Significance
Sodium ion/proton antiporters exchange sodium ions against protons across the membrane and are thereby essential for ion homeostasis inside living cells. The bacterial sodium ion/proton antiporter NhaA is the most important working model for this class of proteins. However, the structure of NhaA has only been determined at low pH where it is inactive. Nonetheless, different models were proposed for the mechanism of antiport. Here, we analyzed the conformational changes of unmodified NhaA upon ligand binding at physiological pH. We utilized hydrogen/deuterium exchange mass spectrometry to monitor conformational changes of the entire protein. We observed that, upon ligand binding, NhaA undergoes a conformational change consisting of two movements, and propose a model that reconciles apparently conflicting data in the field.
Keywords: NhaA, HDX-MS, conformational change, antiporter, membrane protein
Abstract
Na+/H+ antiporters comprise a family of membrane proteins evolutionarily conserved in all kingdoms of life and play an essential role in cellular ion homeostasis. The NhaA crystal structure of Escherichia coli has become the paradigm for this class of secondary active transporters. However, structural data are only available at low pH, where NhaA is inactive. Here, we adapted hydrogen/deuterium-exchange mass spectrometry (HDX-MS) to analyze conformational changes in NhaA upon Li+ binding at physiological pH. Our analysis revealed a global conformational change in NhaA with two sets of movements around an immobile binding site. Based on these results, we propose a model for the ion translocation mechanism that explains previously controversial data for this antiporter. Furthermore, these findings contribute to our understanding of related human transporters that have been linked to various diseases.
Na+/H+ antiporters found in the membranes of nearly all eukaryotic and prokaryotic cells (1) are essential for cell homeostasis (2, 3), as they regulate intracellular pH, sodium ion concentration, and volume. They represent drug targets as mutations in the human NHA2 have been implied in essential hypertension and diabetes (4–6), and a change in the activity of NHE1 plays a role in heart failure (4). Due to their evolutionary conservation, studies of prokaryotic antiporters can contribute to the understanding of human homologs important in health and disease (6).
The Escherichia coli protein NhaA is one of the central model proteins for the class of Na+/H+ antiporters (7). It exchanges one Na+ (or Li+) for two protons (8) and is thereby responsible for intracellular Na+ and H+ homeostasis. The antiport activity is strongly pH dependent, as NhaA is inactive below pH 6.5 and reaches its activity maximum at pH 8.5 (9). The functional unit of NhaA is the monomer (10), although in membranes and in detergent micelles it typically forms dimers (11, 12).
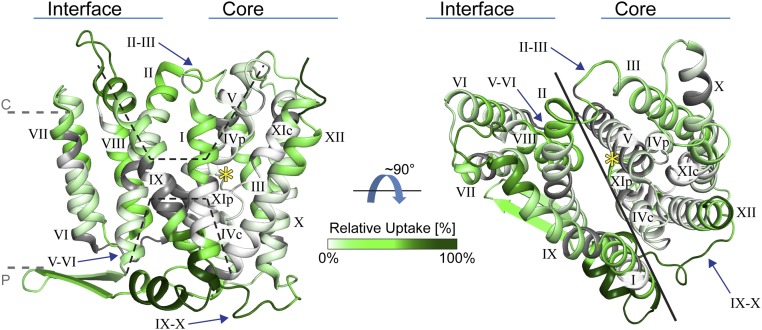
The crystal structure of the ligand-free, inactive NhaA monomer and dimer were both determined at acidic pH (13, 14). Each monomer contains 12 transmembrane helices (TMs) organized in two domains (Fig. 1, Left): the core domain (the functional unit, TMs III–V and X–XII) and the interface domain (connecting the two monomers, TMs I, II, VI, VII, VIII, and IX). The two domains are connected by three loops (II–III, V–VI, and IX–X) and form the cytoplasmic and the periplasmic funnel (Fig. 1, Left).
Fig. 1.
Deuterium uptake of NhaA. NhaA (PDB ID 4au5) is viewed parallel and normal to the membrane (dashed gray lines), and the TMs are numbered. Dashed lines show the cytoplasmic (c) and periplasmic (p) funnels (Left), and a solid line shows the separation between the interface and core domain (Right). The loops connecting the two domains (arrows) and the unique NhaA-fold in proximity to the binding site (yellow star) are indicated. The structure is colored according to the relative deuterium uptake measured after 30 min of incubation in D2O in the presence of K+, from white to dark green (zero to maximum deuterium uptake), and gray indicates no sequence coverage.
Two structurally related helix bundles (TMs III, IV, and V, and TMs X, XI, and XII) are topologically inverted to each other (13) (Fig. 1, Left). In each repeat, one TM (TM IV/XI assembly) is interrupted by an extended chain. They cross each other in the middle of the membrane, leaving two short helices oriented either toward the cytoplasm (c) or toward the periplasm (p), IVc/IVp and XIc/XIp, respectively (Fig. 1, Left). These interrupted helices contribute to a delicately balanced electrostatic environment at the ion binding site (Asp163 and Asp164) (15) and play a critical role in cation translocation (13). This general architecture, the “NhaA-fold” (16), is found in a steadily increasing number of secondary active transporters (17–21).
The Na+/H+ antiport cycle (22–24) is in principle compatible with the canonical “alternating-access” mechanism of secondary active transporters (25, 26). This model describes a shift between at least two conformations, alternatively exposing the substrate binding site either to the cytoplasm or to the periplasm. To date, no crystal structure of NhaA in its active, ligand-bound conformation is available. However, many complementary techniques (in silico and in situ) to analyze conformational changes in membrane proteins were applied to NhaA. Most of these approaches focus on the conformational change during pH activation (23, 27–29), and very little information has been obtained regarding conformational changes induced by substrate binding (29–31). Although these studies provided important information to understand the regulation of NhaA, only movements in specific regions of the protein were analyzed. Specifically, in contrast to a small number of other transporters [e.g., LeuT (32)], no global dynamic data for active NhaA as well as for other family members are available to date.
Here we used hydrogen–deuterium exchange mass spectrometry (HDX-MS) (33, 34) to directly analyze Li+-induced conformational changes of native, unmodified NhaA at physiological pH. In HDX-MS, the exchange rate of the protein backbone amide hydrogen atoms with the surrounding D2O is analyzed. At constant pH and temperature, this process is a function of the local accessibility of surrounding water and the strength of intramolecular hydrogen bonding (33). A combination of proteolytic digestion with HDX-MS enables the determination of deuterium uptake into specific peptides. Therefore, changes in deuterium uptake due to ligand binding can be correlated to specific areas of the protein (34, 35). Hence, HDX-MS has become an important tool for structural and functional analyses of soluble proteins.
However, to date only a limited number of membrane proteins have been analyzed using HDX-MS (36), as they represent a particular challenge for this technique (37) (Discussion). Here, we report the successful implementation of HDX-MS for the analysis of an integral membrane protein, NhaA. Our study revealed a global, coordinated conformational change in the architecture of NhaA upon Li+ binding.
Results
HDX-MS of a Membrane Protein in a Detergent Micelle.
