Abstract
Intestinal bacteria can affect xenobiotic metabolism through both direct bacterial enzyme-catalyzed modification of the xenobiotics and indirect alterations of the expression of host genes. To determine how intestinal bacteria affect the expression of host xenobiotic-processing genes (XPGs), the mRNA profiles of 303 XPGs were characterized by RNA sequencing in four intestinal sections and compared with that in the liver from adult male conventional (CV) and germ-free (GF) mice. Fifty-four XPGs were not expressed in the intestine of either CV or GF mice. The GF condition altered the expression of 116 XPGs in at least one intestinal section but had no effect on 133 XPGs. Many cytochrome P450 family members such as Cyp1a, Cyp2b10, Cyp2c, and most Cyp3a members, as well as carboxylesterase (Ces) 2a were expressed lower in the intestine of GF than CV mice. In contrast, GF mice had higher intestinal expression of some phase I oxidases (alcohol dehydrogenase 1, aldehyde dehydrogenase a1l1 and 4a1, as well as flavin monooxygenase 5) and phase II conjugation enzymes (UDP-glucuronosyltransferase 1a1, and sulfotransferase 1c2, 1d1, and 2b1). Several transporters in the intestine, such as bile acid transporters (apical sodium-dependent bile acid transporter, organic solute transporter α and β), peptide transporter 1, and multidrug and toxin extrusion protein 1, exhibited higher expression in GF mice. In conclusion, lack of intestinal bacteria alters the expression of a large number of XPGs in the host intestine, some of which are section specific. Cyp3a is downregulated in both the liver and intestine of GF mice, which probably contributes to altered xenobiotic metabolism.
Introduction
Trillions of microbes inhabit the human intestine, collectively known as the intestinal microbiota, and form a complex ecological community that influences normal physiology and disease susceptibility (Lozupone et al., 2012). The intestinal bacteria are beneficial for host metabolism, aid in digestion, produce vitamins B and K, and contribute to normal immune function, thereby creating a symbiotic relationship with the host (Tremaroli and Backhed, 2012). It has been increasingly recognized that intestinal bacteria are implicated in several diseases, such as obesity, diabetes, neurologic diseases, inflammatory bowel disease, cancer, and liver diseases (Fu and Cui, 2017).
Metabolism of orally administered drugs by intestinal bacteria can alter the efficacy and toxicity of drugs (Klaassen and Cui, 2015). A wide range of metabolic reactions are performed by intestinal bacteria, such as hydrolysis, reduction, dehydroxylation, decarboxylation, dealkylation, dehalogenation, deamination, heterocyclic ring fission, aromatization, nitrosamine formation, acetylation, esterification, isomerization, and oxidation (Mikov, 1994). Intestinal bacteria-mediated metabolism of biliary-excreted metabolites is often crucial for the enterohepatic circulation of xenobiotics. Bacterial enzymes in the intestine often make drugs more lipophilic by deconjugating conjugated drug metabolites, which favor intestinal uptake and increase the half-life of drugs (Stojančević et al., 2014). According to the PharmacoMicrobiomics database, approximately 60 drugs are known to be affected by intestinal microbes (Rizkallah et al., 2012). Some well-known examples include the analgesic acetaminophen (by Clostridium difficile), cardiotonic drug digoxin (by Eggerthella lenta), antiviral drug sorivudine (by Bacteroides), hypnotic nitrazepam (by Clostridium leptum), and anthelminitic levamisole (by Bacteroides and Clostridium species) (Klaassen and Cui, 2015; Jourova et al., 2016). The wide yet less controlled use of intestinal bacteria modulators (prebiotics, probiotics, synbiotics, and antibiotics) can be problematic in polypharmacy due to bacteria-mediated alteration of the pharmacokinetics and pharmacodynamics of concomitant orally administered drugs. Humans have marked variation in microbiota inhabiting their intestine, as evidenced by metagenomic studies (Human Microbiome Project Consortium, 2012), and may help to explain interindividual variations in the pharmacokinetics of xenobiotics. Thus, understanding the functions of individual microbial populations and how the intestinal microbiota varies across age, sex, ethnicity, life styles, health or disease states, and medications of humans is emerging as a novel component of personalized medicine.
In addition to the direct bacterial enzyme–catalyzed metabolism, intestinal bacteria can also affect the biotransformation of xenobiotics via an indirect mechanism of regulating the expression of host XPGs. Modulation of intestinal bacteria has been shown to alter the hepatic and renal expression of genes involved in drug metabolism and disposition, evidenced by GF, probiotic-, or antibiotic-treated animals (Bjorkholm et al., 2009; Toda et al., 2009; Selwyn et al., 2015b, 2016; Kuno et al., 2016). Given the crucial role of intestinal tissue in xenobiotic absorption and metabolism, as well as its close proximity to the microbiota in the lumen, it is of great importance to determine the effect of intestinal bacteria on the expression of xenobiotic metabolism–related genes in the intestine.
In this study, GF mice that do not have microbes, as well as CV counterparts, were used to investigate the role of intestinal bacteria on host gene expression. RNA sequencing was applied to provide comprehensive and unbiased mRNA profiles of various XPGs, including phase I– and phase II drug–metabolizing enzymes, drug uptake, and efflux transporters, as well as related transcription factors (TFs). These results will advance our knowledge on the regulation of host drug metabolism by intestinal bacteria and shed light on the underlying mechanisms for food-drug and drug-drug interactions, as well as interindividual differences in xenobiotic metabolism.
Materials and Methods
Animals.
All mice used in the studies were male C57BL/6 mice at 2–3 months of age (n = 3) due to the known effect of age and sex on XPG expression (Cui et al., 2012a; Fu et al., 2012). All mice were housed in an Association for Assessment and Accreditation of Laboratory Animal Care International–accredited facility at the University of Kansas Medical Center (KUMC), with a 14-hour light/10-hour dark cycle, in a temperature- and humidity-controlled environment. All mice were given ad libitum access to autoclaved rodent chow (catalog #5K67; LabDiet, St. Louis, MO) and autoclaved water. The initial breeding colony of GF C57BL/6J mice was established with mice purchased from the National Gnotobiotic Rodent Resource Center (University of North Carolina at Chapel Hill). GF mice were born and raised inside sterile isolators and received sterile feed, water, and bedding. All CV mice were purchased from The Jackson Laboratory (Bar Harbor, ME). All animal experiments were approved by the Institutional Animal Care and Use Committee at KUMC.
Tissue Collection.
Animals were not fasted due to the known effect of nutritional status on XPG expression (Fu and Klaassen, 2014). All tissues were harvested between 9:00 AM and noon. Intestinal contents were flushed with ice-cold phosphate-buffered saline, and intestinal tissues were separated into various sections, namely duodenum (duo), jejunum (jej), ileum (ile), and large intestine (LI). Liver and intestinal sections were snap frozen in liquid nitrogen and stored at −80°C before further analysis.
Total RNA Isolation.
Total RNA was isolated from tissues using RNA-Bee reagent (Tel-Test Inc., Friendswood, TX) following the manufacturer's protocol. The concentration of total RNA in each sample was quantified spectrophotometrically at 260 nm using a NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA). RNA integrity was confirmed using a dual Agilent 2100 Bioanalyzer (Agilent Technologies Inc., Santa Clara, CA), and the samples with RNA integrity numbers above 7.0 were used for the following experiments.
cDNA Library Preparation and RNA-Seq.
The cDNA library preparation and sequencing of the transcriptome were performed in the KUMC Genome Sequencing Facility. The cDNA libraries from total RNA samples (n = 3/group) were prepared using an TruSeq RNA Sample Prep Kit (Illumina, San Diego, CA). Three micrograms of total RNA were used as the RNA input according to recommendations of the manufacturer protocol. The mRNAs were selected from the total RNAs by purifying the poly-A–containing molecules using poly-T primers. The RNA fragmentation, first- and second-strand cDNA syntheses, end repair, adaptor ligation, and PCR amplification were performed according to the manufacturer's protocol. The average size of the cDNAs was approximately 160 base pairs (excluding the adapters). The cDNA libraries were validated for RNA-integrity and quantity using an Agilent 2100 Bioanalyzer (Agilent Technologies Inc.) before sequencing. The cDNA libraries were clustered onto a TruSeq paired–end flow cell and sequenced (2 × 50 base pairs) using a TruSeq SBS kit (Illumina) on an Illumina HiSeq2000 sequencer with a multiplexing strategy of four samples per lane.
RNA-Seq Data Analysis.
After the sequencing platform generated the sequencing images, the pixel-level raw data collection, image analysis, and base calling were performed using the Illumina Real-Time Analysis (RTA) software on a Dell (Round Rock, TX) personal computer attached to a HiSeq2000 Sequencer (Illumina). The base call files (*.BCL) were converted to qseq files by the Illumina BCL Converter, and the qseq files were subsequently converted to Fastq files for downstream analysis. The RNA-Seq reads from the Fastq files were mapped to the mouse reference genome (UCSC mm10), and the splice junctions were identified by TopHat2. The output files in BAM (binary sequence alignment) format were analyzed by Cufflinks to estimate the transcript abundance. Differential expression (DE) analysis between CV and GF mice was determined using Cuffdiff [significant at a false discovery rate with Benjamini-Hochberg false discovery rate adjustment (FDR-BH) < 0.05]. The mRNA abundance was expressed as fragments per kilobase of exon per million reads mapped (FPKM). RNA-Seq data are uploaded to the National Center for Biotechnology Information Gene Expression Omnibus database with accession number GSE102867.
