Significance
Epigenetically programmed stress adaptation may be a conduit for informing offspring of environmental challenge. We employed ChIP-sequencing to examine effects of early environment on epigenetic regulation using hippocampal samples from macaques exposed to disruption in maternal care. We found decreased H3K4me3 binding at genes critical to behavioral stress response, the most robust being the oxytocin receptor gene (OXTR), for which we observed a corresponding decrease in RNA expression. Post hoc analysis showed that a gain-of-function OXTR SNP rescued behavioral differences in early stress-exposed subjects. Our data suggest that epigenetic down-modulation of OXTR in brain could contribute to behavioral differences observed in early stress-exposed subjects and that functional genetic variation plays a role. These could have translational implications for human psychiatric disease and personality disorders.
Keywords: primate, epigenetic, stress, maternal care, oxytocin
Abstract
Adaptations to stress can occur through epigenetic processes and may be a conduit for informing offspring of environmental challenge. We employed ChIP-sequencing for H3K4me3 to examine effects of early maternal deprivation (peer-rearing, PR) in archived rhesus macaque hippocampal samples (male, n = 13). Focusing on genes with roles in stress response and behavior, we assessed the effects of rearing on H3K4me3 binding by ANOVA. We found decreased H3K4me3 binding at genes critical to behavioral stress response, the most robust being the oxytocin receptor gene OXTR, for which we observed a corresponding decrease in RNA expression. Based on this finding, we performed behavioral analyses to determine whether a gain-of-function nonsynonymous OXTR SNP interacted with early stress to influence relevant behavioral stress reactivity phenotypes (n = 194), revealing that this SNP partially rescued the PR phenotype. PR infants exhibited higher levels of separation anxiety and arousal in response to social separation, but infants carrying the alternative OXTR allele did not exhibit as great a separation response. These data indicate that the oxytocin system is involved in social-separation response and suggest that epigenetic down-modulation of OXTR could contribute to behavioral differences observed in PR animals. Epigenetic changes at OXTR may represent predictive adaptive responses that could impart readiness to respond to environmental challenge or maintain proximity to a caregiver but also contribute to behavioral pathology. Our data also demonstrate that OXTR polymorphism can permit animals to partially overcome the detrimental effects of early maternal deprivation, which could have translational implications for human psychiatric disorders.
Stress is a universal condition of life, but if it is chronic, severe, or occurs during critical developmental windows, it contributes to a variety of disease vulnerabilities, particularly disorders of the brain (1–3). In humans, there are known links between prenatal or early stress and a variety of psychiatric and developmental disorders, including depression, schizophrenia, autism spectrum disorders, posttraumatic stress disorder, anxiety disorders, and substance use disorders (4–6). A major advantage in using animal models is the ability to follow animals prospectively from or before birth and then to control environmental exposure. As such, studies aimed at determining the mechanisms through which in utero or postnatal environments induce long-lasting differences in neurophysiology and behavior have been performed using animal models.
Early infant development is a time of high brain plasticity as well as intense mother–infant interaction. For an infant, the mother’s behaviors and other cues (i.e., hormones in the milk, pheromones are among the main sources of information about the environment to which an infant must adapt. An early period of enhanced environmental sensitivity has been documented in wild, laboratory, and domestic animals alike. In 1937, the ethologist Lorenz (7) defined a “critical period” for the social bonding that occurs during early development. Bateson later modified the nomenclature, instead referring to a “sensitive period,” which he described as a developmental phase during which events are particularly likely to produce prolonged effects on an individual (8).
Years later, rodent studies performed by Levine showed that early experience, as determined by the extent and quality of maternal care, produced long-lasting alterations in hypothalamic–pituitary–adrenal (HPA) axis activity, fearfulness, and social behaviors (9). These studies were rapidly replicated and expanded, demonstrating that epigenetic factors played a significant role, especially in stress-sensitive regions of brain such as the hippocampus (10–15). However, while rodent studies were foundational and have been critical to our understanding of the epigenetic mechanisms driving behavioral effects related to variation in maternal care, they are not without limitations. For example, the relative levels of expression and distributions of key mediators of stress responses differ between catarrhine primates (Old World monkeys, apes, and humans) and other animal species. Of particular relevance to studies modeling effects of early adversity, other laboratory animal species (e.g., rodents) are hyporesponsive to stress during the first several weeks of development, with delayed maturation of the endocrine stress response. This is not the case in humans and other catarrhine primates, such as the rhesus macaque (Macaca mulatta) (16, 17).
As with many other primate species, rhesus macaque mothers typically have singleton births and invest much of their energy into defending, comforting, and caring for their infants. This maternal “buffering” appears to be critical to normal infant development (18). A manipulation that has been demonstrated to produce long-lasting effects on behavior in nonhuman primates (rhesus macaques) is disruption of maternal care in the form of nursery or peer rearing. In this condition, subjects are removed from their parents at birth and reared with other age-matched infants so that they develop in the absence of adult influence (19, 20). Peer-reared (PR) monkeys develop strong bonds with their age mates and often use them as a base from which to explore. Compared with their mother-reared (MR) counterparts, however, PR subjects exhibit evidence of insecure attachment, higher levels of anxiety, and lower levels of exploration in novel settings (18). Macaques exposed to early adversity in the form of peer rearing show long-lasting differences in brain function and behavior, appearing to be more reactive and less able to adapt to stress (20–23).
The aim of the current study was to identify early-stress–mediated epigenetic regulation (24, 25) of DNA derived from hippocampus in adult rhesus macaques. Rather than using a candidate gene-centric approach, we wanted to perform a study that was hypothesis driven but that cast a wider net (Table S1 for gene list). The emergence of next-generation sequencing technologies has broadened our potential for discovery of epigenetic effects. DNA modifications and location relative to the gene promoter can produce varied effects (silencing vs. activation). One commonly studied epigenetic mark that has clear and well-accepted mechanistic implications is the histone 3 protein trimethylated at lysine 4 (modified histone, H3K4me3). This is a histone modification that marks active promoters and which therefore would be expected to facilitate gene expression (26–28). In addition, ChIP-sequencing (ChIP-seq) for the H3K4me3 modification requires relatively low sequencing depth, and, more importantly, because it is a point-source factor that produces localized peaks, it can be more easily associated with a particular gene (29). As such, we believed it would increase accuracy and reliability for comparisons of peak size between treatment groups. Here, we employed ChIP-seq to examine effects of early peer rearing on epigenetic regulation in adult rhesus hippocampus. We describe the effects of rearing condition at the OXTR and perform post hoc analyses to show that a gain-of-function nonsynonymous OXTR SNP partially rescues behavioral differences observed among PR subjects.
Table S1.
