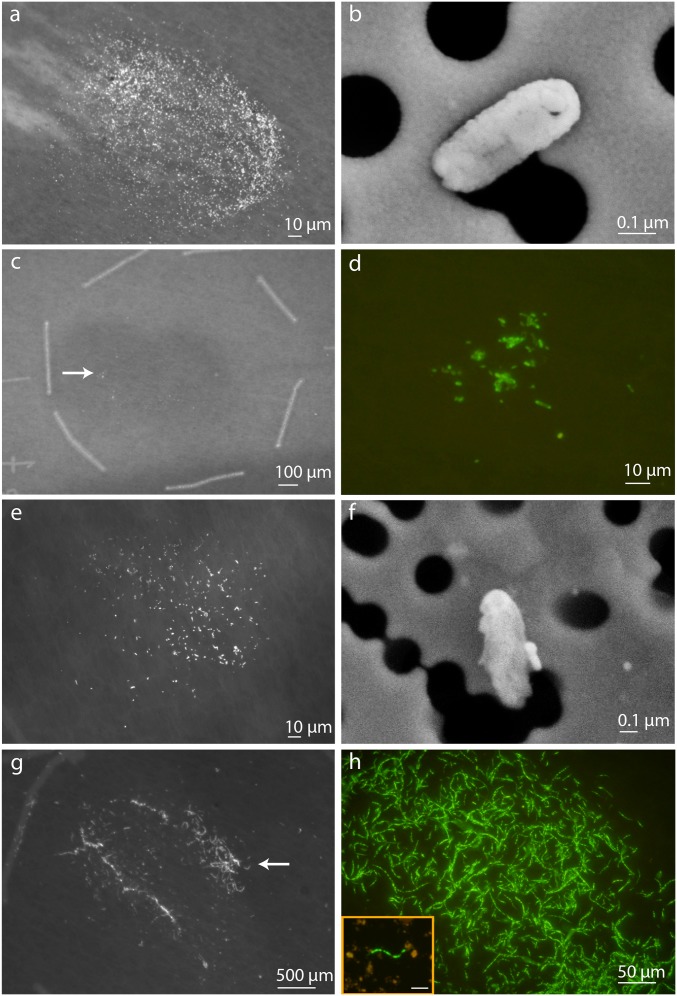
Significance
Microbial cells are widespread in diverse deep subseafloor environments; however, the viability, growth, and ecophysiology of these low-abundance organisms are poorly understood. Using single-cell–targeted stable isotope probing incubations combined with nanometer-scale secondary ion mass spectrometry, we measured the metabolic activity and generation times of thermally adapted microorganisms within Miocene-aged coal and shale bed samples collected from 2 km below the seafloor during Integrated Ocean Drilling Program Expedition 337. Microorganisms from the shale and coal were capable of metabolizing methylated substrates, including methylamine and methanol, when incubated at their in situ temperature of 45 °C, but had exceedingly slow growth, with biomass generation times ranging from less than a year to hundreds of years as measured by the passive tracer deuterated water.
Keywords: subseafloor life, coal bed biosphere, NanoSIMS, stable isotope probing, microbial generation time
Abstract
The past decade of scientific ocean drilling has revealed seemingly ubiquitous, slow-growing microbial life within a range of deep biosphere habitats. Integrated Ocean Drilling Program Expedition 337 expanded these studies by successfully coring Miocene-aged coal beds 2 km below the seafloor hypothesized to be “hot spots” for microbial life. To characterize the activity of coal-associated microorganisms from this site, a series of stable isotope probing (SIP) experiments were conducted using intact pieces of coal and overlying shale incubated at in situ temperatures (45 °C). The 30-month SIP incubations were amended with deuterated water as a passive tracer for growth and different combinations of 13C- or 15N-labeled methanol, methylamine, and ammonium added at low (micromolar) concentrations to investigate methylotrophy in the deep subseafloor biosphere. Although the cell densities were low (50–2,000 cells per cubic centimeter), bulk geochemical measurements and single-cell–targeted nanometer-scale secondary ion mass spectrometry demonstrated active metabolism of methylated substrates by the thermally adapted microbial assemblage, with differing substrate utilization profiles between coal and shale incubations. The conversion of labeled methylamine and methanol was predominantly through heterotrophic processes, with only minor stimulation of methanogenesis. These findings were consistent with in situ and incubation 16S rRNA gene surveys. Microbial growth estimates in the incubations ranged from several months to over 100 y, representing some of the slowest direct measurements of environmental microbial biosynthesis rates. Collectively, these data highlight a small, but viable, deep coal bed biosphere characterized by extremely slow-growing heterotrophs that can utilize a diverse range of carbon and nitrogen substrates.
Advances in deep-sea drilling technology and analytical methods for cored samples have led to the discovery of microbial life in a range of deep subseafloor habitats, from Earth’s most oligotrophic sediments (1) to its largest aquifer (2). To probe the limits for microbial life, Integrated Ocean Drilling Program (IODP) Expedition 337 sampled a deeply buried [2 km below seafloor (kmbsf)] coal bed system of terrestrial origin and low thermal alteration at Site C0020. In addition to increased depth, temperature, and organic carbon input from coal beds, Site C0020 lacked the dominant electron acceptors (oxygen and sulfate) that have been the focus of other deep-biosphere–focused IODP expeditions (3, 4). Initial results from Expedition 337 showed cell abundances no longer tracked the global depth trend based on previous cruises (5), suggesting that life-limiting conditions may have been reached at these extreme depths (6). The coal bed, however, contained comparatively high concentrations of microbial cells, 10- to 1,000-fold higher than in the adjacent shale and sandstone lithologies, which is a feature common to lignite-containing terrestrial sediments, where higher organic substrate concentrations sourced from the lignite can sustain higher cell numbers (7, 8). Recovered 16S rRNA gene diversity from the coal beds revealed an assemblage that phylogenetically resembled modern terrestrial environments (e.g., peat or forest soil), which was interpreted as representing indigenous microorganisms within Miocene-aged coal, rather than contaminants or overprinting of commonly observed marine sediment microbes (6).
While the concept of coal beds hosting microbial life for millions of years has existed for almost a century (9), microbial activity and metabolic potential through time remain largely unknown in these systems. Measuring activity in the deep biosphere requires a balance between stimulating life enough to observe a metabolic signal and overstimulation of the system such that experimental conditions no longer approximate in situ conditions (10). Subseafloor microbial metabolic activity, such as bacterial sulfate reduction, is often interpreted through geochemical profiles or microcosm incubations to determine rates of substrate utilization (11–14). Other approaches include traditional (13) and subseafloor-adapted (15) cultivation-based methods, analysis of actively transcribed genes (16), and biomolecule degradation modeling (17) from recovered core materials. These bulk methods have been valuable in developing a picture of the deep subseafloor biosphere characterized by viable, but extremely slow-growing, microorganisms with a range of metabolisms. There are limitations to these methods in some subseafloor habitats like the Shimokita coal bed, off Japan, where biomass is extremely low (1–1,000 cells per cubic centimeter), and bulk analyses may lack the sensitively to detect microbial activity and growth. Stable isotope probing (SIP) coupled to single-cell–targeted nanometer-scale secondary ion mass spectrometry (NanoSIMS) analysis is well suited for direct activity measurements in low biomass environments and has been applied in select deep subseafloor biosphere studies (18). This technique additionally offers a unique perspective on the cell-to-cell variation in growth and metabolic potential of co-occurring microorganisms in nature.
Here we used SIP-NanoSIMS to determine the viability and metabolic capability of microorganisms associated with the Shimokita Peninsula coal bed and surrounding lithologies since burial over 20 Mya. We focused specifically on methyl substrate metabolism, which could be hypothetically sourced from coal breakdown products, and rates of microbial growth in long-term, high-temperature microcosm experiments. Taking advantage of the ability to measure three different stable isotope tracers in parallel with NanoSIMS, we applied combinations of 13C- and 15N-labeled substrates in tandem with deuterated water (2H2O) as a passive tracer for growth. The use of 2H2O with NanoSIMS is an effective method for measuring single-cell microbial biosynthesis with high sensitivity, capable of detecting extraordinarily slow rates of growth (19) predicted to be characteristic of subseafloor biosphere ecosystems (20). This tracer has been successfully combined with lipid biomarker and NanoSIMS analyses for assessing rates of growth in pure culture and mammalian microbiome studies (19, 21, 22), soils (23), and seabed methane seep environments (24, 25), but it has not yet been applied to study microbial growth in low-biomass, deep subsurface ecosystems. This multiple-isotope SIP-NanoSIMS study of microbial life in deep subseafloor environments recovered evidence for methyl substrate metabolism in deep coal and shale beds and exceedingly slow growth within this low-abundance, high-temperature microbial community.
Materials and Methods
SIP Incubation Preparation.
IODP Expedition 337 operations commenced July 26 and continued through September 30, 2012, on the drilling research vessel Chikyu. Utilizing riser drilling, a sedimentary sequence was recovered down to 2,466 mbsf at Site C0020 Hole A (41°10′36′′N, 142°12′02′′E) in a water depth of 1,180 m off the Shimokita Peninsula, Japan. The drilled sequence transitioned from open marine (youngest; late Pliocene, ∼5 Ma) to terrestrial (oldest; late Oligocene, ∼30 Ma) with depth. Models for maximum temperature reached by Expedition 337 coring report 63.7 °C (26). Shipboard sedimentological, geochemical, and microbiological data and methods are available through IODP publications (27). Additional coal petrography is available in a report by Gross et al. (28).
A total of 52 incubation amendment conditions were prepared onboard to interrogate a range of potential deep biosphere metabolic strategies, and then incubated back in the laboratory at temperatures approximating in situ measurements (27). In this study, incubations from shale (core 8L4, 1,606 mbsf, 37 °C incubation temperature), coal (core 15R3, 1,921 mbsf, 45 °C incubation temperature), and mixed (homogenized mixture from multiple cores 19R1, 19R5, 19R7, 20R3, 23R6, 23R8, 24R3, 25R1, 25R2, and 25R3; 1,950–1,999 mbsf; 45 °C incubation temperature) with methanol, methylamine, and ammonium substrate additions were analyzed. Age estimates of these samples are early to middle Miocene (6). In situ temperatures ranged from 38–48 °C at these sample depths, with pressures of ∼30 MPa (27). Two coal beds were included in these incubations: a shallower coal-only sample deposited under more marine-influenced conditions (∼1,921 mbsf, core 15R3) and a deeper coal bed deposited under more limnic conditions included in the mixed lithology sample (∼2,000 mbsf, cores 24R3 and 25R1).
