Abstract
DNA double-strand breaks (DSBs) are a particularly deleterious class of DNA damage that threatens genome integrity. DSBs are repaired by three pathways: nonhomologous-end joining (NHEJ), homologous recombination (HR), and single-strand annealing (SSA). Drosophila melanogaster Blm (DmBlm) is the ortholog of Saccharomyces cerevisiae SGS1 and human BLM, and has been shown to suppress crossovers in mitotic cells and repair mitotic DNA gaps via HR. To further elucidate the role of DmBlm in repair of a simple DSB, and in particular recombination mechanisms, we utilized the Direct Repeat of white (DR-white) and Direct Repeat of white with mutations (DR-white.mu) repair assays in multiple mutant allele backgrounds. DmBlm null and helicase-dead mutants both demonstrated a decrease in repair by noncrossover HR, and a concurrent increase in non-HR events, possibly including SSA, crossovers, deletions, and NHEJ, although detectable processing of the ends was not significantly impacted. Interestingly, gene conversion tract lengths of HR repair events were substantially shorter in DmBlm null but not helicase-dead mutants, compared to heterozygote controls. Using DR-white.mu, we found that, in contrast to Sgs1, DmBlm is not required for suppression of recombination between diverged sequences. Taken together, our data suggest that DmBlm helicase function plays a role in HR, and the steps that contribute to determining gene conversion tract length are helicase-independent.
Keywords: homologous recombination, homeologous recombination, gene conversion tracts, suppression of recombination between diverged sequences, Drosophila
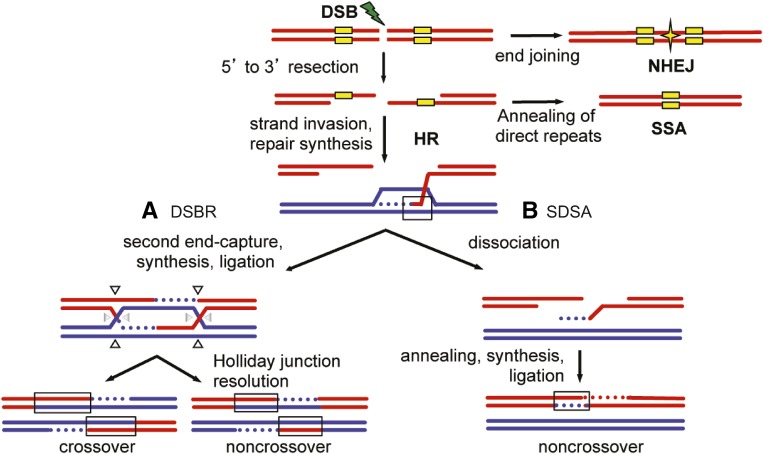
DNA double-strand breaks (DSBs) must be accurately and efficiently repaired to maintain genome integrity. DSBs can be repaired through homologous recombination (HR), nonhomologous end-joining (NHEJ), or single-strand annealing (SSA) (Heyer et al. 2010) (Figure 1). The choice between NHEJ, HR, and SSA is determined by a variety of inputs, including cell cycle regulation (Delacote and Lopez 2008; Shrivastav et al. 2008; Kass and Jasin 2010) and chromatin context (Ryu et al. 2015; Janssen et al. 2016), and can also be species- and reporter-specific (Paques and Haber 1999; Do et al. 2014). The HR repair mechanism involves utilizing a homologous donor sequence as a template for synthesis to accurately repair the DSB. HR is initiated by 5′–3′ resection to reveal single-stranded DNA 3′ overhangs, followed by Rad51-dependent strand invasion into a homologous donor sequence (Sugawara et al. 1995; McIlwraith et al. 2000). Strand invasion results in heteroduplex DNA (hDNA) and forms a D-loop to allow for nascent strand synthesis (Figure 1, black boxes). Following DNA synthesis, there are two current models that describe alternative HR mechanisms: DSB repair (DSBR) and synthesis-dependent strand annealing (SDSA) models. In the DSBR mechanism (Figure 1A), the second end is captured after synthesis to create a double Holliday Junction (dHJ) (Szostak et al. 1983). Resolution of the dHJ can result in a crossover or noncrossover product. Alternatively, the SDSA mechanism involves dissociation of the newly-synthesized strand before ligation to the other DNA end (Figure 1B) (Nassif et al. 1994). SDSA always provides a noncrossover gene conversion product and is the preferred pathway in mitotically-dividing cells (Nassif et al. 1994; Rong and Golic 2003; LaRocque and Jasin 2010), while DSBR is essential for meiotically-dividing cells to generate crossover products.
Figure 1.
Models of double-strand break repair (DSBR). Double-strand breaks (DSBs) can be repaired by homologous recombination (HR), single-strand annealing (SSA), or nonhomologous end-joining (NHEJ). In NHEJ, processed ends are joined by ligation (star). HR repair is initiated by 5′–3′ resection at the DSB. If the DSB occurs between direct repeats (yellow boxes), extensive resection followed by annealing of the direct repeats results in SSA, resulting in loss of the intervening sequence. Alternatively, the resected 3′ overhang invades the homologous template (blue) to initiate repair synthesis (blue dotted line). The invaded strand may result in heteroduplex DNA (hDNA) between the red and blue sequences (black box). DNA repair synthesis is then initiated. (A) In the DSBR model, the second strand of the DSB is captured, followed by repair synthesis, and then the newly synthesized strands are ligated to form a double Holliday junction (dHJ). Depending on how the dHJ is cleaved (arrow heads), resolution can result in a crossover or a noncrossover. (B) In synthesis-dependent strand annealing (SDSA), the newly synthesized strand dissociates, anneals to the other end, the gap is filled in, and nicks ligated to result in a noncrossover product. The newly synthesized strands in both DSBR and SDSA also form hDNA. hDNA in these later HR intermediates can be repaired by mismatch repair, resulting in gene conversion (data not shown). Direct repeats are shown only for SSA for simplicity.
Human BLM is one of five members of the evolutionarily-conserved RecQ helicase family and is important for facilitating accurate DSB repair. Mutations in human BLM result in Bloom syndrome (BS), which can cause cancer susceptibility, sterility, immunodeficiency, dwarfism, and premature aging (Bloom 1954; Ellis et al. 2008). Furthermore, BS is characterized at the cellular level by chromosomal instability leading to an elevated rate of sister chromatid exchanges and vast structural rearrangements (German et al. 1965; Chaganti et al. 1974). Mechanistically, previous studies have demonstrated BLM to have a role in DSB repair pathway choice (Grabarz et al. 2013), single-strand DNA resection (Gravel et al. 2008; Nimonkar et al. 2011), D-loop dissociation (Bachrati et al. 2006), dHJ branch migration (Karow et al. 2000), and dissolution (Wu and Hickson 2003; Wu et al. 2006). Taken together, it is evident that human BLM is involved in multiple components of both the DSBR and SDSA HR mechanisms.
Human BLM has also been shown to directly interact with mismatch repair (MMR) proteins MLH1 (Langland et al. 2001) and MSH6 (Pedrazzi et al. 2003; Yang et al. 2004). MMR machinery is important for maintaining genome stability by suppressing recombination of diverged sequences (i.e., homeologous recombination), and repairing nucleotide mismatches in hDNA (Figure 1, black boxes) [reviewed in Spies and Fishel (2015)]. Both functions are initiated by highly conserved MSH2–MSH6 heterodimers that locate and bind to nucleotide mismatches in hDNA generated from strand invasion (Drummond et al. 1995). Studies in Saccharomyces cerevisiae have suggested that if the MMR machinery rejects the hDNA, Sgs1, the sole RecQ helicase in S. cerevisiae, unwinds the DNA strands (Sugawara et al. 2004). Additionally, similar observations between bacterial and yeast systems suggest that the Msh2–Msh3 heterodimer and Sgs1 may reject the hDNA following nascent strand annealing (Spies and Fishel 2015).
Sgs1 facilitates suppression of recombination between diverged sequences in S. cerevisiae (Myung et al. 2001), however it remains unclear whether this functionality is conserved in other organisms. Similar to Sgs1 (Sugawara et al. 2004), Blm was demonstrated to facilitate suppression of SSA between diverged sequences in Drosophila (Kappeler et al. 2008). However, BLM-deficient murine cells and BS cells sufficiently suppress recombination between sequences with divergence up to 1.5% (LaRocque and Jasin 2010). Interestingly, BS cells fail to suppress recombination when the sequence divergence is increased to 19% (Wang et al. 2016).
