Abstract
Recent studies have highlighted great challenges of transgene silencing for transgenic plants facing climate change. In order to understand the impacts of elevated CO2 on exogenous Bacillus thuringiensis (Bt) toxins and transgene expression in transgenic rice under different levels of N-fertilizer supply, we investigated the biomass, exogenous Bt toxins, Bt-transgene expression and methylation status in Bt rice exposed to two levels of CO2 concentrations and nitrogen (N) supply (1/8, 1/4, 1/2, 1 and 2 N). It is elucidated that the increased levels of global atmospheric CO2 concentration will trigger up-regulation of Bt toxin expression in transgenic rice, especially with appropriate increase of N fertilizer supply, while, to some extent, the exogenous Bt-transgene expression is reduced at sub-N levels (1/4 and 1/2N), even though the total protein of plant tissues is reduced and the plant growth is restricted. The unpredictable and stochastic occurrence of transgene silencing and epigenetic alternations remains unresolved for most transgenic plants. It is expected that N fertilization supply may promote the expression of transgenic Bt toxin in transgenic Bt rice, particularly under elevated CO2.
Introduction
Global atmospheric CO2 concentrations have been increasing at an accelerating rate from 280 ppm before industrialization to 402 ppm in recent years (Mauna Loa Observatory: NOAA-ESRL), and are anticipated to reach at least 550 ppm by the year 20501. Also, CO2 has been attracting people’s attention owing to its “greenhouse effects”, which is closely related to more frequent extreme weather events2. Typically, the effects of elevated CO2 on plants are generally characterized by increases in the photosynthetic rate, biomass, and carbon (C): nitrogen (N) ratio, especially in C3 crops3–5. Elevated CO2 can alter plant phenotype and chemistry by inducing changes in assimilation and re-assignment of C and N resources to primary and secondary metabolites in plant tissues, which, in turn, affects life-history and feeding responses of herbivorous insects6–10.
Bacillus thuringiensis (Bt) is a ubiquitous gram-positive and spore-forming bacterium, which produces specific insecticidal crystal proteins known as δ-endotoxins11,12. Based on amino acid sequence similarity and protein function, Bt δ-endotoxins are classified into two major groups, that is, Cry and Crt proteins13. The Cry toxins have been shown toxic towards a wide variety of larval stages of the Lepidoptera, Diptera, Coleoptera, Hymenoptera, Homoptera, Orthoptera, Mallophaga, Nematoda and Acarina14,15. Bt toxins are proven alternatives or supplement to synthetic chemical pesticides in commercial agriculture, forest management, and public health. However, directly transferring wild-type Bt genes into plants resulted in poor expression and potency, with the toxin comprising less than 0.005% of the total proteins in the plant, making transgenic plants still susceptible to insects even if the transgenes were driven by a strong plant promoter16–18. Thus, the usage of synthetic genes to express hybrid Bt toxins is regarded as a promising strategy in genetic engineering. A chimeric Cry1Ab/Ac gene, a fusion of Cry1Ab (GenBank Accession No. ×54939) and Cry1Ac (GenBank Accession No. Y09787) into a single gene, is highly toxic to three important rice lepidopteran pests, including Chilo suppressalis, Scirpophaga incertulas, and Cnaphalocrocis medinalis 19,20. Because of the excellent efficiency against lepidopteran pests and its environmental friendliness, China’s Ministry of Agriculture granted safety certificates in 2009 to two transgenic varieties of rice with hybrid Bt toxins for commercialization: a restorer line (cv. Bt Huahui-1) and a hybrid line (cv. Bt Shanyou-63), both of which expressed fused Cry1Ab/Cry1Ac genes20.
It is speculated that global agriculture will face numerous challenges owing to climate change1. Elevated CO2 can have various effects on different trophic levels, plants, herbivores, and predators/parasitoids3,6,7,9,10,21. Under these circumstances, transgenic plants may become a significantly greater component of cropping systems for sustainable agriculture. However, the performance of transgenic plants, the stability of the transgenic traits, and their ecological interactions have been studied under atmospheres with elevated CO2 22–25. Several studies suggested that the expression of exogenous gene in transgenic plants diverts some nutrients from the normal physiological pathways which could alter the C–N balance, especially under changed abiotic conditions23–26. Coviella and Trumble22 and Tsutsumi et al.27 reported that applied N fertilization can relieve such nutrient diversion. N fertilization is, thus, regarded as an attractive strategy to improve the C-N balance in transgenic plants under elevated CO2 in the future22,23.
Methylation of cytosines in DNA is a common eukaryotic DNA modification that functions as a powerful mechanism to regulate gene expression through gene silencing28–30. Napoli et al.31 and Van der Krol et al.32 introduced CHS (chalcone synthase) gene and DFR (dihydroflavonol-4-reductase) gene into petunia which was expected to overexpress the floral pigmentation. Unexpectedly, the introduced transgene created a blockage in anthocyanin biosynthesis. Napoli et al.31 and Reddy et al.33 demonstrated that the transgene silencing occurred in transgenic plant, and suggested that it might be connected to methylation. Subsequent studies found much more transgene silencing in transgenic plants34,35 as well as in fungi36 and higher order animal taxa37,38. Numerous researches have indicated that transgenes are susceptible to silencing in all studied plant species39,40, including rice41. Silencing resulting from interactions among multiple copies of transgenes or when additional copies of an endogenous gene are expressed ectopically involves homology dependent gene silencing (HDGS). The mechanism of HDGS is not yet clearly understood, though a number of speculative hypotheses have been put forward and extensively reviewed42–44. Basically two forms of transgene silencing have been described, transcriptional gene silencing (TGS), in which gene expression is directly blocked, and posttranscriptional gene silencing (PTGS) in which mRNA is degraded45. Both types of silencing are associated with de novo methylation of cognate sequences: in TGS, DNA methylation occurs in the promoter region46,47, and in PTGS, it is associated with DNA methylation in the coding sequences48–50.
In this study, we simulated the future global atmospheric CO2 concentrations to determine how elevated CO2 affects the Bt-transgene methylation and Bt toxins expression in transgenic Bt rice under different rates of N-fertilizer augmentation. Two hypotheses were issued: (1) The expression of Bt transgene should be down-regulation by hyper-methylation of transgenic rice facing elevated CO2 levels, and (2) N fertilization should relieve or remove the N limitation for transgenic Bt rice under elevated CO2.
Materials and Methods
Plant materials and growth conditions
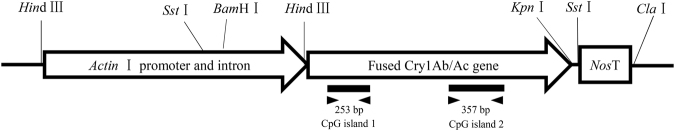
The generalized modified rice used in this experiment was the transgenic restorer line, Huahui-1, which was developed by using Minghui 63 as recipient to harbor the fusion gene Cry 1Ab/Cry1Ac from transgenic event TT51-1 (GenBank Accession Number: EU880444.1), provided by Huazhong Agricultural University (Wuhan, China). The transgene is regulated by the rice actin 1 promoter and the nopaline synthase (NOS) gene terminator (Fig. 1).
Figure 1.
Schematic diagram of the fused Cry1Ab/Ac gene and its plasmid.
Experiments were conducted at Nanjing Agricultural University in two electronically controlled growth incubators (GDN-400D-4/ CO2; Ningbo Southeast Instrument CO., LTD, Ningbo, China) with a gas-tank system that maintained the desired CO2 concentration. The CO2 concentrations in these two growth incubators were set at the current atmospheric CO2 level (400 ± 20 ppm) and at an elevated level (800 ± 50 ppm, slightly higher than the predicted level at the end of this century)1. Within each CO2 level, N-fertilizer sub-plot treatment was set at five levels, including 1/8, 1/4, 1/2, 1, and 2 N; the standard N-fertilizer supply or 1 N was 1.25 mM NH4NO3), delivered through NH4NO3. The entire experiment, thus, consisted of 2 CO2 concentrations ×5 N-fertilizer rates (total 10 treatment combinations). The two growth incubators were alternated by switching CO2 concentration rates as well as swapping the entire content of each chamber every five days in order to equalize the possible bias in plant growth due to incubator-specific growth conditions.
