Alcohol abuse is among the top causes of preventable death, generating considerable financial, health, and societal burdens. Paradoxically, alcohol...
Keywords: alcohol, HSF1, optogenetics, protein kinase A, UNC-18
Abstract
Alcohol is a potent pharmacological agent when consumed acutely at sufficient quantities and repeated overuse can lead to addiction and deleterious effects on health. Alcohol is thought to modulate neuronal function through low-affinity interactions with proteins, in particular with membrane channels and receptors. Paradoxically, alcohol acts as both a stimulant and a sedative. The exact molecular mechanisms for the acute effects of ethanol on neurons, as either a stimulant or a sedative, however remain unclear. We investigated the role that the heat shock transcription factor HSF-1 played in determining a stimulatory phenotype of Caenorhabditis elegans in response to physiologically relevant concentrations of ethanol (17 mM; 0.1% v/v). Using genetic techniques, we demonstrate that either RNA interference of hsf-1 or use of an hsf-1(sy441) mutant lacked the enhancement of locomotion in response to acute ethanol exposure evident in wild-type animals. We identify that the requirement for HSF-1 in this phenotype was IL2 neuron-specific and required the downstream expression of the α-crystallin ortholog HSP-16.48. Using a combination of pharmacology, optogenetics, and phenotypic analyses we determine that ethanol activates a Gαs-cAMP-protein kinase A signaling pathway in IL2 neurons to stimulate nematode locomotion. We further implicate the phosphorylation of a specific serine residue (Ser322) on the synaptic protein UNC-18 as an end point for the Gαs-dependent signaling pathway. These findings establish and characterize a distinct neurosensory cell signaling pathway that determines the stimulatory action of ethanol and identifies HSP-16.48 and HSF-1 as novel regulators of this pathway.
Alcohol is one of the most prevalent addictive substances and its overuse produces a severe burden on society (WHO 2011). Acute effects of alcohol include motor incoordination, sedation, and anesthesia. Alcohol also acts as a stimulant, activating neurotransmitter release within the brain’s reward circuitry, increasing heart rate, and inducing aggression and risk-taking (Hendler et al. 2013). The cellular mechanisms underlying either the anesthetic or stimulatory effects of ethanol are only now being elucidated. Ethanol was initially hypothesized to act through indirect perturbations of the lipid environment thereby affecting broadly all membrane protein function (Harris et al. 2008). Recent work instead implicates more direct effects on specific protein targets, particularly membrane proteins such as ion channels and receptors (Howard et al. 2014; Trudell et al. 2014). Ethanol has a very simple molecular structure and is thought to exert its intoxicating effects through interactions with these target proteins with very low affinity (∼0.1 M) to alter the natural dynamics of protein function (Howard et al. 2011; Olsen et al. 2014). A complete understanding of both the acute and chronic effects of alcohol within the nervous system is an important unresolved question in physiology.
Invertebrates are excellent genetic models for the identification of cellular/molecular mechanisms governing the sedative properties of ethanol and other anesthetics (Barclay et al. 2010; Devineni and Heberlein 2013; Bettinger and Davies 2014). In Drosophila, studies of acute ethanol intoxication have implicated a large number of proteins and signaling pathways (Kaun et al. 2012). In Caenorhabditis elegans, proteins such as Munc18 and Rab3 (Kapfhamer et al. 2008; Graham et al. 2009; Johnson et al. 2013), the BK channel slo-1 (Davies et al. 2003), the leak channel regulator Lightweight (Speca et al. 2010), chloride intracellular channels (Bhandari et al. 2012), and the α-crystallin ortholog HSP-16.48 (Johnson et al. 2016) all affect sensitivity to high levels of ethanol (400 mM). The use of very high external ethanol in C. elegans behavioral analysis is thought to be a consequence of poor penetration through the nematode cuticle (Alaimo et al. 2012). Indeed, internal concentrations in response to 400 mM external ethanol are estimated between 20 and 60 mM (Alaimo et al. 2012; Johnson et al. 2016), although not all studies are in agreement (Mitchell et al. 2007). In addition to sedation, at lower concentrations ethanol acts as a stimulant (Phillips and Shen 1996; Wolf et al. 2002; Balino et al. 2016). In C. elegans, ethanol-induced stimulation occurs at external concentrations (17 mM, 0.1%) whose absolute values would be physiologically consistent with blood alcohol limits for impaired driving (House of Commons Transport Committee 2010). Acute exposure of nematodes to 17 mM ethanol causes a small, but characteristic increase in locomotion rate (Graham et al. 2009; Johnson et al. 2013). Virtually nothing is known about the cellular and molecular basis underlying C. elegans phenotypes at this ethanol concentration.
Heat shock activates a transcriptional response to toxic insults whereby protective cellular chaperone (heat shock protein) expression is increased under the control of the heat shock transcription factor (HSF1) (Anckar and Sistonen 2011). The HSF1-HSP pathway is also ubiquitously involved in stress-independent cellular functions such as polypeptide folding, protein–protein interactions (Kampinga and Craig 2010), and proteostasis (Morimoto 2011), as well as contributing to diseases like cancer (Mendillo et al. 2012) and neurodegeneration (Kondo et al. 2013). In C. elegans, RNA interference (RNAi) or loss-of-function mutations of hsf-1 increase stress sensitivity, but also accelerate ageing (Hsu et al. 2003; Prahlad et al. 2008; Baird et al. 2014). We have recently shown that hsf-1 loss-of-function increases sensitivity to 400 mM ethanol (Johnson et al. 2016). This hypersensitivity was partially the result of the downstream basal expression of HSP-16.48, an ortholog of the human small heat shock protein α-crystallin, in a process unrelated to an HSF-1-dependent heat shock stress response. Here, we identify that the stimulatory ethanol phenotype also required HSF-1, but specifically in six IL2 chemosensory neurons and could be completely rescued by transgenic expression of HSP-16.48 in IL2 neurons of the hsf-1 mutant. Using a combination of pharmacology, genetics, and optogenetics, we determined further that this ethanol-dependent stimulation of motility acts via a Gαs-cAMP-protein kinase A (PKA) signaling pathway within the IL2 sensory neurons and identifies the exocytotic protein UNC-18 as a downstream effector for PKA. Although individual components of the Gαs pathway have been linked previously to the neuronal effects of ethanol, this study uniquely characterizes the entire signaling pathway in ethanol-dependent stimulation.
Materials and Methods
Nematode culturing, strains, and genetics
C. elegans were cultured under standard conditions at 20° on Nematode Growth Media (NGM) agar plates with OP50 Escherichia coli as a food source as previously described (Brenner 1974; Graham et al. 2009; Edwards et al. 2012). These experiments used the following strains: Bristol N2 (wild-type), PS3551 hsf-1(sy441), KG524 gsa-1(ce94), KG421 gsa-1(ce81), MT363 goa-1(n363), NM1380 egl-30(js126), KG1180 lite-1(ce314), and NL2099 rrf-3(pk1426). To investigate effects of single-copy transgenic rescue of hsf-1(sy441) mutants, we analyzed the OG532 (hsf-1(sy441);drSi13[hsf-1p::hsf-1::GFP::unc-54 3′UTR + Cbr-unc-119(+)]) and OG580 (hsf-1(sy441);drSi28[hsf-1p::hsf-1(R145A)::GFP::unc-54 3′UTR + Cbr-unc-119(+)]) strains (Morton and Lamitina 2013), which are single-copy mos1-mediated Single Copy Insertion (MosSCI)-produced rescues of the hsf-1(sy441) mutant. Basal locomotion rates for each strain can be found in Supplemental Material, Table S1 in File S1. Transgenic animals were derived by germline injection as previously described (Mello et al. 1991; Graham et al. 2009; Edwards et al. 2012). All injections were performed with 10 ng/µl (indicated construct) and 30 ng/µl (indicated co-injection marker), and made up to 100 ng/µl in total with empty filler DNA (either pUC19 or pBluescript). For each transgenic strain, three independent lines were isolated and tested phenotypically. The results presented were consistent for all individual lines; however, individual line results can be found in Table S2 in File S1. Transgenic lines used in this study include the following: N2;ulvEx[Phsf-1::hsf-1], N2;ulvEx[Prab-3::hsb-1], N2;ulvEx[Pklp-6::hsb-1], N2;ulvEx[Prab-3::hsp-16.48]; N2;ulvEx[Prab-3::hsp-16.48 Δ38-44], N2;ulvEx[Pklp-6::hsp-16.48], N2;ulvEx[Punc-18::unc-18 S322A], N2;ulvEx[Pklp-6::unc-18 S322A], N2;ulvEx[Pklp-6::kin-1::Pklp-6], hsf-1(sy441);ulvEx[Phsf-1::hsf-1], hsf-1(sy441);ulvEx[Prab-3::hsf-1], hsf-1(sy441);ulvEx[Pmyo-3::hsf-1], hsf-1(sy441);ulvEx[Pglr-1::hsf-1], hsf-1(sy441);ulvEx[Punc-17::hsf-1], hsf-1(sy441);ulvEx[Posm-6::hsf-1], hsf-1(sy441);ulvEx[Pgcy-8::hsf-1], hsf-1(sy441);ulvEx[Pklp-6::hsf-1], hsf-1(sy441);ulvEx[Prab-3::hsp-16.48], hsf-1(sy441);ulvEx[Prab-3::hsp-16.48 Δ38-44], and hsf-1(sy441);ulvEx[Pklp-6::hsp-16.48]. Transgenic constructs in Bristol N2 were coexpressed with either a Psur-5::GFP or a Prab-3::GFP marker. To indicate appropriate cellular expression for additional cell–tissue-specific promoters, each of these transgenics used a GFP coexpression marker under the control of the same promoter. IL2 neurons were visualized with the promoter::GFP reporter hsf-1(sy441);ulvEx[Pklp-6::GFP; Pmyo-2::mCherry]. To examine cell-specific RNAi, Bristol N2 worms were injected with the plasmid Pklp-6::kin-1::Pklp-6, which drove cell-specific expression of kin-1 (catalytic subunit of C. elegans protein kinase A) in both the forward and reverse direction (Esposito et al. 2007).
RNAi experiments
RNAi experiments were performed using the rrf-3(pk1426) strain. RNAi was induced by feeding (Kamath and Ahringer 2003) using the ORFeome-based RNAi library (Rual et al. 2004) as described previously (Johnson et al. 2016). HT115 RNAi bacteria were cultured in LB media with 100 μg/ml ampicillin and spotted onto 60 mm diameter NGM plates supplemented with 1 mM isopropyl β-1-thiogalactopyranoside and 25 μg/ml carbenicillin. NGM plates were dried for 4 days before spotting. Five L3–L4 worms were added to each RNAi plate and cultured at 20°. Phenotypic analysis was performed on first-generation progeny fed with the indicated RNAi bacterial clones. For the negative control for the RNAi screen, worms were fed with an empty feeding vector.
Cloning
C. elegans genes of interest were amplified from either Bristol N2 genomic DNA (hsp-16.48) or cDNA (hsf-1), cloned into pDONR201 and recombined into DEST vectors to create tissue-specific expression vectors, as previously described (Johnson et al. 2016). For hsb-1, the gene was amplified from Bristol N2 genomic DNA using the following primers:
hsb-1 attB forward: 5ʹ-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGTCCGATGAGAAGTCTACC-3ʹ.
hsb-1 AttB reverse:
5ʹ-GGGGACCACTTTGTACAAGAAAGCTGGGTCTTATTGAGCGCTTGGCGGATGTTC-3ʹ.
Mutagenesis of the hsp-16.48 gene was performed using a Q5 Site-Directed Mutagenesis Kit (New England Biolabs, Beverly, MA) as per the manufacturer’s instructions. The unc-18 S322A expression vector was created using the Gene Tailor mutagenesis kit (Invitrogen, Carlsbad, MA) from a vector carrying the unc-18 cDNA under the control of the unc-18 genomic flanking regions (gift from H. Kitayama, Kyoto University, Japan) (Gengyo-Ando et al. 1996). To create the Pklp-6::unc-18 construct, the existing unc-18 promoter was excised by EcoRI and BamHI restriction digest and replaced with a klp-6 promoter using the NEBuilder DNA assembly (New England Biolabs). For glutathione-S-transferase (GST) fusion protein production, unc-18 was subcloned into pGEX-6p-1 as previously described (Edwards et al. 2012).