Statistical Analysis.
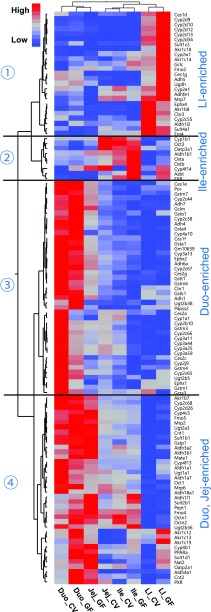
Data are presented as the mean FPKM ± S.E.M. XPGs with mean FPKM values per tissue less than 1 in each of the four sections of the intestine were defined as not expressed (NE) in the intestine. Differences between CV and GF mice that were significant by Cuffdiff (FDR-BH < 0.05) were represented with asterisks. Hepatic expression of the XPGs in CV and GF mice was reported previously (Selwyn et al., 2015b) and is included in this article to compare the effect of the GF condition in the liver to that in the various sections of the intestine. To compare the expression of XPGs in intestines of CV and GF mice, a two-way hierarchical clustering dendrogram (Ward’s minimum variance method, distance scale) was generated by JMP version 12.1.0 software (SAS Institute, Inc., Cary, NC) on the mean mRNA expression of XPGs with DE in at least one section of the intestine between CV and GF mice (FDR-BH < 0.05, Cuffdiff). The expression of each individual gene was standardized among eight experimental groups to present the DE pattern of multiple genes between CV and GF mice in various sections of the intestine. Relatively high mRNA abundance is represented in red, whereas relatively low mRNA abundance is in blue. Relative color intensities are not comparable among genes.
Results
mRNA Expression of All XPGs in Intestines of CV and GF Mice
In total, 303 XPGs with known important functions in xenobiotic metabolism and transport were analyzed. These genes belong to various categories as reported previously (Fu et al., 2016), namely 158 phase I enzymes, 78 phase II enzymes, 35 uptake transporters, 21 efflux transporters, and 11 TFs. Among these 303 XPGs, 116 XPGs had DE between CV and GF mice in at least one section of the intestine (FDR-BH < 0.05, Cuffdiff). This includes 69 phase I enzymes, 27 phase II enzymes, 17 transporters, and 3 TFs (Table 1). Table 2 lists 54 XPGs that are NE in the intestine of CV or GF mice (FPKM < 1 in all sections of the intestine). Table 3 lists 133 XPGs with no DE (NDE) between CV and GF mice in the intestine (FDR-BH ≥ 0.05, Cuffdiff).
TABLE 1.
List of 116 XPGs that have differential expression (DE) between CV and GF mice in at least one section of the intestine (FDR-BH < 0.05, Cuffdiff)
| DE XPGs # | Duodenum (Duo) | Jejunum (Jej) | Ileum (Ile) | Large intestine (LI) | |
|---|---|---|---|---|---|
| 8 Cess | Ces1d, 2a, | Ces1e, 1f, 2a, 2b | Ces2a, 2g | Ces1e, 1g, 2a, 2c, 2g | |
| 3 Ephxs | Ephx1 | Ephx1, 2 | Ephx2 | Ephx2, 4 | |
| 7 Akrs | Akr1c19 | Akr1b7, 1c12, 1c13, 1c14, 1c19 | Akr1c12, 1c13, 1c14, 1c19 | Akr1b8, 1c12, 1c13, 1c14, 1c18, 1c19 | |
| 2 Cbrs | Cbr1 | Cbr3 | |||
| 69 phase I enzymes | 30 P450s + Por | Cyp1a1, 2b10, 2c55, 2c66, 2d10, 3a11, 3a25, 3a44, 3a59, 4b1 | Cyp1a1, 1b1, 2c38, 2c44, 2c55, 2c66, 2c67, 2c68, 2d26, 2e1, 2w1, 3a13, 4a10, 4b1, 4f13, 4f14, 4v3, Por | Cyp1a1, 2b10, 2c29, 2c55, 2c65, 2c66, 2j9, 3a11, 3a25, 3a59, 4b1 | Cyp2d13, 2c55, 2c65, 2c66, 2d9, 2d10, 2d12, 2d26, 2d34, 2w1, 3a13, 4b1, 4f14, 4v3 |
| 6 Adhs | Adh1l2 | Adh1, 4, 6a, 7, fe1, | Adh1, 4, 6a | Adh5 | |
| 9 Aldhs | Aldh4a1, 18a1 | Aldh1a1, 1l1, 3a2, 3b1, 4a1 | Aldh1a1, 1a7, 1b1, 1l2 | ||
| 3 Fmos | Fmo2 | Fmo4, 5 | Fmo5 | ||
| 5 Ugts | Ugt2b38 | Ugt2a3 | Ugt1a1, 2b5, 2b36 | ||
| 5 Sults | Sult1c2 | Sult1b1, 1c2, 1d1, 2b1, 4a1 | Sult2b1 | Sult1c2 | |
| 27 phase II enzymes | 12 Gsts | Gsta1, m1, m3, m4 | Gsta1, a4, k1, m6, m7, o1, p1, t1 | Gstk1, m1, m3, m4, m6, t1 | Gsta3, o1 |
| 1 Nat | Nat2 | ||||
| 4 enzymes for cofactors | Ugdh, Papss2, Gclm | Gclc | Ugdh, Gclc | ||
| 17 transporters | 11 Uptake | Cnt2 | Asbt, Pept1, Oct1, Oct3, Octn2, Cnt1, Cnt2, Ent4, Oatp2a1, Oatp3a1 | Asbt, Cnt1, Cnt2 | Asbt, Pept1, Octn1, Cnt2, Oatp2a1 |
| 6 Efflux | Mate1 | Mrp2, 7, Mate1 | Mrp6 | Mrp6, Ostα, Ostβ | |
| 3 TFs | FXR, PPARα, PXR, | FXR |
TABLE 2.
List of 54 XPGs that are not expressed (NE) in the intestine of CV or GF mice (FPKM <1 in all sections of the intestine)
| NE XPGs # | XPGs | |
|---|---|---|
| 4 Cess | Ces1a, 2f, 4a, 5a | |
| 1 Ephx | Ephx3 | |
| 21 phase I enzymes | 1 Akr | Akr1c21 |
| 2 Nqos | Nqo1, Nqo2 | |
| 11 P450s | Cyp2b9, 2b19, 2b23, 2c39, 2g1, 2j5, 2j8, 2j11, 2t4, 4f39, 4x1 | |
| 1 Aox | Aox3l1 | |
| 1 Fmo | Fmo3 | |
| 4 Ugts | Ugt1a2, 1a10, 2a1, 2a2 | |
| 10 Sults | Sult1c1, 1e1, 2a1, 2a2, 2a3, 2a4, 2a5, 2a7, 3a1, 6b1 | |
| 18 phase II enzymes | 2 Gsts | Gsto2, t4 |
| 2 Nats | Nat1, 3 | |
| 15 transporters | 13 Uptake | Ntcp, Oct2, Oat1, Oat3, Urat1, Oatp1a5, 1a6, 1c1, 4c1, 5a1, 6b1, 6c1, 6d1 |
| 2 Efflux | Mrp9, Mate2 |
TABLE 3.
List of 133 XPGs with no differential expression (NDE) in the intestine between CV and GF mice (FDR-BH ≥ 0.05, Cuffdiff)
| NDE XPGs # | XPGs | |
|---|---|---|
| 6 Cess | Ces1b, 1c, 2e, 2h, 3a, 3b | |
| 3 Pons | Pon1, 2, 3 | |
| 8 Akrs | Akr1a4, 1b3, 1b10, 1c6, 1c20, 1d1, 1e1, 7a5 | |
| 2 Cbrs | Cbr2, 4 | |
| 68 phase I enzymes | 35 P450s | Cyp1a2, 2a4, 2a5, 2a12, 2a22, 2b13, 2c37, 2c40, 2c50, 2c54, 2c69, 2c70, 2d11, 2d22, 2d40, 2f2, 2j6, 2j13, 2r1, 2s1, 2u1, 3a16, 3a41a, 3a57, 4a12a, 4a12b, 4a14, 4a31, 4a32, 4f15, 4f16, 4f17, 4f18, 4f37, 4f40 |
| 11 Aldhs | Aldh1a2, 1a3, 2, 3a1, 3b2, 5a1, 6a1, 7a1, 8a1, 9a1, 16a1 | |
| 2 Aoxs | Aox1, 3 | |
| 1 Fmo | Fmo1 | |
| 9 Ugts | Ugt1a5, 1a6a, 1a6b, 1a7c, 1a9, 2b1, 2b34, 2b35, 2b37 | |
| 2 Sults | Sult1a1, 5a1 | |
| 11 Gsts | Gsta2, cd, m2, m5, p2, t2, t3, z1, Mgst1, 2, 3 | |
| 3 methyl transferase | Comt, As3mt, Tpmt | |
| 33 phase II enzymes | 3 Amino acid-conjugation enzymes | Bal, Bat, Glyat |
| 5 enzymes for cosubstrates | Ugp2, Papss1, Mat1a, 2a, 2b | |
| 24 transporters | 12 Uptake | Pept2, Octn3, Oat2, Cnt3, Ent1, 2, 3, Oatp1a1, 1a4, 1b2, 2b1, 4a1 |
| 12 Efflux | Abca1, Mdr1a, 1b, Mdr2, Bsep, Mrp1, 3, 4, 5, Bcrp, Abcg5, g8 | |
| 8 TFs | AhR, CAR, HNF1α, 4α, RXRα, β, γ, Nrf2 |
Bcrp, breast cancer resistance protein; Bsep, bile salt export pump.
