Significance
Host lysis and virion assembly are essential processes during the infection cycle of single-stranded RNA (ssRNA) viruses. Using single-particle cryoelectron microscopy, we visualized how the ssRNA virus, Qβ, uses its single-molecule “tail protein,” A2, to inhibit MurA, a bacterial enzyme essential for cell wall biosynthesis, leading to lysis of the host cell. We also revealed an extra coat protein dimer, which instead of being a part of the viral capsid, is sequestered within the virion, binding to an RNA hairpin from a five-way junction in the genomic RNA. The same five-way junction also presents hairpins to bind A2 and other coat protein dimers in the capsid, potentially supporting a nucleation event for virion assembly.
Keywords: single-particle cryo-EM, ssRNA virus, genome packaging, maturation protein, lysis protein
Abstract
In single-stranded RNA bacteriophages (ssRNA phages) a single copy of the maturation protein binds the genomic RNA (gRNA) and is required for attachment of the phage to the host pilus. For the canonical Allolevivirus Qβ the maturation protein, A2, has an additional role as the lysis protein, by its ability to bind and inhibit MurA, which is involved in peptidoglycan biosynthesis. Here, we determined structures of Qβ virions, virus-like particles, and the Qβ–MurA complex using single-particle cryoelectron microscopy, at 4.7-Å, 3.3-Å, and 6.1-Å resolutions, respectively. We identified the outer surface of the β-region in A2 as the MurA-binding interface. Moreover, the pattern of MurA mutations that block Qβ lysis and the conformational changes of MurA that facilitate A2 binding were found to be due to the intimate fit between A2 and the region encompassing the closed catalytic cleft of substrate-liganded MurA. Additionally, by comparing the Qβ virion with Qβ virus-like particles that lack a maturation protein, we observed a structural rearrangement in the capsid coat proteins that is required to package the viral gRNA in its dominant conformation. Unexpectedly, we found a coat protein dimer sequestered in the interior of the virion. This coat protein dimer binds to the gRNA and interacts with the buried α-region of A2, suggesting that it is sequestered during the early stage of capsid formation to promote the gRNA condensation required for genome packaging. These internalized coat proteins are the most asymmetrically arranged major capsid proteins yet observed in virus structures.
Single-stranded RNA (ssRNA) viruses infect all domains of life and are important pathogens (1–4). Bacteriophages or phages, especially the canonical Leviviridae Qβ and MS2, have been model systems for studying ssRNA viral gene regulation (5–8), genome replication (9, 10), and encapsidation (11–14). They have a near-icosahedral capsid, encapsidating genomic RNAs (gRNAs) of ∼3,500–4,300 nucleotides. Recent advances in single-particle cryoelectron microscopy (cryo-EM) have enabled characterization of asymmetric features in ssRNA phages. Of note, the gRNA has been found to adopt a single, well-folded, 3D structure within the virion (15–18).
The 4,217-nucleotide gRNA of Qβ has genes for the maturation protein (A2), the coat protein, and the replicase (the viral subunit of the RNA-dependent RNA polymerase) (19–21). The gene for the Qβ coat protein contains a “leaky” stop codon that occasionally results in a readthrough by the host ribosome producing a minor coat protein A1. A1 is essential and consists of the full-length coat domain connected by a flexible linker to the readthrough domain, a 196-amino acid C-terminal extension (22). These four proteins have essential functions at the various stages of the Qβ life cycle, from genome replication and packaging to host lysis and the subsequent adsorption to the F pilus of a new host.
Unlike double-stranded DNA phages, which use specialized machinery to pump their genetic material into a preformed capsid, ssRNA phages assemble their capsid proteins around their gRNA. In Qβ, the maturation protein, A2, along with 89 coat protein dimers, directly binds the gRNA, encapsidating it in a near-icosahedral (T = 3) capsid (15). Around 10 copies of A1 are incorporated into the capsid, replacing monomers of the coat protein, but the positions of these substitutions and the basis for the requirement for A1 are unknown (23, 24). The composition of ribonucleoprotein assembly intermediates of the related phage MS2 was estimated to be 1:15:1:1 for the replicase subunit, coat protein, maturation protein, and RNA, including both gRNA and ribosomal RNAs (25). This suggests that the maturation proteins of ssRNA phages bind before the vast majority of coat proteins, as there are ∼180 coat proteins in the fully assembled virion. However, the detailed interactions between the components of Qβ were not clear, due to the lack of high-resolution structures of the Qβ virion.
One major difference between Qβ and other ssRNA phages, such as MS2, is that the maturation protein of Qβ, A2, has an additional role. While MS2 dedicates a unique protein, L, for host lysis (26), Qβ uses the maturation protein A2 to bind and inhibit MurA (27), the enzyme for the first committed step in bacterial cell wall biosynthesis (28). The inhibition of cell wall biosynthesis leads to lysis at the next cell division, releasing the progeny Qβ virions (29). Therefore, A2, as well as the mature Qβ virion, has been regarded as a “protein antibiotic” (27). Although it has been shown that in vitro A2 binds MurA in the presence of its first substrate, uridine 5′-diphosphate-N-acetylglucosamine (UDP-GlcNAc) (19), the basis of how A2 inhibits MurA was unknown, again largely due to the lack of structural information.
Here we present cryo-EM structures of Qβ both free and bound with MurA, to determine the structural details of bacterial cell lysis by Qβ. Moreover, by comparing the structures of the Qβ virion and noninfectious Qβ virus-like particles (VLPs), which are copurified with the Qβ virions, we reveal a structural rearrangement in the capsid coat proteins that is required for the assembly of viable phages. These results are discussed in terms of the pathway of Qβ assembly and the genetics of resistance to Qβ propagation.
Results
Qβ Virion at Near-Atomic Resolution.
Using single-particle cryo-EM, 2,370 movie stacks of the Qβ phages were recorded on a direct detection camera in the superresolution electron counting mode (30), yielding 248,434 high-quality phage particles. Classification of these particles revealed that ∼20% of them contain A2 in the capsid and have a single, dominant gRNA conformation (SI Appendix, Fig. S1). These particles were then used to generate a symmetry-free reconstruction of the mature Qβ virion at an overall resolution of 4.7 Å, an improvement from the 7-Å resolution reconstruction in our previous work (15). The local resolution of the virion is not uniform, with the gRNA in the center of the virion having the lowest resolution, presumably due to the intrinsic flexibility of the RNA helices that are not directly bound to the capsid. The resolution for the asymmetric reconstruction of capsid components, including the coat proteins and A2, was further improved to 4.4 Å after excluding the gRNA during the map refinement (SI Appendix, Fig. S2 A and C).