All C. elegans promoter fragments were amplified from Bristol N2 genomic DNA and cloned into pPD117.01 (kind gift of A. Fire, Stanford University) in place of Pmec-7. These vectors were converted into Gateway DEST vectors using a conversion cassette (Life Technologies). Prab-3 (kind gift of M. Nonet, Washington University in St. Louis), Pmyo-3 (kind gift of A. Fire, Stanford University), and Phsf-1 are previously described (Edwards et al. 2012; Johnson et al. 2016). The following primers were used for additional promoter cloning:
Punc-17 forward: 5ʹ-AGTCGGCGCGCCATCCGTTCCCATCCGCTTCATC-3ʹ.
Punc-17 reverse: 5ʹ-AGGAGGATCCGGTTACTATTTTGAACAAGAGATGCGG-3ʹ.
Pglr-1 forward: 5ʹ-AGTCGGCGCGCCCTGTAGCCGGTATGCACTGATAAC-3ʹ.
Pglr-1 reverse: 5ʹ-AGTCGGATCCTGTGAATGTGTCAGATTGGGTG-3ʹ.
Posm-6 forward: 5ʹ-ATGTGGCGCGCCCAGTGGAATCACCATTGGGTATCCAG-3ʹ.
Posm-6 reverse: 5ʹ-GGGTGGATCCGAAGGTAATAGCTTGAAAGAGATATAAGCCC-3ʹ.
Pgcy-8 forward: 5ʹ-AGTCGGCGCGCCAACTACCTTCCTCCGCGTCC-3ʹ.
Pgcy-8 reverse: 5ʹ-AGTCGGATCCTTTGATGTGGAAAAGGTAGAATCG-3ʹ.
Pklp-6 forward: 5ʹ-CCCCGGCGCGCCAACGTCCCAGACAATTTCAAC-3ʹ.
Pklp-6 reverse: 5ʹ-CTACGGATCCGGAGTCACCCTTTCCCCTTATTCTG-3ʹ.
The Pklp-6::kin-1::Pklp-6 construct was made as follows. A 750-bp fragment of kin-1 was amplified from the Vidal library clone (Rual et al. 2004) and subcloned into the Pklp-6::GFP expression vector, downstream of Pklp-6 in place of GFP, using the NEBuilder cloning kit (New England Biolabs). The Pklp-6::kin-1 PCR fragment was then amplified from this construct and again subcloned into the same Pklp-6 construct as before, but in reverse orientation. Correct construction was confirmed by PCR and sequencing. Primers used were as follows:
Protein phosphorylation and mass spectrometry
For in vitro biochemistry, recombinant proteins (GST, GST-UNC-18) were produced as described previously (Edwards et al. 2012). For phosphorylation experiments, 2 μg of substrate protein was incubated with 2 units of PKA catalytic subunit (Sigma [Sigma Chemical], St. Louis, MO), 100 μM ATP, and 2 μCi [γ-32P]ATP (GE Healthcare) in a 50 μl final reaction volume of 2-(N-morpholino)ethanesulfonic acid (MES) buffer (50 mM MES, 10 mM MgCl2, 1 mM DTT, and 0.5 mM EDTA, pH 6.9). Reactions were incubated at 30° for 3 hr before termination. To determine phosphorylation, 20 μl of the kinase reaction was separated by SDS-PAGE, stained with Coomassie Blue dye, destained overnight in a destainer [35% ethanol, 2% glycerol (v/v)], air-dried in Hoeffer Easy Breeze plastic frames (Thermo Fisher Scientific), exposed to a phosphor screen for 2–4 hr, and scanned by a PhosphorImager 425 (Molecular Dynamics). For in vitro phosphorylation site determination, phosphorylated samples were separated on NuPAGE 4–12% Bis-Tris precast gels (Invitrogen) and stained with Coomassie Blue dye before excision of protein bands. Gel plugs were destained in 50% acetonitrile (v/v)/50 mM ammonium bicarbonate and dried before incubation in trypsin (5 ng/μl in 50 mM ammonium bicarbonate) for 16 hr at 37°. Peptides were then extracted by sonication of gel plugs in 60% (v/v) acetonitrile/1% (v/v) trifluoroacetic acid. Extracts were thoroughly dried and ZipTipped (Millipore, Bedford, MA) before electrospray ionization mass spectrometry (MS). Residual peptides were resuspended in 50% (v/v) acetonitrile/0.05% trifluoroacetic acid, and 5 μl of suspension was delivered into a QStar Pulsar I hybrid quadrupole time-of-flight MS (AB Sciex) by automated in-line liquid chromatography [integrated LCPackings System, 5 mm C18 nano-precolumn, and 75 μm × 15 cm C18 PepMap column (Dionex)]. A gradient from 5 to 48% acetonitrile/0.05% trifluoroacetic acid (v/v) in 60 min was applied at a 300 nl/min flow rate and survey scans of 1 sec were acquired for m/z 400–2000. The most intense ions were selected for tandem MS with 2 sec accumulation times and a dynamic exclusion of 30 sec. Identification and analysis was performed using MASCOT software (Matrix Science).
Optogenetics
JellyOp (Bailes et al. 2012) and hRh1 (Bailes and Lucas 2013) coding domain sequences were PCR amplified from pcDNA3.1 and pcDNA3.5 expression vectors, respectively, and subcloned downstream of Pklp-6 in pPD117.01 (detailed above) via Gibson Cloning techniques (New England Biolabs). The optogenetic vectors were then transformed by germline injection into C. elegans as described above. The vectors were expressed in the lite-1(ce314) background to prevent any potential photophobic responses (Husson et al. 2013). All optogenetic worm strains were then grown in the dark at 20° on standard NGM plates with OP50 bacteria as a food source. One day prior to assays, late L4 worms were picked onto standard 60 mm NGM plates with 50 μl OP50 supplemented with 100 μM 9-cis-Retinal, an active chromophore for opsin. All plates and worms were handled either in the dark or under a dim red light (> 630 nm). Thrashing assays were performed under the dim red light in Dent’s solution (140 mM NaCl, 6 mM KCl, 1 mM CaCl2, 1 mM MgCl2, and 5 mM HEPES, pH 7.4, with bovine serum albumin at 0.1 mg/ml), in the presence or absence of 17 mM ethanol, using a Leica MZ10F-stereomicroscope (Leica, UK) equipped with a pE-300 light-emitting diode (LED) fluorescence light source (CoolLED, UK). To activate opsin, worms were illuminated with a single flash of a 100% power green LED through the ET GFP filter set for 5 sec. For hRh1, opsin activation occurred as soon as they were placed in the Dent’s solution and thrashing was quantified after 10 min acclimation (as below). For JellyOp, worms were illuminated following the 10 min acclimation in Dent’s solution just prior to thrashing quantification, as the JellyOp activation is more transient (Bailes et al. 2012). Assays were repeated on three separate days to confirm replicability.
Behavioral assays
Phenotypic analysis was performed in a temperature-controlled room on young adult hermaphrodites from sparsely-populated plates grown at 20°. Unless otherwise indicated, experiments were conducted at an ambient temperature of 20°. Here, locomotion rate was measured as thrashing (Gjorgjieva et al. 2014) in 200 μl Dent’s solution as previously described (Graham et al. 2009; Johnson et al. 2013). One thrash was defined as a complete movement from maximum to minimum amplitude and back again. For acute ethanol experiments, ethanol was diluted to 17 mM in Dent’s solution, and locomotion was quantified following 10 min exposure and normalized as a percentage of the mean thrashing rate of untreated worms measured each day (at least 10 control worms per strain). For forskolin (Sigma) experiments, forskolin was diluted in dimethyl sulfoxide (DMSO) and added to Dent’s solution in the indicated concentration. For direct comparison in these experiments, control (untreated) worms were exposed to Dent’s with an equal concentration of DMSO. H-89 (Sigma) was diluted in water and added to the Dent’s/DMSO solution in the indicated concentration. For chemotaxis assays (Kashyap et al. 2014), experimental assay plates (100 mm Petri dishes) contained 2% (w/v) agar, 5 mM KH2PO4, 1 mM CaCl2, and 1 mM MgSO4. Plates were poured 3 days prior to use and allowed to dry. Worms were washed in M9 buffer and placed on the center of the plate. Next, 1 μl of the attractant (100% ethanol) and a negative control (distilled water) were pipetted on opposite sides of the plate. The number of worms on either side of the plate was calculated following 90 min. The chemotaxis index (C.I.) was calculated as C.I. = (# worms at attractant − # worms at control)/total number of worms. For worm avoidance assays, a 0.9-cm ring of a substance was made by a stamping procedure on unseeded NGM plates. Immediately after stamping, 30 worms were placed within the center of the ring, completed within 2 min. Following 10 min exposure, the number of worms that had crossed the ring was counted. In preliminary experiments, 0.9 cm was found to be sufficient to permit most worms to cross a neutral substance (distilled water) within 10 min. All data are expressed as mean ± SE. As indicated, significance was tested by Student’s t-test, Mann–Whitney U-test, or ANOVA with Tukey post hoc comparisons where appropriate.
Data availability
Strains are available upon request. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
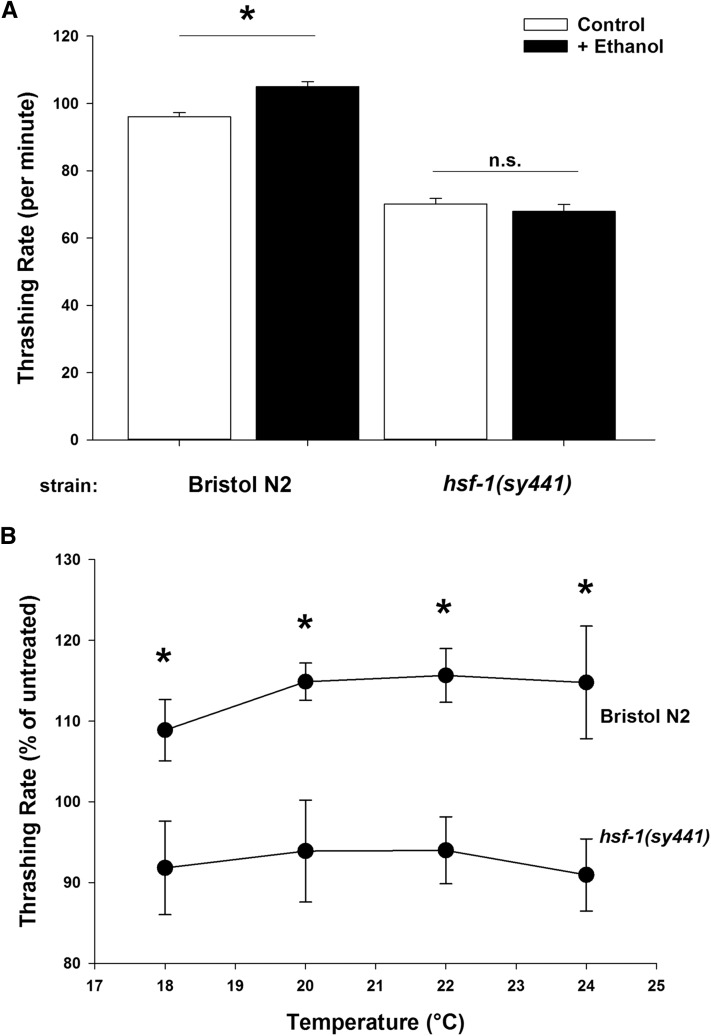
HSF-1, the heat shock transcription factor, is involved in a plethora of cellular functions (Anckar and Sistonen 2011; Morimoto 2011; Vihervaara and Sistonen 2014). In C. elegans, the hsf-1(sy441) allele is a viable loss-of-function point mutation in the hsf-1 gene that acts as an inhibitor of HSF-1 transcriptional activity (Hajdu-Cronin et al. 2004), increasing temperature sensitivity and decreasing lifespan (Baird et al. 2014). Nematodes respond to high external ethanol concentration (400 mM) by a dose-dependent decrease in coordinated locomotion (Davies et al. 2003; Graham et al. 2009) and we have recently characterized a novel role for HSF-1 in determining this phenotype (Johnson et al. 2016). However, worms additionally respond phenotypically to low levels of external ethanol (17 mM) with a reproducible enhancement in locomotor activity (Graham et al. 2009; Johnson et al. 2013). In Bristol N2 wild-type worms, this stimulation is a 5–10% increase in basal locomotion rate, as quantified by thrashing (Figure 1A). We tested whether the hsf-1(sy441) mutation would also hypersensitize worms to the effects of low concentrations of ethanol. Surprisingly, we found that the stimulatory effect of 17 mM external ethanol was completely absent in worms containing the hsf-1(sy441) mutation (Figure 1A). The hsf-1(sy441) mutant is well established to have a temperature-sensitive phenotype and we verified that the low-dose ethanol phenotype was not affected by the ambient temperature (Figure 1B); however, all other described experiments were conducted at 20° standard temperature.