To determine the tissue-specific expression patterns of the 116 differentially expressed XPGs, a two-way hierarchical clustering dendrogram was generated using their standardized mean FPKM values (Fig. 1). These XPGs partitioned into four distinctive expression patterns, namely LI enriched (cluster 1), ile enriched (cluster 2), duo enriched (cluster 3), as well as duo enriched and jej enriched (cluster 4). In general, XPGs within the same intestinal section clustered together regardless of phenotype, indicating that sections of the intestine were a more predominant regulatory factor in XPG expression than phenotype. Among all four sections of the intestine, duo was the section where most XPGs were most highly expressed in both CV and GF mice. The effect of lack of intestinal bacteria on XPG expression was specific to intestinal sections. Certain XPGs within the same family also exhibited similar expression alteration patterns by lack of intestinal bacteria. For example, multiple Cyp2d subfamily members (Cyp2d9, 2d10, 2d12, 2d13, and 2d34) had higher expression in LI of GF mice (cluster 1); multiple Cyp3a subfamily members (Cyp3a11, 3a25, 3a44, and 3a59) had lower expression in the duo of GF mice (cluster 3); and multiple Akr1c subfamily members (Akr1c12, 1c13, and 1c19) were expressed higher in the duo and LI of GF mice (cluster 4). The FPKMs of individual XPGs of each gene family are further described below.
Fig. 1.
Two-way hierarchical clustering of XPGs with DE between CV and GF. Various sections of the intestine (duo, jej, ile, as well as LI) from C57BL/6J CV and GF male mice 2–3 months of age were used for RNA-Seq quantification. The heatmap dendrogram describes XPG expression profiles between CV and GF mice in various intestine sections (FDR-BH < 0.05 by Cuffdiff analysis, in at least one section of the intestine). Average FPKM values of three mice per tissue are presented by colored squares: red, relatively high expression; blue, relatively low expression.
Intestinal Expression of Phase I Enzymes in GF Mice
Phase I Enzymes Involved in Hydrolysis Reactions.
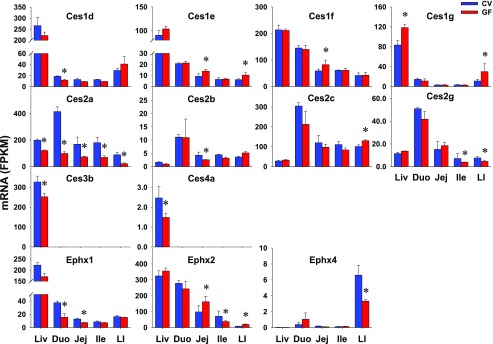
There are three major families of xenobiotic hydrolytic enzymes, including Cess for esters and amides; paraoxonases (Pons) for organophosphates, aromatic carboxylic acid esters, cyclic carbonates, and lactones; as well as epoxide hydrolases (Ephxs) for electrophilic epoxides. In total, 18 Cess, 3 Pons, and 4 Ephxs were investigated in this study. Ces1a, 2f, 4a, and 5a and Ephx3 were NE in intestines of CV or GF mice (Table 2). Ces1b, 1c, 2e, 2h, 3a, and 3b and all three Pons (Pon1–3) did not show differential intestinal expression between CV and GF mice (Table 3). Eight Cess and three Ephxs had DE between CV and GF mice in at least one section of the intestine (Table 1). As shown in Fig. 2, four hydrolases showed higher intestinal expression in GF mice, such as Ces1e in jej (51%) and LI (67%), Ces1f in jej (40%), as well as Ces1g (168%) and Ces2c (29%) in LI. In contrast, six hydrolases, such as Ces1d in duo (36%), Ephx1 in duo (58%) and jej (42%), Ces2a in all four sections of the intestine (duo 76%; jej 56%; ile 61%; LI 74%), Ces2b in jej (39%), Ces2g in ile (45%) and LI (40%), and Ephx4 in LI (50%), were lower in GF mice. Ephx2 had higher expression in jej (64%) and LI (118%) but lower expression in ile (46%) of GF mice. Similar to the liver data, Ces1g had higher expression in LI of GF mice. Probably of greatest significance is that Ces2a had lower expression in intestine as well as liver of GF mice. In contrast to the similar expression of some Cess and Ephxs in livers of CV and GF mice, some genes had higher (Ces1e, 1f, 2c) or lower (Ces1d, 2b, 2g, Ephx1, 4) expression in intestines of GF mice. Some genes (Ces3b and 4a) with DE in liver remained unchanged in intestines of GF mice.
Fig. 2.
The intestinal expression of Cess and Ephxs in GF mice. Data are represented as the mean FPKM ± S.E.M. of three individual animals. Differences between CV and GF mice that were significant by Cuffdiff (FDR-BH < 0.05) were represented with asterisks.
Phase I Enzymes Involved in Reduction Reactions.
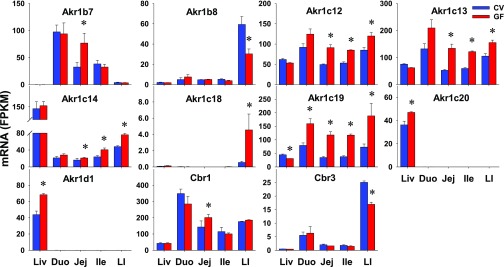
Three major families of xenobiotic reductases were investigated, namely 16 aldo-keto reductases (Akrs) from six subfamilies that metabolize aldehydes and ketones, 4 carbonyl reductases (Cbrs) that metabolize carbonyl-containing xenobiotics, and 2 NAD(P)H:quinone oxidoreductases (Nqos) that metabolize quinones. Akr1c21 and both Nqos (Nqo1 and Nqo2) were NE in intestines of CV or GF mice (Table 2). The intestinal expression of eight Akrs (1a4, 1b3, 1b10, 1c6, 1c20, 1d1, 1e1, and 7a5) and two Cbrs (Cbr2 and Cbr4) showed no differences in expression between CV and GF mice (Table 3). Seven Akrs and two Cbrs had DE between CV and GF mice in at least one section of the intestine (Table 1). Seven of nine reductases showed higher intestinal expression in GF mice, such as Akr1b7 in jej (136%); Akr1c12 (84%; 60%; 41%), Akr1c13 (152%; 104%; 48%), and Akr1c14 (29%; 71%; 58%) in jej, ile, and LI; Akr1c18 in LI (741%); Akr1c19 in all four sections of the intestine (101%; 241%; 211%; 158%); and Cbr1 in jej (41%); whereas a couple of reductases, namely Akr1b8 (49%) and Cbr3 (32%) in LI, had lower expression in GF mice (Fig. 3). In contrast to the similar expression of some Akrs in the livers of CV and GF mice, some genes had higher expression (Akr1b7, 1c12, 1c13, 1c14, 1c18, Cbr1) or lower expression (Akr1b8, Cbr3) in intestines of GF mice. Substrates of Akr1c12, 1c13, and 1c19 were likely to be affected the most by the lack of intestinal bacteria because the expression of these genes was higher in the intestine, and there was an increase in the expression of these genes in multiple intestinal sections.
Fig. 3.
The intestinal expression of Akrs and Cbrs in GF mice. Data are represented as the mean FPKM ± S.E.M. of three individual animals. Differences between CV and GF mice that were significant by Cuffdiff (FDR-BH < 0.05) were represented with asterisks.
Phase I enzymes Involved in Oxidation Reactions
Cytochrome P450s.