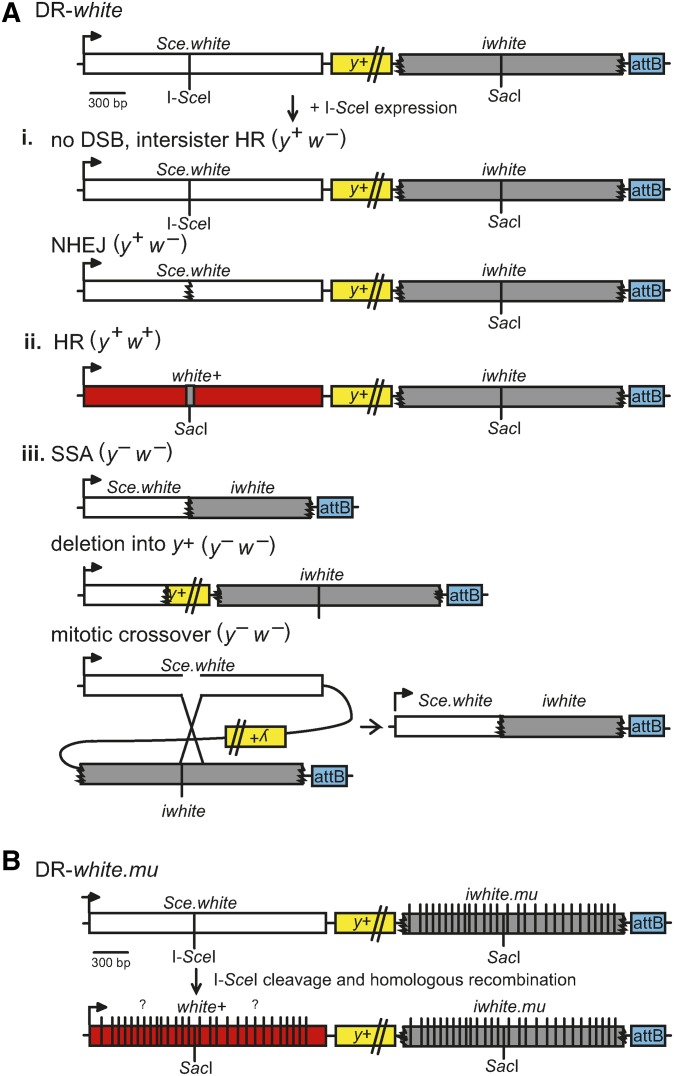
Drosophila has orthologs of four of the five human RecQ helicases: DmBlm, WRNexo, DmRecQ4, and DmRecQ5 (Supplemental Material, Figure S1 in File S1). DmBlm functionality is largely conserved across species, thus is highly involved in DSB repair by HR (Kusano et al. 1999, 2001; Adams et al. 2003; McVey et al. 2007). In this study, we used the Direct Repeat of white (DR-white) assay to determine how DmBlm functions in DSB repair pathway usage by measuring the relative frequencies of noncrossover HR, SSA, and NHEJ repair outcomes of an I-SceI-induced DSB in both null and helicase-dead DmBlm mutants (Figure 2A) (Do et al. 2014). The Direct Repeat of white with mutations (DR-white.mu) assay, which contains 28 silent nucleotide polymorphisms (SNPs) in the donor sequence increasing sequence divergence by 1.4% (Figure 2B), was used to analyze the structures of gene conversion tracts (GCTs) and determine if DmBlm plays a role in suppressing recombination between diverged sequences in multicellular systems (Do and LaRocque 2015). Our results further elucidate the role of DmBlm in multiple aspects of HR repair, and how impaired recombination mechanisms may impact the length of GCTs.
Figure 2.
DR-white and DR-white.mu DSB repair assays. (A) The DR-white assay contains two nonfunctional direct repeats of the white gene. The first repeat, Sce.white, is nonfunctional due to the insertion of an I-SceI recognition sequence into the wild-type SacI recognition sequence of white cDNA resulting in a defective white gene. The second repeat, iwhite, is nonfunctional due to 5′ and 3′ truncations and serves as a homologous donor sequence for repair. DR-white is targeted using the attB sequence (blue) and integration is confirmed using yellow (y+) transgene expression. DR-white flies are crossed with flies containing an I-SceI transgene, in which expression results in DSB formation at the I-SceI recognition sequence. Repair events are observed by crossing these males to y w tester females; progeny of this cross represent single DSB repair events of the male germline. One of three phenotypes will result depending on the repair. (i) White-eyed progeny (y+ w−) suggest no DSB, intersister HR, or repair by NHEJ with processing, resulting in loss of the I-SceI recognition sequence. NHEJ with processing can be identified through molecular analysis. (ii) Repair by HR results in restoration of the wild-type SacI site from the iwhite donor sequence and a red-eyed fly (y+ w+). (iii) Yellow-bodied, white-eyed (y− w−) progeny indicates repair by SSA, mitotic crossover event (indistinguishable from SSA), or an aberrant repair event that impedes y+ expression, such as a deletion into the y+ transgene. (B) The DR-white.mu assay includes the incorporation of 28 silent polymorphisms on the iwhite.mu donor sequence, resulting in a 1.4% increase of sequence divergence between the two direct repeats. HR gene conversion of each of the polymorphisms varies from one repair product to the next (indicated by “?”), and can be determined by molecular analyses. DR-white; Direct Repeat of white; DR-white.mu; Direct Repeat of white with mutations; DSB, double-strand break; HR, homologous recombination; NHEJ, nonhomologous end-joining; SSA, single-strand annealing.
Materials and Methods
Drosophila stocks and maintenance
Drosophila were maintained on standard Nutri-fly Bloomington Formulation medium (Genesee Scientific, San Diego, CA) at 25°. DR-white and DR-white.mu transgenic stocks were previously described (Do et al. 2014; Do and LaRocque 2015; Delabaere et al. 2016) and are available upon request. I-SceI transgenic stocks included either a Drosophila Ubiquitin promoter for constitutive expression (Preston et al. 2006) or Drosophila hsp70 promoter for heat shock induction (Rong and Golic 2000; Wei and Rong 2007). Blm mutant stocks were as previously described: DmBlmN1 null allele (McVey et al. 2007), DmBlmD2 null allele (Boyd et al. 1981), and DmBlmD3 helicase-dead allele (Boyd et al. 1981).
DSB repair assay
To induce DSBs, females heterozygous for BlmD2 containing DR-white or DR-white.mu were crossed to males heterozygous for BlmN1 or BlmD3 containing either the heat-inducible or the constitutively-active I-SceI transgene. After 3 days, flies were removed. For the heat shock-inducible I-SceI transgene, 0–3-day-old embryos were then heat-shocked in a 38° water bath for 1 hr. Single F1 males of either heterozygote (balanced with TM6B), null mutant, or helicase-dead mutant Blm status containing both DR-white (or DR-white.mu) and heat-inducible (or constitutively-active) I-SceI transgene were crossed to five to eight y w females in vials. For each experiment, F2 progeny from 31 to 125 individual F1 male germlines were scored for phenotypes, as described in Figure 2 and summarized in Table S1 in File S1. For suppression of recombination between diverged sequences analysis, only the experiments in which DR-white and DR-white.mu assays were performed side-by-side were included for comparison (see Table S1 in File S1). All data from individual male germlines from these DR-white.mu experiments were combined by genotype to determine HR frequencies relative to DR-white.
Molecular analyses
For molecular analyses, one or two F2 progeny from each male germline were analyzed to avoid frequency biases attributable to potential germline jackpot events (Luria and Delbruck 1943). Genomic DNA was isolated from individual flies using 50 μl Squishing Buffer (10 mM Tris-Cl, 25 mM NaCl, and 1 mM EDTA) and Proteinase K (0.2 mg/ml). Samples were incubated at 37° for 30 m, followed by inactivation of Proteinase K by heating to 95° for 2 min (Gloor et al. 1993). To determine the proportion of “NHEJ with processing” events in y+ w− flies, Sce.white was PCR amplified using Sce.white-specific primers (forward, DR-white1, 5′-GTGTGAAAAATCCCGGCA-3′; reverse, DR-white2a 5′-TGGCAACCATCGTTGTCTG-3′) and SapphireAmp Fast PCR Master Mix (Clontech, Mountain View, CA). Sce.white PCR products were directly digested with I-SceI restriction enzyme (New England Biolabs, Beverly, MA) and visualized on a 1% agarose gel. PCR products that failed to cleave with I-SceI restriction enzyme were classified as NHEJ with processing and sent for sequencing using the DR-white2 primer (5′-ATGCAGGCCAGGTGCGCCTATG-3′) (Genewiz, South Plainfield, NJ) to determine the sequence of repair junctions including the presence of microhomologies. Sequences were analyzed using 4Peaks software (Nucleobytes, Aalsmeer, The Netherlands).
