Significance
We question a central paradigm: namely, that the protein domain is the “atomic unit” of evolution. In conflict with the current textbook view, our results unequivocally show that duplication of protein segments happens both above and below the domain level among amino acid segments of diverse lengths. Indeed, we show that significant evolutionary information is lost when the protein is approached as a string of domains. Our finer-grained approach reveals a far more complicated picture, where reused segments often intertwine and overlap with each other. Our results are consistent with a recursive model of evolution, in which segments of various lengths, typically smaller than domains, “hop” between environments. The fit segments remain, leaving traces that can still be detected.
Keywords: protein space, protein evolutionary patterns, protein function annotation, ancestral segments
Abstract
Proteins share similar segments with one another. Such “reused parts”—which have been successfully incorporated into other proteins—are likely to offer an evolutionary advantage over de novo evolved segments, as most of the latter will not even have the capacity to fold. To systematically explore the evolutionary traces of segment “reuse” across proteins, we developed an automated methodology that identifies reused segments from protein alignments. We search for “themes”—segments of at least 35 residues of similar sequence and structure—reused within representative sets of 15,016 domains [Evolutionary Classification of Protein Domains (ECOD) database] or 20,398 chains [Protein Data Bank (PDB)]. We observe that theme reuse is highly prevalent and that reuse is more extensive when the length threshold for identifying a theme is lower. Structural domains, the best characterized form of reuse in proteins, are just one of many complex and intertwined evolutionary traces. Others include long themes shared among a few proteins, which encompass and overlap with shorter themes that recur in numerous proteins. The observed complexity is consistent with evolution by duplication and divergence, and some of the themes might include descendants of ancestral segments. The observed recursive footprints, where the same amino acid can simultaneously participate in several intertwined themes, could be a useful concept for protein design. Data are available at http://trachel-srv.cs.haifa.ac.il/rachel/ppi/themes/.
Studying segment reuse across proteins can reveal the mechanics of protein evolution (1–9). Identifying reuse also has many practical applications, and thus, many tools have been designed to harvest the reuse signal (e.g., for prediction of structure and function) by comparing protein structures (10, 11) and sequences [e.g., the hidden Markov model (HMM) aligners HHSearch (12) and HMMER (13)]. However, even if one has a sensitive and accurate tool to compare proteins, the challenge of quantifying reuse across the entirety of protein space remains. In the context of structure reuse, several studies have attempted to evaluate the extent to which particular components recur among proteins; these studies have addressed several different scales, ranging from a few residues or fragments (14, 15) to sets of secondary structure elements (16–18) and full domains, or folds (19, 20). The reuse of protein segments is not uniform. Some segments are reused extensively, whereas others recur only rarely; scholars have described the reuse patterns of protein segments of various lengths by the power law distribution (15, 21–27). Structure reuse may reflect the recurrence of biophysically favorable conformations (18, 27–30). However, sequence reuse and in particular, reuse of substantial segments hints at an evolutionary relationship among proteins.
Domains are the prime example of protein segments that are reused across multiple proteins. Domains duplicate, diverge, and recombine to form protein chains (21). Thus, a dominant view among protein scholars is that domains are the “atomic” evolutionary units (4, 31–33). The exact definition of a domain is not fully agreed on, because the term is used to refer to several things that often, but not necessarily, coincide: (i) a reused element, (ii) an independently folded unit, and even, (iii) a structural region (34). Thus, the de facto definition of a domain is an entity in one of the domain databases [e.g., Class Architecture Topology Homology (CATH), Structural Classification of Proteins (SCOP), and Evolutionary Classification of Protein Domains (ECOD) (9, 35, 36)], and these were generally selected for inclusion based on reuse. The interpretation of the definition of a domain varies among the databases. For example, only 60% of CATH domains have a similar SCOP counterpart (34, 37, 38). The lengths of the domains are assumed to be around 100 residues (39), and indeed, the domain lengths in CATH, SCOP, and ECOD follow very similar, and narrow, distributions around this mean (refs. 35, figure 8B and 40).
The view that domains are the only atomic evolutionary unit is challenged by evidence of substantial cross-protein similarities at the subdomain level (1, 3, 6, 41, 42) dating back to the work of Eck and Dayhoff (43), which was carried out even before the concept of domains had been introduced. Such similarities were identified for segments of different lengths based on similar sequences, structures, or both and described in the literature under different names. For example, several investigations identified cross-fold similarities of short and long segments (27, 44–47) and attributed subdomain similarities to “domain atrophy” (48). Such similarities have also been described as protein motifs (16, 49–52). To better understand the evolution of proteins, Lupas and coworkers (1, 3) documented shared segments, referred to as “antecedent domain segments.” Following a similar reasoning, Frenkel and coworker (53, 54) described the reuse of (very) short protein segments, referred to as “modalities,” and Goncearenco and Berezovsky (55, 56) examined reuse of closed rings referred to as “elementary functional loops.”
Clearly, reuse is common in protein space, but many details of the overall pattern of reuse are yet to be described. Here, we focus on the relationship between the length (in residues) of the reused segments and the extent of reuse. Our goal is to differentiate between two alternative evolutionary scenarios: (i) domains are the minimal evolutionary units vs. (ii) shorter (and longer) amino acid segments duplicate and mutate as well. By studying the properties and extent of reuse patterns, we attempt to elucidate whether a simple segmentation of protein chains into domains (i.e., nonoverlapping segments of fixed length) is a sufficiently detailed abstraction of the data.
To this end, we present an automated pipeline to identify segments of various lengths of reused sequences that also have similar structures; these segments are referred to as “themes” (as in “variations on a theme”). To derive a set of candidate themes, we introduce an efficient dynamic programming algorithm that finds an optimal segmentation of each protein based on all of its HHSearch alignments. Using this framework, we identify themes of lengths ranging from 35 to 200 residues. Reuse across protein space can then be quantified: we show that, when considering increasingly shorter themes (i.e., of fewer residues), reuse increases. The results indicate that duplication had happened for a range of segment lengths and is not limited to entities around 100 residues (i.e., domains). We show examples where elaborate patterns of reuse offer hints regarding the evolutionary process. Consequently, we argue that one must take a more holistic view and consider reuse of elements of diverse sizes rather than rely only on the segmentation of protein chains into a set of nonoverlapping domains.