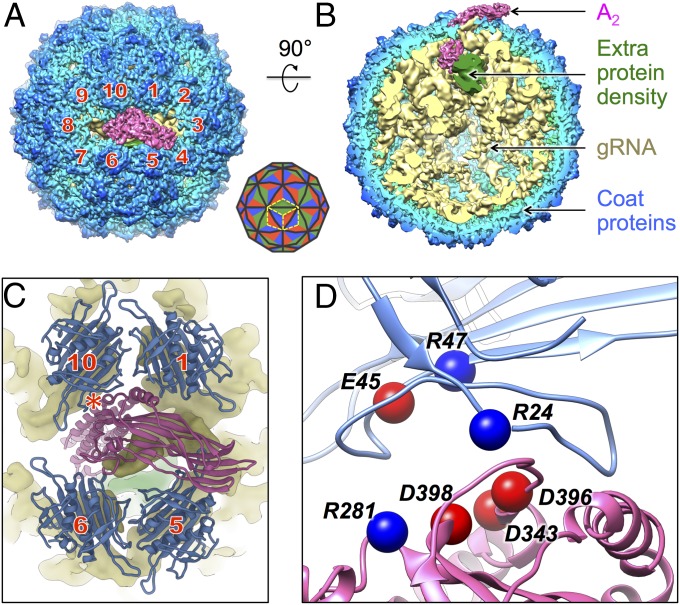
The cryo-EM density of the maturation/lysis protein, A2, matches the recent crystal structure of purified A2, in the absence of the capsid and gRNA (31). A2 consists of an α-region and a β-region. It replaces a coat protein dimer at a twofold axis, with its α-region inserted into the capsid, bound to the gRNA, and its β-region exposed outside the virion. By labeling the coat protein dimers surrounding A2 from 1 to 10 in a clockwise fashion, the β-region of A2 is seen to lean toward the interface of neighboring coat protein dimers 3 and 4 (Fig. 1A). These results are consistent with our previous observations at a lower resolution (15). A cutaway side view (Fig. 1B) shows the components of the mature Qβ including A2, the gRNA, and the coat proteins. Surprisingly, an extra protein density is observed inside the virion close to the bottom of A2, around the α-region. This density does not resemble any RNA secondary structures and is not proximal to any coat proteins on the capsid. The identity of this density will be revealed in the present study.
Fig. 1.
The structure of the Qβ virion shows interactions between the maturation protein and the capsid coat proteins. (A) Near-atomic asymmetric structure of Qβ shows the maturation protein surrounded by 10 coat protein dimers, which are labeled clockwise 1–10. Diagram in the Lower Right corner shows the triangulation pattern for T = 3 and the placement of 3 coat protein dimers labeled by three yellow dashed diamond boxes. (B) A 90° turn and cutaway view of Qβ shows the coat proteins (radially colored from light blue to blue), the gRNA (yellow, low-pass filtered to 6-Å resolution), A2 (hot pink), and the electron density of the previously uncharacterized proteins (green). (C) Close-up image of A2 (hot pink) in proximity to the 4 coat protein dimers (blue) neighboring it; the red star labels the tightest interaction. The densities for the gRNA (yellow) and the previously uncharacterized proteins (green) are also shown. (D) A zoomed-in view of the tightest interaction between the maturation protein and coat protein dimer 10 with the clusters of charged residues labeled and shown as spheres (blue and red spheres for positively and negatively charged residues, respectively).
The improved resolution of the capsid components in our cryo-EM reconstruction, from 6.5 Å in previous work (15) to 4.4 Å, allowed us to build the entire backbone model of the Qβ protein capsid into the cryo-EM density map and examine the detailed interactions between the A2 and neighboring coat protein dimers. Four surrounding coat protein dimers (dimers 1, 5, 6, and 10 in Fig. 1 A and C) present loops to interact with A2 (Fig. 1C and SI Appendix, Fig. S3), wherein the tightest interaction occurs from coat protein dimer 10. A zoomed-in view of the interface between coat protein dimer 10 and A2 reveals that several amino acid residues from coat protein dimer 10 and A2 carry opposite charges (Arg24, Glu45, and Arg47 on the coat protein and Arg281, Asp343, Asp396, and Asp398 from A2, Fig. 1D), suggesting that salt bridges are involved in the structure.
Comparison of the Qβ Virion and Qβ Virus-Like Particles.
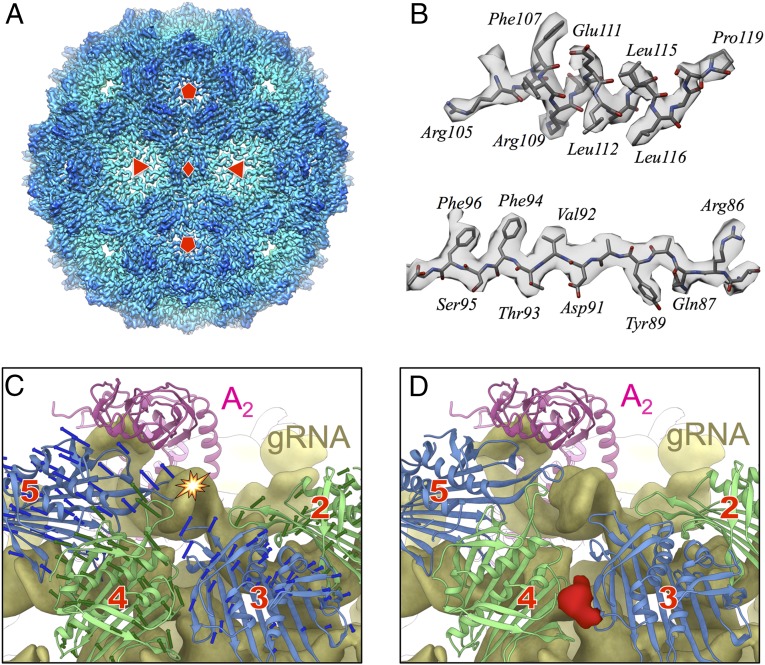
The capsids of 70–80% of the particles are perfectly icosahedral and do not have A2 incorporated into the surface of the capsid. Among these particles, the RNA density within the capsid does not adopt a dominant conformation (SI Appendix, Fig. S1). These particles are regarded as noninfectious VLPs. The electron density of the capsids for Qβ VLPs was resolved to 3.3-Å resolution with icosahedral symmetry applied (Fig. 2A and SI Appendix, Fig. S2A). Sidechains for the coat proteins in the Qβ VLP are clearly seen (Fig. 2B).