GCT analyses
Sce.white fragments from DR-white.mu HR events (y+ w+) were amplified with DR-white1 and DR-white1a (5′-AGACCCACGTAGTCCAGC-3′) as described above, and Sce.white PCR products were directly sequenced (Genewiz) with primers DR-white 2, DR-white 2a, DR-white1, or DR-white1.3 (5′-GTTTTGGGTGGGTAAGCAGG-3′) and DR-white1a to detect incorporations of any of the 28 silent polymorphisms from the iwhite donor sequence. Sequences were analyzed using 4Peaks software (Nucleobytes, Aalsmeer, The Netherlands).
Data availability
All strains and reporter constructs used in this study are available upon request. Table S1 in File S1 contains the raw data from experiments presented in Figure 3, Figure 5, and Figure S2 and Figure S5 in File S1. Table S2 in File S1 contains raw data presented in the Results. Figure S3 in File S1 contains the individual GCTs that are presented in Figure 4.
Figure 3.
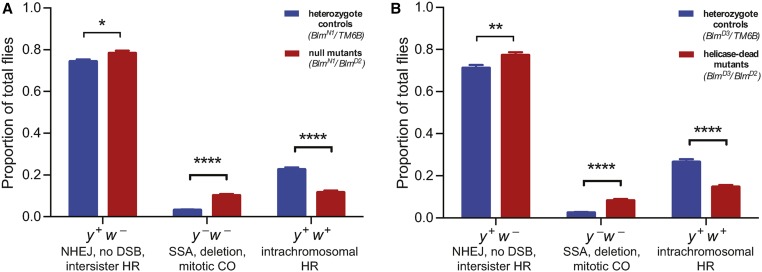
DmBlm impacts DSB repair pathway usage. (A) I-SceI heat shock-induced DSB repair events in a DmBlmN1/D2 null mutant background (red; n = 94) compared to DmBlmN1 heterozygote controls (blue; n = 125). Results shown are averages and SEM of individual male germline events compiled from five independent experiments. * P < 0.05 and **** P < 0.0001 by unpaired Student’s t-test. (B) I-SceI heat shock-induced DSB repair events in a DmBlmD3/D2 helicase-dead mutant background (red; n =52) compared to DmBlmD3 heterozygote controls (blue; n = 59). Results shown are averages and SEM of individual male germline events compiled from three independent experiments. ** P < 0.01 and **** P < 0.0001 by unpaired Student’s t-test. CO, crossover; DSB, double-strand break; HR, homologous recombination; NHEJ, nonhomologous end-joining; SSA, single-strand annealing.
Figure 5.
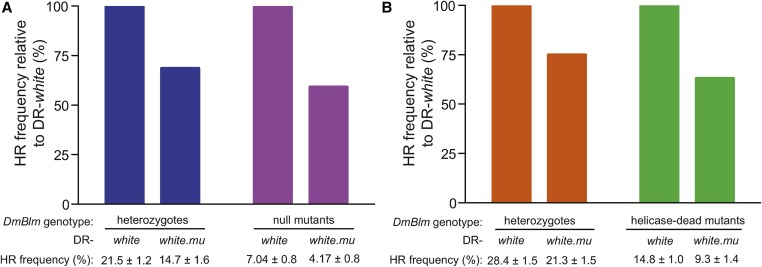
Relative recombination frequencies between homologous and diverged sequences are unaltered in DmBlm mutants. Relative recombination frequencies between homologous and diverged sequences were determined using DR-white and DR-white.mu, respectively, in (A) DmBlmN1 heterozygote controls and DmBlmN1/D2 null mutants, and (B) DmBlmD3 heterozygote controls and DmBlmD3/D2 helicase-dead mutants. Average HR frequencies (with SEM) were compiled from individual germlines from two independent experiments and are presented below the graphs (and in Table S1 in File S1). Average HR frequencies relative to DR-white are plotted. DR-white; Direct Repeat of white; DR-white.mu; Direct Repeat of white with mutations; HR, homologous recombination.
Figure 4.
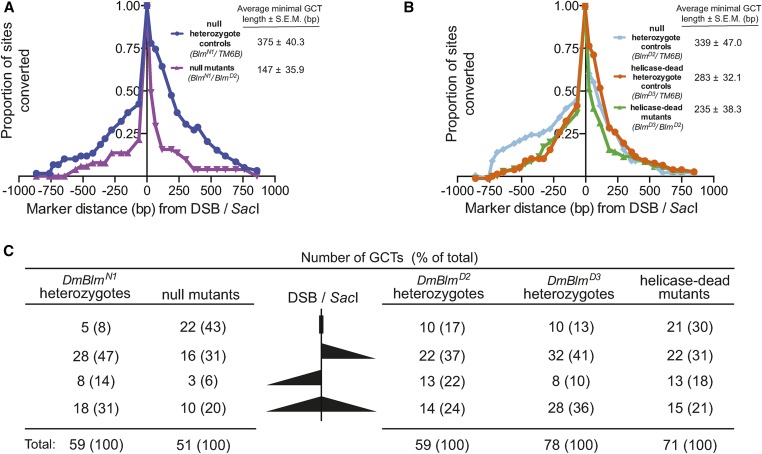
DmBlm impacts gene conversion tract length independently of helicase function. Intrachromosomal noncrossover HR events using the DR-white.mu assay were isolated and the GCT direction and length were determined. (A) The proportions of SNP sites converted are displayed for DmBlmN1 heterozygote control HR events (blue; n = 59) and DmBlmN1/D2 null mutant HR events (purple; n = 51). The average distance converted and SEM (base pair) to the left and to the right of the SacI site/DSB (0) is given for both genotypes. Data are from two independent experiments. (B) The proportions of SNP sites converted are displayed for DmBlmD2 heterozygote control HR events (light blue; n = 59), DmBlmD3 heterozygote control HR events (orange; n = 78) and DmBlmD3/D2 helicase-dead mutant HR events (green; n = 71). The average distance and SEM converted (base pair) to the left and to the right of the SacI site/DSB (0) is given for both genotypes. Data are from four independent experiments. (C) Classes of gene conversion tracts from combined DR-white.mu assay experiments in DmBlmN1/D2 null mutants (left) and DmBlmD3/D2 helicase-dead mutants (right) and the respective heterozygote control. Each tract was grouped into one of four classes (represented graphically in descending order): conversion of only the DSB/SacI site, conversion to the right of the DSB/SacI site (> 32 bp; unidirectional), conversion to right of the DSB/SacI site (> 63 bp; unidirectional), and conversion to both sides of the break (bidirectional). DR-white.mu; Direct Repeat of white with mutations; DSB, double-strand break; GCT, gene conversion tract; HR, homologous recombination; SNP, single nucleotide polymorphism.
Results
DmBlm is involved in accurate intrachromosomal HR repair of a single break
The DR-white reporter determines the relative distribution of three phenotypes that represent one or more DSB repair events (Do et al. 2014). Briefly, DSBs are induced using an I-SceI enzyme that cleaves at the I-SceI recognition sequence of Sce.white. After I-SceI expression, cleavage, and DSB repair, males are crossed to tester females to isolate and measure the relative frequencies of individual repair products in germline cells. No DSB formation, repair by intersister HR, or repair by NHEJ result in brown-bodied and white-eyed (y+ w−) progeny (Figure 2A, i). NHEJ with processing and microhomology-mediated end joining (MMEJ) can be detected from this group by molecular analysis at the I-SceI site; loss of the I-SceI recognition sequence suggests repair by NHEJ with processing (Figure 2A, i) and annealing of short homologous sequences suggests MMEJ (McVey and Lee 2008). Accurate repair by intrachromosomal noncrossover HR restores the wild-type SacI site and w+ expression, resulting in brown-bodied, red-eyed (y+ w+) progeny (Figure 2A, ii). NHEJ with processing leading to a perfect deletion of the 23 bp I-SceI recognition sequence could result in a wild-type SacI sequence and y+ w+ progeny. However, NHEJ with processing (this study, Table S2 in File S1), particularly deletions > 10 bp, are exceptionally rare (Do et al. 2014), suggesting that the vast majority of y+ w+ events are due to noncrossover HR. Yellow-bodied, white-eyed (y− w−) progeny may result from loss of y + transgene expression by extensive end resection followed by SSA of the direct repeat sequence, a mitotic crossover event, or any aberrant repair event in which the y+ transgene expression is lost, such as a deletion (Figure 2A, iii).
