Abstract
Peripheral serotonin (5-hydroxytryptamine, 5-HT) regulates cell growth and differentiation in numerous cell types through engagement of seven types of cell surface receptors (HTR1–7). Deregulated 5-HT/HTR levels contribute to pathology in chronic inflammatory diseases, with macrophages being relevant targets for the physio-pathological effects of 5-HT. In fact, 5-HT skews human macrophage polarization through engagement of 5-HT2BR and 5-HT7R receptors. We now report that 5-HT primes macrophages for reduced pro-inflammatory cytokine production and IFN type I-mediated signaling, and promotes an anti-inflammatory and pro-fibrotic gene signature in human macrophages. The acquisition of the 5-HT-dependent gene profile primarily depends on the 5-HT7R receptor and 5-HT7R-initiated PKA-dependent signaling. In line with the transcriptional results, 5-HT upregulates TGFβ1 production by human macrophages in an HTR7- and PKA-dependent manner, whereas the absence of Htr7 in vivo results in diminished macrophage infiltration and collagen deposition in a mouse model of skin fibrosis. Our results indicate that the anti-inflammatory and pro-fibrotic activity of 5-HT is primarily mediated through the 5-HT7R-PKA axis, and that 5-HT7R contributes to pathology in fibrotic diseases.
Introduction
Serotonin (5-hydroxytryptamine, 5-HT) is a monoamine neurotransmitter derived from L-tryptophan via a rate-limiting reaction catalyzed by tryptophan hydroxylases (TPH1 in periphery, TPH2 in brain)1,2. Brain-derived 5-HT controls mood, behavior, sleep, blood pressure and thermoregulation3 whereas peripheral 5-HT regulates vascular and heart functions4 and gastrointestinal mobility5. Enterochromaffin cells in the gastrointestinal tract produce 90% of the human body’s 5-HT6, which is actively taken up by blood platelets and stored in dense granules. Upon platelet activation, released 5-HT modifies vascular smooth muscle tone, promotes proliferation of smooth muscle cells7, hepatocytes8 and endothelial cells9, and critically contributes to wound healing. All 5-HT actions are exerted through engagement of seven types of receptors (5-HT1-7R) which, except for 5-HT3R, belong to the G protein-coupled superfamily of receptors10. 5-HT also functions as a regulator of immune and inflammatory responses11. 5-HT modulates T-cell activation, proliferation and differentiation12 and modifies cytokine production in a cell type-dependent manner13–16. The regulatory role of 5-HT in inflammation is illustrated by the pathological consequences of its altered production or absence in chronic inflammatory diseases. 5-HT contributes to Pulmonary Arterial Hypertension (PAH)17, atopic dermatitis18 and systemic sclerosis19, and modifies the outcome of inflammatory gut disorders20–25. 5-HT also favors colon cancer angiogenesis26 and neuroendocrine neoplasms proliferation27, and its absence increases pathologic scores in collagen-induced arthritis28. The close link between 5-HT and chronic inflammatory pathologies29 is in line with the anti-inflammatory actions of selective 5-HT reuptake inhibitors (SSRI) like fluoxetine30,31. Further supporting the 5-HT/inflammation link, 5-HT2BR has been shown to mediate the effects of 5-HT on tissue fibrosis19 and PAH17, 5-HT7R mediates the 5-HT contribution to gut inflammation in IBD models20,22,23, and 5-HT3R or 5-HT4R ligands reduce inflammatory reactions during postoperative ileus32,33.
Macrophages are critical for maintaining tissue homeostasis and promoting the initiation and resolution of inflammatory processes. The balance between pro- and anti-inflammatory (resolving) macrophages is required for restoring tissue homeostasis34–36, and its deregulation leads to chronic inflammatory diseases37,38. Since macrophages rapidly adapt their functions to micro-environmental stimuli (e.g., cytokines, growth factors, pathogen- and damage-associated molecular patterns)34–38, targeting macrophages is currently proposed as a therapeutic approach for chronic inflammatory diseases37. Not surprisingly, some of the effects of 5-HT on inflammation are mediated through direct and indirect actions on myeloid cells17,26,32,33. Further supporting this relationship, bone marrow-derived cells are responsible for the contribution of 5-HT and 5-HT7R to intestinal inflammation22, and macrophages mediate the anti-inflammatory action of SSRI30,39.
We have previously demonstrated that human pro-inflammatory and anti-inflammatory macrophages40–43 exhibit a distinct profile of 5-HT receptors, and that 5-HT2BR and 5-HT7R shape macrophage effector functions towards the anti-inflammatory side44. To dissect the molecular mechanisms underlying the inflammation-modulating action of 5-HT, we undertook the determination of the 5-HT-dependent transcriptome of human macrophages. 5-HT rapidly altered the human macrophage transcriptome towards a growth-promoting, anti-inflammatory and pro-fibrotic gene profile, whose acquisition was dependent on the 5-HT7R -PKA signaling axis. Moreover, and in line with these findings, Htr7 −/− mice exhibited significantly reduced macrophage accumulation and collagen deposition in a bleomycin-induced model of skin fibrosis.
Results
5-HT promotes the expression of an anti-inflammatory gene profile and inhibits pro-inflammatory cytokine production
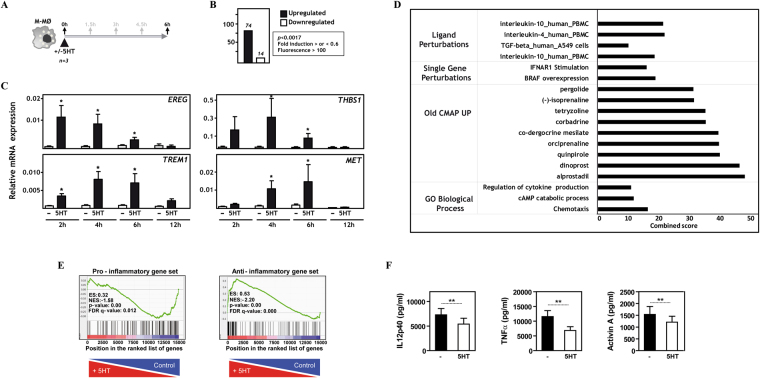
To determine the 5HT-dependent transcriptome of human macrophages, a global gene expression analysis was performed on human monocyte-derived macrophages (M-MØ) exposed to 5-HT for 6 hours (Fig. 1A), a time at which exposure to 5-HT modifies the LPS-induced production of inflammatory cytokines (see below). Transcriptional profiling revealed that 5-HT increases the expression of 170 annotated genes (p < 0.01; log2 ratio 5-HT/untreated ≥0.6) and downregulated the expression of 41 genes (p < 0.01; log2 ratio 5-HT/untreated ≤ −0.6) (Supplementary Table I). Further filtering (normalized expression levels higher than 100 in untreated or 5-HT-treated M-MØ, and p < 0.0017 for the M-MØ vs. M-MØ + 5-HT comparison) identified 74 genes upregulated and 14 genes downregulated upon 5-HT exposure (Table 1) (Fig. 1B). Analysis of independent 5-HT-treated M-MØ samples confirmed the microarray data and revealed distinct kinetics for the 5-HT-upregulated genes. As shown in Fig. 1C, EREG expression was maximally upregulated only two hours after exposure to 5-HT, while other genes (TREM1, MET, THBS1) exhibited maximal level of up-regulation 4–6 hours after 5-HT treatment. Therefore, 5-HT modifies the gene signature of human macrophages and its effects can be detected as early as 2 hours after exposure to the neurotransmitter.
Figure 1.
Serotonin promotes the acquisition of an anti-inflammatory gene profile and conditions human macrophages for diminished LPS-stimulated proinflammatory cytokine production. (A) Experimental design of the gene profiling experiment. (B) Number of annotated genes whose expression is upregulated or downregulated in M-MØ exposed to 5-HT for 6 hours (p < 0.0017; Upregulated, log2 M-MØ + 5-HT/M-MØ > 0.6; Downregulated, log2 M-MØ + 5-HT/M-MØ < −0.6). (C) Relative expression of the indicated genes in non-treated M-MØ (-) or M-MØ treated with 5-HT (5HT) for 2, 4, 6 and 12 hours. Results are expressed as the mRNA level of each gene relative to the GAPDH mRNA level in the same sample (n = 3; *p < 0.05). (D) Gene ontology analysis of the set of genes upregulated by 5-HT in M-MØ, as determined using the ENRICHR tool and the indicated databases [combined score = log(p-value) x z-score]. (E) GSEA on the “t statistic-ranked” list of genes obtained from the 5-HT-treated M-MØ versus M-MØ limma analysis, using the proinflammatory (left panel) and anti-inflammatory (right panel) gene sets previously defined48. Vertical black lines indicate the position of each of the genes comprising the “Pro-inflammatory” and “Anti-inflammatory” gene sets. (F) Production of IL-12p40, TNFα and Activin A by non-treated (-) or 5-HT-pretreated (6 h) LPS-stimulated (18 h) M-MØ, as determined by ELISA (n = 12; *p < 0.05; **p < 0.01).
Table 1.
Gene expression analysis on untreated (M-MØ) and 5-HT-treated (10 μM, 6h) M-MØ (M-MØ+5HT), where the normalized fluorescence of each probe (probeid) and the corresponding gene symbol (genesymbol) is indicated. Probes are ordered according to the t-value (t) of the M-MØ+5HT/ M-MØ ratio, and the p values obtained after adjusting for multiple hypotheses testing (adj.pval) are shown.
