Abstract
The lipid droplet (LD) is a unique multi-functional organelle that contains a neutral lipid core covered with a phospholipid monolayer membrane. The LDs have been found in almost all organisms from bacteria to humans with similar shape. Several conserved functions of LDs have been revealed by recent studies, including lipid metabolism and trafficking, as well as nucleic acid binding and protection. We summarized these findings and proposed a hypothesis that the LD is a conserved organelle.
KEYWORDS: lipid droplet, conserved organelle, lipid metabolism, nucleic acid handling
INTRODUCTION
The lipid droplet (LD) is a multi-functional organelle with unique structure that distinguishes it from other cellular organelles (Murphy and Vance, 1999; Martin and Parton, 2006; Farese and Walther, 2009; Welte, 2015). Since discovered in 1674 by Van Leeuwenhoek, the LD has been found to be an organelle necessary for many cellular functions that are essential for the organismic energy homeostasis, and more importantly for human health and aging. In addition to its role in lipid storage and metabolism (Cao et al., 2008; Cohen et al., 2011), recent studies have revealed that the LD is critical for membrane trafficking (Liu et al., 2004; Bartz et al., 2007), protein storage (Li et al., 2012) and degradation (Ploegh, 2007), and has a key role in hepatitis C virus (HCV) replication and assembly (Miyanari et al., 2007) and neurodegeneration (Liu et al., 2015). As important sites of neutral lipid storage and metabolism, the ectopic storage of lipids in LDs is a key cellular component in many diseases. On other hand, LDs in plants and oleaginous microorganisms provide not only food oil but also feedstock for biodiesel and industrial oil (Murphy, 2001; Alvarez and Steinbuchel, 2002; Murphy, 2012; Chen et al., 2014).
LIPID DROPLETS EXIST FROM BACTERIA TO HUMANS
LDs are found in almost all organisms from bacteria to humans (Murphy, 2012; Waltermann et al., 2005). So far, except for knowing that LDs exist in all eukaryotic cells, it is also reported that some actinobacteria and cyanobacteria contain LDs, such as the genera Micromonospora, Dietzia, Nocardia, Rhodococcus, Mycobacterium, Gordonia, some streptomycetes (Murphy, 2001; Murphy, 2012), Nostoc punctiforme (Peramuna and Summers, 2014), Synechococcus lividus (Edwards et al., 1968), Anabaena variabilis (Wolk, 1973), and Synechocystis sp. PCC 6803 (Van de Meene et al., 2006). In addition, in comparison with other bacterial microcompartments including protein-based and lipid-bilayer membrane-based (Cornejo et al., 2014; Bobik et al., 2015), the LD is an unique organelle due to its particular structure and composition: neutral lipid core, phospholipid monolayer membrane, and peripheral proteins (Martin and Parton, 2006; Ding et al., 2012). This unique property is conserved from bacteria to humans.
THE STRUCTURE AND COMPOSITION OF LIPID DROPLETS ARE CONSERVED
The core content of LDs in bacteria and eukaryotic cells is neutral lipid. Although some LDs contain retinyl ester (O’Mahony et al., 2015), polyhydroxyalkanoate or wax ester (Murphy, 2012), triacylglycerol (TAG) and cholesterol ester (CE) are the major neutral lipids of LDs in most cells (Waltermann and Steinbuchel, 2005; Barbosa and Siniossoglou, 2017). The neutral lipid core is coated by a phospholipid monolayer membrane in bacteria and eukaryotes (Martin and Parton, 2006; Farese and Walther, 2009; Waltermann and Steinbuchel, 2005), although the phospholipid composition may be different (Chitraju et al., 2012). In addition to the conserved lipid contents, the resident proteins of the organelle, including microorganism lipid droplet small (MLDS) and eukaryotic PERILIPIN (PLIN) family proteins (Kimmel et al., 2010), display conserved properties including the ability to target the phospholipid monolayer membrane and by the fact that they are all belong to apolipoprotein-like protein family (Yang et al., 2012).