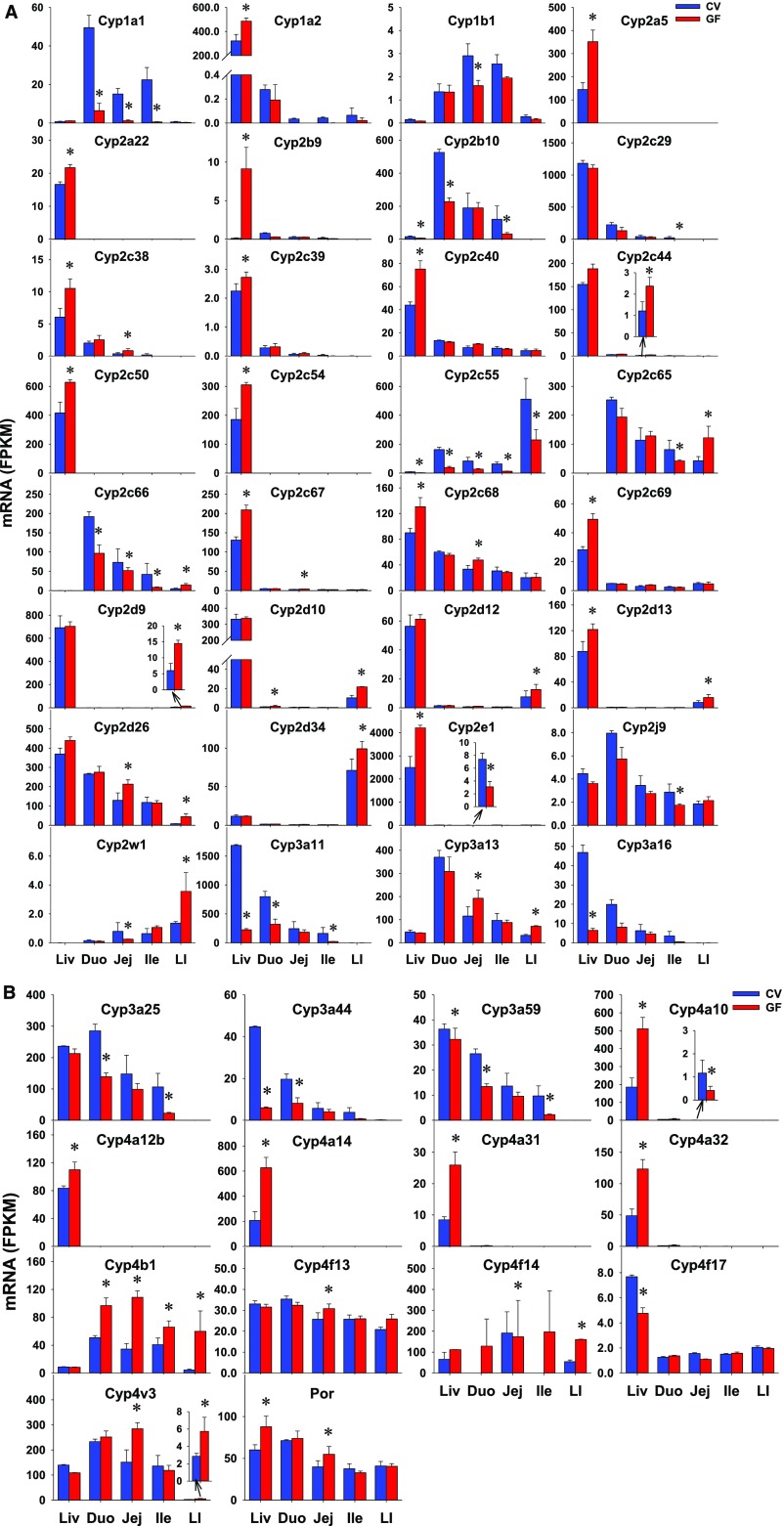
Cytochrome P450s (P450s), a class of heme-containing monooxygenases, are major enzymes for the biotransformation of numerous endobiotics as well as the detoxification or bioactivation of various xenobiotics (e.g., drugs, chemical carcinogens, and environmental chemicals) (Danielson, 2002; Parkinson et al., 2013). P450 oxidoreductase (Por) is an important electron donor for P450s for facilitation of their catalytic functions. The intestinal expression of 76 P450s in the Cyp1–4 families as well as Por were investigated. There were 11 P450s that were NE in the intestine of CV or GF mice (Table 2). As detailed in Table 3, 35 P450s did not show any differences in intestinal expression between CV and GF mice. Thirty P450s and Por had DE between CV and GF mice in at least one section of the intestine (Table 1). 11 of 30 P450s had lower intestinal expression in GF mice, including Cyp1a1 in all three sections of small intestine (87%; 92%; 97%), Cyp1b1 in jej (45%), Cyp2b10 in duo (57%) and ile (74%), Cyp2c29 in ile (90%), Cyp2c55 in all four sections of intestine (76%; 66%; 79%; 55%), Cyp2e1 in jej (58%), Cyp2j9 in ile (39%), Cyp3a11 in duo (60%) and ile (86%), Cyp3a25 in duo (51%) and ile (78%), Cyp3a44 in duo (58%), and Cyp3a59 in duo (49%) and ile (77%) (Fig. 4). Some genes showed higher expression in GF mice, such as Cyp2c38 (141%), Cyp2c44 (97%), Cyp2c67 (43%), and Cyp2c68 (43%) in jej; Cyp2d9 (142%) and Cyp2d34 (39%) in LI; Cyp2d10 in duo (69%) and LI (114%); Cyp2d12 (65%) and Cyp2d13 (87%) in LI; Cyp2d26 (65%; 163%) and Cyp3a13 (67%; 124%) in jej and LI; Cyp4b1 in all four sections of intestine (91%; 216%; 62%; 127%); Cyp4v3 in jej (88%) and LI (100%); as well as Por in jej (38%). In addition, Cyp2c65 and Cyp2c66 had section-specific DE between CV and GF mice, which is an increase in one section and a decrease in another section. In contrast to the similar expression of some P450s in livers of CV and GF mice, some genes had higher expression (Cyp2c44, 2d9, 2d10, 2d12, 2d26, 2d34, 3a13, 4b1, 4f13, and 4v3) or lower expression (Cyp1a1, 1b1, 2c29, 2j9, and 3a25) in intestines of GF mice. Some genes with DE in liver (Cyp1a2, 2a5, 2a22, 2b29, 2c39, 2c40, 2c50, 2c54, 2c69, 3a16, 4a12b, 4a14, 4a31, 4a32, and 4f17) remained unchanged in intestines of GF mice. Some genes (Cyp2b10, 2c38, 2c55, 2c67, 2c68, 2d13, 3a11, 3a44, and 3a59, and Por) had similar alterations of expression in livers and intestines of GF mice. Probably the most important change is the decrease in Cyp3a subfamily, because the decrease was observed in the intestine and liver, and the Cyp3a subfamily metabolizes over 50% of drugs. Substrates of Cyp1a1, 2b10, and 2c55 were likely to be metabolized more slowly with the lack of intestinal bacteria because they were primarily expressed in the intestine, where they were expressed at lower levels in GF mice. Most of the Cyp4 enzymes were expressed at a higher level in the liver than in the intestine (Cyp4a10, 4a12b, 4a14, 4a31, and 4a32), whereas Cyp4b1 was expressed at a higher level in the intestine, and expression was also increased in GF mice.
Fig. 4.
The intestinal expression of some P450s (Cyp1 to Cyp4 family) and Por (A and B) in GF mice. Data are represented as mean FPKM ± S.E.M. of three individual animals. Differences between CV and GF mice that were significant by Cuffdiff (FDR-BH < 0.05) were represented with asterisks.
Non–P450 Oxidation.
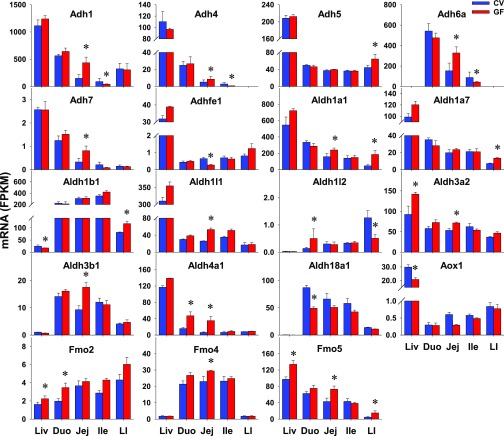
Four families of non-P450 enzymes that mediate xenobiotic oxidation were investigated, including 6 alcohol dehydrogenases (Adhs) for ethanol and other aliphatic alcohols, 20 aldehyde dehydrogenases (Aldhs) for aldehydes, 3 aldehyde oxidases (Aoxs) for aromatic aldehydes, and 5 flavin monooxygenases (Fmos) for xenobiotics with nucleophilic atoms. Aox3l1 and Fmo3 were NE in intestine of CV or GF mice (Table 2). As detailed in Table 3, 11 Aldhs did not show any differences in intestinal expression between CV and GF mice, in addition to Aox1, Aox3, and Fmo1. Six Adhs, nine Aldhs, and three Fmos had DE between CV and GF mice in at least one section of the intestine (Table 1). As shown in Fig. 5, 12 of 18 oxidases had higher intestinal expression in GF mice, such as Adh5 in LI (45%); Adh7 in jej (148%); Aldh1a1 in jej (52%) and LI (291%); Aldh1a7 (88%) and Aldh1b1 (43%) in LI; Aldh1l1 (103%), Aldh3a2 (34%), and Aldh3b1 (89%) in jej; and Aldh4a1 in duo (195%) and jej (458%); Fmo2 in duo (75%); Fmo4 in jej (28%); and Fmo5 in jej (70%) and LI (205%). Some genes, such as Adhfe1 in jej (58%) and Aldh18a1 in duo (44%), had lower expression in GF mice. In addition, expressions of Adh1, 4, and 6a were higher in jej (192%; 64%; 114%) but lower in ile (53%; 73%; 52%); Aldh1l2 expression was higher in duo (261%) but lower in LI (60%). In contrast to the similar expression in livers of CV and GF mice, some genes had higher expression (Adh5, 7, Aldh1a1, 1a7, 1l1, 3a2, 3b1, and 4a1, and Fmo4) or lower expression (Adhfe1 and Aldh18a1) in intestines of GF mice. Aox1 had DE in the liver but remained unchanged in intestines of GF mice. Some genes (Aldh3a2, Fmo2, and Fmo5) had similar alterations of expression in livers and intestines of GF mice. Aldh1b1 had lower expression in the liver but higher expression in the LI of GF mice. Because none of the Adhs, Aldhs, or Fmos had consistently higher or lower expression in all sections of the GF mice, it is not likely that these changes will have marked changes on the metabolism of their substrates.
Fig. 5.
The intestinal expression of Adhs, Aldhs, Aox1, and Fmos in GF mice. Data are represented as the mean FPKM ± S.E.M. of three individual animals. Differences between CV and GF mice that were significant by Cuffdiff (FDR-BH < 0.05) were represented with asterisks.