| probeid | M-MØ | M-MØ + 5HT | log2.fold M-MØ + 5HT/M-MØ | t | pval | adj.pval | genesymbol |
|---|---|---|---|---|---|---|---|
| A_23_P19333 | 80.05 | 344.74 | 2.0767 | 16.5371 | 0 | 0.192 | TREM1 |
| A_23_P401106 | 154.79 | 2069.16 | 3.6833 | 14.6249 | 0 | 0.192 | PDE2A |
| A_23_P416711 | 51.15 | 137.60 | 1.4367 | 14.5716 | 0 | 0.192 | ST6GALNAC3 |
| A_23_P208900 | 275.45 | 686.62 | 1.3333 | 14.3717 | 0 | 0.192 | SEMA6B |
| A_23_P32404 | 410.56 | 963.29 | 1.2367 | 13.1219 | 1.0000E-04 | 0.2092 | ISG20 |
| A_23_P132515 | 102.95 | 300.13 | 1.55 | 12.9583 | 1.0000E-04 | 0.2092 | SIDT1 |
| A_23_P257649 | 331.87 | 745.56 | 1.1767 | 12.2964 | 1.0000E-04 | 0.2138 | RBP1 |
| A_23_P373724 | 55.60 | 115.94 | 1.0967 | 11.7738 | 1.0000E-04 | 0.2138 | PPFIBP1 |
| A_24_P49260 | 120.84 | 254.30 | 1.14 | 11.5085 | 1.0000E-04 | 0.2138 | SPTLC3 |
| A_23_P154605 | 3414.30 | 7447.51 | 1.1567 | 10.6682 | 2.0000E-04 | 0.2138 | SULF2 |
| A_23_P85453 | 45.04 | 102.33 | 1.2633 | 10.2964 | 2.0000E-04 | 0.2138 | CD244 |
| A_24_P32085 | 269.48 | 651.45 | 1.2633 | 10.2497 | 2.0000E-04 | 0.2138 | MOB3B |
| A_33_P3352578 | 74.65 | 140.30 | 0.93 | 10.043 | 2.0000E-04 | 0.2138 | CLEC4D |
| A_23_P321354 | 389.53 | 744.31 | 0.9467 | 10.037 | 2.0000E-04 | 0.2138 | TMEM71 |
| A_33_P3333436 | 78.19 | 169.81 | 1.1033 | 9.9911 | 2.0000E-04 | 0.2138 | SGSM3 |
| A_23_P69109 | 1470.47 | 2684.51 | 0.8667 | 9.957 | 2.0000E-04 | 0.2138 | PLSCR1 |
| A_24_P349547 | 218.04 | 750.66 | 1.9733 | 9.8847 | 2.0000E-04 | 0.2138 | |
| A_33_P3322288 | 101.13 | 372.80 | 1.95 | 9.6901 | 2.0000E-04 | 0.2138 | AZI2 |
| A_24_P132383 | 491.79 | 927.06 | 0.94 | 9.3149 | 3.0000E-04 | 0.2138 | GIMAP8 |
| A_24_P93703 | 371.19 | 656.60 | 0.8167 | 8.9764 | 3.0000E-04 | 0.2138 | TMEM198B |
| A_24_P324674 | 492.95 | 846.92 | 0.7767 | 8.9556 | 3.0000E-04 | 0.2138 | LY9 |
| A_23_P414654 | 56.39 | 113.19 | 0.9633 | 8.9112 | 3.0000E-04 | 0.2138 | RAB37 |
| A_24_P245379 | 51.84 | 148.73 | 1.4133 | 8.8935 | 3.0000E-04 | 0.2138 | SERPINB2 |
| A_23_P72117 | 446.16 | 1126.78 | 1.2967 | 8.8485 | 4.0000E-04 | 0.2138 | SMPDL3A |
| A_24_P296508 | 702.06 | 1395.79 | 0.9533 | 8.7928 | 4.0000E-04 | 0.2138 | SLC43A2 |
| A_32_P161762 | 69.56 | 124.60 | 0.8667 | 8.7471 | 4.0000E-04 | 0.2138 | RUNX2 |
| A_23_P166297 | 227.26 | 450.70 | 0.9833 | 8.6544 | 4.0000E-04 | 0.2138 | ABCG1 |
| A_23_P428129 | 407.37 | 1022.43 | 1.32 | 8.6251 | 4.0000E-04 | 0.2138 | CDKN1C |
| A_23_P102731 | 127.71 | 316.19 | 1.2833 | 8.6233 | 4.0000E-04 | 0.2138 | SMOX |
| A_23_P404481 | 116.49 | 243.45 | 1.1133 | 8.5894 | 4.0000E-04 | 0.2138 | S1PR1 |
| A_33_P3226050 | 548.01 | 953.06 | 0.8 | 8.4776 | 4.0000E-04 | 0.2138 | GATSL3 |
| A_23_P10559 | 264.88 | 447.64 | 0.75 | 8.4521 | 4.0000E-04 | 0.2138 | AATK |
| A_33_P3240843 | 170.64 | 314.95 | 0.8833 | 8.4384 | 4.0000E-04 | 0.2138 | TMEM71 |
| A_33_P3382560 | 7622.06 | 15383.62 | 1.06 | 8.424 | 4.0000E-04 | 0.2138 | RPL23A |
| A_23_P25503 | 2952.57 | 6154.31 | 1.0633 | 8.4164 | 5.0000E-04 | 0.2138 | FNDC3A |
| A_23_P86653 | 11484.56 | 19458.03 | 0.77 | 8.3019 | 5.0000E-04 | 0.2193 | SRGN |
| A_23_P15108 | 532.44 | 913.10 | 0.77 | 8.1222 | 5.0000E-04 | 0.2193 | YPEL3 |
| A_23_P37375 | 217.10 | 359.90 | 0.7433 | 8.0574 | 5.0000E-04 | 0.2193 | RPS6KA5 |
| A_23_P89570 | 551.51 | 948.42 | 0.7633 | 8.0314 | 6.0000E-04 | 0.2193 | ZMYND15 |
| A_33_P3236177 | 703.17 | 1139.93 | 0.6967 | 7.9894 | 6.0000E-04 | 0.2193 | ANG |
| A_23_P170453 | 4108.14 | 10205.73 | 1.4933 | 7.968 | 6.0000E-04 | 0.2193 | CST5 |
| A_33_P3273885 | 409.11 | 870.16 | 1.0067 | 7.9645 | 6.0000E-04 | 0.2193 | |
| A_33_P3323722 | 2078.85 | 4415.23 | 1.0967 | 7.9589 | 6.0000E-04 | 0.2193 | ARL4C |
| A_23_P59637 | 957.90 | 1874.73 | 0.96 | 7.9163 | 6.0000E-04 | 0.2193 | DOCK4 |
| A_23_P154849 | 87.31 | 145.08 | 0.7267 | 7.9088 | 6.0000E-04 | 0.2193 | OLIG1 |
| A_23_P167920 | 117.43 | 211.91 | 0.8467 | 7.7999 | 6.0000E-04 | 0.2296 | DLL1 |
| A_24_P315184 | 9659.77 | 16932.61 | 0.8333 | 7.7041 | 7.0000E-04 | 0.2387 | NBEAL1 |
| A_32_P117464 | 190.00 | 365.73 | 0.97 | 7.6492 | 7.0000E-04 | 0.2392 | MB21D2 |
| A_33_P3422294 | 101.01 | 288.34 | 1.4233 | 7.6364 | 7.0000E-04 | 0.2392 | |
| A_33_P3317431 | 1153.70 | 2714.24 | 1.3067 | 7.5237 | 8.0000E-04 | 0.2392 | |
| A_23_P110445 | 129.59 | 225.31 | 0.8067 | 7.5107 | 8.0000E-04 | 0.2392 | APBB3 |
| A_23_P23438 | 741.59 | 1400.56 | 0.9633 | 7.4906 | 8.0000E-04 | 0.2392 | SEMA4A |
| A_23_P57760 | 101.41 | 159.07 | 0.6567 | 7.4768 | 8.0000E-04 | 0.2392 | ACPL2 |
| A_33_P3351775 | 79.06 | 130.10 | 0.6867 | 7.4673 | 8.0000E-04 | 0.2392 | |
| A_33_P3227716 | 820.46 | 1369.66 | 0.7333 | 7.4591 | 8.0000E-04 | 0.2392 | GATSL3 |
| A_23_P76969 | 785.27 | 2508.70 | 1.6 | 7.4097 | 8.0000E-04 | 0.2392 | SIPA1L1 |
| A_23_P115011 | 59.66 | 102.35 | 0.8333 | 7.3499 | 8.0000E-04 | 0.2392 | ADAMTSL4 |
| A_32_P87697 | 29475.87 | 46802.86 | 0.6833 | 7.3421 | 8.0000E-04 | 0.2392 | HLA-DRA |
| A_23_P55356 | 279.95 | 489.77 | 0.82 | 7.2414 | 9.0000E-04 | 0.2392 | VMO1 |
| A_23_P214139 | 349.66 | 647.84 | 0.8833 | 7.2258 | 9.0000E-04 | 0.2392 | REV3L |
| A_33_P3259557 | 296.93 | 544.75 | 0.8333 | 7.1249 | 0.001 | 0.2435 | TMEM198B |
| A_23_P79518 | 166.63 | 375.23 | 1.0967 | 7.111 | 0.001 | 0.2435 | IL1B |
| A_24_P370172 | 236.53 | 391.70 | 0.72 | 7.0777 | 0.001 | 0.2457 | LILRA5 |
| A_23_P152791 | 163.03 | 270.51 | 0.75 | 6.851 | 0.0011 | 0.2668 | SLC16A6 |
| A_23_P400378 | 512.19 | 844.84 | 0.8067 | 6.8421 | 0.0011 | 0.2668 | GPBAR1 |
| A_23_P85716 | 2690.03 | 4201.95 | 0.6933 | 6.8414 | 0.0011 | 0.2668 | FCGR2A |
| A_33_P3413840 | 121.69 | 228.70 | 0.9067 | 6.8214 | 0.0012 | 0.2668 | GK |
| A_33_P3372727 | 54.20 | 141.80 | 1.44 | 6.8087 | 0.0012 | 0.2668 | SEMA5A |
| A_23_P38795 | 1232.70 | 2077.44 | 0.8033 | 6.7376 | 0.0012 | 0.2692 | FPR1 |
| A_33_P3352098 | 11435.65 | 17831.64 | 0.6367 | 6.7192 | 0.0012 | 0.2692 | MS4A7 |
| A_33_P3315320 | 70.38 | 106.96 | 0.6 | 6.7149 | 0.0012 | 0.2692 | CNTD1 |
| A_23_P55020 | 656.97 | 1013.28 | 0.6133 | 6.659 | 0.0013 | 0.2725 | CD300LF |
| A_33_P3415191 | 182.14 | 313.76 | 0.78 | 6.6105 | 0.0013 | 0.2733 | ATP8B1 |
| A_33_P3232557 | 119.84 | 206.66 | 0.7833 | 6.5667 | 0.0014 | 0.2733 | DLGAP3 |
| A_23_P41114 | 878.89 | 1323.40 | 0.6167 | 6.4491 | 0.0015 | 0.2733 | CSTA |
| A_23_P201747 | 114.55 | 205.24 | 0.7933 | 6.4307 | 0.0015 | 0.2733 | PADI2 |
| A_33_P3236267 | 228.15 | 690.87 | 1.7367 | 6.4271 | 0.0015 | 0.2733 | KCNQ1OT1 |
| A_23_P380998 | 165.10 | 269.18 | 0.74 | 6.4024 | 0.0015 | 0.2733 | R3HDM1 |
| A_23_P60166 | 395.34 | 627.24 | 0.6367 | 6.3945 | 0.0016 | 0.2733 | DEPTOR |
| A_23_P215744 | 91.56 | 144.62 | 0.7067 | 6.3883 | 0.0016 | 0.2733 | CTTNBP2 |
| A_23_P258108 | 11895.64 | 19363.83 | 0.7267 | 6.3718 | 0.0016 | 0.2733 | |
| A_33_P3386132 | 397.26 | 623.42 | 0.66 | 6.3502 | 0.0016 | 0.2733 | C2orf49 |
| A_24_P110273 | 676.36 | 1044.92 | 0.6333 | 6.3217 | 0.0016 | 0.2733 | |
| A_23_P25566 | 4726.99 | 7713.17 | 0.7067 | 6.3212 | 0.0016 | 0.2733 | GPR183 |
| A_23_P67896 | 269.49 | 137.51 | −0.9233 | −6.3894 | 0.0016 | 0.2733 | SCN3A |
| A_23_P125204 | 492.06 | 317.00 | −0.6133 | −6.4729 | 0.0015 | 0.2733 | OR10G8 |
| A_33_P3232955 | 920.03 | 555.00 | −0.77 | −6.662 | 0.0013 | 0.2725 | F2RL3 |
| A_32_P134290 | 1591.48 | 1022.82 | −0.65 | −6.7639 | 0.0012 | 0.2689 | ZCCHC2 |
| A_23_P62967 | 1743.65 | 1144.21 | −0.6033 | −6.7737 | 0.0012 | 0.2689 | DISC1 |
| A_33_P3512350 | 675.48 | 399.65 | −0.7567 | −6.9789 | 0.0011 | 0.2531 | LOC339807 |
| A_23_P340848 | 1536.35 | 979.80 | −0.6667 | −7.0574 | 0.001 | 0.246 | PTGIR |
| A_23_P103034 | 214.84 | 114.56 | −0.9067 | −7.2742 | 9.0000E-04 | 0.2392 | CRYBA4 |
| A_23_P328545 | 440.40 | 259.98 | −0.7633 | −8.0807 | 5.0000E-04 | 0.2193 | GABRP |
| A_23_P404162 | 100.99 | 46.26 | −1.0767 | −8.2829 | 5.0000E-04 | 0.2193 | HDAC9 |
| A_23_P81441 | 1493.93 | 874.24 | −0.76 | −8.7105 | 4.0000E-04 | 0.2138 | C5orf20 |
| A_33_P3380383 | 4374.93 | 2579.19 | −0.8067 | −8.8739 | 4.0000E-04 | 0.2138 | TIFAB |
| A_23_P91095 | 523.44 | 291.00 | −0.8367 | −9.6298 | 2.0000E-04 | 0.2138 | CD28 |
| A_32_P155666 | 716.76 | 350.13 | −1.0733 | −9.703 | 2.0000E-04 | 0.2138 | ECEL1 |
Gene ontology analysis supported the relevance of the transcriptomic data because the 5-HT-upregulated gene set included a significant percentage of genes whose expression is increased by serotonin receptors agonists like co-dergocrine mesilate (adj p = 2.9 × 10−10) and pergolide (adj p = 3.03 × 10−8) (Fig. 1D). In fact, pergolide is an agonist for 5-HT2BR that causes valvular heart disease45,46 and M-MØ express functional 5-HT2BR receptors44. Besides, 5-HT enhanced the expression of genes positively regulated by prostaglandins (Alprostidil, Dinoprost), dopamine (Quinpirole, co-dergocrine mesilate), and adrenergic (Orciprenaline, Tetryzoline, (-)-isoprenaline) receptor ligands (Fig. 1D). Regarding biological processes, gene ontology analysis revealed that 5-HT-upregulated genes are significantly enriched in genes involved in chemotaxis (adj p = 1.8 × 10−3), cAMP catabolic process (adj p = 1.3 × 10−2) and regulation of cytokine production (adj p = 1.7 × 10−2), as well as in genes regulated by IL-10 (adj p = 8.3 × 10−6 and adj p = 4.2 × 10−5) and TGFβ (adj p = 2.7 × 10−3), and negatively regulated upon IFNAR1 stimulation (adj p = 7.0 × 10−5) (Fig. 1D).
