Abstract
The addition of a new telomere onto a chromosome break, a process termed healing, has been studied extensively in organisms that utilize telomerase to maintain their telomeres. In comparison, relatively little is known about how new telomeres are constructed on broken chromosomes in organisms that do not use telomerase. Chromosome healing was studied in somatic and germline cells of Drosophila melanogaster, a nontelomerase species. We observed, for the first time, that broken chromosomes can be healed in somatic cells. In addition, overexpression of the telomere cap component Hiphop increased the survival of somatic cells with broken chromosomes, while the cap component HP1 did not, and overexpression of the cap protein HOAP decreased their survival. In the male germline, Hiphop overexpression greatly increased the transmission of healed chromosomes. These results indicate that Hiphop can stimulate healing of a chromosome break. We suggest that this reflects a unique function of Hiphop: it is capable of seeding formation of a new telomeric cap on a chromosome end that lacks a telomere.
Keywords: telomere, terminin, Drosophila, double strand break, hiphop, HOAP, FLP, dicentric chromosome
CHROMOSOMES that experience double-strand breaks (DSBs) are usually repaired with great efficiency. However, in some cases a cell may have difficulty achieving proper rejoining of two broken ends. If cells are exposed to significant doses of ionizing radiation, the number of DSBs may overwhelm a cell’s repair capacity and lead to cell death. Another form of damage that presents difficulty for a cell is the occurrence of a single broken chromosome end. Such damage may arise by breakage of a dicentric chromosome during a mitotic or meiotic division, by division of a cell with an unrepaired DSB, or, in some organisms, by telomere shortening in somatic cells owing to a lack of telomerase expression. A chromosome with a single broken end is unrepairable by normal DNA DSB repair mechanisms, which are geared to rejoin two broken ends.
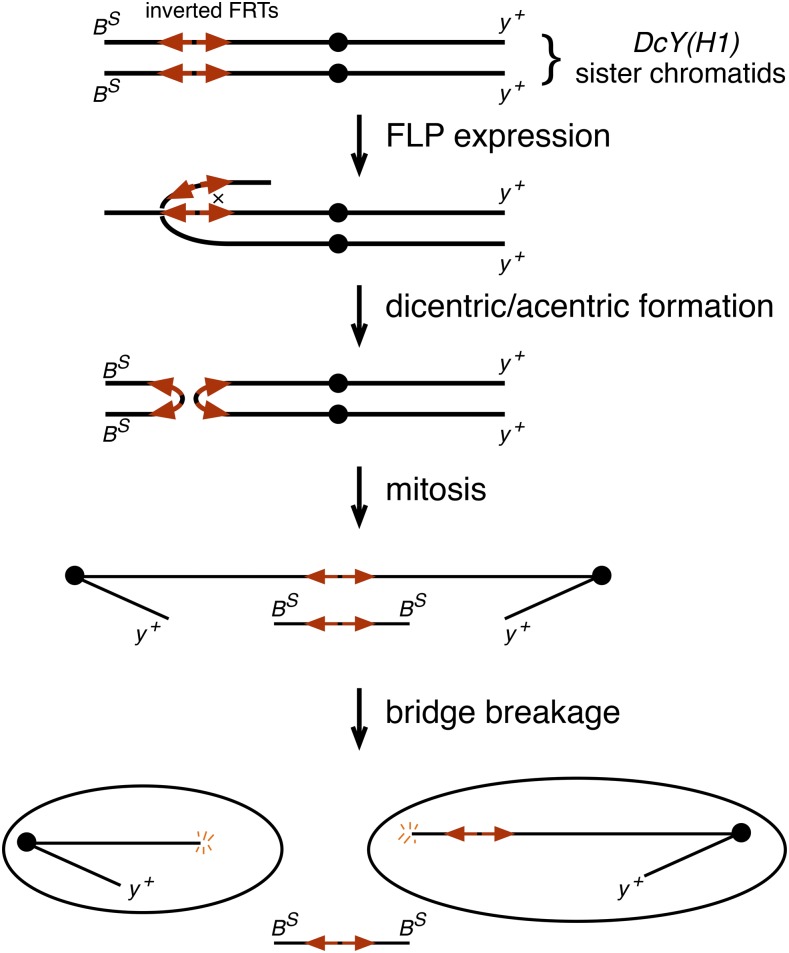
Dicentric chromosomes, having two centromeres rather than the normal single centromere, can be produced experimentally by using the FLP recombinase to mediate recombination between inverted FRTs on sister chromatids (Figure 1) (Falco et al. 1982; Golic 1994). The sister centromeres conjoined in this fashion then separate to opposite poles during anaphase. In Drosophila, the resulting chromatin bridges typically break in mitosis to produce daughter cells that receive a chromosome with a single broken end (Titen and Golic 2008). The predominant consequence of this event in the soma is cell death (Ahmad and Golic 1999; Titen and Golic 2008). Nonetheless, some cells do escape death and continue to divide and differentiate (Golic 1994; Titen and Golic 2008; Kurzhals et al. 2011). In the male germline most cells experiencing dicentric formation are also eliminated, but a small fraction are repaired by de novo telomere addition (Ahmad and Golic 1998; Titen and Golic 2010)—a phenomenon known as healing (McClintock 1939, 1941).
Figure 1.
Dicentric chromosome formation mediated by FLP recombinase with the DcY(H1) chromosome. The DcY(H1) chromosome is a BS Y y+ chromosome carrying inverted FRTs inserted proximal to BS on YL, but distal to all fertility factors. FLP-mediated recombination between inverted FRTs on sister chromatids produces a dicentric Y chromosome, and a small acentric chromosome carrying both copies of BS. In mitosis, the dicentric bridge breaks to produce daughter cells, each carrying a chromosome with one broken end. If these chromosomes are healed and transmitted to progeny they may be recognized as y+ B+ offspring. Dicentric chromosome formation on other chromosomes uses inverted FRTs to similar effect.
Chromosome healing has been observed in many species, from yeast to humans (Gorovsky 1980; Mason et al. 1984; Flint et al. 1994; Melek and Shippen 1996; Sprung et al. 1999; Pennaneach et al. 2006). Healing of broken chromosomes is likely to play a significant role in human health. For instance, healing of broken ends has been observed in tumor cells (Fouladi et al. 2000), and is often associated with constitutional chromosome abnormalities (Fortin et al. 2009). During evolution, new telomeres must be generated when a karyotype changes by chromosome fission to produce an increase in chromosome number. Striking examples of this are seen in butterflies, with some species having evolved karyotypes with >200 chromosomes (Lukhtanov 2015; Šíchová et al. 2015).
A chromosome that is broken in the germline may be considered healed if it is transmissible through generations without incurring lethality or being subject to continuing chromosome rearrangements. Healing events in the germline can be readily detected in crosses using appropriately marked chromosomes (Levis 1989; Tower et al. 1993; Ahmad and Golic 1998; Titen and Golic 2010; Titen et al. 2014). Although healing of broken ends clearly occurs in the Drosophila germline, it is unknown whether healing can occur in somatic cells that experience dicentric breakage.
In organisms that utilize telomerase, healing requires that new telomeric repeats are added to the broken chromosome end, and that these repeats successfully recruit the full array of capping proteins that recognize these repeats, as well as their critical partners, which then prevent the chromosome end from being perceived as damaged DNA. Drosophila are one of a number of organisms that do not use telomerase (Mason et al. 2011). In place of the simple sequence repeats that are normally added by telomerase, Drosophila extend chromosomal DNA by the mobilization of specific retrotransposons to chromosome ends (Biessmann et al. 1990b, 1992; Levis et al. 1993). The precise complement of proteins that are needed to form a functional capping complex in Drosophila differs from that found in organisms that use telomerase, though there is some overlap. The set of proteins that are found uniquely at Drosophila telomeres has been called the terminin complex, in analogy with the shelterin complex found in organisms that use telomerase (de Lange 2005; Palm and de Lange 2008; Raffa et al. 2013; Zhang et al. 2016).
