Abstract
Insecticide resistance is considered a classic model of microevolution, where a strong selective agent is applied to a large natural population, resulting in a change in frequency of alleles that confer resistance. While many insecticide resistance variants have been characterized at the gene level, they are typically single genes of large effect identified in highly resistant pest species. In contrast, multiple variants have been implicated in DDT resistance in Drosophila melanogaster; however, only the Cyp6g1 locus has previously been shown to be relevant to field populations. Here we use genome-wide association studies (GWAS) to identify DDT-associated polygenes and use selective sweep analyses to assess their adaptive significance. We identify and verify two candidate DDT resistance loci. A largely uncharacterized gene, CG10737, has a function in muscles that ameliorates the effects of DDT, while a putative detoxifying P450, Cyp6w1, shows compelling evidence of positive selection.
Keywords: Drosophila Genetic Reference Panel (DGRP), DDT, CG10737, Cyp6w1, triallele
MANY insights into the genetic basis of adaptation have been gained by genetic analyses of resistance to man-made chemicals; whether that be bacteria to antibiotics (Zhang et al. 2011), weeds to herbicides (Baucom 2016), fungi to fungicides (Schoustra et al. 2005), or insects to insecticides (Crow 1957; McKenzie 1996). Typically, the genetics of such traits are thought to be strongly monogenic and thus distinct from most other traits whose variation is governed by the combined effect of multiple loci of small effect (Lande 1983; Roush and McKenzie 1987; Allen et al. 2010). This contention is central to the debate over the genetics of DDT resistance in Drosophila melanogaster. As it is not considered a pest, this model insect may have never been exposed to the high selection intensities thought necessary for the evolution of major-gene-based resistance (Macnair 1991; McKenzie and Batterham 1994). Despite most genetic investigations supporting a polygenic architecture (Crow 1954; King and Sømme 1958; Dapkus and Merrell 1977; Shepanski et al. 1977) or more precisely weak polygenicity (Dapkus 1992; i.e., at most one or two loci per chromosome arm), a gene of major effect, Cyp6g1, was identified and described as “necessary and sufficient” for DDT resistance (Daborn et al. 2002). Proposed solutions to this apparent contradiction between “polygenic” and “monogenic” schools include delineating a difference between field and laboratory evolved resistance (Ffrench-Constant 2013) or suggesting that Cyp6g1 is only inconsistently associated with resistance (Kuruganti et al. 2007).
In the specific case of the long-term DDT-selected strain 91-R, there is a strong argument for the polygenicity of DDT resistance. 91-R is over 1500 times more resistant to DDT than another laboratory strain, Canton-S (Strycharz et al. 2013); levels far beyond those attributable to Cyp6g1 alone. Much of the recent work aimed at identifying polygenes associated with DDT resistance has focused on contrasting these resistant and susceptible laboratory strains and has allowed the generation of transcriptomic (Pedra et al. 2004), proteomic (Festucci-Buselli et al. 2005), physiological (Strycharz et al. 2013), and genomic (Steele et al. 2014, 2015) data sets of differentiated factors. These candidates, however, remain to be validated, and their contribution to either the selection response in the laboratory, or to field resistance, have yet to be quantified. Until now Cyp6g1 has remained the only DDT resistance locus with molecularly defined alleles that contribute to DDT-related genetic variation observed in field populations.
Previously Schmidt et al. (2010) demonstrated that an allelic series at the Cyp6g1 locus is sweeping through D. melanogaster populations. The resistant Cyp6g1 variant described by Daborn et al. (2002), which harbored a partial Accord transposable element insertion upstream of the Cyp6g1 locus, was subsequently found to be invariably associated with a duplication of the Cyp6g1 locus (Schmidt et al. 2010). It is still unclear to what extent the upregulation of Cyp6g1 observed at this allele is due to the Accord element (Chung et al. 2007) or to the presence of multiple Cyp6g1 copies, or both. However, it is clear that the ancestral Cyp6g1 allele (Cyp6g1-M) is at very low frequencies throughout the world and has largely been replaced by haplotypes carrying the duplication and various transposable element insertions. The Cyp6g1-AA allele (which has two copies of the gene both bearing the Accord element) and the Cyp6g1-BA allele (where one of the duplicated copies contains an additional HMS-Beagle transposable element insertion) both confer resistance to DDT relative to the Cyp6g1-M allele, although they show only subtle phenotypic differences between one another. A fourth allele, Cyp6g1-BP, is at high frequencies in populations in northeastern Australia, and provides a further increase in resistance, in both a set of isochromosomal lines and in a field population (Schmidt et al. 2010). In the latter, the Cyp6g1-BP allele accounts for ∼16% of the total variation in the DDT resistance phenotype.
The analysis of field variation at the Cyp6g1 locus provides two arguments suggesting that other loci contribute to DDT resistance in the field. First, as Cyp6g1 allelic variation does not account for all the 20–50% heritability expected for insecticide resistance (McKenzie 2000), there is much genetic variation that is not accounted for (20–68%). Second, as a succession of Cyp6g1 alleles provides a series of adaptive events at one locus, there has been time and selective pressure for other adaptive events arising elsewhere in the genome.
DDT affects the nervous system by targeting the voltage-gated sodium channel resulting in uncontrolled nerve firing. In D. melanogaster, the α-subunit of the voltage-gated sodium channel is encoded by para, and target-site resistance to DDT has been described in para mutants by Pittendrigh et al. (1997) and Lindsay et al. (2008). However, para variation affecting DDT resistance has not yet been identified in outbred populations of D. melanogaster, nor has it been implicated in the phenotypes of well-studied DDT-resistant laboratory strains (Dapkus and Merrell 1977; Steele et al. 2014).
DDT-induced mortality is typically preceded by the loss of peripheral muscle control and manifests as a “knockdown” phenotype in which the fly lies paralyzed with appendages twitching, or exhibits uncontrolled flight. Here we use the Drosophila Genetic Reference Panel (DGRP; Mackay et al. 2012; Mackay and Huang 2017), a resource for the dissection of quantitative traits, to characterize the genetic architecture of both knockdown and mortality caused by DDT exposure using predominantly field-derived genetic variation from a single population. This population has shown great phenotypic diversity for, and furnished genetic associations with, a diverse range of traits including insecticide resistance (Mackay et al. 2012; Huang et al. 2014; Battlay et al. 2016). We performed genome-wide association studies (GWAS) on two DDT-related traits to gain insights into insecticide biology and insecticide-driven selection using a classic insecticide model.
Materials and Methods
Fly stocks
DGRP lines were obtained from the Bloomington Stock Center, and were maintained on corn meal media within a 22–25° temperature range. For the DDT assays, larval density was controlled by setting up vials with 10 mated females that were allowed to lay for 3 days, then transferred to fresh media for a further 3 days to establish an additional brood to provide enough progeny for replication. At this stage, two to three replicates per line were established. Adults were allowed to eclose for 2–3 days, and females were collected to food-containing holding vials after brief CO2 anesthetization. After 2–3 days’ recovery, adult females were assayed, thus the age range was 4–6 days for flies assayed.
DDT assays
Glass scintillation vials were inoculated with 200 μl of Acetone/DDT solution at a concentration of 0.5 μg/ml, giving a final contact assay amount of 100 μg of DDT. Cotton wool moistened with 10% sucrose solution was used to stopper the scintillation vials. Assays were set up between 7 and 8 am, roughly corresponding with the onset of the light phase of the diurnal cycle. Time of setup was recorded, and each vial was scored for the presence of flies exhibiting either knockdown or mortality at ∼1-hr intervals. Knockdown here is defined as either flies permanently seen to be in a prone position with jolting movement of legs or wings, or a prolonged inability to right themselves from a prone position after tapping of the scintillation vial on the laboratory bench. Mortality was determined as the point at which all external movement ceased. Data were recorded in data sheets with time points as columns and DGRP line number as rows. Assay vials contained 10 females, and there were at least three replicates per DGRP line. A total of 184 DGRP lines were assayed. In total, the assay time was 30 hr. Phenotypes were scored through 1–15, 24, and 30 hr.
Estimation of heritability
At least three replicates of ten flies per vial were analyzed per DGRP line, which allowed the calculation of broad-sense heritability (H2). Following the method of Clowers et al. (2010), heritability was estimated from the variance components of a linear model of the form: Phenotype = Population mean + Line effect + error. As these lines are inbred, the components of the Genetic Variance (e.g., additive or dominant), cannot be separated, thus these estimates are of broad-sense (H2) heritability. An ANOVA was performed in Statistics Package in Social Sciences (SPSS), and the variance components were estimated as the Mean Sum of Squares. Total phenotypic variance was estimated as Genetic Variance + Environmental Variance. H2 was thus estimated as Gv/Gv+Ev. This was calculated for each discrete time point.
GWAS
Phenotypes were submitted to the Mackay Laboratory DGRP2 website for GWAS (http://dgrp.gnets.ncsu.edu/; Huang et al. 2014). To perform our own GWAS for DDT resistance traits, we used the PLINK GWAS program (PLINK v1.9, https://www.cog-genomics.org/plink2; Chang et al. 2015). For genotypes, we initially downloaded the DGRP freeze 1 SNP tables (http://www.hgsc.bcm.tmc.edu/projects/dgrp/freeze1_July_2010/snp_calls/). These tables encoded “haploid” variant calls, with heterozygous positions encoded with the relevant ambiguity codes. To make PLINK-compatible files, we used perl scripts to modify these SNP tables into diploid genotypes. We were surprised by a large number of sites that contained more than two alleles. The initial DGRP freeze 1 GWAS tool removed all such sites, but assuming that many of these represented low-frequency sites or errors, we ranked each variant by frequency and kept the two most frequent ones. The one major difference between our initial results and that of the DGRP freeze 1 GWAS tool was the triallelic variant at Cyp6w1.
