2006; Zou et al. 2012). crb3a, on the other hand, is expressed predominantly in the otic vesicle at stages that were investigated thus far (Omori and Malicki 2006). Consistent with these expression patterns, antisense morpholino knockdown of zebrafish crb2b and crb3a reduces cilia size in the pronephros and the ear, respectively (Omori and Malicki 2006). An even stronger crumbs phenotype has been reported in tissue culture; small interfering RNA (siRNA) knockdown of the crumbs 3 gene in Madin-Darby canine kidney (MDCK) cells eliminates cilia entirely (Fan et al. 2004). In agreement with crumbs cilia phenotype, downregulation of other apico-basal polarity determinants, aPKC, Par6, and Par3, also leads to cilia loss (Fan et al. 2004; Sfakianos et al. 2007). To explain these observations in mechanistic terms, it has been postulated that Par proteins bridge transmembrane Crumbs 3 with a subunit of the main ciliary kinesin, Kif3a (Sfakianos et al. 2007).
As morpholino knockdown results are frequently difficult to interpret (Kok et al. 2015), we chose to analyze the role of crumbs in ciliogenesis using mutants of several zebrafish crumbs genes. We found that mutant alleles of oko meduzy (crb2a), crb2b, and crb3a, cause changes in cilia length. This is accompanied by a massive accumulation of other Crumbs proteins and intraflagellar transport (IFT) particle components in the ciliary compartment of ome and ome;crb3a mutants. Consistent with the above, Crb3 knockdown in mammalian inner medullary collecting duct cells (IMCD3) cells affects the dynamics of IFT particle movement. These studies reveal crumbs-dependent mechanisms that affect the subcellular localization of Crumbs proteins and show that crumbs genes affect ciliary protein composition and modulate intraflagellar transport.
Materials and Methods
Zebrafish strains and maintenance
crb3a mutant alleles crb3ash410 and crb3ash346 were generated using transcription activator-like effector nucleases (TALENs) as described previously (Zu et al. 2013; Pooranachandran and Malicki 2016). The crb2am98 mutant allele was described previously in detail (Malicki et al. 1996; Malicki and Driever 1999; Omori and Malicki 2006) and the crb2bsa18042 allele was obtained from the Sanger Institute TILLING project. Zebrafish were maintained in accordance with UK Home Office regulations and the UK Animals (Scientific Procedures) Act 1986. Fish genotypes were determined by fin-clipping adults at 3 months of age or later followed by DNA isolation, PCR amplification of mutant sites, and Sanger sequencing. The following primers were used: 5′-TTCTACACTTCTGGCTTCCG-3′ and 5′-ATTGTGGCCATCGTTGTA-3′ for crb3ash410 and crb3ash346, and 5′-AAACTTCCGACTCCTCTCCG-3′ and 5′-AAAGATGTCCTACCCAGCTT-3′ for crb2bsa18042. During phenotypic analysis, mutants were compared to phenotypically wild-type siblings or to phenotypically wild-type animals derived from common ancesteral generation. Mutations that do not cause lethality (crb3ash410; crb3ash346; and crb2bsa18042) were maintained as homozygous strains. Consequently, analysis of cilia phenotype was performed on maternal/zygotic mutants.
Photography of adult zebrafish
To record adult phenotypes, zebrafish 6 months old or older more were placed in weighing boats (7 ml, 611–9179; VWR) containing E3 medium with tricaine (E10521, 0.2 mg/ml; Sigma). Photographs were obtained using an iPAD Pro digital camera, 12MP, F/2.2, 29mm, phase detection autofocus.
Immunostaining, mounting, and microscopy
Staining of whole zebrafish at 36 h postfertilization (hpf), 72 hpf, and 5 days postfertilization (dpf) was performed as previously described (Leventea et al. 2016). The following primary antibodies and dilutions were used: anti-acetylated tubulin, 1:500–1:1000 (T6793; Sigma [Sigma Chemical], St. Louis, MO); anti-CRB (Omori and Malicki 2006), 1:250; anti-Kif17(ab11261; Abcam), 1:500; anti-IFT88, 1:500; and anti-IFT52, 1:500. Anti-IFT antibodies were kindly provided by Brian Perkins. Embryos were then counterstained with DAPI to visualize nuclei. Stained embryos were placed in imprinted wells created by placing molds onto a liquid 1% agarose layer in 35-mm petri dishes (Leventea et al. 2016). To examine the cilia of the ear, the nasal pit, the pronephros, and the lateral line, embryos were positioned on their sides in the imprinted wells and immobilized by overlaying with 1.5% low-melting point agarose. Images of whole embryos were collected using an Olympus FV1000 confocal microscope with either a 40×/0.8 or 60×/0.9 water dipping lens.
IMCD3 cell culture and siRNA experiments
Mouse inner medullary collecting duct cells stably expressing IFT88 (IMCD3-IFT88-GFP cells, a gift from Hiroaki Ishikawa) were grown in full medium containing Dulbecco’s modified Eagle’s medium (DMEM) and Ham’s F12 nutrient mixture (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS) and 1% Pen/Strep Amphotericin B (100×) (Lonza) at 37° in a tissue culture incubator (Sanyo inCu Safe). Cells were seeded in cell culture flasks (cat no. 156472; Nunc). Medium was changed daily.
ON-TARGET plus SMART pool siRNAs (Dharmacon, GE Healthcare) against mouse Crb3 were used to transfect IMCD3-IFT88 cells using Lipofectamine RNAiMAX transfection reagent (Life Technologies) following the manufacturer’s recommendations. The following siRNA target sequences were used: 5′-GCACCGGACCCUUUCCAA-3′, 5′-AGGCAAGCAGGAUGGGACU-3′, 5′-CAACACCCUCUUUGGGCAA-3′, and 5′-GAUAGGGACAAUAAAGGUU-3′. As a negative control, we used nontargeting pool directed to the following sequences: 5′-UGGUUUACAUGUCGACUAA-3′, 5′-UGGUUUACAUGUUGUGUGA-3′, and 5′-UGGUUUACAUGUUUUCUGA-3′, 5′-UGGUUUACAUGUUUUCCUA-3′.
Immunostaining of IMCD3 cells
For staining with anti-acetylated tubulin and anti-Crumbs (CRB) antibodies, cells were rinsed with PBS, fixed with 4% PFA for 10 min at room temperature (RT), permeabilized with 0.1% Triton X-100 in PBS for 10 min, and blocked with 3% BSA in PBS (1×) for 30 min at RT. Cells were then incubated with appropriate primary and secondary antibodies using standard protocols. Cells were counterstained with DAPI and mounted on glass slides using ProLong Gold antifade reagent (Life Technologies). Imaging was performed on an Olympus FV1000 confocal microscope using a 60×/1.42 Plan Apo N oil lens.
Total Internal Reflection Fluorescence (TIRF) imaging of IFT in IMCD3-IFT88 cells
IMCD3 cells were seeded on transwell cups (Costar, Cambridge, MA; 6.5 mm–0.5 μm pore size) at a density of 2 × 105 cells/ml as previously described (Ott and Lippincott-Schwartz 2012; Ishikawa and Marshall 2015). Upon reaching 60–70% confluency, cells were transfected with siRNAs as above. 48 hr after transfection, cells were serum-starved for an additional 48 hr to induce ciliogenesis. The transwell cups were then placed in glass-bottom dishes (cat. no. 81153; ibidi) and imaged on an Eclipse Ti Microscope (Nikon, Shinagawa, Tokyo, Japan) supplied with a heating chamber (Oko Touch) using the Apo TIRF 100×, 1.49 NA oil lens (Nikon). Images were acquired at 100-msec intervals using the ixOn Ultra EMCCD camera (Andor Technology) and analyzed using the “KymoResliceWide” Fiji plug-in as previously described (Ishikawa and Marshall 2015). The lengths of tracks were measured using the “segmented line tool” in Fiji and expressed as the percentage of cilia length.