The availability of the 5-HT-dependent transcriptome in M-MØ allowed us to address the global effect of 5-HT on the macrophage transcriptome. We have previously determined the gene expression profiles of IL-10-producing anti-inflammatory (M-MØ) and TNF-α-producing pro-inflammatory (GM-MØ) human macrophages47–49, and identified two sets of 150 genes that best define their corresponding transcriptomes (“Anti-inflammatory gene set” for M-MØ, and “Pro-inflammatory gene set” for GM-MØ)47,48. Analysis of the expression of both gene sets in 5-HT-treated macrophages using Gene Set Enrichment Analysis (GSEA) revealed that, compared to untreated cells, the transcriptome of 5-HT-treated macrophages shows a significantly higher expression of the “Anti-inflammatory gene set” (FDR q value = 0.000) together with a significantly lower expression of the “Pro-inflammatory gene set” (FDR q value = 0.012) (Fig. 1E). In fact, genes within the leading edge of the “anti-inflammatory gene set” included CD163L1, HTR2B, and IL10, whose expression is closely linked to M-CSF-driven anti-inflammatory polarization44,48,49 (Fig. 1E, right panel and data not shown). At the functional level, and in line with GSEA results, 5-HT pretreatment (6 hours) led to a significant reduction in the LPS-induced production of IL-12p40, TNFα and Activin A of M-MØ (Fig. 1F). Therefore, 5-HT promotes the acquisition of an anti-inflammatory gene profile and conditions macrophages for a diminished pro-inflammatory response towards pathogenic stimuli.
5-HT modulates macrophage transcriptome and function primarily via the 5-HT7R-PKA signaling axis
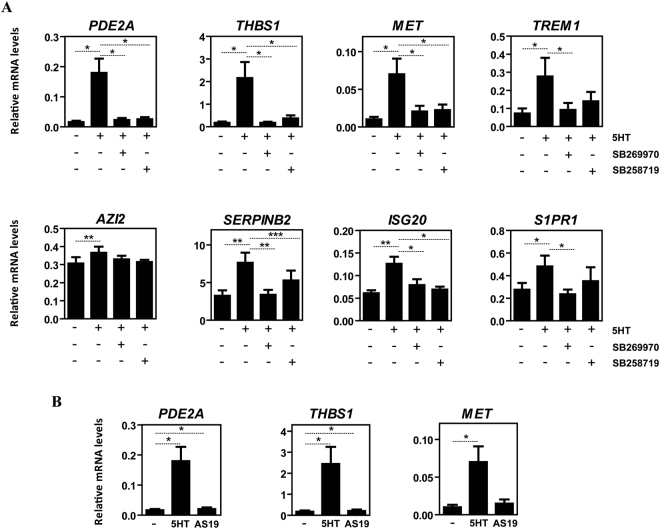
To determine the receptors responsible for the acquisition of the 5HT-dependent transcriptional and functional profile in 5-HT7R+/5-HT2BR+ M-MØ44, macrophages were exposed to 5-HT in the presence of 5-HT7R antagonists SB26997050 or SB25871951, or the 5-HT2BR antagonist SB20474152. Whereas blockade of 5-HT2BR had no effect (Supplementary Figure 1), 5-HT7R antagonists prevented or significantly inhibited the 5-HT-dependent up-regulation of PDE2A, THBS1, MET, TREM1, SERPINB2, ISG20, AZI2 and S1PR1 (Fig. 2A). Although to a lower extent than 5-HT, the 5-HT7R agonist AS19 was capable of enhancing the expression of PDE2A and THBS1 (Fig. 2B), further supporting the involvement of 5-HT7R in the 5-HT-dependent gene expression changes in human macrophages. However, since AS19 had no effect on other 5-HT-regulated genes (MET) (Fig. 2B), additional receptors might also contribute to the acquisition of the 5-HT-dependent gene signature.
Figure 2.
The acquisition of the 5-HT-dependent gene signature of human M-MØ is mediated by the 5-HT7R receptor. (A) Relative expression of the indicated genes in non-treated (-) or 5-HT-treated (5HT, 6 h) M-MØ and either in the absence (-) or in the presence of the 5-HT7R antagonists SB269970 or SB258719. Results are expressed as the mRNA level of each gene relative to the TBP mRNA level in the same sample (n = 6; *p < 0.05; **p < 0.01; ***p < 0.001). (B) Relative expression of the indicated genes in M-MØ either non-treated (-) or treated for 6 h with 5-HT or the 5-HT7R agonist AS19. Results are expressed as the mRNA level of each gene relative to the TBP mRNA level in the same sample (n = 6; *p < 0.05).
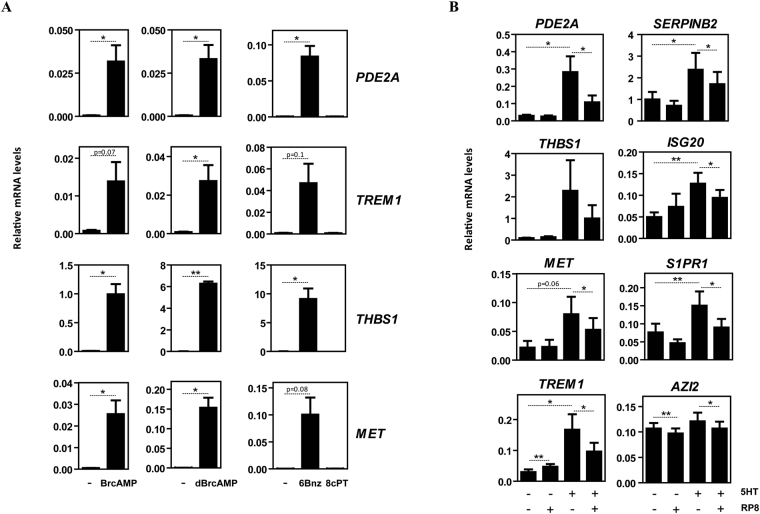
Since 5-HT7R engagement leads to increased intracellular levels of cAMP53, whose effectors include PKA and “Exchange factor directly activated by cAMP” (Epac), we next assessed the effect of cAMP analogs (BrcAMP, dBrcAMP) and modifiers of cAMP-initiated signaling (6Bnz, 8cPT, RP8) on the acquisition of the 5-HT-dependent gene signature in M-MØ. The cAMP analogs BrcAMP and dBrcAMP greatly enhanced the expression of genes upregulated by 5-HT (PDE2A, TREM1, THBS1, MET) (Fig. 3A). Similar changes in the expression of these genes were seen in M-MØ exposed to the PKA-specific activator 6-Bnz-cAMP (Fig. 3A), while the Epac activator 8-pCPT had no effect (Fig. 3A). Moreover, the positive effect of 5-HT on the expression of 5-HT-upregulated genes was significantly blunted or inhibited in the presence of the PKA inhibitor RP8 (Fig. 3B). Therefore, 5-HT shapes M-MØ gene expression primarily via engagement of 5-HT7R and activation of PKA.
Figure 3.
The acquisition of the 5-HT/5-HT7R-dependent gene signature of human M-MØ is mediated by PKA. (A) Relative expression of the indicated genes in M-MØ either untreated (-) or treated for 6 h to the PKA activators BrcAMP, dBrcAMP or 6Bnz, or to the Epac activator 8cPT. Results are expressed as the mRNA level of each gene relative to the GAPDH mRNA level in the same sample (n = 3; *p < 0.05). (B) Relative expression of the indicated genes in non-treated (-) or 5-HT-treated (5HT, 6 h) M-MØ and either in the absence (-) or in the presence of the PKA inhibitor RP8. Results are expressed as the mRNA level of each gene relative to the TBP mRNA level in the same sample (n = 6; *p < 0.05; **p < 0.01).
