Abstract
Phagocytes express multiple phosphatidylserine (PtdSer) receptors that recognize apoptotic cells. It is unknown whether these receptors are interchangeable or if they play unique roles during cell clearance. Loss of the PtdSer receptor Mertk is associated with apoptotic corpse accumulation in the testes and degeneration of photoreceptors in the eye. Both phenotypes are linked to impaired phagocytosis by specialized phagocytes: Sertoli cells and the retinal pigmented epithelium (RPE). Here, we overexpressed the PtdSer receptor BAI1 in mice lacking MerTK (Mertk −/− Bai1 Tg) to evaluate PtdSer receptor compensation in vivo. While Bai1 overexpression rescues clearance of apoptotic germ cells in the testes of Mertk −/− mice it fails to enhance RPE phagocytosis or prevent photoreceptor degeneration. To determine why MerTK is critical to RPE function, we examined visual cycle intermediates and performed unbiased RNAseq analysis of RPE from Mertk +/+ and Mertk −/− mice. Prior to the onset of photoreceptor degeneration, Mertk −/− mice had less accumulation of retinyl esters and dysregulation of a striking array of genes, including genes related to phagocytosis, metabolism, and retinal disease in humans. Collectively, these experiments establish that not all phagocytic receptors are functionally equal, and that compensation among specific engulfment receptors is context and tissue dependent.
Introduction
Cell death is a crucial part of life. Each day, billions of cells in the human body undergo apoptotic cell death and must be cleared1–3. In many tissues, dying cells are cleared by phagocytes that engulf and digest the apoptotic corpse3–5. Impediments to apoptotic cell clearance can lead to chronic inflammation and autoimmunity6–9. Apoptotic cell clearance is stimulated upon apoptotic cell exposure of the ‘eat-me’ signal phosphatidylserine (PtdSer) and its subsequent recognition by PtdSer receptors on phagocytes10–12. Phagocytes express a multitude of PtdSer receptors, yet, despite extensive research on apoptotic cell recognition and clearance, the relative importance of PtdSer receptors in a given phagocyte or tissue context is unclear4,13.
PtdSer receptors include many different families of proteins including integrins, the BAI family of GPCRs, the TIM family, and the TAM family of receptor tyrosine kinases2,3,14–20. These protein families are structurally diverse and are related only by their shared ligand, PtdSer3–5. Given the importance of apoptotic cell clearance to health and homeostasis, it is not surprising that many different receptors have evolved. However, the loss of a single receptor often causes deficits in apoptotic cell clearance, suggesting that the presence of many receptors cannot be explained as a simple redundancy mechanism1,7,9,10,17,18. Thus, the question of whether PtdSer receptors are interchangeable or unique in cell clearance remains unclear. To address this question, we considered designing a mouse model in which one PtdSer receptor is overexpressed in the absence of another in order to assess functional compensation in vivo.
In most tissues, evaluation of phagocytosis in vivo is complicated by the unpredictable timing of apoptosis and phagocytic events. Two exceptions to this rule are tissues where phagocytosis is mediated by ‘specialized’ phagocytes: the testes and the retina5,14,18. Specialized phagocytes are epithelial-derived, mitotically-quiescent cells21–23. Currently, two best known examples of specialized phagocytes are the retinal pigmented epithelium (RPE) of the eye and the Sertoli cells of the testes5. Sertoli cell phagocytosis is easily studied in the testes as apoptosis of developing germ cells occurs with sufficient regularity to quantify apoptotic corpse accumulation. Similarly, RPE phagocytosis is conducive to in vivo analysis as RPE phagocytosis is circadian-regulated and occurs daily around the time of light onset14,21,24,25. Furthermore, loss of a single PtdSer receptor, MerTK, leads to phagocytic defects in both Sertoli cells and the RPE10,14,21,26. Mertk −/− mice exhibit apoptotic corpse accumulation in the testes and profound retinal degeneration10,26,27. Thus, Mertk −/− mice are uniquely suited for in vivo evaluation of compensation among PtdSer receptors. Indeed, an elegant study by Vollrath et. al. determined that enhanced of expression of Tyro3 can rescue the defective RPE phagocytosis in Mertk −/− mice27. Therefore, we asked whether a PtdSer receptor from a completely different family would be capable of the same rescue. To answer this question, we overexpressed the PtdSer receptor BAI1 in Mertk −/− mice (Mertk −/− Bai1 Tg).
Specialized phagocytes perform a multitude of supportive functions within the tissue. Though the testes and retina are highly disparate at first glance, Sertoli cells and RPE share several key features, including: epithelial derivation, mitotic quiescence, formation of a blood-tissue barrier, maintenance of immunological privilege, and as stated above, phagocytosis22,23,26. As, phagocytes, Sertoli cells are responsible for clearing germ cells that undergo apoptosis during spermatogenesis and the residual-body of cytoplasm that is removed from maturing sperm during spermiation25,28. RPE have a slightly different phagocytic function, unlike Sertoli cells which mediate corpse clearance, the RPE ‘trim’ the adjacent photoreceptors in a PtdSer dependent manner13,21,26,29. This RPE-mediated pruning of the photoreceptors occurs daily and is a waste-removal mechanism, removing the photo-oxidative byproducts that accumulate during phototransduction14,29,30. Mertk −/− mice are born with a full complement of photoreceptors but exhibit early-onset photoreceptor degeneration due to impaired phagocytosis (pruning) of photoreceptor outer segments (POS)1,14,21.
Here, we observe that while the BAI1 transgene has the ability to reduce apoptotic corpse accumulation in the testes of Mertk −/− mice following testicular torsion, the phagocytic defect in Mertk −/− RPE is not compensated by transgenic overexpression of BAI1. When we evaluated the visual cycle (a function unique to RPE), we observed that MerTK expression impacted the visual cycle prior to the onset of retinal degeneration. In addition, RNAseq revealed that the expression of many genes, including those related to phagocytosis and metabolism and other forms of retinal disease, were dysregulated in Mertk −/− RPE prior to the onset of retinal degeneration. Collectively, these findings suggest that PtdSer receptor functionality is contingent on tissue and context, and that while they can compensate for each other in certain contexts, they also have unique roles where they are not interchangeable.
Results
BAI1-transgene reduces corpse accumulation in the testes of Mertk−/− mice
To determine whether distinct PtdSer receptors play unique roles in the process of engulfment, we designed a genetic approach to determine whether the overexpression of one receptor could rescue for the loss of another. We crossed MerTK null (Mertk −/−) and BAI1-overexpressing (Bai1 Tg) mice to generate Mertk −/− Bai1 Tg mice. The rationale for choosing Mertk −/− mice is that they have two in vivo phenotypes associated with impaired phagocytosis. First, Mertk −/− mice exhibit accumulation of apoptotic germ cells in the testes due to impaired Sertoli cell phagocytosis10,31. Second, these mice exhibit profound retinal degeneration due to failed clearance of photoreceptor outer segments (POS) by the RPE2,14,32. We elected to overexpress Bai1 in an attempt to rescue the phenotypes in Mertk −/− mice, as BAI1 overexpression can enhance PtdSer-dependent apoptotic cell clearance by multiple phagocytes, including intestinal epithelial cells and Sertoli cells. Furthermore, Bai1 Tg mice had been previously generated and characterized1,5,18,33.
Sertoli cells are the specialized phagocytes of the testes and promote routine phagocytosis of apoptotic germ cells. Sertoli cells utilize both BAI1 and MerTK during the phagocytosis of apoptotic germ cells1,10. Mertk −/− mice exhibit accumulation of apoptotic corpses at baseline10 and Bai1 −/− mice exhibit apoptotic corpse accumulation following testicular torsion1. Furthermore, Bai1 Tg mice exhibit a decrease in corpse accumulation following torsion1. Given the endogenous role for MerTK and BAI1 in the testes, we initially tested PtdSer receptor compensation in Mertk −/− Bai1 Tg mice in the context of Sertoli cells.
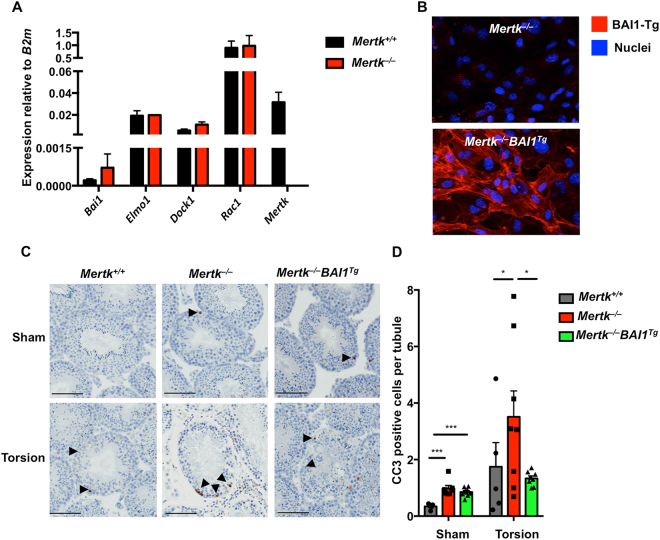
Prior to evaluating apoptotic cell accumulation in the testes, we confirmed that Sertoli cells endogenously express Mertk, Bai1 and components of the BAI1 signaling pathway: Elmo1, Dock180 and Rac1 (Fig. 1A)4. Importantly, expression of Bai1, Elmo1, Dock180 and Rac1 did not differ between Mertk +/+ and Mertk −/− mice (Fig. 1A). In addition, we confirmed that the Bai1 Tg was expressed by Sertoli cells and that the BAI1-Tg properly localized to the surface of Mertk −/− Sertoli cells (Fig. 1B). To determine whether Bai1 overexpression could rescue phagocytic deficits in Mertk −/− Sertoli cells, we evaluated apoptotic corpse accumulation in testes that had undergone surgical torsion and those that underwent sham surgery6,8. Corpse accumulation was quantified by counting cleaved-caspase 3 positive cells in testicular cross-sections (Figs 1C, S1). As was previously reported, Mertk −/− mice exhibited a slight but significant increase in apoptotic corpse accumulation at baseline (Fig. 1C,D ) 10. While the Mertk −/− Bai1 Tg mice trended towards fewer apoptotic corpses at baseline, this was not statistically significant (Fig. 1D). Analysis of injured testes revealed that Mertk −/− mice had substantially more apoptotic cell accumulation than control littermates (Fig. 1C,D). Interestingly, corpse accumulation in Mertk −/− Bai1 Tg mice was significantly decreased compared to Mertk −/− mice. In fact, the corpse numbers in Mertk −/− Bai1 Tg mice were reduced to the number in Mertk +/+ mice (Fig. 1C,D). These data suggest that in the context of testicular torsion, overexpression of Bai1 can reduce the number of apoptotic corpses that seem to accumulate in Mertk −/− Sertoli cells.