To elucidate the impact of DmBlm on DSB repair pathway distribution, we tested hetero-allelic DmBlmN1/D2 null mutants with the DR-white assay and I-SceI expression by heat shock. DmBlmN1 is characterized as a 2480 bp deletion that includes the start codon (McVey et al. 2007), and DmBlmD2 is a nonsense mutation (Kusano et al. 2001). Both the DmBlmN1 and DmBlmD2 alleles are characterized as genetic nulls (McVey et al. 2007). In accordance with trends reported with the Rr3 DSB repair assay in a previous study (Johnson-Schlitz et al. 2007), we measured a ∼50% decrease of noncrossover HR repair events from 22.6% in DmBlmN1 heterozygote controls to 11.7% in DmBlmN1/D2 null mutants (P < 10−12, Student’s t-test) (Figure 3A). Furthermore, there was a threefold increase of y− w− flies from 3.1% in the DmBlmN1 heterozygotes to 10.1% in the DmBlmN1/D2 null mutants (P < 10−13, Student’s t-test) (Figure 3A). Notably, because the study by Johnson-Schlitz et al. (2007) utilized a constitutively-active I-SceI transgene with their Rr3 DSB repair assay, we also tested the constitutively-active I-SceI transgene in DR-white and observed a similar decrease in noncrossover HR and a compensatory increase in y− w− (Figure S2 in File S1). Finally, we detected a relatively small, yet significant, increase in the frequency of the y+ w− class from 74.3% in DmBlmN1 heterozygotes to 78.3% in DmBlmN1/D2 null mutants (P < 0.05, Student’s t-test) (Figure 3A). Within this y+ w− pool, 8.1% of the repair events from DmBlmN1 heterozygotes were due to NHEJ with processing, which was not significantly different from the 5.9% NHEJ with processing repair events in DmBlmN1/D2 null mutants (P = 0.6, Fisher’s exact test) (Table S2 in File S1). Also within the NHEJ with processing events, there were no significant differences in the proportion of events with microhomologies between DmBlmN1 heterozygotes (23/38 NHEJ events contained microhomologies) and DmBlmN1/D2 null mutants (22/34 NHEJ events contained microhomologies) (P = 0.8, Fisher’s exact test) (Table S2 in File S1).
DSB repair pathway distribution is DmBlm helicase-dependent
To specifically test the DmBlm helicase enzymatic function in our system, we used the DR-white assay with DmBlmD2/D3 helicase-dead mutants. The DmBlmD3 allele is characterized as a missense mutation (Kusano et al. 2001) that alters a critical motif for nucleotide cofactor binding and hydrolysis, thus encoding DmBlm protein with impaired helicase function (McVey et al. 2007). Similar to the experiments using null mutants, the noncrossover HR frequency decreased by ∼50% from 26.5% in DmBlmD3 heterozygote controls to 14.6% in DmBlmD2/D3 helicase-dead mutants (P < 10−9, Student’s t-test). Furthermore, the frequency of the y− w− class increased threefold from 2.4% in DmBlmD3 heterozygotes to 8.2% in DmBlmD2/D3 helicase-dead mutants (P < 10−7, Student’s t-test), and the frequency of y+ w− flies slightly increased from 71.1% in DmBlmD3 heterozygotes to 77.2% in DmBlmD2/D3 helicase-dead mutants (P < 0.01, Student’s t-test) (Figure 3B). Within the y+ w− pool, 11.3% of the repair events from DmBlmD3 heterozygotes were due to NHEJ with processing, which was not significantly different from the 8.1% NHEJ with processing repair events in DmBlmD2/D3 helicase-dead mutants (P = 0.6, Fisher’s exact test) (Table S2 in File S1). These results indicate the role of DmBlm in multiple DSB repair pathway distributions to be, at least partially, helicase-dependent.
GCTs are significantly shorter in DmBlm null but not helicase-dead mutants
Gene conversion results from a noncrossover HR repair event and can have significant impacts on genome stability and evolution [reviewed in Chen et al. (2007)]. To further investigate the noncrossover HR deficiencies observed in DmBlm mutants, we quantified HR GCT lengths of DmBlm null and helicase-dead mutants by molecular analysis using the DR-white.mu assay (Do et al. 2014). By sequencing the recipient fragment, Sce.white, we were able to determine the structure and length of GCTs based on the presence or absence of SNPs from the donor sequence, iwhite.mu (Figure 2B).
We determined DmBlmN1 heterozygotes to have a mean GCT length of 374.8 ± 40.3 bp, and this dropped substantially to 146.9 ± 35.9 bp in DmBlmN1/D2 null mutants (P < 10−4, Student’s t-test) (Figure 4A and Figure S3 in File S1). Moreover, of all DmBlmN1 heterozygote GCTs analyzed (n = 59), five (8.5%) of the GCTs were limited to the SacI site, 36 (61.0%) were unidirectional, and 18 (30.5%) were bidirectional (Figure 4C and Figure S3 in File S1). In contrast, of all DmBlmN1/D2 null mutant GCTs analyzed (n = 51), 22 (43.1%) of the GCTs were limited to the SacI site, 19 (37.3%) were unidirectional, and 10 (19.6%) were bidirectional (Figure 4C and Figure S3 in File S1). Interestingly, while the proportion of bidirectional GCTs does not differ between DmBlmN1 heterozygotes and DmBlmN1/D2 null mutants (P = 0.5, Fisher’s exact test), the two genotypes significantly differ in the proportion of GCTs limited to the SacI site (P < 0.001, Fisher’s exact test) and unidirectional GCTs (P < 0.001, Fisher’s exact test). Two (3.4%) discontinuous tracts were observed in DmBlmN1 heterozygote controls and four (7.8%) were observed in DmBlmN1/D2 null mutant flies (P = 0.4, Fisher’s exact test) (Figure S3 in File S1). Finally, neither DmBlmN1 heterozygote (P = 0.1, Student’s t-test) nor DmBlmN1/D2 null mutant GCTs (P = 0.7, Student’s t-test) exhibited significant biases in either direction of the SacI site (Figure S4 in File S1).
To test whether the apparent effects of DmBlm on GCT length are helicase-dependent, we also analyzed GCTs from DmBlm helicase-dead mutants. Interestingly, the mean GCT length for DmBlmD2 heterozygote controls was longer than the DmBlmD3/D2 helicase-dead mutants (338.7 ± 47.0 bp vs. 235.8 ± 38.3 bp), but the difference did not reach significance (P = 0.09) (Figure 4B and Figure S3 in File S1). Similarly, the mean GCT length in DmBlmD3 heterozygote controls was 283.4 ± 32.1 bp, which did not significantly differ from that of DmBlmD3/D2 helicase-dead mutants, 235.8 ± 38.3 bp (P = 0.3, Student’s t-test) (Figure 4B and Figure S3 in File S1). These results suggest that the decrease in GCT lengths in DmBlm mutants is mostly helicase-independent.
Of all DmBlmD2 heterozygote GCTs analyzed (n = 59), 10 (16.9%) of the GCTs were limited to the SacI site, 35 (59.3%) were unidirectional, and 14 (23.7%) were bidirectional (Figure 4C and Figure S3 in File S1). Of all DmBlmD3 heterozygote GCTs analyzed (n = 78), 10 (12.8%) of the GCTs were limited to the SacI site, 40 (51.3%) were unidirectional, and 28 (35.9%) were bidirectional (Figure 4C and Figure S3 in File S1). Of all DmBlmD3/D2 helicase-dead mutant GCTs analyzed (n = 71), 21 (29.6%) of the GCTs were limited to the SacI site, 35 (49.3%) were unidirectional, and 15 (21.1%) were bidirectional (Figure 4C and Figure S3 in File S1). Between DmBlmD2 heterozygotes and DmBlmD3/D2 helicase-dead mutants, the proportions of bidirectional (P = 0.8, Fisher’s exact test) and unidirectional (P = 0.3, Fisher’s exact test) GCTs did not differ. Similarly, between DmBlmD3 heterozygotes and DmBlmD3/D2 helicase-dead mutants, the proportions of bidirectional (P = 0.2, Fisher’s exact test) and unidirectional (P = 0.6, Fisher’s exact test) GCTs did not differ. However, the DmBlmD3 heterozygotes and DmBlmD3/D2 helicase-dead mutants did significantly differ in the proportion of GCTs limited to the SacI site (P < 0.05, Fisher’s exact test), although there was no statistical different between DmBlmD2 heterozygotes and DmBlmD3/D2 helicase-dead mutants (P = 0.1, Fisher’s exact test). One (1.7%) and four (5.1%) discontinuous tracts were observed in DmBlmD2 and DmBlmD3 heterozygote controls, respectively. Two (2.8%) discontinuous tracts were observed in DmBlmD3/D2 helicase-dead mutants (P = 1.0 and 0.7, Fisher’s exact test comparing D2 heterozygotes to helicase-dead mutants and D3 heterozygotes to helicase-dead mutants, respectively) (Figure S3 in File S1). Finally, similar to the DmBlm null mutant experiment, neither DmBlmD2 heterozygote (P = 0.1, Student’s t-test), DmBlmD3 heterozygote (P = 0.1, Student’s t-test), nor DmBlmD3/D2 helicase-dead mutant GCTs (P = 0.9, Student’s t-test) exhibited significant biases in either direction of the SacI site (Figure S4 in File S1).
