Significance
Environmental exposures are among the best predictors of health and educational outcomes. Models that estimate the effect of environmental exposures on developmental outcomes typically ignore genetic factors or focus on gene–environment interaction (whether individuals’ response to environmental exposures depends on their genotype). Here we test gene–environment correlation (whether individuals’ exposure to environments depends on their genotype). Using a method that tests specific genetic effects while controlling for background genetic effects, we estimate covariation between children’s genetic liability/propensity for core developmental outcomes and a wide range of environmental exposures. Findings suggest that genetic variants associated with traits, such as educational attainment, body mass index, and schizophrenia, also capture environmental risk and protective factors.
Keywords: environmental risk, polygenic scores, gene–environment correlation, human complex traits, prediction
Abstract
Although gene–environment correlation is recognized and investigated by family studies and recently by SNP-heritability studies, the possibility that genetic effects on traits capture environmental risk factors or protective factors has been neglected by polygenic prediction models. We investigated covariation between trait-associated polygenic variation identified by genome-wide association studies (GWASs) and specific environmental exposures, controlling for overall genetic relatedness using a genomic relatedness matrix restricted maximum-likelihood model. In a UK-representative sample (n = 6,710), we find widespread covariation between offspring trait-associated polygenic variation and parental behavior and characteristics relevant to children’s developmental outcomes—independently of population stratification. For instance, offspring genetic risk for schizophrenia was associated with paternal age (R2 = 0.002; P = 1e-04), and offspring education-associated variation was associated with variance in breastfeeding (R2 = 0.021; P = 7e-30), maternal smoking during pregnancy (R2 = 0.008; P = 5e-13), parental smacking (R2 = 0.01; P = 4e-15), household income (R2 = 0.032; P = 1e-22), watching television (R2 = 0.034; P = 5e-47), and maternal education (R2 = 0.065; P = 3e-96). Education-associated polygenic variation also captured covariation between environmental exposures and children’s inattention/hyperactivity, conduct problems, and educational achievement. The finding that genetic variation identified by trait GWASs partially captures environmental risk factors or protective factors has direct implications for risk prediction models and the interpretation of GWAS findings.
Environmental exposures are among the best early predictors of developmental outcomes. For instance, maternal smoking during pregnancy, socioeconomic deprivation, and time spent watching television and playing video games are associated with lower academic achievement (1–9). Harsh parental physical discipline such as hitting has been linked to increased emotional and behavioral problems including aggression in adolescence (10–14). Paternal age is a risk factor for a range of disorders and subclinical phenotypes including low academic achievement (15), with the link to autism spectrum disorders and schizophrenia most robustly replicated (16–21). Breastfeeding and higher parental socioeconomic status (education, income, occupation) are protective factors for a range of outcomes including educational achievement (7, 8, 22).
Evidence from many family, twin, and adoption studies converges in showing that individuals’ exposure to environments partially depends on their genotype (i.e., genotype–environment correlation). This includes both parenting characteristics and broad socioeconomic variables; all are partially heritable (23–28). In the past decade, quantitative genetic research of this type has been extended to explore genetic and environmental contributions to correlations between environmental factors and children’s outcomes (29–32). Some new designs such as the children-of-twins designs make it possible to tease apart different types of genotype–environment correlation and identify environmental influences free of genetic confounds (33–37). These designs are limited by the extent to which environmental variables differ between close relatives.
Converging evidence for gene–environment correlation comes more recently from “single nucleotide polymorphism (SNP)-heritability” studies that estimate overall genetic influences from genome-wide DNA differences in unrelated individuals. These studies have shown that variation in individuals’ social deprivation, household income, stressful life events, and family socioeconomic status partially reflects individuals’ differences across genome-wide common genetic variants measured on SNP arrays (38–44). There have also been a few reports of extending SNP heritability analysis to estimate genetic correlations between environmental measures and measures of children’s developmental outcomes (38–40).
Gene–environment correlation is recognized and investigated by family studies and recently by SNP-heritability studies. However, the possibility that genetic effects on traits capture environmental risk factors or protective factors has been neglected by polygenic prediction models, which use trait-associated genetic variants identified by genome-wide association studies (GWASs) to estimate individual-level genetic trait propensities for trait prediction.
Here we tested whether genetic variation identified by trait GWASs captures variation in environmental risk factors or protective factors. Specifically, as children’s environments and genetic propensities are both “provided by” their parents, these are expected to correlate because parents pass on genetic variants to their offspring that influence parents’ environment-providing behaviors. Therefore, we examine to what extent offspring trait-associated alleles covary with parental traits and behaviors previously reported to be environmental risk or protective factors for important child outcomes. We also tested to what extent offspring genetic trait propensities contribute to the correlation between parenting characteristics and children’s developmental outcomes.
First, we conducted a systematic investigation of covariation between children’s genetic propensities for specific developmental outcomes and a wide range of environmental exposures previously shown to be risk or protective factors for these outcomes (SI Appendix, Methods S3). We focus on genetic propensities—that is, individual-specific genomic profiles of trait-associated alleles—for three core developmental outcomes: educational attainment (45), body mass index (BMI) (46), and schizophrenia (47). These traits from three important domains of child development—social-cognitive, mental health, and physical health—each are robust predictors of mortality and life expectancy, with substantial associated societal and personal burden (48–55). They were chosen because of the availability of statistically powerful GWAS summary statistics for these traits (56).
Second, we tested whether the environmental exposures predicted children’s developmental outcomes (as would be expected based on previous literature) and to what extent these associations are captured by children’s polygenic propensities for education, BMI, and schizophrenia. For this, we examined associations between the environmental exposures and three developmental outcomes assessed at age 16 in our sample: educational achievement, inattention-hyperactivity symptoms, and conduct problems (SI Appendix, Methods S3).
We used a sample of 6,710 unrelated individuals, drawn from the Twins Early Development Study (TEDS), for whom genotype data and a wide range of specific environmental exposure measures and developmental outcomes from birth to adolescence are available. TEDS is a multivariate longitudinal study that recruited over 11,000 twin pairs born in England and Wales in 1994, 1995, and 1996 (57, 58), shown to be representative of the UK population (38, 59).