However, our analyses here are based on the newer DGRP freeze 2 release. For this we downloaded the dgrp2.bed file (PLINK format) from the DGRP2 website (http://dgrp2.gnets.ncsu.edu/data/website/dgrp2.bed). Thus, these genotypes are identical to those used in the DGRP webtool, except for choices in variant filtering as indicated in the PLINK command line below. It should also be noted that now no heterozygous genotypes are present, and that the two most common variants at triallelic sites are also present in this bed file.
We used the following base PLINK command to perform GWAS (linear regression of genotype phenotype association assuming an additive model of allelic effects) after initially filtering for SNPs with MAF ≥ 0.05 and at least 70% genotyping rate (given that a triallelic site with equal proportions of all three genotypes would have a rate of ∼66% in the DGRP freeze 2 data):
plink–allow-extra-chr–allow-no-sex–bfile dgrp2–covar dgrp2_ESTRAT_PCA20.txt–linear–map3–no-fid–no-parents–no-pheno–no-sex–aperm 5 1000000
The “aperm” option was used to generate empirical estimates of the P-value for each SNP. To calculate the family-wise error rate (FWER) for a given P-value or level of significance, we generated 5000 random permutations of the 4-hr knockdown (4 hrkd) and 24-hr mortality (24 hrm) phenotype data and performed the same linear association model as for the observed phenotypes. For each permutation we record the lowest observed P-value. The FWER is then the number of random permutations that generated a P-value lower than or equal to that observed for each SNP, i.e., that chance of at least one false positive at this level of significance. After filtering, 1,776,058 SNPs were tested for each PLINK GWAS.
We performed principal components analysis on the DGRP data using PLINK after prefiltering genotypes for minor allele frequency (>0.05), missing rate (<0.70), and linkage disequilibrium (LD) pruning (r2 < 0.2) using the indep pairwise command, with both window and step size of 500 variants. To control for confounding cryptic relatedness in the DGRP, we used the first 20 principal components as covariables in all GWAS, as indicated above with the “–covar dgrp2_ESTRAT_PCA20.txt” option in the PLINK command. As inversions and Wolbachia infection status can also influence the phenotypes of the DGRP lines, we used the phenotypes adjusted for these factors outputted from the DGRP2 website. Thus, our results for the DGRP freeze 1 and DGRP freeze 2 differ due to now correcting for population structure, adjusting phenotypes for inversion and Wolbachia infection status, and the removal of heterozygous genotypes in the DGRP freeze 2. Finally, for annotating variants we used the annotations made available on the DGRP2 website (http://dgrp2.gnets.ncsu.edu/data/website/dgrp.fb557.annot.txt).
To test specific variants at Cyp6g1, we used PCR primers described in Schmidt et al. (2010) and/or local alignments of Illumina and 454 reads to determine the Cyp6g1 genotypes. For Cyp6w1, our DGRP freeze 1 GWAS indicated the presence of a triallelic site at 2R:6,174,944[V6]. We used the reprocessed DGRP genotype data from the Drosophila Genome Nexus (Lack et al. 2015) files to extract genotype data for this position (described further under the iHS and nSL tests methods), as the genotypes for the third variant state are removed from the DGRP freeze 2 data. For both these genes, the sites of interest are triallelic in the DGRP. We generated files with the ancestral allele and derived alleles, both singularly and combined after collapsing to the same allelic code, included to test for associations with DDT phenotypes. In the particular case of Cyp6g1, we introduced a variable site to code Cyp6g1-M, Cyp6g1-AA, or Cyp6g1-BA alleles.
iHS and nSL tests of recent positive selection
We downloaded the haploid FASTA alignments for 205 DGRP2 genomes, described by Lack et al. (2015), that are contained in the Drosophila Genome Nexus (DGN; http://johnpool.net/genomes.html). We applied the supplied masking filter for identity by decent (IBD) regions, but not the masks for admixture. For ease of downstream processing, we converted these FASTA alignments into Variant Call Format (VCF) files using custom bash and perl scripts. Note, these scripts kept the genotype information at triallelic sites.
After IBD masking and the prefiltering of excessively heterozygous regions in the DGN data (using the scripts available from http://johnpool.net/genomes.html), the genomes varied drastically in the proportion of sites with missing data. We excluded chromosomes with >20% missing data, leaving us with 134 lines for chromosome 3L. For other chromosomes, we randomly sampled lines without replacement to n = 134. In the compilation of VCF files, we excluded sites with >15% missing genotypes in the DGRP2. There were of course missing data remaining after these two steps, still rendering them unsuitable for haplotype tests. We filled in missing genotypes by randomly sampling from called genotypes, in proportion to their population frequency. In the main, this procedure will reduce local LD, and thus is conservative vis-a-vis LD-based tests of selection, and is similar to the method employed by Garud et al. (2015).
We used two tests of selection that both utilize signatures of LD around a beneficial allele. This LD can be measured by the extended haplotype homozygosity (EHH) statistic (Sabeti et al. 2002). EHH summarizes the probability that two chromosomes bearing the beneficial allele are also identical by state at a nearby neutral variant. Increased LD will thus also increase the EHH statistic. The integrated haplotype score (iHS; Voight et al. 2006) extends EHH by calculating the area under the EHH curve (iHH, and thus integrated) for both the derived and ancestral variant, and then finding the ratio of iHHDerived/iHHAncestral. If LD for both the derived and ancestral allele is approximately the same, this ratio should be close to 1, whereas large departures could indicate the action of recent positive selection. As LD is also influenced by the age of mutations, iHS scores are standardized in bins of derived allele frequency, as under a neutral model frequency is indicative of allele age (on average). nSL (number of segregating sites by length; Ferrer-Admetlla et al. 2014) is similar in spirit to iHS, but instead of EHH, the base statistic is the average count of SNPs for which two haplotypes are identical. We use it here because this statistic may be less biased toward regions of low recombination and has an increased ability to detect soft sweeps (i.e., the beneficial allele is segregating on more than one distinct haplotype at the time of selection).
To calculate both iHS and nSL, we used the versions implemented in selscan with default parameters for each (Szpiech and Hernandez 2014). We kept biallelic SNPs with 5% < DAF < 95%. The ancestral state was estimated as the homologous D. simulans reference genome allele (aligned to D. melanogaster, also downloaded from http://johnpool.net/genomes.html). Both statistics were normalized in 1% frequency bins per chromosome, and a P-value per site was calculated from the empirical distribution of normalized scores (|iHS| |nSL|) per chromosome. Genetic map positions were estimated using the recombination maps generated by Comeron et al. (2012).
For the triallelic sites at Cyp6g1 and Cyp6w1, we manually constructed haplotype files containing the set of one of the derived haplotypes and the ancestral haplotypes. These sets are a subset of those for biallelic sites, and P-values for each of these were calculated as above.
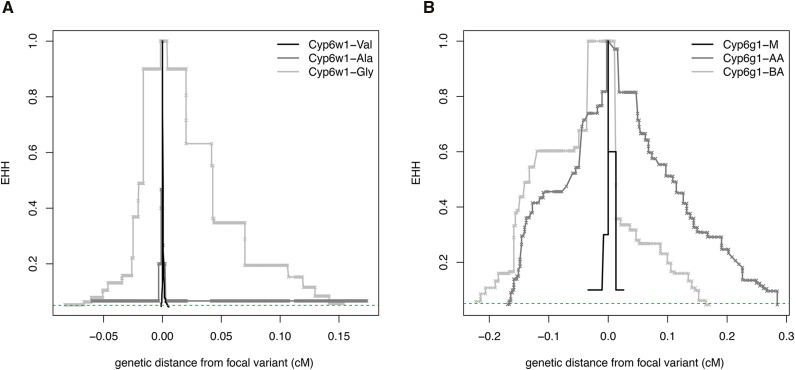
For the extended haplotype structure for the Cyp6w1_GLY allele, the EHH curve does not decay below 0.05 within the default window size, a reflection of both increased haplotype structure but also a region of increased heterozygosity downstream of Cyp6w1, which is filtered out in most DGRP2 individuals and introduces a gap >200 kb. To compute an iHS value for this variant we relaxed the default parameters, so that the max gap is 250 kb. Manual inspection of the EHH curves (Figure 4) suggests that the high iHS statistic for this variant is due to haplotype structure and not this gap in the estimated haplotypes.
Figure 4.
Extended Haplotype Homozygosity (EHH) plots: (A) around the Cyp6w1 locus using the triallelic second site of codon 370 as the focal variant and (B) around the Cyp6g1 locus using transposable element insertion sites as the focal variant.
CG10737 RNAi
Line CG10737-KK [Vienna Drosophila Resource Centre (VDRC) ID 106383] that has an upstream activator sequence (UAS) followed by a hairpin sequence matching CG10737 with no recorded off-targets was obtained from the VRDC. It was crossed to Mef2-GAL4 (Bloomington ID 27390). Line y, w[1118]; P(attP,y+,w[3′]) (VDRC ID 60100) was used as control and also crossed to the Mef2-GAL4 driver. The offspring of these crosses were evaluated for the DDT resistance phenotypes, 3hrkd, and 24hrm. The screens for each cross were performed over a range of DDT concentrations using replicates of 10 females aged 4–6 days old. For every DDT concentration, a minimum of 80 females were screened for 3hrkd and a minimum of 50 females were screened for 24hrm. The relationship between the 3hrkd resistance phenotypes and DDT concentration was evaluated using XLSTAT to perform a Probit analysis, a log normal regression of the knockdown data (Sakuma 1998). Dosage mortality curves were constructed for the relevant crosses and their controls and the ED50, the dose that will theoretically knockdown 50% of the population, was estimated. To ascertain that the gene had indeed been knocked down, total RNA was extracted from 20 to 30 adult females using the TRIsure-reagent. The extracted RNA was treated with RNA-free DNase (New England Biolabs, Ipswich, Massachusetts) and residual genomic DNA contamination was assessed for each sample by attempting to amplify a PCR product from the RNA. The samples were converted to complementary DNA (cDNA) using 1 µg of RNA in 20 µl reaction with Anchored Oligo (dT)20 and MuLV Reverse Transcriptase (NEB) according to the manufacturer’s instructions.