These apolipoprotein-like proteins have also the ability to target LDs in diverse organisms, for example, mammalian LD proteins (PLINs) are targeted to LDs in yeast (Rowe et al., 2016) and bacteria (Hanisch et al., 2006). The LD resident proteins in C. elegans, DHS-3 and MDT-28/PLIN1 (Chughtai et al., 2015) behave similarly to target mammalian LDs (Na et al. 2015). In addition, a Drosophila LD resident protein, LSD1/PLIN1 localizes to LDs in C. elegans (Liu et al., 2014). The LD resident proteins, human adipose differentiation-related protein (ADRP)/PLIN2, C. elegans MDT-28, and bacterial MLDS are all able to bind to adiposomes that contain a TAG core with a phospholipid (DOPC) monolayer to mimic LDs in vitro (Wang et al., 2016). The ability of these proteins to target LDs of other organisms indicates that this fundamental process is highly conserved.
THE LIPID DROPLET IS A FUNCTIONALLY CONSERVED ORGANELLE FROM BACTERIA TO HUMANS
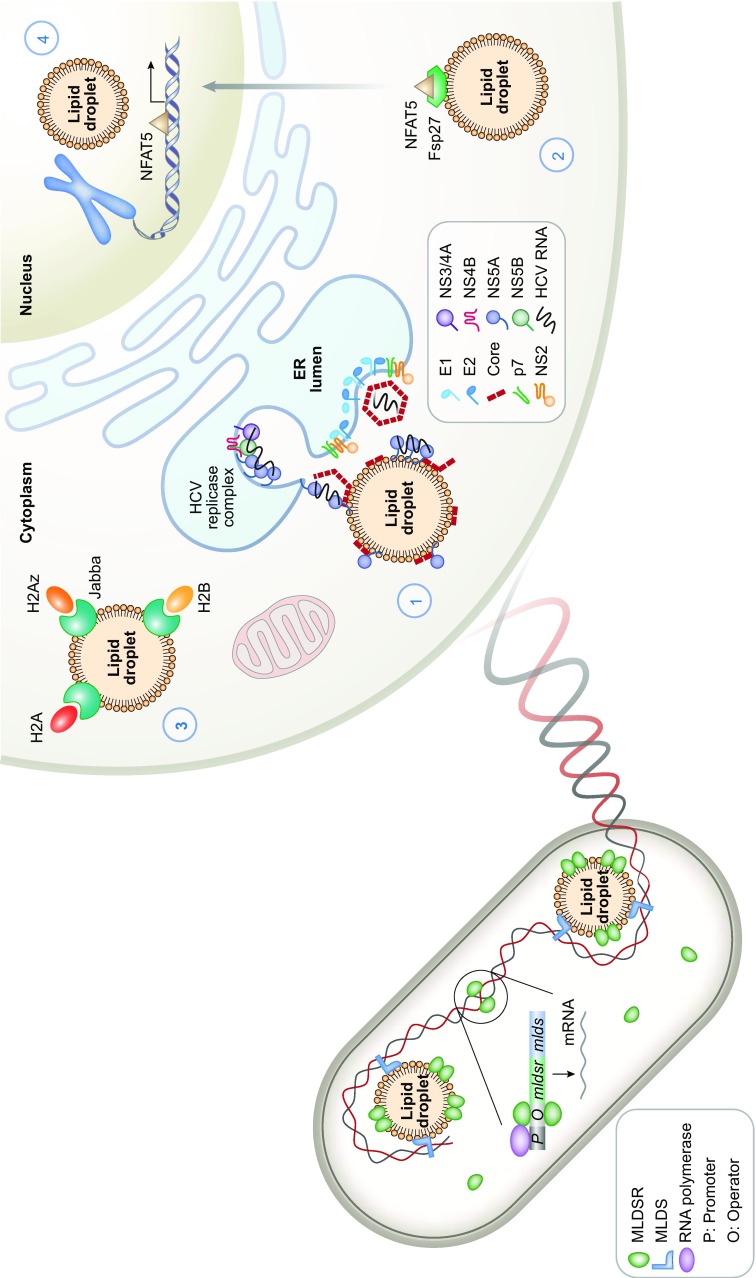
Several functions of LDs are common through bacteria to humans, such as lipid storage and metabolism. However, the study of other functions of LDs, especially in bacteria, is insufficient. Recently, we found that the LDs in a bacterium, Rhodococcus jostii RHA1 (RHA1), bind to genomic DNA (Fig. 1) (Zhang et al., 2017) and protect it via their major protein, MLDS, which promotes bacterial survival under stress (Zhang et al., 2017). Furthermore, the study also reports that LDs are involved in transcriptional regulation via a LD-associated transcriptional regulator, MLDSR (Zhang et al., 2017). These two newly identified functions in bacteria suggest that LDs are unique endomembrane organelles involved in nucleic acid handling and facilitate bacterial survival in and adaptation to extreme environments (Zhang et al., 2017).