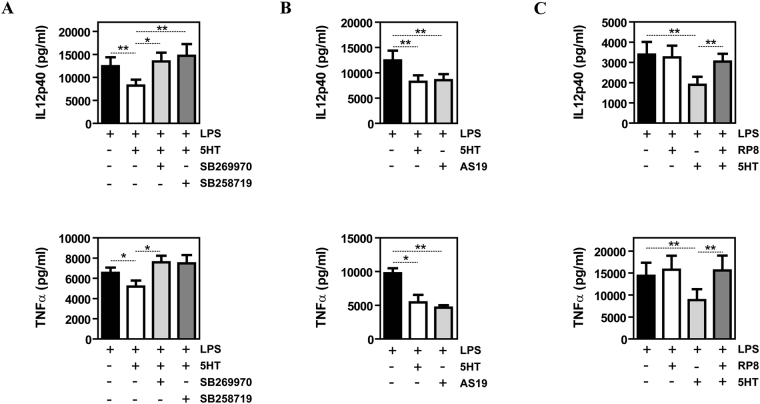
Since the 5-HT7R-PKA axis mediates the expression of the 5-HT-dependent M-MØ transcriptome, we next evaluated whether this signaling axis contributes to the inhibitory effect of 5-HT on the LPS-induced production of inflammatory cytokines by M-MØ, which produce undetectable levels of TNFα and IL-12p40 in the absence of stimulation44. Both 5-HT7R antagonists (SB269970 and SB258719) dose-dependently reversed the inhibitory action of 5-HT on the production of TNFα and IL-12p40 induced by LPS (Fig. 4A), while the 5-HT7R agonist AS19 (1 μM) mimicked the effect of 5-HT on the LPS-induced expression of TNFα and IL-12p40 (Fig. 4B). Furthermore, the inhibitory effect of 5-HT on the LPS-induced production of TNFα and IL-12p40 was significantly reduced in the presence of the PKA inhibitor RP8 (Fig. 4C). Thus, 5-HT conditions macrophages for a diminished production of pro-inflammatory cytokines primarily via engagement of 5-HT7R and activation of PKA. As a whole, this set of results demonstrates that the 5-HT7R-PKA axis mediates the acquisition of the 5-HT-dependent gene and cytokine profile in human macrophages.
Figure 4.
The inhibitory effect of 5-HT on the LPS-induced pro-inflammatory cytokine production of human macrophages is dependent on 5-HT7R and PKA. (A) Production of LPS-stimulated IL-12p40 and TNFα by M-MØ non-treated (-) or pretreated with 5-HT (6 h) in the presence or absence of the 5-HT7R antagonists SB269970 or SB258719 (n = 6; *p < 0.05; **p < 0.01; ***p < 0.001). (B) Production of LPS-stimulated IL-12p40 and TNFα by M-MØ non-treated (-) or pretreated (6 h) with either 5-HT (5HT) or the 5-HT7R agonist AS19 (n = 6; *p < 0.05; **p < 0.01). (C) Production of LPS-stimulated IL-12p40 and TNFα by M-MØ non-treated (-) or pretreated with 5-HT (5HT, 6 h) in the presence or absence of the PKA inhibitor RP8 (n = 6; *p < 0.05; **p < 0.01).
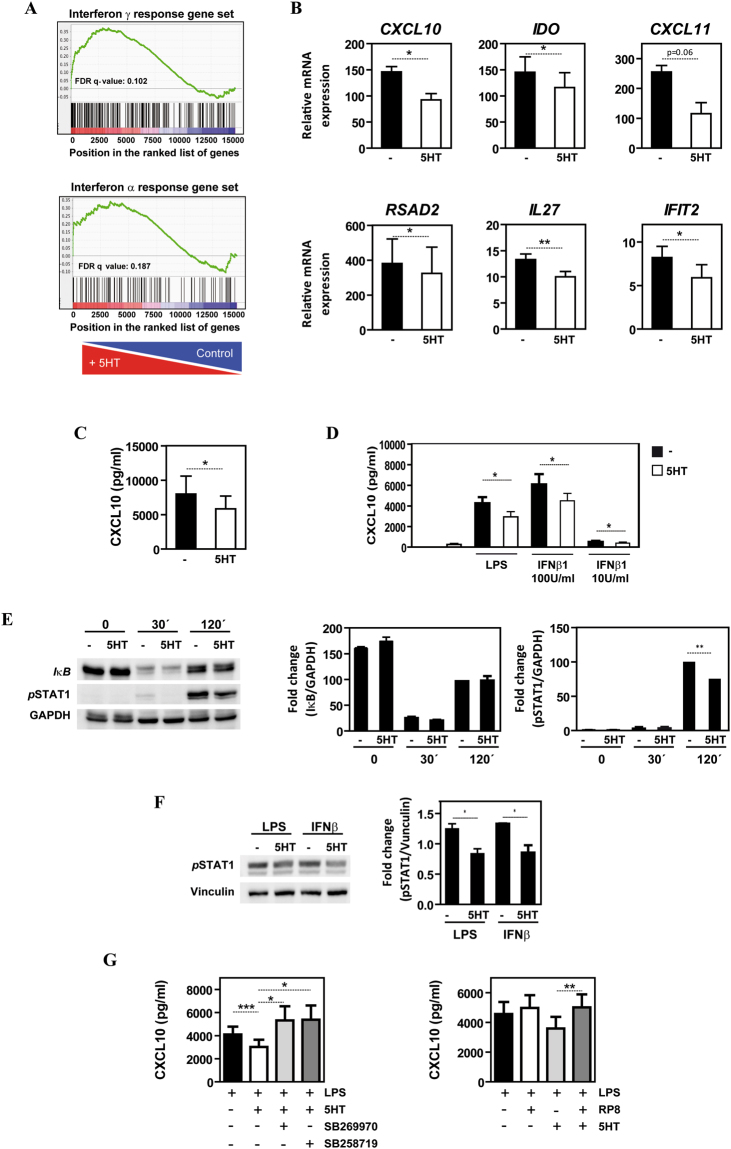
5-HT impairs type I IFN-dependent gene and chemokine expression through the 5-HT7R-PKA axis
To gain further biological insights from the 5-HT-dependent M-MØ transcriptome, we performed GSEA on the 5-HT-induced gene profile using gene sets contained in the Molecular Signature databases available at the GSEA Website. GSEA on the 5-HT-dependent transcriptome using the gene sets revealed a significant enrichment of the “hallmark IFNγ response” (FDR q value = 0.102) and “hallmark IFNα response” (FDR q value = 0.187) gene sets within the genes upregulated by 5-HT (Fig. 5A). In agreement with the GSEA data, the mRNA levels of type I IFN-dependent genes (CXCL10, CXCL11, IDO1, RSAD2, IL27 and IFIT2) (Fig. 5B) and the production of the type I IFN-dependent chemokine CXCL10 (Fig. 5C) were significantly reduced in LPS-treated M-MØ that had been pre-treated for 6 hours with 5-HT. Moreover, M-MØ also produced reduced levels of CXCL10 in response to IFNβ if previously exposed to 5-HT (Fig. 5D). The decrease in LPS- or IFNβ-induced CXCL10 production caused by 5-HT exposure correlated with a reduced activation of STAT1 in response to either LPS or IFNβ (Fig. 5E,F), further supporting the 5-HT´s ability to limit macrophage responses to type I IFN. Importantly, the ability of 5-HT to limit the expression of type I IFN-responsive genes is also mediated by the 5-HT7R-PKA axis, as it was abolished by either 5-HT7R antagonists (SB269970 and SB258719) or the PKA inhibitor RP8 (Fig. 5G).
Figure 5.
Serotonin modifies the type I IFN-dependent gene profile and impairs the response of human macrophages to type I IFN through 5-HT7R and PKA. (A) GSEA on the “t statistic-ranked” list of genes obtained from the 5-HT-treated M-MØ versus M-MØ limma analysis, using the “Hallmark_Interferon_gamma_response” (left panel) and the “Hallmark_Interferon_alpha_response” (right panel) gene sets. (B) Expression of the indicated genes in untreated (-) or 5-HT-pretreated (5HT, 6 h) LPS-stimulated (4 h) M-MØ, as determined by qRT-PCR. Results are expressed as the mRNA level of each gene relative to the level of TBP mRNA in the same sample (n = 3; *p < 0.05; **p < 0.01). (C) Production of LPS-stimulated CXCL10 by untreated (-) M-MØ and 5-HT-treated M-MØ (5HT, 6 h) exposed to LPS for 18 h (n = 12; *p < 0.05). (D) Production of IFNβ1-stimulated CXCL10 by M-MØ non-treated (-) or pretreated with 5-HT (5HT, 6 h), using the indicated concentrations of IFNβ1 (n = 13; *p < 0.05). (E) Levels of IkBα and phosphorylated STAT1 in untreated (-) M-MØ and 5-HT-treated M-MØ (5HT, 6 h), after stimulation with LPS for the indicated periods of time (left panel). Protein loading was normalized using a monoclonal antibody against GAPDH. Densitometric analysis of three independent experiments is shown in the right panels (n = 3; **p < 0.01). (F) Levels of phosphorylated STAT1 in untreated (-) M-MØ and 5-HT-treated M-MØ (5HT, 6 h), after stimulation with LPS or IFNβ1 (for 2 h). Protein loading was normalized using a monoclonal antibody against Vinculin. Densitometric analysis of three independent experiments is shown in the right panels (n = 3; *p < 0.05). (G) Production of LPS-stimulated CXCL10 by non-treated (-) M-MØ or M-MØ pretreated with 5-HT (5HT, 6 h) in the presence or absence of the 5-HT7R antagonists SB269970 or SB258719 (n = 6; *p < 0.05; ***p < 0.001) (left panel) or in the presence or absence of the PKA inhibitor RP8 (n = 6; **p < 0.01) (right panel).
5-HT also promotes pro-fibrotic gene expression in human macrophages
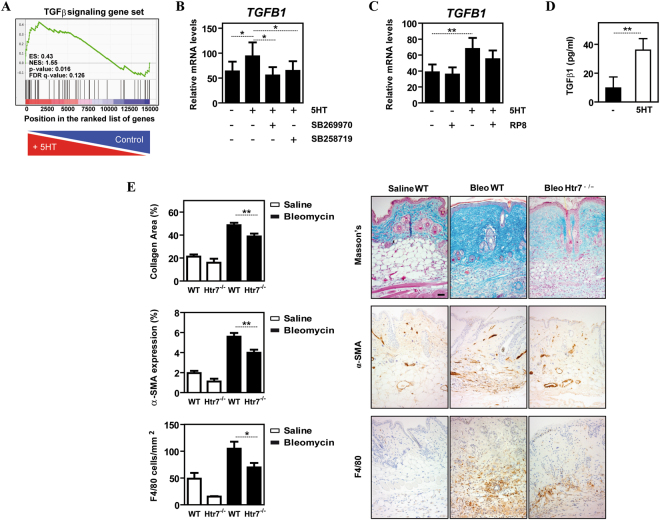
Additional GSEA results evidenced that a short-term (6 h) exposure to 5-HT causes a global upregulation of the “Angiogenesis” gene sets, as well as a very significant downregulation of the “Cholesterol homeostasis” and “Fatty Acid Metabolism” genes sets (Supplementary Figure 2). Furthermore, 5-HT treatment led to a significant upregulation of the “TGFβ signaling” gene set (FDR q-val = 0.126) (Fig. 6A). In line with the GSEA data, 5-HT was found to induce a significant increase in TGFB1 mRNA (Fig. 6B). Like most 5-HT-upregulated genes, the enhanced expression of TGFB1 mRNA was prevented by 5-HT7R antagonists (Fig. 6B) and did not occur in the presence of the PKA inhibitor RP8 (Fig. 6C). More importantly, 5-HT treatment of M-MØ resulted in a significantly increase production of TGFβ1 (Fig. 6D). Therefore, engagement of 5-HT7R by 5-HT promotes the production of TGFβ1 as well as the acquisition of a pro-fibrotic gene signature in human macrophages.
