Significance
Neurons are highly polarized cells critical to the processing of signals within the brain, and are distinguished by morphologically and functionally distinct axonal and somatodendritic compartments. Understanding mechanisms dictating neuronal morphological specialization will advance our understanding of neuronal function and neurodegenerative diseases. This report reveals that the spectrin-actin cytoskeleton contributes to the origin of dendritic arbor morphology. We show that this cytoskeleton is required for formation and stabilization of complex dendritic arbors in Drosophila. A human spinocerebellar ataxia type 5 mutation causes β-III-spectrin to bind actin with high affinity and disrupts expansion of the spectrin-actin cytoskeleton into distal regions of the arbor. Our data suggest that loss of the spectrin-actin cytoskeleton reduces stabilization of distal dendrites required for arbor outgrowth.
Keywords: spinocerebellar ataxia type 5, SCA5, dendritic arborization, spectrin, ankyrin
Abstract
A spinocerebellar ataxia type 5 (SCA5) L253P mutation in the actin-binding domain (ABD) of β-III-spectrin causes high-affinity actin binding and decreased thermal stability in vitro. Here we show in mammalian cells, at physiological temperature, that the mutant ABD retains high-affinity actin binding. Significantly, we provide evidence that the mutation alters the mobility and recruitment of β-III-spectrin in mammalian cells, pointing to a potential disease mechanism. To explore this mechanism, we developed a Drosophila SCA5 model in which an equivalent mutant Drosophila β-spectrin is expressed in neurons that extend complex dendritic arbors, such as Purkinje cells, targeted in SCA5 pathogenesis. The mutation causes a proximal shift in arborization coincident with decreased β-spectrin localization in distal dendrites. We show that SCA5 β-spectrin dominantly mislocalizes α-spectrin and ankyrin-2, components of the endogenous spectrin cytoskeleton. Our data suggest that high-affinity actin binding by SCA5 β-spectrin interferes with spectrin-actin cytoskeleton dynamics, leading to a loss of a cytoskeletal mechanism in distal dendrites required for dendrite stabilization and arbor outgrowth.
Spinocerebellar ataxia type 5 (SCA5) is a human neurodegenerative disease that causes gait and limb ataxia, slurred speech, and abnormal eye movements (1). SCA5 stems from autosomal dominant mutations in the SPTBN2 gene that encodes β-III-spectrin (2), a cytoskeletal protein predominantly expressed in the brain and enriched in cerebellar Purkinje cells (3). A necessary function of β-III-spectrin in Purkinje cells was demonstrated by β-III-spectrin–null mice, which show ataxic phenotypes and decreased Purkinje cell dendritic arborization (4–6). β-III-spectrin consists of an N-terminal actin-binding domain (ABD) followed by 17 spectrin-repeat domains and a C-terminal pleckstrin homology domain. SCA5 mutations that result in single amino acid substitutions or small in-frame deletions have been identified in the ABD and neighboring spectrin-repeat domains (2, 7–10). In a SCA5 mouse model, expression in Purkinje cells of a β-III-spectrin transgene containing a spectrin-repeat domain mutation, E532_M544del, causes ataxic phenotypes and thinning of the cerebellar molecular layer that contains Purkinje cell dendrites (11). This suggests that the cellular mechanism underlying SCA5 pathogenesis is a Purkinje cell deficit linked to the loss of dendritic arborization.
The functional unit of β-III-spectrin is considered to be a heterotetrameric complex containing two β-spectrin subunits and two α-spectrin subunits. Through the β-spectrin subunits the spectrin heterotetramer binds and cross-links actin filaments. Multiple β-spectrin protein isoforms have been shown to form a spectrin-actin cytoskeletal structure that lines the plasma membrane of axons and dendrites. The spectrin-actin lattice is a highly conserved neuronal structure identified in the axons of a broad range of neuron types in mammals (12–14) and in invertebrates, including Drosophila (14, 15). A spectrin-actin lattice containing β-III-spectrin, or the homolog β-II-spectrin, was identified in the dendrites of hippocampal neurons (16). Recent studies suggest that the dendritic spectrin-actin cytoskeleton is a ubiquitous feature of neurons, prominent in both dendritic shafts and spines (17–19). The widespread localization of β-III-spectrin within the Purkinje cell dendritic arbor (3) suggests that similar spectrin–actin interactions are important for Purkinje cell dendritic function.
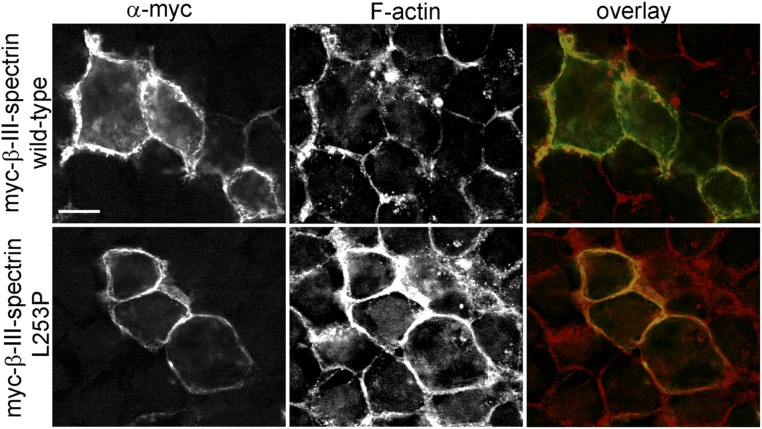
The spectrin-actin cytoskeleton functions to organize integral membrane proteins through the spectrin adaptor ankyrin (12) and provides mechanical stability to neuronal processes (20, 21). A form of erythrocyte ankyrin, ankyrin-R, is expressed in Purkinje cells and appears to be required for Purkinje cell health and normal motor function. A hypomorphic ankyrin-R mutation, termed “normoblastosis” (22, 23), causes Purkinje cell degeneration and ataxia in mice (24). The subcellular localization of ankyrin-R in the Purkinje cell soma and dendrites mirrors the distribution of β-III-spectrin (25–27), and recently β-III-spectrin was shown to physically interact with ankyrin-R (27). In β-III-spectrin–null mice, ankyrin-R is present in the soma but absent in Purkinje cell dendrites (27), suggesting that Purkinje cell degeneration and ataxic phenotypes observed in the absence of β-III-spectrin may be linked to a loss of ankyrin-R function in dendrites. A SCA5 mutation that results in a leucine 253-to-proline (L253P) substitution in the ABD of β-III-spectrin causes ectopically expressed β-III-spectrin and ankyrin-R to colocalize internally in HEK293T cells, in contrast to control cells where wild-type β-III-spectrin colocalizes with ankyrin-R at the plasma membrane (27). This previous study suggests that neurotoxicity caused by the L253P mutation may be connected to spectrin mislocalization and the concomitant mislocalization of ankyrin-R. However, it has not been established whether the L253P mutation affects the dendritic localization of β-spectrin or ankyrin proteins in any neuronal system.
This report extends our analysis of the β-III-spectrin L253P mutation, which we recently demonstrated causes an ∼1,000-fold increase in the binding affinity of the β-III-spectrin ABD for actin filaments in vitro (28). The mutation is also destabilizing in vitro, causing the ABD to begin to unfold near physiological temperature. Given these results, a key question with important implications for the SCA5 disease mechanism is whether the previously described mislocalization of L253P β-III-spectrin in mammalian cells is driven by a loss of ABD-binding activity, as originally proposed (29), or instead is the consequence of increased ABD-binding activity. To address the mechanistic basis of β-III-spectrin dysfunction, we have characterized the L235P mutant protein behavior in mammalian cells. In addition, we generated a Drosophila SCA5 model in which a Drosophila β-spectrin transgene containing the equivalent mutation is conditionally expressed in dendritic arborization sensory neurons. We use the Drosophila model to analyze the impact of the mutation on dendritic morphology, an aspect of Purkinje cell dysfunction that potentially underlies SCA5 pathology. In living, fully intact larvae, we examine the consequence of the ABD mutation on dendritic arborization, β-spectrin subcellular localization, and the functional interaction of β-spectrin and ankyrin in dendrites.
Results
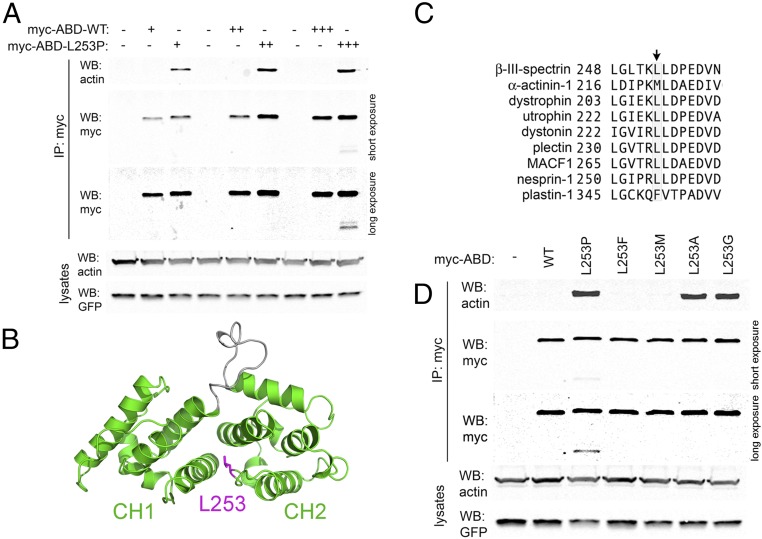
Biochemical analyses demonstrated that a SCA5 missense mutation resulting in a L253P substitution in the ABD of human β-III-spectrin causes high-affinity F-actin binding (Kd = 75.5 nM vs. 75.8 µM for wild-type) (28). Here we report on the properties of the SCA5 mutant β-III-spectrin in mammalian cells and consider potential mechanisms underlying SCA5 pathology. First, using a coimmunoprecipitation (co-IP) assay in HEK293T cells, we demonstrate that mutant L253P ABD also binds actin with increased affinity in mammalian cells at physiological temperature (Fig. 1A and Fig. S1). The large difference in binding affinities characterized in vitro is reflected in the co-IP assay, which readily detects the interaction of the mutant ABD with actin but does not detect binding of the wild-type ABD to actin, a dynamic, low-affinity interaction probably below the sensitivity of the assay. Our previous thermal denaturation studies also revealed that in vitro the L253P mutant ABD begins to unfold at 42 °C, raising the question of whether the L253P β-III-spectrin ABD is prone to denaturation in living cells at physiological temperature. This might explain the previously reported loss of binding to an actin-related protein, actin-related protein 1 (ARP1), by L253P β-III-spectrin (29). However, in the co-IP assay only minor degradation of the L253P ABD is observed (Fig. 1A), and only at the highest level of ABD expression, suggesting that the mutant ABD is largely well-folded in mammalian cells at 37 °C. Moreover, the mutation does not cause degradation of full-length β-III-spectrin in mammalian cells (Fig. S2), suggesting that full-length L253P β-III-spectrin is also well-folded. Importantly, these results in mammalian cells corroborate our in vitro analyses showing that a well-folded L253P ABD binds actin with high affinity.
Fig. 1.
The L253P mutation causes high-affinity actin binding in mammalian cells by disrupting leucine 253 hydrophobic contacts. (A) Co-IP assay in HEK293T cells transiently transfected with varying amounts of myc-epitope–tagged wild-type (WT) β-III-spectrin ABD or L253P. Actin coimmunoprecipitataes with L253P ABD but not with wild-type ABD. In the long-exposure α-myc Western blot (WB) panel, image contrast settings were adjusted to better observe low-intensity L253P ABD degradation products. (B) Structural homology model of the β-III-spectrin ABD (amino acids 56–284) (28). The left and right globular domains represent the CH1 and CH2 domains, respectively. Leucine 253, colored magenta, is positioned at the interface of the two CH domains. (C) ClustalW alignment of human ABD proteins showing the conservation of leucine or a similarly hydrophobic residue at the position equivalent to β-III-spectrin leucine 253. (D) Co-IP assay in HEK293T cells expressing myc-ABD proteins with leucine 253 substitutions to vary hydrophobicity. Loss of hydrophobicity caused by alanine or glycine substitutions increases actin binding, as does proline substitution. Unlike proline substitution, alanine and glycine substitutions do not cause ABD degradation.
Fig. S1.
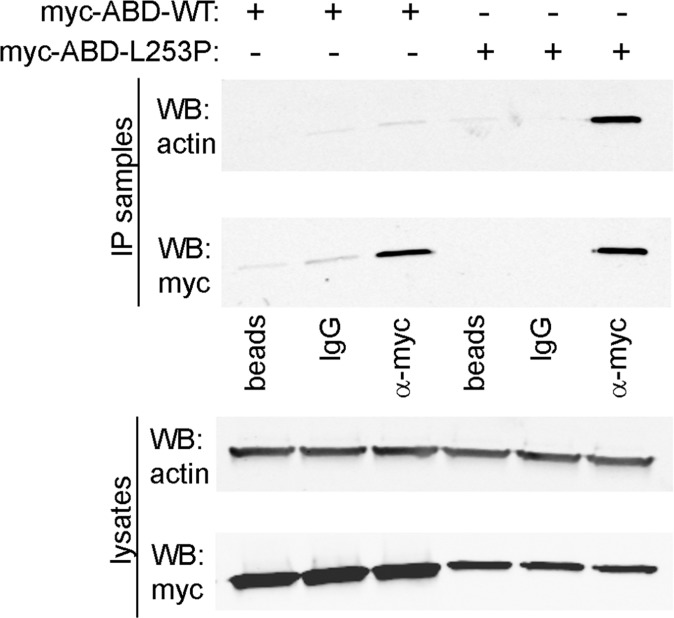
The interaction of the L253P β-III-spectrin ABD with actin by co-IP is specific. HEK293T cells were transiently transfected with wild-type (WT) or L253P myc-epitope–tagged β-III-spectrin ABD. The myc-ABD proteins were isolated from lysates using 1 µg rabbit α-myc antibody or, as controls, 1 µg normal rabbit IgG or protein A Sepharose (beads) only. Myc-ABD proteins are greatly enriched in α-myc antibody IP samples compared with control antibody IP samples. Actin is enriched only in the myc-L253P ABD α-myc antibody IP sample. In lysates, the reduced amount of L253P ABD compared with wild-type ABD reflects the accumulation of L253P ABD in the clarified Triton X-100–insoluble fraction, which contains about ∼50% of total cellular actin (Fig. S2).
