Significance
The metazoan protein BCL-2–associated athanogene cochaperone 6 (Bag6) acts as a central hub for several essential cellular processes, including immunoregulation, gene regulation, apoptosis, and proteostasis. These roles are in both the nucleus and the cytosol, but the mechanism by which Bag6 traffics between these compartments remains elusive. Here we present the crystal structure of Bag6 in complex with its cytoplasmic retention factor transmembrane domain recognition complex 35 (TRC35) and suggest a mechanism of regulation for the nucleo-cytoplasmic transport of Bag6.
Keywords: tail-anchor targeting, X-ray crystallography, tail-anchor recognition complex, proteasome-dependent degradation, GET pathway
Abstract
The metazoan protein BCL2-associated athanogene cochaperone 6 (Bag6) forms a hetero-trimeric complex with ubiquitin-like 4A and transmembrane domain recognition complex 35 (TRC35). This Bag6 complex is involved in tail-anchored protein targeting and various protein quality-control pathways in the cytosol as well as regulating transcription and histone methylation in the nucleus. Here we present a crystal structure of Bag6 and its cytoplasmic retention factor TRC35, revealing that TRC35 is remarkably conserved throughout the opisthokont lineage except at the C-terminal Bag6-binding groove, which evolved to accommodate Bag6, a unique metazoan factor. While TRC35 and its fungal homolog, guided entry of tail-anchored protein 4 (Get4), utilize a conserved hydrophobic patch to bind their respective partners, Bag6 wraps around TRC35 on the opposite face relative to the Get4–5 interface. We further demonstrate that TRC35 binding is critical not only for occluding the Bag6 nuclear localization sequence from karyopherin α to retain Bag6 in the cytosol but also for preventing TRC35 from succumbing to RNF126-mediated ubiquitylation and degradation. The results provide a mechanism for regulation of Bag6 nuclear localization and the functional integrity of the Bag6 complex in the cytosol.
Tail-anchored (TA) membrane proteins contain a single, C-terminal, hydrophobic transmembrane-helix domain (TMD) (1). The TMD, which acts as the endoplasmic reticulum (ER) targeting signal, emerges from the ribosome after translation termination necessitating posttranslational targeting and insertion. Transmembrane domain recognition complex (TRC) proteins in mammals and guided entry of tail-anchored proteins (GET) in fungi are the best-characterized molecular players in TA targeting (2). In the TRC pathway, TA proteins are captured by the small glutamine-rich tetratricopeptide-repeat protein A (SGTA) after release from the ribosome then transferred to the ATPase TRC40, a process facilitated by the hetero-trimeric Bag6–TRC35–Ubl4A complex (3, 4). TA binding stimulates hydrolysis of ATP by TRC40 (5) leading to release of the TRC40–TA complex that then localizes to the ER membrane via its interaction with calcium-modulating ligand (CAML) (6). Release of the TA substrate for insertion is facilitated by CAML and tryptophan rich basic protein (WRB) (7). Notably, loss of WRB in mouse cardiomyocytes and hepatocytes results in partial mislocalization of TA proteins (8), suggesting that other pathways, including the signal-recognition particle (SRP) (9), SRP-independent targeting (SND) proteins (10), the Hsc70 family of chaperones (11), and ubiquilins (12), can compensate to target TA in vivo.
Structural information for the mammalian proteins is sparse; however, homology to the equivalent fungal counterparts, Get1–5 and Sgt2, allow for parallels. Bag6 is unique among the mammalian proteins, and its introduction alters the molecular architecture of the central hand-off complex. For TA targeting, the heterotrimeric Bag6 complex is equivalent to the fungal heterotetrameric Get4–Get5 complex (3, 13), but the addition of Bag6 leads to different interactions.
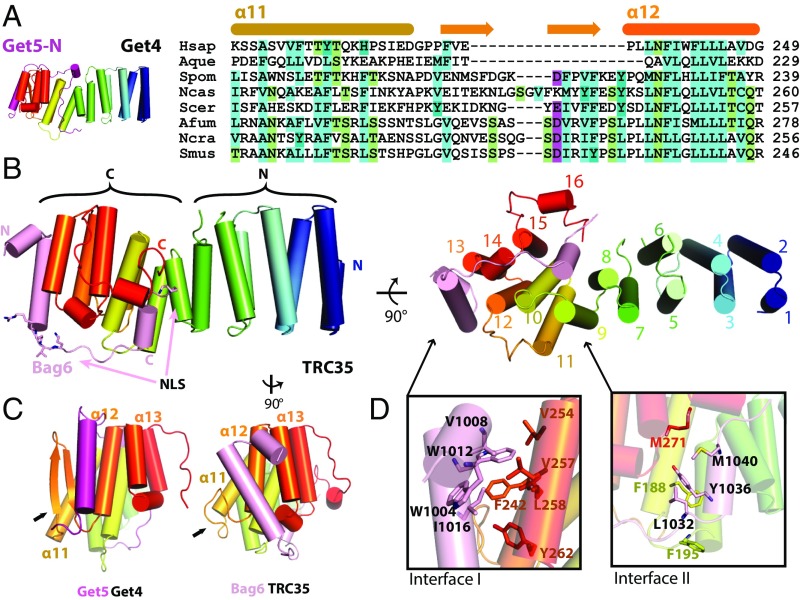
Crystal structures of fungal Get4 revealed 14 α-helices that can be divided into N-terminal domains (NTD) and C-terminal domains (CTD) (Fig. 1A and Fig. S1A) (14–16). The NTD contains conserved residues (Fig. S1C) involved in binding and regulating Get3 (13, 15), while the CTD forms a stable interaction with the Get5 N terminus (Get5-N) (15). The Get5 interface includes a hydrophobic cleft in Get4 formed by a β-hairpin between α13 and α14 that binds the helix in Get5-N (15). Sequence alignment of Get4/TRC35 and Get5/Ubl4A led to predictions that the Get4 β-hairpin and Get5 N-terminal extension are absent in metazoans (Fig. 1 A and B) (15), which suggested a different structural organization for the metazoan Bag6–TRC35–Ubl4A complex.
Fig. 1.
The structure of the Bag6NLS–TRC35 heterodimer. (A) The structure of Saccharomyces cerevisiae Get4–Get5N complex [Protein Data Bank (PDB) ID code: 3LKU], Get4 in rainbow and Get5 in magenta. Sequence alignment of TRC35/Get4 from Homo sapiens, Amphimedon queenslandica, Schizosaccharomyces pombe, Naumovozyma castelli, Saccharomyces cerevisiae, Aspergillus fumigatus, Neurospora crassa, and Sphaerulina musiva. The secondary structure based on fungal Get4s is highlighted above the text. (B, Left) The overall structure of Bag6–TRC35 heterodimer in cylinder representation with Bag6 in light pink and TRC35 in rainbow. The nuclear localization sequence is highlighted in sticks on Bag6. (B, Right) A 90° in plane rotated ‘bottom’ view. The 16 helices of TRC35 are labeled from N to C terminus. (C) Comparison of the C-terminal faces of TRC35 and Get4 that bind Bag6 and Get5, respectively. The arrows highlight the significant structural difference in the residues between α11 and α12. (D) Zoomed in view of the regions, defined as interface I and II. Hydrophobic residues that are involved in Bag6–TRC35 dimerization are shown as sticks.
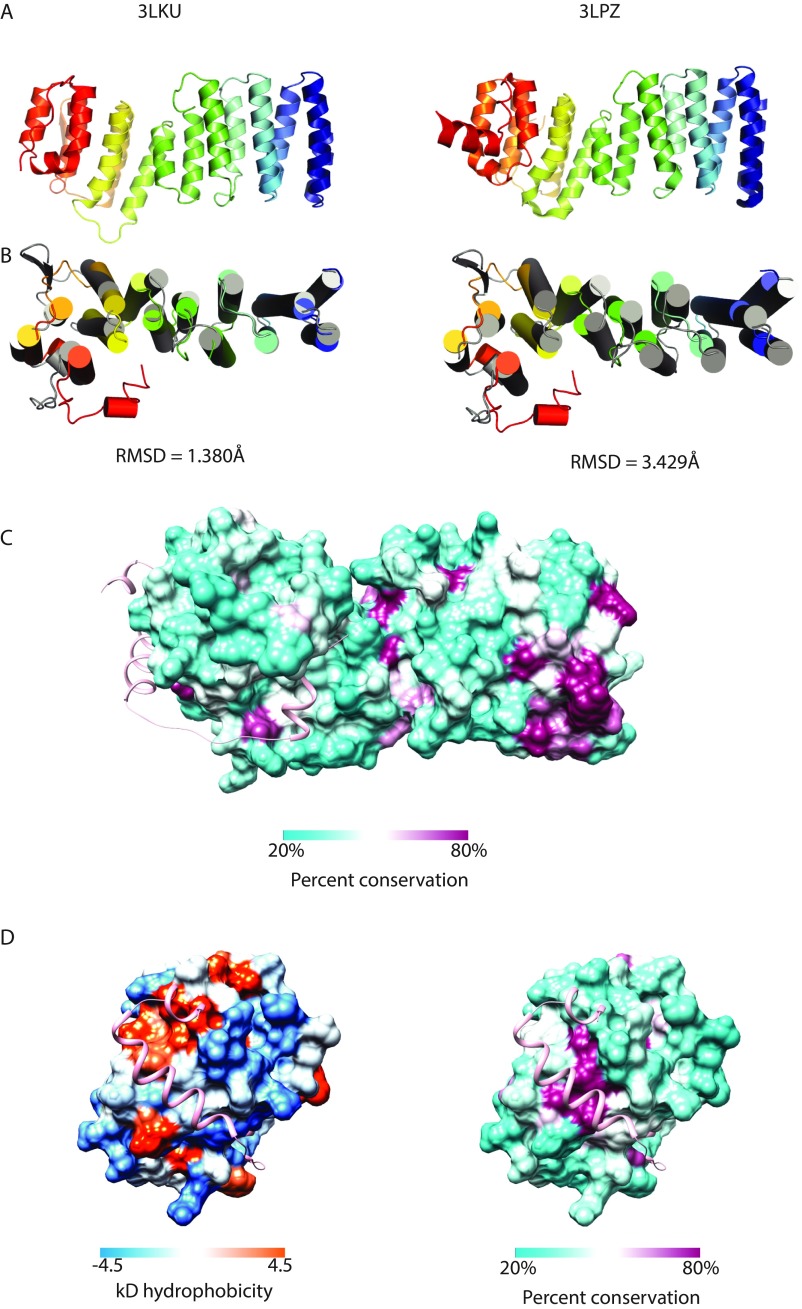
Fig. S1.
Comparison of TRC35 and Get4 structures. (A) Representative structures of fungal Get4 homologs. 3LKU is from Saccharomyces cerevisiae (Sc) and 3LPZ is from Chaetomium thermophilum. (B) Aligned human TRC35 (color ramped) and ScGet4 (3LKU, gray) using Pymol (55) super for sequence-independent structural alignment. (Left) Structures aligned to the six N-terminal helices. (Right) Structures aligned to the seven C-terminal helices. (C) Accessible surface representation of TRC35 colored based on percent conservation as implemented in Chimera (56). Conservation based on a MAFFT (57) alignment of TRC35/Get4 sequences from Homo sapiens, Xenopus laevis, Danio rerio, Drosophila melanogaster, Nematostella vectensis, Monosiga brevicollis, Schizosaccharomyces pombe, and Saccharomyces cerevisiae. Bag6 is in ribbons representation in pink. (D) The Bag6-binding surface of TRC35 colored based on hydrophobicity (Kyle–Doolittle scale) and percent conservation as implemented in Chimera (56).
In metazoans, Bag6 abrogates the interaction of TRC35 with Ubl4A by separately binding these factors (3, 17) resulting in a heterotrimer. The interface between TRC35 and Bag6 had previously been mapped to the CTD of TRC35 and the region on Bag6 that contains the bipartite nuclear localization sequence (NLS) (3, 18). Overexpression of TRC35 results in Bag6 retention in the cytosol (18), suggesting that TRC35 plays a role in determining the nucleo-cytoplasmic distribution of Bag6. Potential roles for Bag6 in the nucleus are varied and include modulation of p300 acetylation (19, 20), facilitation of histone methylation (21), and regulation of DNA damage signaling-mediated cell death (22). How Bag6 localization is regulated remains unclear.