Seed germination was induced on a moist filter paper for 48 h and then the seeds were sown into plastic foam (0.7 cm in thickness) covering the plastic cups (9 cm in diameter and 6.5 cm in height). There were five holes in each piece of plastic foam and two seeds were transferred into each hole (2 rice plants per hole ×5 holes per piece of plastic foam ×1 piece of plastic foam per plastic cup ×4 plastic cups = 40 rice plants per treatment). The plastic cups were filled with modified culture solutions51, and the nutrient solution was renewed daily. The composition of modified culture solutions was as follows (per liter): (1) Macronutrient solution: NH4NO3, 1.25 mM; KH2PO4, 0.3 mM; K2SO4, 1 mM; CaCl2·2H2O, 1 mM; MgSO4·7H2O, 1 mM; Na2SiO3·9H2O, 0.5 mM. (2) Micronutrient solution: MnCl2·4H2O, 9 μM; Na2MoO4·2H2O, 0.39 μM; H3BO3, 20 μM; ZnSO4·7H2O, 0.77 μM; CuSO4·5H2O, 0.32 μM; FeSO4·7H2O + Na2-EDTA, 20 μM. All rice plants were grown at 26.5 ± 1.0 °C with 70 ± 10% humidity at a photoperiod of 14 h: 10 h (light/dark).
The measurement of transgenic Bt rice biomass
At 35 days after planting (DAP), twenty rice seedlings were randomly selected to determine the biomass of aboveground (shoot) and belowground (root) plant tissues. Fresh weights of each of these two plant parts were recorded by using precision scales with an accuracy of ± 0.1 mg (Mettler Toledo AL104). These post-fresh weight samples were collected in the self-sealing bags separately and stored at −80 °C in an ultra-cold storage freezer (Thermo Scientific Forma 702, USA) for the bioassay of relative expression and methylation status of Bt transgene.
The measurement of foliar Bt toxins and total soluble proteins of transgenic Bt rice
On 35 DAP, ten seedlings were randomly selected from the remaining twenty plants in each treatment combination for the measurement of foliar Bt toxins and total soluble proteins. Three leaves from each seedling were separately collected (approximately 30–40 mg total fresh weight), and were then placed into 1.5 ml microtube. All samples were homogenized in a TissueLyser II (Qiagen) by shaking for 2 min at 28 Hz with one steel ball per tube. After homogenization, the extraction buffer PBST was added into the tube at a ratio of 1:10 (tissue weight in g: buffer volume in ml) for Bt toxins test, and 0.9% saline was used as an extraction buffer at a ratio of 1:9 (tissue weight in g: buffer volume in ml) for total soluble proteins test, according to the specification respectively. The supernate of extraction buffer was used for the following test as protein solution. The foliar content of total soluble proteins was determined following the corresponding diagnostic kit A045-2 (Nanjing Jiancheng Bioengineering Institute), and the Bt toxins content was analyzed with an ELISA test (Bt-Cry1Ab/Ac ELISA Kit, Agdia, Elkhart, IN, USA) according to the manufacturer protocol and with Cry1Ab/Ac standards for quantitative determination. Each sample of the above two foliar content tests for Bt toxins and total soluble proteins was replicated three times.
RNA preparation and reverse transcription
Total RNA was extracted from leaf tissues of the sampled seedlings by using TRIzol® reagent (Invitrogen). The concentration and quality of samples was determined by NanoDropTM spectrophotometer (Thermo Scientific) and 1.5% agarose gel electrophoresis. The first-strand complementary cDNA templates were synthesized with 100 ng total RNA by using PrimeScriptTM RT reagent Kit with gDNA Eraser (TaKaRa, Japan). Reverse transcriptase reactions were performed in a 20 μl final volume reaction.
Real-time PCR analysis
Each cDNA product was diluted from 20 µl to 200 µl with RNase-free dH2O, in order to make the Ct value within the suitable range of 15–35 based on preliminary experiments. For fluorescence-based quantitative real-time PCR (qRT-PCR), 2 µl cDNA dilution and 0.2 μM primers were used in 1 × SYBR® Premix Ex Taq TM (TaKaRa, Japan) with 7500 Real-Time PCR Detection System (Applied Biosystems) following the supplier’s instructions. Reactions were performed in a 20 µl final volume. Specific primers for the fusion Cry1Ab/Ac gene were designed by Beacon DesignerTM 7.9 software, and the housekeeping genes actin1 and ubiquitin were used as the internal standard to analyze target gene expression (Primers for qRT-PCR were as shown in Table 1). Quantification of the transcript level of target gene was conducted following the 2−ΔΔCt normalization method52. The expression levels of internal control genes were examined in every PCR plate to eliminate systematic error. Three biological replicates were made for each treatment in the qRT-PCR analysis, and each biological replicate contained three technical repeats.
Table 1.
Primers for qRT-PCR.
| Primer | Sequence (5′-3′) | GeneBank Accession | Description |
|---|---|---|---|
| Cry1Ab/Ac-F | TAGAGTTCGTGTGAGGTA | EU816953 | Bt protein gene |
| Cry1Ab/Ac-R | CTGTATTGGAGAAGATGGAT | ||
| actin1-F | ATGGCAACATTGTGCTCAGTG | Bt13042791 | Rice housekeeping gene |
| actin1-R | CCTCCGATCCAGACGCTGTA | ||
| ubiquitin-F | GCTCCGTGGCGGTATCAT | NC_02925892 | Rice housekeeping gene |
| ubiquitin-R | CGGCAGTTGACAGCCCTAG |
Genomic DNA isolation of transgenic Bt rice leaves
Genomic DNA was isolated and purified from 40 mg leaf tissue samples prepared in foliar Bt toxin and total protein estimation section by using DNAsecure Plant Kit (TIANGEN, Beijing, China), following its manufacturer’s instructions. DNA was quantified by using NanoDropTM spectrophotometer (Thermo Scientific) and stored at −20 °C before use.
Bisulfite genomic sequencing
DNA methylation analysis was performed by the bisulfite sequencing method. Because bisulfite treatment can convert unmethylated cytosines to uracils, it was used to determine the methylation status of cytosines in CG, CHG, and CHH sequences (where H could be A, T, or C). In order to obtain detailed information on the methylation status of cytosine which could represent the integral level of the transgene in silence performance, we used bisulfite sequencing, which allowed detection of mostly methylated cytosine residues within a DNA region of the Bt-transgene body. Two pairs of primers were designed to amplify methylation and non-methylation copies of the transgene, for acquiring major information on methylation of the transgene. Two PCR fragments (CpG island) were analyzed: P1 (CpG island 1) was amplified from the top strand of the transgene, and P2 (CpG island 2) was amplified from the bottom strand (Fig. 1).
Bisulfite treatment of genomic DNA was performed by using the DNA Bisulfite Conversion Kit (TIANGEN, Beijing, China). A total of 0.6 µg genomic DNA was converted for 70 min with the following program: 95 °C for 10 min, 64 °C for 60 min, and reactions were then maintained at 4 °C until the next step. The target sequences were amplified by PCR from the bisulfite-treated DNA (150 ng) by using the Methylation-specific Kit (TIANGEN, Beijing, China). The optimized PCR program consisted of 95 °C for 5 min, 35 cycles of 94 °C for 20 sec, 55 °C for 30 sec, 72 °C for 20 sec, and a final step with 72 °C for 5 min. The PCR products were excised and purified from the gel with AxyPrep DNA Gel Extraction Kit (Axygen, Union City, USA), and cloned into pEASY ®-T3 Cloning Vector using pEASY-T3 Cloning Kit (TransGen, Beijing, China) with Trans 1-T1 Phage Resistant Chemically Competent Cell (TransGen, Beijing, China). At least ten independent clones from each PCR product were sequenced (Table 2). Primers for bisulfite sequencing were designed using Methyl Primer Express Software® (Applied Biosystems). The primer sequences and products are summarized in Fig. 1 and Table 3.
Table 2.
The number of sequenced clones from each PCR product.
| Number (clones) | Nitrogen concentration treatments | |||||
|---|---|---|---|---|---|---|
| 1/8 N | 1/4 N | 1/2 N | 1 N | 2 N | ||
| CO2 treatments | Ambient | 15 | 14 | 19 | 16 | 14 |
| Elevated | 14 | 13 | 10 | 12 | 10 | |
Table 3.