Cores used for incubations were prepared by removal of outer drill-fluid–contaminated layers by means of a sterile ceramic knife as soon as possible after core recovery and stored at 4 °C until incubation preparation, while maintaining an anaerobic atmosphere during the entire process. For preparation of the SIP incubations, the interior portion of the core was manually crushed into centimeter-sized pieces under sterile, anaerobic conditions and distributed evenly into sterile 50-mL glass vials with butyl rubber stoppers and screw caps (Nichidenrika-Glass Co. Ltd.). Vials were flushed with argon and pressurized to 1 atm argon headspace. Sterile C-, N-, and S-free media (1% PBS, 30 g/L NaCl, 12 g/L MgCl2, and 3 g/L KCl) was prepared anaerobically with 2H2O [20 atom % (at. %) 2H2O]. This value (20 at. % 2H2O) was selected as the highest level of enrichment with little to no effect on the activity of microorganisms in pure culture (21, 22). Time point 1, time point 2, and autoclaved treatments were prepared for each substrate condition. Time point 1 incubations lasted for 6 mo, while time point 2 and autoclaved treatments were maintained at the in situ incubation temperature for 2.5 y. Due to low levels of activity ascertained from geochemical measurements, all NanoSIMS analyses were conducted on time point 2 and autoclaved samples. Amendments and incubation conditions for the methyl-substrate subset analyzed in this study are listed in Table 1. Equimolar amounts of substrate (30 μmol C, 1.5 mM final; 3 μmol N, 0.15 mM final) were added across incubation conditions at 50 at. % enrichment (Cambridge Isotopes). Hydrogen was added as 5 mL of 100% H2 overpressure to incubations (∼0.15 atm H2). A full list of the additional incubation conditions prepared onboard is provided in cruise methods (29). Alkalinity (34.39–9.68 mM) and ammonium (2.80–1.83 mM) concentrations from formation fluid samples collected onboard exceed concentrations of C and N amendments (30). Concentrations of methylamine (0.05 mM) and methanol (1 mM) measured from another study of lignite coal (31) also suggested our substrate additions were environmentally relevant.
Table 1.
Geochemical and cell enumeration for SIP-NanoSIMS incubations
| DIC, mM | δ13C-DIC, ‰ | CH4/Ar, ‰ | δ13C-CH4, ‰ | δ2H-CH4, ‰ | Cells, g (rock) | Total cells | Percent active | |||||||||||
| Lithology | 13C source, 1.5 mM | 15N source, 0.15 mM | H2, 15% | A | − | A | − | A | − | A | − | − | A | − | A | − | A | − |
| Shale - 8L4 (1,606 mbsf) | — | — | — | 0.36 | nm | −0.46 | nm | nm | nm | nm | nm | nm | nm | 770 | nm | nm | nm | nm |
| — | MeAm. | Y | 0.28 | 0.10 | 0.66 | −1.24 | 0.2 | 0.3 | nm | nm | nm | 851 | 1,548 | 1E + 05 | 1E + 05 | nd | nd | |
| — | MeAm. | — | 0.30 | 2.31 | 4.62 | 4.80 | 0.1 | 0.3 | nm | nm | nm | 153 | 5,873 | 2E + 04 | 6E + 05 | 5 | 26 | |
| MeAm. | Am. | Y | 0.51 | 0.72 | 8.32 | 5.72 | 0.1 | 0.2 | nm | nm | nm | 1,205 | 95 | 1E + 05 | 1E + 04 | 0 | 61 | |
| MeAm. | Am. | — | 0.28 | 0.40 | −2.86 | 2.45 | 0.1 | 0.2 | nm | nm | nm | 26,717 | 1,664 | 3E + 06 | 2E + 05 | nd | 33 | |
| MeOH | Am. | Y | 0.74 | 0.69 | 123.87 | 430.03 | 0.1 | 0.2 | nm | nm | nm | 539 | 7,503 | 5E + 04 | 1E + 06 | nd | 78 | |
| MeOH | — | — | 0.09 | 1.16 | 6.43 | 261.94 | 0.2 | 0.1 | nm | nm | nm | 1,948 | 1,967 | 2E + 05 | 2E + 05 | 0 | 25 | |
| Coal - 15R3 (1,921 mbsf) | — | — | — | 0.16 | nm | −10.33 | nm | nm | nm | nm | nm | nm | nm | 60 | nm | nm | nm | nm |
| — | MeAm. | Y | 0.06 | 0.07 | −6.00 | −3.77 | 1.1 | 5.5 | −42.8 | −46.1 | −170.4 | 69 | 263 | 7E + 02 | 3E + 03 | 5 | 6 | |
| — | MeAm. | — | 0.04 | 0.23 | −16.19 | −7.20 | 0.8 | 8.6 | −57.2 | −55.7 | −198.2 | 21* | 70,363 | 1E + 02 | 9E + 05 | 0 | 61 | |
| MeAm. | Am. | Y | 0.02 | 0.03 | 88.77 | 20.10 | 0.8 | 4.5 | −42.3 | −55.0 | −144.1 | 37* | 529 | 3E + 02 | 7E + 03 | 14 | 19 | |
| MeAm. | Am. | — | 0.11 | 0.03 | 108.06 | 55.79 | 1.0 | 8.1 | −58.0 | −58.8 | −158.2 | 4,864 | 34* | 4E + 04 | 4E + 02 | 7 | 4 | |
| MeOH | Am. | Y | 0.06 | 0.08 | 19.09 | −6.46 | 2.4 | 5.4 | −49.7 | −52.9 | −96.5 | 32* | 268 | 4E + 02 | 4E + 03 | nd | nd | |
| MeOH | — | — | 0.07 | 0.05 | −15.14 | 26.67 | 1.2 | 3.8 | −57.5 | −56.6 | bd | 7,693 | 11,046 | 6E + 04 | 1E + 05 | 0 | 33 | |
| Mixed - 19:25R (1,950–2,000 mbsf) | — | — | — | 0.16 | nm | 4.29 | nm | nm | nm | nm | nm | nm | nm | 22,200 | nm | nm | nm | nm |
| — | MeAm. | Y | 1.48 | 1.38 | 5.63 | 3.35 | 0.2 | 1.4 | nm | nm | nm | 32* | 24* | 8E + 02 | 7E + 02 | nd | nd | |
| — | MeAm. | — | 0.90 | 1.97 | 5.72 | 6.85 | 0.1 | 2.0 | nm | nm | nm | 24 | 43 | 7E + 02 | 1E + 03 | nd | 8 | |
| MeAm. | Am. | Y | 0.98 | 0.97 | 38.45 | 37.73 | 0.1 | 0.5 | nm | nm | nm | 17* | 13* | 6E + 02 | 5E + 02 | 7 | 11 | |
| MeAm. | Am. | — | 1.73 | 1.79 | 48.63 | 327.56 | 0.1 | 0.1 | nm | nm | nm | 42 | 130 | 9E + 02 | 5E + 03 | nd | nd | |
| MeOH | Am. | Y | 1.54 | 1.76 | 6.32 | 14.87 | 0.2 | 2.2 | nm | nm | nm | 16* | 271 | 5E + 02 | 1E + 04 | nd | 37 | |
| MeOH | — | — | 1.83 | 1.96 | 9.42 | 44.64 | 0.4 | 2.2 | nm | nm | nm | 14* | 2,692 | 5E + 02 | 1E + 05 | 15 | 100 | |
Geochemical summary of incubations by lithology and substrates added [MeAm., methylamine (tan); MeOH, methanol (gray); Am., ammonium], with 2H2O only (no C or N added) provided as a background sample, after 864 d incubation. Addition of hydrogen to incubation is indicated by Y (yes). Samples are in paired columns of autoclaved (A) and untreated (−). DIC concentration, 13C-carbon isotopic composition of DIC, ratio of methane (mass 16) to argon (mass 40), and 13C-carbon and 2H-hydrogen isotopic composition of methane in incubation headspace in permil are shown. Due to low yields, methane isotopic analyses were only performed on coal incubations. Only nonautoclaved samples had enough methane to determine 2H enrichment, except for the last condition, which was below detection (bd). Cell density was measured from incubation separates before NanoSIMS sorting after 864 d for all substrate-added conditions and after 1,140 d for no-substrate–added control (2H2O only). Total cells were calculated from estimates of grams of rock per incubation. The active cell percentage is the number of ROIs above 10-fold natural abundance for 2H. nd, not detected (specifically, no cells were detected on the NanoSIMS membrane); nm, not measured.
Indicates a sample of lower statistical reliability that was below the quantification limit (negative control) but cells were detected. Errors on DIC measurements are ±0.5‰ δ13C-DIC and ±0.0086 μmol DIC.
After 30 months of incubation (March 2014), all treatments were sampled for geochemical analyses before preparation for NanoSIMS. Three milliliters of headspace gas was removed to a vial filled with 0.1 M NaOH for methane analysis. About 1 mL of liquid was filtered through a 0.1-μm 13-mm Whatman Polycarbonate Nuclepore Track-Etched Membrane (110405) for dissolved inorganic carbon (DIC) analysis. A detailed description of methane and DIC analyses is provided in Supporting Information.
Sample Preparation for NanoSIMS Analysis.
To overcome technical challenges for NanoSIMS analysis of low-biomass samples, cell separation and fluorescence-activated cell sorting (FACS) were used to directly concentrate cells in a small analysis area, ∼1–0.5 mm2 (32). NanoSIMS samples were prepared from paraformaldehyde (PFA)-fixed cell separates after 30 months of incubation. Cell preservation, separation, enumeration, and FACS were all conducted in the clean booth and clean room facilities at the Kochi Institute for Core Sample Research, Japan Agency for Marine-Earth Science and Technology. Half of the solid portion and half of the liquid portion of each sample were fixed overnight in a solution of 2% PFA, 3× PBS. Samples were then subjected to two washes, incubating in 3× PBS for 6 h and then for 2 h after each wash. Samples were centrifuged (3,500 × g), and supernatant was decanted after each wash. PFA-fixed samples were stored in 50% ethanol, 3× PBS. The other half of the sample was preserved in glyTE (70% glycerol, 100 mM Tris, and 10 mM EDTA; Bigelow Single Cell Genomics Center preservation protocol), frozen by a cell alive system (CAS; ABI Corporation, Ltd.), and stored at −80 °C (33).
One milliliter of liquid and ∼1 g of sediment chips were subsampled by pipette and sterile cell culture loop, respectively, from the PFA-fixed sample. Cell separation, microscopy, and sorting procedures followed the method of Morono et al. (32), with the following modifications to improve cell recovery: (i) Samples were sonicated (Bioruptor UCD-250; COSMO BIO) in an ice bath for 20 cycles of 30 s at 200 W on and 30 s off, and (ii) samples were incubated in hydrofluoric acid after initial sonication, rather than after first-density gradient separation. The cell detection limit was determined by no-sample–added controls run in parallel with samples. The same cell enumeration method was used for incubations and initial shipboard samples.