Cilia length measurements and statistical analysis
Zebrafish cilia were measured on TIFF files of Z-stack projections of cristae and the nasal pit confocal images using the “segmented line tool” in ImageJ software. At least 10 animals (mutants and wild-types each) from two to three independent experiments were used. Measurements from each crista were averaged before performing comparisons. Cilia of IMCD3 cells were measured on TIFF images of cells stained with anti-acetylated tubulin antibody by tracing their length using ImageJ/FIJI software as above. Statistical analysis was carried out using the Student’s t-test, and the Mann–Whitney test included in GraphPad Prism 7.0 software (http://www.graphpad.com/). Data are presented as mean ± 95% C.I. Statistical significance is indicated as follows: * for P < 0.05, ** P < 0.01, *** P < 0.001, and **** P < 0.0001.
Data availability
All animal strains and reagents will be distributed through international stock centers or directly by the Malicki laboratory.
Results
crb3a affects cilia length in vestibular system cristae
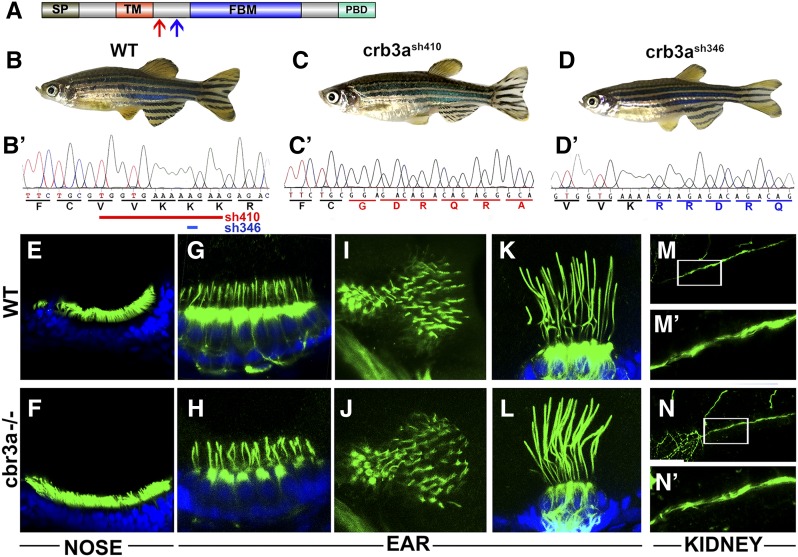
Knockdown of the CRB3 gene in MDCK cells was shown to block cilia formation (Fan et al. 2004). Similar phenotypes were seen following knockdowns of other apico-basal polarity determinants: aPKC, Par6, and Par3 (Fan et al. 2004; Sfakianos et al. 2007). Moreover, morpholino knockdown of crb3a in zebrafish was shown to reduce cilia length (Omori and Malicki 2006); However, these ciliary phenotypes have not been investigated in mutants. To address this deficiency, we generated several crb3a mutant alleles using TALEN nucleases as previously described (Zu et al. 2013). Two alleles were analyzed in this study, crb3ash410 and crb3ash346. The former introduces a deletion of 13 bp, causing a frameshift between the TM and FERM domains (Figure 1A, red arrow and Figure 1C’). The latter contains a 1-bp deletion that also causes a frameshift at another site between the TM and FERM domains (Figure 1A, blue arrow and Figure 1D’). Homozygous carriers of either allele do not display any external abnormalities, survive to adulthood, and are fertile (Figure 1, B–D). This is also true for homozygous animals that originate from homozygous mothers and thus did not receive maternal contribution during embryogenesis.
Figure 1.
crb3a mutant phenotype. (A) Schematic of Crb3a protein domain structure. Signal peptide (SP); transmembrane domain (TM); FERM-binding motif (FBM); and PDZ-binding domain (PBD) are indicated. Red arrow indicates the start of the frameshift in crb3a−/−sh410 mutant allele; blue arrow, the start of frameshift in crb3a−/−sh346 allele. (B–D) External phenotypes of wild-type (WT) (B), crb3a−/−sh410 homozygous mutant (C), and crb3a−/−sh346 homozygous mutant (D) adult zebrafish. (B’–D’) Sequences of wild-type (B’), and two mutant alleles: crb3a−/−sh410 (C’), and crb3a−/−sh346 (D’). Deletions in crb3a−/−sh410 (red line) and crb3a−/−sh346 (blue line) mutants are indicated in (B’). (E–N’) Images of wild-type and crb3a−/− mutant embryos stained with anti-acetylated tubulin antibody (in green) and counterstained with DAPI (in blue) at 5 days postfertilization: olfactory placode (E and F); anterior macula (G and H); posterior macula (I and J); lateral crista (K and L); and pronephros (M and N). (M’ and N’) are enlarged images of pronephric cilia shown in (M and N).
To assess cilia morphology in these mutants, we immunostained embryos at 5 dpf using anti-acetylated tubulin antibody. Mutant homozygotes for either allele do not manifest any gross ciliary phenotypes in nasal pits (Figure 1, E and F), anterior and posterior maculae (Figure 1, G–J), lateral cristae (Figure 1, K and L), or pronephroi (Figure 1, M–N’). However, thorough measurements of cilia length in vestibular system cristae revealed that crb3a mutant cilia are somewhat longer compared to those of the wild type (Figure 2P). These observations lead to the conclusion that crb3a function contributes to cilia length.
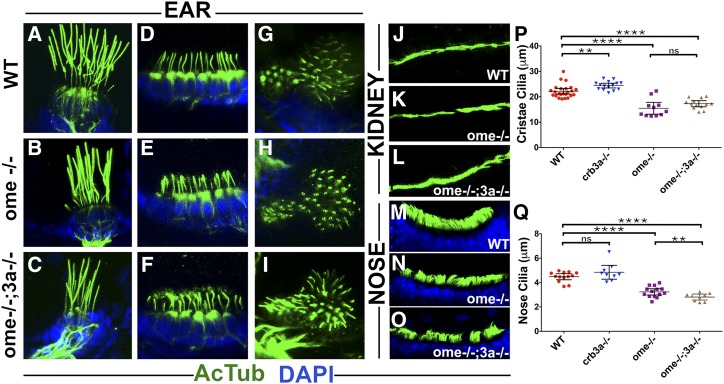
Figure 2.