Primers for bisulfite sequencing.
| Primer | Sequence (5′-3′) | GeneBank Accession | Description | Designed by |
|---|---|---|---|---|
| P1-F | TTGGTGTAAATTGAGTAGTTGAT | EU880444.1 | CpG island 1 | Methyl Primer Express Software v1.0 |
| P1-R | ACACRAACAAAAAAAAAACTTA | |||
| P2-F | GGAGAGTATTATTGGTTTGGATA | EU880444.1 | CpG island 2 | Methyl Primer Express Software v1.0 |
| P2-R | CAACCTATAAAAAAATCCTTACCT |
Data Analysis
Statistical analysis of data was performed by using SPSS 20.0 (SPSS Inc., Chicago IL, USA). The biomass of different rice plant parts, total soluble proteins and Bt toxins in foliar contents, and foliar Bt-transgene (i.e., Cry1Ab/Ac) expression levels were analyzed by a two-way analysis of variance (ANOVA) with CO2 concentration as main factor (ambient CO2 vs. elevated CO2) and N-fertilizer supply (5 levels: 1/8, 1/4, 1/2, 1 and 2 N) as sub-factor. If significant effects of CO2 concentration, N-fertilizer supply, and their interactions were found, the two-tailed Student’s t-test was used to compare the means between ambient CO2 and elevated CO2 at P < 0.05, and the least significant difference (LSD) test was used to separate the treatment means. Sequences were manually trimmed and the data analysis performed with the online tool CyMATE (http://cymate.org/).
Results
Aboveground and belowground biomass
The results showed that the aboveground and belowground biomasses of transgenic Bt rice were significantly affected by N supply rates (Table 4). Also, the augmentation of 1 N (1.25 mM NH4NO3) as the standard level of N fertilization was the optimal N condition for the biomass production regardless of the CO2 level. The aboveground and belowground biomass of transgenic Bt rice was increased by elevated CO2 in comparison with ambient CO2 at the 2 N level, but the effect of elevated CO2 on rice seedling biomass decreased at sub-optimal levels of N (i.e., 1/8, 1/4 and 1/2 N), with a conspicuous sharp decrease at 1/2 N (Fig. 2).
Table 4.
F-values and P-values from two-way ANOVA for the effects of CO2 levels and N levels on the fresh weight of rice plants, total soluble protein, Bt toxin content, and Bt gene expression.
| Parameters | df | CO2 | N | CO2 × N | |||
|---|---|---|---|---|---|---|---|
| F-values | P-values | F-values | P-values | F-values | P-values | ||
| Fresh weight of aboveground parts (g) | 199 | 0.148 | 0.701 | 11.677 | 0.000*** | 1.578 | 0.182 |
| Fresh weight of belowground parts (g) | 199 | 0.003 | 0.953 | 9.252 | 0.000*** | 1.422 | 0.228 |
| Total soluble protein (mg/g leaf fresh weight) | 99 | 0.661 | 0.419 | 26.577 | 0.000*** | 14.250 | 0.000*** |
| Bt protein content (mg/g leaf fresh weight) | 99 | 13.937 | 0.000*** | 11.820 | 0.000*** | 2.475 | 0.050* |
| Bt gene expression | 29 | 0.002 | 0.969 | 26.130 | 0.000*** | 10.597 | 0.000*** |
*P < 0.05; ***P < 0.001.
Figure 2.
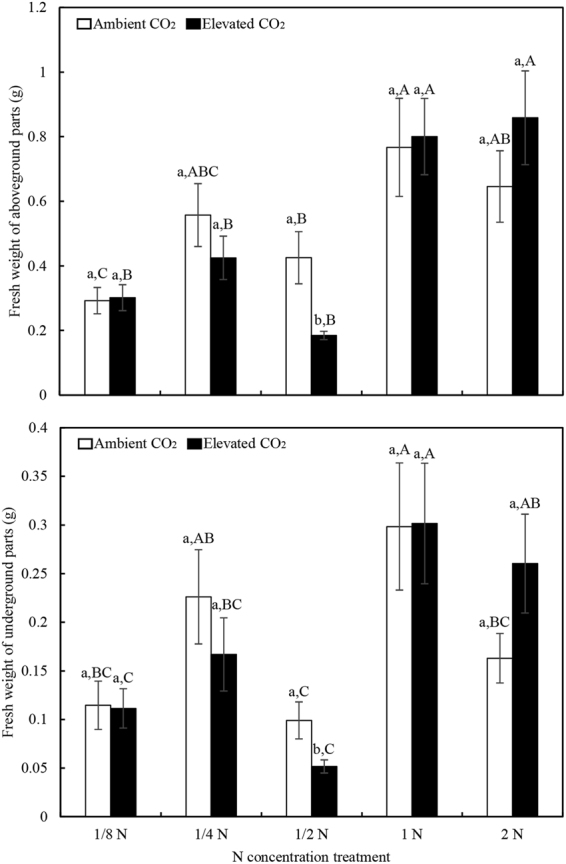
Aboveground (A) and belowground (B) biomass of transgenic Bt rice plants grown under ambient and elevated CO2 at different N-fertilizer augmentation rates. Different lowercase and uppercase letters indicate significant differences between the ambient CO2 and elevated CO2 within N-fertilizer rates, and between the different N-fertilizer rates within CO2 level by LSD test at P < 0.05.
Foliar Bt toxins and total soluble proteins
CO2 level, N augmentation rates, and CO2 × N interactions all significantly influenced foliar Bt toxin content on transgenic rice while N augmentation rates and CO2 × N interactions significantly affected the total soluble proteins (Table 4). Transgenic Bt rice grown under ambient CO2 contained 32.89 μg Bt toxins/g foliar fresh weight at 1/8 N level and showed rising tendency until about 1/2 N level, but the further increase in N rates did not correspondingly increase the Bt toxin (Fig. 3). Compared with ambient CO2, elevated CO2 increased foliar Bt toxin content on transgenic Bt rice with increased N-fertilizer augmentation, and there was significant increase at 2 N level (Fig. 3). However, elevated CO2 treatment showed no significant influence on the foliar total soluble proteins at optimal (standard) and sub-optimal N levels, except at 1/4 N, but significantly increased total soluble proteins was observed at 2 N level (Fig. 4).
Figure 3.
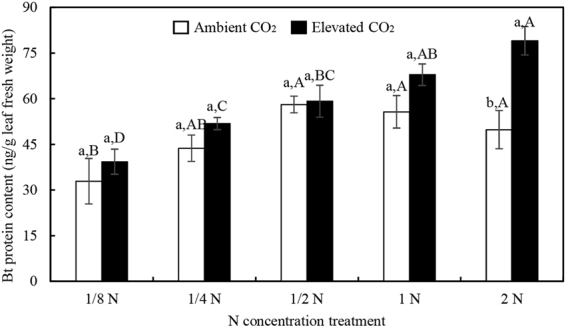
The foliar Bt protein contents of transgenic Bt rice grown under ambient and elevated CO2 with different N-fertilizer augmentation rates. Different lowercase and uppercase letters indicate significant differences between the ambient CO2 and elevated CO2 within N-fertilizer rate, and between the different N-fertilizer rates within CO2 level by LSD test at P < 0.05.
Figure 4.
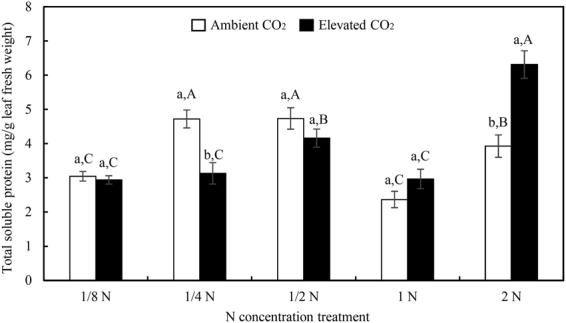
The foliar total soluble protein of transgenic Bt rice grown under ambient and elevated CO2 with different N-fertilizer augmentation rates. Different lowercase and uppercase letters indicate significant differences between the ambient CO2 and elevated CO2 within N-fertilizer rate, and between the different N-fertilizer rates within CO2 level by LSD test at P < 0.05.
