Abstract
Ferrochelatase-1 as a terminal enzyme of heme biosynthesis regulates many essential metabolic and physiological processes. Whether FC1 is involved in plant response to salt stress has not been described. This study shows that Arabidopsis overexpressing AtFC1 displays resistance to high salinity, whereas a T-DNA insertion knock-down mutant fc1 was more sensitive to salt stress than wild-type plants. AtFC1 conferred plant salt resistance by reducing Na+ concentration, enhancing K+ accumulation and preventing lysis of the cell membrane. Such observations were associated with the upregulation of SOS1, which encodes a plasma membrane Na+/H+ antiporter. AtFC1 overexpression led to a reduced expression of several well known salt stress-responsive genes such as NHX1 and AVP1, suggesting that AtFC1-regulated low concentration of Na+ in plants might not be through the mechanism for Na+ sequestration. To investigate the mechanism leading to the role of AtFC1 in mediating salt stress response in plants, a transcriptome of fc1 mutant plants under salt stress was profiled. Our data show that mutation of AtFC1 led to 490 specific genes up-regulated and 380 specific genes down-regulated in fc1 mutants under salt stress. Some of the genes are involved in salt-induced oxidative stress response, monovalent cation-proton (Na+/H+) exchange, and Na+ detoxification.
Introduction
Soil salinity is one of the major environmental problems that seriously cause osmotic stress and ionic toxicity, thus limiting the productivity of crops. Plants have to develop remarkable capabilities for adapting themselves to adverse environments1. Plants also evolve various strategies to cope with the environmental challenge via perceiving stressful signals and transmitting them through diverse metabolic pathways; and upon receipt of the signal, a number of molecular and cellular responses are initiated2.
Plant growth responds to salt stress in three phases: Phase 0: initial response to NaCl; Phase I: osmotic phase that physiologically inhibits growth of young leaves; and Phase II: ionic phase that facilitates senescence of mature leaves3. Phase 0 occurs during the first minutes to hours after salt exposure. There are transient changes in turgor, growth, membrane potential4. During the first phase, ion accumulation in root medium may reduce the water potential and water availability to plants5. The second phase occurs when vacuoles no longer sequester incoming salts, but the concentration of toxic ions rises rapidly in leaves6. Na+ is the main toxic ion. Accumulation of excessive Na+ in cytosol is detrimental to many metabolic and physiological processes. Thus, maintaining a low concentration of cytoplasmic Na+ under salt stress is critical for plant growth and development2. Plants prevent Na+ accumulation in symplast through multiple ways such as extrusion, influx restriction and vacuolar sequestration7. Na+ efflux is catalyzed by a plasma membrane Na+/H+ antiporter encoded by Salt Overly Sensitive 1 (SOS1)8. SOS1 is mainly expressed in epidermis of root tip regions and is responsible for the long-distance Na+ transport from roots to shoots in xylem parenchyma to protect cells from Na+ toxicity8. Sequestration of Na+ into vacuoles is catalyzed by a vacuolar Na+/H+ antiporter, by which energy-consuming H+ transporting pumps such as H+-ATPase and H+-PPase are involved9. Reducing salt-induced toxicity in plants needs numerous resistant genes working together. Recent genome-wide profiling of transcriptome results in identification of a large number of genes in response to salt stress in plants10.
Ferrochelatase 1 (FC1, EC4.99.1.1) is the terminal enzyme of heme biosynthesis, catalyzing the insertion of ferrous iron into protoporphyrin IX11. Protoporphyrin IX is the branch point of the tetrapyrrole biosynthesis pathway of heme and chlorophyll. While chlorophyll is well known for its functions associated with solar energy absorption, transfer and photosynthesis in plants, heme is one of the important tetrapyrroles with functions associated with fundamental cellular aspects12. It is apparent that FC1 is a critical enzyme in the tetrapyrrole biosynthesis pathway for coordination of heme and apoprotein production13. FCs are conserved in organisms. In Arabidopsis and several other plants such as barley and cucumber, only two isoform genes of FC are available11,14–18. While AtFC1 (At5g26030) is expressed in both mitochondria and chloroplasts, AtFC2 (At2g30390) is only present in chloroplasts11,16. Although AtFC1 and AtFC2 share high identify of nucleotides (83%) and amino acids (69%), both did originate from a segmental duplication event in the Arabidopsis genome and thus belong to two distinct groups of plant ferrochelatases11,12,18. AtFC1 and AtFC2 have different expression patterns11,13,15,16,19. Compared to AtFC1 that ubiquitously expresses in whole plants, AtFC2 tends to express in stems, flowers and leaves, but not in roots15,16. Identifying promoters in response to biotic and abiotic stresses shows that the leaf AtFC1 promoter activity increased in response to norflurazon (an inhibitor of carotenoid biosynthesis), wounding and viral infection, whereas AtFC2 promoter activity was in most instances repressed19. Furthermore, expression of AtFC1 could be triggered by reagents generating reactive oxygen species (ROS), inhibitors of cytoplasmic protein synthesis and other environmental stresses13,18. These results suggest that FC1 is mainly involved in stress responses, whereas FC2 is thought of generating heme for photosynthetic cytochromes15,16,18,20. In this paper, we showed that AtFC1 could positively regulate Arabidopsis resistance to salinity stress. AtFC1 overexpression improved seed germination and primary root elongation and reduced Na+ accumulation under salt stress, whereas the loss of function fc1 mutant had adverse phenotypes. AtFC1 overexpression altered expression of a set of genes responsible for Na+ uptake, efflux and detoxification in plants.
Results
Expression of AtFC1 is upregulated by salt stress
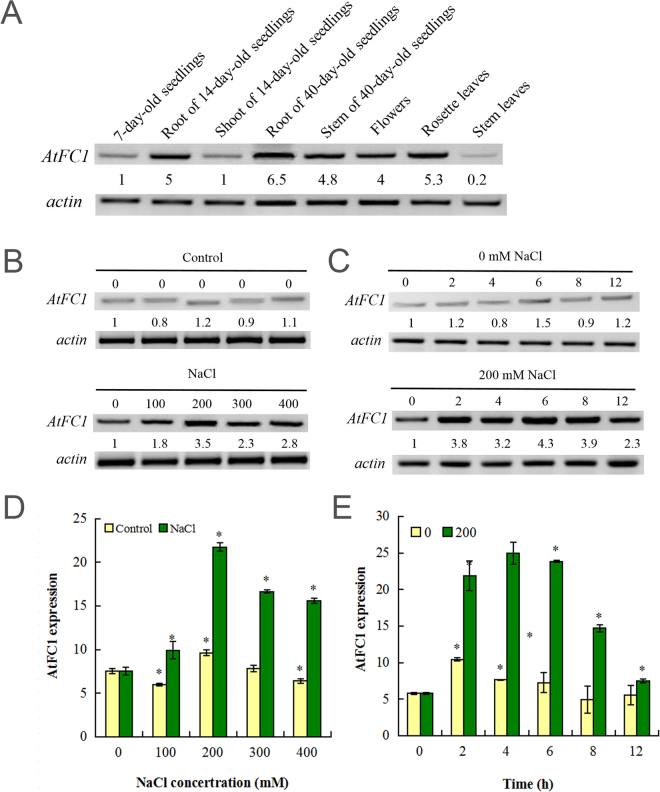
The transcriptional expression pattern of AtFC1 was analyzed during plant growth and development. RT-PCR analysis showed that AtFC1 was ubiquitously expressed, including early or late developing cotyledons, roots, stems, shoots, leaves and flowers, but the abundance of their transcripts varied considerably (Fig. 1A). Roots showed a high abundance of AtFC1 transcripts. Compared to young seedlings, mature stems and flowers also had a higher AtFC1 expression level.
Figure 1.
Expression pattern of AtFC1 in Arabidopsis and its regulation by NaCl treatment. (A) Semi-quantitative RT-PCR (sqRT-PCR) analysis of AtFC1 expression at different developmental stages. B-E: sqRT-PCR and qRT-PCR analyses of AtFC1 expression in response to salt stress. Two week-old seedlings growing in MS media were exposed to 0–400 mM NaCl for 5 h (B,D) or 200 mM NaCl for 0–12 h (C,E). Total RNA was extracted from whole plants and transcripts were analyzed by sqRT-PCR (B,C) and qRT-PCR (D,E). Actin primers were used in PCR as an internal control. The number below the band indicates the relative abundance of the corresponding transcripts with respect to the loading control. Vertical bars represent mean values ± SE. Asterisk indicates the significant difference in expression between the treatments and control (p < 0.05). The color of images for sqRT-PCR was inversed. The uncropped images are shown in Supplementary Data 8.
Several studies have indicated that expression of AtFC1 was induced by wounding, ozone and oxidative stress13,19,21,22. To investigate the expression pattern of AtFC1 under salt stress, transcript levels of AtFC1 were assessed by RT-PCR. As shown in Fig. 1B, exposure of two-week old Arabidopsis seedlings to 100–400 mM NaCl induced AtFC1 expression by 1.8–3.5 fold. The time-kinetics study, in which seedlings were treated with 200 mM NaCl for 12 hours, also showed 2.3–4.3 fold enhanced expression of AtFC1 under salt stress (Fig. 1C). These results were well confirmed by qRT-PCR (Fig. 1D,E).