Cyp6w1 transgenic overexpression
UAS-Cyp6w1 constructs were designed using the y; cn bw sp; reference strain Cyp6w1 coding sequence as a template. Constructs containing each state of Cyp6w1 triallele identified in the 24 hrm GWAS were created by mutating the VAL370 codon in the reference sequence to ALA370 and GLY370. All three constructs were manufactured by Biomatik USA (Wilmington, DE) and were supplied cloned into plasmid pUASTattB.
The UAS-Cyp6w1 constructs were transformed into the y1 M{vas-int.Dm}ZH- 2A w*; (EPS) M{36P3-RFP.attP}ZH-86Fb recipient strain, which has a defined integration site on Chromosome 3, using the attP–attB system and QC31 integrase (Bischof et al. 2007). To generate heterozygous Cyp6w1/tubulin-GAL4 or serrate flies, virgin female homozygous UAS-Cyp6w1 flies were collected on light CO2, stored at 22–25° for 4 days to ensure virginity, and then 10 females per vial were crossed to tubulin-GAL4/serrate males. At the same time, vials with either 10 UAS-Cyp6w1 or tubulin- GAL4/serrate females were established. Adults for all these fly lines were allowed to eclose for 2–3 days, and females were collected to food-containing holding vials after brief CO2 anesthesia.
Each line was tested on a minimum of five doses of DDT. A minimum of five replicates of 20 individuals per dose per line were treated in the bio-assay. Glass scintillation vials were inoculated with 200 μl of Acetone/DDT solution with concentrations ranging from 5 × 10−5 to 1.0 μg/μl, giving a final contact assay range of 0.01–200 μg of DDT. Cotton wool moistened with 10% sucrose solution was used to stopper the scintillation vials. Flies were assayed at 25° for 24 hr. Data for each tested line were individually analyzed using PriProbit (Sakuma 1998) to estimate the LC50 and generate data for plotting dosage mortality curves and 95% confidence intervals.
91-R and 91-C genome examination
bam files containing alignments of 91-R and 91-C sequencing reads to the y; cn bw sp; reference genome were obtained from the sequence read archive (Leinonen et al. 2011; SRR1237973; SRR1237974). Alignments at relevant loci were visualized using IGV software (Thorvaldsdóttir et al. 2013) to determine presence of SNPs and discordant reads consistent with structural variation.
Data availability
Raw phenotypic data are available in the Supplemental Material, File S1.
Results
GWAS
One hundred eighty-four lines of the DGRP were exposed to a dose of 100 μg of DDT and monitored over 30 hr. Two phenotypes were recorded; percentage knocked down and percentage dead. The median of the 50% knockdown time for the DGRP lines was 15 hr, while the median of 50% mortality time was 18 hr.
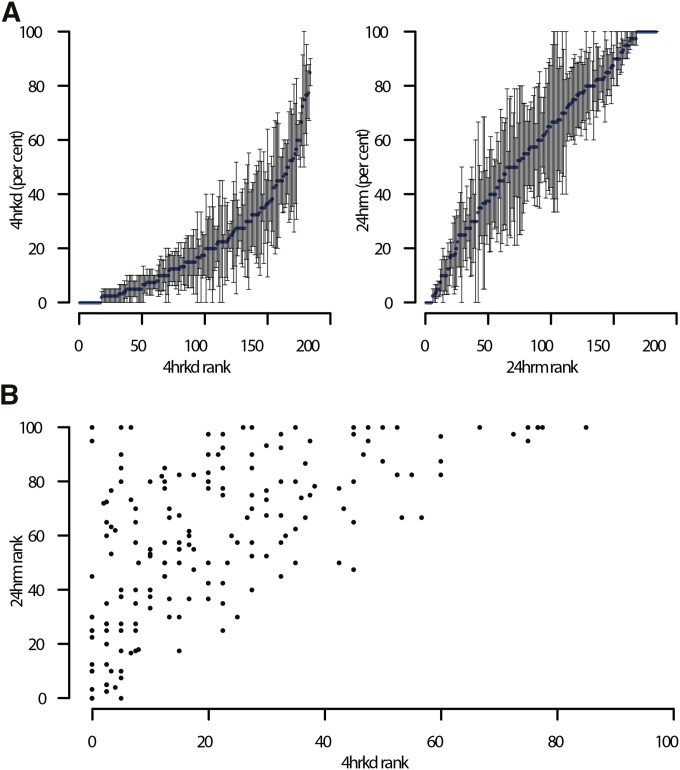
We chose to focus on 24hr mortality (24hrm), as it is a standard assay in D. melanogaster insecticide resistance literature (Daborn et al. 2001; Schmidt et al. 2010). We also observed minimal zero-dose control mortality and maximal broad-sense heritability (H2 = 0.8) at this time point. Of our knockdown timepoints, we chose 4 hrkd for further analysis, as the H2 of our assay reached an early peak at this time point and it thus provides a robust yet contrasting data set to the 24hrm (Figure 1A and File S1). Based on the number of lines phenotyped and genotyped (n = 179), the number of replicates (r = 3), and the broad-sense heritability of the trait (H2 = 0.8), the method of Mackay and Huang (2017) was used to estimate the power to detect variants for a range of effect sizes. Variants with an effect of 12% of H2 (∼0.1 units of the trait’s SD) would be detected with 50% chance and variants with an effect of 22% of H2 (∼0.18 units of the trait’s SD) with 95% chance, respectively (File S4).
Figure 1.
DDT-Knockdown and mortality. (A) The mean percentage 4-hr knockdown (4hrkd) and 24-hr mortality (24hrm) for each DGRP line (based on at least 3 replicates of 10 individuals each) with lines ordered from lowest to highest for each trait. (B) The scatterplot shows the correlation between 24-hr mortality and 4-hr knockdown. Flies that are knocked down early can recover and exhibit late mortality.
The 4 hour knockdown (4hrkd) phenotype may reveal the intrinsic variation in insecticide uptake routes and nervous system sensitivities, while the later 24hrm phenotype may encompass variation in the physiological response to the insecticide such as inducible detoxification mechanisms. Four-hour knockdown and 24hrm show distinctly different distributions within the DGRP. The population mean for 4 hrkd is 21% with SD of 19%, while 24 hrm has a mean mortality of 58%, with SD 29%. The genetic correlation between the two traits was estimated to be 0.42 (Figure 1B).
We performed GWAS on our 4hrkd and 24hrm phenotypes using both the DGRP2 webtool (Huang et al. 2014) and a custom implementation using PLINK (version 1.9). We did not observe a significant (ANOVA P > 0.05) effect on DDT-induced knockdown or mortality of Wolbacchia infection (knockdown: r2 = 0.013 and mortality: r2 = 0.010), inversions with a frequency >5% (knockdown: r2 range = −0.145–0.176 and mortality: r2 range = −0.264–0.136), or genome size (knockdown: r2 = 0.005 and mortality: r2 = 0.004). All four GWAS detected variants with P-values less than the arbitrary genome-wide significance threshold (1 × 10−5; File S2; Mackay et al. 2012). None of the variants associated with 24 hrm were also associated with the 4 hrkd phenotype. Only 24 hrm with the PLINK pipeline showed a variant with FWER <0.05, in a cytochrome P450 gene Cyp6w1. Using a less conservative cutoff of P < 1 × 10−6, 4hrkd showed eight variants. Of note, one of these is in CG10737, a gene that has previously been implicated in DDT resistance.
CG10737
Two SNPs separated by 30 nucleotides occur in the intron of CG10737 and are highly associated with 4hrkd in both PLINK and DGRP2 GWAS; they are in complete LD with each other and have slightly different P-values of association because of missing data (2R:19,169,399[V6], 2R:19,169,429[V6]; File S2). CG10737 is predicted to encode a protein with C1 and C2 domains that are capable of diacylglycerol binding and calcium-dependent targeting, respectively, and is therefore postulated to be involved in intracellular signal transduction (Mitchell et al. 2015; Attrill et al. 2016). A previous study highlighted that CG10737 is one of five “lipid metabolism” genes shown to be differentially expressed among two DDT-resistant lines and a susceptible laboratory line. Specifically, CG10737 was expressed at significantly lower levels in the DDT-resistant Wisc-1 line than the susceptible Canton-S line (Pedra et al. 2004). This led us to hypothesize that a reduction of CG10737 transcript would lead to increased resistance to DDT-knockdown. To test this hypothesis, we crossed a VDRC UAS-CG10737-KK line to knockdown this gene using a Mef2-GAL4 driver, which targets the muscles. The justification for this driver is fourfold: (i) according to the modENCODE data sets CG10737 is highly expressed in the muscle-associated tissues (crop, hindgut, heart, carcass, and male accessory gland) and is considered muscle-enriched in two microarray analyses (Zhan et al. 2007; Weake et al. 2008); (ii) genes that are coexpressed with CG10737 are known muscle genes (Myosin heavy chain, myosin light chain 1 and 2, troponin, and myosuppressins); (iii) chromatin co-immunoprecipitation experiments show that MEF2 binds to the CG10737 cis regulatory region (Sandmann et al. 2006; Cunha et al. 2010); and (iv) Bonn (2010) demonstrated that antibodies to CG10737 stained less intensely in the somatic and visceral mesoderm when in a mef2 null background.