H3K4me3 binding differing as a function of rearing condition at genes relating to stress response and behavior
| Gene symbol | Mean peak AUC | aveCount | Highest | High50Ave | RPKM transposed |
| ADRA2C | — | — | — | — | — |
| AVPR1A | — | — | — | — | — |
| AVPR1B | — | — | — | — | — |
| BDNF | — | P ≤ 0.01 | P ≤ 0.05 | P ≤ 0.05 | - |
| MR = 2.25 ± 0.23 vs. PR = 1.38 ± 0.16 | MR = 10.5 ± 1.1 vs. PR = 6.87 ± 0.9 | MR = 9.52 ± 1.1 vs. PR = 6.19 ± 0.99 | |||
| COMT | — | — | — | — | — |
| CRH | P ≤ 0.05 | P ≤ 0.05 | — | — | — |
| MR = 3,393 + 736 vs. PR = 1,735.7 ± 245.5 | MR = 5.62 ± 1.2 vs. PR = 2.8 ± 0.4 | ||||
| CRHBP | — | — | — | — | — |
| CRHR1 | — | — | — | — | — |
| CRHR2 | — | — | — | — | — |
| DGKH | — | — | — | — | — |
| DISC1 | — | — | — | — | — |
| DRD1 | — | — | — | — | — |
| DRD2 | — | — | — | — | — |
| DRD3 | P ≤ 0.02 | P ≤ 0.03 | — | — | — |
| MR = 7,170.0 + 1,321 vs. PR = 4,052.1 ± 524.5 | MR = 2.25 ± 0.23 vs. PR = 1.38 ± 0.16 | ||||
| DRD4 | — | — | — | — | — |
| ESR1 | — | — | — | — | — |
| ESR2 | P ≤ 0.03 | — | — | — | — |
| MR = 3,135.2 ± 460 vs. PR = 1,866.7 ± 263.2 | |||||
| FKBP5 | — | — | — | — | — |
| FOXP1 | — | — | — | — | — |
| GALR1 | — | — | — | — | — |
| GALR3 | — | P ≤ 0.02 | P ≤ 0.02 | P ≤ 0.04 | - |
| MR = 4.6 ± 0.6 vs. PR = 2.44 ± 0.4 | MR = 15.5 ± 3.5 vs. PR = 7.4 ± 1.4 | MR = 14.08 ± 3.1 vs. PR = 6.8 ± 1.2 | |||
| GMIP | — | P ≤ 0.02 | — | — | — |
| MR = 3.8 ± 0.6 vs. PR = 1.36 ± 0.3 | |||||
| HTR1A | — | — | — | — | — |
| HTR1B | P ≤ 0.04 | P ≤ 0.04 | — | — | — |
| MR = 3.8 ± 0.6 vs. PR = 1.36 ± 0.3 | MR = 3.9 ± 0.7 vs. PR = 2.2 ± 0.28 | ||||
| HTR2A | P ≤ 0.02 | P ≤ 0.03 | — | — | — |
| MR = 6,988.3 ± 365 vs. PR = 5,599.6 ± 332.7 | MR = 1.7 ± 0.08 vs. PR = 1.39 ± 0.08 | ||||
| HTR6 | — | — | — | — | — |
| MTHFR | — | — | — | — | — |
| NGFR | — | — | — | — | — |
| NPY | P ≤ 0.05 | P ≤ 0.04 | — | — | — |
| MR = 6,988.3 ± 365 vs. PR = 5,599.6 ± 340.7 | MR = 2.5 + 0.5 vs. PR = 1.27 + 0.26 | ||||
| NR3C1 | P ≤ 0.03 | P ≤ 0.04 | — | — | — |
| MR = 23,892.5 ± 3,615 vs. PR = 14,471.4 ± 1,952.7 | MR = 6.3 + 9.4 vs. PR = 3.8 + 0.51 | ||||
| NR3C2 | — | — | — | — | — |
| OPRD1 | — | — | — | — | — |
| OPRK1 | — | — | — | — | — |
| OPRL1 | — | — | — | — | — |
| OPRM1 | — | — | — | — | — |
| OXTR | P ≤ 0.01 | P ≤ 0.02 | P ≤ 0.05 | P ≤ 0.04 | P ≤ 0.05 |
| MR = 2,745 ± 420 vs. PR = 1,416.5 ± 202.6 | MR = 3.82 + 0.58 vs. PR = 1.9 ± 0.28 | MR = 19.2 + 3.9 vs. PR = 9.8 ± 1.7 | MR = 17.4 ± 3.5 vs. PR = 8.6 ± 1.5 | MR = 8.9 ± 1.4 vs. PR = 5.6 ± 0.6 | |
| SLCA2 | — | — | — | — | — |
| TACR1 | — | — | — | — | — |
Results
Effects of Early Rearing on H3K4me3 Binding in Hippocampus.
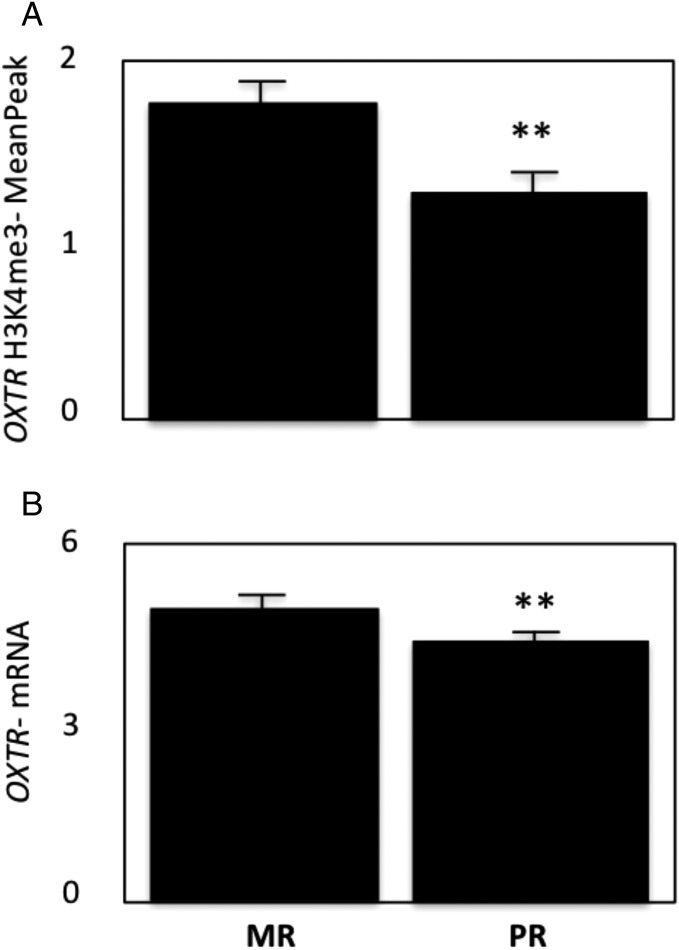
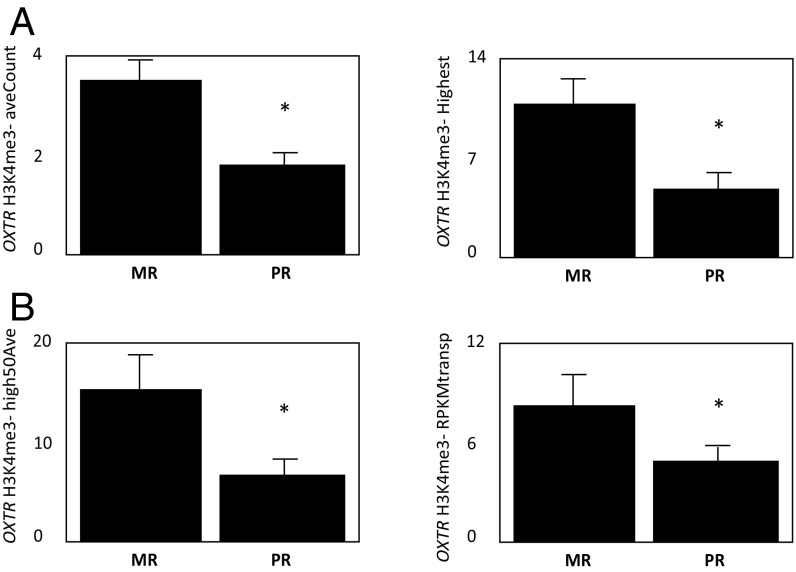
There were effects of rearing condition on H3K4me3 binding, with a diminution noted in PR hippocampus. Depending on which analytical method/sampling window was employed, decreased H3K4me3 binding was observed in PR hippocampus at the BDNF, CRH, DGKH, DRD3, ESR1/2, FKBP5, GALR3, GMIP, HTR1B, HTR2A, NPY, NR3C1, OPRM1, and OXTR promoters (Table S1 for means and SEMs). H3K4me3 binding at the OXTR promoter was significantly decreased in PR hippocampus for all windows and transformations used in the analysis [mean peak F(1,11) = 12.2, P = 0.005 (Fig. 1); average count in the peak (aveCount) F(1,11) = 8.8, P = 0.012; the highest count in the peak (highest) F(1,11) = 9.3, P = 0.01; the average count of 50 bases with highest count in this peak (high50Ave) F(1,11) = 5.8, P = 0.03; reads per kilobase of transcript per million reads mapped (RPKM) transposed F(1,11) = 6.8, P = 0.02 (Fig. 1 and Fig. S1)]. Expression microarray analysis indicated a small but significant corresponding decrease in hippocampal OXTR mRNA expression levels (Welsh t test, P = 0.008) (Fig. 1B), which was not observed for the other genes (Table S1) for which we had RNA expression data (BDNF, P > 0.94; CRH, P > 0.79; GMIP, P > 0.88; HTR1B, P > 0.34, and NPY, P > 0.33).