Bt transgene expression on leaves
Two-way ANOVA indicated that N-fertilizer augmentation and CO2 × N interactions had significant effects on Bt transgene expression (Table 4). Under ambient CO2 growing conditions, the expression of Bt transgene was up-regulated with increased N-fertilizer supply from 1/8 to 1/2 N, but the transgene expression stabilized beyond the 1/4 N and 1/2 N levels. However, increased N supply significantly up-regulated the Bt-transgene expression beyond optimum supply of nitrogen under elevated CO2, especially at 2 N (Fig. 5). Compared with ambient CO2, elevated CO2 significantly down-regulated the Bt-transgene expression (35.31%) at the lowest N-fertilizer rate (i.e., 1/8 N) while significantly up-regulated the Bt-transgene expression (75.13%) at the highest N-fertilizer rate (i.e., 2 N) (Fig. 5).
Figure 5.
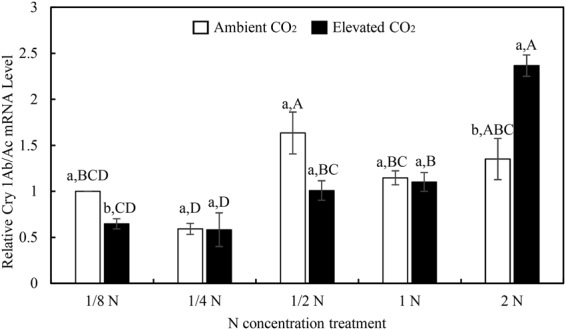
The relative transcript levels of fused Cry 1Ab/Ac gene in leaves of transgenic Bt rice grown under ambient and elevated CO2 with different N-fertilizer rates. Different lowercase and uppercase letters indicate significant differences between the ambient CO2 and elevated CO2 within N-fertilizer rate, and between the different N-fertilizer rates within CO2 level by LSD test at P < 0.05.
Methylation status of Bt-transgene of transgenic Bt rice leaves
The methylation status of Bt transgene (i.e., Cry1Ab/Ac fusion gene) transferred to rice plant showed uneven methylation under different combination treatments of CO2 concentration and N-fertilizer supply (Fig. 6A). Different N supply showed various effects towards the methylation of Bt-transgene body. Under ambient CO2, lowest and highest methylation percentages were observed at 1/8 N (1.28% of cytosines) and 1/4 N (4.01% of cytosines), respectively; methylation was stabilized at ½ N to 2 N range (2.06% at 1 N level to 2.38% at ½ N level of cytosines). In contrast, under elevated CO2, the methylation status showed declining tendency at sub-optimal levels of N (3.18% to 0.64% of cytosines), and then it moved to a hypermethylation status at 2 N level (2.67% of cytosines) (Fig. 6A). Furthermore, at severe nitrogen-deficit (1/8 N) and excessive nitrogen fertility (2 N) conditions, Bt-transgene body appeared to undergo hypermethylation under elevated CO2 (3.18% and 2.67% of cytosines) compared to ambient CO2 (1.28% and 2.12% of cytosines), respectively. In contrast, elevated CO2 (2.44%, 2.04%, and 0.64% of cytosines) led lower methylation in Bt-transgene body than ambient CO2 (4.01%, 2.38%, and 2.06% of cytosines) at intermediate N levels (i.e., 1/4, 1/2, 1 N) (Fig. 6A).
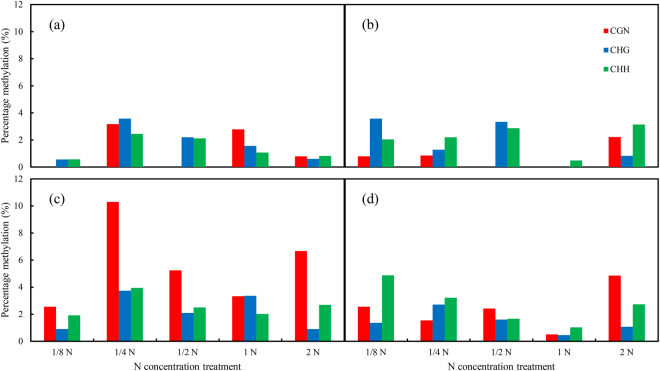
Figure 6.
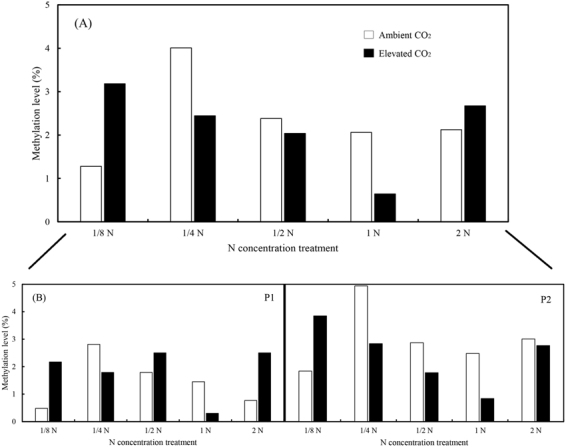
The cytosine methylation of fused Cry 1Ab/Ac transgene in leaves of transgenic Bt rice grown under ambient and elevated CO2 with different N-fertilizer rates. (n = minimum of 10 clones for each treatment; (A) Percentage methylation of the fused Cry 1Ab/Ac transgene (both two fragments); (B) Percentage methylation of P1 and P2 fragments in the fused Cry 1Ab/Ac transgene, respectively).
The integral methylation level of fragment P1 was lower than P2 under same treatments, except for 1/2 N level under elevated CO2. At 1/8, 1/4 and 1 N levels, the methylation status of P1 or P2 manifested a similar tendency as the integral level of this Bt-transgene body under CO2 treatment (Fig. 6B). However, at 1/2 and 2 N levels, elevated CO2 increased methylation frequency in fragment P1, and it had inverse effects on methylation frequency in fragment P2 (Fig. 6B). Moreover, within the 253 bp fragment P1, a total of 9 CG, 12 CHG, and 35 CHH sites were found as potential targets for methylation, while in 357 bp fragment P2, there were 15 CG, 17 CHG and 60 CHH sites as potential targets for methylation. Also, the methylation of cytosines located at CHG and CHH sites appeared to be higher in fragment P1 (Fig. 7, Supplementary Figs 1 and 2). Conversely, the CG site preference was more apparent in fragment P2 compared with a low level of methylation in fragment P1 (Fig. 7, Supplementary Figs 3 and 4).
Figure 7.
Percentage methylation of different methylation patterns (CGN, CHG and CHH) in P1 (A,B) and P2 (C,D) fragments of the fused Cry 1Ab/Ac transgene in leaves of transgenic Bt rice grown under ambient (A,C) and elevated (B,D) CO2 with different N-fertilizer rates.
Discussion
Global climate change has introduced a new challenge to the current plant genetic improvement programs53,54. Previous studies focused on plants grown under elevated CO2 have suggested that there is a link between ‘up-regulation’ and ‘down-regulation’ of plant growth traits55. Moutinho-Pereira et al.56 reported increased leaf tissue thickness in grape plants (Vitis vinifera L.) as a growth response to elevated CO2. Guo et al.57 and Reich et al.58 noted that the elevated CO2 resulted in increased plant biomass in Medicago truncatula (63.3%) and grassland (33%), respectively. While the occurrence of down-regulation of photosynthesis has been shown in several free-air CO2 enrichment (FACE) experiments, it has not been a consistent phenomenon55. A short-term effect of elevated CO2 may be misleading when attempting to predict longer-term effects59. Moreover, the effect of elevated CO2 on plant biomass of transgenic plants is quite complex, e.g., increased leaf biomass, increased leaf area and plant height26,60, in contrast, there was different conjecture based on their results, because heterologous protein produced by transgenic plant, as a kind of toxic molecule, could impair the plant tissues, which in turn progressively damaged host cells61. In this study, we first investigated the interactive effect of various levels of N fertilizer augmentation and elevated CO2 on transgenic rice biomass. The results suggested that there is a positive effect of elevated CO2 on biomass at increased N augmentation (>1 N level), but on the contrary, elevated CO2 resulted in decreased plant biomass under N-deficit production regimes (1/2 and 1/4 N levels). Based on these results, we conclude that elevated CO2 and N-fertilizer supply both make an effect on foliar biomass. It appears that the increased CO2 puts a higher N demand on transgenic rice to achieve the same level of biomass.