Figure 1.
Bai1 Tg reduces accumulation of apoptotic corpses in Mertk −/− mice post-torsion. (A) Sertoli cell expression of Bai1, BAI1 signaling pathway genes, and Mertk were analyzed by quantitative RT-PCR. Sertoli cells were isolated from Mertk +/+ (n = 4) and Mertk −/− (n = 2) mice and were cultured for 3 days to expand them prior to RNA isolation. Error bars are standard error of mean (SEM). (B) Representative images of isolated Sertoli cells from Mertk −/− and Mertk −/− Bai1 Tg mice were stained for BAI1 to confirm surface expression of the Bai1 Tg. (C) Mice (8–12 weeks-old) underwent testicular torsion surgery to induce ischemic injury. Testicular cross sections from sham and torsion testes were stained for cleaved caspase 3 (CC3) (black arrowheads). Images are of representative tubule cross sections from matched sham and torsion testes. (D) The number of CC3 positive cells per tubule cross section was determined by analyzing the entire testicular cross section. Each mouse is represented by individual data points within the bars. Mertk +/+ (n = 5) Mertk −/− (n = 8) Mertk −/− Bai1 Tg (n = 9). Error bars represent SEM. Statistical analysis was performed with a Wilcoxon rank-sum test. *p < 0.05, ***p < 0.001.
RPE express the Bai1Tg and BAI1 signaling pathway in Mertk−/−Bai1Tg mice
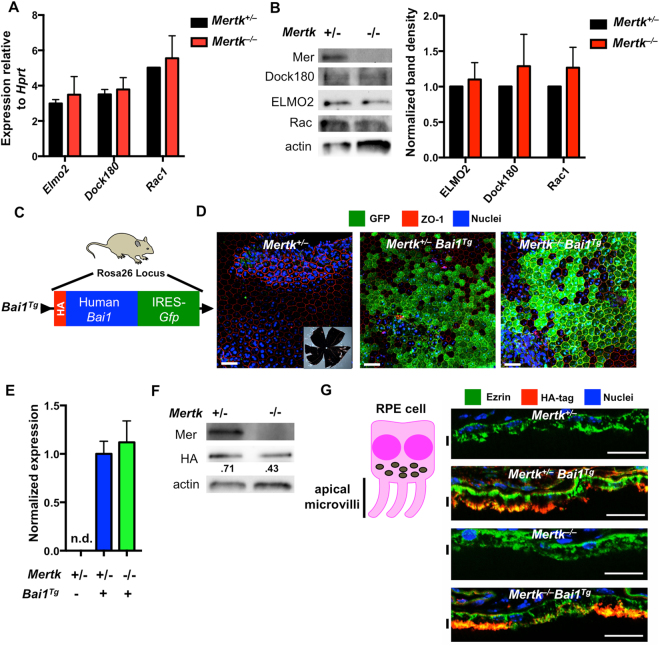
Previous studies have established that POS expose PtdSer that is subsequently recognized by the RPE13 and Mertk −/− mice exhibit profound retinal degeneration linked to failed clearance of POS by the RPE14,15. While Mertk is highly expressed by the RPE14,15,21, RPE do not express BAI1 endogenously (Figure S2A). We first confirmed that downstream components of the BAI1 signaling pathway are present within the RPE. Isolated RPE from Mertk +/− and Mertk −/− mice were analyzed by RT-PCR and immunoblotting. These analyses showed expression of the BAI1 signaling components ELMO, Dock180 and Rac1 by RT-PCR (Fig. 2A,B). Importantly, we found that ELMO2, Dock180 and Rac1 were all expressed. Furthermore expression of BAI1 signaling components did not differ between RPE from Mertk +/− and Mertk −/− mice as assessed by both RNA and protein (Fig. 2A,B), suggesting that BAI1 could theoretically function in the RPE.
Figure 2.
RPE express components of BAI1-signaling pathway and Bai1 Tg. (A) Expression of the BAI1 signaling pathway was analyzed by quantitative RT-PCR in RPE isolated from P14 Mertk +/− and Mertk −/− mice (n = 2) Error bars represent SEM. (B) Immunoblot analysis of BAI1 signaling pathway in RPE whole cell lysates isolated from P14 mice. Left panel shows representative immunoblots for Mer, Dock180, Elmo2 and Rac. Right panel shows combined densitometry analysis of immunoblots from n = 3 mice per genotype. For densitometry analysis, band volumes were normalized to an actin loading control and band densities in Mertk −/− were then normalized to Mertk +/− for comparison across multiple blots. (C) Schematic of Bai1 Tg insertion in the Rosa26 locus indicating the N-terminal HA-tag in red and IRES-GFP in green. Mouse image licensed from Motifolio Inc. (D) GFP expression was analyzed in RPE flat mount preparations at 20x magnification. Inset image in the left panel shows a representative flat mount at 1.2x magnification. White scale bars in images are 50 μm. Images are representative of n = 2 mice per genotype. (E) qRT-PCR analysis of Bai1 Tg expression in Mertk +/− Bai1 Tg (n = 9) and Mertk −/− Bai1 Tg (n = 3) RPE isolated on P14. n.d. = not detected. Error bars represent SEM. (F) Immunoblot analysis of HA-tag (Bai1 Tg) in RPE whole cell lysates isolated from P13-P14 mice. Immunoblot shown is representative of n = 2 experiments. (G) HA staining and localization was analyzed in eyecups at 40x magnification. Neural retinas were removed prior to fixation and staining. Black lines next to the figure align with apical microvilli of RPE. Scale bars are 20 μm. Images shown are representative of n = 2 mice per genotype.
We next assessed Bai1 Tg expression and localization within the RPE. The Bai1 Tg mouse construct contains three separate methods to detect expression in situ (Fig. 2C ). First, the Bai1 Tg construct includes an IRES-driven GFP, which allows visualization of the transcriptional activity at the Bai1 Tg locus. To assess GFP expression in RPE, we prepared RPE flat mounts from Mertk +/− Bai1 Tg and Mertk −/− Bai1 Tg mice and imaged GFP by confocal microscopy (Fig. 2D). The GFP expression pattern appears uniform across the RPE layer in both Mertk +/− Bai1 Tg and Mertk −/− Bai1 Tg mice. On average, 45% of the area in each 20x field is GFP positive (data not shown). Importantly, no GFP signal was detected in mice lacking the Bai1 Tg (Fig. 2D). Second, the Bai1 Tg is derived from the human Bai1 cDNA1. Despite being highly homologous to mouse BAI1 at the protein level and indistinguishable in functional assays14,18, human Bai1 transcript can be distinguished from the murine transcript by RT-PCR. Human Bai1 was readily detected in the RPE of Mertk +/− Bai1 Tg and Mertk −/− Bai1 Tg but not littermate control mice (Fig. 2E). Third, the Bai1 Tg construct has an N-terminal HA-tag, which facilitates detection of the Bai1 Tg protein. The HA-BAI1 protein was readily detected in the extracts of isolated RPE cells by immunoblotting (Fig. 2F).
RPE cells are highly polarized and the apical microvilli mediate the phagocytosis of photoreceptor outer segments. Therefore, to assess whether the Bai1 Tg protein was properly localized on the apical surface in a location similar to MerTK21, we stained for the HA-tag in eyecup cross sections. HA-BAI1 showed apical localization and co-localized with ezrin, a cytoskeletal protein enriched in microvilli (Fig. 2G). Furthermore, HA-BAI1 was detected on the apical RPE surface of both Mertk +/− Bai1 Tg and Mertk −/− Bai1 Tg mice. These data suggested that RPE cells in the Mertk −/− Bai1 Tg mice express the Bai1 Tg, and that the BAI1 protein localizes to the region of the RPE that mediates phagocytosis.
To examine whether there are signals initiated by the BAI1 transgene within the RPE, we performed RNAseq on Mertk +/+ and Mertk +/+ Bai1 Tg mice to identify genes likely altered by the Bai1 Tg. Significant transcriptional changes were noted in 63 genes from the Bai1 Tg mice. When we focused on processes previously associated with BAI1, such as cholesterol homeostasis, axonal growth and synaptogenesis33–35, we identified 10 genes with functions that are associated with the aforementioned processes (Figure S2B). These data suggest that BAI1 is not only expressed by RPE cells at the correct location, but can also signal to induce transcriptional changes in RPE cells.