oko meduzy and crb3a genes modulate cilia length. (A–O) Whole-mount immunostaining of wild-type (WT), ome−/−, and ome−/−;3a−/− double mutant cilia in several tissues at 5 days postfertilization. (A–C) lateral crista, (D–F) anterior macula, (G–I) posterior macula, (J–L) pronephros, and (M–O) nasal pits. Zebrafish larvae were immunostained using anti-acetylated tubulin antibody (in green) and counterstained with DAPI (in blue) to visualize nuclei. (P) Graph of cilia length in the cristae of WT and crumbs mutants as indicated. Each dot represents the average length of all cilia in one crista. (Q) Graph of cilia length in the olfactory placodes of WT and crumbs mutants as indicated. In (P and Q), data were collected from at least two independent experiments using at least five animals per experiment. The mean and 95% C.I. are indicated. Based on Student’s t-tests; ** P < 0.01, *** P < 0.001, and **** P < 0.0001; not significant, ns. All differences were also significant based on Mann–Whitney test.
oko meduzy (ome) mutations affect ciliogenesis
ome (crb2a) functions in the apico-basal polarity of the eye neuroepithelium and retinal neurogenesis. ome mutants are characterized by abnormal body axis curvature, edema, nonuniform eye pigmentation, grossly disorganized retinal neurons, and lethality by 7 dpf (Malicki et al. 1996; Malicki and Driever 1999; Omori and Malicki 2006). Previous studies did not evaluate ome function in cilia. To investigate the cilia phenotype, we analyzed vestibular cristae in ome mutants and found that cilia are shortened by ∼30% (Figure 2B, quantified in Figure 2P). To test functional relationship between crb3a and ome, we examined cilia of ome;crb3a double mutants. This analysis revealed that cilia of double mutants display similar length reduction to ome cristae, revealing that ome is epistatic to crb3a. Similarly, nose cilia are significantly shorter in both ome homozygotes and crb3a;ome double mutants when compared to those of the wild-type or crb3a homozygotes, (Figure 2, M–O, quantified in Figure 2Q). Moreover, olfactory placode cilia of crb3a;ome double mutants are significantly shorter than those of ome mutants (Figure 2Q). Cilia in kidney (Figure 2, K and L, compare to Figure 2J) and maculae (Figure 2, E, F, H, and I compare to Figure 2, D and G) are not obviously affected in ome or double mutants. These observations reveal that ome/crb2a and crb3a are necessary for proper cilia formation and control their lengths in a subset of tissues. It appears that ome functions downstream or in parallel to crb3a in cristae. These two genes also display some functional redundancy in nasal cilia.
Crumbs proteins accumulate in cilia of oko meduzy mutants
Crumbs 3 localizes to cilia in mammalian cell culture and is enriched at the base of cilia in zebrafish (Fan et al. 2004; Omori and Malicki 2006). To investigate how crumbs mutations affect Crumbs protein localization, we immunostained embryos with anti-acetylated tubulin and anti-Crumbs antibodies. The anti-Crumbs antibody used in these experiments is directed to the cytoplasmic tail and recognizes all zebrafish Crumbs proteins on western blots of Crumbs-GST fusions (Hsu et al. 2006; Omori and Malicki 2006).
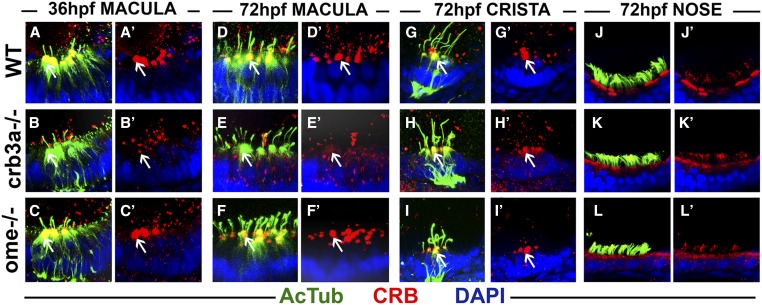
In the wild-type, Crumbs proteins are found at the base of hair cell kinocilia in ear maculae at 36 and 72 hpf (Figure 3, A, A’, D, and D’). This is no longer the case in crb3a mutant homozygotes (Figure 3, B, B’, E, and E’). However, in contrast to crb3a mutants, Crumbs localization in ome mutants is not affected (Figure 3, C, C’, F, and F’). These observations are in agreement with our previous report that crb3a, but not other crumbs genes, is strongly transcribed in the otic vesicle between 24 and 72 hpf (Omori and Malicki 2006). Crumbs proteins are also present at the apical surface of cells in olfactory placodes (Figure 3, J and J’). The apical localization is not obviously affected in either crb3a or ome mutants (Figure 3, K–L’).
Figure 3.
Crumbs expression in crb3a mutants at early stages of development. Confocal images of whole-mount cilia staining with anti-acetylated tubulin (AcTub) (green) and anti-CRB antibody (red). (A–C’) Staining of the otic vesicle at 36 h postfertilization (hpf). Crumbs proteins localize to the cilia base in the wild-type (WT) (A and A’) and ome−/− mutants (C and C’), but are absent in crb3a−/− mutant homozygotes (B and B’). At 72 hpf, Crumbs proteins still localize to the cilia base in maculae of WT animals (D and D’) and ome−/− mutants (F and F’), but very little signal is seen in crb3a−/− mutants (E and E’). No obvious differences are found in the localization of Crumbs proteins in the cristae of ome−/− (I and I’), crb3a−/− (H and H’), and WT individuals (G and G’). The localization patterns of Crumbs proteins in nasal pits of WT (J and J’), crb3a−/− mutant (K and K’), and ome−/− mutant (L and L’) animals do not show any obvious differences either. All samples were counterstained with DAPI to visualize nuclei. Arrows point to Crumbs signal at the apical surface of hair cells.
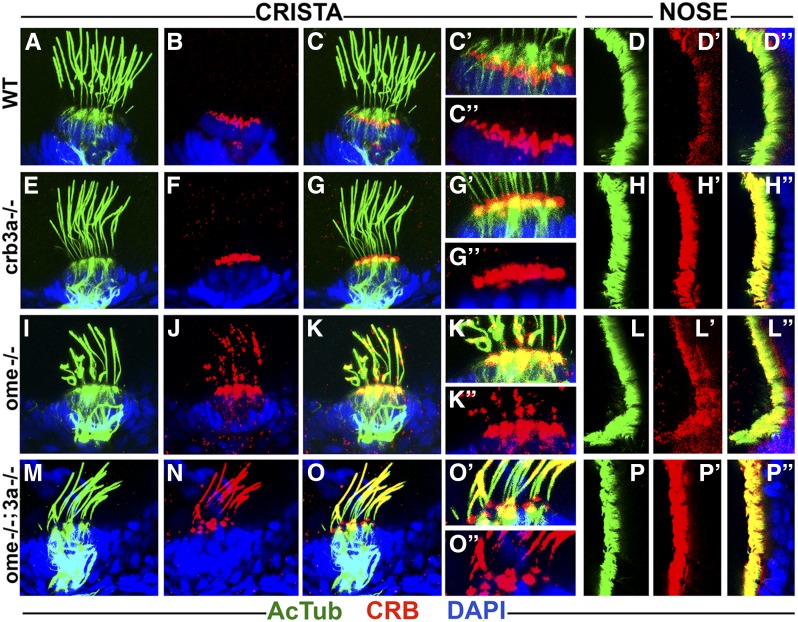
In contrast to ear maculae, hair cells of semicircular canals contain normal levels of Crumbs in crb3a and ome mutant homozygotes at 3 dpf (Figure 3, H–I’ compare to Figure 3, G and G’). This is also the case for crb3a mutants at 5 dpf (Figure 4, E–G’’, compare to Figure 4, A–C’’). However, we did observe strong enrichment of Crumbs proteins in olfactory placode cilia of these mutants (Figure 4, H–H’’, compare to Figure 4, D–D’’, 10/10 olfactory placodes). Interestingly, in ome mutant homozygotes, Crumbs proteins are mislocalized into cilia of both ear cristae and olfactory placodes (Figure 4, I–K’’, 23/23 cristae and Figure 4, L–L’’, 11/14 olfactory placodes). These findings reveal that ome, and to a lesser extent crb3a, strongly affect the subcellular localization of other Crumbs proteins. An enrichment of Crumbs proteins is also found in ome;crb3a double mutants both in cristae and olfactory placodes (Figure 4, M–O’’ 33/33 cristae and Figure 4, P–P’’ 6/6 olfactory placodes). While Crumbs staining forms puncta in ome mutant cilia, ome;crb3a double mutants display a uniform Crumbs signal along most of the ciliary axoneme, with the exception of the proximal region (Figure 4, I–K’’, compare to Figure 4, M–O’’). These findings reveal regulatory relationships between crumbs genes.