In this study, we present a crystal structure of a complex that includes TRC35 from Homo sapiens (Hs) and the TRC35-binding domain of HsBag6. The structure of TRC35 reveals the conserved architecture relative to fungal Get4 homologs. Surprisingly, Bag6 utilizes the same conserved pockets on TRC35 that are recognized by Get5 on Get4; yet the extended domain wraps around the opposite face of the α-solenoid. The resulting interaction leads to masking of the bipartite NLS preventing nuclear trafficking of Bag6 by blocking its binding to karyopherin α (KPNA), which is demonstrated experimentally. The interaction also prevents RNF126-mediated degradation of TRC35. These results suggest a mechanism for regulation of Bag6 nucleo-cytoplasmic distribution.
The Crystal Structure of the TRC35–Bag6–NLS Complex
A complex between TRC35 (residues 23–305), shown to be competent for TA transfer (3), and a minimal TRC35-binding domain of Bag6 that contains the NLS (residues 1,000–1,054) was coexpressed, purified and crystallized. A dataset from a single crystal was collected to 1.8 Å resolution and phases were obtained using single wavelength anomalous dispersion from a rubidium derivative. The final model refined to 1.8–39 Å had good statistics with an Rfree = 20.1%. Full crystallographic statistics are provided in Table S1. All TRC35 residues in the crystallized construct were visible in the electron density except for S137 and G138 that are disordered in the loop between α6 and α7. For Bag6, residues 1,002–1,043 were resolved with only a few terminal residues ambiguous in the electron density.
Table S1.
Data collection and refinement statistics
| Statistics | Native | Rubidium |
| Data collection | ||
| Protein | Bag6–TRC35 | Bag6–TRC35 |
| Synchrotron | SSRL | SSRL |
| Beamline | 12-2 | 12-2 |
| Space group | P 21 21 21 | P21 21 21 |
| Cell dimensions | ||
| a, b, c, Å | 41.7, 84.6, 102.6 | 42.0, 84.3, 102.5 |
| α, β, γ, ° | 90.0 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Wavelength | 0.9200 | 0.8154 |
| Resolution, Å | 39.1–1.80 (1.92–1.80) | 50.0–1.99 (2.11–1.99) |
| Rmeas*, % | 9.3 (64.3) | 16.9 (118.8) |
| I/σ(I) | 16.9 (3.8) | 14.9 (2.1) |
| CC1/2, % | 99.9 (95) | 99.9 (88.0) |
| Completeness*, % | 98.7 (92.7) | 99.2 (95.6) |
| No. of observations | 384,047 (60,190) | 655,714 (96,962) |
| No. of unique reflections* | 40,776 (6,194) | 47,793 (7,417) |
| Redundancy* | 9.4 (9.7) | 13.7 (13.1) |
| Refinement | ||
| Resolution, Å | 39.1–1.80 | |
| No. of reflections | 34,334 | |
| No. of reflections test set | 1,998 | |
| Rwork/Rfree | 15.8/20.0 | |
| No. atoms (nonhydrogen) | 2,738 | |
| Protein | 2,574 | |
| Water | 152 | |
| Ligand/Ions | 12 | |
| B-factors | ||
| Protein | 31.7 | |
| Water | 39.0 | |
| RMSD | ||
| Bond lengths, Å | 0.010 | |
| Bond angles, ° | 1.16 | |
| Ramachandran plot | ||
| Favored, % | 98 | |
| Additionally allowed, % | 2.0 | |
| Outliers, % | 0.0 |
Highest-resolution shell is shown in parentheses.
TRC35 has the same overall architecture as fungal Get4 (Fig. S1A), revealing that the Getfourfold has been conserved across Opisthokonta. Opisthokonta includes metazoan, choanozoan, and fungal lineages (Fig. S2), which share common ancestry as determined by analysis of 16S-like rRNA (23) and protein sequences (24). The first 15 α-helices form an α-solenoid fold that can be divided into N- and C-terminal halves (NTD and CTD) (Fig. 1B). Alignment of the NTD between TRC35 and Get4 results in an RMSD = 1.380 Å, while the equivalent alignment in the CTD results in an RMSD = 3.429 Å (Fig. S1B). As seen in Get4 (15), there is likely some flexibility between the two halves based on differences across crystal forms. TRC35 and TRC40 are conserved throughout opisthokont evolution and seem to occur as a pair, suggesting the essentiality of the two proteins in the pathway (Fig. S2). Accordingly, residues at the TRC35–TRC40 interface (13) and the TRC35–Bag6 interface in TRC35 are well conserved (Fig. S1 C and D). As predicted from sequence alignment, the β-hairpin in Get4 is replaced by a shorter loop in TRC35 (Fig. 1C, arrows). In fact, sequence alignment of the predicted Get4 proteins of selected opisthokonts revealed that the β-hairpin is unique to the fungal lineage (Fig. S2 and Table S3). The C-terminal α16 is flanked by two extended stretches that cover part of Bag6 (Fig. 1B).
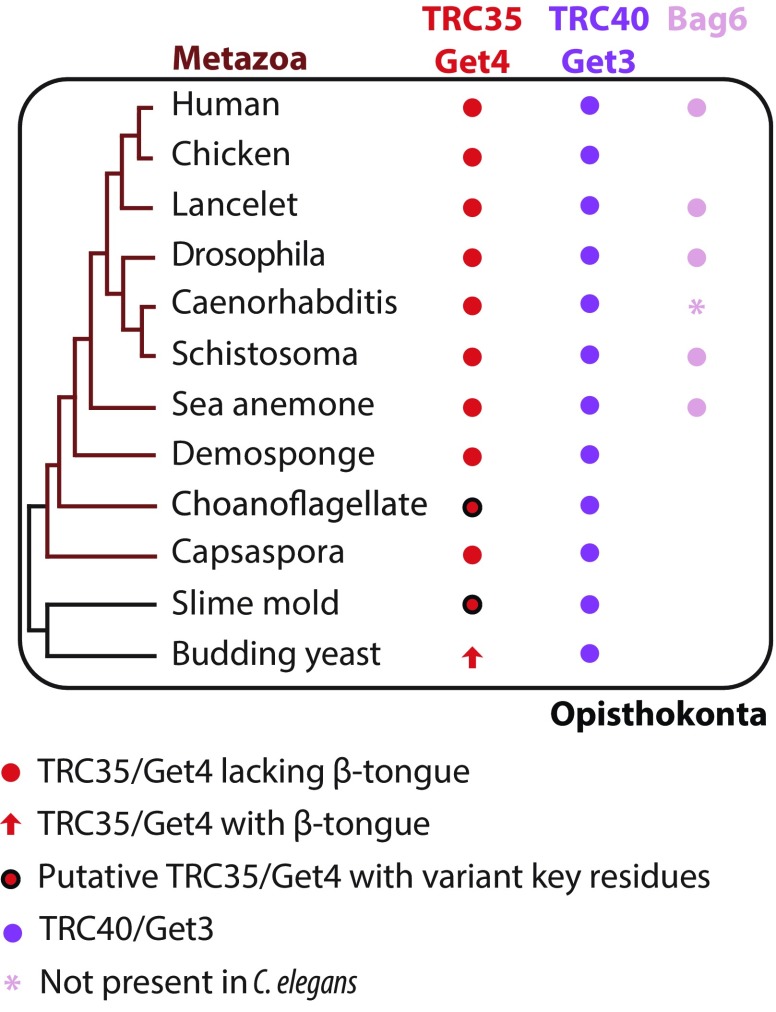
Fig. S2.
Survey of factors involved in the TRC pathway in eukaryotes (Table S3). A condensed phylogenetic tree of representative eukaryotes was built based on the maximum likelihood phylogenetic tree of eukaryotes by Eme et al. (51). The genome of each organism was searched for the presence of Get4/TRC35, Get3/TRC40, and Bag6 using MEME suite motif discovery tool (MEME) (43) and motif-scanning tool (MAST) (58) in addition to NCBI protein BLAST (58). Proteins are color coded and sequence elements are highlighted. Black circles indicate homologs that are missing residues demonstrated to be critical in fungal TA-targeting studies. Pink asterisk indicates that although a clear Bag6 homology could not be identified in the genome of Caenorhabditis elegans, Bag6-like proteins have been identified in genomes of other nematodes: Brugia malayi, Loa loa, Wuchereria bancrofti, and Toxocara canis.
Table S3.
Statistics used for data quantification in Fig. 4
| Bag6 variant | Cytosolic Bag6, no. of cells | Nuclear Bag6, no. of cells | Total no. of cells |
| wtBag6 | 77 | 7 | 84 |
| Bag6(W1004A) | 50 | 30 | 80 |
| Bag6(W1012A) | 35 | 65 | 100 |
| Bag6(Y1036A) | 32 | 56 | 88 |
| Bag6(W1004A/1036A) | 18 | 113 | 131 |
| Bag6(W1012A/1036A) | 15 | 137 | 152 |
The Bag6 NLS region wraps around the TRC35 CTD (Fig. 1B, light pink) at an interface that resembles that of Get5–Get4 (Fig. 1C). The interaction is stabilized by two hydrophobic interface sites. In interface I, Bag6 α1 and α2 dock into a conserved pocket created by α12 and α13 of TRC35 (Fig. S1D) and include W1004, V1008 and W1012 from Bag6 and V254, V257, F242, L258 and Y262 from TRC35 (Fig. 1D). In fungal Get4–5, the Get5 N-terminal helix docks into a groove formed by α12, α13, and the β-hairpin of Get4 (Fig. 1C). The missing β-hairpin in TRC35 results in the Bag6 helix shifting away from α11 toward α12 and α13 near the bottom of TRC35 (Fig. 1C, arrows). The differences in the interface result in rearrangement of α11, α13, and α15 of TRC35 (Fig. 1C) relative to those of Get4. Interface II is less extensive involving fewer residues, L1032, Y1036, M1040 of Bag6 and F195, M271 of TRC35 (Fig. 1D). While the connecting loops between the two interfaces are well-ordered in both contexts, the Bag6 loop makes fewer contacts to TRC35, buried surface area of 2,841 Å2, than the extensive interactions in the Get5-loop to Get4, buried surface area of 5,554 Å2 (Fig. 1B).
The structure reveals how TRC35 masks the Bag6 NLS (Fig. 1B). The conserved first basic cluster of the predicted NLS (K1024RVK) is sequestered between interfaces I and II (Fig. 1B, sticks). Only the first lysine residue of the second cluster (K1043RRK) is resolved in our model (Fig. 1B, sticks). The truncated C-terminal residues of TRC35 in our construct are predicted to be disordered and include conserved multiple negative charges (five Glu and three Asp) (Fig. S3) that would be in position to mask the second NLS basic cluster, K1043RRK, through charge-mediated interactions.
Fig. S3.
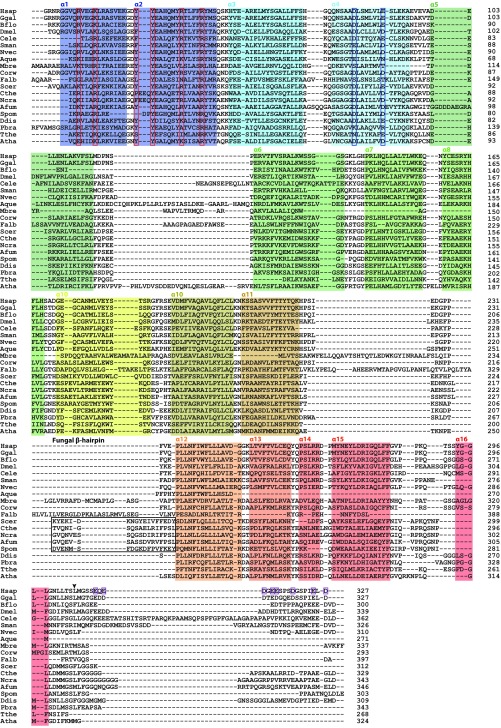
Representative aligned sequences of eukaryotic TRC35/Get4. Species selected based on the eukaryotic phylogenetic tree by Eme et al. (51). Sequences were aligned with MAFFT (57). α-Helices, based on the TRC35 structure, are highlighted in colors that correspond to the crystal structure on Fig. 1B and numbered on top. Residues highlighted in red boxes were identified as critical to fungal Get4 binding Get3 (13). Residues highlighted in blue boxes are critical to Get4/TRC35 regulating Get3/TRC40 (4, 13). The residues that comprise the fungal β-hairpin are highlighted in a black box. The arrow indicates the end of the crystallization construct. Purple boxes highlight glutamate and aspartate residues in the C terminus of TRC35.