Fig. 1.
H3K4me3 binding and OXTR mRNA expression levels are decreased in hippocampus from PR macaques. Shown are H3K4me3 values assessed by ChIP-seq (A) and OXTR mRNA values generated by expression microarray (B). Data are shown as mean readings ± SEM; **P ≤ 0.01.
Fig. S1.
H3K4me3-binding levels are decreased in hippocampus from PR macaques. Shown are the H3K4me3 values assessed by ChIP-seq: aveCount and highest (A) and high50Ave and RPKM transposed (RPKMtransp) (B). Data are shown as mean readings ± SEM; *P < 0.05.
Functional Analysis of the OXTR Gene.
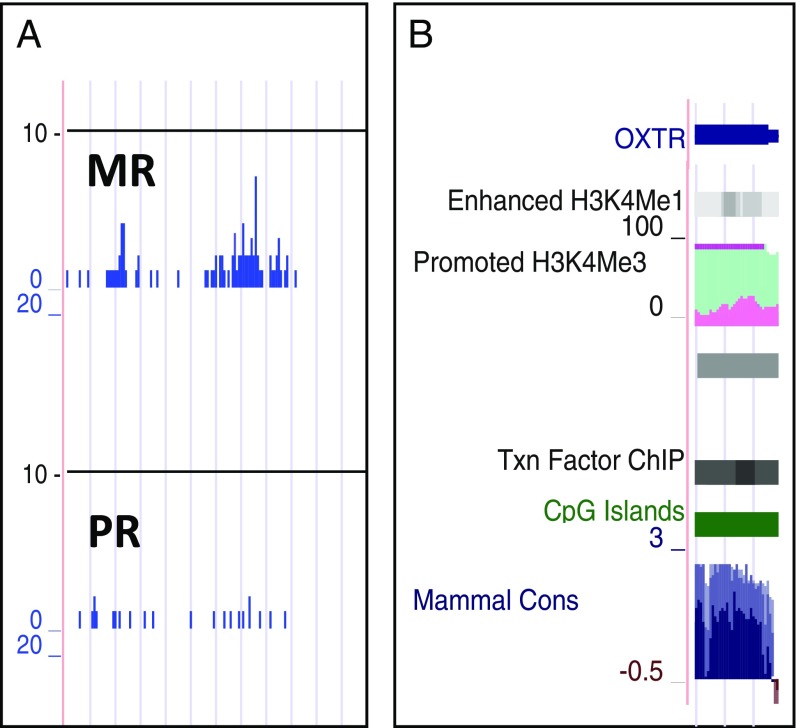
We identified a region of histone binding that spanned the first exon of the rhesus OXTR gene (Chr2: 52,233,200–52,234,100, rhMac2), with peak binding observed in a central 700-nt region (Fig. 2A). In silico analysis (University of California, Santa Clara Genome Browser, hereafter “UCSC”) indicated there are sites of epigenetic regulation (CpG islands and histone binding) in the corresponding region in the human OXTR gene (exon 1, hg19, Chr3: 8,808,733–8,810,082) (Fig. 2B).
Fig. 2.
Region of H3Kfme3 binding found in rhesus hippocampus and potential for epigenetic and transcriptional regulation in the corresponding region at human OXTR. (A) Output showing H3K4me3-binding peaks at the OXTR promoter in MR vs. PR macaque hippocampus. (B) UCSC output for human OXTR showing potential sites for epigenetic regulation (H3K4me1, H3K4me3, and CpG islands) and transcription factor (Txn Factor)-binding sites [DNase hypersensitivity clusters and transcription factor ChIP-seq] for human OXTR. Also shown are regions of conservation across species (Mammal Cons).
OXTR and Social-Separation Stress Responses in PR Macaques.
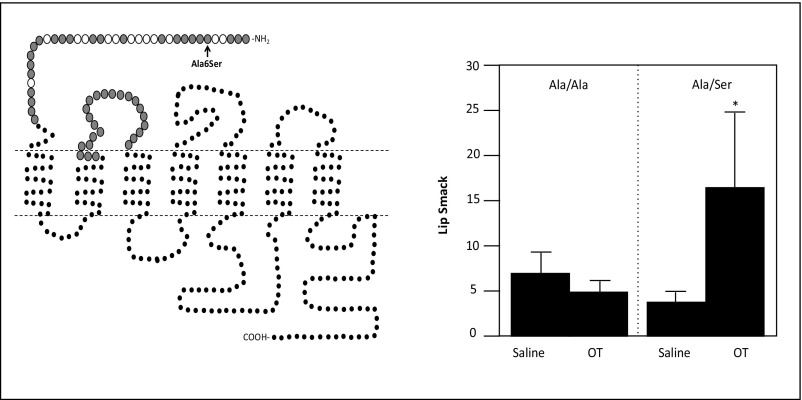
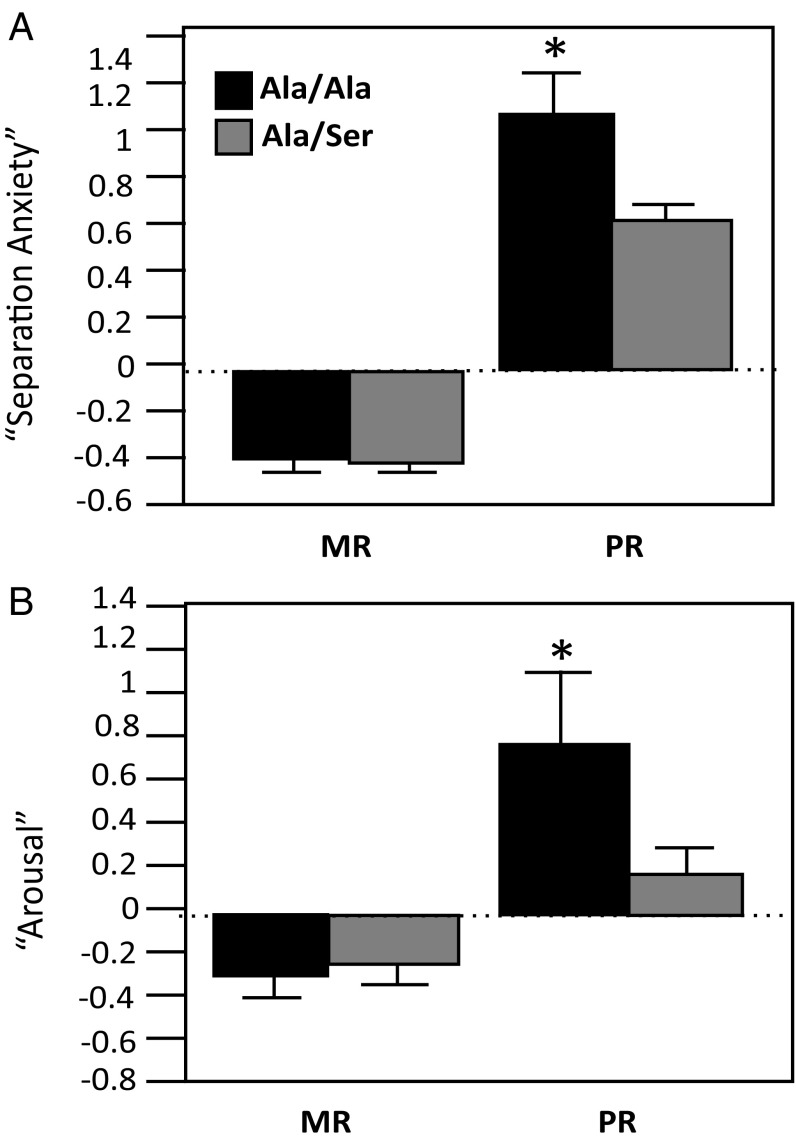
In attempts to validate a role for OXTR in behavioral differences known to occur in PR macaques (30), we investigated whether a naturally occurring gain-of-function nonsynonymous OXTR polymorphism could partially rescue the PR phenotype in a larger sample. Using archived social-separation response factors from studies in which 6-mo-old animals were separated from their social attachment sources (peers for PR infants and social group including mother for MR infants; see ref. 30 for prior factor analysis), we found that a newly discovered gain-of-function polymorphism, Ala6Ser (SI Materials and Methods for discovery and functional characterization and Fig. S2), partially rescued the PR behavioral phenotype (Fig. 3). We found main effects of rearing on separation anxiety and arousal. In both cases, PR monkeys exhibited higher response levels [F(1,186) = 284.1, P ≤ 0.001 and F(1,186) = 14.5, P ≤ 0.001, respectively]. For both types of separation response, there were also rearing × OXTR genotype interactions [separation anxiety, F(1,186) = 5.29, P ≤ 0.03; arousal, F(1,186) = 7.3, P ≤ 0.008]. PR monkeys that were carriers of the Ser allele did not exhibit the same levels of separation anxiety or arousal as their PR Ala/Ala counterparts (P ≤ 0.05, Tukey–Kramer test). There were no effects of sex or any interactive effects between sex and rearing and/or OXTR genotype.