In Drosophila, the end-replication solution and capping functions are not interdependent as they are in organisms that use telomerase. Thus, Drosophila may heal a broken chromosome end by the addition of the cap structure without the presence of retrotransposon sequences that are normally found at chromosome ends (Levis 1989; Biessmann et al. 1990a). In Drosophila, chromosomes with functional caps may terminate at a variety of locations, and within a variety of sequences (Levis 1989; Biessmann et al. 1990a; Ahmad and Golic 1998; Titen and Golic 2010; Beaucher et al. 2012). Although a fully functional telomere will carry the proteins that form a functional cap, and the retrotransposons that solve the end replication problem, the latter are only necessary in the long term so that vital genes are not lost as chromosomes shorten over many generations. In the short term, possession of a functional cap is sufficient to allow survival, and we consider such a chromosome to be healed.
Flies carrying loss-of-function mutations in terminin complex proteins show end-to-end chromosome fusions and activate the DNA Damage Response (DDR) (Musarò et al. 2008; Cipressa and Cenci 2013). The terminin proteins include HOAP (encoded by cav), Tea (tea), Modigliani (moi), Verocchio (ver), and the paralogous pair of proteins Hiphop (hiphop) and Ms(3)K81 (or simply K81; ms(3)K81) (Rong 2008; Raffa et al. 2011; Zhang et al. 2016). A number of other proteins that are conserved in telomerase and nontelomerase organisms, such as HP1 (HP1), ATM (tefu), and the MRN complex (mre11, rad50, nbs), function in telomere maintenance in both types of organisms (Fanti et al. 1998; Pandita 2002; Rong 2008; Sabourin and Zakian 2008; Lamarche et al. 2010; Canudas et al. 2011). A common characteristic of members of this latter group is that they also play roles at nontelomeric locations.
In spite of the identification of many genes required for telomere function, little is known of the mechanisms that mediate chromosome healing in Drosophila. Genes involved in the DDR and DNA repair are known to influence the frequency of chromosome healing. Females that carry the mu-2 mutation allow healing of terminally truncated chromosomes in the female germline (Mason et al. 1984), and mu-2 is homologous to a mammalian DNA damage checkpoint protein (Dronamraju and Mason 2009). The lok (encoding Chk2) and p53 genes also strongly influence the recovery of healed chromosomes (Titen et al. 2014). However, it is probable that mu-2, lok and p53 all act indirectly, via cell cycle checkpoint delays or control of cell growth and viability, rather than participating directly in the addition of a new telomere to a broken end. DNA repair proteins influence the healing of chromosomes that have been cut by the meganuclease I-SceI, but this effect may also be indirect—when mutants eliminate the normal DSB repair pathways the frequency of healing may increase simply because the preferred repair mechanisms are unavailable (Beaucher et al. 2012). The influence of the telomere capping complex proteins on healing has yet to be investigated.
In the work reported here we examined the fate of a broken chromosome in somatic cells and found strong evidence for occasional healing. Additionally, we identified one component of Drosophila telomeres, Hiphop, whose overexpression promotes the survival of cells with a broken chromosome and the transmission of broken-and-healed chromosomes through the male germline. We propose that Hiphop acts to seed telomere cap formation on broken chromosome ends, and that an increased frequency of chromosome healing with hiphop overexpression is the basis for these somatic and germline effects.
Materials and Methods
Karyotype examination
DAPI-stained larval brain chromosomes were prepared as described (Fanti and Pimpinelli 2004). To obtain metaphase preparations for the immunostaining of Figure 2, the larval brains were dissected in 0.7% sodium chloride and treated with hypotonic solution (0.5% sodium citrate) for 8 min. Brains were then fixed for 8 min with 2% formaldehyde and 45% acetic acid, and squashed in the same fixative. Slides were frozen in liquid nitrogen, and, after flipping off the coverslip, were immersed in PBS, washed in PBS containing 1% Triton-X, and incubated in 1× PBS with dried nonfat milk for 30 min (∼1 spoonful of milk in 40 ml of 1× PBS). The slides were then incubated with HOAP antibody for 1 hr at room temperature, and then overnight at 4°. The HOAP antibodies, obtained in guinea pig, were diluted 1:100 in PBS 1% bovine serum albumin (BSA). Cy3-conjugated Goat Anti-Guinea Pig IgG (H+L) was used as a secondary antibody (diluted 1:100; Life Technologies). For examination by DAPI staining only, squashes were prepared as described (Gatti and Pimpinelli 1983)
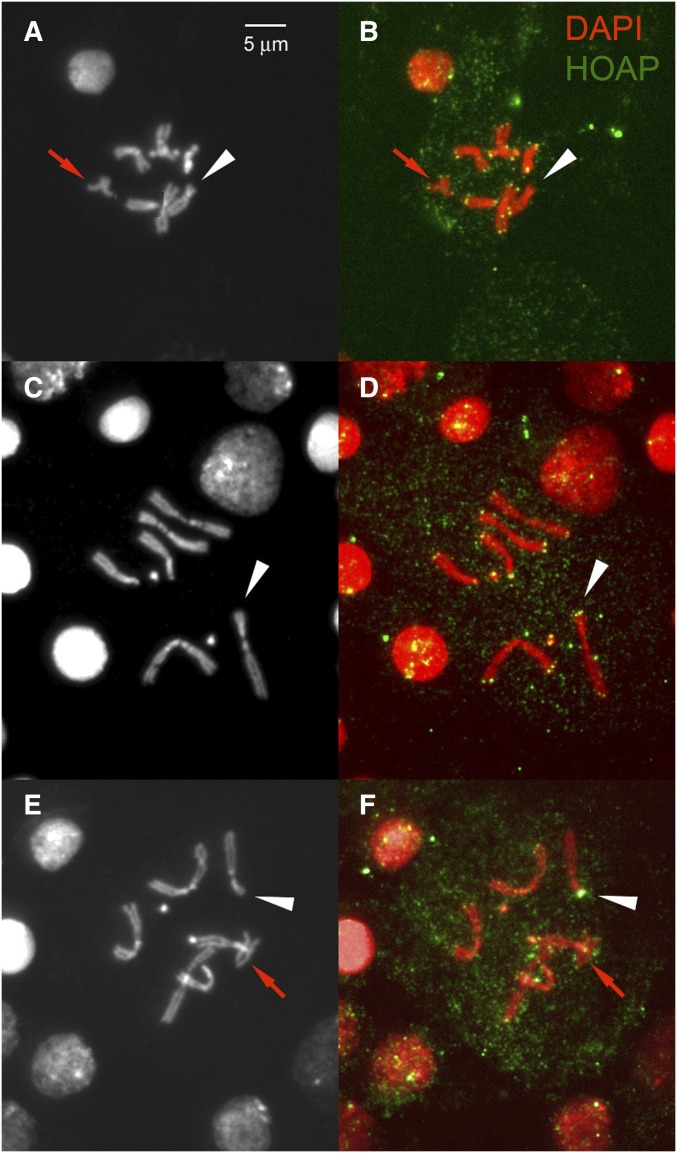
Figure 2.
Detection of HOAP on chromosome breaks. Acentric chromosomes are indicated with a red arrow, and are expected to have telomeres on both ends. In each panel the broken end of chromosome 3 is indicated with a white arrowhead. (A, C, E) DAPI staining of chromosomes; (B, D, F) HOAP staining shown in green with chromosomes in red. (A and B) A nucleus with no HOAP staining on the broken end, but HOAP staining clearly visible on the acentric portion and on the remaining chromosomes. (C and D) A nucleus lacking the acentric chromosome, but with clear HOAP staining detected on the broken chromosome end. (E and F) A nucleus with very strong HOAP signal on the broken chromosome end.