We created genome-wide polygenic scores for trait-associated genetic variants for each individual in the sample using summary statistics from the independent GWAS of years of education (EDU) (45), BMI (46), and schizophrenia (SCZ) (47). We used a Bayesian approach (60) that estimates posterior mean effect size of each marker by using a point-normal mixture prior on effect sizes and linkage disequilibrium (LD) information (Materials and Methods).
Because of the salience of possible population stratification when investigating the genetic effect on differences in environmental exposures, we estimated the effect of the polygenic scores while controlling for overall genetic relatedness in the form of a genomic relatedness matrix restricted maximum likelihood model. Specifically, we fit the effects of all SNPs as random effects, while estimating the fixed effects of the polygenic scores (Materials and Methods).
Results
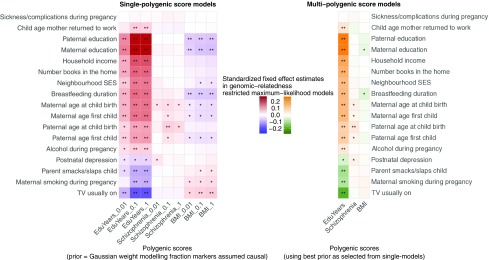
To estimate the univariate effect of each polygenic score on the environmental exposures, we fit a series of single-score models, which reveal significant trait-associated polygenic effects across a wide range of environmental exposures. Fig. 1, Left (and SI Appendix, Table S1) shows the estimated variance explained by each polygenic score for each of the environmental measures. Environmental factors varied significantly as a function of trait-associated polygenic variation, independently of population stratification. This provides evidence for trait-associated genotype–environment correlation. However, given the robust evidence for extensive pleiotropy across complex traits (61), we aimed to isolate the effects of each trait-associated polygenic score using a multiscore model. To test the trait specificity of the polygenic effects on environmental exposures, we jointly modeled the three scores for years of education, BMI, and schizophrenia, allowing us to estimate the effects of each polygenic score while statistically adjusting for the effects of the others. Fig. 1, Right (and SI Appendix, Table S2) shows that the multiscore models revealed some attenuation of the polygenic score effects compared with the single-score models, suggesting that the effects of the three scores on environmental exposures are nonindependent. Specifically, the effects of BMI-associated polygenic variation on several environmental measures (including watching television and parental education) were no longer significant.
Fig. 1.
(Left) Single-polygenic score models: associations between polygenic scores and environmental exposures and single-predictor effects of polygenic scores for years of education, BMI, and schizophrenia on the environmental exposures. (Right) Multipolygenic score models: joint estimation of effects of polygenic scores on environmental exposures and effects of polygenic scores for years of education, BMI, and schizophrenia on the environmental exposures while adjusting for other predictors, respectively. Color gradients represent effect sizes as standardized coefficients—that is, SDs change in the environmental exposure, per SD increase in the polygenic predictor (see SI Appendix, Tables S1–S3 for full statistics). *Uncorrected P < 0.05, **multiple testing corrected P < 0.05 (see Materials and Methods).
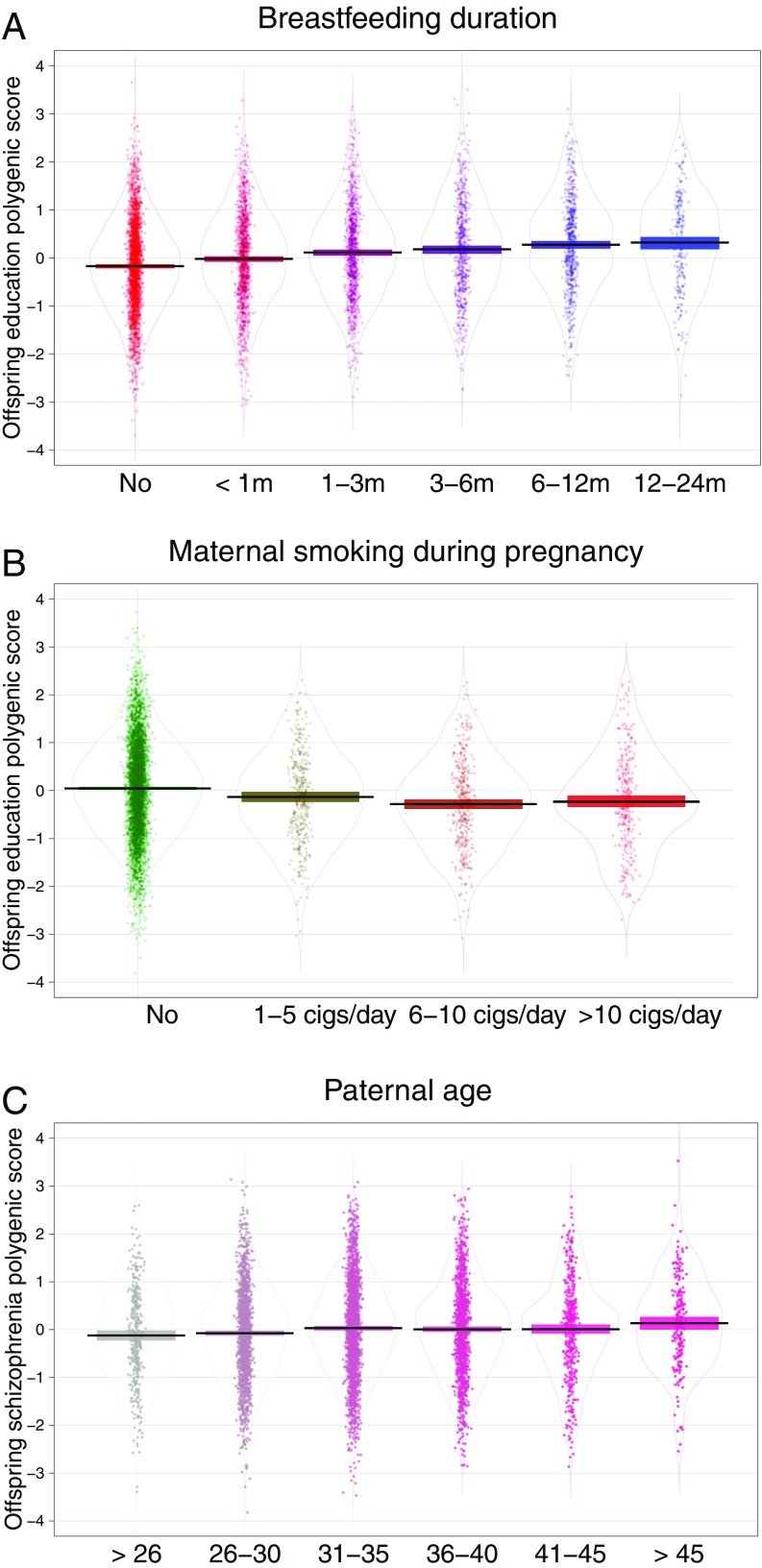
Breastfeeding duration was positively associated with offspring education polygenic score, adjusted for BMI and schizophrenia polygenic scores (R2 = 0.021, beta = 0.144; P = 7e-30). Fig. 2A displays children’s adjusted education polygenic score as a function of whether and for how long they were breastfed. Children who were breastfed had, on average, an education polygenic score approximately one third SD higher (Hedges’ g = 0.30) than children who were not breastfed (t = −11.55, df = 5,664.2, P = 1.6e-30).