Fig. S2.
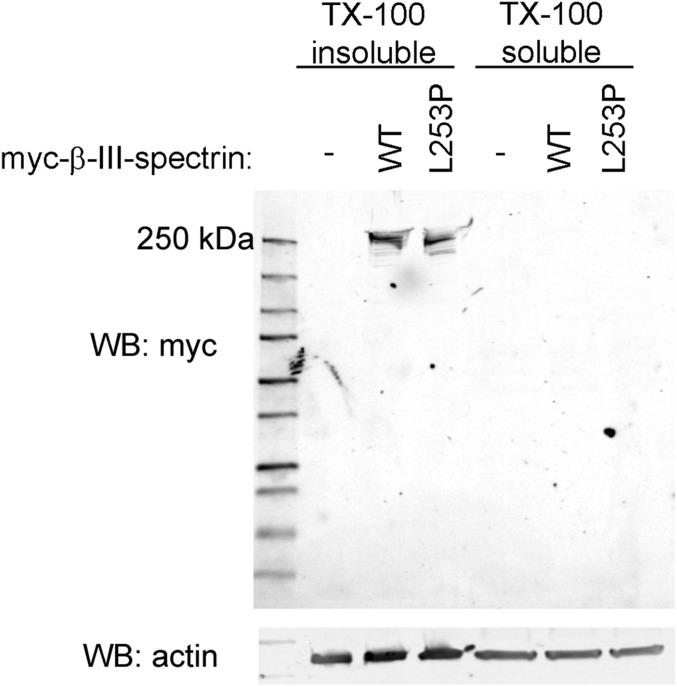
The L253P mutation does not induce degradation of full-length β-III-spectrin in mammalian cells at physiological temperature. HEK293T cells were transiently transfected with myc-epitope–tagged full-length wild-type (WT) or L253P β-III-spectrin or no β-III-spectrin protein. Wild-type and L253P full-length β-III-spectrin proteins are ∼97% and 100%, respectively, contained in the Triton X-100–insoluble fraction that also contains plasma membrane material and ∼50% of total cellular actin. Protein bands for wild-type and L253P β-III-spectrin run near the predicted 271-kDa mass. Only minor degradation products are observed for either wild-type and L253P protein.
To further examine the role of leucine 253 in regulating actin-binding affinity and protein stability, we characterized additional leucine 253 mutations. The ABD consists of two calponin-homology (CH) domains in tandem, with leucine 253 located in the second CH domain (CH2) (Fig. 1B). An alignment of β-III-spectrin with other human tandem-CH ABD proteins shows a preference for highly hydrophobic amino acids at the position equivalent to leucine 253 (Fig. 1C). Most of the aligned ABD proteins contain a leucine residue at this position, while α-actinin and plastin contain methionine and phenylalanine residues, respectively. Leucine 253 is predicted to be in a loop structure at the interface of the two CH domains (28), and our previous in vitro characterization of the L253P ABD indicated that the mutant ABD can attain a well-folded state, suggesting that high-affinity actin binding caused by the L253P substitution is due to a localized disruption of hydrophobic interactions normally mediated by leucine 253. To further test this mechanistic hypothesis for elevated actin-binding affinity, we performed additional mutagenesis to substitute leucine 253 with phenylalanine or methionine, which have hydrophobicity similar to that of the native leucine, or with alanine or glycine, which are of low hydrophobicity comparable to that of the mutant proline. A co-IP assay in HEK293T cells shows that substitution with alanine or glycine, like proline, causes increased actin binding, while substitution with phenylalanine or methionine does not (Fig. 1D). Thus, these data support the model that the L253P substitution increases actin binding by causing a loss of hydrophobic interactions normally mediated by leucine 253 at the CH1–CH2 interface. In this experiment, we emphasize that for both the alanine and glycine substitutions, no ABD degradation products were detected, further supporting our inference that, in vivo, the elevated actin binding is associated with a well-folded ABD.
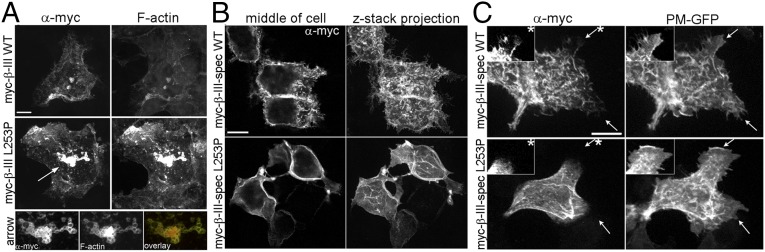
We next examined the impact of altered actin binding on the cellular distribution of L253P β-III-spectrin. A previous study examined the effect of the L253P mutation on the subcellular localization of full-length rat β-III-spectrin in HEK293T cells and reported that L253P β-III-spectrin localizes internally in cells, in contrast to wild-type β-III-spectrin, which localizes to the plasma membrane (29). In our studies, the mutant human L253P β-III-spectrin is also observed to accumulate internally in HEK293T cells, and at higher spatial resolution our analysis shows that internal L253P β-III-spectrin localizes to vesicular structures (Fig. 2A). We further demonstrate, using a fluorescent conjugate of phalloidin, that the L253P β-III-spectrin vesicles stain intensely positive for F-actin, suggesting that L253P β-III-spectrin on vesicles is bound to actin filaments. We speculate that colocalization of β-III-spectrin and actin on vesicular structures derives from the internalization of plasma membrane resulting from the high affinity of L253P β-III-spectrin for actin.
Fig. 2.
L253P β-III-spectrin accumulates on F-actin–positive vesicular membranes and is excluded from plasma membrane protrusions. (A) Confocal images of HEK293T cells transiently transfected with myc-epitope–tagged full-length human wild-type or L253P β-III-spectrin. Immunofluorescence staining showing accumulation of L253P β-III-spectrin vesicular membranes that also stain positive for F-actin, labeled with a phalloidin fluorescent conjugate. To a lesser extent, wild-type β-III-spectrin is observed on vesicular membranes. (Scale bar, 10 µm.) (B) Immunofluorescence staining showing the localization of β-III-spectrin proteins at the plasma membrane of HEK293T cells. (Left) Single z-stack slices through the middle of cells showing abundant plasma membrane localization of L253P β-III-spectrin, similar to wild-type β-III-spectrin. (Right) Corresponding z-stack projections showing that the cell surface, revealed by staining for wild-type β-III-spectrin, is highly textured with filopodium-like protrusions. Staining for L253P β-III-spectrin reveals, by comparison, a smooth cell surface in which filopodium-like extensions are not detected. (Scale bar, 10 µm.) (C) Immunofluorescence staining of HEK293T cells showing the cell surface localization of β-III-spectrin proteins relative to a GFP plasma membrane marker (PM-GFP). (Upper) Immunofluorescence staining showing that wild-type β-III-spectrin–positive filopodium-like extensions are similarly revealed by PM-GFP. Arrows indicate lamellipodium-like extensions that are also colabeled by wild-type β-III-spectrin and PM-GFP. (Lower) L253P β-III-spectrin is not present in filopodium-like or lamellipodium-like plasma membrane extensions revealed by PM-GFP. (Insets) Lamellipodium-like extensions, indicated by asterisks, in which contrast settings were adjusted to better show β-III-spectrin localization. (Scale bar, 10 µm.)
Unexpectedly, we also observed a striking difference in the localization pattern of wild-type versus L253P β-III-spectrin at the plasma membrane. In cells expressing wild-type β-III-spectrin, the spectrin protein is enriched in numerous, short filopodium-like structures that decorate the surface of HEK293T cells (Fig. 2B). Intriguingly, in cells transfected with the mutant β-III-spectrin, the distribution of spectrin at the plasma membrane is relatively “smooth” by comparison. This suggests that L253P β-III-spectrin either inhibits the formation of filopodium-like structures or is restricted from localizing to these membrane protrusions. To distinguish between these possibilities, a plasma membrane marker consisting of GFP fused to conserved myristoylation and palmitoylation sequences (PM-GFP) (30) was cotransfected with β-III-spectrin. In cells expressing wild-type β-III-spectrin, PM-GFP reveals a textured cell surface with filopodium-like structures that stain positive for β-III-spectrin (Fig. 2C). In cells expressing the mutant L253P β-III-spectrin, PM-GFP reveals that the cell surface is also textured with filopodium-like structures. However, L253P β-III-spectrin, while present at the plasma membrane, does not localize to these plasma membrane extensions. In addition, PM-GFP shows the presence of lamellipodium-like structures that contain wild-type β-III-spectrin but not L253P β-III-spectrin. The absence of L253P β-III-spectrin in these membrane protrusions suggests that high-affinity actin binding restricts β-III-spectrin from localizing to regions of the plasma membrane that are structurally dynamic. Instead, the mutation appears to cause preferential localization of β-III-spectrin to stable membrane regions, such as sites of cell–cell contact, where a robust colocalization with a cortical actin network is observed (Fig. S3).
Fig. S3.
Wild-type and L253P β-III-spectrin colocalize with F-actin at the cell cortex. Confocal images cutting through the middle of HEK293T cells transiently transfected with myc-epitope–tagged, full-length human wild-type or L253P β-III-spectrin. Immunofluorescence staining shows localization of full-length β-III-spectrin proteins at the plasma membrane boundaries between cells. Phalloidin staining reveals intense F-actin colocalization with L253P β-III-spectrin at the plasma membrane. (Scale bar, 10 µm.)
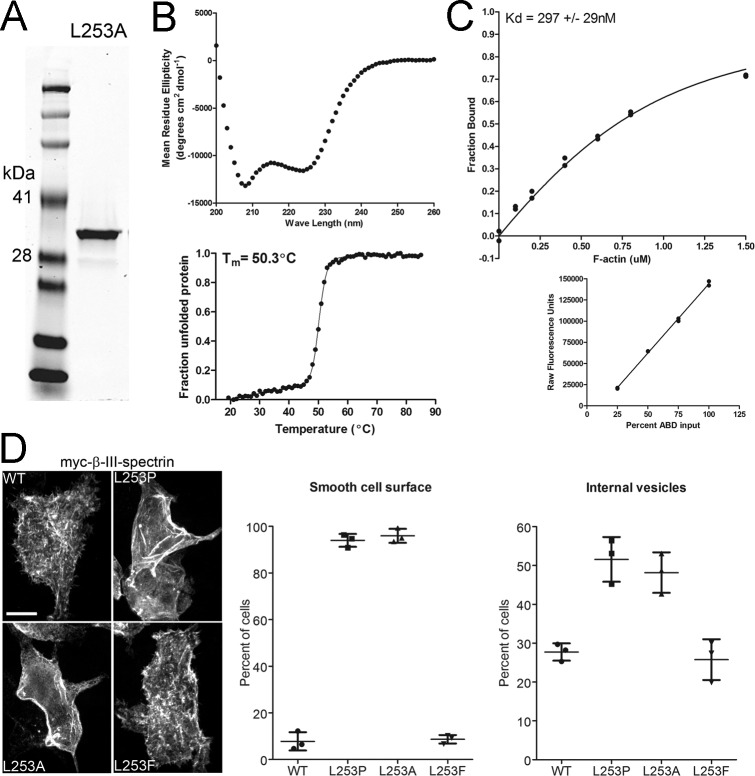
To further interrogate the mechanism(s) driving the altered subcellular localization of L253P β-III-spectrin, we also characterized the alanine variant of the ABD, L253A, in greater detail. First, we quantified the actin-binding affinity and stability of the variant. As performed previously for the wild-type and L253P ABD proteins, the L253A ABD was expressed in bacteria and purified (Fig. S4A). CD spectroscopy was performed to assess secondary structure; the CD spectrum for the L253A ABD shows a pronounced α-helical profile (Fig. S4B, Upper), as observed previously for the wild-type and L253P ABD proteins (28). CD spectroscopy was further employed to assess the thermal stability of the L253A ABD. Thermal melting curve analysis shows that the L253A ABD unfolds with a melting temperature (Tm) of 50.3 °C (Fig. S4B, Lower). The Tm of the L253A ABD is intermediate between the Tms of the wild-type (59.5 °C) and L253P (44.6 °C) ABD proteins, thus confirming that the alanine substitution is less destabilizing than the proline substitution. Further, F-actin cosedimentation reveals that the L253A ABD binds actin with a Kd of 297 nM (Fig. S4C), which is approximately fourfold weaker than the actin-binding affinity of the L253P ABD (Kd = 75.5 nM) but is still ∼250-fold higher than the actin-binding affinity of the wild-type ABD (75.8 µM). These biochemical analyses together with the cell-based binding assay (Fig. 1D) demonstrate that the L253A substitution causes high-affinity F-actin binding without perturbing protein folding at a physiological temperature.
Fig. S4.