The Intestinal Expression of Phase II Enzymes in GF Mice
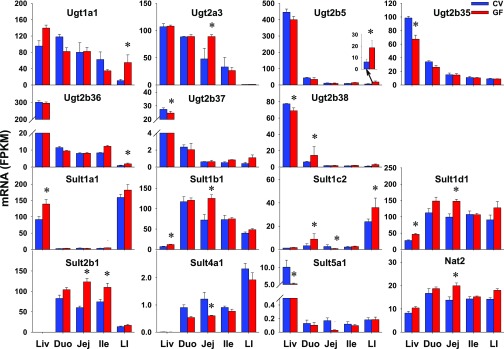
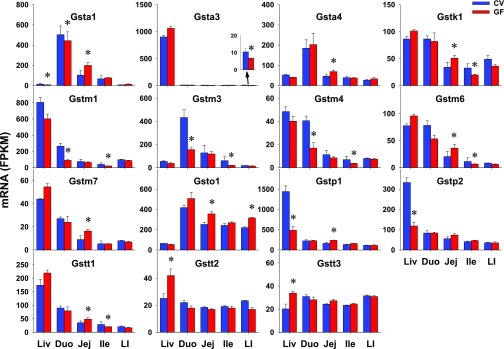
Xenobiotics can be conjugated by phase II enzymes with glucuronic acid [by UDP-glucuronosyltransferases (Ugts)], sulfate [by sulfotransferases (Sults)], glutathione [by glutathione S-transferases (Gsts)], or amino acids (by amino acid-conjugation enzymes), as well as methyl (by methyltransferases) or acetyl group [N-actyltransferases (Nats)]. In this study, 18 Ugts, 17 Sults, 25 Gsts, 3 methyltransfersases [catechol O-methyltransferase (Comt), arsenic (+3 oxidation state) methyltransferase (As3mt), thiopurine S-methyltransferase (Tpmt)], 3 Nats, and 3 amino acid-conjugation enzymes [bile acid-CoA ligase (Bal), bile acid-CoA:amino acid N-acyltransferase (Bat), and glycine N-acyltransferase (Glyat)] were investigated. Four Ugts, 10 Sults, 2 Gsts, as well as Nat1 and 3 were NE in the intestines of CV or GF mice (Table 2). As detailed in Table 3, some genes did not show any differences in intestinal expression between CV and GF mice, including 9 Ugts, 2 Sults, 11 Gsts, 3 methyltransferases (Comt, As3mt, and Tpmt), as well as 3 amino acid-conjugation enzymes (Bal, Bat, and Glyat). Five Ugts, 5 Sults, 12 Gsts, and Nat2 had DE between CV and GF mice in at least one section of the intestine (Table 1). As shown in Figs. 6 and 7, 13 of 23 phase II enzymes, such as Ugt1a1 (415%), Ugt2b5 (197%), and Ugt2b36 (106%) in LI; Ugt2a3 in jej (85%); Ugt2b38 in duo (129%); Sult1b1 (74%) and Sult1d1 (49%) in jej; Sult2b1 in jej (105%) and ile (47%); Gsta4 in jej (49%); Gstm7 in jej (82%); Gsto1 in jej (41%) and LI (44%); Gstp1 in jej (46%); and Nat2 in jej (45%), had higher expression in distinct intestinal sections of GF mice. Some genes, such as Sult4a1 in jej (50%); Gsta3 in LI (35%); Gstm1, m3, and m4 in duo (66%; 64%; 58%); and ile (49%; 68%; 45%), had lower expression in GF mice. In addition, Sult1c2, and Gsta1, k1, m6, and t1 had section-specific DE between CV and GF mice. In contrast to similar expression in livers of CV and GF mice, some genes (Ugt1a1, 2a3, 2b5, and 2b36; Sult2b1; Nat2; and Gsta4, m7, and o1) had higher expression whereas some genes had lower expression (Sult4a1, and Gsta3, m1, m3, m4) in intestines of GF mice. Some genes (Ugt2b35 and 2b37; Sult1a1 and 5a1; and Gstp2, t2, and t3) had DE in the livers of the CV and GF mice but remained unchanged in intestines of GF mice. Some genes (Sult1b1 and 1d1) had similar alterations of expression in livers and intestines of GF mice. Ugt2b38 had lower expression in the liver but higher expression in the duo of GF mice. Some Ugts (Ugt1a1 and 2a3) had comparable expression in the intestine and liver, whereas most Ugts (Ugt2b5, 2b35, 2b36, 2b37, and 2b38) had much lower expression in the intestine than the liver. Substrates of some Sults (Sult1b1, 1d1, and 2b1) and Gsts (Gsta1, m3, and o1) are likely to be affected by the lack of intestinal bacteria, because they were expressed at higher levels in the intestine than the liver, and the expressions of these genes were altered in the intestines of GF mice.
Fig. 6.
The intestinal expression of Ugts, Sults, and Nat2 in GF mice. Data are represented as the mean FPKM ± S.E.M. of three individual animals. Differences between CV and GF mice that were significant by Cuffdiff (FDR-BH < 0.05) were represented with asterisks. Nat, N-acetyltransferase.
Fig. 7.
The intestinal expression of Gsts in GF mice. Data are represented as the mean FPKM ± S.E.M. of three individual animals. Differences between CV and GF mice that were significant by Cuffdiff (FDR-BH < 0.05) were represented with asterisks.
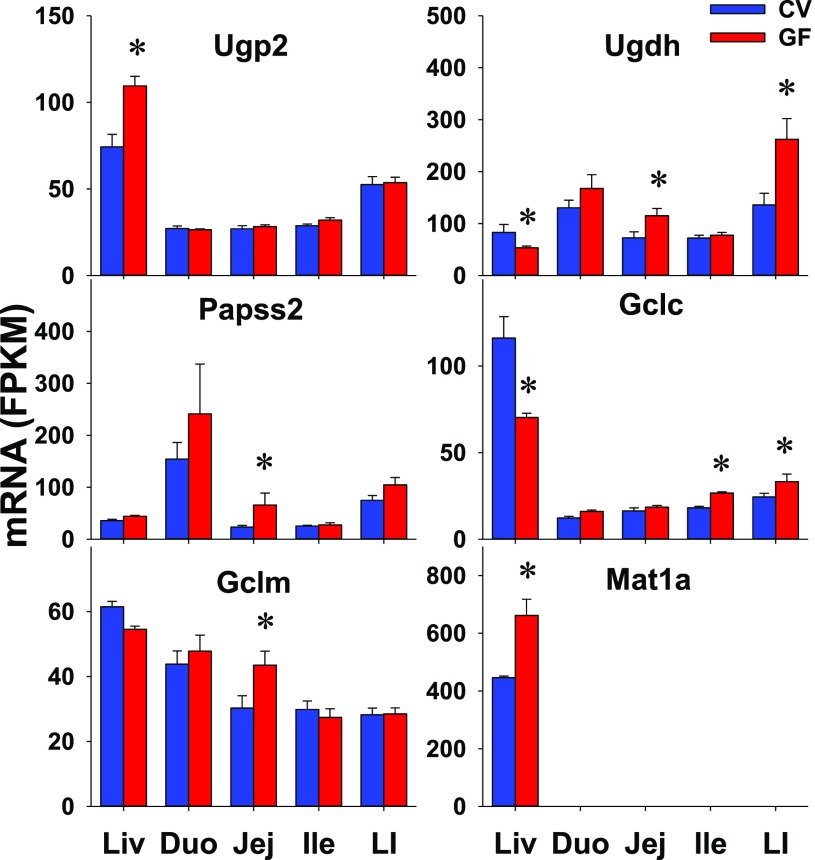
Several enzymes involved in the synthesis of phase II conjugation cosubstrates were also investigated, including UDP-glucose pyrophosphorylase 2 (Ugp2) (for UDP-glucose synthesis), UDP-glucose 6-dehydrogenase (Ugdh) (for the synthesis of the glucuronidation cosubstrate uridine diphosphate glucuronic acid), 3′-phosphoadenosine 5′-phosphosulfate synthases/Papss (for the synthesis of the sulfation cosubstrate 3′-phosphoadenosine 5′-phosphosulfate), glutamate-cysteine ligase catalytic subunit (Gclc) and glutamate-cysteine ligase modifier subunit (Gclm) (for the synthesis of Gst cosubstrate glutathione), and methionine adenosyltransferases (Mats) (for the synthesis of the common methyl donor S-adenosylmethionine). The intestinal expression of five of these enzymes, namely Ugp2, Papss1, and Mat1a, 2a, and 2b, was not different between CV and GF mice (Table 3). Compared with CV mice, GF mice had higher intestinal expression of other genes, including Ugdh in jej (59%) and LI (93%), Papss2 in jej (182%), Gclc in ile (47%) and LI (36%), and Gclm in jej (44%) (Fig. 8). In contrast to similar expression in livers of CV and GF mice, Papss2 and Gclm had higher expression in intestines of GF mice. Some genes (Ugp2, Mat1a) had DE in the liver but remained unchanged in the intestine of GF mice. Ugdh and Gclc had opposite patterns of altered expression in the liver and intestine of GF mice. Because none of these genes had constant higher or lower expression in all sections of the GF mice, it is not likely that these changes will have marked changes on the synthesis of these cosubstrates for phase II conjugation.
Fig. 8.
The intestinal expression of phase II conjugation cosubstrate synthetic enzymes in GF mice. Data are represented as the mean FPKM ± S.E.M. of three individual animals. Differences between CV and GF mice that were significant by Cuffdiff (FDR-BH < 0.05) were represented with asterisks.