Cells were stained with SYBR Green I [1:40 dilution of SYBR Green in TE (10 mM Tris and 1 mM EDTA)] and sorted following the flow cytometry protocol of Morono et al. (32). Sorted cells were concentrated directly from the sorter onto NanoSIMS-compatible 0.2-μm polycarbonate filters coated with indium tin oxide (ITO) as described by Morono et al. (18) and Inagaki et al. (6). ITO coating on polycarbonate membranes (Isopore GTBP02500; Millipore) was prepared by a sputtering deposition technique at Astellatech Co. Ltd. Scanning electron microscopy of the filters was done on a Zeiss 1550 VP Field Emission Scanning Electron Microscope at the Geological and Planetary Sciences Division Analytical Facility at the California Institute of Technology (Caltech), and SYBR-stained cells were imaged with a BX51 epifluorescence microscope (Olympus) using 20× (UPlanFL N) dry, 60× (PlanApo N), and 100× (UPlanFL N) oil immersion objectives (Fig. S1).
Fig. S1.
SYBR-stained cells after separation and FACS concentration on ITO-coated 0.2-μm polycarbonate NanoSIMS membranes from the two highest cell abundance conditions in 15R3 coal and mixed lithology. SYBR and SEM images highlight some distinctions in cell morphology between the two methanol amended samples. White arrows indicate region of SYBR image on larger membrane target area. The 15R3 coal incubation amended with methylamine (A), SEM image of A (B), 15R3 coal amended with methanol (C), C magnified with false color (D), mixed lithology amended with methanol + H2 + ammonium (E), SEM image of E (F), mixed lithology amended with methanol (G), and mixed lithology amended with methanol magnified with false color (H; Inset, the same sample filtered before cell sorting to demonstrate cell density without concentration). (Scale bar: H, Inset, 20 μm.)
NanoSIMS Instrument Tuning and Analysis.
Cell targets were identified (by SYBR stain) and marked on NanoSIMS membranes with a laser dissection microscope (LMD6000; Leica Microsystems) for ease of rediscovery on the NanoSIMS. Samples were analyzed by raster ion imaging with a CAMECA NanoSIMS 50L at the Caltech Microanalysis Center in the Division of Geological and Planetary Sciences. A focused primary Cs+ beam of ∼1 pA was used for sample collection, with rasters of 256 × 256 or 512 × 512 pixels. Ions 1H– (EM 1), 2H– (EM 2), 12C2– (EM 3), 13C12C– (EM 4), 12C14N– (EM 5), and 12C15N– (EM 6) were measured simultaneously. Technical development has been described by Kopf et al. (22). Collection began after a presputtering of equal intensity to one collection frame (∼45 min). Recorded images and data were processed using Look@NanoSIMS software (34). Images were deadtime-corrected, and individual ion image frames were merged and aligned using the 12C14N ion image to correct for drift during acquisition. Cell-based regions of interest (ROIs) were determined by “interactive thresholding” with the 12C14N ion image. Final ion images and counts per ROI were calculated by summation of ion counts for each pixel over all scans. Calculation of cell size from Look@NanoSIMS software was based on the diameter of a circle (in micrometers) derived from the equivalent number of pixels from the ROI drawn on the NanoSIMS 14N12C ion image. We also confirmed that cell ROIs had a total C-to-total N ratio that was distinct from the background correction ROIs or coal to ensure drawn ROIs only included biomass targets.
Sample Preparation for DNA and iTag Processing.
Cells for gDNA extraction were recovered from unfixed samples stored in glycerol using a modified version of the NanoSIMS protocol by omitting detergent and methanol treatments and only shaking (60 min at 500 rpm using a Shake Master; Bio Medical Science) before centrifugation. Nycodenz was also omitted from the gradient separation solution to prevent PCR inhibition (linear gradients of 10%, 30%, and 67% sodium polytungstate solutions were utilized instead). After separation, the staining and flow cytometry sorting protocol was identical to NanoSIMS processing, with the exception that the cells were collected into a UV sterilized 0.5-mL PCR tube rather than on the ITO filter. Sorted cells were stored at −20 °C until DNA extraction. A proteinase K digestion mixture consisting of 20 mM Tris⋅HCl (pH 8.0), 5 mM MgCl2, 2 mM DTT, 4 U/mL heat-labile double-strand specific DNase (HL-dsDNase; ArcticZymes), and 0.4 mg/mL proteinase K was utilized for cell lysis. Before proteinase K application, potential endogenous DNA contamination in the proteinase K mixture was removed by digestion using HL-dsDNase. The HL-dsDNase extracellular DNA digestion was carried out at 25 °C for 2 h, followed by an inactivation step at 60 °C for 1 h. The sorted cells and negative controls (sheath fluid) were then lysed using the pretreated proteinase K. One-third volume of the HL-dsDnase–treated proteinase K solution was added to one volume of sheath fluid, which contained the sorted cells (total volume varied by cell sort time). Proteinase K digestion was performed at 60 °C for 8 h, followed by proteinase K inactivation at 95 °C for 10 min. The resultant DNA extract was then used directly for first-round PCR amplification with universal primers targeting the V4 region of the 16S rRNA gene (U515F: 5′-TGY CAG CMG CCG CGG TAA-3′, U806R: 5′-GGA CTA CHV GGG TWT CTA AT-3′). First-round PCR amplicons were quantified and purified via gel electrophoresis and then indexed and barcoded in the second PCR assay. The iTag sequencing was performed on a MiSeq Illumina platform following the methods outlined by Hoshino and Inagaki (35). Resultant paired-end reads were processed and assigned taxonomy using Qiime (36) and a custom version of the URef_NR99_119_SILVA database (37, 38) optimized for marine sedimentary microorganisms. The total sequences and relative abundance of most abundant taxa are provided in Dataset S1, and all taxa are provided in Dataset S2.
Results
Evaluation of Incubation Cellular Abundances and Sample Purity.
Stringent contamination monitoring during Expedition 337 suggested sandstone lithologies were the most susceptible to drilling mud contamination (27). Therefore, only shale and coal lithologies were used in this study. While cells were detected in the drilling fluids [2.66 × 108 cells per milliliter (30)] and have the potential to contaminate downstream analyses (6), two lines of evidence suggest exogenous cell contamination was minimal in our long-term stable isotope incubation study. First, molecular screening of the 16S rRNA sequences recovered from our 37 °C and 45 °C incubations was distinct from that of sequences associated with common drilling mud contaminants from Expedition 337 (6). Second, cultivation attempts to enrich for bacteria (39) or fungi (40) from drilling fluids were unsuccessful, suggesting that contaminant cells had low viability and were unlikely to overgrow the indigenous microorganisms in our moderately thermophilic, low-nutrient coal and shale incubations. The low microbial viability of exogenous microorganisms may be due to the chemical composition of the drilling mud, containing bentonite and detergent chemicals, as well as the extreme physical conditions experienced during circulation of the high pH (10.5–11) drilling fluids between the 2-kmbsf drilling formation and the ship, with repeated cycling between oxic and anoxic conditions and temperatures ranging from 2–60 °C.
Cell abundances were calculated from concentrates after cell separation from the rock matrix (Table 1). Cell numbers measured at the end of the incubations, including no amendment controls, were all elevated relative to cell concentrations calculated from shipboard-processed samples, reported as 7 cells per gram of shale, 200 cells per gram of coal, and 942 cells per gram of mixed lithology (6). The elevated cell concentration in the incubated no-amendment controls suggested that the addition of water alone could have been enough to stimulate growth in this system using substrates native to the coal and shale. Microbial cells stained with SYBR Green I appeared as small cocci (single or doublets) or rods, with the exception of the mixed incubation, which also contained filamentous microorganisms (Fig. S1). The range of microbial cell sizes determined from NanoSIMS ion images yielded median cell lengths ranging from 0.7–1.2 μm in shale, 0.3–0.9 μm in coal, and 0.4–1.0 μm in the mixed incubations, consistent with previous reports from deep subseafloor sediments (41).
Assessing Methylated Substrate Metabolism by Bulk 13C and 2H Geochemical Analysis of Methane and DIC Measurements.
Total methane production at the end of the incubation period was higher in coal incubations, relative to shale and mixed samples, and highest overall in nonautoclaved coal samples (Table 1). Lower, but detectable, levels of methane production were also measured in the mixed incubations. The δ13C-CH4 was measured for coal incubations, which showed minimal 13C enrichment from the added 13C-methanol or methylamine (−42.3 to −58.8‰) relative in situ methane values [approximately −62‰ in both formation water and core gas sampling (6)]. Kinetic isotope effects during methane desorption could account for a portion of the methane enrichment [<5‰ (42, 43)], but the higher δ13C-CH4 enrichments (∼15–20‰) are unlikely explained by kinetic fractionation alone, and most likely contain some 13C-substrate incorporation. If all isotopic enrichment in these heaviest samples were from 13C substrate, it would correspond to ∼0.04% of total methane being sourced from the added 13C-methylamine or 13C-methanol. Since H2 in the incubations is predicted to exchange with 2H2O (44), any H2-derived methane produced over the course of the incubations should also be enriched in δ2H-CH4. As observed with 13C measurements, the values of δ2H-CH4 were also minimally enriched in deuterium (−96.5 to −198‰) relative to the in situ δ2H-CH4 [approximately −200‰ in both formation water and core gas sampling (6)]. The most enriched δ2H-CH4 was observed in incubations amended with 13C-methanol + 15N-ammonium + H2 (−96.5‰). Isotope mass balance indicates this level of enrichment could at most account for ∼0.007% of the total methane recovered in the incubation. Therefore, both isotopic methods demonstrate consistently low, but detectable, levels of biogenic methane production in the coal incubations, with carbon isotope data supporting the presence of some methylotrophic methanogenesis.
Methylotrophic methanogenesis conversion of the 13C-labeled methanol or methylamine should also lead to the production of 13C-DIC in our incubations. Although shale incubations had overall higher DIC concentrations than incubations with only coal, DIC concentrations varied widely within each lithology. Methanol-amended shale incubations produced enriched δ13C-DIC values, with the highest values observed in the 13C-methanol + 15N-ammonium + H2 amendment (+430.03‰). In contrast, methylamine amendments to the shale incubations did not show any appreciable enrichment (−2.86 to +8.32‰). The opposite trend was observed in the coal incubations, where 13C-DIC production from 13C-methanol was minimal (−15.14 to +26.67‰), but some of the 13C-methylamine incubations (autoclaved treatment) produced enriched δ13C-DIC (+108.06‰). The 13C-DIC trends with the mixed incubations were similar to what was observed in the coal incubations, with the highest enrichment in 13C-methylamine amendments (+327.56‰) and lower activity observed with the addition of 13C-methanol (+6.32 to +44.64‰). By comparison, the in situ gaseous 13C-CO2 ranged from −19 to +1‰, with the most 13C-enriched CO2 recovered from the coal beds (6). Assuming steady-state production during the incubation, the highest δ13C-DIC enrichment in coal would correspond to 2–0.2 pmol DIC per cm3⋅d−1 or 0.2–0.1% of total moles of DIC generated from 13C-methylamine and 13C-methanol, respectively. In summary, the observed minor contributions of labeled methane and DIC production in our incubations indicated the potential for both methane- and DIC-generating metabolisms (methylotrophic methanogenesis) in addition to DIC-only–generating metabolisms (methyl fermentation).