Figure 4.
Crumbs proteins accumulate in cilia of ome mutants at 5 days postfertilization (dpf). Confocal images of whole-mount cilia staining with anti-acetylated tubulin antibody (AcTub) (green) and anti-CRB antibody (red) at 5 dpf. Shown are cristae and olfactory placodes of wild-types (WT), crb3a−/−, ome−/−, and ome−/−;crb3a−/− mutants as indicated. No obvious difference is seen in the localization of Crumbs proteins in cristae cilia of crb3a−/− mutants (E–G’’) when compared to WT (A–C’’). Crumbs proteins are strongly enriched in the cilia of nasal pits in crb3a−/− mutants (H–H’’) when compared to the WT (D–D’’). ome−/− and ome−/−;crb3a−/− double mutants show massive accumulation of Crumbs proteins inside cilia of cristae (I–K’’ and M–O’’) and nasal pits (L–L’’ and P–P’’). All samples were counterstained with DAPI to visualize nuclei (in blue).
IFT proteins are highly enriched in cilia of ome;crb3a double mutants
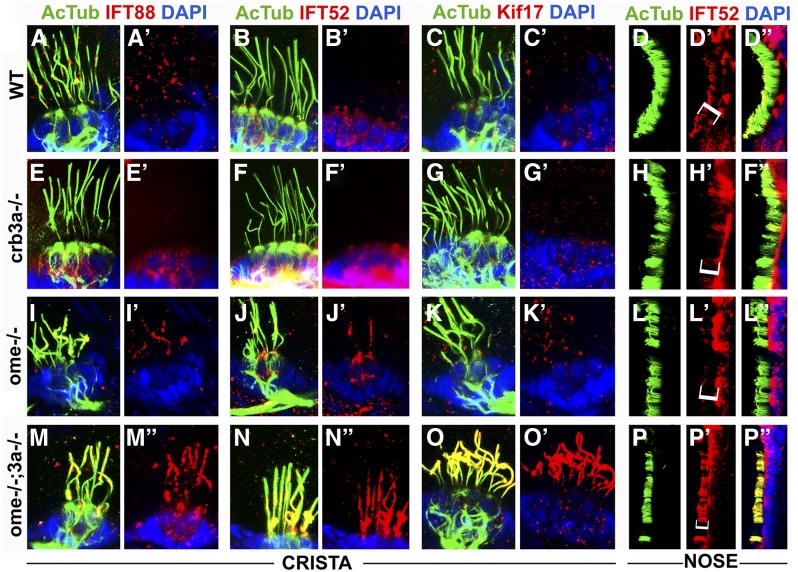
Par3, a key regulator of apico-basal cell polarity, is required for ciliogenesis in cell culture conditions (Sfakianos et al. 2007). Furthermore, the Par3 C-terminal region binds directly to Kif3a, the main anterograde motor of IFT particles (Nishimura et al. 2004). This led to the hypothesis that the Par3/Par6/aPKC complex bridges Crumbs proteins and Kif3a (Sfakianos et al. 2007). If true, Crumbs proteins could affect IFT by competing for the Kif3A motor. To test whether the loss of ome and/or crb3a function and the accompanying accumulation of Crumbs proteins in cilia affects IFT, we stained ome mutants and ome;crb3a double mutants with antibodies to IFT particle components: IFT88, IFT52, and Kif17. In crb3a mutants, IFT protein localization in cilia is largely unchanged (Figure 5, E–H’’, compare to Figure 5, A–D’’). However, weak accumulation of IFT proteins is observed in cristae cilia of ome mutants (Figure 5, I–K’). The olfactory cilia in both crb3a and ome single mutants do not show obvious differences in IFT distribution when compared to the wild type (Figure 5, H–H’’ and L–L’’). Strikingly, in ome;crb3a double mutants, IFT proteins massively accumulate inside cilia of ear cristae (Figure 5, M–O’) and the olfactory placode (Figure 5, P–P’’). In a control experiment, we have not observed any enrichment of γ-tubulin in ome;crb3a double mutants (data not shown). These observations indicate that ome and crb3a genes function redundantly in the ciliary localization of IFT proteins.
Figure 5.
Intraflagellar transport (IFT) particle components accumulate in the ciliary compartment of crumbs mutants. Confocal images of whole-mount cilia staining with antibodies to acetylated tubulin (AcTub) (green), and to IFT proteins (in red): IFT88, IFT52, and Kif17 at 5 days postfertilization. Shown are cristae and olfactory placodes of wild-type (WT), ome−/−, crb3a−/−, and ome−/−;crb3a−/− double mutants as indicated. IFT proteins are not detected in the ciliary shaft of WT cristae using this staining method (A–C’) and a low amount of IFT52 is found in WT olfactory cilia (D–D’’). Similarly, in crb3a−/− mutants, IFT proteins are not detected in cilia (E–H’’). Low levels of some IFT proteins are found in cristae cilia of ome−/− mutants (I–K’). Compared to WT, IFT52 localization is not affected in olfactory placode cilia of ome−/− mutants (L–L’’). In contrast to that, IFT proteins, including Kif17, strongly accumulate in cilia of ome−/−;crb3a−/− double mutants (M–P’’). All samples were counterstained with DAPI to visualize nuclei (in blue). Brackets in (D’, H’, L’, and P’) indicate nasal cilia.