Overexpression of AtFC1 in Arabidopsis enhances salt resistance
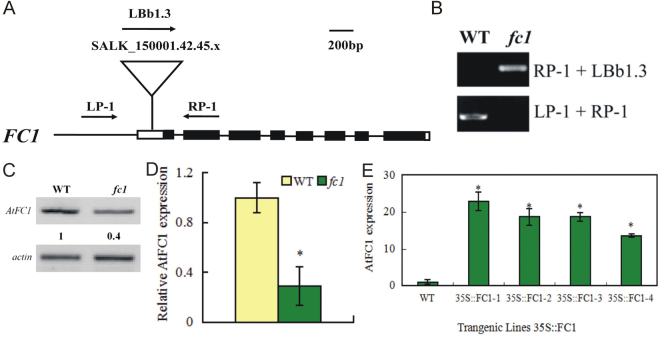
To test the biological function of AtFC1 in Arabidopsis in response to salt stress, a T-DNA insertion mutant of AtFC1 (SALK-150001.42.45x, fc1) was identified. The mutant fc1 was verified by diagnostic PCR using gene-specific primers and found with a T-DNA insertion in the 5’-untranslated region (Fig. 2A). The RT-PCR analysis proved the right T-DNA insertion in the homozygous mutant (Fig. 2B). AtFC1 transcripts in fc1 were assessed by RT-PCR, showing substantial decrease compared to the wild-type (Fig. 2C,D), which was in a good agreement with the recent report18. We further generated transgenic lines overexpressing AtFC1 driven by the cauliflower mosaic virus (CaMV) 35 S promoter. The homozygous transgenic lines were filtered in. The AtFC1 mRNA level of the 35 S::AtFC1 transgenic lines was confirmed by qRT-PCR. The transgenic plants carrying 35 S::AtFC1 showed 13.5 to 22.8-fold higher transcripts of AtFC1 than the wild-type (WT) plants (Fig. 2E).
Figure 2.
Identification of fc1 mutant and 35 S::AtFC1 transgenic lines in Arabidopsis. (A) Schematic diagram of AtFC1 structure and T-DNA diagnostic PCR and RT-PCR. The coding region and 5’ and 3’ untranslated regions are illustrated by the black and white boxes, respectively; introns are indicated by lines. The position of the T-DNA insertion is indicated by a triangle. The locations of the primer pairs used to analyze the mutation by RT-PCR are indicated by arrows. (B) RT-PCR analysis of the fc1 insertion mutant. The reverse transcription products were PCR-amplified using primer pairs LP-1 + RP-1, LP-1 + LBb1.3. (C) RT-PCR analysis of AtFC1 transcript levels in fc1 mutant. (D) qRT-PCR analysis of AtFC1 transcript levels in fc1 mutant. E: qRT-PCR analysis of AtFC1 transcript levels in 35 S::AtFC1 lines. Fourteen day-old seedlings were used for RT-PCR analysis. The number below the band indicates the relative abundance of the corresponding proteins with respect to the loading control. Vertical bars represent mean values ± SE. Asterisk indicates the significant difference in expression between the fc1 mutant/35 S::AtFC1 lines and wild type (p < 0.05). The color of image in C was inversed. The uncropped images are shown in Supplementary Data 9.
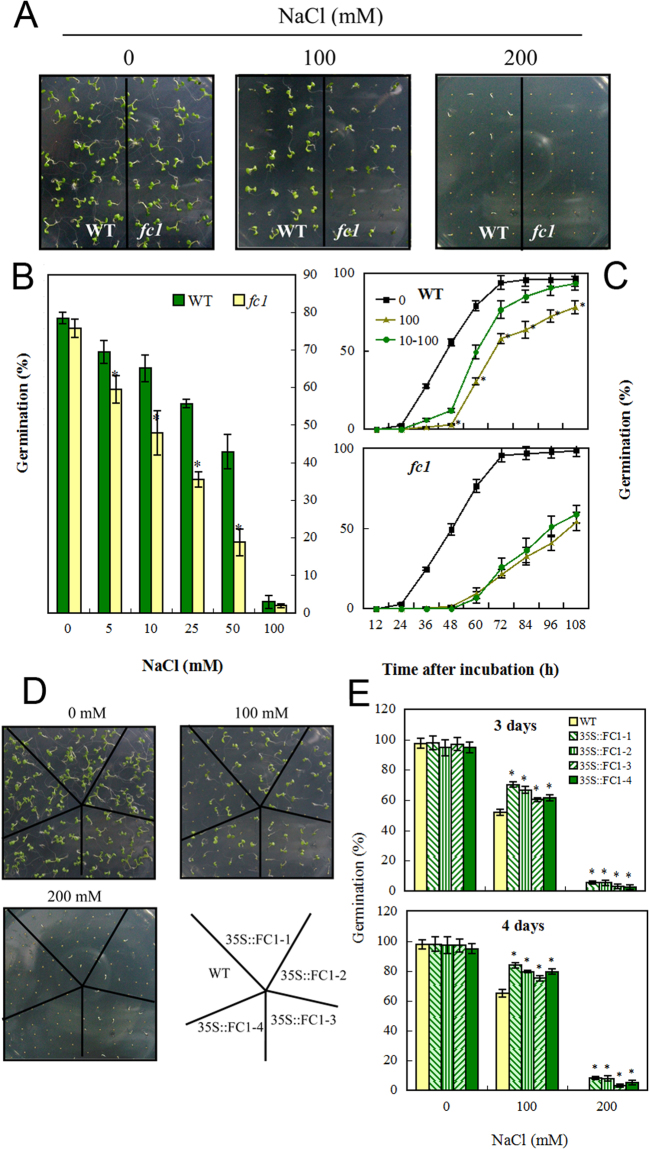
Seed germination in response to salt stress was determined in wild-type, fc1 mutants and AtFC1-overexpression plants. Seeds were placed on the solid 1/2 MS medium supplemented with 0, 100 and 200 mM NaCl. First, the response of wild-type and fc1 mutants to salt stress was compared. The fc1 mutants always had a lower germination rate under salt stress (Fig. 3A,B). Further, a salt acclimation experiment was made with WT and fc1 mutant seeds before exposure to 100 mM NaCl. Seeds were pretreated with 10 mM NaCl for 48 h. The WT seeds had a rapid germination after the 48 h salt acclimation, whereas fc1 seeds had no significant increase in germination after that (Fig. 3C), indicating that loss of AtFC1 function impaired seed germination. We then studied the effect of AtFC1 overexpression on seed germination under salt stress. The seed germination responded differently to NaCl stress. The 35 S::AtFC1 lines always had higher germination rates than WT under salt stress (Fig. 3D,E).
Figure 3.
Germination responses of fcl mutants and 35 S::AtFC1 plants to salt stress. (A) Phenotypes of wild-type (WT) and fc1 mutant seeds exposed to 0, 100 and 200 mM NaCl for 7 d. (B) Quantification of germination rates of WT and fc1 seeds with 0–100 mM NaCl for 2 d. C: Quantification of germination rates of WT and fc1 seeds in an acclimation way of 10 mM NaCl pre-treatment for 48 h (circles) and then 100 mM NaCl to the end of treatment (108 h) (triangles). Both 0 (square) and 100 (circles) mM NaCl treatments were set as control. D: Phenotypes of wild-type (WT) and 35 S::AtFC1 lines exposed the indicated concentrations of NaCl for 7 d. E: Quantification of germination rates of WT and fc1 seeds with 0, 100 and 200 mM NaCl for 3 or 4 d. Vertical bars represent mean values ± SE. Asterisk indicates the significant difference in germination rate between fc1 mutants/35 S::AtFC1 lines and wild type (p < 0.05).
