Significance
Cholesterol and its transformation into cholesterol-5,6-epoxides (5,6-EC) was long suspected as contributing to breast cancer (BC) pathogenesis, before it was found that 5,6-EC metabolism controls BC development and is deregulated in breast cancers. Herein, we studied in tumor cells and human samples how 5,6-EC metabolism deregulation promotes tumor progression. We have discovered a pathway in BCs producing an oncometabolite derived from 5,6-EC, through the action of the cortisol-inactivating enzyme, and identified the glucocorticoid receptor (GR) as the target mediating its proliferative effects. Inhibition of its production or GR significantly blocked its action on BC progression. Thus, targeting this oncometabolism and GR represents a new opportunity for therapeutic intervention in BCs and potentially other cancers presenting such deregulations.
Keywords: breast cancer, oncometabolism, dendrogenin A, therapy, nuclear receptor
Abstract
Breast cancer (BC) remains the primary cause of death from cancer among women worldwide. Cholesterol-5,6-epoxide (5,6-EC) metabolism is deregulated in BC but the molecular origin of this is unknown. Here, we have identified an oncometabolism downstream of 5,6-EC that promotes BC progression independently of estrogen receptor α expression. We show that cholesterol epoxide hydrolase (ChEH) metabolizes 5,6-EC into cholestane-3β,5α,6β-triol, which is transformed into the oncometabolite 6-oxo-cholestan-3β,5α-diol (OCDO) by 11β-hydroxysteroid-dehydrogenase-type-2 (11βHSD2). 11βHSD2 is known to regulate glucocorticoid metabolism by converting active cortisol into inactive cortisone. ChEH inhibition and 11βHSD2 silencing inhibited OCDO production and tumor growth. Patient BC samples showed significant increased OCDO levels and greater ChEH and 11βHSD2 protein expression compared with normal tissues. The analysis of several human BC mRNA databases indicated that 11βHSD2 and ChEH overexpression correlated with a higher risk of patient death, highlighting that the biosynthetic pathway producing OCDO is of major importance to BC pathology. OCDO stimulates BC cell growth by binding to the glucocorticoid receptor (GR), the nuclear receptor of endogenous cortisol. Interestingly, high GR expression or activation correlates with poor therapeutic response or prognosis in many solid tumors, including BC. Targeting the enzymes involved in cholesterol epoxide and glucocorticoid metabolism or GR may be novel strategies to prevent and treat BC.
A role for cholesterol in the etiology of cancers has long been suspected, and 5,6-cholesterol epoxides (5,6-EC) were initially thought to be the causative agents. However, recent data have now shown that it is the metabolism of 5,6-EC that is deregulated in breast cancer (BC), and that this metabolism controls BC development (1–4). To develop novel precision therapeutic strategies, a deeper understanding of 5,6-EC metabolism is required. Indeed, BC remains the most frequent cause of death from cancer among women worldwide, despite the development of targeted therapies, such as Tamoxifen (Tam) for treating tumors expressing the estrogen receptor (ER), or agents that target the overexpressed growth factor receptor HER2 (human epidermal growth factor receptor). These failures are explained by the fact that many BC do not respond to these therapies or develop resistance, and there are currently no effective targeted therapies to treat tumors that express neither ER nor HER2. In addition to its role as a competitive inhibitor for ER, Tam also impacts on cholesterol metabolism by targeting cholesterol epoxide hydrolase (ChEH), an enzymatic complex formed by two cholesterogenic enzymes, DHCR7 and D8D7I (also known as EBP) (5–7). In normal tissues, ChEH catalyzes the hydrolysis of the cholesterol 5,6-epoxides α and β (5,6α-EC and 5,6β-EC) into cholestane-3β,5α,6β-triol (CT) (5, 6, 8). In normal mammalian tissues, 5,6α-EC reacts with histamine via an enzyme to produce a tumor suppressor metabolite named dendrogenin A (DDA), whose levels are significantly decreased in BC compared with normal adjacent tissues (NAT), indicating a deregulation of this pathway during BC development (3, 4). These data combined suggest a potential role of ChEH activation in BC progression that we explored in the present study. Herein, we describe how this led us to discover an oncometabolite, 6-oxo-cholestan-3β,5α-diol (OCDO), that drives BC progression through the glucocorticoid receptor (GR). Furthermore, we show that OCDO is derived from 5,6-ECs through the action of ChEH and the cortisol-inactivating enzyme, 11βHSD2. Inhibiting this oncometabolic pathway or GR significantly reduced BC proliferation, suggesting that targeting the actors of this pathway could represent new strategies in BC therapy and prevention.
Results
Kinetic Analysis of 5,6-EC and CT Metabolism in MCF7 Cells.
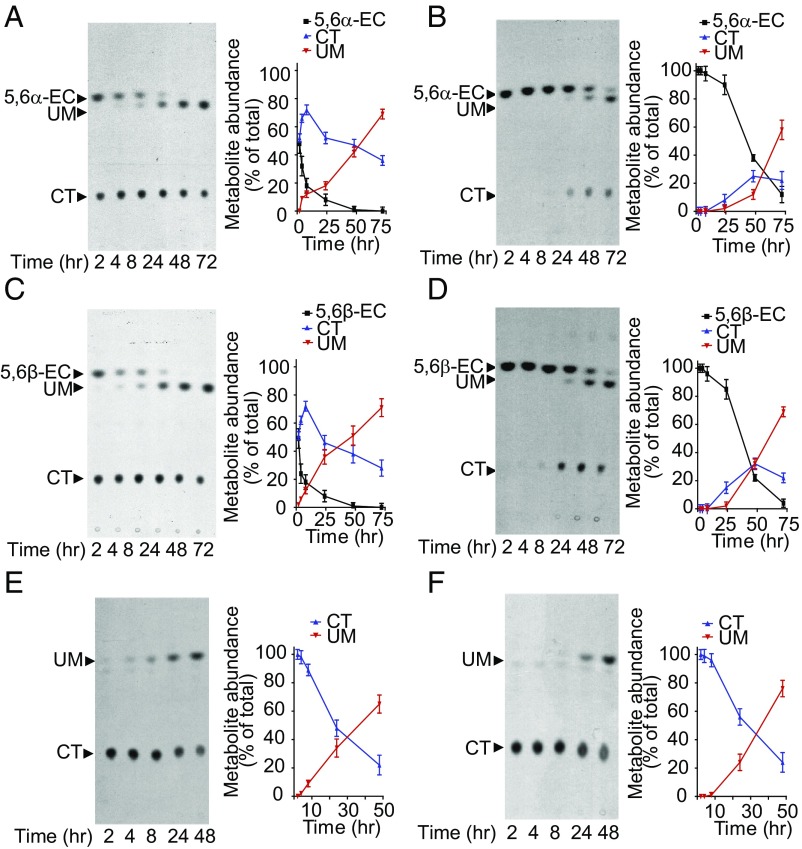
We first examined the metabolism of [14C]5,6α-EC, [14C]5,6β-EC, and [14C]CT over a 72-h period in the breast cancer cell line MCF7. Thin-layer chromatography (TLC) analyses of cell and media extracts showed that 5,6α-EC (Fig. 1 A and B) and 5,6β-EC (Fig. 1 C and D) were first converted into CT (retention factor, Rf = 0.21) as a result of ChEH activity. However, over time, an unknown metabolite (UM), Rf = 0.60, appeared and its level increased at the expense of the 5,6-ECs and CT. Similar experiments performed using [14C]CT alone revealed that the UM was a metabolite of CT (Fig. 1 E and F).
Fig. 1.
UM is a metabolite of CT in tumor cells. (A–F, Left) Representative TLC autoradiograms (n = 5) showing time-dependent production of UM in MCF7 cells treated with (A and B) 600 nM [14C]5,6α-EC, (C and D) 600 nM [14C]5,6β-EC, and (E and F) 1 µM [14C]-CT. (Right) Quantitative analyses of the metabolites extracted from cells (A, C, and E) and media (B, D, and F). The regions corresponding to the radioactive metabolites of interest (arrows) were recovered and counted using a β-counter. Results are the mean (±SEM) of five independent experiments.
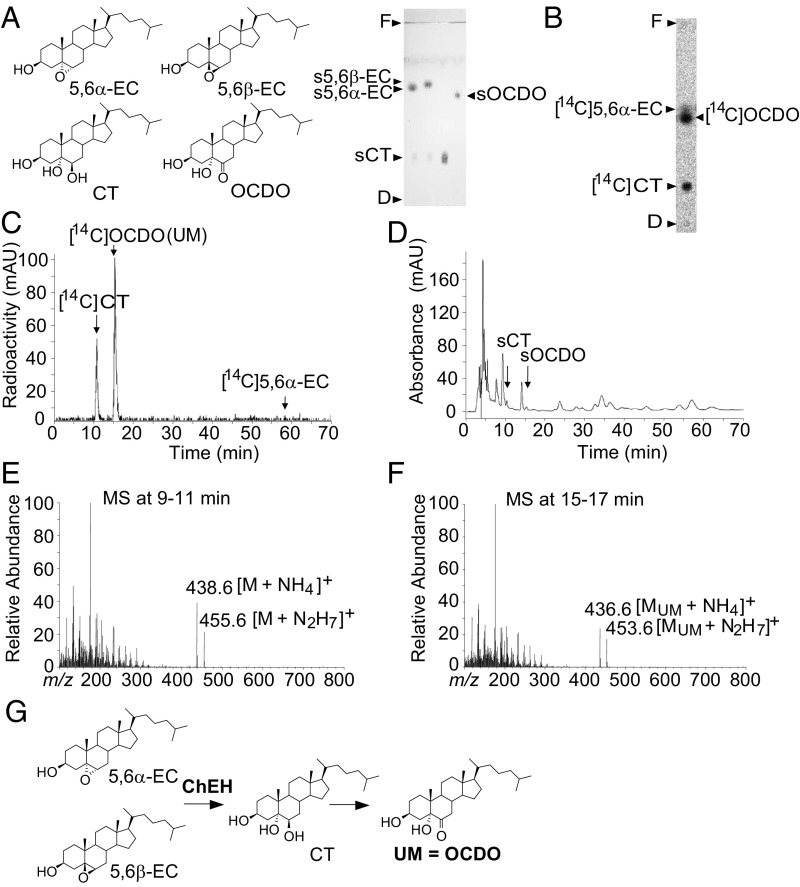
Characterization of UM as OCDO.