Figure 1.
Low concentrations of ethanol stimulate C. elegans locomotion in an hsf-1-dependent fashion. (A) In comparison to untreated controls, exposure to 17 mM ethanol (+ Ethanol) significantly increased the locomotion of Bristol N2 wild-type worms. The hsf-1(sy441) loss-of-function mutant demonstrated a reduced untreated locomotion rate that was unaffected by ethanol. Locomotion rate was quantified by thrashing after 10 min immersion in Dent’s solution. * P < 0.001 (one-way ANOVA with Tukey post hoc comparisons); n.s. = not significant. N = 100 for each condition. (B) The absence of ethanol stimulation in hsf-1(sy441) worms is independent of temperature. Locomotion rate was quantified at the indicated temperature and expressed here as a percentage of mean thrashing rate of untreated worms. At all temperatures, ethanol stimulated Bristol N2 locomotion, but had no effect on hsf-1(sy441) loss-of-function worms. * P < 0.001 (two-way ANOVA with Tukey post hoc comparisons); N = 20 for each condition.
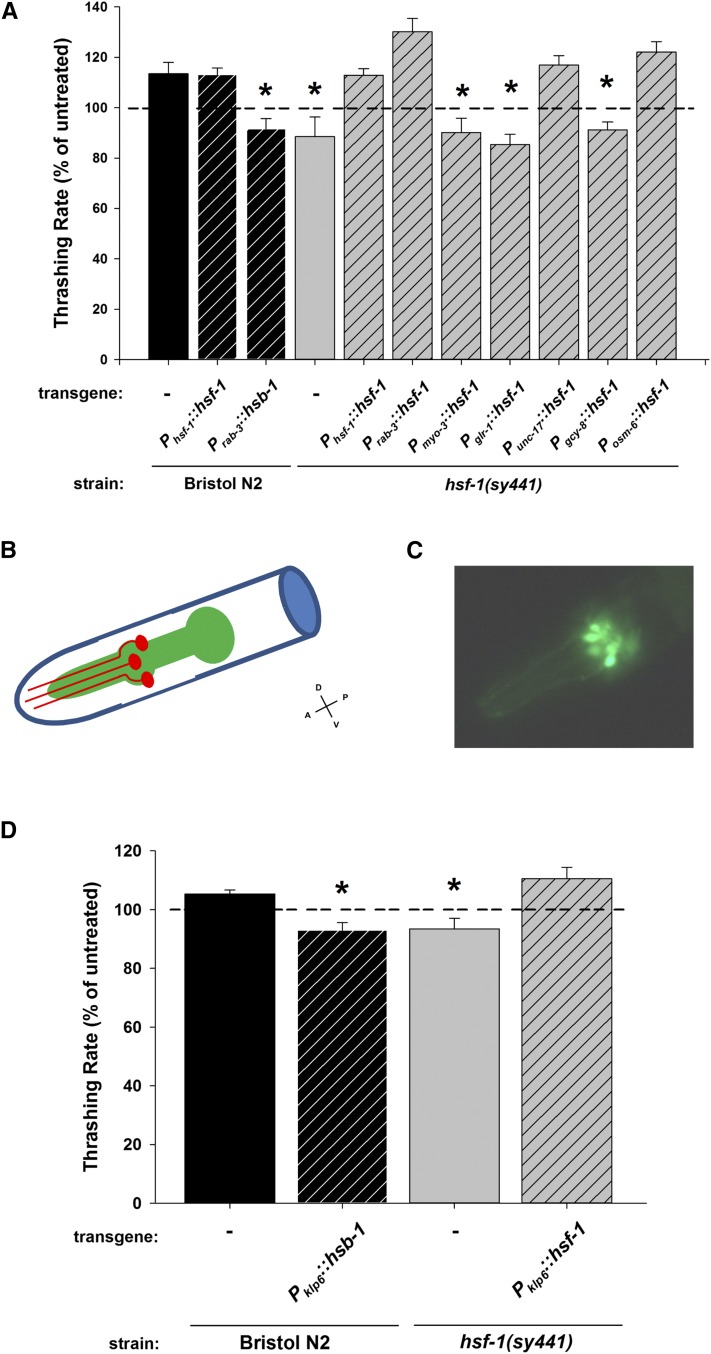
HSF-1 is a ubiquitously expressed transcription factor that is active in all cell types; however, the role of hsf-1(sy441) in the hypersensitivity to 400 mM external ethanol is dependent upon pan-neuronal hsf-1 expression (Johnson et al. 2016). Therefore, we next investigated next whether the ablation of the low (17 mM) ethanol stimulation of locomotion phenotype observed in the hsf-1(sy441) mutant was also pan-neuronal in nature by tissue-specific transgenic rescue. To allow for a direct comparison of the stimulatory effects of ethanol independent of basal locomotor rates, the data are presented as thrashing rate normalized to untreated worms of the same strain as done in previous studies (Davies et al. 2003; Graham et al. 2009; Johnson et al. 2016). However, basal rates for all strains can be compared in Table S1 in File S1. While overexpression of hsf-1 in Bristol N2 controls had no additional effect, transgenic rescue of hsf-1(sy441) under its own promoter (Phsf-1::hsf-1) or specifically under the control of a pan-neuronal promoter (Prab-3::hsf-1) completely rescued the stimulatory ethanol phenotype (Figure 2A). However, transgenic rescue of hsf-1(sy441) using a body wall muscle promoter (Pmyo-3::hsf-1) was insufficient to rescue. As a complementary approach to investigate hsf-1 function in the ethanol phenotype, we cloned and overexpressed hsb-1 in Bristol N2. hsb-1 is a negative regulator for hsf-1 transcriptional function and its overexpression in C. elegans results in similar phenotypes to loss-of-function of hsf-1 (Satyal et al. 1998). Overexpression of hsb-1 pan-neuronally in Bristol N2 worms (Prab-3::hsb-1) also resulted in a loss of ethanol stimulation of locomotion similar to that seen in the hsf-1(sy441) mutant (Figure 2A). From these transgenic rescue experiments, we conclude that the loss of the ethanol stimulation phenotype is a consequence of a loss in neuronal hsf-1 function.
Figure 2.
The hsf-1 dependence of the low ethanol stimulation of locomotion requires hsf-1 expression in IL2 neurons. (A) In hsf-1(sy441) worms, the low ethanol stimulation of locomotion was restored by transgenic rescue of wild-type hsf-1 under the control of its endogenous (Phsf-1), pan-neuronal (Prab-3), cholinergic neuron (Punc-17), or ciliated sensory neuron promoter (Posm-6). Tissue-specific expression in muscle (Pmyo-3), interneurons (Pglr-1), or the AFD thermosensory neurons (Pgcy-8) was unable to rescue the hsf-1(sy441) mutant. In Bristol N2 worms, overexpression of hsf-1 under its endogenous promoter (Phsf-1) had no additional effect on ethanol stimulation of locomotion, whereas neuronal overexpression of the hsf-1 inhibitor, hsb-1, blocked the ethanol effect. (B) Cartoon schematic of the C. elegans head region indicating the location of the IL2 neurons (one neuron of each of the three pairs are depicted) and their projections (in red) adjacent to the pharynx (in green). The approximate anterior–posterior (A–P) and ventral–dorsal (V–D) axes are indicated and apply to part C as well. (C) Expression of green fluorescent protein (GFP) under the control of the klp-6 promoter, showing expression in the IL2 neurons of hsf-1(sy441) mutants. Expression in Bristol N2 worms was anatomically similar. (D) In comparison to hsf-1(sy441) worms, low ethanol stimulation of locomotion was restored by transgenic expression of wild-type hsf-1 using an IL2-specific promoter (Pklp-6). In Bristol N2 worms, IL2 neuron overexpression of the hsf-1 inhibitor, hsb-1, blocked the ethanol effect. In both (A) and (D), data are expressed normalized to untreated controls. For each experiment, exposure to ethanol enhanced the locomotion rate of Bristol N2 worms (Mann–Whitney U-test; P < 0.05). * indicates significant difference in comparison to treated Bristol N2. Comparisons were made by one-way ANOVA with Tukey post hoc comparisons (P < 0.001; N = 30 for each condition). Bristol N2 worms are depicted in black, hsf-1(sy441) in gray. Hatching indicates transgenic expression (transgene and promoter indicated below graph).
As the rab-3 promoter drives expression throughout the nervous system, we next determined whether the stimulatory ethanol phenotype could be localized more precisely in the nervous system. Intriguingly, the 17-mM ethanol stimulatory phenotype of hsf-1(sy441) worms could be reestablished by transgenic expression of wild-type hsf-1 specifically in cholinergic neurons (Punc-17::hsf-1) or in ciliated sensory neurons (Posm-6::hsf-1), but not in interneurons (Pglr-1::hsf-1) or the AFD thermosensory neurons (Pgcy-8::hsf-1) (Figure 2A). The localization of the ethanol-induced stimulation of locomotion to either cholinergic neurons or ciliated sensory neurons was particularly serendipitous, as these two promoters are predicted to overlap in only one nematode cell type, the inner labial sensilla neurons, IL2 (Figure 2B). There are three pairs of IL2 neurons located in the head of the animal, all with ciliated endings (White et al. 1986; Burket et al. 2006). IL2 neurons are cholinergic (Zhang et al. 2014) and are predicted to be chemosensory as their dendrites are exposed to the external environment. Very little has been published ascribing function to the IL2 neurons; however, they do regulate nictation, a behavior whereupon a nematode waves its head in three dimensions (Lee et al. 2012). We tested whether the absence of the stimulatory ethanol phenotype of hsf-1(sy441) could be reversed by transgenic expression of wild-type hsf-1 specifically in the IL2 neurons. For this, we cloned the promoter region of the klp-6 gene, which in hermaphrodites is expressed specifically in the IL2 neurons (Peden and Barr 2005). Localization was verified by examination of a promoter::GFP transgenic worm, confirming that the IL2 neuronal structures appeared anatomically intact even in the hsf-1(sy441) mutant (Figure 2C), although an in-depth ultrastructural analysis of the cilia in this mutant remains to be fully investigated. We then determined that expression of wild-type hsf-1 in the IL2 neurons (Pklp-6::hsf-1) alone could restore the stimulatory ethanol phenotype of the hsf-1(sy441) mutant to a level indistinguishable from wild-type worms (Figure 2D). The complementary approach also confirmed that IL2-specific overexpression of hsb-1 (Pklp-6::hsb-1), the negative regulator for hsf-1, was able to block the ethanol-induced stimulation in Bristol N2 (Figure 2D).
hsf-1(sy441) worms have a slightly lower basal locomotor rate (Figure 1A); however, previous transgenic rescue experiments indicate that the basal locomotion rate is uncorrelated with the high ethanol phenotype (Johnson et al. 2016). Our data here also support a lack of correlation between basal locomotor rate and the low-ethanol phenotype (Table S1 in File S1). For example, IL2 neuron overexpression of hsb-1 in Bristol N2 does not reduce basal locomotion rate at all, but does block the low-ethanol phenotype. Alternatively, overexpression of wild-type hsf-1 in IL2 neurons in the hsf-1(sy441) mutant background does not rescue the locomotor defect of the mutant, but did restore the low-ethanol phenotype. Notwithstanding this lack of correlation, we were interested in determining whether the reduced locomotor rate in the hsf-1(sy441) mutant was a consequence of the point mutation. The hsf-1(sy441) strain was originally isolated in a genetic screen of suppressors of heat shock-induced expression, where it was both backcrossed and outcrossed (Hajdu-Cronin et al. 2004). We have demonstrated here (Table S1 in File S1) and elsewhere (Johnson et al. 2016) that transgenic rescue can alter many hsf-1-dependent phenotypes, without altering the basal locomotor rates, consistently indicating that the defect in locomotion may be independent of the sy441 mutation or possibly incomplete rescue by the transgene. To address this question more directly we analyzed the OG532 strain, which is a single-copy MosSCI-integrated rescue of hsf-1(sy441) (Morton and Lamitina 2013). The OG532 strain demonstrated a significant, yet incomplete, rescue of the locomotion defect in comparison to Bristol N2 (Table S1 in File S1). The incomplete rescue of the locomotion defect was reminiscent of a reported incomplete rescue of the hsf-1(sy441) temperature-sensitivity (Morton and Lamitina 2013). Reassuringly, the single-copy rescue did restore the ethanol stimulatory phenotype (Figure S1) to a level similar to that seen with transgenic overexpression. In contrast, the OG580 strain, a single-copy rescue of hsf-1(sy441) with a DNA-binding defective mutant hsf-1 (Morton and Lamitina 2013), impaired locomotion further (Table S1 in File S1) and did not rescue the ethanol phenotype (Figure S1). These data indicate a potentially complex role for hsf-1 in basal locomotor rate, which is perhaps unsurprising given the broad cellular functions of the HSF-1 transcription factor and its downstream transcriptional targets. Critically, however, our results show that the basal locomotor rate is unrelated to the ethanol stimulatory effect and is instead a consequence of the hsf-1(sy441) mutation.