Data Compilation and Processing
We introduce an algorithm to optimally identify themes of different lengths and use it to analyze reuse in two datasets of proteins of known structure: (i) a set of 28,223 [70% nonredundant (NR)] domains referred to as the ECOD dataset and (ii) a set of 31,417 (70% NR; Sep6_2014) chains referred to as the Protein Data Bank (PDB) dataset. Using HHSearch (12), we align all-vs.-all within these sets and calculate for each significant alignment the percentage of similarity of aligned residues and the rmsd of the Cα atoms of the aligned residues. We construct similarity networks, where the nodes are the dataset proteins and the edges are alignments, for which the calculated sequence similarity is greater than 40% and the calculated rmsd is lower than 3.5 Å (45). Because we identify reuse based on alignments, reuse can only be found among proteins in the same connected component in the similarity network. Hence, within the ECOD network, we focus on the single largest connected component of 15,016 domains, and in the PDB network, we focus on the 20,398 chains in the connected components of at least 10 chains.
Our procedure has two steps. (i) The split step identifies reused nonoverlapping themes. We run this step for each protein and all of its alignments. (ii) In the search-and-join step, for each theme identified in the first step, we collect all variations of that theme by traversing the network. For the ECOD dataset, we also further group themes on the basis of their structural similarity.
The split step identifies the most extensively reused pieces in each protein (regardless of whether they are domains or chains) that are longer than some prespecified threshold (Fig. 1); these are the candidate themes. Given a protein and the set of its significant alignments, we can calculate for each range of residues s-e a reuse score. The reuse score is the sum of all of the substitution matrix BLOSUM-62 scores for the residues s-e and the residues aligned to them (including gap penalty) in any of the alignments that are matched to the protein. Searching for the highest scoring combination of nonoverlapping themes by enumerating all possible combinations is a very expensive computation, because there are many such combinations. Instead, we use a dynamic programming algorithm to efficiently identify the highest scoring combination (SI Appendix, Methods has details). The split step ends by providing a list of candidate themes described as protein ranges [e.g., P(s-e)]. We ran the split step repeatedly, with different minimal theme lengths. For the ECOD set, we considered themes of at least 35–65 residues, and for the PDB set, we considered themes of at least 35–200 residues.
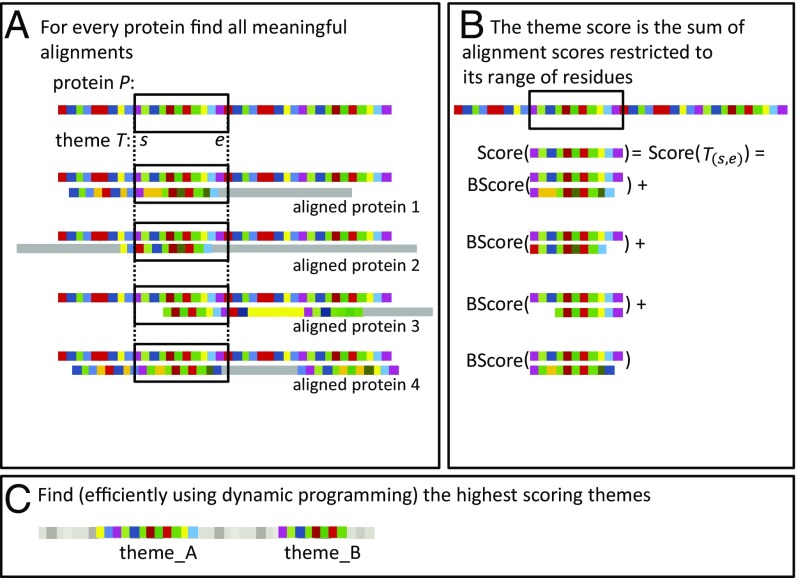
Fig. 1.
(A) The most reused themes in a protein P are derived from the set of meaningful alignments of P and other proteins: in this example, proteins 1–4. For any possible theme (for example, theme T that spans residues s-e), we can consider the parts in the alignments that are restricted to these residues, which are marked here by black rectangles. (B) We assign a score for every theme within the protein P based on the scores of these restricted parts, which is the sum over the BLOSUM-62 scores for the aligned parts. (C) Our goal is to identify the largest set of nonoverlapping themes (for example, theme_A and theme_B), such that the sum of these scores is optimal. Rather than exhaustively scoring all possible theme end points to find the optimal one, we find it more efficiently using dynamic programming (SI Appendix, Methods has details).
The search-and-join step groups themes to reveal overall reuse in protein space. Given a candidate theme T (protein P, residues s-e), we identify all of the variations of T in the network. A variation of T, denoted T′, is a range of residues in another protein that are aligned to T and similar: namely, there is a meaningful alignment to more than 85% of the residues of T, with a sequence similarity of more than 50% and structure similarity lower than 3.5 Å rmsd over the aligned residues. When we find a variation T′, we join it to the set of variations initiated by T and repeat the process to include in the set any additional themes that are variations of a theme already in that set (T, T′, or any other theme that was added in the process). If an added variation is also one of the candidate themes identified in the split step described above, we merge its set of variations into the set of T. We denote by the set of all identified themes.
Results
Reuse Is Prevalent in Protein Space.
Focusing on theme reuse, we count, for every theme , the number of unique proteins (domains in the ECOD set, chains in the PDB set) that it appears in or equivalently, its number of variations; we refer to this value as the “size” of the theme (a concept that is distinct from the length of the theme or the number of residues that it contains) and denote it by . SI Appendix, Fig. S1A, and Fig. 2 show, for the ECOD and PDB datasets, the number of themes detected (for different minimal residue numbers or “theme lengths”) as a function of . The reuse pattern, sorted by along the x axis, is similar for all lengths and in both datasets: many themes are used only occasionally, and a few are used extensively. We further cluster the themes of the ECOD dataset to create structurally similar “superthemes” (rmsd lower than 4.5 Å). SI Appendix, Fig. S1B shows the number of superthemes vs. the number of variations in (or the size of) the supertheme: we see that, when considering increasingly shorter themes, there are more themes and that they are of larger size, but the distributions stay qualitatively the same.