At high N levels, CO2 concentration may prove to be a limiting factor affecting plant photosynthesis. Yamori et al.62 and Adachi et al.63 reported that the photosynthesis of rice leaves is limited by CO2 concentration, which showed positive correlation, at carboxylation sites inside chloroplast when nitrogen fertilizer is controlled. During photosynthesis, CO2 molecules in the air diffuse through the stomata into the substomatal cavities and then move via cell walls, plasmalemma, cytosol, and chloroplast envelope membranes to the carboxylation sites in the stroma64. While there is a different response pattern at low N levels, N supply may play a more important factor than CO2 concentration towards plant growth. The effects of CO2 concentration may be weaker when plant growth is limited by nutrient availability58. Several studies have shown that N promotes photosynthesis which has a strong and positive correlation with biomass accumulation by increasing Rubisco content and CO2 diffusion conductance in rice leaves62,65,66. Sub-optimal levels of N may lead to reduction in total fresh, as well as dry weight of leaf blades, leaf sheaths plus stems, and total shoot weight in rice, but the cultivar response to N supply varies67. The N limitation feedback hypothesis suggests that the negative impacts of elevated CO2 on N cycling can constrain responses of plants to elevated CO2 and it may also be induced by the low supply of N67–69. Nguyen et al.67 reported that nitrogen use efficiency (NUE) increased gradually when lowering the N supply from 1 N to 1/8 N, while the NUE was maximized at the 1/2 or 1/4 N levels. Elevated CO2 appeared to have varying effects on the N concentrations in different plant types, with greater reduction of N concentration in C3 plants than in C4 plants. Cotrufo et al.70 reported that plants exposed to elevated CO2 altered their N allocation between above- and below-ground components: root N concentrations were reduced by ~9% compared to ~14% reduction for above-ground tissues. Results of this study are in agreement with prior studies related to theory of multiple-resource-limitation69–72 as we demonstrated the separate effect of CO2 and N, as limiting factors, on plant biomass accumulation in transgenic rice.
Our results indicated that Bt protein content of rice plants under elevated CO2 was higher than that under ambient CO2 regardless of N treatment. In plants, low concentrations of reactive oxygen species (ROS) functions in signal transduction leading to activation of defense responses73, while higher levels of ROS lead to oxidative damage of lipids, DNA, and proteins74. Two plausible pathways exist for elevated CO2 to decrease oxidative stress via reduction in cellular production of ROS. First, increased CO2:O2 ratios within chloroplasts would decrease electron leakage from PSI to O2, thereby attenuating ·O2 − formation, while decreased photorespiration would reduce cellular H2O2 production associated with glycolate metabolism75. Second, growth at elevated CO2 often improves plant water status, which would indirectly decrease antioxidant activities that are stimulated by water stress76. So, the decreased oxidative stress may display less oxidative damage to proteins including Bt protein, as observed in our study. Leaf age may have many problematic effects to plant growth77. Leaf-specific differences were observed in chlorophyll and protein content and starch and metabolite accumulation, and elevated CO2 caused both suppression and promotion of CO2 assimilation within the same plant depending on leaf age78. Thus, the effects of elevated CO2 strongly depend on the developmental state of the leaves. As mentioned, both the aging of leaves and the adaptation towards elevated CO2 contributed to the changes in profiles of chlorophyll, protein, and several plant metabolites79, which might contribute to the results in our study.
In the current study, expression levels and methylation status of Cry1 Ab/Ac fusion gene were determined to estimate the potential mechanism of Bt toxins in response to elevated CO2 and N fertilization treatments. The expression of Bt transgene was up-regulated when rice plants were exposed to excessively sub-optimal N fertility (<1/2 N) and ambient CO2, whereas elevated CO2 significantly up-regulated the Bt-transgene expression only under an excessively high N regime (i.e., 2 N). Previous studies have noted that the expression of protein is controlled by not only mRNA through transcriptional level but also RNA-binding proteins and microRNAs through post-transcriptional level80,81, likewise, the expression of exogenous Bt gene was also regulated through this way. Post-transcriptional regulation is increasingly recognized as a complex system that controls every aspect of RNA metabolism, while it is mediated by the interactions of trans-factors such as RNA-binding proteins and microRNAs with cis-acting elements located in mRNAs. RNA-binding proteins critically regulate mRNA splicing, localization, degradation and translation by binding to short sequences and/or structure motifs in target mRNAs81,82. Likewise, microRNAs can be sequestered and neutralized by the target mRNAs in addition to competition between binding sites on different mRNAs, which shows the fundamental principle of post-transcriptional regulation80,83. We speculate that the difference in Bt expression at various N augmentation rates as modulated by CO2 level may be due to the variation in RNA-binding proteins. Jens and Rajewsky80 reported that mRNA un-translated regions flanked the coding sequence and were bound by post-transcriptional regulators (RNA-binding proteins and microRNAs), which collectively controlled mRNA stability, mRNA localization and protein production. Melanson et al.84 demonstrated that the synthetic novel cis-acting elements from 3′ un-translated region of DNA damage-binding protein 2 can lead to more rapid induction of the reporter mRNA, export of the message to the cytoplasm, and the subsequent accumulation of the encoded reporter protein, which also can affect transcriptional and post-transciptional processes84. Thus, we argue that the post-transcriptional regulation resulted in the difference between ambient CO2 and elevated CO2 treatments within a given N level.
Several explanations have been given that DNA methylation plays an important role in regulating gene expression in transgenic plants as well as plants in general34,35,85. Weinhold et al.34 noted that three independently transformed tobacco lines rapidly lost the expression of the resistance marker and down-regulated transgene expression by more than 200 fold after only one plant generation, while at the same time, the hypermethylation status was displayed within the 35 S and NOS promoters of these lines. Additionally, different methylation patterns (CG, CHG and CHH) may also play an important role in regulating gene expression34,35,86. Previous studies have also indicated that the methylation status may be influenced by external factors, including virus infection35 or nutrition-induced87 plant response factors. Furthermore, environmental factors are also considered as potential inducers to change the methylation status. For example, a sudden cold environment has been shown to trigger the demethylation of hemi-methylated or internally full methylated cytosine in cotton, and this change could be reversed following a subsequent normal temperature88. However, unlike the promoter region, loss of body methylation does not appear to trigger a systematic and drastic over-expression of body-methylated genes to the same extent as transposon reactivation, whereas moderate regulation of body-methylated genes has been observed, suggesting that body methylation may be involved in fine-tuning transcription levels89,90. Our results seem to concur with this phenomenon, which may show a moderate regulation to the expression of Bt gene, because the methylation remain at relatively low level in this study.
In summary, transgene expression in plants is characteristically unpredictable, and depends on many internal and external factors. In this study, the expression of Bt protein toxin transferred in rice was relatively stable under elevated CO2 at different N levels, except at 2 N level. It means there are no significant effects on exogenous Bt toxin affected by elevated CO2 under same N level (below standard N level), but under high N level, elevated CO2 shows promotive effect. In the foreseeable future, it is not expected that the global atmospheric CO2 concentrations will trigger a significant down-regulation of transgene expression in transgenic Bt rice and appropriate increase of N fertilization may be prudent for its expression. It is noteworthy that the foreign protein expression is not reduced under N limitation production scenarios, although the plant growth is restricted and total protein content of the plant is reduced. Ruiz et al.65 showed that the subjecting of genetically modified plants to induction conditions drives the production and accumulation of the recombinant phytotoxic molecule, which in turn progressively damages host cells. This damage depends on the concentration and the length of exposure to the newly synthesized molecule, consequently negatively impacting the biomass and yield. With the development of genetic engineering, the mechanism of exogenous gene expression and post-transcriptional regulation will be further studied. And it is expected that N fertilization supply may promote the expression of Bt toxin in transgenic Bt rice, particularly under elevated CO2.
Electronic supplementary material
Acknowledgements
We thank Prof. Donglei Yang, National Key Laboratory of Crop Genetics and Germplasm Enhancement, Agricultural College, Nanjing Agricultural University, for his help in guiding experiments. This research was supported by the National Basic Research Program of China ‘973’ (2010CB126200), the National Nature Science Foundations of China (31272051 and 31470454), and the Qing Lan Project of Jiangsu Province of China.