Figure 6.
5-HT7R engagement promotes the acquisition of a pro-fibrotic gene signature in human macrophages and contributes to pathology-associated parameters in a mouse model of skin fibrosis. (A) GSEA on the “t statistic-ranked” list of genes obtained from the 5-HT-treated M-MØ versus M-MØ limma analysis, using the “TGFβ signaling” gene set. Black vertical lines indicate the position of each of the genes included within the “TGFβ signaling” Hallmark gene set. (B) Relative expression of TGFB1 mRNA in non-treated M-MØ (-) or M-MØ exposed to 5-HT (5HT, 6 h) in the absence or in the presence of the 5-HT7R antagonists SB269970 or SB258719. Results are expressed as the TGFB1 mRNA level relative to the TBP mRNA level in the same sample (n = 6; *p < 0.05). (C) Relative expression of TGFB1 mRNA in non-treated M-MØ (-) or M-MØ exposed to 5-HT (5HT, 6 h) in the absence or in the presence of the PKA inhibitor RP8. Results are expressed as the TGFB1 mRNA level relative to the TBP and HPRT1 mRNA level in the same sample (n = 6; *p < 0.05; **p < 0.01). (D) Production of TGFβ1 by non-treated M-MØ (-) or M-MØ treated with 5-HT (5HT) for 24 h. (n = 3; **p < 0.01). (E) Fibrosis was measured as the collagen Masson stained area (upper panel), immunohistochemistry analysis of activated fibroblasts (α-SMA+, middle panel) and number of F4/80+ cells per area (lower panel), in lesional skin from saline- or bleomycin-treated control and Htr7 −/− mice. Shown are the mean and SEM of three independent experiments with 10 mice per group. Statistical significance was evaluated using Mann-Whitney U-test, (*p < 0.05; **p < 0.01). Representative skin sections stained are shown (bar, 50 μM).
Lack of Htr7 results in diminished macrophage infiltration and pathology in a mouse model of skin fibrosis
Macrophages exert critical functions during tissue repair after injury but their deregulated polarization can also result in excessive scarring and chronic fibrosis54,55. In fact, macrophages are important cells for the onset of scleroderma56,57 and pulmonary fibrosis58,59, and their deregulated polarization results in fibrosis in muscle60. Given the 5-HT/5-HT7R-upregulated expression of TGFβ1 in human macrophages, and to analyze the contribution of 5-HT7R to skin fibrosis development, we assessed the effect of Htr7 gene ablation in the mouse model of bleomycin-induced dermal fibrosis that mimics histological features of human scleroderma61. To this end we first determined Htr7 expression of in mouse macrophages in vitro and in vivo. Htr7 mRNA was readily detected in liver Kupffer cells and peritoneal F4/80+ macrophages, with Htr7a being the predominant splicing isoform in both cases (Supplementary Figure 3A). Unlike human monocyte-derived macrophages, Htr7 mRNA was only detected in murine pro-inflammatory GM-MØ, where also Htr7a was the predominant isoform (Supplementary Figure 3B). Also in marked contrast with human macrophages, where HTR7 mRNA is greatly reduced in response to LPS44, macrophage Htr7 mRNA was greatly upregulated after LPS stimulation (Supplementary Figure 3C).
Once the presence of Htr7 mRNA had been demonstrated in murine macrophages, the effect of Htr7 gene deletion in a mouse model of fibrosis was assessed. Whereas no histological differences in collagen stained area were observed between saline-treated Htr7 −/− and WT mice, a significant increase in skin collagen content was observed after bleomycin injection in WT mice (Fig. 6E). Conversely, Htr7 −/− mice appeared protected from bleomycin-induced fibrosis since significantly reduced collagen area content was observed in bleomycin-treated Htr7 −/− skin compared to WT skin (Fig. 6E). In addition, myofibroblast differentiation evaluated as α-SMA expression, was also significantly reduced in Htr7 −/− compared to WT mice (Fig. 6E). Likewise, a significantly lower infiltration of F4/80+ cells was found in bleomycin-treated Htr7 −/− mice (Fig. 6E). In addition, bleomycin treatment significantly enhanced α-SMA expression in WT mice but not in Htr7 −/− mice (Fig. 6E). Therefore, and in line with the transcriptional results in human macrophages, 5-HT7R expression contributes to macrophage accumulation and fibrosis in the bleomycin model of skin fibrosis.
Discussion
Macrophages exhibit a huge phenotypic and functional heterogeneity, and their effector functions (“polarization state”) are determined by the integration of the intracellular signals initiated by the surrounding extracellular cues and stimuli. Elimination of inflammatory insults requires the balanced and sequential dominance of pro-inflammatory and anti-inflammatory/resolving macrophages62 whose deregulation leads to chronic inflammatory diseases63–65. Given their critical role in the initiation and resolution of inflammation, modulation of the macrophage polarization state has been proposed as a therapeutic approach for numerous chronic inflammatory pathologies37. While the physiological processes regulated by 5-HT (cell proliferation, tissue repair, inflammation) are also critically modulated by macrophages66, the influence of 5-HT on macrophage plasticity is not yet completely understood11. Based on the ability of 5-HT to modulate the macrophage cytokine profile44, we undertook the determination of the 5-HT-dependent human macrophage transcriptome. Our results indicate that 5-HT conditions macrophages for impaired production of pro-inflammatory cytokines and type I IFN-inducible cytokines, and also shapes the macrophage gene signature towards the acquisition of an anti-inflammatory and pro-fibrotic gene profile, with all these effects being primarily mediated by the 5-HT7R-PKA axis.
The link between 5-HT and fibroblast proliferation/fibrosis has been known to be primarily mediated by the 5-HT2BR receptor, which induces extra-cellular matrix synthesis in fibroblasts19 and whose over-activation leads to excessive proliferation of cardiac valves fibroblasts and severe cardiac pathologies67–69. Our results reveal that 5-HT7R is a primary mediator of the pro-fibrotic action of 5-HT on human macrophages because 5-HT7R antagonists and inhibitors of 5-HT7R-initiated signaling block the acquisition of the pro-fibrotic gene signature as well as the 5-HT-upregulated production of TGFβ1 in human macrophages. The pro-fibrotic action of 5-HT7R is further supported by in vivo results, as Htr7 −/− mice exhibit diminished pathology (lower collagen deposition and F4/80+ cell infiltration) in the bleomycin-induced model of skin fibrosis. Macrophages play a critical role in fibrotic processes54. In fact, elimination of macrophages expressing Folr2, a myeloid-specific protein exclusively expressed by anti-inflammatory M-MØ70, greatly diminishes pathology in the bleomycin-induced experimental skin fibrosis71. Therefore, it can be speculated that the absence of 5-HT7R in mouse macrophages contributes to the diminished skin pathology we have observed. Alternatively, other macrophage 5-HT7R-dependent functions might contribute to the reduced pathology seen in Htr7 −/− mice. Specifically, impaired migration of myeloid cells to the bleomycin-treated tissue might explain the reduced accumulation of F4/80+ cells in the damaged skin of Htr7 −/− mice. This explanation would fit with the impaired migration of mouse bone marrow-derived dendritic cells from Htr7 −/− mice72 and is supported by the significant 5-HT-dependent upregulation of genes involved in cell chemotaxis (Fig. 1D). However, these interpretations should be taken cautiously, because expression of 5-HT7R appears to be differentially regulated in mouse and human macrophages: 5-HT7R in human macrophages is greatly downregulated by pathogenic stimuli like LPS44 whereas Ht7r expression in mouse myeloid cells is greatly upregulated by LPS72. In line with our findings, the 5-HT7R agonist LP-44 has been reported to reduce pro-inflammatory cytokine production in vivo in a carbon tetrachloride-induced rat model of liver fibrosis73, where the agonist was, however, also capable of inhibiting Tgfb1 mRNA73. The latter discrepancy between these results and ours might be explained by the use of different 5-HT7R agonists (5HT versus a chemical agonist) and animal models of fibrosis (bleomycin-induced mouse skin fibrosis versus carbon tetrachloride-induced rat liver fibrosis), but indicate a significant involvement of 5-HT7R in fibrotic responses.
In peripheral tissues, 5-HT7R is expressed in smooth muscle cells of blood vessels and the gastrointestinal tract, as well as in kidney, liver, pancreas and spleen74–76. Within the immune system, the functional consequences of 5-HT7R ligation are far from clear. 5-HT7R on mouse naive T cells contributes to early T-cell activation12, whereas 5-HT7R on human monocytes has been reported to inhibit77 or enhance78 LPS-induced pro-inflammatory cytokine release. A similar controversy appears to exist regarding the in vivo role of Htr7, whose deletion has been reported to improve22 or exacerbate23 mouse gut inflammation. Our transcriptional results clarify this issue and demonstrate that 5-HT7R, acting through PKA, conditions macrophages towards the acquisition of a more anti-inflammatory and pro-fibrotic polarization state and for an impaired production of pro-inflammatory functions. As a consequence, our results place 5-HT7R as a potentially relevant molecule for modulation of macrophage effector functions under physiological and pathological settings.
The anti-inflammatory gene profile promoted by 5-HT/5-HT7R is compatible with the known signaling ability of 5-HT7R. Although 5-HT7R has been shown to activate NFκB in monocytes12,78, and ERK1/279, Akt80, p38MAPK and protein kinase Cε81, or the Cdc42-Gα12-SRF axis82 in various cell types, 5-HT7R couples positively to adenylate cyclase through activating Gαs, leading to increased cAMP levels and activation of PKA and Epac1/274. These cell-specific differences in 5-HT7R signaling might derive from the presence of distinct splicing isoforms or heterodimerization with other receptors83,84. In the case of human macrophages, where the three splicing isoforms can be detected at the mRNA level (data not shown), our results clearly establish PKA, and not Epac1/2, as a major effector of 5-HT7R, as most of the transcriptional actions of 5-HT7R engagement by 5-HT can be abolished by PKA inhibitors and mimicked by PKA activators. Furthermore, the connexion between 5-HT7R and PKA activation fits well with the global anti-inflammatory skewing induced by 5-HT via 5-HT7R because PKA leads to CREB activation, which favours the acquisition of a “M2 polarization state”62. In addition, cAMP-initiated signaling limits the effector functions of pro-inflammatory stimuli85,86, which is consistent with the reduced production of proinflammatory cytokines seen with 5-HT7R activation.