Figure 1.
The conserved lipid droplet functions of binding and regulating nucleic acids from bacterial to human cells. In bacteria (left), LDs bind and protect genomic DNA via the major LD-associated protein, MLDS, which enhances the survival and adaptation of bacteria in extreme environments. Furthermore, a LD-associated transcriptional regulator, MLDSR, whose gene is in the same operon as mlds, induces or reduces the expression of MLDS when its cytosolic concentration is low or high, respectively. LDs have key role in transcriptional regulation by recruiting MLDSR to control its cytosolic concentration. Similar functions of LDs are also found in mammalian cells. In liver cells, hepatitis C virus is assembled around the LD surface and viral RNA is located to LDs through NS5A and core proteins. A hypothesis is proposed that after replication of viral RNA on the ER membrane, the newly synthesized RNA is moved by NS5A to the core protein on LDs, which triggers the initial viral assembly (right, part 1). In adipocytes, moreover, a transcriptional factor NFAT5 can be sequestered to LDs by Fsp27, which prevents its nuclear importation to initiate transcription (right, part 2). Several histones such as H2A, H2B, and H2Av are localized to LDs via the anchor protein Jabba in Drosophila (right, part 3). In addition, LDs are also present in the liver cell nucleus (right, part 4). The facts that both bacterial and mammalian LDs possess the function of nucleic acid handling indicate that LDs in living cells on earth are evolutionary conserved from prokaryotes to humans
In eukaryotic and prokaryotic cells, LD proteomic analysis has revealed that RNA-binding proteins, ribosomal subunits, and/or translation factors are present on LDs (Ding et al., 2012; Sato et al., 2006; Zhang et al., 2012). Ribosomes and RNA are also found on mammalian LDs (Dvorak et al., 2003; Dvorak, 2005; Wan et al., 2007). In addition, HCV localizes and assembles around the LD surface (Fig. 1) (Miyanari et al., 2007; Shi et al., 2002; Gentzsch et al., 2013; Fiches et al., 2016). Furthermore, a mammalian homologue of the most abundant LD resident protein in C. elegans, MDT-28, is a mediator of RNA polymerase II (Zhang et al., 2012; Li et al., 2015). LDs in Drosophila store histones via the Jabba protein (Fig. 1) (Li et al., 2012, 2014; Cermelli et al., 2006). Interestingly, several recent studies identified LDs in the nuclei of mammalian cells (Fig. 1) (Layerenza et al. 1831; Wang et al., 2013; Ohsaki et al., 2016). LDs inhibit the translocation of NFAT5 to the nucleus via the LD-associated protein FSP27 and reduce NFAT5 transcriptional activity (Fig. 1) (Ueno et al., 2012). Altogether, these reports suggest that eukaryotic LDs partially mimic some nuclear functions, which is similar to bacterial LDs.
According to these previous studies, both bacterial and eukaryotic LDs are involved in nucleic acid handling, suggesting that the LD is a functionally conserved organelle. In the evolution from prokaryotes to eukaryotes, the most important feature is the protection of hereditary material (nuclear emergence). Thus, the function of bacterial LDs to protect and regulate nucleic acids indicates that they are analogous to the eukaryotic nuclear membrane.
Based on the extensive distribution, as well as the conservation of structure, composition, and functions of LDs from almost all living organisms, we propose a hypothesis that the LD is a conserved organelle from bacteria to humans (Fig. 1).