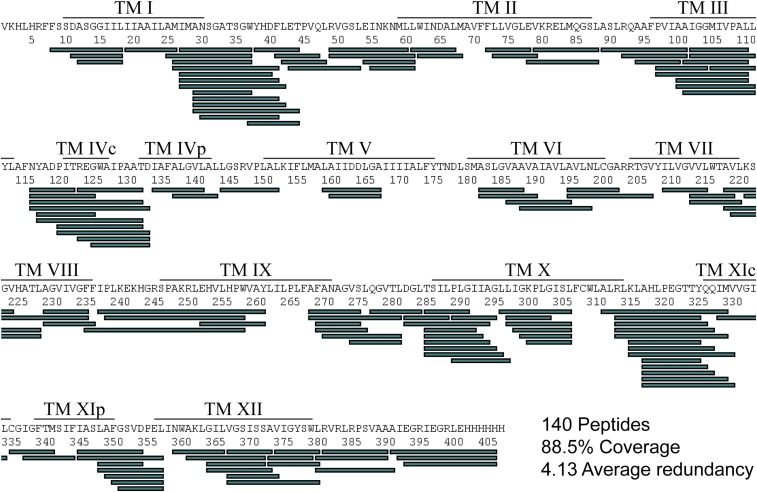
Affinity-purified dimeric NhaA in n-dodecyl β-d-maltoside (DDM) (10) was prepared at pH 7.5 in two buffers containing either Li+ or K+. We used Li+ as a ligand because it has a 10-fold higher affinity compared with Na+, and K+ served as a negative control as it does not bind to NhaA (8, 15). The samples were incubated in an excess of D2O at 20 °C for various times (0, 1, 5, 10, 30, and 60 min), and the exchange reaction was rapidly quenched by shifting pH and temperature (2 °C, pH 2.6). Subsequently, the protein was digested online on a pepsin column and analyzed using liquid chromatography-coupled ion mobility time-of-flight mass spectrometry (SI Methods). Using this setup, we identified 193 peptides across both conditions (Li+ and K+) covering 92% of the protein. To determine back-exchange levels, predigested NhaA was incubated in D2O for 4 h at room temperature (RT) before the HDX-MS measurements. Back-exchange levels could be assessed for 140 of the 193 identified peptides, covering 88.5% of the protein, with an average redundancy of four peptides per amino acid (Fig. 1 and Fig. S1). On average, peptides showed 47% back-exchange, with about 80% of the peptides within 35% and 55% deuterium loss.
Fig. S1.
Sequence coverage of NhaA. The peptides of NhaA obtained via peptic digestion and LC-IMS/MS analysis are shown as green bars along the sequence. TMs are indicated by black lines above the respective sequence. Only peptides identified with high confidence (score < 6) in four independent experiments per condition were retained for further analysis.
Next, we determined the relative deuterium uptake for each peptide and time point by normalizing the observed mass shift to the maximum of exchangeable backbone amides (excluding N terminus and prolines) and corrected for back-exchange (SI Methods). Only peptides with back-exchange correction were retained for statistical evaluation. However, the behavior of all identified peptides was taken into consideration when interpreting the data. We observed significant uptake for most parts of the protein, with a SD for the relative uptake below 2% on average (Fig. 1 and Dataset S1A).
We further analyzed the distribution of the deuterium uptake across the protein structure. The measured relative deuterium uptake after 30 min of incubation in D2O in the presence of K+ was projected onto the latest NhaA crystal structure (Fig. 1 and Movie S1). This projection demonstrated a clear correlation between high deuterium uptake and protein areas exposed to water, including the periplasmic and cytoplasmic surfaces of the protein (Fig. 1 and Movie S1). In contrast, the majority of the membrane-embedded segments showed lower deuterium uptake. This strong overlap clearly shows that solvent accessibility in the crystal structure correlates with the relative deuterium uptake in our experiments. In addition, our data suggest that no major structural changes occur during pH activation, as also suggested by electrophysiology (22) and observed by cryo-electron microscopy (38). In summary, our analysis yielded a global and comprehensive deuterium uptake map in strong agreement with the crystal structure, and we subsequently analyzed the differences in deuterium uptake induced by Li+.
Li+-Induced Differences in Deuterium Uptake.
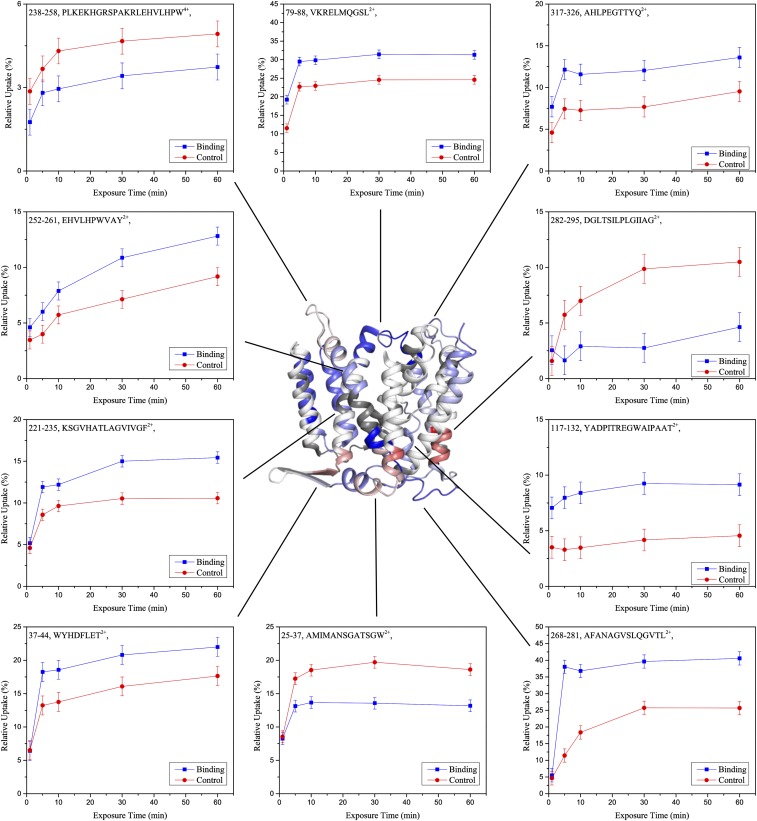
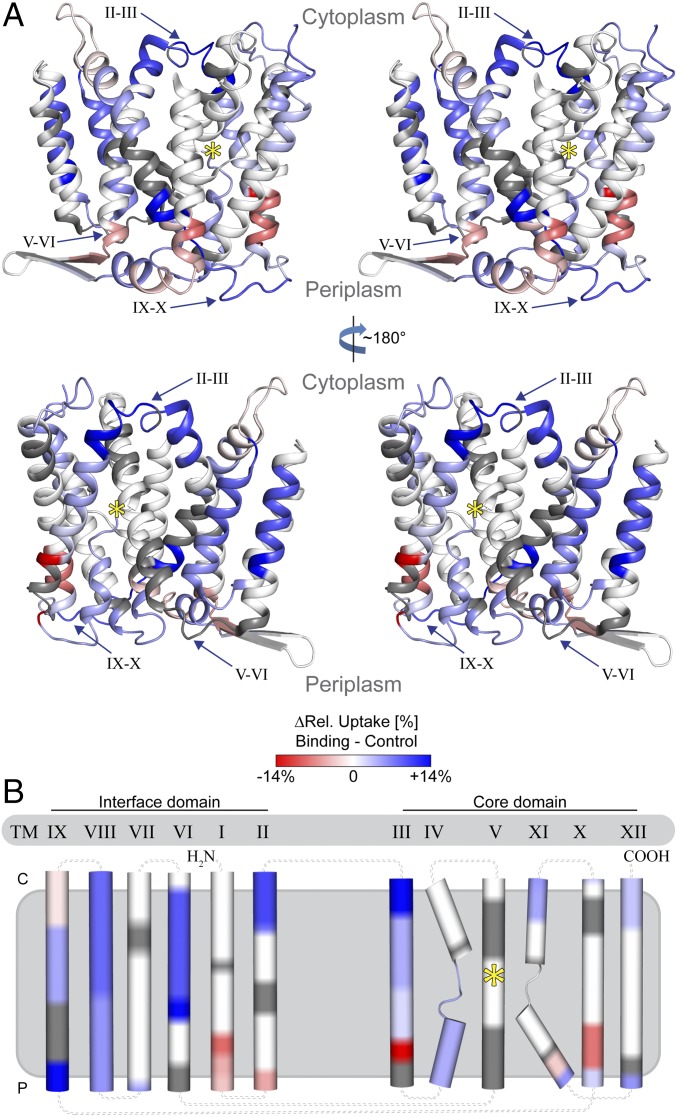
We compared relative deuterium uptake into NhaA in the presence of Li+ and in the presence of K+ by calculating the uptake differences for each individual peptide and time point. For statistical data evaluation, we performed a two-stage t test (P ≤ 0.05 and P ≤ 0.01) and only retained peptides with significant uptake differences between the conditions for further analyses (Fig. S2 and Dataset S1A). These uptake differences were plotted onto the NhaA crystal structure (Fig. 2 and Movie S2). Li+-induced differences in deuterium uptake were observed in different segments of the protein (Fig. 2 and Fig. S3A). In several areas, the presence of Li+ induced an increase in the deuterium uptake (Fig. 2, shades of blue), while in other areas it induced a decrease (Fig. 2, shades of red). Surprisingly, no differences in deuterium uptake were observed at the cation binding site (15), located at TM V (Fig. 2, yellow star), and in the adjacent loops (IV–V and V–VI). However, from this area, we observed changes in HDX spreading toward the N and the C termini (Fig. 2).