The ability of 5-HT to promote the acquisition of a pro-fibrotic and anti-inflammatory signature in human macrophages has relevant pathophysiological implications. While normal peripheral blood levels of 5-HT range between 0.7 and 2.5 μM87–91, 5-HT concentrations at the neuronal synapse have been estimated to reach the millimolar range92, and the available platelet serotonin is close to 20 μM93. Since platelets release serotonin during inflammation as a means to activate endothelial cells and promote leukocyte adhesion and recruitment93, our findings on the macrophage polarizing effects of 10 μM 5-HT are physiologically relevant, especially at the initial stages of inflammatory responses. Besides, and regarding pathology, the serum levels of 5-HT detected in metastatic carcinoid tumors exceed 30 μM91, thus pointing to 5-HT as a factor that contributes to polarization of macrophages in serotonin-producing neuroendocrine tumors. Therefore, 5HT7R-regulated genes should be considered as potential targets to modify macrophage polarization under pathological settings.
Materials and Methods
Ethical statement
Ethical approvals for all blood sources and processes used in this study have been approved by the Centro de Investigaciones Biológicas Ethics Committees. Subjects gave written informed consent in accordance to the Declaration of Helsinki. All experiments were carried out in accordance with the approved guidelines and regulations.
All experiments on mice were conducted according to the Spanish and European regulations on care and protection of laboratory animals and were approved by the Consejo Superior de Investigaciones Científicas ethics committee.
Generation of human monocyte-derived macrophages and cell isolation and culture
Human peripheral blood mononuclear cells (PBMC) were isolated from buffy coats from normal donors over a Lymphoprep (Nycomed Pharma, Oslo, Norway) gradient according to standard procedures. Monocytes were purified from PBMC by magnetic cell sorting using CD14 microbeads (Miltenyi Biotech, Bergisch Gladbach, Germany). Monocytes (>95% CD14+ cells) were cultured at 0.5 × 106 cells/ml for 7 days in RMI supplemented with 10% fetal calf serum (FCS) (completed medium), at 37 °C in a humidified atmosphere with 5% CO2, and containing M-CSF (10 ng/ml) (ImmunoTools GmbH, Friesoythe, Germany) to generate monocyte-derived macrophages (M-MØ). Cytokine was added every two days. Before treatment with 5HT, M-MØ macrophages were maintained in serum-free medium for 48 hours, without a significant change in the level of expression of M-MØ-specific markers. Macrophage activation was accomplished with either ultrapure LPS (E. coli 055:B5, 10 ng/ml, Invivogen, San Diego, CA), synthetic triacylated lipoprotein (Pam3CSK4, 10 μg/ml, Invivogen) or IFNγ (10–100 IU/ml, Miltenyi Biotech). Bone marrow-derived mouse macrophages were also generated using GM-CSF (mouse GM-MØ) or M-CSF (mouse M-MØ) as described previously94,95. RNA from mouse liver cells was obtained as previously described44. Isolation of mouse peritoneal mouse macrophages was done by magnetic cell sorting using F4/80-biotin and Streptavidin microbeads (Miltenyi Biotech). For activation, macrophages were treated with Escherichia coli 055:B5 LPS (100 ng/ml for mouse macrophages; 10 ng/ml for human macrophages) for 24 h. 5-HT was obtained from Sigma Aldrich and used at 10μM. 5-HT7R agonists AS19 was obtained from Tocris Bioscience and used at 1μM. 5-HT7R antagonists (SB269970 and SB258719 were purchased from Sigma Aldrich and Tocris Bioscience, respectively, and added at 10μM 1 hour before 5-HT treatment. cAMP analogues (BrcAMP and dBrcAMP) were used at 200μM and 50μM, respectively. The PKA activator 6BNZ-cAMP (6Bnz) and the EPAC-specific activator 8-pCPT-2′-O-Me-cAMP (8cPT) were obtained from Sigma Aldrich and used at 200μM and 100μM, respectively. The PKA-specific inhibitor RP-8-CPT-cAMPs (RP8) was obtained from Biolog and used at 100μM.
ELISA
Culture supernatants from untreated or LPS-treated (24 h) human macrophages were assayed for the presence of cytokines using commercially available ELISA for human TNFα, IL-12p40 (BD Pharmingen), CXCL10, IL-10 (Biolegend), activin A and TGFβ (R&D Systems). ELISA was performed following the protocols supplied by the manufacturers.
Quantitative real-time RT-PCR
Oligonucleotides for selected genes were designed according to the Universal Probe Roche library system (Roche Diagnostics) for quantitative real time PCR (qRT-PCR). Total RNA was extracted using the RNeasy kit (Qiagen), retrotranscribed, and amplified in triplicates. Results were expressed relative to the expression level of TBP RNA. When indicated, results were expressed relative to the mean of the expression level of endogenous reference genes HPRT1, TBP and RPLP0. In all cases the results were expressed using the ΔΔCT method for quantitation.
Microarray analysis
Global gene expression analysis was performed on RNA obtained from untreated or 5-HT-treated (10 μM, 6 h) M-MØ from three independent healthy donors. RNA was isolated using the RNeasy Mini kit (Qiagen, Germantown, MD) and labeled RNA used as a hybridization probe on Whole Human Genome Microarrays (Agilent Technologies, Palo Alto, CA). Only probes with signal values > 60% quantile in at least one condition were considered for the differential expression and statistical analysis. Statistical analysis for differential gene expression was carried out using empirical Bayes moderated t test implemented in the limma package (http://www.bioconductor.org). The p values were further adjusted for multiple hypotheses testing using the Benjamini-Hochberg method to control the false discovery rate96. All the above procedures were coded in R (http://www.r-project.org). Microarray data were deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession GSE94608. Differentially expressed genes were analysed for annotated gene sets enrichment using the online tool ENRICHR (http://amp.pharm.mssm.edu/Enrichr/)97,98. Enrichment terms were considered significant when they had a Benjamini-Hochberg-adjusted p value < 0.05. For gene set enrichment analysis (GSEA)99, the gene sets contained in the Molecular Signature databases available at the GSEA website, and the previously defined “Pro-inflammatory gene set” and “Anti-inflammatory gene set”48, which contain the top and bottom 150 probes from the GM-MØ versus M-MØ limma analysis of the microarray data in GSE68061 (ranked on the basis of the value of the t statistic), were used.
Western blot
Cell lysates were obtained in RIPA buffer (10 mM Tris-HCl pH 8, 150 mM NaCl, 1% NP-40, 2 mM Pefabloc, 2 mg/ml aprotinin/antipain/leupeptin/pepstatin, 10 mM NaF, and 1 mM Na3VO4). 20 µg of cell lysate was subjected to SDS-PAGE and transferred onto an Immobilon polyvinylidene difluoride membrane (Millipore). Protein detection was carried out using antibodies against phosphorylated STAT1 (BD Biosciences, CA, USA) and a monoclonal antibody against GAPDH (sc-32233, Santa Cruz, CA, USA).
Bleomycin-induced skin fibrosis mouse model
Skin fibrosis was induced in 6–8 week-old, pathogen-free WT or Htr7 −/− 100 female C57BL/6NJ (The Jackson Laboratory) by subcutaneous injections of 100 μg of bleomycin (1 mg/ml, Mylan Pharmaceuticals, Barcelona, Spain) or 0.9% saline control into the shaved back skin every day for 4 weeks as previously described61,101. Skin was harvested and frozen for mRNA/protein studies or paraffin embedded for histological studies. Fibrosis was determined by Masson’s trichrome staining, and the presence of α-SMA+ or F4/80+ determined using anti-α Smooth Muscle Actin antibody (α-SMA; 1A4 clone, Sigma-Aldrich, Spain) and anti-F4/80 antibody (BM8 clone; eBioscience, San Diego, CA, USA).
Statistical analysis
For comparison of means, and unless otherwise indicated, statistical significance of the generated data was evaluated using the Student t test. In all cases, p < 0.05 was considered as statistically significant.
Data Availability
All data generated or analysed during this study are included in this published article. Moreover, microarray data were deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession GSE94608.
Electronic supplementary material
Acknowledgements
This work was supported by grants from Ministerio de Economía y Competitividad (SAF2011-23801 and SAF2014-52423-R to MAV and ALC, and PI I12/439 to JLP), “Programa de Actividades de I + D” from the Comunidad de Madrid/FEDER (RAPHYME S2010/BMD-2350 to JLP and ALC), and RIER (Red de Investigación en Inflamación y Enfermedades Reumáticas, RD12/09 to ALC and JLP) from the Instituto de Salud Carlos III, Ministerio de Economía y Competitividad, Spain (co-financed by FEDER, European Union). MCE was supported by an FPI predoctoral fellowship (BES-2009-021465) from Ministerio de Economía e Innovación.