It has been reported that CT can be chemically oxidized into OCDO (9), so we hypothesized that the UM could be OCDO (Fig. 2A). In normal-phase TLC, we found that synthetic OCDO (sOCDO) and [14C]OCDO had a similar retention factor (Rf = 0.60) to that of the UM (Fig. 2 A and B, respectively). Reverse-phase HPLC (RP-HPLC) also showed a similar retention time between UM and sOCDO or [14C]OCDO (Rt = 15 min) (Fig. 2 C and D, respectively), while synthetic CT (sCT) and [14C]CT had a Rt of 10 min (Fig. 2 C and D, respectively). To confirm the identity of the UM, MCF7 cells were treated with 5,6α-EC for 72 h, then lipids were extracted and submitted to RP-HPLC. Mass spectrometric analysis of the 9- to 11-min RP-HPLC fractions (Fig. 2D) showed peaks of m/z [M+NH4]+ = 438.6 and m/z [M+N2H7]+ = 455.6, corresponding to the mass of CT and consistent with the transformation of 5,6α-EC into CT by ChEH (Fig. 2E). Analysis of the 15- to 17-min RP-HPLC fractions (Fig. 2D) showed peaks of m/z [MUM+NH4]+ = 436.6 and m/z [MUM+N2H7]+ = 453.6 (Fig. 2F), corresponding to the mass of sOCDO. This confirmed that the UM was OCDO (Fig. 2G).
Fig. 2.
Structural characterization of UM. (A, Left) Chemical structure of the metabolites of interest. (Right) Representative migration performed by silica gel TLC (n = 5) of the synthetic (s) metabolites of interest, indicated by arrows. (B) MCF7 cells were incubated for 72 h with [14C]5,6α-EC and analyzed as described for Fig. 1 (n = 5). Analysis of cell extracts by (B) polar silica gel TLC or (C) hydrophobic RP-HPLC. (D) RP-HPLC profile of the metabolites extracted from MCF-7 cells that had been treated for 72 h with 5,6α-EC. Arrows indicate peaks corresponding to the authentic standards: sCT and sOCDO. (E) CI-MS spectra of the RP-HPLC peak eluted between 9 and 11 min in D. (F) CI-MS spectra of the RP-HPLC peak eluted between 15 and 17 min in D. (G) Scheme describing the formation of OCDO.
ChEH Inhibition Abrogates OCDO Formation in BC Cells.
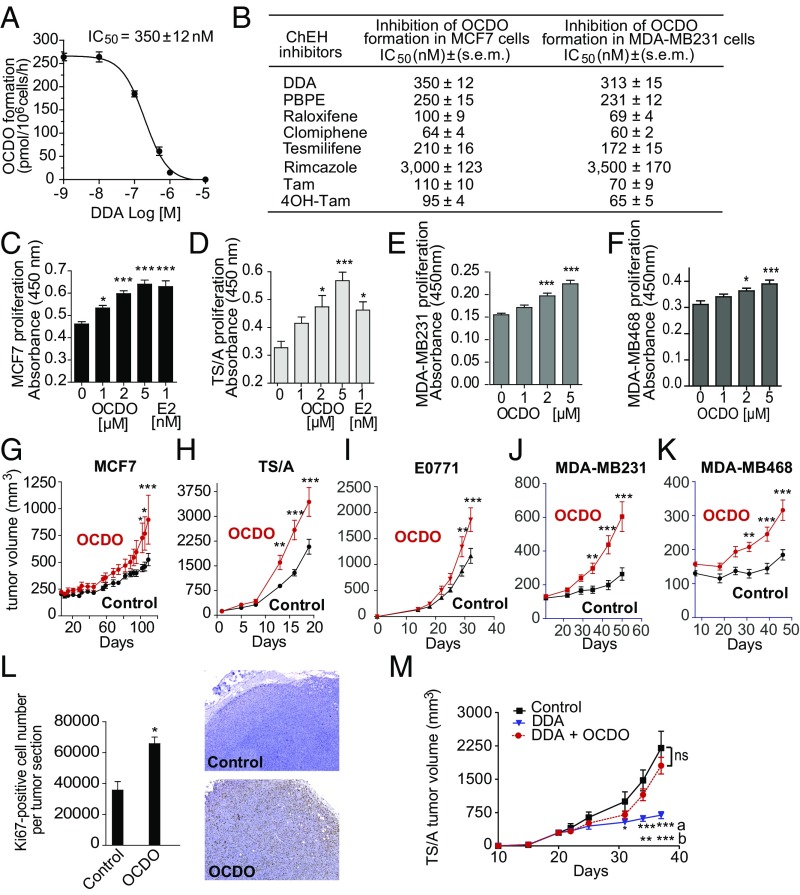
We next measured the impact of DDA and other ChEH inhibitors (3, 5) on OCDO formation by incubating human MCF7 or MDA-MB-231 tumor cells (expressing or not the ERα, respectively) with [14C]5,6α-EC and the indicated ChEH inhibitors. As shown in Fig. 3 A and B, in both cell lines DDA and the ChEH inhibitors tested inhibited OCDO production, including ChEH inhibitors known to have antitumor activity and to inhibit ERα, such as Tam or raloxifene, or to act independently of the ERα, such as DDA, tesmilifene, or PBPE (1-[2-[4-(phenylmethyl)phenoxy]ethyl]-pyrrolidine) (3, 5, 10). These data indicate that all of these ChEH inhibitors target the OCDO pathway independently of ERα expression.
Fig. 3.
OCDO is a tumor promoter and its inhibition contributes to the antitumor effects of DDA. (A) Representative inhibition curve showing the inhibition of OCDO formation by increasing concentrations of DDA in human MCF7 cells treated with 600 nM [14C]5,6α-EC for 72 h. OCDO formation was quantified after TLC analysis as described for Fig. 1 and IC50 value was calculated from the concentration-inhibition curve. (B) Analysis of the inhibition of OCDO formation in MCF7 and MDA-MB231 cells treated as in A with increasing concentrations of the indicated ChEH inhibitors. OCDO formation and IC50 values were measured as in A. (C–F) Tumor cell proliferation using a colorimetric BrDU immunoassay, n = 8. *P < 0.05, ***P < 0.001, one-way ANOVA, Tukey’s posttest. (A–F) Data are the mean (±SEM) of five separate experiments. (G–K) Mice (10 per group) implanted with the indicated cell lines were treated with the solvent vehicle or OCDO and monitored for tumor growth over time. (L, Left) Mean (±SEM) of Ki67-positive cells quantified from IHC staining of MCF7 tumor sections from G, n = 8. *P < 0.05, Student’s t test, two-tailed; ***P < 0.001. (Right) Representative Ki67 staining of TS/A tumor sections from H. (M) Mice (10 per group) implanted with TS/A cells were treated every day, starting at day 1, with either the solvent vehicle, DDA (0.37 µg/kg), or DDA (0.37 µg/kg) + OCDO (50 µg/kg). (G–K and M) Data are representative of three independent experiments. Mean tumor volumes (±SEM) are shown, two-way ANOVA, Bonferroni posttest, *P < 0.05, **P < 0.01, ***P < 0.001. In M, letters indicate the comparison between: a: DDA vs. control; b: DDA vs. DDA + OCDO. ns: not significant at each time.
OCDO Stimulates BC Cell Proliferation in Vitro and in Vivo.
Treatment with OCDO or 17β-estradiol (E2) significantly increased the growth rate of ER+ cell lines (Fig. 3 C and D). OCDO also increased the proliferation of the ER− cells lines MDA-MB231 and MDA-MB468 (Fig. 3 E and F). OCDO significantly promoted the growth of both ER+ and ER− tumors grafted onto mice (Fig. 3 G and K). Histological analysis of tumors revealed that the proliferative marker Ki67 was increased following OCDO treatment (Fig. 3L).
Inhibition of OCDO Production Contributes to the Antitumor Activities of DDA in Mice.
We then determined whether the antiproliferative effects of DDA involved the inhibition of OCDO production. As shown in Fig. 3M, DDA significantly inhibited the growth of tumors compared with vehicle-treated mice. However, when OCDO was added back by treating engrafted animals with DDA plus OCDO, the growth inhibitory action of DDA was reversed, indicating that the inhibition of OCDO production contributed to the antitumor activity of this compound.
Identification of the Enzymes That Regulate the Production of OCDO from CT.
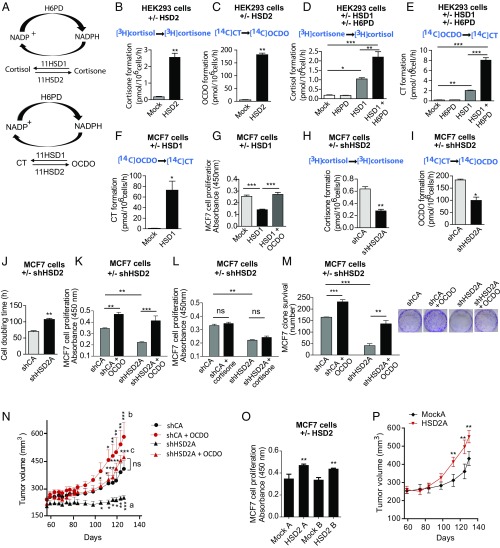
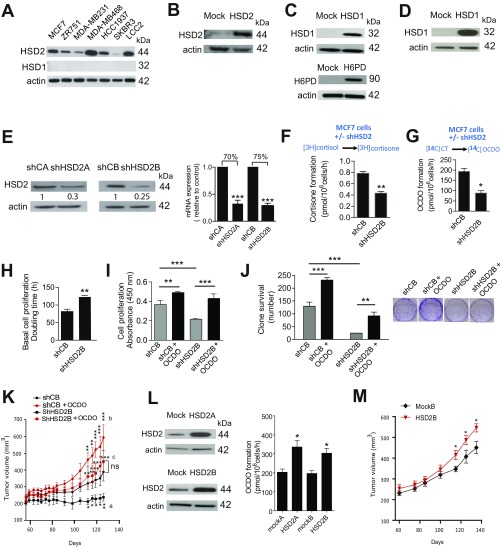
Since OCDO is produced from CT, we hypothesized the existence of an enzyme distinct from ChEH that would realize this reaction. We considered that a hydroxysteroid dehydrogenase (HSD) could catalyze the dehydrogenation of the alcohol function in position 6 of CT into a ketone in OCDO. A local symmetry axis on the steroid backbone makes positions 11β and 7α equivalent (11), which suggested to us that an 11βHSD enzyme was a good candidate for catalyzing this reaction. 11βHSD exist as two enzymes, 11βHSD type 2 (11HSD2), which catalyzes the dehydrogenation of cortisol into the inactive cortisone, and 11βHSD type 1 (11HSD1), which performs the reverse reaction (12) (Fig. 4A). In accordance with this hypothesis, 11HSD2 mRNA and protein expression was measured in a panel of human BC cell lines expressing or not ERα, while 11HSD1 expression was not detectable and all of the cell lines tested produced OCDO (Fig. S1A and Table S1).
Fig. 4.