Fig. S2.
Uptake curves of selected peptides. Exemplary uptake curves of peptides displaying significantly altered deuterium uptake in the presence of Li+ and their location on the NhaA crystal structure are shown. The observed relative deuterium uptake for each peptide, time point, and condition were calculated and plotted against the labeling time. Error bars represent the average SD observed across time points and replicates. The difference between these curves were accessed by a two-stage t test (P < 0.05, P < 0.01). Peptides which passed this test were accounted to show significantly altered deuterium uptake in the presence of Li+. Color code as in Fig. 2.
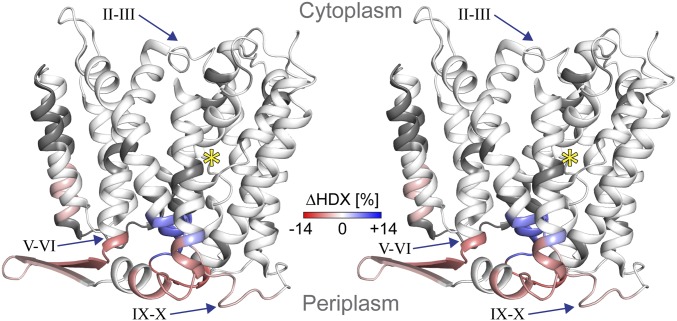
Fig. 2.
Differences in deuterium uptake upon Li+ binding observed for NhaA. Relative deuterium uptake into NhaA was measured after 30 min of incubation in the presence of Li+ (binding) or K+ (control). The uptake difference (binding minus control) was projected on the crystal structure (PDB ID code 4au5) and indicated using a color gradient from red (decreased) via white (unchanged) to blue (increased). (A) NhaA is viewed in stereo parallel to the membrane as ribbon representation (indicators as in Fig. 1). (B) Topology map of NhaA. All TMs are grouped according to the crystal structure into either the core or the interface domain (loop simplified and uncolored for clarity).
Fig. S3.
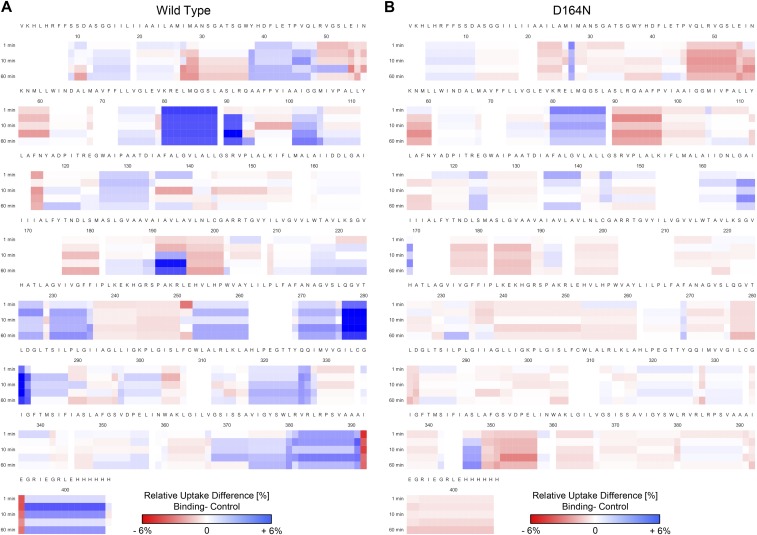
Heat maps of differential deuterium uptake for WT NhaA and variant D164N. The differential deuterium uptake, observed in the presence of Li+, is shown as heat map representation for the WT (A) and variant D164N (B). No back-exchange correction and statistical filtering were performed for the heat map generation. For the filtered results, refer to Fig. 2 and Dataset S1.
Toward the N terminus, differences in deuterium uptake were observed in all helices from TM IVp to TM I. TM IVp and loop IV–III showed a low but significant increase in deuterium uptake (Fig. 2). In TM III, a distinct pattern of deuterium uptake differences was observed, which repeated itself in several other helices; at the C terminus of TM III, there was a decreased deuterium uptake in the presence of Li+ (Fig. 2B, red). In the middle of the helix, the deuterium uptake gradually increased and reached the highest value at the N terminus, loop II–III (Fig. 2A) and the C terminus of TM II (Fig. 2B, dark blue). In the middle of TM II, no uptake difference was observed, but at its N terminus a decrease in deuterium uptake was detected. Hence, TM II showed a pattern of uptake differences similar to TM III (Fig. 2B). The C terminus of TM I showed a decrease in deuterium uptake like the N terminus of TM II, but no further changes were observed in TM I.
Toward the C terminus of NhaA, Li+-induced changes in deuterium uptake were observed in all TMs from VI–XII (Fig. 2A). TMs IX, X, and XI showed the distinct pattern of uptake differences at their termini, as the pattern observed for TMs II and III: an increase in deuterium uptake at one terminus was accompanied by a decrease at the other terminus (Fig. 2B). A high Li+-induced increase in deuterium uptake was observed at the C terminus of TM IX and part of loop IX–X (Fig. 2).
The Li+-Induced Deuterium Uptake Differences Only Occur in Functional NhaA.
As mentioned previously, the deuterium uptake rate into the protein backbone is mainly a function of its local accessibility to water and its structural rigidity. Both may also be perturbed by different ionic strengths or buffer conditions. Even though we used identical salt and buffer concentrations in all experiments, the different ions (Li+ or K+) could cause unspecific effects or produce nonphysiological peptide adducts.
To address this question, we made use of an inactive variant (D164N) (39), which does not bind Li+ (24). HDX-MS measurements of this variant under identical conditions showed an overall deuterium uptake pattern like the wild type (WT) in the absence of Li+. However, only minor differences in deuterium uptake were observed in the protein upon addition of Li+ (Fig. 3; Fig. S3B and Dataset S1B). In particular, no global changes as found for the WT were detected in this variant. Taken together, these results imply that the deuterium uptake differences observed in the presence of Li+ for WT NhaA represent conformational dynamics induced by ion binding to functional NhaA at physiological pH.
Fig. 3.
HDX-MS results for NhaA variant D164N. Differences in deuterium uptake measured after 30 min of incubation in the presence of Li+ (binding) or K+ (control) for NhaA variant D164N are projected onto the crystal structure viewed in stereo parallel to the membrane (color coding and indicators as in Fig. 2A).
SI Methods
Plasmid, Bacterial Strains, and Culture Conditions.
Plasmid pAXH is a pET20b (Novagen; Merck) derivative encoding wild-type (WT) His-tagged NhaA (56). Plasmid pAXH-D164N encodes the variant D164N (39). Bacterial strain RK20 (melBLid, ΔlacZY, thr1, and lacIQ) is an Escherichia coli K-12 derivative (9). Cells were grown in modified Luria broth media (LBK) in which NaCl is substituted by KCl, in the presence of tetracycline (6.5 µg/mL) and carbenicillin (100 µg/mL).
Production and Purification of NhaA Protein Variants.