Fig. 2.
(A) Offspring adjusted education polygenic score (standardized) by level of breastfeeding. Education polygenic score was adjusted for schizophrenia and BMI polygenic scores. Positive association (R2 = 0.021, beta = 0.144; P = 7e-30). Children who were breastfed had, on average, an education polygenic score approximately one third SD higher (Hedges’ g = 0.30) than children who were not breastfed (t = −11.55, df = 5,664.2; P = 1.6e-30). (B) Offspring adjusted education polygenic score (standardized) by level of maternal smoking during pregnancy. Education polygenic score was adjusted for schizophrenia and BMI polygenic scores. Negative association (R2 = 0.008, beta = −0.090; P = 5e-13). Children exposed to maternal smoking prenatally had, on average, an education polygenic score approximately one quarter SD lower (Hedges’ g = 0.26) than children whose mothers did not smoke (t = 7.93, df = 1,556.3; P = 4e-15). (C) Offspring adjusted schizophrenia polygenic score (standardized) by paternal age at birth of offspring. Genetic risk for schizophrenia was adjusted for education and BMI polygenic scores. Positive association (R2 = 0.002, beta = 0.049; P = 1e-04). Children whose father was aged over 45 at their birth had, on average, a genetic risk score for schizophrenia over one quarter SD (Hedges’ g = 0.26) higher than children whose father was under the age of 26 at their birth (t = −3.01, df = 411.91; P = 3e-03). Horizontal lines and bars represent means and 95% confidence intervals. Violin shapes represent probability densities.
Maternal smoking during pregnancy was negatively associated with offspring education polygenic score adjusted for BMI and schizophrenia polygenic scores (R2 = 0.008, beta = −0.090; P = 5e-13; Fig. 2B). Children exposed to maternal smoking prenatally had, on average, an education polygenic score approximately one quarter SD lower (Hedges’ g = 0.26) than children whose mothers did not smoke (t =7.93, df = 1,556.3; P = 4e-15).
Other effects of education-associated polygenic variation on environmental exposures included 3.3% in household income (beta = 0.181, P = 1e-22), 6.5% in maternal education level (beta = 0.255, P = 3e-96), 1% in parental smacking (beta = −0.10, P = 4e-15), and 3.4% in television watching in the household (beta = −0.184, P = 5e-47).
Offspring genetic risk for schizophrenia was positively associated with paternal age, even when adjusting for education and BMI-associated polygenic variation (R2 = 0.002, beta = 0.049; P = 1e-04). Fig. 2C shows children’s adjusted genetic risk for schizophrenia as a function of paternal age. Children whose father was aged over 45 at their birth had, on average, a genetic risk score for schizophrenia over one quarter SD (Hedges’ g = 0.26) higher than children whose father was under the age of 26 at their birth (t = −3.01, df = 411.91; P = 3e-03).
Next, we examined the extent to which associations between environmental exposures and developmental outcomes were explained by trait-associated polygenic variation for education, BMI, and schizophrenia (SI Appendix, Fig. S3). We examined associations between environmental exposures and three developmental outcomes: educational achievement, inattention-hyperactivity symptoms, and conduct problems. Of the three polygenic scores, only the education polygenic score captured covariation between environmental exposures and the three developmental outcomes (SI Appendix, Table S3).
On average education-associated polygenic variation explained 15% of the associations between the environmental measures and children’s developmental outcomes. For example, the education polygenic score explained 23% (P = 1.2e-18) of the beta = 0.19 covariance between child educational achievement and breastfeeding. Education-associated polygenic variation also captured 6% (P = 1.9e-05) and 7% (P = 4.4e-06) of the associations between parental slapping/smacking and conduct problems and hyperactivity/inattention problems (beta = 0.20 for both).
Discussion
We report evidence for covariation between trait-associated polygenic variation and early environmental exposures independently of population stratification. We show that a wide range of parental, neighborhood, and parent–child perinatal characteristics, representing key early life “environmental” influences, present at birth or early in life, correlate with offspring genetic propensity—specifically, with the allele frequency at loci associated with education, BMI, and schizophrenia. We also demonstrate that covariance between environments and important developmental outcomes are partially captured by education-associated polygenic variation.
The present study combines family and molecular data. In addition to replicating the general finding that individuals’ environmental exposures vary as a function of their genotype, the current findings suggest that trait GWASs are detecting genetic variants associated with parental characteristics and their correlation with child outcomes.