Fig. 2.
A comparison between the capsids of Qβ virions and Qβ virus-like particles. (A) An icosahedral reconstruction of the Qβ VLPs at 3.3-Å resolution with the red diamond, triangles, and pentagons labeling the twofold, threefold, and fivefold axes, respectively. (B) Sidechains visualized on an α-helix (Top) and a β-strand (Bottom) in the coat proteins. (C) Coat protein dimers from the capsid of the Qβ virus-like particles are overlaid onto the gRNA (yellow density) and A2 (pink model) of the Qβ virion. Only four coat protein dimers (green and blue alternating) are shown and numbered as in Fig. 1A. The arrows indicate the conformational difference between these four coat protein dimers in the Qβ virus-like particles and the Qβ virion. The star indicates a severe steric clash where the gRNA density collides with the EF loop of a coat protein in dimer 5 of the Qβ virus-like particles. (D) The same view as in C but with the four coat protein dimers in their conformations within a mature Qβ virion. An extra density (red) was observed at the interface between the models of coat protein dimers 3 and 4.
Compared with our previous cryo-EM structures (15), these improved resolutions for both the Qβ virion and the VLP allowed us to more accurately delineate the structural differences between these two types of capsids, which are particularly striking for coat dimers 2–6, with deviations in certain regions larger than 10 Å (SI Appendix, Fig. S4). When replacing the conformations of the coat proteins in the virion capsid with their corresponding conformations in the VLP (Fig. 2C), there are apparent steric clashes between the VLP capsid and the gRNA, particularly between a gRNA hairpin loop and one FG loop from coat protein dimer 5 (labeled by a star in Fig. 2C). A large structural rearrangement, expanding the capsid locally around these coat protein dimers, is necessary to avoid these collisions in the mature Qβ virions. The expansion also leaves a gap between the models of coat protein dimers 3 and 4. Interestingly, at this gap between the coat protein models, some extra density (colored red in Fig. 2D) is observed, protruding from the C termini of the coat proteins (SI Appendix, Fig. S5). The identity of this extra density is yet to be determined. One explanation is that one or a few coat proteins in dimers 3 and 4 are replaced by A1 and this extra density belongs to part of the flexible linker of the readthrough domain in A1.
MurA Binds A2 in Mature Qβ Virions.
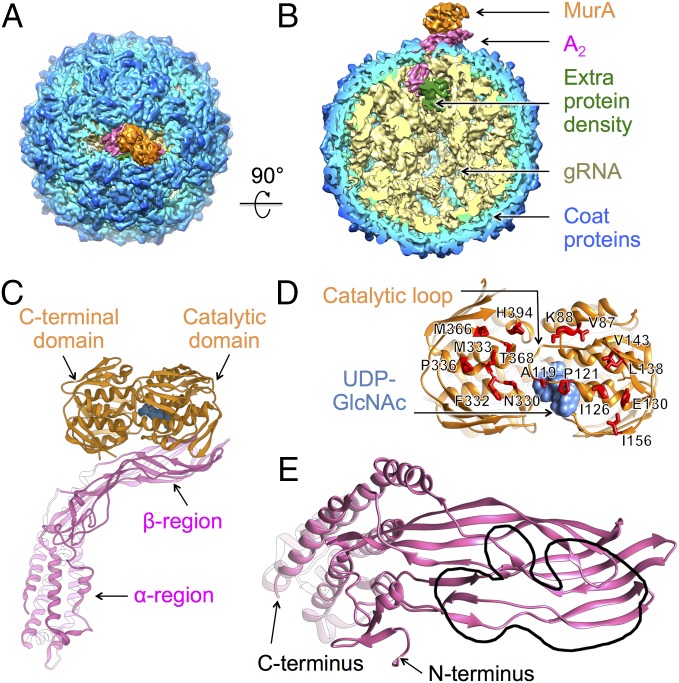
During Qβ infection, A2 inhibits MurA, resulting in host lysis. In addition, the mature Qβ virion has been shown to be a stoichiometric inhibitor of MurA, via its single molecule of A2 (27). To reveal the interactions between A2 and MurA, we collected 2,871 movie stacks of the Qβ–MurA complex on a direct detection camera in the superresolution electron counting mode using single-particle cryo-EM. Approximately 20% of total 130,013 particles correspond to Qβ–MurA complexes. The resolution was determined to 6.1 Å for the Qβ–MurA complex and improved to 5.7 Å for the protein components by excluding gRNA during map refinement (SI Appendix, Fig. S2 B and D). The architecture of the Qβ–MurA complex is almost identical to that of the Qβ alone (Fig. 3 A and B), except for the additional density from MurA binding to A2. The extra protein density sequestered inside the virion, seen in the structure of Qβ alone, is still observed. No significant conformational rearrangement of the gRNA is seen (SI Appendix, Fig. S6).
Fig. 3.
The structure of Qβ bound with MurA reveals the structural mechanism of A2-mediated host lysis. (A) The structure of Qβ bound to MurA colored as in Fig. 1 A and B with MurA colored orange. (B) A 90° turn and cutaway view of Qβ shows MurA bound to the maturation protein. (C) The ribbon model of A2 bound to MurA with uridine diphosphate N-acetylglucosamine (UDP-GlcNAc) in the active site (cornflower blue). (D) The ribbon model of MurA viewed from the MurA–A2 interface. The point mutations that render MurA resistant to A2 are labeled and shown as red stick models. Locations of the catalytic loop and the UDP-GlcNAc are indicated by black arrows. (E) Ribbon model of A2 as viewed from the MurA–A2 interface. The region interacting with MurA, encompassing the N-terminal β-sheet region of residues 30–120, is outlined by a black lasso. The N and C termini are indicated by black arrows.