ACKNOWLEDGEMENTS
The authors thank Dr. Mark Christian for his critical reading and useful suggestions. The authors also thank Ms. Libing Mu for the graphical summary. This work was supported by the Ministry of Science and Technology of China (Grant No. 2016YFA0500100), National Natural Science Foundation of China (Grant Nos. U1402225, 31571388, 61273228, and 81270932), Chinese Academy of Sciences (Grant No. XDA12030201).
ABBREVIATIONS
CE, cholesterol ester; HCV, hepatitis C virus; LD, lipid droplet; MLDS, microorganism lipid droplet small; TAG, triacylglycerol
COMPLIANCE WITH ETHICS GUIDELINES
Congyan Zhang declares that he has no conflict of interest. Pingsheng Liu declares that he has no conflict of interest. This article does not contain any studies with human or animal subjects performed by the any of the authors.
References
- Alvarez HM, Steinbuchel A. Triacylglycerols in prokaryotic microorganisms. Appl Microbiol Biotechnol. 2002;60:367–376. doi: 10.1007/s00253-002-1135-0. [DOI] [PubMed] [Google Scholar]
- Barbosa AD, Siniossoglou S (2017) Function of lipid droplet-organelle interactions in lipid homeostasis. Biochimica et Biophysica Acta. [DOI] [PubMed]
- Bartz R, Zehmer JK, Zhu M, Chen Y, Serrero G, Zhao Y, Liu P. Dynamic activity of lipid droplets: protein phosphorylation and GTP-mediated protein translocation. J Proteome Res. 2007;6:3256–3265. doi: 10.1021/pr070158j. [DOI] [PubMed] [Google Scholar]
- Bobik TA, Lehman BP, Yeates TO. Bacterial microcompartments: widespread prokaryotic organelles for isolation and optimization of metabolic pathways. Mol Microbiol. 2015;98:193–207. doi: 10.1111/mmi.13117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Gerhold K, Mayers JR, Wiest MM, Watkins SM, Hotamisligil GS. Identification of a lipokine, a lipid hormone linking adipose tissue to systemic metabolism. Cell. 2008;134:933–944. doi: 10.1016/j.cell.2008.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermelli S, Guo Y, Gross SP, Welte MA. The lipid-droplet proteome reveals that droplets are a protein-storage depot. Curr Biol. 2006;16:1783–1795. doi: 10.1016/j.cub.2006.07.062. [DOI] [PubMed] [Google Scholar]
- Chen Y, Ding YF, Yang L, Yu JH, Liu GM, Wang XM, Zhang SY, Yu D, Song L, Zhang HX, Zhang CY, Huo LH, Huo CX, Wang Y, Du YL, Zhang HN, Zhang P, Na HM, Xu SM, Zhu YX, Xie ZS, He T, Zhang Y, Wang GL, Fan ZH, Yang FQ, Liu HL, Wang XW, Zhang XG, Zhang MQ, Li YD, Steinbuchel A, Fujimoto T, Cichello S, Yu J, Liu PS. Integrated omics study delineates the dynamics of lipid droplets in Rhodococcus opacus PD630. Nucleic Acids Res. 2014;42:1052–1064. doi: 10.1093/nar/gkt932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitraju C, Trotzmuller M, Hartler J, Wolinski H, Thallinger GG, Lass A, Zechner R, Zimmermann R, Kofeler HC, Spener F. Lipidomic analysis of lipid droplets from murine hepatocytes reveals distinct signatures for nutritional stress. J Lipid Res. 2012;53:2141–2152. doi: 10.1194/jlr.M028902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chughtai AA, Kassak F, Kostrouchova M, Novotny JP, Krause MW, Saudek V, Kostrouch Z, Kostrouchova M. Perilipin-related protein regulates lipid metabolism in C. elegans. PeerJ. 2015;3:e1213. doi: 10.7717/peerj.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JC, Horton JD, Hobbs HH. Human fatty liver disease: old questions and new insights. Science. 