Importantly, the association between exposures and outcomes was by no means entirely captured by offspring trait-associated polygenic variation. There are three likely, nonmutually exclusive, explanations for this. First, a substantial proportion of the exposure–outcome associations is likely due to nongenetic factors. Second, polygenic scores intrinsically underestimate the total genetic effects on the exposure–outcome associations because they are limited to the additive effects of common variants on a particular trait that the discovery GWAS was powered to detect. Third, we only measure offspring polygenic variation, but offspring phenotype can be influenced not only by transmitted but also by nontransmitted parental alleles via parental phenotype (i.e., child exposure).
The education-associated polygenic variation showed the strongest and most consistent correlations with environmental exposures. This is consistent with research showing associations between educational attainment and many parental behaviors and characteristics (e.g., refs. 12, 31, 62, and 63). Moreover, the multipolygenic score models showed that the association between BMI-associated polygenic variation and environmental exposures such as television watching and parental education is explained by education-associated genetic variations. This suggests the potential for multipolygenic models for isolating polygenic effects, provided the underlying discovery GWASs are similarly powered. The finding of an association between paternal age and offspring genetic risk for schizophrenia is consistent with previous evidence for older fathers’ elevated risk for conceiving a child who will go on to develop schizophrenia (18, 19, 63). Although the current findings provide evidence for the relevance of gene–environment correlation for polygenic trait prediction methods, they are not informative about the mechanisms involved.
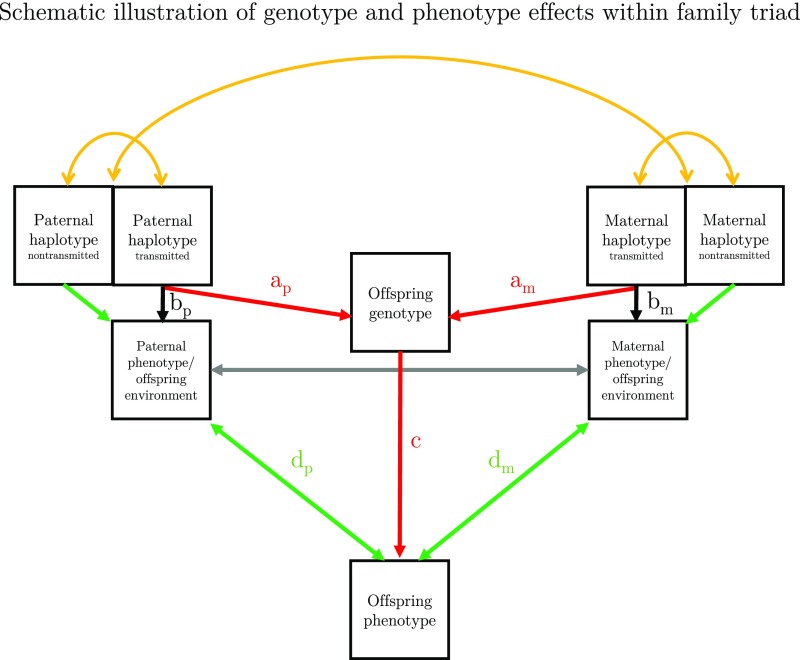
The observed associations could arise from passive or active gene–environment correlation or via environmentally mediated genetic effects, all of which are nonmutually exclusive. Fig. 3 illustrates these possibilities schematically. Many of the observed associations between offspring genotype and environment-providing parental characteristics are outside of the offspring’s influence (e.g., parental age and education level at child birth) and are therefore likely to result from passive gene–environment correlation. That is, parental genetic propensities that were passed down to offspring are also associated with environment-providing parental behavior (through both path a and b, Fig. 3). However, some of the investigated parental behaviors could partially be evoked by offspring genetic propensities (through paths c and d in Fig. 3; e.g., breastfeeding, watching television). Finally, genetic correlations could arise as a result of environmentally mediated genetic effects (e.g., if education-associated genetic variation influenced mothers’ predisposition to smoke during pregnancy and prenatal exposure to nicotine had an environmental effect on offspring attention problems, this could result in offspring education-associated polygenic variation being associated with maternal smoking pregnancy as well as capturing part of its correlation with offspring attention problems).
Fig. 3.
Schematic illustration of cross-generational effects within family triad. Because of the lack of parental genotype data, the present study was unable to distinguish passive and evocative gene–environment correlation. Passive gene–environment correlation: am,p × bm,p. Evocative gene–environment correlation: c × bm,p. Offspring phenotype can be influenced by both the transmitted paternal and maternal alleles (red arrows) and by nontransmitted alleles via parental phenotype (green arrows). Provided that paternal, maternal, and offspring genotype and phenotype data were available in a single sample, the effect of parental trait-associated alleles on offspring phenotype independently of genetic sharing between parents and offspring (green arrows) could be estimated (70–72). A testable assumption for investigating these mechanisms is there is no correlation between parental genotypes and between each parent’s haplotypes (i.e., assortative mating) (yellow arrows).
The design of the current study is unable to distinguish environmentally mediated genetic effects, passive, and evocative gene–environment correlations. One way to investigate the contributions of these different mechanisms would be to use samples incorporating parental genotype data. In analyses of such samples, confounding of offspring genotype by parental genotypes could be accounted for. Provided that paternal, maternal, and offspring genotype and phenotype data were available in a single sample, cross-generational effects of genetics and environment could be further disentangled (see Fig. 3 for schematic illustration).
Nurture has a genetic component; trait-associated alleles in the offspring explain variation in environment-providing parental behaviors and their covariation with offspring developmental outcomes. This provides evidence that the observed effects from GWASs are not only reflecting direct trait effects. This evidence resonates with the hypothesis that trait GWASs capture variation in risk factors as well as direct genetic effects on the trait (64). Here we showed that polygenic scores derived from trait GWASs predict variation in variables beyond the target trait, including variables often presumed to be environmental in origin such as parenting. This suggests incorporating genetic variants associated with environmental risk or predictive factors into polygenic prediction models might improve trait prediction.