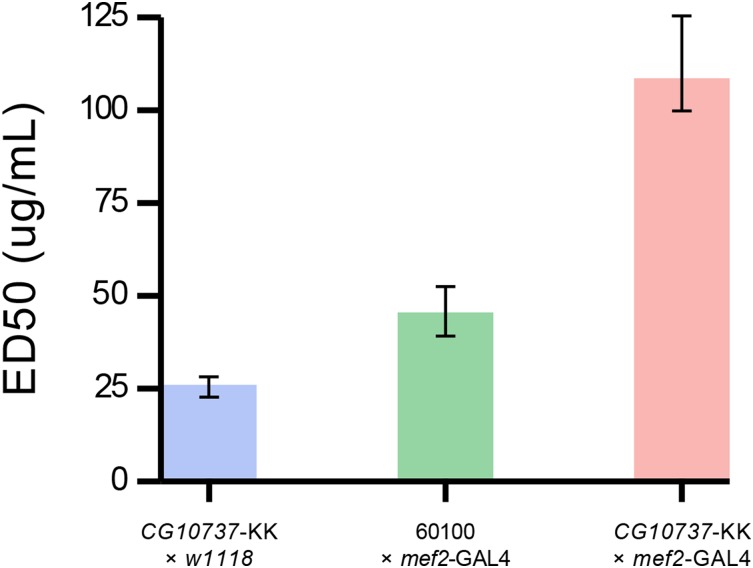
The progeny of the Mef2-GAL4 × UAS-CG10737-KK cross were assayed for DDT-knockdown resistance over a range of doses. The dose of DDT required to knockdown 50% of flies (ED50) was more than twice as high for UAS-CG10737-KK lines as it was for the control lines (109 μg/vial vs. <47 μg/vial; Figure 2). These findings support the hypothesis that CG10737 transcript reduction increases resistance to DDT-knockdown.
Figure 2.
RNAi knockdown confirms that lowering CG10737 transcript abundance increases DDT resistance and that this effect can be mediated by manipulating muscle expression. Flies in which CG10737 is knocked down in muscles using the mef2-GAL4 driver crossed to the VDRC CG10737-KK line are significantly more resistant to DDT than control flies, which are the result of substituting the relevant genetic background line for either mef2-GAL4 (w1118) or the CG10737-KK line (60100) in the cross.
Cyp6w1
This gene is the standout candidate as judged by significance of association with variation in 24hrm. The associated SNP (2R:6,174,944[V6]) results in a coding sequence change in an enzyme belonging to a family capable of detoxification and which is most abundantly expressed in the fly fat body, a known detoxification tissue. It is also expressed in the spermatheca, heart, head, and carcass (Chintapalli et al. 2007), and Pedra et al. (2004) found it to be overexpressed in the DDT-resistant 91-R line relative to the Canton-S susceptible line. The variant site associated with DDT 24hrm in Cyp6w1 is particularly interesting as three different allelic states at this site are observed within the DGRP, and each state encodes a different amino acid. Such triallelic sites, which represent 2.5% of variable sites in the DGRP, get excluded by many analyses (either explicitly, by excluding sites with more than two states, or implicitly, because the most abundant two states combined are not enough to pass abundance thresholds).We surmise that this latter factor is the reason we do not recover this variant using the Mackay DGRP2 webtool (Huang et al. 2014; http://dgrp.gnets.ncsu.edu/), as <75% of lines have a called genotype for this position. In contrast, we used a lower missing genotype threshold of 60% in our PLINK GWAS pipeline. We initially identified 2R:6,174,944[V6] as a biallelic TC transition resulting in a Val370Ala substitution. By investigating this site in the Drosophila genome DGRP2 data, we found a third state, G, that is at a frequency of 14% in the DGRP, and results in a Val370Gly substitution. Combining the effect of the C and G alleles, the P-value for association with DDT mortality is 7.88 × 10−7. Individually the G allele is insignificant genome-wide (P = 0.015), but taken together these results suggest that it too could influence DDT resistance.
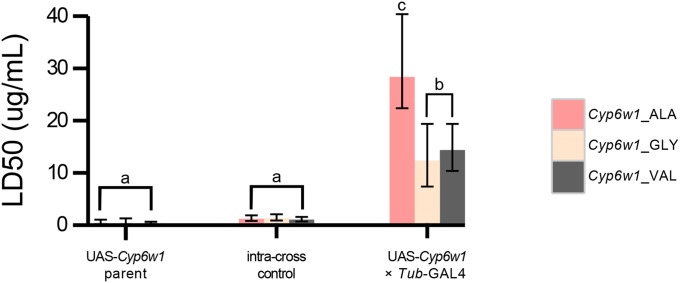
To test what effect the three amino acid states at site 370 of Cyp6w1 have on DDT resistance, three transgenic lines were generated such they that only differed at that single site. The three lines were crossed to a GAL4 driver line under the control of tubulin promoter sequence, so that the UAS-Cyp6w1 isoforms were expressed in a ubiquitous manner. All three of the Cyp6w1 transgenes were at least 12 times more resistant to DDT than their intracross sibs when under the control of the tubulin-GAL4 driver [Figure 3; LD50 (the dose predicted to result in 50% mortality) for Cyp6w1 overexpression lines: Cyp6w1_VAL = 14 μg/vial, Cyp6w1_GLY = 12 μg/vial and Cyp6w1_ALA = 28 μg/vial]. This indicates that overexpression of Cyp6w1 in itself can result in a DDT resistance. However, an additional effect is seen for the Ala370 amino acid substitution, as flies overexpressing this isoform are at least two times more resistant than either Val370 or Gly370 counterparts (Figure 3). Thus, the Ala370 substitution appears to increase DDT resistance, supporting the results of our 24hrm GWAS.
Figure 3.
Toxicology of transgenic lines overexpressing the three allelic forms of Cyp6w1 (far right) and various controls. All three allelic forms of Cyp6w1 confer DDT resistance relative to controls, and Cyp6w1_ALA confers a further significant increase in DDT resistance relative to the other two alleles.
As our transgenic analyses showed that Cyp6w1 overexpression can yield resistance and Cyp6w1 transcripts are more abundant in Pedra et al. (2004), we examined Cyp6w1 transcription in females from a DGRP transcriptome data set (Huang et al. 2015). However, there was no strong correlation between Cyp6w1 transcription level and DDT mortality or knockdown data, nor between Cyp6w1 transcription level and the triallelic states of Cyp6w1. Similarly, we did not find any of the other eight detoxification or four lipid metabolism genes that were focused on by Pedra et al. (2004) to have strong correlations between the DGRP transcription data sets and either DDT traits we examined (File S3).
Selective sweep tests at DDT-associated loci
To assess whether 4 hrkd- and 24 hrm-associated (P < 1 × 10−5) variants from the PLINK GWAS exhibited evidence of being the targets of selection, we used the integrated Haplotype Score (iHS) test (Sabeti et al. 2002; Voight et al. 2006). We also calculated the related statistic nSL (number of segregating sites by length; Ferrer-Admetlla et al. 2014), which defines haplotype lengths via pairwise differences and may have higher power to detect soft selective sweeps. The only derived variant yielding a significant iHS or nSL score was the Glycine allele of Cyp6w1 (|iHS| = 3.82, P-value = 0.00085; Figure 4; |nSL| = 3.04, P-value = 0.0028). The other derived variants did not show significant departures from expectation, including the second derived allele at Cyp6w1 – the alanine allele that confers increased DDT resistance in our transgenic experiments.
To ascertain the power of these iHS and nSL approaches with this data set we also examined Cyp6g1, the genomic region around which Catania et al. (2004), Schlenke and Begun (2004), and Garud et al. (2015) noted a reduction in genetic diversity indicative of a selective sweep. While there is an allelic series at this locus (Schmidt et al. 2010), the two predominate alleles in the DGRP are Cyp6g1-AA and Cyp6g1-BA, and the EHH for both of these alleles and for the ancestral Cyp6g1-M allele was determined. Clearly both derived Cyp6g1 alleles exhibit large extended haplotypes. Both of these are significant compared to ancestral Cyp6g1-M allele (Figure 4; Cyp6g1-AA: |iHS| = 2.45, P-value = 0.018; |nSL| = 2.61, P-value = 0.011; Cyp6g1-BA: |iHS| = 1.96, P-value = 0.027; |nSL| = 2.47, P-value = 0.012).
Further evidence that insecticide resistance alleles may be adaptive comes from elevated levels of population differentiation (Taylor et al. 1995). We examined Cyp6w1 allele frequencies in African, European, Australian, and North American populations (Pool et al. 2012; Martins et al. 2014; Reinhardt et al. 2014; Bergman and Haddrill 2015; Table 1); most populations indicate segregation of each of the triallelic states, except for Portugal, which does not exhibit Ala370. The frequency of the variants, however, shows marked population differentiation, perhaps indicating geographical variation in selection intensity by DDT or other insecticides.