DmBlm mutants suppress noncrossover recombination between diverged sequences
A previous study found that DmBlmD2/D3 helicase-dead mutants failed to suppress SSA between diverged sequences, suggesting a function for the DmBlm helicase in SSA hDNA rejection (Kappeler et al. 2008). With the DR-white and DR-white.mu assays, we tested suppression of noncrossover HR between diverged sequences by directly measuring the proportion of HR between homologous (DR-white) and diverged (DR-white.mu) sequences. Our earlier study demonstrated that wild-type DmBlm+/+ flies suppressed HR frequency measured with DR-white.mu by 31.5% relative the HR frequency measured with DR-white (Do et al. 2014). Furthermore, msh6 mutants failed to suppress recombination with the DR-white.mu assay relative to that with DR-white, suggesting this effect to be MMR machinery-dependent (Do and LaRocque 2015).
To determine if DmBlm suppresses recombination of diverged sequences similar to S. cerevisiae Sgs1 (Myung et al. 2001) and Drosophila Msh6, we tested these effects in DmBlmN1/D2 null mutants. Nearly identical to DmBlm+/+ flies, the noncrossover recombination frequency of DmBlmN1 heterozygote controls with diverged sequences was suppressed by 31.5% relative to HR with homologous sequences (21.5 ± 1.2% HR with DR-white and 14.7 ± 1.6% HR with DR-white.mu; P < 0.001, Student’s t-test) (Figure 5A). Similarly, DmBlmN1/D2 null mutants suppressed noncrossover HR with diverged sequences by 40.8% compared to HR with homologous sequences (7.0 ± 0.8% HR with DR-white and 4.2 ± 0.8% HR with DR-white.mu; P < 0.05, Student’s t-test) (Figure 5A). The relative frequencies of the other phenotypic classes were also consistent regardless of DmBlm status or the presence of a homologous or diverged donor sequence (Figure S5 in File S1).
Since the previously mentioned study by Kappeler et al. (2008) tested DmBlmD3/D2 helicase-dead mutants, we repeated this assay in a DmBlmD3/D2 helicase-dead genetic background. The recombination frequency in DmBlmD3 heterozygote controls with diverged sequences was suppressed by 25.1% relative to HR with homologous sequences (28.4 ± 1.5% HR with DR-white and 21.3 ± 1.5% HR with DR-white.mu; P < 0.01, Student’s t-test) (Figure 5B). Similar to DmBlmN1/D2 null mutants, DmBlmD3/D2 helicase-dead mutants also suppressed noncrossover recombination frequency by 36.9% relative to HR with homologous sequences (14.8 ± 1.0% HR with DR-white and 9.3 ± 1.4% HR with DR-white.mu; P < 0.01, Student’s t-test) (Figure 5B).
Discussion
DmBlm impacts DSB repair pathway distribution by its involvement in HR
DmBlm promotes efficient and accurate repair of DNA gaps by the SDSA HR pathway (Adams et al. 2003). Johnson-Schlitz et al. (2007) previously found that DmBlm mutants suppress interhomolog HR and compensate by increasing SSA repair pathway usage in their Rr3 DSB repair assay. In this current study, we utilize the DR-white assay to demonstrate consistent results with a DSB detector assay that measures intrachromosomal noncrossover HR repair. Furthermore, DmBlm null and helicase-dead mutants both exhibited a threefold increase of y− w− flies, which may be explained by a shift from noncrossover HR to crossover HR, as observed by the increase in crossovers and flanking deletions associated with DmBlm deficiencies (Johnson-Schlitz and Engels 2006; McVey et al. 2007), although these events cannot be precisely identified molecularly in our assay (see Figure 2A, iii).
The statistically significant increase in y+ w− repair events in DmBlm mutants could be due to a shift from noncrossover HR to NHEJ events. However, neither the relative proportion of NHEJ with processing events nor NHEJ with microhomology events differ between DmBlm null and helicase-dead mutants and the respective heterozygote controls (Table S2 in File S1). Given the sparse evidence connecting DmBlm functionally to the NHEJ pathway, this increase is more likely due to cell death of failed intrachromosomal HR repair events in the germline, thus causing a proportional increase in this class (y+ w−). It is also possible that the increase in y+ w− flies reflects a shift from intrachromosomal noncrossover HR to intersister HR using the Sce.white sequence as a donor sequence. However, these events cannot be molecularly distinguished from “no DSB” events, which retain the I-SceI recognition sequence. Overall, these data demonstrate the ability of DR-white to simultaneously capture multiple types of repair events and identify compensatory pathway shifts in mutant backgrounds, as observed previously in mutants required for HR (e.g., DmRad51) (Do et al. 2014).
In general, DmBlmN1/D2 null and DmBlmD3/D2 helicase-dead genetic backgrounds had similar effects on DSB repair pathway usage detected with the DR-white assay (Figure 3). These results are consistent with a previous study in which DmBlmD3/D2 helicase-dead flies exhibited increased ionizing radiation sensitivity, although interestingly not to the extent of DmBlmN1/N1 null mutants (McVey et al. 2007). Furthermore, BLM helicase-deficient human cells were recently shown to suppress the elongation of branch migration and dHJ crossovers (Suzuki et al. 2016). Taken together, these results indicate that the helicase enzymatic activity is necessary for many functions of DmBlm in DSB repair.
A helicase-independent role of DmBlm in homologous recombination impacts GCT lengths
DmBlmN1/D2 null mutants exhibit a striking decrease in GCT lengths compared to DmBlmN1 heterozygote controls (Figure 4A). However, DmBlmD3/D2 helicase-dead mutant GCT lengths do not differ significantly from DmBlmD2 heterozygote controls or DmBlmD3 heterozygote controls, suggesting this effect to be mostly helicase-independent (Figure 4B). Others have reported the DmBlmD3 allele to be antimorphic in SDSA gap repair assays (McVey et al. 2007). In this study, the average GCT lengths in DmBlmD3 heterozygote controls were lower than in both DmBlmN1 and DmBlmD2 heterozygote controls, although this difference does not reach statistical significance (P = 0.08 and P = 0.33, respectively; Student’s t-test). Additionally, we see no statistically significant difference in noncrossover HR frequency in DmBlmN1/D2 and DmBlmD3/D2 mutants (P = 0.06). However, although our data does not reach statistical significance, the trends we report here suggest that the DmBlmD3 allele may be a semidominant phenotype, which may be more prevalent when assaying for gap repair (McVey et al. 2007).
Considering the impact of DmBlm on GCT length, there are multiple mechanisms that could contribute to GCT tract length, of which DmBlm may play a role. Based on the current SDSA and DSBR pathway models, GCT lengths may be affected by: (1) end resection, (2) branch migration, (3) strand synthesis, and/or (4) DNA mismatch repair. First, following a DSB, 5′–3′ resection determines how much sequence information is available for invasion into the homologous donor sequence. If the resection to GCT length relationship was isolated, less resection could result in shorter GCT lengths, and vice versa. Indeed, S. cerevisiae exo1 mutants, which have reduced end resection, are associated with crossovers and shorter GCTs (Yin and Petes 2014). However, contrary to this model, S. cerevisiae yku70 mutants with increased resection and mre11 mutants with reduced resection have GCTs similar to wild-type in a chromosomal context (Krishna et al. 2007). A previous study outlined two DNA end resection machinery complexes in human cells that both involve BLM (Nimonkar et al. 2011). One pathway requires BLM helicase functionality to unwind DNA to allow for resection by DNA2, but the other pathway involves EXO1 end resection that is stimulated by BLM independently of the helicase functionality (Nimonkar et al. 2008, 2011). Our results corroborate a helicase-independent role for DmBlm in end resection, which could ultimately impact GCT length.