Fly husbandry, stocks, and transgenes
All flies were raised at 25° on standard medium. Heat shocks were performed by partial submersion of culture vials in a circulating water bath. The eyGal4 UASFLP combination used was P{Gal4-ey.H}4–8 P{USFLP.D}JD1; the eyFLP transgene was P{ey-FLP.n}2; the brain Gal4 driver was P{GawB}6011A. These stocks were obtained from the Bloomington (IN) Drosophila stock center. Use of the eyGal4 UASFLP and eyFLP combinations has been described previously (Kurzhals et al. 2011). An Epgy2 element (EY09894, obtained from the Bloomington stock center) was used for overexpression of hiphop in Figure 3D. The UAShiphop (fused to mCherry) and UASK81 (fused to GFP) transgenes and hiphopL14 and hiphopL32 mutant alleles have been described elsewhere (Gao et al. 2011). Figure 3E used the HRH008-A1 insertion on 2; Figure 3F used the insertion HRH008-E on chromosome 3; Figure 3G used the insertion HGK-4B insertion on chromosome 3. The UAScav transgene construct (fused to GFP) has been described elsewhere (Raffa et al. 2010). We used two different insertions of this element on chromosome 2, #1 and #3, for Figure 3, H and I, respectively. The HS-HP1 construct has been described elsewhere (Eissenberg and Hartnett 1993; Fanti et al. 1998). The 70FLP3F line was previously described (Golic et al. 1997). For cytological experiments, insertions of the P{FrTr} element, carrying inverted FRTs and an adjacent mini-white gene (Titen and Golic 2010), were used to generate dicentric chromosomes. P{FrTr)1D at 72D1 (Figure 2 and Table 1) and P{FrTr)1B at 46F3 (Figure 4) were used. The DcY(H1) chromosome, carrying y+ near the tip of the short arm, BS near the tip of the long arm, and P{iw} with inverted FRTs proximal to BS has been described elsewhere, along with its use to assay germline chromosome healing (Kurzhals et al. 2011; Titen et al. 2014). The nosGal4 and UASFLP transgenes have been described elsewhere (Van Doren et al. 1998; Beumer 1997).
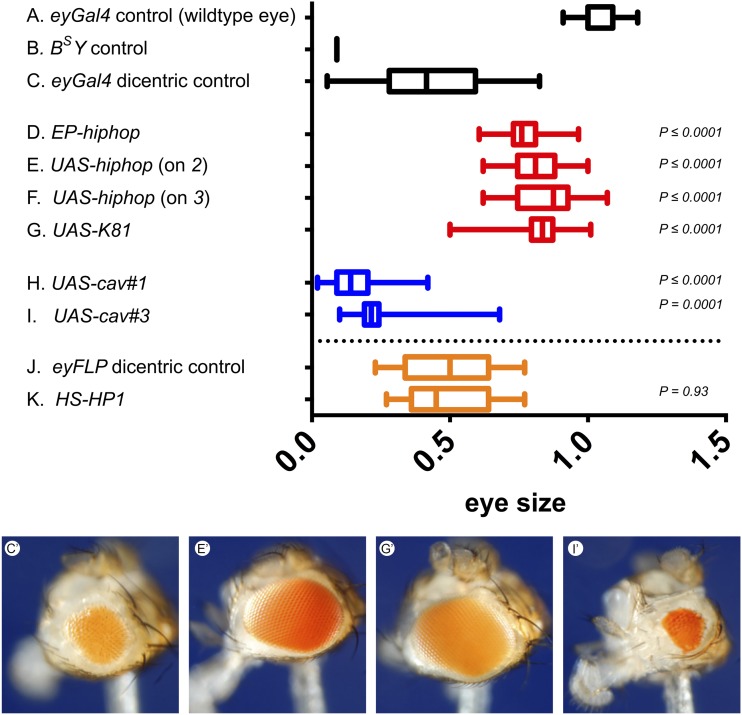
Figure 3.
Effects of telomere capping genes in the BARTL assay. Dicentric chromosome formation was induced in the developing eyes of males carrying the DcY(H1), y+ BS chromosome, using eyGal4 UASFLP (C–I) or eyFLP (J and K), and eye sizes were measured for each genotype. (A and B) Wild-type and BS controls, respectively. (C) The effect of DcY(H1) dicentric production. The eyes are larger than in B, owing to survival and differentiation of some cells that lost BS. The effect of hiphop (D–F), ms(3)K81 (G), and cav (H and I) overexpression. P values are for comparison with C. (J and K) Eye size when FLP is expressed from eyFLP transgene (J), and when Su(var)205 (encoding HP1) is also overexpressed with an early developmental heat shock (K). Typical phenotypes are shown below [i.e., (C′) eyGal4, (E′) UAS-hiphop, (G′) UAS-K81, and (I′) UAS-cav#3]. The numbers of eyes measured for each genotype (A–K) were 20, 43, 110, 35, 16, 16, 14, 18, 18, 30, and 30, respectively. The plots show 5th, 25th, median, 75th, and 95th percentiles.
Table 1. Frequency of HOAP staining on broken chromosomes.
| Time after heat shock (hr) | Nuclei with acentric | Nuclei lacking acentric | |||
|---|---|---|---|---|---|
| Total | Acentric positive | Broken end positive | Total | Broken end positive | |
| 8 | 28 | 18 | 7 | 3 | 1 |
| 16 | 138 | 102 | 16 | 74 | 7 |
| 24 | 58 | 39 | 14 | 30 | 3 |
| 40 | 23 | 15 | 5 | 19 | 2 |
| 72 | 13 | 12 | 1 | 27 | 1 |
| Total | 260 | 186 | 43 | 153 | 14 |
Nuclei that carried a broken chromosome were scored for HOAP staining on the broken chromosome end, and on the acentric chromosome (if present) as a control for staining efficiency. The larvae that were examined carried 70FLP3F on their X and the inverted FRT-bearing element FrTr1D (at 72D1 on 3L). They were heat shocked at 38° for 1 hr, and dissected at various times after heat shock.
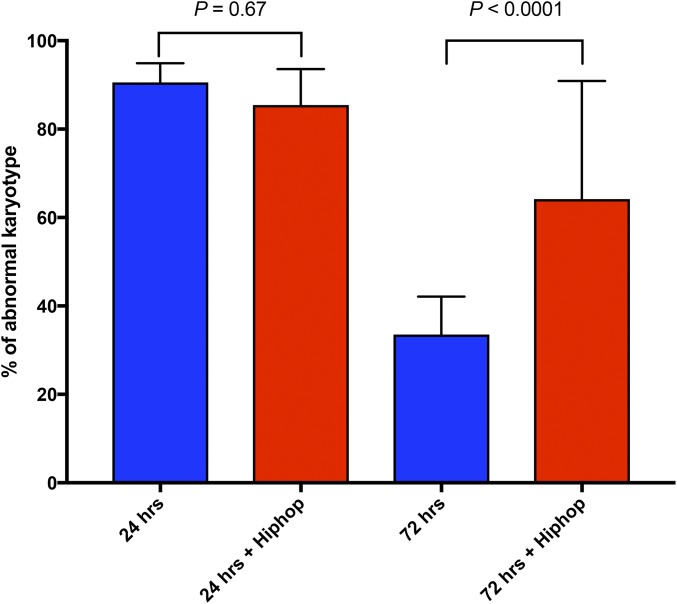
Figure 4.
Survival of cells that experienced dicentric formation. Dicentric chromosome formation was induced in larvae of all stages by heat shock (38°, 1 hr) using 70FLP3F. P{GawB}6011A drove expression of UAShiphop in the brain. Metaphase nuclei from third instar larval brains were examined at 24 and 72 hr after heat shock. Cells carrying broken chromosomes (visibly shortened) and/or acentric chromosomes were scored as abnormal. The numbers of nuclei examined for each time point and genotype were 115 (5 brains), 99 (4 brains), 133 (4 brains), and 33 (3 brains), respectively. Significance was calculated with a 2 × 2 contingency test. SD are indicated.
BARTL assay
The BARTL assay was performed as previously described (Kurzhals et al. 2011).
Transmission of healed chromosomes
To test for the influence of hiphop overexpression on healing in the male germline, we generated males of the following genotypes: (1) y w/DcY(H1); nosGal4 UASFLP/+; (2) y w/DcY(H1); UAShiphop/+; nosGal4 UASFLP/+ ; (3) y w/DcY(H1); lokP6; nosGal4 UASFLP/+; (4) y w/DcY(H1); lokP6 UAShiphop/lokP6; nosGal4 UASFLP/+. Three different nosGal4 UASFLP combinations (1, 40, and 95) with different efficiencies were tested for each genotype. The UAShiphop insertion HRH008-A1 on 2 was used in these experiments. The males were individually testcrossed to y w females, and the progeny were counted through day 18 of the cross.