Fig. S2.
(Left) OXTR typology showing rhesus nonsynonymous changes. Larger circles indicate residues directly involved in ligand–receptor interactions. Gray filled circles indicate residues conserved across species (human, rhesus monkey, pig, bovine, sheep, rat, and mouse). Open circles indicate residues that vary among species, indicating that nonsynonymous changes may be better tolerated without influencing functionality of the receptor. This figure was modified from ref. 63. (Right) A nonsynonymous SNP (Ala6Ser) in the ligand-binding domain of the rhesus macaque OXTR confers increased sensitivity to exogenous intranasal oxytocin. Shown are infants’ lip-smacks during a face-to-face interaction with a human caretaker at 1 h after administration of saline or oxytocin (OT) to infants with the Ala/Ala and Ala/Ser genotypes. *P < 0.05.
Fig. 3.
A gain-of-function OXTR polymorphism partially rescues the PR phenotype. Shown are results from ANOVA performed using as dependent variables factors derived from factor analysis of social-separation data averaged from macaques at 6–8 mo of age. In both separation anxiety (A) and arousal (B), PR monkeys that are homozygous for the Ala allele exhibit higher reactivity than the other groups studied, including PR Ala/Ser monkeys; *P ≤ 0.05.
SI Materials and Methods
Sequencing of the OXTR Gene and Assessment of in Vivo Function.
Genomic DNA was extracted from whole blood from rhesus macaques from the NIH Animal Center, and direct sequencing was performed. We used primers designed from published human sequences and rhesus sequences (genome.ucsc.edu/cgi-bin/hgGateway) to sequence the regions encoding amino acid residues known to be involved in receptor–oxytocin interactions (61). Using standard AmpliTaq Gold (Applied Biosystems) parameters, a 521-bp amplicon was sequenced to screen for genetic variants in regions encoding the N-terminal arm of the receptor (primers: forward: GGGGAGGTGGTTTGGTTTTA; reverse: GAGTTGACTGCCTCGGTGCT, 57 °C annealing temperature) (Fig. S1A). Since oxytocin also binds to the first extracellular loop of the receptor, a second 496-bp amplicon was also generated for screening DNA encoding that region for nonsynonymous variation (primers: forward: GCCTCTACACCCTCCGACAC; reverse: CAGGGACATGAGTAGCAGCA, 62 °C annealing temperature). Cycle sequencing was performed using the Big Dye Terminator Version 3.1 reaction (Applied Biosystems, Inc.) in 96-well optical plates. Variants were detected by visualization of electropherograms generated by ABI Sequencing Analysis software.
We identified a nonsynonymous SNP that caused an amino acid change in the ligand-binding domain of the oxytocin receptor. Given this location, we predicted that the most likely functional effect of this SNP would be in the modulation of oxytocin binding to its receptor. To determine if this was the case, we administered exogenous oxytocin to animals and examined genotype effects on individuals’ response to oxytocin challenge. Prior studies have shown that intranasal administration of exogenous oxytocin to infant rhesus macaques increases their affiliativeness during an interaction with a human caregiver, as measured by levels of lip-smacking. We sequenced infant macaques (PR, n =24, 9 females, 14 males) that were nebulized intranasally with aerosolized oxytocin to assess oxytocin-induced effects on attending to social cues and affiliativeness during early infancy (62). Because we were specifically interested in polymorphisms that would increase oxytocin response, we focused on searching for nonsynonymous changes in the ligand-binding domains of the oxytocin receptor. We identified a G/T SNP, Ala6Ser, at position chr2:52233200 (rhMac2) that met this criterion (Fig. S2, Left). The effects of polymorphism on oxytocin-induced lip-smacking were assessed using repeated-measures ANOVA, with genotype (Ala/Ala and Ala/Ser) and sex (male and female) as between-subjects variables and treatment condition (saline vs. oxytocin) as the within-subjects variable (Fig. S2, Right).
Use of Microarray Expression Data to Determine OXTR Expression in PR and MR Macaques.
Microarray was performed in the Laboratory of Clinical and Translational Sciences of the NIAAA. Total RNA from the hippocampus from the subjects used in the ChIP-seq experiment was extracted using the TRIzol (Gibco BRL) protocol. Samples were cleaned using spin-columns with an additional DNase step to ensure there was no DNA contamination. cDNA synthesis was performed using reverse-transcription reagents (starting material, 100 ng RNA), and samples were stored at −80 °C until use. A quality check for samples (260/280 nm ≤ 1.9–2.1) was performed on an Agilent 2100 BioAnalyzer (Agilent Technologies). Samples were then analyzed using the Affymetrix GeneChip Rhesus Macaque Genome Array (Thermo Fisher Scientific). Data were log-transformed, and expression levels for the OXTR gene (Affy Chip gene ID UB2440, mmu: 11995) were compared using the Welsh t test.
OXTR Ala6Ser Genotyping.
Genotyping for Ala6Ser was performed with a TaqMan DME Genotyping Assay according to the manufacturer’s instructions (Applied Biosystems, Life Technologies). The forward primer was 5′-GCCGCTCCAAGCCGTAA-3′, and the reverse primer was 5′-CGGCGCTGGAGTTGACT-3′, with reporter sequences AGCTCGCAGCCAAC and CGAGCTCTCAGCCAAC (female: MR G/G, n = 19; PR G/G, n = 6; MR T carrier, n = 43; PR T carrier, n = 22; male: MR G/G, n = 12; PR G/G, n = 9; MR T carrier, n = 48; PR T carrier, n = 35). The frequency of the 6Ser allele was 0.48, and genotype frequencies did not deviate from Hardy–Weinberg predictions (n = 194).
Discussion
Many forms of psychopathology and psychiatric illness can occur through the pathways of altered environmental sensitivity, social functioning, and anxious responding, and, while these traits are also heritable, environmental conditions are known to play a critical role (31, 32). The adaptation to unpredictable or stressful environmental conditions can occur at both the species and the individual levels. While the former occurs through genetic polymorphism, the latter can occur through epigenetic processes (33–35) and may be a conduit for informing offspring of the potential for environmental challenge (36).
One prominent example of such an epigenetic phenomenon as it relates to early-life stress exposure and maternal care comes from work by Meaney and Szyf (10). This body of work demonstrates that, among other epigenetic effects, early-life stress in both rodents and humans results in long-lasting, epigenetically driven decreases in hippocampal expression of glucocorticoid receptor (GR, or NR3C1) (13, 37–39) via alterations in DNA methylation. This is of relevance to stress-related disorders, since hippocampal GR is thought to be involved in engaging the fast-feedback “brake” on the HPA axis and therefore to be important for termination of the endocrine stress response (40). Disruption of this brake and a resultant persistence of elevated levels of corticosteroids have been observed in a number of stress-related psychiatric disorders (41, 42).