MurA consists of two globular domains: a catalytic domain and a C-terminal domain, resulting in a clamshell shape (32). The catalytic domain has an 11-amino acid loop (catalytic loop, residues 111–121), creating a “lid” over the active site, where the substrates UDP-GlcNAc and phosphoenolpyruvate (PEP) bind. By refining a model of the complex based on the crystal structures of MurA (32) and A2 (31) into our cryo-EM density map, we are able to determine the overall interaction pattern (Fig. 3C). Both the C-terminal and the catalytic domains of MurA are seen to interact with the β-region of A2, with the outer surface of A2 binding at the concave side of MurA, where the catalytic loop is positioned. This interface between MurA and A2, spanning ∼170 Å2, agrees with previous studies that mapped murA mutations blocking A2 lysis in vivo (19). These MurA mutants are fully functional but cannot be inhibited by A2. The positions of the rat (resistant to A-two) mutations, labeled as red stick models in Fig. 3D, are clearly capable of disrupting the interactions between A2 and MurA (Fig. 3D). Of particular interest is Leu138 in MurA, the site of the first rat mutation (L138Q) (27). Leu138 is positioned directly at the interface where a three-stranded β-sheet (residues 106–110, 135–139, and 142–146) from MurA comes in contact with a five-stranded β-sheet (residues 38–45, 49–55, 91–103, 107–114, and 367–379) from A2 (SI Appendix, Fig. S7). Mutating this residue from a hydrophobic leucine to a hydrophilic glutamine may prevent the proper association between the β-sheets from the two proteins. Further examination of the MurA–A2 interface reveals that most of the interactions with MurA occur in the N-terminal β-sheet region of A2 (between residues 30 and 120), as outlined in Fig. 3E.
To verify that the imaged interaction involving the N-terminal β-sheet region of A2 with MurA had biological significance, we tested different truncations and deletions of A2 for their effect on cell lysis caused by induction of plasmid-borne clones of A2 (27) (SI Appendix, Fig. S9). As expected from the structure, deletions of N-terminal domains were uniformly nonlytic. The shortest N-terminal fragment that was capable of lysis comprised residues 1–180, which spans the entire interface observed by cryo-EM.
During catalysis, MurA undergoes a large conformational change between the unliganded (open) and liganded (closed) states (33). Binding of UDP-GlcNAc triggers the catalytic loop to close, before the association of the second substrate PEP (34, 35). Previous research has shown that the interactions between A2 and MurA are conformation dependent, specifically favoring the conformation when MurA binds UDP-GlcNAc (19). To stabilize MurA in the UDP-GlcNAc bound state, we pretreated MurA with both UDP-GlcNAc and fosfomycin before incubating with Qβ. Fosfomycin covalently binds to Cys115 in the catalytic loop of MurA to lock it in the closed state, and the structure of the UDP-GlcNAc/fosfomycin bound state (PDB ID: 1UAE) is similar to that of the UDP-GlcNAc bound state (PDB ID: 3KQJ). In our cryo-EM map of Qβ–MurA complex, the density of UDP-GlcNAc is clearly visible in the active site of MurA (Fig. 3 C and D and SI Appendix, Fig. S8). Fosfomycin is not visualized, most likely due to its small mass and the limited resolution of the electron density map. In the unliganded open conformation of MurA, the catalytic loop flips out from the active site and may collide with the β-region of A2 (Movie S1), which prevents their association. In fact, even a slight flipping out of the catalytic loop for the UDP-GlcNAc/PEP bound state could inhibit its binding to A2 (19).
An Extra Coat Protein Dimer Is Sequestered Inside the Qβ Virion.
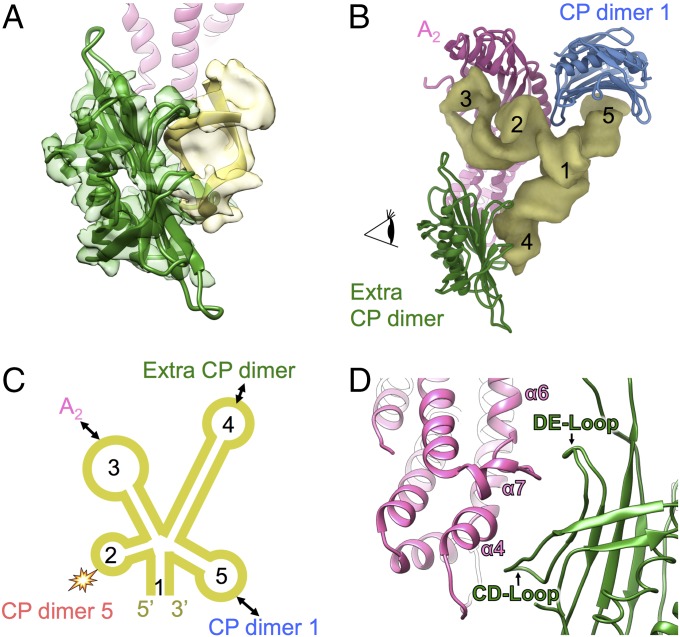
In both the cryo-EM density maps of Qβ alone and the Qβ–MurA complex a strong protein density was observed inside the virion between the bottom of A2 and the gRNA. To resolve this density better, we combined both datasets and subjected them to local 3D classification in the region including A2, the extra protein density, and the interacting gRNA hairpins (Experimental Procedures). The resultant map shows much better resolvability, with the improved extra density able to fit a coat protein dimer, which interacts with a gRNA hairpin in a mode that is similar to the interaction between a coat protein dimer and the RNA hairpin “operator” at the start of the replicase gene (36) (Fig. 4A). When compared at the local resolution (8 Å) in this region, the cross-correlation score between this density and the fitted crystal structure of the Qβ coat protein dimer, in complex with the operator RNA (PDB ID 4L8H), is 0.91.
Fig. 4.
A coat protein dimer is sequestered inside the capsid proximal to the bottom of A2 but away from the rest of the capsid. (A) The additional protein density (transparent green) inside the virion fits the model of a coat protein dimer (green), which interacts with a gRNA hairpin (yellow ribbon model in transparent yellow electron density). The ribbon model of A2 is colored pink and shown in the back. (B) A gRNA domain (yellow density), containing a five-way junction, interacts with A2 (pink), the sequestered coat protein (CP) dimer (green) and CP dimer 1 (blue). The helices on this gRNA five-way junction are labeled 1–5 from the 5′ to the 3′ ends. (C) A scheme showing the secondary structures of the gRNA five-way junction domain with helices labeled from 1 to 5. Double-headed arrows indicate the interacting component of each hairpin on the corresponding helix. The star collision sign indicates where hairpin 2 displaces coat protein dimer 5 as in Fig. 2C. (D) Zoomed-in region as viewed by the eye cartoon in B showing the interactions between the extra coat protein dimer and the bottom of the α-region in A2. The CD loop and DE loop from the sequestered CP dimer and the α-helices of α4, α6, and α7 from A2 are labeled.