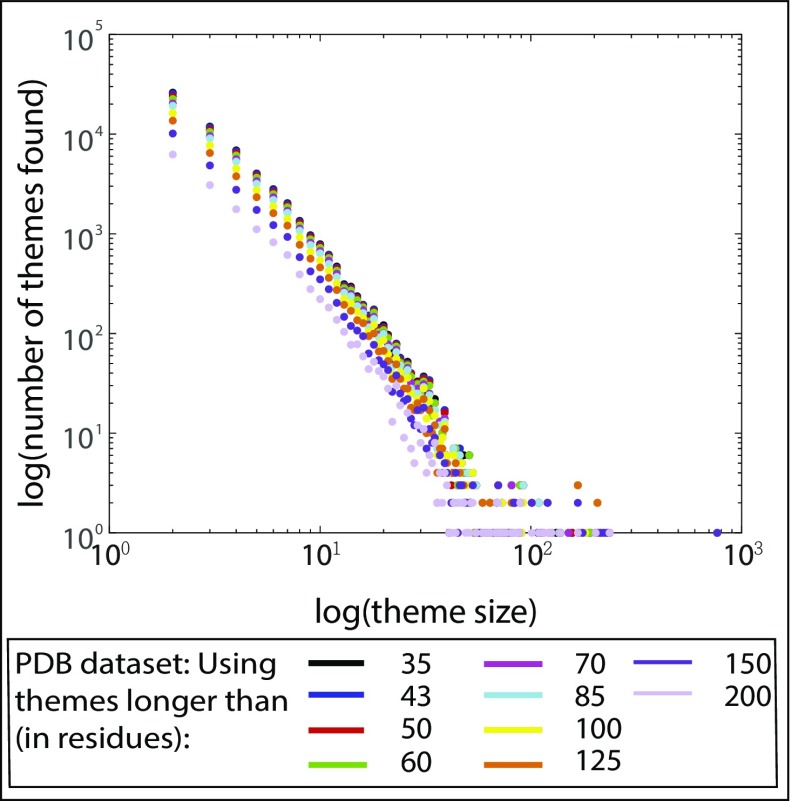
Fig. 2.
Usage of the protein themes. Log–log plot of the number of themes vs. theme size [i.e., number of variations per theme for the PDB datasets (for ECOD dataset see SI Appendix, Fig. S1A)]. Results for themes of different minimal lengths are presented using different colors. In all cases, we see that there are many themes with a small size and few large-sized themes; we also see that reuse increases as the minimal theme length decreases.
Reuse Increases with the Decrease in Theme Length.
Focusing on each protein and fixing a set of detected themes S, we calculate for each residue r the maximal size of a theme that it is part of: (i.e., the theme with greatest number of variations). Then, we use values to calculate the total number of residues that appear in any theme in [i.e., all residues for which ] (Eq. 1). For a set of proteins , the total number of residues in that participate in any theme is
| [1] |
To show another perspective of reuse, we also calculate the number of unique residues in the dataset (Eq. 2) (57). Residues in a theme T are variations of the same theme and should be counted only once rather than times in a straightforward count. Thus, we sum for each residue in T: the contributions from variants of T will total one for each residue along T, as it should. Because a residue r may be part of more than one theme, we sum :
| [2] |
If there is no reuse, the number of unique residues will equal the total number of residues in all proteins in the set. Every theme that we detect shows reuse and can decrease this value by the number of residues that are similar to ones that were already counted. SI Appendix, Fig. S2 shows three toy examples of a small protein dataset, its varying lengths themes, and how changes in reuse as a function of the minimal theme length are manifested by the measures in Eqs. 1 and 2.
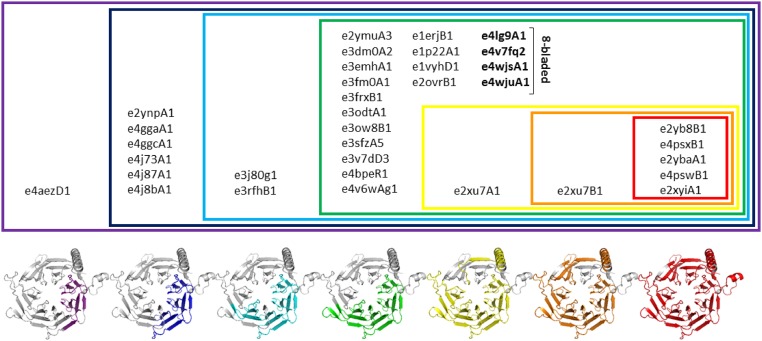
Fig. 3 shows the reuse pattern of themes in the seven-blade propeller domain e2xyiA1 from the ECOD dataset. When considering alignments that conform to the abovementioned conservative thresholds, we find the most reused theme in 34 neighboring domains; these residues of e2xyiA1 are marked in purple in Fig. 3, and the list of domains is bounded in a purple box in Fig. 3. A larger (encompassing) part of the domain e2xyiA1 appears in a smaller set of 33 neighbors (shown in blue in Fig. 3). Similarly, increasingly larger (encompassing) parts of the domain appear in decreasingly smaller sets of neighbors; we color these in light blue, green, yellow, orange, and red in Fig. 3. The largest part (red in Fig. 3), which covers almost the whole domain, appears in four other domains. Using our notation, the residues marked in purple in Fig. 3 are part of a theme of = 35 (e2xyiA1 + 34 additional domains): hence, these residues contribute 1/35 to the total unique count. At the other extreme, the residues marked in red in Fig. 3 (part of the longest theme) do not overlap with any of the shorter themes and are only part of a theme of = 5, and thus, they each contribute 1/5 to the total unique count. We use the per residue measures of reuse in Eqs. 1 and 2 to capture the details of the reuse pattern as manifested in this example. In SI Appendix, we survey common themes and provide an online interface to explore them (trachel-srv.cs.haifa.ac.il/rachel/ppi/themes/).
Fig. 3.