Probing the Bag6–TRC35 Interface
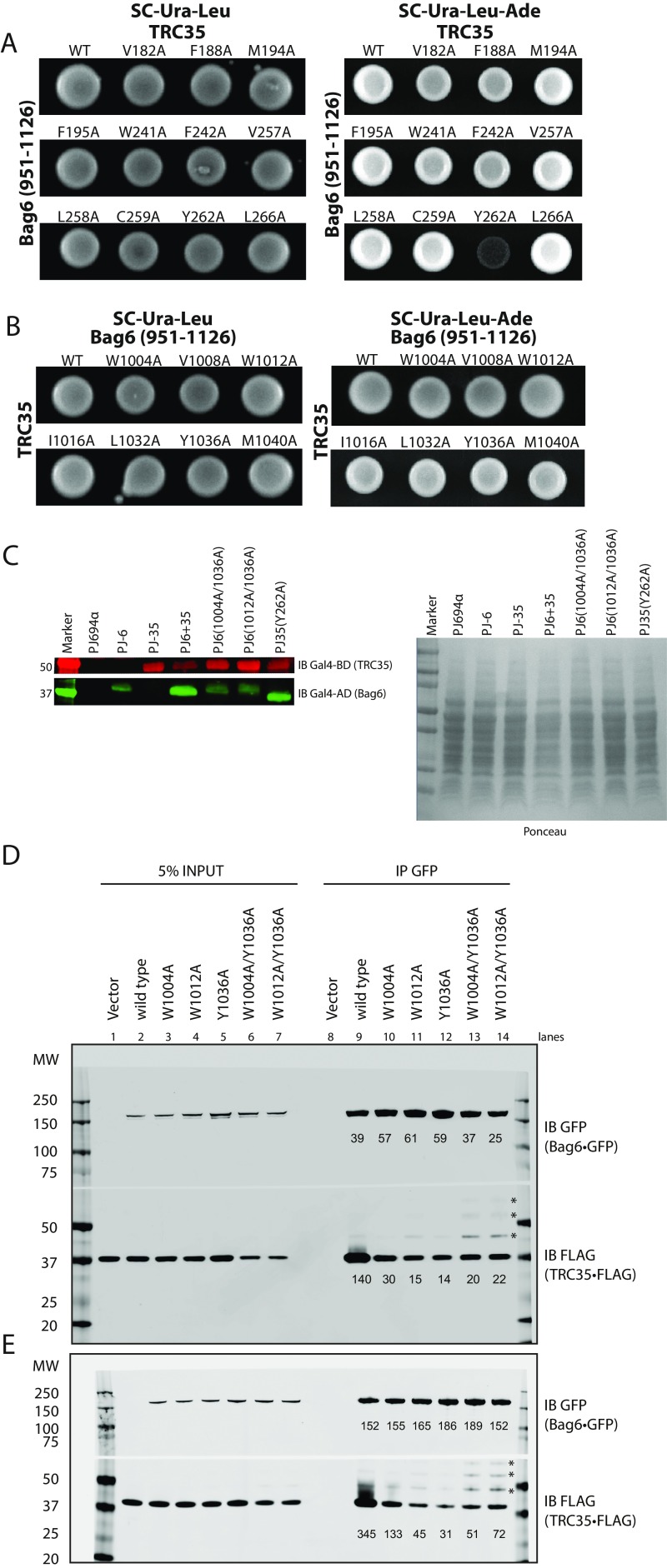
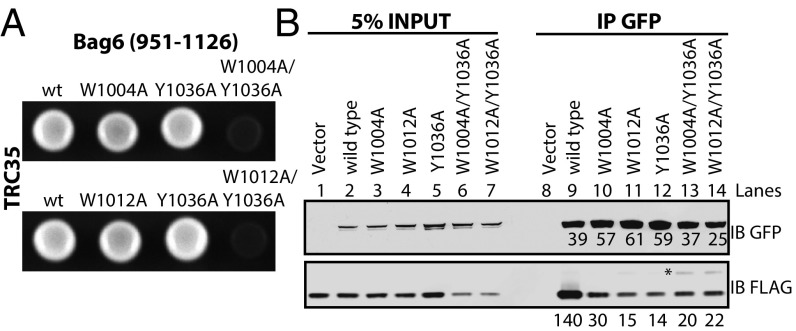
To validate the interfaces observed in the structure, we generated alanine mutants for analysis by yeast two-hybrid (Y2H) analysis. A fragment of Bag6 (residues 951–1,126) was attached to the GAL4 transcription activating domain and full-length TRC35 was attached to the GAL4 DNA-binding domain. The transformation of both vectors was confirmed by cell growth on SC-Ura-Leu medium (Fig. S4 A and B). Of the single amino acid substitutions (Fig. 2A and Fig. S4 A and B), only one residue TRC35 (Y262A) at the end of interface I (Fig. 1D) disrupted the Y2H interaction (Fig. S4A). The Bag6 mutations W1004A and W1012A localize at interface I and Y1036A localizes at interface II. While the individual mutations did not show a phenotype, the combination of the two mutations (W1004A/Y1036A or W1012A/Y1036A) synthetically disrupted the interaction (Fig. 2A). For mutants that failed to grow, expression was confirmed by immunoblotting (Fig. S4C). The results confirm that both interfaces are critical for forming a stable complex between Bag6 and TRC35.
Fig. S4.
Validation of the Bag6–TRC35 interface. (A) Yeast two-hybrid assay to validate the interface identified in the crystal structure. WT or mutant full-length TRC35 conjugated to the DNA binding domain was expressed with WT Bag6(951–1126) conjugated to the transcription activating domain. Transformation was confirmed by ability to grow on SC-Ura-Leu plates. Interaction was determined by ability to grow on SC-Ura-Leu-Ade medium. (B) Western blot analysis of yeast whole-cell lysate for proper expression of prey and bait proteins. Whole-cell lysates of PJ694α (without plasmid), PJ6 (PJ694α transformed with wtBag6), PJ-35 (PJ694α transformed with wtTRC35), PJ6+35 (PJ694α transformed with WT Bag6 and wtTRC35), PJ6(1004A/1036A) (PJ694α transformed with wtTRC35 and Bag6(W1004A/Y1036A), PJ6(1012A/1036A) (PJ694α transformed with wtTRC35 and Bag6(W1012A/Y1036A), and PJ35(Y262A) (PJ694α transformed with wtBag6 and TRC35(Y262A) were analyzed by immunoblotting with indicated Abs. (C) WT full-length TRC35 conjugated to the DNA binding domain was expressed with WT or mutant Bag6(951–1126) conjugated to the transcription activating domain. (D and E) WT or mutant Bag6⋅GFP was coexpressed in Bag6−/− 293T cells with TRC35⋅FLAG and immunoprecipitated using anti-GFP Ab. Amount of TRC35 retrieved by Bag6 was assessed by blotting with anti-FLAG Ab. The positions of the higher molecular weight TRC35⋅FLAG are indicated by asterisks.
Fig. 2.
Validation of the Bag6–TRC35 interface. (A) Yeast two-hybrid assay to validate the interface identified in the crystal structure. WT TRC35 conjugated to the DNA binding domain was expressed with WT or mutant Bag6(951–1126) conjugated to the transcription activating domain. Interaction was determined by ability to grow on adenine-negative medium. (B) WT or mutant Bag6•GFP was coexpressed in Bag6−/− 293T cells with TRC35•FLAG and immunoprecipitated using anti-GFP Ab. Amount of TRC35 retrieved by Bag6 was assessed by blotting with anti-FLAG Ab. The position of the higher molecular weight TRC35⋅FLAG is indicated by an asterisk.
We further validated the interaction of TRC35 with full length Bag6 in the context of a mammalian cellular environment. GFP-tagged WT Bag6 or the mutants Bag6(W1004A), Bag6(W1012), Bag6(Y1036A), Bag6(W1004A/Y1036A), and Bag6(W1012A/Y1036A) were coexpressed with FLAG-tagged TRC35 in CRISPR-mediated Bag6-knockout 293T cells (25). Bag6 and associated proteins were captured from cell extracts by immunoprecipitation (IP) using a GFP-Ab. Viewed by immunoblot (Fig. 2B and Fig. S4 D and E), each Bag6 mutation resulted in a reduction of the captured TRC35 relative to wtBag6 (lane 9 vs. lanes 10–12). The double mutants (lanes 13 and 14) also had reduced capture of TRC35, but they unexpectedly appeared to capture more TRC35 than the single mutants. Furthermore, higher molecular weight TRC35 bands appeared that became more pronounced in the double mutants (Fig. 2, asterisk and Fig. S4 D and E). This will be discussed below. Overall, the general effects of mutations at the Bag6 interface are consistent between the Y2H and the mammalian pulldowns.
Improperly Assembled TRC35 Is Ubiquitylated by RNF126
As noted, IP results (Fig. 2B and Fig. S4 D and E) revealed two interesting observations: (i) an increase in TRC35 binding to Bag6 double mutants relative to single mutants (Fig. 2B and Fig. S4 D and E, lanes 11 and 12 vs. lanes 13 and 14) and (ii) the appearance of higher molecular weight products of TRC35 captured by Bag6 double mutants (Fig. 2B and Fig. S4 D and E, asterisk). Evidence supports that the stability of TRC35 requires forming a proper complex with Bag6 (17, 22). Given the implication of Bag6 as a chaperone holdase in protein quality control processes such as mislocalized protein degradation (26) and ER-associated protein degradation pathway (18), we postulated that TRC35 mutants that fail to bind Bag6 at its physiological binding site are unstable and become a target for Bag6-mediated degradation. In this case, unassembled TRC35 would bind to the Bag6 substrate-binding site. Because the Bag6 construct used in our Y2H studies lacks the substrate-binding domain, increased binding is not observed in the double mutant experiments. If this model is correct, the higher molecular weight bands should be ubiquitylated TRC35 bound to the substrate-binding site on Bag6.