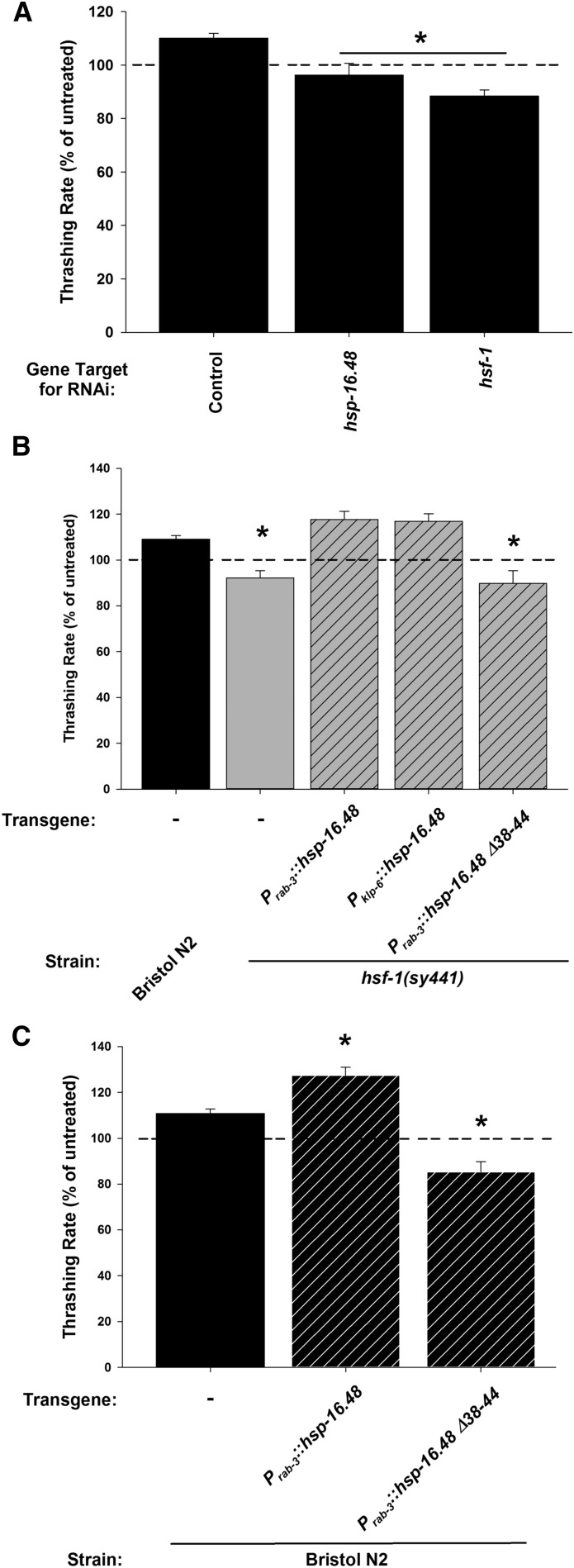
Our previous work on HSF-1 and its effect on sensitivity to 400 mM external ethanol indicated that the main effector for HSF-1 was the small heat shock protein HSP-16.48 (Johnson et al. 2016). This work showed that RNAi of hsp-16.48 phenocopied RNAi of hsf-1 and that transgenic expression of hsp-16.48 partially rescued the high-dose ethanol sensitivity of the hsf-1(sy441) mutants. Finally, overexpression of hsp-16.48 greatly reduced wild-type worm sensitivity to high external ethanol in a manner dependent upon an intact seven amino acid region of the N-terminus (amino acids 38–44) (Johnson et al. 2016). We next performed a series of experiments to test for functional similarities with the stimulatory ethanol phenotype. There are unfortunately no available hsp-16.48 null mutations as the gene has undergone evolutionary genetic duplication, rendering null mutations prohibitively difficult to isolate (Johnson et al. 2016). Our results showed that RNAi knockdown of hsf-1 also resulted in a loss of the 17 mM ethanol phenotype (Figure 3A), to an extent similar to that seen with the sy441 point mutation. Reminiscent of the sedative ethanol phenotype results (Johnson et al. 2016), RNAi of hsp-16.48 also blocked the stimulatory ethanol phenotype to an equivalent level to hsf-1 RNAi (Figure 3A). However, distinct from the sedative ethanol phenotype (Johnson et al. 2016), transgenic expression of hsp-16.48 either pan-neuronally (Prab-3::hsp-16.48) or in IL2 neurons specifically (Pklp-6::hsp-16.48) was sufficient to restore completely the stimulatory ethanol phenotype in the hsf-1(sy441) mutant (Figure 3B). Finally, we determined that overexpression of hsp-16.48 in Bristol N2 wild-types (Prab-3::hsp-16.48) increased the effect of 17 mM external ethanol, whereas the N-terminal truncation mutant (Prab-3::hsp-16.48 Δ38-44) appeared to act as neomorphic or as dominant-negative (Figure 3C). However, expression of the truncation mutant in the hsf-1(sy441) background did not result in an enhancement of the ethanol-dependent reduction in locomotion in comparison to hsf-1(sy441) alone (Figure 3B). Therefore, we hypothesize that the truncation mutant is possibly acting here as a dominant-negative. These experiments implicate HSP-16.48 in the stimulatory ethanol phenotype, acting downstream of HSF-1 in a manner requiring the seven amino acid region of the N-terminus.
Figure 3.
HSP-16.48 acts downstream of HSF-1 in IL2 neurons. (A) RNA interference (RNAi) knockdown of the small heat shock protein hsp-16.48 statistically phenocopied the ethanol sensitivity of hsf-1 RNAi knockdown in comparison to control. RNAi was performed by feeding where controls were fed empty vector. n.s. = not significant. (B) Either pan-neuronal (Prab-3) or IL2-specific (Pklp-6) expression of hsp-16.48 rescued the hsf-1(sy441) mutant phenotype. Expression of an hsp-16.48 truncation mutant (+Δ38-44) was unable to rescue. (C) Overexpression of hsp-16.48 in Bristol N2 worms enhanced the ethanol stimulation of locomotion to a significantly greater extent, whereas the hsp-16.48 truncation mutant (+Δ38-44) blocked the ethanol enhancement of locomotion. For (A–C), data are expressed normalized to untreated controls. For each experiment, exposure to ethanol enhanced the locomotion rate of RNAi control or Bristol N2 worms (Mann–Whitney U-test; P < 0.05). * indicates significant difference in comparison to treated RNAi control or Bristol N2. Comparisons were made by one-way ANOVA with Tukey post hoc comparisons [P < 0.001; N = 10 (A), 30 (B) and 20 (C) for each condition]. Bristol N2 worms are depicted in black, hsf-1(sy441) in gray. Hatching indicates transgenic expression (transgene and promoter indicated below graph).
As HSF-1/HSP-16.48 was acting in a chemosensory neuron to increase locomotion, we were interested to determine whether this increase in motility was reflective of either positive or negative chemotaxis. Indeed, the performed thrashing assays immersed the animals directly into ethanol where it would be impossible to determine any directionality to movement. We tested for this possibility by a performing a chemotaxis assay on wild-type animals toward or away from ethanol. Ethanol has been previously reported to have mild to negative effects on chemotaxis only at very high concentrations (Bargmann et al. 1993; Lee et al. 2009). We found that, in our hands, ethanol did not act as either a chemoattractant or repellent whereas butanol, a longer-chain alcohol, did act as a strong attractant (Figure S2A) as has been previously described (Bargmann et al. 1993). As an alternative approach, we adapted an osmotic aversion assay (Solomon et al. 2004) to investigate whether populations of worms would be averse to crossing a ring of ethanol. We found that if the ethanol concentration of the ring was increased to 100%, we could induce avoidance in comparison to a neutral (distilled water) control; however, 17 mM (0.1%) ethanol had no effect (Figure S2B). In contrast, worms were averse to octanol at concentrations as low as 1%.
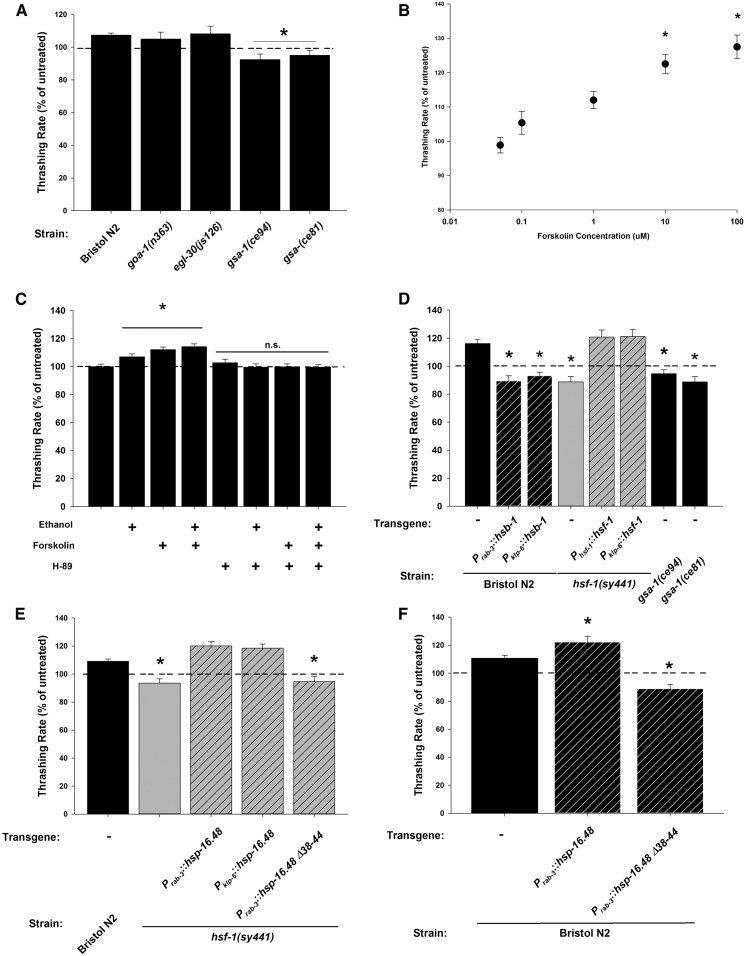
We next investigated whether the low concentration of external ethanol was instead activating the IL2 chemosensory neurons to stimulate nematode motility through a G-protein-coupled receptor (GPCR)-dependent signaling mechanism. Physiological effects of ethanol are known to be modulated by various GPCRs, for example the GABAB or G-protein-coupled corticotropin-releasing factor (CRH) 1 receptors (Nie et al. 2004; Kelm et al. 2011; Agabio and Colombo 2014). However, to screen GPCRs directly would be prohibitively difficult as there are > 1000 GPCRs in the C. elegans genome, very few of which have been functionally characterized (Frooninckx et al. 2012). Therefore, for simplicity, we instead tested available G-protein mutants. Many loss-of-function nematode G-protein mutants are lethal or have severe locomotor defects, which would interfere with quantification using our assay. Therefore, we screened gain-of-function mutants instead and determined that the stimulatory effect of 17 mM external ethanol remained intact for both Gαo (goa-1) and Gαq (egl-30) mutants, but was absent specifically for mutants of Gαs, gsa-1(ce81) and gsa-1(ce94) (Figure 4A).
Figure 4.