11βHSD2 and 11βHSD1 interconvert OCDO and CT. (A, Upper) 11HSD2 catalyzes the dehydrogenation of cortisol into cortisone. 11HSD1 realizes the reverse reaction. H6PD is the enzyme that produces the cofactor NADPH necessary for the reductase activity of 11HSD1. (A, Lower) 11HSD2 produces OCDO from CT and 11HSD1 produces CT from OCDO. (B–M and O) HEK293T (HEK) or MCF7 cells were transfected with plasmids encoding the indicated enzyme or the empty vector (mock) or shRNA. (B–F) The production of the indicated metabolites in transfected HEK (B–E) or MCF7 cells (F) was analyzed as in Fig. 1, n = 5. (G) The proliferation of transfected MCF7 cells was analyzed as in Fig. 3 C–F, n = 8. (H–M) shCA- and shHSD2A-MCF7 cells were assayed for: (H) cortisone or (I) OCDO production, as measured in Fig. 1, n = 5; (J–L) cell proliferation by (J) counting cell numbers, n = 5, or (K–L) as in G, n = 8; or (M) cell clonogenicity (n = 3), with or without 5 µM OCDO or cortisone. (N) shCA or shHSD2A-MCF7 tumors engrafted into mice (10 per group) were treated with solvent vehicle (control) or OCDO (16 µg/kg, 5 d/wk). Data are representative of three independent experiments. (O) Control (mock) or HSD2 overexpressing MCF7 cell proliferation was analyzed as in G. (P) The growth of control (mock) or 11HSD2 overexpressing MCF7 tumors engrafted into mice (10 mice per group) were compared over time. Data are representative of three independent experiments. (B–M and O) Data are the mean (±SEM) of five separate experiments and were analyzed (B, C, F, and H–J) by a Student’s t test, two-tailed, or (D, E, G, and K–O) by one-way ANOVA, Tukey’s posttest. *P < 0.05, **P < 0.01, ***P < 0.001. (N and P) Mean tumor volumes (±SEM) are shown, and data were analyzed by two-way ANOVA, Bonferroni posttest. *P < 0.05, **P < 0.01, ***P < 0.001. In N, letters indicate the comparison between: a: shCA vs. shHSD2A; b: shCA vs. shCA + OCDO; c: shHSD2A vs. shHSD2A + OCDO.
Fig. S1.
Measuring the expression or extinction of the proteins of interest and their activity. (A) The expression of endogenous HSD2 and HSD1 was analyzed by immunoblotting in the indicated mammary tumor cells (related to Table S1). (B) HEK293T cells were transfected with a plasmid encoding HSD2 or the empty vector (mock) and the expression of the enzyme was confirmed by immunoblotting. (C) HEK293 cells were transfected with a plasmid encoding HSD1, H6PD, or the empty vector and the expression of the enzyme was confirmed by immunoblotting. (D) MCF7 cells were transfected with a plasmid encoding 11HSD1 and the expression of the enzyme was confirmed as in C. (E) MCF7 cells were transfected with two different shRNA plasmids targeting HSD2 (shHSD2), or with a control shRNA (shC) (SureSilencing ShRNA plasmid; Qiagen). The decreased expression of the enzyme was confirmed by immunoblotting and by qRT-PCR. Two clones (shHSD2A and shHSD2B) were selected. (F–J) shCB-MCF7 and shHSD2B-MCF7 cells were assayed for (F and G) cortisone or OCDO production, respectively, as described in Fig. 4 B and C; cell proliferation (H and I) by counting cell numbers (n = 5) or using a colorimetric BrDu assay, respectively (n = 8), or (J) cell clonogenicity (n = 3); with or without 5 µM OCDO, as indicated. Data were analyzed as described in Fig. 4 H–K and M and are the mean (±SEM) of five separate experiments. (K) The growth of shCB or shHSD2B-MCF7 tumors engrafted into mice (10 per group) were compared. OCDO treatment (16 µg/kg, 5 d/wk) increased shCB-MCF7 and reversed the growth inhibition of shHSD2-MCF7 tumors. Data are representative of three independent experiments. (L and M) MCF7 cells were transfected with a plasmid encoding 11HSD2 or the empty vector (mock). (L) Two stable clones were selected for the overexpression of 11HSD2 (HSD2A and HSD2B) and were compared with control cells (mockA and mockB) by immunoblotting. Mock-MCF7 or HSD2-MCF7 cells were incubated with [14C]CT and the production of OCDO was analyzed as in Fig. 1. Data are the mean (±SEM) of three experiments. (M) The growth of Mock-MCF7 or HSD2-MCF7 tumors engrafted into mice (10 per group) were compared. Mean tumor volumes (±SEM) are shown. Data are representative of three independent experiments. (E and H) Data were analyzed by a Student’s t test, two-tailed, or (I and J) by one-way ANOVA, Tukey’s posttest. *P < 0.05, **P < 0.01, ***P < 0.001. (K and M) Mean tumor volumes (±SEM) are shown, and data were analyzed by two-way ANOVA, Bonferroni posttest. *P < 0.05, **P < 0.01, ***P < 0.001. In K, letters indicate the comparison between: a: shCB vs. shHSD2B; b: shCB vs. shCB + OCDO; c: shHSD2B vs. shHSD2B + OCDO. ns, not significant.
Table S1.
Analysis of a panel of BC cell lines for 11HSD1 and 11HSD2 expression and OCDO production
| Cells | 11HSD1 Relative mRNA expression | 11HSD1 protein expression | 11HSD2 Relative mRNA expression | 11HSD2 protein expression | OCDO production pmol/106 cells/h ± SEM |
| MCF7 (ER+/PR+/HER2−) | — | — | 10.5 ± 0.1 | + | 254.4 ± 48 |
| ZR751 (ER+/PR−/HER2−) | — | — | 10.3 ± 0.4 | + | 223.2 ± 24 |
| SKBr3 (ER−/PR−/HER2+) | — | — | 12.1 ± 0.4 | + | 84.0 ± 7.2 |
| MDA-MB468 (ER−/PR−/HER2−) | — | — | 8.0 ± 0.3 | + | 573.6 ± 81.6 |
| HCC1937 (ER−/PR−/HER2−) | — | — | 11.0 ± 0.4 | + | 175.2 ± 7.2 |
| MDA-MB231 (ER−/PR−/HER2−) | — | — | 13.5 ± 0.2 | + | 48.0 ± 9.6 |
| LCC2 (TamR) (ER+/PR+/HER2−) | — | — | 7.6 ± 0.3 | + | 703.2 ± 216 |
| TS/A (ER+) | — | — | 10.7 ± 0.2 | ND | 228.0 ± 2.4 |
| E0771 (ER+) | — | — | 8.2 ± 0.3 | ND | 540.0 ± 96 |
Different subtypes of BC cells were analyzed for the expression of 11HSD1 and 11HSD2 by immunoblotting and qRT-PCR and assessed for OCDO production by incubating tumor cells with [14C]5,6α-EC for 24 h, as described for Fig. 1. Relative mRNA expression was expressed as ΔCT (HSD2CT-cycloA1CT and HSD1CT-cycloA1CT). Expression was considered as negative (−) when the CT of the mRNA of interest was >35. Results are the mean (±SEM) of three experiments. (+) and (−) indicate that the protein of interest was detected or not, respectively, by immunoblotting (n = 3); see immunoblot in Fig. S1A; TamR: cells derived from MCF7 cells with acquired resistance to Tam. Primers used: cycloA1: forward: GCA-TAC-GGG-TCC-TGG-CAT-CTT-GTC-C; reverse: ATG-GTG-ATC-TTC-TTG-CTG-GTC-TTG-C. 11βHSD1: forward: GA-CAG-CGA-GGT-CAA-AAG-AAA; reverse: GTC-CTC-CCA-TGA-GCT-TTC-CTG. 11βHSD2: forward: CCA-CCG-TAT-TGG-AGT-TGA-ACA; reverse: CGC-GGC-TAA-TGT-CTC-CTG-G.
To confirm a role for 11HSD2 in the production of OCDO from CT, we transfected HEK293 cells, a cell model that has previously been used to study 11HSD1 and 11HSD2 (13), with a plasmid encoding either 11HSD2 or the empty vector (mock) (Fig. S1B). When incubated with [3H]cortisol ([3H]-CRT) (Fig. 4B) or [14C]CT (Fig. 4C), 11HSD2-transfected cells produced significantly higher levels of cortisone or OCDO, respectively, than mock-transfected cells, indicating that 11HSD2 is able to produce OCDO in addition to cortisone. To study the involvement of 11HSD1 in the reverse transformation of OCDO into CT (Fig. 4A), HEK293T cells were transfected with a plasmid encoding 11HSD1 (HSD1) or the empty vector (mock), and with or without a plasmid encoding H6PD, the enzyme that produces the NADPH cofactor that is necessary for the reductase activity of 11HSD1 (14) (Fig. S1C). When cells were incubated with [3H]cortisone (Fig. 4D) or [14C]OCDO (Fig. 4E), 11HSD1-transfected cells produced significantly more cortisol or CT, respectively, than mock- or H6PD-transfected cells. This production was further increased by two- and fourfold, respectively, by cotransfecting 11HSD1 with H6PD. Thus, 11HSD1 produces significant levels of CT in addition to cortisol.
Ectopic 11HSD1 Expression in MCF-7 Cells Produces CT and Decreases Cell Proliferation, and OCDO Treatment Reverses This Effect.
The expression of 11HSD1 in MCF7 cells (Fig. S1D) significantly stimulated the conversion of OCDO to CT compared with controls (Fig. 4F). In addition, 11HSD1 expression in MCF7 cells also significantly decreased cell proliferation, an effect that was reversed by OCDO treatment (Fig. 4G). This indicates that 11HSD1 inhibits tumor cell proliferation through the transformation of OCDO into CT.
11HSD2 Controls Cell Proliferation and Tumor Growth Through OCDO Production.
To confirm that 11HSD2 produces OCDO and stimulates tumor cell proliferation, we knocked down 11HSD2 expression in MCF7 cells using shRNA. Two stable clones (shHSD2A and shHSD2B) were selected in which the expression of 11HSD2 was significantly decreased at both the protein and mRNA levels compared with shRNA control clones (shCA and shCB) (Fig. S1E). A significant decrease in cortisone (Fig. 4H) and OCDO (Fig. 4I) production was measured in shHSD2A cells compared with shCA cells, and their doubling time increased by 150% (Fig. 4J). In addition, OCDO significantly increased ShCA cell proliferation and rescued the decreased proliferation of shHSD2A cells (Fig. 4K), while no such effect was observed with cortisone (Fig. 4L). Moreover, OCDO also significantly increased ShCA clonogenecity and rescued the decreased clonogenicity of 11HSD2A cells (Fig. 4M). Together, these results indicate that 11HSD2 controls cell proliferation and clonogenicity through OCDO production. We then tested the impact of 11HSD2 knockdown in vivo by implanting shHSD2A cells into mice. As shown in Fig. 4N, the growth of shHSD2A tumors was significantly reduced (by 50%) compared with shCA tumors. Importantly, OCDO significantly stimulated the growth of ShCA tumor and rescued the decrease growth of shHSD2A tumors, indicating that 11HSD2 controls tumor growth through OCDO production. Similar results, both in vitro and in vivo, were obtained with the other shHSD2B and shCB clones (Fig. S1 F–K). To confirm the involvement of 11HSD2 in cell proliferation, MCF7 cells were stably transfected with a plasmid expressing 11HSD2 or the control plasmid. Two clones stably overexpressing 11HSD2 and two control clones (mock) were selected (Fig. S1L). The clones overexpressing 11HSD2 showed a greater capacity to produce OCDO when incubated with [14C]CT (Fig. S1L) and proliferated significantly faster in vitro, indicating an effect on tumor cells (Fig. 4O), and in vivo when implanted into mice, compared with control clones (Fig. 4P and Fig. S1M).