Antibody staining of Hiphop:mCherry fusion in testis
Testes were dissected from young males (0- to 3-day-old adults) in 1× PBS, fixed for 30 min in 1× PBS + 4% paraformaldehyde, then washed in PBS-T (1× PBS + 0.1% Triton-X) for 60 min at room temperature (RT). The testes were incubated overnight with the primary antibody (1:200 dilution of rabbit polyclonal antibody against mCherry; Thermo-Fisher PA5-34974) at 4° in 1× PBS-T + 3% BSA, then 1 hr at RT. They were then washed three times, 20 min each, in PBS-T at RT, then incubated with the secondary antibody (1:400 dilution of goat anti-rabbit coupled to AlexaFluor 488; Thermo-Fisher A-11008) in PBS-T + 3% BSA at RT for 2 hr. The testes were then washed 3× for 20 min each, in PBS-T at RT. DAPI was added to the final wash at a concentration of 0.4 µg/ml. Testes were then transferred to fresh 1× PBS, and stored at 4° until examined. Testes were mounted in Vectashield and examined with an Olympus DSU disc scanning microscope using 60× and 100× PlanApo oil immersion objectives.
Data availability
Supplemental Material, Table S1 (see File S1 for legend) contain complete progeny counts used for testing the effect of hiphop overexpression in the germline. Fly strains are available from the authors upon request.
Results
Experimental system
In these experiments, we used FLP-mediated recombination between inverted FRTs on sister chromatids to produce a dicentric and an acentric chromosome. This process is very efficient, and can be induced to occur in nearly 100% of cells. In some experiments, inverted FRTs inserted at a medial location on an autosomal arm were used because this generates dicentric and acentric chromosomes that are easily detected in metaphase chromosome preparations. In other experiments we used a BS Y y+ chromosome, carrying the dominant eye shape marker BarStone on the long arm distal to the inverted FRTs, and the dominant body color marker yellow+ on the other arm (Figure 1). Because the Y is not required for cell viability, the generation of dicentric and acentric chromosomes does not produce deleterious aneuploidy as it does on a large autosome. The placement of BS distal to the inverted FRTs makes this system especially useful to assess survival of cells in the eye after dicentric formation (Kurzhals et al. 2011). We also used this chromosome to assess breakage and healing in the germline, as described below.
Healing of broken chromosomes in somatic cells
To determine whether chromosome healing may occur in the soma, we generated dicentric chromosomes using inverted FRTs at a medial site of the left arm of chromosome 3. At different times, between 8 and 72 hr following heat shock induction of FLP expression, we examined the broken centric fragments of chromosome 3 for the presence of the vital telomere cap component HOAP (Cenci et al. 2003) as an indicator of whether a broken end had been healed (Figure 2). We used the frequency of HOAP staining on the acentric fragment (which carries the normal telomeres of 3L sister chromatids) as a measure of staining efficiency (186/260 = 0.715), and normalized the frequency of staining on the broken chromosome accordingly (Table 1). Overall, ∼19% (57/413/0.715) of broken chromosome ends exhibited HOAP staining. HOAP is normally only found at telomeres. Its presence at sites of chromosome breakage, far from its normal telomeric location, strongly suggests that healing occurs in a significant, though minor, fraction of somatic cells. Thus, the persistence of somatic cells that have experienced chromosome breakage is likely the result, in part, of healing the broken chromosome end. Curiously, we observed that in a few instances (at the 16 hr time point), the strongest HOAP signal by far was to be found on the broken chromosome end (Figure 2F), though the reason for this is not known.
Most cells did not add a new telomere to the broken end. The persistence of cells with an unhealed chromosome break is not unprecedented. Cells may also survive through other mechanisms such as adaptation, wherein a cell with an unrepaired DSB can survive for an extended period, though it may ultimately succumb to cell death (Sandell and Zakian 1993; Titen and Golic 2008). It is likely that the surviving cells with uncapped broken chromosomes utilize such a mechanism.
Hiphop increases survival of somatic cells with a broken chromosome
These experiments strongly suggest that chromosome healing can occur in somatic cells. We therefore investigated whether overexpression of telomere cap components might offer additional protection from the lethal effect of a broken chromosome. The BARTL (Bar and Telomere Loss) assay relies on Y chromosome dicentric formation (Figure 1) produced by constitutive FLP synthesis during eye development, and uses adult eye size as a metric for survival of cells with broken chromosomes (Kurzhals et al. 2011). The broken end activates the DDR, frequently resulting in apoptosis. Flies generated in this screen have eyes that are approximately half the size of wildtype eyes (Figure 3C). Although some cells do survive and differentiate, many clearly do not. An eyGal4 UASFLP combination, or eyFLP, was used to drive FLP. Telomere cap genes were driven simultaneously with Gal4, or by heat shock induction. In this assay we found that overexpression of hiphop (Gao et al. 2010), or the closely related paralog ms(3)K81 (Loppin et al. 2005; Dubruille et al. 2010; Gao et al. 2011), produced flies with larger eyes (Figure 3, D–G). However, overexpression of cav (encoding HOAP; Figure 3, H and I) or Su(var)205 [encoding HP1 (Eissenberg et al. 1990); Figure 3K] did not produce an increase in eye size. In fact, overexpression of cav in this assay had the opposite effect, producing significantly smaller eyes. Overexpression of hiphop, K81, cav or HP1 had no effect on otherwise wildtype eyes (not shown). These results suggest that the terminin component Hiphop has a unique role in protecting cells from the deleterious effect of a broken chromosome.
To further test whether hiphop overexpression ameliorates the effect of a broken chromosome, we examined cells by looking at metaphase chromosome spreads after dicentric chromosome induction, using heat shock to induce FLP expression. Previously we showed that, in wildtype flies, cells with broken chromosomes are very frequent shortly after FLP induction, but are eliminated over time, so that, after 3 days, few such cells remain (Titen and Golic 2008). When comparing wildtype with hiphop overexpressing cells, at the initial assessment 24 hr after heat shock, the majority of cells in each genotype exhibit abnormal karyotypes, characterized by dicentric chromosomes, broken centric fragments, or single or multiple copies of acentric fragments. At this early time point, hiphop overexpression had no discernable effect (Figure 4, columns 1, 2). However, when cells were examined 72 hr after heat shock, there was a significant difference between wildtype and hiphop overexpressing cells. In wildtype flies, few cells with abnormal karyotypes remain (Figure 4, column 3). In contrast, we found that hiphop overexpression rescued cells with abnormal karyotypes, allowing them to persist at high frequency, even 3 days after FLP induction (Figure 4, column 4). This provides further evidence that cells with a broken chromosome can be rescued by hiphop overexpression.
Hiphop promotes chromosome healing
The BARTL assay and karyotype examinations both show that hiphop overexpression can save cells that would otherwise die following dicentric formation and breakage. The presence of the telomere cap component HOAP on a fraction of broken chromosome ends in somatic cells is a probable indicator that such chromosomes are healed. It is tempting to speculate that hiphop overexpression rescues cells with broken chromosomes by promoting formation of new telomeres on the broken chromosome ends. However, it is also conceivable that hiphop promotes cell survival entirely apart from chromosome healing. One may argue whether or not HOAP association to broken ends represents true healing (see Discussion), but a definitive test of healing is possible. When a chromosome is transmitted through the germline, and is then recovered in normal viable offspring, by definition, that chromosome must carry a functional telomere. Thus, the transmission and recovery of broken chromosomes through the germline provides a solid and quantifiable test of chromosome healing. Accordingly, we asked whether hiphop overexpression in the male germline could increase the transmission of healed chromosomes.
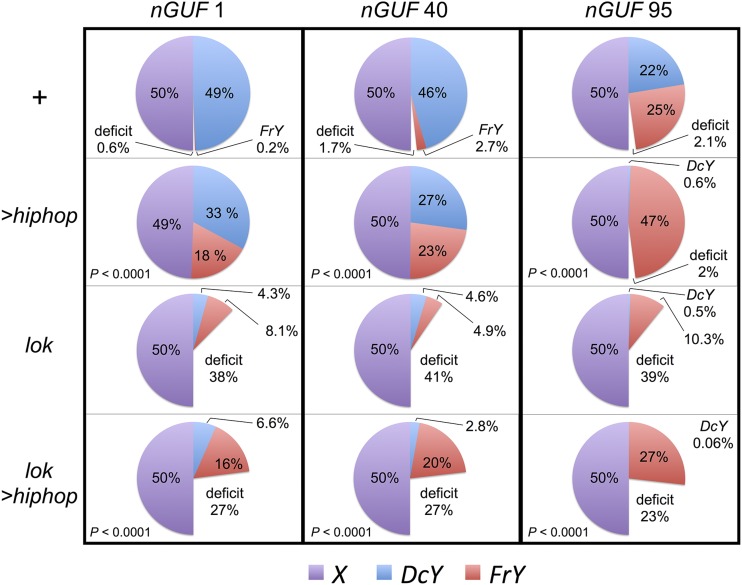
To assay germline breakage and healing we use the same DcY(H1) (Dicentric-forming Y) chromosome that was used in the BARTL assay. FLP-mediated recombination generates a dicentric chromosome and an acentric chromosome, with BS on the acentric fragment, which is subsequently lost. Breakage and healing of the dicentric portion generates y+ B+ sons (denoted as FrY, for Fragment Y) in testcrosses to y w females. To drive expression of FLP in the germline we used chromosomes carrying nosGal4 and UASFLP (indicated as nGUF). The transmission of the healed FrY chromosomes, as a fraction of all sex chromosomes, varied between 0.2 and 25%, depending on whether a weak or strong nGUF combination was used (Figure 5, top row; complete data in Table S1).