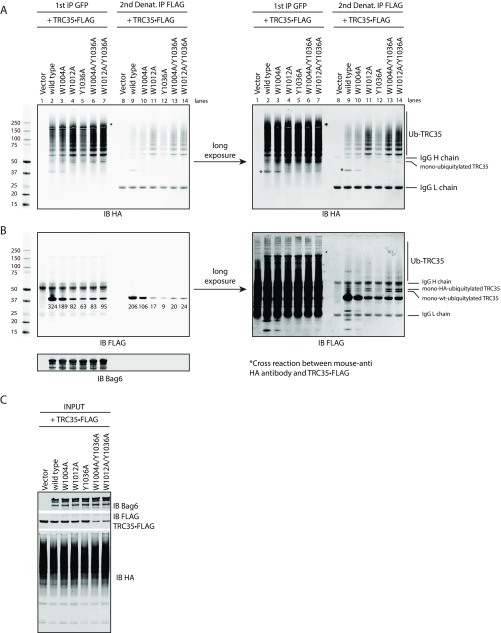
To verify a role for Bag6 in TRC35 ubiquitylation, Bag6 deficient cells were used to coexpress TRC35⋅ holdase FLAG, Bag6 holdase GFP variants and HA holdase ubiquitin (Ub). Proteins bound to Bag6 were first immunoprecipitated with GFP Ab (Fig. S5 A and B, lanes 2–7). To remove other ubiquitylated Bag6 substrates (18, 27), the samples obtained from the GFP IP were subject to a second round of IP using anti-FLAG beads under denaturing conditions (Fig. S5 A and B, lanes 9–14). Immunoblotting showed that all Bag6 variants pulled down ubiquitylated proteins, suggesting that the mutations did not affect its substrate-binding activity (Fig. S5 A and B, lanes 2–7). After the second IP with FLAG Ab, TRC35 associated with wtBag6 carried a small amount of Ub conjugates (Fig. S5 A and B, lane 9), while those associated with Bag6 variants that disrupted physiological association with TRC35 carried significantly more Ub conjugates (Fig. S5 A and B, lanes 10–14). Compared with single Bag6 mutations, TRC35 bound to the double mutants had the highest Ub-modified to unmodified TRC35 ratio (Fig. S5 A and B, lanes 10–12 vs. lanes 13 and 14). Furthermore, the level of TRC35 in cells expressing Bag6 double mutants was consistently lower than in cells expressing wtBag6 (Fig. S5C), which could be rescued by treatment with the proteasome inhibitor MG132 (Fig. 3A, lane 2 vs. lane 4). Treating cells with MG132 also increased ubiquitylated TRC35 that was captured by either wtBag6 or a Bag6 double mutant (Fig. 3A). The accumulation of unmodified TRC35 (Fig. 3A, lanes 9 and 10 vs. lanes 11 and 12) is probably due to depletion of free Ub in the cell (28). Collectively, these data support the idea that the TRC35 molecules unable to associate with Bag6 at the NLS domain become targets for Ub-dependent degradation through a client–chaperone interaction with Bag6. This suggests that the ubiquitylated TRC35 associates with the quality control module (QC) of Bag6 (27).
Fig. S5.
Ubiquitylation of TRC35 in cells expressing Bag6 mutants. (A) IP of TRC35 in Bag6−/− 293T cells cotransfected with plasmids encoding TRC35⋅FLAG (WT), Bag6⋅GFP (WT or mutants), and HA⋅ubiquitin. Anti-GFP Ab was used for the first IP. Anti-FLAG Ab was used for the second IP in denaturing conditions. TRC35 ubiquitylation was assessed by immunoblotting with anti-HA Ab. The exposure times were adjusted to improve visibility of the reactive bands. (B) IP was carried out as in A in Bag6−/− 293T cells cotransfected with plasmids encoding TRC35⋅FLAG (WT) and Bag6⋅GFP (WT or mutants). TRC35 ubiquitylation was assessed by immunoblotting with anti-FLAG Ab. The exposure times were adjusted to improve visibility of the reactive bands. (C) The cell extract used for IP was immunoblotted for Bag6, TRC35, and HA⋅ubiquitin with Bag6 Ab, FLAG Ab, and HA Ab, respectively.
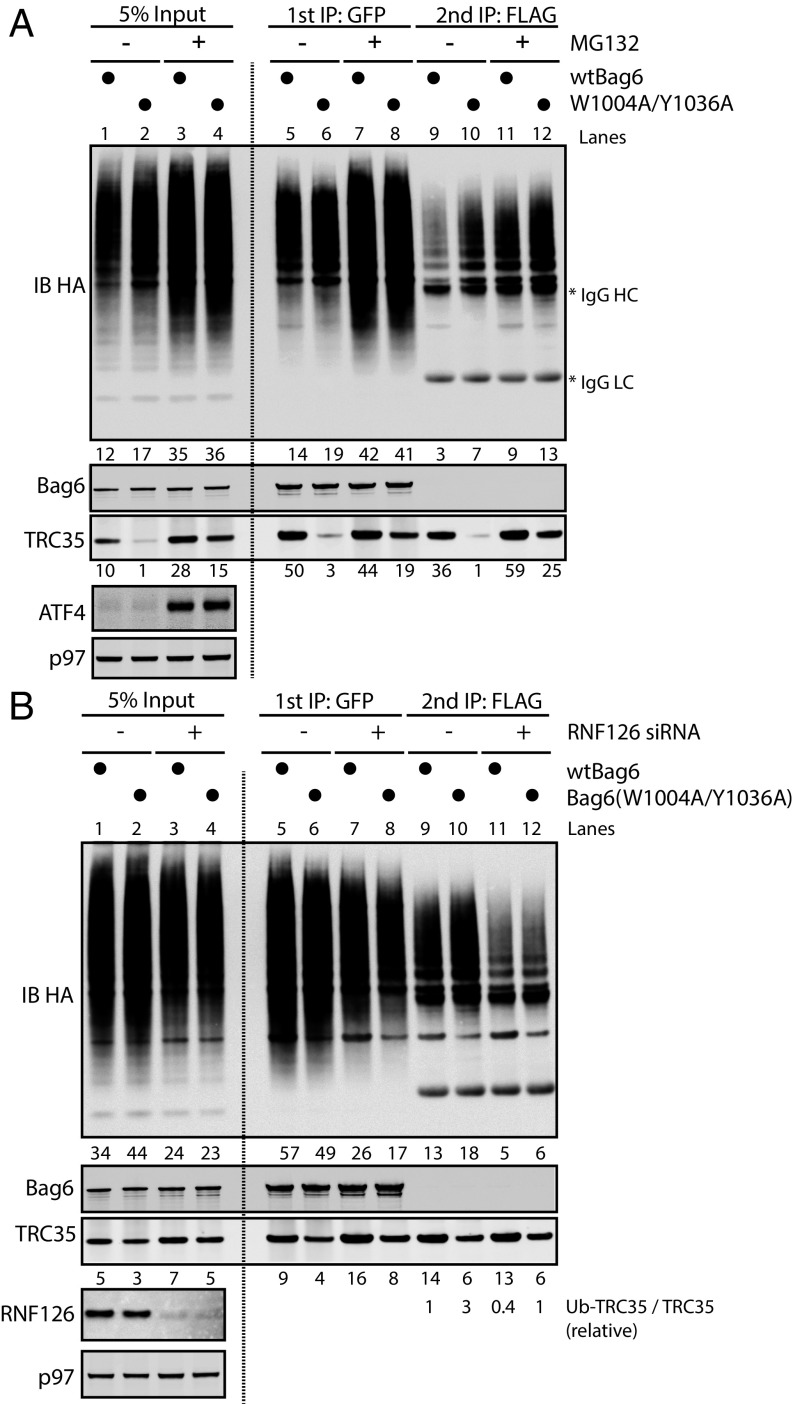
Fig. 3.
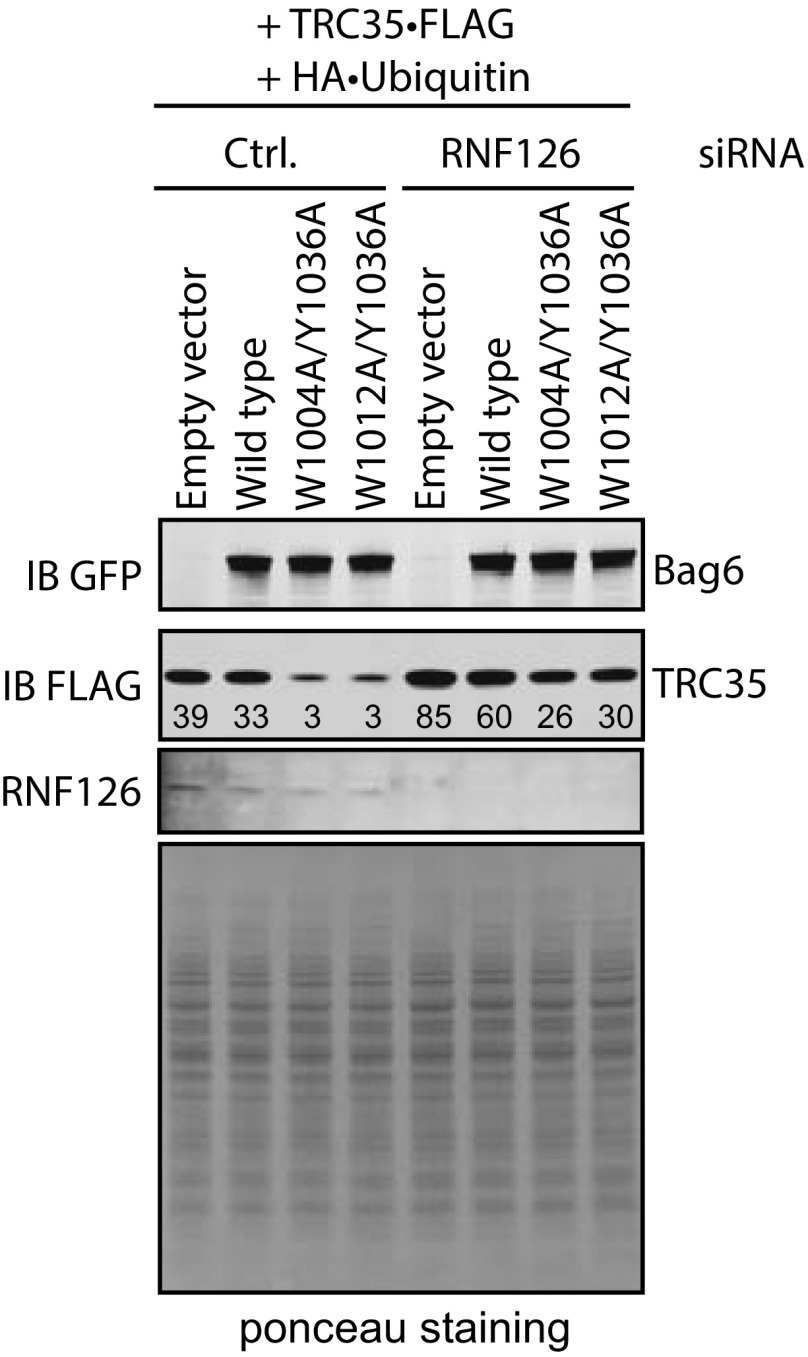
Ubiquitylation of TRC35 in cells expressing Bag6 mutants is dependent on RNF126. (A) Bag6−/− 293T cells cotransfected with plasmids encoding TRC35•FLAG (WT), Bag6⋅GFP (WT or mutant), and HA•Ub and treated with 10 µM MG132. The cell extracts were subject to two rounds of IP with anti-GFP Ab then with anti-FLAG Ab in denaturing conditions. Ubiquitylation was assessed by immunoblotting with HA Ab. (B) Bag6−/− 293T cells cotransfected with plasmids encoding TRC35•FLAG (WT), Bag6•GFP (WT or mutants), and HA⋅Ub. The cells were simultaneously treated with 10 µM MG132 and siRNAs against RNF126. The cell extracts were subject to two rounds of denaturing IP with anti-GFP Ab and then with anti-FLAG Ab. TRC35 ubiquitylation was assessed by immunoblotting with HA Ab.
To further ensure that unassembled TRC35 is a Bag6-QC substrate, we investigated the effect of the Ub-ligase RNF126 on TRC35 ubiquitylation. RNF126 is a Bag6-associated Ub-ligase utilized by the Bag6-QC for ubiquitylation of Bag6-associated clients in the cytosol (27). If TRC35 ubiquitylation is mediated by Bag6-QC, knocking down RNF126 in cells expressing Bag6 mutants would result in reduced ubiquitylation and increased stability of TRC35. To test this hypothesis, we examined the effect of siRNA-mediated RNF126 knockdown (KD) on TRC35 ubiquitylation. 293T cells coexpressing HA⋅Ub, TRC35⋅FLAG, and either WT or mutant Bag6⋅GFP (W1004A/Y1036A or W1012A/Y1036A) were treated with siRNA against RNF126 followed by 6-h treatment with MG132. Bag6-associated TRC35 was isolated by two rounds of IP. Immunoblotting was used to analyze the relative ratio between ubiquitylated TRC35 and unmodified TRC35 (Ub–TRC35/TRC35). The ratio in cells expressing both wtTRC35 and wtBag6 without RNF126 was defined as 1. In cells expressing Bag6 mutants, ubiquitylation of TRC35 bound to Bag6 is moderately increased (Fig. 3B, lane 9 vs. lane 10); RNF126 KD reduces TRC35 ubiquitylation in cells expressing either wtBag6 or Bag6 double mutants, resulting in approximately two- to threefold reduction in Ub–TRC35/TRC35 (Fig. 3B, lanes 9 and 10 vs. lanes 11 and 12). Importantly, like MG132 treatment, RNF126 KD rescued the expression of TRC35 in cells expressing the double mutant (Fig. S6), presumably due to increased stability. Together these results demonstrate that ubiquitylation of unassembled TRC35 is modulated by the QC role of Bag6 and RNF126.