The altered subcellular localization of L253P β-III-spectrin is a consequence of increased ABD-binding activity. (A–C) In vitro characterization of the L253A ABD. (A) A Coomassie blue-stained gel containing 2 µg of purified L253A ABD protein resolved by SDS/PAGE. (B) CD characterization of L253A ABD. (Upper) CD spectra between 200 and 260 nm showing a pronounced α-helical absorption profile. (Lower) CD melting curve analysis at 222 nm. The Tm is 50.3 °C. (C, Upper) Results of an individual F-actin cosedimentation assay performed using 0.8 µM ABD. The L253A ABD binds actin with a Kd of 297 ± 29 nM. The Kd is the average Kd ± SD from three cosedimentaton assays. (C, Lower) Standard curve to convert Coomassie blue fluorescence ABD band intensities to the amount of ABD protein in the F-actin cosedimentation assay. (D) The effect of different leucine 253 substitutions on the subcellular localization of β-III-spectrin in HEK293T cells. (Left) Confocal images of cells transfected with myc-epitope–tagged full-length human wild-type β-III-spectrin or the L253P, L253A, or L253F mutant. Immunofluorescence staining for wild-type or L253F β-III-spectrin proteins reveals a textured cell surface in which β-III-spectrin proteins are present in filopodium-like extensions. Staining for L253P and L253A β-III-spectrin proteins reveals a smooth cell surface in which β-III-spectrin proteins are not detected in filopodium-like extensions. (Middle and Right) Percentage of cells showing smooth cell surface (Middle) or β-III-spectrin vesicles (Right). For the cell-surface phenotype, each data point represents a single experiment in which 138–328 cells were scored for a smooth or textured cell surface. For the β-III-spectrin vesicle phenotype, each point represents a single experiment in which 221–336 cells were scored for the presence of β-III-spectrin vesicles. Error bars represent SD.
Next, we tested whether the L253A substitution results in the same β-III-spectrin subcellular localization changes in HEK293T cells as observed for the L253P mutation. In addition, the subcellular localization of full-length β-III-spectrin containing the L253F substitution, which did not cause increased actin binding (Fig. 1D), was included as a control. As observed for L253P β-III-spectrin, staining for β-III-spectrin in cells transfected with L253A β-III-spectrin reveals that nearly all (∼95%) transfected cells exhibit the smooth cell surface phenotype in which β-III-spectrin is present at the plasma membrane but not in filopodium-like structures (Fig. S4D). In contrast, most cells (>90%) transfected with wild-type or L253F β-III-spectrin show a textured cell surface. Further, ∼50% percent of cells expressing L253P or L253A β-III-spectrin exhibit β-III-spectrin internal vesicles, compared with ∼27% wild-type or L253F β-III-spectrin cells showing internal vesicles. The similarity in subcellular localization phenotypes between the L253P and L253A substitutions in β-III-spectrin strongly supports the conclusion that increased ABD-binding activity underlies the altered behavior of L253P β-III-spectrin in cells. We emphasize that, since the L253A β-III-spectrin is well-folded and binds actin with high affinity, the altered localization of mutant L253P β-III-spectrin in cells is not readily explained by a loss in ABD-binding activity. Our results point to the interesting possibility that high-affinity actin binding acts dominantly as a driver of L253P β-III-spectrin neurotoxicity and SCA5 pathology.
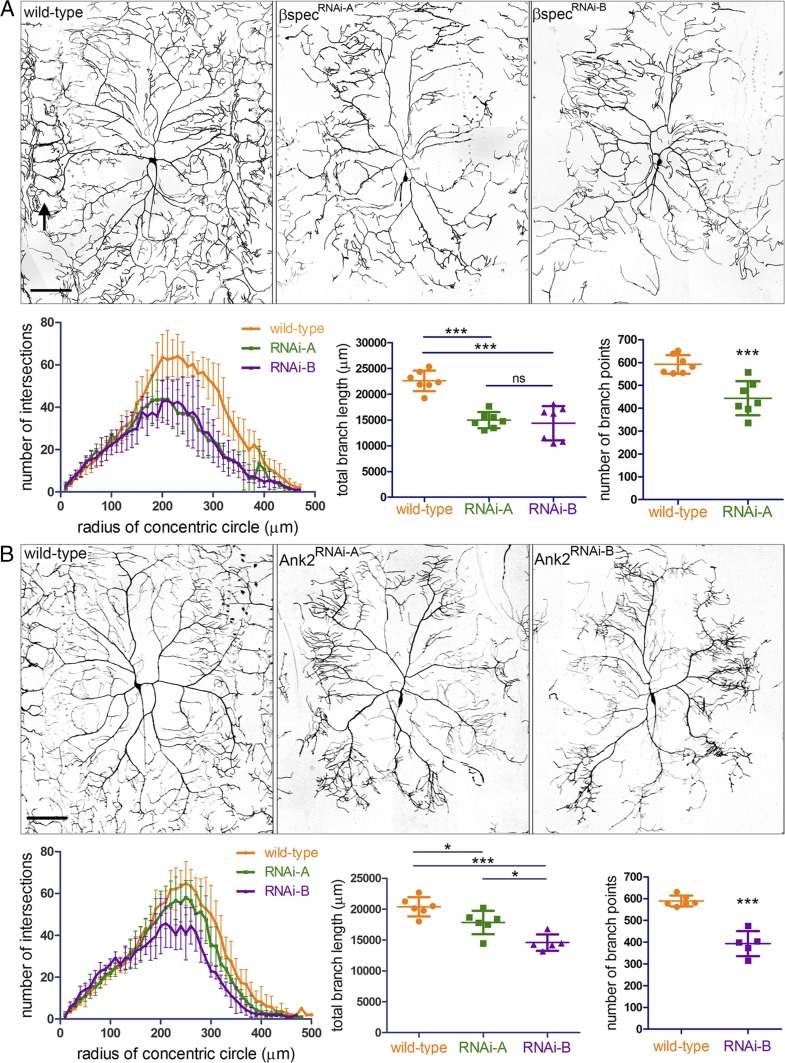
The neuronal population targeted in SCA5 pathogenesis is cerebellar Purkinje cells, which extend elaborate dendritic arbors required for control of body movements. To model how the L253P mutation impacts dendritic arborization, we performed studies in Drosophila class IV dendritic arborization (da) neurons that, like Purkinje cells, form complex dendritic arbors. We first determined if the endogenous Drosophila spectrin cytoskeleton is required for dendritic arborization of class IV da neurons. Using the ppk-Gal4 driver (31), UAS-RNAi transgenes targeting distinct sequences of β-spectrin mRNA were specifically expressed in class IV da neurons. The da neuron arbor morphologies were documented by live imaging of fully intact larvae at late third instar, a developmental stage when arbors are fully formed (Fig. 3A). Wild-type da neurons extend dendrites that reach body segment boundaries, where distal dendrites intertwine with dendrites of neighboring da neurons to form ring-like patterns (Fig. 3A, arrow). In contrast, neurons expressing β-spectrin RNAi show a clear loss of distal dendrites near segmental boundaries. This reduction in branches is reflected in Sholl analysis plots showing that β-spectrin RNAi causes decreased distal dendritic branch complexity and is demonstrated further in analyses showing that β-spectrin RNAi decreases total dendritic branch length and the number of branch points. In addition, we tested whether the neuron-specific spectrin adaptor protein ankyrin-2 is required for arborization. Ankyrin-2 is expressed as multiple splice forms (S, M, L, and XL) (32). Ankyrin-2 RNAi targeting all splice forms (RNAi-A) or targeting M, L, and XL splice forms (RNAi-B) causes reduced dendritic arborization (Fig. 3B). Like β-spectrin RNAi, ankyrin-2 RNAi decreases distal dendritic branch complexity and reduces total branch length and the number of branch points. Together, these data establish that the spectrin cytoskeleton is required for class IV da neuron dendritic arborization.
Fig. 3.
The Drosophila spectrin cytoskeleton is required for dendritic arborization. (A) Effect of β-spectrin RNAi on dendritic arborization of class IV da neurons. (Upper) Reconstructions of wild-type and β-spectrin RNAi dendritic arbors imaged by fluorescence confocal microscopy in third-instar larvae. In the wild-type arbor, the arrow at left indicates ring-like patterns formed by distal dendrites at body segment boundaries. β-Spectrin RNAi causes a loss of distal dendrites near segment boundaries. (Scale bar, 100 µm.) (Lower Left) Sholl analysis plots showing β-spectrin RNAi decreases distal dendritic complexity. For all samples n = 7 neurons. (Lower Middle) β-Spectrin RNAi decreases total branch length. ***P < 0.001; ns, not significant. (Lower Right) β-Spectrin RNAi decreases number of branch points. P = 0.0006. (B) Effect of ankyrin-2 RNAi on dendritic arborization. (Upper) Reconstructions of wild-type and ankyrin-2 RNAi dendritic arbors. (Scale bar, 100 µm.) (Lower Left) Sholl analysis plots showing that ankyrin-2 RNAi decreases distal dendritic complexity. For wild-type or RNAi-A, n = 6 neurons. For RNAi-B, n = 5 neurons. (Lower Middle) Ankyrin-2 RNAi decreases total branch length. *P < 0.05. ***P < 0.001. (Lower Right) Ankyrin-2 RNAi deceases the number of branch points. P < 0.0001. Error bars represent SD in all graphs.
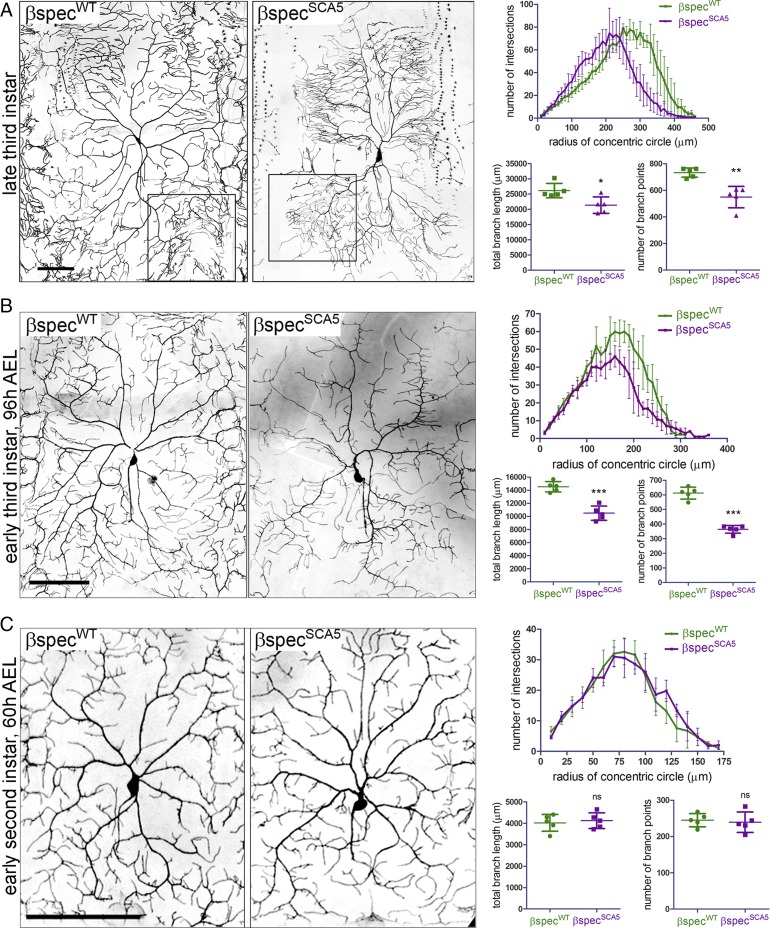
To examine the effect of the L253P SCA5 mutation on dendritic arborization, we generated transgenic Drosophila lines in which UAS-transgenes containing wild-type β-spectrin or L246P β-spectrin (L246P is equivalent to the human L253P, and herein is termed “SCA5”) are inserted into the same genomic locus to control for equal transgene expression. The 477-Gal4 driver (33), a weaker driver than ppk-Gal4, was employed to express wild-type or SCA5 β-spectrin specifically in class IV da neurons. In late-third-instar larvae, neurons expressing SCA5 β-spectrin (βspecSCA5) show a pronounced reduction in dendritic arbor size, with a complete loss of distal dendrites near segmental boundaries (Fig. 4A). As for β-spectrin RNAi and ankyrin-2 RNAi, this loss in distal dendrites is reflected in Sholl analysis plots showing that SCA5 β-spectrin causes decreased branch complexity at locations far from the soma. Also, similar to β-spectrin RNAi and ankyrin-2 RNAi, SCA5 β-spectrin reduces total branch length and the number of branch points. We further noted that while SCA5 β-spectrin causes a loss of dendrites at distal segmental boundaries and an overall reduction in branch points, high-complexity terminal branching is present near the ends of primary dendrites where complex terminal branching is observed in control neurons (Fig. 4A, boxed regions). Intriguingly, this feature distinguishes the SCA5 da neuron phenotype from the SCA1 and SCA3 phenotypes in which all terminal branching in the arbor is lost (34). Similar to the SCA5 arbor phenotype, complex terminal branching is also observed following ankryin-2 RNAi but is not pronounced after depletion of β-spectrin by RNAi.
Fig. 4.