The afore-mentioned studies were foundational in the field of epigenetics for psychiatric research, but, although controlled rodent experiments have been invaluable in furthering the field, examining early environmental influences on behavior, and pointing to underlying epigenetic processes, some of the key mediators of stress response differ between catarrhine primates and other animal species (16, 17, 43). As stated above, there are papers reporting epigenetic variation as it relates to environmental exposure and to psychiatric disease burden in humans, although environmental factors are less controlled and many are performed using peripheral samples. For this reason, studies performed using primate brain are important additions to this body of work (44). For the present study, we had access to brain samples that had been archived from a terminal study performed in 2002 (43). We wanted to make use of this resource to determine whether we could observe epigenetic effects of the disruption of early maternal care in hippocampal tissue from adult rhesus macaques. Here, we performed H3K4me3 ChIP-seq and examined the effects of early peer rearing on epigenetic regulation at genes in hippocampus thought to moderate the risk for human psychopathology. Depending on the analytical window used for sampling, we found decreased H3K4me3 binding in a number of genes that are critical to behavioral stress response, including the GR gene, NR3C1 (Table S1), although most results did not stand up after correcting for the false discovery rate (FDR). It should be noted that one potential confound of examining the effects of early adversity on molecular changes occurring in the hippocampus is that studies have indicated a potential for stress-induced alteration in neuron number, a sensitivity which differs across hippocampal cell fields (45, 46). Structural MRI studies performed in MR and PR monkeys do not indicate that early rearing history influences hippocampal volume (21); however, because each of the H3K4me3 marks that we observed to differ between the two rearing conditions was down-regulated in PR subjects, we were concerned about the potential for this observation being secondary to there being fewer neurons. We wanted to try to rule out the possibility that decreased H3K4me3 binding in PR brain was merely a reflection of a different degree of cellular heterogeneity between the two treatments. We therefore performed a post hoc examination to determine whether levels of expression for six neuronal or glial markers (microtubule-associated protein, glial fibrillary acidic protein, neuronal nitric oxide synthase, myelin basic protein, neural cell adhesion molecule, and calbindin) differed in PR and MR hippocampus, using expression microarray data. As none of these markers was differentially expressed (Fig. S3), we do not believe the potential for neuron loss secondary to early peer rearing was a confounding variable for the differences in H3K4me3 binding observed in this study.
Fig. S3.
Effects of rearing condition on levels of mRNA expression of six neuronal or glial markers: microtubule-associated protein (MAP), glial fibrillary acidic protein (GFAP), neuronal nitric oxide synthase (NOS1), myelin basic protein (MBP), neural cell adhesion molecule (NCAM), and calbindin (CALB1).
The most robust and consistent result from this study was that obtained for OXTR (Fig. 1 and Table S1). Results for OXTR held with all five different ChIP-seq analytical transformations/windows (Fig. 1 and Fig. S1), and it was the only gene at which FDR correction consistently resulted in P values ≤ 0.05. H3K4me3-binding levels were decreased in hippocampus from PR animals, with a corresponding decrease in OXTR RNA expression. Another potential confound for this study is that the oxytocin system may both drive and be modulated by ethanol consumption (47). Since PR animals have been shown to exhibit higher levels of ethanol preference (22), and animals used for the epigenetics study had been given access to ethanol at some point before being killed, we therefore performed follow-up ANCOVA with ethanol consumption (expressed as grams per kilogram per hour) included as a continuous covariable. Since there were no effects of individual differences in ethanol intake (P > 0.4) and because rearing history had similar effects on H3K4me3 binding with inclusion of this covariate (P < 0.05), we do not believe ethanol exposure is a confound for this study.
Oxytocin is a neuropeptide critically involved in parturition and milk letdown but also one that plays roles in stress response, learning and memory, social affiliation, and care-giving (48–51). As such, there are certainly broad implications for behavioral differences that might result from a persistent decrease in epigenetic regulation and brain expression of OXTR in PR monkeys. The ability to remanipulate the system to demonstrate that the observed molecular changes contribute to the PR phenotype would contribute significantly to these findings. One of the major disadvantages of using a catarrhine primate model, however, is that the molecular tools that can be relatively easily employed in live rodents for interrogation of gene or protein function are less feasible in rhesus macaques. In lieu of this, we investigated whether a naturally occurring nonsynonymous OXTR polymorphism, which predicted increased affiliative responses to intranasal oxytocin in PR infants (Fig. S2), could partially rescue the PR phenotype in a larger sample. We predicted that, despite decreased epigenetic priming and expression at OXTR in PR monkeys, for PR subjects expressing a receptor that is more sensitive or effective would be less severely affected than those with the wild-type receptor. This study and others have demonstrated that PR infants show higher levels of behavioral reactivity to social separation (18, 30). Here, we show that PR infants carrying a gain-of-function OXTR allele, OXTR Ala6Ser (Fig. S2), exhibit attenuated social-separation responses relative to PR infants homozygous for the ancestral allele. These data indicate that the oxytocin system is involved in driving behavioral responses to social separation and suggest that epigenetic down-modulation of OXTR could be involved in the behavioral differences observed between PR and MR animals.
The performance of this study relied on the use of archived brain tissue samples, DNA samples, and behavioral datasets. While having access to these resources was an enormous benefit and allowed us to perform directed analyses without needing to use additional animals, one limitation is that the ChIP-seq experiment was performed only in male brains. Given the role of oxytocin in maternal behavior and sexually dichotomous effects of genetic variation and stress exposure on behavioral responses to stress, and because disorders in which social cognition is severely impaired (autism spectrum disorders and antisocial personality disorder) are more common among males than females, demonstration of diminished H3K4me3 binding at the OXTR promoter in female macaque brain would be of great interest (52, 53). This being said, when we examined the role of the OXTR Ala6Ser genotype in social-separation responses in infants and included sex as a variable, we saw no demonstration of sexually dichotomous effects or genotype/rearing × sex interactions. Furthermore, it could be that diminution in H3K4me3 binding is observed only at more advanced developmental stages, in other words, as a result of chronic social stress/adversity. Future studies should examine whether early disruption of maternal caregiving results in effects on the maternal behavior in female offspring, the developmental stage at which epigenetic effects emerge, whether endocrine regulation of OXTR plays a significant role, and whether any potential deficits are rescued by functional genetic variation. The study of primate species in which paternal caregiving plays a significant role would also be of great interest and may have additional translational implications for the human condition.
To see if the epigenetic findings reported here had potential for translating to humans, we queried the human OXTR gene (UCSC) and found indication of both the regulation of OXTR by epigenetic processes (CpG islands and H3K4me3-binding peaks) and between-species conservation. These findings indicate that H3K4me3 binding may have a role in determining the effects on oxytocin response in humans. Further, they suggest that genetic selection is acting at this region. Because oxytocin plays critical roles in parturition and milk letdown in mammals, it is not surprising that this gene would be both under purifying selection and environmentally sensitive, in part through epigenetic mechanisms. Oxytocin is also a key player in social recognition, affiliation, and attachment. For species, such as Homo sapiens, in which mothers give birth to particularly helpless and altricial offspring, the development of the social bond between parent and infant is critical for infant survival and fitness. The importance of social bonding extends not only to other species but also to other types of social relationships within species. As a result of the importance of social connections and coalitions for mammalian survival and fitness, the neurobiological and behavioral systems driving sociality are thought to be highly conserved (54). These motivations are adaptive through driving caregiving behavior and the development of the parent–infant bond, pair-bonding/monogamy, and empathy and can even extend to phenomena such as parochial altruism. The oxytocin system is known to be involved in the evolution of these processes (55). If epigenetic regulation in human brain at OXTR occurs via disturbances in parental care during early development, this suggests one mechanism by which early experience could moderate the risk for later-developing psychopathology. It is also possible that epigenetic mechanisms at OXTR in brain could drive sensitivity to environmental effects as they relate to individual variation in temperament styles and the balance of alternative strategies, social/sexual orientation, and even political preference in human societies (2, 56).