Single-Cell Substrate Uptake and Generation Time Estimates of Deep Subseafloor Microorganisms by 15N and 2H2O.
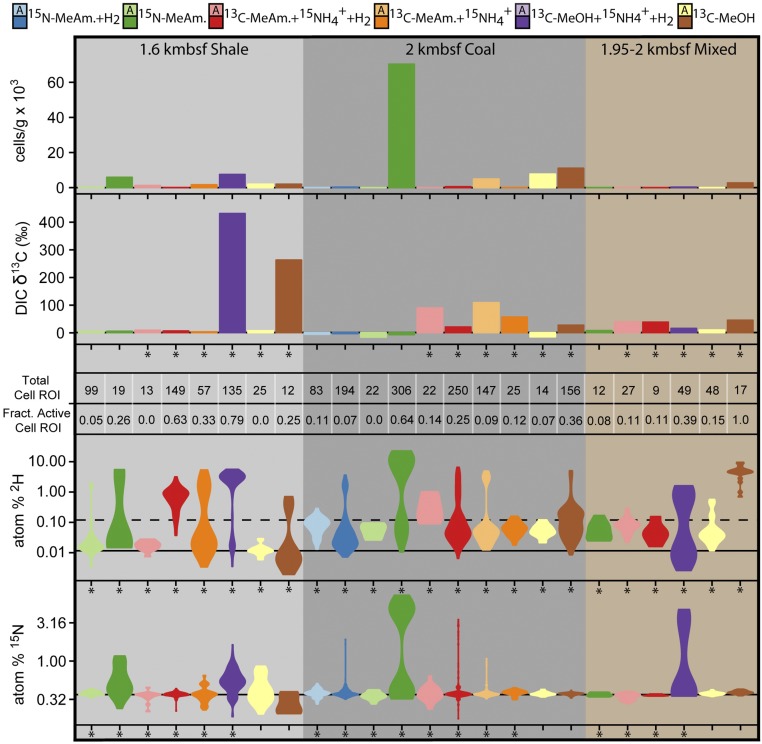
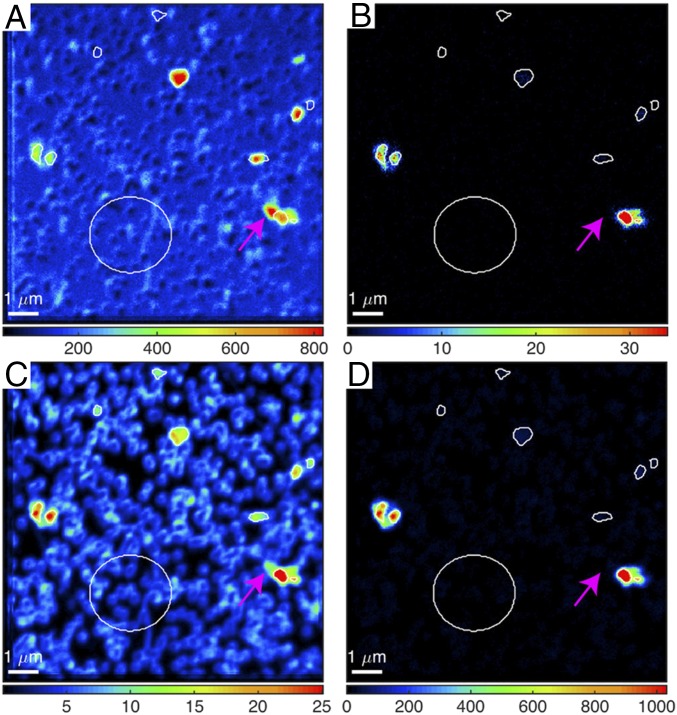
Of the 12 different sample conditions prepared for each lithology, eight shale, 10 coal, and six mixed lithology incubations had cells detectible by NanoSIMS after the cell separation protocol. The number of cellular ROIs analyzed per incubation ranged from nine to 306 (Fig. 1). Due to low cellular 13C enrichment relative to carbon background on NanoSIMS filters, single-cell 13C-carbon enrichment was within instrument error of natural abundance, and was therefore not included. Although it cannot be determined if active cells were from the regeneration of existing biomass or the generation of new cells through division, at least one instance of potential cell division was observed via NanoSIMS from the coal incubation amended with 15N-methylamine (Fig. 2).
Fig. 1.
NanoSIMS cellular ROIs are visualized by kernel density distribution in violin plots of enrichment in 2H from 2H2O and 15N from methylamine and ammonium in log-scale atom percent, along with incubation cellular abundance and bulk δ13C-DIC values from Table 1 [13C-methanol (MeOH) and 13C-methylamine (MeAm.)]. Incubation lithology is indicated by the shaded background for 1.6 kmbsf 8L4 shale (light gray), 2 kmbsf 15R3 coal (dark gray), and mixed 1.95–2 kmbsf (brown). Incubation conditions are indicated by color, where the lighter color of each pair is the autoclave-treated sample. An asterisk indicates that isotope label was added. The number of ROIs identified in NanoSIMS analysis and the “active” fraction of those ROIs (atom % 2H greater than 10-fold natural abundance) are recorded in tabular format. Solid line indicates natural abundance for each isotope, and the dashed line indicates the 2H threshold for active ROIs.
Fig. 2.
1H– ion (A), 2H– ion (B), 14N12C– ion (C), and 15N12C– ion (D) counts of ROIs from 15R3 coal with 15N-methylamine and 2H2O. The pink arrow indicates biomass that is only present in 1H and 14N12C ion maps and connected to biomass present in all four ion maps, indicative of new biomass growth. Large circles are used for nitrogen background standard correction. There are also six other ROIs drawn on the 14N12C map that do not show 15N12C or 2H enrichment for comparison.
The cellular isotopic enrichment in 2H and 15N was highest in the 45 °C 15N-methylamine coal incubations, with both isotopes showing a similar bimodal distribution of ROIs (Fig. 1). This sample also had the highest cell number of all incubations. When compared with the paired 15N-methylamine sample set with H2 added, cellular activities (2H and 15N enrichment) were lower, suggesting the methylamine-utilizing microorganisms in the coal could be inhibited by H2. Although not as high as 15N-methylamine, 2H and 15N enrichment was observed in treatments with 13C-methylamine + 15N-ammonium and 2H enrichment was seen with 13C-methanol in coal samples. In shale incubations, the highest enrichment in bulk δ13C-DIC and cellular 2H and 15N was seen in treatments amended with 13C-methanol + 15N-ammonium + H2. Bulk δ13C-DIC was not enriched in 13C-methylamine–amended shale incubations, yet 2H and 15N (from 15N-ammonium) enrichment was still observed in cells from these incubations. The 15N-methylamine–amended shale incubations without hydrogen also showed high 2H and 15N incorporation. Finally, mixed lithology incubations had the highest δ13C-DIC (+327‰) in the 13C-methylamine + 15N-ammonium treatment, although this incubation did not yield enough cells to analyze by NanoSIMS. From the mixed incubations that could be analyzed by NanoSIMS, the 13C-methanol amendment had the highest cellular 2H enrichment (no 15N was added to this incubation).
In summary, both methanol and methylamine treatments produced 13C-DIC in coal and mixed incubations, but only methanol produced 13C-DIC in the shale incubations. Cells enriched with 15N were recovered from both 15N-methlyamine– and 15N-ammonium–amended incubations in shale and coal incubations, but only 15N-ammonium incubations showed cellular 15N uptake for mixed lithology incubations. The 2H-enriched cells were recovered across all substrate combinations and all lithologies. While bulk δ13C-DIC suggested methanol and shale, and methylamine and coal amendments and lithologies were correlated, single-cell incorporation suggests that viability and activity in these incubations were not uniformly linked to substrate addition or lithology.
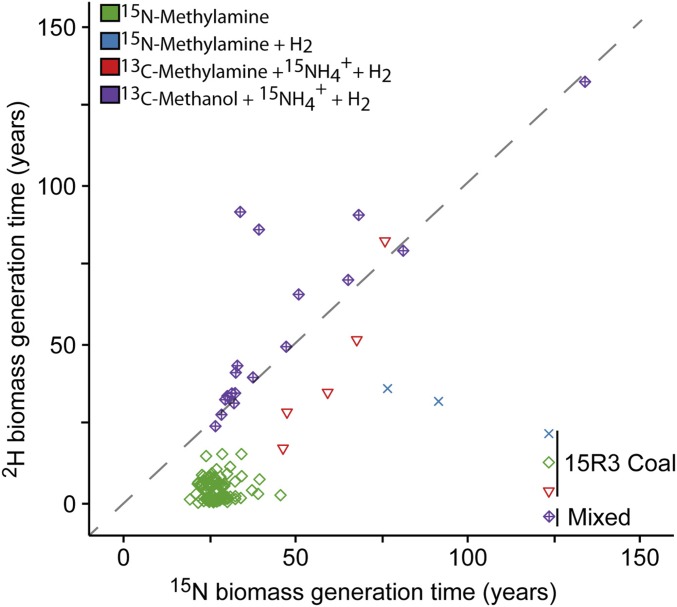
Biomass generation times were calculated from the most robust NanoSIMS cellular ROI enrichments in 2H and 15N (Materials and Methods). As such, our estimates of microbial generation times represent values for the fastest growing cells within our incubations (Fig. 3), yet still result in generation time estimates from a few months to more than 100 y. The greatest overall 2H and 15N incorporation was from coal amended with 15N-methylamine, with much longer generation times for the rest of the incubations, which all had hydrogen added. This concurs with 13C-DIC production results, where samples containing coal had depressed activity when H2 was added. This sample also had the greatest offset between 2H and 15N enrichment, where generation time estimates calculated from cellular 2H2O enrichment were shorter than 15N-nitrogen enrichment from methylamine. The mixed lithology samples amended with 15N-ammonium or 15N-methylamine as the nitrogen source had more comparable uptake of 15N and 2H, with slightly higher enrichment observed in the 15N-ammonium treatment. This suggests that while ammonium and methylamine can both be assimilated into biomass, ammonium was more readily used for anabolism compared with methylamine.
Fig. 3.
2H biomass generation time versus 15N biomass generation time, in years, calculated from ROIs where the 15N12N ion counts were above the Poisson error (Materials and Methods). The dashed line is where 2H and 15N generation time is at parity.