Drosophila SCA5 β-spectrin decreases dendritic arborization. Reconstructions of class IV da neuron dendritic arbors imaged by fluorescence confocal microscopy in living, fully intact larvae at different developmental stages. Da neurons expressed wild-type Drosophila β-spectrin (βspecWT) or SCA5 (L246P) β-spectrin (βspecSCA5), together with the fluorescent membrane marker CD4tdGFP. (A, Left) Reconstructions of class IV da neuron dendritic arbors imaged by fluorescence confocal microscopy in late-third-instar larvae. Boxed regions highlight complex branching at the ends of primary dendrites observed in both SCA5 and wild-type β-spectrin neurons. (Scale bar, 100 µm.) (Right) Quantitation of arbor phenotypes. (Upper) Sholl analysis plots showing that SCA5 β-spectrin causes a loss of distal branch complexity. (Lower) SCA5 β-spectrin decreases total branch length (*P = 0.0172) and the number of branch points (**P = 0.0053). (B, Left) Reconstructions of class IV da neuron dendritic arbors imaged by fluorescence confocal microscopy in early-third-instar larvae, 96 h AEL. (Scale bar, 100 µm.) (Right) Quantitation of arbor phenotypes. (Upper) Sholl analysis plots showing that SCA5 β-spectrin causes a loss of distal branch complexity. (Lower) SCA5 β-spectrin decreases total branch length and the number of branch points (***P < 0.0001). (C, Left) Early second instar larvae, 60 h AEL. (Scale bar, 100 µm.) (Right) Quantitation of arbor phenotypes. (Upper) Sholl analysis plots showing a similar distribution in branch complexity for neurons expressing wild-type and SCA5 β-spectrin. (Lower) Wild-type and SCA5 β-spectrin–expressing neurons are not different in total branch length [P = 0.6757, not significant (ns)] or in number of branch points (P = 0.7287, ns). For all samples, n = 5 (one neuron imaged in each of five larvae). Error bars represent SD in all graphs.
To gain insight into whether the SCA5 arbor phenotype reflects a degenerative or a developmental defect, we documented neuronal morphologies at earlier stages of development. At the earliest larval stage investigated, second instar [60 h after egg laying (AEL)], the dendritic arbors in larvae expressing βspecWT and βspecSCA5 appear identical, with no significant difference in Sholl analysis plots, total branch length, or the number of branch points (Fig. 4C). The dendritic arbors in βspecWT and βspecSCA5 second-instar larvae are similar to the arbors of βspecWT late-third instar-larvae, covering the full body segment with dendrites extending to the segmental boundaries. By comparison, at an intermediate developmental stage, early third instar (96 h AEL), the arbor fields for larvae expressing βspecWT and βspecSCA5 are clearly distinguishable (Fig. 4B). In βspecSCA5 early-third-instar larvae arbors show decreased dendrites near segmental boundaries, accounting for the reduced distal complexity in the Sholl analyses. Moreover, in βspecSCA5 early-third-instar larvae the arbors have reduced total branch length and number of branch points. Progressing from early to late third instar, arbors continue to grow in the presence of βspecSCA5, indicated by increases in total branch length and branch points. However, growth appears restricted to regions proximal to the soma. Further, progression from early to late third instar appears to be associated with a continued loss of distal dendrites near segmental boundaries. Taking these findings together, we conclude that the SCA5 arbor phenotype results from both a developmental defect, initiating after early second instar, in which branch growth is restricted to regions close to the soma, and a degenerative defect, in which distal dendrites present at segmental boundaries are eliminated.
To test whether the SCA5 mutation alters the subcellular localization of Drosophila β-spectrin in da neurons, additional transgenic lines containing wild-type or SCA5 β-spectrin fused to the fluorescent protein mEos3.2 (35) were generated. Wild-type Drosophila β-spectrin with the N-terminal mEos3.2 fusion retains function in neurons, as expression of the mEos3.2-βspecWT transgene using the elav-Gal4 pan-neuronal driver rescues the lethality of a loss-of-function allele, βspecem21 (36) to a similar extent as the untagged wild-type transgene βspecWT (Table S1). This is consistent with prior work demonstrating that pan-neuronal expression of β-spectrin containing an N-terminal myc-epitope tag likewise rescues lethality caused by loss of β-spectrin function (37). In contrast, neither the mEos3.2-βspecSCA5 nor βspecSCA5 transgene rescues lethality caused by the βspecem21 allele. Furthermore, animals heterozygous for the βspecem21 allele are viable, with or without βspecWT or mEos3.2-βspecWT expression. Importantly, in βspecem21 heterozygous animals, expression of either βspecSCA5 or mEos3.2-βspecSCA5 causes lethality. These results indicate that Drosophila β-spectrin is not haploinsufficient and that mutant L253P β-III-spectrin acts dominantly. Moreover, the mEos3.2 tag does not interfere with the dominant neurotoxic property conferred by the SCA5 mutation.
Table S1.
Rescue of em21 lethal mutation by untagged and mEos3.2-tagged β-spectrin transgenes
| Progeny class: total number of adults | ||||
| Transgene | em21, elav-Gal4/Y; transgene/+ | FM7/Y; transgene/+ | em21, elav-Gal4/+; transgene/+ | FM7/+; transgene/+ |
| attP154 | 0 | 99 | 100 | 107 |
| UAS-βspecWT | 17 | 145 | 150 | 144 |
| UAS-mEos3.2-βspecWT | 26 | 119 | 108 | 130 |
| UAS-βspecSCA5 | 0 | 152 | 2 | 177 |
| UAS-mEos3.2-βspecSCA5 | 0 | 196 | 4 | 206 |
Males homozygous for UAS-βspec transgenes or the third chromosome attP154 landing site, into which UAS-βspec transgenes were inserted, were crossed to females carrying the em21, elav-Gal4 recombinant chromosome balanced over the FM7 chromosome.
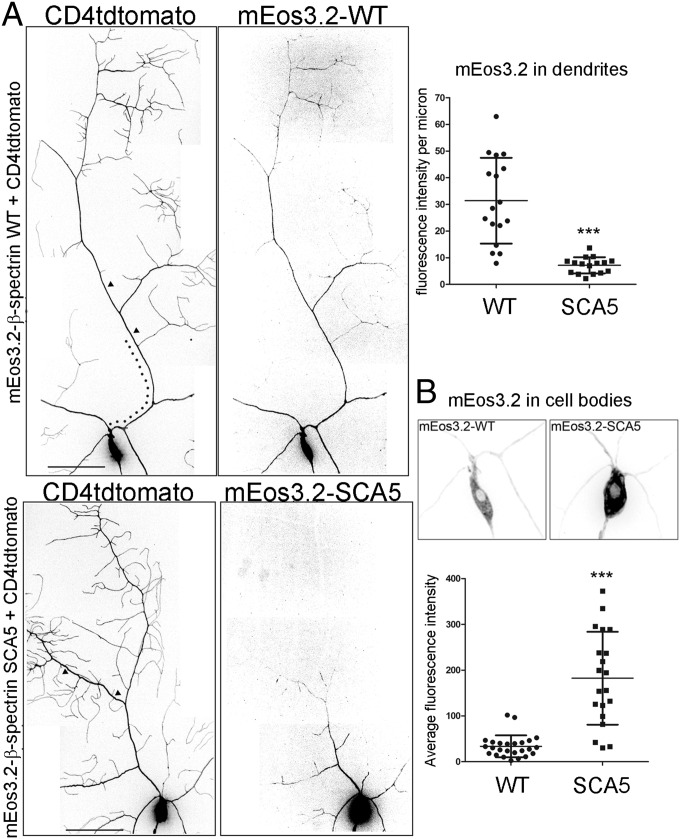
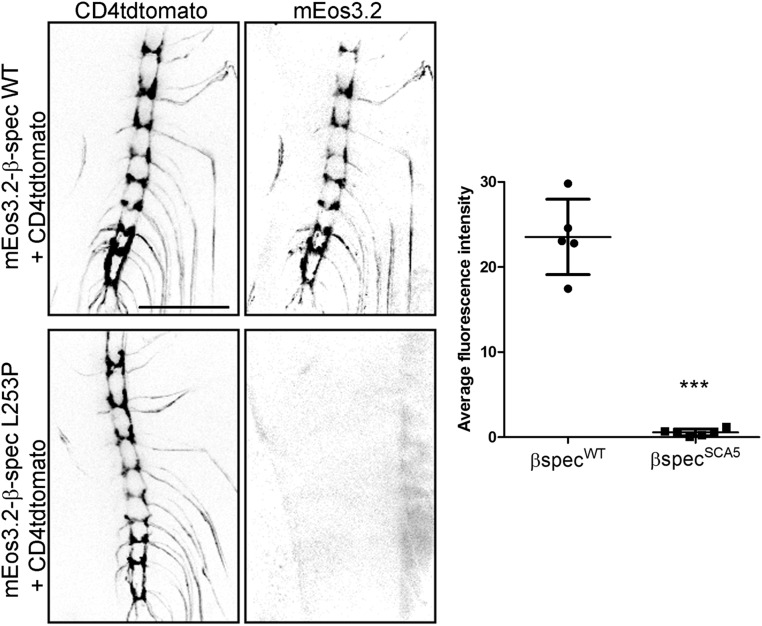
To assess the effect of the mutation on β-spectrin subcellular localization, live imaging of mEos3.2-β-spectrin was performed in da neurons. Wild-type β-spectrin is present throughout the dendritic arbor, in addition to localizing to the soma and axon (Fig. 5). In contrast, SCA5 β-spectrin is strikingly absent at distal dendritic locations and is detectable only within proximal branches of the arbor and in the soma, where it shows approximately sixfold accumulation compared with wild-type. Significantly, the SCA5 mutation not only restricts β-spectrin localization near the soma but also generates a proximal shift in dendritic branching. This suggests that the spectrin cytoskeleton in distal dendrites is critical for normal arborization. Moreover, the impact of the SCA5 mutation on β-spectrin localization is not confined to the somatodendritic compartment but extends to the axon, where SCA5 spectrin is localized to the proximal axon but is absent from the axon terminal (Fig. S5). The altered subcellular distribution of SCA5 β-spectrin in da neurons suggests that the high-affinity actin binding constrains the expansion of the spectrin-actin cytoskeleton from the soma into the distal axon and distal dendritic branches.
Fig. 5.
SCA5 β-spectrin is absent in distal dendrites and accumulates in the soma. (A) Confocal-image arbor reconstructions of class IV da neurons expressing mEos3.2-β-spectrin wild-type (WT) or SCA5 proteins, together with the fluorescent membrane marker CD4tdtomato. (Left) Fluorescence signals of CD4tdtomato and mEos3.2-β-spectrin proteins. Wild-type mEos3.2-β-spectrin localizes throughout arbor and soma. mEos3.2-β-spectrin SCA5 is absent in distal dendrites but is present in proximal dendrites. (Scale bar, 50 µm.) (Right) mEos3.2-β-spectrin fluorescence intensities were quantified in 50-µm segments of a primary dendrite (arrowheads in Left) located 100 µm (dotted line in Left) from the soma. mEos3.2-β-spectrin SCA5 localization is significantly reduced in the distal regions of primary branches. For wild type, n = 17 neurons (one to three neurons imaged per larva). For SCA5, n = 16 neurons (one to three neurons imaged per larva). ***P < 0.0001. (B) Images of wild-type mEos3.2-β-spectrin and SCA5 fluorescence signals in somata of neurons shown in A. Somata are magnified 2× and contrast settings were adjusted to compare signals in somata. mEos3.2-β-spectrin SCA5 accumulates in the soma. For wild type, n = 26 neurons (one to three neurons per larva). For SCA5, n = 21 neurons (one to three neurons per larva). ***P < 0.0001. Error bars represent SD in all graphs.
Fig. S5.
SCA5 β-spectrin is absent at da neuron axon terminals. (Left) Confocal images of ventral nerve cords showing the axon terminals of class IV da neurons expressing mEos3.2-β-spectrin proteins and the fluorescent membrane marker CD4tdtomato. Wild-type β-spectrin is present in axon terminals, but SCA5 β-spectrin is not. (Scale bar, 100 µm.) (Right) Quantitation of mEos3.2 signals at the ventral nerve cord. ***P = 0.0003; n = 5 for wild-type; n = 6 for SCA5. Error bars represent SD.
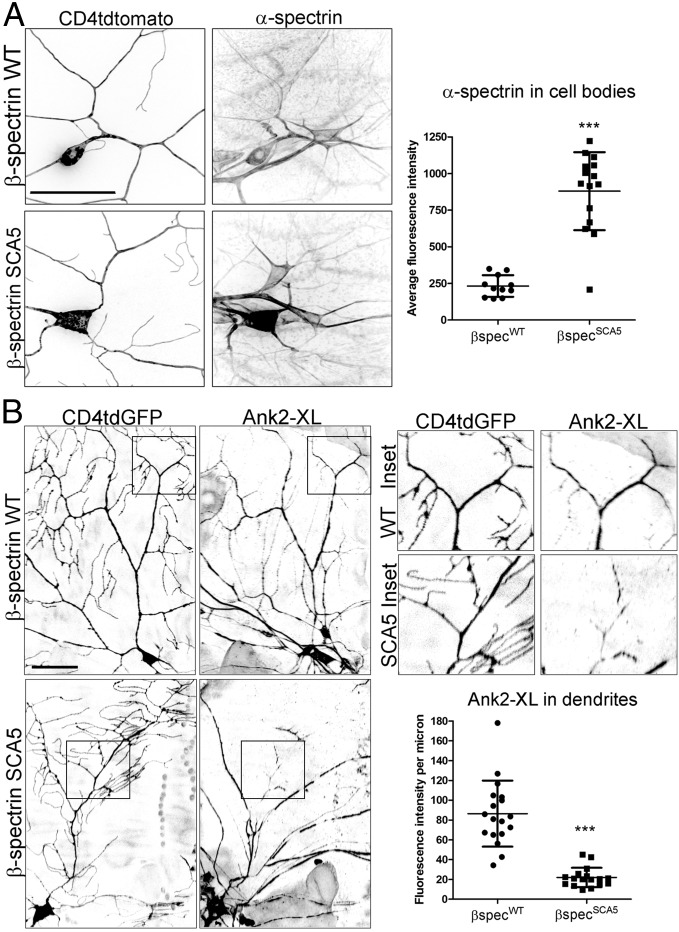
We next tested if SCA5 β-spectrin acts dominantly to mislocalize the endogenous spectrin cytoskeleton. We assessed the subcellular localization of α-spectrin, taking advantage of a Drosophila line containing a GFP insertion in the endogenous α-Spec locus (38), resulting in a functional α-spectrin-GFP fusion protein. In control neurons, α-spectrin is present in the soma and dendrites of class IV da neurons labeled with CD4tdtomato (Fig. 6A). However, neurons expressing SCA5 β-spectrin show a striking accumulation of α-spectrin in the soma, mirroring the mislocalization of mutant β-spectrin itself. High expression of endogenous α-spectrin in epidermal cells, innervated by da neuron dendrites, prevented quantitation of α-spectrin in dendrites. To determine how SCA5 β-spectrin affects the localization of the endogenous spectrin cytoskeleton in dendrites, we assessed the localization of neuron-specific ankyrin-2 using an antibody that specifically recognizes the XL splice form. In control class IV da neurons, ankyrin-2 XL is present in both proximal and distal regions of the arbor (Fig. 6B). In neurons expressing SCA5 β-spectrin, ankyrin-2 XL localization in distal dendrites is significantly decreased.