CRB3 affects IFT train dynamics in IMCD3 cells
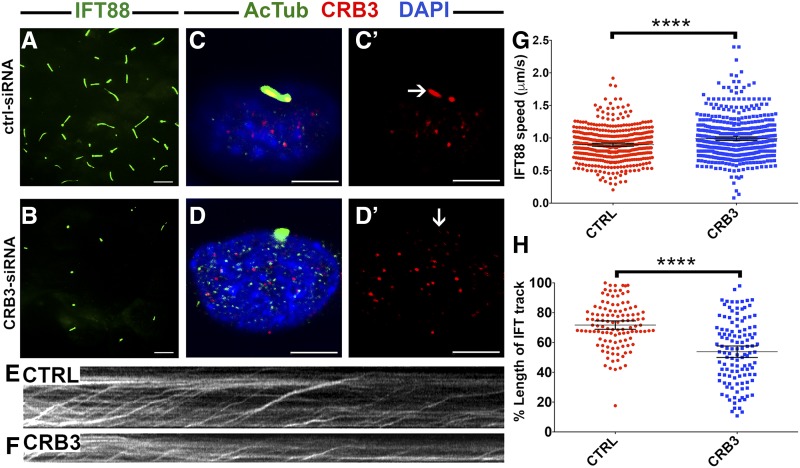
To further the understanding of the relationship between Crumbs proteins and intraflagellar transport, we decided to test whether IFT train movement is affected by crumbs genes. Imaging of IFT movement is difficult in zebrafish but can be efficiently performed in mammalian cells (Jin et al. 2014; Ishikawa and Marshall 2015). To this end, we knocked down CRB3 in an IMCD3 cell line stably expressing an IFT88-GFP fusion (Ishikawa and Marshall 2015). As reported previously, CRB3-knockdown cells display fewer and shorter cilia compared to controls (Figure 6, A–D) and the level of CRB3 proteins is reduced (Figure 6, C’ and D’). Imaging of IFT particle movement using IFT88 fluorescence (Figure 6, E and F) revealed that IFT particle speed is somewhat faster in knockdown cells, compared to controls (Figure 6G). Moreover, when adjusted for cilia length, IFT tracks are 25% shorter in knockdown cells when compared to control cells (Figure 6H). These observations are consistent with the idea that Crumbs affects IFT processivity and speed.
Figure 6.
CRB3 knockdown in mammalian cells affects intraflagellar transport (IFT) dynamics. (A and B) Maximum projections of total internal reflection fluorescence (TIRF) time-lapse recordings of IMCD3 cells grown on transwells and transfected with scrambled Ctrl-small interfering RNA (siRNA) (A) or CRB3-siRNA (B). These cells are stably transfected with an IFT88-GFP construct to visualize intraflagellar transport (green signal). (C–D’) Confocal images of control (CTRL) siRNA- (C and C’) and CRB3 siRNA-treated (D and D’) IMCD3 cells. Cilia are stained with antibodies to acetylated tubulin (AcTub) (green) and Crumbs (red). Samples are counterstained with DAPI to mark nuclei (in blue). (E and F) Kymographs of IFT movement in an IMCD3-IFT88 cell line transfected with CTRL or CRB3 siRNA as indicated. (G) Graph showing the speed of IFT particle movement in cilia of IFT88-GFP IMCD3 cells. Date collected from three independent experiments. (H) Lengths of IFT tracks in CTRL siRNA- and CRB3 siRNA-transfected cells expressed as percentages of total cilia length. Data are collected from two independent experiments. In (G and H), the mean and 95% C.I. are indicated. P < 10−4 based on Student’s t-tests. Bar, 10 μm (A and B) and 5 μm (C–D’).
crb2b mutation increases cilia length in a subset of tissues
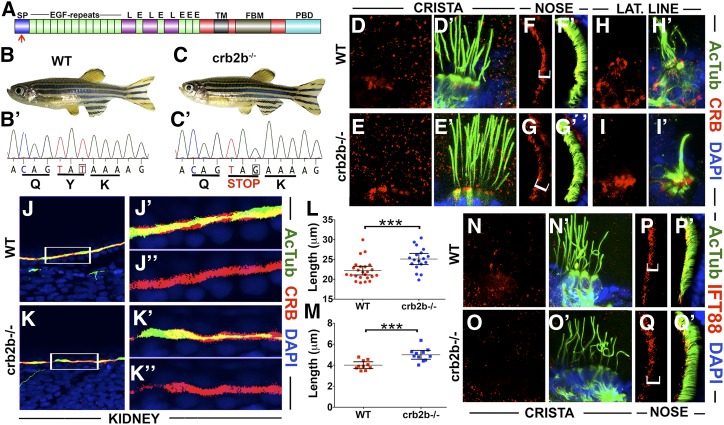
Morpholino knockdown studies of crb2b revealed that this gene is necessary for the elongation and motility of pronephric cilia (Omori and Malicki 2006). To gain further insight into crb2b function in cilia formation, we analyzed homozygous carriers of the crb2bsa18042 allele. The zebrafish crumbs 2b gene encodes two polypeptides that share most of the amino acid sequence. The shorter polypeptide does not include 11 N-terminal fibroblast growth factor-like repeats present in the long form and features a separate signal sequence (Zou et al. 2012). The crb2bsa18042 allele that we chose to use contains a stop codon at amino acid 10 of the long form, and thus is likely to eliminate the function of the long form (red arrow in Figure 7, A and B’–C’). A possible use of an alternative initiation codon at position 19 of the open reading frame could lead to protein expression, but it would eliminate most of the signal sequence rendering the long form of Crb2b dysfunctional. crb2bsa18042 homozygotes have normal external appearance and are fertile (Figure 7, B and C). This is also true for the offspring of homozygous mothers. To analyze cilia morphology in these mutants, we stained them using anti-acetylated tubulin antibody as above. No gross abnormalities were seen in the cilia of most tissues, including the inner ear (Figure 7E’, compare to Figure 7D’), the olfactory placode (Figure 7G’, compare to Figure 7F’), and the lateral line (Figure 7I’, compare to Figure 7H’). Similarly, contrary to the phenotype observed following morpholino knockdown (Omori and Malicki 2006) (Table 1), pronephric cilia were not obviously affected in crb2b mutants when compared to the wild type (Figure 7, K and K’, compare to Figure 7, J and J’). Staining of mutant homozygotes with anti-CRB antibody did not reveal differences in Crumbs protein localization in cristae (Figure 7E, compare to Figure 7D), olfactory placodes (Figure 7G, compare to Figure 7F), the lateral line (Figure 7I, compare to Figure 7H), and the pronephros (Figure 7K’’, compare to Figure 7J’’). However, a statistically significant difference of cilia length was observed between crb2b mutants and the wild-type in cristae and the olfactory placode (Figure 7, L and M). Contrary to cilia shortening seen in ome and ome;crb3a double mutants, and similar to the crb3a cilia phenotype (Figure 2P), cilia of crb2b mutant homozygotes are longer than those of their wild-type siblings. To test whether IFT protein localization is affected in these mutants, we immunostained homozygous crb2b mutant embryos for IFT88 at 5 dpf. No detectable difference was seen in the localization of IFT88 between crb2b mutants and the wild-type in inner ear cilia (Figure 7O, compare to Figure 7N) and the nose (Figure 7Q, compare to Figure 7P). These results further confirm that crumbs genes modulate cilia lengths in several tissues. In contrast to ome mutants, crb2bsa18042 mutant homozygotes do not display crumbs upregulation in cristae cilia.