An RNA domain that contains a five-way junction was observed close to A2 and the extra coat protein dimer, with the five RNA helices around the junction labeled from 1 to 5 (Fig. 4 B and C). This local RNA domain connects back to other parts of the gRNA through helix 1. The hairpin loop of RNA helix 2 displaces the FG loop of one coat protein on coat protein dimer 5. RNA helices 3–5, each containing a hairpin loop, interact with A2, the sequestered coat protein dimer and coat protein dimer 1, respectively. The sequestered coat protein dimer interacts with A2 via its CD and DE loops, which clamp the bottom of the α-region of A2 (Fig. 4D). Along with the interaction between coat protein dimer 1 and A2, the interactions between these protein components may further compact the RNA five-way junction.
Discussion
Low Infectivity for Qβ May Be Due to Either VLP Formation or MurA Binding.
Qβ, and many ssRNA phages, have low viability, as only a fraction of the purified particles are actually infectious (37, 38). In our cryo-EM data analysis, around 20% of the total number of particles contain A2 and have a defined gRNA conformation inside the capsid (SI Appendix, Fig. S1). After further classification of the remaining dataset, we determined that the remaining particles are VLPs, inasmuch as they lack A2, leading to a perfectly icosahedral capsid. The RNA in these VLPs could be viral RNA, which does not have A2 bound; or host RNA, since it has been demonstrated that ssRNA viruses can encapsidate host RNAs (39). By testing coat protein binding to purified RNAs from Qβ and MS2, it was found that the Qβ coat protein has a higher affinity for both homologous and heterologous assembly than the MS2 coat protein (40). The higher affinity for the coat proteins of Qβ to bind RNA could be due to its positively charged interior EF loop (15), which may facilitate the nonspecific binding to the negatively charged RNA backbones. Indeed, the ability to encapsidate foreign RNA is the basis on which the Qβ coat protein is commercially used to protect heterologous RNAs (41). It is also possible that just as there is a sequestered coat protein dimer within the virion, the maturation protein may also bind to the Qβ gRNA in these VLPs but be internalized due to mispackaging. The low infectivity of Qβ could further be due to the cellular MurA binding A2, wherein it prevents efficient assembly of the mature virion or occupies the interface of A2 that is required for attachment to the host F pilus.
Potential Role of the Extra Coat Protein Dimer.
The extra coat protein dimer sequestered in the interior of Qβ has not been seen in other ssRNA viruses. Its presence lends credence to the role of “packaging signals” (11) in virion morphogenesis. As the viral gRNA is replicated and translated, coat proteins are synthesized and form dimers to bind to packaging signals, mainly RNA hairpins, such as the operator, a hairpin at the start of the replicase gene that is known to bind coat proteins to repress translation of the replicase (42). The current resolutions of the Qβ RNA do not allow the assignment of the genome sequence solely based on its cryo-EM density. It needs to be determined whether the RNA sequence that the internalized coat protein dimer binds corresponds to the operator of the replicase gene or a sequence similar to the operator. It also cannot be ruled out that there may be a small fraction of Qβ particles that contain more coat protein dimers, internalized at different locations in the virion and bound to other gRNA hairpins facing inwards. The internalized coat protein dimer, observed in our Qβ virion, may participate in a nucleation event wherein the combination of the maturation protein, the sequestered coat protein dimer and the capsid coat protein dimers condense the gRNA locally around the RNA five-way junction, compacting the gRNA and enabling it to fit into the dimensions of the capsid.
Such an internalized coat protein dimer and its associated compact gRNA domain may also affect the genome release during viral infection. Helix 3 from this gRNA domain directly associates with A2 (Fig. 4) and is likely the first portion of the RNA to exit the capsid during the genome release. The expansion of the capsid around A2 (Fig. 2 and SI Appendix, Fig. S4) may facilitate the initial release of this ordered gRNA domain. It would be interesting to determine if the sequestered coat protein dimer remains inside the capsid or just simply diffuses away after the maturation protein and gRNA are removed from the virion. Alternatively, the sequestered coat protein dimer may recap the opening that is left after A2 is removed from the capsid, leaving an empty but icosahedral VLP.
Structural Comparison of the Maturation Proteins from Two Model ssRNA Phages.
A2 from Qβ and the A protein from MS2 share high structural similarity within the α-region (SI Appendix, Fig. S10A). However, the connections between the α- and β-regions of these two proteins are very different. A2 has a long α-helix (residues 128–177) from the α-region to the β-region, while the A protein has a zigzag helix–loop–helix (residues 117–169) motif to connect the two regions (SI Appendix, Fig. S10B). The different angles between the α- and β-regions for these two proteins lead to altered insertions into the capsid, at varying angles relative to the opening in the capsid (SI Appendix, Fig. S10C). When viewing A and A2 in their corresponding virions, we see that the α-region of A2 inserts at an orientation that is more perpendicular to the capsid opening (SI Appendix, Fig. S10D). Even though the coat proteins and α-region of the maturation proteins from MS2 and Qβ are structurally similar, the interactions between the α-region of the maturation protein and its surrounding coat proteins are different between MS2 and Qβ (SI Appendix, Fig. S10C), indicating these interactions are not conserved between the two model phages. The maturation protein may be loosely anchored in the capsid while being held in place by the neighboring coat proteins that wrap around it. The weak interactions between the maturation protein and the coat proteins could further facilitate the release of the maturation protein, along with the attached gRNA, from the capsid during infection.
Even though folded differently, the overall geometries of the β-regions in A and A2 are similar, both presenting a flat outer surface that is angled ∼30° to the opening of the capsid (SI Appendix, Fig. S10D). Based on a low-resolution density map of MS2 bound to the F pilus obtained through cryoelectron tomography (43), the outer surface of the β-region of the MS2 A protein binds the pilus. Presumably, the similar outer surface of Qβ would also bind to the pilus. It has also been reported that treating Qβ with MurA abolishes its ability to infect Escherichia coli (29), suggesting that the F pilus and MurA bind the same or overlapping locations on the β-region of A2. However, a detailed picture of the interaction between the F pilus and the maturation protein requires a high-resolution structure of the phage–pilus complex.