Fig. 6.
SCA5 β-spectrin dominantly mislocalizes the endogenous spectrin cytoskeleton. (A, Left) Confocal images showing the localization of endogenous α-spectrin in the somata and dendrites of class IV da neurons expressing wild-type (WT) or SCA5 β-spectrin together with the fluorescent membrane marker CD4tdtomato. α-Spectrin can also be seen in the somata and dendrites of neighboring sensory neurons not expressing β-spectrin transgenes. (Scale bar, 50 µm.) (Right) Quantitation of α-spectrin in cell bodies. P < 0.0001. For wild-type, n = 11 neurons. For SCA5, n = 15. (B, Left) Confocal images showing the dendritic localization of endogenous ankyrin-2 XL in class IV da neurons expressing wild-type (WT) or SCA5 β-spectrin together with CD4tdGFP. Ankyrin-2 XL localization can also be seen in the dendrites of neighboring sensory neurons. Enlarged views (2.16× magnification) of boxed areas are shown at Upper Right. (Lower Right) In control class IV da neurons, ankyrin-2 XL is present in proximal and distal regions of the arbor. In neurons expressing SCA5 β-spectrin, ankyrin-2 localization is reduced in distal dendrites. (Scale bar, 50 µm.) P < 0.0001. For wild-type, n = 18 neurons. For SCA5, n = 17. Error bars represent SD in all graphs.
Discussion
The morphology of dendritic arbors dictates the connectivity of neuronal networks, integrating inputs and propagating signals (39, 40). The question of how neurons modulate dendritic morphology is of keen interest in the study of neuronal function and neurodegeneration (41–43). For example, the molecular and cell biological mechanisms that control branch stability and remodeling within a dendritic field remain largely elusive (44, 45). In this report, we determined the consequence of a SCA5 mutation on the binding of β-III-spectrin to actin in mammalian cells and leveraged the Drosophila model system to reveal the impact of the SCA5 disease mutation on the neuronal spectrin-actin cytoskeleton and dendritic arborization. This work identifies an important cytoskeletal mechanism in distal dendrites required for formation of large, complex arbors, critical to the function of Purkinje cells targeted in hereditary ataxias.
Our data suggest that high-affinity actin binding acts dominantly as a driver of L253P β-III-spectrin neurotoxicity by impacting the dynamics of the spectrin-actin network. We observed that Drosophila SCA5 β-spectrin containing the equivalent L253P mutation accumulates in the da neuron soma and is absent in distal dendritic regions, in contrast to wild-type β-spectrin that localizes throughout the arbor. In the axons of mammalian neurons the spectrin-actin lattice initially forms near the soma and propagates distally (16), suggesting that the loss of Drosophila SCA5 β-spectrin in distal dendrites reflects a defect in expansion of the spectrin-actin cytoskeleton from the soma into dendrites. Such an expansion defect may be a consequence of a slow dissociation rate that is typical of high-affinity molecular interactions (46). Specifically, high-affinity actin binding caused by the mutation may limit the pool of free β-spectrin molecules available to be recruited to an expanding cytoskeleton. Like the loss of Drosophila SCA5 β-spectrin in da neuron dendritic extensions, we observed a reduction of human L253P β-III-spectrin in HEK293T cell plasma membrane protrusions. The absence of L253P β-III-spectrin in filopodium-like and lamellipodium-like extensions, despite abundant localization elsewhere at the plasma membrane, suggests a partitioning between structurally dynamic and stable membrane regions. This partitioning supports the idea that high-affinity actin binding reduces the availability of β-III-spectrin to be recruited to newly formed membrane structures. Our data predict that high-affinity binding of L253P β-III-spectrin to actin filaments within the neuronal spectrin-actin lattice negatively impacts Purkinje cell arborization and function by impeding the expansion of the spectrin-actin cytoskeleton in dynamically growing or remodeling dendritic branches and spines.
In addition to increasing actin-binding affinity, the L253P mutation destabilizes β-III-spectrin, causing the ABD to begin to unfold near physiological temperature in vitro. This denaturation raised the possibility that cellular phenotypes in mammalian cells are the consequence of ABD protein unfolding and loss of ABD-binding activity rather than elevated ABD-binding activity. Our experiments do not support this interpretation, showing instead through co-IP assays that the mutant ABD retains high-affinity actin binding in cells. Indeed, in cultured mammalian cells, protein unfolding reflected in minor degradation products was detected only when the mutant ABD was highly overexpressed. Significantly, the high-affinity actin binding observed for the L253P mutation is mimicked by the alternative substitution, L253A, which, like the L253P mutation, is predicted to disrupt the normal hydrophobic contacts of leucine 253 in the β-III-spectrin ABD. In this case, no degradation of the L253A mutant is detected, and we confirmed in vitro the increased protein stability of the L253A mutation. In light of these results, our observation that the L253A and L253P mutations cause the same β-III-spectrin subcellular localization phenotypes in HEK293T cells supports the conclusion that the behavior of L253P β-III-spectrin is driven by increased ABD-binding activity. These results support the hypothesis that high-affinity actin binding contributes to the L253P β-III-spectrin neurotoxicity that underlies SCA5 pathology.
How do the L253P and L253A substitutions account for elevated actin-binding affinity? The location of the mutations in the CH2 domain is consistent with a suspected regulatory role for the domain in mediating actin binding. Biochemical studies of the isolated CH domains of β-spectrin or of the related α-actinin ABD previously documented actin-binding activity for the CH1 domain but not for the CH2 domain (47–49). Confinement of binding activity to the CH1 domain is further supported by a structural model for α-actinin ABD-actin complexes in which only a single CH domain is bound to actin filaments (50). Consistent with the idea that the L253P mutation disrupts a CH2 domain regulatory function, leucine 253 in the CH2 domain is predicted to interface with the CH1 domain and physically bridge the two domains through hydrophobic contacts (28). The decrease in hydrophobicity introduced by the L253P or L253A substitution is thus predicted to disrupt inter-CH domain contacts and relieve CH2 inhibition. Significantly, disease-causing mutations located in the CH2 domain of α-actinin (51, 52) or filamin (53–55) also increase actin-binding affinity.
In addition to binding filaments of conventional actin, the β-III-spectrin ABD also interacts with ARP1 (56), a component of the dynactin complex that facilitates transport mediated by microtubule motors. Consistent with an ARP1 interaction, we previously reported that expression of SCA5 β-spectrin in Drosophila motoneurons impairs axonal transport (57). Given the ∼75% similarity in actin and ARP1 primary structures, we predict the L253P mutation will similarly enhance the binding of β-III-spectrin to ARP1. Our current studies have not directly addressed this prediction. However, in a previous study, Clarkson et al. (29) conducted a bimolecular fluorescence complementation assay in HEK293T cells overexpressing ARP1, concluding that the L253P mutation reduces the interaction of β-III-spectrin ABD with ARP1. Nonetheless, a direct test of how L253P β-III-spectrin impacts ARP1 binding is lacking. Indeed, ARP1-binding studies are not straightforward; the native ARP1 filament is difficult to purify, ARP1-specific antibodies are not available, and there is a strong propensity for ARP1 to form nonnative structures when overexpressed in cells (58). We believe further experiments are needed to fully understand the impact of the L253P mutation on ARP1 binding and intracellular vesicular transport.
In Drosophila, the lone homolog of β-III-spectrin, β-spectrin, localizes to both dendrites and axons, where the function of the spectrin cytoskeleton has been extensively studied (32, 59, 60). We previously reported the impact of SCA5/L246P β-spectrin expression on synaptic organization at the neuromuscular junction (NMJ) (57). Spectrin RNAi also disrupts synaptic bouton organization but further leads to NMJ retraction (59), a phenotype not observed in SCA5 motoneurons (57). In the current work, we show that SCA5 β-spectrin is absent not only in distal dendrites but also at the axon terminus of da neurons. Thus, SCA5 β-spectrin may dominantly mislocalize the spectrin cytoskeleton not only in dendrites but also in distal axons. In light of these findings, the reduced size of the Drosophila NMJ reported in motor neurons expressing SCA5 β-spectrin (57) may in part reflect a disruption of the spectrin cytoskeleton and associated ankyrin-2.
This work points to a molecular mechanism in the somatodendritic compartment of neurons that enables the formation of large, complex dendritic arbors. As the dendritic arbor grows, dynamic actin assembly in the distal dendrites drives the formation of new terminal branches (61). We suggest that during arbor growth expansion of the spectrin-actin cytoskeleton is required to stabilize terminal branches and allow for continued expansion of the arbor. To explain the mutant SCA5 arbor phenotypes, we propose that high-affinity binding of the mutant β-spectrin decreases spectrin-actin dynamics and consequently constrains expansion of the spectrin-actin cytoskeleton and stabilization of growing dendrites. The spectrin cytoskeleton has similarly been implicated in axonal growth (16) and stabilization of synaptic junctions (59). In support of this cytoskeletal mechanism regulating dendritic arbor stability and potentially underlying SCA5 pathology, we show that the loss of β-spectrin, as well as ankyrin-2, in the distal dendrites of Drosophila da neurons correlates with a proximal shift in dendritic branching. Importantly, expression of mutant SCA5 β-spectrin and ankyrin-2 RNAi resulted in similar dendritic arborization defects, and SCA5 β-spectrin causes a loss of ankyrin-2 XL in distal dendrites. We characterize a progressive elimination of distal dendrites at segmental boundaries in SCA5 arbors. Moreover, lacking expansion of the spectrin-actin cytoskeleton in terminal dendrites, dynamic actin-based assembly drives complex terminal branching at the periphery of SCA5 arbors. However, the stability of the SCA5 terminal branching is compromised, and the outward growth of the arbor field is defective. One possibility is that impaired expansion of the spectrin-actin cytoskeleton and loss of ankyrin-2 in dendrites impacts localization of neuroglian, a cell-adhesion molecule required for arborization (62) and which may mediate stability of dendritic branching in Drosophila da neurons (59). In Purkinje cells, we predict that L253P β-III-spectrin will similarly impair expansion of the spectrin-actin lattice, disrupting dendritic localization of critical membrane proteins ankyrin-R (27), EAAT4 (4, 6), and mGluR1α (11), and in consequence promoting defects in arborization and postsynaptic signaling that characterize SCA5 pathology.
Significantly, our model for the impact of a SCA5 mutation on cytoskeletal dynamics and distal arborization is similar to a disease model proposed for autosomal recessive spastic ataxia of Charlevoix–Saguenay (ARSACS) in which a decrease in mitochondrial dynamics is suggested to disrupt distal Purkinje cell arborization (63). Like the mislocalization of SCA5 β-spectrin in da neurons, loss of function of the ARSACS disease protein sacsin, a mitochondrial protein, causes mitochondria to accumulate in the soma and proximal dendrites but fail to reach distal dendrites in mammalian neurons. Neurons such as Purkinje cells and da neurons that extend complex arbors appear to be especially vulnerable to disruptions to pathways in distal dendrites that support arborization, and this sensitivity possibly explains the cerebellar specificity of SCA5 pathology.
Materials and Methods
Experimental procedures for mammalian cell culture studies, in vitro ABD protein characterization, and Drosophila studies are described in detail in SI Materials and Methods. Experiments in these studies were approved by the Institutional Biosafety Committee at the University of Minnesota. The DNA constructs for human β-III-spectrin used in our experiments are based on a wild-type human cDNA clone from an available collection and were not derived from patients.
SI Materials and Methods
Experiments in these studies were approved by the Institutional Biosafety Committee at the University of Minnesota. The DNA constructs for human β-III-spectrin used in our experiments are based on a wild-type human cDNA clone from an available collection and were not derived from patients.
Mammalian Cell Culture Studies.
Expression constructs.