Table 1. Population frequencies of Cyp6w1 codon 370 trialleles.
| Continent | Population | Data Source | Sample Size | Allele Frequency | ||
|---|---|---|---|---|---|---|
| VAL | ALA | GLY | ||||
| Africa | Combined | Pool et al. (2012) | 139 | 0.9 | 0.07 | 0.3 |
| Australia | Queensland | Reinhardt et al. (2014) | 17 | 0.53 | 0.03 | 0.46 |
| Tasmania | Reinhardt et al. (2014) | 15 | 1 | 0 | 0 | |
| Europe | France – A | Bergman and Haddrill 2015 | 50 | 0.78 | 0.02 | 0.2 |
| France – B | Pool et al. (2012) | 8 | 0.5 | 0 | 0.5 | |
| Portugal | Martins et al. (2014) | 12 | 0.95 | 0 | 0.05 | |
| North America | Maine | Reinhardt et al. (2014) | 16 | 0.69 | 0.25 | 0.06 |
| North Carolina | Mackay et al. (2012) | 162 | 0.54 | 0.32 | 0.14 | |
| Florida | Reinhardt et al. (2014) | 16 | 0.63 | 0.31 | 0.06 | |
Discussion
The DGRP is a powerful resource for identifying natural variation contributing to a range of phenotypes (Mackay and Huang 2017). However, it is of particular interest to the study of insecticide resistance due to the important role insecticides have apparently played in the evolutionary history of this population; two of the strongest peaks of selection in the DGRP, genome wide, are Ace and Cyp6g1 (Garud et al. 2015).
Ace is the molecular target of organophosphate and carbamate insecticides, and within the DGRP segregate three of the four substitutions in Ace known to confer resistance to these insecticide classes in D. melanogaster (Menozzi et al. 2004; Battlay et al. 2016). Cyp6g1 has likewise been demonstrated to provide resistance to insecticides, including the neonicotinoids imidacloprid and nitenpyram (Daborn et al. 2001, 2007; Joußen et al. 2008) and the organophosphate azinphos-methyl (Battlay et al. 2016). There is also evidence that Cyp6g1 may contribute to resistance in other organophosphates including malathion, parathion, and diazinon (Kikkawa 1961; Pyke et al. 2004). But the best studied of Cyp6g1’s insecticide substrates remains DDT (Daborn et al. 2002, 2007; Joußen et al. 2008; Schmidt et al. 2010); derived alleles of Cyp6g1 have previously been shown to contribute significantly to DDT resistance, with the Cyp6g1-BP allele explaining ∼16% of the total variance in DDT resistance in a field population from Queensland, Australia (Schmidt et al. 2010).
While DDT is not used in much of the world today, DDT resistance is a trait of interest for two important reasons: first, it may help to explain the strong and recent sweeps at the Cyp6g1 locus (Schmidt et al. 2010; Kolaczkowski et al. 2011; Garud et al. 2015), and second it was the subject of the classic work by Crow (1956), Sokal and Hunter (1954), and others on microevolution. We performed GWAS of two DDT resistance phenotypes in the hope of dissecting the quantitative nature of a selective response to DDT exposure.
Cyp6g1 was not identified in the GWAS
The association of Cyp6g1 allelic variation with either the 4 hrkd or 24 hrm DDT phenotypes is not significant, even at the genome-wide significance threshold. This is partly attributable to the Cyp6g1 sweep itself; resistant Cyp6g1-AA and Cyp6g1-BA alleles are present in all but nine DGRP lines (all nine were phenotyped in this study), while the Cyp6g1-BP allele, which confers the most drastic increase in DDT resistance (Schmidt et al. 2010), is absent from the population. Although much of the power to detect the effect of Cyp6g1 resistance is gone from the DGRP, Battlay et al. (2016) have previously shown that a haplotype in LD with the ancestral Cyp6g1-M allele was strongly associated with susceptibility to a low dose of the organophosphate insecticide azinphos-methyl in DGRP larvae. One reason Cyp6g1 was associated with the azinphos-methyl phenotype and not our DDT phenotypes could be the relative contributions of Cyp6g1 overexpression to these traits: Daborn et al. (2007) demonstrated a threefold increase in adult DDT LD50 when Cyp6g1 was overexpressed using the GAL4-UAS system, while Battlay et al. (2016) observed a 6.5-fold increase in larval azinphos-methyl LD50 in the same crosses. It is also possible that the dose of the DDT used in this study did not maximize the chances of recovering the Cyp6g1 variation present in the DGRP. Steele et al. (2014) also reported an absence of Cyp6g1 signal in their comparison of 91-R and 91-C genome sequences. This is due, however, to Steele et al. (2014) limiting their study to analysis of open reading frames. We revisited genome sequence alignments generated by Steele et al. (2014) and found that evidence of the Accord LTR insertion into the 5′ UTR of Cyp6g1 (Daborn et al. 2002) as well as copy number variation in both Cyp6g1 and Cyp6g2 (Schmidt et al. 2010), both identifiers of Cyp6g1 resistance alleles, are present in 91-R but not 91-C.
The two detoxification enzymes implicated in DDT mortality, Cyp6g1 and Cyp6w1, could interact in parallel or in series. Epistatic interactions between alleles at the two loci were examined but, unfortunately, we did not have the power to test for such interaction in the DGRP because there is only one DGRP line (Ral-486) that is homozygous for the susceptible Cyp6g1-M allele and homozygous for the resistant Cyp6w1-Ala allele. We have, however, assessed pairwise epistatic interactions of the GWAS nominally associated variants listed in the combined mortality and knockdown lists, and none were significantly extreme to pass the Bonferroni correction threshold (∼0.000015) for either the knockdown (lowest value: 0.0002) or the mortality (lowest value: 0.0015) traits.
Variation at CG10737 contributes to 4-hrkd but not 24-hrm
The GWAS presented here demonstrate that the two DDT-associated traits under examination have distinct genetic architecture, and for each trait we have generated transgenic evidence to support the involvement of a top candidate in DDT phenotype variation. CG10737 has previously been implicated as a DDT resistance candidate, being one of 158 genes shown to be significantly differentially regulated between DDT-resistant 91-R and a standard susceptible laboratory stock (Canton-S; Pedra et al. 2004). Moreover, inspection of the 91-R and 91-C genome sequences generated by Steele et al. (2014) shows that both CG10737 GWAS SNPs, associated with increased knockdown susceptibility, are present in 91-C but absent in 91-R. Using a completely independent approach we have found that these noncoding variants in CG10737, present in ∼10% of the DGRP, are strongly associated with 4 hrkd. Our RNAi knockdown of CG10737 increased DDT resistance, consistent with Pedra et al.’s (2004) observation that the CG10737 transcript was less abundant in the resistant 91-R line. We note that the significantly associated GWAS variants also lie in a region that modENCODE found to bind the Trithorax-like (Trl) GAGA factor, suggesting an untested mechanism linking the naturally occurring polymorphisms to transcriptional abundance.
An important insight gained through these experiments is that they demonstrate a role of CG10737 in the muscles of flies, and a relationship between DDT and muscle biology. CG10737 knockdown in the muscles resulted in a two- to fourfold increase in resistance, demonstrating that DDT perturbation can be ameliorated by genetic manipulation limited to the muscles.
The molecular function of CG10737 has not been well characterized. It is a gene with 14 coding exons and produces at least 12 alternatively spliced mRNA isoforms. It putatively encodes a transmembrane protein with C1 and C2 calcium/lipid-binding domains and it has been annotated as “protein kinase c-like” although it lacks a catalytic domain. C1 domains have cysteine motifs that form zinc fingers and typically bind diacylglycerol (Colón-González and Kazanietz 2006), while C2 domains typically bind calcium ions and are associated with protein–protein and protein–membrane interactions (Ochoa et al. 2001). Given the important role that calcium plays in synapse and muscle signaling, perhaps a decrease in the expression level of CG10737 makes flies less sensitive to Ca2+ ion signaling. It thereby may reduce the expression of the DDT-knockdown phenotype by mediating the effect of overstimulation of muscle cells caused by the uncontrolled firing of nerves due to DDT’s effect on the voltage-gated sodium channel, Para, rather than influencing nerve functionality directly.
While CG10737 is associated with DDT-knockdown in the GWAS, it was not significantly associated with 24hrm GWAS. Furthermore, a reexamination of the DDT-knockdown phenotype for DGRP lines over the initial time course of the experiment found that variants in this gene were only strongly associated with knockdown over a narrow time window (3–5 hr), suggesting that the gene’s role in knockdown resistance is transient. Additionally, in the RNAi experiments, the dose that effectively interrogated the temporally defined knockdown was so high that the flies did not live long enough to accurately score 24-hrm. This is attributable to genetic background differences between the lines going into the RNAi crosses and the DGRP lines. These issues highlight the differences in genetic architecture of subtly different DDT-related traits. While the knockdown trait may give us insight into the effect of perturbations of DDT on insect physiology, and the function of previously uncharacterized genes, one has to ask how relevant this variation is to the field exposure to DDT. We were unable to detect any signature of selection around variants in this gene, or indeed any high-frequency derived knockdown-associated variants, that would indicate microevolutionary change in the recent history of the Raleigh population of D. melanogaster – the source of the DGRP. However, this may reflect an issue of power to detect subtle changes in frequency of preexisting variants rather than an irrelevance of the phenotype to survivorship in the field. We also note that in early DDT selection experiments in D. melanogaster by Sokal and Hunter (1954), pupation height was correlated with DDT resistance and that Riedl et al. (2007) mapped a pupal height QTL to a narrow region encompassing CG10737.
Triallelic variation at Cyp6w1 separates DDT-resistant allele from a selective sweep signal
Like CG10737, Cyp6w1 was previously identified as a DDT resistance candidate by Pedra et al. (2004), who found it had high transcriptional abundance in 91-R relative to the susceptible Canton-S. Again, inspection of the Steele et al. (2014) genomes revealed that 91-R also carries the DDT resistance-associated Cyp6w1_ALA allele, whereas 91-C carries the Cyp6w1_GLY allele. While our transgenic manipulation of this gene shows that increased expression of three different alleles of Cyp6w1 can indeed lead to increased resistance to DDT, it is amino acid variants that appear to explain the difference in resistance between DGRP lines. The associated variant is a rare triallelic site where each state encodes a different amino acid (Valine GTG, Alanine GCG, or Glycine GGG). No sites are in LD with it that are also significantly associated with 24hrm, and the sharp and immediate decrease in EHH for both the Val370 and Ala370 variants makes it highly unlikely that this variant is a proxy for an undetected, linked regulatory change polymorphism. Amino acid site 370 of Cyp6w1 is predicted to occur in Substrate Recognition Domain SRS5 (Gotoh 1992; Zawaira et al. 2011), of an enzyme in a gene family frequently associated with insecticide resistance. It is therefore completely credible that it acts to metabolize DDT, perhaps in a similar way to its paralog Cyp6g1 (Joußen et al. 2008; Hoi et al. 2014).