Author Contributions
S.L.J. and F.J.C. designed the study; S.L.J., Y.Q.L., Y.D. and T.L. performed the experiments; S.L.J. and F.J.C. analysed the data; S.L.J. wrote the manuscript; S.L.J., F.J.C., M.N.P., G.J.W., L.Q. and A.B.M. reviewed and polished the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-15321-9.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.IPCC. Climate Change 2007: The Physical Science Basis. Contribution of Working Group I to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change. (eds, Solomon, S., Qin, D., Manning, M., Chen, Z., Marquis, M., Averyt, K. B., Tignor, M. & Miller, H. L.). Cambridge University Press, Cambridge, UK and New York, NY, USA (2007).
- 2.Tubiello FN, Donatelli M, Rosenzweig C, Stockle CO. Effects of climate change and elevated CO2 on cropping systems: Model predictions at two Italian locations. Eur. J. Agron. 2000;13(2):179–189. doi: 10.1016/S1161-0301(00)00073-3. [DOI] [Google Scholar]
- 3.Ainsworth EA, Long SP. What have we learned from 15 years of free-air CO2 enrichment (FACE)? A meta-analytic review of the responses of photosynthesis, canopy properties and plant production to rising CO2. New Phytol. 2005;165(2):351–372. doi: 10.1111/j.1469-8137.2004.01224.x. [DOI] [PubMed] [Google Scholar]
- 4.Ainsworth EA, et al. Does elevated atmospheric [CO2] alter diurnal C uptake and the balance of C and N metabolites in growing and fully expanded soybean leaves? J. Exp. Bot. 2007;58(3):579–591. doi: 10.1093/jxb/erl233. [DOI] [PubMed] [Google Scholar]
- 5.Long SP, Ainsworth EA, Leakey AD, Morgan PB. Global food insecurity. Treatment of major food crops with elevated carbon dioxide or ozone under large-scale fully open-air conditions suggests recent models may have overestimated future yields. Philosophical Transactions of the Royal Society of London B: Bilon. Sci. 2005;360(1463):2011–2020. doi: 10.1098/rstb.2005.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbehenn RV, Chen Z, Karowe DN, Spickard A. C3 grasses have higher nutritional quality than C4 grasses under ambient and elevated atmospheric CO2. Global Change Biol. 2004;10(9):1565–1575. doi: 10.1111/j.1365-2486.2004.00833.x. [DOI] [Google Scholar]
- 7.Reich PB, Hungate BA, Luo Y. Carbon-nitrogen interactions in terrestrial ecosystems in response to rising atmospheric carbon dioxide. Annu. Rev. Ecol. Evol. S. 2006;37:611–636. doi: 10.1146/annurev.ecolsys.37.091305.110039. [DOI] [Google Scholar]
- 8.Couture JJ, Servi JS, Lindroth RL. Increased nitrogen availability influences predator–prey interactions by altering host-plant quality. Chemoecology. 2010;20(4):277–284. doi: 10.1007/s00049-010-0058-y. [DOI] [Google Scholar]
- 9.Robinson EA, Ryan GD, Newman JA. A meta-analytical review of the effects of elevated CO2 on plant–arthropod interactions highlights the importance of interacting environmental and biological variables. New Phytol. 2012;194(2):321–336. doi: 10.1111/j.1469-8137.2012.04074.x. [DOI] [PubMed] [Google Scholar]
- 10.Guo H, et al. Elevated CO2 alters the feeding behaviour of the pea aphid by modifying the physical and chemical resistance of Medicago truncatula. Plant Cell Environ. 2014;37(9):2158–2168. doi: 10.1111/pce.12306. [DOI] [PubMed] [Google Scholar]
- 11.Schnepf E, et al. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. R. 1998;62(3):775–806. doi: 10.1128/mmbr.62.3.775-806.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.García-Robles I, et al. Combining hexanoic acid plant priming with Bacillus thuringiensis insecticidal activity against Colorado potato beetle. Int. J. Mol. Sci. 2013;14(6):12138–12156. doi: 10.3390/ijms140612138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crickmore N, et al. Revision of the nomenclature for the Bacillus thuringiensis pesticidal crystal proteins. Microbiol. Mol. Biol. R. 1998;62(3):807–813. doi: 10.1128/mmbr.62.3.807-813.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beegle CC, Yamamoto T. Invitation paper (CP Alexander Fund): History of Bacillus thuringiensis Berliner research and development. Can. Entomol. 1992;124(04):587–616. doi: 10.4039/Ent124587-4. [DOI] [Google Scholar]
- 15.Feitelson JS, Payne J, Kim L. Bacillus thuringiensis: Insects and beyond. Nat. Biotechnol. 1992;10(3):271–275. doi: 10.1038/nbt0392-271. [DOI] [Google Scholar]
- 16.Barton KA, Whiteley HR, Yang NS. Bacillus thuringiensis §-endotoxin expressed in transgenic Nicotiana tabacum provides resistance to lepidopteran insects. Plant Physiol. 1987;85(4):1103–1109. doi: 10.1104/pp.85.4.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vaeck M, et al. Transgenic plants protected from insect attack. Nature. 1987;328:33–37. doi: 10.1038/328033a0. [DOI] [Google Scholar]
- 18.Perlak FJ, Fuchs RL, Dean DA, McPherson SL, Fischhoff DA. Modification of the coding sequence enhances plant expression of insect control protein genes. P. Natl. Acad. Sci. 1991;88(8):3324–3328. doi: 10.1073/pnas.88.8.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng X, Sardana R, Kaplan H, Altosaar I. Agrobacterium-transformed rice plants expressing synthetic cryIA (b) and cryIA (c) genes are highly toxic to striped stem borer and yellow stem borer. P. Natl. Acad. Sci. 1998;95(6):2767–2772. doi: 10.1073/pnas.95.6.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu C. The first approved transgenic rice in China. GM Crops. 2010;1(3):113–115. doi: 10.4161/gmcr.1.3.12377. [DOI] [PubMed] [Google Scholar]
- 21.Chen FJ, Wu G, Parajulee MN, Ge F. Impact of elevated CO2 on the third trophic level: A predator Harmonia axyridis (Pallas) and a parasitoid Aphidius picipes (Nees) Biocontrol Sci. Tec. 2007;17(3):313–324. doi: 10.1080/09583150701211814. [DOI] [Google Scholar]
- 22.Coviella CE, Trumble JT. Effect of elevated atmospheric carbon dioxide on the use of foliar application of Bacillus thuringiensis. Biocontrol. 2000;45(3):325–336. doi: 10.1023/A:1009947319662. [DOI] [Google Scholar]
- 23.Coviella CE, Stipanovic RD, Trumble JT. Plant allocation to defensive compounds: Interactions between elevated CO2 and nitrogen in transgenic cotton plants. J. Exp. Bot. 2002;53(367):323–331. doi: 10.1093/jexbot/53.367.323. [DOI] [PubMed] [Google Scholar]
- 24.Chen F, Wu G, Ge F, Parajulee MN, Shrestha RB. Effects of elevated CO2 and transgenic Bt cotton on plant chemistry, performance, and feeding of an insect herbivore, the cotton bollworm. Entomol. Exp. Appl. 2005;115(2):341–350. doi: 10.1111/j.1570-7458.2005.00258.x. [DOI] [Google Scholar]
- 25.Chen F, Wu G, Ge F, Parajulee MN. Relationships between exogenous-toxin quantity and increased biomass of transgenic Bt crops under elevated carbon dioxide. Ecotox. Environ. Safe. 2011;74(4):1074–1080. doi: 10.1016/j.ecoenv.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 26.Himanen SJ, et al. Interactions of elevated carbon dioxide and temperature with aphid feeding on transgenic oilseed rape: Are Bacillus thuringiensis (Bt) plants more susceptible to nontarget herbivores in future climate? Global Change Biol. 2008;14(6):1437–1454. doi: 10.1111/j.1365-2486.2008.01574.x. [DOI] [Google Scholar]