For myc-ABD co-IP experiments, the human wild-type β-III-spectrin ABD or L253P coding sequence (amino acids 1–284) containing an N-terminal myc-epitope tag fusion was PCR amplified using the pUASp-hSPWT or pUASp-hSPGM template (57) and the oligonucleotides GCG GGT ACC ACC ATG GAA CAA AAA CTC ATC and AAA TCT AGA CTA CTT CAT CTT GGA GAA GTA ATG GTA GTA AG. PCR products were digested with Kpn1 and Xba1 restriction enzymes and ligated into pcDNA3.1(+) digested with the same enzymes, resulting in the wild-type pcDNA3.1-myc-ABD or L253P constructs. Additional leucine 253 substitutions were generated by site-directed mutagenesis using wild-type pcDNA3.1-myc-ABD as the template and the oligonucleotides (L253M sense) CTG GGA CTT ACC AAG ATG CTG GAT CCC GAA GAC G, (L253M antisense) CGT CTT CGG GAT CCA GCA TCT TGG TAA GTC CCA G; (L253F sense) CTG GGA CTT ACC AAG TTT CTG GAT CCC GAA GAC G, (L253F antisense) CGT CTT CGG GAT CCA GAA ACT TGG TAA GTC CCA G; (L253G sense) CTG GGA CTT ACC AAG GGG CTG GAT CCC GAA GAC G, (L253G antisense) CGT CTT CGG GAT CCA GCC CCT TGG TAA GTC CCA G; (L253A sense) CTG GGA CTT ACC AAG GCG CTG GAT CCC GAA GAC G, and (L253A antisense) CGT CTT CGG GAT CCA GCG CCT TGG TAA GTC CCA G. Resulting constructs were pcDNA3.1-myc-ABD L253M, L253F, L253G, and L253A. For subcellular localization experiments, the coding sequence for full-length human wild-type β-III-spectrin or L253P, with an N-terminal myc-epitope tag, was isolated from pUASp-hSPWT or pUASp-hSPGM using Kpn1 and Xba1 restriction enzymes. β-III-Spectrin DNAs were ligated into pcDNA3.1+ using the same restriction enzymes to generate the final wild-type pcDNA3.1-myc-β-III-spectrin and L253P constructs. To generate L253A and L253F variants, site-directed mutagenesis was performed using template DNA consisting of a 5′ Kpn1-Age1 β-III-spectrin DNA fragment, which was contained in pcDNA3.1, and the L253A sense/antisense or L253F sense/antisense oligonucleotides. Following mutagenesis, the 5′ β-III-spectrin fragments were isolated from pcDNA3.1 using Kpn1 and Age1 restriction enzymes and were ligated into wild-type pcDNA3.1-myc-β-III-spectrin, digested with the same restriction enzymes. All DNA sequences were verified. PM-GFP was a gift from Tobias Meyer, Stanford University, Stanford, CA (plasmid #21213; Addgene). pcDNA3.1-eGFP was generously provided by the G. W. Gant Luxton laboratory at the University of Minnesota-Twin-Cities, Minneapolis.
Cell culture.
HEK293T cells were grown in DMEM (Thermo Fisher Scientific) containing 4.5 g/mL glucose and 0.862 g/L l-alanyl-glutamine and supplemented with 10% FBS and penicillin-streptomycin. Cells were maintained at 37 °C and 5% CO2. For all experiments, HEK293T cells were dissociated with TrypLE (Thermo Fisher Scientific) and plated into tissue culture-treated six-well plates at a density of 0.5 × 106 cells per well. Twenty-four hours after plating, at 60–70% cell confluency, transient transfections were performed using Lipofectamine 3000 (Thermo Fisher Scientific). Following the manufacturer’s recommendations, transfection reactions for single wells contained 2.5 µg total DNA, 5 µL Lipofectamine 3000, and 5 µL P3000 reagent, diluted in Opti-MEM (Gibco) to a total volume of 250 µL.
Coimmunoprecipitation assays.
In Fig. 1A, HEK293T cells in six-well microplates were transfected with 125, 375, or 1.25 µg of wild-type pcDNA3.1-myc-ABD or L253P, 250 ng pcDNA3.1-eGFP as transfection control, and pcDNA3.1(+) filler to a total of 2.5 µg DNA per well. In Fig. 1D, cells were transfected with 750 ng wild-type pcDNA3.1-myc-ABD or L253 variant, 250 ng pcDNA3.1-eGFP, and pcDNA3.1(+)to a total of 2.5 µg DNA per well. In Fig. S1, cells were transfected with 750 ng wild-type pcDNA3.1-myc-ABD or L253P, 250 ng pcDNA3.1-eGFP, and pcDNA3.1(+)to a total of 2.5 µg DNA per well. At 24 h posttransfection, the medium was aspirated, and cells were washed once with 1 mL ice-cold PBS. Cells were lysed for 15 min in 750 µL ice-cold Triton immunoprecipitation (IP) buffer containing 20 mM Tris (pH 7.5), 150 mM NaCl, 1% Triton X-100, and 5% glycerol, supplemented with protease inhibitors (cOmplete mini protease inhibitor tablet, EDTA-free; Roche). Lysates were transferred to prechilled microtubes and clarified at 16,000 × g for 10 min at 4 °C. Supernatants were transferred to prechilled microtubes and clarified for 30 min at 4 °C, with inversion, with Sepharose 4B beads (Sigma) equilibrated in Triton IP buffer. Sepharose beads were pelleted at 750 × g, 4 °C, and supernatants were transferred to prechilled microtubes. Samples (75 µL) of clarified lysates were combined with 75 µL 2× Laemmli buffer for the SDS/PAGE and Western blot analyses described below. One microgram of α-myc A-14 antibody (Santa Cruz Biotechnology) was added to remaining lysates. In Fig. S1, control IP samples received 1 µg normal rabbit IgG (Cell Signaling Technology) or no antibody. Lysate samples were incubated with antibody for 1 h at 4 °C with inversion. Protein A agarose (Roche) equilibrated in Triton IP buffer was added to lysates and incubated for 1 h at 4 °C with inversion. Beads were pelleted at 750 × g, 4 °C, and were washed four times in 1 mL Triton IP buffer. After the final washing, 110 µL of 1× Laemmli buffer was added to the beads, and samples were boiled for 2 min. Proteins were resolved by SDS/PAGE and transferred to Immobilon-FL membranes (Millipore). Membranes were blocked in 1× PBS-L (10× PBS-L: 90 g NaCl, 20 g Na2HPO4, 8.3 g NaH2PO4⋅H2O, dissolved in 1 L H2O) with 1% casein (Hammersten; Affymetrix). Membranes were probed overnight at 4 °C with primary antibodies diluted in PBS-L plus 1% casein plus 0.1% Tween 20. Primary antibodies were α-myc 9E10 (1:1,000; Santa Cruz Biotechnology), α-actin JLA20 (1:2,000; Developmental Studies Hybridoma Bank), and α-GFP (1:2,000; Millipore). Primary antibodies were removed by washing with PBS-L plus 0.1% Tween 20. Membranes were probed for 1 h at room temperature with secondary antibodies diluted in PBS-L plus 1% casein plus 0.1% Tween 20 plus 0.01% SDS. Secondary antibodies were α-mouse IRDye 800CW (1:5,000; LI-COR Biosciences) and α-rabbit IRDye 680RD (1:5,000; LI-COR Biosciences). Secondary antibodies were removed by washing with PBS-L plus 0.1% Tween 20. Membranes were imaged using 700- and 800-nm channels in an Odyssey Imager (LI-COR Biosciences).
Solubility and degradation analysis of full-length β-III-spectrin proteins.
In Fig. S2, HEK293T cells were plated at 0.5 × 106 cells per well in a six-well microplate and 24 h later were transfected with 750 ng pcDNA3.1-myc-β-III-spectrin, 250 ng PM-GFP, and pcDNA3.1(+),to a total of 2.5 µg DNA. Twenty-four hours posttransfection, the medium was aspirated, and cells were washed once with 1 mL ice-cold PBS. Cells were lysed for 15 min in 750 µL ice-cold Triton IP buffer. To separate Triton X-100–soluble and –insoluble fractions, lysates were centrifuged at 16,000 × g for 20 min at 4 °C. Supernatants were transferred to new prechilled microtubes. For lysate samples, 150 µL of supernatant was combined with 50 µL of 4× Laemmli buffer and was boiled for 2 min. The Triton X-100–insoluble pellets were resuspended in 750 µL 1× Laemmli buffer by passing the pellets through an 18-gauge needle 20 times, followed by boiling for 2 min. For Triton X-100–insoluble samples, 150 µL of resuspended pellet was combined for with 50 µL 1× Laemmli buffer. SDS/PAGE and Western blotting were performed as described for co-IP assays.
Immunofluorescence studies.
HEK293T cells were plated onto glass coverslips (22 × 22 mm; #1.5; Thermo Fisher Scientific) in six-well microplates. Coverslips were cleaned with sodium hydroxide, sterilized, and coated with polylysine as described (64). For coating, coverslips were incubated with poly-d-lysine, 70–150 kDa (Sigma) solution, 0.1 mg/mL in water, for 20 min at room temperature. Cells in individual wells were transfected with 150 ng pcDNA3.1-myc-β-III-spectrin and pcDNA3.1(+) filler to a total of 2.5 µg DNA per well. In some experiments, 250 ng PM-GFP was included. Twenty-four hours after transfection, cells were rinsed once in PBS and then were fixed for 15 min in PBS containing 4% paraformaldehyde. Fixed cells were permeabilized in PBS containing 0.1% saponin and were blocked in PBS containing 0.1% saponin and 5% goat serum. The primary antibody, α-myc 9E10 (Santa Cruz Biotechnology) at 1:500 in PBS plus 0.1% saponin plus 5% goat serum, was applied to the coverslips and incubated overnight at 4 °C. Primary antibodies were removed by washing with PBS plus 0.1% saponin. Secondary antibodies, α-mouse Alexa-488 (donkey) or α-mouse Alexa-594 (donkey) (Jackson ImmunoResearch), diluted 1:500 in PBS plus 0.1% saponin plus 5% goat serum, were incubated with coverslips for 1 h at room temperature. Secondary antibodies were removed by washing with PBS plus 0.1% saponin. For labeling F-actin, coverslips were incubated with phalloidin-iFluor 594 (Abcam) diluted 1:1,000 in PBS plus 1% BSA plus 0.1% saponin for 1 h at room temperature. Phalloidin was removed by washing with PBS plus 0.1% saponin. Coverslips were mounted with ProLong Gold (Molecular Probes). Cell images were captured using a Plan-APOCHROMAT 63×/1.40 oil objective and an ORCA-ER camera (Hamamatsu) on a spinning-disk confocal microscope (Zeiss). In Fig. S4, cells stained for myc-β-III-spectrin were visually scored for a smooth or textured cell surface. For each L253 variant, 138–328 cells were scored in each of three replicate experiments. Similarly, 221–336 cells for each L253 variant were scored for the presence of β-III-spectrin vesicles in each of three experiments.
L253A ABD in Vitro Characterization.
PCR site-directed mutagenesis was performed to introduce the L253A substitution in the human β-III-spectrin ABD coding sequence (amino acids 1–284) contained in the pET-30a bacterial expression vector (wild-type pET-30a-ABD) (28). L253A sense/antisense oligonucleotides, described above, were used. The resulting construct, pET-30a-ABD L253A, was sequence verified and then was transformed into Escherichia coli BL21(DE3) cells (Novagen). Subsequent bacterial expression and purification were performed as described previously (28). Using an ABD concentration of 247 ng/µL, CD absorption spectra were measured as described (28). To determine the Tm, thermal melting curve data at 222 nm were fit to an equation for a two-state transition (65) using nonlinear regression in Prism 5. For F-actin cosedimentation assays, actin was prepared from rabbit skeletal muscle as reported (66). Binding assays were carried out using 0.8 μM ABD and varying concentrations of phalloidin-stabilized F-actin, as previously reported (28). Nonlinear regression analysis was performed in Prism 5 (GraphPad) to fit binding data to the equation Y = {(X + 0.8 + Kd) – [(X + 0.8 + Kd)2 – 4*X*0.8]0.5}/(2*0.8), where Y equals the ABD-bound fraction, X equals the total F-actin concentration, and 0.8 equals total ABD concentration in micromolar units.
Drosophila Studies.
Fly stocks.
Flies were raised on standard food. For experiments using UAS-β-spectrin or UAS-mEos3.2-β-spectrin flies, crosses were performed at 22 °C. For RNAi experiments, crosses were performed at 25 °C or 29 °C, as described below. The following stocks were obtained from the Bloomington Drosophila Stock Center: 477-Gal4/CyO, ppk-Gal4.G(2), ppk-CD4-tdGFP(1b), ppk-CD4-tdGFP(8)/TM6,Tb, ppk-CD4-tdTom(10a)/TM6,Tb, UAS-Dcr-2, UAS-βspecRNAi-A (βspecGL01174), UAS-Ank2RNAi-B (Ank2JF03374), and elav-Gal4. The following stocks were obtained from the Vienna Drosophila Resource Center: UAS-βspecRNAi-B (βspecGD11790) (42054) and UAS-Ank2RNAi-A (Ank2GD12247) (40638). The βspecem21 (36) stock was generously provided by Ronald Dubreuil, University of Illinois at Chicago, Chicago. The attP154 stock (67) was generously provided by Norbert Perrimon, Harvard Medical School, Boston. The weeP386 α-spectrin-GFP line (38) was from Melissa Rolls, The Pennsylvania State University, University Park, PA. OreR was used as a wild-type control.
Generation of UAS-βspecWT and UAS-βspecSCA5 flies.
The wild-type and SCA5 (L246P) Drosophila β-spectrin coding sequences were isolated from the constructs pUAST-FSPWT and pUAST-FSPGM (57), respectively, using Not1 and Xba1 restriction enzymes and were ligated into the pUASTattB transformation vector using the same restriction enzymes. The resulting constructs, pUASTattB-β-spectrin wild-type and pUASTattB-β-spectrin SCA5, were sequence verified. PhiC31 integrase-mediated transgenesis using the attP154 docking site was carried out by BestGene, Inc.
Generation of UAS-mEos3.2-βspecWT and UAS-mEos3.2-βspecSCA5 flies.