Variation in maternal care as it relates to oxytocin system functioning has been demonstrated in rodents. Our results show that disrupted maternal care produces decreased binding of an activating histone and lower OXTR mRNA expression levels in adult macaque brain. Stress-induced epigenetic changes within the oxytocin system may represent predictive adaptive responses (e.g., increased sensitivity to environmental stress) that, in the face of prolonged stress exposure, could contribute to attachment disorders, stress-induced pathology, and allostatic load. Oxytocin dysregulation has been implicated in disorders such as social phobia, mood disorders, the addictions, and posttraumatic stress disorder (57–59). The predominant role of the oxytocin system in mediating social behaviors, learning, and anxiolytic effects, together with the potential for epigenetic regulation at the OXTR, make OXTR regulation × stress interactions good candidates for elucidating the etiology and/or pathogenesis of such disorders (60, 61). Given the critical role of oxytocin in social behaviors and empathic responses, our findings may also have implications for vulnerability to the autism spectrum disorders, sociopathy, and other disorders in which disrupted social cognition or empathy are observed.
Materials and Methods
Early Rearing Conditions.
Rearing conditions have been previously described (62). Briefly, animals were reared either in social groups composed of 8–14 females (about half of which had same-aged infants) and two adult males (MR group) or were separated from their mothers at birth and hand-reared by human caregivers in a neonatal nursery for the first 37 d of life (PR group). For the first 14 d, PR infants were kept in an incubator and hand-fed. From day 15 until day 37, PR infants were placed alone in a nursery cage and provided a blanket and a terrycloth-covered, rocking surrogate. A bottle from which the infants fed was fixed to the surrogate. At 37 d of age, they were placed in a cage with three other age-mates with whom they had continuous contact. At ∼8 mo of age, animals were placed into age-matched (birth year) social groups and housed in large indoor–outdoor runs through late adolescence, at which point the cohorts were divided into same-sex groups under similar housing conditions. The macaques in this colony were maintained in an outbred state, with frequent introduction of new breeding stock, so that genetic diversity approached that occurring in free-ranging populations. Studies were approved by the National Institute on Alcohol Abuse and Alcoholism (NIAAA) and the Eunice Shriver Kennedy National Institute of Child Health and Human Development Animal Care and Use Committees.
Subjects and Tissue Collection for Epigenetic Analysis.
The tissues used in the present study had been archived from a prior experiment (43). The subjects were 13 young adult male rhesus macaques (M. mulatta) (MR, n = 6, PR, n = 7), previously members of a longitudinal study that investigated genetic and environmental influences on neurobiology, behavior, and alcohol consumption (62). At the time animals were killed, animals’ ages ranged from 4 to 7 y. There were no age differences between the rearing groups (unpaired t test, t = −0.81, P = 0.43). The mean age (+ SEM) of the MR group was 58 + 6.7 mo (range = 47–91 mo), and the mean age of the PR group was 66.7 + 7.3 mo. All subjects had been killed during the morning hours on four consecutive days. Animals were administered ketamine (15 mg/kg, i.m.) anesthesia and were perfused with ice-cold normal saline. Craniotomies were performed, brains were rapidly removed, and the left and right cerebral hemispheres were separated by a midsagittal incision. Both hemispheres were blocked into 0.5- to 0.8-cm-thick coronal sections, frozen in liquid isopentane (−42 °C), and stored at −80 °C until ready for processing. Brain tissue collections occurred between 08:00 and 11:30 h to control for diurnal variation, making sure that animals from both experimental groups were represented each morning. The hippocampus was identified in the right hemisphere slabs using gross anatomical landmarks and was excised using a fine-toothed saw. The entire dissection was done with the slab sitting on a sheet of dry ice. Given the relative ease of identification and accuracy of dissection, and because prior in situ studies demonstrated the rostral hippocampus to be particularly stress sensitive in rhesus macaques (43), blocks with the rostral hippocampus (at the level of the red nucleus) were identified, and the hippocampal region was further trimmed at −20 °C on a Peltier cooling plate using a razor.
ChIP.
Postmortem hippocampal tissue samples (50 mg) were cut into slices less than 1 mm in thickness and were fixed in 3 mL of 1% formaldehyde/PBS solution for 10 min at room temperature to cross-link chromatin DNA and proteins. After being washed with PBS, the tissue samples were homogenized in a glass-Teflon homogenizer. Following homogenization, chromatin was isolated using the Magna ChIP G kit (Millipore) according to the Millipore protocol. Briefly, cells were lysed in cell lysis buffer in the presence of protease inhibitor mixture (Magna ChIP G, cat. no. 17-611; Millipore). Nuclei were isolated from the lysed cells by centrifugation and were resuspended in nuclear lysis buffer. The chromatin DNA was then fragmented into the 150- to 500-bp range by sonication using a Branson Sonifier.
To immunoprecipitate specific genomic regions of chromatin DNA, 25 mg of the isolated chromatin fragments were incubated with 0.5 mg of an antibody directed against HeK4me3 (anti-H3K4me3; Abcam) and magnetic protein G beads (Millipore) at 4 °C for 2.5 h. Following incubation, the beads were washed with low-salt, high-salt, LiCl salt, and Tris-EDTA buffers, and chromatin fragments were reverse cross-linked by proteinase K digestion at 62 °C for 2 h. The eluted DNA was purified after reverse cross-linking by column purification. The H3k4me3-specific enrichment was validated by PCR amplification of select positive and negative genomic regions, and amplification signals were normalized to the input DNA samples, which were extracted from the same pools of chromatin fragments without enrichment by immunoprecipitation.
Sequencing with the Illumina Genome Analyzer.
Sample preparation and sequencing on an Illumina’s Genome Analyzer (Illumina) were carried out according to Illumina protocols with some modifications. Briefly, double-stranded ChIP-enriched genomic DNA fragments were treated with T4 DNA polymerase and Klenow fragments for end repair. The 5′ ends of DNA fragments were then phosphorylated by T4 polynucleotide kinase, and an adenosine base was added to the 3′ end of the fragments by Klenow (3′–5′ exonuclease). A universal adaptor was added to the both ends of the DNA fragments by A-T ligation. Following 18 cycles of PCR with Phusion DNA polymerase (New England Biolabs), the DNA library was purified on a 2% agarose gel, and fragments 170- to 350-bp in size were recovered. Approximately 10 ng of the prepared DNA was then used for cluster generation on a grafted Illumina Flow Cell and sequenced on the Genome Analyzer for 36 cycles using the sequencing-by-synthesis method.
Sequence Base Calling, Mapping to Genome, and Data Normalization.
Sequences were called from image files with the Illumina’s Genome Analyzer Pipeline (GApipeline) version 0.3.0 and aligned to the reference genome (UCSC rheMac2) using Extended Eland in the GApipeline. The unique mapped reads varied from 4.4 million to 10 million. To control the variation due to total mapped reads, a total of 4,436,000 uniquely mapped H3K4me3 reads for each sample were retrieved from export.txt files (output of Extended Eland) based on their physical locations on the sequencing slides. Based on their mapping locations on the reference genome, these selected reads were parsed with in-house Perl scripts to generate base coverage in WIG file format. The raw sequence depths in the WIG files were smoothed with a 201-base moving window. The chromosome locations of enrichment peaks were identified from pooled WIG files using in-house Perl scripts. For peak identification from the pooled WIG files, the minimum height, width, and border distances between two peaks (two peaks were merged if they were within this minimum distance) were 15, 80, and 1,000 bp, respectively, for H3K4me3 ChIP-seq. The numbers of reads (aveCount, the average count in the peak), peak height (highest, the highest count in the peak and high50Ave, the average count of 50 bases with highest count in this peak), and area under the curve (AUC) (mean peak AUC, the sum of counts in the peak) of each individual peak were determined for each sample. Read counts were then log2 transformed and normalized using quantile normalization (RPKM transposed). ChIP-seq data have been deposited in the National Center for Biotechnology Information Sequence Read Archive database with the accession number SRP004886. They can be accessed at https://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi?study=SRP004886.
Statistical Analysis.
We elected to examine epigenetic effects within regulatory regions for genes with known or suspected effects on stress response and behavior. The candidates on which we focused are listed in Table S1. Effects of rearing condition on H3K4me3 binding (across promoter and within peak) were assessed by one-way ANOVA with rearing condition (MR vs. PR) included as a nominal independent variable.