The Intestinal Expression of Xenobiotic Transporters in GF Mice
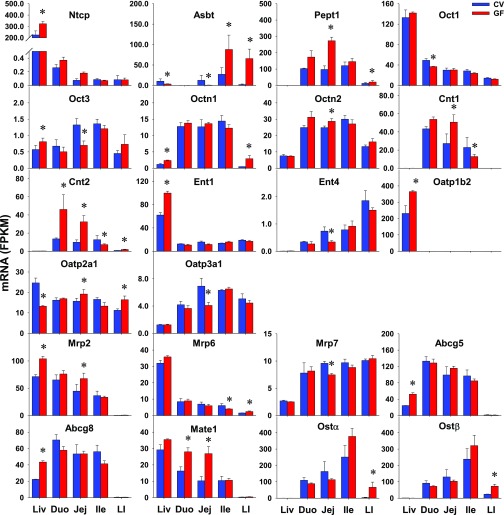
Transporters with importance in xenobiotic transport were investigated, including 36 uptake transporters and 20 efflux transporters. Thirteen uptake transporters and two efflux transporters were NE in intestines of CV or GF mice (Table 2). Twelve uptake transporters and 12 efflux transporters did not have DE between CV and GF mice in the intestine (Table 3). As shown in Fig. 9, 8 of 17 transporters, such as peptide transporter (Pept) 1/solute carrier (Slc) 15a1 in jej (179%) and LI (65%); organic cation/carnitine transporter (Octn) 1/Slc22a4 in LI (507%) and Octn2/Slc22a5 in jej (16%); organic anion-transporting polypeptide (Oatp) 2a1/Slco2a1 in jej (22%) and LI (46%); multidrug resistance–associated protein (Mrp) 2/ATP-binding cassette (Abc) c2 in jej (51%); multidrug and toxin extrusion protein (Mate) 1/Slc47a1 in duo (72%) and jej (162%); and organic solute transporter (Ost) α/Slc51a and Ostβ/Slc51b in LI (18-fold; 208%), had higher expression in the intestines of GF mice. Some transporters had lower intestinal expression in GF mice, such as organic cation transporter (Oct) 1 (Oct1/Slc22a1; 25%) in duo; and Oct3/Slc22a3 (47%), equilibrative nucleoside transporter (Ent) 4/Slc29a4 (54%), Oatp3a1/Slco3a1 (41%), and Mrp7/Abcc10 (22%) in jej. In contrast to similar expression in the liver of CV and GF mice, some transporters had higher (Pept1, Octn2, Mate1, Ostα, Ostβ) or lower (Oct1, Ent4, Otap3a1, Mrp7) expression in intestines of GF mice. Some transporters (Ntcp, Ent1, Oatp1b2, Abcg5, and Abcg8) had altered expression in livers of GF mice but remained unchanged in intestines of GF mice. Some genes (Octn1, Mrp2) had similar alterations of expression in livers and intestines of GF mice. Some genes (Oct3 and Oatp2a1) had opposite patterns of altered expression in livers and intestines of GF mice. Because none of the transporters had constant higher or lower expression in all sections of the GF mice, it is not likely these changes will have marked changes on the transport of their substrates.
Fig. 9.
The intestinal expression of uptake and efflux xenobiotic transporters in GF mice. Data are represented as the mean FPKM ± S.E.M. of three individual animals. Differences between CV and GF mice that were significant by Cuffdiff (FDR-BH < 0.05) were represented with asterisks. Abst, apical sodium-dependent bile acid transporter; Cnt, concentrative nucleoside transporter.
The Intestinal Expression of Xenobiotic-Related TFs in GF Mice
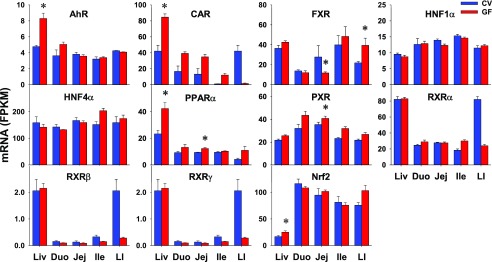
Various TFs that are involved in the transcriptional regulation of XPGs were also investigated in this study, including the aryl hydrocarbon receptor (AhR), constitutive androstane receptor (CAR)/NR1i3, farnesoid X receptor (FXR)/NR1h4, hepatocyte nuclear factor (HNF) 1α and HNF4α/NR2a1, peroxisome proliferator–activated receptor α (PPARα)/NR1c1, pregnane X receptor (PXR)/NR1i2), three retinoid X receptors (RXRs) (RXRα/NR2b1, RXRβ/NR2b2, and RXRγ/NR2b3), and nuclear factor erythroid 2–related factor 2 (Nrf2). As shown in Fig. 10, some TFs showed higher expression in GF mice, such as PPARα (31%) and PXR (15%) in jej. CAR appeared to have higher expression in GF than CV mice in all three sections of the small intestine (although not statistically significant), but lower expression in the LIs of GF mice. In contrast to the similar expression in livers of CV and GF mice, FXR had lower expression in jejs (57%), but higher expression in LIs (79%) of GF mice. Some TFs appeared to have lower expression in LIs of GF mice but did not achieve statistical significance. HNF1α and HNF4α did not have DE in intestines of CV and GF mice. In contrast to the higher expression in livers of GF mice, AhR and Nrf2 expression in the intestine remained similar between CV and GF mice.
Fig. 10.
The intestinal expression of xenobiotic-related TFs in GF mice. Data are represented as mean FPKM ± S.E.M. of three individual animals. Differences between CV and GF mice that were significant by Cuffdiff (FDR-BH < 0.05) were represented with asterisks.
Discussion
The present study was the first to investigate how a lack of intestinal bacteria regulates the expression of host genes involved in drug metabolism and transport in the intestine. Comparing GF mice with CV mice, this study has demonstrated that the lack of intestinal bacteria alters the gene expression of a number of phase I and phase II drug–metabolizing enzymes as well as transporters in the intestine (Table 1), and the altered pattern of XPG expression in the intestine induced by the lack of intestinal bacteria differs from that in the liver.
To provide a condensed picture focusing on the genes altered the most, XPGs with over 5-fold DE in GF versus CV mice in at least one intestinal section are listed in Table 4, which includes eight phase I enzymes, one phase II enzyme, and three transporters. Six of these are P450s that are responsible for the biotransformation of approximately 75% of drugs (Guengerich, 2008). Noticeably, Cyp3A, which is present in critical drug-metabolic tissues such as liver and intestine, metabolizes more drugs than any other P450 (Wilkinson, 1996). In this study, the majority of Cyp3a subfamily members have lower intestinal expression in GF mice compared with CV mice, including Cyp3a11, 3a16, 3a25, 3a44, and 3a59 (Fig. 4). Similarly, these Cyp3a members have markedly lower expression in the livers of GF mice (Fig. 4), which is consistent with previous reports (Toda et al., 2009; Kuno et al., 2016). In contrast, Cyp3a13 is the only Cyp3a member whose expression is higher in the intestines of GF mice (Fig. 4). It is known that the Cyp3a13 gene locates in a distinct position away from the Cyp3a locus on chromosome 5, where all other Cyp3a members cluster (Zaphiropoulos, 2003; Cui et al., 2012b). It is likely that Cyp3a13 and the other Cyp3a members may have a different regulatory mechanism for expression due to this distinct gene location. Moreover, many other P450s show lower expression in GF mice compared with CV mice, such as the Cyp1a, Cyp2b10, and Cyp2c subfamilies (Cyp2c29, 2c55, and 2c66), whereas a couple of P450s (Cyp2d26, 4b1, and 4v3) have higher expression in GF mice (Fig. 4). The current finding of altered P450 expression in the intestine of GF mice suggests that intestinal bacteria play an important role in regulating P450-mediated first-pass metabolism by the host intestine.
TABLE 4.
List of XPGs that have over 5-fold DE in GF vs. CV mice in at least one section of the intestine
| DE XPGs # | Increase | Decrease |
|---|---|---|
| Akr1c18 (LI) | Cyp1a1 (Duo, Jej, Ile) | |
| 8 phase I enzymes | Cyp2d26 (LI) | Cyp2c29 (Ile) |
| Aldh4a1 (Jej) | Cyp2c55 (Ile) | |
| Cyp2c66 (Ile) | ||
| Cyp3a11 (Ile) | ||
| 1 phase II enzyme | Ugt1a1 (LI) | |
| 3 transporters | Asbt (LI) | Asbt (Jej) |
| Octn1 (LI) | ||
| Ostα (LI) |
In addition to P450s, several other phase I drug–metabolizing enzymes are regulated by intestinal bacteria. Intestine-enriched Ces2a are Cess for large alcohol or small acyl groups, such as the cancer prodrug irinotecan (Humerickhouse et al., 2000; Zhang et al., 2012). This study shows that Ces2a has markedly lower expression in intestines of GF mice compared with CV mice (Fig. 2), indicating a possible role of intestinal bacteria in the hydrolysis and activation of prodrugs mediated by host Cess of the intestine. The Akr1c subfamily members are ketosteroid reductases that play crucial roles in the metabolism of steroid hormones and prostaglandins, as well as metabolic activation of the polycyclic aromatic hydrocarbon carcinogens and inactivation of nicotine-derived nitrosaminoketones (Penning and Drury, 2007). This study reveals that several Akr1c subfamily members (Ark1c12, 1c13, 1c14, 1c18, and 1c19) have higher expression in intestines of GF mice than CV mice (Fig. 3), indicating that the lack of intestinal bacteria may trigger the upregulation of the metabolism of steroid hormones and xenobiotics by the intestine. Aldh4a1 is a host enzyme involved in proline conversion to glutamate (Marchitti et al., 2008). Proline has been shown to be an important carbon and nitrogen source for the growth of Gram-negative bacteria, which can convert proline to glutamate entering the tricarboxylic acid cycle (Krishnan and Becker, 2006). This study identifies increased intestinal expression of Aldh4a1 in GF mice (Fig. 5), which suggests that the proline metabolism pathway of the host intestine is induced as a result of a lack of intestinal bacteria.