Recursive reuse of parts of e2xyiA1 in the ECOD dataset. Reuse manifests a Russian nested dolls effect (in sequence; not to be confused with the structural one described in refs. 8 and 9). The themes are marked on the e2xyiA1 domain. The shortest theme, shown in purple, appears in the largest set of domains (listed within the purple box). A longer (encompassing) theme, shown in blue, appears in fewer domains. Similarly, increasingly longer themes of e2xyiA1, shown in light blue, green, yellow, orange, and red, are found in increasingly smaller sets of domains. This example manifests the complexity of the reuse pattern in evolution, where the same amino acid can appear in more than one theme, and shows that, to accurately describe reuse of a domain, we must consider a per residue resolution.
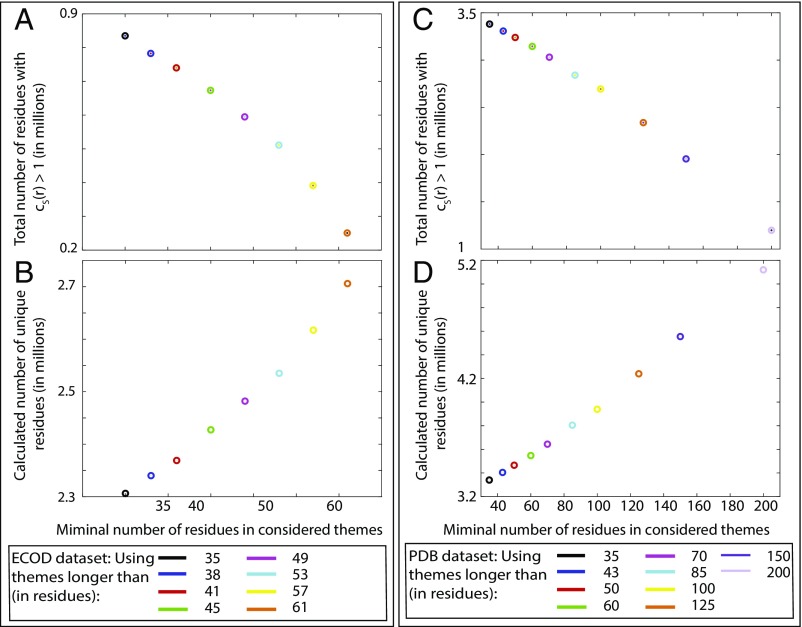
Fig. 4 plots the number of residues detected in themes (Fig. 4 A and C) and the calculated number of unique residues (Fig. 4 B and D) vs. the minimal number of residues in the set of themes S for the ECOD and PDB datasets. In both cases, when considering sets of themes with minimal lengths that are increasingly shorter, the total number of residues in detected themes increases (Fig. 4 A and C), and the calculated number of unique residues decreases (Fig. 4 B and D). This quantifies the fact that reuse in protein space is more prevalent when considering increasingly shorter themes. For the PDB dataset, the minimal lengths of the themes range from 35 to 200, including themes of 100 residues, or the average lengths of domains, which do not show any unique or singular pattern. Actually, relying only on a limited range of theme lengths to describe reuse in protein space leads to detecting far less reuse as quantified by a greater calculated number of unique residues: SI Appendix, Fig. S3 compares the calculated number of unique residues when considering only themes of limited lengths, showing that, in all cases, considering all theme lengths leads to detection of significantly more reuse.
Fig. 4.
Reuse in protein space is greater when considering sets of themes of increasingly shorter minimal lengths. Reuse in the ECOD (A and B) and PDB (C and D) datasets. (A and C) The number of recurring residues (i.e., amino acids that appear in any theme) (Eq. 1) using different sets of themes. (B and D) The number of unique residues (Eq. 2) obtained using sets of themes with different minimal lengths.
As the minimal threshold for theme length decreases, the calculated number of unique residues also decreases. The first reason for this is the increase in the total number of residues with (Fig. 4 A and C). SI Appendix, Fig. S4 shows that this increase is because of a twofold effect: (i) as theme length decreases, themes are detected in more proteins in the dataset, and (ii) the coverage (i.e., the percentage of residues in a protein described by themes) increases. SI Appendix, Fig. S5 shows an example for the second effect, where new (shorter) themes emerge in a different region of domain e3nytA1 (SI Appendix, Fig. S5, orange) and in different domains; a nested pattern also appears (SI Appendix, Fig. S5, red, yellow, and green). SI Appendix, Fig. S4 A and B shows that, in the ECOD dataset, the number of domains containing themes increases from 2,927 to 7,468 as the minimal theme length decreases from 60 to 35 residues, and coverage within these domains increases from 55 to 81%. Similarly, SI Appendix, Fig. S4 C and D shows that, in the PDB dataset, the number of chains containing themes increases from 4,080 to 15,998 as the minimal theme length decreases from 200 to 35 residues, and coverage within these domains increases from 83 to 91%.
Fig. 3 shows another factor contributing to the decrease in the calculated number of unique residues: within the set of residues in the seven-blade propeller domain e2xyiA1 that satisfies , the value of increases as the minimal theme length decreases. This is typical: SI Appendix, Fig. S6A shows that, in the ECOD dataset, as the minimal theme length decreases from 60 to 35 residues, the average increases from 3.8 to 26. Likewise, SI Appendix, Fig. S6B shows that, in the PDB dataset, as the minimal theme length decreases from 200 to 35 residues, the average increases from 13.7 to 33.1. That is, many amino acids are included in more than one theme.
Divergent vs. Convergent Evolution in Our Dataset.
The sequence similarity threshold needs to be sufficiently high, so that pairs marked as evolutionarily related are (mostly) caused by divergent evolution. The concern here is that a sequence similarity threshold that is too small may lead to many false positives: cases which are marked as if they diverged from a common ancestor, although they have not. In support of the 50% threshold being sufficiently high is that it corresponds to ∼40% sequence identity on average, and almost all pairs in the dataset have a sequence identity greater than 30% (SI Appendix, Fig. S7). Another concern is that the sequences may be similar, because they have converged because of their similar structures (recall that we enforce structural similarity). To address this, we consider 23 helix–strand–helix–strand themes of similar structures (grouped together in the second level of clustering), which we identified with the 50% threshold. This supertheme includes 75 variations from 62 domains of the a/b barrels and a/b three-layered sandwiches classes (in five X-level classifications). We use the global Needleman–Wunch sequence alignment to compare all-vs.-all 75 segments sequences and collect the sequence similarity, identity, and raw scores. SI Appendix, Fig. S8 plots the distributions of these measures for the 146 pairs of segments that are in the same theme (that is, have similar sequences and structures) and the 2,629 pairs that are in different themes that share only a similar common structure. We see that the similarity of the sequences of similar structures (or similar biophysical properties) is markedly lower than that of the ones within our themes (P value < 10−15 in all three cases, Wilcoxon test), implying that the similarity within themes is not merely a consequence of similar structures.