To provide validation for a role of epigenetic regulation occurring at OXTR, we performed set of post hoc analyses to determine whether a gain-of-function nonsynonymous SNP (Fig. S2) would interact with rearing to influence social-separation–related reactivity phenotypes, assessed at 6 mo of age (30). We performed three-way ANOVA with rearing (PR vs. MR), OXTR genotype (Ala/Ala vs. Ser allele carriers; SI Materials and Methods), and sex (female vs. male) as nominal independent variables. For dependent variables, we used social-separation response factors that had previously been demonstrated to be increased among PR animals: separation anxiety (positive loading for vocalization and self-directed behavior) and arousal (positive loading for environmental exploration and locomotion) (30). Analyses were performed using StatView 5.01 statistical software. The criterion for significance was set at P < 0.05.
Acknowledgments
This work was supported by the Intramural Research Program of the National Institute on Alcohol Abuse and Alcoholism, NIH.
Footnotes
The authors declare no conflict of interest.
Data deposition: The ChIP-seq data reported in this paper have been deposited in National Center for Biotechnology Information Sequence Read Archive database (accession no. SRP004886). They can be accessed at https://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi?study=SRP004886.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1706206114/-/DCSupplemental.
References
- 1.McEwen BS. Physiology and neurobiology of stress and adaptation: Central role of the brain. Physiol Rev. 2007;87:873–904. doi: 10.1152/physrev.00041.2006. [DOI] [PubMed] [Google Scholar]
- 2.Korte SM, Koolhaas JM, Wingfield JC, McEwen BS. The Darwinian concept of stress: Benefits of allostasis and costs of allostatic load and the trade-offs in health and disease. Neurosci Biobehav Rev. 2005;29:3–38. doi: 10.1016/j.neubiorev.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 3.Maren S, Phan KL, Liberzon I. The contextual brain: Implications for fear conditioning, extinction and psychopathology. Nat Rev Neurosci. 2013;14:417–428. doi: 10.1038/nrn3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neigh GN, Gillespie CF, Nemeroff CB. The neurobiological toll of child abuse and neglect. Trauma Violence Abuse. 2009;10:389–410. doi: 10.1177/1524838009339758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sinha R. Chronic stress, drug use, and vulnerability to addiction. Ann N Y Acad Sci. 2008;1141:105–130. doi: 10.1196/annals.1441.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kessler RC, Davis CG, Kendler KS. Childhood adversity and adult psychiatric disorder in the US National Comorbidity Survey. Psychol Med. 1997;27:1101–1119. doi: 10.1017/s0033291797005588. [DOI] [PubMed] [Google Scholar]
- 7.Lorenz K. The companion in the bird’s world. Der Kumpan in der Urnwelt des Vogels (Der Artgenosse als aus16sendes Moment sozialer Verhaltungsweisen)'. Auk. 1937;54:245–273. [Google Scholar]
- 8.Bateson P. How do sensitive periods arise and what are they for? Anim Behav. 1979;27:470–486. [Google Scholar]
- 9.Levine S. The pituitary-adrenal system and the developing brain. Prog Brain Res. 1970;32:79–85. doi: 10.1016/S0079-6123(08)61521-6. [DOI] [PubMed] [Google Scholar]
- 10.Meaney MJ, (37m not 10 M. Environmental programming of stress responses through DNA methylation: Life at the interface between a dynamic environment and a fixed genome. Dialogues Clin Neurosci. 2005;7:103–123. doi: 10.31887/DCNS.2005.7.2/mmeaney. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roman E, Gustafsson L, Berg M, Nylander I. Behavioral profiles and stress-induced corticosteroid secretion in male Wistar rats subjected to short and prolonged periods of maternal separation. Horm Behav. 2006;50:736–747. doi: 10.1016/j.yhbeh.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 12.Roman E, Gustafsson L, Hyytiä P, Nylander I. Short and prolonged periods of maternal separation and voluntary ethanol intake in male and female ethanol-preferring AA and ethanol-avoiding ANA rats. Alcohol Clin Exp Res. 2005;29:591–601. doi: 10.1097/01.alc.0000158933.70242.fc. [DOI] [PubMed] [Google Scholar]
- 13.Meaney MJ, et al. Early postnatal handling alters glucocorticoid receptor concentrations in selected brain regions. Behav Neurosci. 2013;127:637–641. doi: 10.1037/a0034187. [DOI] [PubMed] [Google Scholar]
- 14.Szyf M, Weaver I, Meaney M. Maternal care, the epigenome and phenotypic differences in behavior. Reprod Toxicol. 2007;24:9–19. doi: 10.1016/j.reprotox.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 15.Litvin Y, et al. Fibroblast growth factor 2 alters the oxytocin receptor in a developmental model of anxiety-like behavior in male rat pups. Horm Behav. 2016;86:64–70. doi: 10.1016/j.yhbeh.2016.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sánchez MM, Young LJ, Plotsky PM, Insel TR. Autoradiographic and in situ hybridization localization of corticotropin-releasing factor 1 and 2 receptors in nonhuman primate brain. J Comp Neurol. 1999;408:365–377. [PubMed] [Google Scholar]
- 17.Barr CS. Non-human primate models of alcohol-related phenotypes: The influence of genetic and environmental factors. Curr Top Behav Neurosci. 2013;13:223–249. doi: 10.1007/7854_2011_142. [DOI] [PubMed] [Google Scholar]
- 18.Suomi SJ. Abnormal behavior in and primate models of psychopathology. In: Fobes JL, King JE, editors. Primate Behavior. Academic; New York: 1982. pp. 171–215. [Google Scholar]
- 19.Chamove AS, Rosenblum LA, Harlow HF. Monkeys (Macaca mulatta) raised only with peers. A pilot study. Anim Behav. 1973;21:316–325. doi: 10.1016/s0003-3472(73)80073-9. [DOI] [PubMed] [Google Scholar]
- 20.Harlow HF, Suomi SJ. Induced depression in monkeys. Behav Biol. 1974;12:273–296. doi: 10.1016/s0091-6773(74)91475-8. [DOI] [PubMed] [Google Scholar]
- 21.Spinelli S, et al. Early-life stress induces long-term morphologic changes in primate brain. Arch Gen Psychiatry. 2009;66:658–665. doi: 10.1001/archgenpsychiatry.2009.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Higley JD, Hasert MF, Suomi SJ, Linnoila M. Nonhuman primate model of alcohol abuse: Effects of early experience, personality, and stress on alcohol consumption. Proc Natl Acad Sci USA. 1991;88:7261–7265. doi: 10.1073/pnas.88.16.7261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spinelli S, et al. Effects of early-life stress on serotonin(1A) receptors in juvenile Rhesus monkeys measured by positron emission tomography. Biol Psychiatry. 2010;67:1146–1153. doi: 10.1016/j.biopsych.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Millan MJ. An epigenetic framework for neurodevelopmental disorders: From pathogenesis to potential therapy. Neuropharmacology. 2013;68:2–82. doi: 10.1016/j.neuropharm.2012.11.015. [DOI] [PubMed] [Google Scholar]
- 25.Toth M. Mechanisms of non-genetic inheritance and psychiatric disorders. Neuropsychopharmacology. 2015;40:129–140. doi: 10.1038/npp.2014.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dincer A, et al. Deciphering H3K4me3 broad domains associated with gene-regulatory networks and conserved epigenomic landscapes in the human brain. Transl Psychiatry. 2015;5:e679. doi: 10.1038/tp.2015.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maćkowiak M, Bator E, Latusz J, Mordalska P, Wędzony K. Prenatal MAM administration affects histone H3 methylation in postnatal life in the rat medial prefrontal cortex. Eur Neuropsychopharmacol. 2014;24:271–289. doi: 10.1016/j.euroneuro.2013.05.013. [DOI] [PubMed] [Google Scholar]