Enzymes Involved in the Production of OCDO Are Overexpressed in Human BC Compared with Matched Normal Adjacent Breast Tissue.
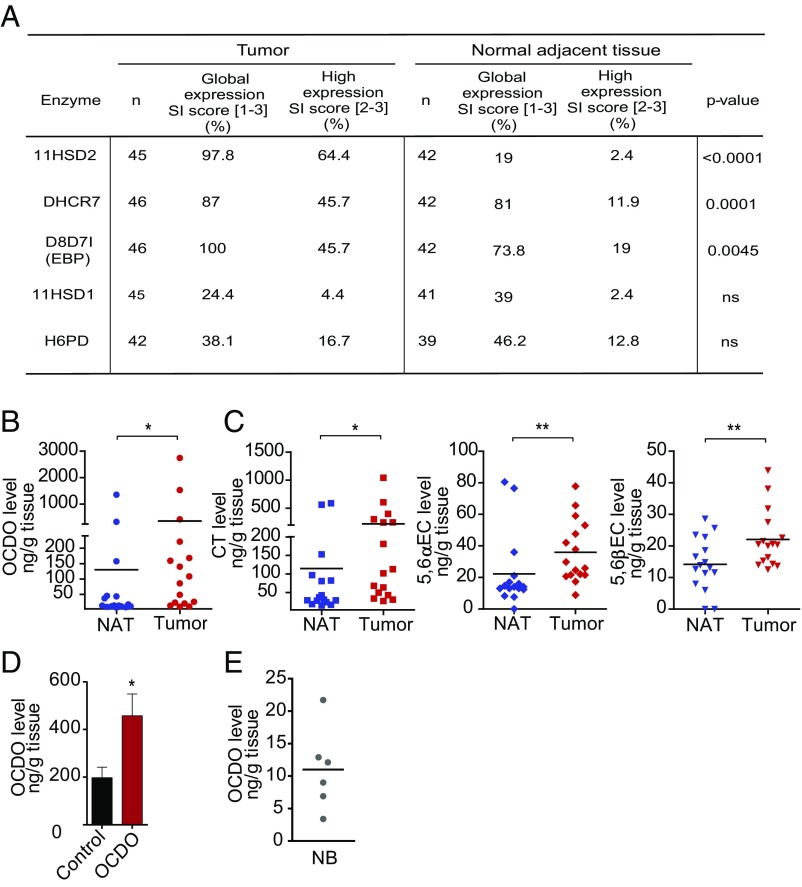
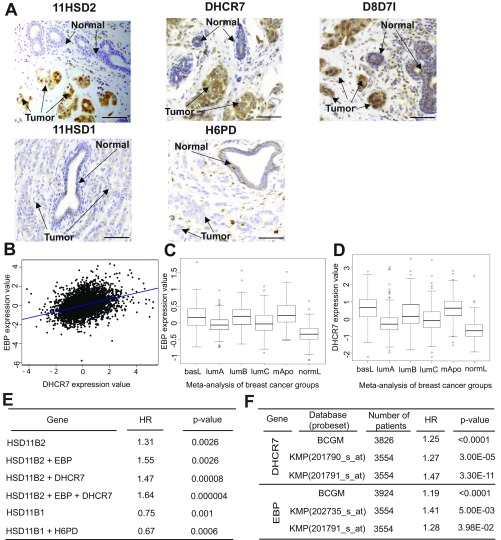
We then compared the expression of enzymes regulating OCDO production (namely 11HSD2, DHCR7, D8D71/EBP, 11HSD1, H6PD) in 50 patient BC samples with matched NAT, by immunohistochemistry (IHC). Patient characteristics are summarized in Table S2. As shown in Fig. 5A and Fig. S2A, 11HSD2 expression was significantly higher in BC than in matched NAT. In addition, a positive correlation was found between the expression of 11HSD2 in BC and grade III tumors (Table S3). In contrast, 11HSD1 and H6PD were weakly present in both BC and NAT samples, with no significant difference between their expression levels in the two tissues (Fig. 5A and Fig. S2A). D8D7I (EBP) and DHCR7, which together make up the ChEH, had a significantly higher expression in tumors compared with NAT (Fig. 5A and Fig. S2A) and their expression positively correlated with each other in tumors (r = 0.43; P = 0.004). The expression of the two ChEH subunits was also highly significantly correlated at the mRNA level (r = 0.37; P < 0.0001) (Fig. S2B). These data are in agreement with the fact that the two enzymes work together for an optimal ChEH activity (5). A positive correlation was also found between the expression of DHCR7 and grade III tumors. In addition, ER−/PR− patients had a significant increased proportion of tumors expressing high levels of DHCR7 and D8D7I. In contrast, a high level of 11HSD2 appeared independent of hormone receptor status (Table S3). At the mRNA level, both ChEH subunits were also expressed in the more aggressive molecular subtypes (i.e., basal, luminal B, and mApo) (15) (Fig. S2 C and D and Table S4). Taken together, these results indicate that the enzymes involved in the production of OCDO are more highly expressed in BC relative to NAT, while those involved in its conversion are weakly expressed.
Table S2.
Patient and tumor characteristics
| Clinicopathological parameters | Level | n (%) |
| Age at diagnostis | ≤60 y | 30 (56.6) |
| >60 y | 23 (43.4) | |
| Body mass index | ≤25 y | 31 (58.5) |
| >25 y | 22 (41.5) | |
| HR expression | ER+/PR+ | 31 (59.6) |
| ER+/PR− | 14 (26.9) | |
| ER−/PR− | 7 (13.5) | |
| Missing | 1 | |
| TN | No | 45 (86.5) |
| Yes | 7 (13.5) | |
| Missing | 1 | |
| HER2+ expression | No | 41 (80.4) |
| Yes | 10 (19.6) | |
| Missing | 2 | |
| Tumor stage | 0 | 2 (3.8) |
| I | 17 (32.7) | |
| II | 24 (46.2) | |
| III | 6 (11.5) | |
| IV | 3 (5.8) | |
| Missing | 1 | |
| Pathological nodal stage, pN | pN0 | 26 (49.1) |
| pN+ | 27 (50.9) | |
| Pathological tumor size, pT | ≤2 cm | 22 (44) |
| 2–5 cm | 23 (46) | |
| >5 cm | 5 (10) | |
| Missing | 3 |
HR, Hormone receptor; TN, triple negative (ER−/PR−/HER2−).
Fig. 5.
Expression levels of the enzymes regulating OCDO production and dosage of their metabolites in patient samples. (A) IHC analyses using specific antibodies against the enzymes of interest (Table S5). SI, Staining intensity score. Enzyme expression in BC and NAT was analyzed using the McNemar test for paired samples. (B and C) The indicated endogenous metabolites levels were quantified by GC/MS in matched patient tumors and NAT (n = 16). *P < 0.05, **P < 0.01, Wilcoxon test for paired samples, two-tailed. (D) OCDO level was quantified by GC/MS in TS/A tumors implanted into mice treated with 50 µg/kg OCDO for 19 d or treated with solvent vehicle (control) (n = 10 mice per group). Mean OCDO levels (±SEM), n = 10, are shown, *P < 0.05, Mann–Whitney test, two-tailed. Data are representative of three independent experiments. (E) Endogenous OCDO level was quantified by GC/MS in normal breast (NB) samples (n = 6). (B, C, and E) Each point represents the mean level of the metabolite of interest analyzed twice.
Fig. S2.
Expression of the enzymes regulating OCDO production and patient survival (related to Fig. 5A and Table S3). (A) Representative immunostaining images illustrating the expression of the indicated enzymes (in brown) in tumor breast samples. (Scale bars, 50 µm.) (B) Pearson’s pairwise correlation plot for EBP (D8D7I) vs. DHCR7 expression in 5,164 samples using the Breast Cancer Gene-Expression Miner v3 targeted correlation analysis module. Box-plots show the expression of (C) EBP (D8D7I) and (D) DHCR7 genes according to tumor molecular subtypes. This analysis was performed by the Carte d’Identité des Tumeurs from the Ligue contre le Cancer on a cohort of 726 tumors (dataset E-MTAB-365) organized in six subtypes (13). ANOVA tests were performed to calculate P values. DHCR7 and EBP levels were significantly different in the subtypes (P = 4.17e-48 and P = 1.02e-53, respectively). Tukey’s post hoc tests performed after one-way ANOVA (function Tukey’s HSD, stats R package) confirmed that the DHCR7 and EBP mean expression in basL, lumB, and mApo subtypes was significantly different from in normL and lumA subtypes (Table S4). (E) The BreastMark mining tool was used to calculate HR and P values, taking into account patient overall survival and median cut-off values for the genes indicated in the first column. (F) Correlation of DHCR7 and EBP (D8D7I) gene expression with survival using the Breast Cancer Gene-Expression Miner v3 targeted prognosis analysis module BCGM (breast cancer gene-expression miner) or the Kaplan–Meier Plotter online software (KMP). The probeset numbers are indicated, together with hazard ratios and logrank P values.
Table S3.