Experimental Procedures
Sample and Cryo-EM Grid Preparation.
Qβ and MurA were purified as previously described with minor modifications (19, 44). After separation using a cesium chloride gradient, Qβ particles were further purified by a 200-mL hand-packed S500 gel-filtration column. The sample of MurA-bound Qβ was prepared by pretreating MurA with 1 mM UDP-GlcNAc and 1 mM fosfomycin on ice for 30 min, and then incubating with Qβ on ice for another 30 min. The molar ratio of MurA to Qβ was ∼5–1. Cryo-EM specimens were prepared by applying 3 µL of the sample solution to a C-Flat 1.2/1.3 or 2/1 400-mesh Holey Carbon Grid (Protochips) at 22 °C with 100% relative humidity and vitrified using a Vitrobot Mark III (FEI Company).
Cryo-EM Data Collection.
Qβ alone and some of the Qβ–MurA complex images were recorded under a JEM3200FSC cryoelectron microscope with a field emission gun (JEOL) operated at 300 kV. Data were collected using SerialEM (45) on a K2 Summit direct detection camera (Gatan) in the superresolution electron counting mode (30). A nominal magnification of 30,000× was used, yielding a subpixel size of 0.608 Å. The beam intensity was adjusted to a dose rate of 7e− per pixel per second on the camera. A 50-frame movie stack was recorded for each exposure with 0.2 s per frame for a total exposure time of 10 s. An in-column energy filter was used with a slit width of 29 eV.
Most of the Qβ–MurA complex images were obtained under an FEI Tecnai F20 cryoelectron microscope with a field emission gun (FEI Company) operated at 200 kV. Data were recorded using SerialEM on a K2 Summit direct detection camera (Gatan) in the superresolution electron counting mode. A nominal magnification of 29,000× was used, yielding a subpixel size of 0.625 Å. The beam intensity was adjusted to a dose rate of 8.7e− per pixel per second on the camera. A 33-frame movie stack was recorded for each picture, with 0.2 s per frame for a total exposure time of 6.6 s.
Image Preprocessing.
The collected superresolution movie stacks were first binned by 2, then aligned, summed, and filtered using Unblur (46). The defocus value of each summed micrograph was determined using CTFFIND4 (47). Micrographs with strong power spectrums were used to semiautomatically pick particles with e2boxer.py in EMAN2 (48). The 2D and 3D classifications and 3D refinement of the image data were performed in Relion (49).
The data processing for the Qβ alone sample was as follows (SI Appendix, Fig. S1): Particles were selected from 2,370 micrographs and subjected to 2D classifications in Relion to remove bad particles. After 2D classifications, 248,445 particles were chosen for three independent rounds of unsupervised 3D classifications, each for 30 interactions, requesting three, four, and five classes, respectively. Each round of the 3D classification produced a single class containing particles with defined densities for A2 and the gRNA. These three single classes were combined to yield 76,843 particles without the duplication of particles. To improve the resolution for A2, 50 iterations of a masked classification around A2 with signal subtraction was performed (50). A total of 46,471 particles with good A2 density were selected for the 3D refinement of the Qβ virion, giving a 4.7-Å resolution map of the Qβ virion. The resolution of the capsid for the Qβ virion was improved to 4.4 Å after a masked refinement excluding the gRNA density. The 3D classification of the remaining 171,602 particles revealed no A2 density and the capsid of these particles was perfectly icosahedral. These particles were subjected to icosahedral refinement, giving a 3.3-Å resolution map of the Qβ VLP capsid.
The structure of the Qβ–MurA complex was determined by combining 1,117 and 1,754 images collected using the JEM3200FSC and the FEI Tecnai F20 microscopes, respectively. By carefully comparing the two maps calculated from icosahedral refinement in those two datasets, images collected on the JEM3200FSC microscope were downscaled to match the pixel size of those collected on the FEI Tecnai F20 microscope. After 2D and 3D classifications, 25,597 of the 130,013 total particles were used for the final reconstruction to yield a 5.7-Å map after using an outer shell mask, which excludes the gRNA.
To achieve better map quality for the extra coat protein dimer density near the bottom of A2, we combined all 5,241 images of Qβ alone and Qβ–MurA complex by the same procedure as we combined the two Qβ–MurA datasets. With a masked classification around A2 and the extra coat protein dimer, 47,621 good particles were selected for the final refinement. A 6.2-Å map of A2 and the coat protein dimer was obtained after masking.
Resolution Estimation.
The overall resolutions of all these reconstructed maps were assessed using the gold-standard criterion of Fourier shell correlation, with a cutoff at 0.143, between two half-maps from two independent half-sets of data (51). Local resolutions were estimated using Resmap (52).
Modeling and Visualization.
To build the atomic model for the Qβ VLP capsid, our previous icosahedral Qβ structure (PDB ID: 5KIP) was used as the initial model and subsequently refined into the 3.3-Å icosahedral map of the Qβ VLP by the real-space refinement from PHENIX (53). To build the backbone model for the Qβ virion capsid, the crystal structure of A2 (PDB ID: 5MNT) was used to replace one coat protein dimer on the Qβ VLP capsid from the previous step. The model for the internalized coat protein dimer was then added to the capsid of the Qβ virion by fitting into its corresponding density as a rigid body. The backbone of the Qβ virion capsid was then refined into the 4.4-Å asymmetric map using the real-space refinement with the secondary structure restraints in PHENIX. To model the interaction between A2 and MurA, the density of the A2–MurA complex was firstly segmented out from the 5.7-Å map of the Qβ–MurA complex using University of California San Francisco (UCSF) Chimera (54). The crystal structures of MurA (PDB ID: 1UAE) and A2 were then rigidly fitted into the segmented density and further refined by the molecular dynamics flexible fitting method (55) followed by the real-space refinement with the secondary structure restraints in PHENIX. The model statistics are summarized in SI Appendix, Table S1.
The model of the E. coli MurA in its open conformation was built using homology modeling by SWISS-MODEL (56), based on the crystal structure of the Enterobacter cloacae MurA (PDB ID: 1EJD). All of the figures and movies were made using UCSF Chimera.