Mertk-linked retinal degeneration is not rescued by BAI1
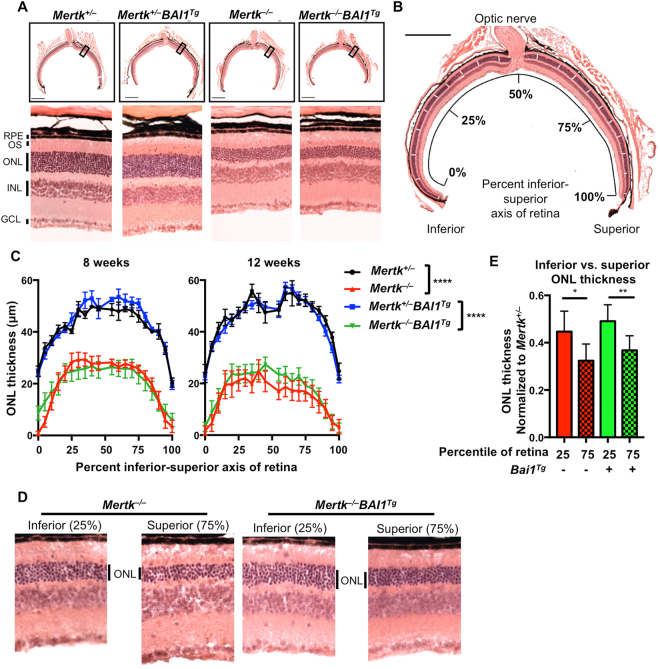
We next assessed the retinal degeneration in Mertk −/− Bai1 Tg mice. Photoreceptors in Mertk −/− mice begin to show signs of overgrowth at post-natal day 17 and exhibit highly disorganized POS by 35 days after birth14,21. Degeneration of the photoreceptor layer begins soon afterwards and previous reports have demonstrated that most photoreceptors are lost by 12 weeks of age14,21,26. To assess retinal degeneration, we collected sagittal sections of eyes that transect the optic cup (where the optic nerve meets the retina) (Fig. 3A,B). Inspection of central retinas from 12 week-old Mertk −/− and Mertk −/− Bai1 Tg revealed equivalent degeneration of the outer nuclear layer (ONL) consisting of photoreceptor nuclei. Importantly, Mertk +/− Bai1 Tg retinal cross sections exhibited normal retinal architecture suggesting that the retinal structure was not adversely affected by transgenic Bai1 expression (Fig. 2A).
Figure 3.
Bai1 Tg does not rescue retinal degeneration in Mertk −/− mice. (A) H&E stained eyecup sections that transect the optic cup were imaged from the different genotypes at 20x magnification. (B) An image mask was applied to the images in Photoshop marking the regions to be measured (white dashes overlaid on ONL). Image is a representative eyecup with the overlaid image mask. Scale bar is 500 μm. (C) ONL measurements were taken at the indicated points (according to the image mask) along the inferior-superior axis of the retina. The left panel includes measurements from 8 week-old Mertk +/− (n = 3) Mertk +/− Bai1 Tg (n = 5), Mertk −/− (n = 10) and Mertk −/− Bai1 Tg (n = 13) mice. Right panel shows measurements from 12 week-old Mertk +/− (n = 6) Mertk +/− Bai1 Tg (n = 8), Mertk −/− (n = 6) and Mertk −/− Bai1 Tg (n = 10) mice. Statistical analysis by 2-way ANOVA showed no difference between Mertk +/− and Mertk +/− Bai1 Tg mice or Mertk −/− and Mertk −/− Bai1 Tg mice at any time point. Asterisks indicate the difference between genotypes as measured by 2-way ANOVA. Error bars represent SEM. ****p < 0.0001. (D) Images of the ONL in the inferior retina (25% measurement mark) and the superior retina (75% measurement mark) from representative mice. (E) Quantification of the difference in ONL thickness in inferior retina (25% measurement mark) and the superior retina (75% measurement mark) at 12 weeks of age. Mertk −/− (n = 6) Mertk −/− Bai1 Tg (n = 10). Significance was determined using a Wilcoxon rank-sum test. *p < 0.05, **p < 0.01. Error bars represent SEM.
ONL thickness is not equivalent across the retina and degeneration is not necessarily homogeneous26,27. To standardize the measurement of ONL across the entire section, we adapted a previously described technique26 and measured the ONL thickness at 20 standardized points across the ‘inferior-superior’ axis of the retinal section (Fig. 3B). We measured ONL thickness in mice at 8 and 12 weeks of age (Fig. 3C). At 8 weeks of age, both Mertk −/− and Mertk −/− Bai1 Tg mice exhibited moderate degeneration across the entire inferior-superior axis (Fig. 3C). By 12 weeks of age, retinal degeneration had progressed in both Mertk −/− and Mertk −/− Bai1 Tg mice (Fig. 3C). As previously reported26,27, degeneration was more severe in the superior than the inferior retina (Fig. 3D,E). Importantly, degeneration was equivalent between Mertk −/− and Mertk −/− Bai1 Tg animals. Collectively, these data indicate that overexpression of Bai1 did not rescue retinal degeneration due to loss of Mertk.
For analysis of ONL degeneration we facilitated littermate comparison by using Mertk +/− and Mertk −/−mice as previous studies demonstrated that the retinas of Mertk +/− mice are indistinguishable from Mertk +/+ mice21,26. Importantly, we also confirmed that Mertk +/− mice exhibited no retinal degeneration as late as 16 weeks of age (Figure S3A), despite decreased expression of MerTK in Mertk +/− relative to Mertk +/+ animals (Figure S3B,C ).
Phagocytosis defect in MerTK-null mice is not rescued by the BAI1 transgene
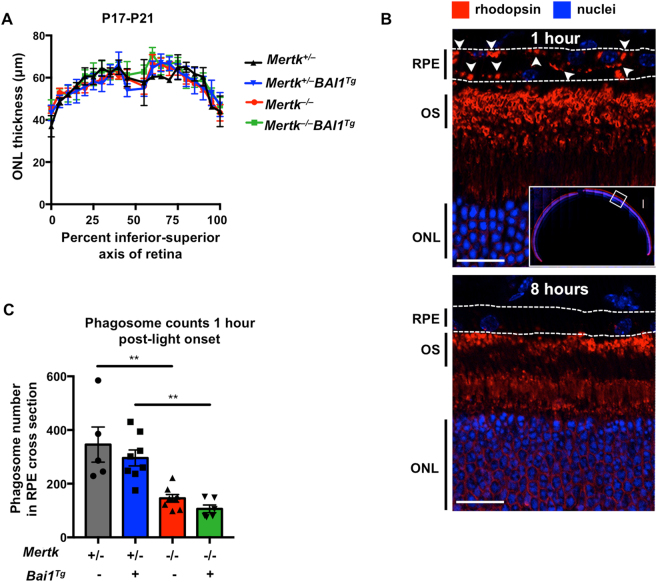
Retinal degeneration in Mertk −/− mice is attributed to decreased phagocytosis of POS. Therefore, we analyzed RPE phagocytosis of POS in Mertk −/− and Mertk −/− Bai1 Tg mice. Phagocytosis by RPE in vivo is readily analyzed due to its temporal regulation (around light onset) and because the content of the phagosomes (rhodopsin) can be visualized14,30. The rate and amount of phagocytosis is a dual function of phagocyte efficiency and the ratio of targets to phagocytes. Since Mertk −/− mice exhibit photoreceptor degeneration, the number of POS targets decreases as degeneration progresses. Therefore, decreases in RPE phagocytosis after the onset of degeneration could be due to the inefficient uptake by the RPE or a decreased target to phagocyte ratio. To ensure that any observed changes in phagocytosis were due to RPE phagocytic efficiency, we analyzed mice prior to the onset of retinal degeneration, at P17-P21 days of age. Importantly, analysis of ONL thickness at this age confirmed that degeneration in Mertk −/− and Mertk −/− Bai1 Tg mice had not yet begun (Fig. 4A).
Figure 4.
Bai1 Tg is unable to restore RPE phagocytosis. (A) Eyes were isolated from P17-P21 mice 1 h after light onset. ONL measurements were completed as described above. Measurements were taken along the inferior-superior axis at the indicated points from Mertk +/− (n = 2), Mertk +/− Bai1 Tg (n = 5), Mertk −/− (n = 5), and Mertk −/− Bai1 Tg (n = 4) mice. Error bars represent SEM. No significant difference was found between any genotypes by 2-way ANOVA. (B) Eyecups isolated 1 h after light onset (top panel) were stained for rhodopsin and nuclei (Hoechst). The entire eyecup was imaged at 40x (top panel, inset). Rhodopsin staining is robust in the outer segments (OS) of the photoreceptors. Rhodopsin puncta are apparent in the RPE layer (white arrowheads). Bottom panel shows representative staining from an eyecup isolated 8 h after light onset. (C) Rhodopsin puncta in the RPE, referred to as phagosomes, were quantified in ImageJ by an automated particle count. Particle size was constrained from 0.5 μm to ∞ and minimum circularity was restricted to 0.2. Symbols within the bars represent the average particle count from the left and right eyes of one mouse. Mertk +/− (n = 5) Mertk +/− Bai1 Tg (n = 8) Mertk −/− (n = 8) Mertk −/− Bai1 Tg (n = 6). Error bars represent SEM. Significance was determined by one-way ANOVA. Multiple comparison analysis was corrected with a post-hoc Tukey’s test.
To quantify phagocytosis, we harvested eyes 1 h after light onset and immunostained sagittal-eye sections for rhodopsin (Fig. 4B, white arrows). Importantly, rhodopsin puncta are degraded over time and thus are mostly absent 8 h after light onset, suggesting that these puncta are not artifacts of sectioning or staining (Fig. 4B). Quantification of puncta at 1 h post-light onset revealed a striking decrease in rhodopsin puncta in the RPE of Mertk −/− mice relative to Mertk +/− mice. However, neither Mertk +/− Bai1 Tg nor Mertk −/− Bai1 Tg mice exhibited differences in the number of puncta compared to their respective controls (Fig. 4C). Furthermore, neither the loss of Mertk nor overexpression of BAI1 affected phagosome trafficking in RPE, as indicated by basolateral localization of phagosomes (Figure S4 ). BAI1 overexpression was previously shown to enhance phagocytosis in WT epithelial cells (both in vitro and in vivo)1. However, BAI1 was unable to enhance phagocytosis in Mertk +/+ mice and importantly, the number of puncta in Mertk +/+ mice were equivalent to the number seen in Mertk +/− (Figure S3D). Collectively, these data suggest that Bai1 Tg expression is unable to rescue the RPE phagocytic defect observed in Mertk −/− mice.