Correlation between the expression of the enzymes generating OCDO and the histological grade and subtypes of patient tumors
| Enzyme | Histological grade | HER2 and hormone receptor status | ||||||||||||
| I | II | III | P | HER2− | HER2+ | P | ER−/PR− | ER+/PR− | ER+/PR+ | P | TN | P | ||
| No | Yes | |||||||||||||
| 11HSD2 | n = 7 | n = 17 | n = 21 | n = 36 | n = 9 | n = 11 | n = 7 | n = 27 | n = 40 | n = 5 | ||||
| Low IRS (0–6) | 4/34 | 16/34 | 14/34 | 0.0417 | 30/34 | 4/34 | 0.0275 | 8/34 | 4/34 | 22/34 | ns | 30/34 | 4/34 | ns |
| n = 34, % | (11.8) | (47.1) | (41.2) | (88.2) | (11.8) | (23.5) | (11.8) | (64.7) | (88.2) | (11.8) | ||||
| High IRS (7–12) | 3/11 | 1/11 | 7/11 | 6/11 | 5/11 | 3/11 | 3/11 | 5/11 | 10/11 | 1/11 | ||||
| n = 11, % | (27.3) | (9.1) | (63.6) | (54.5) | (45.5) | (27.3) | (27.3) | (45.5) | (90.9) | (9.1) | ||||
| DHCR7 | n = 7 | n = 18 | n = 20 | n = 36 | n = 9 | n = 12 | n = 7 | n = 26 | n = 39 | n = 6 | ||||
| Low IRS (0–6) | 7/37 | 18/37 | 12/37 | 0.0021 | 35/37 | 2/37 | <0.0001 | 7/37 | 5/37 | 25/37 | 0.0064 | 32/37 | 5/37 | ns |
| n = 37, % | (18.9) | (48.6) | (32.4) | (94.6) | (5.4) | (18.9) | (13.5) | (67.6) | (86.5) | (13.5) | ||||
| High IRS (7–12) | 0/8 | 0/8 | 8/8 | 1/8 | 7/8 | 5/8 | 2/8 | 1/8 | 7/8 | 1/8 | ||||
| n = 8, % | (0) | (0) | (100) | (12.5) | (87.5) | (62.5) | (25) | (12.5) | (87.5) | (12.5) | ||||
| D8D7I | n = 7 | n = 19 | n = 20 | n = 36 | n = 10 | n = 12 | n = 7 | n = 27 | n = 41 | n = 5 | ||||
| Low IRS (0–6) | 7/38 | 17/38 | 14/38 | ns | 31 | 7 | ns | 6 | 7 | 25 | 0.0038 | 36 | 2 | 0.0307 |
| n = 38, % | (18.4) | (44.7) | (36.8) | (81.6) | (18.4) | (15.8) | (18.4) | (65.8) | (94.7) | (5.3) | ||||
| High IRS([7–12) | 0/8 | 2/8 | 6/8 | 5/8 | 3/8 | 6/8 | 0/8 | 2/8 | 5/8 | 3/8 | ||||
| n = 8, % | (0) | (25) | (75) | (62.5) | (37.5) | (75) | (0) | (25) | (62.5) | (37.5) | ||||
IHC analyses were scored as described in SI Experimental Procedures and Table S4. ER+ and PR+ tumors were defined by at least 10% of tumor cells being positive for these receptors by IHC analysis. HER2+ tumors were defined by at least 10% of tumor cells being positive for HER2 expression by IHC or a positive in situ hybridation assay. ns, not significant; TN, triple negative (ER−, PR−, and HER2−). Correlations between tumor sample immunoreactive score (IRS) and pathological characteristics were analyzed using χ2 test or Fisher’s exact test.
Table S4.
Statistical analysis of DHCR7 and EBP (D8D7I) in BC subtypes
| DHCR7 or EBP | basL | lumA | lumB | lumC | mApo |
| DHCR7 | |||||
| lumA | 0.01 | ||||
| lumB | 0.02 | <0.001 | |||
| lumC | <0.001 | 0.01 | 0.22 | ||
| mApo | 0.82 | <0.001 | 0.99 | 0.11 | |
| normL | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
| EBP | |||||
| lumA | 0.02 | ||||
| lumB | 0.51 | <0.001 | |||
| lumC | 0.58 | 0.61 | <0.001 | ||
| mApo | 0.94 | <0.001 | 0.99 | 0.11 | |
| normL | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
The DHCR7 and EBP (D8D7I) mean expression in the BC subtypes was compared using Tukey’s post hoc tests after one-way ANOVA (function TukeyHSD, stats R package).
Levels of OCDO and Its Precursors Are Higher in Patient BC Samples Compared with Normal Tissues.
We then quantified the levels of OCDO and its precursors in 16 paired patient BC and NAT by gas chromatography coupled to mass spectrometry (GC/MS). The levels of OCDO (Fig. 5B) and its precursors CT, 5,6α-EC, and 5,6β-EC (Fig. 5C) were significantly higher in patient tumors compared with NAT (P = 0.0245, 0.0162, 0.0121, and 0.0077, respectively; Wilcoxon test for paired samples, two-tailed), indicating that the increased expression of 11HSD2 and the ChEH subunits in BC favors the production of OCDO. To determine whether OCDO concentration used to treat mice was relevant to the pathological conditions, we measured OCDO levels in TS/A tumors treated or not with 50 µg/kg OCDO for 19 d (Fig. 3H) by GC/MS. The mean levels of OCDO measured in control tumors were of 197 ± 44 ng/g tissue (∼0.5 µM) and significantly increased to 458 ± 91 ng/g tissue in OCDO-treated tumors (∼1 µM) (Fig. 5D). These levels are within the pathological levels found in human tumors (mean: 357 ± 183 ng/g tissue, ∼0.85 µM) (Fig. 5B). We also measured the physiological levels of OCDO in six normal breast samples (Fig. 5E). The median level of OCDO measured in these normal samples was 10.6 ng/g tissue (range: 3.4–21.7; ∼25 nM) and was not significantly different from the median level of OCDO measured in NAT (Fig. 5B), 14.1 ng/g tissue (range: 7.5–1,350; P = 0.0768, Mann–Whitney test; ∼33 nM).
Expression of the Enzymes Producing OCDO in BC Correlates with Patient Survival.
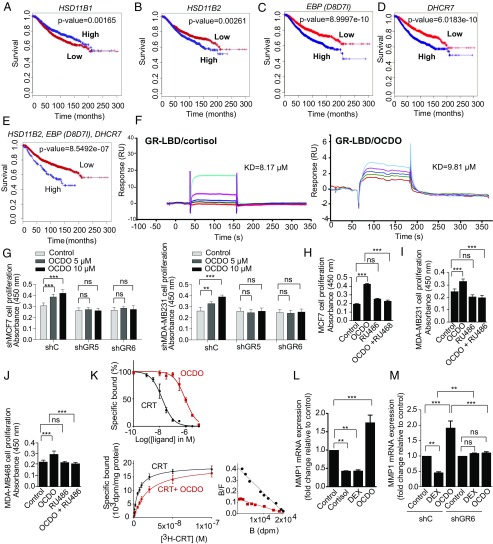
The BreastMark algorithm was used to perform Kaplan–Meier analysis on several datasets (16). Low levels of 11HSD1 (HSD11B1) mRNA and high levels of 11HSD2 (HSD11B2) mRNA were significantly associated with a poor prognosis (decreased overall survival rate) (Fig. 6 A and B). As shown in Fig. 6 C and D and Fig. S2E, high levels of EBP (D8D7I) and DHCR7 mRNA were also significantly associated with a lower survival rate of patients. Interestingly, the discrimination between the overall survival rates of patients using DHCR7 or EBP (D8D7I) mRNA levels was also found using other online databases, such as Breast Cancer Gene-Expression Miner and Kaplan–Meier Plotter (Fig. S2F). When the expression levels of 11HSD2, DHCR7, and EBP (D8D7I) were taken into account, the risk of death was the highest [hazard ratio (HR) = 1.849] (Fig. 6E). Altogether, these data demonstrate that a high expression level of the enzymes involved in the pathway producing OCDO correlates with lower patient overall survival rates.
Fig. 6.
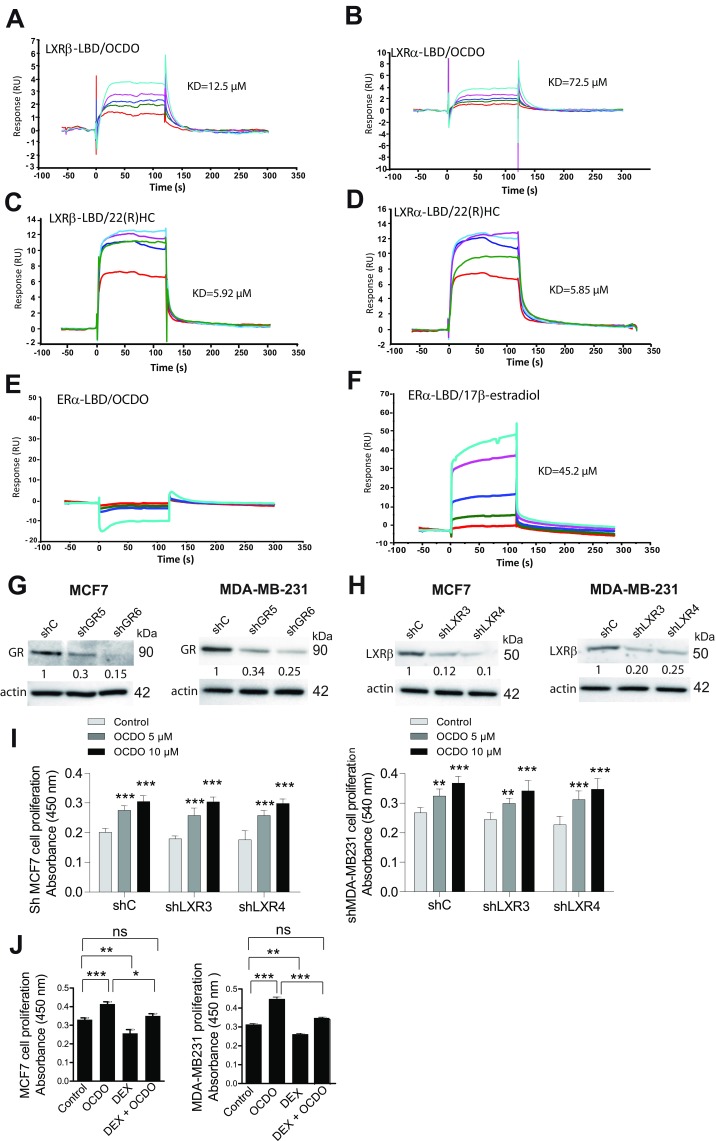
Expression of the enzymes regulating OCDO production and patient survival, and evidence that OCDO binds and stimulates cell proliferation through the GR and regulates GR transcriptional activity. (A–D) Kaplan–Meier representation of patient overall survival according to the indicated enzyme expression (median cut-off) using the BreastMark mining tool on 21 individual datasets (4,738 samples). Survival curves are based on Kaplan–Meier estimates and log-rank P values were calculated for differences in survival. Cox regression analysis was used to calculate HRs. (E) Kaplan–Meier representation of patient overall survival taking into account the expression of the HSD11B2, EBP (D8D7I), and DHCR7 genes using the BreastMark mining tool. (F) Representative SPR sensorgrams from three experiments showing the binding of a series of concentrations of cortisol or OCDO (µM) to the GR-LBD captured on a Biacore sensor chip: 6.25 (red); 12.5 (green); 25 (dark blue); 50 (pink); 100 (light blue). (G–J) Proliferation of the indicated tumor cells was analyzed as in Fig. 3C, n = 8. (H–J) The indicated tumor cells was treated with either the solvent vehicle (control), 5 µM OCDO, 1 µM RU486, or 5 µM OCDO plus 1 µM RU486. Data are the means (±SEM) of five separate experiments, n = 8, **P < 0.01, ***P < 0.001, one-way ANOVA, Tukey’s posttest. (K, Upper) Cell cytosols were incubated with 10 nM [3H]-CRT and increasing concentrations of unlabeled CRT or OCDO for competition binding assays. (K, Lower) Saturation and scatchard plots analyses were performed with cell cytosols incubated with increasing concentration of [3H]-CRT in the absence or in the presence of 1 µM unlabeled CRT (nonspecific binding) or 1 µM OCDO for competitivity studies. Data are the mean (±SEM) of triplicate and are representative of three experiments. (L and M) qRT-PCR analysis of MMP1 gene expression in MDA-MB231 (L) or shC and shGR MDA-MB231 (M) cells treated either with the solvent vehicle (control), 0.5 µM cortisol, 0.1 µM DEX or 5 µM OCDO. (L and M) Data are the means ± SEM of three experiments performed in triplicate, **P < 0.01, ***P < 0.001, one-way ANOVA, Tukey’s posttest. ns, not significant.