Fig. S6.
The effect of RNF126 on TRC35 stabilization. Immunoblot analysis of the effect of RNF126 on TRC35 levels in wt293T cells cotransfected with plasmids encoding TRC35⋅FLAG (WT), Bag6⋅GFP (WT or mutants), and HA⋅ubiquitin. A combination of three siRNAs against RNF126 was used. Bag6 and TRC35 levels were assessed by immunoblotting with anti-GFP and anti-FLAG Abs.
TRC35 Masks the Bipartite Nuclear Localization Sequence of Bag6 and Retains Bag6 in the Cytosol
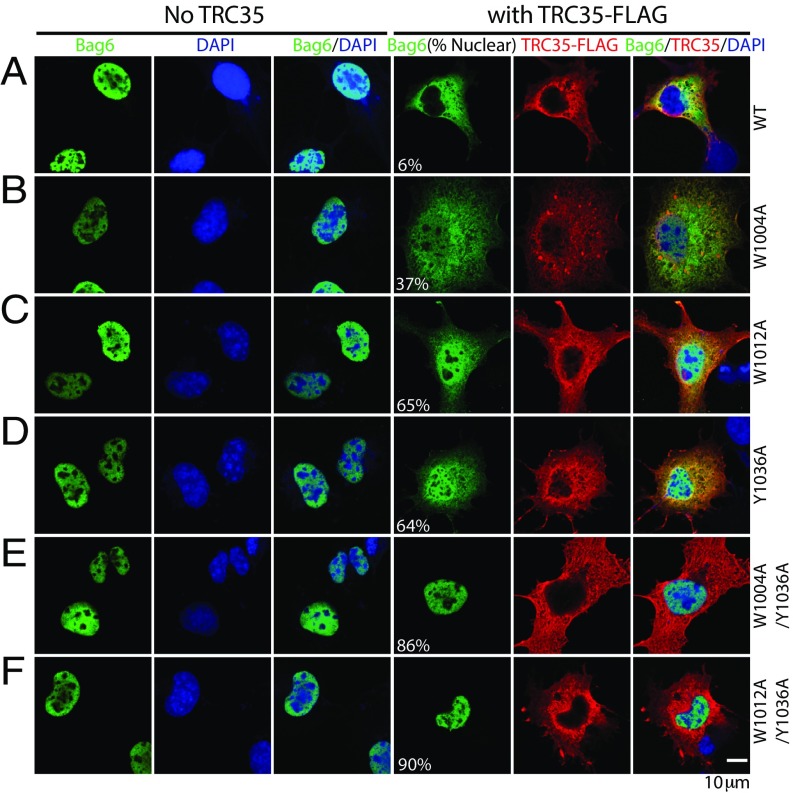
To investigate the Bag6–TRC35 interface in the context of Bag6 localization, the mutants identified from Y2H and IP were used for localization studies. Overexpression of TRC35 has been shown to retain Bag6 in the cytosol (18), suggesting that TRC35 binding is required for cytosolic localization of Bag6. Because the mutations identified in Y2H experiments specifically prevent TRC35 binding at its native binding site, exogenously expressed Bag6 mutants defective in TRC35 binding would localize primarily in the nucleus regardless of TRC35 expression. To test this hypothesis, WT or mutant Bag6•GFP were expressed in Cos7 cells with or without TRC35•FLAG. The localization of Bag6 and TRC35 was examined by immunofluorescence.
As expected (18), overexpressed wtBag6 and various Bag6 mutants were localized to the nucleus (Fig. 4A). This is likely due to excess Bag6 that cannot be retained in the cytosol by endogenous TRC35. When TRC35 is coexpressed with wtBag6, the increased cytosolic pool of TRC35 captures wtBag6 and both proteins stain primarily in the cytosol (Fig. 4A). In accordance with the IP results (Fig. 2B), introduction of a single mutation—W1004A, W1012A, or Y1036A—results in some Bag6 localization to the nucleus (Fig. 4 B–D and Table S3). Mutations at both interface I (W1004A or W1012A) and interface II (Y1036A) further disrupt the binding between Bag6 and TRC35 (Fig. 2), and Bag6 localizes primarily to the nucleus (Fig. 4 E and F and Table S3). These results unequivocally establish TRC35 as a cytoplasmic retention factor of Bag6.
Fig. 4.
Bag6 mutations at the TRC35 binding site results in nuclear localization of Bag6. (A–F) Representative images of Cos7 cells expressing WT TRC35⋅FLAG and WT or mutant Bag6⋅GFP. Cos7 cells were transfected either with Bag6⋅GFP (WT or mutant) expressing plasmid alone or cotransfected with TRC35•FLAG (WT) expressing plasmid. Cells were stained with anti-GFP (green) and/or anti-FLAG (red) Abs. DNA was stained with DAPI where indicated (blue). In the Bag6 column, the percent of cells that contain nuclear Bag6 is noted in parenthesis. For each Bag6 variant, ∼100 cells were counted and categorized as having nuclear or cytosolic Bag6.
TRC35 Has Higher Affinity for Bag6 than Karyopherin-α 2
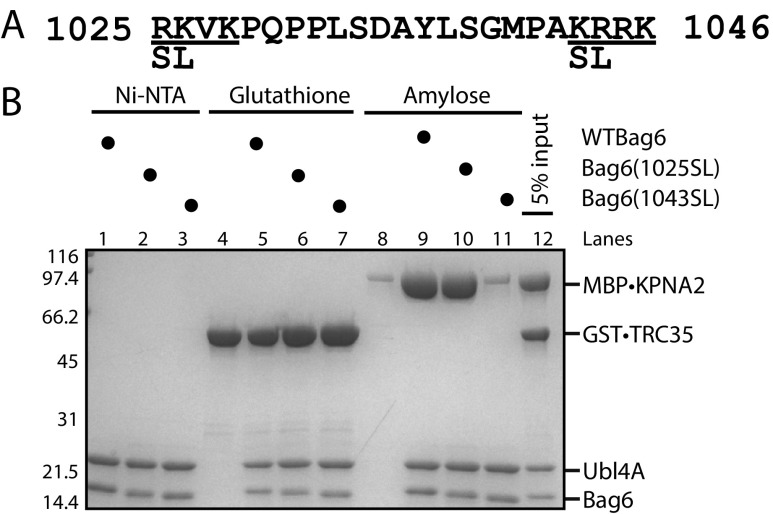
All molecules destined for the nucleus must move through the nuclear pore complex, which spans the nuclear envelope and facilitates nucleo-cytoplasmic traffic (29). Macromolecules larger than ∼40 kDa cannot freely diffuse through and require carrier proteins, such as the karyopherin-α (KPNA) family of transporters (30). The two basic clusters, R1024KVK and K1043RRK (Fig. S7A), in Bag6 are thought to act as a bipartite NLS by specifically recognizing the acidic substrate-binding surface of karyopherins (31). In HeLa cells, the K1045R to S1045L mutation has been shown to abrogate nuclear localization of Bag6 (31). Therefore, TRC35 and KPNA likely bind Bag6 at the fragment that contains the NLS.
Fig. S7.
The second basic cluster of Bag6 is necessary and sufficient for binding karyopherin α 2. (A) The putative bipartite nuclear localization sequence of Bag6. The serine and leucine mutations introduced in this study are highlighted. (B) Recombinantly purified Bag6C131–6xHis⋅Ubl4A (500 pmol) was incubated with twofold excess GST⋅TRC35 or MBP⋅KPNA2 for 20 min at room temperature. Ni-NTA beads were used to capture purified Bag6C131–6xHis⋅Ubl4A and bound factors.
To confirm that the Bag6-NLS is a KPNA-binding site and to define the residues involved in KPNA binding, 131 C-terminal residues of Bag6 (Bag6C131) (32) in complex with hexahistidine-tagged full-length Ubl4A (Bag6C131–6xHis⋅Ubl4A or Bag6–Ubl4A) was purified from E. coli. KPNA2 has been shown to interact with Bag6 (33) and could be stably purified. Mutations previously shown to abrogate Bag6 nuclear localization (31) were introduced at either the first basic cluster (1024SL) or the second basic cluster (1045SL) (Fig. S7A). The purified Bag6–Ubl4A variants were incubated with either GST⋅TRC35 or MBP⋅KPNA2 lacking the importin β binding domain (58–529), and the resulting complexes were isolated using Ni-NTA beads. WT, 1024SL, and 1045SL Bag6-Ubl4A pulled down GST⋅TRC35 with similar efficiency (Fig. S7B, lanes 5–7), demonstrating that these residues are not involved in binding TRC35. MBP⋅KPNA2 formed a complex with Bag6 that can be visualized in a pulldown (Fig. S7B, lane 9). Surprisingly, only mutation of the second basic cluster led to disruption of the interaction between Bag6–Ubl4A and MBP⋅KPNA2 (Fig. S7B, lane 10 vs. lane 11). These results show that the residues required for binding KPNA2 are distinct from those required for binding TRC35. Moreover, unlike what had been previously suggested (31), the Bag6 NLS is monopartite with the second basic cluster both necessary and sufficient for binding KPNA2. The overlap of the two binding sites provides a mechanism for Bag6 retention in the cytoplasm.
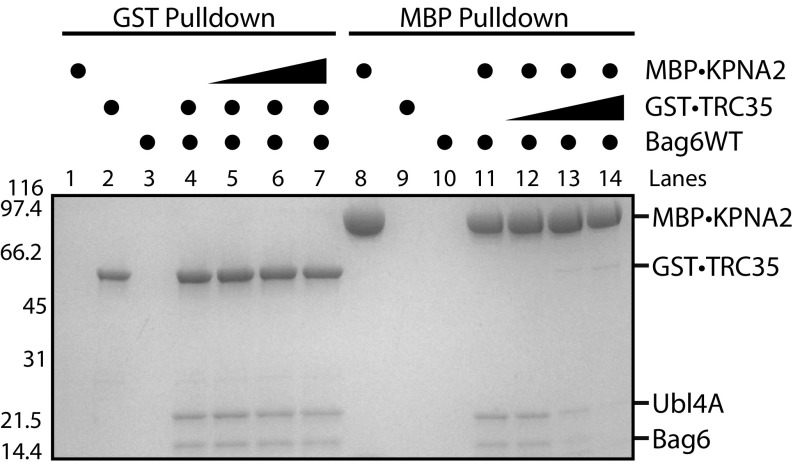
If binding of TRC35 to Bag6 prevents KPNA-mediated translocation, only TRC35-free Bag6 should be able to bind KPNA and be translocated to the nucleus. One way this could be achieved is if KPNAs have a higher affinity for Bag6 than TRC35, up-regulation of KPNA expression would lead to displacement of TRC35 from Bag6 and formation of a Bag6–KPNA complex. To test this hypothesis, we compared the relative binding affinities of TRC35 and KPNA2 to Bag6 using an exchange assay. Bag6–Ubl4A complexes with GST•TRC35 were generated and bound to glutathione affinity resin. After washing, increasing amounts of KPNA2 were added. The ability of KPNA2 to displace Bag6–Ubl4A from TRC35 was determined by the amount of Bag6–Ubl4A eluted from the resin after incubation. In this case, even at the highest concentration tested (twofold molar excess), there was no significant displacement of Bag6 from TRC35 by KPNA2 (Fig. 5, lanes 4–7). When starting with MBP⋅KPNA2–Bag6–Ubl4A on amylose beads, adding excess TRC35 resulted in the dissociation of the KPNA2–Bag6–Ubl4A complex (Fig. 5, lanes 11–14). These results highlight the stability of the TRC35–Bag6 complex and argue against the ability of KPNA regulation as a means for modulating the nuclear pool of Bag6.
Fig. 5.