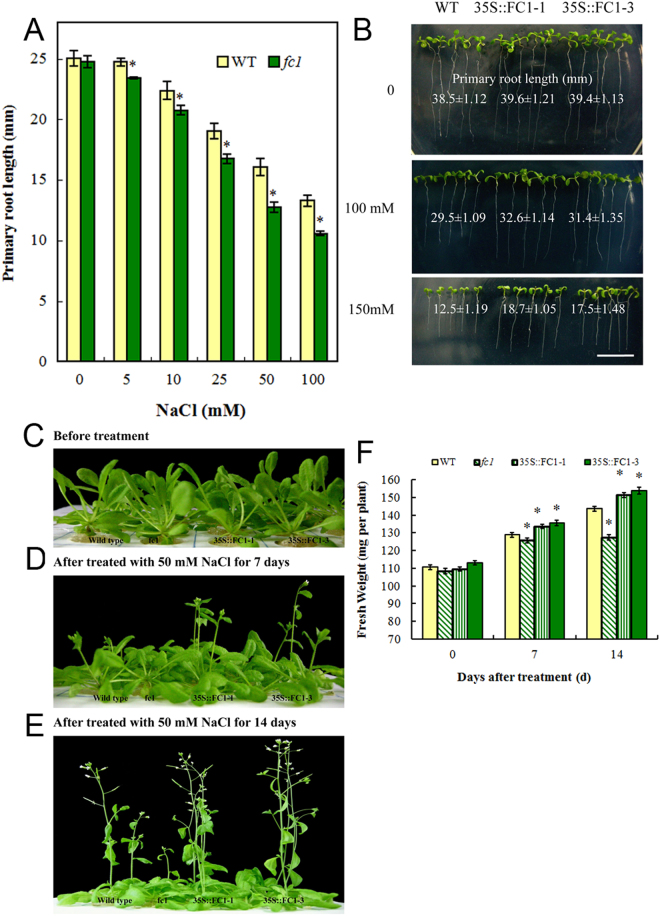
Root growth is sensitive to NaCl and is often used as a biomarker of salt stress23. Under 5–100 mM NaCl exposure for 7 d, the primary root elongation of fc1 mutants was relatively weak compared to WT (Fig. 4A). In contrast, 35 S::FC1 plants displayed strong root growth compared to the wild-type under salt stress (Fig. 4B). Under 150 mM NaCl stress, elongation of 35 S::AtFC1 roots was 1.4–1.5 fold higher than that of wild-type. The AtFC1-improved plant growth was also found in shoots. Three-week-old WT, fc1 and 35 S::AtFC1 seedlings were treated with 50 mM NaCl for 7 and 14 d. Thereafter, the fresh weight was measured. Compared to WT, the total fresh weight of fc1 mutants was lower, whereas the fresh weight of 35 S::AtFC1 plants was higher (Fig. 4C–E). The fresh weight of 35 S:FC1-1 and 35 S:FC1-3 seedlings with 150 mM NaCl was increased 27–34% compared to WT (Fig. 4F).
Figure 4.
Growth responses of fc1 and 35 S::AtFC1 plants to salt stress. (A) Primary root growth of wild-type and fc1 germinating seedlings with 0–100 mM NaCl for 7 d. (B) Phenotype of elongation of primary roots of 35 S::AtFC1 plants with 0, 100 and 200 mM NaCl for 10 d. (C–E) Phenotype of shoots of three week-old WT, fc1 and 35 S::AtFC1 plants exposed to 0, 100 and 150 mM NaCl for 7 and 14 d. (F) Quantification of fresh weight of plants treated with NaCl under the same condition of (C–E). Vertical bars represent mean values ± SE. Asterisk indicates the significant difference in fresh weight between the fc1/35 S::AtFC1 lines and wild type (p < 0.05).
Overexpression of AtFC1 attenuates electrolyte leakage and proline accumulation
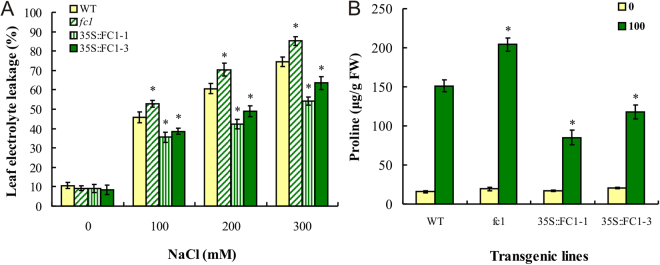
The electrolyte leakage represents the damage of plasma membrane in plant cells and the degree towards abiotic stress response24. Compared to WT, 100–300 mM NaCl induced a higher level of electrolyte leakage in fc1 mutants, whereas a lower level was observed in 35 S::AtFC1 plants, (Fig. 5A).
Figure 5.
Physiological responses of fc1 and 35 S::AtFC1 plants to salt stress. (A) Leaf tissues from three week-old WT, fc1 and 35 S::AtFC1 plants were carefully excised 4 h after 0–300 mM NaCl treatments and used for electrolyte leakage (see Materials and Methods). (B) Three week-old WT, fc1 and 35 S::AtFC1 plants were exposed 0 and 100 mm NaCl for 4 h before determination of free proline concentration. Vertical bars represent mean values ± SE. Asterisk indicates the significant difference between fc1/35 S::AtFC1 and WT plants (p < 0.05).
Proline acts as an osmotprotectant which plays an important role in osmotic balancing and increasing the turgor necessary for cell expansion under salt stress; in addition, it is serves as a redox potential regulator, protecting macromolecules (e.g. proteins or enzymes) from damage or denaturation caused by heat, NaCl, and other stresses2. Similarly, a higher level of proline was found in the fc1 mutants than in WT under NaCl stress; in contrast, the concentration of proline in 35 S::AtFC1 plants was lower than WT (Fig. 5B).
AtFC1 overexpression maintains Na+ and K+ homeostasis under salt stress
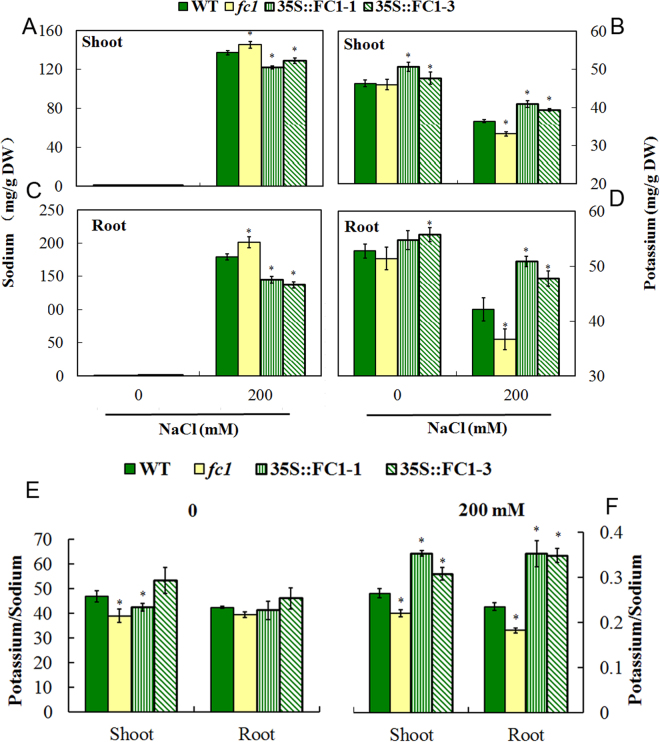
Excessive Na+ is toxic for plants, whereas K+ is an opponent against Na+ under salt stress25. To figure out the physiological role of AtFC1 in resistance to salt stress, three week-old WT, fc1 mutant and 35 S::AtFC1 plants were treated with 200 mM NaCl for 3 d, and the concentrations of Na+ and K+ in roots and shoots were determined by coupled plasma-atomic emission spectrometry. There was no difference of Na+ concentrations in WT, fc1 mutant and 35 S::AtFC1 plants grown in absence of NaCl (Fig. 6A,C). However, when exposed to 200 mM NaCl, the fc1 mutants accumulated more Na+ in shoots and roots, whereas the Na+ concentration in the tissues was relatively lower in 35 S::AtFC1 plants compared to wild-type. In absence of NaCl, no difference of shoot K+ concentrations was observed between WT and fc1 mutants (Fig. 6B); although fc1 mutants accumulated less K+ in roots, there was no significant difference (Fig. 6D). Under control conditions, 35 S::AtFC1 plants accumulated a litter higher level of K+ in plants. However, the 35 S::AtFC1 plants accumulated more K+ in their shoots and roots under salt stress, whereas the K+ concentration was significantly lower in fc1 mutants than in WT. We further analyzed the K+/Na+ ratio which represents the balance of the two ions under salt stress26. It was unclear for K+/Na+ ratio between fc1, 35 S::AtFC1 and WT plants under normal condition. But the imbalance of Na+ and K+ uptake led to an increase in K+/Na+ ratio in 35 S::AtFC1 plants and a decrease in K+/Na+ ratio in fc1 mutants (Fig. 6E,F).
Figure 6.
Effects of AtFC1 overexpression on Na and K accumulation. Three week-old wild type, fc1 mutant and over-expression transgenic plants were treated with 200 mM NaCl for 3 d. Sodium (A,C) and potassium (B,D) contents in shoots and roots were quantified. K+/Na+ ratio in shoots and roots of the 35 S::AtFC1 transgenic lines and wild type after treatment with 0 (E) or 200 mM NaCl (F) for 3 d. Vertical bars represent mean values ± SE. Asterisk indicates the significant difference between fc1/35 S::AtFC1 and WT plants (p < 0.05).