OCDO Binds to the GR, Liver X-Receptors, but Not ERα.
Since OCDO and cortisol production are regulated by the same enzymes, we tested whether OCDO can bind to the GR, the cortisol receptor. In addition, the oxysterol 27-hydroxycholesterol has been shown to act through the ERα and the liver-X-receptors (LXRs) (17, 18); therefore, we investigated whether OCDO interacts with these receptors. Surface plasmon resonance (SPR) assays indicated that OCDO binds to the ligand binding domain (LBD) of the GR, as observed with the positive control cortisol (Fig. 6F), and to the LBD of the LXR subtypes α and β (LXRα and LXRβ), as observed with the LXR ligand, 22(R)hydroxycholesterol [22(R)HC] (Fig. S3 A–D). In contrast, we did not detect any interaction between OCDO and the LBD of ERα, whereas 17β-estradiol binds to the ERα (Fig. S3 E–F). These data indicate that OCDO binds to the GR and the LXRs but not ERα.
Fig. S3.
Binding of the indicated ligands to ERα and LXRs, extinction of GR and LXRβ expression in the indicated BC cells and evidence that OCDO does not stimulate cell proliferation through the LXRβ. (A–F) Representative SPR sensorgrams from three experiments showing the binding of a series of concentrations of OCDO, 22(R)HC, or 17β-estradiol to the indicated receptor-LBD, captured on a Biacore sensor chip. Concentrations: 6.25 µM (red), 12.5 µM (green), 25 µM (dark blue), 50 µM (pink), 100 µM (light blue). Each sensorgram (expressed in RUs as a function of time in seconds) represents a differential response where the response on an empty reference channel (Fc1) was subtracted. The affinity constant (KD) is determined at equilibrium by the BIAevaluation software. (G and H) GR expression (G) or LXRβ expression (H) in MCF7 or MDA-MB-231 cells was knocked down using either shRNA against the GR (clones shGR5 and shGR6), or the LXRβ (clones shLXR3 and LXR4), or control shRNA (shC) and the expression of receptors was analyzed by immunoblotting. The blots are representative of three experiments. (I) Effect of solvent vehicle (control) or OCDO (5 or 10 µM) on the proliferation of the indicated tumor cells using a colorimetric BrDU immunoassay. (J) Effects of solvent vehicle (control) or OCDO (5 µM) or DEX 0.1 µM, or OCDO (5 µM) + DEX 0.1 µM on the proliferation of the indicated tumor cells using a colorimetric BrDU immunoassay. (I and J) Data are the means (±SEM) of five separate experiments, (n = 8), **P < 0.01, ***P < 0.001, one-way ANOVA, Tukey’s posttest. ns, not significant.
OCDO Mediates Tumor Cell Proliferation Through the GR and Regulates GR Transcriptional Activity.
To determine the involvement of GR and LXRs in OCDO-induced tumor cell proliferation, we knocked down the expression of GR or LXRβ, the only isoform present in MCF7 and MDA-MB231 cells (19), using shRNA. Two stable clones with significantly decreased GR and LXRβ expression (shGR or shLXR) compared with control clones (shC) were selected (Fig. S3 G and H). GR knockdown in MCF7 and MDA-MB231 cells abolished the cell proliferation induced by OCDO (Fig. 6G) and RU486 (Mifepristone), a GR antagonist, completely abolished OCDO-induced cell proliferation in MCF7, MDA-MB231, and MDA-MB468 tumor cell lines (Fig. 6 H–J). In contrast, the knockdown of LXRβ did not affect OCDO-mediated cell proliferation in either tumor cell lines (Fig. S3I). These data indicate that the proliferative effect of OCDO is dependent of the GR, but not of the LXRβ. We then tested whether the synthetic agonist dexamethasone (DEX) could inhibit tumor cell proliferation induced by OCDO. DEX inhibited basal and OCDO-induced MCF7 and MDA-MB231 cell proliferation at 1 µM concentration but had no effect at lower concentrations (Fig. S3J). To determine whether OCDO is or not a competitive inhibitor of cortisol on the GR, we performed competition binding experiments on lysates of GR-transfected cells using [3H]-CRT (Fig. 6K). OCDO fully inhibited [3H]-CRT binding to the GR in a concentration-dependent manner with an IC50 of 0.91 ± 0.12 µM and a Ki of 0.3 ± 0.07 µM (Fig. 6K, Upper). Saturation analyses showed that cortisol bound to the GR with a Kd of 5.9 ± 0.5 nM (Bmax of 0.92 ± 0.18 pmol/mg proteins). OCDO, at 1 µM, decreased the affinity of cortisol (Kd = 17.8 ± 3.1 nM) without changing the Bmax, indicating that OCDO is a competitive inhibitor of cortisol on the GR (Fig. 6K, Lower).
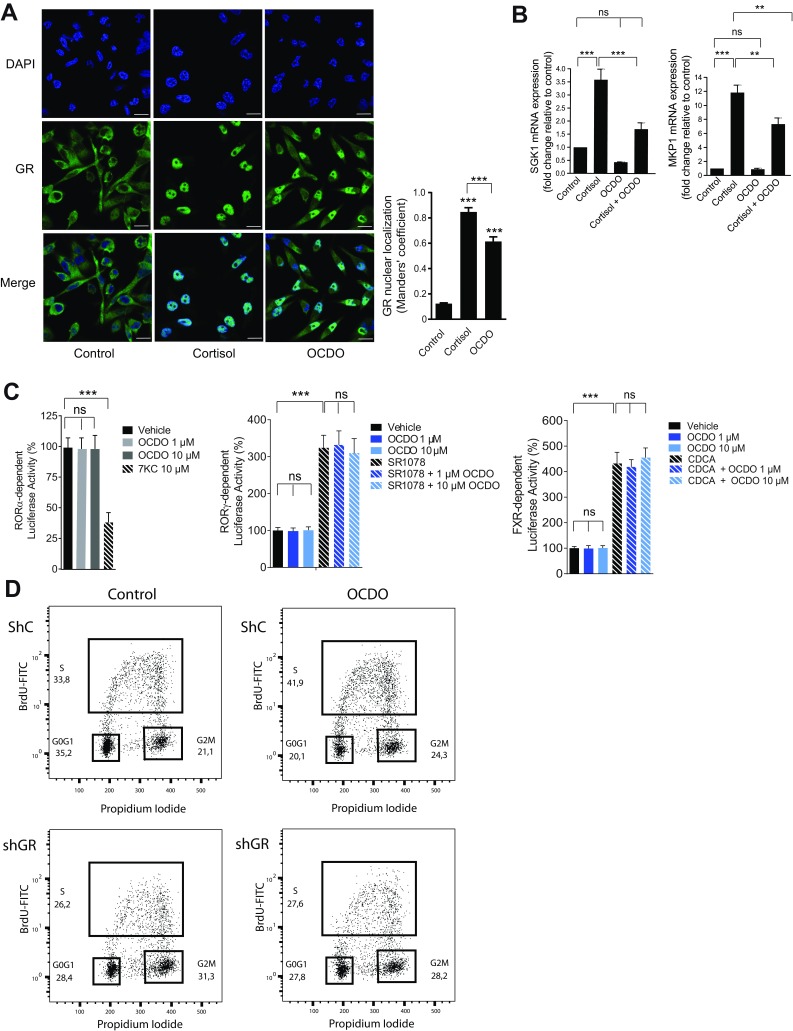
After binding to active glucocorticoids, GR translocates from the cytoplasm to the nucleus, where it positively or negatively regulates the expression of glucocorticoid-responsive genes via different mechanisms, including transactivation via the binding of liganded-GR to glucocorticoid-response elements (GREs) or repression via the binding of a negative GRE (nGRE) or transrepression via its interaction with other transcription factors, such as activator protein-1 or NFκB (20, 21). To determine whether, OCDO regulates GR transcriptional activity, OCDO was first tested for its ability to translocate GR into the nucleus in comparison with glucocorticoids (cortisol). As observed in Fig. S4A, in vehicle-treated MDA-MB231 cells, GR is mostly localized in the cytoplasm in an inactive form. Addition of either cortisol or OCDO to MDA-MB231 cells caused a significant eightfold and sixfold increase in GR nuclear localization. We next determined whether OCDO regulates the transcriptional activity of GR in MDA-MB231 cells by evaluating the transcription of canonical endogenous genes regulated by GR, such as SGK1 and MKP1, which are activated (22, 23), or MMP1 (collagenase), which is repressed by cortisol or dexamethasone (24). As shown in Fig. S4B, cortisol significantly increased SGK1 and MKP1 gene expression compared with vehicle-treated cells while OCDO had no significant effect on the transcription of these genes. When cells were treated with OCDO plus cortisol, SGK1 and MKP1 expression was significantly decreased compared with cortisol alone. In contrast, cortisol and DEX significantly inhibited the transcription of the MMP1 gene, while OCDO significantly stimulated its transcription (Fig. 6L). The induction of MMP1 gene transcription by OCDO through the GR was confirmed in control or GR-knockdown MDA-MB231 cells (ShC and shGR). The up-regulation of MMP1 mRNA by OCDO measured in shC cells was abolished in shGR cells and the inhibition of MMP1 mRNA by DEX in shC cells was reversed to basal in shGR cells (Fig. 6M). Taken together, these data indicate that OCDO regulates positively or negatively GR transcriptional activity by binding to the GR and inducing its nuclear localization. OCDO was next evaluated for its capacity to regulate the transcriptional activity of other nuclear receptors known to be regulated by oxidation products of cholesterol, such as the retinoid orphan-related receptors α and γ (RORα and RORγ) (25, 26) and the farnesoid X receptor (FXR) (27). OCDO did not regulate positively or negatively the transcriptional activity of these receptors as observed with their ligands (Fig. S4C).
Fig. S4.