Figure 5.
Effect of Hiphop on transmission of healed chromosomes. The transmission of X, DcY, and FrY chromosomes was measured in testcrosses. FLP expression was driven by three combinations of nosGal4 UASFLP (nGUF 1, 40, or 95) on chromosome 3 in males that were: y w/DcY(H1); nGUF/+, or y w/DcY(H1); UAS-hiphop/+; nGUF/+, or y w/DcY(H1); lok/lok; nGUF/+, or y w/DcY(H1); lok UAS-hiphop/lok; nGUF/+. It was assumed that sons should represent half of all progeny. In crosses that produced <50% males, the size of that deficit is indicated. Results were corrected for the slightly reduced viability of the Y-bearing progeny. Between 41 and 115 fertile males, with an average of ∼80 progeny each, were tested for each combination. P values are given for comparisons of UAShiphop flies with corresponding genotypes that lack UAShiphop (i.e., the group immediately above). P values were determined by comparing the FrY/X ratios produced by the individual males in each group using the Mann-Whitney test. Complete summary results for each experiment are presented in Table S1.
We tested whether hiphop overexpression would affect the recovery of healed chromosomes by using the same combinations with the addition of UAShiphop. The combination of nosGal4 with UAShiphop drives robust expression of hiphop in the germline (Figure 6). When this is added to the normal level of Hiphop already present in these hiphop+/+ animals, the result is significant overexpression of hiphop. With UAShiphop the transmission of FrY chromosomes increased significantly, ranging from 18% (representing an increase of ∼100-fold) to 47% (representing ∼99% of all Y chromosomes transmitted by these males; Figure 5, second row). The increased transmission of healed chromosomes is consistent with the promotion of new telomere formation by Hiphop.
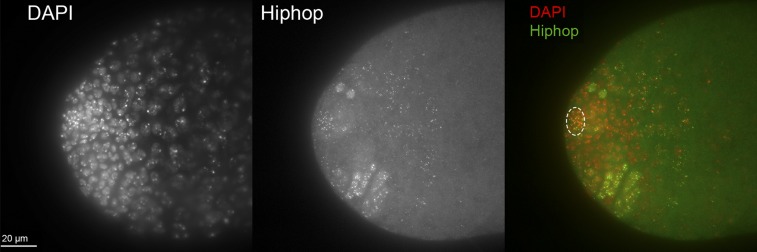
Figure 6.
Expression of UAShiphop in the male germline. Testes of y w/Y; UAShiphop/+; nosGal4 UASFLP/+ males were dissected and stained with DAPI and an antibody to mCherry (to which Hiphop is fused in this construct). The HRH008-A1 insertion and the nGUF95 combination were used (the same combination as in Figure 5, third column, second and fourth rows). Seven testes were examined—all showed strong expression in germline stem cells at the hub (circled in panel at right), and variable expression at later stages. Expression was in the form of bright puncta, as expected for telomeric staining, and a diffuse and dimmer cytoplasmic staining.
In this experiment UASFLP and UAShiphop expression were driven in mitotically dividing cells, at a time when Hiphop normally functions. Although ms(3)K81 does substitute for hiphop late in spermiogenesis (Dubruille et al. 2010; Gao et al. 2011), that is at a much later stage than the nos driven expression used here.
A concern with the proposal that Hiphop promotes telomere cap formation derives from our previous finding that lok (encoding Chk2) mutant males also transmit healed FrY chromosomes at a higher rate (Titen et al. 2014). In principle, Chk2 might act to block chromosome healing, and in lok mutants this block would be relieved to allow an increased rate of healing. But with lok this is more likely an indirect effect of allowing cells with broken chromosomes to continue to survive and divide, thus providing an extended time to heal, rather than a direct regulation of chromosome healing. In lok mutant males, premeiotic germline cells with unhealed chromosomes can continue to divide for at least 9–10 days. When these cells reach meiosis, the unhealed sister chromatids undergo end-to-end fusion and generate Meiosis II dicentric bridges. These bridges lead to the elimination of spermatid nuclei that carry them, resulting in a large deficit of sons when the Y chromosome is involved. Thus, if overexpression of hiphop increased transmission of healed chromosomes simply by permitting extended survival of cells with broken Y chromosomes, rather than by healing broken ends, we would expect to see a deficit of sons. This was not observed, and instead we saw a male:female ratio that was essentially equivalent to the control (Figure 5, second row). In confirmation of our previous findings, when we drove FLP expression in lok mutant males, we saw an increase in transmission of FrY chromosomes relative to intact DcY chromosomes, and a large deficit of sons (Figure 5, third row). Moreover, when hiphop overexpression was driven in lok males, the deficit of sons was partially alleviated, with a concomitant increase in the recovery of FrY-bearing sons (Figure 5, fourth row). Very similar results were obtained using all three nosGal4 UASFLP chromosomes. In all cases, UAShiphop produced a strong increase in FrY recovery in lok+ males, and UAShiphop rescued Y-bearing sperm in lok mutant males, producing FrY offspring from sperm that would otherwise have been eliminated. These results argue strongly that Hiphop does not merely promote survival of cells that carry broken chromosomes, but that such cells now survive because Hiphop promotes healing the broken chromosome ends.
We also tested the effect of hiphop null mutations. Since hiphop is a vital gene it was not possible to test homozygous mutants, but we did test two mutant alleles for their effect in heterozygotes. The rate of fragment transmission was measured as the fragment ratio [FR = FrY/(FrY + DcY)]. FrY chromosomes were recovered more frequently in the controls of these crosses than in the previous crosses, likely indicating an effect of genetic background. Nonetheless, in every case, the hiphop+/− heterozygotes showed significantly reduced recovery of FrY relative to their hiphop+/+ siblings with matched genetic background (apart from chromosome 3 where hiphop is located; Table 2), supporting the hypothesis that Hiphop is a critical player in the process of de novo telomere formation.
Table 2. The recovery of healed chromosomes from males carrying a hiphop mutant allele.
| Genotype | Fathers | Progeny | |||||
|---|---|---|---|---|---|---|---|
| Fertile | Sterile | DcY | FrY | X | FR | P value | |
| nGUF40/TM6 | 20 | 31 | 523 | 99 | 686 | 0.16 | |
| nGUF40/hiphopL14 | 18 | 12 | 705 | 16 | 796 | 0.02 | <0.0001 |
| nGUF40/ hiphopL32 | 38 | 22 | 1487 | 21 | 1583 | 0.01 | <0.0001 |
| nGUF95/TM6 | 11 | 20 | 30 | 283 | 386 | 0.90 | |
| nGUF95/ hiphopL14 | 7 | 20 | 174 | 95 | 341 | 0.35 | <0.0001 |
| nGUF95/ hiphopL32 | 15 | 16 | 113 | 369 | 505 | 0.77 | <0.0001 |
y w/DcY males carrying nosGal4 UASFLP (40 or 95) and homozygous for hiphop+ (one copy on the nGUF chromosome 3 and one on TM6), or heterozygous for hiphop+ and a mutant allele (either hiphopL14 or hiphopL32) were testcrossed to y w females. Progeny were scored as DcY (BS y+ sons), FrY (B+ y+ sons) or X (y w daughters). Fragment ratio is calculated as FrY/total Y progeny. Heterozygous mutants were compared with their respective +/+ controls using a 2 × 2 contingency test comparing DcY and FrY progeny from each, with P values shown.