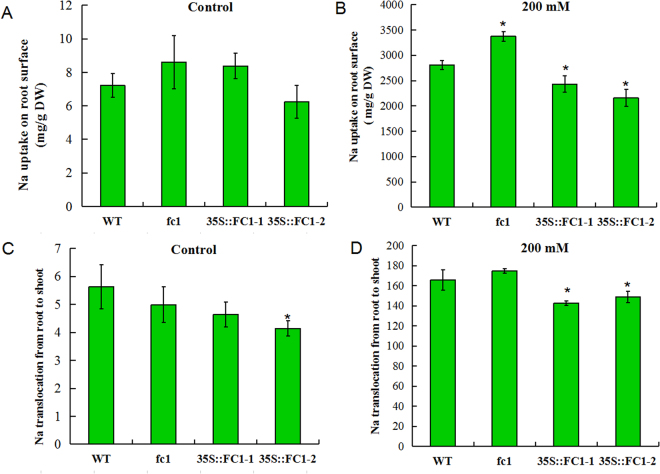
To get an insight into the Na+ location, we analyzed the Na+ uptake on root surface and Na+ translocation from roots to shoots based on the method described previously27. Under normal condition, there was no significant difference of the Na+ uptake on root surface between WT and 35 S::AtFC1 or fc1 mutant plants; however under salt stress (200 mM NaCl), the concentration of Na+ on the root surface was lower in 35 S::AtFC1 plants but higher in fc1 mutant compared to WT (Fig. 7). Also, the AtFC1 overexpressing plants showed less Na+ transfer from roots to shoots. For fc1 mutants, no much difference was found.
Figure 7.
Effect of AtFC1 over-expression on Na+ uptake and Na+ translocation. (A,B) Na+ uptake at root surface under control (A) and 200 mM NaCl (B) conditions. C and D: Na+ translocation from roots to shoots under control (C) and 200 mM NaCl (D) conditions. Vertical bars represent mean values ± SE. Asterisk indicates the significant difference between fc1/35 S::AtFC1 and WT plants (p < 0.05).
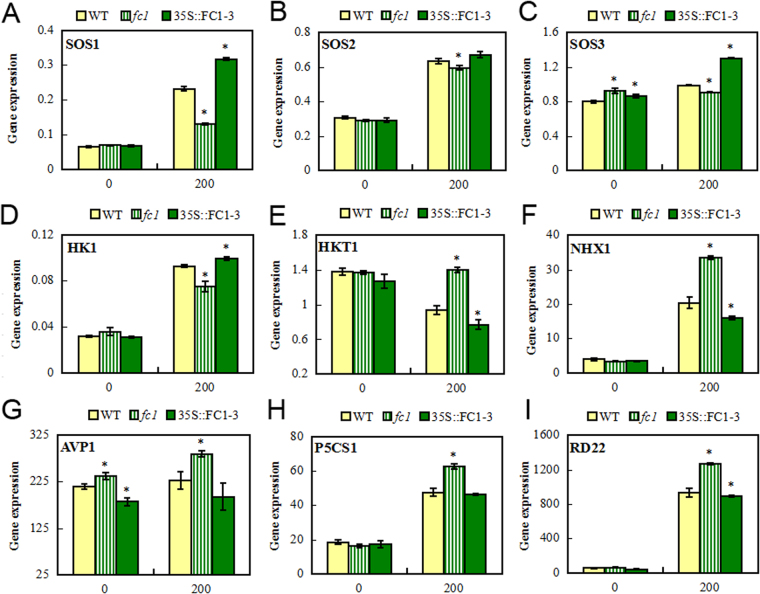
The altered Na+ accumulation due to the AtFC1 expression prompted us to analyze genes responsible for Na+ and K+ acquisition and homeostasis. The SOS (Salt Overly Sensitive) pathway plays an important role in Arabidopsis to regulate Na+ concentration and maintain ionic homeostasis2. SOS1 encodes a plasma membrane-localized Na+/H+ antiporter, functioning in extrusion of Na+ from cells and contributes to salt resistance in Arabidopsis8. SOS2 encodes a member of the CBL-interacting protein kinase family, which combines with SOS328. SOS3 encodes a protein that shares significant sequence similarity with the calcineurin B subunit from yeast and neuronal calcium sensors from animals; this kind of intracellular calcium signaling through a calcineurin-like pathway mediates the beneficial effect of calcium on plant salt tolerance2. qRT-PCR analyses showed that compared to the controls, expression of SOS1 and SOS3 was lower in fc1 mutants, whereas it was higher in 35 S::AtFC1 plants under salt stress (Fig. 8A,C). SOS2 showed a similar expression pattern, but the SOS2 transcription was very weak in 35 S::FC1-3 plants under salt stress (Fig. 8B).
Figure 8.
qRT-PCR analysis of salt stress-responsive genes in fc1 mutant and 35 S::AtFC1 plants. Two week-old wild type, fc1 mutant and 35 S::AtFC1 plants were treated with 0 and 200 mM NaCl for 4 h. Total RNA was isolated from the plants and analyzed by qRT-PCR. The graphs (y-axis) indicate the induction fold of the genes with 200 mM NaCl as compared with the control (0 mM NaCl) (x-axis). Vertical bars represent mean values ± SE. Asterisks indicate that mean values are significantly different between the fc1/35 S:AtFC1 and WT (p < 0.05).
HK1 encodes a member of the histidine kinase family, and is considered as an osmosensor29. Analysis of HK1 transcripts showed that expression of HK1 in salt-treated fc1 mutants was lower than in WT, accounting for about 70% of the WT (Fig. 8D). We further assessed the expression of HKT1, NHX1 and AVP1. HKT1 encodes a sodium transporter expressed in xylem parenchyma cells, but SOS3 and other salt-related protein in the SOS pathway can inhibit the expression of HKT1 to reduce the intracellular concentration of Na+ 2,30. NHX1 encodes a vacuolar Na+/H+ antiporter involved in salt resistance and transports Na+ into the vacuole using the electrochemical gradient of protons generated by the vacuolar H+-translocating enzymes, H+-adenosine triphosphatase and H+-inorganic pyrophosphatase31. AVP1 encodes an H+-translocating (pyrophosphate-energized) inorganic pyrophosphatase located in the vacuolar membrane32,33. Under salt stress, expression of HKT1, NHX1 and AVP1 was higher in fc1 mutants, while their expression in 35 S::AtFC1 plants was lower (Fig. 8E–G). AVP1 was an exception because it had a similar expression pattern under both –NaCl and + NaCl conditions (Fig. 8G). P5CS1 (Delta-1-pyrroline-5-carboxylate synthase 1) is a rate-limiting enzyme in proline biosynthesis, whose mRNA is induced by drought and salinity34. RD22 belongs to the DRE/CRT (drought responsive/C-repeat) elements-containing class of stress-responsive genes35. qRT-PCR analysis showed that both genes had a expression pattern similar to NHX1 in fc1 mutants and 35 S::AtFC1 plants under salt stress (Fig. 8H,I).
Exogenous hematin alleviates tissue damage of fc1 mutant under salt stress
Knowing that AtFC1 was involved in plant salt stress response, we asked whether feeding fc1 with heme, the product of AtFC1 could restore the impaired seed germination caused by NaCl stress. To validate the assumption, the fc1 mutant plants were treated with hematin (protoheme), a highly stable heme substitute36,37. Under salt stress (100 mM NaCl), the fc1 seeds showed a reduced germination rate, but adding 2 μM hematin to the growth media increased the number of germinating seeds (Fig. 9A–C). Addition of hematin led to a 43% recovery in germination rate compared to the control in which no hematin was added. The exogenous hematin could also recover 47% fresh weight of fc1 mutant plants (Fig. 9D). These results indicate that the heme produced by FC1 is partially responsible for resistance of plant to salt stress. Further functional analysis of hematin was performed by assessing Na+ accumulation in fc1 mutant plants. Three week-old WT and fc1 plants were exposed to 100 mM NaCl with or without 2 μM hematin for 3 d. Compared to NaCl treatment alone, concomitant supply of hematin resulted in a significant decrease of Na+ accumulation in fc1 mutants (Fig. 9E). Furthermore, qRT-PCR analysis showed that expression of SOS1 with NaCl was enhanced by hematin (Fig. 9F).
Figure 9.
Effect of exogenous hematin on growth, Na+ concentration and SOS1 expression in fc1 mutants under salt stress. (A,B) Wild-type and fc1 mutant plants after germination were grown in MS medium containing 0 or 2 μM hematin with or without 100 mM NaCl for 14 d. Exogenous hematin effect was assayed as the whole plants fresh weight when treated with or without hematin. C: Germination rate of WT and fc1 seeds with 100 mM NaCl and/or 2 μM hematin for 7 d. D: Fresh weight of WT and fc1 seeds with 100 mM NaCl and/or 2 μM hematin for 7 d. E: Three-week-old wild type and fc1 mutant plants were treated with 100 mM NaCl with and/or 2 μM hematin for 3 days. Then sodium ion contents in whole plants were detected. F: Two week-old WT and fc1 mutant plants were treated with 0 and 200 mM NaCl with and/or 2 μM hematin for 4 h. Total RNA was isolated from the plants and analyzed by qRT-PCR. SOS1 transcript level was analyzed by qRT-PCR. Vertical bars represent Values represent mean values ± SE. Asterisk indicates the significant difference between fc1 mutant and wild type (p < 0.05).