Figure 7.
crb2b affects cilia length. (A) Schematic of Crb2b protein domain structure (not to scale). Indicated are the signal peptide (SP), EGF-like repeats (E), Laminin G domains (L), transmembrane domain (TM), FERM-binding motif (FBM), and PDZ-binding domain (PBD). Red arrow shows the position of mutation in the crb2b−/−sa18042 mutant allele. (B and C) Phenotypes of wild-type (WT) (B) and crb2b−/−sa18042 (C) homozygous mutant adult zebrafish. (B’ and C’) Sequences of WT and crb2b−/−sa18042 mutant alleles. (D–I’) Whole-mount staining of WT and crb2b−/− mutants using anti-acetylated tubulin (AcTub) (green), and anti-CRB (red) antibodies at 5 days postfertilization. Samples were counterstained with DAPI to visualize nuclei (in blue). Crumbs proteins are not detected in the cilia of cristae (D–E’) and the lateral (LAT.) line (H–I’) of crb2b−/− mutants or their WT siblings. No differences in Crumbs signal are found between WT and mutants in the cilia of olfactory placodes (F–G’) and the pronephric duct (J–K’’). (L) Graph of cilia length in WT and crb2b−/− mutants. Each dot represents the average length of all cilia in one crista. Data were collected from three independent experiments using at least five animals per experiment. (M) Graph of cilia length in olfactory placodes of WT and crb2b−/− mutants. (N–O’) IFT proteins are not detected in the cristae cilia of WT (N and N’) and crb2b−/− mutants (O and O’). (P–Q’) IFT88 localization is not obviously different in nasal pit cilia of WT (P and P’) and crb2b−/− mutant (Q and Q’) animals. Brackets in (F, G, P, and Q) indicate nasal cilia. In (L and M), the mean and 95% C.I. are indicated. P < 0.001 based on Student’s t-test and Mann–Whitney test.
Table 1. Summary of mutant and morphant crumbs phenotypes in cilia.
| Genotype | Tissue Examined | Cilia Length Phenotype | Crumbs Localization (3 dpf) | Crumbs Localization (5 dpf) | IFT in Cilia (5 dpf) |
|---|---|---|---|---|---|
| Wild-type | Ear macula | — | Cilia base | Cilia base | n.d |
| Ear cristae | — | Cilia base | Cilia base | None | |
| Nasal placode | — | Cilia base | Weak in ciliary shaft | ? | |
| Kidney | — | Cilia base/apical surface (36 hpf) | n.d. | n.d. | |
| crb3a−/− | Ear macula | n.d. | Absent | Cilia base | n.d. |
| Ear cristae | Longer | Cilia base | Cilia base | None | |
| Nasal placode | No change | Cilia base | Strong ciliary shaft | ? | |
| crb3a MO | Ear macula | Shortera | Reduced (2 dpf)a | n.d. | n.d. |
| ome−/− | Ear macula | n.d. | Cilia base | Cilia base | n.d. |
| Ear cristae | Shorter | Cilia base | Cilia base, puncta in ciliary shaft | Weak | |
| Nasal placode | Shorter | Cilia base | Ciliary shaft | Cilia base | |
| crb3a−/−; ome−/− | Ear cristae | Shorter | n.d. | Cilia base, ciliary shaft, weaker proximally | Strong |
| Nasal placode | Shorter | n.d. | Strong ciliary shaft | Ciliary shaft | |
| crb2b−/− | Ear cristae | Longer | n.d. | Cilia base | None |
| Nasal placode | Longer | n.d. | Weak in ciliary shaft | ? | |
| Kidney | No obvious change | cilia base/apical surface | n.d. | n.d. | |
| crb2b MO | Kidney | Shorter and disorganizeda | reduced (1 dpf)a | n.d. | n.d. |
Cilia length relative to wild-type cilia. dpf, days postfertilization; IFT, intraflagellar transport; n.d., not determined; ?, weak signal comparable to background; hpf, hours postfertilization. MO, morpholino.
Discussion
Our studies reveal that crumbs genes function in three interconnected aspects of ciliogenesis: the regulation of protein composition in the ciliary shaft, IFT movement dynamics, and cilia length determination (summarized in Table 1). The absence of some crumbs genes, either singly and/or in double mutants, results in a massive accumulation of other Crumbs proteins and IFT particle components inside the ciliary shaft in some tissues. In a subset of cilia, the increase in ciliary Crumbs localization correlates with a decrease of cilia length. Interestingly, IFT dynamics is affected following Crb3 knockdown in mammalian cells; IFT trains are somewhat faster and IFT tracks are markedly shorter. As discussed below, this may be related to a global role of Crumbs proteins in the morphogenesis of the apical surface of the cell.
An increase in ciliary Crumbs content in ome mutants is counterintuitive and reveals that crumbs genes or their protein products may negatively regulate each other. Such regulation could occur at the level of transcript or protein expression. It could also be mediated by protein degradation pathways. Previous studies suggested that crumbs expression may be regulated post-transcriptionally. The zebrafish Crb3a protein is enriched in mechanosensory hair cells while its transcript is uniformly expressed throughout the otic vesicle, suggesting a regulatory mechanism that affects translation or protein stability (Omori and Malicki 2006). Since mouse studies of the retina did not detect changes in the transcriptome of the Crb2 mutant during development, cross talk between crumbs genes on the level of transcriptional regulation appears less likely (Alves et al. 2013). Consistent with the above, zebrafish studies did not reveal compensatory Crumbs protein upregulation in ome mutants and similarly did not detect transcriptional upregulation of Crb2b in the same mutants (Hsu et al. 2006). Alternatively, as discussed below, changes in Crumbs protein level in cilia may reflect the function of this group of genes in gating mechanisms at the cilia base.
Equally unexpected is the enrichment of IFT proteins in the cilia of crumbs mutants. It could be partially explained by the trapping of the heterotrimeric IFT kinesin by the mislocalized Crumbs in the ciliary shaft (see below). In addition, the regulation of IFT protein content by Crumbs could occur at the level of gating mechanisms that regulate trafficking into the ciliary compartment. This is suggested by observations that a Crumbs 3 isoform interacts with Importin β-1 in a RAN-regulated manner (Fan et al. 2007). A related importin, importin β-2 localizes to the proximal region of the ciliary axoneme and was proposed to mediate the ciliary entry of kif17, one of the two IFT kinesins, also in a RAN-regulated fashion (Dishinger et al. 2010). It is thus possible that Crumbs mutations affect RAN–Importin-mediated gating mechanisms at the cilia base that regulate IFT entry into the ciliary compartment. This possibility is also supported by observations that Crumbs is enriched at the base hair cell kinocilia, where it could function in regulating cilia-directed traffic (Omori and Malicki 2006).
crumbs mutants display cilia abnormalities only in some organs. One possible reason is that crumbs genes, crb2b and crb3a in particular, are expressed in a subset of tissues. Another and perhaps more intriguing possibility is that the crumbs cilia phenotype varies across tissues due to intrinsic differences in cilia assembly mechanisms. Cristae cilia in particular are genetically different from most other cilia. The most striking indication of their unique genetic characteristics is that they are unaffected in mutants of kif3b, a subunit of the major ciliary kinesin, while most other cilia, including kinocilia of ear maculae, are absent in kif3b mutants (Zhao et al. 2012). Similarly, the kif3a mutant phenotype of cristae cilia differs from that of other cilia. Short-cristae cilia form in the absence of kif3a function and, in contrast to maculae for example, IFT88 protein persists at the base of these cilia in kif3a mutants (Pooranachandran and Malicki 2016). Although morphological abnormalities of cilia in crumbs mutants are fairly subtle, the accumulation of Crumbs and IFT proteins in the ciliary shaft may have profound functional consequences, such as malfunction of cilia-mediated signal transduction cascades. This may account for the severity of the oko meduzy phenotype in many organs including the central nervous system, the cardiovascular system, and the pronephros (Malicki and Driever 1999; Omori and Malicki 2006).