- 27.Tsutsumi K, Konno M, Miyazawa SI, Miyao M. Sites of action of elevated CO2 on leaf development in rice: Discrimination between the effects of elevated CO2 and nitrogen deficiency. Plant Cell Physiol. 2014;55(2):258–268. doi: 10.1093/pcp/pcu006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bird AP, Wolffe AP. Methylation induced repression—belts, braces and chromatin. Cell. 1999;99(5):451–454. doi: 10.1016/S0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 29.Chan SWL, Henderson IR, Jacobsen SE. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat. Rev. Genet. 2005;6(5):351–360. doi: 10.1038/nrg1601. [DOI] [PubMed] [Google Scholar]
- 30.Klose RJ, Bird AP. Genomic DNA methylation: The mark and its mediators. Trends Biochem. Sci. 2006;31(2):89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 31.Napoli C, Lemieux C, Jorgensen R. Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell. 1990;2(4):279–289. doi: 10.1105/tpc.2.4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van der Krol AR, Mur LA, Beld M, Mol JN, Stuitje AR. Flavonoid genes in petunia: Addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell. 1990;2(4):291–299. doi: 10.1105/tpc.2.4.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reddy MSS, Dinkins RD, Collins GB. Gene silencing in transgenic soybean plants transformed via particle bombardment. Plant Cell Rep. 2003;21(7):676–683. doi: 10.1007/s00299-002-0567-4. [DOI] [PubMed] [Google Scholar]
- 34.Weinhold A, Kallenbach M, Baldwin IT. Progressive 35S promoter methylation increases rapidly during vegetative development in transgenic Nicotiana attenuata plants. BMC. Plant Biol. 2013;13(1):99. doi: 10.1186/1471-2229-13-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao M, San León D, Delgadillo M, García JA, Simón-Mateo C. Virus-induced gene silencing in transgenic plants: Transgene silencing and reactivation associate with two patterns of transgene body methylation. Plant J. 2014;79(3):440–452. doi: 10.1111/tpj.12579. [DOI] [PubMed] [Google Scholar]
- 36.Cogoni C, Macino G. Conservation of transgene-induced post-transcriptional gene silencing in plants and fungi. Trends Plant Sci. 1997;2(11):438–443. doi: 10.1016/S1360-1385(97)90028-5. [DOI] [Google Scholar]
- 37.Robl JM, Wang Z, Kasinathan P, Kuroiwa Y. Transgenic animal production and animal biotechnology. Theriogenology. 2007;67(1):127–133. doi: 10.1016/j.theriogenology.2006.09.034. [DOI] [PubMed] [Google Scholar]
- 38.Yan J, et al. Expression of human transferrin can be regulated effectively by rabbit transferrin regulatory elements in transgenic mice. Biotechnol. Lett. 2014;36(6):1209–1216. doi: 10.1007/s10529-014-1484-7. [DOI] [PubMed] [Google Scholar]
- 39.Jeong BR, Wu-Scharf D, Zhang C, Cerutti H. Suppressors of transcriptional transgenic silencing in Chlamydomonas are sensitive to DNA-damaging agents and reactivate transposable elements. P. Natl. Acad. Sci. 2002;99(2):1076–1081. doi: 10.1073/pnas.022392999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang H, Zhu JK. RNA-directed DNA methylation. Curr. Opin. Plant Biol. 2011;14(2):142–147. doi: 10.1016/j.pbi.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kohli A, Gahakwa D, Vain P, Laurie DA, Christou P. Transgene expression in rice engineered through particle bombardment: Molecular factors controlling stable expression and transgene silencing. Planta. 1999;208(1):88–97. doi: 10.1007/s004250050538. [DOI] [Google Scholar]
- 42.Cogoni C, Macino G. Homology-dependent gene silencing in plants and fungi: A number of variations on the same theme. Curr. Opin. Microbiol. 1999;2(6):657–662. doi: 10.1016/S1369-5274(99)00041-7. [DOI] [PubMed] [Google Scholar]
- 43.Matzke M, Matzke AJ, Kooter JM. RNA: Guiding gene silencing. Science. 2001;293(5532):1080–1083. doi: 10.1126/science.1063051. [DOI] [PubMed] [Google Scholar]
- 44.Waterhouse PM, Wang MB, Lough T. Gene silencing as an adaptive defense against viruses. Nature. 2001;411(6839):834–842. doi: 10.1038/35081168. [DOI] [PubMed] [Google Scholar]
- 45.Sijen T, et al. Transcriptional and posttranscriptional gene silencing are mechanistically related. Curr. Biol. 2001;11(6):436–440. doi: 10.1016/S0960-9822(01)00116-6. [DOI] [PubMed] [Google Scholar]
- 46.Mette MF, Aufsatz W, Van der Winden J, Matzke MA, Matzke AJM. Transcriptional silencing and promoter methylation triggered by double-stranded RNA. Embo. J. 2000;19(19):5194–5201. doi: 10.1093/emboj/19.19.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang X, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell. 2006;126(6):1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 48.Dalakouras A, Dadami E, Zwiebel M, Krczal G, Wassenegger M. Transgenerational maintenance of transgene body CG but not CHG and CHH methylation. Epigenetics. 2012;7(9):1071–1078. doi: 10.4161/epi.21644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lister R, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133(3):523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vermeersch L, et al. Transitive RNA silencing signals induce cytosine methylation of a transgenic but not an endogenous target. Plant J. 2013;74(5):867–879. doi: 10.1111/tpj.12172. [DOI] [PubMed] [Google Scholar]
- 51.Kumar A, Silim SN, Okamoto M, Siddiqi MY, Glass ADM. Differential expression of three members of the AMT1 gene family encoding putative high-affinity NH4+ transporters in roots of Oryza sativa subspecies indica. Plant Cell Environ. 2003;26(6):907–914. doi: 10.1046/j.1365-3040.2003.01023.x. [DOI] [PubMed] [Google Scholar]
- 52.Wan G, et al. Rice stripe virus counters reduced fecundity in its insect vector by modifying insect physiology, primary endosymbionts and feeding behavior. Sci. Rep-UK. 2015;5:12527. doi: 10.1038/srep12527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiang SL, et al. Damage of maize borer and maize weevil on the yield of transgenic phytase maize. Agron. J. 2015;107(1):25–32. doi: 10.2134/agronj14.0366. [DOI] [Google Scholar]
- 54.Han L, Liu P, Wu K, Peng Y, Wang F. Population dynamics of Sesamia inferens on transgenic rice expressing Cry1Ac and CpTI in southern China. Environ. Entomol. 2008;37(5):1361–1370. doi: 10.1093/ee/37.5.1361. [DOI] [PubMed] [Google Scholar]
- 55.Nowak RS, Ellsworth DS, Smith SD. Functional responses of plants to elevated atmospheric CO2– do photosynthetic and productivity data from FACE experiments support early predictions? New Phytol. 2004;162(2):253–280. doi: 10.1111/j.1469-8137.2004.01033.x. [DOI] [Google Scholar]
- 56.Moutinho-Pereira J, et al. Effects of elevated CO2 on grapevine (Vitis vinifera L.): Physiological and yield attributes. VITIS-J. Grapevine Res. 2009;48(4):159. [Google Scholar]
- 57.Guo H, et al. Pea aphid promotes amino acid metabolism both in Medicago Truncatula, and bacteriocytes to favor aphid population growth under elevated CO2. Global Change Biol. 2013;19(10):3210–3223. doi: 10.1111/gcb.12260. [DOI] [PubMed] [Google Scholar]
- 58.Reich PB, Hobbie SE, Lee TD. Plant growth enhancement by elevated CO2 eliminated by joint water and nitrogen limitation. Nat. Geosci. 2014;7(12):920–924. doi: 10.1038/ngeo2284. [DOI] [Google Scholar]
- 59.Niklaus PA, Leadley PW, Schmid B. A long-term field study on biodiversity x elevated CO2 interactions in grassland. Ecol. Monogr. 2001;71(3):341–356. [Google Scholar]
- 60.Wu G, Chen FJ, Ge F, Sun YC. Effects of elevated carbon dioxide on the growth and foliar chemistry of transgenic Bt cotton. J. Integr. Plant Biol. 2007;49(9):1361–1369. doi: 10.1111/j.1744-7909.2007.00472_1.x. [DOI] [Google Scholar]