Second, branch migration of recombination intermediates may also affect GCT length. DmBlm is known to promote SDSA by facilitating D-loop branch migration in order for the invading strand to dissociate (McVey et al. 2004; Weinert and Rio 2007). Given this function, a common phenotype associated with DmBlm mutants is increased mitotic crossovers (Johnson-Schlitz and Engels 2006; McVey et al. 2007), which could result from an inability to resolve recombination intermediates. In S. cerevisiae, mitotic crossovers are more often associated with longer GCTs (Jinks-Robertson et al. 1993; Symington et al. 2000). Therefore, it is plausible that a subset of noncrossover HR events associated with longer GCTs due to extensive branch migration are resolved as y− w− crossover products, which would explain our results of a decrease in noncrossover HR and a loss of longer GCTs in DmBlm null mutants.
Third, repair synthesis may impact GCT length. In S. cerevisiae, DSB repair requires both leading and lagging DNA polymerase functionality (Holmes and Haber 1999). S. cerevisiae lacking DNA polymerase δ have shorter GCT lengths, suggesting this effect to be correlated with primed DNA synthesis lengths (Maloisel et al. 2008). DmBlm lacks polymerase enzymatic properties, thus is unlikely to directly impact repair synthesis. However, the proposed DmBlm DNA end resection function may indirectly affect subsequent nascent strand DNA synthesis and therefore GCT length.
Lastly, GCT lengths may also be affected if MMR machinery is unable to correct base pair mismatches in hDNA [reviewed in Spies and Fishel (2015)]. Failure to convert mismatches in hDNA to the recipient sequence strand would result in longer GCTs. However, our result that DmBlm null mutants have shorter GCTs suggests that DmBlm is not involved in conversion of hDNA to the recipient strand sequence. This result may be consistent with previous studies that have shown S. cerevisiae sgs1 mutants (Lo et al. 2006) and BS human cells (Langland et al. 2001) to have fully functional base pair mismatch repair capabilities. Given the complexity of mechanisms that drive GCT length and the versatility of the DmBlm protein, more work is needed to isolate and quantify the determinants of GCT length and structure in multicellular organisms.
The GCTs analyzed in DmBlm mutants are a subset of DSB repair events that are able to complete HR (∼50% of the events in heterozygote controls). It is possible that the detectable mean GCT lengths with the DR-white.mu assay are skewed in DmBlm mutants since we are only measuring viable y+w+ progeny. For example, mechanisms that lead to longer GCT HR repair events in the absence of DmBlm may cause cell death, or be more susceptible to becoming the aberrant repair products frequently observed in DmBlm null mutants (Johnson-Schlitz and Engels 2006). These mechanisms would result in the y− w− phenotype as opposed to the y+ w+ HR phenotype. However, if we were unable to detect the longer GCT lengths due to the HR impairment in DmBlm mutants, we would expect DmBlmD3 helicase-dead mutants to similarly exhibit shorter mean GCT lengths compared to DmBlmD3 or DmBlmD2 heterozygote controls. Additionally, extensive gene conversion beyond the iwhite donor sequence may also skew the relative phenotype distribution by resulting in a y+ w− phenotype that contains a converted SacI sequence at the repair junction, instead of a y+ w+ HR event. Importantly, we did not observe a SacI conversion event in any repair events from y+ w− flies, suggesting that extensive gene conversion is unlikely.
DmBlm is not involved in suppression of noncrossover recombination between diverged sequences
Our DR-white and DR-white.mu assays can directly measure the ratio of noncrossover HR between homologous and 1.4% diverged sequences, respectively (Do et al. 2014). In both DmBlmN1/D2 null and DmBlmD2/D3 helicase-dead mutants, the ratio of noncrossover HR between homologous and diverged sequences was not significantly affected, suggesting that DmBlm is not involved in the suppression of recombination between diverged sequences. At similar levels of sequence divergence, BLM is dispensable for suppressing recombination between diverged sequences in both human and murine cells as well (LaRocque and Jasin 2010), but the sole RecQ helicase in S. cerevisiae, Sgs1, is necessary to suppress recombination of sequences with ≤ 9% divergence (Myung et al. 2001).
Whereas S. cerevisiae and Escherichia coli bacteria have only one known RecQ helicase, fruit flies, humans, and mice all have at least four RecQ helicase paralogs (Figure S1 in File S1), suggesting duplicate gene evolution in metazoans. Two possible outcomes of duplicate gene evolution are: (1) subfunctionalization, the duplicated genes each retain different subfunctions of the ancestral gene, and (2) neofunctionalization, one of the duplicated genes acquires a novel functionality while the other copy retains the ancestral functionality (Lynch and Conery 2000). Either of these gene evolutionary histories can explain why the BLM ortholog in species with one RecQ helicase suppresses recombination of sequences with nucleotide divergence < 9%, but not in species with multiple RecQ helicases. Additionally, high conservation of the helicase domain of RecQ helicases within and across species may allow for partial functional redundancy in species with multiple RecQ helicases (Kitao et al. 1998). To elucidate the collective roles of the RecQ helicase protein family in suppressing recombination of diverged sequences, future experiments are needed in other RecQ helicases within Drosophila, as well as in other genetically-tractable model organisms with multiple RecQ helicases, such as Caenorhabditis elegans (Jung et al. 2014; Ryu and Koo 2016), Danio rerio (Xie et al. 2007), and Arabadopsis thaliana (Knoll and Puchta 2011).
In contrast to our data quantifying recombination between diverged sequences, Kappeler et al. (2008) demonstrated that DmBlmD2/D3 helicase-dead mutants failed to suppress SSA with sequence divergences < 0.5%. Sequence divergence used in these two assays are similar, thus this discrepancy may most likely be attributed to the differences in the SSA and HR mechanisms. HR and SSA are both initiated by 5′ to 3′ resection and utilize similar components of the MMR machinery to ensure sufficient homology before undergoing recombination. Furthermore, the SDSA HR model and SSA share the S. cerevisiae Rad1-Rad10 nuclease-dependent 3′ flap processing step to stabilize hDNA (Fishman-Lobell and Haber 1992; Ivanov and Haber 1995; Mazon et al. 2012). However, multiple intermediate steps in the HR pathway that differ from the SSA pathway may consequently affect hDNA rejection mechanisms. For example, Rad51 coats the 3′ single-strand DNA and facilitates invasion into a homologous donor sequence (McIlwraith et al. 2000), and is then disassembled by a complex including S. cerevisiae Rad54 (Wright and Heyer 2014), the C. elegans Rad51 ortholog RFS-1, and DNA helicase HELQ-1 (Ward et al. 2010). These upstream steps of hDNA rejection are not necessary for SSA, and it is unclear how these differences between HR and SSA pathways may alter the respective recombination suppression mechanisms.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.300285/-/DC1.
Acknowledgments
We thank the Sekelsky and McVey laboratories for reagents and members of the LaRocque laboratory for discussion. This research was supported by National Institutes of Health grant 1R15-GM-110454-01 (to J.R.L.) and in part by the Georgetown Undergraduate Research Opportunities Program (J.T.B. and N.S.).