Comparison of 16S rRNA Gene Profiles to Geochemically Predicted Metabolisms.
Microbial diversity surveys using 16S rRNA gene iTag sequencing were generated from FAC sorting of the seven most active shale (n = 2), coal (n = 3), and mixed lithology (n = 2) SIP incubations after 2.5 y of incubation at 37 °C and 45 °C (Datasets S1 and S2). The recovered microbial diversity was broadly similar to the in situ microbial assemblage reported from the same coal and shale beds described by Inagaki et al. (6), with an abundance of anaerobic heterotrophic groups that more closely resembled terrestrial and wetland microbial lineages than marine sediments, many of which are generally associated with heterotrophs described from coal bed methane systems (45–47). The fact that the microbial taxa recovered were derived from intact FACS cells rather than environmental sample DNA extracts suggests that the diversity within the deep coal and shale SIP incubations is likely associated with viable microorganisms that were capable of persisting or growing in the long-term incubations at elevated temperatures and anoxic or microoxic conditions.
The 16S rRNA gene sequences within the bacterial domain were dominant in our diversity surveys of the seven most active incubations, with rare archaeal sequences affiliated with marine benthic group B detected in only two of the shale incubations incubated at 37 °C (Dataset S2). Some recovered taxa in the 45 °C incubation were related to moderately thermophilic microorganisms [e.g., Thermomonas within the Gammaproteobacteria (48), Acidothermus within the Acinetobacteria (49)], although the majority of the lineages recovered are not known for being thermotolerant. Thermophilic and spore-forming microorganisms often show enhanced cultivation and growth after high-pressure temperature treatment (50), consistent with the dominance of Firmicutes from the autoclaved coal incubation amended with methylamine and ammonium, reaching 76% of the total diversity, relative to 2–3% Firmicutes within the parallel nonautoclaved incubation (Dataset S1).
Dominant bacterial groups in the incubations included Actinobacteria, Firmicutes, Bacteroidetes, Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, and Deltaproteobacteria, with additional representatives belonging to the Chloroflexi (Dehalococcoidia), Deinococcus, Verucomicrobia (Opitatue), and Epsilonproteobacteria (Campylobacterales). At the genus level, microbial assemblages differed between samples, with no genus shared across all seven samples, and 37% of genera were found only in one of the seven samples. These differences were not statistically correlated with substrate amendment or lithology, despite bulk geochemistry analysis suggesting a preference for methanol utilization in shale and methylamine utilization in coal. There were also no statistically supported differences in the recovered diversity associated with lithology between the shale, coal, and mixed samples. When observed at lower taxonomic resolution and grouped by assigned taxonomic phylum, the microbial assemblages share the same dominant phyla across all incubations (Dataset S1). Therefore, while microdiversity exists at the genus level, the dominant phyla maintained over the 2.5-y incubation period were predominantly heterotrophic microorganisms (e.g., Gammaproteobacteria were at ≥10% relative abundance in all incubations).
Within these heterotrophic bacteria, there was some correlation between relative abundance of specific microbial taxa and hydrogen addition. For example, both shale and mixed lithology samples with 13C-methanol had a higher relative abundance of Firmicutes and Alphaproteobacteria in the absence of hydrogen addition to the headspace. In comparison, parallel hydrogen-amended incubations contained a higher percentage of Actinobacteria, suggesting potential utilization. While commonly described from deep subseafloor biosphere ecosystems, sequences affiliated with the Chloroflexi were only abundant (28% relative abundance) in the 13C-methanol mixed lithology incubation. Many members of the Chloroflexi have a characteristic filamentous morphology, making them likely candidates for the abundant filamentous cells recovered from this incubation (Fig. S1H).
While we did not recover sequences for methylotrophic methanogens, some of the bacterial lineages have cultured representatives reported to use methanol and/or methylamine as the sole carbon or energy source, including members of the Nocardia within the Actinobacteria (51), Rhizobiales [e.g., Rhodopseudomonas acidophilus (52)], Desulfovibrionales (Desulfovibrio fructosovorans) (53), and Firmicutes (54), especially many of the thermophilic spore-forming bacilli (55, 56). Other detected bacteria may contribute to methanol and methylamine production through breakdown of organic compounds, a process that has also been documented in marine sediments (57). For example, Verrucomicrobia (Opitutus sp.) recovered in some of the coal and shale incubations has cultured representatives that are capable of producing methanol during pectin degradation (58). Select members of the Clostridia also produce methanol from pectin and can degrade N-methyl compounds, such as betaine, to methylamines (54), while others within this class consume methanol and methoxylated compounds as sole carbon and energy sources.
Discussion
IODP Expedition 337 off Shimokita Peninsula explored a pristine, deeply buried, Miocene-aged coal bed system in an attempt to characterize the deep subseafloor biosphere within this unique environment. The initial site characterization from this expedition, described by Inagaki et al. (6), demonstrated higher cell concentrations in the coal bed compared with surrounding lithologies but reported in situ cell counts for this site that were far below the predicted depth-based trends recorded in previous ocean drilling studies (5). Successful cultivation of a hydrogenotrophic methanogen (Methanobacterium sp.), combined with geochemical evidence for sulfate reducers and methanogens in isotopically enriched pyrite (14) and 13C-depleted methane, as well as detection of coenzyme F420 (6), indicated the potential for biogenic methane formation in the organic carbon- and H2-replete coal bed environment (6). The calculated Gibbs free energies of methylotrophic methanogenesis and methyl fermentation under in situ conditions indicate that both methylotrophic methanogenesis (approximately −300 kJ/mol) and fermentation (approximately −10 kJ/mol) were possible, but methylotrophic methanogenesis is more energetically favorable. Given these energetic predictions, it was unexpected that the coal and shale SIP experiments with 13C-labeled methylated substrates did not show significant biological methanogenic activity during the 2.5-y incubation period. Instead, our isotopic data suggested the majority of methane detected during these long-term experiments was associated with the coal before incubation and subsequently released from the sample matrix over time, with minimal contributions from methylotrophic methanogenesis via the 13C-labeled substrates (at most, 0.04% of total methane from labeled substrate). This could also explain why the autoclaved coal samples had lower methane production, as the heat and pressure treatment during autoclaving, followed by flushing with argon, may have accelerated degassing of methane compared to treatments without autoclaving. Our finding that the majority of methane produced was not enriched in 13C may be attributed to other carbon substrates native to the organic-rich coal bed. However, the methane produced in the incubations also did not show enrichment in 2H, which is predicted to occur during active biogenic methane production in the presence of 2H2O, as biogenic methane production requires water-derived hydrogen (59). From incubation δ2H-CH4 values, we calculated a maximum of 0.007% of the total methane could have been biologically produced during the coal incubations from utilization of any carbon substrate available in the coal, or added methylated substrate. The combined 13C and 2H data point to the majority of biogenic methane recovered over the 2.5-y experiment being produced before the start of the incubation, highlighting an apparent temporal offset between in situ generation and subsequent release from the coal matrix (60).
While we have evidence for low levels of methanogenesis occurring in our incubations that could be attributed to the amended substrates, the in situ geochemistry at this site indicates a potential temporal succession in the microbial community over geological time from a methanogenic period to a now quiescent period that has retained the geochemical record [∼30 mM CH4 (30)] of a once more active in situ methanogenic assemblage. Since this Miocene-aged coal bed has experienced a wide range of temperatures, pressures, and geochemical conditions during burial over the past 20 Mya, it is possible that one or more of these variables changed at a rate, or to a degree, where the less metabolically versatile methanogens were impacted more than the comparatively metabolically versatile bacterial assemblage.
Overall, the microbial diversity recovered from our incubations was consistent with the bulk microbial diversity recovered in situ (6), including a dominance of Gammaproteobacteria, Betaproteobacteria, Alphaproteobacteria, Actinobacteria, Firmicutes, Chloroflexi, and Bacteroidetes. The combined multidisciplinary results from our stable isotope incubation experiments suggest that these deep subseafloor microorganisms are metabolic generalists that appear to be resistant to the unique stressors of the in situ environment beyond what has been described of their better characterized surface relatives in culture. While total cell numbers and sample sizes were low, as expected given this environment’s inherent constraints, there were general trends in relative abundance of specific taxonomic groups aligning with hydrogen amendment inhibition (Firmicutes, Alphaproteobacteria) or stimulation (Actinobacteria), as well as observations of increased numbers of putative spore-forming Firmicutes in the autoclave-treated sample. Gibbs free energies calculated for our incubation conditions with hydrogen addition (∼10-fold in situ hydrogen, +10 kJ/mol) and no hydrogen addition (near zero hydrogen approaches favorability of methanogenesis) concur with the coupling of fermentation and hydrogen addition, where incubations without hydrogen amendment should be much more favorable for fermentation. It is also notable that methanol anabolism (modeled via conversion to formaldehyde, approximately −30 kJ/mol at standard temperature and pressure) is more energetically favorable than catabolism, which may have elevated importance in subsurface environments, where most, if not all, cellular energy is likely devoted to cell maintenance (61, 62).
The highest rate of 13C-DIC production from methyl substrate amendments in our coal incubations (2 pmol of DIC per cm3⋅d−1) was 25-fold faster than potential sulfate reduction rates from the same coal bed with radiolabeled sulfate (14). Using a conversion factor to translate radiolabeled sulfate rates to DIC production rates [2 mol of C from 1 mol of acetate, per 1 mol of sulfate (61)], other subseafloor systems yield rates ranging from ∼10−4 fmol of C per cell per day in Peru Margin sediments (13) to shallow sediment rates of ∼10−2 fmol of C per cell per day (12). Converting 13C-DIC production in our coal incubations to cellular rates using final cell concentrations, DIC production was ∼0.02 fmol of C per cell per day for both methylamine and methanol. Assuming a minimal extraction efficiency of only 10%, this would decrease rates to ∼0.002 fmol of C per cell per day, which is still above Peru Margin sediment rates. Our NanoSIMS data also indicate that it is unlikely all cells are active in our incubations (Table 1), and when assuming only the active portion of cells in these coal incubations produced the labeled 13C-DIC, rates are estimated to be 30–60% higher. Accepting that both ex situ stable-isotope and radioisotope incubation methods likely caused some perturbation to the natural system, either through the addition of micromolar concentrations of carbon and nitrogen (this study) or through homogenization of the sample and shaking during 35S-sulfate incubation (14), these rates are unlikely exact representations of in situ subseafloor activity, but remain consistent with the multiple independent indications of slow rates of metabolism and growth of deep biosphere microorganisms relative to pure culture studies.