The mEos3.2 coding sequence was PCR amplified using the forward oligonucleotide CGG GGT ACC GCG GCC GCA TGA GTG CGA TTA AGC CAG ACA TGA AG and a reverse oligonucleotide CTG GGA TCC CAG CGA ACA ATC GAA ATG TCC GTC GTC ATG GGT CGT CTG GCA TTG TCA GGC AAT CCA GAA TG that includes a 5′ β-spectrin coding sequence. The template DNA was pmEos3.2-C1, a gift from the Michael W. Davidson laboratory at Florida State University, Tallahassee, FL. The PCR product was digested with Kpn1 and BamHI restriction enzymes and was ligated into pBluescript II KS(+) digested with the same enzymes. The resulting intermediate construct, pBS-mEos3.2–5′β-spec, consisted of the mEos3.2 coding sequence fused to the 5′ β-spectrin coding sequence containing an internal BamHI site. The 3′ region of β-spectrin was isolated from the pUASTattB-β-spectrin wild-type or SCA5 construct, described above, using BamHI and Xba1 restriction enzymes and was ligated into pBS-mEos3.2–5′β-spec digested with the same enzymes. The resulting intermediate construct, wild-type or L246P pBS-mEos3.2-β-spectrin, contained mEos3.2 fused to full-length wild-type or SCA5 β-spectrin. The mEos3.2-β-spectrin fusion coding sequence was isolated from wild-type or SCA5 pBS-mEos3.2-β-spectrin using Not1 and Xba1 restriction enzymes and was ligated into pUASTattB digested with the same enzymes. The resulting constructs, wild-type and SCA5pUASTattB-mEos3.2-β-spectrin, were sequenced to verify the mEos3.2 sequence and DNA junctions. PhiC31 integrase-mediated transgenesis using the attP154 docking site was carried out by BestGene, Inc.
Da neuron analyses.
In Fig. 3A, ppk-Gal4, ppk-CD4-tdGFP; UAS-dcr2/TM6,Tb flies were crossed to OreR, UAS-βspecRNAi-A, or UAS-βspecRNAi-B flies at 29 °C. ddaC class IV da neurons in abdominal segments A3 and A4 in crawling, late-third-instar larvae were imaged using a Plan-APOCHROMAT 20×/0.75 objective and an ORCA-ER camera (Hamamatsu) on a spinning-disk confocal microscope (Zeiss). Arbor image reconstructions were performed in Photoshop CS2 (Adobe). For analysis, arbors were traced using the Simple Neurite Tracer plug-in in ImageJ. Sholl analysis on arbor traces was performed in ImageJ. A step-by-step arbor analysis protocol was reported (68). Branch points were counted in ImageJ using the Cell Counter plug-in. Statistical analysis of total branch length was performed in Prism 5 by one-way ANOVA followed by Tukey’s multiple comparison test. Statistical analysis of number of branch points was performed in Prism 5 by two-tailed, unpaired t test. For all samples, n = 7 neurons (one neuron imaged in each of seven larvae).
In Fig. 3B, ppk-Gal4; ppk-CD4-tdGFP, UAS-dcr2/TM6,Tb flies were crossed to OreR, UAS-Ank2RNAi-A, or UAS-Ank2RNAi-B flies at 25 °C. ddaC class IV da neurons in abdominal segments A3 and A4 in crawling, late-third-instar larvae were imaged, and arbors were analyzed as in Fig. 3A. Statistical analysis of total branch length was performed in Prism 5 by one-way ANOVA followed by Tukey’s multiple comparison test. Statistical analysis of total branch points was performed in Prism 5 by two-tailed, unpaired t test. For wild-type or Ank2 RNAi-A, n = 6 neurons (one neuron imaged in each of six larvae). For Ank2 RNAi-B, n = 5 neurons (one neuron in each of five larvae).
In Fig. 4A, the fly-line 477-Gal4; ppk-CD4-tdGFP was crossed to UAS-βspecWT or UAS-βspecSCA5 flies at 22 °C. ddaC class IV da neurons in abdominal segments A3 and A4 were imaged in crawling, late-third-instar larvae as described for Fig. 3A. Because βspecSCA5 arborization defects were stronger in females than in males, only female larvae were imaged for UAS-βspecWT and UAS-βspecSCA5. For Fig. 4 B and C, eggs were collected for 4 h on grape juice plates containing yeast paste. ddaC class IV da neurons in abdominal segments A3 and A4 were imaged in second-instar larvae at 60 h AEL or in early-third-instar larvae at 96 h AEL. The sex of second-instar larvae was determined after imaging by preparing genomic DNA and performing PCR with oligos against the Ppr-y gene as previously described (69). Dendritic arbors were analyzed as described for Fig. 3A. Statistical analysis of total branch length or branch points was performed in Prism 5 by two-tailed, unpaired t test. For wild-type or SCA5 sample, n = 5 neurons (one neuron imaged in each of five larvae).
In Fig. 5, 477-Gal4; ppk-CD4-tdTom flies were crossed to UAS-mEos3.2-βspecWT and UAS-mEos3.2-βspecSCA5 flies at 22 °C. ddaC class IV da neurons in abdominal segments A3 and A4 were imaged in female, crawling third-instar larvae. CD4tdTom and mEos3.2-β-spectrin fluorescence signals were imaged in cell bodies and dendrites using a Plan-APOCHROMAT 63×/1.40 oil objective and an ORCA-ER camera (Hamamatsu) on a spinning-disk confocal microscope (Zeiss). Quantification of mEos3.2-β-spectrin fluorescence signals was performed on Z-stack maximum intensity projections using Metamorph (Molecular Devices). For cell bodies, mEos3.2 average fluorescence intensity was measured by drawing a region around the cell body and background subtracting to an equivalent region positioned adjacent to the cell body. For dendrites, mEos3.2 fluorescence intensities were measured in a 50-µm segment of a primary dendrite extending toward the dorsal midline. The 50-µm segment was located at a dendritic distance 100 µm away from the cell body (Fig. 5). Using a three-pixel-wide line scan, maximum mEos3.2 fluorescence intensities were measured along the 50-µm segment. For an individual segment, maximum intensity values were summed and background subtracted using a line scan of equivalent length positioned adjacent to the dendrite. Statistical analyses were performed in Prism 5 using two-tailed, unpaired t tests with Welch’s correction. For dendrites, n = 17 neurons (one to three neurons imaged per larva) for wild type, and n = 16 neurons (one to three neurons imaged per larva) for SCA5. For cell bodies, n = 26 (one to three neurons imaged per larva) for wild type, and n =21 (one to three neurons imaged per larva) for SCA5.
In Fig. 6A, the 477-Gal4; ppk-CD4-tdTom, UAS-βspecWT or 477-Gal4; ppk-CD4-tdTom, UAS-βspecSCA5 flies were crossed to weeP386 flies expressing α-spectrin-GFP from the endogenous α-Spec locus and were grown at 22 °C. CD4tdTom and α-spectrin-GFP fluorescence signals in ddaC class IV da neuron cell bodies in abdominal segments A3 and A4 of female larvae were imaged and quantified as described for Fig. 5. Statistical analysis was performed in Prism 5 by two-tailed, unpaired t test with Welch’s correction. For wild-type, n = 11 neurons; for SCA5, n = 15 neurons.
In Fig. 6B, the 477-Gal4; ppk-CD4-tdGFP flies were crossed to UAS-βspecWT or UAS-βspecSCA5 flies at 22 °C. Female, crawling third-instar larvae were dissected as described (70). Larval fillets were fixed in PBS containing 4% paraformaldehyde. Fixed larvae were permeabilized in PBS containing 0.1% saponin and were blocked in PBS containing 0.1% saponin and 5% goat serum. Primary antibody, α-Ank2XL (32) at 1:500 in PBS plus 0.1% saponin plus 5% goat serum, was incubated with larvae overnight at 4 °C. The primary antibody was removed by washing with PBS plus 0.1% saponin. Secondary antibodies, α-rabbit Alexa-594 (donkey) (Jackson ImmunoResearch), diluted 1:500 in PBS plus 0.1% saponin plus 5% goat serum, was incubated with larvae for 1 h at room temperature. Secondary antibody was removed by washing with PBS plus 0.1% saponin. Larvae were mounted with ProLong Gold (Molecular Probes). Dendrites were imaged using a Plan-APOCHROMAT 20×/0.75 objective and an ORCA-ER camera (Hamamatsu) on a spinning-disk confocal microscope (Zeiss). Dendritic signals were quantified as described for Fig. 5. Statistical analysis was performed in Prism 5 by two-tailed, unpaired t test with Welch’s correction. For wild type, n = 18 neurons; for SCA5, n = 17.
In Fig. S5, ppk-Gal4; ppk-CD4-Tom flies were crossed to UAS-mEos3.2-βspecWT and UAS-mEos3.2-βspecSCA5 flies at 22 °C. Axon terminals at the ventral nerve cord were imaged in female, crawling third-instar larvae. CD4tdTom and mEos3.2-β-spectrin fluorescence signals were imaged using a Plan-APOCHROMAT 20×/0.75 objective and an ORCA-ER camera (Hamamatsu) on a spinning-disk confocal microscope (Zeiss). Quantification of mEos3.2-β-spectrin fluorescence signals was performed on Z-stack maximum intensity projections using Metamorph (Molecular Devices). Statistical analyses were performed in Prism 5 using a two-tailed, unpaired t test with Welch’s correction; n = 5 wild-type larvae; n = 6 SCA5larvae.
UAS-βspec transgene rescue of em21 lethality.
In Table S1, male flies homozygous for UAS-βspec transgenes or the third chromosome attP154 landing site, into which UAS-βspec transgenes are inserted, were crossed to females carrying the βspecem21, elav-Gal4 recombinant chromosome balanced over the FM7 chromosome. Crosses were performed at 22 °C. Adult progeny were scored for all possible genotypes.
Acknowledgments
We thank Dr. Harry T. Orr at the University of Minnesota for critical review of the manuscript; Michael E. Fealey for assistance with protein purification and circular dichroism; Amanda L. Neisch for assistance with dissections and critical input on experiments; R. Grace Owens-Kurtz and Madison L. Irwin, undergraduates who helped with the dendritic arbor analyses; fellow laboratory members Min-Gang Li and Madeline Serr for intellectual input and critical reading; Dr. Sean D. Conner for advice on the mammalian cell culture work; and Dr. Hermann Aberle for the ankyrin-2 XL antibody. This work was supported by NIH Grant R01 GM44757 (to T.S.H.) and NIH Training Grant T32 AR007612 (to D.D.T.). A.W.A. received fellowship support from the National Ataxia Foundation and the Bob Allison Ataxia Research Center.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1707108114/-/DCSupplemental.