Regulation of FC1 involves extensive transcriptome remodeling
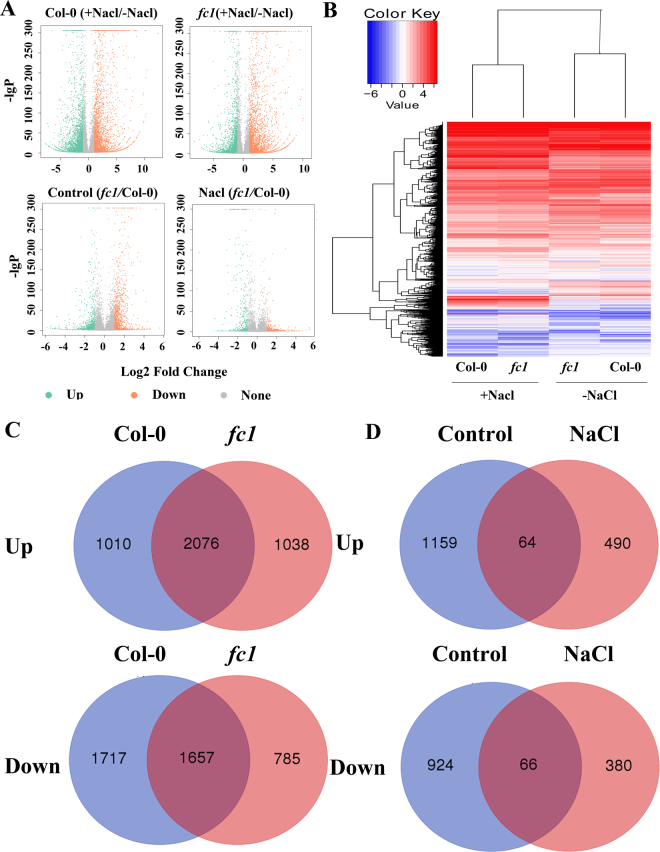
To identify putative FC1 downstream genes associated with the salt stress response in Arabidopsis, we comparatively analyzed the global transcripts of fc1 mutant and wild-type plants by RNA-sequencing. In total, 30.0–31.6 million clean reads were generated in four libraries using Illumina sequencing technology (Supplementary Data 1). Mapping the reads to Arabidopsis genome led to identification of 6460 (3086 up and 3374 down) and 5556 (3114 up and 2442 down) genes in wild-type and fc1 mutant plants (> two fold change, p< 0.05) under salt stress, respectively (Supplementary Data 2–5). Figure 9A shows the distribution of the differentially expressed genes (DEGs) represented by color dots (blue, down, and red, up). Compared to the wild-type Col-0, fc1 mutants under salt stress showed more red dots (more DEGs up-regulated) and less blue dots (less DEGs down-regulated). Under the control (-NaCl) and NaCl treatment, more red dots were detected than blue dots for fcl / Col-0 ratio, indicating that in the fc1 mutant plants more genes were induced relative to wide-type (Col-0) (Fig. 10A). This observation was reflected by the heat-map graph (Fig. 10B), from which the same results could be figured out. To detail the gene expression, Venn diagrams were plotted and showed that 1010 genes were specifically induced and 1717 were repressed in Col-0, while 1038 genes were specifically induced and 785 genes were repressed in fc1 mutant plants under salt stress (Fig. 10C). By comparative analysis of transcripts between fc1 mutant and wild-type plants, 1159 genes were found to be specifically induced and 924 genes were repressed under control condition, whereas 490 genes were specifically induced and 380 genes were repressed under salt stress (Fig. 10D). The expression patterns of some randomly selected genes were well validated by qRT-PCR (Supplementary Data 6)
Figure 10.
Differentially expressed genes in wild-type (WT, Col-0) and fc1 mutant plants under salt stress as determined by mRNA-seq. (A) Differential transcript abundance of NaCl-free and NaCl-treated WT and fc1 mutant plants. The x axis represents the log2 fold change under the mean normalized expression of all transcripts (y axis). Green dots indicate the down-regulated genes and red dots indicate the up-regulated genes. (B) Heatmap representation of a one-dimensional hierarchical clustering of differential gene expression for the NaCl-exposed seedlings (WT and fc1) relative to the control (NaCl-free). (C) Venn diagrams showing up- and down-regulated genes in WT and fc1 mutant seedlings under salt stress relative to control conditions. (D) Venn diagrams showing up- and down-regulated genes in fc1 seedlings relative to WT seedlings grown under the control (0 mM) and 200 mM NaCl treatment.
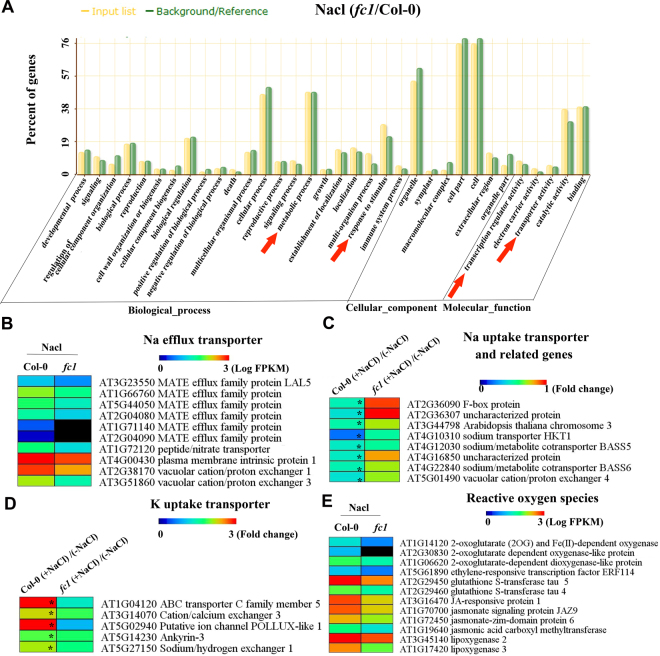
By BGI WEGO (Web Gene Ontology Annotation Plotting), the DEGs from salt-exposed fc1/Col-0 samples were functionally classified. Based on their functional specificity, these DEGs were subdivided into three major groups including biological process, cellular component and molecular function (Fig. 11A). Several categories such as metabolic process, response to stimulus, transcription regulation activity and transporter activity were presented. We further specified some DEGs based on the GO categories. The first group contains genes encoding Na efflux transporters (Fig. 11B), which are responsible for Na exclusion in plants when exposed to excess salt2. The second group comprised genes encoding Na uptake transporters and Na tolerant-related proteins, such as sodium transporter HKT1 and sodium/metabolite co-transporter (Fig. 11C). Compared to WT, these genes in fc1 mutants were induced under salt stress. Potassium is an important ion balancing excessive sodium during plants subjected to salt stress. Examination of K+ uptake transporter genes revealed that five were transcriptionally repressed in fc1 mutants (Fig. 11D). Finally, expression of a group of ROS (reactive oxygen species)-responsive genes was also found to be lower in fc1 mutants than in wild-type (Fig. 11E). These results indicate that disruption of FC1 expression could modify transcription of genes responsible for salt exclusion and detoxification.
Figure 11.
Differential gene expression as determined by mRNA-seq for fc1 seedlings relative to the wild-type (WT, Col-0) under NaCl stress. (A) GO enrichment analysis of specific genes in fc1 mutants which come from the 490 up-expression and 380 down-expression genes of DEGs in fc1 seedlings relative to Col-0 under NaCl stress. (B) Heat map represents the transcript levels of genes encoding Na efflux transporter genes of fc1 mutant and Col-1. (C,D): Heat map represents the DEGs encoding sodium (C) and potassium (D) uptake transporters/related genes in Col-0 and fc1 mutant plants under normal and NaCl stress. Asterisks mean significantly different expressed genes (p < 0.05). (E) Heat map represents the gene expression levels of ROS (Reactive oxygen species)-related genes in Col-0 and fc1 mutant plants.
Discussion
Although several biological and defense functions of FC1 in Arabidopsis and other plant species have been identified in recent years11,12,18,19,38,39, the functional regulation of plant response to salt stress by FC1 has not been described. A recent study using ATH1 microarray chip demonstrated that some genes (e.g. SALT-INDUCIBLE ZINC FINGER 2 and WRKY33) associated with salt stress could be co-regulated by FC1 18, suggesting that FC1 would be involved in the salt stress response. The present study provided genetic and physiological evidence that AtFC1 was able to regulate plant resistance to salt stress. AtFC1 was transcriptionally induced by NaCl. AtFC1 overexpression resisted salt stress by promoting seed germination (Fig. 3E), root and shoot growth (Fig. 4), accumulating less Na+ and more K+ (Fig. 6) in plants. By contrast, AtFC1 loss of function led to adverse phenotypes. Importantly, several genes responsible for Na+/H+ antiporter were up-regulated in 35 S::AtFC1 plants. These results indicate that expression of AtFC1 is required for plant resistance to salt stress.