For production of NhaA variants, strains RK20/pAXH or RK20/pAXH-D164N expressing WT his-tagged NhaA or its variant D164N were grown in minimal medium A, as previously described (57). The NhaA variants were affinity purified (Ni2+-nitrilotriacetic acid-agarose; Qiagen) as described (57). The protein was eluted in a buffer (pH 4) containing 300 mM imidazole, 25 mM sodium citrate, 100 mM KCl, 5 mM MgCl2, and 0.03% n-dodecyl β-d-maltopyranoside (DDM). The eluted protein solution was dialyzed overnight at 4 °C in acidic elution buffer (56) containing 20% sucrose, 100 mM KCl, 25 mM sodium citrate, 5 mM MgCl2, and 0.03% DDM (pH 4.0), and was then frozen at −80 °C in aliquots (10–50 µL) of 10–12 mg/mL.
Preparation of Samples for the HDX-MS Experiments.
To increase pH, to get rid of the sucrose, and to eliminate Na+, samples were dialyzed against a buffer containing 25 mM K-acetate (pH 5) (titrated with KOH), 100 mM Na-free KCl (31248; Sigma-Aldrich), 5 mM MgCl2, and 0.03% DDM, and dialyzed in a small dialysis cartridge (69570; Thermo Scientific) at 4 °C for 16 h. Then, the dialysis buffer was changed, to either the control buffer comprised of 10 mM NH4Ac, 100 mM KCl, 0.03% DDM, or the binding buffer in which KCl was replaced by 100 mM LiCl, and dialysis continued at 4 °C for 4 h. The experimental samples were diluted (∼0.6 mg/mL protein) and immediately analyzed by HDX-MS.
HDX-MS.
HDX-MS experiments were carried out on a fully automated and integrated platform based on the Waters HDX Automation setup, as previously described (35). In brief, the exchange reaction was performed by a temperature-controlled sample-handling system (PAL HTX-xt; CTC Analytics). The protein samples were equilibrated at 2 °C and labeled by 15-fold dilution with the corresponding fully deuterated buffer (control/binding) at 20 °C, for various times (0, 1, 5, 10, 30, and 60 min). Subsequently, the reaction was rapidly quenched by 1:1 dilution in precooled (2 °C) quench buffer containing 50 mM KH2PO4, 50 mM K2PO4, and 0.03% DDM at pH 2.6 (adjusted with HCl). Then, 50 µL of sample (∼23-pmol column load) were immediately injected into a temperature-controlled chromatography system (HDX nanoAcquity UPLC; Waters). The sample was digested online using an immobilized pepsin column (Enzymate BEH pepsin column; 2.1 × 30 mm; Waters) kept at 20 °C, and eluting peptides were subsequently trapped and washed on a C18 precolumn (at 0 °C, VanGuard precolumn, 1.7-μm C18, 2.1 mm × 5 mm; Waters) over a total period of 3 min with 100 µL/min H2O plus 0.1% formic acid (FA). The retained proteolytic peptides were separated on an analytical C18 column (Acquity UPLC 1.7-µm C18, 1 × 100 mm; Waters) using a linear gradient ranging from 8–40% acetonitrile (ACN) plus 0.1% FA in 7 min with a flow rate of 40 µL/min. The two columns were connected in back flush mode, and all parts, except the pepsin column, were kept at 0 °C and pH 2.5 to reduce hydrogen back-exchange to a minimum. The chromatographic system was coupled via Z-spray–positive electrospray ionization (alternating lock mass infusion: GluFib, 786 m/z) to a Synapt G2 Si HDMS QTOF mass spectrometer. Mass spectra were obtained in Waters HDMSe mode (50–2,000 m/z), which includes ion mobility separation (IMS) for 3D (LC, IM, m/z) peptide separation.
For back-exchange correction, WT NhaA or variant D164N prepared in the control buffer were predigested using the same setup as for the HDX experiments. Eluting peptides of multiple digestions were desalted and combined, and the solvent evaporated using vacuum centrifugation. The peptides were reconstituted in the fully deuterated control buffer and incubated for 4 h at RT to reach full deuteration. Peptide samples were processed and analyzed using the full HDX-MS workflow as described above with the only exception that an inactivated pepsin column was utilized to avoid double digestion. All HDX-MS experiments were conducted in at least triplicate measurements.
Data Evaluation and Statistical Analysis.
Peptides were identified from undeuterated control measurements by ProteinLynx Global Server (PLGS) 3.0.2 (Waters). Only peptides identified with high confidence (score > 6) in three out of four replicates measured per condition (control, binding) were retained for HDX analysis. Peak picking of all corresponding peaks was performed by DynamX 3.0 (Waters). Centroids were extracted for all assigned isotopic peaks for each peptide, and center masses were calculated. All assigned ion spectra were manually inspected and revised. The deuterium uptake for each peptide was calculated by the difference in the center masses at each time point compared with the undeuterated control. The observed uptake was corrected for the D2O content and the observed peptide-specific back-exchange. Next, the relative deuterium uptake was calculated by normalizing the corrected deuterium uptake to the maximum of exchangeable backbone amides (excluding N terminus and prolines). Deuterium uptake curves were generated by plotting the relative deuterium uptake against the labeling time (Fig. S2).
To test for significant differences in deuterium uptake between the conditions over time, an in-house R script was written (35) based on a two-stage t test as described by Houde et al. (58). In brief, from the mean ± average SEM observed for each peptide, a t test (n = 3; P ≤ 0.05, two-sided, unpaired) had to be passed by at least three time points before the summed difference was submitted again to a t test (n = 5; P ≤ 0.01, two-sided, unpaired), for overall difference in the dynamic uptake curve. Peptides which passed the two stages of these t tests were accounted to show significantly altered uptake rates in the presence of Li+ (Fig. S2 and Dataset S1). These peptides were then used to generate global, dynamic uptake difference maps, which were illustrated using the most recent crystal structure of inactive NhaA (PDB ID code 4AU5) and PyMOL.
Discussion
HDX-MS of a Membrane Protein.
HDX-MS as a tool for the structural elucidation of proteins has first been described by Zhang and Smith (33) in the early 1990s. Since then, it has developed to a well-established method, mainly for soluble proteins (36). Integral membrane proteins represent a challenge for this method (40) as their hydrophobicity leads to low deuterium uptake rates and insufficient proteolytic digestion efficiencies (40). In addition, the required detergents or lipids interfere with chromatography and mass spectrometry (37).
In the past 5 years, the number of HDX-MS–based studies on membrane proteins steadily increased (36, 40). While most of these studies focused only on soluble domains (41–43), determined the average deuterium uptake of the entire proteins, or showed low peptide coverage (44, 45), advances in instrumentation and methodology enabled the comprehensive analysis of nearly entire integral membrane proteins (46–48).
Here, we report an HDX-MS–based analysis of an integral membrane protein, NhaA, in a detergent micelle at physiological pH. In our setup, we achieved exceptionally high sequence coverage for a membrane protein (88.5%) with robust deuterium uptake (Fig. 1; Movie S1 and Dataset S1). Back-exchange levels were assessed using predigested protein to ensure complete unfolding and deprotection of the peptide backbone. We observed a comparatively high deuterium loss of 47% on average. This value is considerably higher than the ∼30% back-exchange frequently observed in other HDX-MS experiments. However, it is in the range of up to 50% deuterium loss reported in literature (49), and recent studies using similar experimental conditions and digestion temperatures also reported back-exchange values of about 40% (50). The observed deuterium loss can be attributed to the elevated digestion temperature of 20 °C (compared with 0 °C used regularly) and to the working speed of the auto sampler. For NhaA, lower digestion temperatures significantly lowered the sequence coverage and the use of an automated system greatly reduced workload and sample variation. We hence decided to continue with these experimental parameters in favor of the higher sequence coverage and reproducibility, being aware that thereby the sensitivity of our HDX-MS setup was lower. The observed uptake for each time point and condition was corrected for the peptide specific back-exchange level to still ensure a viable sensitivity and proper data interpretation.