OCDO causes GR nuclear localization, regulates GR transcriptional activity and induces cell cycle progression. (A) Representative immunofluorescence image of three different experiments of GR nuclear localization. MDA-MB231 cells were cultured in complete medium with dextran-coated charcoal-stripped FBS for 24 h and then treated for 24 h with either the solvent vehicle (control), 0.5 µM cortisol, or 5 µM OCDO and analyzed by indirect immunofluorescence with a confocal microscope (LSM 780; Zeiss) confocal microscope, 63×, using anti-GR (1:100) primary rabbit monoclonal antibody and a secondary rabbit Alexa fluor-488 secondary goat polyclonal antibody. The nuclei were stained with DAPI. Cells (100 per slide) were analyzed for nuclear GR localization using Jacop plugin in ImageJ v1.51. (Scale bars, 10 µm.) The experiments were performed in triplicate. (B) qRT-PCR analysis of SGK1 and MKP1 mRNA expression in MDA-MB231 tumor cells treated either with solvent vehicle (control), 0.5 µM cortisol, 5 µM OCDO, or 0.5 µM cortisol + 5 µM OCDO for 24 h. Data are the means ± SEM of three experiments performed in triplicate, **P < 0.01, ***P < 0.001, one-way ANOVA, Tukey’s posttest. (C) Effect of OCDO on the transcriptional modulatory activity of RORα, RORγ, and FXR. (Left) HEK293T cells were cotransfected with pSG5-RORα and a RORE-Luc plasmid and treated with or without 10 µM 7-ketocholestrol (7KC) or 1 or 10 µM OCDO for 24 h. (Center) Cells were transfected with pCMV-RORγ and a RORE-Luc plasmid and treated with the solvent-vehicle, 1 or 10 µM OCDO with or without 10 µM SR1078 for 24 h. (Right) Cells were transfected with pSG5-FXR, pSG5-RXR, and the tk-EcRE-Luc plasmid and incubated with either the solvent-vehicle, 1 or 10 µM OCDO with or without 10 µM chenodeoxycholic acid (CDCA). Luciferase activity was measured and normalized per microgram of proteins then expressed as percent activity relative to vehicle-treated cells and represented the mean of three independent experiments performed in triplicate (±SEM), ***P < 0.001, one-way ANOVA, Tukey’s posttest. ns, not significant. (D) Representative cell cycle analysis by flow cytometry of three independent experiments. ShC and shGR6 MDA-MB231 cells cultured for 48 h in serum-free medium were treated with the solvent vehicle (control) or 5 µM OCDO for 7 h. Cells were stained with BrdU and propidium iodide. The percentages of cells within the G0/G1, S, and G2/M phases of the cycle were calculated using FlowJo software. Numbers in the panels indicate the percentages of cells within G0/G1, S, and G2/M phases of the cell cycle.
We then analyzed the effects of OCDO on cell cycle progression in shC and ShGR MDA-MB231 cells. As shown in Fig. S4D, OCDO induced cell cycle progression by decreasing the percent of the G0/G1 phase and increasing the S and G2/M phases in shC cells. These effects were not observed in shGR cells treated with OCDO. These data confirm the mechanism of GR-dependent promotion of tumor cell proliferation by OCDO.
Discussion
Our study reveals that 5,6-EC can be metabolized into an oncometabolite, identified by MS as the oxysterol OCDO, and that it is found at significant greater levels in BC compared with normal tissues. We have shown that several enzymes are involved in the biosynthetic pathway that leads to OCDO production: ChEH, which is formed by D8D7I and DHCR7, and which mediates the transformation of 5,6-EC into CT (5, 6); and 11HSD2, which is involved in the final step in the transformation of CT into OCDO. We found that 11HSD2 controls BC cell proliferation both in vitro and in vivo through OCDO production. 11HSD2 is known to regulate glucocorticoid metabolism by converting active cortisol into inactive cortisone (12). In this study, we have shown that OCDO binds to the GR and LXRs but not to ERα. Moreover, OCDO stimulates the growth of BC cells and cell cycle progression irrespective of their ERα expression status by acting through the GR. Thus, the properties and mechanism of action of OCDO are distinct from another oxysterol, 27-hydroxycholesterol (27-HC), which increases the growth of ERα+ BC cells by interacting with ERα (17, 18). 27-HC also interacts with the LXRβ in BC cells to mediate tumor cell invasion, but not proliferation as observed with OCDO (17, 18). Therefore, the effect of OCDO on tumor cell invasion and the involvement of LXRβ in this event should be explored in the future. Competitive binding experiments indicate that OCDO inhibits cortisol binding to the GR with a Ki of 0.30 ± 0.07 µM and that it is a competitive inhibitor of cortisol. The concentration of OCDO in mouse xenografts or in patient tumors was estimated to be around 0.5 and 0.85 µM, respectively. These concentrations are close to the Ki value of OCDO for the GR.
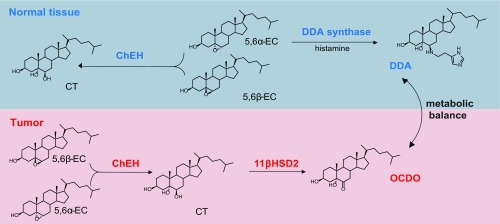
OCDO, like the agonist cortisol, causes the translocation of the GR from the cytoplasm to the nucleus to regulate positively or negatively GR target gene transcription (20, 21). OCDO does not stimulate the transcription of SGK1 or MKP1/DUSP1 genes, as observed with cortisol, while OCDO inhibits the induction of the transcription of these genes by cortisol. In addition, OCDO increases MMP1 gene expression by acting through the GR, while cortisol or DEX represses MMP1 gene transcription. We have found that DEX inhibits the basal and OCDO-induced MCF7 and MDA-MB231 cell proliferation, but only at 1 µM, consistent with data from the literature showing that DEX inhibits tumor cell proliferation at high concentrations (28). Combined, these data suggest that OCDO and glucocorticoids, while acting through the GR, have distinct mechanisms of action on GR transcription and cell proliferation. In a nonexhaustive way, this could be explained by the recruitment of different cofactors, which will determine GR activation or repression activity, or by posttranslational modifications of the GR, such as phosphorylation, since ligand-selective GR phosphorylation has been shown to correlate with levels of GR transcriptional activity (29). Interestingly, a recent study comparing the pharmacodynamics of different GR ligands in activating gene expression showed that ligands elicit differential activation of distinct genes, where ligand-intrinsic efficacy and GR density were essential determinants (30). Thus, additional work will be required to elucidate the molecular mechanisms underlying the proliferative effect of OCDO mediated by the GR and the profile of the genes involved. The effects of OCDO are opposite to those of DDA, which inhibits BC progression (3, 31). Both compounds arise from 5,6-ECs; however, only 5,6α-EC generates DDA (3), while both 5,6α-EC and 5,6β-EC produce OCDO (Fig. S5). We have previously reported that DDA levels are decreased in human BC samples relative to normal tissues (3), whereas here we show that OCDO levels are increased in human BC samples compared with normal tissues. This indicates the existence of a metabolic balance between these two 5,6-EC derivatives in normal breast and BC that may either control or stimulate BC progression (Fig. S5). Interestingly, DDA inhibits the production of OCDO in BC cells through its action on ChEH, independently of ERα expression, and the inhibition of OCDO production contributes to the efficacy of DDA in vivo, indicating that the targeting of this oncometabolism could be relevant for BC treatment, and that treatment with DDA may compensate for its deficiency in BC. In addition, other ChEH inibitors, such as Tam or raloxifene (5), known to inhibit ERα and used as antiestrogen therapy in BC, block ChEH activity and OCDO production in ER+ MCF7 and ER− MDA-MB231 cells, indicating that these molecules target the OCDO pathway independently of ERα expression. We, and other laboratories, have previously reported that Tam inhibits the proliferation of MDA-MB231 cells and other ER− BC cell lines (2, 32–34). These data suggest that the inhibition of the OCDO pathway may contribute to the antiproliferative effect of Tam in these ER− BC cells, and thus that this molecule may be useful in the treatment of certain ER− BC. This opens new avenues of research that deserve further study.
Fig. S5.
Scheme of the metabolic balance between OCDO and DDA.
OCDO was previously described as a derivative of 5,6β-EC when it was found in the lungs of mice exposed to extreme conditions of oxidation, such as ozone exposure, but not under normal conditions (35). Consistent with our data, Pulfer et al. postulated the existence of an enzymatic mechanism to produce OCDO, since its formation requires living cells (35). 5,6-ECs were reported to be produced from cholesterol either by an enzyme not yet identified or via the autoxidation and lipoperoxidation of cholesterol to yield a mixture of 5,6-ECs, but with predominantly 5,6β-EC (1, 2). Since OCDO is a derivative of these compounds, conditions that generate oxidative stress and lipid peroxidation, such as chronic inflammatory processes, may therefore lead to OCDO production and be detrimental to DDA biosynthesis. Taking into account the fact that OCDO binds to the GR, its impact on inflammation deserves further study.
In the present study, IHC analyses indicated that the expression of all of the enzymes involved in the pathway producing OCDO, namely 11HSD2, D8D7I (EBP), and DHCR7, is higher in BC compared with NAT, while the enzymes that convert OCDO into CT (11HSD1 and H6PD) are weakly expressed in BC and NAT. Thus, the expression of these enzymes is consistent with both a higher production of OCDO in BC relative to NAT and with OCDO having tumor promoting properties. Accordingly, the analysis of several human BC mRNA databases also indicated that a high 11HSD2 expression or a low 11HSD1 expression correlates with a bad prognosis in BC patients. In addition, the elevated expression of DHCR7 and/or D8D7I (EBP) was associated with a bad survival outcome, and the risk of death was the highest when the expression levels of 11HSD2, DHCR7, and D8D7I (EBP) were taken into account, highlighting that the biosynthetic pathway producing OCDO is of major importance to BC pathology. Moreover, different studies revealed that high GR expression or activation correlated with poor therapeutic response or prognosis in ERα− breast cancers, as well as in many other solid tumors (36–40). Glucocorticoids exert complex and opposite effects and can enhance chemotherapy sensitivity by inhibiting cell proliferation or chemotherapeutic resistance by inducing cell survival and resistance to apoptosis (28). Interestingly, treatment with the GR antagonist, mifepristone (RU486), potentiates the antitumor efficacy of chemotherapy in ER− BC mouse models (38). In ERα+ breast cancer, GR expression has been associated with good clinical outcomes (37). The fact that glucocorticoids antagonize E2-induced gene expression in BC cells (41) and up-regulate the enzyme involves in estrogen inactivation, may explain the better outcome in ER+ BC patients expressing a high level of GR (42). Recently, liganded-GR was shown to repress a large ERα-activated transcriptional program by binding in trans to ERα-occupied enhancers. This event was associated with poorer metastasis-free outcomes in BC patients (43). These data highlight the important role of GR in BC and other tumors.
In conclusion, the discovery of OCDO, its proliferative effects through the GR, and the identification of the enzymes inducing or regulating its production are important findings, which should have major implications in the biology and diagnosis of BC and in the development of new therapeutic approaches. The targeting of the enzymes involved in cholesterol epoxide and glucocorticoid metabolism, as well as of the receptor mediating the proliferative effects of OCDO, represent new opportunities for therapeutic intervention in different BC subtypes, particularly in case of resistance to conventional therapies.