In summary, we show that genetic variation identified by trait GWASs partially captures environmental risk or protective factors, indicating that some of the same genetic variation underlies both traits and environments. In contrast to the conceptual dichotomy often imposed between traits and environments, this finding implies that the pleiotropy widely found in phenome–genome associations also crosses over to the realm of environments and manifests across generations. Findings illustrate the relevance of gene–environment correlation for polygenic prediction models and that combining family and molecular data might help reveal mechanisms by which genetic variation is translated into phenotypic variation.
Materials and Methods
We used genome-wide SNP and environment-wide phenotype data from 6,710 unrelated individuals drawn from the UK-representative TEDS (57, 58). TEDS data can be accessed in accordance with the Data Access Policy, which can be viewed at www.teds.ac.uk/research/collaborators-and-data/teds-data-access-policy. We processed the 6,710 genotypes using stringent quality control procedures followed by imputation of SNPs to the Haplotype Reference Consortium reference panel (65) (SI Appendix, Methods S1). This included removing one individual from any pair of individuals with an estimate SNP marker relatedness > 0.05. After quality control, 7,581,516 genotyped or well-imputed (info > 0.70) variants remained.
Polygenic Scores.
For each individual in the sample, we created polygenic scores for years of education, schizophrenia, and BMI. After coordinating overlapping markers between each of the three GWA summary statistics and the target data by excluding markers due to nucleotide inconsistencies or low minor allele frequency (<1%), we retained 5,690,632 for the years of education (45), 5,781,731 for schizophrenia (47), and 1,810,667 for BMI (46). We constructed polygenic scores as the effect-size weighted sums of individuals’ trait-associated alleles across all SNPs. We used LDpred (60), which places a prior on the markers’ effect sizes and adjusts summary statistics for LD between markers. For each trait, we created the score using three different priors on the fraction of causal markers—0.01, 0.1, and 1.0—from which the one yielding the largest R2 in the single-polygenic score models was then entered into the multipolygenic score model. For details on the polygenic score construction, see SI Appendix, Methods S2.
To account for population stratification, we adjusted the polygenic predictors by the first 30 principal components (PCs) generated from genotype data before the analysis. We used the top 30 PCs as well as genotyping array and plate to create a N × P matrix Z of eigenvectors across the P selected PCs. We then regressed the genetic polygenic predictor onto the eigenvectors as S = μ + Zβ + e, where μ is the mean and β is a P × 1 vector of the regression coefficients and e is the residual error.
Single-Score and Multiscore Genomic-Relatedness Matrix Restricted Maximum-Likelihood Models.
When estimating genetic effects on environmental exposures, the possibility of population stratification is especially salient. This is because genetic and common environment effects, even if uncorrelated, may be confounded as close relatives share both genes and their environment to a greater extent than other individuals. We control this type of confounding because, under only population stratification, we would not expect an association between polygenic predictors and environmental measures within the mixed effect model of Eqs. 1 and 2. This is because they account for population stratification by both regressing PCs from the polygenic predictors (see above) and fitting a relationship matrix estimated from all SNP markers (see below).
To estimate the degree to which trait-associated polygenic variation captures variation in environmental measures, we estimated the relationship between the polygenic scores and the environmental measures, while controlling for net genetic relatedness by fitting the effects of all of the SNPs as random effects by a mixed linear model:
| [1] |
| [2] |
y is an n × 1 vector containing the level of environmental exposure, with n being the sample size. β is a vector of fixed effects estimating the effects of the polygenic predictor, independently of overall genetic relatedness g.
In the single-score model (Eq. 1), Si is a vector containing individuals’ polygenic score for one of i ∈ [years of education (EDU) (45), BMI (46), schizophrenia (SCZ) (47)], adjusted for 30 PCs, genotyping array, and plate (see section above). g is an n × 1 vector of the total genetic effects of the individuals, independently of β, with g ∼ N(0,Aσ2g), and A is interpreted as the genetic relationship matrix (GRM) between individuals (MAF > 0.01; relatedness < 0.05 as described above). The genomic relationship of each pair of subjects j and k is calculated as Ajk = 1N∑Ni = 1(xij − 2pi)(xik − 2pi)/2pi(1 − pi) with xij being the number of copies of the reference allele for the ith SNP of the jth individual and pi being the frequency of the reference allele (66).
In the multiscore model (Eq. 2), the effects of the three polygenic predictors are being estimated jointly, thereby allowing the effect of each polygenic predictor independently of each other and of overall genetic relatedness g.
The genetic relatedness matrix accounts for population stratification in the environmental exposure, because it is equivalent to fitting all of the PCs within the model. Eqs. 1 and 2 were estimated using the restricted maximum likelihood (REML) approach implemented in the reml function in GCTA v1.26.0 (67).
Decomposition of Covariance Between Environmental Exposures and Developmental Outcomes.
We fit structural equation models to decompose the covariance between environmental exposures and developmental outcomes into effects of the three polygenic scores and residual covariance (SI Appendix, Fig. 3). The total covariance estimated as was decomposed into the effect of the education score , that of the BMI score:, that of the schizophrenia score , and residual covariance g. We used maximum likelihood estimation with robust (Huber–White) SEs. The analyses were conducted using the lavaan package in R (68).
Multiple Testing Correction.
P values obtained for each statistic were corrected for multiple testing using the Šidák correction (69). The Šidák adjusted alpha level is equal to1 − (1 − α)(1/k), where k is the number of tests. The total number of tests was: 357, with 153 (3 scores × 3 priors × 17 exposures) tests for the single-polygenic score models, 51 (3 scores × 17 exposures) tests for the multipolygenic score model, and 153 (3 scores × 17 exposures × 3 outcomes) test for the decomposition of covariance models. The multiple comparison adjustments were applied to α = 0.05. Hence, the corrected “experimentwise” α level was 1 − (1 − 0.05)(1/357) = 1.44e-04.