MerTK regulates the gene expression program in the RPE
The lack of rescue by BAI1 prompted us to ask whether MerTK regulates the phagocytic capacity of RPE in ways beyond PtdSer binding. To address this question, we chose an unbiased transcriptomics approach, and performed RNAseq analysis of RPE from Mertk +/+ and Mertk −/− mice. RPE were isolated at day 14 after birth, a time when the mouse pups have opened their eyes but Mertk −/− mice do not yet exhibit photoreceptor overgrowth that could impact RPE gene expression14,21.
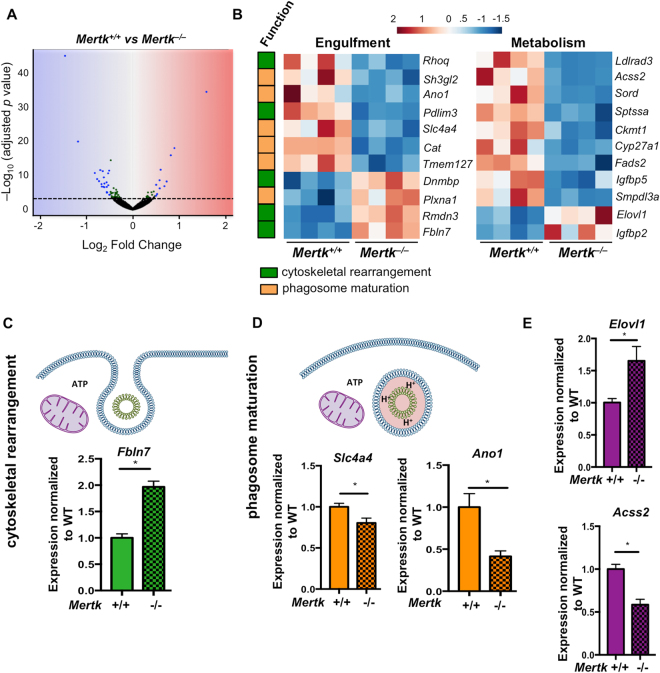
RNAseq analysis identified 60 genes that were differentially regulated in Mertk −/− RPE (Fig. 5A). Further analysis revealed that 11 of these genes have annotated functions related to either cytoskeletal rearrangement or endosomal maturation, two processes that are essential for phagocytosis (Fig. 5B). The gene fibulin 7 (Fbln7), important for extracellular matrix adhesion and cytoskeletal rearrangement36–38, has a SNP associated with age-related macular degeneration and its expression is altered in patients with retinoschisis, a form of inherited retinal dystrophy39,40. qRT-PCR validation of the original library and an additional cohort of Mertk +/+ and Mertk −/− mice confirmed that Fbln7 is upregulated in Mertk −/− RPE (Fig. 5C). Phagosome maturation culminates in acidification, which promotes the breakdown of the phagosome content41. Chloride flux across the lysosome membrane regulates lysosome acidification42,43. Therefore we further assessed the chloride channel anoctamin 1 (Ano1) and the bicarbonate transporter Slc4a4 for further validation. Although Ano1 has yet to be implicated in the chloride flux across lysosomal membranes, it is a well-established calcium-activated chloride transporter44 and RPE phagocytosis is accompanied by substantial calcium accumulation in the RPE45. Slc4a4 is a bicarbonate transporter that is thought to regulate intracellular pH by electrogenic flux of H+ and HCO3 − 46. Interestingly, it has been proposed that these ion transporters work together in certain cellular processes47. We validated that Slc4a4 and Ano1 were reproducibly downregulated in both the original RPE samples as well as in an additional cohort of RPE (Fig. 5D). These data suggest that the presence of MerTK can regulate the expression of various components of the phagocytic machinery, either directly or indirectly.
Figure 5.
Mertk regulates multiple genes linked to phagocytosis and metabolism. (A) RNAseq was performed on RPE isolated from P14 Mertk +/+ (n = 4) and Mertk −/− (n = 4) mice 2 h after light-onset. DEseq. 2 analysis identified 60 genes that were differentially expressed according to p values adjusted for multiple comparisons. The log2 fold change and –Log10padj values are plotted for all hits. Green dots represent genes that had a padj value <0.05. Blue dots indicate genes with a padj <0.05 and log2 fold change >0.5. (B) Further annotation of the differentially regulated genes revealed 11 linked to pathways important in phagocytosis and 11 linked to metabolic pathways. These genes and their expression changes are depicted in heat maps. Functional associations with regard to phagocytosis are indicated on the left. (C) Genes related to cytoskeletal reorganization (graphs in green), (D) genes related to phagosome maturation (graphs in yellow) and (E) genes related to metabolism (graphs in purple) were selected for further validation by qRT-PCR. Validation studies were done with fresh RPE samples from P14 Mertk +/+ (n = 4) and Mertk −/− (n = 4) and original library preparations for a total of 8 samples per genotype. Significance was calculated by student’s T-test. *p < 0.05.
Phagocytosis is a metabolically demanding process for the phagocyte that requires significant ATP generation to mediate cytoskeletal rearrangement and digestion of cargo48,49. In addition to annotating multiple genes involved in phagocytosis, we identified an additional 11 genes related to cell metabolism (Fig. 5B). Although these genes we identified have yet to be directly linked to phagocytic processes, the number of genes that were differentially regulated was intriguing. We validated the fatty acid elongase, Elovl1 and the acetyl CoA synthetase, Acss2, and found that the expression changes were reproducible, in both the original library and a fresh cohort of Mertk +/+ and Mertk −/− mice (Fig. 5E). Overall, these data suggest that MerTK might coordinate the phagocytic process at multiple levels from PtdSer binding, to cytoskeletal reorganization and phagosome maturation, as well as coordinating metabolic changes during phagocytosis (and potentially the visual cycle).
In addition, 9 genes identified from the RNAseq screen were linked to human retinal disease (Table 1). Five of these genes were also annotated under the metabolic subset (marked with*) and three were annotated in the phagocytosis subset (marked with #) (Fig. 5B, Table 1). Two genes, Gja1 and Cyp27a1 have a striking monogenic association with oculodentodigital dysplasia and cerebrotendinous xanthomatosis respectively, both of which are associated with retinal abnormalities50,51. Three genes had SNP associations to retinal disease: Ldlrad3, Fads2, and Fbln7 (Table 1). Five of the genes we annotated as being linked to retinal disease, were identified in the comparative toxicogenomics database, which infers gene-disease associations by examining curated associations between pharmacological agents, known diseases and the gene of interest52,53. These associations were further validated by experimental links to retinal disease or RPE function. Slc4a4 has also been linked to retinitis pigmentosa (RP) type 17 and Elovl1 is an endogenous inhibitor of the visual cycle enzyme RPE65, defects in which cause RP type 2054–56. Overall, our findings suggest that the loss of MerTK (directly or indirectly) perturbs multitude of genes, which might influence additional RPE functions beyond phagocytic pruning of photoreceptors.
Table 1.
The absence of Mertk alters expression of genes linked to human retinal diseases.
| Gene | Log2 fold change | Reported retinal disease association in humans | Experimental evidence |
|---|---|---|---|
| Ldlrad3* | −1.08 | SNP66, inferred52,53 | Intronic SNP associated with pathological myopia, a disease associated with degeneration of several eye structures including RPE67–69 |
| Gja1 | −0.54 | Monogenic51,70, inferred52,53 | Mutations in Gja1 have a monogenic association with oculodentodigital dysplasia, which has multiple manifestations including retinal dysplasia51. Gja1 is also critical in RPE differentiation and communication between RPE cells71,72. |
| Cyp27a1* | −0.48 | Monogenic50, inferred52,53 | Mutations in Cyp27a1 have a monogenic association with cerebrotendinous xanthamatosis, which is associated with premature retinal senescence50,73. Mice with mutations in Cyp27a1 exhibit abnormal retinal vascularization and cholesterol deposits in the RPE74,75. |
| Slc4a4 # | −0.48 | inferred52–54,76 | Retinitis pigmentosa 17 (RP17) is characterized by mutations in Ca4 77,78. Some variants of RP17 have Ca4 mutations that prevent interaction and activation of Slc4a4 54. |
| Cat # | −0.48 | inferred76,79–81 | Increased expression of Cat in RPE prevents oxidative damage to photoreceptors80. Age-related macular degeneration (AMD) is associated with decreased catalase activity in RPE79. |
| Fads2* | −0.36 | SNP82, inferred52,53 | SNPs in intronic and regulatory regions of Fads2 have been linked to AMD82. Fads2 −/− mice exhibit structural changes in interphase between RPE and photoreceptors83. |
| Igfbp5* | −0.36 | inferred52,53 | Altered Igfbp5 expression is associated with myofibroblastic changes in RPE84. |
| Elovl1* | 0.45 | inferred52,53 | ELOVL1 is an endogenous inhibitor of the visual cycle enzyme, RPE6556. Mutations in Rpe65 cause retinitis pigmentosa55. |
| Fbln7 # | 0.82 | SNP39, inferred52,53 | Intronic SNP associated with reduced severity of AMD39. |
*Indicates gene function is associated with metabolism.
#Indicates gene function is associated with phagocytosis.
