Abstract
Candida albicans is a diploid fungus that is a frequent cause of mucosal and systemic infections in humans. This species exhibits an unusual parasexual cycle in which mating produces tetraploid cells that undergo a nonmeiotic program of concerted chromosome loss to return to a diploid or aneuploid state. In this work, we used a multipronged approach to examine the capacity of parasex to generate diversity in C. albicans. First, we compared the phenotypic properties of 32 genotyped progeny and observed wide-ranging differences in fitness, filamentation, biofilm formation, and virulence. Strikingly, one parasexual isolate displayed increased virulence relative to parental strains using a Galleria mellonella model of infection, establishing that parasex has the potential to enhance pathogenic traits. Next, we examined parasexual progeny derived from homothallic, same-sex mating events, and reveal that parasex can generate diversity de novo from identical parental strains. Finally, we generated pools of parasexual progeny and examined resistance of these pools to environmental stresses. Parasexual progeny were generally less fit than control strains across most test conditions, but showed an increased ability to grow in the presence of the antifungal drug fluconazole (FL). FL-resistant progeny were aneuploid isolates, often being diploid strains trisomic for both Chr3 and Chr6. Passaging of these aneuploid strains frequently led to loss of the supernumerary chromosomes and a concomitant decrease in drug resistance. These experiments establish that parasex generates extensive phenotypic diversity de novo, and that this process has important consequences for both virulence and drug resistance in C. albicans populations.
Keywords: Sexual reproduction, aneuploidy, antifungal, fluconazole, pathogenesis
SEXUAL reproduction is a strategy that evolved early in the eukaryotic lineage and has since been retained by most extant species. The benefits of sex include the ability to combine beneficial alleles while eliminating detrimental mutations from the population (Burt 2000; Heitman 2015). Sex can also increase population diversity that may be particularly advantageous for adaptation to stressful or fluctuating environments (Goddard et al. 2005; McDonald et al. 2016). In addition, sex can ensure that organisms keep pace with coevolving pathogens that would otherwise drive the host to extinction, as envisaged in the Red Queen hypothesis (Jokela et al. 2009; Lively 2010; Morran et al. 2011). Despite numerous advantages, the sexual program is also associated with significant costs so that asexual reproduction is favored under many situations, particularly as a short-term reproductive strategy (Sun and Heitman 2011; Lehtonen et al. 2012).
Fungal species exhibit enormous diversity in their reproductive strategies and, historically, have been classified by their mode of sexual reproduction. Ascomycetes, named for their ability to generate sexual ascospores, represent the largest phylum in the fungal kingdom, and include the model species Saccharomyces cerevisiae. Here, pheromone signaling between haploid a and α cells leads to cell fusion and formation of diploid a/α cells (Jones and Bennett 2011; Merlini et al. 2013). Diploid cells can propagate asexually, or can be induced to undergo meiosis and sporulation, leading to the formation of highly recombinant haploid progeny (Neiman 2011; van Werven and Amon 2011). While similar sexual cycles have been defined for many fungal species, a large number of ascomycetes undergo cryptic sexual programs, or have yet to be observed to undergo any form of sexual reproduction (Heitman 2010; Ni et al. 2011).
Candida albicans is an ascomycete that diverged from S. cerevisiae 200–800 MYA (Heckman et al. 2001; Taylor and Berbee 2006). This species is a common component of the human microbiome, estimated to be present in more than half of the world’s population (Mukherjee et al. 2015), but is also an important opportunistic pathogen capable of causing both superficial and systemic infections. C. albicans is the fourth most common cause of bloodstream infections in hospitalized patients in the United States, and infections can lead to death in close to 50% of these cases (Gudlaugsson et al. 2003; Wisplinghoff et al. 2004), making an understanding of its biology of pressing importance.
Originally labeled an obligate asexual species, it is now established that C. albicans can undergo an unusual parasexual cycle. Natural isolates are diploid or aneuploid strains that display extensive heterozygosity between chromosome homologs (Jones et al. 2004; Selmecki et al. 2006; Rustchenko 2007; Butler et al. 2009; Ford et al. 2015; Hirakawa et al. 2015). As in S. cerevisiae, mating occurs between a and α cells of opposite mating types, but C. albicans cells must also undergo a phenotypic switch from the sterile “white” phenotypic state to the mating-competent “opaque” state (Miller and Johnson 2002). Pheromone signaling between opaque a and α cells leads to cell and nuclear fusion, thereby generating tetraploid a/α cells (Bennett et al. 2003; Lockhart et al. 2003; Panwar et al. 2003; Bennett et al. 2005). Tetraploid cells can be stably propagated or can undergo concerted chromosome loss (CCL) to return to a diploid, or near diploid, state (Bennett and Johnson 2003; Hickman et al. 2015).
Same-sex mating has also been documented in C. albicans, in which diploid a-a or α-α fusion events generate MTL homozygous tetraploid cells that can use CCL to complete a homothallic parasexual cycle (Alby et al. 2009; Alby and Bennett 2010). Same-sex mating may serve as an alternative reproductive mechanism to promote adaptation, as unisexual mating in the fungal pathogen Cryptococcus neoformans has been shown to generate progeny with de novo genetic and phenotypic diversity (Ni et al. 2013). Finally, the process of CCL has been documented in C. albicans diploid cells to produce rare haploid cells, which can then mate or auto-diploize to reform diploid cells (Hickman et al. 2013).
Analyses of C. albicans CCL progeny demonstrates that parasex generates diverse genotypes via (i) the shuffling of different chromosome homologs; (ii) recombination between homologs; and (iii) the generation of cells in various ploidy states, including aneuploid cells with one or more supernumerary chromosomes (Forche et al. 2008). Interestingly, the “meiosis-specific” gene SPO11 is implicated in promoting recombination during CCL, suggesting mechanistic parallels between CCL and that of a more conventional meiosis (Forche et al. 2008). It is also notable that multiple steps in the parasexual cycle are stimulated by stress, and that parasex may therefore enhance diversity under conditions where diversity is particularly beneficial for adaptation and survival (Berman and Hadany 2012; Zhang et al. 2015).
C. albicans mating has been observed in multiple murine infection models (Hull et al. 2000; Magee and Magee 2000; Dumitru et al. 2007; Zhang et al. 2015), and there is evidence for recombination in natural isolates despite the population structure being largely clonal (Graser et al. 1996; Anderson et al. 2001; Tavanti et al. 2004; Odds et al. 2007; Zhang et al. 2015). Even in the absence of sex, C. albicans strains exhibit considerable karyotypic plasticity (Chibana et al. 2000; Berman 2016; Gerstein et al. 2017). Multiple types of genome rearrangements have been described including chromosome translocations, chromosome length polymorphisms and chromosome truncations, as well as the gain or loss of whole chromosomes (Rustchenko 2007; Selmecki et al. 2010; Bennett et al. 2014). Aneuploid forms can promote fitness, including the discovery that a segmental aneuploidy involving isochromosome 5L mediates azole resistance in a number of clinical C. albicans isolates (Selmecki et al. 2006, 2009). In addition, C. albicans strains experience frequent loss of heterozygosity (LOH) events, some of which also can contribute to increased drug resistance (Coste et al. 2006; Dunkel et al. 2008).
In this study, we investigated the potential for the C. albicans parasexual cycle to promote phenotypic diversity and enhance adaptation, two tenets of sexual reproduction. Through analysis of large numbers of parasexual progeny, we establish that this program generates highly diverse forms following both heterothallic and homothallic mating. The latter establishes that parasex can produce diversity de novo following mating of identical parental strains. Aneuploidy was common among parasexual progeny and contributed to important phenotypic differences, including whether a strain was hyper- or hypo-virulent. Finally, we tested large, heterogeneous pools of parasexual progeny for their ability to resist different environmental stresses and show that parasex supports the emergence of strains able to grow in the presence of fluconazole (FL). All of these FL-resistant strains contained one or more supernumerary chromosomes, and passaging in the absence of drug led to loss both of supernumerary chromosomes and drug resistance phenotypes. Overall, our study establishes that parasex introduces extensive phenotypic diversity into the population, and that this program impacts important processes relevant to C. albicans pathogenesis and drug resistance.
Materials and Methods
Strains and media
Synthetic complete dextrose medium (SCD) and yeast extract peptone dextrose medium (YPD) were prepared as described (Guthrie and Fink 1991). Presporulation medium (“pre-spo”) contained 0.8% yeast extract, 0.3% peptone, 10% glucose, and 2% agar (glucose added prior to autoclaving medium). Casitone medium; 0.9% bacto casitone, 0.5% yeast extract, 1% sodium citrate, and 1.5% agar. Spider medium; 1% nutrient broth, 1% mannitol, 0.4% K2HPO4, and 1.35% agar, and was adjusted to pH 7.2. CHROMagar was made as per the manufacturer’s instructions (CHROMagar, #CA222).
Tetraploid strain RBY18 was constructed as described (Bennett and Johnson 2003). The set of 32 genotyped progeny were generated previously (Forche et al. 2008), with the exception of Ss2a (CAY3269) and Ss2b (CAY3268) which were isolated in this study. MTLa (DSY219) and MTLα (DSY220) derivatives of SC5314 were generated by deletion of the MTLα or MTLa locus using plasmids pRB102 or pRB101, respectively, as described (Alby and Bennett 2009). Heterothallic mating was performed by mixing MTLa and MTLα cells on Spider medium at 22° for 2–5 days followed by selection or picking zygotes using a micromanipulator. Strains used in this study are listed in Supplemental Material, Table S1 in File S1.
Growth assays
C. albicans cells were grown overnight in YPD at 30°. The following day, cells were diluted to a final concentration of ∼0.1 OD600 in 450 µl fresh YPD in 48-well plates. Plates were shaken continuously on a low setting, and OD600 readings recorded every 15 min for 18 hr using a BioTek Synergy HT plate reader/incubator (BioTek, Winooski, VT). Doubling times were calculated during exponential growth.
Colony morphology and agar invasion assays
To quantify colony morphology variations, cells were grown overnight at 30° in YPD. The following day, ∼50–100 cells were plated onto Spider medium and incubated at 30 or 37° for 7 days. Colony images were collected with a Chemidoc XRS+ and ImageLab software and contrast adjusted using Adobe Photoshop. Morphology scores (M-scores) were calculated relative to an SC5314 diploid control strain using a previously described method (Noble et al. 2010). In brief, the divergence of colony wrinkling (C) and extent of peripheral filamentation (P) was measured relative to the control from −3 to +3. The M-score is the sum of the absolute values of C and P:
To assay for colony invasion into the agar, cells were grown overnight at 30° in YPD. The next day, cells were washed once in sterile water, and 50–100 cells plated onto YPD and incubated for 3 days at 30°. Following incubation, colonies were washed from the surface of the YPD using water and gentle agitation, and images of colonies were taken before and after washing using a Chemidoc XRS+ and ImageLab software.