2011;332:1519–1523. doi: 10.1126/science.1204265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornejo E, Abreu N, Komeili A. Compartmentalization and organelle formation in bacteria. Curr Opin Cell Biol. 2014;26:132–138. doi: 10.1016/j.ceb.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Yang L, Zhang S, Wang Y, Du Y, Pu J, Peng G, Chen Y, Zhang H, Yu J, Hang H, Wu P, Yang F, Yang H, Steinbuchel A, Liu P. Identification of the major functional proteins of prokaryotic lipid droplets. J Lipid Res. 2012;53:399–411. doi: 10.1194/jlr.M021899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak AM. Mast cell secretory granules and lipid bodies contain the necessary machinery important for the in situ synthesis of proteins. Chem Immunol Allergy. 2005;85:252–315. doi: 10.1159/000086520. [DOI] [PubMed] [Google Scholar]
- Dvorak AM, Morgan ES, Weller PF. RNA is closely associated with human mast cell lipid bodies. Histol Histopathol. 2003;18:943–968. doi: 10.14670/HH-18.943. [DOI] [PubMed] [Google Scholar]
- Edwards MR, Berns DS, Ghiorse WC, Holt SC. Ultrastructure of the thermophilic blue-green alga, synechococcus lividus copeland(1) J Phycol. 1968;4:283–298. doi: 10.1111/j.1529-8817.1968.tb04697.x. [DOI] [PubMed] [Google Scholar]
- Farese RV, Walther TC. Lipid droplets finally get a little R-E-S-P-E-C-T. Cell. 2009;139:855–860. doi: 10.1016/j.cell.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiches GN, Eyre NS, Aloia AL, Van Der Hoek K, Betz-Stablein B, Luciani F, Chopra A, Beard MR. HCV RNA traffic and association with NS5A in living cells. Virology. 2016;493:60–74. doi: 10.1016/j.virol.2016.02.016. [DOI] [PubMed] [Google Scholar]
- Gentzsch J, Brohm C, Steinmann E, Friesland M, Menzel N, Vieyres G, Perin PM, Frentzen A, Kaderali L, Pietschmann T. Hepatitis C virus p7 is critical for capsid assembly and envelopment. PLoS Pathogens. 2013;9:e1003355. doi: 10.1371/journal.ppat.1003355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanisch J, Waltermann M, Robenek H, Steinbuchel A. Eukaryotic lipid body proteins in oleogenous actinomycetes and their targeting to intracellular triacylglycerol inclusions: Impact on models of lipid body biogenesis. Appl Environ Microbiol. 2006;72:6743–6750. doi: 10.1128/AEM.00584-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel AR, Brasaemle DL, McAndrews-Hill M, Sztalryd C, Londos C. Adoption of PERILIPIN as a unifying nomenclature for the mammalian PAT-family of intracellular lipid storage droplet proteins. J Lipid Res. 2010;51:468–471. doi: 10.1194/jlr.R000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layerenza JP, Gonzalez P, Garcia de Bravo MM, Polo MP, Sisti MS, Ves-Losada A. Nuclear lipid droplets: a novel nuclear domain. Biochem Biophys Acta. 1831;2013:327–340. doi: 10.1016/j.bbalip.2012.10.005. [DOI] [PubMed] [Google Scholar]
- Li Z, Thiel K, Thul PJ, Beller M, Kuhnlein RP, Welte MA. Lipid droplets control the maternal histone supply of Drosophila embryos. Curr Biol. 2012;22:2104–2113. doi: 10.1016/j.cub.2012.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Johnson MR, Ke Z, Chen L, Welte MA. Drosophila lipid droplets buffer the H2Av supply to protect early embryonic development. Curr Biol. 2014;24:1485–1491. doi: 10.1016/j.cub.2014.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Walsh RM, Wagh V, James MF, Beauchamp RL, Chang YS, Gusella JF, Hochedlinger K, Ramesh V. Mediator subunit Med28 is essential for mouse peri-implantation development and pluripotency. PLoS ONE. 2015;10:e0140192. doi: 10.1371/journal.pone.0140192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Ying Y, Zhao Y, Mundy DI, Zhu M, Anderson RG. Chinese hamster ovary K2 cell lipid droplets appear to be metabolic organelles involved in membrane traffic. J Biol Chem. 2004;279:3787–3792. doi: 10.1074/jbc.M311945200. [DOI] [PubMed] [Google Scholar]