MerTK−/−mice exhibit diminished retinyl ester accumulation prior to the onset of degeneration
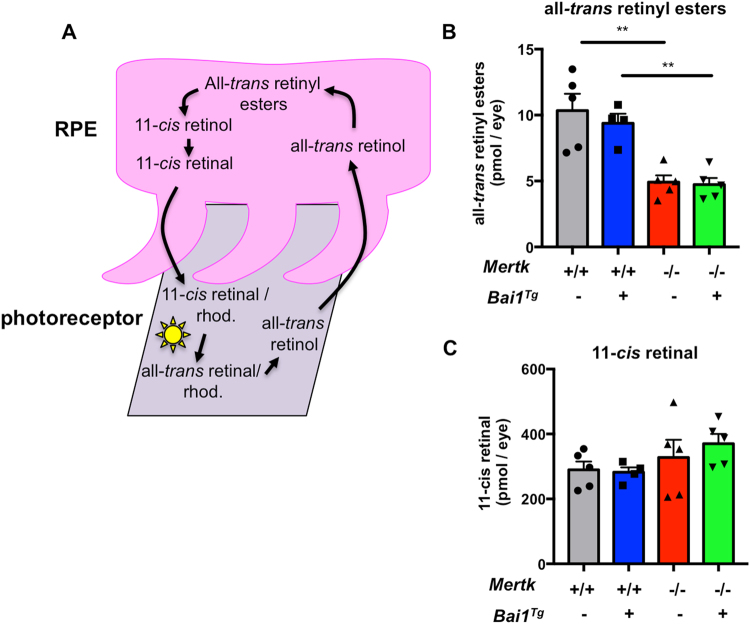
In addition to promoting photoreceptor survival, RPE contribute indirectly to phototransduction by participating in the visual cycle, which supplies photoreceptors with the chromophore 11-cis-retinal. The visual or retinoid cycle is the recycling of vitamin A (all-trans-retinol) to the light-reactive chromophore 11-cis-retinal (Fig. 6A)32. Interestingly, one of the first characterizations of Mertk −/− mice revealed that vision is affected in mice as early as P20, prior to the onset of retinal degeneration14. Given the broader changes in gene expression that we observed in Mertk −/− RPE, we investigated whether Mertk expression influenced the visual cycle.
Figure 6.
Mertk −/− mice have reduced retinyl ester accumulation. (A) Schematic of the visual (retinoid) cycle that occurs in RPE (pink) and POS (gray). (B) P20-P21 mice were dark adapted overnight and HPLC analysis was performed to quantify all-trans-retinyl esters and (C) 11-cis-retinal. Individual points within bars are the average value for both eyes from one mouse: Mertk +/+(n = 5) Mertk +/+ Bai1 Tg (n = 4) Mertk −/− (n = 5) Mertk −/− Bai1 Tg (n = 5). Error bars represent SEM. Statistical analysis was performed by one-way ANOVA. Multiple comparisons were corrected with a post-hoc Tukey’s test. **p < 0.01.
High-performance liquid chromatography was used in a blinded assay to quantify all-trans-retinyl esters and 11-cis-retinal in the eyes of dark-adapted mice at P21 (prior to degeneration but after significant retinal maturation)14,21,57,58. Retinyl esters are notoriously difficult to extract and quantify, thus we developed a novel procedure that allows extraction and quantification of the esters with more than 90 percent accuracy59. Interestingly, we found that all-trans-retinyl ester levels were significantly lower in Mertk −/− relative to WT mice (Fig. 6B). However, there was no difference in the levels of the chromophore 11-cis-retinal at this time point (Fig. 6C). Retinyl esters are a form of stored vitamin A; esterification to fatty acids prevents their release in to the extracellular space60,61. The shifts we observed in metabolic gene expression could influence the availability of fatty acids for esterification, possibly affecting retinyl ester accumulation. Diminished retinyl ester levels in Mertk −/− mice prior to the loss of photoreceptors support the hypothesis that Mertk can influence critical processes beyond phagocytosis, and that MerTK is uniquely critical for RPE function.
Discussion
Phagocytes express multiple PtdSer receptors and it has been unclear whether these receptors are functionally redundant or if they have tissue and context specific functions. In an attempt to determine if PtdSer receptors are functionally unique, we asked if PtdSer receptors are capable of compensating for one another in specialized phagocytes in vivo. Our analysis of the Mertk −/− Bai1 Tg mice led to two major findings. First, Bai1 Tg expression appears to rescue phagocytic deficits in Mertk −/− Sertoli cells. Second, Bai1 Tg expression was not able to prevent retinal degeneration or rescue phagocytic deficits in the Mertk −/− RPE. These findings provide the first evidence for functional compensation between two different PtdSer phagocytosis receptors from two distinct receptor families: The adhesion family GPCR, BAI1 and the receptor tyrosine kinase MerTK. Our results also suggest that the role of a given PtdSer receptor in cell clearance may differ depending on the tissue context. Overall, this indicates that PtdSer receptors are not fully interchangeable, which could, in part, explain why phagocytes express a variety of PtdSer receptors on their cell surface. It also suggests that expression of MerTK is non-redundant in the eye/retina.
To understand what unique role(s) MerTK might have in the RPE, we performed RNAseq on Mertk −/− and Mertk +/+ RPE and identified 60 genes that were differentially expressed in Mertk −/− RPE cells. From those genes, three critical subsets were dysregulated: genes linked to retinal disease, phagocytosis, and metabolism. Genes linked to human retinal disease have associations that are either monogenic, SNP, or inferred based on curated experimental and pharmacological data. Interestingly, 8 of the 9 genes associated with retinal disease were linked to either metabolic or phagocytic functions. These perturbations could explain why Mertk −/− mice exhibit visual defects (as measured by electroretinogram recordings) at P20, prior to the onset of retinal degeneration14. Furthermore, the observed transcriptional changes may help to explain the decreased retinyl ester accumulation in Mertk −/− mice as our data revealed significant changes in metabolic genes that might perturb the availability of fatty acids for esterification.
The finding that Bai1 overexpression can reduce apoptotic corpse accumulation in the testes is intriguing. Though RPE phagocytosis of POS is PtdSer dependent, it is not ‘traditional apoptotic corpse clearance’ and could instead be analogous to the cell pruning observed in the nervous system. It is possible that the mechanism of action of BAI1 in phagocytosis is not conducive to pruning-type events, but rather that BAI1 is more suited for larger corpse uptake. Alternatively, BAI1-mediated phagocytosis could require an as-of-yet unidentified co-factor that RPE cells do not express. This might explain why the Bai1 Tg can enhance phagocytosis in Mertk −/− Sertoli cells, where it is known to function endogenously1,62.
Overall, this work suggests that the variety of PtdSer receptors did not evolve as a simple redundancy mechanism. Rather, these receptors likely play unique roles beyond PtdSer recognition, including the regulation of genes crucial to the process of phagocytosis and biological processes essential for general homeostasis and even specialized tissue functions. Overall, this suggests that PtdSer receptors should not be regarded as having homogenous functionality. Further research into the downstream functions of PtdSer receptors is critical to understanding how the phagocytic process is coordinated in different tissues and contexts.
Experimental Procedures
Mice
Mertk −/− mice were purchased from Jackson Laboratories (stock no: 011122 – B6;129-Mertk tm1Grl/J). Bai1 Tg mice were previously generated by our lab on a C57Bl/6 N background and then backcrossed with C57Bl/6 J mice1. The Bai1 Tg mice were screened for the RD8 mutation and were confirmed to be RD8 negative63 Mertk −/− mice were crossed to Bai1 Tg mice to generate the first generation of Mertk +/− Bai1 Tg mice. Progeny from this initial cross were bred to the original Mertk −/− line from Jackson Laboratories. For line maintenance, Mertk +/− Bai1 Tg progeny were crossed to the original Mertk −/− line from Jackson Laboratories. Mertk +/+ and Mertk +/+ Bai1 Tg mice were generated by crossing Mertk +/− and Mertk +/− Bai1 Tg siblings. Mice were maintained on a 14–10 h light-dark cycle. Animals for analysis were euthanized by CO2 asphyxiation. All animal procedures were approved by and performed according to guidelines of the Institutional Animal Care and Use Committee (IACUC) at the University of Virginia.
RPE isolation for gene expression analyses
The RPE isolation protocol was previously described64. Eyes were enucleated from P14 neonatal mice 2 hours after light onset. Globes were incubated in serum free DMEM (Corning) with 2% dispase (Worthington). Following incubation, eyes were washed 3 times in DMEM supplemented with 10% fetal bovine serum (FBS) (Gemini). Cornea, iris and lens were removed from each eyecup. The eyecups were incubated for 20 minutes at 37 °C in DMEM supplemented with 10% FBS. Following incubation, the neural retina was removed from each eyecup. The eyecup was tugged at opposite ends to release the RPE layer. RPE sheets were washed in Ca2+/Mg2+ free HBSS (Gibco). RPE to be used for RT-PCR or RNAseq were lysed in 350 μL of RLT buffer (Qiagen). RPE to be used for protein analysis were lysed in 40 μL of RIPA buffer.
Immunoblotting
Crude RPE lysates in RIPA buffer were sonicated 2 times for 10 s to shear DNA. Sonicated lysates were incubated at 37 °C in Laemmli sample buffer for 30 min. Lysates were run on Any-kD stain free gels (Bio-Rad) and transferred to PVDF membranes. Membranes were blocked in Tris buffered saline with Tween 20 (TBS-T) with 5% milk for 1 h at room temperature. The following primary antibodies were used at 1:1000 dilution unless otherwise indicated: MerTK (R&D #AF591), HA (Cell Signaling Technology, clone C29F4), Rac1 (Millipore, clone 23A8), Dock180 1:200 (Santa Cruz #6043 and #6167), ELMO2 (in-house65), and β-actin 1:100,000 (Sigma clone AC-15).
RT-PCR
RNA was purified from cell lysates with RNeasy kit (Qiagen). cDNA was prepared with Superscript III kit (Thermo Fisher). The following Taqman probes (Thermo Fisher) were used for qPCR: Bai1 (Mm00558144_m1), Bai1 human (Hs01105174_m1), Bai2 (Mm00557365_m1), Bai3 (Mm00657451_m1), Mertk (Mm00434920_m1), Dock1 (Mm01269874_m1), Elmo2 (Mm01248046_m1) Elmo1 (Mm00519109_m1), Rac1 (Mm01201657_m1), Ano1 (Mm00724407_m1), Elovl1 (Mm01188316_g1), Acss2 (Mm00480101_m1), Fbln7 (Mm01336227_m1) β2 m (Mm00437762_m1), Hprt (Mm00446968_m1), and Slc4a4 (Mm01347935_m1).