Inspection of CYP6W1 site 370 in related species suggests that Val370 is the ancestral state of this site; the Ala370 and Gly370 substitutions are therefore both derived. The Ala370 variant is strongly associated (P = 5 × 10−10) with reduced 24hrm in our GWAS, and this is supported by our transgenic work, which found overexpression of Cyp6w1_ALA increased DDT resistance relative to Cyp6w1_VAL and Cyp6w1_GLY. Calculation of iHS and nSL did identify a significant signal of selection at Cyp6w1_ALA, but it was associated with the other derived allele, Gly370. This allele is not significantly associated with 24-hrm in our GWAS, and transgenic overexpression of Cyp6w1_GLY does confer resistance to DDT relative to Cyp6w1_VAL. There is no evidence, therefore, that DDT is responsible for the selective footprint identified at Cyp6w1, and we propose that the Gly370 mutation increases fitness under a different selective pressure, possibly by increasing the affinity of Cyp6w1 to another insecticide. The extreme population differentiation observed (Table 1) is also consistent with multiple selective agents acting on this site.
GWAS-associated variants in 91-R and 91-C
Interestingly, resistance-associated variants in CG10737 and Cyp6w1 from our GWAS were present in 91-R but not 91-C. To see if any of our other DGRP resistance candidates were differentiated between 91-R and 91-C, we manually searched the genome sequences of these lines generated by Steele et al. (2014) for each of our GWAS candidates (P < 1 × 10−5; File S2). Fifty distinct GWAS candidate variants varied between 91-R and 91-C. Of these, 17 (including the two SNPs in CG10737 and the Cyp6w1 triallele) varied in the expected direction, with the “resistant” allele in 91-R and the “susceptible” allele in 91-C (Table 2). Thus, these variants may have been segregating in the population used to found 91-R and 91-C and potentially contributed to the strong selection response observed in 91-R.
Table 2. GWAS resistance variants enriched in 91-R but not 91-C.
| Phenotype | Pipeline | Location [V6] | P-value | Gene | Site class | 91-C | 91-R |
|---|---|---|---|---|---|---|---|
| 24 hrm | PLINK | 2R:6,174,944 | 1.72E−09 | Cyp6w1 | Nonsynonymous | T | C |
| 4 hrkd | DGRP2, PLINK | 2R:19,169,429 | 4.28E−07, 7.58E−07 | CG10737 | Intron | T | G |
| 4 hrkd | DGRP2 | 2L:14,188,350 | 1.64E−06 | smi35A | Intron | G | G/C |
| 4 hrkd | PLINK | 3L:4,059,748 | 1.75E−06 | — | Intergenic | T | C |
| 4 hrkd | PLINK | 2L:5,600,056 | 1.97E−06 | — | Intergenic | C | A/C |
| 4 hrkd | PLINK | 3L:12,404,415 | 3.00E−06 | CG32103 | Synonymous | C | C/T |
| 4 hrkd | DGRP2 | 3R:30,968,896 | 3.70E−06 | — | Intergenic | A | ATA |
| 4 hrkd | DGRP2, PLINK | 2R:19,169,399 | 5.45E−06; 7.62E−06 | CG10737 | Intron | T | C |
| 24 hrm | PLINK | 2L:10,546,377 | 5.68E−06 | Trim9 | Intron | A | A/G |
| 4 hrkd | DGRP2 | 3L:7,102,962 | 6.79E−06 | form3 | Intron | C | C/T |
| 4 hrkd | PLINK | 3L:6,935,976 | 7.88E−06 | — | Intergenic | A | A/G |
| 24 hrm | PLINK | 3L:15,854,279 | 8.76E−06 | pHCl | Intron | G | A |
| 4 hrkd | DGRP2 | 2L:8,232,517 | 9.49E−06 | Pvr | Intron | C/A | C |
| 4 hrkd | DGRP2 | 2L:4,086,206 | 9.57E−06 | ed | Intron | T | G |
| 4 hrkd | DGRP2 | 2L:19,231,491 | 1.66E−05 | — | Intergenic | A | G |
| 4 hrkd | DGRP2, PLINK | 3L:7,495,781 | 2.02E−05; 7.38E−06 | Cyp4d8 | Intron | G | T |
| 4 hrkd | DGRP2 | 3R:25,716,964 | 2.15E−05 | LpR2 | Intron | A | C |
Other DDT resistance genes
Three substitutions in Cyp6a2 increasing its DDT metabolism capacity have been described in RalDDTR (Amichot et al. 2004), a laboratory strain selected for DDT resistance over 50 generations (Cùany et al. 1990). The first two substitutions (R335S and L336V) are absent from the DGRP, as well as population samples from Africa Drosophila Population Genomics Project; (DPGP; Pool et al. 2012), France and Portugal in Europe, Florida and Maine in the USA, and Queensland and Tasmania in Australia (Pool et al. 2012; Martins et al. 2014; Reinhardt et al. 2014; Bergman and Haddrill 2015). The third substitution, V476L, is found in around a quarter of DGRP and DPGP sequences (24.1% of DGRP, 26.5% of screened DGRP, 32.2% of DPGP), but this variant does not affect DDT metabolism by itself (Amichot et al. 2004), and was not associated with either 4 hrkd or 24 hrm at P < 1 × 10−5. Similarly, the resistance-linked Cyp6a2 promoter variant described by Wan et al. (2014) is present in the DGRP (68.8% of DGRP, 68.6% of screened DGRP), but was not associated with either of our phenotypes at P < 1 × 10−5.
Another P450, Cyp12d1, is overexpressed in the DDT-resistant Wisc-1 strain relative to Canton-S (Pedra et al. 2004) and has been shown to increase DDT resistance when overexpressed transgenically (Daborn et al. 2007), as well as reducing resistance when knocked down with RNAi (Gellatly et al. 2015). We did not find associations with Cyp12d1 in any of our four GWAS, nor did we find strong correlations between our phenotypes and Cyp12d1 copy number variation or transcription level (File S3). Steele et al. (2014) likewise found no differentiation in Cyp12d1 coding regions between 91-R and 91-C, and our inspection of these genomes found no differentiation in copy number between the two strains.
Cyp12d1 has been shown to be inducible by DDT (Brandt et al. 2002; Festucci-Buselli et al. 2005; Willoughby et al. 2006), as well as other xenobiotics (Willoughby et al. 2006; Misra et al. 2011). Our results do not rule out the involvement of this gene in DDT resistance in the DGRP, but show that genomic variation at Cyp12d1 and its transcription level without induction do not correlate with the DDT resistance phenotypes measured in this study (File S5).
In the case of para, neither the kdr nor super-kdr resistance mutations are present in the DGRP, nor any of the analogous sites conferring resistance in mutagenized lines characterized by Pittendrigh et al. (1997) or Lindsay et al. (2008).
Conclusions
There are three main lines of evidence used in the literature to associate genes with DDT resistance in D. melanogaster: differentiation between resistant and susceptible laboratory strains, association of natural alleles with resistance in outbred populations, and transgenic manipulation in a controlled genetic background. Until now, only Cyp6g1 has bridged the divides between these approaches. With this study, Cyp6w1 and CG10737 can be added to this list. These results alone support a polygenic model and are reinforced by the possibility that other genes from our GWAS, or other studies, also contribute to DDT resistance but have yet to be verified. We have focused on two specific DDT-related traits; 4hrkd and 24hrm, both in adult females with a single dose of DDT, and they give only partially correlated results. Which of these has greater relevance in the field, however, is not clear. Furthermore, there are many alternate parameter values and indeed parameters (e.g., temperature; Fournier-Level et al. 2016) that may make laboratory-based DDT assays more accurately reflect the variation most relevant to field survivorship. Perhaps one of the most compelling diagnostics for a relevant assay would be if the loci uncovered showed the molecular signs of selection. Previous studies have revealed that the major DDT resistance locus of some populations, Cyp6g1, shows the hallmarks of a selective sweep. We did not, however, observe signs of selection around our DGRP GWAS candidates. While we did detect signs of a sweep at Cyp6w1, it was associated with a second derived variant, not with the variant associated with DDT resistance. Given that Cyp6g1 is also capable of providing resistance to other insecticides, the failure to find sweeps at DDT-specific resistance loci strengthens the possibility that other insecticides have contributed to driving the Cyp6g1 selective sweeps in field populations of D. melanogaster. Finally, it is also a possibility that statistics such as iHS and nSL are insensitive to the slight changes in allele frequency that might occur if a trait is largely polygenic.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.300310/-/DC1.
Acknowledgments
Jason Somers contributed to the cloning and transgenic delivery of the Cyp6w1 constructs. We thank Trudy Mackay for her advice and support for this work, and the anonymous reviewers for suggestions that improved this manuscript. This work was supported by Australian Research Council (ARC) Discovery grant DP0985013.