TRC35 binding precludes karyopherin α binding to Bag6. Recombinantly purified Bag6C131–6xHis⋅Ubl4A (500 pmol) was first incubated with either GST⋅TRC35 or MBP⋅KPNA2 (500 pmol). The resulting GST⋅TRC35–Bag6–Ubl4A complex was incubated with glutathione resin beads, and increasing amounts of MBP⋅KPNA2 (50, 250, or 1,000 pmol) was added. The ability of MBP⋅KPNA2 to displace Bag6–Ubl4A from GST⋅TRC35 was examined by eluting the GST⋅TRC35 and bound Bag6–Ubl4A from the glutathione resin. The opposite experiment was also carried out using amylose resin beads.
Discussion
Bag6 is a critical scaffolding factor that forms a stable complex with TRC35 and Ubl4A and is involved in protein targeting, protein quality control, and transcription regulation. While there is ample evidence that Bag6 plays both cytosolic and nuclear roles, it is unclear how the localization of Bag6 is regulated. Here we report the structure of the Bag6 NLS region bound to its cytosolic retention factor TRC35, and suggest a mechanism for the regulation of Bag6 localization.
The remarkable conservation of TRC35 both in sequence (Fig. S3) and at the structural level (Fig. S1) from yeast to human can be attributed to the role of TRC35 as a hub of protein–protein interactions in the TRC pathway. The most important interaction, based on its conservation, is between TRC35 and TRC40 (Fig. S2). This interaction is critical for the regulation of TA protein transfer (3, 13) and, as shown in fungal Get4, for release of Get3/TRC40 from the ER membrane to restore the cytosolic pool of complex ready for TA protein transfer (34). The importance of linking TRC35 homologs to Ubl4A homologs is sustained in humans despite the differences in structural features (15). In metazoans, the component Bag6 acts as a scaffold to link TRC35 to Ubl4A. The Bag6–TRC35 structure reveals that TRC35 binds this binding partner utilizing conserved patches in a distinct fashion (Fig. 1). This led to the expansion of the TRC35 protein–protein interaction network while maintaining its crucial interaction with TRC40.
Our results also show that TRC35 acts as an intermolecular mask to Bag6 NLS in a role similarly performed in other pathways. Examples of other cytosolic retention factor pairs include IκB and NF-κB (35) and BRAP2 that retains HMG20A (36). Unlike other cytoplasmic retention factors, which have only been shown to bind their target NLS-containing proteins for occlusion of the NLS, TRC35 also plays a distinct role in the cytoplasmic TA targeting and protein quality control when in complex with Bag6. This dual functionality may be evolutionarily conserved. In yeast, fungal Get4 binds the NTD of Get5, which appears to contain a functional NLS that directs Get5 to the nucleus during a Get5-mediated stress response (37).
These results allow speculation of possible regulatory mechanism for Bag6 nuclear localization. First, disrupting the Bag6–TRC35 interface by introducing alanine mutations led to ubiquitylation of TRC35 (Fig. S5), which is mediated by RNF126. Knocking down Bag6 has been shown to reduce levels of TRC35 in HeLa cells (22), suggesting that Bag6 is required for TRC35 stability in vivo. We also showed that KPNA2 has a lower affinity for Bag6 than TRC35 (Fig. 5), which suggests that for Bag6 to bind KPNA and translocate into the nucleus, it needs to be free of TRC35. The most likely explanation is that cells regulate Bag6 localization by modulating Bag6 levels or decreasing TRC35 levels. In humans, the Bag6 rs3117582 single nucleotide polymorphism found in the promoter region of Bag6 likely affects expression levels and is associated with higher incidence of lung cancer (38–40) and osteoarthritis (41). The localization could also be pretranslationally regulated with differential splicing. In brain and breast tissue, for instance, Bag6 isoforms that lack the NLS are expressed at higher levels than isoforms with the NLS (42).
Our findings establish TRC35 as a universally conserved protein in the opisthokont lineage in both structure and function. These findings support the model in which TRC35 is an important hub of a protein–protein interaction network with its interaction with TRC40 at the core. Higher eukaryotes expanded on this network by the addition of Bag6. Our biochemical studies unequivocally establish TRC35 as a cytosolic retention factor of Bag6 and suggest that Bag6 needs to be in excess for nuclear localization. However, how different cell types regulate Bag6 localization, whether the regulation can be mediated by specific stress, and the biological implications of differential distribution of Bag6 are important questions for future studies.
Materials and Methods
Detailed descriptions of experiments are provided in SI Materials and Methods. Briefly, for crystallization and in vitro binding assays human genes were subcloned and expressed in Escherichia coli. Proteins were purified using affinity chromatography, anion-exchange, and size-exclusion chromatography. Truncated fragments of Bag6 and TRC35 were crystallized, and the structure was determined using molecular-replacement single-wavelength anomalous dispersion. The genomes were surveyed using MEME suite (43). The IP assays were carried out using 293T cell extracts. A combination of three siRNAs against RNF126 was added to 293T cells. MG132 (10 µM) was added to inhibit the proteasome. Cos7 cells were used for localization assays.
SI Materials and Methods
Cloning, Expression, and Purification.
All constructs and specifically the major Bag6 isoform 2 were derived from human cDNA. For crystallization, both Bag6(1,000–1,054) and TRC35(23–305) were inserted into the multiple cloning site of the pACYCDuet plasmid (Novagen) with an N-terminal hexahistidine tag on Bag6(1000–1054). The plasmid was transformed into E. coli NiCo21(DE3) (New England Biolabs). The plate was scraped to inoculate 12 2L baffled flasks containing 2×YT medium then grown by shaking at 37 °C in a shaking incubator (Multitron Standard; Infors HT) at 250 rpm. Cell growth was monitored to an OD600 = 0.6 then protein expression was induced for 3 h by the addition of 500 μM isopropyl-β-d-thiogalactopyranoside (IPTG) (Affymetrix). Cells were harvested by centrifugation in a TLA 8.1 rotor at 4,000 ×g for 20 min. The pellet was resuspended in 50 mM Mops (pH 7.2), 300 mM K⋅glutamate, 5 mM MgOAc, 20 mM imidazole, 5 mM β-mercaptoethanol (1 g cell/10 mL lysis buffer) and supplemented with 0.1 mM PMSF and 1 mM benzamidine. Cells were lysed using an M-110L microfluidizer (Microfluidics) by two passes at ∼17,500 psi. The lysate was clarified by centrifugation at 235,000 ×g in a Beckman Ti45 rotor for 30 min at 4 °C. The clarified lysate was incubated for 1 h with 3 mL of a 50% (vol/vol) slurry of nickel-nitrilotriacetic acid (Ni-NTA) agarose (Qiagen) by rocking. The mix was poured into a gravity column then washed with 100 mL lysis buffer. The protein was eluted with 12 mL elution buffer (20 mM Mops (pH 7.2), 150 mM K⋅glutamate, 300 mM Imidazole, 5 mM β-mercaptoethanol). The eluent was mixed with ∼0.5 mg of TEV protease in snakeskin dialysis tubing with 10 kDa molecular weight cutoff (Thermo Fisher Scientific) and dialyzed overnight at room temperature in (20 mM Mops (pH 7.2), 100 mM K⋅glutamate, and 10 mM β-mercaptoethanol). Precipitate was removed by centrifugation with Beckman SX4750A rotor at 3,000 ×g for 5 min at 4 °C and filtered with a 0.22 µm syringe filter. The sample was concentrated and loaded onto a 5 mL UnoQ ion-exchange column (Bio-Rad) then eluted with a 60 mL 20 mM Mops (pH 7.2), 50–500 mM K⋅glutamate gradient, 5 mM β-mercaptoethanol. The fractions containing the protein (∼5–23 mL) were dialyzed in snakeskin dialysis bag in 20 mM Mops (pH 7.2), 50 mM K⋅glutamate, 5 mM β-mercaptoethanol for 2 h at 4 °C. The sample was concentrated to 2 mL, filtered with a 0.22-µm syringe filter, then further purified by size-exclusion chromatography over a 120 mL Superdex 75 column (GE Healthcare) (20 mM Mops (pH 7.2), 50 mM K⋅glutamate, 5 mM β-mercaptoethanol). The fractions containing the heterodimer (61 mL–72 mL) and monomeric Bag6 (77–82 mL) were pooled and concentrated to ∼10 mg/mL using centrifugal filter units with 10 kDa molecular weight cutoff (Merck Millipore).
The plasmid containing human KPNA2 was obtained from Addgene (#26677). A truncated KPNA2 (58–529) was subcloned into pMAL-C2 vector (New England Biolabs) and transformed into E. coli NiCo21(DE3). The cells were grown in 2×YT medium and induced at 37 °C until OD600 = 0.1 and then were cooled on ice for 1 h. Expression was induced with 500 μM IPTG for 18 h at 16 °C in a shaking incubator at 200 rpm. The cells were harvested as above and resuspended in 50 mM Mops (pH 7.2), 300 mM K⋅glutamate, 5 mM β-mercaptoethanol, supplemented with cOmplete EDTA-free protease inhibitor mixture (Roche) and lysed using the M-110L microfluidizer by two passes at ∼17,500 psi. The lysate was clarified by centrifugation at 235,000 ×g in a Beckman Ti45 rotor for 30 min at 4 °C. The lysate was incubated for 1 h with 3 mL of a 50% (vol/vol) slurry of amylose resin (New England Biolabs) by rocking. The mix was poured into a gravity column then washed with 100 mL lysis buffer. The protein was eluted with 12 mL elution buffer (20 mM Mops (pH 7.2), 150 mM K⋅glutamate, 10 mM maltose, 5 mM β-mercaptoethanol). The sample was placed in snakeskin dialysis bag (10 kDa cutoff, Thermo Fisher) and dialyzed overnight at 4 °C in 20 mM Mops (pH 7.2), 50–800 mM K⋅glutamate gradient, 5 mM β-mercaptoethanol. The fractions containing the protein (∼36–70 mL) were pooled and dialyzed in snakeskin dialysis bag in 20 mM Mops (pH 7.2), 50 mM K⋅glutamate, 5 mM β-mercaptoethanol for 2 h at 4 °C. The sample was concentrated to 2 mL, filtered with a 0.22-µm syringe filter, and purified by size-exclusion chromatography over a 120-mL Superdex 200 column (GE Healthcare) (20 mM Mops (pH 7.2), 50 mM K⋅glutamate, 5 mM β-mercaptoethanol). Fractions containing the sample (60 mL–90 mL) were pooled and concentrated with centrifugal filtration units with a 50-kDa cutoff.
For expression and purification of GST⋅TRC35, full-length TRC35 was subcloned into pGEX6P-1 (GE Healthcare) and transformed into NiCo21(DE3). The cells were grown at 37 °C until OD600 = 0.1, chilled on ice for 1 h, then induced with 500 μM IPTG at 16 °C for 18 h in a shaking incubator at 200 rpm. Cells were lysed using 10 mL lysis buffer [50 mM Hepes (pH 7.2), 400 mM KCl, 5 mM mercaptoethanol] per g cell pellet and were incubated with 3 mL 50% (vol/vol) slurry of glutathione resin (GE Healthcare) for 2 h at 4 °C by rocking. The resin was washed with 100 mL lysis buffer, and the protein was eluted with 12 mL of freshly prepared 20 mM Hepes (pH 7.2), 150 mM KCl, 33 mM glutathione, 5 mM β-mercaptoethanol. The sample was placed in snakeskin dialysis bag (10 kDa cutoff) (Thermo Fisher) and dialyzed overnight at 4 °C in 20 mM Hepes (pH 7.2), 100 mM K⋅glutamate, 5 mM β-mercaptoethanol, before concentration with centrifugal filter unit (50-kDa cutoff) (Millipore).
Crystallization.
Crystallization screening was performed using the sitting-drop vapor-diffusion method with the commercially available PEG Ion screen (Hampton Research) and MRC 2 Well Crystallization plate (Hampton Research). Initial screening was performed by a Mosquito robot (TTP Labtech) with 100 nL protein:100-nL drops of well solution and 50-uL wells. No refinement was necessary. Crystals grew to full size after 5 d as rectangular prisms in 20% PEG-3350 (Wt/vol), 0.2 M DL-malic acid (pH 7.0). For cryoprotection, crystals were transferred into 100 uL of well solution with 20% (vol/vol) glycerol for 15 s and then were frozen in liquid nitrogen. Rubidium derivatives were generated by transferring crystals into 40 μL of well solution plus 350 mM rubidium iodide for 2–10 s before cryopreservation with glycerol.