Biofilms
Biofilm assays were performed as previously described (Nobile et al. 2012). In brief, nonreinforced silicone elastomer (0.06 inch thickness, Bentec Medical, Woodland, CA) was cut into squares of 1.5 cm × 1.5 cm, autoclaved and weighed. Silicone squares were treated with adult bovine serum (ABS) in a polystyrene 12-well plate by incubation for 24 hr at 37° on a shaking platform at 150 rpm. ABS-treated squares were washed with sterile phosphate-buffered saline (PBS) and placed in liquid Spider medium. C. albicans cells grown overnight in YPD at 30° were diluted to an OD600 of 0.2 into wells containing silicone squares in Spider medium. Cells were incubated at 37° shaking at 150 rpm for 90 min to allow for cells to adhere. Next, squares were washed with sterile PBS and placed into a new 12-well dish containing fresh Spider medium. These were incubated at 37° shaking for 60 hr to allow for biofilm formation. After the 60 hr incubation, silicone squares were removed, allowed to dry for 24 hr, and weighed to determine biofilm mass.
FL susceptibility assays
To measure minimum inhibitory concentration (MIC) values, C. albicans cells were grown overnight in YPD at 30°, cells diluted in sterile water and ∼105 colony forming units (CFU) plated onto casitone agar. Plates were allowed to dry for ∼20 min and antifungal Etest strips (BioMérieux, France) added to the center of each plate. Etest plates were incubated at 37° for 48 hr, and MIC values read at the point where cell growth was inhibited.
FL disc assays were also performed to determine the susceptibility of C. albicans strains to this drug. C. albicans cells were grown overnight in YPD at 30°, diluted in sterile water and ∼105 CFU plated onto YPD. Plates were allowed to dry and a 25 mg FL disc (Liofilchem, Italy) added to the center of the plate. Cells were incubated at 30° for 48 hr and imaged using a Chemidoc XRS+ system with Image Lab software (Bio-Rad Laboratories, Hercules, CA). FL disc assays were quantified by measuring radii of inhibition using RAD50 measurements in the RScript DiskImageR program (Gerstein et al. 2016).
Galleria mellonella virulence assay
C. albicans cells were grown overnight in YPD at 30°, and washed three times with 5 ml sterile PBS. Cell density was determined using a hemocytometer and inocula prepared in PBS. Cell suspensions were plated onto YPD plates and incubated overnight at 30° to establish the inoculum; 2.5 × 105 CFU were injected into the last proleg of G. mellonella larvae in a 10 µl volume using a 26 gauge Hamilton fixed needle syringe (Hamilton, No. 80300). Totals of 30 larvae were injected per strain. Infected larvae were incubated in a sterile Petri plate for 7 days at 37° and monitored daily. Worms were scored as dead when they had turned black and could no longer move when stimulated. Dead worms were removed daily. Survival curves were plotted using Prism software and statistical significance determined with log-rank Mantel-Cox survival analysis.
Murine model of systemic infection
C. albicans cells were grown overnight in liquid YPD at 30°, washed three times with 5 ml PBS and cell density quantified using a hemocytometer. Cells were diluted in PBS to a concentration of 1 × 106 CFU/ml. Mouse tail veins were injected with 2 × 105 CFU in a 0.2 ml volume using 27 G needles (BD Precision Glide). Female Balb/C mice (8 weeks old; n = 9 mice per group, Charles River Laboratories, Wilmington, MA) were used, and monitored daily for 7 days. Mice were killed when morbid or at the end of the experiment. Systemic organs were harvested and processed through a 70 µl nylon filter into PBS containing antibiotics. Dilutions of these organ suspensions were plated onto YPD plates and incubated at 30° for 48 hr to quantify fungal burden.
Light microscopy
Differential interference contrast (DIC) microscopy was performed using a Zeiss Inverted Microscope (Axio Observer. Z1) fitted with an AxioCam HR. Images were processed with AxioVision Rel. 4.8 (Zeiss, Germany).
DNA staining and flow cytometry
The ploidy of C. albicans isolates was determined by cytometric analysis as previously described (Hull et al. 2000). Cells were grown overnight in liquid YPD at 30° and diluted to OD600 ≈ 2.0 in 500 µl H2O. Cell suspensions were fixed with 1.1 ml 100% ethanol and incubated at 4° overnight. Fixed cells were washed twice with 250 μl Buffer A (50 mM Tris-HCl, pH 8.0 and 5 mM EDTA) and sonicated for 5 sec on low power (Branson #4C15). Cells were incubated with RNase A (2 mg/ml) in Buffer A for 3 hr at 37°. Cells were pelleted, resuspended in 250 µl 55 mM HCl with pepsin (5 mg/ml), and incubated at 37° for 45 min. Cells were washed with 250 µl Buffer B (50 mM Tris-HCl, pH 7.5 and 5 mM EDTA), sonicated at low power for 5 sec, and resuspended in 250 µl Buffer B. Fifty microliters of each cell solution was diluted into 500 µl of 1 µM Sytox Green and incubated overnight at 4°. A total of 10,000 cells from each sample was run on a FACSAria (BD) and data analyzed using FlowJo (Tree Star, Ashland, OR).
Homothallic mating
To generate an MTLa tetraploid, ∼2 × 107 cells of an MTLa/Δα nourseothricin resistant (NAT+) strain (DSY220) were mixed with ∼2 × 107 cells of an MTLa/Δα hygromycin B-resistant (HYG+) strain (CAY5297) in sterile water. The mating mixture was pelleted, resuspended in 10 µl sterile water and spotted onto Spider medium. The mating mixture was incubated at 22° for 5 days and 2.5 µl of synthetic C. albicans α pheromone (10 mg/ml, Lifetein, Somerset, NJ) added daily to the cell spot to induce unisexual a-a mating (Alby and Bennett 2011). After 5 days, dilutions of the mating mixture were plated onto YPD supplemented with NAT (200 µg/ml) and HYG (1 mg/ml) to select for mating products. Mating products were analyzed by Sytox staining and flow cytometry to establish tetraploidy.
Generation of pools of parasexual progeny
To generate heterogeneous pools of parasexual progeny, an MTLa/α tetraploid strain, RBY18 (Bennett and Johnson 2003), was induced to undergo CCL by plating onto S. cerevisiae pre-spo agar and incubating at 37° for 10 days. After 10 days, cells were removed and resuspended in liquid YPD + 25% glycerol. The number of viable cells in parasexual pools was determined by plating onto YPD agar. Three independent parasexual pools were generated and aliquots stored at −80°.
Stress assays
C. albicans diploid, tetraploid, and parasexual progeny cells were compared for their resistance to six different stressors. Wildtype SC5314 (diploid a/α control), RBY18 (tetraploid a/α control), and three independently generated parasexual pools were evaluated. Conditions tested were 60° heat shock for 60 min, calcofluor white (70 µg/ml), congo red (200 µg/ml), hydrogen peroxide (7 mM), thiomersol (0.00001%), and FL (4 µg/ml). Chemical stress agents were included in YPD medium with the exception of FL, which was added to Candida CHROMagar medium. This was necessary as FL was not effective when dissolved in YPD (all isolates grew at >256 µg/ml FL). For each stress, we aimed to utilize conditions that inhibited growth of at least 50% of the SC5314 control cells plated. Aliquots of frozen diploid/tetraploid controls and parasexual progeny were thawed and ∼1000 CFU inoculated onto stress and YPD plates, and incubated at 30°. The percentage growth of each strain was determined by dividing the average number of colonies that grew on stress media by the number of colonies on YPD medium.
Genomic DNA and library preparation
High molecular weight genomic DNA (gDNA) was isolated from ∼109 cells grown overnight at 30°. SC5314 was grown overnight in liquid YPD and FL-selected parasexual progeny were grown in liquid YPD + FL (256 μg/ml). gDNA was isolated using a Qiagen Genomic Buffer Set and Qiagen Genomic-Tip 100G. Lyticase (Sigma-Aldrich, L2524, St. Louis, MO) was used to enzymatically digest fungal cell walls during genomic preparations. gDNA was sheared using a Bioruptor (Diagenode, Cat. No. UCD-200) on medium power for 2 × 10 min cycles of 15 sec on/15 sec off. Genomic DNA libraries were prepared as described by the Illumina TruSeq DNA Sample Preparation Guide (Illumina, San Diego, CA). gDNA libraries were indexed with Illumina TruSeq DNA HT adapters for multiplexed sequencing. gDNA libraries were pooled and sequenced using an Illumina HiSeq 2500 on high output mode (V4 chemistry) with single-end 50 bp reads or paired-end 100 bp reads (Genewiz, Cambridge, MA).
Genomic analysis
Illumina sequencing reads were aligned to C. albicans reference genome assembly 21 (van het Hoog et al. 2007) using the Bowtie2 aligner (Langmead and Salzberg 2012). Aligned reads were indexed and sorted using SAMtools (Li et al. 2009). Copy numbers for each chromosome were estimated by calculating read depth per chromosome using the idxstats program in the SAMtools package and normalizing to diploid levels. Reads from the rDNA locus (ChrR:1880000–1900000) were excluded from coverage analysis due to extensive expansion and contraction of this locus that interfered with chromosome copy number estimates. To plot coverage across the genome, the average read depth across 10 kb tiling windows was performed using the coverage tool in the BEDTools suite (Quinlan and Hall 2010). Genome coverage plots and karyotype heatmaps were generated in R. To identify potential SNP differences between Ss2a and Ss2b, sequence variants were called using both GATK HaplotypeCaller (Van der Auwera et al. 2013) and Pilon (Walker et al. 2014), and manually inspected using the Integrative Genomics Viewer (Robinson et al. 2011; Thorvaldsdottir et al. 2013).
Data availability
Genome sequencing data has been uploaded to the National Center for Biotechnology Information Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra). Data can be accessed using the study accession #SRP113728 or the BioProject #PRJNA394762.
Results
C. albicans parasexual progeny exhibit diverse phenotypic properties
This study consists of three parts. In the first part of the study, we examine the phenotypic properties of 32 parasexual progeny derived from the “laboratory” strain of C. albicans, SC5314. Progeny were generated via heterothallic mating between MTLa and MTLα cells and tetraploid a/α cells induced to undergo CCL by growth on S. cerevisiae pre-spo medium or sorbose medium (Figure 1A and Figure S1A in File S1). Most of these progeny were previously genotyped (Forche et al. 2008), and include both diploid strains that have a euploid complement of chromosomes, as well as those containing one to three supernumerary chromosomes (Figure S1B in File S1). In the second part of the study, we address whether the parasexual cycle can generate diversity following mating between different clinical isolates, and if diversity is generated de novo from same-sex mating events. The third part of the paper examines large pools consisting of diverse parasexual progeny, and evaluates the ability of these pools to grow in the presence of different forms of environmental stress.