Footnotes
Communicating editor: C. Jones
Literature Cited
- Allen H. L., Estrada K., Lettre G., Berndt S. I., Weedon M. N., et al. , 2010. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 467: 832–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amichot M., Tares S., Brun‐Barale A., Arthaud L., Bride J. M., et al. , 2004. Point mutations associated with insecticide resistance in the Drosophila cytochrome P450 Cyp6a2 enable DDT metabolism. Eur. J. Biochem. 271: 1250–1257. [DOI] [PubMed] [Google Scholar]
- Attrill H., Falls K., Goodman J. L., Millburn G. H., Antonazzo G., et al. , 2016. FlyBase: establishing a gene group resource for Drosophila melanogaster. Nucleic Acids Res. 44: D786–D792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battlay P., Schmidt J. M., Fournier-Level A., Robin C., 2016. Genomic and transcriptomic associations identify a new insecticide resistance phenotype for the selective sweep at the Cyp6g1 locus of Drosophila melanogaster. G3 (Bethesda) 6: 2573–2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baucom R. S., 2016. The remarkable repeated evolution of herbicide resistance. Am. J. Bot. 103: 181–183. [DOI] [PubMed] [Google Scholar]
- Bergman C. M., Haddrill P. R., 2015. Strain-specific and pooled genome sequences for populations of Drosophila melanogaster from three continents. F1000Res. 4: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J., Maeda R. K., Hediger M., Karch F., Basler K., 2007. An optimized transgenesis system for Drosophila using germ-line-specific φC31 integrases. Proc. Natl. Acad. Sci. USA 104: 3312–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonn, B., 2010 Myosin heavy chain like (Mhcl) agiert während der Embryonalentwicklung und Myogenese von Drosophila melanogaster in Redundanz zu Zipper, die Funktion des C2-Domänen-Proteins CG10737-P während der Muskelentwicklung bleibt unklar. Ph.D. Thesis, Philipps-Universität, Marburg. [Google Scholar]
- Brandt A., Scharf M., Pedra J. H. F., Holmes G., Dean A., et al. , 2002. Differential expression and induction of two Drosophila cytochrome P450 genes near the Rst (2) DDT locus. Insect Mol. Biol. 11: 337–341. [DOI] [PubMed] [Google Scholar]
- Catania F., Kauer M. O., Daborn P. J., Yen J. L., Ffrench-Constant R. H., et al. , 2004. World‐wide survey of an Accord insertion and its association with DDT resistance in Drosophila melanogaster. Mol. Ecol. 13: 2491–2504. [DOI] [PubMed] [Google Scholar]
- Chang C. C., Chow C. C., Tellier L. C., Vattikuti S., Purcell S. M., et al. , 2015. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintapalli V. R., Wang J., Dow J. A., 2007. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 39: 715–720. [DOI] [PubMed] [Google Scholar]
- Chung H., Bogwitz M. R., McCart C., Andrianopoulos A., Batterham P., et al. , 2007. Cis-regulatory elements in the Accord retrotransposon result in tissue-specific expression of the Drosophila melanogaster insecticide resistance gene Cyp6g1. Genetics 175: 1071–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clowers K. J., Lyman R. F., Mackay T. F., Morgan T. J., 2010. Genetic variation in senescence marker protein-30 is associated with natural variation in cold tolerance in Drosophila. Genet. Res. 92: 103–113. [DOI] [PubMed] [Google Scholar]
- Colón-González F., Kazanietz M. G., 2006. C1 domains exposed: from diacylglycerol binding to protein–protein interactions. Biochim. Biophys. Acta 1761: 827–837. [DOI] [PubMed] [Google Scholar]
- Comeron J. M., Ratnappan R., Bailin S., 2012. The many landscapes of recombination in Drosophila melanogaster. PLoS Genet. 8: e1002905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow J. F., 1954. Analysis of a DDT-resistant strain of Drosophila. J. Econ. Entomol. 47: 393–398. [Google Scholar]
- Crow J. F., 1956. Genetics of DDT resistance in Drosophila. Cytologia Proceedings of the International Genetics Symposia, pp. 408–409. Tokyo, Japan. [Google Scholar]
- Crow J. F., 1957. Genetics of insect resistance to chemicals. Annu. Rev. Entomol. 2: 227–246. [Google Scholar]
- Cùany A., Pralavorio M., Pauron D., Berge J. B., Fournier D., et al. , 1990. Characterization of microsomal oxidative activities in a wild-type and in a DDT resistant strain of Drosophila melanogaster. Pestic. Biochem. Physiol. 37: 293–302. [Google Scholar]
- Cunha P. M., Sandmann T., Gustafson E. H., Ciglar L., Eichenlaub M. P., et al. , 2010. Combinatorial binding leads to diverse regulatory responses: Lmd is a tissue-specific modulator of Mef2 activity. PLoS Genet. 6: e1001014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daborn P., Boundy S., Yen J., Pittendrigh B., 2001. DDT resistance in Drosophila correlates with Cyp6g1 over-expression and confers cross-resistance to the neonicotinoid imidacloprid. Mol. Genet. Genomics 266: 556–563. [DOI] [PubMed] [Google Scholar]
- Daborn P. J., Yen J. L., Bogwitz M. R., Le Goff G., Feil E., et al. , 2002. A single P450 allele associated with insecticide resistance in Drosophila. Science 297: 2253–2256. [DOI] [PubMed] [Google Scholar]
- Daborn P. J., Lumb C., Boey A., Wong W., Batterham P., 2007. Evaluating the insecticide resistance potential of eight Drosophila melanogaster cytochrome P450 genes by transgenic over-expression. Insect Biochem. Mol. Biol. 37: 512–519. [DOI] [PubMed] [Google Scholar]
- Dapkus D., 1992. Genetic localization of DDT resistance in Drosophila melanogaster (Diptera: Drosophilidae). J. Econ. Entomol. 85: 340–347. [DOI] [PubMed] [Google Scholar]
- Dapkus D., Merrell D. J., 1977. Chromosomal analysis of DDT-resistance in a long-term selected population of Drosophila melanogaster. Genetics 87: 685–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer-Admetlla A., Liang M., Korneliussen T., Nielsen R., 2014. On detecting incomplete soft or hard selective sweeps using haplotype structure. Mol. Biol. Evol. 31: 1275–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festucci-Buselli R. A., Carvalho-Dias A. S., Oliveira-Andrade D., Caixeta-Nunes C., Li H. M., et al. , 2005. Expression of Cyp6g1 and Cyp12d1 in DDT resistant and susceptible strains of Drosophila melanogaster. Insect Mol. Biol. 14: 69–77. [DOI] [PubMed] [Google Scholar]
- Ffrench-Constant R. H., 2013. The molecular genetics of insecticide resistance. Genetics 194: 807–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier-Level A., Neumann-Mondlak A., Good R. T., Green L. M., Schmidt J. M., et al. , 2016. Behavioural response to combined insecticide and temperature stress in natural populations of Drosophila melanogaster. J. Evol. Biol. 29: 1030–1044. [DOI] [PubMed] [Google Scholar]
- Garud N. R., Messer P. W., Buzbas E. O., Petrov D. A., 2015. Recent selective sweeps in North American Drosophila melanogaster show signatures of soft sweeps. PLoS Genet. 11: e1005004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellatly K. J., Yoon K. S., Doherty J. J., Sun W., Pittendrigh B. R., et al. , 2015. RNAi validation of resistance genes and their interactions in the highly DDT-resistant 91-R strain of Drosophila melanogaster. Pestic. Biochem. Physiol. 121: 107–115. [DOI] [PubMed] [Google Scholar]
- Gotoh O., 1992. Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J. Biol. Chem. 267: 83–90. [PubMed] [Google Scholar]
- Hoi K. K., Daborn P. J., Battlay P., Robin C., Batterham P., et al. , 2014. Dissecting the insect metabolic machinery using twin ion mass spectrometry: a single P450 enzyme metabolizing the insecticide imidacloprid in vivo. Anal. Chem. 86: 3525–3532. [DOI] [PubMed] [Google Scholar]
- Huang W., Massouras A., Inoue Y., Peiffer J., Ràmia M., et al. , 2014. Natural variation in genome architecture among 205 Drosophila melanogaster genetic reference panel lines. Genome Res. 24: 1193–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Carbone M. A., Magwire M. M., Peiffer J. A., Lyman R. F., et al. , 2015. Genetic basis of transcriptome diversity in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 112: E6010–E6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joußen N., Heckel D. G., Haas M., Schuphan I., Schmidt B., 2008. Metabolism of imidacloprid and DDT by P450 CYP6G1 expressed in cell cultures of Nicotiana tabacum suggests detoxification of these insecticides in Cyp6g1‐overexpressing strains of Drosophila melanogaster, leading to resistance. Pest Manag. Sci. 64: 65–73. [DOI] [PubMed] [Google Scholar]
- Kikkawa H., 1961. Genetical studies on the resistance to parathion in Drosophila melanogaster. Annu. Rep. Sci. Wks Osaka Univ. 9: 1–20. [Google Scholar]
- King J. C., Sømme L., 1958. Chromosomal analyses of the genetic factors for resistance to DDT in two resistant lines of Drosophila melanogaster. Genetics 43: 577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolaczkowski B., Kern A. D., Holloway A. K., Begun D. J., 2011. Genomic differentiation between temperate and tropical Australian populations of Drosophila melanogaster. Genetics 187: 245–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuruganti S., Lam V., Zhou X., Bennett G., Pittendrigh B. R., et al. , 2007. High expression of Cyp6g1, a cytochrome P450 gene, does not necessarily confer DDT resistance in Drosophila melanogaster. Gene 388: 43–53. [DOI] [PubMed] [Google Scholar]
- Lack J. B., Cardeno C. M., Crepeau M. W., Taylor W., Corbett-Detig R. B., et al. , 2015. The Drosophila genome nexus: a population genomic resource of 623 Drosophila melanogaster genomes, including 197 from a single ancestral range population. Genetics 199: 1229–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R., 1983. The response to selection on major and minor mutations affecting a metrical trait. Heredity 50: 47–65. [Google Scholar]