Ethanol enhances locomotion through an IL2 neuron Gαs-dependent signaling pathway. (A) Stimulation of locomotion by ethanol was blocked in Gαs mutants (gsa-1(ce94) and gsa-1(ce81)), but not in Gao (goa-1 (n363)) or Gaq (egl-30(js126)) mutants. (B) Forskolin-stimulated C. elegans Bristol N2 locomotion. Increasing concentrations of forskolin significantly (*) stimulated locomotion at 10 and 100 μM in comparison to untreated worms. (C) Ethanol and forskolin stimulate locomotion of Bristol N2 worms to an equivalent level and this stimulation can be blocked by addition of the PKA inhibitor H-89. * indicates significant increase from untreated. n.s = not significant. (D) Forskolin-dependent stimulation also requires IL2 neuron expression of hsf-1. The forskolin effect was absent in hsf-1(sy441) mutants but could be rescued by reexpression of wild-type hsf-1 under its endogenous (Phsf-1) or an IL2-specific (Pklp-6) promoter. Like ethanol, the forskolin effect was blocked in Bristol N2 worms by pan-neuronal (Prab-3) or IL2 neuron-specific (Pklp-6) expression of the hsf-1 inhibitor, hsb-1. The forskolin effect was also absent in Gαs mutants. (E) Forskolin-dependent stimulation also involved IL2 neuron expression of hsp-16.48. Pan-neuronal (Prab-3) or IL2-specific (Pklp-6) overexpression of hsp-16.48 rescued the absence of the forskolin phenotype in hsf-1(sy441) mutant worms. Expression of the hsp-16.48 truncation mutant (+Δ38-44) was unable to rescue. (F) Overexpression of hsp-16.48 in Bristol N2 worms enhanced the forskolin stimulation of locomotion, whereas the truncation mutant (+Δ38-44) acted in a dominant-negative fashion. For each experiment (A–F), data are expressed normalized to untreated controls. Exposure to ethanol or forskolin enhanced the locomotion rate of Bristol N2 worms (Mann–Whitney U-test; P < 0.05). * indicates significant difference in comparison to treated Bristol N2. Comparisons were made by one-way ANOVA with Tukey post hoc comparisons [P < 0.001; N = 20 (A and B), 25 (C, D, and F) or 30 (E) for each condition]. Bristol N2 worms are depicted in black, hsf-1(sy441) in gray. Hatching indicates transgenic expression (transgene and promoter indicated below graph).
Gαs is a G-protein that, upon activation, stimulates adenylyl cyclase to synthesize the cAMP signaling molecule from ATP (Dorsam and Gutkind 2007). We addressed whether we could replicate the stimulatory effect of 17 mM ethanol by applying the chemical forskolin, which directly activates adenylyl cyclase pharmacologically. A dose-response curve of varying forskolin concentrations demonstrated that the stimulatory ethanol phenotype could indeed be phenocopied by forskolin (Figure 4B). Addition of ethanol and forskolin together did not have additive effects on nematode motility (Figure 4C), suggestive that ethanol and forskolin acted in the same pathway. One downstream cellular effector for cAMP is the cAMP-dependent protein kinase PKA and we tested for its role in the stimulatory ethanol phenotype by the addition of the specific PKA inhibitor H-89. On its own, H-89 had no effect on locomotion; however, it could completely block the stimulatory effects of ethanol and forskolin, either separately or both together (Figure 4C).
Next, as previously shown for the stimulatory effects of ethanol, we verified that stimulation by forskolin was also dependent upon IL2 neuron expression of HSF-1 and HSP-16.48. In comparison to Bristol N2 wild-types, the effect of forskolin was absent in hsf-1(sy441) mutants, but was completely restored in both the full (Phsf-1::hsf-1) or IL2 neuron-specific transgenic rescues (Pklp-6::hsf-1) (Figure 4D). Complementary to these results, stimulation by forskolin was blocked by the hsf-1 inhibitor hsb-1 expressed either pan-neuronally (Prab-3::hsb-1) or in IL2 neurons specifically (Pklp-6::hsb-1) in Bristol N2 worms. As seen with ethanol, the effect of forskolin was restored in the OG532 single-copy rescue of hsf-1(sy441), but not in the OG580 DNA-binding defective rescue (Figure S1). The forskolin effect was also blocked in the Gαs gain-of-function mutants gsa-1(ce81) and gsa-1(ce94) (Figure 4D). Furthermore, pan-neuronal (Prab-3::hsp-16.48) or IL2-specific (Pklp-6::hsp-16.48) overexpression of hsp-16.48 alone could completely rescue the forskolin phenotype of the hsf-1(sy441) mutation (Figure 4E). Also identical to that seen with ethanol, overexpression of hsp-16.48 in Bristol N2 controls (Prab-3::hsp-16.48) enhanced the effect of forskolin, whereas the N-terminal truncation mutant (Prab-3::hsp-16.48 Δ38-44) appeared to act negatively (Figure 4F). The truncation mutant again had no additional negative effects when expressed in the hsf-1(sy441) background (Figure 4E). Taken together, these data indicate that both the 17 mM ethanol and forskolin stimulatory phenotypes are likely acting through the same pathway. Therefore, ethanol appears to activate a Gαs-cAMP-PKA signaling pathway that requires hsf-1 and hsp-16.48 expression within the IL2 neurons. Additionally, the requirement of HSF-1/HSP-16.48 is likely to be acting downstream of adenylyl cyclase activation as the effect of forskolin is absent in the hsf-1(sy441) mutant.
The data thus far pointed to an activation of Gαs in IL2 neurons by 17 mM external ethanol; however, our evidence was obtained by pharmacological activation of the entire nematode. We next wanted to activate Gαs in the IL2 neurons directly. To accomplish this, we genetically expressed a photoactivatable Gαs-linked jellyfish opsin, JellyOp (Bailes et al. 2012), in the IL2 neurons specifically and demonstrated that photoactivation could enhance the nematode locomotion rate to a level statistically indistinguishable from ethanol exposure (Figure 5). In contrast, control worms demonstrated no enhancement of locomotion by photostimulation; however, their ethanol effect remained intact. Importantly, photoactivation of the Gαs-linked JellyOp with the addition of ethanol did not produce a greater summative effect on locomotion rate, supporting the hypothesis that the Gαs-linked JellyOp and ethanol were acting in the same pathway (Figure 5). Next, we antagonized Gαs signaling in IL2 neurons specifically by genetic expression of a photoactivatable Gαi-linked human rod opsin (hRh1) (Cao et al. 2012; Bailes and Lucas 2013), which acts to counter Gαs by inhibiting the production of cAMP. On its own, photoactivation of the Gαi-linked hRh1 again had no effect on nematode locomotion; however, it was able to block completely the stimulatory effect of low concentrations of external ethanol (Figure 5). As an alternative approach to optogenetic activation or inhibition of the Gαs signaling pathway, we produced a transgenic worm with IL2 neuron-specific RNAi against the catalytic subunit of C. elegans PKA (Pklp-6::kin-1::Pklp-6). The created transgenic worms appeared qualitatively normal and had only a small quantitative reduction in basal locomotion rate (Table S1 in File S1). While we cannot quantify definitive cell specificity of the RNAi, if the PKA knockdown had spread greatly to other cells it would be expected to be lethal (Rual et al. 2004). Therefore, we are reasonably confident in a degree of cellular restriction of the knockdown. Despite the qualitatively wild-type appearance of the worms, the stimulation of locomotion by either ethanol or forskolin was absent (Figure S3). These data support the interpretation that the effects of ethanol to stimulate C. elegans locomotion are regulated in IL2 neurons by a Gαs signaling pathway.
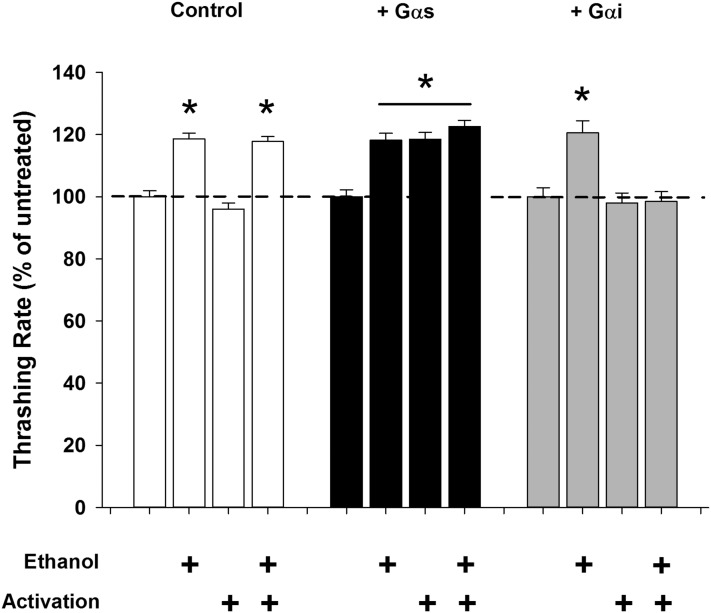
Figure 5.
Ethanol enhancement of locomotion requires Gαs protein activation in IL2 neurons. Left: in control animals (lite-1(ce314)), addition of ethanol enhanced locomotion; however, photostimulation has no stimulatory or depressive effects. Middle: in comparison, IL2-specific optogenetic activation of Gαs stimulated C. elegans locomotion to an equivalent level to low ethanol exposure (lite-1(ce314);Ex[Pklp-6::JellyOp]). Simultaneous ethanol exposure and Gαs photoactivation had no additional effect on locomotion. Right: inhibition of Gαs by IL2-specific optogenetic activation of Gαi blocked ethanol-dependent enhancement of locomotion (lite-1(ce314);Ex[Pklp-6::hRh1]). Activation of Gαi alone had no depressive effect on nematode locomotion. Addition of ethanol without Gαi activation exhibited normal locomotor enhancement. All data are expressed normalized to untreated controls. * indicates significant enhancement in comparison to untreated control. Comparisons were made by one-way ANOVA with Tukey post hoc comparisons (P < 0.001; N = 30 for each condition). Control worms (lite-1(ce314)) are depicted in white, lite-1(ce314);Ex[Pklp-6::JellyOp] in black, and lite-1(ce314);Ex[Pklp-6::hRh1] in gray.
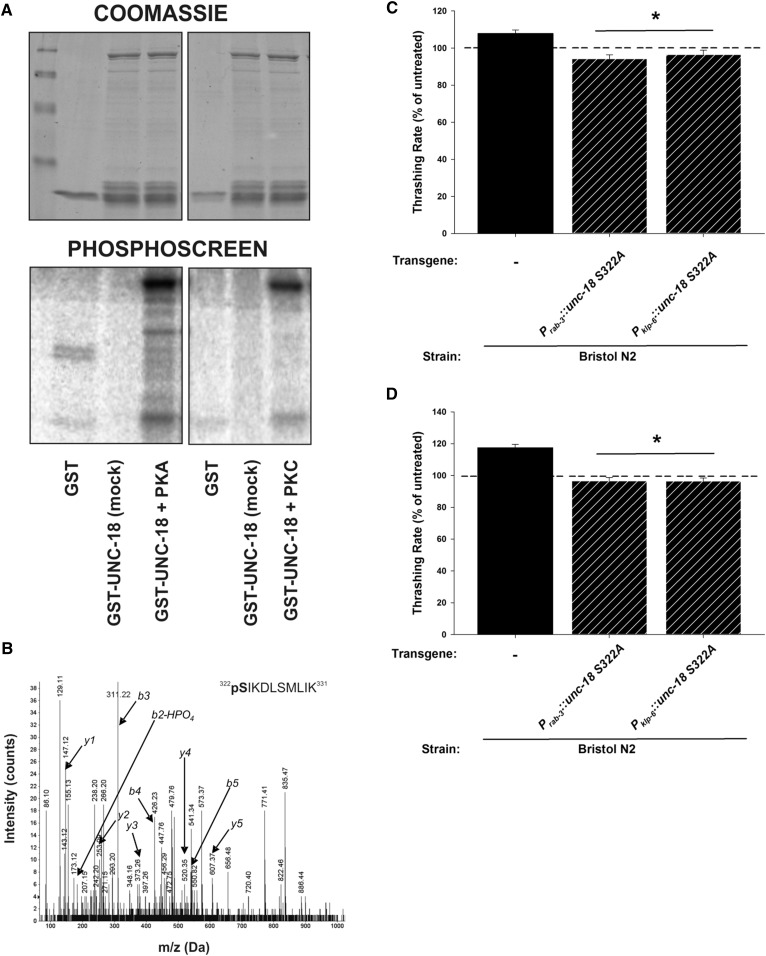
Finally we were interested in identifying what was potentially acting downstream of the Gαs-cAMP-PKA signaling pathway. PKA is a serine/threonine kinase that would be expected to phosphorylate many downstream targets in response to activation. Sec1-Munc18 (SM) proteins are essential components of the exocytotic machinery thought to function primarily through interactions with soluble N-ethylmaleimide-sensitive factor receptor proteins (Rizo and Sudhof 2012). We have previously shown that expression of a single point mutation in the C. elegans SM protein UNC-18 (D214N) was also able to block the low-ethanol phenotype (Graham et al. 2009). Therefore, we suspected that UNC-18 could be one potential downstream target for PKA phosphorylation. Both mammalian Munc18 and the nematode ortholog UNC-18 are known to be phosphorylated by Protein Kinase C (Barclay et al. 2003; Edwards et al. 2012); however, PKA phosphorylation has not been previously demonstrated. We expressed recombinant UNC-18 and exposed it to PKA, determining that it could be phosphorylated in vitro (Figure 6A). We then determined in vitro phosphorylation sites for PKA by MS, positively identifying Ser322 as an in vitro PKA target (Figure 6B). We tested the relevance of this putative phosphorylation site in vivo by expressing phospho-null versions of UNC-18 (S322A), which act in a dominant fashion to endogenous unc-18 (Edwards et al. 2012). In our assays, expression of this single point mutation of UNC-18 pan-neuronally (Punc-18::unc-18 S322A) or in IL2 neurons specifically (Pklp-6::unc-18 S322A) was able to block completely the stimulatory effects on motility of either ethanol or forskolin (Figure 6, C and D).