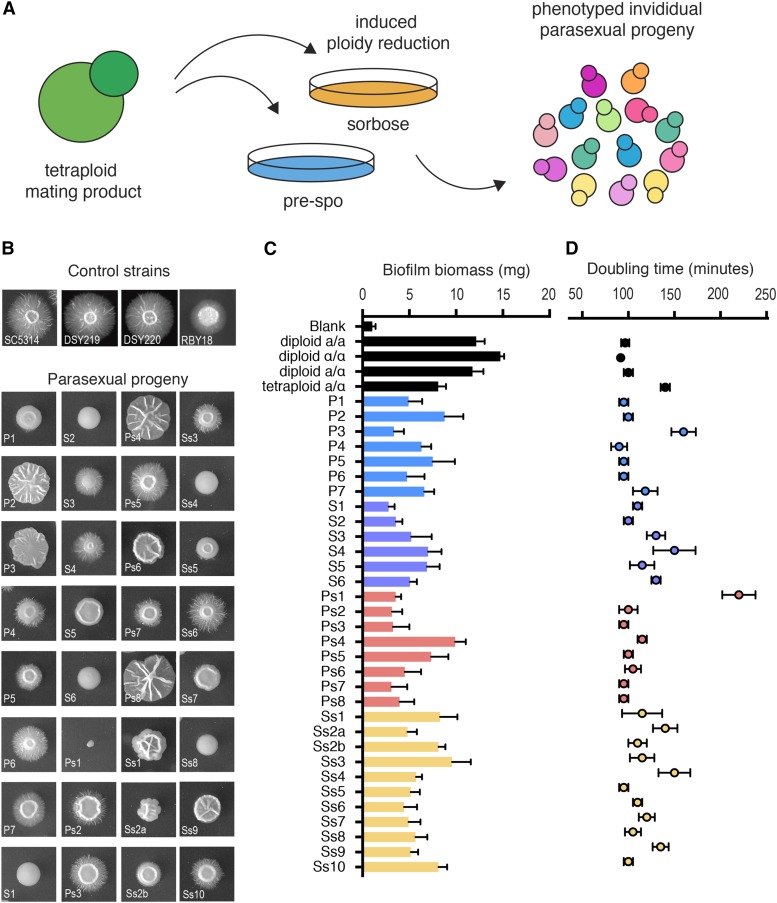
Figure 1.
Phenotypic diversity among 32 parasexual progeny of C. albicans. (A) Overview indicating how parasexual progeny were generated for this study. (B) Colony morphologies of progeny grown for 7 days on Spider medium at 37°. (C) Biofilm formation of parasexual progeny on silicone elastomer (n = 5 biological replicates per strain, error bars = SEM). (D) Doubling times of progeny during exponential growth in YPD medium at 30° (n = 3 biological replicates, error bars = SEM). Strain abbreviations and grouping are based on the media used to induce CCL and the genotype of the parental tetraploid (P, grown on pre-spo; S, grown on sorbose; Ps, spo11Δ cells grown on pre-spo; Ss, spo11Δ cells grown on sorbose).
Parasexual phenotypes: colony morphologies
The ability to undergo a dimorphic transition between yeast and hyphal forms is closely linked to C. albicans pathogenesis (Saville et al. 2003; Sudbery 2011). The 32 parasexual progeny were compared for their ability to undergo filamentation on Spider medium for 7 days at both 30 and 37°. Parental SC5314 cells formed colonies with a wrinkled central region surrounded by extensive peripheral filamentation, whereas parasexual progeny exhibited diverse morphologies that ranged from extensive colony wrinkling and agar invasion to colonies that showed a complete lack of these properties (Figure 1B). The colony morphology was quantified using a morphology “M-score” (Noble et al. 2010) that measures phenotypic variation relative to a control, and includes parameters measuring both colony wrinkling and peripheral filamentation (Figure S2, A and B in File S1). M-score measurements were generated relative to parental SC5314 strains (MTLa/a, MTLα/α, and MTLa/α) that exhibited indistinguishable colony morphologies (Figure 1B and Figure S2, A and B in File S1). The latter result establishes that MTL configuration does not affect the morphology of C. albicans colonies under these conditions. In general, parasexual progeny formed colonies with a wide range of M-scores, and these scores were higher (indicating more divergent phenotypes) at 37° than at 30°. Thus, at 37°, the average M-score across the 32 progeny was ∼4, whereas at 30° the average M-score was ∼3 (P < 0.0001, t-test) (Figure S2C in File S1). In addition, aneuploid isolates (22 strains) gave rise to colonies with an average M-score of ∼3, whereas colony morphologies of euploid progeny (10 strains) produced an M-score of ∼2 (Figure S2D in File S1, growth at 30°, P = 0.005, t-test). This indicates that aneuploid progeny, on average, display more divergent colony phenotypes than euploid progeny, consistent with a role for aneuploidy in augmenting phenotypic diversity.
Parasexual phenotypes: biofilm formation
C. albicans readily generates biofilms that present a major challenge in the clinic (Finkel and Mitchell 2011; Nobile and Johnson 2015) and involve a regulated network of ∼1000 genes (Nobile et al. 2012). Here, the 32 parasexual progeny were compared for biofilm formation using a standard assay involving growth on silicone elastomer, a material commonly used for catheters (Figure 1C). The most robust biofilms were formed by diploid control strains, producing biofilms weighing 11.8–14.7 mg for MTLa/a, MTLα/α, or MTLa/α strains. A tetraploid a/α strain generated a slightly smaller biofilm mass of 8.1 mg. Parasexual progeny produced biofilms with highly variable weights, although all were smaller than those of diploid controls; progeny biofilms ranged from 2.8 mg (S1) to 9.9 mg (Ps4), with a median weight of 5.1 mg. In general, euploid progeny trended toward forming biofilms of greater mass (mean of 6.7 mg) than aneuploid progeny (mean of 5.2 mg; P = 0.051, t-test).
Parasexual phenotypes: growth rates
The in vitro fitness of parasexual progeny was examined by measuring doubling times during exponential growth in liquid YPD medium at 30 and 37° (Figure 1D and Figure S3A in File S1). The diploid parent doubled every ∼100 min at 30°, whereas the tetraploid parent grew more slowly, doubling every ∼140 min. The mean doubling time of parasexual isolates was ∼115 min, with the fastest-growing isolate (P4) doubling every 90 min, and the slowest-growing isolate (Ps1) doubling every 220 min. Aneuploid isolates grew, on average, more slowly than euploid isolates (average doubling time of euploid strains was ∼100 min, whereas that of aneuploid strains was ∼120 min, P = 0.036, t-test, Figure S3B in File S1). Doubling times were also measured at 37° where the diploid control doubled every 85 min, and the tetraploid strain every 120 min. The mean doubling time of parasexual progeny was ∼100 min, with the fastest growing strain (Ps7) doubling every 80 min and the slowest growing strain (Ps1) doubling every 130 min. However, unlike growth at 30°, there was no significant differences in doubling times between aneuploid and euploid groups at 37° (both groups doubled on average every ∼100 min, P = 0.28, Figure S3B in File S1).
Parasexual phenotypes: susceptibility to antifungal drugs
The 32 parasexual progeny were assayed for their ability to grow in the presence of different classes of antifungal drugs. Minimum inhibitory concentrations (MIC) were defined using Etest strips for FL, voriconazole (VO), amphotericin B (AP), flucytosine (FC), and caspofungin (CS) (Table S2 in File S1). The median MIC values for the 32 parasexual strains were 0.125 μg/ml (FL), 0.44 μg/ml (FC), 0.004 μg/ml (VO), 0.125 μg/ml (AP), and 0.035 μg/ml (CS). Although variable MICs were observed among the group, none of the parasexual isolates reached clinical breakpoints for the tested drugs.
Parasexual phenotypes: virulence of parasexual progeny
Virulence is a complex trait that is impacted by multiple aspects of C. albicans biology (Calderone and Fonzi 2001; Mayer et al. 2013). Here, we used a standard G. mellonella larval model of fungal infection (Cotter et al. 2000; Fuchs et al. 2010b) to compare the virulence of parasexual progeny with that of control strains. Previous studies observed a correlation between the virulence of C. albicans strains in G. mellonella with that in a mouse tail-vein model of systemic infection, although there were also isolates for which this correlation did not hold (Brennan et al. 2002; Fuchs et al. 2010a; Hirakawa et al. 2015).
Each of the 32 parasexual isolates and control strains were injected into G. mellonella and their survival monitored over the course of 7 days at 37°. Larvae infected with a control diploid a/α strain had a median survival of 3 days, and 93% of infected larvae had died by 7 days (Figure 2A and Table S3 in File S1). Previous studies showed that MTL heterozygous strains exhibited increased virulence relative to MTL homozygous a/a or α/α strains in the murine tail-vein model of infection (Wu et al. 2007). To test if MTL configuration influences virulence in G. mellonella, we compared diploid a/α strains with isogenic strains deleted for MTLa or MTLα, and found that all three genotypes produced equivalent levels of virulence (Figure S4 in File S1). The majority of parasexual strains were less virulent than the control diploid strain, with 20 of the 32 strains being significantly reduced for virulence (Figure 2A and Table S3 in File S1, P < 0.05, log-rank test). Both euploid and aneuploid strains were virulent in the insect model, and no obvious correlations were found between virulence and different karyotypes, or between virulence and filamentation.
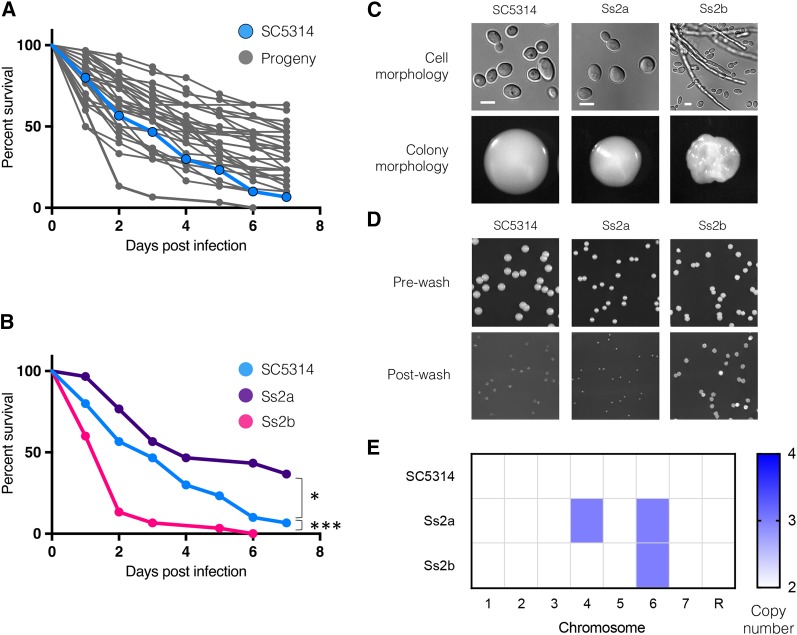
Figure 2.