What mechanism could account for the role of Crumbs in cilia elongation? It was previously reported that the C-terminus of Par3, a key regulator of apico-basal polarity, binds directly to the C-terminal coiled coil region of Kif3a, the main anterograde motor of IFT particles (Nishimura et al. 2004). This, combined with observations that Crb3 and Par3 function in ciliogenesis, led to the idea that the Par3/Par6/aPKC complex bridges Crumbs proteins to Kif3a (Fan et al. 2004; Sfakianos et al. 2007). A genetic interaction between crumbs and kinesin-1 was also reported in the fly eye (League and Nam 2011). It is then tempting to hypothesize that Crumbs proteins compete for the Kif3a motor and, as a consequence, slow down IFT. In this model, cilia shortening in ome mutants is explained by the accumulation of other Crumbs proteins in cilia.
The results of crumbs function analysis in tissue culture are difficult to reconcile with cilia elongation in the zebrafish model. In contrast to fish phenotypes, RNAi knockdown in tissue culture has the opposite effect and causes cilia loss. This could be due to a global role of crumbs in cell polarity. Although cell junctions are largely intact in Crb3 siRNA knockdown cells and in Crb3 mutant mice, analysis of epithelia in Crb3 mouse mutants reveals substantial abnormalities, such as the appearance of prominent blebs on the apical surface of lung cells and a shortening and fusion of apical villi in the intestine (Fan et al. 2004; Whiteman et al. 2014). These defects reveal a role of Crb3 in apical surface morphogenesis, which could account for cilia loss in tissue culture studies. Nonetheless, the Crb3 mouse mutant phenotype is inconsistent with tissue culture studies as it does not affect cilia morphology (Whiteman et al. 2014).
crb3 mutant phenotypes also differ between fish and mice; the mouse knockout phenotype is lethal whereas the zebrafish crb3a phenotype is not (Whiteman et al. 2014). This is most likely due to the duplication of the crb3 gene in the zebrafish genome. It has been argued for quite a while now that gene duplication frequently leads to a subfunctionalization of duplicates relative to the ancestral gene (Force et al. 1999; Braasch et al. 2016). This is likely to have happened in the case of crb3 genes: zebrafish crb3a is mainly expressed in the otic vesicle and only weakly in the digestive system, while the crb3b transcript is found strongly expressed in the digestive system and not at all in the ear (Omori and Malicki 2006). It is thus likely that zebrafish crb3a mutants are viable because, in contrast to mouse Crb3 mutants, they do not affect essential digestive organ functions.
Differences between the outcome of tissue culture studies and genetic analysis in animal models are not uncommon and are frequently difficult to explain. Although HDAC6 and Rab8 appear to function as potent regulators of ciliogenesis in tissue culture studies (Nachury et al. 2007; Pugacheva et al. 2007), mice mutant for these genes do not display cilia defects (Zhang et al. 2008; Sato et al. 2014; Ying et al. 2016). Similarly, substantial differences are frequently seen between morphant and mutant phenotypes in zebrafish (Kok et al. 2015). In our study of crumbs mutants, we also found phenotypic differences in comparison to morpholino knockdowns performed previously (Omori and Malicki 2006) (summarized in Table 1). Such differences could be explained by compensatory mechanisms that become active in mutants, such as the upregulation of paralogous genes. Increased presence of Crumbs proteins in the cilia of crumbs mutants may represent such a compensatory mechanism. Such mechanisms may account for some of the differences seen between tissue culture, morphant, and mutant analyses. Taken together, our data show that some crumbs genes affect the subcellular localization of protein products expressed by other crumbs genes, either through direct regulatory relationships or indirectly by affecting the function of gating mechanisms at the cilia base. crumbs genes function in multiple interrelated aspects of ciliogenesis, including intraflagellar transport, the determination of cilia length, and the protein composition of the ciliary shaft.
Acknowledgments
The authors are thankful to Brian Perkins for providing anti-IFT antibodies, and Hiroaki Ishikawa for the IFT88-GFP cell line. We thank Stone Elworthy for the help in designing the TALENs. Stone Elworthy, Colin Johnson, Iain Drummond, Tomer Avidor-Reiss, and Natalia Bulgakova provided helpful comments on earlier versions of this manuscript. Imaging work was performed at the University of Sheffield Wolfson Light Microscopy Facility, funded in part by Medical Research Council (MRC) grant MR/K015753/1. This project was supported by funding from the National Eye Institute/National Institutes of Health (R01EY018176) and the MRC (MR/N000714/1).
Footnotes
Communicating editor: M. Halpern
Literature Cited
- Alves C. H., Bossers K., Vos R. M., Essing A. H. W., Swagemakers S., et al. , 2013. Microarray and morphological analysis of early postnatal CRB2 mutant retinas on a pure C57BL/6J genetic background. PLoS One 8: e82532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braasch I., Gehrke A. R., Smith J. J., Kawasaki K., Manousaki T., et al. , 2016. The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat. Genet. 48: 427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chih B., Liu P., Chinn Y., Chalouni C., Komuves L. G., et al. , 2011. A ciliopathy complex at the transition zone protects the cilia as a privileged membrane domain. Nat. Cell Biol. 14: 61–72. [DOI] [PubMed] [Google Scholar]
- Craige B., Tsao C. C., Diener D. R., Hou Y., Lechtreck K. F., et al. , 2010. CEP290 tethers flagellar transition zone microtubules to the membrane and regulates flagellar protein content. J. Cell Biol. 190: 927–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hollander A. I., ten Brink J. B., de Kok Y. J., van Soest S., van den Born L. I., et al. , 1999. Mutations in a human homologue of Drosophila crumbs cause retinitis pigmentosa (RP12). Nat. Genet. 23: 217–221. [DOI] [PubMed] [Google Scholar]
- Dishinger J. F., Kee H. L., Jenkins P. M., Fan S., Hurd T. W., et al. , 2010. Ciliary entry of the kinesin-2 motor KIF17 is regulated by importin-β2 and RanGTP. Nat. Cell Biol. 12: 703–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan S., Hurd T. W., Liu C. J., Straight S. W., Weimbs T., et al. , 2004. Polarity proteins control ciliogenesis via kinesin motor interactions. Curr. Biol. 14: 1451–1461. [DOI] [PubMed] [Google Scholar]
- Fan S., Fogg V., Wang Q., Chen X. W., Liu C. J., et al. , 2007. A novel Crumbs3 isoform regulates cell division and ciliogenesis via importin interactions. J. Cell Biol. 178: 387–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force A., Lynch M., Pickett F. B., Amores A., Yan Y. L., et al. , 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151: 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosens I., den Hollander A. I., Cremers F. P. M., Roepman R., 2008. Composition and function of the Crumbs protein complex in the mammalian retina. Exp. Eye Res. 86: 713–726. [DOI] [PubMed] [Google Scholar]
- Grawe F., Wodarz A., Lee B., Knust E., Skaer H., 1996. The Drosophila genes crumbs and stardust are involved in the biogenesis of adherens junctions. Development 122: 951–959. [DOI] [PubMed] [Google Scholar]
- Hirokawa N., Tanaka Y., Okada Y., 2012. Cilia, KIF3 molecular motor and nodal flow. Curr. Opin. Cell Biol. 24: 31–39. [DOI] [PubMed] [Google Scholar]