We also consider a less strict threshold for identifying evolutionarily related segments. SI Appendix, Fig. S9 shows how reuse in the ECOD dataset varies with minimal theme length when using a lower threshold of 40% sequence similarity. The lower threshold identifies more segments as evolutionarily related, leading to a greater total number of residues in any theme and a smaller total number of unique residues. However, regardless of the threshold used, the fundamental observation holds: when considering increasingly shorter minimal theme lengths, we detect more reuse.
Because we use conservative similarity thresholds (a shared segment length of at least 35 residues), most of the theme reuse that we observe emerges among similar proteins. Nonetheless, some of our themes appear to “hop contexts.” In the example in Fig. 3, most of the domains that share similar themes with e2xyiA1 are close neighbors and have seven-blade propellers; however, among the domains that share four blades with e2xyiA1, six are eight-blade propellers (orange-colored nodes in SI Appendix, Fig. S10). SI Appendix, Fig. S11 A and B shows the number of themes that we found in the ECOD dataset that span more than a single fold (X-level classification) as a function of the minimal theme length: among the 862 themes that are found in more than five domains each, 84 span at least two ECOD X classifications. The themes found are in many different folds: SI Appendix, Fig. S11C shows that the total number of folds spanned by any theme is significant. However, as the themes are characterized not only by similar structures (like folds) but also, by similar sequence (and especially since we are using strict thresholds), most themes span only a small part (less than 10%) of their folds (SI Appendix, Fig. S11D). These measures depend on the sequence similarity threshold: SI Appendix, Fig. S11 E–H shows that, using the laxer threshold of 40%, we find more themes in general and more themes that span multiple folds or a greater portion of their folds. Notice that domains from the same X-level classification in ECOD often share only similar geometry and might not necessarily share a common ancestor. In contrast, variations of the same theme observed here share very high sequence similarity and are likely to have emerged from a common ancestor.
Discussion
Reuse patterns in protein space reflect 3.7 billion y of evolution (58), restrained by the underlying physicochemical qualities of covalently linked amino acids. When analyzing segment reuse in present day proteins, it is difficult to differentiate between evolutionary traces and effects of physicochemical constraints resulting from, for example, limitations on backbone conformations, the tendency to satisfy backbone hydrogen bonds, or amino acids qualities. As a rule of thumb, instances of structural reuse—or segments of similar shape—are likely attributable to physicochemical constraints and convergent evolution, whereas sequence reuse may be more indicative of divergence from a common ancestor. Overall, it is unclear how one can definitively separate between divergent and convergent evolution when relying on present day data. Our premise is that relatively long protein segments (≥35 residues) with similar sequences that recur in evolutionarily unrelated contexts provide support to the idea of divergent evolution, because it is unlikely that two such low-probability events happened independently of each other. Thus, our starting point is a set of sequence alignments (we enforce structural similarity only as a precaution against including segments that are too divergent). To study evolution, we investigate reuse patterns of protein segments with similar sequences and of diverse lengths, referred to as themes, on a large scale.
To automatically identify sequence reuse from a set of all-vs.-all alignments, we rely on a computational framework introduced here. Given a set of all alignments of a protein (or domain), we can use this framework to derive an optimal set of nonoverlapping themes corresponding to that protein. Then, we repeatedly follow alignment relationships to identify additional variations of each theme. This way, we collect sets of themes that include many variations. Rather strict similarity thresholds were used here, but one can use our tools for similar analyses with even lower probability events (enforcing longer segments and higher sequence similarity thresholds).
Herein, we identified sets of themes from the ECOD and PDB databases and used them to quantify overall reuse in protein space. We showed that, when considering increasingly shorter themes, reuse is more prevalent. Using conservative thresholds for theme length, we identified themes that appear in multiple domains/chains in the connected regions of protein space. For the ECOD dataset, we detected themes in 19–50% of the domains when considering minimal theme lengths ranging from 61 to 35; for the PDB dataset, we detected themes in 20–78% of the chains when considering minimal theme lengths ranging from 200 to 35. This reuse is significant, because it is observed after removing redundancy (up to 70% sequence identity).
The themes are not reused uniformly. Rather, a few are heavily reused, whereas many recur only sparsely. This pattern is observed across all theme lengths and in both datasets. Our analysis of the ECOD dataset suggests a similar pattern for second-level clustering (based on structural similarity) as well. This observation is in agreement with previous studies investigating both shorter (15) and longer segments (21–27). Our study completes the picture of segment reuse for the missing lengths in the midrange, showing that, indeed, this usage pattern is a ubiquitous feature of the protein universe (25). Consequently, any conclusions emanating from it about the underlying evolutionary process hold for segments of all lengths. The fact that reuse increases when shorter themes are considered is consistent with models of evolution by duplication and divergence. The recurrent themes (of diverse lengths) might have served as recursively used “evolutionary building blocks.”
By using the same automated methodology systematically, we can compare results obtained for themes of different minimal lengths. We see that a given protein chain may include both shorter themes that are reused more extensively in protein space and longer ones that are reused more sparsely. This is illustrated in the propeller domain, where short themes that are shared by many domains overlap with longer themes shared by only some of these domains (Fig. 3). This pattern of reuse may be indicative of shared evolutionary ancestry ranging from further back to more recent. In this context, it is interesting to notice that, of 26 domains that share the four-blade theme, 6 are from eight-blade propellers and 20 from seven-blade propellers, suggesting an evolutionary link between these two classes of domains. Evolutionary analysis (59) and directed evolution studies (60) support this scenario for the emergence of the propeller domain by amplification from single blades followed by function differentiation. Our analysis shows that such recursive patterns are common in protein space (SI Appendix, Fig. S4 A and B) for themes both longer and shorter than the typical domain, suggesting that many proteins emerged through similar processes (data are available in SI Appendix).