Virulence of parasexual progeny and morphological traits of a hyper-virulent strain. (A) Survival curves of G. mellonella larvae injected with C. albicans parasexual progeny (n = 30 larvae per strain, SC5314 parental control is highlighted in blue). (B) Survival curves highlighting the hyper-virulent isolate Ss2b, the hypo-virulent isolate Ss2a, and the control diploid SC5314 strain (* P < 0.05, *** P < 0.001; Log-rank test). (C) Representative cell and colony morphologies of parasexual progeny Ss2a and Ss2b after growth on YPD agar at 30° for 3 days (Bar, 5 μm). (D) Invasive growth of isolate Ss2b on YPD agar at 30°. Cells were grown for 3 days and imaged before and after washing colonies from the agar surface. (E) Heatmap depicting karyotypes of Ss2a and Ss2b as defined by whole-genome sequencing.
Interestingly, we identified one parasexual isolate, Ss2b, that was significantly more virulent than control diploid strain(s), with a median survival rate of 2 days and killing of 100% of larvae after 6 days (Figure 2B) (P = 0.0007, log-rank test). The in vitro properties of Ss2b, an MTLα strain, revealed that it grew more slowly than a control diploid MTLα strain (doubling time of 110 min vs. 97 min in YPD) and produced smaller biofilms (8.1 mg vs. 14.7 mg). The filamentation phenotypes of Ss2b were complex, as it was largely defective for peripheral filamentation on Spider medium, yet exhibited a more wrinkled and invasive colony morphology when grown on YPD at 30°, and cells from these wrinkled colonies were hyper-filamentous when compared to those from control colonies (Figure 2, C and D).
Isolate Ss2b was isolated together with a “sister” progeny, Ss2a, that both arose during propagation of a single parasexual isolate and were distinguished by contrasting colony phenotypes. Thus, Ss2b produced highly wrinkled colonies on YPD, whereas Ss2a formed smooth, nonfilamentous colonies (Figure 2, C and D). We found that Ss2b was hyper-virulent when tested in G. mellonella, whereas Ss2a showed decreased virulence relative to control strains (Figure 2B) (P = 0.02, log-rank test). To further compare these two strains, genome sequencing was performed to determine their genotypes (see Materials and Methods). Sequencing revealed that there were no SNP differences between the two progeny, and the isolates displayed identical karyotypes except for the presence or absence of a Chr4 trisomy in Ss2a and Ss2b, respectively (Figure 2E). In contrast, both Ss2a and Ss2b were trisomic for Chr6. These results imply that simple changes in aneuploidy involving the presence or absence of a single trisomic chromosome can make profound differences to phenotypes such as filamentation and virulence in C. albicans.
Given the elevated virulence of isolate Ss2b in G. mellonella, we also tested its virulence in the standard murine tail-vein model of systemic infection. We found that the control SC5314 diploid strain killed ∼90% of infected mice, whereas all mice infected with Ss2b survived infection (P = 0.0001, log-rank test), displayed no disease symptoms, and did not lose any body weight (Figure S5 in File S1). In line with these results, diploid control cells colonized mouse organs to higher fungal burdens than Ss2b; control cells were present at 5.8 × 104 CFU/g of kidney tissue, whereas Ss2b cells were present at an order of magnitude lower (Figure S5 in File S1, P = 0.006, t-test). Control cells were also recovered from the brain and liver (∼40 CFU/g and ∼70 CFU/g, respectively), whereas no Ss2b cells were recovered from either organ. These results establish that Ssb2 displays significant differences in virulence between G. mellonella and mice, and, while this isolate is hyper-virulent in the insect larval model, it is defective for virulence in the murine model.
Parasex generates diverse progeny following outcrossing of C. albicans isolates
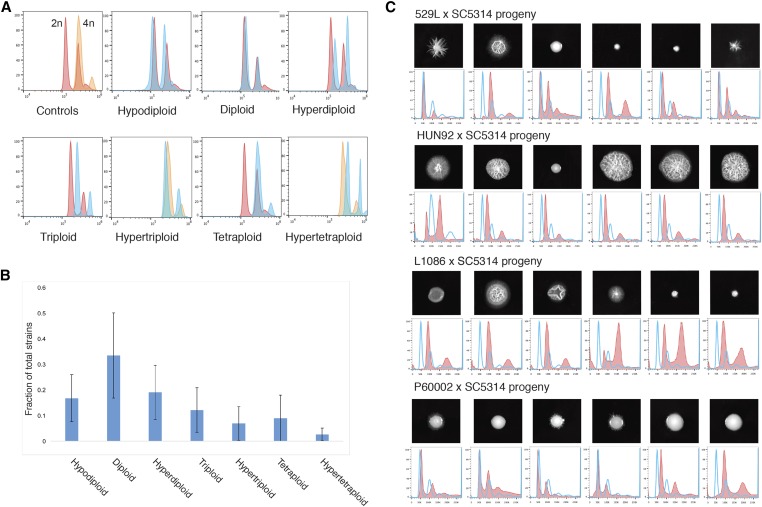
The potential for parasex to generate diversity between strains from different backgrounds was determined by mating SC5314 to one of six different clinical isolates (HUN92, YsU751, L1086, J981315, 529L, and P60002) representing four different C. albicans clades. A gal1/gal1 SC5314-derived strain was used in these crosses so that tetraploid products are heterozygous (gal1/gal1/GAL1/GAL1) at this locus. Following induction of CCL, cells that had undergone a reduction in ploidy were selected for using 2-deoxygalactose, which counter-selects against GAL1 function (Bennett and Johnson 2003). The ploidy of ∼200 parasexual progeny from each tetraploid strain were analyzed by Sytox staining and flow cytometry, and revealed a wide range of ploidy states (Figure 3A). Progeny were categorized as being hypo-diploid, diploid, hyper-diploid, triploid, hyper-triploid, tetraploid, or hyper-tetraploid. The most common ploidy state, with ∼1/3 of progeny, was diploid, while the second most common state was hyper-diploid, as ∼20% of isolates exhibited DNA content between that of diploid and triploid (Figure 3B). This is consistent with previous studies showing that many parasexual progeny are aneuploid due to the presence of one or more supernumerary chromosomes (Bennett and Johnson 2003; Forche et al. 2008; Hickman et al. 2015). None of the progeny analyzed were haploid despite the recent discovery of viable haploid forms of SC5314 (Hickman et al. 2013), although several hypodiploid isolates with a ploidy less than that of diploid controls were obtained (Figure 3B).
Figure 3.
Ploidy outcomes during parasex across diverse strain backgrounds. (A) Representative plots of ploidy states observed by flow cytometric measurements of DNA content. Red profiles are those of diploid control cells, and orange profiles are tetraploid control cells. (B) Cumulative breakdown of ploidy states observed among ∼200 progeny derived from CCL induced from six diverse strain backgrounds (error bars = SD). (C) Colony morphologies of parasexual progeny from different clinical strain backgrounds (529L, HUN92, L1086, and P60002) grown on Spider medium for 10 days at 37° (top), and their corresponding ploidy measurements (bottom; red lines depict parasexual progeny, and blue lines represent diploid control).
To establish that parasexual progeny derived from different isolates exhibit phenotypic diversity, progeny were assayed for colony morphologies on Spider medium. We observed diverse phenotypes at the macroscopic level, from smooth, nonfilamentous colonies to highly wrinkled, filamentous colonies (Figure 3C). These experiments therefore establish that mating and parasex can generate progeny with diverse karyotypes from different clinical backgrounds, and that the progeny exhibit a plethora of distinct phenotypic forms.
Parasex can generate diversity de novo
Conventional mating involves conjugation between opposite sex a and α cells (Hull et al. 2000; Magee and Magee 2000), but C. albicans can also undergo homothallic a-a or α-α mating (Alby et al. 2009; Alby and Bennett 2011). Homothallic mating between a cells can be induced by α-pheromone from MTLα cells or by synthetic α-pheromone added exogenously (Alby and Bennett 2011). Here, we addressed if parasexual progeny from a same-sex mating event exhibit diverse phenotypes, thereby establishing that parasex can generate diversity de novo. To test this, homothallic mating of a cells was performed to generate an MTLa tetraploid that was then induced to undergo CCL and the resulting progeny examined (see Materials and Methods).
DNA content of parasexual progeny from the MTLa tetraploid was determined by flow cytometry and revealed a range of ploidy states, from diploid to tetraploid, similar to that of heterothallic progeny (Figure S6 in File S1). Colony morphologies were compared by growth on Spider medium at 37°, and revealed diverse phenotypes that included different colony sizes as well as both smooth and wrinkled colonies (Figure S6 in File S1). These results establish that the parasexual cycle in C. albicans can create extensive phenotypic diversity through either opposite-sex or same-sex mating events. In the case of same-sex mating, the parental strains are genetically identical, so that diversity is not pre-existing in the population but is generated de novo.
Generation of parasexual pools and evaluation of resistance to environmental stress
The experiments described above establish that parasexual progeny include multiple aneuploid forms, and that progeny accordingly exhibit a wide range of phenotypic properties. Previous studies have shown that aneuploidy is an effective mechanism to increase population diversity and to promote adaptation to environmental stress (Chen et al. 2012a,b; Santaguida and Amon 2015; Wertheimer et al. 2016). We therefore examined whether the extensive genetic diversity present in C. albicans parasexual populations facilitates adaptation. To this end, we generated large, heterogeneous pools of parasexual isolates and examined their ability to generate forms resistant to different in vitro stressors.
Three independent pools of parasexual progeny were generated from tetraploid SC5314 a/α cells by induction of CCL and collection of random progeny (see Materials and Methods). These cell populations were challenged with different stressors and survival compared to that of control diploid/tetraploid cells (Figure 4A). We examined the response to heat shock (60° for 1 hr), cell wall stress (calcofluor white and congo red), oxidative stress (H2O2), and antifungal compounds (FL and thimersol); ∼1000 cells of each cell type were subjected to the test condition, and to growth on nonselective medium, and the fraction of stress-resistant isolates in each population determined (Figure 4B).