Figure 6.
UNC-18 is phosphorylated by protein kinase A, and phosphorylation of UNC-18 Ser322 is required for both ethanol and forskolin-dependent stimulation of locomotion. (A) Recombinant UNC-18 protein (GST-UNC-18) was incubated with 32P-labeled ATP in the absence (mock) or presence of either protein kinase C (+PKC) or protein kinase A (+PKA). Proteins were separated by SDS-PAGE and phosphorylation was determined by PhosphorImager (Phosphoscreen). Coomassie staining indicates equal protein loading. (B) Liquid chromatography-tandem mass spectrometry data positively indicating phosphorylation of Ser322 of UNC-18 in PKA-phosphorylated protein samples. (C) Pan-neuronal (Punc-18) or IL2-specific (Pklp-6) expression of a phospho-null mutation of a PKA-phosphorylation site of unc-18 (S322A) in Bristol N2 blocked the ethanol-dependent stimulation of locomotion. (D) Pan-neuronal (Punc-18) or IL2-specific (Pklp-6) expression of a phospho-null mutation of a PKA-phosphorylation site of unc-18 (S322A) in Bristol N2 blocked the forskolin-dependent stimulation of locomotion. For (C and D), data are expressed normalized to untreated controls. Exposure to ethanol or forskolin enhanced the locomotion rate of Bristol N2 worms (Mann–Whitney U-test; P < 0.05). * indicates significant difference in comparison to treated Bristol N2. Comparisons were made by one-way ANOVA with Tukey post hoc comparisons (P < 0.001; N = 30 for each condition). Bristol N2 worms are depicted in black. Hatching indicates transgenic expression (transgene and promoter indicated below graph).
Discussion
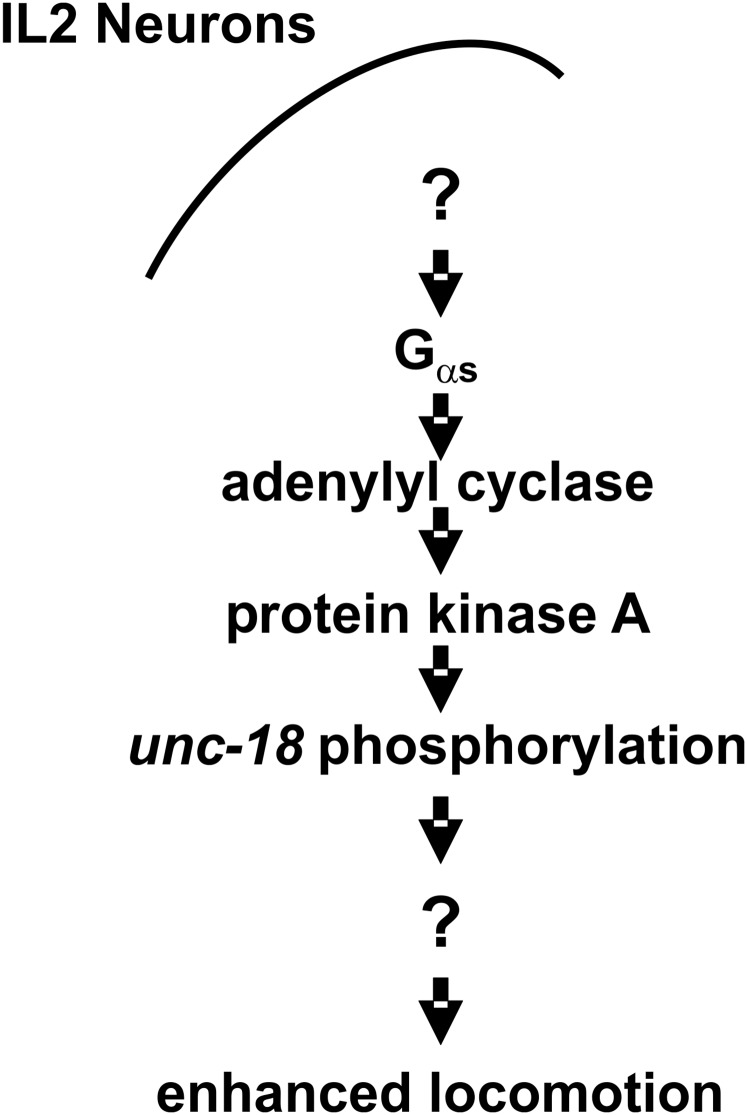
In this manuscript, we demonstrate that a Gαs-signaling pathway is stimulated by the addition of ethanol at external concentrations (17 mM; 0.1%) whose absolute values are physiologically consistent with blood alcohol limits for impaired driving. Through a combination of genetics, pharmacology, and optogenetic manipulation, we have uniquely delineated the entire cell signaling pathway in response to ethanol specifically in IL2 chemosensory neurons (Figure 7). Our model for ethanol stimulation is that ethanol, in some way, activates Gαs to stimulate adenylyl cyclase to produce cAMP. The cAMP in turn activates PKA to phosphorylate various downstream proteins, one possibility of which is the synaptic SM protein UNC-18. The ultimate downstream effect of this cellular signaling pathway is an enhancement in nematode locomotion, as quantified by thrashing rate. This cell-specific signaling pathway requires the transcription factor HSF-1, most likely through its transcriptional control of the α-crystallin ortholog, small heat shock protein HSP-16.48.
Figure 7.
A model for ethanol-dependent enhancement of locomotion in C. elegans. The addition of ethanol at a low external concentration (17 mM) activates a Gαs-dependent signaling pathway in the IL2 chemosensory neurons, likely through an as yet unidentified G-protein coupled receptor. Whether ethanol directly activates or modulates an existing signal remains to be determined. The Gαs-dependent signaling activates adenylyl cyclase and protein kinase A (PKA). PKA-dependent phosphorylation of the exocytotic protein UNC-18 at Ser322 in some way alters signaling from IL2 neurons, which feeds into the neurons controlling nematode locomotion.
Our data support the interpretation that a Gαs-signaling pathway is directly activated or modulated by ethanol. There is ample evidence in other systems for ethanol-induced activation of G-protein-coupled signaling pathways, at least in vitro. Ethanol can enhance GABA release and the extent of this enhancement can be regulated by various GPCRs, mostly via Gαi-coupled receptors (Kelm et al. 2011). Ethanol activation of GABAergic release in the central amygdala, an area of the brain prominently associated with alcohol dependence, requires the G-protein-coupled CRH 1 receptor (Nie et al. 2004). Downstream of GPCRs, ethanol can directly activate specific forms of adenylyl cyclase (Yoshimura and Tabakoff 1995; Yoshimura et al. 2006), affect cellular cAMP levels (Rex et al. 2008; Gupta et al. 2013), activate PKA (Kelm et al. 2008; Wang et al. 2015), and induce protein phosphorylation (Conti et al. 2009). Genetic or pharmacological manipulation of individual components of this pathway has measurable effects on complex alcohol dependency phenotypes, as adenylyl cyclase mutants and knockouts affect ethanol sensitivities and consumption in both mice and Drosophila (Moore et al. 1998; Maas et al. 2005; Xu et al. 2012). Indeed, the role of cAMP in the ethanol-induced stimulation of locomotion of mice was recently reported using a selective phosphodiesterase-4 inhibitor (Balino et al. 2016). Despite evidence of the Gαs-signaling pathway in other models, it has been relatively unexplored in C. elegans. In C. elegans, the CRH1 homolog seb-3 has a characterized high-alcohol phenotype; however, seb-3 is specifically expressed in head ganglia and nerve cords (Jee et al. 2013) and not sensory neurons such as the IL2 neurons. Our data use combinations of optogenetic, genetic, and pharmacological manipulation to trace the involvement of the entire Gαs -cAMP-PKA signaling pathway in the IL2 neurons in ethanol-dependent stimulation. Additionally, we extend this pathway to the downstream phosphorylation of the exocytotic protein UNC-18 and implicate HSP-16.48 as a novel regulatory protein.
Phosphorylation of synaptic proteins is a dynamic mechanism for the regulation of vesicle fusion and synaptic transmission. Many phosphorylation targets have been described for serine/threonine kinases, although mostly in vitro. Phosphorylation of Munc18 has been demonstrated in vivo, primarily by Protein Kinase C (de Vries et al. 2000; Craig et al. 2003). PKA has not previously been shown to phosphorylate Munc18 or UNC-18, despite strong evidence that it does phosphorylate other synaptic proteins such as cysteine string protein (Evans et al. 2001). Both protein kinase C and PKA phosphorylation consensus sequences have similar requirements of basic amino acids upstream of the target serine, and indeed the region upstream of Ser322 is rich in lysine residues (Edwards et al. 2012). Intriguingly, in rat pancreatic acini, PKC-dependent phosphorylation of Munc18C occurs in response to acute exposure to ethanol (Cosen-Binker et al. 2007). Physiologically, phosphorylation alters the kinetics of amperometric spikes, in particular reducing the quantal size of individual release events (Barclay et al. 2003). Munc18 phosphorylation contributes to short-term plasticity at the synapse by controlling post-tetanic potentiation (Genc et al. 2014) and UNC-18 phosphorylation contributes to thermosensory behaviors of C. elegans (Edwards et al. 2012). Biochemically, phosphorylation of this region of either UNC-18 or Munc18 reduces the binding affinity for closed-conformation syntaxin (Barclay et al. 2003; Edwards et al. 2012). However, the classically characterized closed-conformation mutation of Munc18, R39C, has no effect on the stimulatory alcohol phenotype (Johnson et al. 2013), arguing against the physiological effects of low ethanol concentrations being achieved through regulation of that particular interaction. The IL2 neurons release acetylcholine as a fast neurotransmitter (Lee et al. 2012) and connect into the main locomotor circuits via intermediary neurons (White et al. 1986). It is therefore possible that phosphorylation is altering synaptic vesicle fusion in the IL2 neurons, thereby shaping the patterns of electrical activity governing nematode motility.
Our data demonstrate that the α-crystallin ortholog HSP-16.48 acts directly downstream of HSF-1, indicating that the effects of HSF-1 in this phenotype are likely simply as a transcription factor driving constitutive HSP-16.48 expression. HSF-1 controls the expression of heat-inducible stress proteins such as the small HSPs, and HSP-16.48 specifically, as evidenced by numerous experiments (Hsu et al. 2003; Prahlad et al. 2008; Larance et al. 2011; Kourtis et al. 2012). Exposure to very high concentrations of ethanol can induce the expression of many heat shock proteins (Kwon et al. 2004; Pignataro et al. 2007; Urquhart et al. 2016) and overexpression of some HSPs can alter sensitivity to the sedative effects of ethanol (Awofala et al. 2011). In C. elegans, basal expression of hsp-16.48 alters the sedative effects of high doses of ethanol, but its overexpression only partially rescues the hsf-1(sy441) mutant phenotype (Johnson et al. 2016). In contrast, our results indicate that hsp-16.48 can completely restore the stimulatory phenotype for the hsf-1(sy441) mutant. Although there is little constitutive HSP-16.48 expressed, even in response to 400 mM ethanol, our RNAi experiments show that these small quantities are sufficient to affect whole-animal behavior. Small HSPs can be directly phosphorylated by PKA (Garrido et al. 2012), but it is currently unknown how HSP-16.48 regulates the Gαs -cAMP-PKA signaling pathway. It may be that HSP-16.48 is acting simply as a chaperone to preserve either structural conformation of individual proteins, protein–protein interactions, or possibly to assist in the phosphorylation event itself. Alternatively, HSP-16.48 may directly interact with specific components of the signaling pathway independent of its chaperone ability. Nonetheless, the correct functioning of HSP-16.48 in ethanol-dependent phenotypes explicitly required a seven amino acid region of the protein’s N-terminus, a region not found in other small HSPs linked to temperature stress tolerance (Kourtis et al. 2012), and it will be intriguing to determine the mechanistic role for this protein region.