Both 15N-ammonium and 2H2O have been used to assess anabolic activity and generation times of diverse microorganisms in culture and the environment (18, 19, 21, 63, 64). In the context of this single-cell–targeted multiple-isotope labeling study, the passive tracer 2H2O was observed to be a more sensitive measure of overall anabolic activity compared with 15N assimilation from 15N-ammonium or methylamine within the deep subseafloor biosphere. However, when these assimilation rates were converted to biomass generation times, the paired 2H-based and 15N-based generation time estimates were consistent (Fig. 3). This suggests that the most active cells were not selectively producing different types of biomass, such as repairing lipids over generating new proteins, but were generally producing all forms of biomass.
Comparison of cellular enrichment from 2H2O incorporation across different incubations revealed a notable difference in cellular anabolic activity (generation time) in incubations supplied with H2 relative to those without. H2 concentrations have been reported as low as 10−2 μM in subsurface coal beds with active hydrogenotrophic methanogenesis (65), but in situ levels were much higher in the Shimokita coal bed (1–500 μM) (6), suggesting that methanogenesis was not the dominant control over environmental hydrogen concentrations (66). In contrast to many deep biosphere systems that are hypothesized to be fueled by hydrogen (67), the addition of hydrogen (0.15 atm H2) depressed microbial anabolic activity relative to those incubations without hydrogen for the coal incubations (Fig. 3), and suggests that fermentative microorganisms suggested by 16S rRNA gene sequencing are active in coal bed incubations. Microorganisms in the shale incubations from an overlying horizon produced the opposite trend. In this set of incubations, a larger fraction of active cells (ROIs) was detected with H2 amendment, relative to the paired incubation without H2, in both 13C-methylamine and 13C-methanol treatments (Fig. 1). These findings suggest the influence of H2 is complex in organic-rich systems, and as observed in the deep coal horizon, H2 may not always be a beneficial source of energy and, in fact, may depress microbial activity and growth.
Patterns of nitrogen source utilization (ammonia or methylamine) by the active microbial assemblage also differed between the coal and shale incubations. The 15N-ammonium incorporation was observed in the majority of incubations, independent of lithology. Methylotrophy via the microbial assimilation of 15N-methylamine, however, was predominately observed by the coal-associated microbial assemblage. It is expected that most anaerobes can use ammonium as a nitrogen source, but assimilation of methylamine is less common, and the observed difference in methylamine utilization between the coal and shale incubations is notable. Supporting these observations, the diversity recovered in the coal incubations included microbial taxa affiliated with bacterial lineages shown to be capable of methylamine metabolism. These findings may point to the potential use of methylated amines in lignite coal as a source of nitrogen in this deep biosphere ecosystem.
We compared the 15N-estimated generation times from previously published SIP-NanoSIMS data of 15N-ammonium uptake in cells from deep marine sediments (219 mbsf) that overlay the coal and shale beds studied here (18). In the study by Morono et al. (18), 15N-ammonium–based estimates of doubling times ranged from 63 to 252 d depending on added carbon source (e.g., glucose, amino acids), whereas the fastest 15N generation times calculated from our deep coal bed incubations were ∼30-fold slower (∼20 y; Fig. 3). The study by Morono et al. (18) did not include 2H2O, precluding direct comparison; however, estimates of 2H-based generation times from coal bed microorganisms did show some overlap with the slowest rates measured within the overlying sediment microbial assemblage (1–2 y) and other marine sediment studies using 2H2O [∼20 y, 2H-lipid incorporation rates (25)]. Our inability to detect 13C-labeled biomass despite indication of carbon utilization from the bulk geochemical data also supports the findings of the study by Morono et al. (18), where nitrogen was shown to be assimilated to a greater extent than carbon by microorganisms in their sediment incubations.
Overall, the single-cell generation times from SIP-NanoSIMS data for carbon, nitrogen, and 2H2O in our study, as well as those reported by Morono et al. (18), yield estimates that are one to two orders of magnitude faster than deep biosphere turnover times determined from bulk methods, such as amino acid racemization and lipid degradation (17, 65). The simultaneous use of 15N-labeled substrates in combination with the passive 2H2O tracer offered two independent means to track single-cell anabolic activity within our incubations. Although our study focused on the most active cells in our generation time estimates, this targeted single-cell analysis illustrates the high level of heterogeneity in anabolic activity between cells within the subseafloor microbial community, with the active fraction of the community ranging between no cells above the enrichment threshold (10-fold natural abundance) to 76% of the total cells with values above the threshold. This suggests that bulk methods estimating average cellular activity through the incorporation of environmental cell counts are likely to underestimate microbial activity and generation times. The complementarity in these independent multiscale approaches offers an important frame of reference for setting minimum and maximum bounds for growth and metabolic activity estimates within deep subseafloor ecosystems and demonstrates that even at the fringes of microbial activity, survival is not a simple story of dead or alive.
Conclusion
Despite cell abundances that were substantially lower than expected from global depth trends and sediment surface concentrations, this single-cell–targeted study successfully detected active microorganisms in terrestrial-sourced shale and coal buried at 2 kmbsf for millions of years. Incubations of intact shale and coal pieces maintained at in situ temperatures and amended with methyl compounds, ammonium, and 2H2O enabled the detection of some of the slowest measurements of growth using SIP-NanoSIMS. With biomass generation times of years to hundreds of years, these rates were much faster than deep biosphere activity estimates via other bulk methods (approximately thousands of years). Although methanogenic activity could be detected via geochemical analysis, methanogenic archaea were not recovered by 16S rRNA gene iTag sequencing. Instead, heterotrophy appears to be the predominant microbial metabolism for this system, which has been found in many deep subseafloor sedimentary environments (8, 68–70). Although variation in activity among incubations associated with methyl substrate amendment, presence/absence of hydrogen, high temperature/pressure (autoclave), and 16S rRNA diversity depict a heterogeneous and diverse heterotrophic community. Through this unique snapshot into a pristine coal bed, results from Expedition 337 have also opened new avenues for conceptualizing the residence time of carbon, nitrogen, and sulfur in coals that have never reached sterilization conditions. With global lignite reserves estimated at 839 gigatons (71), understanding what portion of this potential microbial energy source, assumed biologically inaccessible in the lithosphere, may be mobilized and returned to the surface biosphere is important for understanding both deep life and global biogeochemical cycles.
Geochemical Analyses
DIC samples were injected into preweighed, He-flushed Exetainer vials (Labco Limited) containing 100 μL of 40% phosphoric acid. Concentrations and stable carbon isotopes of DIC were measured on a Gasbench II (Thermo Scientific) coupled to a Delta V Plus IRMS instrument (Thermo Scientific) in the Caltech Stable Isotope Facility, following the methods of Torres et al. (72). Briefly, Exetainers were sampled using a Gas Chromatograph Prep and Load autosampler (CTC Analytics) equipped with a doubled-holed needle that transferred headspace using a 0.5 mL⋅min−1 continuous flow of He to a 50-μm sample loop before separation by a PoraPlotQ fused silica column (25 m, i.d. = 0.32 mm) at 72 °C. A 20 mM NaHCO3 solution was used to establish a DIC concentration standard curve by comparing the average of the total peak area (masses 44, 45, and 46) of replicate sample injections with the volume of NaHCO3 added. Using the estimated error in the volume measurement and the SD of the peak area for the total carbon of bicarbonate standards, the estimated error of DIC concentrations was ±0.009 μmol. The δ13C values were corrected for sample size dependency and then normalized to the Vienna Pee Dee Belemnite scale with a three-point correction (73) using National Bureau of Standards standard 19, a previously calibrated internal laboratory standard, and the average of all of the NaHCO3 standards. Accuracy (0.11‰, n = 79) was determined by considering independent standards as samples, and precision (0.5‰) was determined from replicate measurements of all standard solutions.
Methane headspace concentrations were measured relative to argon (added to ca. equal partial pressure in all incubations) using a Hewlett Packard 5972 Series Mass Selective Detector and Hewlett Packard 5890 Series II Plus Gas Chromatograph at the Caltech Environmental Analysis Center. The δ13C-CH4 was analyzed as per the methods of Tsunogai et al. (74), and δ2H-CH4 was analyzed as per the methods of Kikuchi et al. (75) at the Kochi Institute for Core Sample Research, Japan Agency for Marine-Earth Science and Technology (JAMSTEC).
Isotope Calculations
Isotope mass balance was calculated using Eqs. S1 and S2, where is the number of moles, is the fractional abundance of the rare isotope (Eq. S3), and is the ratio of ion counts of the rare isotope over ion counts of the more abundant isotope (76). , , , and were known and used to solve for . Fractional abundances are displayed as atom percent, which is 100-fold the fractional abundance (Eq. S4). DIC production rates were calculated by dividing the moles of DIC from substrate () by sample volume (4 cm3 for coal samples) and days of incubation (864 d). The percent molar methane from substrate was calculated from Eq. S5, a rearrangement of Eqs. S1 and S2:
| [S1] |
| [S2] |
| [S3] |
| [S4] |
| [S5] |
Data analysis and display as violin plots of the kernel density function were done using R (77) with the “ggplot2” (78), “dplyr” (79), “gridExtra” (80), and “RColorBrewer” (81) packages. Cell ROIs with deuterium enrichment at least 10-fold above natural abundance were deemed “active.” This threshold was chosen as a number that was sufficiently higher than natural abundance to account for remnant isotope from hydrogen exchange and error in the NanoSIMS measurement. Our chosen threshold (0.115 at. %) is about half of that used by Kopf et al. (22) and a third of that reported by Berry et al. (21) for deuterium background in fixed pure culture studies.