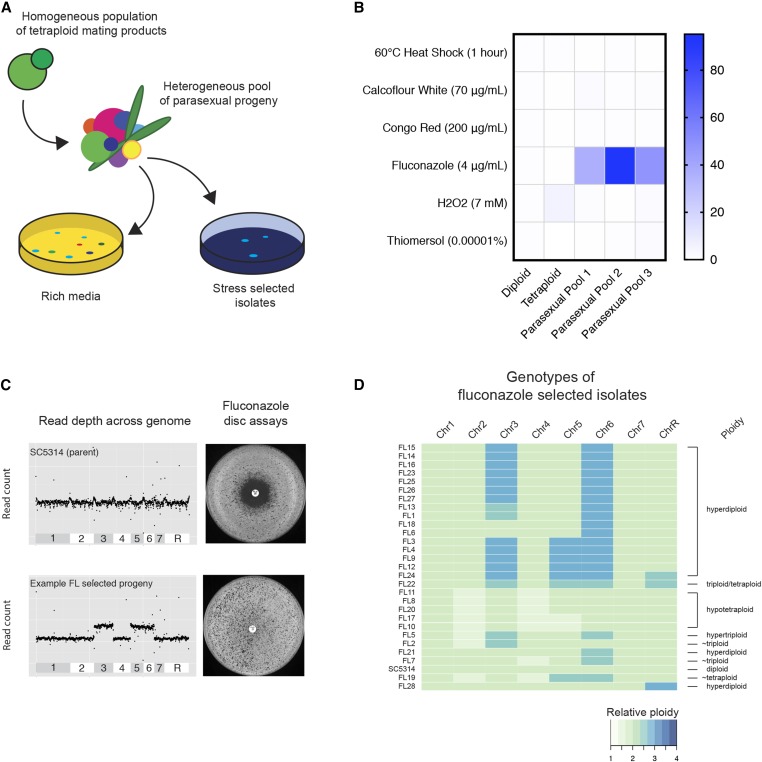
Figure 4.
Stress resistance among pools of parasexual isolates. (A) Schematic of the approach for stress selection on pools of heterogeneous parasexual progeny. (B) Heatmap showing cell viability of three independently generated parasexual pools, and tetraploid parents relative to the SC5314 diploid strain under different test conditions. (C) Example of genome sequencing coverage plots and corresponding FL disc diffusion assays. Top panel shows the euploid control and bottom panel depicts a FL-resistant isolate that exhibits elevated chromosomal coverage for Chr3, Chr5, and Chr6. (D) Heatmap summarizing karyotypes of FL-selected parasexual progeny determined by full-genome sequencing (coverage normalized to diploid levels).
Pools of parasexual progeny were morphologically heterogeneous and generally showed reduced fitness relative to control diploid and tetraploid strains, as evidenced by the formation of smaller colonies on YPD medium (Figure S7 in File S1). This is consistent with the fact that parasex often generates aneuploid progeny with slower growth rates than euploid strains (Figure 1D). Cell populations exposed to a variety of stresses revealed that control diploid cells were the most resistant to heat shock stress, and were also more resistant than tetraploid cells to cell wall damaging agents such as calcofluor white and congo red (Figure 4B and Table S4 in File S1). In contrast, tetraploid cells exhibited the highest levels of resistance to H2O2, with 26% of tetraploid cells producing viable colonies, which was fivefold higher than that of diploid cells and 4- to 29-fold higher than that of the parasexual pools.
Notably, cells from parasexual pools produced significantly more colonies than control populations when grown in the presence of FL (P < 0.05, t-test). Thus, whereas only 0.05% of diploid cells and 0.02% of tetraploid cells produced colonies on medium containing 4 µg/ml FL, the three parasexual pools produced colonies at frequencies of 1.9, 5.0, and 2.5%, respectively, on this medium. This result indicates that parasexual progeny are either intrinsically more resistant to FL or they rapidly adapt to the drug at high frequency. Individual FL-resistant colonies arising from the parasexual pools were further examined for genetic and phenotypic properties to understand the mechanism of resistance, as discussed below.
Genomic analysis of FL-resistant parasexual isolates
Drug resistance levels were defined for 28 parasexual progeny that arose on FL plates by determination of MIC values (see Materials and Methods, Table S5 in File S1). None of the progeny had MIC values that reached clinical breakpoints, although all clones exhibited higher MIC values than parental diploid or tetraploid strains. FL-selected progeny exhibited a range of ploidies between diploid and tetraploid, suggesting that many were aneuploid (Figure S8 in File S1). To more precisely define karyotypes, isolates were genome sequenced (∼20× coverage) and relative read depth measured across all eight chromosomes (Figure 4C and Figure S9 in File S1). All of the parasexual isolates from the FL exposure experiment were aneuploid, with the majority of isolates being close to diploid in content (18/28), although strains with triploid (3/28) and tetraploid (6/28) DNA content were also observed (Figure 4D and Figure S9 in File S1). The most common cause of aneuploidy in the set of FL-selected isolates involved diploid strains that were trisomic for Chr6 (17/28 strains), of which 14 were also trisomic for Chr3. Five of the diploid isolates trisomic for Chr3 and Chr6 were also trisomic for Chr5, and one of these strains included an additional ChrR trisomy. This reveals that particular trisomies were shared among the set of azole-resistant strains, and these patterns were observed despite isolates being derived from three independent pools of parasexual progeny.
We also note that three strains, FL2, FL5, and FL7, were close to triploid, yet all were also aneuploid due to imbalances in the copy number of one or more chromosomes. FL2 and FL5 were disomic for Chr2 and tetrasomic for Chr3, while FL5 included an additional tetrasomy for Chr6. The genome of one isolate, FL22, appeared halfway between that of triploid and tetraploid, with Chrs 3, 5, 6, and R present at tetrasomic levels. Finally, six isolates were close to tetraploid, although all six appeared to have lost one copy of both Chr2 and Chr4, which were therefore present at trisomic levels. Two of the tetraploid isolates were also disomic for Chr5, while one isolate, FL19, appeared pentasomic for this chromosome as well as Chr6. Together, these results reveal that all FL-selected parasexual isolates were aneuploid, and, although a number of complex karyotypes were observed, half the isolates included elevated copy numbers of Chr3 and Chr6.
Stability of FL-resistance phenotypes and karyotypes
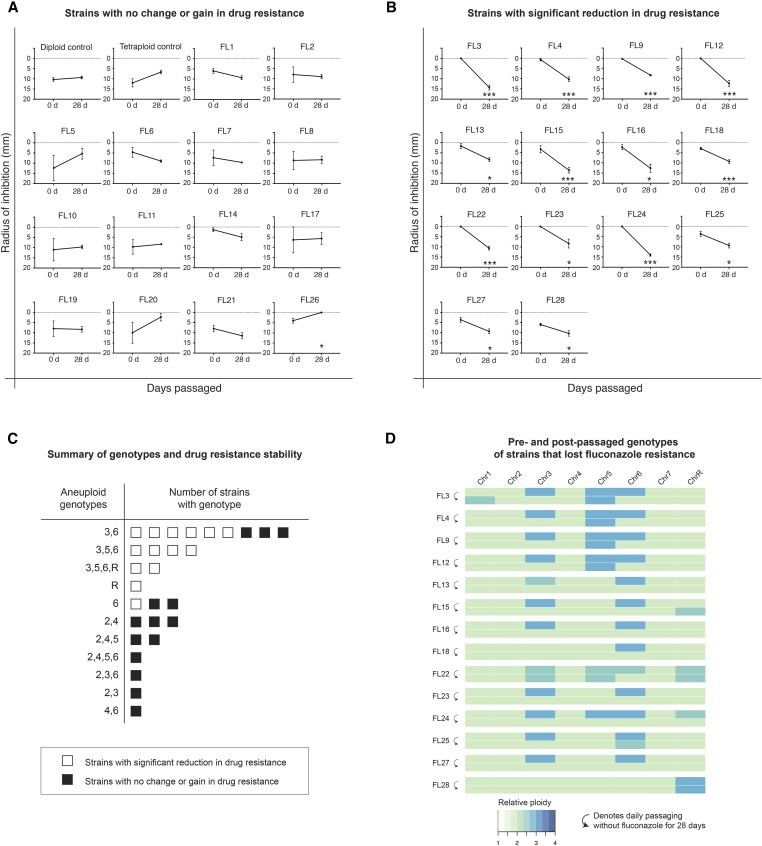
Aneuploid states can promote adaptation to stress in multiple fungal species, particularly in response to antifungal drugs (Chen et al. 2012b; Kwon-Chung and Chang 2012; Morrow and Fraser 2013; Bennett et al. 2014; Santaguida and Amon 2015; Wertheimer et al. 2016). During exposure to stress, selection ensures that fitness-enhancing karyotypes are maintained in the population, but in the absence of selection cells may revert to the parental euploid state (Gerstein et al. 2017). We therefore evaluated whether drug susceptibility in FL-selected progeny changed after extended passaging in the absence of the drug. Progeny were passaged daily for 4 weeks (∼200 generations) and retested for their ability to grow in the presence of FL. For these assays, drug responses were quantified by measuring the radial distance (RAD50) between the disc and the area where fungal growth was inhibited by 50% (Gerstein et al. 2016). Based on these criteria, all FL-selected strains were initially more resistant (smaller RAD50 values) than control diploid and tetraploid strains (Figure 5, A and B). However, following passaging, 14 out of 28 FL-selected progeny showed a significant reduction in FL resistance (P < 0.05, t-test) (Figure 5B). We note that 13/14 isolates that showed reduced drug resistance after passaging had originally been trisomic for Chr6, and that 12 of the 14 isolates had also started out as trisomic for Chr3 (Figure 5C).
Figure 5.
Analysis of the stability of FL-resistant phenotypes in parasexual progeny. FL-resistant isolates were passaged daily in YPD medium without FL for 28 days. FL disc diffusion assays were performed on original resistant strains, and on the corresponding strains following passaging (n = 3 biological replicates per time point). FL resistance was quantified by measuring the radius of inhibition (RAD50) surrounding the FL disc as determined by diskImageR software (Gerstein et al. 2016). (A) Strains showed increased or no change in FL resistance after passaging. (B) Strains showed decreased resistance after passaging (* P < 0.05, *** P < 0.001). (C) Summary of karyotypes observed in FL-selected isolates and their associated drug resistance phenotypes following passaging in the absence of the drug. (D) Heatmap depicting karyotypes determined by genome sequencing of original FL-resistant strains and after passaging of these isolates.