References
- 1.Dick KA, Ikeda Y, Day JW, Ranum LP. Spinocerebellar ataxia type 5. Handb Clin Neurol. 2012;103:451–459. doi: 10.1016/B978-0-444-51892-7.00028-0. [DOI] [PubMed] [Google Scholar]
- 2.Ikeda Y, et al. Spectrin mutations cause spinocerebellar ataxia type 5. Nat Genet. 2006;38:184–190. doi: 10.1038/ng1728. [DOI] [PubMed] [Google Scholar]
- 3.Ohara O, Ohara R, Yamakawa H, Nakajima D, Nakayama M. Characterization of a new beta-spectrin gene which is predominantly expressed in brain. Brain Res Mol Brain Res. 1998;57:181–192. doi: 10.1016/s0169-328x(98)00068-0. [DOI] [PubMed] [Google Scholar]
- 4.Stankewich MC, et al. Targeted deletion of betaIII spectrin impairs synaptogenesis and generates ataxic and seizure phenotypes. Proc Natl Acad Sci USA. 2010;107:6022–6027. doi: 10.1073/pnas.1001522107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perkins EM, et al. Loss of beta-III spectrin leads to purkinje cell dysfunction recapitulating the behavior and neuropathology of spinocerebellar ataxia type 5 in humans. J Neurosci. 2010;30:4857–4867. doi: 10.1523/JNEUROSCI.6065-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao Y, et al. β-III spectrin is critical for development of purkinje cell dendritic tree and spine morphogenesis. J Neurosci. 2011;31:16581–16590. doi: 10.1523/JNEUROSCI.3332-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacob FD, Ho ES, Martinez-Ojeda M, Darras BT, Khwaja OS. Case of infantile onset spinocerebellar ataxia type 5. J Child Neurol. 2013;28:1292–1295. doi: 10.1177/0883073812454331. [DOI] [PubMed] [Google Scholar]
- 8.Cho E, Fogel BL. A family with spinocerebellar ataxia type 5 found to have a novel missense mutation within a SPTBN2 spectrin repeat. Cerebellum. 2013;12:162–164. doi: 10.1007/s12311-012-0408-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, et al. A Japanese SCA5 family with a novel three-nucleotide in-frame deletion mutation in the SPTBN2 gene: A clinical and genetic study. J Hum Genet. 2014;59:569–573. doi: 10.1038/jhg.2014.74. [DOI] [PubMed] [Google Scholar]
- 10.Liu LZ, et al. A novel missense mutation in the spectrin beta nonerythrocytic 2 gene likely associated with spinocerebellar ataxia type 5. Chin Med J (Engl) 2016;129:2516–2517. doi: 10.4103/0366-6999.191834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Armbrust KR, et al. Mutant β-III spectrin causes mGluR1α mislocalization and functional deficits in a mouse model of spinocerebellar ataxia type 5. J Neurosci. 2014;34:9891–9904. doi: 10.1523/JNEUROSCI.0876-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu K, Zhong G, Zhuang X. Actin, spectrin, and associated proteins form a periodic cytoskeletal structure in axons. Science. 2013;339:452–456. doi: 10.1126/science.1232251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D’Este E, et al. Subcortical cytoskeleton periodicity throughout the nervous system. Sci Rep. 2016;6:22741. doi: 10.1038/srep22741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He J, et al. Prevalent presence of periodic actin-spectrin-based membrane skeleton in a broad range of neuronal cell types and animal species. Proc Natl Acad Sci USA. 2016;113:6029–6034. doi: 10.1073/pnas.1605707113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qu Y, Hahn I, Webb SE, Pearce SP, Prokop A. Periodic actin structures in neuronal axons are required to maintain microtubules. Mol Biol Cell. 2017;28:296–308. doi: 10.1091/mbc.E16-10-0727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhong G, et al. Developmental mechanism of the periodic membrane skeleton in axons. Elife. 2014;3:e04581. doi: 10.7554/eLife.04581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D’Este E, Kamin D, Göttfert F, El-Hady A, Hell SW. STED nanoscopy reveals the ubiquity of subcortical cytoskeleton periodicity in living neurons. Cell Rep. 2015;10:1246–1251. doi: 10.1016/j.celrep.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 18.Sidenstein SC, et al. Multicolour multilevel STED nanoscopy of actin/spectrin organization at synapses. Sci Rep. 2016;6:26725. doi: 10.1038/srep26725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bär J, Kobler O, van Bommel B, Mikhaylova M. Periodic F-actin structures shape the neck of dendritic spines. Sci Rep. 2016;6:37136. doi: 10.1038/srep37136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hammarlund M, Jorgensen EM, Bastiani MJ. Axons break in animals lacking beta-spectrin. J Cell Biol. 2007;176:269–275. doi: 10.1083/jcb.200611117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krieg M, et al. Genetic defects in β-spectrin and tau sensitize C. elegans axons to movement-induced damage via torque-tension coupling. Elife. 2017;6:e20172. doi: 10.7554/eLife.20172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bodine DM, 4th, Birkenmeier CS, Barker JE. Spectrin deficient inherited hemolytic anemias in the mouse: Characterization by spectrin synthesis and mRNA activity in reticulocytes. Cell. 1984;37:721–729. doi: 10.1016/0092-8674(84)90408-2. [DOI] [PubMed] [Google Scholar]
- 23.Birkenmeier CS, Gifford EJ, Barker JE. Normoblastosis, a murine model for ankyrin-deficient hemolytic anemia, is caused by a hypomorphic mutation in the erythroid ankyrin gene Ank1. Hematol J. 2003;4:445–449. doi: 10.1038/sj.thj.6200307. [DOI] [PubMed] [Google Scholar]
- 24.Peters LL, et al. Purkinje cell degeneration associated with erythroid ankyrin deficiency in nb/nb mice. J Cell Biol. 1991;114:1233–1241. doi: 10.1083/jcb.114.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kordeli E, Bennett V. Distinct ankyrin isoforms at neuron cell bodies and nodes of Ranvier resolved using erythrocyte ankyrin-deficient mice. J Cell Biol. 1991;114:1243–1259. doi: 10.1083/jcb.114.6.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lambert S, Bennett V. Postmitotic expression of ankyrinR and beta R-spectrin in discrete neuronal populations of the rat brain. J Neurosci. 1993;13:3725–3735. doi: 10.1523/JNEUROSCI.13-09-03725.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clarkson YL, et al. β-III spectrin underpins ankyrin R function in purkinje cell dendritic trees: Protein complex critical for sodium channel activity is impaired by SCA5-associated mutations. Hum Mol Genet. 2014;23:3875–3882. doi: 10.1093/hmg/ddu103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Avery AW, Crain J, Thomas DD, Hays TS. A human β-III-spectrin spinocerebellar ataxia type 5 mutation causes high-affinity F-actin binding. Sci Rep. 2016;6:21375. doi: 10.1038/srep21375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clarkson YL, Gillespie T, Perkins EM, Lyndon AR, Jackson M. Beta-III spectrin mutation L253P associated with spinocerebellar ataxia type 5 interferes with binding to Arp1 and protein trafficking from the Golgi. Hum Mol Genet. 2010;19:3634–3641. doi: 10.1093/hmg/ddq279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teruel MN, Blanpied TA, Shen K, Augustine GJ, Meyer T. A versatile microporation technique for the transfection of cultured CNS neurons. J Neurosci Methods. 1999;93:37–48. doi: 10.1016/s0165-0270(99)00112-0. [DOI] [PubMed] [Google Scholar]
- 31.Grueber WB, Ye B, Moore AW, Jan LY, Jan YN. Dendrites of distinct classes of Drosophila sensory neurons show different capacities for homotypic repulsion. Curr Biol. 2003;13:618–626. doi: 10.1016/s0960-9822(03)00207-0. [DOI] [PubMed] [Google Scholar]
- 32.Koch I, et al. Drosophila ankyrin 2 is required for synaptic stability. Neuron. 2008;58:210–222. doi: 10.1016/j.neuron.2008.03.019. [DOI] [PubMed] [Google Scholar]
- 33.Grueber WB, Jan LY, Jan YN. Different levels of the homeodomain protein cut regulate distinct dendrite branching patterns of Drosophila multidendritic neurons. Cell. 2003;112:805–818. doi: 10.1016/s0092-8674(03)00160-0. [DOI] [PubMed] [Google Scholar]
- 34.Lee SB, Bagley JA, Lee HY, Jan LY, Jan YN. Pathogenic polyglutamine proteins cause dendrite defects associated with specific actin cytoskeletal alterations in Drosophila. Proc Natl Acad Sci USA. 2011;108:16795–16800. doi: 10.1073/pnas.1113573108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang M, et al. Rational design of true monomeric and bright photoactivatable fluorescent proteins. Nat Methods. 2012;9:727–729. doi: 10.1038/nmeth.2021. [DOI] [PubMed] [Google Scholar]
- 36.Dubreuil RR, Wang P, Dahl S, Lee J, Goldstein LS. Drosophila beta spectrin functions independently of alpha spectrin to polarize the Na,K ATPase in epithelial cells. J Cell Biol. 2000;149:647–656. doi: 10.1083/jcb.149.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mazock GH, Das A, Base C, Dubreuil RR. Transgene rescue identifies an essential function for Drosophila beta spectrin in the nervous system and a selective requirement for ankyrin-2-binding activity. Mol Biol Cell. 2010;21:2860–2868. doi: 10.1091/mbc.E10-03-0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khanna MR, et al. Spectrin tetramer formation is not required for viable development in Drosophila. J Biol Chem. 2015;290:706–715. doi: 10.1074/jbc.M114.615427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.London M, Häusser M. Dendritic computation. Annu Rev Neurosci. 2005;28:503–532. doi: 10.1146/annurev.neuro.28.061604.135703. [DOI] [PubMed] [Google Scholar]
- 40.Branco T, Häusser M. The single dendritic branch as a fundamental functional unit in the nervous system. Curr Opin Neurobiol. 2010;20:494–502. doi: 10.1016/j.conb.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 41.Wong RO, Ghosh A. Activity-dependent regulation of dendritic growth and patterning. Nat Rev Neurosci. 2002;3:803–812. doi: 10.1038/nrn941. [DOI] [PubMed] [Google Scholar]
- 42.Gao FB. Molecular and cellular mechanisms of dendritic morphogenesis. Curr Opin Neurobiol. 2007;17:525–532. doi: 10.1016/j.conb.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parrish JZ, Emoto K, Kim MD, Jan YN. Mechanisms that regulate establishment, maintenance, and remodeling of dendritic fields. Annu Rev Neurosci. 2007;30:399–423. doi: 10.1146/annurev.neuro.29.051605.112907. [DOI] [PubMed] [Google Scholar]
- 44.Scott EK, Luo L. How do dendrites take their shape? Nat Neurosci. 2001;4:359–365. doi: 10.1038/86006. [DOI] [PubMed] [Google Scholar]
- 45.Jan YN, Jan LY. Branching out: Mechanisms of dendritic arborization. Nat Rev Neurosci. 2010;11:316–328. doi: 10.1038/nrn2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pollard TD. A guide to simple and informative binding assays. Mol Biol Cell. 2010;21:4061–4067. doi: 10.1091/mbc.E10-08-0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karinch AM, Zimmer WE, Goodman SR. The identification and sequence of the actin-binding domain of human red blood cell beta-spectrin. J Biol Chem. 1990;265:11833–11840. [PubMed] [Google Scholar]
- 48.Way M, Pope B, Weeds AG. Evidence for functional homology in the F-actin binding domains of gelsolin and alpha-actinin: Implications for the requirements of severing and capping. J Cell Biol. 1992;119:835–842. doi: 10.1083/jcb.119.4.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bañuelos S, Saraste M, Djinović Carugo K. Structural comparisons of calponin homology domains: Implications for actin binding. Structure. 1998;6:1419–1431. doi: 10.1016/s0969-2126(98)00141-5. [DOI] [PubMed] [Google Scholar]
- 50.Galkin VE, Orlova A, Salmazo A, Djinovic-Carugo K, Egelman EH. Opening of tandem calponin homology domains regulates their affinity for F-actin. Nat Struct Mol Biol. 2010;17:614–616. doi: 10.1038/nsmb.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaplan JM, et al. Mutations in ACTN4, encoding alpha-actinin-4, cause familial focal segmental glomerulosclerosis. Nat Genet. 2000;24:251–256. doi: 10.1038/73456. [DOI] [PubMed] [Google Scholar]
- 52.Weins A, et al. Disease-associated mutant alpha-actinin-4 reveals a mechanism for regulating its F-actin-binding affinity. Proc Natl Acad Sci USA. 2007;104:16080–16085. doi: 10.1073/pnas.0702451104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Clark AR, Sawyer GM, Robertson SP, Sutherland-Smith AJ. Skeletal dysplasias due to filamin A mutations result from a gain-of-function mechanism distinct from allelic neurological disorders. Hum Mol Genet. 2009;18:4791–4800. doi: 10.1093/hmg/ddp442. [DOI] [PubMed] [Google Scholar]
- 54.Sawyer GM, Clark AR, Robertson SP, Sutherland-Smith AJ. Disease-associated substitutions in the filamin B actin binding domain confer enhanced actin binding affinity in the absence of major structural disturbance: Insights from the crystal structures of filamin B actin binding domains. J Mol Biol. 2009;390:1030–1047. doi: 10.1016/j.jmb.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 55.Duff RM, et al. Mutations in the N-terminal actin-binding domain of filamin C cause a distal myopathy. Am J Hum Genet. 2011;88:729–740. doi: 10.1016/j.ajhg.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Holleran EA, et al. beta III spectrin binds to the Arp1 subunit of dynactin. J Biol Chem. 2001;276:36598–36605. doi: 10.1074/jbc.M104838200. [DOI] [PubMed] [Google Scholar]
- 57.Lorenzo DN, et al. Spectrin mutations that cause spinocerebellar ataxia type 5 impair axonal transport and induce neurodegeneration in Drosophila. J Cell Biol. 2010;189:143–158. doi: 10.1083/jcb.200905158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Holleran EA, Tokito MK, Karki S, Holzbaur EL. Centractin (ARP1) associates with spectrin revealing a potential mechanism to link dynactin to intracellular organelles. J Cell Biol. 1996;135:1815–1829. doi: 10.1083/jcb.135.6.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pielage J, Fetter RD, Davis GW. Presynaptic spectrin is essential for synapse stabilization. Curr Biol. 2005;15:918–928. doi: 10.1016/j.cub.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 60.Pielage J, et al. A presynaptic giant ankyrin stabilizes the NMJ through regulation of presynaptic microtubules and transsynaptic cell adhesion. Neuron. 2008;58:195–209. doi: 10.1016/j.neuron.2008.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Soba P, et al. The Ret receptor regulates sensory neuron dendrite growth and integrin mediated adhesion. Elife. 2015;4:e05491. doi: 10.7554/eLife.05491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamamoto M, Ueda R, Takahashi K, Saigo K, Uemura T. Control of axonal sprouting and dendrite branching by the Nrg-Ank complex at the neuron-glia interface. Curr Biol. 2006;16:1678–1683. doi: 10.1016/j.cub.2006.06.061. [DOI] [PubMed] [Google Scholar]
- 63.Girard M, et al. Mitochondrial dysfunction and purkinje cell loss in autosomal recessive spastic ataxia of Charlevoix-Saguenay (ARSACS) Proc Natl Acad Sci USA. 2012;109:1661–1666. doi: 10.1073/pnas.1113166109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fischer AH, Jacobson KA, Rose J, Zeller R. Preparation of slides and coverslips for microscopy. CSH Protoc. 2008;2008:pdb.prot4988. doi: 10.1101/pdb.prot4988. [DOI] [PubMed] [Google Scholar]
- 65.Legardinier S, et al. A two-amino acid mutation encountered in Duchenne muscular dystrophy decreases stability of the rod domain 23 (R23) spectrin-like repeat of dystrophin. J Biol Chem. 2009;284:8822–8832. doi: 10.1074/jbc.M805846200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Prochniewicz E, Zhang Q, Howard EC, Thomas DD. Microsecond rotational dynamics of actin: Spectroscopic detection and theoretical simulation. J Mol Biol. 1996;255:446–457. doi: 10.1006/jmbi.1996.0037. [DOI] [PubMed] [Google Scholar]
- 67.Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat Genet. 2008;40:476–483. doi: 10.1038/ng.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Neisch AL, Avery AW, Machamer JB, Li MG, Hays TS. Methods to identify and analyze gene products involved in neuronal intracellular transport using Drosophila. Methods Cell Biol. 2016;131:277–309. doi: 10.1016/bs.mcb.2015.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Testa ND, Ghosh SM, Shingleton AW. Sex-specific weight loss mediates sexual size dimorphism in Drosophila melanogaster. PLoS One. 2013;8:e58936. doi: 10.1371/journal.pone.0058936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tenenbaum CM, Gavis ER. Removal of Drosophila muscle tissue from larval fillets for immunofluorescence analysis of sensory neurons and epidermal cells. J Vis Exp. 2016:e54670. doi: 10.3791/54670. [DOI] [PMC free article] [PubMed] [Google Scholar]