Environmental Exposures and Child Outcome Measures.
For a detailed description of all measures, see SI Appendix, Methods S3.
Supplementary Material
Acknowledgments
We gratefully acknowledge the ongoing contribution of the participants in TEDS and their families. TEDS is supported by UK Medical Research Council Program Grant MR/M021475/1 (and previously Grant G0901245) (to R.P.), with additional support from National Institutes of Health Grant AG046938. The research leading to these results has also received funding from the European Research Council under the European Union’s Seventh Framework Programme (FP7/2007-2013)/Grant Agreement 602768 and ERC Grant Agreement 295366. R.P. is supported by Medical Research Council Professorship Award G19/2. E.K. is supported by the MRC/IoPPN Excellence Award. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. This study presents independent research supported by the National Institute for Health Research (NIHR) Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London. The views expressed are those of the authors and not necessarily those of the NHS, NIHR, Department of Health or King’s College London. We gratefully acknowledge capital equipment funding from Maudsley Charity Grant Ref. 980 and Guy’s and St Thomas’s Charity Grant Ref. STR130505. S.J.N. is also supported by the NIHR University College London Hospitals Biomedical Research Centre, and by awards establishing the Farr Institute of Health Informatics Research at UCLPartners, from the Medical Research Council, Arthritis Research UK, British Heart Foundation, Cancer Research UK, Chief Scientist Office, Economic and Social Research Council, Engineering and Physical Sciences Research Council, NIHR, National Institute for Social Care and Health Research, and Wellcome Trust (Grant MR/K006584/1). P.F.O. receives funding from the UK Medical Research Council (Grant MR/N015746/1) and the Wellcome Trust (Grant 109863/Z/15/Z). L.J.H. is supported by an Economic and Social Research Council multidisciplinary studentship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. C.F.C. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1707178114/-/DCSupplemental.
References
- 1.Danner FW. A national longitudinal study of the association between hours of TV viewing and the trajectory of BMI growth among US children. J Pediatr Psychol. 2008;33:1100–1107. doi: 10.1093/jpepsy/jsn034. [DOI] [PubMed] [Google Scholar]
- 2.Jago R, Baranowski T, Baranowski JC, Thompson D, Greaves KA. BMI from 3-6 y of age is predicted by TV viewing and physical activity, not diet. Int J Obes. 2005;29:557–564. doi: 10.1038/sj.ijo.0802969. [DOI] [PubMed] [Google Scholar]
- 3.Anderson CA, et al. Violent video game effects on aggression, empathy, and prosocial behavior in eastern and western countries: A meta-analytic review. Psychol Bull. 2010;136:151–173. doi: 10.1037/a0018251. [DOI] [PubMed] [Google Scholar]
- 4.Gentile DA, Lynch PJ, Linder JR, Walsh DA. The effects of violent video game habits on adolescent hostility, aggressive behaviors, and school performance. J Adolesc. 2004;27:5–22. doi: 10.1016/j.adolescence.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Räsänen P, et al. Maternal smoking during pregnancy and risk of criminal behavior among adult male offspring in the Northern Finland 1966 Birth Cohort. Am J Psychiatry. 1999;156:857–862. doi: 10.1176/ajp.156.6.857. [DOI] [PubMed] [Google Scholar]
- 6.Huizink AC, Mulder EJH. Maternal smoking, drinking or cannabis use during pregnancy and neurobehavioral and cognitive functioning in human offspring. Neurosci Biobehav Rev. 2006;30:24–41. doi: 10.1016/j.neubiorev.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 7.White KR. The relation between socioeconomic status and academic achievement. Psychol Bull. 1982;91:461–481. [Google Scholar]
- 8.Sirin SR. Socioeconomic status and academic achievement: A meta-analytic review of research. Rev Educ Res. 2005;75:417–453. [Google Scholar]
- 9.Caspi A, et al. Childhood forecasting of a small segment of the population with large economic burden. Nat Hum Behav. 2016;1:5. doi: 10.1038/s41562-016-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor CA, Manganello JA, Lee SJ, Rice JC. Mothers’ spanking of 3-year-old children and subsequent risk of children’s aggressive behavior. Pediatrics. 2010;125:e1057–e1065. doi: 10.1542/peds.2009-2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bender HL, et al. Use of harsh physical discipline and developmental outcomes in adolescence. Dev Psychopathol. 2007;19:227–242. doi: 10.1017/S0954579407070125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Afifi TO, Mota NP, Dasiewicz P, MacMillan HL, Sareen J. Physical punishment and mental disorders: Results from a nationally representative US sample. Pediatrics. 2012;130:184–192. doi: 10.1542/peds.2011-2947. [DOI] [PubMed] [Google Scholar]
- 13.Knox M. On hitting children: A review of corporal punishment in the United States. J Pediatr Health Care. 2010;24:103–107. doi: 10.1016/j.pedhc.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 14.Gershoff ET. Corporal punishment by parents and associated child behaviors and experiences: A meta-analytic and theoretical review. Psychol Bull. 2002;128:539–579. doi: 10.1037/0033-2909.128.4.539. [DOI] [PubMed] [Google Scholar]
- 15.D’Onofrio BM, et al. Paternal age at childbearing and offspring psychiatric and academic morbidity. JAMA Psychiatry. 2014;71:432–438. doi: 10.1001/jamapsychiatry.2013.4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reichenberg A, et al. Advancing paternal age and autism. Arch Gen Psychiatry. 2006;63:1026–1032. doi: 10.1001/archpsyc.63.9.1026. [DOI] [PubMed] [Google Scholar]
- 17.Sandin S, et al. Autism risk associated with parental age and with increasing difference in age between the parents. Mol Psychiatry. 2016;21:693–700. doi: 10.1038/mp.2015.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malaspina D. Paternal factors and schizophrenia risk: De novo mutations and imprinting. Schizophr Bull. 2001;27:379–393. doi: 10.1093/oxfordjournals.schbul.a006882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Byrne M, Agerbo E, Ewald H, Eaton WW, Mortensen PB. Parental age and risk of schizophrenia: A case-control study. Arch Gen Psychiatry. 2003;60:673–678. doi: 10.1001/archpsyc.60.7.673. [DOI] [PubMed] [Google Scholar]