- Liu Z, Li X, Ge Q, Ding M, Huang X. A lipid droplet-associated GFP reporter-based screen identifies new fat storage regulators in C. elegans. J Genet Genomics. 2014;41:305–313. doi: 10.1016/j.jgg.2014.03.002. [DOI] [PubMed] [Google Scholar]
- Liu L, Zhang K, Sandoval H, Yamamoto S, Jaiswal M, Sanz E, Li Z, Hui J, Graham BH, Quintana A, Bellen HJ. Glial lipid droplets and ROS induced by mitochondrial defects promote neurodegeneration. Cell. 2015;160:177–190. doi: 10.1016/j.cell.2014.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S, Parton RG. Lipid droplets: a unified view of a dynamic organelle. Nat Rev Mol Cell Biol. 2006;7:373–378. doi: 10.1038/nrm1912. [DOI] [PubMed] [Google Scholar]
- Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, Zayas M, Bartenschlager R, Wakita T, Hijikata M, Shimotohno K. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol. 2007;9:1089–1097. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- Murphy DJ. The biogenesis and functions of lipid bodies in animals, plants and microorganisms. Prog Lipid Res. 2001;40:325–438. doi: 10.1016/S0163-7827(01)00013-3. [DOI] [PubMed] [Google Scholar]
- Murphy DJ. The dynamic roles of intracellular lipid droplets: from archaea to mammals. Protoplasma. 2012;249:541–585. doi: 10.1007/s00709-011-0329-7. [DOI] [PubMed] [Google Scholar]
- Murphy DJ, Vance J. Mechanisms of lipid-body formation. Trends Biochem Sci. 1999;24:109–115. doi: 10.1016/S0968-0004(98)01349-8. [DOI] [PubMed] [Google Scholar]
- Na H, Zhang P, Chen Y, Zhu X, Liu Y, Liu Y, Xie K, Xu N, Yang F, Yu Y, Cichello S, Mak HY, Wang MC, Zhang H, Liu P. Identification of lipid droplet structure-like/resident proteins in Caenorhabditis elegans. Biochem Biophys Acta. 2015;1853:2481–2491. doi: 10.1016/j.bbamcr.2015.05.020. [DOI] [PubMed] [Google Scholar]
- Ohsaki Y, Kawai T, Yoshikawa Y, Cheng J, Jokitalo E, Fujimoto T. PML isoform II plays a critical role in nuclear lipid droplet formation. J Cell Biol. 2016;212:29–38. doi: 10.1083/jcb.201507122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Mahony F, Wroblewski K, O’Byrne SM, Jiang H, Clerkin K, Benhammou J, Blaner WS, Beaven SW. Liver X receptors balance lipid stores in hepatic stellate cells through Rab18, a retinoid responsive lipid droplet protein. Hepatology. 2015;62:615–626. doi: 10.1002/hep.27645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peramuna A, Summers ML. Composition and occurrence of lipid droplets in the cyanobacterium Nostoc punctiforme. Arch Microbiol. 2014;196:881–890. doi: 10.1007/s00203-014-1027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploegh HL. A lipid-based model for the creation of an escape hatch from the endoplasmic reticulum. Nature. 2007;448:435–438. doi: 10.1038/nature06004. [DOI] [PubMed] [Google Scholar]
- Rowe ER, Mimmack ML, Barbosa AD, Haider A, Isaac I, Ouberai MM, Thiam AR, Patel S, Saudek V, Siniossoglou S, Savage DB. Conserved amphipathic helices mediate lipid droplet targeting of perilipins 1-3. J Biol Chem. 2016;291:6664–6678. doi: 10.1074/jbc.M115.691048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Fukasawa M, Yamakawa Y, Natsume T, Suzuki T, Shoji I, Aizaki H, Miyamura T, Nishijima M. Proteomic profiling of lipid droplet proteins in hepatoma cell lines expressing hepatitis C virus core protein. J Biochem. 2006;139:921–930. doi: 10.1093/jb/mvj104. [DOI] [PubMed] [Google Scholar]