Footnotes
Communicating editor: J. Surtees
Literature Cited
- Adams M. D., McVey M., Sekelsky J., 2003. Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science 299: 265–267. [DOI] [PubMed] [Google Scholar]
- Bachrati C. Z., Borts R. H., Hickson I. D., 2006. Mobile D-loops are a preferred substrate for the bloom’s syndrome helicase. Nucleic Acids Res. 34: 2269–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom D., 1954. Congenital telangiectatic erythema resembling lupus erythematosus in dwarfs; probably a syndrome entity. AMA Am. J. Dis. Child. 88: 754–758. [PubMed] [Google Scholar]
- Boyd J. B., Golino M. D., Shaw K. E. S., Osgood C. J., Green M. M., 1981. Third-chromosome mutagen-sensitive mutants of Drosophila melanogaster. Genetics 97: 607–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaganti R. S., Schonberg S., German J., 1974. A manyfold increase in sister chromatid exchanges in bloom’s syndrome lymphocytes. Proc. Natl. Acad. Sci. USA 71: 4508–4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Cooper D. N., Chuzhanova N., Ferec C., Patrinos G. P., 2007. Gene conversion: mechanisms, evolution and human disease. Nat. Rev. Genet. 8: 762–775. [DOI] [PubMed] [Google Scholar]
- Delabaere L., Ertl H. A., Massey D. J., Hofley C. M., Sohail F., et al. , 2016. Aging impairs double-strand break repair by homologous recombination in Drosophila germ cells. Aging Cell 16: 320–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delacote F., Lopez B. S., 2008. Importance of the cell cycle phase for the choice of the appropriate DSB repair pathway, for genome stability maintenance: the trans-S double-strand break repair model. Cell Cycle 7: 33–38. [DOI] [PubMed] [Google Scholar]
- Do A. T., LaRocque J. R., 2015. The role of Drosophila mismatch repair in suppressing recombination between diverged sequences. Sci. Rep. 5: 17601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do A. T., Brooks J. T., Le Neveu M. K., LaRocque J. R., 2014. Double-strand break repair assays determine pathway choice and structure of gene conversion events in Drosophila melanogaster. G3 (Bethesda) 4: 425–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond J. T., Li G. M., Longley M. J., Modrich P., 1995. Isolation of an hMSH2-p160 heterodimer that restores DNA mismatch repair to tumor cells. Science 268: 1909–1912. [DOI] [PubMed] [Google Scholar]
- Ellis N. A., Sander M., Harris C. C., Bohr V. A., 2008. Bloom’s syndrome workshop focuses on the functional specificities of RecQ helicases. Mech. Ageing Dev. 129: 681–691. [DOI] [PubMed] [Google Scholar]
- Fishman-Lobell J., Haber J. E., 1992. Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science 258: 480–484. [DOI] [PubMed] [Google Scholar]
- German J., Archibald R., Bloom D., 1965. Chromosomal breakage in a rare and probably genetically determined syndrome of man. Science 148: 506–507. [DOI] [PubMed] [Google Scholar]
- Gloor G. B., Preston C. R., Johnson-Schlitz D. M., Nassif N. A., Phillis R. W., et al. , 1993. Type I repressors of P element mobility. Genetics 135: 81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabarz A., Guirouilh-Barbat J., Barascu A., Pennarun G., Genet D., et al. , 2013. A role for BLM in double-strand break repair pathway choice: prevention of CtIP/Mre11-mediated alternative nonhomologous end-joining. Cell Rep. 5: 21–28. [DOI] [PubMed] [Google Scholar]
- Gravel S., Chapman J. R., Magill C., Jackson S. P., 2008. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 22: 2767–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer W. D., Ehmsen K. T., Liu J., 2010. Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 44: 113–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes A. M., Haber J. E., 1999. Double-strand break repair in yeast requires both leading and lagging strand DNA polymerases. Cell 96: 415–424. [DOI] [PubMed] [Google Scholar]
- Ivanov E. L., Haber J. E., 1995. RAD1 and RAD10, but not other excision repair genes, are required for double-strand break-induced recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 15: 2245–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen A., Breuer G. A., Brinkman E. K., van der Meulen A. I., Borden S. V., et al. , 2016. A single double-strand break system reveals repair dynamics and mechanisms in heterochromatin and euchromatin. Genes Dev. 30: 1645–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinks-Robertson S., Michelitch M., Ramcharan S., 1993. Substrate length requirements for efficient mitotic recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 13: 3937–3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson-Schlitz D., Engels W. R., 2006. Template disruptions and failure of double holliday junction dissolution during double-strand break repair in Drosophila BLM mutants. Proc. Natl. Acad. Sci. USA 103: 16840–16845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson-Schlitz D. M., Flores C., Engels W. R., 2007. Multiple-pathway analysis of double-strand break repair mutations in Drosophila. PLoS Genet. 3: e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H., Lee J. A., Choi S., Lee H., Ahn B., 2014. Characterization of the Caenorhabditis elegans HIM-6/BLM helicase: unwinding recombination intermediates. PLoS One 9: e102402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappeler M., Kranz E., Woolcock K., Georgiev O., Schaffner W., 2008. Drosophila bloom helicase maintains genome integrity by inhibiting recombination between divergent DNA sequences. Nucleic Acids Res. 36: 6907–6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karow J. K., Constantinou A., Li J. L., West S. C., Hickson I. D., 2000. The bloom’s syndrome gene product promotes branch migration of holliday junctions. Proc. Natl. Acad. Sci. USA 97: 6504–6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass E. M., Jasin M., 2010. Collaboration and competition between DNA double-strand break repair pathways. FEBS Lett. 584: 3703–3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitao S., Ohsugi I., Ichikawa K., Goto M., Furuichi Y., et al. , 1998. Cloning of two new human helicase genes of the RecQ family: biological significance of multiple species in higher eukaryotes. Genomics 54: 443–452. [DOI] [PubMed] [Google Scholar]
- Knoll A., Puchta H., 2011. The role of DNA helicases and their interaction partners in genome stability and meiotic recombination in plants. J. Exp. Bot. 62: 1565–1579. [DOI] [PubMed] [Google Scholar]
- Krishna S., Wagener B. M., Liu H. P., Lo Y. C., Sterk R., et al. , 2007. Mre11 and Ku regulation of double-strand break repair by gene conversion and break-induced replication. DNA Repair (Amst.) 6: 797–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano K., Berres M. E., Engels W. R., 1999. Evolution of the RECQ family of helicases. A Drosophila homolog, DmBlm, is similar to the human bloom syndrome gene. Genetics 151: 1027–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano K., Johnson-Schlitz D. M., Engels W. R., 2001. Sterility of Drosophila with mutations in the bloom syndrome gene complementation by Ku70. Science 291: 2600–2602. [DOI] [PubMed] [Google Scholar]
- Langland G., Kordich J., Creaney J., Goss K. H., Lillard-Wetherell K., et al. , 2001. The bloom’s syndrome protein (BLM) interacts with MLH1 but is not required for DNA mismatch repair. J. Biol. Chem. 276: 30031–30035. [DOI] [PubMed] [Google Scholar]
- LaRocque J. R., Jasin M., 2010. Mechanisms of recombination between diverged sequences in wild-type and BLM-deficient mouse and human cells. Mol. Cell. Biol. 30: 1887–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Y. C., Paffett K. S., Amit O., Clikeman J. A., Sterk R., et al. , 2006. Sgs1 regulates gene conversion tract lengths and crossovers independently of its helicase activity. Mol. Cell. Biol. 26: 4086–4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luria S. E., Delbruck M., 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28: 491–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Conery J. S., 2000. The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155. [DOI] [PubMed] [Google Scholar]
- Maloisel L., Fabre F., Gangloff S., 2008. DNA polymerase δ is preferentially recruited during homologous recombination to promote heteroduplex DNA extension. Mol. Cell. Biol. 28: 1373–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazon G., Lam A. F., Ho C. K., Kupiec M., Symington L. S., 2012. The Rad1-Rad10 nuclease promotes chromosome translocations between dispersed repeats. Nat. Struct. Mol. Biol. 19: 964–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlwraith M. J., Van Dyck E., Masson J. Y., Stasiak A. Z., Stasiak A., et al. , 2000. Reconstitution of the strand invasion step of double-strand break repair using human Rad51 Rad52 and RPA proteins. J. Mol. Biol. 304: 151–164. [DOI] [PubMed] [Google Scholar]
- McVey M., Lee S. E., 2008. MMEJ repair of double-strand breaks (director’s cut): deleted sequences and alternative endings. Trends Genet. 24: 529–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey M., LaRocque J. R., Adams M. D., Sekelsky J. J., 2004. Formation of deletions during double-strand break repair in Drosophila DmBlm mutants occurs after strand invasion. Proc. Natl. Acad. Sci. USA 101: 15694–15699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey M., Andersen S. L., Broze Y., Sekelsky J., 2007. Multiple functions of Drosophila BLM helicase in maintenance of genome stability. Genetics 176: 1979–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung K., Datta A., Chen C., Kolodner R. D., 2001. SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nat. Genet. 27: 113–116. [DOI] [PubMed] [Google Scholar]