We obtained a comprehensive map for NhaA of ligand-induced significant uptake differences, including data on transmembrane segments that have not been described before. In addition, we demonstrated that the observed differences (Fig. 2) were induced by Li+ binding to active WT NhaA using an inactive variant (D164N, Fig. 3). In summary, we revealed the global conformational changes in NhaA upon Li+ binding and highlighted the power of HDX-MS for analyzing the conformational dynamics of membrane proteins.
Deuterium Uptake Changes Reveal Helix Movements in Active NhaA.
The observed differences in deuterium uptake in NhaA can be due to two main factors, which are not mutually exclusive: changes in the structural rigidity of the protein backbone and/or changes in the accessibility of the amide hydrogens to water. The observation that, in six helices (I, II, III, IX, X, XI), the change in deuterium uptake at one end of the helix was inverted with respect to the change at the other end (Fig. 2B and Movie S2) implies a movement of the TMs relative to the detergent micelle. Thereby, one helix terminus becomes more accessible to the solvent and the opposite terminus becomes less accessible. It should also be noted that the tightly folded α-helices are unlikely to change their structural rigidity upon ion binding.
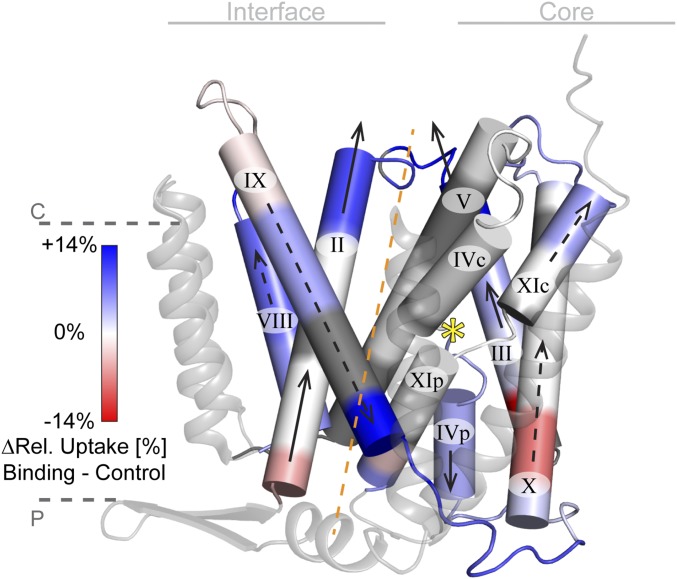
Following the conformational changes in neighboring helices and loops, we traced the spread of the movements from one helix to another (Figs. 2 and 4). We therefore propose that the Li+-induced movements represent two conformational changes originating at the Li+-binding site: one toward the N terminus and the other toward the C terminus of the protein, movement 1 and 2, respectively, as outlined below.
Fig. 4.
Schematic representation of movement 1 and 2. Cylindrical representation of the TMs and loops involved in the conformational changes upon Li+ binding viewed parallel to the membrane (color coding and indicators as in Fig. 2A). Arrows indicate the general movements of TMs upon Li+ binding in movement 1 (solid arrows) and movement 2 (dashed arrows) around the binding site (yellow star). The movements proceed along a central symmetry axis (dashed orange line) running through both funnel centers. TMs I, VI, VII, and XII are shown as transparent ribbons for clarity.
Movement 1 (binding site toward the N terminus) showed a high increase in deuterium uptake upon Li+ binding in the cytoplasmic segment of loop II–III (Lys80–Gln93, Fig. S2), the N-terminal section of TM II, and the C-terminal part of TM III (Figs. 2 and 4). These findings imply that this segment was displaced from the micelle and became more exposed to the solvent at the cytoplasmic face of NhaA (Fig. 4). Consistently, this displacement toward the cytoplasm extended to the periplasmic face of the protein via the membrane-embedded parts of TMs II and III. Although the latter segments did not show any uptake difference, their ends (C terminus of TM II, flanking loop I–II, and N terminus of TM III), which had been exposed to the periplasmic side, now became less exposed or occluded (Figs. 2 and 4). Toward the N terminus of NhaA, movement 1 weakened but extended to TM I (Fig. 2). Toward the C terminus of NhaA, the displacement of TM III toward the cytoplasmic side extended to TM IVp, exposing its end on the periplasmic face and therefore displacing TM IVc away from the cytoplasm. As mentioned above, movement 1 did not affect the N terminus of TM VI, loop IV–V, and TM V, as no differential uptake was observed in this area (Figs. 2 and 4; Fig. S3).
Movement 2 (binding site toward the C terminus) showed a strong increase in deuterium uptake upon Li+ binding in the periplasmic facing segment (Phe269–Leu287, Fig. 2 and Fig. S3A) of loop IX–X and its flanking regions. This result suggests that the segment was displaced away from the micelle into the periplasm. Accordingly, the corresponding segments at the cytoplasmic side of TM IX and loop VIII–IX became less exposed to the solvent (Figs. 2 and 4). This displacement toward the periplasmic side extended to loop X–XI and TM XIc, which became more exposed to the cytoplasm. TM XIp, including its extended chain (Gly336–Leu348), became slightly less exposed to the periplasm, followed by movements in loop XIp–XII and the C terminus of TM XII (Fig. 2). Toward the N terminus of NhaA, movement 2 included movements in TMs VI and VII and finally, like movement 1, abated toward TM V (Figs. 2 and 4).
Movement 1 and 2 Overlap with Structurally and Functionally Important Sites of NhaA.
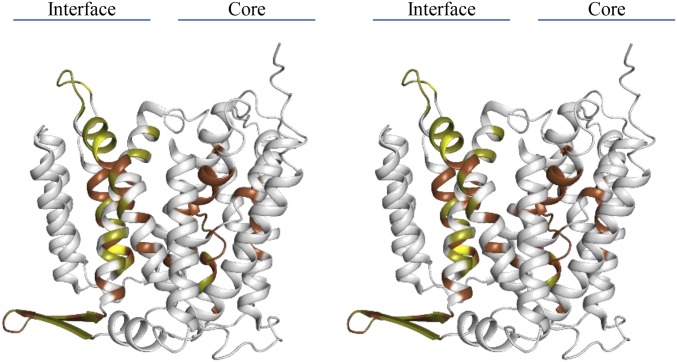
Many indirect biochemical and biophysical approaches have been applied to get insights into the structural dynamics of active NhaA at physiological pH and to identify the amino acid residues involved. In these experiments, specific sites and amino acid residues were identified using proteolysis, binding of various probes, or point mutations (Dataset S2). For comparison with our HDX-MS results, it is important to consider the differences between these indirect techniques and the direct HDX-MS approach. Most of the indirect techniques are conducted in situ (in intact cells or isolated membrane vesicles), are dependent on various reagents that can affect conformation or activity, and are site-directed. In contrast, HDX-MS experiments were performed with purified NhaA in detergent micelles and were neither dependent on any reagent nor limited to specific sites. Remarkably, despite the differences between these techniques, all results overlap regarding the localization of the conformational changes in NhaA (Fig. S4 and Dataset S2). Furthermore, most of the functionally important residues in NhaA activity were found in the helices displaying changes in deuterium uptake (TMs II, IVp, IX, X, XIp, and XIc). These matching results, obtained by different approaches, strongly support our interpretation of the HDX-MS data.
Fig. S4.
TMs and loops comprising movements 1 and 2 include key residues for structure and function of NhaA. The location of NhaA mutations affecting function/structure of NhaA are shown on the crystal structure by coloring the respective amino acid location; brown, mutations affecting the apparent Km for Li+/Na+; yellow, mutations affecting the pH dependence of the NhaA exchange activity; olive, mutation affecting both the apparent Km and the pH response. For source data, refer to Dataset S2.
Notably, deletion of TMs VI and VII and the β-sheet segments of loop I–II revealed that they are not involved in the antiporter activity but contribute to protein stability (51). We therefore assume that the conformational changes observed by HDX-MS in these areas are a consequence of the movements induced by Li+ binding to NhaA.