Eyecup dissection and ONL analysis
Central corneas of enucleated eyes were punctured with a 25-gauge needle. Eyes were submerged in Hartman’s fixative (Sigma) and incubated for 1–3 h at room temperature. Following initial fixation, the cornea, iris and lens were removed. Eyecups were returned to fixative and incubated overnight at 4 °C. Eyecups were moved to 30% sucrose for cryo-protection and incubated at 4 °C until they sank. Eyecups were embedded in O.C.T. (Tissue-Tek) and flash frozen in an isobutane dry ice bath. Consistent orientation of the ‘nasal-notch’ during embedding was maintained to control eyecup orientation. Sagittal eyecup sections were cut at 10 μm thickness and sections transecting the optic cup were collected. Sections for ONL analysis were stained with Mayer’s haematoxylin (Sigma) and eosin (Fisher) and tiled at 20x magnification on an Axio Imager.z1 (Zeiss) with Stereo Investigator software (MBF Biosciences). An image mask with 20 fixed measurement points was applied to eyecup images in Photoshop (Adobe). The ONL was measured at points indicated by the mask in ImageJ software (NIH).
In situ rhodopsin analysis
P17-P21 mice were euthanized 1 h after light onset. Eyes were dissected, fixed and sectioned as described above. Eye sections were blocked in PBS (Corning) with 10% normal goat serum (Jackson Immunoresearch) for 1 h at room temperature. Sections were stained overnight with rhodopsin antibody diluted 1:500 (Abcam clone Rho 4D2). AF647 conjugated secondary antibody was used to detect rhodopsin. Tiled images were acquired at 40x magnification on an Axio Observer.z1 microscope (Zeiss). The RPE layer was isolated in Photoshop (Adobe). Quantification of puncta was performed by automated particle count in ImageJ (NIH).
Flat mount preparation and staining
Mice were euthanized between 4 and 6 weeks of age. Eyes were enucleated and the cornea, iris, lens and neural retina were removed. Eyes were cut at four points for flat mount ‘clover’ preparation and fixed for 1 h in PBS 4% paraformaldehyde (PFA) at room temperature. Flat mounts were blocked and permeabilized in PBS 10% normal horse serum (Hyclone) and 0.1% Tween 20 then stained over night at 4 °C with anti-GFP antibody at 1:50 (Abcam #6673). Secondary antibody conjugated to AF488 was used to detect GFP staining. Flat mounts were then stained with anti-ZO-1 at 1:250 (Thermo Fisher #61-7300) for 1 h at room temperature. Secondary antibody conjugated to AF647 was used to detect ZO-1 staining. Images were acquired at 20x magnification on a confocal Axio Observer.z1 microscope (Zeiss).
Eyecup staining for HA
Mice were euthanized between 4 and 6 weeks of age. Eyes were enucleated and the cornea, iris, lens, and neural retinas were removed. Eyecups were fixed for 1 h in PBS 4% PFA at room temperature then moved to 30% sucrose for cryo-protection and incubated at 4 °C until the eyecups sank. Eyes were embedded in O.C.T. Eyecups were sectioned at 10 μm and sections bisecting the optic cup were collected. Sections were blocked in PBS with 10% normal goat serum for 1 h at room temperature. Sections were stained overnight at 4 °C with anti-HA antibody 1:100 (Cell Signaling Technology clone C29F4). AF647 conjugated secondary antibody was used to detect staining (ThermoFisher). Sections were then stained with biotin conjugated anti-ezrin antibody 1:100 (ThermoFisher clone 3C12) overnight at 4 °C. AF488 conjugated streptavidin was used to detect ezrin staining (ThermoFisher). Images were acquired at 40x magnification on an Axio Imager.z2 microscope with Apotome (Zeiss).
Testicular torsion
Torsion surgery was performed as previously described8. The studies were performed in accordance with the ‘Guiding Principles of the Care and Use of Research Animals’ promulgated by the Society for the Study of Reproduction. The male mice were anesthetized with an intraperitoneal (IP) injection of a mixture of 6 mg /100 g of ketamine and 0.5 mg/100 g of xylazine. After the testes were exteriorized through a low ventral midline incision, the testes were released from the epididymo-testicular membrane through incising the gubernaculums. For torsion, the testis of right side was rotated 720° for 2 h, during which time it remained in the abdomen with a closed incision. For sham control, the testis was freed of the epididymo-testicular membrane and left in the abdomen. After 2 h, the incision was reopened, the testis was counter-rotated to the natural position, the gubernaculum was rejoined, the testes were reinserted into the scrotum, and then the incision was closed. 24 h after operation, mice were euthanized. The testes were removed and fixed for 6 h in Bouin’s fixative for paraffin embedding. Testicular cross sections were stained for cleaved caspase-3 at the University of Virginia Biorepository and Tissue Research Facility. Testicular cross sections were imaged at 20x with an Aperio Scanscope at the University of Virginia Biomolecular Analysis Facility and Shared Instrumentation Core. The number of apoptotic cells per tubule was determined for the entire cross section.
Sertoli cell isolation and staining
Testes from P11-P21 day-old mice were isolated and decapsulated. Tubules were dispersed in a solution of HBSS, 0.0625% Trypsin (Corning) 10 μg/mL DNase (Sigma) for 20 min. at 37 °C then 0.00625% Soybean trypsin inhibitor (SBTI) (Sigma) was added and the supernatants were decanted. Tubules were re-suspended in HBSS with 1 M glycine, 2 mM EDTA, 0.0625% SBTI, and 10 μg/mL DNase and incubated for 10 min. at room temperature. Suspensions were spun at 1000 RPM. Tubules were minced and re-suspended in HBSS with 1 mg/mL collagenase and 10 μg/mL DNase in a shaking water bath at 37 °C for 30 minutes. Tubule fragments were allowed to sediment at room temperature and re-suspended in HBSS with 15 U/mL hyaluronidase (Sigma) and 10 μg/mL DNase and incubated in a shaking water bath at 37 °C for 30 min. Tubule fragments were centrifuged at 1000 RPM. Pellets are washed and re-suspended in F12/DMEM (Corning) with 10% FBS, 1% PSQ 1% (Corning), sodium pyruvate (Corning), 10 mM HEPES (Corning), 5 μg/mL Transferrin (Sigma), 2.5 ng/mL epidermal growth factor (Gibco), 10 μg/mL insulin (Gibco). Media was changed after 24 h of culture to remove floating germ cells. Sertoli cell cultures were passaged after 3–5 days to chamber slides for staining. Sertoli cells were fixed in PBS with 4% PFA for 30 min. at room temperature, blocked in PBS with 10% normal goat serum (Jackson Immunoresearch) and stained overnight in anti-BAI1 antibody 1:100 (R&D #AF4969).
Retinoid analysis
P20-P21 mice were dark adapted overnight. Eyes were collected under dim red light and stored at −80 °C until they were analyzed. Whole globes were then homogenized in 10 mM sodium phosphate buffer, pH 8.0, containing 50% methanol (v/v) and 100 mM hydroxylamine. The resulting mixture was extracted twice with 4 ml of ethyl acetate. The combined organic layers were dried in vacuo, reconstituted in 400 μl of hexanes, and 100 μl of the extract was injected on to a normal-phase high-performance liquid chromatography (HPLC) (Agilent Sil, 5 μm, 4.6 × 250 mm; Agilent Technologies, Santa Clara, CA) in a stepwise gradient of ethyl acetate in hexanes (0–17 min, 0.6%; 17.01–42 min. 10%) at a flow rate of 1.4 ml·min−1. Retinoids were detected by monitoring absorbance at 325 nm and quantified based on a standard curve representing the relationship between the amount of synthetic retinoid standard and the area under the corresponding chromatographic peak.
RNAseq
RPE were isolated from littermate P14 mice 2 h after light onset. RNA was isolated with an RNeasy kit (Qiagen). An mRNA library was prepared using an Illumina TruSeq platform. The transcriptome was sequenced on a NextSeq. 500 cartridge. The statistical software package R (version 3.3.2) was used for all analyses. The Bioconductor package DESeq. 2 was used for differential gene expression analysis of RNA-seq data. Heatmaps were created using the R package gplots via the heatmap.2 package. The R code used for bioinformatics analysis and heatmap generation is available upon request.
Statistics
Statistics analysis was performed in GraphPad Prism 7.0. Statistical test used as indicated in the figure legends. A p-value < 0.05 was considered statistically significant.
Data availability
All data is available upon request. RNAseq datasets are publicly accessible on GEO.
Electronic supplementary material
Acknowledgements
We would like to thank all of the members of the Ravichandran Lab for their advice and support. We would also like to thank Tal Burstyn-Cohen for her assistance in developing dissection techniques. We would like to acknowledge the support of the University of Virginia Research Histology Core and Biological Tissue Repository Facility for preparing testicular sections and cleaved-caspase 3 staining, respectively. We would like to thank the University of Virginia Genomics Core Facility and AnhThu Nguyen for her preparation of the cDNA libraries for RNAseq. We would like to thank Emily Farber for running the Illumina NextSeq analysis. We would like to thank Alex Koeppel and the University of Virginia Bioinformatics Core for performing the DESeq. 2 analysis of the raw RNAseq data. The stereology data described in this manuscript was gathered on an “MBF Bioscience and Zeiss microscope system for stereology and tissue morphology” funded by National Institutes of Health grant 1S10RR026799-01. This work is supported by grants to KSR from NIGMS (GM064709), NHLBI (P01HL120840), and the Center for Cell Clearance/University of Virginia School of Medicine. KKP is supported by an NHLBI F30 award (F30 HL126385) and previously by the NIH T32 Immunology Training Grant (T32 AI007496).