For generation time calculations, a conservative approach was used that only included ROIs where 15N12C counts were above the Poisson error provided by Look@NanoSIMS. Poisson error is based on the theoretical precision of the mean for the minor ion. Eqs. S6 and S7 were used to calculate the rate of biomass generation via fractional abundance of isotope label in cellular biomass over the incubation period, where is the generation rate (encompassing both cell maintenance and generation of new cells), is the length of the incubation, is the labeling strength, is the single-cell NanoSIMS measurement, and is the natural abundance. We used Eq. S8 to estimate generation time, as the reciprocal of the rate, per Zilversmit et al. (82). NanoSIMS and values were multiplied by a conversion factor determined for single-cell to bulk isotope measurements by Kopf et al. (22) of 0.67 (2H) and 0.94 (15N). While lipid water assimilation values are not known for this system, the maximal value (aw of 0.83) from Zhang et al. (83) was used as an upper estimate of assimilation based on findings that lipid water assimilation increases as growth rate decreases (84). Nitrogen-based rate calculations assumed all nitrogen was derived from the substrate (methylamine or ammonium); therefore, the assimilation constant was excluded from Eq. S7:
| [S6] |
| [S7] |
| [S8] |
Supplementary Material
Acknowledgments
We thank the IODP for providing access and samples from the deep coalbed biosphere off Shimokita during Expedition 337. We thank the crew, drilling team members, laboratory technicians, and scientists on the drilling vessel Chikyu for supporting core sampling and onboard measurements. We also thank S. Fukunaga, S. Hashimoto, and A. Imajo [Japan Agency for Marine-Earth Science and Technology (JAMSTEC)] and T. Terada (Marine Works Japan, Ltd) for assistance in microbiological analyses; Y. Guan, F. Wu, C. Ma, and N. Dalleska (Caltech) for assistance with geochemical analyses; and A. L. Sessions, G. L. Chadwick, K. S. Metcalfe, M. K. Lloyd, and S. Kopf (Caltech) and H. Imachi (JAMSTEC) for feedback and valuable discussions. We appreciate the comments of two reviewers that also improved this manuscript. Funding for this work was provided by the Center for Dark Energy Biosphere (C-DEBI), NASA Astrobiology-Life Underground (NAI-LU; Award NNA13AA92A), the Gordon and Betty Moore Foundation Grant GBMF3780 (to V.J.O.), and Post Expedition Award (to E.T.-R. and V.J.O.), the Japan Society for the Promotion of Science (JSPS) Strategic Fund for Strengthening Leading-Edge Research and Development (F.I. and JAMSTEC), the JSPS Funding Program for Next Generation World-Leading Researchers (NEXT Program, Grant GR102 to F.I.), and JSPS Grants-in-Aid for Science Research (Grant 26251041 to F.I.; Grant 15K14907 to T.H.; and Grants 24687004, 15H05608, 24651018, 2665169, and 16K14817 to Y.M.). E.T.-R. was additionally supported, in part, by a Schlanger Ocean Drilling Fellowship, a C-DEBI travel grant for sample processing at the JAMSTEC Kochi Institute for Core Sample Research, and the Deep Life Cultivation Internship Program from the Deep Carbon Observatory (DCO). This is C-DEBI Grant contribution no. 389 and NAI-LU no. 314.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Data are available at National Center for Biotechnology Information (NCBI) BioProject, https://www.ncbi.nlm.nih.gov/bioproject/ (PRJNA381552) and NCBI BioSample, https://www.ncbi.nlm.nih.gov/biosample/ (SAMN06676442-48). BioSamples are identified in Dataset S2. Nanometer-scale secondary ion mass spectrometry and geochemical data are available at Biological and Chemical Oceanography Data Management Office (https://www.bco-dmo.org/projects; Project 672592).
See Commentary on page 11568.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1707525114/-/DCSupplemental.
References
- 1.D’Hondt S, et al. Subseafloor sedimentary life in the South Pacific Gyre. Proc Natl Acad Sci USA. 2009;106:11651–11656. doi: 10.1073/pnas.0811793106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Expedition 336 Scientists (2012) Mid-Atlantic Ridge microbiology: Initiation of long-term coupled microbiological, geochemical, and hydrological experimentation within the seafloor at North Pond, western flank of the Mid-Atlantic Ridge (Integrated Ocean Drilling Program Management International, La Jolla, CA), Technical Report 336.
- 3.D’Hondt S, et al. Presence of oxygen and aerobic communities from sea floor to basement in deep-sea sediments. Nat Geosci. 2015;8:299–304. [Google Scholar]
- 4.D’Hondt S, et al. Distributions of microbial activities in deep subseafloor sediments. Science. 2004;306:2216–2221. doi: 10.1126/science.1101155. [DOI] [PubMed] [Google Scholar]
- 5.Parkes RJ, et al. A review of prokaryotic populations and processes in sub-seafloor sediments, including biosphere:geosphere interactions. Mar Geol. 2014;352:409–425. [Google Scholar]
- 6.Inagaki F, et al. DEEP BIOSPHERE. Exploring deep microbial life in coal-bearing sediment down to ∼2.5 km below the ocean floor. Science. 2015;349:420–424. doi: 10.1126/science.aaa6882. [DOI] [PubMed] [Google Scholar]
- 7.Detmers J, Schulte U, Strauss H, Kuever J. Sulfate reduction at a lignite seam: Microbial abundance and activity. Microb Ecol. 2001;42:238–247. doi: 10.1007/s00248-001-1014-8. [DOI] [PubMed] [Google Scholar]
- 8.Fry JC, et al. Prokaryotic populations and activities in an interbedded coal deposit, including a previously deeply buried section (1.6–2.3 km) above ∼150 Ma basement rock. Geomicrobiol J. 2009;26:163–178. [Google Scholar]
- 9.Lipman CB. Living microörganisms in ancient rocks. J Bacteriol. 1931;22:183–198. doi: 10.1128/jb.22.3.183-198.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Postgate JR, Hunter JR. Acceleration of bacterial death by grown substrates. Nature. 1963;198:273. doi: 10.1038/198273a0. [DOI] [PubMed] [Google Scholar]
- 11.D’Hondt S, Rutherford S, Spivack AJ. Metabolic activity of subsurface life in deep-sea sediments. Science. 2002;295:2067–2070. doi: 10.1126/science.1064878. [DOI] [PubMed] [Google Scholar]
- 12.Leloup J, et al. Sulfate-reducing bacteria in marine sediment (Aarhus Bay, Denmark): Abundance and diversity related to geochemical zonation. Environ Microbiol. 2009;11:1278–1291. doi: 10.1111/j.1462-2920.2008.01855.x. [DOI] [PubMed] [Google Scholar]
- 13.Parkes RJ, et al. Bacterial biomass and activity in deep sediment layers from the Peru Margin [and discussion] Philos Trans R Soc A. 1990;331:139–153. [Google Scholar]
- 14.Glombitza C, et al. Microbial sulfate reduction potential in coal-bearing sediments down to ∼2.5 km below the seafloor off Shimokita Peninsula, Japan. Front Microbiol. 2016;7:1576. doi: 10.3389/fmicb.2016.01576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imachi H, et al. Cultivation of methanogenic community from subseafloor sediments using a continuous-flow bioreactor. ISME J. 2011;5:1913–1925. doi: 10.1038/ismej.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orsi WD, Edgcomb VP, Christman GD, Biddle JF. Gene expression in the deep biosphere. Nature. 2013;499:205–208. doi: 10.1038/nature12230. [DOI] [PubMed] [Google Scholar]
- 17.Lomstein BA, Langerhuus AT, D’Hondt S, Jørgensen BB, Spivack AJ. Endospore abundance, microbial growth and necromass turnover in deep sub-seafloor sediment. Nature. 2012;484:101–104. doi: 10.1038/nature10905. [DOI] [PubMed] [Google Scholar]
- 18.Morono Y, et al. Carbon and nitrogen assimilation in deep subseafloor microbial cells. Proc Natl Acad Sci USA. 2011;108:18295–18300. doi: 10.1073/pnas.1107763108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kopf SH, et al. Trace incorporation of heavy water reveals slow and heterogeneous pathogen growth rates in cystic fibrosis sputum. Proc Natl Acad Sci USA. 2016;113:E110–E116. doi: 10.1073/pnas.1512057112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Starnawski P, et al. Microbial community assembly and evolution in subseafloor sediment. Proc Natl Acad Sci USA. 2017;114:2940–2945. doi: 10.1073/pnas.1614190114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berry D, et al. Tracking heavy water (D2O) incorporation for identifying and sorting active microbial cells. Proc Natl Acad Sci USA. 2015;112:E194–E203. doi: 10.1073/pnas.1420406112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kopf SH, et al. Heavy water and (15) N labelling with NanoSIMS analysis reveals growth rate-dependent metabolic heterogeneity in chemostats. Environ Microbiol. 2015;17:2542–2556. doi: 10.1111/1462-2920.12752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eichorst SA, et al. Advancements in the application of NanoSIMS and Raman microspectroscopy to investigate the activity of microbial cells in soils. FEMS Microbiol Ecol. 2015;91:fiv106. doi: 10.1093/femsec/fiv106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kellermann MY, Yoshinaga MY, Wegener G, Krukenberg V, Hinrichs K-U. Tracing the production and fate of individual archaeal intact polar lipids using stable isotope probing. Org Geochem. 2016;95:13–20. [Google Scholar]
- 25.Wegener G, et al. Assessing sub-seafloor microbial activity by combined stable isotope probing with deuterated water and 13C-bicarbonate. Environ Microbiol. 2012;14:1517–1527. doi: 10.1111/j.1462-2920.2012.02739.x. [DOI] [PubMed] [Google Scholar]
- 26.Tanikawa W, et al. Thermal properties and thermal structure in the deep-water coalbed basin off the Shimokita Peninsula, Japan. Mar Pet Geol. 2016;73:445–461. [Google Scholar]
- 27.Inagaki F, Hinrichs K-U, Kubo Ys. Expedition 337 Scientists (2012) Deep coalbed biosphere off Shimokita: Microbial processes and hydrocarbon system associated with deeply buried coalbed in the ocean. Available at http://publications.iodp.org/preliminary_report/337/index.html. Accessed September 22, 2017.
- 28.Gross D, Bechtel A, Harrington GJ. Variability in coal facies as reflected by organic petrological and geochemical data in Cenozoic coal beds offshore Shimokita (Japan) - IODP Exp. 337. Int J Coal Geol. 2015;152:63–79. [Google Scholar]
- 29. Expedition 337 Scientists (2013) Methods. Available at http://publications.iodp.org/proceedings/337/102/102_.htm. Accessed September 22, 2017.
- 30. Expedition 337 Scientists (2013) Site C0020. Available at http://publications.iodp.org/proceedings/337/103/103_.htm. Accessed September 22, 2017.