To determine if loss of FL resistance following passaging was associated with karyotypic changes, the 14 strains that lost resistance were reanalyzed by genome sequencing. We found that 13 out of 14 strains exhibited substantial karyotypic alterations; the 12 strains that were initially trisomic for Chr3 all lost this trisomy, and 12/13 strains that were trisomic for Chr6 also lost this trisomy during passaging (Figure 5D). In contrast, however, out of the six strains that were trisomic for Chr5, five strains stably maintained this trisomy during passaging. Only one isolate, FL28, did not undergo a change in karyotype during passaging, and remained trisomic for ChrR. Additionally, two strains appeared to acquire new forms of aneuploidy during passaging; strain FL3 gained a segmental aneuploidy of the right arm of Chr1, and FL15 gained a segmental aneuploidy for the right arm of ChrR (Figure S10 in File S1).
Discussion
In this study, we examined the capacity of the parasexual cycle of C. albicans to increase phenotypic diversity and promote adaptation. Unlike most sexually reproducing eukaryotes, C. albicans has not been observed to undergo a traditional meiosis. Instead, a parasexual cycle has been defined that involves CCL to reduce the ploidy of tetraploid mating products to that of diploid or aneuploid cells. Here, we establish that the parasexual cycle generates extensive phenotypic diversity, and that this diversity can impact traits relevant to the clinic.
We first evaluated the properties of a genotyped set of 32 parasexual progeny (Forche et al. 2008), and compared growth rates, filamentation, biofilm formation, and virulence between progeny and parental strains. Diverse phenotypes were observed in each of these assays, establishing that parasex is capable of introducing extensive variation into a population. In most cases, parasexual progeny exhibited reduced fitness relative to parental control strains. For example, all 32 progeny grew more slowly than the parental diploid when cultured in replete conditions, and many also showed a decreased ability to form biofilms. We observed that aneuploid progeny (diploid cells carrying 1–3 supernumerary chromosomes) exhibited more variable phenotypes than euploid progeny. These results are consistent with studies in multiple species where aneuploidy is associated with a negative impact on fitness levels under replete conditions yet can also extend phenotypic variation within a population (Torres et al. 2010; Chen et al. 2012b; Tan et al. 2013; Wertheimer et al. 2016).
Analysis of the virulence of parasexual progeny in G. mellonella produced an unexpected result. While most progeny were less virulent than control strains in this infection model, we found one isolate, Ss2b, to be significantly more virulent than any other strain, including the parental SC5314 strain. As these progeny were derived from crosses between SC5314 a and α cells, this result indicates that mating between similar parents (differing only at the mating-type-like locus) can produce cells with enhanced virulence. The Ss2b hyper-virulent isolate was closely related to a second isolate, Ss2a, that both derived from the same parasexual product. However, whereas Ss2b formed highly filamentous, invasive colonies on rich medium, and was hyper-virulent in G. mellonella, Ss2a formed nonfilamentous, noninvasive, colonies and was defective for virulence in this model. Genotyping of the two isolates revealed that they were diploid and isogenic, apparently differing only by the presence or absence of a single trisomy. Thus, whereas Ss2a was trisomic for both Chr4 and Chr6, Ss2b was trisomic for Chr6 alone. We infer that this single chromosome disparity is responsible for the phenotypic differences between the two progeny, and that this could be due to an imbalance in gene expression levels between Chr4 and Chr6. For example, the FLO8 gene resides on Chr6 and acts to promote filamentation (Cao et al. 2006), and this gene is likely to be expressed at a higher level in cells trisomic for this chromosome. However, isolate Ss2a is also trisomic for Chr4 which contains negative regulators of filamentation such as RFX2 that has been shown to bind to the FLO8 promoter (Hao et al. 2009; Fox et al. 2015). In this manner, cells that are trisomic for Chr4 could potentially negate the increased filamentation tendencies of cells that are trisomic for Chr6. A previous study also implicated a chromosomal trisomy as being responsible for differences in virulence; Chen et al. showed that C. albicans strains trisomic for Chr1 were avirulent as compared to closely related euploid strains (Chen et al. 2004). Our findings suggest a new form of aneuploidy is associated with virulence differences in C. albicans, and extend the evidence that changes in chromosomal copy number can alter pathogenic attributes.
In contrast to its hyper-virulence in G. mellonella, isolate Ss2b exhibited reduced virulence relative to the parental strains when tested in a murine model of systemic infection. It is possible that changes in the propensity to undergo the yeast-hyphal transition contribute to the increased pathogenicity of Ss2b in G. mellonella, but negatively impact its ability to disseminate in mice. This is consistent with increased filamentation in Candida species being linked to pathogenesis in G. mellonella (Jacobsen 2014). However, it is also evident that the relationship between filamentation and virulence in C. albicans is a complex one (Fuchs et al. 2010a; Noble et al. 2010; Sudbery 2011), so that additional experiments will be necessary to define the mechanism by which Ssb2 shows contrasting virulence properties in G. mellonella and mice. Interestingly, parallels exist with studies in C. neoformans, whereby isolates evolved mutations in a pseudohyphal pathway that promoted virulence in an amoeba host, but attenuated virulence in mice (Magditch et al. 2012). These studies reveal limitations in trying to make direct correlations from simpler infection models to the more complex mammalian system.
While the majority of C. albicans research has utilized the “laboratory” strain SC5314, different clinical isolates often exhibit traits that differ both from SC5314 and from one another (Odds et al. 2007; MacCallum et al. 2009; Hirakawa et al. 2015; Schonherr et al. 2017). We therefore examined whether strain background impacts the ability of parasexual reproduction to generate phenotypic diversity. Mating crosses were performed between SC5314 and six different clinical isolates, and, in all cases, mating products were competent to undergo CCL. A wide variety of ploidy states were observed in these parasexual progeny, with many existing in states between that of diploid and tetraploid. Notably, no haploid parasexual progeny were recovered, regardless of the strains involved, which could be due to the fact that C. albicans haploids are unstable, and often auto-diploidize (Hickman et al. 2013; Zeng et al. 2014). We conclude that diverse strains are competent to undergo CCL, and that the resulting progeny display extensive karyotypic and phenotypic diversity.
In addition to heterothallic mating between opposite-sex cells, it is now established that C. albicans cells can undergo same-sex mating, and that the resulting tetraploids can reduce their ploidy via CCL (Alby et al. 2009). We analyzed a number of homothallic parasexual progeny and found that they demonstrated a wide range of ploidy states and colony morphologies, indicating that parasex can produce diversity de novo even when there is no pre-existing diversity in the population. Studies in Cryptococcus neoformans have similarly revealed that unisexual reproduction can generate diversity de novo, and that variation is frequently the result of aneuploid forms (Ni et al. 2013). In C. albicans, aneuploidy is one important mechanism contributing to parasexual diversity, although additional mechanisms include the shuffling of chromosome homologs as well as recombination between homologs (Forche et al. 2008; Berman and Hadany 2012; Bennett et al. 2014).
Given the diversity produced by parasex, we examined whether this variation could generate progeny that were better adapted for survival when exposed to environmental stress. This was relevant given that aneuploid states have been shown to expand the phenotypic landscape, and to provide cells with high adaptive capacities to stress (Rancati et al. 2008; Chen et al. 2012a,b; Yona et al. 2012). We found that C. albicans parasexual progeny generally exhibited lower fitness levels than control strains under replete conditions. In contrast, however, cells from parasexual populations produced 10- to 100-fold more colonies with elevated resistance to the antifungal FL than equivalent numbers of euploid control cells. These results establish that parasex could be particularly effective in promoting the emergence of azole resistance in C. albicans populations.
The karyotypes of 28 parasexual progeny that formed colonies in the presence of FL (4 µg/ml) revealed that many were diploid except for trisomies of Chr6 (19 strains) and/or Chr3 (14 strains). Strains trisomic for Chr3 have been observed following exposure to FL in vitro, as well as in persistent isolates during FL treatment in patients (Perepnikhatka et al. 1999; Ford et al. 2015). In contrast, Chr6 trisomies have not been associated with FL resistance, although resistance was observed in strains that were monosomic for Chr6 (Perepnikhatka et al. 1999). Interestingly, both Chr3 and Chr6 include well-documented genes involved in FL resistance. Chr3 harbors the ABC family transporter genes CDR1 and CDR2 that are involved with drug efflux out of the cell (Prasad et al. 1995; Sanglard et al. 1997), as well as the transcription factor MRR1 that regulates MDR1, a gene on Chr6 that encodes another multidrug efflux pump involved in FL resistance (Wirsching et al. 2000; Morschhauser et al. 2007). It is therefore possible the increased copy number of one or more of these genes mediates the elevated levels of FL resistance observed in many parasexual progeny.
We also addressed whether FL-selected progeny maintained resistance levels following passaging in the absence of the drug. We found that half of the 28 progeny showed a significant decrease in FL resistance following passaging. Of these strains, 13 were initially trisomic for Chr6 and 12 were trisomic for Chr3, and both of these trisomies were lost in virtually all of the isolates following passaging. These results are consistent with trisomies for Chr3/Chr6 contributing to FL resistance in C. albicans. Interestingly, we note that not all trisomic chromosomes were unstable during passaging; strains trisomic for Chr5 often retained this trisomy, indicating that supernumerary chromosomes can show remarkable differences in their levels of stability.
It is worth highlighting that FL treatment of C. albicans diploid cells was recently shown to promote the formation of “trimeras”; three interconnected cells produced by abnormal cell divisions that frequently gave rise to tetraploid and aneuploid cell types (Harrison et al. 2014). The high frequency with which trimeras generate aneuploidy could underlie how drug resistance arises in natural populations of C. albicans. Indeed, approximately half of all clinical FL-resistant isolates exhibit some form of aneuploidy (Selmecki et al. 2009, 2010). Our studies further establish a close link between aneuploidy and FL susceptibility, and that the parasexual program can readily produce progeny that can grow in the presence of the drug. Perhaps surprisingly, we did not observe any resistant progeny carrying isochromosome i5L, a chromosome abnormality frequently associated with FL resistance in the clinic (Selmecki et al. 2006, 2008), which could indicate that this form of aneuploidy is not readily generated by the parasexual mechanism. However, it would be interesting to perform parasexual CCL in the presence of FL (as against using selection on the products of parasex) to determine if the same or different karyotypes emerge.
In conclusion, this study establishes that the C. albicans parasexual cycle is a potent mechanism by which phenotypic diversity can be introduced into the population. This diversity can be generated de novo following mating between identical isolates and affects multiple phenotypes, including those directly relevant to disease. Much of this diversity is due to different karyotypic configurations arising from CCL, and extends the evidence that aneuploidy can expand the phenotypic landscape that can be sampled. Together, these results reveal that parasexual reproduction provides an effective alternative to conventional sexual programs for promoting population diversity, and enabling adaptation to environmental challenges.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.300295/-/DC1.