Movements 1 and 2 Affect the Cytoplasmic and Periplasmic Funnel.
Although NhaA forms dimers, its functional unit is the monomer (10). Thus, we projected the HDX-MS data onto the crystal structure of the NhaA monomer to analyze the conformational change in NhaA upon Li+ binding (Fig. 4). Movement 1 (TM I–IV) involves both the cytoplasmic and periplasmic funnels (Figs. 1 and 2). TM II, showing the distinct, inverted uptake difference pattern at its termini, is the only TM that lines both funnels and crosses the hydrophobic barrier between them (13). TM IVc also lines the cytoplasmic funnel and TM IVp lines the periplasmic funnel. Movement of TM IVp upon ligand binding has been observed by 2D electron crystallography (38), directly matching our findings (Figs. 2 and 4). These small helices are part of the interrupted TMs of the unique NhaA fold (13, 16). The Li+-induced movement 2 (TM VI–XII) also involves both funnels; TM IX lines the cytoplasmic funnel and TM XIp lines the periplasmic funnel (13). TMs XIc and XIp and the extended chain between them form the other part of the unique NhaA fold (16). Taken together, we conclude that the movements play an important function in opening and closing the NhaA funnels.
No changes were observed in loops IV–V, V–VI, and TM V (Figs. 2 and 4), where the most conserved and essential residues of the binding site, Asp163 and Asp164 (15), are located. We thus conclude that this segment does not move upon Li+ binding. Notably, HDX-MS monitors differential deuterium uptake of the backbone amides and not of the side chains (52). Therefore, our results do not exclude movements of the side chains at or around the active site.
Taken together, the HDX analysis revealed Li+-induced movements, occurring on both sides of the active site, proceeding almost parallel to an axis running through both funnels and between the two NhaA domains (Fig. 4). Indeed, the most pronounced Li+-induced deuterium uptake changes occurred in loops II–III and IX–X that connect the two domains (Figs. 2 and 4).
A Model for NhaA Na+/H+ Antiport Based on the Movements Observed by HDX-MS.
The central alternating-access mechanism proposed for secondary active transporters has first been visualized as a “rocker switch” (25). In its simplest form, this model describes a shift between two conformations allowing access to the active site either from the cytoplasm or periplasm while the rest of the protein, including the binding site, remains immobile relative to the membrane (25). However, recent studies on distinct transporters described an “elevator-like” mechanism (53). Therein the protein domain harboring the substrate performs a vertical translation of 5–16 Å relative to the membrane plane (53).
For NhaA, all known crystal structures have been determined of an inactive conformation at low pH, and studies of these structures favored a rocker switch mechanism for ion translocation (13, 14). However, recent studies on the related Na+/H+ antiporters, NapA from Thermus thermophiles and NhaP from Methanocaldococcus jannaschii, described an elevator-like movement upon ion binding (20, 54) including a 5- to 10-Å translation of the core domain relative to the membrane plane.
Our HDX data neither provide exact dimensions and orientations of the observed movements nor temporal resolution. Thus, no information on the specific spatial translocation and the timing of the observed conformational changes is given. The Li+-induced movements can either occur simultaneously or consecutively, leading to the final global conformation. Based on our findings, we propose the following model for NhaA antiport: an immobile binding site is alternatively exposed to either the cytoplasmic or the periplasmic side by a translational motion of the core domain relative to the membrane plane (Fig. 5). We assume a translation of the core domain, because the interface domain is anchored in the membrane by its twin of the NhaA dimer (14), a connection critical for NhaA stability (51). Indeed, in the core domain, the movements span entire TMs (III, X, and XII) (Figs. 2 and 4), while in the interface domain they mainly affect the C terminus of TM II and the N terminus of TM IX (Figs. 2 and 4). These observations suggest a more pronounced translation of the core domain, directly matching our assumption.
Fig. 5.
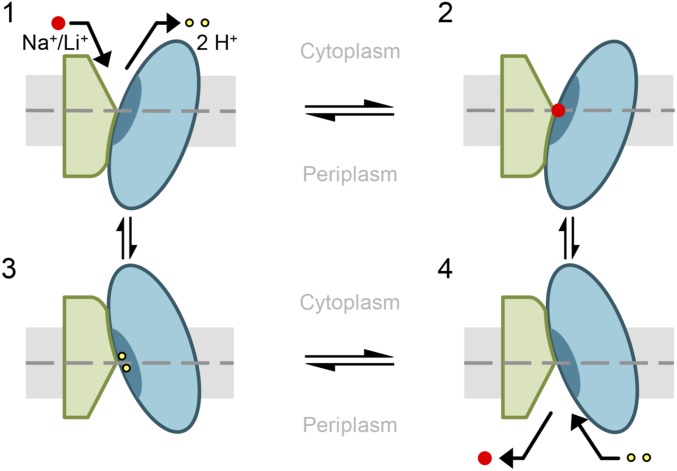
Proposed alternating-access mechanism of NhaA. Schematic representation of NhaA viewed parallel to the membrane. The two domains are shown in green (interface) and blue (core). TM V including the binding site is indicated as a dark blue segment in the core domain. The membrane is shown in gray, and a dashed line indicates its center. NhaA in the “cytoplasmic open” conformation releases two protons (yellow dots) (1) and binds Na+/Li+ (red dots) (2). After Na+/Li+ binding, the core domain translocates by a rotational upward translation, closing the cytoplasmic and opening the periplasmic funnel (3). Thereby the Li+/Na+ is released into the periplasm and two protons can bind (4). Due to the proton binding, NhaA switches back into the cytoplasmic open conformation and a new transport cycle starts (1). TM V including the binding site remains immobile relative to the membrane throughout the entire cation binding and translocation process.
In the antiport cycle, two protons are released upon Li+ binding (Fig. 5, steps 1–2). Then, the core domain shifts toward the cytoplasm into closer proximity of the interface domain, leading to a closure of the cytoplasmic funnel and opening of the periplasmic funnel (Fig. 5, step 3). Notably, the observed movements in the middle of NhaA (TMs IV and XI) (Figs. 2 and 4) would certainly cause a disruption of the charge balance delicately maintained at the ion binding pocket (13). Thus, even though TM V does not move, this effect may trigger the release of the substrate ion via the open periplasmic funnel (Fig. 5, step 3) in exchange for two protons (Fig. 5, step 4) to complete the antiport cycle.
Our model is directly supported by two computational approaches that have been used to model the periplasmic open conformation of NhaA (23). Both analyses predicted similar types of conformational changes. Overall, the two NhaA domains move around a putative symmetry axis at the funnel centers, perpendicular to the membrane plane. This motion results in the closure of the cytoplasmic funnel and the opening of the periplasmic funnel. Remarkably, the TMs involved were predicted to be TM II, TM IX, TM IVc, and TM XIp, and thus directly match with the HDX-MS results in this study.
In summary, our HDX-MS–based analysis revealed a global conformational change in NhaA mediated by two movements that form the basis of the alternating-access mechanism. Our findings highlight the value of this powerful technique and can contribute critically to driving structure–function-based experiments in NhaA. On the basis of the NhaA crystal structure, we successfully modeled human NHE1 and NHA2 (23, 55). Therefore, the results presented here can guide experiments to improve our understanding of the human antiporters of the cation/proton antiporter (CPA) family, which play essential roles in health and disease (1).