Our experiments have used thrashing as a measure of locomotion (Gjorgjieva et al. 2014). The main rationale for this selection was the simplicity and reproducibility of the assay. It is unknown experimentally whether other measures of locomotion, such as body bends or locomotor speed, would be similarly affected; however, that speculation would be considered likely. High alcohol concentrations reduce all aspects of locomotion by similar extents including thrashing (Mitchell et al. 2007; Graham et al. 2009; Johnson et al. 2013), body bends (Johnson et al. 2016), and locomotor speed (Davies et al. 2003; Kapfhamer et al. 2008) and there is no reason to suspect differences with low alcohol concentrations. Exposure to high or low ethanol has no reported effect on reversals or ω-turns of C. elegans on agar (Mitchell et al. 2010), but high alcohol did increase spontaneous reversals in solution by ∼1 reversal per minute (Topper et al. 2014). While uncharacterized in this study, it is unlikely that a change in reversal frequency would be significant enough to influence rates as high as 110–120 thrashes per minute.
The internal concentration of ethanol in nematodes is reported to be ∼10% of external experimental levels due to poor penetration through the cuticle (Davies et al. 2003; Alaimo et al. 2012; Johnson et al. 2016). However, the structure of the IL2 neurons, with dendritic projections directly to the exterior of the body (White et al. 1986), would be in contact with the exact level of ethanol seen in solution. Therefore, it is unclear whether this low-alcohol phenotype represents a phenotypic response to 17 mM or a substantially reduced concentration. However, in either case this phenotype does indicate a neuronal cell signaling effect in C. elegans in response to ethanol at levels that would be seen in intoxicated humans. The stimulation of locomotion has been reproducibly demonstrated in C. elegans (Graham et al. 2009; Johnson et al. 2013) but is smaller than that seen in Drosophila (Wolf et al. 2002). It is difficult to determine whether there could be a further enhancement of locomotion with exposure to higher levels of ethanol as competing depressive and stimulatory effects of ethanol may begin to cancel each other out as concentration is increased. Indeed, there is no net effect of 100 mM external ethanol on locomotion rate (Davies et al. 2003; Mitchell et al. 2007; Graham et al. 2009; Johnson et al. 2016). It remains possible that the stimulatory effects of 17 mM ethanol are actually underrepresented here due to the antagonistic sedative effects of higher alcohol concentrations. Why would C. elegans increase their movement in response to 17 mM ethanol? The ecology of the species is not fully understood; however, some evidence points to C. elegans feeding on bacteria associated with decomposing material. However, unlike longer chain alcohols, evidence for ethanol acting as a chemotactic cue signaling a potential nutrient source is not well supported. Although we were unable to reproduce the results, some data point to ethanol acting as a low-level chemorepellent (Bargmann et al. 1993; Lee et al. 2009). It is therefore conceivable that the stimulus in locomotor rate is enhancing escape or avoidance of a potentially poisonous substance. Alternatively, the locomotor stimulus is merely reflective of an indirect activation of a neuronal G-protein signaling pathway that feeds into the circuitry governing nematode locomotion (Gjorgjieva et al. 2014).
Alcohol acts paradoxically as both a depressant and as a stimulant. These stimulatory effects in humans are thought to be more rewarding than sedative effects and thus may play a more prominent role in determining addiction. The differentiator model for risk to alcoholism is associated with a trade-off between stimulant and sedative effects (Hendler et al. 2013). We have shown a unique genetic link between the α-crystallin homolog HSP-16.48 and both the sedative (Johnson et al. 2016) and now also the stimulatory effects of alcohol, where an increase in HSP-16.48 expression biphasically both enhances ethanol-induced stimulation and decreases ethanol-induced sedation. Importantly, the expression of the α-crystallin chaperone is upregulated in mice strains with a high alcohol intake preference (Mulligan et al. 2006) as well as human alcohol addicts (Iwamoto et al. 2004), indicative that the phenotypic effects presented here may indeed reflect neuronal pathways and have biomedical relevance in higher organisms. This identification of an ethanol-dependent signaling pathway, from G-protein to the phosphorylation target of UNC-18, therefore presents novel targets for future pharmacological intervention that could be exploited to control the devastating physiological effects of alcohol.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.300119/-/DC1.
Acknowledgments
We thank A. Morgan (University of Liverpool) for critical comments on a previous draft of this manuscript. This work was supported by a research grant to J.W.B. from the Wellcome Trust (088779/Z/09/Z) and a grant to R.J.L. from the Biotechnology and Biological Sciences Research Council (BB//K002252/1). M.R.E. was supported by a Medical Research Council Studentship. Some strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health National Office of Research Infrastructure Program (P40 OD010440).
Footnotes
Communicating editor: P. Sengupta
Literature Cited
- Agabio R., Colombo G., 2014. GABAB receptor ligands for the treatment of alcohol use disorder: preclinical and clinical evidence. Front. Neurosci. 8: 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alaimo J. T., Davis S. J., Song S. S., Burnette C. R., Grotewiel M., et al. , 2012. Ethanol metabolism and osmolarity modify behavioral responses to ethanol in C. elegans. Alcohol. Clin. Exp. Res. 36: 1840–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anckar J., Sistonen L., 2011. Regulation of HSF1 function in the heat stress response: implications in aging and disease. Annu. Rev. Biochem. 80: 1089–1115. [DOI] [PubMed] [Google Scholar]
- Awofala A. A., Jones S., Davies J. A., 2011. The heat shock protein 26 gene is required for ethanol tolerance in Drosophila. J. Exp. Neurosci. 5: 31–44. [Google Scholar]
- Bailes H. J., Lucas R. J., 2013. Human melanopsin forms a pigment maximally sensitive to blue light (λmax ≈ 479 nm) supporting activation of G(q/11) and G(i/o) signalling cascades. Proc. Biol. Sci. 280: 20122987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailes H. J., Zhuang L. Y., Lucas R. J., 2012. Reproducible and sustained regulation of Gαs signalling using a metazoan opsin as an optogenetic tool. PLoS One 7: e30774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird N. A., Douglas P. M., Simic M. S., Grant A. R., Moresco J. J., et al. , 2014. HSF-1-mediated cytoskeletal integrity determines thermotolerance and life span. Science 346: 360–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balino P., Ledesma J. C., Aragon C. M., 2016. Role of phosphodiesterase-4 on ethanol elicited locomotion and narcosis. Neuropharmacology 101: 271–278. [DOI] [PubMed] [Google Scholar]
- Barclay J. W., Craig T. J., Fisher R. J., Ciufo L. F., Evans G. J., et al. , 2003. Phosphorylation of Munc18 by protein kinase C regulates the kinetics of exocytosis. J. Biol. Chem. 278: 10538–10545. [DOI] [PubMed] [Google Scholar]
- Barclay J. W., Graham M. E., Edwards M. R., Johnson J. R., Morgan A., et al. , 2010. Presynaptic targets for acute ethanol sensitivity. Biochem. Soc. Trans. 38: 172–176. [DOI] [PubMed] [Google Scholar]
- Bargmann C. I., Hartwieg E., Horvitz H. R., 1993. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell 74: 515–527. [DOI] [PubMed] [Google Scholar]
- Bettinger J. C., Davies A. G., 2014. The role of the BK channel in ethanol response behaviors: evidence from model organism and human studies. Front. Physiol. 5: 346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandari P., Hill J. S., Farris S. P., Costin B., Martin I., et al. , 2012. Chloride intracellular channels modulate acute ethanol behaviors in Drosophila, Caenorhabditis elegans and mice. Genes Brain Behav. 11: 387–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burket C. T., Higgins C. E., Hull L. C., Berninsone P. M., Ryder E. F., 2006. The C. elegans gene dig-1 encodes a giant member of the immunoglobulin superfamily that promotes fasciculation of neuronal processes. Dev. Biol. 299: 193–205. [DOI] [PubMed] [Google Scholar]
- Cao P., Sun W., Kramp K., Zheng M., Salom D., et al. , 2012. Light-sensitive coupling of rhodopsin and melanopsin to G(i/o) and G(q) signal transduction in Caenorhabditis elegans. FASEB J. 26: 480–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti A. C., Maas J. W., Jr, Moulder K. L., Jiang X., Dave B. A., et al. , 2009. Adenylyl cyclases 1 and 8 initiate a presynaptic homeostatic response to ethanol treatment. PLoS One 4: e5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosen-Binker L. I., Lam P. P., Binker M. G., Reeve J., Pandol S., et al. , 2007. Alcohol/cholecystokinin-evoked pancreatic acinar basolateral exocytosis is mediated by protein kinase C alpha phosphorylation of Munc18c. J. Biol. Chem. 282: 13047–13058. [DOI] [PubMed] [Google Scholar]
- Craig T. J., Evans G. J., Morgan A., 2003. Physiological regulation of Munc18/nSec1 phosphorylation on serine-313. J. Neurochem. 86: 1450–1457. [DOI] [PubMed] [Google Scholar]
- Davies A. G., Pierce-Shimomura J. T., Kim H., VanHoven M. K., Thiele T. R., et al. , 2003. A central role of the BK potassium channel in behavioral responses to ethanol in C. elegans. Cell 115: 655–666. [DOI] [PubMed] [Google Scholar]
- Devineni A. V., Heberlein U., 2013. The evolution of Drosophila melanogaster as a model for alcohol research. Annu. Rev. Neurosci. 36: 121–138. [DOI] [PubMed] [Google Scholar]
- de Vries K. J., Geijtenbeek A., Brian E. C., de Graan P. N. E., Ghijsen W., et al. , 2000. Dynamics of munc18–1 phosphorylation/dephosphorylation in rat brain nerve terminals. Eur. J. Neurosci. 12: 385–390. [DOI] [PubMed] [Google Scholar]
- Dorsam R. T., Gutkind J. S., 2007. G-protein-coupled receptors and cancer. Nat. Rev. Cancer 7: 79–94. [DOI] [PubMed] [Google Scholar]
- Edwards M. R., Johnson J. R., Rankin K., Jenkins R. E., Maguire C., et al. , 2012. PKC-2 phosphorylation of UNC-18 Ser322 in AFD neurons regulates temperature dependency of locomotion. J. Neurosci. 32: 7042–7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito G., Di Schiavi E., Bergamasco C., Bazzicalupo P., 2007. Efficient and cell specific knock-down of gene function in targeted C. elegans neurons. Gene 395: 170–176. [DOI] [PubMed] [Google Scholar]
- Evans G. J., Wilkinson M. C., Graham M. E., Turner K. M., Chamberlain L. H., et al. , 2001. Phosphorylation of cysteine string protein by PKA: implications for the modulation of exocytosis. J. Biol. Chem. 276: 47877–47885. [DOI] [PubMed] [Google Scholar]
- Frooninckx L., Van Rompay L., Temmerman L., Van Sinay E., Beets I., et al. , 2012. Neuropeptide GPCRs in C. elegans. Front. Endocrinol. 3: 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido C., Paul C., Seigneuric R., Kampinga H. H., 2012. The small heat shock proteins family: the long forgotten chaperones. Int. J. Biochem. Cell Biol. 44: 1588–1592. [DOI] [PubMed] [Google Scholar]
- Genc O., Kochubey O., Toonen R. F., Verhage M., Schneggenburger R., 2014. Munc18–1 is a dynamically regulated PKC target during short-term enhancement of transmitter release. Elife 3: e01715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gengyo-Ando K., Kitayama H., Mukaida M., Ikawa Y., 1996. A murine neural specific homolog corrects cholinergic defects in Caenorhabditis elegans unc-18 mutants. J. Neurosci. 16: 6695–6702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjorgjieva J., Biron D., Haspel G., 2014. Neurobiology of Caenorhabditis elegans locomotion: where do we stand? Bioscience 64: 476–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham M. E., Edwards M. R., Holden-Dye L., Morgan A., Burgoyne R. D., et al. , 2009. UNC-18 modulates ethanol sensitivity in Caenorhabditis elegans. Mol. Biol. Cell 20: 43–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R., Qualls-Creekmore E., Yoshimura M., 2013. Real-time monitoring of intracellular cAMP during acute ethanol exposure. Alcohol. Clin. Exp. Res. 37: 1456–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdu-Cronin Y. M., Chen W. J., Sternberg P. W., 2004. The L-type cyclin CYL-1 and the heat-shock-factor HSF-1 are required for heat-shock-induced protein expression in Caenorhabditis elegans. Genetics 168: 1937–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris R. A., Trudell J. R., Mihic S. J., 2008. Ethanol’s molecular targets. Sci. Signal. 1: re7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendler R. A., Ramchandani V. A., Gilman J., Hommer D. W., 2013. Stimulant and sedative effects of alcohol. Curr. Top. Behav. Neurosci. 13: 489–509. [DOI] [PubMed] [Google Scholar]
- House of Commons Transport Committee, 2010 Drink and drug driving law: first report of session 2010–2011. Available at: https://publications.parliament.uk/pa/cm201011/cmselect/cmtran/460/460.pdf.