Acknowledgments
We would like to thank Iuliana Ene, Joshua Wang, Matthew Anderson, and Kevin Alby for assistance with experiments. We also thank Christina Cuomo (Broad Institute) for advice regarding genome sequencing experiments and Donna MacCallum (University of Aberdeen) for sharing clinical isolates of C. albicans. This work was supported by National Institutes of Health grants AI081704 and AI112363 (to R.J.B.), F31 DE023726 (to M.P.H.), and by a Pathogenesis of Infectious Disease (PATH) award from the Burroughs Wellcome Fund (to R.J.B.).
Footnotes
Communicating editor: A. Mitchell
Literature Cited
- Alby K., Bennett R. J., 2009. Stress-induced phenotypic switching in Candida albicans. Mol. Biol. Cell 20: 3178–3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alby K., Bennett R. J., 2010. Sexual reproduction in the Candida clade: cryptic cycles, diverse mechanisms, and alternative functions. Cell. Mol. Life Sci. 67: 3275–3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alby K., Bennett R. J., 2011. Interspecies pheromone signaling promotes biofilm formation and same-sex mating in Candida albicans. Proc. Natl. Acad. Sci. USA 108: 2510–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alby K., Schaefer D., Bennett R. J., 2009. Homothallic and heterothallic mating in the opportunistic pathogen Candida albicans. Nature 460: 890–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J. B., Wickens C., Khan M., Cowen L. E., Federspiel N., et al. , 2001. Infrequent genetic exchange and recombination in the mitochondrial genome of Candida albicans. J. Bacteriol. 183: 865–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett R. J., Johnson A. D., 2003. Completion of a parasexual cycle in Candida albicans by induced chromosome loss in tetraploid strains. EMBO J. 22: 2505–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett R. J., Uhl M. A., Miller M. G., Johnson A. D., 2003. Identification and characterization of a Candida albicans mating pheromone. Mol. Cell. Biol. 23: 8189–8201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett R. J., Miller M. G., Chua P. R., Maxon M. E., Johnson A. D., 2005. Nuclear fusion occurs during mating in Candida albicans and is dependent on the KAR3 gene. Mol. Microbiol. 55: 1046–1059. [DOI] [PubMed] [Google Scholar]
- Bennett R. J., Forche A., Berman J., 2014. Rapid mechanisms for generating genome diversity: whole ploidy shifts, aneuploidy, and loss of heterozygosity. Cold Spring Harb. Perspect. Med. 4: a019604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman J., 2016. Ploidy plasticity: a rapid and reversible strategy for adaptation to stress. FEMS Yeast Res. 16: pii: fow020. [DOI] [PubMed] [Google Scholar]
- Berman J., Hadany L., 2012. Does stress induce (para)sex? Implications for Candida albicans evolution. Trends Genet. 28: 197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan M., Thomas D. Y., Whiteway M., Kavanagh K., 2002. Correlation between virulence of Candida albicans mutants in mice and Galleria mellonella larvae. FEMS Immunol. Med. Microbiol. 34: 153–157. [DOI] [PubMed] [Google Scholar]
- Burt A., 2000. Perspective: sex, recombination, and the efficacy of selection–was Weismann right? Evolution 54: 337–351. [DOI] [PubMed] [Google Scholar]
- Butler G., Rasmussen M. D., Lin M. F., Santos M. A., Sakthikumar S., et al. , 2009. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459: 657–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderone R. A., Fonzi W. A., 2001. Virulence factors of Candida albicans. Trends Microbiol. 9: 327–335. [DOI] [PubMed] [Google Scholar]
- Cao F., Lane S., Raniga P. P., Lu Y., Zhou Z., et al. , 2006. The Flo8 transcription factor is essential for hyphal development and virulence in Candida albicans. Mol. Biol. Cell 17: 295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Bradford W. D., Seidel C. W., Li R., 2012a Hsp90 stress potentiates rapid cellular adaptation through induction of aneuploidy. Nature 482: 246–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Rubinstein B., Li R., 2012b Whole chromosome aneuploidy: big mutations drive adaptation by phenotypic leap. Bioessays 34: 893–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Magee B. B., Dawson D., Magee P. T., Kumamoto C. A., 2004. Chromosome 1 trisomy compromises the virulence of Candida albicans. Mol. Microbiol. 51: 551–565. [DOI] [PubMed] [Google Scholar]
- Chibana H., Beckerman J. L., Magee P. T., 2000. Fine-resolution physical mapping of genomic diversity in Candida albicans. Genome Res. 10: 1865–1877. [DOI] [PubMed] [Google Scholar]
- Coste A., Turner V., Ischer F., Morschhauser J., Forche A., et al. , 2006. A mutation in Tac1p, a transcription factor regulating CDR1 and CDR2, is coupled with loss of heterozygosity at chromosome 5 to mediate antifungal resistance in Candida albicans. Genetics 172: 2139–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter G., Doyle S., Kavanagh K., 2000. Development of an insect model for the in vivo pathogenicity testing of yeasts. FEMS Immunol. Med. Microbiol. 27: 163–169. [DOI] [PubMed] [Google Scholar]
- Dumitru R., Navarathna D. H., Semighini C. P., Elowsky C. G., Dumitru R. V., et al. , 2007. In vivo and in vitro anaerobic mating in Candida albicans. Eukaryot. Cell 6: 465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkel N., Blass J., Rogers P. D., Morschhauser J., 2008. Mutations in the multi-drug resistance regulator MRR1, followed by loss of heterozygosity, are the main cause of MDR1 overexpression in fluconazole-resistant Candida albicans strains. Mol. Microbiol. 69: 827–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel J. S., Mitchell A. P., 2011. Genetic control of Candida albicans biofilm development. Nat. Rev. Microbiol. 9: 109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forche A., Alby K., Schaefer D., Johnson A. D., Berman J., et al. , 2008. The parasexual cycle in Candida albicans provides an alternative pathway to meiosis for the formation of recombinant strains. PLoS Biol. 6: e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford C. B., Funt J. M., Abbey D., Issi L., Guiducci C., et al. , 2015. The evolution of drug resistance in clinical isolates of Candida albicans. Elife 4: e00662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox E. P., Bui C. K., Nett J. E., Hartooni N., Mui M. C., et al. , 2015. An expanded regulatory network temporally controls Candida albicans biofilm formation. Mol. Microbiol. 96: 1226–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs B. B., Eby J., Nobile C. J., El Khoury J. B., Mitchell A. P., et al. , 2010a Role of filamentation in Galleria mellonella killing by Candida albicans. Microbes Infect. 12: 488–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs B. B., O’Brien E., Khoury J. B., Mylonakis E., 2010b Methods for using Galleria mellonella as a model host to study fungal pathogenesis. Virulence 1: 475–482. [DOI] [PubMed] [Google Scholar]
- Gerstein A. C., Rosenberg A., Hecht I., Berman J., 2016. diskImageR: quantification of resistance and tolerance to antimicrobial drugs using disk diffusion assays. Microbiology 162: 1059–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein A. C., Lim H., Berman J., Hickman M. A., 2017. Ploidy tug-of-war: evolutionary and genetic environments influence the rate of ploidy drive in a human fungal pathogen. Evolution 71: 1025–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard M. R., Godfray H. C., Burt A., 2005. Sex increases the efficacy of natural selection in experimental yeast populations. Nature 434: 636–640. [DOI] [PubMed] [Google Scholar]
- Graser Y., Volovsek M., Arrington J., Schonian G., Presber W., et al. , 1996. Molecular markers reveal that population structure of the human pathogen Candida albicans exhibits both clonality and recombination. Proc. Natl. Acad. Sci. USA 93: 12473–12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudlaugsson O., Gillespie S., Lee K., Vande Berg J., Hu J., et al. , 2003. Attributable mortality of nosocomial candidemia, revisited. Clin. Infect. Dis. 37: 1172–1177. [DOI] [PubMed] [Google Scholar]
- Guthrie C., Fink G. R., 1991. Guide to yeast genetics and molecular biology. Methods Enzymol. 194: 1–863. [PubMed] [Google Scholar]
- Hao B., Clancy C. J., Cheng S., Raman S. B., Iczkowski K. A., et al. , 2009. Candida albicans RFX2 encodes a DNA binding protein involved in DNA damage responses, morphogenesis, and virulence. Eukaryot. Cell 8: 627–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison B. D., Hashemi J., Bibi M., Pulver R., Bavli D., et al. , 2014. A tetraploid intermediate precedes aneuploid formation in yeasts exposed to fluconazole. PLoS Biol. 12: e1001815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckman D. S., Geiser D. M., Eidell B. R., Stauffer R. L., Kardos N. L., et al. , 2001. Molecular evidence for the early colonization of land by fungi and plants. Science 293: 1129–1133. [DOI] [PubMed] [Google Scholar]
- Heitman J., 2010. Evolution of eukaryotic microbial pathogens via covert sexual reproduction. Cell Host Microbe 8: 86–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J., 2015. Evolution of sexual reproduction: a view from the fungal kingdom supports an evolutionary epoch with sex before sexes. Fungal Biol. Rev. 29: 108–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman M. A., Zeng G., Forche A., Hirakawa M. P., Abbey D., et al. , 2013. The ‘obligate diploid’ Candida albicans forms mating-competent haploids. Nature 494: 55–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman M. A., Paulson C., Dudley A., Berman J., 2015. Parasexual ploidy reduction drives population heterogeneity through random and transient aneuploidy in Candida albicans. Genetics 200: 781–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa M. P., Martinez D. A., Sakthikumar S., Anderson M. Z., Berlin A., et al. , 2015. Genetic and phenotypic intra-species variation in Candida albicans. Genome Res. 25: 413–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull C. M., Raisner R. M., Johnson A. D., 2000. Evidence for mating of the “asexual” yeast Candida albicans in a mammalian host. Science 289: 307–310. [DOI] [PubMed] [Google Scholar]
- Jacobsen I. D., 2014. Galleria mellonella as a model host to study virulence of Candida. Virulence 5: 237–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jokela J., Dybdahl M. F., Lively C. M., 2009. The maintenance of sex, clonal dynamics, and host-parasite coevolution in a mixed population of sexual and asexual snails. Am. Nat. 174(Suppl. 1): S43–S53. [DOI] [PubMed] [Google Scholar]