The expression of some phase II enzymes in the intestine is also affected by the intestinal bacteria. Ugt1a1 catalyzes the glucuronidation of bilirubin and the chemotherapeutic drug irinotecan, and Ugt1a1 polymorphism is implicated in Gilbert syndrome, Crigler-Najjar syndrome, and irinotecan toxicity (Sugatani, 2013). Glucuronidates can be deconjugated by β-glucuronidases in the intestinal bacteria and then enter the enterohepatic circulation. Of note, Ugt1a1 expression in LI is markedly higher in GF mice compared with CV mice (Fig. 6). It is possible that when the intestinal bacteria are absent, the LI increases its glucuronidation to reduce the recycling of chemicals in the enterohepatic circulation.
Intestinal bacteria are responsible for producing secondary bile acids from primary bile acids through deconjugation, dehydroxylation, epimerization, and oxidation (Fu et al., 2014; Wahlstrom et al., 2016; Li et al., 2017). It is known that GF mice have diminished secondary bile acids but more total and conjugated bile acids in the intestine (Sayin et al., 2013; Selwyn et al., 2015a). The current study shows that bile acid transporters have higher expression in the intestines of GF mice compared with CV mice, specifically Asbt in ile and LI and Ostα/Ostβ in the LI (Fig. 9). This finding suggests an upregulation of bile acid intestinal reabsorption when intestinal bacteria are absent.
The expression of TFs in the intestine appears to be differentially regulated by intestinal bacteria. PXR and CAR are two master regulators of drug metabolism, and their target drug-metabolizing genes in the intestine have been reported (Hartley et al., 2004; Park et al., 2016). The current study shows that intestinal bacteria–mediated changes in the expression profiles of PXR and CAR in the intestine are not consistent with the changes of their prototypical target genes Cyp3a11 and Cyp2b10, respectively (Figs. 4 and 10). Other xenobiotic-sensing TFs, AhR and Nrf2, have similar expression in the intestine between CV and GF mice, which is inconsistent with decreased expression of the AhR target gene Cyp1a1 and increased expression of some Gsts that are targets of Nrf2. Therefore, the XPG alterations by lack of intestinal bacteria may not mediated by a single TF.
Previous publications indicate that microbial metabolites (e.g., short-chain fatty acids and tryptophan metabolites), sensing through xenobiotic receptors that PXR and AhR can regulate host intestinal barrier function and innate immunity (Jin et al., 2014; Venkatesh et al., 2014; Ranhotra et al., 2016; Lanis et al., 2017). The present study has compared the intestinal expression profiles between CV and GF mice of target genes of PXR (Cui and Klaassen, 2016) and those of AhR (Sartor et al., 2009) as previously reported. The marked lower expression of PXR target genes Cyp3a11, Cyp2b10, as well as Gstm1, 3, and 4 in the intestines of GF mice than in CV mice (Figs. 4 and 7) theoretically could be due to less activation of PXR. However, an exception to this includes some PXR target genes (such as Oatp1a4 and Mrp3) with similar intestinal expression between CV and GF mice (Table 3). Similarly, AhR target gene Cyp1a1 expression in the intestine of GF mice is lower than that in CV mice, whereas other target genes Cyp1a2 and Aldh3a1 have similar intestinal expression between CV and GF mice (Fig. 4; Table 3). Therefore, it does not appear that alterations in the PXR or AhR signaling pathway are responsible for the upregulation and downregulation of XPGs in GF mice. The exact mechanism for XPG alterations by intestinal bacteria remains elusive.
Lack of intestinal bacteria leads to similar alterations of some XPGs in liver and intestine, but different alterations of other XPGs in the intestine from that in the liver. Compared with CV mice, GF mice have lower expression of Cyp3a11 and Cyp2b10 in the liver (Selwyn et al., 2015b) and similarly lower expression in duo and ile (Fig. 4), but their expressions in jej and LI are similar in CV and GF mice (Fig. 4). Several phase II enzymes that are not differentially expressed in liver of GF and CV mice have higher intestinal expression in GF mice, such as Ugt1a1, 2a3, Sult1c2, 2b1, and Gsto1 (Figs. 6 and 7). In contrast, some enzymes with higher (Sult1a1, Gstt2, Gstt3) or lower (Gstp1, Gstp2) hepatic expression in GF mice are not differentially expressed in intestines of CV and GF mice (Figs. 6 and 7).
One should be cautious that mRNA level does not necessarily correlate with protein function. The protein levels and enzyme/transporter activities of XPGs were not determined in this study due to technique limitations. Specific antibodies and substrates of many isoforms of drug-metabolizing enzymes and transporters are not available. Technical breakthroughs in proteomics and metabolomics are essential to determine the protein levels and activities of these enzymes and transporters. Furthermore, additional studies are needed in the future to specify which genera of intestinal microflora and which bacterial metabolites are responsible for the regulation of host gene expression.
In conclusion, the present study has demonstrated that despite many exceptions, the lack of intestinal bacteria decreases the expression of numerous P450s (Table 4) but increases phase II enzymes and transporters in the intestine, both of which help to decrease the formation of reactive metabolites and elimination of chemicals. This reveals the important role of intestinal bacteria on host drug metabolism in an indirect manner and provides insights into the interaction between intestinal bacteria and pharmaceuticals. Furthermore, this finding paves a new path for exploring individual differences in pharmacokinetics from a metagenomic perspective.
Acknowledgments
We thank Clark Bloomer of the KUMC genomic sequencing facilities for his technical assistance on RNA-Seq.
Abbreviations
- Abc
ATP-binding cassette
- Adh
alcohol dehydrogenase
- AhR
aryl hydrocarbon receptor
- Akr
aldo-keto reductase
- Aldh
aldehyde dehydrogenase
- Aox
aldehyde oxidase
- Asbt
apical sodium-dependent bile acid transporter
- As3mt
arsenic (+3 oxidation state) methyltransferase
- Bal
bile acid-CoA ligase
- Bat
bile acid-CoA:amino acid N-acyltransferase
- CAR
constitutive androstane receptor
- Cbr
carbonyl reductase
- Ces
carboxylesterase
- Cnt
concentrative nucleoside transporter
- Comt
catechol O-methyltransferase
- CV
conventional
- DE
Differential expression
- duo
duodenum
- Ephx
epoxide hydrolase
- Ent
equilibrative nucleoside transporter
- FDR-BH
Benjamini-Hochberg false discovery rate adjustment
- Fmo
flavin monooxygenase
- FPKM
fragments per kilobase of exon per million reads mapped
- FXR
farnesoid X receptor
- Gclc
glutamate-cysteine ligase catalytic subunit
- Gclm
glutamate-cysteine ligase modifier subunit
- GF
germ-free
- Glyat
glycine N-acyltransferase
- Gst
glutathione S-transferase
- HNF
hepatocyte nuclear factor
- ile
ileum
- jej
jejunum
- KUMC
University of Kansas Medical Center
- LI
large intestine
- liv
liver
- Mat
methionine adenosyltransferase
- Mate
multidrug and toxin extrusion protein
- Mdr
multidrug resistance protein
- Mrp
multidrug resistance–associated protein
- Nat
N-acetyl transferase
- NDE
no differential expression
- NE
not expressed
- Nqo, NAD(P)H
quinone oxidoreductase
- NR
nuclear receptor
- Nrf2
nuclear factor erythroid 2–related factor 2
- Ntcp
Na+-taurocholate cotransporting polypeptide
- Oat
organic anion transporter
- Oatp
organic anion-transporting polypeptide
- Oct
organic cation transporter
- Octn
organic cation/carnitine transporter
- Ost
organic solute transporter
- P450
cytochrome P450
- Paps
3′-phosphoadenosine 5′-phosphosulfate synthase
- Pept
peptide transporter
- Pon
paraoxonase
- Por
cytochrome P450 oxidoreductase
- PPARα
peroxisome proliferator–activated receptor α
- PXR
pregnane X receptor
- RNA-Seq
RNA-sequencing
- RT-qPCR
reverse transcription-quantitative real-time polymerase chain reaction
- RXR
retinoid X receptor
- Slc
solute carrier
- Slco
solute carrier organic anion
- Sult
sulfotransferase
- TF
transcription factor
- Tpmt
thiopurine S-methyltransferase
- Ugdh
UDP-glucose 6-dehydrogenase
- Ugp2
UDP-glucose pyrophosphorylase 2
- Ugt
UDP-glucuronosyltransferase
- XPG
xenobiotic-processing gene
Authorship Contributions
Participated in research design: Selwyn, Cui, Klaassen.
Conducted experiments: Selwyn and Cui.
Performed data analysis: Fu, Cui, and Klaassen.
Wrote or contributed to the writing of the manuscript: Fu, Cui, and Klaassen.