Discussion
The ability of organisms with linear chromosomes to heal broken chromosome ends appears to be universal, occurring in organisms that use telomerase, and in those that do not (Haber and Thorburn 1984; Mason et al. 1984; Matsumoto et al. 1987; Pologe and Ravetch 1988; Levis 1989; Flint et al. 1994; Melek and Shippen 1996; Sprung et al. 1999; Fortin et al. 2009). Healing in organisms with telomerase most often involves telomerase-mediated addition of telomere repeats to broken ends, and utilizes microhomologies to the normal telomeric repeat (Greider and Blackburn 1985; Mangahas et al. 2001; Pennaneach et al. 2006; Gao et al. 2008; Murnane 2012). It is thought that the newly added repeats are then capable of recruiting the full array of shelterin components and associated proteins. In organisms that do not use telomerase, such as Drosophila, among others (Mason et al. 2011), the mechanism of healing is unknown.
In most cases, it seems that healing is a repair of last resort, used only after normal repair mechanisms, which conserve the genome, fail. However, there are also well-known examples of developmentally programmed chromosome breakage and healing in particular cells or particular compartments of the cell (Baroin et al. 1987; Forney and Blackburn 1988; Spangler et al. 1988; Müller et al. 1991; Magnenat et al. 1999). In some plants, chromosome healing is also differentially regulated between tissues (McClintock 1939). Thus, it is not a foregone conclusion that healing may occur in all tissues of an organism once it has been demonstrated in one.
In previous work, we showed that somatic cells can occasionally survive and differentiate as adult tissue despite the occurrence of a broken chromosome (Golic 1994; Kurzhals et al. 2011). Such ends cannot be repaired by normal mechanisms, and, most often, but not always, lead cells into apoptosis (Ahmad and Golic 1999; Titen and Golic 2008). How some cells survive DNA damage that is normally lethal has been an open question. By staining for the unique telomere component HOAP, we find that ∼20% of these cells show evidence of chromosome healing. The addition of a new telomere to the broken end would allow these cells to repress the DDR and escape apoptosis. It has been known for some time that mammalian somatic cells may heal broken ends by de novo telomere addition (Murnane and Yu 1993). Our results indicate that this is also true for Drosophila.
It might be argued that HOAP staining is not an absolute indication of a functional telomere. For example, moi, tefu, and tea mutants have dysfunctional telomeres, yet still show localization of HOAP at chromosome ends (Bi et al. 2004; Raffa et al. 2009; Zhang et al. 2016). However, this appears to result from an epistatic relationship in which moi, tefu, and tea lie downstream of HOAP (discussed further below). Since all telomere cap components were wildtype in the experiment of Figure 2, HOAP association likely reflects an actual healing event. Furthermore, HOAP is normally found exclusively at telomeres, and in our experiments HOAP appeared at sites that are normally found in the middle of a chromosome arm. The occurrence of HOAP at these newly broken ends must represent, at a minimum, an attempt by the cell to heal that end by cap addition.
In spite of this strong evidence for chromosome healing in somatic cells, only a minor fraction of the somatic cells with a broken chromosome survive by chromosome healing; the majority show no evidence of healing. Most of these cells must survive by other means. In the soma, the Chk2 and P53-mediated apoptotic response to DNA damage is largely responsible for eliminating cells with damaged genomes. Any mechanisms that interfere with this apoptotic response, such as the P53 negative regulator corp (Chakraborty et al. 2015), could contribute to continued survival of the remaining cells.
In somatic cells, we tested overexpression of three telomere cap components: HOAP, HP1, and Hiphop/K81. Of these, only Hiphop and its paralog K81 [which is capable of substituting for Hiphop in somatic cells (Gao et al. 2011)] increased the survival of cells with a broken chromosome. Moreover, in germline cells, hiphop overexpression increased the transmission of broken chromosomes by healing chromosomes that would otherwise have been eliminated. This result indicates that Hiphop is capable of promoting chromosome healing, and this likely represents the mechanism of its action in somatic cells as well. We propose that this reflects a unique role for Hiphop at broken ends: Hiphop “seeds” the formation of a new cap structure.
It was surprising to find that overexpression of cav (Raffa et al. 2010) in somatic cells had a negative effect on survival of cells with a broken chromosome, leading to smaller eyes in the BARTL assay. This may reflect a real effect of excess HOAP, and possible interference with the stoichiometry needed for de novo telomere capping. Alternatively, since HOAP physically interacts with Hiphop (Gao et al. 2010), excess HOAP may prevent Hiphop from reaching chromosome ends and seeding telomere formation. Finally, it is possible that this reflects a deleterious effect that is unique to this HOAP-GFP fusion construct.
It is interesting to note that, in a few nuclei, the HOAP signal on the broken chromosome end was far stronger than HOAP signals at normal chromosome ends. We speculate this may reflect an early stage in the healing process, with either a quantity or a configuration of HOAP that differs from that found at normal “mature” telomeres. Either possibility suggests that chromosome healing occurs in stages, with the cap progressing from an immature to mature structure. Zhang et al. (2016) examined telomeric localization of the known terminin components in mutants that lacked one of these components, and elucidated a hierarchy of telomere associations. Hiphop and HOAP lie at the top of this hierarchy, followed by Tea, which is followed by Ver and Moi. Altogether, these results suggest a model for healing in which Hiphop is the first terminin component to recognize a nontelomeric chromosome end, perhaps in cooperation with components of the DDR, such as ATM or the MRN complex, which are also required for functioning telomeres and for the normal localization of Hiphop to telomeres (Ciapponi et al. 2004; Ciapponi 2006; Gao et al. 2009, 2010). Hiphop then recruits HOAP and HP1, the Hiphop-HOAP-HP1 complex recruits Tea, and this assembly then recruits Ver and Moi.
Such a role for Hiphop fits perfectly with what is known about the Hiphop and K81 proteins. K81 is required during spermiogenesis when histones that package chromosomal DNA are replaced with protamines (Jayaramaiah-Raja and Renkawitz-Pohl 2005). The telomeric cap is also remodeled at this time, with K81 replacing Hiphop and HOAP at chromosome ends (Dubruille et al. 2010; Gao et al. 2011). This replacement is vital, because sperm without K81 fail to construct functional telomeres in the male pronucleus after fertilization, leading to chromosome fusions and zygotic lethality. Gao et al. (2011) suggested that a role for K81 might be to recruit other cap components to chromosome ends in the male pronucleus. Our findings, that hiphop or ms(3)K81 overexpression promotes survival of cells with broken chromosomes, and, more critically, that hiphop overexpression promotes healing of broken chromosome ends in the male germline, provide strong support for the model that one role of Hiphop (and K81) is to seed telomere formation. The hypothesis that Hiphop plays a critical role in recruiting other telomere capping proteins is further supported by recent results showing that overexpression of Hiphop in the male germline is accompanied by increased localization of HOAP and HP1 to telomeres (Dubruille and Loppin 2015).
One striking aspect of the germline results is the revelation that the mechanism which preserves a functional germline with undamaged chromosomes is extremely robust. This is apparent when considering the results with lok; nGUF1 males. Sons with an intact DcY chromosome accounted for only ∼4% of all their progeny, indicating that dicentric chromosome formation must occur in at least ∼90% of germline cells with this least effective driver combination. Nevertheless, nearly all of the sons of nGUF1 wildtype males carry the intact DcY chromosome. When using the lok; nGUF95 combination, sons with an intact DcY were nearly nonexistent, indicating that dicentric formation must occur in nearly 100% of germ cells, but the intact chromosome still accounted for almost half of the sons of nGUF95 wildtype males. The fact that these males are reasonably fertile (∼80%, Table S1) points to the existence of an extremely effective mechanism to preserve the germline, even in the presence of pervasive DNA damage. Repair of broken chromosomes by de novo telomere addition must be one part of this mechanism, but is clearly not preferred. The favored mechanism must involve removing the cells with a broken chromosome and replacing them with undamaged cells. Chk2 and P53 are involved in the former and latter aspects, respectively (Titen et al. 2014; Wylie et al. 2014; Ma et al. 2016).
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.300317/-/DC1.
Acknowledgments
We thank Mary Golic, Ho-Chen Lin, and Katelyn Froehlich for technical assistance. Stocks obtained from the Bloomington Drosophila Stock Center [National Institutes of Health (NIH) P40OD018537] were used in this study. This work was supported by the National Institute of General Medical Sciences of the NIH under award number RO1GM065604 (K.G.G.), by Southeast Missouri State University (R.L.K.), by the Epigenomics Flagship Project EpiGen, the Italian Ministry of Education and Research, National Research Council (L.F. and S.P.), and by the Intramural Program of the National Cancer Institute (Y.S.R.).