- Nassif N., Penney J., Pal S., Engels W. R., Gloor G. B., 1994. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol. Cell. Biol. 14: 1613–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimonkar A. V., Ozsoy A. Z., Genschel J., Modrich P., Kowalczykowski S. C., 2008. Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc. Natl. Acad. Sci. USA 105: 16906–16911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimonkar A. V., Genschel J., Kinoshita E., Polaczek P., Campbell J. L., et al. , 2011. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 25: 350–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques F., Haber J. E., 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63: 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrazzi G., Bachrati C. Z., Selak N., Studer I., Petkovic M., et al. , 2003. The bloom’s syndrome helicase interacts directly with the human DNA mismatch repair protein hMSH6. Biol. Chem. 384: 1155–1164. [DOI] [PubMed] [Google Scholar]
- Preston C. R., Flores C. C., Engels W. R., 2006. Differential usage of alternative pathways of double-strand break repair in Drosophila. Genetics 172: 1055–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong Y. S., Golic K. G., 2000. Gene targeting by homologous recombination in Drosophila. Science 288: 2013–2018. [DOI] [PubMed] [Google Scholar]
- Rong Y. S., Golic K. G., 2003. The homologous chromosome is an effective template for the repair of mitotic DNA double-strand breaks in Drosophila. Genetics 165: 1831–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu J. S., Koo H. S., 2016. Roles of Caenorhabditis elegans WRN helicase in DNA damage responses, and a comparison with its mammalian homolog: a mini-review. Gerontology 62: 296–303. [DOI] [PubMed] [Google Scholar]
- Ryu T., Spatola B., Delabaere L., Bowlin K., Hopp H., et al. , 2015. Heterochromatic breaks move to the nuclear periphery to continue recombinational repair. Nat. Cell Biol. 17: 1401–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastav M., De Haro L. P., Nickoloff J. A., 2008. Regulation of DNA double-strand break repair pathway choice. Cell Res. 18: 134–147. [DOI] [PubMed] [Google Scholar]
- Spies M., Fishel R., 2015. Mismatch repair during homologous and homeologous recombination. Cold Spring Harb. Perspect. Biol. 7: a022657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N., Ivanov E. L., Fishman-Lobell J., Ray B. L., Wu X., et al. , 1995. DNA structure-dependent requirements for yeast RAD genes in gene conversion. Nature 372: 84–86. [DOI] [PubMed] [Google Scholar]
- Sugawara N., Goldfarb T., Studamire B., Alani E., Haber J. E., 2004. Heteroduplex rejection during single-strand annealing requires Sgs1 helicase and mismatch repair proteins Msh2 and Msh6 but not Pms1. Proc. Natl. Acad. Sci. USA 101: 9315–9320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Yasui M., Honma M., 2016. Mutator phenotype and DNA double-strand break repair in BLM helicase-deficient human cells. Mol. Cell. Biol. 36: 2877–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington L. S., Kang L. E., Moreau S., 2000. Alteration of gene conversion tract length and associated crossing over during plasmid gap repair in nuclease-deficient strains of Saccharomyces cerevisiae. Nucleic Acids Res. 28: 4649–4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak J. W., Orr-Weaver T. L., Rothstein R. J., Stahl F. W., 1983. The double-strand-break repair model for recombination. Cell 33: 25–35. [DOI] [PubMed] [Google Scholar]
- Wang Y., Li S., Smith K., Waldman B. C., Waldman A. S., 2016. Intrachromosomal recombination between highly diverged DNA sequences is enabled in human cells deficient in bloom helicase. DNA Repair (Amst.) 41: 73–84. [DOI] [PubMed] [Google Scholar]
- Ward J. D., Muzzini D. M., Petalcorin M. I., Martinez-Perez E., Martin J. S., et al. , 2010. Overlapping mechanisms promote postsynaptic RAD-51 filament disassembly during meiotic double-strand break repair. Mol. Cell 37: 259–272. [DOI] [PubMed] [Google Scholar]
- Wei D. S., Rong Y. S., 2007. A genetic screen for DNA double-strand break repair mutations in drosophila. Genetics 177: 63–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert B. T., Rio D. C., 2007. DNA strand displacement, strand annealing and strand swapping by the Drosophila bloom’s syndrome helicase. Nucleic Acids Res. 35: 1367–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright W. D., Heyer W. D., 2014. Rad54 functions as a heteroduplex DNA pump modulated by its DNA substrates and Rad51 during D loop formation. Mol. Cell 53: 420–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Hickson I. D., 2003. The bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature 426: 870–874. [DOI] [PubMed] [Google Scholar]
- Wu L., Bachrati C. Z., Ou J., Xu C., Yin J., et al. , 2006. BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proc. Natl. Acad. Sci. USA 103: 4068–4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J., Bessling S. L., Cooper T. K., Dietz H. C., McCallion A. S., et al. , 2007. Manipulating mitotic recombination in the zebrafish embryo through RecQ helicases. Genetics 176: 1339–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Zhang R., Wang X. W., Linke S. P., Sengupta S., et al. , 2004. The mismatch DNA repair heterodimer, hMSH2/6, regulates BLM helicase. Oncogene 23: 3749–3756. [DOI] [PubMed] [Google Scholar]
- Yin Y., Petes T. D., 2014. The role of Exo1p exonuclease in DNA end resection to generate gene conversion tracts in Saccharomyces cerevisiae. Genetics 197: 1097–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All strains and reporter constructs used in this study are available upon request. Table S1 in File S1 contains the raw data from experiments presented in Figure 3, Figure 5, and Figure S2 and Figure S5 in File S1. Table S2 in File S1 contains raw data presented in the Results. Figure S3 in File S1 contains the individual GCTs that are presented in Figure 4.
Figure 3.
DmBlm impacts DSB repair pathway usage. (A) I-SceI heat shock-induced DSB repair events in a DmBlmN1/D2 null mutant background (red; n = 94) compared to DmBlmN1 heterozygote controls (blue; n = 125). Results shown are averages and SEM of individual male germline events compiled from five independent experiments. * P < 0.05 and **** P < 0.0001 by unpaired Student’s t-test. (B) I-SceI heat shock-induced DSB repair events in a DmBlmD3/D2 helicase-dead mutant background (red; n =52) compared to DmBlmD3 heterozygote controls (blue; n = 59). Results shown are averages and SEM of individual male germline events compiled from three independent experiments. ** P < 0.01 and **** P < 0.0001 by unpaired Student’s t-test. CO, crossover; DSB, double-strand break; HR, homologous recombination; NHEJ, nonhomologous end-joining; SSA, single-strand annealing.
Figure 5.
Relative recombination frequencies between homologous and diverged sequences are unaltered in DmBlm mutants. Relative recombination frequencies between homologous and diverged sequences were determined using DR-white and DR-white.mu, respectively, in (A) DmBlmN1 heterozygote controls and DmBlmN1/D2 null mutants, and (B) DmBlmD3 heterozygote controls and DmBlmD3/D2 helicase-dead mutants. Average HR frequencies (with SEM) were compiled from individual germlines from two independent experiments and are presented below the graphs (and in Table S1 in File S1). Average HR frequencies relative to DR-white are plotted. DR-white; Direct Repeat of white; DR-white.mu; Direct Repeat of white with mutations; HR, homologous recombination.
Figure 4.
DmBlm impacts gene conversion tract length independently of helicase function. Intrachromosomal noncrossover HR events using the DR-white.mu assay were isolated and the GCT direction and length were determined. (A) The proportions of SNP sites converted are displayed for DmBlmN1 heterozygote control HR events (blue; n = 59) and DmBlmN1/D2 null mutant HR events (purple; n = 51). The average distance converted and SEM (base pair) to the left and to the right of the SacI site/DSB (0) is given for both genotypes. Data are from two independent experiments. (B) The proportions of SNP sites converted are displayed for DmBlmD2 heterozygote control HR events (light blue; n = 59), DmBlmD3 heterozygote control HR events (orange; n = 78) and DmBlmD3/D2 helicase-dead mutant HR events (green; n = 71). The average distance and SEM converted (base pair) to the left and to the right of the SacI site/DSB (0) is given for both genotypes. Data are from four independent experiments. (C) Classes of gene conversion tracts from combined DR-white.mu assay experiments in DmBlmN1/D2 null mutants (left) and DmBlmD3/D2 helicase-dead mutants (right) and the respective heterozygote control. Each tract was grouped into one of four classes (represented graphically in descending order): conversion of only the DSB/SacI site, conversion to the right of the DSB/SacI site (> 32 bp; unidirectional), conversion to right of the DSB/SacI site (> 63 bp; unidirectional), and conversion to both sides of the break (bidirectional). DR-white.mu; Direct Repeat of white with mutations; DSB, double-strand break; GCT, gene conversion tract; HR, homologous recombination; SNP, single nucleotide polymorphism.