- Hsu Y. C., Willoughby J. J., Christensen A. K., Jensen A. M., 2006. Mosaic Eyes is a novel component of the crumbs complex and negatively regulates photoreceptor apical size. Development 133: 4849–4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q., Milenkovic L., Jin H., Scott M. P., Nachury M. V., et al. , 2010. A septin diffusion barrier at the base of the primary cilium maintains ciliary membrane protein distribution. Science 329: 436–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H., Marshall W. F., 2015. Efficient live fluorescence imaging of intraflagellar transport in mammalian primary cilia. Methods Cell Biol. 127: 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins P. M., McEwen D. P., Martens J. R., 2009. Olfactory cilia: linking sensory cilia function and human disease. Chem. Senses 34: 451–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin D., Ni T. T., Sun J., Wan H., Amack J. D., et al. , 2014. Prostaglandin signalling regulates ciliogenesis by modulating intraflagellar transport. Nat. Cell Biol. 16: 841–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurgens G., Wieschaus E., Nusslein-Volhard C., Kluding C., 1984. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. Rouxs Arch. Dev. Biol. 193: 283–295. [DOI] [PubMed] [Google Scholar]
- Kennedy B., Malicki J., 2009. What drives cell morphogenesis: a look inside the vertebrate photoreceptor. Dev. Dyn. 238: 2115–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok F. O., Shin M., Ni C. W., Gupta A., Grosse A. S., et al. , 2015. Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev. Cell 32: 97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer-Zucker A. G., Olale F., Haycraft C. J., Yoder B. K., Schier A. F., et al. , 2005. Cilia-driven fluid flow in the zebrafish pronephros, brain and Kupffer’s vesicle is required for normal organogenesis. Development 132: 1907–1921. [DOI] [PubMed] [Google Scholar]
- League G. P., Nam S.-C., 2011. Role of kinesin heavy chain in crumbs localization along the rhabdomere elongation in Drosophila photoreceptor. PLoS One 6: e21218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leventea E., Hazime K., Zhao C., Malicki J., 2016. Analysis of cilia structure and function in zebrafish. Methods Cell Biol. 133: 179–227. [DOI] [PubMed] [Google Scholar]
- Malicki J., Driever W., 1999. oko meduzy mutations affect neuronal patterning in the zebrafish retina and reveal cell-cell interactions of the retinal neuroepithelial sheet. Development 126: 1235–1246. [DOI] [PubMed] [Google Scholar]
- Malicki J., Neuhauss S. C., Schier A. F., Solnica-Krezel L., Stemple D. L., et al. , 1996. Mutations affecting development of the zebrafish retina. Development 123: 263–273. [DOI] [PubMed] [Google Scholar]
- Malicki J. J., Johnson C. A., 2017. The cilium: cellular antenna and central processing unit. Trends Cell Biol. 27: 126–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourão A., Christensen S. T., Lorentzen E., 2016. The intraflagellar transport machinery in ciliary signaling. Curr. Opin. Struct. Biol. 41: 98–108. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S., Wen X., Chih B., Nelson C. D., Lane W. S., et al. , 2010. TULP3 bridges the IFT-A complex and membrane phosphoinositides to promote trafficking of G protein-coupled receptors into primary cilia. Genes Dev. 24: 2180–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury M. V., Loktev A. V., Zhang Q., Westlake C. J., Peranen J., et al. , 2007. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell 129: 1201–1213. [DOI] [PubMed] [Google Scholar]
- Nishimura T., Kato K., Yamaguchi T., Fukata Y., Ohno S., et al. , 2004. Role of the PAR-3–KIF3 complex in the establishment of neuronal polarity. Nat. Cell Biol. 6: 328–334. [DOI] [PubMed] [Google Scholar]
- Omori Y., Malicki J., 2006. oko meduzy and related crumbs genes are determinants of apical cell features in the vertebrate embryo. Curr. Biol. 16: 945–957. [DOI] [PubMed] [Google Scholar]
- Ott, C., and J. Lippincott-Schwartz, 2012 Visualization of live primary cilia dynamics using fluorescence microscopy. Curr. Protoc. Cell Biol. 57: 4.26.1–4.26.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellikka M., Tanentzapf G., Pinto M., Smith C., McGlade C. J., et al. , 2002. Crumbs, the Drosophila homologue of human CRB1/RP12, is essential for photoreceptor morphogenesis. Nature 416: 143–149. [DOI] [PubMed] [Google Scholar]
- Pooranachandran N., Malicki J. J., 2016. Unexpected roles for ciliary kinesins and intraflagellar transport proteins. Genetics 203: 771–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugacheva E. N., Jablonski S. A., Hartman T. R., Henske E. P., Golemis E. A., 2007. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell 129: 1351–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Iwano T., Kunii M., Matsuda S., Mizuguchi R., et al. , 2014. Rab8a and Rab8b are essential for several apical transport pathways but insufficient for ciliogenesis. J. Cell Sci. 127: 422–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schou K. B., Pedersen L. B., Christensen S. T., 2015. Ins and outs of GPCR signaling in primary cilia. EMBO Rep. 16: 1099–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfakianos J., Togawa A., Maday S., Hull M., Pypaert M., et al. , 2007. Par3 functions in the biogenesis of the primary cilium in polarized epithelial cells. J. Cell Biol. 179: 1133–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepass U., Theres C., Knust E., 1990. crumbs encodes an EGF-like protein expressed on apical membranes of Drosophila epithelial cells and required for organization of epithelia. Cell 61: 787–799. [DOI] [PubMed] [Google Scholar]
- van den Hurk J. A., Rashbass P., Roepman R., Davis J., Voesenek K. E., et al. , 2005. Characterization of the Crumbs homolog 2 (CRB2) gene and analysis of its role in retinitis pigmentosa and Leber congenital amaurosis. Mol. Vis. 11: 263–273. [PubMed] [Google Scholar]
- Wei X., Malicki J., 2002. nagie oko, encoding a MAGUK-family protein, is essential for cellular patterning of the retina. Nat. Genet. 31: 150–157. [DOI] [PubMed] [Google Scholar]
- Whiteman E. L., Fan S., Harder J. L., Walton K. D., Liu C. J., et al. , 2014. Crumbs3 is essential for proper epithelial development and viability. Mol. Cell. Biol. 34: 43–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarz A., Hinz U., Engelbert M., Knust E., 1995. Expression of crumbs confers apical character on plasma membrane domains of ectodermal epithelia of Drosophila. Cell 82: 67–76. [DOI] [PubMed] [Google Scholar]
- Ying G., Gerstner C. D., Frederick J. M., Boye S. L., Hauswirth W. W., et al. , 2016. Small GTPases Rab8a and Rab11a are dispensable for rhodopsin transport in mouse photoreceptors. PLoS One 11: e0161236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Kwon S., Yamaguchi T., Cubizolles F., Rousseaux S., et al. , 2008. Mice lacking histone deacetylase 6 have hyperacetylated tubulin but are viable and develop normally. Mol. Cell. Biol. 28: 1688–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Omori Y., Brodowska K., Kovach P., Malicki J., 2012. Kinesin-2 family in vertebrate ciliogenesis. Proc. Natl. Acad. Sci. USA 109: 2388–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J., Wang X., Wei X., 2012. Crb apical polarity proteins maintain zebrafish retinal cone mosaics via intercellular binding of their extracellular domains. Dev. Cell 22: 1261–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu Y., Tong X., Wang Z., Liu D., Pan R., et al. , 2013. TALEN-mediated precise genome modification by homologous recombination in zebrafish. Nat. Methods 10: 329–331. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All animal strains and reagents will be distributed through international stock centers or directly by the Malicki laboratory.