Supplementary Material
Acknowledgments
We thank Karthik Chamakura for his expertise in ssRNA viruses and comments on the paper, and the Microscopy and Imaging Center at Texas A&M University and the National Center for Macromolecular Imaging (NCMI) at Baylor College of Medicine for the cryo-EM data collection. NCMI is supported by NIH Grants P41GM103832 and U24GM116787. We also thank the Texas A&M High Performance Research Computing Center for providing the computational resources for the data processing. J.Z. is supported by startup funding from the Department of Biochemistry and Biophysics at Texas A&M University and Welch Foundation Grant A-1863. R.Y. is supported by Public Health Service Grant GM27099. J.Z. and R.Y. acknowledge the support of the Center for Phage Technology jointly sponsored by Texas A&M AgriLife and Texas A&M University.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the EMDataBank (www.emdatabank.org) with accession IDs: EMD-8708 (Qβ virus-like particle), EMD-8709 (Qβ virion), EMD-8711 (Qβ virion bound with MurA), and EMD-8710 (local map including A2, the internalized coat protein dimer, and the interacting gRNA hairpins), and Protein Data Bank (https://www.wwpdb.org) with PDB IDs: 5VLY (capsid for the Qβ virus-like particle), 5VLZ (capsid for the Qβ virion), and 5VM7 (complex between A2 and MurA).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1707102114/-/DCSupplemental.
References
- 1.Koonin EV, Dolja VV, Krupovic M. Origins and evolution of viruses of eukaryotes: The ultimate modularity. Virology. 2015;479–480:2–25. doi: 10.1016/j.virol.2015.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roossinck MJ. Plant RNA virus evolution. Curr Opin Microbiol. 2003;6:406–409. doi: 10.1016/s1369-5274(03)00087-0. [DOI] [PubMed] [Google Scholar]
- 3.Kolakofsky D. A short biased history of RNA viruses. RNA. 2015;21:667–669. doi: 10.1261/rna.049916.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barré-Sinoussi F, Ross AL, Delfraissy JF. Past, present and future: 30 years of HIV research. Nat Rev Microbiol. 2013;11:877–883. doi: 10.1038/nrmicro3132. [DOI] [PubMed] [Google Scholar]
- 5.Kajitani M, Kato A, Wada A, Inokuchi Y, Ishihama A. Regulation of the Escherichia coli hfq gene encoding the host factor for phage Q β. J Bacteriol. 1994;176:531–534. doi: 10.1128/jb.176.2.531-534.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poot RA, Tsareva NV, Boni IV, van Duin J. RNA folding kinetics regulates translation of phage MS2 maturation gene. Proc Natl Acad Sci USA. 1997;94:10110–10115. doi: 10.1073/pnas.94.19.10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Smit MH, van Duin J. Translational initiation at the coat-protein gene of phage MS2: Native upstream RNA relieves inhibition by local secondary structure. Mol Microbiol. 1993;9:1079–1088. doi: 10.1111/j.1365-2958.1993.tb01237.x. [DOI] [PubMed] [Google Scholar]
- 8.Klovins J, Berzins V, van Duin J. A long-range interaction in Qbeta RNA that bridges the thousand nucleotides between the M-site and the 3′ end is required for replication. RNA. 1998;4:948–957. doi: 10.1017/s1355838298980177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown D, Gold L. RNA replication by Q β replicase: A working model. Proc Natl Acad Sci USA. 1996;93:11558–11562. doi: 10.1073/pnas.93.21.11558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gytz H, et al. Structural basis for RNA-genome recognition during bacteriophage Qβ replication. Nucleic Acids Res. 2015;43:10893–10906. doi: 10.1093/nar/gkv1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rolfsson Ó, et al. Direct evidence for packaging signal-mediated assembly of bacteriophage MS2. J Mol Biol. 2016;428:431–448. doi: 10.1016/j.jmb.2015.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stockley PG, et al. Packaging signals in single-stranded RNA viruses: Nature’s alternative to a purely electrostatic assembly mechanism. J Biol Phys. 2013;39:277–287. doi: 10.1007/s10867-013-9313-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peabody DS. Role of the coat protein-RNA interaction in the life cycle of bacteriophage MS2. Mol Gen Genet. 1997;254:358–364. doi: 10.1007/s004380050427. [DOI] [PubMed] [Google Scholar]
- 14.Horn WT, et al. Structural basis of RNA binding discrimination between bacteriophages Qbeta and MS2. Structure. 2006;14:487–495. doi: 10.1016/j.str.2005.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorzelnik KV, et al. Asymmetric cryo-EM structure of the canonical Allolevivirus Qβ reveals a single maturation protein and the genomic ssRNA in situ. Proc Natl Acad Sci USA. 2016;113:11519–11524. doi: 10.1073/pnas.1609482113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dai X, et al. In situ structures of the genome and genome-delivery apparatus in a single-stranded RNA virus. Nature. 2017;541:112–116. doi: 10.1038/nature20589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koning RI, et al. Asymmetric cryo-EM reconstruction of phage MS2 reveals genome structure in situ. Nat Commun. 2016;7:12524. doi: 10.1038/ncomms12524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhong Q, et al. Genetic, structural, and phenotypic properties of MS2 coliphage with resistance to ClO2 disinfection. Environ Sci Technol. 2016;50:13520–13528. doi: 10.1021/acs.est.6b04170. [DOI] [PubMed] [Google Scholar]
- 19.Reed CA, Langlais C, Kuznetsov V, Young R. Inhibitory mechanism of the Qβ lysis protein A2. Mol Microbiol. 2012;86:836–844. doi: 10.1111/mmi.12021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garwes D, Sillero A, Ochoa S. Virus-specific proteins in Escherichia coli infected with phage Qb. Biochim Biophys Acta. 1969;186:166–172. doi: 10.1016/0005-2787(69)90499-7. [DOI] [PubMed] [Google Scholar]
- 21.Blumenthal T, Carmichael GG. RNA replication: Function and structure of Qbeta-replicase. Annu Rev Biochem. 1979;48:525–548. doi: 10.1146/annurev.bi.48.070179.002521. [DOI] [PubMed] [Google Scholar]