Author Contributions
K.K.P., C.R., L.S.S., K.P., S.O., J.L. and K.S.R. designed research. K.K.P., C.R., M.H.R., J.Z., C.H., J.L. performed experiments. C.S.L. generated Bai1 Tg mice. K.K.P. and J.S.A.P. analyzed and visualized data. K.K.P. and K.S.R. wrote the manuscript with input from all other authors.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-15191-1.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lee CS, et al. Boosting Apoptotic Cell Clearance by Colonic Epithelial Cells Attenuates Inflammation In Vivo. Immunity. 2016;44:807–820. doi: 10.1016/j.immuni.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henson PM, Hume DA. Apoptotic cell removal in development and tissue homeostasis. Trends in Immunology. 2006;27:244–250. doi: 10.1016/j.it.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 3.Ravichandran KS, Lorenz U. Engulfment of apoptotic cells: signals for a good meal. Nature reviews Immunology. 2007;7:964–974. doi: 10.1038/nri2214. [DOI] [PubMed] [Google Scholar]
- 4.Penberthy KK, Ravichandran KS. Apoptotic cell recognition receptors and scavenger receptors. Immunological Reviews. 2016;269:44–59. doi: 10.1111/imr.12376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arandjelovic S, Ravichandran KS. Phagocytosis of apoptotic cells in homeostasis. Nature Immunology. 2015;16:907–917. doi: 10.1038/ni.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith-Harrison, L. I. & Koontz, W. W. Torsion of the testis: changing concepts. (AUA update Series, 1990).
- 7.Cohen PL, et al. Delayed Apoptotic Cell Clearance and Lupus-like Autoimmunity in Mice Lacking the c-mer Membrane Tyrosine Kinase. Journal of Experimental Medicine. 2002;196:135–140. doi: 10.1084/jem.20012094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lysiak JJ, Turner SD, Turner TT. Molecular pathway of germ cell apoptosis following ischemia/reperfusion of the rat testis. Biology of Reproduction. 2000;63:1465–1472. doi: 10.1095/biolreprod63.5.1465. [DOI] [PubMed] [Google Scholar]
- 9.Poon IKH, Lucas CD, Rossi AG, Ravichandran KS. Apoptotic cell clearance: basic biology and therapeutic potential. Nature reviews Immunology. 2014;14:166–180. doi: 10.1038/nri3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lew, E. D. et al. Differential TAM receptor-ligand-phospholipid interactions delimit differential TAM bioactivities. eLife3, (2014). [DOI] [PMC free article] [PubMed]
- 11.Fadok VA, et al. Different populations of macrophages use either the vitronectin receptor or the phosphatidylserine receptor to recognize and remove apoptotic cells. The Journal of Immunology. 1992;149:4029–4035. [PubMed] [Google Scholar]
- 12.Fadok VA, et al. Exposure of phosphatidylserine on the surface of apoptotic lymphocytes triggers specific recogntion of removal by macrophages. The Journal of Immunology. 1992;148:2207–2216. [PubMed] [Google Scholar]
- 13.Ruggiero L, Connor MP, Chen J, Langen R, Finnemann SC. Diurnal, localized exposure of phosphatidylserine by rod outer segment tips in wild-type but not Itgb5−/− or Mfge8−/− mouse retina. PNAS. 2012;109:8145–8148. doi: 10.1073/pnas.1121101109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duncan JL, et al. An RCS-like retinal dystrophy phenotype in mer knockout mice. Investigative Ophthalmology & Visual Science. 2003;44:826–838. doi: 10.1167/iovs.02-0438. [DOI] [PubMed] [Google Scholar]
- 15.D’Cruz PM, et al. Mutation of the receptor tyrosine kinase gene Mertk in the retinal dystrophic RCS rat. Hum. Mol. Genet. 2000;9:645–651. doi: 10.1093/hmg/9.4.645. [DOI] [PubMed] [Google Scholar]
- 16.Lemke G, Burstyn-Cohen T. TAM receptors and the clearance of apoptotic cells. Annals of the New York Academy of Sciences. 2010;1209:23–29. doi: 10.1111/j.1749-6632.2010.05744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyanishi M, et al. Identification of Tim4 as a phosphatidylserine receptor. Nature. 2007;450:435–439. doi: 10.1038/nature06307. [DOI] [PubMed] [Google Scholar]
- 18.Park D, et al. BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature. 2007;450:430–434. doi: 10.1038/nature06329. [DOI] [PubMed] [Google Scholar]
- 19.Albert ML, Kim J-I, Birge RB. avB5 integrin recruits the CrkII–Dock180–Rac1 complex for phagocytosis of apoptotic cells. Nature Cell Biology. 2000;2:899–905. doi: 10.1038/35046549. [DOI] [PubMed] [Google Scholar]
- 20.Akakura S, et al. The opsonin MFG-E8 is a ligand for the αvβ5 integrin and triggers DOCK180-dependent Rac1 activation for the phagocytosis of apoptotic cells. Experimental Cell Research. 2004;292:403–416. doi: 10.1016/j.yexcr.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 21.Prasad D, et al. TAM receptor function in the retinal pigment epithelium. Molecular and Cellular Neuroscience. 2006;33:96–108. doi: 10.1016/j.mcn.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 22.Strauss O. The retinal pigment epithelium in visual function. Physiol. Rev. 2005;85:845–881. doi: 10.1152/physrev.00021.2004. [DOI] [PubMed] [Google Scholar]
- 23.Griswold MD. The central role of Sertoli cells in spermatogenesis. Semin. Cell Dev. Biol. 1998;9:411–416. doi: 10.1006/scdb.1998.0203. [DOI] [PubMed] [Google Scholar]
- 24.Nandrot EF, et al. Essential role for MFG-E8 as ligand for avb5 integrin in diurnal retinal phagocytosis. PNAS. 2007;104:12005–12010. doi: 10.1073/pnas.0704756104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang H, et al. Evaluation on the phagocytosis of apoptotic spermatogenic cells by Sertoli cells in vitro through detecting lipid droplet formation by Oil Red O staining. Reproduction. 2006;132:485–492. doi: 10.1530/rep.1.01213. [DOI] [PubMed] [Google Scholar]
- 26.Burstyn-Cohen T, et al. Genetic Dissection of TAM Receptor-Ligand Interaction in Retinal Pigment Epithelial Cell Phagocytosis. Neuron. 2012;76:1123–1132. doi: 10.1016/j.neuron.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vollrath D, et al. Tyro3 Modulates Mertk-Associated Retinal Degeneration. PLoS Genet. 2015;11:e1005723. doi: 10.1371/journal.pgen.1005723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Breucker H, Schäfer E, Holstein AF. Morphogenesis and fate of the residual body in human spermiogenesis. Cell Tissue Res. 1985;240:303–309. doi: 10.1007/BF00222339. [DOI] [PubMed] [Google Scholar]
- 29.Kevany BM, Palczewski K. Phagocytosis of Retinal Rod and Cone Photoreceptors. Physiology. 2010;25:8–15. doi: 10.1152/physiol.00038.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sethna, S. & Finnemann, S. C. In Methods in Molecular Biology935, 245–254 (Humana Press, 2012).
- 31.Lu Q, et al. Tyro-3 family receptors are essential regulators of mammalian spermatogenesis. Nature. 1999;398:723–728. doi: 10.1038/19554. [DOI] [PubMed] [Google Scholar]
- 32.Kiser PD, Palczewski K. Retinoids and Retinal Diseases. Annu. Rev. Vis. Sci. 2016;2:197–234. doi: 10.1146/annurev-vision-111815-114407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fond, A. M., Lee, C. S., Schulman, I. G., Kiss, R. S. & Ravichandran, K. S. Apoptotic cells trigger a membrane-initiated pathway to increase ABCA1. Journal of Clinical Investigation, 10.1172/JCI80300DS1 (2015). [DOI] [PMC free article] [PubMed]
- 34.Zhu D, et al. BAI1 regulates spatial learning and synaptic plasticity in the hippocampus. Journal of Clinical Investigation. 2015;125:1497–1508. doi: 10.1172/JCI74603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duman JG, et al. The adhesion-GPCR BAI1 regulates synaptogenesis by controlling the recruitment of the Par3/Tiam1 polarity complex to synaptic sites. Journal of Neuroscience. 2013;33:6964–6978. doi: 10.1523/JNEUROSCI.3978-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sarangi, P., Wahl, L. & Vega, S. Fibulin-7, a member of the extracellular matrix fibulin family, regulates immune cell migration and function (P5124). The Journal of Immunology (2013).