Author Contributions
A.D.S., M.C.E., M.S.F., A.U., C.N. and V.D.C. performed research; A.D.S., M.A.V., J.L.P. and A.L.C. designed the research; A.D.S. analyzed data and prepared figures; A.L.C. wrote the paper.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Ángeles Domínguez-Soto, Alicia Usategui, Mateo de las Casas-Engel, José Luis Pablos and Ángel L. Corbí contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-15348-y.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lesurtel M, Soll C, Graf R, Clavien PA. Role of serotonin in the hepato-gastroIntestinal tract: an old molecule for new perspectives. Cell Mol Life Sci. 2008;65:940–952. doi: 10.1007/s00018-007-7377-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amireault P, Sibon D, Cote F. Life without peripheral serotonin: insights from tryptophan hydroxylase 1 knockout mice reveal the existence of paracrine/autocrine serotonergic networks. ACS Chem Neurosci. 2013;4:64–71. doi: 10.1021/cn300154j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger M, Gray JA, Roth BL. The expanded biology of serotonin. Annu Rev Med. 2009;60:355–366. doi: 10.1146/annurev.med.60.042307.110802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaumann AJ, Levy FO. 5-hydroxytryptamine receptors in the human cardiovascular system. Pharmacology & therapeutics. 2006;111:674–706. doi: 10.1016/j.pharmthera.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 5.Spiller R. Serotonin and GI clinical disorders. Neuropharmacology. 2008;55:1072–1080. doi: 10.1016/j.neuropharm.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 6.Yano JM, et al. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. 2015;161:264–276. doi: 10.1016/j.cell.2015.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nemecek GM, Coughlin SR, Handley DA, Moskowitz MA. Stimulation of aortic smooth muscle cell mitogenesis by serotonin. Proceedings of the National Academy of Sciences of the United States of America. 1986;83:674–678. doi: 10.1073/pnas.83.3.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lesurtel M, et al. Platelet-derived serotonin mediates liver regeneration. Science. 2006;312:104–107. doi: 10.1126/science.1123842. [DOI] [PubMed] [Google Scholar]
- 9.Pakala R, Willerson JT, Benedict CR. Mitogenic effect of serotonin on vascular endothelial cells. Circulation. 1994;90:1919–1926. doi: 10.1161/01.CIR.90.4.1919. [DOI] [PubMed] [Google Scholar]
- 10.Barnes NM, Sharp T. A review of central 5-HT receptors and their function. Neuropharmacology. 1999;38:1083–1152. doi: 10.1016/S0028-3908(99)00010-6. [DOI] [PubMed] [Google Scholar]
- 11.Ahern, G. P. 5-HT and the immune system. Curr Opin Pharmacol11, 29–33. [DOI] [PMC free article] [PubMed]
- 12.Leon-Ponte M, Ahern GP, O’Connell PJ. Serotonin provides an accessory signal to enhance T-cell activation by signaling through the 5-HT7 receptor. Blood. 2007;109:3139–3146. doi: 10.1182/blood-2006-10-052787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arzt E, Costas M, Finkielman S, Nahmod VE. Serotonin inhibition of tumor necrosis factor-alpha synthesis by human monocytes. Life sciences. 1991;48:2557–2562. doi: 10.1016/0024-3205(91)90612-F. [DOI] [PubMed] [Google Scholar]
- 14.Cloez-Tayarani I, Petit-Bertron AF, Venters HD, Cavaillon JM. Differential effect of serotonin on cytokine production in lipopolysaccharide-stimulated human peripheral blood mononuclear cells: involvement of 5-hydroxytryptamine2A receptors. International immunology. 2003;15:233–240. doi: 10.1093/intimm/dxg027. [DOI] [PubMed] [Google Scholar]
- 15.Durk T, et al. 5-Hydroxytryptamine modulates cytokine and chemokine production in LPS-primed human monocytes via stimulation of different 5-HTR subtypes. International immunology. 2005;17:599–606. doi: 10.1093/intimm/dxh242. [DOI] [PubMed] [Google Scholar]
- 16.Menard G, Turmel V, Bissonnette EY. Serotonin modulates the cytokine network in the lung: involvement of prostaglandin E2. Clinical and experimental immunology. 2007;150:340–348. doi: 10.1111/j.1365-2249.2007.03492.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Launay JM, et al. Serotonin 5-HT2B receptors are required for bone-marrow contribution to pulmonary arterial hypertension. Blood. 2012;119:1772–1780. doi: 10.1182/blood-2011-06-358374. [DOI] [PubMed] [Google Scholar]
- 18.Morita T, et al. HTR7 Mediates Serotonergic Acute and Chronic Itch. Neuron. 2015;87:124–138. doi: 10.1016/j.neuron.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dees C, et al. Platelet-derived serotonin links vascular disease and tissue fibrosis. The Journal of experimental medicine. 2011;208:961–972. doi: 10.1084/jem.20101629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim JJ, Khan WI. 5-HT7 receptor signaling: improved therapeutic strategy in gut disorders. Front Behav Neurosci. 2014;8:396. doi: 10.3389/fnbeh.2014.00396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levin AD, van den Brink GR. Selective inhibition of mucosal serotonin as treatment for IBD? Gut. 2014;63:866–867. doi: 10.1136/gutjnl-2013-305283. [DOI] [PubMed] [Google Scholar]
- 22.Kim JJ, et al. Targeted inhibition of serotonin type 7 (5-HT7) receptor function modulates immune responses and reduces the severity of intestinal inflammation. J Immunol. 2013;190:4795–4804. doi: 10.4049/jimmunol.1201887. [DOI] [PubMed] [Google Scholar]
- 23.Guseva D, et al. Serotonin 5-HT7 receptor is critically involved in acute and chronic inflammation of the gastrointestinal tract. Inflamm Bowel Dis. 2014;20:1516–1529. doi: 10.1097/MIB.0000000000000150. [DOI] [PubMed] [Google Scholar]
- 24.Ghia JE, et al. Serotonin has a key role in pathogenesis of experimental colitis. Gastroenterology. 2009;137:1649–1660. doi: 10.1053/j.gastro.2009.08.041. [DOI] [PubMed] [Google Scholar]
- 25.Li N, et al. Serotonin activates dendritic cell function in the context of gut inflammation. The American journal of pathology. 2011;178:662–671. doi: 10.1016/j.ajpath.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nocito A, et al. Serotonin regulates macrophage-mediated angiogenesis in a mouse model of colon cancer allografts. Cancer research. 2008;68:5152–5158. doi: 10.1158/0008-5472.CAN-08-0202. [DOI] [PubMed] [Google Scholar]
- 27.Svejda B, et al. Serotonin and the 5-HT7 receptor: the link between hepatocytes, IGF-1 and small intestinal neuroendocrine tumors. Cancer Sci. 2013;104:844–855. doi: 10.1111/cas.12174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chabbi-Achengli Y, et al. Serotonin Is Involved in Autoimmune Arthritis through Th17 Immunity and Bone Resorption. The American journal of pathology. 2016;186:927–937. doi: 10.1016/j.ajpath.2015.11.018. [DOI] [PubMed] [Google Scholar]
- 29.Shajib MS, Khan WI. The role of serotonin and its receptors in activation of immune responses and inflammation. Acta Physiol (Oxf) 2015;213:561–574. doi: 10.1111/apha.12430. [DOI] [PubMed] [Google Scholar]
- 30.Nazimek, K. et al. The role of macrophages in anti-inflammatory activity of antidepressant drugs. Immunobiology, 10.1016/j.imbio.2016.07.001 (2016). [DOI] [PubMed]
- 31.Su HC, et al. Glycogen synthase kinase-3beta regulates anti-inflammatory property of fluoxetine. Int Immunopharmacol. 2012;14:150–156. doi: 10.1016/j.intimp.2012.06.015. [DOI] [PubMed] [Google Scholar]
- 32.Tsuchida Y, et al. Neuronal stimulation with 5-hydroxytryptamine 4 receptor induces anti-inflammatory actions via alpha7nACh receptors on muscularis macrophages associated with postoperative ileus. Gut. 2011;60:638–647. doi: 10.1136/gut.2010.227546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maehara T, et al. Therapeutic action of 5-HT3 receptor antagonists targeting peritoneal macrophages in post-operative ileus. British journal of pharmacology. 2015;172:1136–1147. doi: 10.1111/bph.13006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jantsch J, Binger KJ, Muller DN, Titze J. Macrophages in homeostatic immune function. Frontiers in physiology. 2014;5:146. doi: 10.3389/fphys.2014.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. The Journal of clinical investigation. 2012;122:787–795. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinez FO, Sica A, Mantovani A, Locati M. Macrophage activation and polarization. Front Biosci. 2008;13:453–461. doi: 10.2741/2692. [DOI] [PubMed] [Google Scholar]
- 37.Schultze JL. Reprogramming of macrophages–new opportunities for therapeutic targeting. Current opinion in pharmacology. 2016;26:10–15. doi: 10.1016/j.coph.2015.09.007. [DOI] [PubMed] [Google Scholar]
- 38.Schultze JL, Freeman T, Hume DA, Latz E. A transcriptional perspective on human macrophage biology. Seminars in immunology. 2015;27:44–50. doi: 10.1016/j.smim.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 39.Kalkman HO, Feuerbach D. Antidepressant therapies inhibit inflammation and microglial M1-polarization. Pharmacology & therapeutics. 2016;163:82–93. doi: 10.1016/j.pharmthera.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 40.Verreck FA, et al. Human IL-23-producing type 1 macrophages promote but IL-10-producing type 2 macrophages subvert immunity to (myco)bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:4560–4565. doi: 10.1073/pnas.0400983101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li G, Kim YJ, Broxmeyer HE. Macrophage colony-stimulating factor drives cord blood monocyte differentiation into IL-10(high)IL-12absent dendritic cells with tolerogenic potential. J Immunol. 2005;174:4706–4717. doi: 10.4049/jimmunol.174.8.4706. [DOI] [PubMed] [Google Scholar]
- 42.Akagawa KS. Functional heterogeneity of colony-stimulating factor-induced human monocyte-derived macrophages. Int J Hematol. 2002;76:27–34. doi: 10.1007/BF02982715. [DOI] [PubMed] [Google Scholar]
- 43.Hamilton JA. Colony-stimulating factors in inflammation and autoimmunity. Nature reviews. Immunology. 2008;8:533–544. doi: 10.1038/nri2356. [DOI] [PubMed] [Google Scholar]
- 44.de las Casas-Engel, M. et al. Serotonin skews human macrophage polarization through HTR2B and HTR7. Journal of immunology190, 2301–2310, 10.4049/jimmunol.1201133 (2013). [DOI] [PubMed]
- 45.Roth BL. Drugs and valvular heart disease. The New England journal of medicine. 2007;356:6–9. doi: 10.1056/NEJMp068265. [DOI] [PubMed] [Google Scholar]
- 46.Schade R, Andersohn F, Suissa S, Haverkamp W, Garbe E. Dopamine agonists and the risk of cardiac-valve regurgitation. The New England journal of medicine. 2007;356:29–38. doi: 10.1056/NEJMoa062222. [DOI] [PubMed] [Google Scholar]
- 47.Cuevas, V. D. et al. MAFB Determines Human Macrophage Anti-Inflammatory Polarization: Relevance for the Pathogenic Mechanisms Operating in Multicentric Carpotarsal Osteolysis. J Immunol, 10.4049/jimmunol.1601667 (2017). [DOI] [PubMed]
- 48.Gonzalez-Dominguez E, et al. Atypical Activin A and IL-10 Production Impairs Human CD16+Monocyte Differentiation into Anti-Inflammatory Macrophages. J Immunol. 2016;196:1327–1337. doi: 10.4049/jimmunol.1501177. [DOI] [PubMed] [Google Scholar]
- 49.Sierra-Filardi E, et al. Activin A skews macrophage polarization by promoting a proinflammatory phenotype and inhibiting the acquisition of anti-inflammatory macrophage markers. Blood. 2011;117:5092–5101. doi: 10.1182/blood-2010-09-306993. [DOI] [PubMed] [Google Scholar]
- 50.Lovell PJ, et al. A novel, potent, and selective 5-HT(7) antagonist: (R)-3-(2-(2-(4-methylpiperidin-1-yl)ethyl)pyrrolidine-1-sulfonyl) phen ol (SB-269970) J Med Chem. 2000;43:342–345. doi: 10.1021/jm991151j. [DOI] [PubMed] [Google Scholar]