Minimizing the concentration of cytoplasmic Na+ is critical for plant growth and development under salt stress. Strategies for fighting against excess Na+ in plant cells were proposed1,2. Na+ exclusion is one of the most efficiently resistant mechanisms7. The SOS pathway is one of the most extensively studied mechanisms for controlling Na+ accumulation in plants2. During salt stress, a myristoylated calcium-binding protein encoded by SOS3 presumably senses the salt-elicited calcium signal and translates it to downstream responses40. SOS3 interacts with and activates SOS2, a serine/threonine protein kinase28. Both SOS2 and SOS3 regulate the expression of SOS1, a plasma membrane Na+/H+ antiporter which mediates Na+ efflux8. SOS1 can act as a Na+ sensor of adjusting Na+ long-distance transport from roots to shoots and protecting cells from Na+ toxicity8,41. AtFC1-improved resistance to salt stress was associated with activation of SOSs because these genes, particularly for SOS1 were induced in 35 S::AtFC1 plants. The upregulation of SOS1 might be responsible for the low level of Na+ in the 35 S::AtFC1 plants. We examined cellular and tissue Na+ distribution in transgenic plants and found that AtFC1 overexpression led to a lower concentration of Na+ on the root cellular surface and less Na+ translocated from roots to shoots (Fig. 7). This is consistent with the result that 35 S::AtFC1 plants accumulated less Na in roots and shoots. In addition, RNA-seq profiled many Na efflux transporter genes repressed, while some Na uptake transporter genes were enhanced in fc1 mutant plants (Fig. 11). Several other salt stress-responsive genes, such as NHX1 and AVP1 were examined. Both genes are responsible for sequestration of Na+ into vacuoles31,33. However, expression of NHX1 and AVP1 was down-regulated in 35 S::AtFC1 plants, suggesting that sequestration of Na+ into vacuoles by NHX1 and AVP1 was not be involved in AtFC1-mediated salt tolerance mechanism.
Heme is one of the most important tetrapyrroles in plants as it serves as a cofactor for many enzymes, transporters and proteins involved in the essential biological processes36,39,42. Furthermore, FC1 was shown to be induced with demand for heme for respiratory cytochromes and heme-proteins as part of general defense responses13,18,19. To address the question whether heme is also involved in salt stress response, its regulatory function in this regard was tested by supplying hematin to fc1 mutants and WT plants. Exogenous hematin could partially improve seed germination rate and fresh weight of fc1 mutant plants under salt stress, and addition of hematin could also reduce the Na content in fc1 mutants. These results suggest that AtFC1-improved plant resistance to salt stress not only depended on AtFC1 itself but on heme as well. Although AtFC1 generates heme in plastids, AtFC1 could co-induced with heme-proteins outside plastids, which consequently coordinate the defense response to wounding and ozone-induced oxidative stresses13. One of the striking heme-containing enzymes is heme oxygenase-1 (HO1 or HY1) that regulates biosynthesis of phytochrome by taking advantage of heme as substrate to yields biliverdin IXα, carbon monoxide (CO) and iron43. HO1 was induced by abiotic stresses including salinity and heavy metals36,42. In Arabidopsis, transgenic ho1 mutants overexpressing AtHO1 resisted salt stress by limiting K+ efflux and facilitating H+ efflux44. The enhanced H+ efflux was associated with activation of plasma membrane H+-ATPases (AHA1/2/3) in root epidermis and Na + /H + antiporter (SOS1) in the plasma membrane of AtHO overexpressors44. In this way, AtHO1 acted as an essential component of the salt acclimation signaling pathway36. In addition to HO1, many other proteins such as ascorbate peroxidases and cytochrome P450 reductases with heme as co-enzyme are also involved in plant response to abiotic stress42,43. Thus, AtFC1-improved salt stress resistance likely depends on heme through interacting with heme-proteins.
It is mentionable that AtFC1-regulated salt stress resistance involved potassium homeostasis. Our data showed that the 35 S::AtFC1 plants had a higher concentration of tissue K+, whereas the fc1 mutant plants had a lower concentration of K+ than WT under salt stress. Overload of Na+ can dramatically depolarize the plasma membrane, leading to K+ efflux via depolarization-activated outward rectifying K+ (KOR) channels45. AtFC1 overexpression prevented the loss of K+ in salt-stressed roots. This was reinforced by our RNA-seq datasets, from which a group of K+ uptake transporter genes in response to salt stress was profiled. For example, the cation/calcium exchanger 3 (AtCCX3) identified here has been well characterized as an endomembrane H+-dependent K+ transporter46. ABC transporter C family member 5 (AtABCC5 or AtMRP5) mediating guild cell opening was characterized involving ABA signaling47. Both AtCCX3 and AtABCC5 are involved in abiotic stress responses, but both genes showed significantly lower abundance in fc1 mutants than in WT under salt stress (Fig. 11D). These results suggest that the increased K+ which accompanied the reduced Na+ in 35 S::AtFC1 plants would be mediated by a mechanism for K+ influx and Na+ efflux. Further investigation will be required to highlight the essential role of AtFC1 in mediating K+ and Na+ homeostasis in plants.
To figure out the impact of AtFC1 on its downstream genes and the mechanism for AtFC1 regulation, we profiled transcriptomes of fc1 mutant plants exposed to –NaCl and + NaCl. Our data show that mutation of AtFC1 led to 1038 specific genes up-regulated in fc1 mutants under salt stress; by contrast there were 785 specific genes down-regulated in fc1 mutants. These results indicate that mutation of AtFC1 was able to modify the transcriptional pattern of more genes under salt stress. By profiling the specifically up- or down-regulated genes in fc1 mutants, we show that many genes were involved in biological pathways including cellular components and molecular functions. A fairly number of genes that are responding to or mediate plant response to salt stress under FC1 pathway have been identified. For example, the NaCl-specific responsive genes such as cytochrome P45010, was repressed in fc1 mutants under salt stress, and many other genes related to salt stress response were also found to be altered. The vacuolar-type H + -ATPase (V-ATPase) is a multi-subunit endomembrane proton pump involved in ion vesicle trafficking and adaptation to salt stress42. A V-ATPase (AT4G23710) in salt-exposed fc1 mutant plants was found to be down-regulated (< 2 fold change, p<0.05). Simultaneously, several other types of monovalent cation-proton antiporters including vacuolar cation/proton exchangers or sodium/hydrogen exchangers were found to be transcriptionally repressed. These cation-proton antiporters have been identified to be responsible for cellular Na+ efflux and maintenance of ion homeostasis and excessive Na+ detoxification48.
Taken together, this study identified a new function of AtFC1 that involves plant response to salt stress. Overexpression of AtFC1 conferred plant resistance to NaCl stress. The strong expression of AtFC1, which ensures the supply of the cofactor for a variety of heme-proteins and other putative metabolism, is required in plant resistance to salt stress. The salt stress-induced AtFC1 expression in the tetrapyrrole pathway was associated with activation or suppression of many genes responsible for Na+ exclusion or sequestration, antioxidative stress, phytohormones regulation and other defense components, suggesting a cross-talk between the FC1 pathway and salt-responsive resistance pathway. Thus, our data broaden our understanding of a new role of FC1 in mediating plant resistance to salt stress.
Materials and Methods
Plant materials and growth condition
Arabidopsis thaliana (ecotype Col-0) was used throughout the study. The FC1 (At5g26030) T-DNA insertion mutant fc1 (SALK_15000.142.45.X, Col background) were obtained from the Arabidopsis Biological Resource Center. Seeds were surface sterilized and germinated on half-strength MS medium containing 1 to 3% sucrose and 0.8% phytoagar (pH 5.7) in a growth chamber at 22 °C with 100 μE m−2 s−1 photosynthetically active radiation and a 16 h light/8 h dark cycle. Two week-old seedlings were used for NaCl (100–400 mM) treatments for 0–12 h depending on the experiment conducted.
Mutant analysis
The T-DNA insertion position and homozygous lines were verified according to the instructions on the Salk Signal website (http://signal.salk.edu/isects.html). Primers were designed using the SALK T-DNA verification primer design program (http://signal.salk.edu/tdnaprimers.2.html). For qRT-PCR analysis of the insertion mutants, total RNA was isolated and reverse-transcribed. The reverse transcription products were then PCR-amplified. The internal control was normalized with Actin.
RT-PCR analysis
Total RNA was extracted using column plant RNAout Kit (Tiandz). A 1% agarose gel, stained by ethidium bromide, was run to check the integrity of the RNA. All RNA samples were quantified and examined for protein contamination (A260 nm/A280 nm ratios) and reagent contamination (A260 nm/A230 nm ratios) by a Nanodrop ND 1000 spectrophotometer.
The first strand cDNA was synthesized from 1.0 µg total RNA by Moloney Murine Leukemia Virus Reverse Transcriptase (Promega) using oligo (dT) primers. Transcription of genes was analyzed by quantitative real-time RT-PCR (qRT-PCR) using the fluorescent intercalating dye SYBR-Green in a detection system (MJ Research, Opticon 2) in a final volume of 20 µL containing 2 µL of a 1/10 dilution of cDNA in water, 10 µL of the 2 × SYBR Premix Ex Taq (TaKaRa) and 200 nM of forward and reverse primers (Supplementary Data 7). The thermal cycling conditions were 40 cycles of 95 °C for 5 s for denaturation and 60 °C for 30 s for annealing and extension. All reactions were run in triplicate by monitoring the dissociation curve to control the dimers. PCR efficiency was determined by a series of 2-fold dilutions of cDNAs. The calculated efficiency of all primer pairs was 0.9 to 1.0. Gene Actin 2 was used as a reference and relative expression levels of genes were presented by 2−ΔCT. For semi-quantitative RT-PCR (sqRT-PCR), the gene specific primer (Supplementary Data 7) were used for PCR reactions under the following conditions: pre-denaturation at 94 °C for 5 min, followed by 30 cycles of 30 s at 94 °C, 30 s at a specific annealing temperature (57 °C), and 30 s at 72 °C. As an internal control and to exclude genomic contamination, Actin was amplified (same cycling conditions as above for 28 cycles) from the same cDNA samples.