- 61.Ruiz C, Pla M, Riudavets J, Nadal A. High CO2 concentration as an inductor agent to drive production of recombinant phytotoxic antimicrobial peptides in plant biofactories. Plant Mol. Biol. 2016;90(4–5):329–343. doi: 10.1007/s11103-015-0419-6. [DOI] [PubMed] [Google Scholar]
- 62.Yamori W, Nagai T, Makino A. The rate-limiting step for CO2 assimilation at different temperatures is influenced by the leaf nitrogen content in several C3 crop species. Plant Cell Environ. 2011;34(5):764–777. doi: 10.1111/j.1365-3040.2011.02280.x. [DOI] [PubMed] [Google Scholar]
- 63.Adachi S, et al. The mesophyll anatomy enhancing CO2 diffusion is a key trait for improving rice photosynthesis. J. Exp. Bot. 2013;64(4):1061–1072. doi: 10.1093/jxb/ers382. [DOI] [PubMed] [Google Scholar]
- 64.Evans JR, Kaldenhoff R, Genty B, Terashima I. Resistances along the CO2 diffusion pathway inside leaves. J. Exp. Bot. 2009;60(8):2235–2248. doi: 10.1093/jxb/erp117. [DOI] [PubMed] [Google Scholar]
- 65.Li Y, Gao Y, Xu X, Shen Q, Guo S. Light-saturated photosynthetic rate in high-nitrogen rice (Oryza sativa L.) leaves is related to chloroplastic CO2 concentration. J. Exp. Bot. 2009;60(8):2351–2360. doi: 10.1093/jxb/erp127. [DOI] [PubMed] [Google Scholar]
- 66.Xiong D, et al. Rapid responses of mesophyll conductance to changes of CO2 concentration, temperature and irradiance are affected by N supplements in rice. Plant Cell Environ. 2015;38(12):2541–2550. doi: 10.1111/pce.12558. [DOI] [PubMed] [Google Scholar]
- 67.Nguyen HTT, Van Pham C, Bertin P. The effect of nitrogen concentration on nitrogen use efficiency and related parameters in cultivated rices (Oryza sativa L. subsp. indica and japonica and O. glaberrima Steud.) in hydroponics. Euphytica. 2014;198(1):137–151. [Google Scholar]
- 68.Schneider MK, et al. Ten years of free-air CO2 enrichment altered the mobilization of N from soil in Lolium perenne L. swards. Global Change Biol. 2004;10(8):1377–1388. doi: 10.1111/j.1365-2486.2004.00803.x. [DOI] [Google Scholar]
- 69.Reich PB, et al. Nitrogen limitation constrains sustainability of ecosystem response to CO2. Nature. 2006;440(7086):922–925. doi: 10.1038/nature04486. [DOI] [PubMed] [Google Scholar]
- 70.Cotrufo MF, Ineson P, Scott A. Elevated CO2 reduces the nitrogen concentration of plant tissues. Global Change Biol. 1998;4(1):43–54. doi: 10.1046/j.1365-2486.1998.00101.x. [DOI] [Google Scholar]
- 71.Luo Y, Field CB, Mooney HA. Predicting responses of photosynthesis and root fraction to elevated [CO2]a: interactions among carbon, nitrogen, and growth. Plant Cell Environ. 1994;17(11):1195–1204. doi: 10.1111/j.1365-3040.1994.tb02017.x. [DOI] [Google Scholar]
- 72.Rastetter EB, Ågren GI, Shaver GR. Responses of n-limited ecosystems to increased CO2: A balanced-nutrition, coupled-element-cycles model. Ecol. Appl. 1997;7(7):444–460. [Google Scholar]
- 73.Mittler R. Oxidative stress, antioxidants and stress tolerance. Trends in Plant Science. 2002;7(9):405–410. doi: 10.1016/S1360-1385(02)02312-9. [DOI] [PubMed] [Google Scholar]
- 74.Ghezzi P, Bonetto V. Redox proteomics: Identification of oxidatively modified proteins. Proteomics. 2003;3(7):1145–1153. doi: 10.1002/pmic.200300435. [DOI] [PubMed] [Google Scholar]
- 75.Pritchard SG, et al. The influence of elevated CO2 on the activities of antioxidative enzymes in two soybean genotypes. Funct. Plant Biol. 2000;27(11):1061–1068. doi: 10.1071/PP99206. [DOI] [Google Scholar]
- 76.Long SP, Ainsworth EA, Rogers A, Ort DR. Rising atmospheric carbon dioxide: Plants FACE the Future*. Annu. Rev. Plant Biology. 2004;55:591–628. doi: 10.1146/annurev.arplant.55.031903.141610. [DOI] [PubMed] [Google Scholar]
- 77.Sun Z, Niinemets U, Hueve K, Rasulov B, Noe SM. Elevated atmospheric CO2 concentration leads to increased whole-plant isoprene emission in hybrid aspen (Populus tremula × Populus tremuloides) New Phytol. 2013;198(3):788–800. doi: 10.1111/nph.12200. [DOI] [PubMed] [Google Scholar]
- 78.Backhausen JE, Scheibe R. Adaptation of tobacco plants to elevated CO2: Influence of leaf age on changes in physiology, redox states and NADP-malate dehydrogenase activity. J. Exp. Bot. 1999;50(334):665–675. doi: 10.1093/jxb/50.334.665. [DOI] [Google Scholar]
- 79.Webber AN, Nie GY, Long SP. Acclimation of photosynthetic proteins to rising atmospheric CO2. Photosynth. Res. 1994;39(3):413–25. doi: 10.1007/BF00014595. [DOI] [PubMed] [Google Scholar]
- 80.Jens M, Rajewsky N. Competition between target sites of regulators shapes post-transcriptional gene regulation. Nat. Rev. Genet. 2015;16(2):113–126. doi: 10.1038/nrg3853. [DOI] [PubMed] [Google Scholar]
- 81.Romanowski A, Yanovsky MJ. Circadian rhythms and post-transcriptional regulation in higher plants. Front. Plant Sci. 2015;6:437. doi: 10.3389/fpls.2015.00437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li X, Kazan H, Lipshitz HD, Morris QD. Finding the target sites of RNA-binding proteins. WIRES. RNA. 2014;5(1):111–130. doi: 10.1002/wrna.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.HafezQorani S, et al. Modeling the combined effect of RNA-binding proteins and microRNAs in post-transcriptional regulation. Nucleic. Acids. Res. 2016;44(9):e83. doi: 10.1093/nar/gkw048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Melanson, B. D. et al. A novel cis-acting element from the 3′ UTR of DNA damage-binding protein 2 mRNA links transcriptional and post-transcriptional regulation of gene expression. Nucleic. Acids. Res. gkt279 (2013). [DOI] [PMC free article] [PubMed]
- 85.Ikeuchi M, Iwase A, Sugimoto K. Control of plant cell differentiation by histone modification and DNA methylation. Curr. Opin. Plant Biol. 2015;28:60–67. doi: 10.1016/j.pbi.2015.09.004. [DOI] [PubMed] [Google Scholar]
- 86.Okumura A, et al. CaMV-35S promoter sequence-specific DNA methylation in lettuce. Plant Cell Rep. 2016;35(1):43–51. doi: 10.1007/s00299-015-1865-y. [DOI] [PubMed] [Google Scholar]
- 87.Ohkama-Ohtsu N, Kasajima I, Fujiwara T, Naito S. Isolation and characterization of an Arabidopsis mutant that overaccumulates O-Acetyl-L-Ser. Plant Physiol. 2004;136(2):3209–3222. doi: 10.1104/pp.104.047068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fan HH, et al. DNA methylation alterations of upland cotton (Gossypium hirsutum) in response to cold stress. Acta. Physiol. Plant. 2013;35(8):2445–2453. doi: 10.1007/s11738-013-1278-x. [DOI] [Google Scholar]
- 89.Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 2007;39(1):61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
- 90.Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328(5980):916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- 91.Sun G, et al. Quick and accurate detection and quantification of Magnaporthe oryzae in rice using real-time quantitative polymerase chain reaction. Plant Dis. 2015;99(2):219–224. doi: 10.1094/PDIS-05-14-0485-RE. [DOI] [PubMed] [Google Scholar]
- 92.Zhou K, et al. Young Leaf Chlorosis 1, a chloroplast-localized gene required for chlorophyll and lutein accumulation during early leaf development in rice. Planta. 2013;237(1):279–292. doi: 10.1007/s00425-012-1756-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.