Data Collection, Structure Solution, and Refinement.
X-ray diffraction data were collected on Beam line BL12-2 at the Stanford Synchrotron Radiation Laboratory (SSRL). A complete native dataset was collected from a single crystal to 1.8 Å resolution, and a single rubidium derivative crystal dataset was collected from a single crystal to 2.0 Å with a Dectris Pilatus 6M detector. Data were integrated, scaled, and merged using XDS (44) and SCALA (45, 46). Phases were determined by molecular replacement single-wavelength anomalous dispersion (MRSAD) using SHARP (47) and PHASER-MR (48) on PHENIX (49). The initial structure was built by PHASER as implemented by PHENIX (49). The model was further built and refined against the native dataset over several rounds using COOT (50) and PHENIX (49). Statistics are provided in Table S1.
Phylogenetic Tree of GET/TRC Components.
The phylogenetic tree was modified from the maximum-likelihood phylogenetic tree of eukaryotes by Eme et al. (51) The MEME suite (43) was used to determine the presence or absence of the GET/TRC components in the genomes presented in the tree. The MEME motif discovery tool was used to make motifs for Ubl4A, Get5, Get4/TRC35, Sgt2, SGTA, and Bag6. The motifs were then used to search the genomes using the motif-scanning tool MAST (52). Identified proteins were confirmed using BLAST (53) and the corresponding reference ID is provided on Table S2.
Table S2.
Gene IDs used in Fig. S2
| Species | TRC35 | TRC40 | Bag6 |
| Homo sapiens | NP_057033.2 | NP_004308.2 | NP_004630.3 |
| Gallus gallus | XP_015149638.1 | XP_015158190.1 | |
| Branchiostoma floridae | XP_002607071.1 | XP_002605952.1 | XP_002594881.1 |
| Drosophila melanogaster | NP_649464.1 | NP_610296.2 | NP_648132.2 |
| Caenorhabditis elegans | NP_498034.1 | NP_498965.1 | |
| Schistosoma mansoni | XP_018645653.1 | XP_018648676.1 | XP_018648249.1 |
| Nematostella vectensis | XP_001640454.1 | XP_001638254.1 | XP_001635433.1 |
| Amphimedon queenslandica | XP_011404841.1 | XP_003385895.1 | |
| Monosiga brevicollis | XP_001750773.1 | XP_001743630.1 | |
| Capsaspora orwazaki | XP_004342754.1 | XP_004363354.1 | |
| Fonticula alba | KCV72828.1 | XP_009492379.1 | |
| Saccharomyces cerevisiae | NP_014807.3 | NP_010183.1 |
Yeast Two-Hybrid.
The PJ69-4α strain was obtained from the Yeast Resource Center at the University of Washington. Bag6 isoform 2 residues 951–1,126 were cloned into the pGAD-C1 vector, and TRC35 was cloned into pGBDU-C1 vector. Alanine mutations were made using Q5 site-directed mutagenesis (New England Biolabs). pGAD-C1–TRC35 (WT or mutant) and pGBDU-C1–Bag6 (WT or mutant) were cotransformed into PJ69-4α using previously described methods (54) and then were plated onto SC-Ura-Leu plates and incubated at 30 °C. Single colonies from the transformants were cultured in 5 mL SC-Ura-Leu liquid medium and grown overnight in a shaking incubator (Multitron Standard Infors HT) at 200 rpm at 30 °C. Then 2 × 107 cells were transferred into a total of 5 mL SC-Ura-Leu medium and grown in a shaking incubator at 30 °C at 200 rpm for 6 h. Cells were harvested by centrifugation with a Beckman SX4750A rotor at 3,000 × g at 25 °C. The cells were washed twice by resuspension in 5 mL of sterile water followed by centrifugation at 3,000 × g at 25 °C. After the second wash, cells were resuspended in 1 mL sterile water. Concentration was measured, and 1 × 107 cells were resuspended in a total volume of 40 µL of sterile water. Four microliters of this resuspended sample were spot-plated onto a SC-Ura-Leu-Ade plate and incubated for 72 h at 30 °C.
Yeast Two-Hybrid Expression Controls.
Cells were grown overnight in SC-Ura-Leu medium. Then 2.5 × 107 cells were transferred to a total volume of 5 mL medium and grown for ∼6–8 h until OD600 = 2.0. Cells were harvested by centrifugation at 4,000 × g for 5 min at 4 °C. Cells were resuspended in 2 mL 10 mM TE buffer [Tris⋅HCl (pH 8.0), 0.1 mM EDTA]. Cells were centrifuged in a microcentrifuge at 13,500 × g at 4 °C for 2 min. Approximately 37.5 µL of glass beads (415–600 µm) for each OD unit were added to the sample. The same volume of 1× SDS/PAGE loading buffer was added. The cells were vortexed and immediately plunged into a boiling water bath. After boiling for 3 min, tubes were immersed in an ice bucket. The tubes were placed into FastPrep (MP Biomedicals) homogenizer at 6 m/s for 45 s. Cells were boiled for 3 min and centrifuged in a microcentrifuge for 2 min at 13,500 × g at room temperature. Supernatant was transferred to a clean microfuge tube and loaded onto 5–20% gradient gel (Bio-Rad). The protein was transferred onto PVDF membrane. Membrane was blocked with Odyssey TBS blocking buffer (LI-COR) for 1 h. The membrane was incubated with a primary Ab against the Gal4 DNA-binding domain (Santa Cruz) for TRC35 detection or Gal4 transactivating domain (Santa Cruz) for Bag6 detection. After washing three times with TBST [20 mM Tris⋅HCl (pH 7.5), 150 mM NaCl, 0.2% Tween-20] for 10 min , membrane was incubated with anti-mouse secondary Ab (IRDye 680RD or 800CW; LI-COR) for 1 h at room temperature. Membrane was washed three times with TBST for 10 min each. Proteins were detected using an Odyssey Imaging System (LI-COR).
IP from 293T Cells.
On day 0, 5 × 105 Bag6−/− cells were seeded in plates. On day 1, TRC35 with a C-terminal FLAG tag (TRC35⋅FLAG), Bag6 with a C-terminal GFP tag (WT or mutant Bag6⋅GFP), and HA⋅ubiquitin were cotransfected. On day 3, cells were collected and washed with 1 mL of ice-cold PBS. The cells were lysed with Nonidet P-40 lysis buffer containing 1 mM N-ethylmaleimide to inhibit the ubiquitin–proteasome system. The detergent soluble fraction was used for IP using GFP Ab. After IP, the beads were divided into two fractions. Half was analyzed directly with SDS/PAGE and immunoblotting with indicated Abs to assess the interaction between TRC35 and Bag6 and Bag6 mutants. The other half was used for denaturing IP to detect TRC35 ubiquitylation. The beads were resuspended in 150 µL denaturing buffer (1× PBS, 1% SDS, 5 mM DTT) and heated at 95 °C for 10 min. Then 1.35 mL Nonidet P-40 lysis buffer was added into the tube and incubated at 4 °C for 30 min. The supernatant was used for IP with FLAG A-conjugated M2 beads (Sigma-Aldrich). The eluate was analyzed with SDS/PAGE and immunoblotting.
To assay TRC35 ubiquitylation after proteasome inhibition, 5 × 105 WT 293T cells were seeded in plates on day 0. On day 1, cells were transfected as above. Twenty-four hours post transfection, proteasome inhibitor, MG132, was added to the concentration of 10 µM and incubated overnight. Cells were collected and analyzed with sequential IP and immunoblotting as above.
To investigate the effect of simultaneous RNF126 down-regulation and proteasomal inhibition on TRC35 ubiquitylation, 5 × 105 Bag6 293T cells were seeded in plates on day 0. On day 1, cells were transfected with RNF126 siRNA or control siRNA. On day 2, cells were split in half into new plates. After ∼9 h, plasmids encoding TRC35⋅FLAG (WT), Bag6⋅GFP (WT or mutant) and HA⋅ubiquitin were transfected. On day 3, medium was replaced with fresh medium containing either 10 µM MG132 or DMSO. On day 4, cells were collected, and sequential IP was performed as described above.
Localization Assay.
On day 0, 1 × 105 Cos7 cells were seeded onto a 12-well plate with a poly-d-lysine–coated cover glass. After ∼8 h, the cells were cotransfected with TRC35⋅FLAG and Bag6•GFP (WT or mutant). Twenty hours after transfection, the cells were washed with 1× PBS, fixed with 4% (vol/vol) paraformaldehyde for 15 min, then washed with 1× PBS before permeabilization with staining solution [1× PBS, 5% FBS (vol/vol), 0.1% (vol/vol) Nonidet P-40]. Cells were then stained with staining solution containing primary Ab and incubated at room temperature for 1 h. After washing the cells with 1× PBS, the cells were incubated with staining solution with secondary Ab and then were washed with 1× PBS. Cells were counterstained with a mounting medium containing DAPI to illuminate the nucleus for visualization with Axiovert 200M microscope (Zeiss). Confocal microscope analyses were performed with a Zeiss LSM 780 system.
To assay the localization of each Bag6 mutant, three view fields were evaluated with a 40× oil immersion objective. Approximately 100 cells were evaluated for each Bag6 variant, and cells were counted as having Bag6 primarily in either the nucleus or the cytosol.
In Vitro Binding Assay with Purified Proteins.
1 nmol hexahistidine-tagged Ubl4A–Bag6C131 (WT, 1025SL, or 1043SL) was incubated with 1 nmol GST⋅TRC35 or 1 nmol MBP⋅KPNA2 in a 100-µL total volume of binding buffer [20 mM Mops (pH 7.2), 100 mM K⋅glutamate 20 mM imidazole, 5 mM β-mercaptoethanol] at room temperature for 30 min. Then 30 µL 50% slurry of Ni-NTA beads (Qiagen) equilibrated with binding buffer were added to the reaction and incubated at room temperature for 30 min. The beads were resuspended using a pipet every 10 min. The beads were then washed twice with 100 µL of room-temperature binding buffer. Samples bound to the resin were eluted with 25 µL of elution buffer [20 mM Mops (pH 7.2), 100 mM K⋅glutamate, 300 mM imidazole, 5 mM β-mercaptoethanol] and evaluated with Coomassie-stained SDS/PAGE gel.
Exchange Assay.
One nanomole of hexahistidine-tagged Ubl4A–Bag6C131 (WT, 1024SL, or 1043SL) was incubated with 1 nmol GST⋅TRC35 in 100 μL total volume of binding buffer [20 mM Mops (pH 7.2), 100 mM K⋅glutamate, 20 mM imidazole, 5 mM β-mercaptoethanol]. Thirty microliters of a 50% (vol/vol) slurry of glutathione beads (GE Healthcare) equilibrated with binding buffer were added to the reaction and incubated at room temperature for 30 min. The beads were resuspended every 10 min. The beads were washed twice with 100 µL of room-temperature binding buffer [20 mM Mops (pH 7.2), 100 mM K⋅glutamate, 5 mM β-mercaptoethanol]; then 0.5, 1, or 2 nmol of MBP⋅KPNA in 100 µL was added to the resin and incubated for 30 min at room temperature. The beads were washed twice with 100 µL binding buffer and then were eluted with 25 µL elution buffer [20 mM Mops (pH 7.2), 100 mM K⋅glutamate, 33 mM glutathione, 5 mM β-mercaptoethanol]. The reverse experiment starting with hexahistidine-tagged Ubl4A–Bag6C131 and MBP⋅KPNA2 was carried out as above but using 30 µL of a 50% (vol/vol) slurry of amylose beads (New England Biolabs) in 20 mM Mops (pH 7.2), 100 mM K⋅glutamate, and 5 mM β-mercaptoethanol and was eluted with 25 µL of 20 mM Mops (pH 7.2), 100 mM K⋅glutamate, 10 mM maltose, and 5 mM β-mercaptoethanol.