- 37.de Vega, S. et al. Identification of Peptides Derived from the C-terminal Domain of Fibulin-7 Active for Endothelial Cell Adhesion and Tube Formation Disruption. Biopolymers, 10.1002/bip.22754 (2015). [DOI] [PMC free article] [PubMed]
- 38.de Vega S, et al. TM14 is a new member of the fibulin family (fibulin-7) that interacts with extracellular matrix molecules and is active for cell binding. J. Biol. Chem. 2007;282:30878–30888. doi: 10.1074/jbc.M705847200. [DOI] [PubMed] [Google Scholar]
- 39.Sardell RJ, et al. Whole exome sequencing of extreme age-related macular degeneration phenotypes. Molecular Vision. 2016;22:1062–1076. [PMC free article] [PubMed] [Google Scholar]
- 40.Barrett T, et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41:D991–5. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kinchen JM, Ravichandran KS. Phagosome maturation: going through the acid test. Nat. Rev. Mol. Cell Biol. 2008;9:781–795. doi: 10.1038/nrm2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steinberg BE, et al. A cation counterflux supports lysosomal acidification. The Journal of Cell Biology. 2010;189:1171–1186. doi: 10.1083/jcb.200911083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swanson J. CFTR: helping to acidify macrophage lysosomes. Nature Cell Biology. 2006;8:908–909. doi: 10.1038/ncb0906-908. [DOI] [PubMed] [Google Scholar]
- 44.Yang YD, et al. TMEM16A confers receptor-activated calcium-dependent chloride conductance. Nature. 2008;455:1210–1215. doi: 10.1038/nature07313. [DOI] [PubMed] [Google Scholar]
- 45.Zhang L, et al. Calcium overload is associated with lipofuscin formation in human retinal pigment epithelial cells fed with photoreceptor outer segments. Eye (Lond) 2011;25:519–527. doi: 10.1038/eye.2011.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aalkjaer C, Boedtkjer E, Choi I, Lee S. Cation-coupled bicarbonate transporters. Compr Physiol. 2014;4:1605–1637. doi: 10.1002/cphy.c130005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hanzu FA, et al. Expression of TMEM16A and SLC4A4 in human pancreatic islets. Cellular Physiology and Biochemistry. 2012;29:61–64. doi: 10.1159/000337587. [DOI] [PubMed] [Google Scholar]
- 48.Energy metabolism of human neutrophils during phagocytosis. 70, 550–557 (1982). [DOI] [PMC free article] [PubMed]
- 49.Biswas SK, Mantovani A. Orchestration of metabolism by macrophages. Cell Metabolism. 2012;15:432–437. doi: 10.1016/j.cmet.2011.11.013. [DOI] [PubMed] [Google Scholar]
- 50.Dotti MT, Rufa A, Federico A. Cerebrotendinous xanthomatosis: heterogeneity of clinical phenotype with evidence of previously undescribed ophthalmological findings. J. Inherit. Metab. Dis. 2001;24:696–706. doi: 10.1023/A:1012981019336. [DOI] [PubMed] [Google Scholar]
- 51.Gabriel LAR, Sachdeva R, Marcotty A, Rockwood EJ, Traboulsi EI. Oculodentodigital dysplasia: new ocular findings and a novel connexin 43 mutation. Arch. Ophthalmol. 2011;129:781–784. doi: 10.1001/archophthalmol.2011.113. [DOI] [PubMed] [Google Scholar]
- 52.Davis AP, et al. Comparative Toxicogenomics Database: a knowledgebase and discovery tool for chemical-gene-disease networks. Nucleic Acids Res. 2009;37:D786–92. doi: 10.1093/nar/gkn580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Davis AP, et al. The Comparative Toxicogenomics Database’s 10th year anniversary: update 2015. Nucleic Acids Res. 2015;43:D914–20. doi: 10.1093/nar/gku935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alvarez BV, et al. Identification and Characterization of a Novel Mutation in the Carbonic Anhydrase IV Gene that Causes Retinitis Pigmentosa. Investigative Ophthalmology & Visual Science. 2007;48:3459–3468. doi: 10.1167/iovs.06-1515. [DOI] [PubMed] [Google Scholar]
- 55.Cai X, Conley SM, Naash MI. RPE65: role in the visual cycle, human retinal disease, and gene therapy. Ophthalmic Genet. 2009;30:57–62. doi: 10.1080/13816810802626399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li S, et al. Fatty acid transport protein 4 (FATP4) prevents light-induced degeneration of cone and rod photoreceptors by inhibiting RPE65 isomerase. Journal of Neuroscience. 2013;33:3178–3189. doi: 10.1523/JNEUROSCI.2428-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tian N. Visual experience and maturation of retinal synaptic pathways. Vision Res. 2004;44:3307–3316. doi: 10.1016/j.visres.2004.07.041. [DOI] [PubMed] [Google Scholar]
- 58.Dorrell MI, Aguilar E, Weber C, Friedlander M. Global gene expression analysis of the developing postnatal mouse retina. Investigative Ophthalmology & Visual Science. 2004;45:1009–1019. doi: 10.1167/iovs.03-0806. [DOI] [PubMed] [Google Scholar]
- 59.Palczewska G, et al. Receptor MER Tyrosine Kinase Proto-oncogene (MERTK) Is Not Required for Transfer of Bis-retinoids to the Retinal Pigmented Epithelium. J. Biol. Chem. 2016;291:26937–26949. doi: 10.1074/jbc.M116.764563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.O’Byrne, S. M. & Blaner, W. S. Retinol and retinyl esters: Biochemistry and physiology Thematic Review Series: Fat-soluble vitamins: vitamin A. J. Lipid Res. (2013). [DOI] [PMC free article] [PubMed]
- 61.Batten ML, et al. Lecithin-retinol acyltransferase is essential for accumulation of all-trans-retinyl esters in the eye and in the liver. J. Biol. Chem. 2004;279:10422–10432. doi: 10.1074/jbc.M312410200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Elliott MR, et al. Unexpected requirement for ELMO1 in clearance of apoptotic germ cells in vivo. Nature. 2011;467:333–337. doi: 10.1038/nature09356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mattapallil MJ, et al. The Rd8 Mutation of the Crb1 Gene Is Present in Vendor Lines of C57BL/6N Mice and Embryonic Stem Cells, and Confounds Ocular Induced Mutant Phenotypes. Investigative Ophthalmology & Visual Science. 2012;53:2921–2927. doi: 10.1167/iovs.12-9662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gibbs, D. & Williams, D. S. In Retinal Degenerations (eds. LaVail, M. M., Hollyfield, J. G. & Anderson, R. E.) 533, 347–352 (Springer US, 2003).
- 65.Brugnera E, et al. Unconventional Rac-GEF activity is mediated through the Dock180–ELMO complex. Nature Cell Biology. 2002;4:574–582. doi: 10.1038/ncb824. [DOI] [PubMed] [Google Scholar]
- 66.Li MJ, et al. GWASdb: a database for human genetic variants identified by genome-wide association studies. Nucleic Acids Res. 2012;40:D1047–54. doi: 10.1093/nar/gkr1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chan W-M, et al. Choroidal neovascularisation in pathological myopia: an update in management. Br J Ophthalmol. 2005;89:1522–1528. doi: 10.1136/bjo.2005.074716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grossniklaus HE, Green WR. Pathologic findings in pathologic myopia. Retina (Philadelphia, Pa.) 1992;12:127–133. doi: 10.1097/00006982-199212020-00009. [DOI] [PubMed] [Google Scholar]
- 69.Batioğlu F, Ozdek S, Hasanreisoğlu B. An unusual macular involvement in pathological myopia. Eye (Lond) 2003;17:654–656. doi: 10.1038/sj.eye.6700416. [DOI] [PubMed] [Google Scholar]
- 70.Paznekas WA, et al. Connexin 43 (GJA1) mutations cause the pleiotropic phenotype of oculodentodigital dysplasia. Am. J. Hum. Genet. 2003;72:408–418. doi: 10.1086/346090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kojima A, et al. Connexin 43 contributes to differentiation of retinal pigment epithelial cells via cyclic AMP signaling. Biochemical Journal. 2008;366:532–538. doi: 10.1016/j.bbrc.2007.11.159. [DOI] [PubMed] [Google Scholar]
- 72.Malfait M, et al. Effects of hyperglycemia and protein kinase C on connexin43 expression in cultured rat retinal pigment epithelial cells. J. Membr. Biol. 2001;181:31–40. doi: 10.1007/s0023200100082. [DOI] [PubMed] [Google Scholar]
- 73.Verrips A, et al. Clinical and molecular genetic characteristics of patients with cerebrotendinous xanthomatosis. Brain. 2000;123(Pt 5):908–919. doi: 10.1093/brain/123.5.908. [DOI] [PubMed] [Google Scholar]
- 74.Saadane A, et al. Retinal and nonocular abnormalities in Cyp27a1(−/−)Cyp46a1(−/−) mice with dysfunctional metabolism of cholesterol. The American Journal of Pathology. 2014;184:2403–2419. doi: 10.1016/j.ajpath.2014.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Omarova S, et al. Abnormal vascularization in mouse retina with dysregulated retinal cholesterol homeostasis. J. Clin. Invest. 2012;122:3012–3023. doi: 10.1172/JCI63816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pletscher-Frankild S, Pallejà A, Tsafou K, Binder JX, Jensen LJ. DISEASES: text mining and data integration of disease-gene associations. Methods. 2015;74:83–89. doi: 10.1016/j.ymeth.2014.11.020. [DOI] [PubMed] [Google Scholar]
- 77.Rebello G, et al. Apoptosis-inducing signal sequence mutation in carbonic anhydrase IV identified in patients with the RP17 form of retinitis pigmentosa. Proc. Natl. Acad. Sci. USA. 2004;101:6617–6622. doi: 10.1073/pnas.0401529101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Datta R, Waheed A, Bonapace G, Shah GN, Sly WS. Pathogenesis of retinitis pigmentosa associated with apoptosis-inducing mutations in carbonic anhydrase IV. PNAS. 2009;106:3437–3442. doi: 10.1073/pnas.0813178106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liles MR, Newsome DA, Oliver PD. Antioxidant enzymes in the aging human retinal pigment epithelium. Arch. Ophthalmol. 1991;109:1285–1288. doi: 10.1001/archopht.1991.01080090111033. [DOI] [PubMed] [Google Scholar]
- 80.Rex TS, et al. Adenovirus-mediated delivery of catalase to retinal pigment epithelial cells protects neighboring photoreceptors from photo-oxidative stress. Hum. Gene Ther. 2004;15:960–967. doi: 10.1089/hum.2004.15.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Miceli MV, Liles MR, Newsome DA. Evaluation of oxidative processes in human pigment epithelial cells associated with retinal outer segment phagocytosis. Experimental Cell Research. 1994;214:242–249. doi: 10.1006/excr.1994.1254. [DOI] [PubMed] [Google Scholar]
- 82.Meyers KJ, et al. Genetic evidence for role of carotenoids in age-related macular degeneration in the Carotenoids in Age-Related Eye Disease Study (CAREDS) Investigative Ophthalmology & Visual Science. 2014;55:587–599. doi: 10.1167/iovs.13-13216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stoffel W, et al. Delta6-desaturase (FADS2) deficiency unveils the role of omega3- and omega6-polyunsaturated fatty acids. EMBO J. 2008;27:2281–2292. doi: 10.1038/emboj.2008.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mukherjee S, King JL, Guidry C. Phenotype-associated changes in retinal pigment epithelial cell expression of insulin-like growth factor binding proteins. Investigative Ophthalmology & Visual Science. 2009;50:5449–5455. doi: 10.1167/iovs.09-3383. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data is available upon request. RNAseq datasets are publicly accessible on GEO.