- 51.Pouzet B. SB-258741: a 5-HT7 receptor antagonist of potential clinical interest. CNS Drug Rev. 2002;8:90–100. doi: 10.1111/j.1527-3458.2002.tb00217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Forbes IT, Jones GE, Murphy OE, Holland V, Baxter GS. N-(1-methyl-5-indolyl)-N’-(3-methyl-5-isothiazolyl)urea: a novel, high-affinity 5-HT2B receptor antagonist. J Med Chem. 1995;38:855–857. doi: 10.1021/jm00006a001. [DOI] [PubMed] [Google Scholar]
- 53.Raymond JR, et al. Multiplicity of mechanisms of serotonin receptor signal transduction. Pharmacology & therapeutics. 2001;92:179–212. doi: 10.1016/S0163-7258(01)00169-3. [DOI] [PubMed] [Google Scholar]
- 54.Wynn TA, Vannella KM. Macrophages in Tissue Repair, Regeneration, and Fibrosis. Immunity. 2016;44:450–462. doi: 10.1016/j.immuni.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Munoz-Canoves P, Serrano AL. Macrophages decide between regeneration and fibrosis in muscle. Trends Endocrinol Metab. 2015;26:449–450. doi: 10.1016/j.tem.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 56.Christmann RB, et al. Association of Interferon- and transforming growth factor beta-regulated genes and macrophage activation with systemic sclerosis-related progressive lung fibrosis. Arthritis Rheumatol. 2014;66:714–725. doi: 10.1002/art.38288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stifano G, et al. Chronic Toll-like receptor 4 stimulation in skin induces inflammation, macrophage activation, transforming growth factor beta signature gene expression, and fibrosis. Arthritis Res Ther. 2014;16:R136. doi: 10.1186/ar4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Murray LA, et al. TGF-beta driven lung fibrosis is macrophage dependent and blocked by Serum amyloid P. The international journal of biochemistry & cell biology. 2011;43:154–162. doi: 10.1016/j.biocel.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 59.Murray LA, et al. Serum amyloid P therapeutically attenuates murine bleomycin-induced pulmonary fibrosis via its effects on macrophages. PloS one. 2010;5:e9683. doi: 10.1371/journal.pone.0009683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lemos DR, et al. Nilotinib reduces muscle fibrosis in chronic muscle injury by promoting TNF-mediated apoptosis of fibro/adipogenic progenitors. Nature medicine. 2015;21:786–794. doi: 10.1038/nm.3869. [DOI] [PubMed] [Google Scholar]
- 61.Yamamoto T. Animal model of systemic sclerosis. J Dermatol. 2010;37:26–41. doi: 10.1111/j.1346-8138.2009.00764.x. [DOI] [PubMed] [Google Scholar]
- 62.Ruffell D, et al. A CREB-C/EBPbeta cascade induces M2 macrophage-specific gene expression and promotes muscle injury repair. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:17475–17480. doi: 10.1073/pnas.0908641106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lumeng CN, Bodzin JL, Saltiel AR. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. The Journal of clinical investigation. 2007;117:175–184. doi: 10.1172/JCI29881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ruffell B, Coussens LM. Macrophages and therapeutic resistance in cancer. Cancer Cell. 2015;27:462–472. doi: 10.1016/j.ccell.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity. 2010;32:593–604. doi: 10.1016/j.immuni.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 66.Mantovani A, Biswas SK, Galdiero MR, Sica A, Locati M. Macrophage plasticity and polarization in tissue repair and remodelling. The Journal of pathology. 2013;229:176–185. doi: 10.1002/path.4133. [DOI] [PubMed] [Google Scholar]
- 67.Jaffre F, et al. Involvement of the serotonin 5-HT2B receptor in cardiac hypertrophy linked to sympathetic stimulation: control of interleukin-6, interleukin-1beta, and tumor necrosis factor-alpha cytokine production by ventricular fibroblasts. Circulation. 2004;110:969–974. doi: 10.1161/01.CIR.0000139856.20505.57. [DOI] [PubMed] [Google Scholar]
- 68.Nebigil CG, et al. Overexpression of the serotonin 5-HT2B receptor in heart leads to abnormal mitochondrial function and cardiac hypertrophy. Circulation. 2003;107:3223–3229. doi: 10.1161/01.CIR.0000074224.57016.01. [DOI] [PubMed] [Google Scholar]
- 69.Elangbam CS. Drug-induced valvulopathy: an update. Toxicol Pathol. 2010;38:837–848. doi: 10.1177/0192623310378027. [DOI] [PubMed] [Google Scholar]
- 70.Puig-Kroger A, et al. Folate receptor beta is expressed by tumor-associated macrophages and constitutes a marker for M2 anti-inflammatory/regulatory macrophages. Cancer research. 2009;69:9395–9403. doi: 10.1158/0008-5472.CAN-09-2050. [DOI] [PubMed] [Google Scholar]
- 71.Li H, Nagai T, Hasui K, Matsuyama T. Depletion of folate receptor beta-expressing macrophages alleviates bleomycin-induced experimental skin fibrosis. Mod Rheumatol. 2014;24:816–822. doi: 10.3109/14397595.2013.879415. [DOI] [PubMed] [Google Scholar]
- 72.Holst K, et al. The serotonin receptor 5-HT(7)R regulates the morphology and migratory properties of dendritic cells. Journal of cell science. 2015;128:2866–2880. doi: 10.1242/jcs.167999. [DOI] [PubMed] [Google Scholar]
- 73.Polat B, et al. Liver 5-HT7 receptors: A novel regulator target of fibrosis and inflammation-induced chronic liver injury in vivo and in vitro. Int Immunopharmacol. 2017;43:227–235. doi: 10.1016/j.intimp.2016.12.023. [DOI] [PubMed] [Google Scholar]
- 74.Gellynck E, et al. The serotonin 5-HT7 receptors: two decades of research. Exp Brain Res. 2013;230:555–568. doi: 10.1007/s00221-013-3694-y. [DOI] [PubMed] [Google Scholar]
- 75.Bard JA, et al. Cloning of a novel human serotonin receptor (5-HT7) positively linked to adenylate cyclase. The Journal of biological chemistry. 1993;268:23422–23426. [PubMed] [Google Scholar]
- 76.Tuladhar BR, Ge L, Naylor RJ. 5-HT7 receptors mediate the inhibitory effect of 5-HT on peristalsis in the isolated guinea-pig ileum. British journal of pharmacology. 2003;138:1210–1214. doi: 10.1038/sj.bjp.0705184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Idzko M, et al. The serotoninergic receptors of human dendritic cells: identification and coupling to cytokine release. Journal of immunology. 2004;172:6011–6019. doi: 10.4049/jimmunol.172.10.6011. [DOI] [PubMed] [Google Scholar]
- 78.Soga F, Katoh N, Inoue T, Kishimoto S. Serotonin activates human monocytes and prevents apoptosis. The Journal of investigative dermatology. 2007;127:1947–1955. doi: 10.1038/sj.jid.5700824. [DOI] [PubMed] [Google Scholar]
- 79.Norum JH, Hart K, Levy FO. Ras-dependent ERK activation by the human G(s)-coupled serotonin receptors 5-HT4(b) and 5-HT7(a) The Journal of biological chemistry. 2003;278:3098–3104. doi: 10.1074/jbc.M206237200. [DOI] [PubMed] [Google Scholar]
- 80.Johnson-Farley NN, Kertesy SB, Dubyak GR, Cowen DS. Enhanced activation of Akt and extracellular-regulated kinase pathways by simultaneous occupancy of Gq-coupled 5-HT2A receptors and Gs-coupled 5-HT7A receptors in PC12 cells. J Neurochem. 2005;92:72–82. doi: 10.1111/j.1471-4159.2004.02832.x. [DOI] [PubMed] [Google Scholar]
- 81.Lieb K, et al. Serotonin via 5-HT7 receptors activates p38 mitogen-activated protein kinase and protein kinase C epsilon resulting in interleukin-6 synthesis in human U373 MG astrocytoma cells. J Neurochem. 2005;93:549–559. doi: 10.1111/j.1471-4159.2005.03079.x. [DOI] [PubMed] [Google Scholar]
- 82.Kvachnina E, et al. 5-HT7 receptor is coupled to G alpha subunits of heterotrimeric G12-protein to regulate gene transcription and neuronal morphology. J Neurosci. 2005;25:7821–7830. doi: 10.1523/JNEUROSCI.1790-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Renner U, et al. Heterodimerization of serotonin receptors 5-HT1A and 5-HT7 differentially regulates receptor signalling and trafficking. Journal of cell science. 2012;125:2486–2499. doi: 10.1242/jcs.101337. [DOI] [PubMed] [Google Scholar]
- 84.Guseva D, Wirth A, Ponimaskin E. Cellular mechanisms of the 5-HT7 receptor-mediated signaling. Front Behav Neurosci. 2014;8:306. doi: 10.3389/fnbeh.2014.00306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shirshev SV. Role of Epac proteins in mechanisms of cAMP-dependent immunoregulation. Biochemistry (Mosc) 2011;76:981–998. doi: 10.1134/S000629791109001X. [DOI] [PubMed] [Google Scholar]
- 86.Peters-Golden M. Putting on the brakes: cyclic AMP as a multipronged controller of macrophage function. Sci Signal. 2009;2:pe37. doi: 10.1126/scisignal.275pe37. [DOI] [PubMed] [Google Scholar]
- 87.Lieder, H. R., Baars, T., Kahlert, P. & Kleinbongard, P. Aspirate from human stented saphenous vein grafts induces epicardial coronary vasoconstriction and impairs perfusion and left ventricular function in rat bioassay hearts with pharmacologically induced endothelial dysfunction. Physiol Rep4, 10.14814/phy2.12874 (2016). [DOI] [PMC free article] [PubMed]
- 88.Chojnacki C, et al. Serum and ascitic fluid serotonin levels and 5-hydroxyindoleacetic acid urine excretion in the liver of cirrhotic patients with encephalopathy. Adv Med Sci. 2013;58:251–256. doi: 10.2478/ams-2013-0010. [DOI] [PubMed] [Google Scholar]
- 89.Abid S, et al. Inhibition of gut- and lung-derived serotonin attenuates pulmonary hypertension in mice. American journal of physiology. 2012;303:L500–508. doi: 10.1152/ajplung.00049.2012. [DOI] [PubMed] [Google Scholar]
- 90.Comai S, et al. Serum levels of tryptophan, 5-hydroxytryptophan and serotonin in patients affected with different forms of amenorrhea. Int J Tryptophan Res. 2010;3:69–75. doi: 10.4137/IJTR.S3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Onaitis MW, et al. Gastrointestinal carcinoids: characterization by site of origin and hormone production. Ann Surg. 2000;232:549–556. doi: 10.1097/00000658-200010000-00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bunin MA, Wightman RM. Quantitative evaluation of 5-hydroxytryptamine (serotonin) neuronal release and uptake: an investigation of extrasynaptic transmission. J Neurosci. 1998;18:4854–4860. doi: 10.1523/JNEUROSCI.18-13-04854.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Duerschmied D, et al. Platelet serotonin promotes the recruitment of neutrophils to sites of acute inflammation in mice. Blood. 2013;121:1008–1015. doi: 10.1182/blood-2012-06-437392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fleetwood AJ, Dinh H, Cook AD, Hertzog PJ, Hamilton JA. GM-CSF- and M-CSF-dependent macrophage phenotypes display differential dependence on type I interferon signaling. Journal of leukocyte biology. 2009;86:411–421. doi: 10.1189/jlb.1108702. [DOI] [PubMed] [Google Scholar]
- 95.Fleetwood AJ, Lawrence T, Hamilton JA, Cook AD. Granulocyte-macrophage colony-stimulating factor (CSF) and macrophage CSF-dependent macrophage phenotypes display differences in cytokine profiles and transcription factor activities: implications for CSF blockade in inflammation. Journal of immunology. 2007;178:5245–5252. doi: 10.4049/jimmunol.178.8.5245. [DOI] [PubMed] [Google Scholar]
- 96.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 97.Kuleshov MV, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic acids research. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chen EY, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hedlund PB, et al. No hypothermic response to serotonin in 5-HT7 receptor knockout mice. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:1375–1380. doi: 10.1073/pnas.0337340100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Usategui A, Criado G, Del Rey MJ, Fare R, Pablos JL. Topical vitamin D analogue calcipotriol reduces skin fibrosis in experimental scleroderma. Arch Dermatol Res. 2014;306:757–761. doi: 10.1007/s00403-014-1466-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article. Moreover, microarray data were deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession GSE94608.