Transformation of AtFC1 in Arabidopsis
The AtFC1 genomic sequence was inserted into the downstream of the CaMV35s promoter in the binary vector pBI12123. The constructs were then transferred into Agrobacterium tumefaciens for Arabidopsis transformation by the floral dip method. Positive transgenic lines were selected on the 1/2 MS medium with 50 mg/L kanamycin. More than fifteen independent transgenic lines were obtained, and four of them were presented. In this study, the homozygous lines (T4) were used.
Germination assay, root growth and fresh weight measurement
Seeds of wild-type, mutant and transgenic plants were grown on the same plate containing MS medium with or without different concentrations of NaCl or hematin. Plants were grown under conditions of 22 °C with 100 μEm−2s−1 photosynthetically active radiation and a 16 h light/8 h dark cycle. The germination (fully emerged radicle) was recorded. The root length was measured with a ruler, and the fresh mass was weighted at the indicated times.
Analysis of plasma membrane permeability and proline
Plasma membrane permeability of tissues was determined according to the method described previously24. Leaf and root segments were immersed in tubes with deionized water for 30 min, followed by measurement of conductivity of bathing medium (EC1) with a conductivity meter (METTLER TOLEDO FE30-FiveEasy™). Samples were boiled for 20 min and the conductivity of tissues (EC2) was measured. The percent leakage of electrolytes was calculated as the ratio of EC1/EC2. For proline analysis, seedlings were harvested, frozen in liquid nitrogen and dried by lyophilization. Approximately 50 mg of dried seedling tissue was ground in 3% sulfosalicylic acid to extract free proline. Proline concentration was determined as described previously49.
Quantification of ion concentration
Fresh seedlings were harvested and dried at 80 °C, and digested with the mixture of nitric acid and hydrogen peroxide using microwave system (MARS, CEM). The digested samples were used to quantify Na+ and K+ in roots and shoots using inductively coupled plasma-atomic emission spectrometry (ICP-AES) (Optimal 2100DV, Perkin Elmer Instruments). The Na+ uptake (1) and Na+ translocation (2) was calculated as follows27: Na+ uptake = total Na+ content / root dry weight (1); and Na+ translocation = shoot Na+ content / root Na+ content (2).
Preparation of total RNA libraries and mRNA sequencing
Two week-old Arabidopsis seedlings were treated with 0 and 200 mM NaCl and sampled at 1, 2 and 4 h, respectively. Total RNA from NaCl-exposed and NaCl free seedlings was isolated using the TRIzol Reagent (Invitrogen, USA) and pooled for RNA sequencing. The extracted RNA was treated with DNaseI (Qiagen, USA) at 25 °C for 30 min and confirm in quality. mRNA was purified with oligo (dT)-rich magnetic beads and broken into short fragments. The first and second strand cDNAs were synthesized. The cDNAs were end-repaired and phosphorylated using T4 DNA polymerase and Klenow DNA polymerase. The Illumina paired-end solexa adaptors were ligated to these cDNA fragments. The ligated products were purified on a 2% agarose gel. Four libraries (Col-0-NaCl, fc1-NaCl, Col-0 + NaCl and fc1 + NaCl) were sequenced using an Illumina hiseq. 2500 with paired-end of Solexa RNA.
The original image data generated by the sequence providers were transferred into nucleotide sequences data by base calling, defined as raw reads and saved as ‘fastq’ files. All subsequent analyses were performed on the high-quality clean read datasets according to the bioinformatics analysis approach summarized in Supplementary Data 1 50. A rigorous algorithm was used to identify differentially expressed genes (DEGs) between the samples. The expression level for each transcript was calculated as FPKM (fragments per kilobase of exon per million fragments mapped)-derived read counts based on the number of uniquely mapped reads that overlapped with exonic regions51. FDR was used to determine the threshold of the p-value in multiple tests, which corresponded to the differential gene expression test. In this study, FDR ≤ 0.001 and the absolute value of Log2Ratio > 1 were used as a threshold to judge the significant differences of gene expression.
Gene Ontology analysis
The Gene Ontology (GO) category of the DEGs with functional significance was subject to the ultra-geometric test with Benjamini-Hochberg correction (http://www.geneontology.org/). GO terms with corrected p-value < 0.05 were regarded as significant enrichment for the DEGs compared to the genome background.
Statistical analysis
Experiments in the study were independently performed in triplicate. Each result shown in the figures was the mean of three replicated treatments, and each treatment contained at least 15–20 seedlings. Samples for analysis were randomly selected from all transgenic lines. The significant differences between treatments were statistically evaluated by standard deviation and ANOVA methods (p < 0.05).
Electronic supplementary material
Acknowledgements
This research was supported by the Priority Academic Program Development of Jiangsu Higher Education Institutions (200910). We gratefully acknowledge the technique assistance of Annoroad Gene Technology Co., Ltd, Beijing, China.
Author Contributions
Z.M.Y. conceived, designed and wrote the manuscript; F.F. and T.K. revised the manuscript; W.Z., H.L. and N.L.L. carried out the molecular and physiological experiments; S.J.F analyzed the transcriptome data. All authors have read and approved the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Wen Ting Zhao and Sheng Jun Feng contributied equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-13593-9.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Munns. R, Tester M. Mechanisms of salinity tolerance. Annu. Rre. Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- 2.Zhu JK. Regulation of ion homeostasis under salt stress. Curr. Opin. Plant Biol. 2003;6:441–445. doi: 10.1016/S1369-5266(03)00085-2. [DOI] [PubMed] [Google Scholar]
- 3.Munns R. Physiological processes limiting plant growth in saline soils: some dogmas and hypotheses. Plant Cell Environ. 1993;16:15–24. doi: 10.1111/j.1365-3040.1993.tb00840.x. [DOI] [Google Scholar]
- 4.Schubert, S. Salt resistance of crop plants: physiological characterization of a multigenic trait, in Hawkesford, M. J., Barra-clough, P. (eds.): The Molecular and Physiological Basis of Nutrient Use Efficiency in Crops. John Wiley & Sons, Inc., pp. 443–455 (2014).