Acknowledgments
We thank Daniel Lin and Jens Kaiser for help with data processing, Catherine Day for technical support, members of the W.M.C. laboratory for support and useful discussions, Gordon and Betty Moore for support of the Molecular Observatory at California Institute of Technology, and the staff at the Stanford Synchrotron Radiation Lightsource for assistance with synchrotron data collection. W.M.C. is supported by NIH Grant R01GM097572.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.L.B. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors reported in this paper have been deposited in the Protein Data Bank (PDB), https://www.wwpdb.org (PDB ID code 6AU8).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1702940114/-/DCSupplemental.
References
- 1.Kalbfleisch T, Cambon A, Wattenberg BW. A bioinformatics approach to identifying tail-anchored proteins in the human genome. Traffic. 2007;8:1687–1694. doi: 10.1111/j.1600-0854.2007.00661.x. [DOI] [PubMed] [Google Scholar]
- 2.Stefanovic S, Hegde RS. Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell. 2007;128:1147–1159. doi: 10.1016/j.cell.2007.01.036. [DOI] [PubMed] [Google Scholar]
- 3.Mock JY, et al. Bag6 complex contains a minimal tail-anchor-targeting module and a mock BAG domain. Proc Natl Acad Sci USA. 2015;112:106–111. doi: 10.1073/pnas.1402745112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schuldiner M, et al. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell. 2008;134:634–645. doi: 10.1016/j.cell.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rome ME, Rao M, Clemons WM, Shan SO. Precise timing of ATPase activation drives targeting of tail-anchored proteins. Proc Natl Acad Sci USA. 2013;110:7666–7671. doi: 10.1073/pnas.1222054110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamamoto Y, Sakisaka T. Molecular machinery for insertion of tail-anchored membrane proteins into the endoplasmic reticulum membrane in mammalian cells. Mol Cell. 2012;48:387–397. doi: 10.1016/j.molcel.2012.08.028. [DOI] [PubMed] [Google Scholar]
- 7.Vilardi F, Lorenz H, Dobberstein B. WRB is the receptor for TRC40/Asna1-mediated insertion of tail-anchored proteins into the ER membrane. J Cell Sci. 2011;124:1301–1307. doi: 10.1242/jcs.084277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rivera-Monroy J, et al. Mice lacking WRB reveal differential biogenesis requirements of tail-anchored proteins in vivo. Sci Rep. 2016;6:39464. doi: 10.1038/srep39464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abell BM, Pool MR, Schlenker O, Sinning I, High S. Signal recognition particle mediates post-translational targeting in eukaryotes. EMBO J. 2004;23:2755–2764. doi: 10.1038/sj.emboj.7600281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aviram N, et al. The SND proteins constitute an alternative targeting route to the endoplasmic reticulum. Nature. 2016;540:134–138. doi: 10.1038/nature20169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abell BM, Rabu C, Leznicki P, Young JC, High S. Post-translational integration of tail-anchored proteins is facilitated by defined molecular chaperones. J Cell Sci. 2007;120:1743–1751. doi: 10.1242/jcs.002410. [DOI] [PubMed] [Google Scholar]
- 12.Itakura E, et al. Ubiquilins chaperone and triage mitochondrial membrane proteins for degradation. Mol Cell. 2016;63:21–33. doi: 10.1016/j.molcel.2016.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gristick HB, et al. Crystal structure of ATP-bound Get3-Get4-Get5 complex reveals regulation of Get3 by Get4. Nat Struct Mol Biol. 2014;21:437–442. doi: 10.1038/nsmb.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bozkurt G, et al. The structure of Get4 reveals an alpha-solenoid fold adapted for multiple interactions in tail-anchored protein biogenesis. FEBS Lett. 2010;584:1509–1514. doi: 10.1016/j.febslet.2010.02.070. [DOI] [PubMed] [Google Scholar]
- 15.Chartron JW, Suloway CJ, Zaslaver M, Clemons WM., Jr Structural characterization of the Get4/Get5 complex and its interaction with Get3. Proc Natl Acad Sci USA. 2010;107:12127–12132. doi: 10.1073/pnas.1006036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang YW, et al. Crystal structure of Get4-Get5 complex and its interactions with Sgt2, Get3, and Ydj1. J Biol Chem. 2010;285:9962–9970. doi: 10.1074/jbc.M109.087098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mariappan M, et al. A ribosome-associating factor chaperones tail-anchored membrane proteins. Nature. 2010;466:1120–1124. doi: 10.1038/nature09296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Q, et al. A ubiquitin ligase-associated chaperone holdase maintains polypeptides in soluble states for proteasome degradation. Mol Cell. 2011;42:758–770. doi: 10.1016/j.molcel.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sebti S, et al. BAT3 modulates p300-dependent acetylation of p53 and autophagy-related protein 7 (ATG7) during autophagy. Proc Natl Acad Sci USA. 2014;111:4115–4120. doi: 10.1073/pnas.1313618111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sasaki T, et al. HLA-B-associated transcript 3 (Bat3)/Scythe is essential for p300-mediated acetylation of p53. Genes Dev. 2007;21:848–861. doi: 10.1101/gad.1534107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wakeman TP, Wang Q, Feng J, Wang XF. Bat3 facilitates H3K79 dimethylation by DOT1L and promotes DNA damage-induced 53BP1 foci at G1/G2 cell-cycle phases. EMBO J. 2012;31:2169–2181. doi: 10.1038/emboj.2012.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krenciute G, et al. Nuclear BAG6-UBL4A-GET4 complex mediates DNA damage signaling and cell death. J Biol Chem. 2013;288:20547–20557. doi: 10.1074/jbc.M112.443416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wainright PO, Hinkle G, Sogin ML, Stickel SK. Monophyletic origins of the metazoa: An evolutionary link with fungi. Science. 1993;260:340–342. doi: 10.1126/science.8469985. [DOI] [PubMed] [Google Scholar]
- 24.Baldauf SL, Palmer JD. Animals and fungi are each other’s closest relatives: Congruent evidence from multiple proteins. Proc Natl Acad Sci USA. 1993;90:11558–11562. doi: 10.1073/pnas.90.24.11558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu Y, Anderson DE, Ye Y. The HECT domain ubiquitin ligase HUWE1 targets unassembled soluble proteins for degradation. Cell Discov. 2016;2:16040. doi: 10.1038/celldisc.2016.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hessa T, et al. Protein targeting and degradation are coupled for elimination of mislocalized proteins. Nature. 2011;475:394–397. doi: 10.1038/nature10181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodrigo-Brenni MC, Gutierrez E, Hegde RS. Cytosolic quality control of mislocalized proteins requires RNF126 recruitment to Bag6. Mol Cell. 2014;55:227–237. doi: 10.1016/j.molcel.2014.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melikova MS, Kondratov KA, Kornilova ES. Two different stages of epidermal growth factor (EGF) receptor endocytosis are sensitive to free ubiquitin depletion produced by proteasome inhibitor MG132. Cell Biol Int. 2006;30:31–43. doi: 10.1016/j.cellbi.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 29.Hoelz A, Debler EW, Blobel G. The structure of the nuclear pore complex. Annu Rev Biochem. 2011;80:613–643. doi: 10.1146/annurev-biochem-060109-151030. [DOI] [PubMed] [Google Scholar]
- 30.Pumroy RA, Cingolani G. Diversification of importin-α isoforms in cellular trafficking and disease states. Biochem J. 2015;466:13–28. doi: 10.1042/BJ20141186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manchen ST, Hubberstey AV. Human Scythe contains a functional nuclear localization sequence and remains in the nucleus during staurosporine-induced apoptosis. Biochem Biophys Res Commun. 2001;287:1075–1082. doi: 10.1006/bbrc.2001.5701. [DOI] [PubMed] [Google Scholar]
- 32.Wu YH, Shih SF, Lin JY. Ricin triggers apoptotic morphological changes through caspase-3 cleavage of BAT3. J Biol Chem. 2004;279:19264–19275. doi: 10.1074/jbc.M307049200. [DOI] [PubMed] [Google Scholar]
- 33.Rouillard AD, et al. The harmonizome: A collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database (Oxford) 2016;2016:baw100. doi: 10.1093/database/baw100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rome ME, Chio US, Rao M, Gristick H, Shan SO. Differential gradients of interaction affinities drive efficient targeting and recycling in the GET pathway. Proc Natl Acad Sci USA. 2014;111:E4929–E4935. doi: 10.1073/pnas.1411284111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beg AA, et al. I kappa B interacts with the nuclear localization sequences of the subunits of NF-kappa B: A mechanism for cytoplasmic retention. Genes Dev. 1992;6:1899–1913. doi: 10.1101/gad.6.10.1899. [DOI] [PubMed] [Google Scholar]
- 36.Davies RG, Wagstaff KM, McLaughlin EA, Loveland KL, Jans DA. The BRCA1-binding protein BRAP2 can act as a cytoplasmic retention factor for nuclear and nuclear envelope-localizing testicular proteins. Biochim Biophys Acta. 2013;1833:3436–3444. doi: 10.1016/j.bbamcr.2013.05.015. [DOI] [PubMed] [Google Scholar]
- 37.Arhzaouy K, Ramezani-Rad M. Nuclear import of UBL-domain protein Mdy2 is required for heat-induced stress response in Saccharomyces cerevisiae. PLoS One. 2012;7:e52956. doi: 10.1371/journal.pone.0052956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Etokebe GE, et al. Association of the FAM46A gene VNTRs and BAG6 rs3117582 SNP with non small cell lung cancer (NSCLC) in Croatian and Norwegian populations. PLoS One. 2015;10:e0122651. doi: 10.1371/journal.pone.0122651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao J, Wang H, Hu W, Jin Y. Effect of HLA-B-associated transcript 3 polymorphisms on lung cancer risk: A meta-analysis. Med Sci Monit. 2014;20:2461–2465. doi: 10.12659/MSM.891141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen J, Zang YS, Xiu Q. BAT3 rs1052486 and rs3117582 polymorphisms are associated with lung cancer risk: A meta-analysis. Tumour Biol. 2014;35:9855–9858. doi: 10.1007/s13277-014-1912-2. [DOI] [PubMed] [Google Scholar]
- 41.Etokebe GE, et al. Susceptibility to large-joint osteoarthritis (hip and knee) is associated with BAG6 rs3117582 SNP and the VNTR polymorphism in the second exon of the FAM46A gene on chromosome 6. J Orthop Res. 2015;33:56–62. doi: 10.1002/jor.22738. [DOI] [PubMed] [Google Scholar]
- 42.Luce MJ, Akpawu AA, Tucunduva DC, Mason S, Scott MS. Extent of pre-translational regulation for the control of nucleocytoplasmic protein localization. BMC Genomics. 2016;17:472. doi: 10.1186/s12864-016-2854-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bailey TL, et al. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Collaborative Computational Project, Number 4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 46.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W. Generation, representation and flow of phase information in structure determination: Recent developments in and around SHARP 2.0. Acta Crystallogr D Biol Crystallogr. 2003;59:2023–2030. doi: 10.1107/s0907444903017694. [DOI] [PubMed] [Google Scholar]
- 48.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eme L, Sharpe SC, Brown MW, Roger AJ. On the age of eukaryotes: Evaluating evidence from fossils and molecular clocks. Cold Spring Harb Perspect Biol. 2014;6:a016139. doi: 10.1101/cshperspect.a016139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bailey TL, Gribskov M. Combining evidence using p-values: Application to sequence homology searches. Bioinformatics. 1998;14:48–54. doi: 10.1093/bioinformatics/14.1.48. [DOI] [PubMed] [Google Scholar]
- 53.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 54.Gietz RD, Schiestl RH. Large-scale high-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc. 2007;2:38–41. doi: 10.1038/nprot.2007.15. [DOI] [PubMed] [Google Scholar]
- 55. Schrödinger, LLC (2015) The PyMOL Molecular Graphics System (Schrödinger, LLC, New York), Version 2.0.
- 56.Pettersen EF, et al. UCSF Chimera–A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 57.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]