Footnotes
References
- Björkholm B, Bok CM, Lundin A, Rafter J, Hibberd ML, Pettersson S. (2009) Intestinal microbiota regulate xenobiotic metabolism in the liver. PLoS One 4:e6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui JY, Aleksunes LM, Tanaka Y, Fu ZD, Guo Y, Guo GL, Lu H, Zhong XB, Klaassen CD. (2012a) Bile acids via FXR initiate the expression of major transporters involved in the enterohepatic circulation of bile acids in newborn mice. Am J Physiol Gastrointest Liver Physiol 302:G979–G996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui JY, Klaassen CD. (2016) RNA-Seq reveals common and unique PXR- and CAR-target gene signatures in the mouse liver transcriptome. Biochim Biophys Acta 1859:1198–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui JY, Renaud HJ, Klaassen CD. (2012b) Ontogeny of novel cytochrome P450 gene isoforms during postnatal liver maturation in mice. Drug Metab Dispos 40:1226–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielson PB. (2002) The cytochrome P450 superfamily: biochemistry, evolution and drug metabolism in humans. Curr Drug Metab 3:561–597. [DOI] [PubMed] [Google Scholar]
- Fu ZD, Csanaky IL, Klaassen CD. (2012) Effects of aging on mRNA profiles for drug-metabolizing enzymes and transporters in livers of male and female mice. Drug Metab Dispos 40:1216–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu ZD, Cui JY. (2017) Remote sensing between liver and intestine: importance of microbial metabolites. Current Pharmacology Reports 3:101–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu ZD, Cui JY, Klaassen CD. (2014) Atorvastatin induces bile acid-synthetic enzyme Cyp7a1 by suppressing FXR signaling in both liver and intestine in mice. J Lipid Res 55:2576–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu ZD, Klaassen CD. (2014) Short-term calorie restriction feminizes the mRNA profiles of drug metabolizing enzymes and transporters in livers of mice. Toxicol Appl Pharmacol 274:137–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu ZD, Selwyn FP, Cui JY, Klaassen CD. (2016) RNA sequencing quantification of xenobiotic-processing genes in various sections of the intestine in comparison to the liver of male mice. Drug Metab Dispos 44:842–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guengerich FP. (2008) Cytochrome p450 and chemical toxicology. Chem Res Toxicol 21:70–83. [DOI] [PubMed] [Google Scholar]
- Hartley DP, Dai X, He YD, Carlini EJ, Wang B, Huskey SE, Ulrich RG, Rushmore TH, Evers R, Evans DC. (2004) Activators of the rat pregnane X receptor differentially modulate hepatic and intestinal gene expression. Mol Pharmacol 65:1159–1171. [DOI] [PubMed] [Google Scholar]
- Human Microbiome Project Consortium (2012) Structure, function and diversity of the healthy human microbiome. Nature 486:207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humerickhouse R, Lohrbach K, Li L, Bosron WF, Dolan ME. (2000) Characterization of CPT-11 hydrolysis by human liver carboxylesterase isoforms hCE-1 and hCE-2. Cancer Res 60:1189–1192. [PubMed] [Google Scholar]
- Jin UH, Lee SO, Sridharan G, Lee K, Davidson LA, Jayaraman A, Chapkin RS, Alaniz R, Safe S. (2014) Microbiome-derived tryptophan metabolites and their aryl hydrocarbon receptor-dependent agonist and antagonist activities. Mol Pharmacol 85:777–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourova L, Anzenbacher P, Anzenbacherova E. (2016) Human gut microbiota plays a role in the metabolism of drugs. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub 160:317–326. [DOI] [PubMed] [Google Scholar]
- Klaassen CD, Cui JY. (2015) Review: mechanisms of how the intestinal microbiota alters the effects of drugs and bile acids. Drug Metab Dispos 43:1505–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan N, Becker DF. (2006) Oxygen reactivity of PutA from Helicobacter species and proline-linked oxidative stress. J Bacteriol 188:1227–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuno T, Hirayama-Kurogi M, Ito S, Ohtsuki S. (2016) Effect of intestinal flora on protein expression of drug-metabolizing enzymes and transporters in the liver and kidney of germ-free and antibiotics-treated mice. Mol Pharm 13:2691–2701. [DOI] [PubMed] [Google Scholar]
- Lanis JM, Alexeev EE, Curtis VF, Kitzenberg DA, Kao DJ, Battista KD, Gerich ME, Glover LE, Kominsky DJ, Colgan SP. (2017) Tryptophan metabolite activation of the aryl hydrocarbon receptor regulates IL-10 receptor expression on intestinal epithelia. Mucosal Immunol 10:1133–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WK, Li H, Lu YF, Li YY, Fu ZD, Liu J. (2017) Atorvastatin alters the expression of genes related to bile acid metabolism and circadian clock in livers of mice. PeerJ 5:e3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. (2012) Diversity, stability and resilience of the human gut microbiota. Nature 489:220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchitti SA, Brocker C, Stagos D, Vasiliou V. (2008) Non-P450 aldehyde oxidizing enzymes: the aldehyde dehydrogenase superfamily. Expert Opin Drug Metab Toxicol 4:697–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikov M. (1994) The metabolism of drugs by the gut flora. Eur J Drug Metab Pharmacokinet 19:201–207. [DOI] [PubMed] [Google Scholar]
- Park S, Cheng SL, Cui JY. (2016) Characterizing drug-metabolizing enzymes and transporters that are bona fide CAR-target genes in mouse intestine. Acta Pharm Sin B 6:475–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson A, Ogilvie BW, Buckley DB, Kazmi F, Czerwinski M, Parkinson O. (2013) Biotransformation of xenobiotics, in Casarett & Doull’s Toxicology: The Basic Science of Poisons (Klaassen CD. ed), pp 185–367, McGraw-Hill, New York. [Google Scholar]
- Penning TM, Drury JE. (2007) Human aldo-keto reductases: function, gene regulation, and single nucleotide polymorphisms. Arch Biochem Biophys 464:241–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranhotra HS, Flannigan KL, Brave M, Mukherjee S, Lukin DJ, Hirota SA, Mani S. (2016) Xenobiotic receptor-mediated regulation of intestinal barrier function and innate immunity. Nucl Receptor Res 3:101199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizkallah MR, Gamal-Eldin S, Saad R. (2012) The pharmacoMicrobiomics portal: a database for drug-microbiome interactions. Curr Pharmacogenomics Person Med 10:195–203. [Google Scholar]
- Sartor MA, Schnekenburger M, Marlowe JL, Reichard JF, Wang Y, Fan Y, Ma C, Karyala S, Halbleib D, Liu X, et al. (2009) Genomewide analysis of aryl hydrocarbon receptor binding targets reveals an extensive array of gene clusters that control morphogenetic and developmental programs. Environ Health Perspect 117:1139–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayin SI, Wahlström A, Felin J, Jäntti S, Marschall HU, Bamberg K, Angelin B, Hyötyläinen T, Orešič M, Bäckhed F. (2013) Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab 17:225–235. [DOI] [PubMed] [Google Scholar]
- Selwyn FP, Cheng SL, Klaassen CD, Cui JY. (2016) Regulation of hepatic drug-metabolizing enzymes in germ-free mice by conventionalization and probiotics. Drug Metab Dispos 44:262–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selwyn FP, Csanaky IL, Zhang Y, Klaassen CD. (2015a) Importance of large intestine in regulating bile acids and glucagon-like peptide-1 in germ-free mice. Drug Metab Dispos 43:1544–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selwyn FP, Cui JY, Klaassen CD. (2015b) RNA-Seq quantification of hepatic drug processing genes in germ-free mice. Drug Metab Dispos 43:1572–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojančević M, Bojić G, Salami HA, Mikov M. (2014) The influence of intestinal tract and probiotics on the fate of orally administered drugs. Curr Issues Mol Biol 16:55–68. [PubMed] [Google Scholar]
- Sugatani J. (2013) Function, genetic polymorphism, and transcriptional regulation of human UDP-glucuronosyltransferase (UGT) 1A1. Drug Metab Pharmacokinet 28:83–92. [DOI] [PubMed] [Google Scholar]
- Toda T, Saito N, Ikarashi N, Ito K, Yamamoto M, Ishige A, Watanabe K, Sugiyama K. (2009) Intestinal flora induces the expression of Cyp3a in the mouse liver. Xenobiotica 39:323–334. [DOI] [PubMed] [Google Scholar]
- Tremaroli V, Bäckhed F. (2012) Functional interactions between the gut microbiota and host metabolism. Nature 489:242–249. [DOI] [PubMed] [Google Scholar]
- Venkatesh M, Mukherjee S, Wang H, Li H, Sun K, Benechet AP, Qiu Z, Maher L, Redinbo MR, Phillips RS, et al. (2014) Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity 41:296–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlström A, Sayin SI, Marschall HU, Bäckhed F. (2016) Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab 24:41–50. [DOI] [PubMed] [Google Scholar]
- Wilkinson GR. (1996) Cytochrome P4503A (CYP3A) metabolism: prediction of in vivo activity in humans. J Pharmacokinet Biopharm 24:475–490. [DOI] [PubMed] [Google Scholar]
- Zaphiropoulos PG. (2003) A map of the mouse Cyp3a locus. DNA Seq 14:155–162. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Cheng X, Aleksunes L, Klaassen CD. (2012) Transcription factor-mediated regulation of carboxylesterase enzymes in livers of mice. Drug Metab Dispos 40:1191–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]