- Leinonen R., Sugawara H., Shumway M., International Nucleotide Sequence Database Collaboration , 2011. The sequence read archive. Nucleic Acids Res. 39: D19–D21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay H. A., Baines R., Lilley K., Jacobs H. T., O’Dell K. M., 2008. The dominant cold-sensitive Out-cold mutants of Drosophila melanogaster have novel missense mutations in the voltage-gated sodium channel gene paralytic. Genetics 180: 873–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T. F. C., Huang W., 2017. Charting the genotype-phenotype map: lessons from the Drosophila melanogaster genetic reference panel. Wiley Interdiscip. Rev. Dev. Biol. DOI: 10.1002/wdev.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T. F. C., Richards S., Stone E. A., Barbadilla A., Ayroles J. F., et al. , 2012. The Drosophila melanogaster genetic reference panel. Nature 482: 173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnair M. R., 1991. Why the evolution of resistance to anthropogenic toxins normally involves major gene changes: the limits to natural selection. Genetica 84: 213–219. [Google Scholar]
- Martins N. E., Faria V. G., Nolte V., Schlötterer C., Teixeira L., et al. , 2014. Host adaptation to viruses relies on few genes with different cross-resistance properties. Proc. Natl. Acad. Sci. USA 111: 5938–5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie J. A., 1996. Ecological and Evolutionary Aspects of Insecticide Resistance. RG Landes, Austin, TX. [Google Scholar]
- McKenzie J. A., 2000. The character or the variation: the genetic analysis of the insecticide-resistance phenotype. Bull. Entomol. Res. 90(1): 3–7. [DOI] [PubMed] [Google Scholar]
- McKenzie J. A., Batterham P., 1994. The genetic, molecular and phenotypic consequences of selection for insecticide resistance. Trends Ecol. Evol. 9: 166–169. [DOI] [PubMed] [Google Scholar]
- Menozzi P., Shi M. A., Lougarre A., Tang Z. H., Fournier D., 2004. Mutations of acetylcholinesterase which confer insecticide resistance in Drosophila melanogaster populations. BMC Evol. Biol. 4: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra J. R., Horner M. A., Lam G., Thummel C. S., 2011. Transcriptional regulation of xenobiotic detoxification in Drosophila. Genes Dev. 25: 1796–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A., Chang H. Y., Daugherty L., Fraser M., Hunter S., et al. , 2015. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 43: D213–D221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa W. F., Garcia-Garcia J., Fita I., Corbalan-Garcia S., Verdaguer N., et al. , 2001. Structure of the C2 domain from novel protein kinase Cϵ. A membrane binding model for Ca 2+-independent C2 domains. J. Mol. Biol. 311: 837–849. [DOI] [PubMed] [Google Scholar]
- Pedra J. H. F., McIntyre L. M., Scharf M. E., Pittendrigh B. R., 2004. Genome-wide transcription profile of field-and laboratory-selected dichlorodiphenyltrichloroethane (DDT)-resistant Drosophila. Proc. Natl. Acad. Sci. USA 101: 7034–7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittendrigh B., Reenan R., Ganetzky B., 1997. Point mutations in the Drosophila sodium channel gene para associated with resistance to DDT and pyrethroid insecticides. Mol. Gen. Genet. 256: 602–610. [DOI] [PubMed] [Google Scholar]
- Pool J. E., Corbett-Detig R. B., Sugino R. P., Stevens K. A., Cardeno C. M., et al. , 2012. Population genomics of sub-Saharan Drosophila melanogaster: African diversity and non-African admixture. PLoS Genet. 8: e1003080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke F. M., Bogwitz M. R., Perry T., Monk A., Batterham P., et al. , 2004. The genetic basis of resistance to diazinon in natural populations of Drosophila melanogaster. Genetica 121: 13–24. [DOI] [PubMed] [Google Scholar]
- Reinhardt J. A., Kolaczkowski B., Jones C. D., Begun D. J., Kern A. D., 2014. Parallel geographic variation in Drosophila melanogaster. Genetics 197: 361–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl C. A., Riedl M., Mackay T. F., Sokolowski M. B., 2007. Genetic and behavioral analysis of natural variation in Drosophila melanogaster pupation position. Fly (Austin) 1: 23–32. [DOI] [PubMed] [Google Scholar]
- Roush R. T., McKenzie J. A., 1987. Ecological genetics of insecticide and acaricide resistance. Annu. Rev. Entomol. 32: 361–380. [DOI] [PubMed] [Google Scholar]
- Sabeti P. C., Reich D. E., Higgins J. M., Levine H. Z., Richter D. J., et al. , 2002. Detecting recent positive selection in the human genome from haplotype structure. Nature 419: 832–837. [DOI] [PubMed] [Google Scholar]
- Sakuma M., 1998. Probit analysis of preference data. Appl. Entomol. Zool. (Jpn.) 33: 339–347. [Google Scholar]
- Sandmann T., Jensen L. J., Jakobsen J. S., Karzynski M. M., Eichenlaub M. P., et al. , 2006. A temporal map of transcription factor activity: mef2 directly regulates target genes at all stages of muscle development. Dev. Cell 10: 797–807. [DOI] [PubMed] [Google Scholar]
- Schlenke T. A., Begun D. J., 2004. Strong selective sweep associated with a transposon insertion in Drosophila simulans. Proc. Natl. Acad. Sci. USA 101: 1626–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt J. M., Good R. T., Appleton B., Sherrard J., Raymant G. C., et al. , 2010. Copy number variation and transposable elements feature in recent, ongoing adaptation at the Cyp6g1 locus. PLoS Genet. 6: e1000998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoustra S. E., Slakhorst M., Debets A. J., Hoekstra R. F., 2005. Comparing artificial and natural selection in rate of adaptation to genetic stress in Aspergillus nidulans. J. Evol. Biol. 18: 771–778. [DOI] [PubMed] [Google Scholar]
- Shepanski M. C., Glover J., Kuhr R. J., 1977. Resistance of Drosophila melanogaster to DDT. J. Econ. Entomol. 70: 539–543. [DOI] [PubMed] [Google Scholar]
- Sokal R. R., Hunter P. E., 1954. Reciprocal selection for correlated quantitative characters in Drosophila. Science 119: 649–651. [DOI] [PubMed] [Google Scholar]
- Steele L. D., Muir W. M., Seong K. M., Valero M. C., Rangesa M., et al. , 2014. Genome-wide sequencing and an open reading frame analysis of dichlorodiphenyltrichloroethane (DDT) susceptible (91-C) and resistant (91-R) Drosophila melanogaster laboratory populations. PLoS One 9: e98584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele L. D., Coates B., Valero M. C., Sun W., Seong K. M., et al. , 2015. Selective sweep analysis in the genomes of the 91-R and 91-C Drosophila melanogaster strains reveals few of the ‘usual suspects’ in dichlorodiphenyltrichloroethane (DDT) resistance. PLoS One 10: e0123066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strycharz J. P., Lao A., Li H., Qiu X., Lee S. H., et al. , 2013. Resistance in the highly DDT-resistant 91-R strain of Drosophila melanogaster involves decreased penetration, increased metabolism, and direct excretion. Pestic. Biochem. Physiol. 107: 207–217. [Google Scholar]
- Szpiech Z. A., Hernandez R. D., 2014. Selscan: an efficient multithreaded program to perform EHH-based scans for positive selection. Mol. Biol. Evol. 31: 2824–2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M. F. J., Shen Y., Kreitman M. E., 1995. A population genetic test of selection at the molecular level. Science 270: 1497. [DOI] [PubMed] [Google Scholar]
- Thorvaldsdóttir H., Robinson J. T., Mesirov J. P., 2013. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14: 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight B. F., Kudaravalli S., Wen X., Pritchard J. K., 2006. A map of recent positive selection in the human genome. PLoS Biol. 4: e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan H., Liu Y., Li M., Zhu S., Li X., et al. , 2014. Nrf2/Maf‐binding‐site‐containing functional Cyp6a2 allele is associated with DDT resistance in Drosophila melanogaster. Pest Manag. Sci. 70: 1048–1058. [DOI] [PubMed] [Google Scholar]
- Weake V. M., Lee K. K., Guelman S., Lin C. H., Seidel C., et al. , 2008. SAGA‐mediated H2B deubiquitination controls the development of neuronal connectivity in the Drosophila visual system. EMBO J. 27: 394–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willoughby L., Chung H., Lumb C., Robin C., Batterham P., et al. , 2006. A comparison of Drosophila melanogaster detoxification gene induction responses for six insecticides, caffeine and phenobarbital. Insect Biochem. Mol. Biol. 36: 934–942. [DOI] [PubMed] [Google Scholar]
- Zawaira A., Ching L. Y., Coulson L., Blackburn J., Chun Wei Y., 2011. An expanded, unified substrate recognition site map for mammalian cytochrome P450s: analysis of molecular interactions between 15 mammalian CYP450 isoforms and 868 substrates. Curr. Drug Metab. 12: 684–700. [DOI] [PubMed] [Google Scholar]
- Zhan M., Yamaza H., Sun Y., Sinclair J., Li H., et al. , 2007. Temporal and spatial transcriptional profiles of aging in Drosophila melanogaster. Genome Res. 17: 1236–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Lambert G., Liao D., Kim H., Robin K., et al. , 2011. Acceleration of emergence of bacterial antibiotic resistance in connected microenvironments. Science 333: 1764–1767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw phenotypic data are available in the Supplemental Material, File S1.