Footnotes
Communicating editor: B. Calvi
Literature Cited
- Ahmad K., Golic K. G., 1998. The transmission of fragmented chromosomes in Drosophila melanogaster. Genetics 148: 775–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad K., Golic K. G., 1999. Telomere loss in somatic cells of Drosophila causes cell cycle arrest and apoptosis. Genetics 151: 1041–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroin A., Prat A., Caron F., 1987. Telomeric site position heterogeneity in macronuclear DNA of Paramecium primaurelia. Nucleic Acids Res. 15: 1717–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaucher M., Zheng X.-F., Amariei F., Rong Y. S., 2012. Multiple pathways suppress telomere addition to DNA breaks in the Drosophila germline. Genetics 191: 407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beumer K. J., 1997. Which Way Did They Go? Segregation After Mitotic Recombination in Drosophila melanogaster. Doctoral Dissertation, University of Utah, Salt Lake City, UT. [Google Scholar]
- Bi X., Wei S. C., Rong Y. S., 2004. Telomere protection without a telomerase; the role of ATM and Mre11 in Drosophila telomere maintenance. Curr. Biol. 14: 1348–1353. [DOI] [PubMed] [Google Scholar]
- Biessmann H., Carter S. B., Mason J. M., 1990a Chromosome ends in Drosophila without telomeric DNA sequences. Proc. Natl. Acad. Sci. USA 87: 1758–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biessmann H., Mason J. M., Ferry K., d’Hulst M., Valgeirsdottir K., et al. , 1990b Addition of telomere-associated HeT DNA sequences “heals” broken chromosome ends in Drosophila. Cell 61: 663–673. [DOI] [PubMed] [Google Scholar]
- Biessmann H., Valgeirsdottir K., Lofsky A., Chin C., Ginther B., et al. , 1992. HeT-A, a transposable element specifically involved in “healing” broken chromosome ends in Drosophila melanogaster. Mol. Cell. Biol. 12: 3910–3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canudas S., Houghtaling B. R., Bhanot M., Sasa G., Savage S. A., et al. , 2011. A role for heterochromatin protein 1γ at human telomeres. Genes Dev. 25: 1807–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenci G., Siriaco G., Raffa G. D., Kellum R., Gatti M., 2003. The Drosophila HOAP protein is required for telomere capping. Nat. Cell Biol. 5: 82–84. [DOI] [PubMed] [Google Scholar]
- Chakraborty R., Li Y., Zhou L., Golic K. G., 2015. Corp regulates P53 in Drosophila melanogaster via a negative feedback loop. PLoS Genet. 11: e1005400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciapponi L., 2006. The Drosophila Nbs protein functions in multiple pathways for the maintenance of genome stability. Genetics 173: 1447–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciapponi L., Cenci G., Ducau J., Flores C., Johnson-Schlitz D., et al. , 2004. The Drosophila Mre11/Rad50 complex is required to prevent both telomeric fusion and chromosome breakage. Curr. Biol. 14: 1360–1366. [DOI] [PubMed] [Google Scholar]
- Cipressa F., Cenci G., 2013. DNA damage response, checkpoint activation and dysfunctional telomeres: face to face between mammalian cells and Drosophila. Tsitologiia 55: 211–217. [PubMed] [Google Scholar]
- de Lange T., 2005. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 19: 2100–2110. [DOI] [PubMed] [Google Scholar]
- Dronamraju R., Mason J. M., 2009. Recognition of double strand breaks by a mutator protein (MU2) in Drosophila melanogaster. PLoS Genet. 5: e1000473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubruille R., Loppin B., 2015. Protection of Drosophila chromosome ends through minimal telomere capping. J. Cell Sci. 128: 1969–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubruille R., Orsi G. A., Delabaere L., Cortier E., Couble P., et al. , 2010. Specialization of a Drosophila capping protein essential for the protection of sperm telomeres. Curr. Biol. 20: 2090–2099. [DOI] [PubMed] [Google Scholar]
- Eissenberg J. C., Hartnett T., 1993. A heat shock-activated cDNA rescues the recessive lethality of mutations in the heterochromatin-associated protein HP1 of Drosophila melanogaster. Mol. Gen. Genet. 240: 333–338. [DOI] [PubMed] [Google Scholar]
- Eissenberg J. C., James T. C., Foster-Hartnett D. M., Hartnett T., Ngan V., et al. , 1990. Mutation in a heterochromatin-specific chromosomal protein is associated with suppression of position-effect variegation in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 87: 9923–9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falco S. C., Li Y., Broach J. R., Botstein D., 1982. Genetic properties of chromosomally integrated 2 mu plasmid DNA in yeast. Cell 29: 573–584. [DOI] [PubMed] [Google Scholar]
- Fanti, L., and S. Pimpinelli, 2004 Immunostaining of squash preparations of chromosomes of larval brains, pp. 353–361 in Methods in Molecular Biology: Drosophila Cytogenetics Protocols, Vol. 247, edited by D. S. Henderson. Humana Press, New York, NY. [DOI] [PubMed] [Google Scholar]
- Fanti L., Giovinazzo G., Berloco M., Pimpinelli S., 1998. The heterochromatin protein 1 prevents telomere fusions in Drosophila. Mol. Cell 2: 527–538. [DOI] [PubMed] [Google Scholar]
- Flint J., Craddock C. F., Villegas A., Bentley D. P., Williams H. J., et al. , 1994. Healing of broken human chromosomes by the addition of telomeric repeats. Am. J. Hum. Genet. 55: 505–512. [PMC free article] [PubMed] [Google Scholar]
- Forney J. D., Blackburn E. H., 1988. Developmentally controlled telomere addition in wild-type and mutant paramecia. Mol. Cell. Biol. 8: 251–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin F., Beaulieu Bergeron M., Fetni R., Lemieux N., 2009. Frequency of chromosome healing and interstitial telomeres in 40 cases of constitutional abnormalities. Cytogenet. Genome Res. 125: 176–185. [DOI] [PubMed] [Google Scholar]
- Fouladi B., Sabatier L., Miller D., Pottier G., Murnane J. P., 2000. The relationship between spontaneous telomere loss and chromosome instability in a human tumor cell line. Neoplasia 2: 540–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G., Bi X., Chen J., Srikanta D., Rong Y. S., 2009. Mre11-Rad50-Nbs complex is required to cap telomeres during Drosophila embryogenesis. Proc. Natl. Acad. Sci. USA 106: 10728–10733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G., Walser J.-C., Beaucher M. L., Morciano P., Wesolowska N., et al. , 2010. HipHop interacts with HOAP and HP1 to protect Drosophila telomeres in a sequence-independent manner. EMBO J. 29: 819–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G., Cheng Y., Wesolowska N., Rong Y. S., 2011. Paternal imprint essential for the inheritance of telomere identity in Drosophila. Proc. Natl. Acad. Sci. USA 108: 4932–4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q., Reynolds G. E., Wilcox A., Miller D., Cheung P., et al. , 2008. Telomerase-dependent and -independent chromosome healing in mouse embryonic stem cells. DNA Repair (Amst.) 7: 1233–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti M., Pimpinelli S., 1983. Cytological and genetic analysis of the Y chromosome of Drosophila melanogaster. Chromosoma 88: 349–373. [Google Scholar]
- Golic K. G., 1994. Local transposition of P elements in Drosophila melanogaster and recombination between duplicated elements using a site-specific recombinase. Genetics 137: 551–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golic M. M., Rong Y. S., Petersen R. B., Lindquist S. L., Golic K. G., 1997. FLP-mediated DNA mobilization to specific target sites in Drosophila chromosomes. Nucleic Acids Res. 25: 3665–3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorovsky M. A., 1980. Genome organization and reorganization in Tetrahymena. Annu. Rev. Genet. 14: 203–239. [DOI] [PubMed] [Google Scholar]
- Greider C. W., Blackburn E. H., 1985. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43: 405–413. [DOI] [PubMed] [Google Scholar]