- 22.Rumnieks J, Tars K. Crystal structure of the read-through domain from bacteriophage Qβ A1 protein. Protein Sci. 2011;20:1707–1712. doi: 10.1002/pro.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weber K, Konigsberg W. Proteins of the RNA phages. Cold Spring Harb Monogr Arch. 1975;5:51–84. [Google Scholar]
- 24.Hofstetter H, Monstein HJ, Weissmann C. The readthrough protein A1 is essential for the formation of viable Q β particles. Biochim Biophys Acta. 1974;374:238–251. doi: 10.1016/0005-2787(74)90366-9. [DOI] [PubMed] [Google Scholar]
- 25.Richelson E, Nathans D. Association of bacteriophage proteins and RNA in E. coli infected with MS2. Biochem Biophys Res Commun. 1967;29:842–849. doi: 10.1016/0006-291x(67)90296-3. [DOI] [PubMed] [Google Scholar]
- 26.Kastelein RA, Remaut E, Fiers W, van Duin J. Lysis gene expression of RNA phage MS2 depends on a frameshift during translation of the overlapping coat protein gene. Nature. 1982;295:35–41. doi: 10.1038/295035a0. [DOI] [PubMed] [Google Scholar]
- 27.Bernhardt TG, Wang IN, Struck DK, Young R. A protein antibiotic in the phage Qbeta virion: Diversity in lysis targets. Science. 2001;292:2326–2329. doi: 10.1126/science.1058289. [DOI] [PubMed] [Google Scholar]
- 28.Brown ED, Vivas EI, Walsh CT, Kolter R. MurA (MurZ), the enzyme that catalyzes the first committed step in peptidoglycan biosynthesis, is essential in Escherichia coli. J Bacteriol. 1995;177:4194–4197. doi: 10.1128/jb.177.14.4194-4197.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reed CA, Langlais C, Wang IN, Young R. A(2) expression and assembly regulates lysis in Qβ infections. Microbiology. 2013;159:507–514. doi: 10.1099/mic.0.064790-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rumnieks J, Tars K. Crystal structure of the maturation protein from bacteriophage Qβ. J Mol Biol. 2017;429:688–696. doi: 10.1016/j.jmb.2017.01.012. [DOI] [PubMed] [Google Scholar]
- 32.Skarzynski T, et al. Structure of UDP-N-acetylglucosamine enolpyruvyl transferase, an enzyme essential for the synthesis of bacterial peptidoglycan, complexed with substrate UDP-N-acetylglucosamine and the drug fosfomycin. Structure. 1996;4:1465–1474. doi: 10.1016/s0969-2126(96)00153-0. [DOI] [PubMed] [Google Scholar]
- 33.Schönbrunn E, Svergun DI, Amrhein N, Koch MH. Studies on the conformational changes in the bacterial cell wall biosynthetic enzyme UDP-N-acetylglucosamine enolpyruvyltransferase (MurA) Eur J Biochem. 1998;253:406–412. doi: 10.1046/j.1432-1327.1998.2530406.x. [DOI] [PubMed] [Google Scholar]
- 34.Zhu JY, et al. Functional consequence of covalent reaction of phosphoenolpyruvate with UDP-N-acetylglucosamine 1-carboxyvinyltransferase (MurA) J Biol Chem. 2012;287:12657–12667. doi: 10.1074/jbc.M112.342725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skarzynski T, Kim DH, Lees WJ, Walsh CT, Duncan K. Stereochemical course of enzymatic enolpyruvyl transfer and catalytic conformation of the active site revealed by the crystal structure of the fluorinated analogue of the reaction tetrahedral intermediate bound to the active site of the C115A mutant of MurA. Biochemistry. 1998;37:2572–2577. doi: 10.1021/bi9722608. [DOI] [PubMed] [Google Scholar]
- 36.Rumnieks J, Tars K. Crystal structure of the bacteriophage Qβ coat protein in complex with the RNA operator of the replicase gene. J Mol Biol. 2014;426:1039–1049. doi: 10.1016/j.jmb.2013.08.025. [DOI] [PubMed] [Google Scholar]
- 37.Watanabe H, Watanabe M. Comparative biology of five RNA phages, R23, f2, Q β, R34, and R40. Can J Microbiol. 1970;16:859–864. doi: 10.1139/m70-145. [DOI] [PubMed] [Google Scholar]
- 38.Leipold B, Hofschneider PH. Isolation of an infectious RNA-A-protein complex from the bacteriophage M12. FEBS Lett. 1975;55:50–52. doi: 10.1016/0014-5793(75)80954-9. [DOI] [PubMed] [Google Scholar]
- 39.Routh A, Domitrovic T, Johnson JE. Host RNAs, including transposons, are encapsidated by a eukaryotic single-stranded RNA virus. Proc Natl Acad Sci USA. 2012;109:1907–1912. doi: 10.1073/pnas.1116168109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ling CM, Hung PP, Overby LR. Specificity in self-assembly of bacteriophages Q β and MS2. Biochemistry. 1969;8:4464–4469. doi: 10.1021/bi00839a036. [DOI] [PubMed] [Google Scholar]
- 41.Bundy BC, Swartz JR, Chan W. 2010. Encapsidation of heterologous entities into virus-like particles. US Patent 12/621,416.
- 42.Weber H. The binding site for coat protein on bacteriophage Qbeta RNA. Biochim Biophys Acta. 1976;418:175–183. doi: 10.1016/0005-2787(76)90067-8. [DOI] [PubMed] [Google Scholar]
- 43.Dent KC, et al. The asymmetric structure of an icosahedral virus bound to its receptor suggests a mechanism for genome release. Structure. 2013;21:1225–1234. doi: 10.1016/j.str.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strauss JH, Jr, Sinsheimer RL. Purification and properties of bacteriophage MS2 and of its ribonucleic acid. J Mol Biol. 1963;7:43–54. doi: 10.1016/s0022-2836(63)80017-0. [DOI] [PubMed] [Google Scholar]
- 45.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 46.Grant T, Grigorieff N. Measuring the optimal exposure for single particle cryo-EM using a 2.6 Å reconstruction of rotavirus VP6. eLife. 2015;4:e06980. doi: 10.7554/eLife.06980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang G, et al. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 49.Scheres SH. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bai XC, Rajendra E, Yang G, Shi Y, Scheres SH. Sampling the conformational space of the catalytic subunit of human γ-secretase. eLife. 2015;4:e11182. doi: 10.7554/eLife.11182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scheres SH, Chen S. Prevention of overfitting in cryo-EM structure determination. Nat Methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pettersen EF, et al. UCSF Chimera: A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 55.Trabuco LG, Villa E, Mitra K, Frank J, Schulten K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure. 2008;16:673–683. doi: 10.1016/j.str.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.