- 20.de Kluiver H, Buizer-Voskamp JE, Dolan CV, Boomsma DI. Paternal age and psychiatric disorders: A review. Am J Med Genet B Neuropsychiatr Genet. 2017;174:202–213. doi: 10.1002/ajmg.b.32508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janecka M, et al. Paternal age alters social development in offspring. J Am Acad Child Adolesc Psychiatry. 2017;56:383–390. doi: 10.1016/j.jaac.2017.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Victora CG, et al. Association between breastfeeding and intelligence, educational attainment, and income at 30 years of age: A prospective birth cohort study from Brazil. Lancet Glob Health. 2015;3:e199–e205. doi: 10.1016/S2214-109X(15)70002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Plomin R, Bergeman CS. The nature of nurture: Genetic influence on “environmental” measures. Behav Brain Sci. 1991;14:373–386. [Google Scholar]
- 24.Kendler KS, Baker JH. Genetic influences on measures of the environment: A systematic review. Psychol Med. 2007;37:615–626. doi: 10.1017/S0033291706009524. [DOI] [PubMed] [Google Scholar]
- 25.Avinun R, Knafo A. Parenting as a reaction evoked by children’s genotype: A meta-analysis of children-as-twins studies. Pers Soc Psychol Rev. 2014;18:87–102. doi: 10.1177/1088868313498308. [DOI] [PubMed] [Google Scholar]
- 26.Klahr AM, Burt SA. Elucidating the etiology of individual differences in parenting: A meta-analysis of behavioral genetic research. Psychol Bull. 2014;140:544–586. doi: 10.1037/a0034205. [DOI] [PubMed] [Google Scholar]
- 27.Vinkhuyzen AA, van der Sluis S, de Geus EJ, Boomsma DI, Posthuma D. Genetic influences on ‘environmental’ factors. Genes Brain Behav. 2010;9:276–287. doi: 10.1111/j.1601-183X.2009.00554.x. [DOI] [PubMed] [Google Scholar]
- 28.Butcher LM, Plomin R. The nature of nurture: A genomewide association scan for family chaos. Behav Genet. 2008;38:361–371. doi: 10.1007/s10519-008-9198-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larsson H, Sariaslan A, Långström N, D’Onofrio B, Lichtenstein P. Family income in early childhood and subsequent attention deficit/hyperactivity disorder: A quasi-experimental study. J Child Psychol Psychiatry. 2014;55:428–435. doi: 10.1111/jcpp.12140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Colen CG, Ramey DM. Is breast truly best? Estimating the effects of breastfeeding on long-term child health and wellbeing in the United States using sibling comparisons. Soc Sci Med. 2014;109:55–65. doi: 10.1016/j.socscimed.2014.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.D’Onofrio BM, et al. Familial confounding of the association between maternal smoking during pregnancy and offspring criminality: A population-based study in Sweden. Arch Gen Psychiatry. 2010;67:529–538. doi: 10.1001/archgenpsychiatry.2010.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.D’Onofrio BM, et al. Causal inferences regarding prenatal alcohol exposure and childhood externalizing problems. Arch Gen Psychiatry. 2007;64:1296–1304. doi: 10.1001/archpsyc.64.11.1296. [DOI] [PubMed] [Google Scholar]
- 33.Lynch SK, et al. A genetically informed study of the association between harsh punishment and offspring behavioral problems. J Fam Psychol. 2006;20:190–198. doi: 10.1037/0893-3200.20.2.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harden KP, et al. A behavior genetic investigation of adolescent motherhood and offspring mental health problems. J Abnorm Psychol. 2007;116:667–683. doi: 10.1037/0021-843X.116.4.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Narusyte J, et al. Testing different types of genotype-environment correlation: An extended children-of-twins model. Dev Psychol. 2008;44:1591–1603. doi: 10.1037/a0013911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knopik VS, et al. Maternal alcohol use disorder and offspring ADHD: Disentangling genetic and environmental effects using a children-of-twins design. Psychol Med. 2006;36:1461–1471. doi: 10.1017/S0033291706007884. [DOI] [PubMed] [Google Scholar]
- 37.Silberg JL, Maes H, Eaves LJ. Genetic and environmental influences on the transmission of parental depression to children’s depression and conduct disturbance: An extended children of twins study. J Child Psychol Psychiatry. 2010;51:734–744. doi: 10.1111/j.1469-7610.2010.02205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krapohl E, Plomin R. Genetic link between family socioeconomic status and children’s educational achievement estimated from genome-wide SNPs. Mol Psychiatry. 2016;21:437–443. doi: 10.1038/mp.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davies NM, Hemani G, Timpson NJ, Windmeijer F, Davey Smith G. The role of common genetic variation in educational attainment and income: Evidence from the National Child Development Study. Sci Rep. 2015;5:16509. doi: 10.1038/srep16509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trzaskowski M, et al. Genetic influence on family socioeconomic status and children’s intelligence. Intelligence. 2014;42:83–88. doi: 10.1016/j.intell.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benjamin DJ, et al. The genetic architecture of economic and political preferences. Proc Natl Acad Sci USA. 2012;109:8026–8031. doi: 10.1073/pnas.1120666109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marioni RE, et al. Molecular genetic contributions to socioeconomic status and intelligence. Intelligence. 2014;44:26–32. doi: 10.1016/j.intell.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Power RA, et al. Estimating the heritability of reporting stressful life events captured by common genetic variants. Psychol Med. 2013;43:1965–1971. doi: 10.1017/S0033291712002589. [DOI] [PubMed] [Google Scholar]