In contrast to the richness and inherent complexity of the themes described above, where a given amino acid can belong to multiple themes, the entities classified in domain databases, such as SCOP, CATH, and ECOD, aim to provide a much reduced representation of reuse in protein space. That is, domains are reused segments of a particular size—100 residues on average (35). To identify domains for inclusion, the curators of these databases generally relied on the sequence reuse signal (35, 61–63). In particular, this view places every residue in exactly one reused segment (its domain). [Note that here we are referring to the entities classified; indeed, as these are hierarchical classifications, each domain is then classified into several groups (e.g., family, superfamily, fold, class).] This decision regarding database construction has had far-reaching effects. Algorithms that mine the reuse signal depend on the segmented chains in these databases both when aligning a new target chain to a given database and when tuning their parameters (64). Consequently, such algorithms identify domains with lengths similar to those already in the databases (40). This, in turn, influences a host of applications that rely on these data, such as function annotation, which often starts with segmenting the protein chain into domains (65).
Tracing the reuse of themes of minimal lengths ranging from 35 to 200 residues—below and above the length of an average domain—provided us with means for examining whether domains are indeed the basic “building blocks” of evolution. Specifically, we attempted to differentiate between two alternative scenarios: (i) duplication of protein segments occurs (only) at the domain and above-domain levels vs. (ii) duplication occurs among protein segments of diverse lengths and in particular, also among segments that are shorter than domains. In both cases, we expected to observe reuse. The difference is that the first scenario implies that the level of reuse observed with a minimal length threshold of ∼100 residues should be similar to that observed when shorter themes are considered, whereas the second scenario implies that reuse should be more extensive when shorter themes are included. Our results unequivocally support the second scenario. The shorter the segments included, the more reuse we find, and this relationship is monotonic. In other words, contrary to Brenner’s statement that protein evolution acts in units of 100 residues (39), the length of 100 residues is not singular from the perspective of reuse. We, therefore, suggest that, when studying protein function and evolution, it is not sufficient to only consider reuse among substructures of ∼100 residues (66). Rather, researchers must take into account the more complex patterns associated with the reuse of (often overlapping) segments of many different lengths.
Conclusions
Our work suggests two future directions. The first is documenting sequence reuse of segments across a wider range of lengths. We hope that this approach will be incorporated into future versions of existing databases. A holistic and unified view can include reuse of protein themes, motifs, domains, supradomains, chains, and even complexes and represent instances in which residues appear in several (overlapping, occasionally nested) reused segments of different lengths and in (increasingly large) sets of neighboring proteins. The second is identifying sequences of short reused protein segments that fold independently, which can offer powerful hints as to how proteins evolve. Our themes, available at http://trachel-srv.cs.haifa.ac.il/rachel/ppi/themes/, can assist in narrowing the search for such segments. Indeed, our set of themes could be remnants of antecedent peptides from which current proteins emerged. At any rate, the principles presented here and the computational pipeline could facilitate the detection of such peptides. Relating themes to biological functions, such as binding of ligands, nucleic acids, and proteins, may also facilitate this effort, offering a perspective on the evolution of protein function.
Methods
For every protein P, we consider all of its meaningful HHSearch (12) alignments. Based on these alignments, we identify contiguous segments in P (i.e., themes) that have at least minimal theme length residues and that are most reused. To do this, we define a function that quantifies the support that the alignments offer to the idea that a theme is reused and solve the optimization problem of finding the set of themes that are most reused using a dynamic programming algorithm. After a list of all of the candidate themes was found, we used a search-and-join step to join similar themes that were identified in different contexts. This results in a set of bona fide themes: each is a set of segments from different proteins, among which we identified similarity relationships (SI Appendix has details of the algorithms and examples).
Supplementary Material
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Themes Database of Reuse in Proteins, trachel-srv.cs.haifa.ac.il/rachel/ppi/themes/.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1707642114/-/DCSupplemental.
References
- 1.Lupas AN, Ponting CP, Russell RB. On the evolution of protein folds: Are similar motifs in different protein folds the result of convergence, insertion, or relics of an ancient peptide world? J Struct Biol. 2001;134:191–203. doi: 10.1006/jsbi.2001.4393. [DOI] [PubMed] [Google Scholar]
- 2.Söding J, Lupas AN. More than the sum of their parts: On the evolution of proteins from peptides. Bioessays. 2003;25:837–846. doi: 10.1002/bies.10321. [DOI] [PubMed] [Google Scholar]
- 3.Alva V, Söding J, Lupas AN. A vocabulary of ancient peptides at the origin of folded proteins. Elife. 2015;4:e09410. doi: 10.7554/eLife.09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vogel C, Bashton M, Kerrison ND, Chothia C, Teichmann SA. Structure, function and evolution of multidomain proteins. Curr Opin Struct Biol. 2004;14:208–216. doi: 10.1016/j.sbi.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 5.Petrey D, Fischer M, Honig B. Structural relationships among proteins with different global topologies and their implications for function annotation strategies. Proc Natl Acad Sci USA. 2009;106:17377–17382. doi: 10.1073/pnas.0907971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kolodny R, Petrey D, Honig B. Protein structure comparison: Implications for the nature of ‘fold space’, and structure and function prediction. Curr Opin Struct Biol. 2006;16:393–398. doi: 10.1016/j.sbi.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 7.Krishna SS, Grishin NV. Structural drift: A possible path to protein fold change. Bioinformatics. 2005;21:1308–1310. doi: 10.1093/bioinformatics/bti227. [DOI] [PubMed] [Google Scholar]