Methods
WT His-tagged NhaA (56) and variant D164N (39) were produced in E. coli strain RK20 (8) and affinity purified in DDM as previously described (56, 57). Samples were dialyzed against either the control buffer composed of 10 mM NH4Ac, 100 mM KCl, and 0.03% DDM or the binding buffer in which KCl was replaced by 100 mM LiCl and diluted to ∼0.6 mg/mL, before each HDX-MS experiment. The hydrogen/deuterium exchange reaction was performed on a Waters HDX Automation setup, as previously described (35). In brief, samples were 15-fold diluted with the corresponding fully deuterated buffer (control/binding) and incubated at 20 °C for various times (0, 1, 5, 10, 30, and 60 min). Subsequently, the exchange reaction was quenched using acidic conditions (pH 2.6, 0 °C). Peptic digestion was carried out online at 20 °C, and peptides were trapped and separated using reverse-phase chromatography on an Acquity UPLC system (Waters) kept at pH 2.5 and 0 °C. Mass spectra of the eluting peptides were obtained on a Synapt GS-Si (Waters) using HDMSe mode (50–2,000 m/z), which includes ion mobility separation (IMS). Back-exchange levels were accessed using predigested NhaA incubated in fully deuterated control buffer at 20 °C for 4 h. Further details on the experimental procedure and statistical analysis are provided in SI Methods.
Supplementary Material
Acknowledgments
We thank Imke Wüllenweber for excellent technical assistance in protein production. Furthermore, we thank Dr. Alex Muck (Waters) and the Waters HDX team for valuable advice and constructive discussions. This work was supported by the Max Planck Society, Deutsche Forschungsgemeinschaft (Cluster of Excellence Macromolecular Complexes Frankfurt), and the German–Israeli Project Cooperation (Deutsche Forschungsgemeinschaft Grant LA3655/1-1/MI 236/5-1). E.P. also thanks the Israel Science Foundation (Grant 284/12).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1703422114/-/DCSupplemental.
References
- 1.Padan E, Landau M. Sodium-proton (Na+/H+) antiporters: Properties and roles in health and disease. Met Ions Life Sci. 2016;16:391–458. doi: 10.1007/978-3-319-21756-7_12. [DOI] [PubMed] [Google Scholar]
- 2.West IC, Mitchell P. Proton/sodium ion antiport in Escherichia coli. Biochem J. 1974;144:87–90. doi: 10.1042/bj1440087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krulwich TA, Sachs G, Padan E. Molecular aspects of bacterial pH sensing and homeostasis. Nat Rev Microbiol. 2011;9:330–343. doi: 10.1038/nrmicro2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fliegel L. Molecular biology of the myocardial Na+/H+ exchanger. J Mol Cell Cardiol. 2008;44:228–237. doi: 10.1016/j.yjmcc.2007.11.016. [DOI] [PubMed] [Google Scholar]
- 5.Deisl C, et al. Sodium/hydrogen exchanger NHA2 is critical for insulin secretion in β-cells. Proc Natl Acad Sci USA. 2013;110:10004–10009. doi: 10.1073/pnas.1220009110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schushan M, et al. Model-guided mutagenesis drives functional studies of human NHA2, implicated in hypertension. J Mol Biol. 2010;396:1181–1196. doi: 10.1016/j.jmb.2009.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Padan E. Functional and structural dynamics of NhaA, a prototype for Na+ and H+ antiporters, which are responsible for Na+ and H+ homeostasis in cells. Biochim Biophys Acta. 2014;1837:1047–1062. doi: 10.1016/j.bbabio.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 8.Taglicht D, Padan E, Schuldiner S. Proton-sodium stoichiometry of NhaA, an electrogenic antiporter from Escherichia coli. J Biol Chem. 1993;268:5382–5387. [PubMed] [Google Scholar]
- 9.Taglicht D, Padan E, Schuldiner S. Overproduction and purification of a functional Na+/H+ antiporter coded by nhaA (ant) from Escherichia coli. J Biol Chem. 1991;266:11289–11294. [PubMed] [Google Scholar]
- 10.Rimon A, Tzubery T, Padan E. Monomers of the NhaA Na+/H+ antiporter of Escherichia coli are fully functional yet dimers are beneficial under extreme stress conditions at alkaline pH in the presence of Na+ or Li+ J Biol Chem. 2007;282:26810–26821. doi: 10.1074/jbc.M704469200. [DOI] [PubMed] [Google Scholar]
- 11.Williams KA, Geldmacher-Kaufer U, Padan E, Schuldiner S, Kühlbrandt W. Projection structure of NhaA, a secondary transporter from Escherichia coli, at 4.0 Å resolution. EMBO J. 1999;18:3558–3563. doi: 10.1093/emboj/18.13.3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerchman Y, Rimon A, Venturi M, Padan E. Oligomerization of NhaA, the Na+/H+ antiporter of Escherichia coli in the membrane and its functional and structural consequences. Biochemistry. 2001;40:3403–3412. doi: 10.1021/bi002669o. [DOI] [PubMed] [Google Scholar]
- 13.Hunte C, et al. Structure of a Na+/H+ antiporter and insights into mechanism of action and regulation by pH. Nature. 2005;435:1197–1202. doi: 10.1038/nature03692. [DOI] [PubMed] [Google Scholar]
- 14.Lee C, et al. Crystal structure of the sodium-proton antiporter NhaA dimer and new mechanistic insights. J Gen Physiol. 2014;144:529–544. doi: 10.1085/jgp.201411219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maes M, Rimon A, Kozachkov-Magrisso L, Friedler A, Padan E. Revealing the ligand binding site of NhaA Na+/H+ antiporter and its pH dependence. J Biol Chem. 2012;287:38150–38157. doi: 10.1074/jbc.M112.391128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Padan E, Michel H. NhaA: A unique structural fold of secondary active transporters. Isr J Chem. 2015;55:1233–1239. [Google Scholar]
- 17.Shi Y. Common folds and transport mechanisms of secondary active transporters. Annu Rev Biophys. 2013;42:51–72. doi: 10.1146/annurev-biophys-083012-130429. [DOI] [PubMed] [Google Scholar]
- 18.Hu NJ, Iwata S, Cameron AD, Drew D. Crystal structure of a bacterial homologue of the bile acid sodium symporter ASBT. Nature. 2011;478:408–411. doi: 10.1038/nature10450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wöhlert D, Grötzinger MJ, Kühlbrandt W, Yildiz Ö. Mechanism of Na+-dependent citrate transport from the structure of an asymmetrical CitS dimer. Elife. 2015;4:e09375. doi: 10.7554/eLife.09375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paulino C, Wohlert D, Kapotova E, Yildiz O, Kuhlbrandt W. Structure and transport mechanism of the sodium/proton antiporter MjNhaP1. Elife. 2014;3:e03583. doi: 10.7554/eLife.03583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wöhlert D, Kühlbrandt W, Yildiz O. Structure and substrate ion binding in the sodium/proton antiporter PaNhaP. Elife. 2014;3:e03579. doi: 10.7554/eLife.03579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mager T, Rimon A, Padan E, Fendler K. Transport mechanism and pH regulation of the Na+/H+ antiporter NhaA from Escherichia coli: An electrophysiological study. J Biol Chem. 2011;286:23570–23581. doi: 10.1074/jbc.M111.230235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schushan M, et al. A model-structure of a periplasm-facing state of the NhaA antiporter suggests the molecular underpinnings of pH-induced conformational changes. J Biol Chem. 2012;287:18249–18261. doi: 10.1074/jbc.M111.336446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dwivedi M, Sukenik S, Friedler A, Padan E. The Ec-NhaA antiporter switches from antagonistic to synergistic antiport upon a single point mutation. Sci Rep. 2016;6:23339. doi: 10.1038/srep23339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jardetzky O. Simple allosteric model for membrane pumps. Nature. 1966;211:969–970. doi: 10.1038/211969a0. [DOI] [PubMed] [Google Scholar]