- Haber J. E., Thorburn P. C., 1984. Healing of broken linear dicentric chromosomes in yeast. Genetics 106: 207–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaramaiah-Raja S., Renkawitz-Pohl R., 2005. Replacement by Drosophila melanogaster protamines and Mst77F of histones during chromatin condensation in late spermatids and role of sesame in the removal of these proteins from the male pronucleus. Mol. Cell. Biol. 25: 6165–6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurzhals R. L., Titen S. W. A., Xie H. B., Golic K. G., 2011. Chk2 and p53 are Haploinsufficient with dependent and independent functions to eliminate cells after telomere loss. PLoS Genet. 7: e1002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamarche B. J., Orazio N. I., Weitzman M. D., 2010. The MRN complex in double-strand break repair and telomere maintenance. FEBS Lett. 584: 3682–3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis R. W., 1989. Viable deletions of a telomere from a Drosophila chromosome. Cell 58: 791–801. [DOI] [PubMed] [Google Scholar]
- Levis R. W., Ganesan R., Houtchens K., Tolar L. A., Sheen F.-M., 1993. Transposons in place of telomeric repeats at a Drosophila telomere. Cell 75: 1083–1093. [DOI] [PubMed] [Google Scholar]
- Loppin B., Lepetit D., Dorus S., Couble P., Karr T. L., 2005. Origin and neofunctionalization of a Drosophila paternal effect gene essential for zygote viability. Curr. Biol. 15: 87–93. [DOI] [PubMed] [Google Scholar]
- Lukhtanov V. A., 2015. The blue butterfly Polyommatus (Plebicula) atlanticus (Lepidoptera, Lycaenidae) holds the record of the highest number of chromosomes in the non-polyploid eukaryotic organisms. Comp. Cytogenet. 9: 683–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Han Y., Song X., Do T., Yang Z., et al. , 2016. DNA damage-induced Lok/CHK2 activation compromises germline stem cell self-renewal and lineage differentiation. Development 143: 4312–4323. [DOI] [PubMed] [Google Scholar]
- Magnenat L., Tobler H., Müller F., 1999. Developmentally regulated telomerase activity is correlated with chromosomal healing during chromatin diminution in Ascaris suum. Mol. Cell. Biol. 19: 3457–3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangahas J. L., Alexander M. K., Sandell L. L., Zakian V. A., 2001. Repair of chromosome ends after telomere loss in Saccharomyces. Mol. Biol. Cell 12: 4078–4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason J. M., Strobel E., Green M. M., 1984. mu-2: mutator gene in Drosophila that potentiates the induction of terminal deficiencies. Proc. Natl. Acad. Sci. USA 81: 6090–6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason J. M., Reddy H. M., Frydrychova R. C., 2011. Telomere maintenance in organisms without telomerase in DNA Replication - Current Advances, edited by Seligmann H. InTech, Rijeka, Croati. [Google Scholar]
- Matsumoto T., Fukui K., Niwa O., Sugawara N., Szostak J. W., et al. , 1987. Identification of healed terminal DNA fragments in linear minichromosomes of Schizosaccharomyces pombe. Mol. Cell. Biol. 7: 4424–4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B., 1939. The behavior in successive nuclear divisions of a chromosome broken at meiosis. Proc. Natl. Acad. Sci. USA 25: 405–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B., 1941. The stability of broken ends of chromosomes in Zea Mays. Genetics 26: 234–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melek M., Shippen D. E., 1996. Chromosome healing: spontaneous and programmed de novo telomere formation by telomerase. BioEssays 18: 301–308. [DOI] [PubMed] [Google Scholar]
- Müller F., Wicky C., Spicher A., Tobler H., 1991. New telomere formation after developmentally regulated chromosomal breakage during the process of chromatin diminution in Ascaris lumbricoides. Cell 67: 815–822. [DOI] [PubMed] [Google Scholar]
- Murnane J. P., 2012. Telomere dysfunction and chromosome instability. Mutat. Res. Fundam. Mol. Mech. Mutagen. 730: 28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murnane J. P., Yu L. C., 1993. Acquisition of telomere repeat sequences by transfected DNA integrated at the site of a chromosome break. Mol. Cell. Biol. 13: 977–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musarò M., Ciapponi L., Fasulo B., Gatti M., Cenci G., 2008. Unprotected Drosophila melanogaster telomeres activate the spindle assembly checkpoint. Nat. Genet. 40: 362–366. [DOI] [PubMed] [Google Scholar]
- Palm W., de Lange T., 2008. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 42: 301–334. [DOI] [PubMed] [Google Scholar]
- Pandita T. K., 2002. ATM function and telomere stability. Oncogene 21: 611–618. [DOI] [PubMed] [Google Scholar]
- Pennaneach V., Putnam C. D., Kolodner R. D., 2006. Chromosome healing by de novo telomere addition in Saccharomyces cerevisiae. Mol. Microbiol. 59: 1357–1368. [DOI] [PubMed] [Google Scholar]
- Pologe L. G., Ravetch J. V., 1988. Large deletions result from breakage and healing of P. falciparum chromosomes. Cell 55: 869–874. [DOI] [PubMed] [Google Scholar]
- Raffa G. D., Siriaco G., Cugusi S., Ciapponi L., Cenci G., et al. , 2009. The Drosophila modigliani (moi) gene encodes a HOAP-interacting protein required for telomere protection. Proc. Natl. Acad. Sci. USA 106: 2271–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffa G. D., Raimondo D., Sorino C., Cugusi S., Cenci G., et al. , 2010. Verrocchio, a Drosophila OB fold-containing protein, is a component of the terminin telomere-capping complex. Genes Dev. 24: 1596–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffa G. D., Ciapponi L., Cenci G., Gatti M., 2011. Terminin: a protein complex that mediates epigenetic maintenance of Drosophila telomeres. Nucleus 2: 383–391. [DOI] [PubMed] [Google Scholar]
- Raffa G. D., Cenci G., Ciapponi L., Gatti M., 2013. Organization and evolution of Drosophila Terminin: similarities and differences between Drosophila and human telomeres. Front. Oncol. 3: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong Y. S., 2008. Telomere capping in Drosophila: dealing with chromosome ends that most resemble DNA breaks. Chromosoma 117: 235–242. [DOI] [PubMed] [Google Scholar]
- Sabourin M., Zakian V. A., 2008. ATM-like kinases and regulation of telomerase: lessons from yeast and mammals. Trends Cell Biol. 18: 337–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandell L. L., Zakian V. A., 1993. Loss of a yeast telomere: arrest, recovery, and chromosome loss. Cell 75: 729–739. [DOI] [PubMed] [Google Scholar]
- Šíchová J., Voleníková A., Dincă V., Nguyen P., Vila R., et al. , 2015. Dynamic karyotype evolution and unique sex determination systems in Leptidea wood white butterflies. BMC Evol. Biol. 15: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangler E. A., Ryan T., Blackburn E. H., 1988. Developmentally regulated telomere addition in Tetrahymena thermophila. Nucleic Acids Res. 16: 5569–5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprung C. N., Reynolds G. E., Jasin M., Murnane J. P., 1999. Chromosome healing in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA 96: 6781–6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titen S. W. A., Golic K. G., 2008. Telomere loss provokes multiple pathways to apoptosis and produces genomic instability in Drosophila melanogaster. Genetics 180: 1821–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titen S. W. A., Golic K. G., 2010. Healing of euchromatic chromosome breaks by efficient de novo telomere addition in Drosophila melanogaster. Genetics 184: 309–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titen S. W. A., Lin H.-C., Bhandari J., Golic K. G., 2014. Chk2 and p53 regulate the transmission of healed chromosomes in the Drosophila male germline. PLoS Genet. 10: e1004130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tower J., Karpen G. H., Craig N., Spradling A. C., 1993. Preferential transposition of Drosophila P elements to nearby chromosomal sites. Genetics 133: 347–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doren M., Williamson A. L., Lehmann R., 1998. Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr. Biol. 8: 243–246. [DOI] [PubMed] [Google Scholar]
- Wylie A., Lu W.-J., D’Brot A., Buszczak M., Abrams J. M., 2014. p53 activity is selectively licensed in the Drosophila stem cell compartment. eLife 3: e01530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhang L., Tang X., Bhardwaj S. R., Ji J., et al. , 2016. MTV, an ssDNA protecting complex essential for transposon-based telomere maintenance in Drosophila. PLoS Genet. 12: e1006435. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Supplemental Material, Table S1 (see File S1 for legend) contain complete progeny counts used for testing the effect of hiphop overexpression in the germline. Fly strains are available from the authors upon request.