- 31.Mayumi D, et al. Methane production from coal by a single methanogen. Science. 2016;354:222–225. doi: 10.1126/science.aaf8821. [DOI] [PubMed] [Google Scholar]
- 32.Morono Y, Terada T, Kallmeyer J, Inagaki F. An improved cell separation technique for marine subsurface sediments: Applications for high-throughput analysis using flow cytometry and cell sorting. Environ Microbiol. 2013;15:2841–2849. doi: 10.1111/1462-2920.12153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morono Y, et al. Intact preservation of environmental samples by freezing under an alternating magnetic field. Environ Microbiol Rep. 2015;7:243–251. doi: 10.1111/1758-2229.12238. [DOI] [PubMed] [Google Scholar]
- 34.Polerecky L, et al. Look@NanoSIMS–A tool for the analysis of nanoSIMS data in environmental microbiology. Environ Microbiol. 2012;14:1009–1023. doi: 10.1111/j.1462-2920.2011.02681.x. [DOI] [PubMed] [Google Scholar]
- 35.Hoshino T, Inagaki F. Application of stochastic labeling with random-sequence barcodes for simultaneous quantification and sequencing of environmental 16S rRNA genes. PLoS One. 2017;12:e0169431. doi: 10.1371/journal.pone.0169431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quast C, et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yilmaz P, et al. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 2014;42:D643–D648. doi: 10.1093/nar/gkt1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Masui N, Morono Y, Inagaki F. Microbiological assessment of circulation mud fluids during the first operation of riser drilling by the deep-earth research vessel Chikyu. Geomicrobiol J. 2008;25:274–282. [Google Scholar]
- 40.Liu C-H, et al. Exploration of cultivable fungal communities in deep coal-bearing sediments from ∼1.3 to 2.5 km below the ocean floor. Environ Microbiol. 2017;19:803–818. doi: 10.1111/1462-2920.13653. [DOI] [PubMed] [Google Scholar]
- 41.Kallmeyer J, Pockalny R, Adhikari RR, Smith DC, D’Hondt S. Global distribution of microbial abundance and biomass in subseafloor sediment. Proc Natl Acad Sci USA. 2012;109:16213–16216. doi: 10.1073/pnas.1203849109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strapoc D, Schimmelmann A, Mastalerz M. Carbon isotopic fractionation of CH4 and CO2 during canister desorption of coal. Org Geochem. 2006;37:152–164. [Google Scholar]
- 43.Strąpoć D, Mastalerz M, Eble C, Schimmelmann A. Characterization of the origin of coalbed gases in southeastern Illinois Basin by compound-specific carbon and hydrogen stable isotope ratios. Org Geochem. 2007;38:267–287. [Google Scholar]
- 44.Campbell BJ, Li C, Sessions AL, Valentine DL. Hydrogen isotopic fractionation in lipid biosynthesis by H2-consuming Desulfobacterium autotrophicum. Geochim Cosmochim Acta. 2009;73:2744–2757. [Google Scholar]
- 45.Gründger F, et al. Microbial methane formation in deep aquifers of a coal-bearing sedimentary basin, Germany. Front Microbiol. 2015;6:200. doi: 10.3389/fmicb.2015.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Midgley DJ, et al. Characterisation of a microbial community associated with a deep, coal seam methane reservoir in the Gippsland Basin, Australia. Int J Coal Geol. 2010;82:232–239. [Google Scholar]
- 47.Strąpoć D, et al. Biogeochemistry of microbial coal-bed methane. Annu Rev Earth Planet Sci. 2011;39:617–656. [Google Scholar]
- 48.Wang L, Zheng S, Wang D, Wang L, Wang G. Thermomonas carbonis sp. nov., isolated from the soil of a coal mine. Int J Syst Evol Microbiol. 2014;64:3631–3635. doi: 10.1099/ijs.0.063800-0. [DOI] [PubMed] [Google Scholar]
- 49.Mohagheghi A, Grohmann K, Himmel M, Leighton L, Updegraff D. Isolation and characterization of Acidothermus cellulolyticus gen. nov., sp. nov., a new genus of thermophilic, acidophilic, cellulolytic bacteria. Int J Syst Evol Microbiol. 1986;36:435–443. [Google Scholar]
- 50.Hubert C, et al. A constant flux of diverse thermophilic bacteria into the cold Arctic seabed. Science. 2009;325:1541–1544. doi: 10.1126/science.1174012. [DOI] [PubMed] [Google Scholar]
- 51.Goodfellow M. Phylum XXVI: Actinobacteria phyl. nov. In: Whitman W, et al., editors. Bergey’s Manual of Systematics of Archaea and Bacteria. 2nd Ed. Vol 5 Springer; New York: 2012. [Google Scholar]
- 52.Sahm H, Cox RB, Quayle JR. Metabolism of methanol by Rhodopseudomonas acidophila. J Gen Microbiol. 1976;94:313–322. doi: 10.1099/00221287-94-2-313. [DOI] [PubMed] [Google Scholar]
- 53.Ollivier B, Cord-Ruwisch R, Hatchikian EC, Garcia JL. Characterization of Desulfovibrio fructosovorans sp. nov. Arch Microbiol. 1988;149:447–450. [Google Scholar]
- 54.Ludwig W, Schleifer K-H, Whitman WB. Bacilli class. nov. In: Whitman WB, editor. Bergey’s Manual of Systematics of Archaea and Bacteria. Wiley; New York: 2015. [Google Scholar]
- 55.Nicholson WL, Munakata N, Horneck G, Melosh HJ, Setlow P. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol Mol Biol Rev. 2000;64:548–572. doi: 10.1128/mmbr.64.3.548-572.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fang J, et al. Predominance of viable spore-forming piezophilic bacteria in high-pressure enrichment cultures from ∼1.5 to 2.4 km-deep coal-bearing sediments below the ocean floor. Front Microbiol. 2017;8:137. doi: 10.3389/fmicb.2017.00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhuang G-C, Lin Y-S, Elvert M, Heuer VB, Hinrichs K-U. Gas chromatographic analysis of methanol and ethanol in marine sediment pore waters: Validation and implementation of three pretreatment techniques. Mar Chem. 2014;160:82–90. [Google Scholar]
- 58.Chin KJ, Liesack W, Janssen PH. Opitutus terrae gen. nov., sp. nov., to accommodate novel strains of the division ‘Verrucomicrobia’ isolated from rice paddy soil. Int J Syst Evol Microbiol. 2001;51:1965–1968. doi: 10.1099/00207713-51-6-1965. [DOI] [PubMed] [Google Scholar]
- 59.Thauer RK, Kaster A-K, Seedorf H, Buckel W, Hedderich R. Methanogenic archaea: Ecologically relevant differences in energy conservation. Nat Rev Microbiol. 2008;6:579–591. doi: 10.1038/nrmicro1931. [DOI] [PubMed] [Google Scholar]
- 60.Ertefai TF, et al. The biogeochemistry of sorbed methane in marine sediments. Geochim Cosmochim Acta. 2010;74:6033–6048. [Google Scholar]
- 61.Hoehler TM, Jørgensen BB. Microbial life under extreme energy limitation. Nat Rev Microbiol. 2013;11:83–94. doi: 10.1038/nrmicro2939. [DOI] [PubMed] [Google Scholar]
- 62.Jørgensen BB, Marshall IPG. Slow microbial life in the seabed. Annu Rev Mar Sci. 2016;8:311–332. doi: 10.1146/annurev-marine-010814-015535. [DOI] [PubMed] [Google Scholar]
- 63.Musat N, Foster R, Vagner T, Adam B, Kuypers MMM. Detecting metabolic activities in single cells, with emphasis on nanoSIMS. FEMS Microbiol Rev. 2012;36:486–511. doi: 10.1111/j.1574-6976.2011.00303.x. [DOI] [PubMed] [Google Scholar]
- 64.Orphan VJ, House CH. Geobiological investigations using secondary ion mass spectrometry: Microanalysis of extant and paleo-microbial processes. Geobiology. 2009;7:360–372. doi: 10.1111/j.1472-4669.2009.00201.x. [DOI] [PubMed] [Google Scholar]
- 65.Strąpoć D, et al. Methane-producing microbial community in a coal bed of the Illinois basin. Appl Environ Microbiol. 2008;74:2424–2432. doi: 10.1128/AEM.02341-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hoehler TM, Alperin MJ, Albert DB, Martens CS. Thermodynamic control on hydrogen concentrations in anoxic sediments. Geochim Cosmochim Acta. 1998;62:1745–1756. [Google Scholar]
- 67.Nealson KH, Inagaki F, Takai K. Hydrogen-driven subsurface lithoautotrophic microbial ecosystems (SLiMEs): Do they exist and why should we care? Trends Microbiol. 2005;13:405–410. doi: 10.1016/j.tim.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 68.Biddle JF, et al. Heterotrophic Archaea dominate sedimentary subsurface ecosystems off Peru. Proc Natl Acad Sci USA. 2006;103:3846–3851. doi: 10.1073/pnas.0600035103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lipp JS, Morono Y, Inagaki F, Hinrichs K-U. Significant contribution of Archaea to extant biomass in marine subsurface sediments. Nature. 2008;454:991–994. doi: 10.1038/nature07174. [DOI] [PubMed] [Google Scholar]
- 70.Ulrich GA, et al. Sulfur cycling in the terrestrial subsurface: Commensal interactions, spatial scales, and microbial heterogeneity. Microb Ecol. 1998;36:141–151. doi: 10.1007/s002489900101. [DOI] [PubMed] [Google Scholar]
- 71.Killops SD, Killops VJ. Introduction to Organic Geochemistry. Wiley; New York: 2013. [Google Scholar]
- 72.Torres ME, Mix AC, Rugh WD. Precise δ13C analysis of dissolved inorganic carbon in natural waters using automated headspace sampling and continuous-flow mass spectrometry. Limnol Oceanogr Methods. 2005;3:349–360. [Google Scholar]
- 73.Coplen TB, et al. New guidelines for δ13C measurements. Anal Chem. 2006;78:2439–2441. doi: 10.1021/ac052027c. [DOI] [PubMed] [Google Scholar]
- 74.Tsunogai U, Yoshida N, Gamo T. Carbon isotopic evidence of methane oxidation through sulfate reduction in sediment beneath cold seep vents on the seafloor at Nankai Trough. Mar Geol. 2002;187:145–160. [Google Scholar]
- 75.Kikuchi S, et al. Limited reduction of ferrihydrite encrusted by goethite in freshwater sediment. Geobiology. 2016;14:374–389. doi: 10.1111/gbi.12181. [DOI] [PubMed] [Google Scholar]
- 76.Hayes JM. An Introduction to Isotopic Calculations. Woods Hole Oceanographic Institution; Woods Hole, MA: 2004. [Google Scholar]
- 77.R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna: 2015. [Google Scholar]
- 78.Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer; New York: 2009. [Google Scholar]
- 79.Wickham H, Francois R. 2015 dplyr: A Grammar of Data Manipulation, R package Version 0.4.1. Available at https://cran.r-project.org/web/packages/dplyr/index.html. Accessed September 25, 2017.
- 80.Auguie B. 2012 gridExtra: Functions in Grid Graphics, R package Version 0.9.1. Available at https://cran.r-project.org/web/packages/gridExtra/index.html. Accessed September 25, 2017.
- 81.Neuwirth E. 2014 RColorBrewer, R package Version 1.1-2. Available at https://cran.r-project.org/web/packages/RColorBrewer/index.html. Accessed September 25, 2017.
- 82.Zilversmit DB, Entenman C, Fishler MC. On the calculation of “turnover time” and “turnover rate” from experiments involving the use of labeling agents. J Gen Physiol. 1943;26:325–331. doi: 10.1085/jgp.26.3.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang X, Gillespie AL, Sessions AL. Large D/H variations in bacterial lipids reflect central metabolic pathways. Proc Natl Acad Sci USA. 2009;106:12580–12586. doi: 10.1073/pnas.0903030106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kopf SH. 2015. From lakes to lungs: Assessing microbial activity in diverse environments. PhD thesis (California Institute of Technology, Pasadena, CA)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.