- 28.Varma G, Sobolewski M, Cory-Slechta DA, Schneider JS. Sex- and brain region- specific effects of prenatal stress and lead exposure on permissive and repressive post-translational histone modifications from embryonic development through adulthood. Neurotoxicology. 2017;62:207–217. doi: 10.1016/j.neuro.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jung YL, et al. Impact of sequencing depth in ChIP-seq experiments. Nucleic Acids Res. 2014;42:e74. doi: 10.1093/nar/gku178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindell SG, et al. Functional NPY variation as a factor in stress resilience and alcohol consumption in rhesus macaques. Arch Gen Psychiatry. 2010;67:423–431. doi: 10.1001/archgenpsychiatry.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barr C, Becker ML, Suomi SJ, Higley D. Relationships among CSF monoamine metabolite levels, alcohol sensitivity and alcohol-related aggression in rhesus macaques. Aggress Behav. 2003;29:288–301. [Google Scholar]
- 32.Caspi A, Hariri AR, Holmes A, Uher R, Moffitt TE. Genetic sensitivity to the environment: The case of the serotonin transporter gene and its implications for studying complex diseases and traits. Am J Psychiatry. 2010;167:509–527. doi: 10.1176/appi.ajp.2010.09101452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klengel T, Binder EB. Epigenetics of stress-related psychiatric disorders and gene × environment interactions. Neuron. 2015;86:1343–1357. doi: 10.1016/j.neuron.2015.05.036. [DOI] [PubMed] [Google Scholar]
- 34.Labonté B, et al. Genome-wide epigenetic regulation by early-life trauma. Arch Gen Psychiatry. 2012;69:722–731. doi: 10.1001/archgenpsychiatry.2011.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keverne EB. Significance of epigenetics for understanding brain development, brain evolution and behaviour. Neuroscience. 2014;264:207–217. doi: 10.1016/j.neuroscience.2012.11.030. [DOI] [PubMed] [Google Scholar]
- 36.Agrawal AA, Laforsch C, Tollrian R. Transgenerational induction of defences in animals and plants. Nature. 1999;401:60–63. [Google Scholar]
- 37.McGowan PO, et al. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12:342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weaver IC, et al. The transcription factor nerve growth factor-inducible protein a mediates epigenetic programming: Altering epigenetic marks by immediate-early genes. J Neurosci. 2007;27:1756–1768. doi: 10.1523/JNEUROSCI.4164-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suderman M, et al. Conserved epigenetic sensitivity to early life experience in the rat and human hippocampus. Proc Natl Acad Sci USA. 2012;109:17266–17272. doi: 10.1073/pnas.1121260109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Young EA, Vazquez D. Hypercortisolemia, hippocampal glucocorticoid receptors, and fast feedback. Mol Psychiatry. 1996;1:149–159. [PubMed] [Google Scholar]
- 41.Young EA, Haskett RF, Murphy-Weinberg V, Watson SJ, Akil H. Loss of glucocorticoid fast feedback in depression. Arch Gen Psychiatry. 1991;48:693–699. doi: 10.1001/archpsyc.1991.01810320017003. [DOI] [PubMed] [Google Scholar]
- 42.Liberzon I, López JF, Flagel SB, Vázquez DM, Young EA. Differential regulation of hippocampal glucocorticoid receptors mRNA and fast feedback: Relevance to post-traumatic stress disorder. J Neuroendocrinol. 1999;11:11–17. doi: 10.1046/j.1365-2826.1999.00288.x. [DOI] [PubMed] [Google Scholar]
- 43.Lopez JF, Higley JD. The effect of early experience on brain corticosteroid and serotonin receptors in rhesus monkeys. Biol Psychiatry. 2002;51:294. [Google Scholar]
- 44.Kinnally EL, et al. DNA methylation as a risk factor in the effects of early life stress. Brain Behav Immun. 2011;25:1548–1553. doi: 10.1016/j.bbi.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sapolsky RM. 2001. Physiological and pathophysiological implications of social stress in animals. Coping with the Environment: Neural and Endocrine Mechanisms, Handbook of Physiology, eds McEwen BS, Goodman HM, American Physiological Society (American Physiological Society and Oxford Univ Press, New York), Vol 4, pp 517–532.
- 46.Gould E, Woolley CS, McEwen BS. Adrenal steroids regulate postnatal development of the rat dentate gyrus: I. Effects of glucocorticoids on cell death. J Comp Neurol. 1991;313:479–485. doi: 10.1002/cne.903130308. [DOI] [PubMed] [Google Scholar]
- 47.Lee MR, Rohn MC, Tanda G, Leggio L. Targeting the oxytocin system to treat addictive disorders: Rationale and progress to date. CNS Drugs. 2016;30:109–123. doi: 10.1007/s40263-016-0313-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liberzon I, Trujillo KA, Akil H, Young EA. Motivational properties of oxytocin in the conditioned place preference paradigm. Neuropsychopharmacology. 1997;17:353–359. doi: 10.1016/S0893-133X(97)00070-5. [DOI] [PubMed] [Google Scholar]
- 49.Liberzon I, Young EA. Effects of stress and glucocorticoids on CNS oxytocin receptor binding. Psychoneuroendocrinology. 1997;22:411–422. doi: 10.1016/s0306-4530(97)00045-0. [DOI] [PubMed] [Google Scholar]
- 50.Perkeybile AM, Bales KL. Intergenerational transmission of sociality: The role of parents in shaping social behavior in monogamous and non-monogamous species. J Exp Biol. 2017;220:114–123. doi: 10.1242/jeb.142182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnson ZV, Walum H, Xiao Y, Riefkohl PC, Young LJ. Oxytocin receptors modulate a social salience neural network in male prairie voles. Horm Behav. 2017;87:16–24. doi: 10.1016/j.yhbeh.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carter CS. Sex differences in oxytocin and vasopressin: Implications for autism spectrum disorders? Behav Brain Res. 2007;176:170–186. doi: 10.1016/j.bbr.2006.08.025. [DOI] [PubMed] [Google Scholar]
- 53.Barr CS, et al. Sexual dichotomy of an interaction between early adversity and the serotonin transporter gene promoter variant in rhesus macaques. Proc Natl Acad Sci USA. 2004;101:12358–12363. doi: 10.1073/pnas.0403763101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Panksepp J, Nelson E, Bekkedal M. Brain systems for the mediation of social separation-distress and social-reward. Evolutionary antecedents and neuropeptide intermediaries. Ann N Y Acad Sci. 1997;807:78–100. doi: 10.1111/j.1749-6632.1997.tb51914.x. [DOI] [PubMed] [Google Scholar]
- 55.Decety J, Norman GJ, Berntson GG, Cacioppo JT. A neurobehavioral evolutionary perspective on the mechanisms underlying empathy. Prog Neurobiol. 2012;98:38–48. doi: 10.1016/j.pneurobio.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 56.Ebitz RB, Platt ML. An evolutionary perspective on the behavioral consequences of exogenous oxytocin application. Front Behav Neurosci. 2014;7:225. doi: 10.3389/fnbeh.2013.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McQuaid RJ, McInnis OA, Abizaid A, Anisman H. Making room for oxytocin in understanding depression. Neurosci Biobehav Rev. 2014;45:305–322. doi: 10.1016/j.neubiorev.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 58.Cox BM, et al. Oxytocin acts in nucleus accumbens to attenuate methamphetamine seeking and demand. Biol Psychiatry. 2017;81:949–958. doi: 10.1016/j.biopsych.2016.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Horn SR, Charney DS, Feder A. Understanding resilience: New approaches for preventing and treating PTSD. Exp Neurol. 2016;284:119–132. doi: 10.1016/j.expneurol.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 60.Tabak BA. Oxytocin and social salience: A call for gene-environment interaction research. Front Neurosci. 2013;7:199. doi: 10.3389/fnins.2013.00199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McQuaid RJ, McInnis OA, Stead JD, Matheson K, Anisman H. A paradoxical association of an oxytocin receptor gene polymorphism: Early-life adversity and vulnerability to depression. Front Neurosci. 2013;7:128. doi: 10.3389/fnins.2013.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Higley JD, Linnoila M. Low central nervous system serotonergic activity is traitlike and correlates with impulsive behavior. A nonhuman primate model investigating genetic and environmental influences on neurotransmission. Ann N Y Acad Sci. 1997;836:39–56. doi: 10.1111/j.1749-6632.1997.tb52354.x. [DOI] [PubMed] [Google Scholar]
- 63.Gimpl G, Fahrenholz F. The oxytocin receptor system: Structure, function, and regulation. Physiol Rev. 2001;81:629–683. doi: 10.1152/physrev.2001.81.2.629. [DOI] [PubMed] [Google Scholar]