- Jones S. K., Jr, Bennett R. J., 2011. Fungal mating pheromones: choreographing the dating game. Fungal Genet. Biol. 48: 668–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T., Federspiel N. A., Chibana H., Dungan J., Kalman S., et al. , 2004. The diploid genome sequence of Candida albicans. Proc. Natl. Acad. Sci. USA 101: 7329–7334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Chung K. J., Chang Y. C., 2012. Aneuploidy and drug resistance in pathogenic fungi. PLoS Pathog. 8: e1003022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtonen J., Jennions M. D., Kokko H., 2012. The many costs of sex. Trends Ecol. Evol. 27: 172–178. [DOI] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lively C. M., 2010. A review of Red Queen models for the persistence of obligate sexual reproduction. J. Hered. 101(Suppl. 1): S13–S20. [DOI] [PubMed] [Google Scholar]
- Lockhart S. R., Zhao R., Daniels K. J., Soll D. R., 2003. Alpha-pheromone-induced “shmooing” and gene regulation require white-opaque switching during Candida albicans mating. Eukaryot. Cell 2: 847–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCallum D. M., Castillo L., Nather K., Munro C. A., Brown A. J., et al. , 2009. Property differences among the four major Candida albicans strain clades. Eukaryot. Cell 8: 373–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magditch D. A., Liu T. B., Xue C., Idnurm A., 2012. DNA mutations mediate microevolution between host-adapted forms of the pathogenic fungus Cryptococcus neoformans. PLoS Pathog. 8: e1002936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee B. B., Magee P. T., 2000. Induction of mating in Candida albicans by construction of MTLa and MTLalpha strains. Science 289: 310–313. [DOI] [PubMed] [Google Scholar]
- Mayer F. L., Wilson D., Hube B., 2013. Candida albicans pathogenicity mechanisms. Virulence 4: 119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald M. J., Rice D. P., Desai M. M., 2016. Sex speeds adaptation by altering the dynamics of molecular evolution. Nature 531: 233–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlini L., Dudin O., Martin S. G., 2013. Mate and fuse: how yeast cells do it. Open Biol. 3: 130008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. G., Johnson A. D., 2002. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell 110: 293–302. [DOI] [PubMed] [Google Scholar]
- Morran L. T., Schmidt O. G., Gelarden I. A., Parrish R. C., II, Lively C. M., 2011. Running with the Red Queen: host-parasite coevolution selects for biparental sex. Science 333: 216–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow C. A., Fraser J. A., 2013. Ploidy variation as an adaptive mechanism in human pathogenic fungi. Semin. Cell Dev. Biol. 24: 339–346. [DOI] [PubMed] [Google Scholar]
- Morschhauser J., Barker K. S., Liu T. T., Bla B. W. J., Homayouni R., et al. , 2007. The transcription factor Mrr1p controls expression of the MDR1 efflux pump and mediates multidrug resistance in Candida albicans. PLoS Pathog. 3: e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee P. K., Sendid B., Hoarau G., Colombel J. F., Poulain D., et al. , 2015. Mycobiota in gastrointestinal diseases. Nat. Rev. Gastroenterol. Hepatol. 12: 77–87. [DOI] [PubMed] [Google Scholar]
- Neiman A. M., 2011. Sporulation in the budding yeast Saccharomyces cerevisiae. Genetics 189: 737–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M., Feretzaki M., Sun S., Wang X., Heitman J., 2011. Sex in fungi. Annu. Rev. Genet. 45: 405–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M., Feretzaki M., Li W., Floyd-Averette A., Mieczkowski P., et al. , 2013. Unisexual and heterosexual meiotic reproduction generate aneuploidy and phenotypic diversity de novo in the yeast Cryptococcus neoformans. PLoS Biol. 11: e1001653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile C. J., Johnson A. D., 2015. Candida albicans biofilms and human disease. Annu. Rev. Microbiol. 69: 71–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile C. J., Fox E. P., Nett J. E., Sorrells T. R., Mitrovich Q. M., et al. , 2012. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell 148: 126–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble S. M., French S., Kohn L. A., Chen V., Johnson A. D., 2010. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat. Genet. 42: 590–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odds F. C., Bougnoux M. E., Shaw D. J., Bain J. M., Davidson A. D., et al. , 2007. Molecular phylogenetics of Candida albicans. Eukaryot. Cell 6: 1041–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panwar S. L., Legrand M., Dignard D., Whiteway M., Magee P. T., 2003. MFalpha1, the gene encoding the alpha mating pheromone of Candida albicans. Eukaryot. Cell 2: 1350–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perepnikhatka V., Fischer F. J., Niimi M., Baker R. A., Cannon R. D., et al. , 1999. Specific chromosome alterations in fluconazole-resistant mutants of Candida albicans. J. Bacteriol. 181: 4041–4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad R., De Wergifosse P., Goffeau A., Balzi E., 1995. Molecular cloning and characterization of a novel gene of Candida albicans, CDR1, conferring multiple resistance to drugs and antifungals. Curr. Genet. 27: 320–329. [DOI] [PubMed] [Google Scholar]
- Quinlan A. R., Hall I. M., 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rancati G., Pavelka N., Fleharty B., Noll A., Trimble R., et al. , 2008. Aneuploidy underlies rapid adaptive evolution of yeast cells deprived of a conserved cytokinesis motor. Cell 135: 879–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E. S., et al. , 2011. Integrative genomics viewer. Nat. Biotechnol. 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustchenko E., 2007. Chromosome instability in Candida albicans. FEMS Yeast Res. 7: 2–11. [DOI] [PubMed] [Google Scholar]
- Sanglard D., Ischer F., Monod M., Bille J., 1997. Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC transporter gene. Microbiology 143: 405–416. [DOI] [PubMed] [Google Scholar]
- Santaguida S., Amon A., 2015. Short- and long-term effects of chromosome mis-segregation and aneuploidy. Nat. Rev. Mol. Cell Biol. 16: 473–485. [DOI] [PubMed] [Google Scholar]
- Saville S. P., Lazzell A. L., Monteagudo C., Lopez-Ribot J. L., 2003. Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot. Cell 2: 1053–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonherr F. A., Sparber F., Kirchner F. R., Guiducci E., Trautwein-Weidner K., et al. , 2017. The intraspecies diversity of C. albicans triggers qualitatively and temporally distinct host responses that determine the balance between commensalism and pathogenicity. Mucosal Immunol. 10: 1335–1350. [DOI] [PubMed] [Google Scholar]
- Selmecki A., Forche A., Berman J., 2006. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science 313: 367–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmecki A., Gerami-Nejad M., Paulson C., Forche A., Berman J., 2008. An isochromosome confers drug resistance in vivo by amplification of two genes, ERG11 and TAC1. Mol. Microbiol. 68: 624–641. [DOI] [PubMed] [Google Scholar]
- Selmecki A., Forche A., Berman J., 2010. Genomic plasticity of the human fungal pathogen Candida albicans. Eukaryot. Cell 9: 991–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmecki A. M., Dulmage K., Cowen L. E., Anderson J. B., Berman J., 2009. Acquisition of aneuploidy provides increased fitness during the evolution of antifungal drug resistance. PLoS Genet. 5: e1000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudbery P. E., 2011. Growth of Candida albicans hyphae. Nat. Rev. Microbiol. 9: 737–748. [DOI] [PubMed] [Google Scholar]
- Sun S., Heitman J., 2011. Is sex necessary? BMC Biol. 9: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Z., Hays M., Cromie G. A., Jeffery E. W., Scott A. C., et al. , 2013. Aneuploidy underlies a multicellular phenotypic switch. Proc. Natl. Acad. Sci. USA 110: 12367–12372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavanti A., Gow N. A., Maiden M. C., Odds F. C., Shaw D. J., 2004. Genetic evidence for recombination in Candida albicans based on haplotype analysis. Fungal Genet. Biol. 41: 553–562. [DOI] [PubMed] [Google Scholar]
- Taylor J. W., Berbee M. L., 2006. Dating divergences in the fungal tree of life: review and new analyses. Mycologia 98: 838–849. [DOI] [PubMed] [Google Scholar]
- Thorvaldsdottir H., Robinson J. T., Mesirov J. P., 2013. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14: 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres E. M., Dephoure N., Panneerselvam A., Tucker C. M., Whittaker C. A., et al. , 2010. Identification of aneuploidy-tolerating mutations. Cell 143: 71–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera G. A., Carneiro M. O., Hartl C., Poplin R., Del Angel G., et al. , 2013. From FastQ data to high confidence variant calls: the genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinformatics 43: 11.10.1–11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van het Hoog M., Rast T. J., Martchenko M., Grindle S., Dignard D., et al. , 2007. Assembly of the Candida albicans genome into sixteen supercontigs aligned on the eight chromosomes. Genome Biol. 8: R52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Werven F. J., Amon A., 2011. Regulation of entry into gametogenesis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 366: 3521–3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker B. J., Abeel T., Shea T., Priest M., Abouelliel A., et al. , 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9: e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheimer N. B., Stone N., Berman J., 2016. Ploidy dynamics and evolvability in fungi. Philos. Trans. R. Soc. Lond. B Biol. Sci. 371: 20150461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirsching S., Michel S., Morschhauser J., 2000. Targeted gene disruption in Candida albicans wild-type strains: the role of the MDR1 gene in fluconazole resistance of clinical Candida albicans isolates. Mol. Microbiol. 36: 856–865. [DOI] [PubMed] [Google Scholar]
- Wisplinghoff H., Bischoff T., Tallent S. M., Seifert H., Wenzel R. P., et al. , 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39: 309–317. [DOI] [PubMed] [Google Scholar]
- Wu W., Lockhart S. R., Pujol C., Srikantha T., Soll D. R., 2007. Heterozygosity of genes on the sex chromosome regulates Candida albicans virulence. Mol. Microbiol. 64: 1587–1604. [DOI] [PubMed] [Google Scholar]
- Yona A. H., Manor Y. S., Herbst R. H., Romano G. H., Mitchell A., et al. , 2012. Chromosomal duplication is a transient evolutionary solution to stress. Proc. Natl. Acad. Sci. USA 109: 21010–21015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng G., Wang Y. M., Chan F. Y., Wang Y., 2014. One-step targeted gene deletion in Candida albicans haploids. Nat. Protoc. 9: 464–473. [DOI] [PubMed] [Google Scholar]
- Zhang N., Magee B. B., Magee P. T., Holland B. R., Rodrigues E., et al. , 2015. Selective advantages of a parasexual cycle for the yeast Candida albicans. Genetics 200: 1117–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genome sequencing data has been uploaded to the National Center for Biotechnology Information Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra). Data can be accessed using the study accession #SRP113728 or the BioProject #PRJNA394762.