- 44.Hill WD, et al. Molecular genetic contributions to social deprivation and household income in UK Biobank. Curr Biol. 2016;26:3083–3089. doi: 10.1016/j.cub.2016.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okbay A, et al. LifeLines Cohort Study Genome-wide association study identifies 74 loci associated with educational attainment. Nature. 2016;533:539–542. doi: 10.1038/nature17671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Locke AE, et al. LifeLines Cohort Study; ADIPOGen Consortium; AGEN-BMI Working Group; CARDIOGRAMplusC4D Consortium; CKDGen Consortium; GLGC; ICBP; MAGIC Investigators; MuTHER Consortium; MIGen Consortium; PAGE Consortium; ReproGen Consortium; GENIE Consortium; International Endogene Consortium Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schizophrenia Working Group of the Psychiatric Genomics Consortium Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tiihonen J, et al. 11-year follow-up of mortality in patients with schizophrenia: A population-based cohort study (FIN11 study) Lancet. 2009;374:620–627. doi: 10.1016/S0140-6736(09)60742-X. [DOI] [PubMed] [Google Scholar]
- 49.Wang H, et al. GBD 2015 Mortality and Causes of Death Collaborators Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980-2015: A systematic analysis for the Global Burden of Disease Study 2015. Lancet. 2016;388:1459–1544. doi: 10.1016/S0140-6736(16)31012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brown S. Excess mortality of schizophrenia. A meta-analysis. Br J Psychiatry. 1997;171:502–508. doi: 10.1192/bjp.171.6.502. [DOI] [PubMed] [Google Scholar]
- 51.Berrington de Gonzalez A, et al. Body-mass index and mortality among 1.46 million white adults. N Engl J Med. 2010;363:2211–2219. doi: 10.1056/NEJMoa1000367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whitlock G, et al. Prospective Studies Collaboration Body-mass index and cause-specific mortality in 900 000 adults: Collaborative analyses of 57 prospective studies. Lancet. 2009;373:1083–1096. doi: 10.1016/S0140-6736(09)60318-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.OECD 2013 Education at a Glance 2013 (Organisation for Economic Co-operation and Development, Paris). Available at www.oecd-ilibrary.org/content/book/eag_highlights-2013-en. Accessed December 10, 2013.
- 54.Morris JN, Blane DB, White IR. Levels of mortality, education, and social conditions in the 107 local education authority areas of England. J Epidemiol Community Health. 1996;50:15–17. doi: 10.1136/jech.50.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huisman M, et al. Educational inequalities in cause-specific mortality in middle-aged and older men and women in eight western European populations. Lancet. 2005;365:493–500. doi: 10.1016/S0140-6736(05)17867-2. [DOI] [PubMed] [Google Scholar]
- 56.Zheng J, et al. Early Genetics and Lifecourse Epidemiology (EAGLE) Eczema Consortium LD Hub: A centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics. 2017;33:272–279. doi: 10.1093/bioinformatics/btw613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oliver BR, Plomin R. Twins’ Early Development Study (TEDS): A multivariate, longitudinal genetic investigation of language, cognition and behavior problems from childhood through adolescence. Twin Res Hum Genet. 2007;10:96–105. doi: 10.1375/twin.10.1.96. [DOI] [PubMed] [Google Scholar]
- 58.Haworth CMA, Davis OSP, Plomin R. Twins Early Development Study (TEDS): A genetically sensitive investigation of cognitive and behavioral development from childhood to young adulthood. Twin Res Hum Genet. 2013;16:117–125. doi: 10.1017/thg.2012.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kovas Y, Haworth CMA, Dale PS, Plomin R. The genetic and environmental origins of learning abilities and disabilities in the early school years. Monogr Soc Res Child Dev. 2007;72:vii, 1–144. doi: 10.1111/j.1540-5834.2007.00439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vilhjálmsson BJ, et al. Schizophrenia Working Group of the Psychiatric Genomics Consortium, Discovery, Biology, and Risk of Inherited Variants in Breast Cancer (DRIVE) study Modeling linkage disequilibrium increases accuracy of polygenic risk scores. Am J Hum Genet. 2015;97:576–592. doi: 10.1016/j.ajhg.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Visscher PM, Yang J. A plethora of pleiotropy across complex traits. Nat Genet. 2016;48:707–708. doi: 10.1038/ng.3604. [DOI] [PubMed] [Google Scholar]
- 62.Johnson W, et al. Does education confer a culture of healthy behavior? Smoking and drinking patterns in Danish twins. Am J Epidemiol. 2011;173:55–63. doi: 10.1093/aje/kwq333. [DOI] [PubMed] [Google Scholar]
- 63.Janecka M, et al. Advanced paternal age effects in neurodevelopmental disorders-review of potential underlying mechanisms. Transl Psychiatry. 2017;7:e1019. doi: 10.1038/tp.2016.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gage SH, Davey Smith G, Ware JJ, Flint J, Munafò MR. G = E: What GWAS can tell us about the environment. PLoS Genet. 2016;12:e1005765. doi: 10.1371/journal.pgen.1005765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McCarthy S, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Gene. 2016;48:1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang J, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: A tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rosseel Y. lavaan: An R package for structural equation modeling. J Stat Softw. 2012;48:1–36. [Google Scholar]
- 69.Sidak Z. On probabilities of rectangles in multivariate student distributions: Their dependence on correlations. Ann Math Stat. 1971;42:169–175. [Google Scholar]
- 70.Richmond RC, et al. Using genetic variation to explore the causal effect of maternal pregnancy adiposity on future offspring adiposity: A mendelian randomisation study. PLoS Med. 2017;14:e1002221. doi: 10.1371/journal.pmed.1002221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang G, et al. Assessing the causal relationship of maternal height on birth size and gestational age at birth: A mendelian randomization analysis. PLoS Med. 2015;12:e1001865. doi: 10.1371/journal.pmed.1001865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Eaves LJ, Pourcain BS, Smith GD, York TP, Evans DM. Resolving the effects of maternal and offspring genotype on dyadic outcomes in genome wide complex trait analysis (“M-GCTA”) Behav Genet. 2014;44:445–455. doi: 10.1007/s10519-014-9666-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.