- 5.Hütsch BW, Saqib M, Osthushenrich T, Schubert S. Invertase activity limits grain yield of maize under salt stress. J. Plant Nutri.Soil Sci. 2014;177:278–286. doi: 10.1002/jpln.201300345. [DOI] [Google Scholar]
- 6.Fortmeier R, Schubert S. Salt tolerance of maize (Zea mays L.): The role of sodium exclusion. Plant Cell Environ. 1995;18:1041–1047. doi: 10.1111/j.1365-3040.1995.tb00615.x. [DOI] [Google Scholar]
- 7.Tester M, Davenport R. Na+ tolerance and Na+ transport in higher plants. Ann.Bot. 2003;91:503–527. doi: 10.1093/aob/mcg058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi H, Quintero FJ, Pardo JM, Zhu JK. The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell. 2002;14:465–477. doi: 10.1105/tpc.010371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fukuda AK, Chiba M, Maeda A, Nakamura M. Effect of salt and osmotic stresses on the expression of genes for the vacuolar H+ pyrophosphatase, H+-ATPase subunit A, and Na+/H+ antiporter from barley. J Exp Bot. 2004;55:585–594. doi: 10.1093/jxb/erh070. [DOI] [PubMed] [Google Scholar]
- 10.Kreps JA, et al. Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 2002;130:2129–2141. doi: 10.1104/pp.008532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki T, et al. Two types of ferrochelatase in photosynthetic and nonphotosynthetic tissues of cucumber: their difference in phylogeny, gene expression, and localization. J Biol.Chem. 2002;15:4731–4738. doi: 10.1074/jbc.M105613200. [DOI] [PubMed] [Google Scholar]
- 12.Papenbrock J, Grimm B. Regulatory network of tetrapyrrole biosynthesis–studies of intracellular signalling involved in metabolic and developmental control of plastids. Planta. 2001;213:667–681. doi: 10.1007/s004250100593. [DOI] [PubMed] [Google Scholar]
- 13.Nagai S, et al. Induction of isoforms of tetrapyrrole biosynthetic enzymes, AtHEMA2 and AtFC1, under stress conditions and their physiological functions in Arabidopsis. Plant Physiol. 2007;144:1039–1051. doi: 10.1104/pp.107.100065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miyamoto K, et al. Nucleotide sequences of cDNA clones encoding ferrochelatase from barley and cucumber. Plant Physiol. 1994;105:769–777. doi: 10.1104/pp.105.2.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith AG, et al. Isolation of a cDNA encoding chloroplast ferrochelatase from Arabidopsis thaliana by functional complementation of a yeast mutant. J. Biol. Chem. 1994;269:13405–13418. [PubMed] [Google Scholar]
- 16.Chow KS, Singh DP, Walker AR, Smith AG. Two different genes encode ferrochelatase in Arabidopsis: mapping, expression and subcellular targeting of the precursor proteins. Plant J. 1998;15:531–541. doi: 10.1046/j.1365-313X.1998.00235.x. [DOI] [PubMed] [Google Scholar]
- 17.Watanabe S, et al. Mitochondrial localization of ferrochelatase in a red alga Cyanidioschyzon merolae. Plant Cell Physiol. 2013;54:1289–1295. doi: 10.1093/pcp/pct077. [DOI] [PubMed] [Google Scholar]
- 18.Scharfenberg M, et al. Functional characterization of the two ferrochelatases in Arabidopsis thaliana. Plant Cell Environ. 2015;38:280–298. doi: 10.1111/pce.12248. [DOI] [PubMed] [Google Scholar]
- 19.Singh DP, Cornah JE, Hadingham S, Smith AG. Expression analysis of the two ferrochelatase genes in Arabidopsis in different tissues and under stress conditions reveals their different roles in haem biosynthesis. Plant Mol. Biol. 2002;50:773–788. doi: 10.1023/A:1019959224271. [DOI] [PubMed] [Google Scholar]
- 20.Espinas NA, et al. Allocation of heme is differentially regulated by ferrochelatase isoforms in Arabidopsis cells. Front. Plant Sci. 2016;7:1326. doi: 10.3389/fpls.2016.01326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Phung TH, et al. Porphyrin biosynthesis control under water stress: sustained porphyrin status correlates with drought tolerance in transgenic rice. Plant Physiol. 2011;157:1746–1764. doi: 10.1104/pp.111.188276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim JG, et al. Increased expression of Fe-chelatase leads to increased metabolic flux into heme and confers protection against photodynamically induced oxidative stress. Plant Mol. Biol. 2014;86:271–287. doi: 10.1007/s11103-014-0228-3. [DOI] [PubMed] [Google Scholar]
- 23.Song JB, et al. miR394 and LCR are involved in Arabidopsis salt and drought stress responses in an abscisic acid-dependent manner. BMC Plant Biol. 2013;13:210. doi: 10.1186/1471-2229-13-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belkhadi A, et al. Effects of exogenous salicylic acid pre-treatment on cadmium toxicity and leaf lipid content in Linum usitatissimum L. Ecotoxicol. Environ. Saf. 2010;73:1004–1011. doi: 10.1016/j.ecoenv.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 25.Zörb C, et al. Molecular characterization of Na+/H+ antiporters (ZmNHX) of maize (Zea mays L.) and their expression under salt stress. J. Plant Physiol. 2005;162:55–66. doi: 10.1016/j.jplph.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 26.Gao S, et al. A cotton miRNA is involved in regulation of plant response to salt stress. Scientific Reposts. 2016;4:6122. doi: 10.1038/srep19736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Britta P, Abdel M, Anja BN, Sven S. Tonoplast Na+/H+ antiporters of newly developed maize (Zea mays) hybrids contribute to salt resistance during the second phase of salt stress. J. Plant Nutr. Soil Sci. 2013;176:148–156. doi: 10.1002/jpln.201200597. [DOI] [Google Scholar]
- 28.Liu J, Ishitani M, Halfter U, Kim CS, Zhu JK. The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc. Natl. Acad. Sci.USA. 2000;97:3735–3740. doi: 10.1073/pnas.97.7.3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Urao T, Yakubov B, Satoh R, Shinozaki Y. A transmembrane hybrid-type histidine kinase in Arabidopsis functions as an osmosensor. Plant Cell. 1999;11:1743–1754. doi: 10.1105/tpc.11.9.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sunarpi HT, Motoda J, Kubo M, Uozumi N. Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na unloading from xylem vessels to xylem parenchyma cells. Plant J. 2005;44:928–938. doi: 10.1111/j.1365-313X.2005.02595.x. [DOI] [PubMed] [Google Scholar]
- 31.Apse MP, Aharon GS, Snedden WA, Blumwald E. Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science. 1999;20:1256–1258. doi: 10.1126/science.285.5431.1256. [DOI] [PubMed] [Google Scholar]
- 32.Sarafian V, Kim Y, Poole RJ, Rea PA. Molecular cloning and sequence of cDNA encoding the pyrophosphate-energized vacuolar membrane proton pump of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 1992;89:1775–1779. doi: 10.1073/pnas.89.5.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gaxiola RA, Li J, Undurraga S, Dang LM, Allen GJ. Drought-and salt-tolerant plants result from overexpression of the AVPl H+-pump. Proc.Natl. Acad.Sci. USA. 2001;98:11444–11449. doi: 10.1073/pnas.191389398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strizhov N, et al. Differential expression of two P5CS genes controlling proline accumulation during saltstress requires ABA and is regulated by ABA1, ABI1 and AXR2 in Arabidopsis. Plant J. 1997;12:557–569. doi: 10.1046/j.1365-313X.1997.00557.x. [DOI] [PubMed] [Google Scholar]
- 35.Shinozaki K, Yamaguchi-Shinozaki K. Gene expression and signal transduction in water-stress response. Plant Physiol. 1997;115:327–334. doi: 10.1104/pp.115.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cao ZY, et al. BnHO1, a haem oxyenase-1 gene from Brassica napus, is required for salinity and osmotic stress-induced lateral root formation. J. Exp. Bot. 2011;62:4675–4689. doi: 10.1093/jxb/err190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H, Song JB, Zhao WT, Yang ZM. AtHO1 is involved in iron homeostasis in a NO-dependent manner. Plant Cell Physiol. 2013;54:1105–1117. doi: 10.1093/pcp/pct063. [DOI] [PubMed] [Google Scholar]
- 38.Sobotka R, Tichy M, Wilde A, Hunter CN. Functional assignments for the carboxyl-terminal domains of the ferrochelatase from Synechocystis PCC 6803: the CAB domain plays a regulatory role, and region II is essential for catalysis. Plant Physiol. 2011;155:1735–1747. doi: 10.1104/pp.110.167528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Woodson JD, Perez-Ruiz JM, Chory J. Heme synthesis by plastid ferrochelatase I regulates nuclear gene expression in plants. Current Biol. 2011;21:897–903. doi: 10.1016/j.cub.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ishitani M, et al. SOS3 function in plant salt tolerance requires myristoylation and calcium-binding. Plant Cell. 2000;12:1667–1677. doi: 10.1105/tpc.12.9.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pardo JM, Cubero B, Leidi EO. Alkali cation exchangers: role in cellular homeostasis and stress tolerance. J. Exp Bot. 2006;57:1181–1199. doi: 10.1093/jxb/erj114. [DOI] [PubMed] [Google Scholar]
- 42.Shen Q, Jiang M, Li H, Che LL, Yang ZM. Expression of a Brassica napus heme oxygenase confers plant tolerance to mercury toxicity. Plant Cell Environ. 2011;34:752–763. doi: 10.1111/j.1365-3040.2011.02279.x. [DOI] [PubMed] [Google Scholar]
- 43.Kikuchi G, Yoshida T, Noguchi M. Heme oxygenase and heme degradation. Biochem Biophy Res Commun. 2005;338:558–567. doi: 10.1016/j.bbrc.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 44.Bose J, Xie Y, Shen W, Shabala S. Haem oxygenase modifies salinity tolerance in Arabidopsis by controlling K+ retention via regulation of the plasma membrane H+ -ATPase and by altering SOS1 transcript levels in roots. J Exp Bot. 2014;64:471–481. doi: 10.1093/jxb/ers343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen, et al. Root Plasma Membrane Transporters Controlling K1/Na1 Homeostasis in Salt-Stressed Barley. Plant Physiol. 2007;145:1714–1725. doi: 10.1104/pp.107.110262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morris J, et al. AtCCX3 Is an Arabidopsis Endomembrane H+ -Dependent K+ Transporter. Plant Physiol. 2008;148:1474–1486. doi: 10.1104/pp.108.118810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nagy R, et al. The Arabidopsis ATP-binding cassette protein AtMRP5/AtABCC5 is a high affinity inositol hexakisphosphate transporter involved in guard cell signaling and phytate storage. J Biol Chem. 2009;284:33614–33622. doi: 10.1074/jbc.M109.030247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kluge C, et al. New insight into the structure and regulation of the plant vacuolar H+ -ATPase. J Bioenerg Biomembr. 2003;35:377–388. doi: 10.1023/A:1025737117382. [DOI] [PubMed] [Google Scholar]
- 49.Bates LS, Waldren RP, Teare ID. Rapid determination of free proline for water stress studies. Plant Soil. 1973;39:205–207. doi: 10.1007/BF00018060. [DOI] [Google Scholar]
- 50.Feng SJ, et al. Variation of DNA methylation patterns associated with gene expression in rice (Oryza sativa) exposed to cadmium. Plant Cell Environ. 2016;39:2629–2649. doi: 10.1111/pce.12793. [DOI] [PubMed] [Google Scholar]
- 51.Feng SJ, et al. Characterization of long non-coding RNAs involved in cadmium toxic response in Brassica napus. RSC Advances. 2016;6:82157–82173. doi: 10.1039/C6RA05459E. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.