- 8.Swindells MB, Orengo CA, Jones DT, Hutchinson EG, Thornton JM. Contemporary approaches to protein structure classification. Bioessays. 1998;20:884–891. doi: 10.1002/(SICI)1521-1878(199811)20:11<884::AID-BIES3>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 9.Orengo CA, et al. CATH–A hierarchic classification of protein domain structures. Structure. 1997;5:1093–1108. doi: 10.1016/s0969-2126(97)00260-8. [DOI] [PubMed] [Google Scholar]
- 10.Kolodny R, Koehl P, Levitt M. Comprehensive evaluation of protein structure alignment methods: Scoring by geometric measures. J Mol Biol. 2005;346:1173–1188. doi: 10.1016/j.jmb.2004.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koehl P. Protein structure similarities. Curr Opin Struct Biol. 2001;11:348–353. doi: 10.1016/s0959-440x(00)00214-1. [DOI] [PubMed] [Google Scholar]
- 12.Söding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 13.Eddy SR. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009;23:205–211. [PubMed] [Google Scholar]
- 14.Kolodny R, Koehl P, Guibas L, Levitt M. Small libraries of protein fragments model native protein structures accurately. J Mol Biol. 2002;323:297–307. doi: 10.1016/s0022-2836(02)00942-7. [DOI] [PubMed] [Google Scholar]
- 15.Sawada Y, Honda S. Structural diversity of protein segments follows a power-law distribution. Biophys J. 2006;91:1213–1223. doi: 10.1529/biophysj.105.076661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szustakowski JD, Kasif S, Weng Z. Less is more: Towards an optimal universal description of protein folds. Bioinformatics. 2005;21(Suppl 2):ii66–ii71. doi: 10.1093/bioinformatics/bti1111. [DOI] [PubMed] [Google Scholar]
- 17.Fernandez-Fuentes N, Dybas JM, Fiser A. Structural characteristics of novel protein folds. PLoS Comput Biol. 2010;6:e1000750. doi: 10.1371/journal.pcbi.1000750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mackenzie CO, Zhou J, Grigoryan G. Tertiary alphabet for the observable protein structural universe. Proc Natl Acad Sci USA. 2016;113:E7438–E7447. doi: 10.1073/pnas.1607178113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coulson AFW, Moult J. A unifold, mesofold, and superfold model of protein fold use. Proteins. 2002;46:61–71. doi: 10.1002/prot.10011. [DOI] [PubMed] [Google Scholar]
- 20.Orengo CA, Thornton JM. Protein families and their evolution: A structural perspective. Annu Rev Biochem. 2005;74:867–900. doi: 10.1146/annurev.biochem.74.082803.133029. [DOI] [PubMed] [Google Scholar]
- 21.Levitt M. Nature of the protein universe. Proc Natl Acad Sci USA. 2009;106:11079–11084. doi: 10.1073/pnas.0905029106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wuchty S. Scale-free behavior in protein domain networks. Mol Biol Evol. 2001;18:1694–1702. doi: 10.1093/oxfordjournals.molbev.a003957. [DOI] [PubMed] [Google Scholar]
- 23.Unger R, Uliel S, Havlin S. Scaling law in sizes of protein sequence families: From super-families to orphan genes. Proteins. 2003;51:569–576. doi: 10.1002/prot.10347. [DOI] [PubMed] [Google Scholar]
- 24.Vogel C, Berzuini C, Bashton M, Gough J, Teichmann SA. Supra-domains: Evolutionary units larger than single protein domains. J Mol Biol. 2004;336:809–823. doi: 10.1016/j.jmb.2003.12.026. [DOI] [PubMed] [Google Scholar]
- 25.Zeldovich KB, Shakhnovich EI. Understanding protein evolution: From protein physics to Darwinian selection. Annu Rev Phys Chem. 2008;59:105–127. doi: 10.1146/annurev.physchem.58.032806.104449. [DOI] [PubMed] [Google Scholar]
- 26.Wolf YI, Grishin NV, Koonin EV. Estimating the number of protein folds and families from complete genome data. J Mol Biol. 2000;299:897–905. doi: 10.1006/jmbi.2000.3786. [DOI] [PubMed] [Google Scholar]
- 27.Dokholyan NV, Shakhnovich B, Shakhnovich EI. Expanding protein universe and its origin from the biological Big Bang. Proc Natl Acad Sci USA. 2002;99:14132–14136. doi: 10.1073/pnas.202497999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, Hubner IA, Arakaki AK, Shakhnovich E, Skolnick J. On the origin and highly likely completeness of single-domain protein structures. Proc Natl Acad Sci USA. 2006;103:2605–2610. doi: 10.1073/pnas.0509379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skolnick J, Gao M, Zhou H. On the role of physics and evolution in dictating protein structure and function. Isr J Chem. 2014;54:1176–1188. doi: 10.1002/ijch.201400013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skolnick J, Gao M. Interplay of physics and evolution in the likely origin of protein biochemical function. Proc Natl Acad Sci USA. 2013;110:9344–9349. doi: 10.1073/pnas.1300011110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Triant DA, Pearson WR. Most partial domains in proteins are alignment and annotation artifacts. Genome Biol. 2015;16:99. doi: 10.1186/s13059-015-0656-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chothia C, Gough J, Vogel C, Teichmann SA. Evolution of the protein repertoire. Science. 2003;300:1701–1703. doi: 10.1126/science.1085371. [DOI] [PubMed] [Google Scholar]
- 33.Lees JG, Dawson NL, Sillitoe I, Orengo CA. Functional innovation from changes in protein domains and their combinations. Curr Opin Struct Biol. 2016;38:44–52. doi: 10.1016/j.sbi.2016.05.016. [DOI] [PubMed] [Google Scholar]