- Howard R. J., Slesinger P. A., Davies D. L., Das J., Trudell J. R., et al. , 2011. Alcohol-binding sites in distinct brain proteins: the quest for atomic level resolution. Alcohol. Clin. Exp. Res. 35: 1561–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard R. J., Trudell J. R., Harris R. A., 2014. Seeking structural specificity: direct modulation of pentameric ligand-gated ion channels by alcohols and general anesthetics. Pharmacol. Rev. 66: 396–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu A.-L., Murphy C. T., Kenyon C., 2003. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science 300: 1142–1145. [DOI] [PubMed] [Google Scholar]
- Husson S. J., Gottschalk A., Leifer A. M., 2013. Optogenetic manipulation of neural activity in C. elegans: from synapse to circuits and behaviour. Biol. Cell 105: 235–250. [DOI] [PubMed] [Google Scholar]
- Iwamoto K., Bundo M., Yamamoto M., Ozawa H., Saito T., et al. , 2004. Decreased expression of NEFH and PCP4/PEP19 in the prefrontal cortex of alcoholics. Neurosci. Res. 49: 379–385. [DOI] [PubMed] [Google Scholar]
- Johnson J. R., Kashyap S., Rankin K., Barclay J. W., 2013. Rab-3 and unc-18 interactions in alcohol sensitivity are distinct from synaptic transmission. PLoS One 8: e81117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. R., Rajamanoharan D., McCue H. V., Rankin K., Barclay J. W., 2016. Small heat shock proteins are novel common determinants of alcohol and nicotine sensitivity in Caenorhabditis elegans. Genetics 202: 1013–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R. S., Ahringer J., 2003. Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30: 313–321. [DOI] [PubMed] [Google Scholar]
- Kampinga H. H., Craig E. A., 2010. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 11: 579–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapfhamer D., Bettinger J. C., Davies A. G., Eastman C. L., Smail E. A., et al. , 2008. Loss of RAB-3/A in C. elegans and the mouse affects behavioral response to ethanol. Genes Brain Behav. 7: 669–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap S. S., Johnson J. R., McCue H. V., Chen X., Edmonds M. J., et al. , 2014. Caenorhabditis elegans dnj-14, the orthologue of the DNAJC5 gene mutated in adult onset neuronal ceroid lipofuscinosis, provides a new platform for neuroprotective drug screening and identifies a SIR-2.1-independent action of resveratrol. Hum. Mol. Genet. 23: 5916–5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaun K. R., Devineni A. V., Heberlein U., 2012. Drosophila melanogaster as a model to study drug addiction. Hum. Genet. 131: 959–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelm M. K., Criswell H. E., Breese G. R., 2008. The role of protein kinase A in the ethanol-induced increase in spontaneous GABA release onto cerebellar Purkinje neurons. J. Neurophysiol. 100: 3417–3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelm M. K., Criswell H. E., Breese G. R., 2011. Ethanol-enhanced GABA release: a focus on G protein-coupled receptors. Brain Res. Brain Res. Rev. 65: 113–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo N., Katsuno M., Adachi H., Minamiyama M., Doi H., et al. , 2013. Heat shock factor-1 influences pathological lesion distribution of polyglutamine-induced neurodegeneration. Nat. Commun. 4: 1405. [DOI] [PubMed] [Google Scholar]
- Kourtis N., Nikoletopoulou V., Tavernarakis N., 2012. Small heat-shock proteins protect from heat-stroke-associated neurodegeneration. Nature 490: 213–218. [DOI] [PubMed] [Google Scholar]
- Kwon J. Y., Hong M., Choi M. S., Kang S., Duke K., et al. , 2004. Ethanol-response genes and their regulation analyzed by a microarray and comparative approach in the nematode Caenorhabditis elegans. Genomics 83: 600–614. [DOI] [PubMed] [Google Scholar]
- Larance M., Bailly A. P., Pourkarimi E., Hay R. T., Buchanan G., et al. , 2011. Stable-isotope labeling with amino acids in nematodes. Nat. Methods 8: 849–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Choi M. K., Lee D., Kim H. S., Hwang H., et al. , 2012. Nictation, a dispersal behavior of the nematode Caenorhabditis elegans, is regulated by IL2 neurons. Nat. Neurosci. 15: 107–112. [DOI] [PubMed] [Google Scholar]
- Lee J., Jee C., McIntire S. L., 2009. Ethanol preference in C. elegans. Genes Brain Behav. 8: 578–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas J. W., Jr, Indacochea R. A., Muglia L. M., Tran T. T., Vogt S. K., et al. , 2005. Calcium-stimulated adenylyl cyclases modulate ethanol-induced neurodegeneration in the neonatal brain. J. Neurosci. 25: 2376–2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C. C., Kramer J. M., Stinchcomb D., Ambros V., 1991. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10: 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendillo M. L., Santagata S., Koeva M., Bell G. W., Hu R., et al. , 2012. HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell 150: 549–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P., Mould R., Dillon J., Glautier S., Andrianakis I., et al. , 2010. A differential role for neuropeptides in acute and chronic adaptive responses to alcohol: behavioural and genetic analysis in Caenorhabditis elegans. PLoS One 5: e10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. H., Bull K., Glautier S., Hopper N. A., Holden-Dye L., et al. , 2007. The concentration-dependent effects of ethanol on Caenorhabditis elegans behaviour. Pharmacogenomics J. 7: 411–417. [DOI] [PubMed] [Google Scholar]
- Moore M. S., DeZazzo J., Luk A. Y., Tully T., Singh C. M., et al. , 1998. Ethanol intoxication in Drosophila: genetic and pharmacological evidence for regulation by the cAMP signaling pathway. Cell 93: 997–1007. [DOI] [PubMed] [Google Scholar]
- Morimoto R. I., 2011. The heat shock response: systems biology of proteotoxic stress in aging and disease. Cold Spring Harb. Symp. Quant. Biol. 76: 91–99. [DOI] [PubMed] [Google Scholar]
- Morton E. A., Lamitina T., 2013. Caenorhabditis elegans HSF-1 is an essential nuclear protein that forms stress granule-like structures following heat shock. Aging Cell 12: 112–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan M. K., Ponomarev I., Hitzemann R. J., Belknap J. K., Tabakoff B., et al. , 2006. Toward understanding the genetics of alcohol drinking through transcriptome meta-analysis. Proc. Natl. Acad. Sci. USA 103: 6368–6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z., Schweitzer P., Roberts A. J., Madamba S. G., Moore S. D., et al. , 2004. Ethanol augments GABAergic transmission in the central amygdala via CRF1 receptors. Science 303: 1512–1514. [DOI] [PubMed] [Google Scholar]
- Olsen R. W., Li G. D., Wallner M., Trudell J. R., Bertaccini E. J., et al. , 2014. Structural models of ligand-gated ion channels: sites of action for anesthetics and ethanol. Alcohol. Clin. Exp. Res. 38: 595–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden E. M., Barr M. M., 2005. The KLP-6 kinesin is required for male mating behaviors and polycystin localization in Caenorhabditis elegans. Curr. Biol. 15: 394–404. [DOI] [PubMed] [Google Scholar]
- Phillips T. J., Shen E. H., 1996. Neurochemical bases of locomotion and ethanol stimulant effects. Int. Rev. Neurobiol. 39: 243–282. [DOI] [PubMed] [Google Scholar]
- Pignataro L., Miller A. N., Ma L., Midha S., Protiva P., et al. , 2007. Alcohol regulates gene expression in neurons via activation of heat shock factor 1. J. Neurosci. 27: 12957–12966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prahlad V., Cornelius T., Morimoto R. I., 2008. Regulation of the cellular heat shock response in Caenorhabditis elegans by thermosensory neurons. Science 320: 811–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rex E. B., Rankin M. L., Ariano M. A., Sibley D. R., 2008. Ethanol regulation of D(1) dopamine receptor signaling is mediated by protein kinase C in an isozyme-specific manner. Neuropsychopharmacology 33: 2900–2911. [DOI] [PubMed] [Google Scholar]
- Rizo J., Sudhof T. C., 2012. The membrane fusion enigma: SNAREs, Sec1/Munc18 proteins, and their accomplices–guilty as charged? Annu. Rev. Cell Dev. Biol. 28: 279–308. [DOI] [PubMed] [Google Scholar]
- Rual J. F., Ceron J., Koreth J., Hao T., Nicot A. S., et al. , 2004. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 14: 2162–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satyal S. H., Chen D., Fox S. G., Kramer J. M., Morimoto R. I., 1998. Negative regulation of the heat shock transcriptional response by HSBP1. Genes Dev. 12: 1962–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon A., Bandhakavi S., Jabbar S., Shah R., Beitel G. J., et al. , 2004. Caenorhabditis elegans OSR-1 regulates behavioral and physiological responses to hyperosmotic environments. Genetics 167: 161–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speca D. J., Chihara D., Ashique A. M., Bowers M. S., Pierce-Shimomura J. T., et al. , 2010. Conserved role of unc-79 in ethanol responses in lightweight mutant mice. PLoS Genet. 6: e1001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topper S. M., Aguilar S. C., Topper V. Y., Elbel E., Pierce-Shimomura J. T., 2014. Alcohol disinhibition of behaviors in C. elegans. PLoS One 9: e92965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trudell J. R., Messing R. O., Mayfield J., Harris R. A., 2014. Alcohol dependence: molecular and behavioral evidence. Trends Pharmacol. Sci. 35: 317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urquhart K. R., Zhao Y., Baker J. A., Lu Y., Yan L., et al. , 2016. A novel heat shock protein alpha 8 (Hspa8) molecular network mediating responses to stress- and ethanol-related behaviors. Neurogenetics 17: 91–105. [DOI] [PubMed] [Google Scholar]
- Vihervaara A., Sistonen L., 2014. HSF1 at a glance. J. Cell Sci. 127: 261–266. [DOI] [PubMed] [Google Scholar]
- Wang X., Yang Z., Sun Y., Zhou H., Chu G., et al. , 2015. Ethanol activation of PKA mediates single-minded 2 expression in neuronal cells. Mol. Neurobiol. 52: 1234–1244. [DOI] [PubMed] [Google Scholar]
- White J. G., Southgate E., Thomson J. N., Brenner S., 1986. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 314: 1–340. [DOI] [PubMed] [Google Scholar]
- World Health Organization, 2011 Global status report on alcohol and health. Available at: http://www.who.int/substance_abuse/publications/global_alcohol_report/en/index.html. Accessed: October 9, 2017.
- Wolf F. W., Rodan A. R., Tsai L. T., Heberlein U., 2002. High-resolution analysis of ethanol-induced locomotor stimulation in Drosophila. J. Neurosci. 22: 11035–11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S., Chan T., Shah V., Zhang S., Pletcher S. D., et al. , 2012. The propensity for consuming ethanol in Drosophila requires rutabaga adenylyl cyclase expression within mushroom body neurons. Genes Brain Behav. 11: 727–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura M., Tabakoff B., 1995. Selective effects of ethanol on the generation of cAMP by particular members of the adenylyl cyclase family. Alcohol. Clin. Exp. Res. 19: 1435–1440. [DOI] [PubMed] [Google Scholar]
- Yoshimura M., Pearson S., Kadota Y., Gonzalez C. E., 2006. Identification of ethanol responsive domains of adenylyl cyclase. Alcohol. Clin. Exp. Res. 30: 1824–1832. [DOI] [PubMed] [Google Scholar]
- Zhang F., Bhattacharya A., Nelson J. C., Abe N., Gordon P., et al. , 2014. The LIM and POU homeobox genes ttx-3 and unc-86 act as terminal selectors in distinct cholinergic and serotonergic neuron types. Development 141: 422–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains are available upon request. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.