- 26.Smirnova I, Kasho V, Kaback HR. Lactose permease and the alternating access mechanism. Biochemistry. 2011;50:9684–9693. doi: 10.1021/bi2014294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arkin IT, et al. Mechanism of Na+/H+ antiporting. Science. 2007;317:799–803. doi: 10.1126/science.1142824. [DOI] [PubMed] [Google Scholar]
- 28.Olkhova E, Padan E, Michel H. The influence of protonation states on the dynamics of the NhaA antiporter from Escherichia coli. Biophys J. 2007;92:3784–3791. doi: 10.1529/biophysj.106.098269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kozachkov L, Padan E. Conformational changes in NhaA Na+/H+ antiporter. Mol Membr Biol. 2013;30:90–100. doi: 10.3109/09687688.2012.693209. [DOI] [PubMed] [Google Scholar]
- 30.Tzubery T, Rimon A, Padan E. Mutation E252C increases drastically the Km value for Na+ and causes an alkaline shift of the pH dependence of NhaA Na+/H+ antiporter of Escherichia coli. J Biol Chem. 2004;279:3265–3272. doi: 10.1074/jbc.M309021200. [DOI] [PubMed] [Google Scholar]
- 31.Alhadeff R, Warshel A. Simulating the function of sodium/proton antiporters. Proc Natl Acad Sci USA. 2015;112:12378–12383. doi: 10.1073/pnas.1516881112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Penmatsa A, Gouaux E. How LeuT shapes our understanding of the mechanisms of sodium-coupled neurotransmitter transporters. J Physiol. 2014;592:863–869. doi: 10.1113/jphysiol.2013.259051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Z, Smith DL. Determination of amide hydrogen exchange by mass spectrometry: A new tool for protein structure elucidation. Protein Sci. 1993;2:522–531. doi: 10.1002/pro.5560020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Percy AJ, Rey M, Burns KM, Schriemer DC. Probing protein interactions with hydrogen/deuterium exchange and mass spectrometry—a review. Anal Chim Acta. 2012;721:7–21. doi: 10.1016/j.aca.2012.01.037. [DOI] [PubMed] [Google Scholar]
- 35.Demmer JK, Rupprecht F, Eisinger ML, Ermler U, Langer JD. Ligand binding and conformational dynamics in a flavin-based electron bifurcating enzyme complex revealed by HDX-MS. FEBS Lett. 2016;590:4472–4479. doi: 10.1002/1873-3468.12489. [DOI] [PubMed] [Google Scholar]
- 36.Pirrone GF, Iacob RE, Engen JR. Applications of hydrogen/deuterium exchange MS from 2012 to 2014. Anal Chem. 2015;87:99–118. doi: 10.1021/ac5040242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pan Y, Konermann L. Membrane protein structural insights from chemical labeling and mass spectrometry. Analyst (Lond) 2010;135:1191–1200. doi: 10.1039/b924805f. [DOI] [PubMed] [Google Scholar]
- 38.Appel M, Hizlan D, Vinothkumar KR, Ziegler C, Kühlbrandt W. Conformations of NhaA, the Na/H exchanger from Escherichia coli, in the pH-activated and ion-translocating states. J Mol Biol. 2009;386:351–365. doi: 10.1016/j.jmb.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 39.Olkhova E, Kozachkov L, Padan E, Michel H. Combined computational and biochemical study reveals the importance of electrostatic interactions between the “pH sensor” and the cation binding site of the sodium/proton antiporter NhaA of Escherichia coli. Proteins. 2009;76:548–559. doi: 10.1002/prot.22368. [DOI] [PubMed] [Google Scholar]
- 40.Forest E, Man P. Conformational dynamics and interactions of membrane proteins by hydrogen/deuterium mass spectrometry. Methods Mol Biol. 2016;1432:269–279. doi: 10.1007/978-1-4939-3637-3_17. [DOI] [PubMed] [Google Scholar]
- 41.Chung KY, et al. Conformational changes in the G protein Gs induced by the β2 adrenergic receptor. Nature. 2011;477:611–615. doi: 10.1038/nature10488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shukla AK, et al. Visualization of arrestin recruitment by a G-protein-coupled receptor. Nature. 2014;512:218–222. doi: 10.1038/nature13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vahidi S, Bi Y, Dunn SD, Konermann L. Load-dependent destabilization of the γ-rotor shaft in FOF1 ATP synthase revealed by hydrogen/deuterium-exchange mass spectrometry. Proc Natl Acad Sci USA. 2016;113:2412–2417. doi: 10.1073/pnas.1520464113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pan Y, Piyadasa H, O’Neil JD, Konermann L. Conformational dynamics of a membrane transport protein probed by H/D exchange and covalent labeling: The glycerol facilitator. J Mol Biol. 2012;416:400–413. doi: 10.1016/j.jmb.2011.12.052. [DOI] [PubMed] [Google Scholar]
- 45.Hebling CM, et al. Conformational analysis of membrane proteins in phospholipid bilayer nanodiscs by hydrogen exchange mass spectrometry. Anal Chem. 2010;82:5415–5419. doi: 10.1021/ac100962c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang X, et al. Dynamics of the β2-adrenergic G-protein coupled receptor revealed by hydrogen-deuterium exchange. Anal Chem. 2010;82:1100–1108. doi: 10.1021/ac902484p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.West GM, et al. Ligand-dependent perturbation of the conformational ensemble for the GPCR β2 adrenergic receptor revealed by HDX. Structure. 2011;19:1424–1432. doi: 10.1016/j.str.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adhikary S, et al. Conformational dynamics of a neurotransmitter:sodium symporter in a lipid bilayer. Proc Natl Acad Sci USA. 2017;114:E1786–E1795. doi: 10.1073/pnas.1613293114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoofnagle AN, Resing KA, Ahn NG. Protein analysis by hydrogen exchange mass spectrometry. Annu Rev Biophys Biomol Struct. 2003;32:1–25, and erratum (2005) 34:vi. doi: 10.1146/annurev.biophys.32.110601.142417. [DOI] [PubMed] [Google Scholar]
- 50.Jensen PF, et al. Removal of N-linked glycosylations at acidic pH by PNGase A facilitates hydrogen/deuterium exchange mass spectrometry analysis of N-linked glycoproteins. Anal Chem. 2016;88:12479–12488. doi: 10.1021/acs.analchem.6b03951. [DOI] [PubMed] [Google Scholar]
- 51.Padan E, et al. NhaA antiporter functions using 10 helices, and an additional 2 contribute to assembly/stability. Proc Natl Acad Sci USA. 2015;112:E5575–E5582. doi: 10.1073/pnas.1510964112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marcsisin SR, Engen JR. Hydrogen exchange mass spectrometry: What is it and what can it tell us? Anal Bioanal Chem. 2010;397:967–972. doi: 10.1007/s00216-010-3556-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ryan RM, Vandenberg RJ. Elevating the alternating-access model. Nat Struct Mol Biol. 2016;23:187–189. doi: 10.1038/nsmb.3179. [DOI] [PubMed] [Google Scholar]
- 54.Coincon M, et al. Crystal structures reveal the molecular basis of ion translocation in sodium/proton antiporters. Nat Struct Mol Biol. 2016;23:248–255. doi: 10.1038/nsmb.3164. [DOI] [PubMed] [Google Scholar]
- 55.Landau M, Herz K, Padan E, Ben-Tal N. Model structure of the Na+/H+ exchanger 1 (NHE1): Functional and clinical implications. J Biol Chem. 2007;282:37854–37863. doi: 10.1074/jbc.M705460200. [DOI] [PubMed] [Google Scholar]
- 56.Olami Y, Rimon A, Gerchman Y, Rothman A, Padan E. Histidine 225, a residue of the NhaA-Na+/H+ antiporter of Escherichia coli is exposed and faces the cell exterior. J Biol Chem. 1997;272:1761–1768. doi: 10.1074/jbc.272.3.1761. [DOI] [PubMed] [Google Scholar]
- 57.Gerchman Y, Rimon A, Padan E. A pH-dependent conformational change of NhaA Na+/H+ antiporter of Escherichia coli involves loop VIII–IX, plays a role in the pH response of the protein, and is maintained by the pure protein in dodecyl maltoside. J Biol Chem. 1999;274:24617–24624. doi: 10.1074/jbc.274.35.24617. [DOI] [PubMed] [Google Scholar]
- 58.Houde D, Berkowitz SA, Engen JR. The utility of hydrogen/deuterium exchange mass spectrometry in biopharmaceutical comparability studies. J Pharm Sci. 2011;100:2071–2086. doi: 10.1002/jps.22432. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.