- 34.Kelley LA, Sternberg MJ. Partial protein domains: Evolutionary insights and bioinformatics challenges. Genome Biol. 2015;16:100. doi: 10.1186/s13059-015-0663-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng H, et al. ECOD: An evolutionary classification of protein domains. PLoS Comput Biol. 2014;10:e1003926. doi: 10.1371/journal.pcbi.1003926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hubbard TJ, Murzin AG, Brenner SE, Chothia C. SCOP: A structural classification of proteins database. Nucleic Acids Res. 1997;25:236–239. doi: 10.1093/nar/25.1.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Csaba G, Birzele F, Zimmer R. Systematic comparison of SCOP and CATH: A new gold standard for protein structure analysis. BMC Struct Biol. 2009;9:23. doi: 10.1186/1472-6807-9-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Day R, Beck DAC, Armen RS, Daggett V. A consensus view of fold space: Combining SCOP, CATH, and the Dali Domain Dictionary. Protein Sci. 2003;12:2150–2160. doi: 10.1110/ps.0306803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brenner S. The molecular evolution of genes and proteins: A tale of two serines. Nature. 1988;334:528–530. doi: 10.1038/334528a0. [DOI] [PubMed] [Google Scholar]
- 40.Wheelan SJ, Marchler-Bauer A, Bryant SH. Domain size distributions can predict domain boundaries. Bioinformatics. 2000;16:613–618. doi: 10.1093/bioinformatics/16.7.613. [DOI] [PubMed] [Google Scholar]
- 41.Kolodny R, Pereyaslavets L, Samson AO, Levitt M. On the universe of protein folds. Annu Rev Biophys. 2012;42:559–582. doi: 10.1146/annurev-biophys-083012-130432. [DOI] [PubMed] [Google Scholar]
- 42.Harrison A, Pearl F, Mott R, Thornton J, Orengo C. Quantifying the similarities within fold space. J Mol Biol. 2002;323:909–926. doi: 10.1016/s0022-2836(02)00992-0. [DOI] [PubMed] [Google Scholar]
- 43.Eck RV, Dayhoff MO. Evolution of the structure of ferredoxin based on living relics of primitive amino acid sequences. Science. 1966;152:363–366. doi: 10.1126/science.152.3720.363. [DOI] [PubMed] [Google Scholar]
- 44.Friedberg I, Godzik A. Connecting the protein structure universe by using sparse recurring fragments. Structure. 2005;13:1213–1224. doi: 10.1016/j.str.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 45.Nepomnyachiy S, Ben-Tal N, Kolodny R. Global view of the protein universe. Proc Natl Acad Sci USA. 2014;111:11691–11696. doi: 10.1073/pnas.1403395111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alva V, Remmert M, Biegert A, Lupas AN, Söding J. A galaxy of folds. Protein Sci. 2010;19:124–130. doi: 10.1002/pro.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pascual-García A, Abia D, Ortiz ÁR, Bastolla U. Cross-over between discrete and continuous protein structure space: Insights into automatic classification and networks of protein structures. PLoS Comput Biol. 2009;5:e1000331. doi: 10.1371/journal.pcbi.1000331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prakash A, Bateman A. Domain atrophy creates rare cases of functional partial protein domains. Genome Biol. 2015;16:88. doi: 10.1186/s13059-015-0655-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vanhee P, et al. BriX: A database of protein building blocks for structural analysis, modeling and design. Nucleic Acids Res. 2011;39:D435–D442. doi: 10.1093/nar/gkq972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Su QJ, Lu L, Saxonov S, Brutlag DL. eBLOCKs: Enumerating conserved protein blocks to achieve maximal sensitivity and specificity. Nucleic Acids Res. 2005;33:D178–D182. doi: 10.1093/nar/gki060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Henikoff S, Henikoff JG, Pietrokovski S. Blocks+: A non-redundant database of protein alignment blocks derived from multiple compilations. Bioinformatics. 1999;15:471–479. doi: 10.1093/bioinformatics/15.6.471. [DOI] [PubMed] [Google Scholar]
- 52.Bailey TL, et al. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frenkel ZM, Trifonov EN. From protein sequence space to elementary protein modules. Gene. 2008;408:64–71. doi: 10.1016/j.gene.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 54.Trifonov EN, Frenkel ZM. Evolution of protein modularity. Curr Opin Struct Biol. 2009;19:335–340. doi: 10.1016/j.sbi.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 55.Goncearenco A, Berezovsky IN. Prototypes of elementary functional loops unravel evolutionary connections between protein functions. Bioinformatics. 2010;26:i497–i503. doi: 10.1093/bioinformatics/btq374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goncearenco A, Berezovsky IN. Computational reconstruction of primordial prototypes of elementary functional loops in modern proteins. Bioinformatics. 2011;27:2368–2375. doi: 10.1093/bioinformatics/btr396. [DOI] [PubMed] [Google Scholar]
- 57.Yanover C, Vanetik N, Levitt M, Kolodny R, Keasar C. Redundancy-weighting for better inference of protein structural features. Bioinformatics. 2014;30:2295–2301. doi: 10.1093/bioinformatics/btu242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dodd MS, et al. Evidence for early life in Earth’s oldest hydrothermal vent precipitates. Nature. 2017;543:60–64. doi: 10.1038/nature21377. [DOI] [PubMed] [Google Scholar]
- 59.Chaudhuri I, Söding J, Lupas AN. Evolution of the β-propeller fold. Proteins. 2008;71:795–803. doi: 10.1002/prot.21764. [DOI] [PubMed] [Google Scholar]
- 60.Smock RG, Yadid I, Dym O, Clarke J, Tawfik DS. De novo evolutionary emergence of a symmetrical protein is shaped by folding constraints. Cell. 2016;164:476–486. doi: 10.1016/j.cell.2015.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Redfern OC, Harrison A, Dallman T, Pearl FM, Orengo CA. CATHEDRAL: A fast and effective algorithm to predict folds and domain boundaries from multidomain protein structures. PLoS Comput Biol. 2007;3:e232. doi: 10.1371/journal.pcbi.0030232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fox NK, Brenner SE, Chandonia J-M. SCOPe: Structural classification of proteins–Extended, integrating SCOP and ASTRAL data and classification of new structures. Nucleic Acids Res. 2014;42:D304–D309. doi: 10.1093/nar/gkt1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Andreeva A, et al. Data growth and its impact on the SCOP database: New developments. Nucleic Acids Res. 2008;36:D419–D425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nagarajan N, Yona G. Automatic prediction of protein domains from sequence information using a hybrid learning system. Bioinformatics. 2004;20:1335–1360. doi: 10.1093/bioinformatics/bth086. [DOI] [PubMed] [Google Scholar]
- 65.Radivojac P, et al. A large-scale evaluation of computational protein function prediction. Nat Methods. 2013;10:221–227. doi: 10.1038/nmeth.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chothia C. Proteins. One thousand families for the molecular biologist. Nature. 1992;357:543–544. doi: 10.1038/357543a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.