- Shi ST, Polyak SJ, Tu H, Taylor DR, Gretch DR, Lai MMC. Hepatitis C virus NS5A colocalizes with the core protein on lipid droplets and interacts with apolipoproteins. Virology. 2002;292:198–210. doi: 10.1006/viro.2001.1225. [DOI] [PubMed] [Google Scholar]
- Ueno M, Shen WJ, Patel S, Greenberg AS, Azhar S, Kraemer FB. Fat-specific protein 27 modulates nuclear factor of activated T cells 5 and the cellular response to stress. J Lipid Res. 2012;54:734–743. doi: 10.1194/jlr.M033365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Meene AM, Hohmann-Marriott MF, Vermaas WF, Roberson RW. The three-dimensional structure of the cyanobacterium Synechocystis sp. PCC 6803. Arch Microbiol. 2006;184:259–270. doi: 10.1007/s00203-005-0027-y. [DOI] [PubMed] [Google Scholar]
- Waltermann M, Steinbuchel A. Neutral lipid bodies in prokaryotes: recent insights into structure, formation, and relationship to eukaryotic lipid depots. J Bacteriol. 2005;187:3607–3619. doi: 10.1128/JB.187.11.3607-3619.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waltermann M, Hinz A, Robenek H, Troyer D, Reichelt R, Malkus U, Galla HJ, Kalscheuer R, Stoveken T, von Landenberg P, Steinbuchel A. Mechanism of lipid-body formation in prokaryotes: how bacteria fatten up. Mol Microbiol. 2005;55:750–763. doi: 10.1111/j.1365-2958.2004.04441.x. [DOI] [PubMed] [Google Scholar]
- Wan HC, Melo RC, Jin Z, Dvorak AM, Weller PF. Roles and origins of leukocyte lipid bodies: proteomic and ultrastructural studies. FASEB J. 2007;21:167–178. doi: 10.1096/fj.06-6711com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang Y, Liang Y, Li J, Liu Y, Zhang J, Zhang A, Fu J, Jiang G. Specific accumulation of lipid droplets in hepatocyte nuclei of PFOA-exposed BALB/c mice. Sci Rep. 2013;3:2174. doi: 10.1038/srep02174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhou XM, Ma X, Du Y, Zheng L, Liu P. Construction of nano-droplet/adiposome and artificial lipid droplets. ACS Nano. 2016;10:3312–3322. doi: 10.1021/acsnano.5b06852. [DOI] [PubMed] [Google Scholar]
- Welte MA. Expanding roles for lipid droplets. Curr Biol. 2015;25:R470–481. doi: 10.1016/j.cub.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolk CP. Physiology and cytological chemistry blue-green algae. Bacteriol Rev. 1973;37:32–101. doi: 10.1128/br.37.1.32-101.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Ding YF, Chen Y, Zhang SY, Huo CX, Wang Y, Yu JH, Zhang P, Na HM, Zhang HN, Ma YB, Liu PS. The proteomics of lipid droplets: structure, dynamics, and functions of the organelle conserved from bacteria to humans. J Lipid Res. 2012;53:1245–1253. doi: 10.1194/jlr.R024117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Na H, Liu Z, Zhang S, Xue P, Chen Y, Pu J, Peng G, Huang X, Yang F, Xie Z, Xu T, Xu P, Ou G, Zhang SO, Liu P. Proteomic study and marker protein identification of Caenorhabditis elegans lipid droplets. Mol Cell Proteomics. 2012;11:317–328. doi: 10.1074/mcp.M111.016345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Yang L, Ding Y, Wang Y, Lan L, Ma Q, Chi X, Wei P, Zhao Y, Steinbuchel A, Zhang H, Liu P. Bacterial lipid droplets bind to DNA via an intermediary protein that enhances survival under stress. Nat Commun. 2017;8:15979. doi: 10.1038/ncomms15979. [DOI] [PMC free article] [PubMed] [Google Scholar]