Experimental Procedures
For materials, animals, patient samples, and other techniques, see SI Experimental Procedures. All animal procedures concerning the care and use of laboratory animals were conducted according to the ethical guidelines of the Claudius Regaud Institute and followed the general regulations governing animal experimentation. All human samples were collected with the approval of the Institutional Review Board of the Claudius Regaud Institute, Toulouse, France and Tumor Bank Committee of the Claudius Regaud Institute, Institut Universitaire du Cancer Toulouse, France. Normal breast tissue was obtained from six patients who underwent mammoplasty and written informed consent was obtained before inclusion into the study. Patients’ clinical characteristics and tumor pathological features were obtained from their medical reports and followed the standard procedures adopted by the Claudius Regaud Institute.
Metabolism of Sterols in BC Cells.
Cells were plated into six-well plates (1 × 105 cells per well) in the appropriate complete medium. One day after seeding, this medium was replaced with complete medium without Phenol red supplemented with dextran-coated charcoal-stripped FBS, and cells were treated with either 0.6 µM [14C]5,6α-EC for 72 h, 0.6 µM [14C]5,6β-EC for 72 h, 1 µM [14C]CT for 8 h, 1 µM [14C]OCDO for 72 h, 0.2 µM [3H]-CRT for 8 h, or 0.2 µM [3H]cortisone for 72 h. After incubation, cells were washed and scraped, and neutral lipids were extracted with a chloroform-methanol mixture as described in Segala et al. (2), and then separated by TLC using either ethyl acetate as the eluent for [14C]CT and [14C]OCDO (9) or chloroform-methanol [87:13 (vol/vol)] for [3H]-CRT or [3H]cortisone in a method adapted from ref. 44. The radioactive sterols were revealed by autoradiography. For quantification, silica zones at the expected Rf values corresponding to authentic [14C]- or [3H]-labeled standards were scraped and radioactivity was measured using a β-counter, as previously described (5).
Structural Characterization of the Unknown 5,6-EC Metabolite.
MCF-7 cells (8 × 105 cells per dish) were treated with 10 µM 5,6α-EC. After a 72 h treatment, cells were washed twice with 5 mL PBS, then scraped and resuspended in 5 mL PBS and pelleted by centrifugation (800 × g, 5 min, 4 °C). Cells were then prepared for oxysterol analyses as previously described (2). Oxysterols were separated by RP-HPLC isocratically [MeOH:H2O] (95/5) at a flow rate of 0.7 mL/min. CT and OCDO had retention times of 10 and 17 min, respectively, as previously described (9). The 9- to 11- and 15- to 17-min RP-HPLC fractions were dried under vacuum and submitted to chemical ionization-MS (CI-MS) analysis using ammonia as a reagent gas on a Perkin-Elmer SCIEX API 100 spectrometer.
GC/MS Quantification of Oxysterols.
Tissues, tumors and cell homogenates were extracted as previously described (3). In a separate experiment, 0.1 µCi [14C]OCDO was added to determine the yield of OCDO extraction. The samples were then dried under argon, dissolved in 1 mL toluene and passed through a silica cartridge (ISOLUTE SI 100 mg SPE Columns; Biotage). Oxysterol levels were determined by high-performance GC/MS detection using deuterium-labeled internal standards exactly as described in Iuliano et al. (45). The percentage of recovery of [14C]OCDO was 96 ± 8%. The estimation of OCDO concentration in tumor xenografts or in patient tissues was calculated by assuming that 1 g of tissue corresponds to a volume of 1 mL (3).
SI Experimental Procedures
Materials.
[1,2,4,6-3H(N)]-cortisol ([3H]cortisol, specific activity: 82 ci/mmol), [1,2-3H(N)]-cortisone (specific activity: 60 ci/mmol) and [4-14C]Cholesterol (50.8 mCi/mmol) were purchased from Perkin-Elmer. The radiochemical purity of the compounds was verified by TLC and was greater than 98%. [25,26,26,26,27,27,27-d7] cholesterol (99% D) and RU486 (Mifepristone) were from Sigma-Aldrich. CT and OCDO were from Steraloids. Raloxifene was a generous gift from Patrick Van de Velde, Sanofi-Avantis, Paris. HPLC-MS grade acetonitrile was from Pierce. HPLC purifications were carried out using a Gilson HPLC or a LC200 Perkin-Elmer apparatus, equipped with a diode array UV or β-radioactivity detector, and an Ultrasep C18 RP 100 column (Bischoff). Autoradiography experiments were performed with GE Healthcare or Kodak phosphor screens. The NEON Transfection system was purchased from Invitrogen, the BrdU cell proliferation Elisa kit was from Roche Diagnostics or Sigma, and the following plasmids were from OriGene: 11HSD1: sc109325; 11HSD2: sc122552 or RG207796; H6PD: sc117481; DHCR7: sc110871; and D8D7I (EBP): sc116006. pCMV-RORγ plasmid (NBP2-25278) was from Novus biologicals; pSG5-hGR was a gift from H. Richard-Foy, CNRS, Toulouse, France; pSG5-hRORα was a gift from V. Laudet, CNRS, Banyuls-sur-Mer, France; RORE-Luc was from B. Staels, the Pasteur Institute, Lille, France; pSG5-hRXRγ was a gift from M. Oulad-Abdelghani, INSERM, Strasbourg, France; and pSG5-hFXR and tk-EcRE-Luc were gifts from R. M. Evans, The Salk Institute, La Jolla, CA. Other compounds and chemicals, unless specified, were from Sigma-Aldrich, and solvents were from VWR. For all enzymes studied, the same antibodies were used for immunoblotting and IHC analyses and are listed in Table S5. The antibodies against LXRβ and the GR used in immunoblotting (1/1,000) or GR nuclear localization (1/100) were from R&D (PP-K8917-00, clone K8917) and Cell Signaling(12041, clone D6H2L), respectively. The FITC-conjugated BrdU antibody used for cell cycle analysis (1/40) was from BioLegend (364104, clone 3D4).
Table S5.
Antibodies and methods used for IHC analyses
| Antigen | Clone | Dilution | Antigen retrieval | Source | Scoring system | Cut-off |
| ER | M3643, Clone EP1 | RTU | PTM, target retrieval | DAKO | Nuclei staining, % | Positive: >10% |
| Solution, pH high, DAKO | ||||||
| PR | M3569, Clone PgR636 | RTU | PTM, target retrieval | DAKO | Nuclei staining, % | Positive: >10% |
| Solution, pH high, DAKO | ||||||
| Ki67 | M7240, Clone MIB1 | RTU | PTM, target retrieval | DAKO | Nuclei staining, % | <10%: Low |
| Solution, pH high, DAKO | 10–30%: Intermediate | |||||
| >30%: High | ||||||
| HER2 (Hercept test ) | SK001 Polyclonal | RTU | PTM, target retrieval | DAKO | Score 0, 1+, 2+, 3+ following the Hercept test kit scoring system updated according to ASCO/CAP guidelines | Score 0/1: Negative |
| Solution, pH low, DAKO | Score 2+: equivocal | |||||
| Score 3+: Positive | ||||||
| DHCR7 | ab170388 Polyclonal | 1:50 | PTM, target retrieval, solution pH low, DAKO | Abcam | Percentage of cytoplasmic staining × intensity (IRS) | Low or negative IRS: (0–6) |
| High IRS: (7–12) | ||||||
| H6PD | sc-377180, clone C-10 | 1:100 | PTM, target retrieval, solution pH low, DAKO | Santa Cruz, Biotechnologies | Percentage of cytoplasmic staining × intensity (IRS) | Low or negative IRS: (0–6) |
| High IRS: (7–12) | ||||||
| D8D7I (EBP) | AP9004a Polyclonal | 1:500 | PTM, target retrieval, solution pH high, DAKO | Abgent | Percentage of cytoplasmic staining × intensity (IRS) | Low or negative IRS: (0–6) |
| High IRS: (7–12) | ||||||
| 11HSD1 | ab157223, clone EPR9407(2) | 1:50 | PTM, target retrieval, solution pH high, DAKO | Abcam | Percentage of cytoplasmic staining × intensity (IRS) | Low or negative IRS: (0–6) |
| High IRS: (7–12) | ||||||
| 11HSD2 | sc-20176 Polyclonal | 1:50 | — | Santa Cruz, Biotechnologies | Percentage of cytoplasmic staining × intensity (IRS) | Low or negative IRS: (0–6) |
| High IRS: (7–12) |
Description of the antibodies, clones, dilutions, antigen retrieval methods, scoring systems, and cut-offs used in the IHC analyses. ASCO/CAP, American Society of Clinical Oncology/College of American Pathologists; PTM, pretreatment module; RTU, ready to use. See also SI Experimental Procedures.
Animals.
Six-week-old females: C57BL/6 (Charles River Laboratories), BALB/c, and NMRI nude mice (Janvier) were maintained in specific pathogen-free conditions and were included in protocols following a 2-wk quarantine. All animal procedures concerning the care and use of laboratory animals were conducted according to the ethical guidelines of the Claudius Regaud Institute and followed the general regulations governing animal experimentation.
Patient Samples.
All human samples were collected with the approval of the Institutional Review Board of the Institut Claudius Regaud, Toulouse, France, and Tumor Bank Committee of the Claudius Regaud Institute, Institut Universitaire du Cancer Toulouse, France. Normal breast tissue was obtained from six patients who underwent mammoplasty. Written informed consent was obtained before inclusion into the study. Patients’ clinical characteristics and tumor pathological features were obtained from their medical reports and followed the standard procedures adopted by the Claudius Regaud Institute.
Analysis of Tumors in Mice.
MCF7, ShMCF7, E0771, MDA-MB231, MDA-MB468, and TS/A cells growing exponentially were collected, washed twice in PBS, and resuspended in PBS. TS/A and E0771 tumors were prepared by subcutaneous transplantation of 35 × 103 cells or 3 × 105 cells, respectively, in 100 μL PBS into the flank of BALB/c or C57Bl6 mice, respectively. For other tumors, 5 × 106 cells were injected into the flank of NMRI nude mice. Animals were treated subcutaneously once per day, 5 d/wk, with the solvent vehicle or OCDO (16 µg/kg for MCF7, MDA-MB231, and MDA-MB468 or 50 µg/kg for TS/A and E0771) or with DDA, as indicated in the figure legends. Each mouse implanted with MCF7 cells was treated with 20 µL estradiol (10 mM in ethanol), injected three times per week on the scruff of the neck and withdrawn at the time of OCDO treatment. Animals were examined daily, and body weights were measured twice per week. In all experiments, tumor volume was determined by direct measurement with a caliper, and was calculated using the formula (width2 × length)/2. Tumors were fixed in 10% neutral-buffered formalin and embedded in paraffin for histomorphological and IHC analyses. Paraffin sections were stained with H&E for histomorphological analysis. Immunostaining of paraffin sections was preceded by an antigen retrieval technique by heating twice for 10 min with a microwave oven in 10 mM citrate buffer, pH 6. After incubation with a specific rabbit monoclonal Ki67 antibody (clone SP6; Spring Bioscience), 1/200, for 1 h at room temperature, sections were incubated with biotin-conjugated polyclonal anti-rabbit Ig antibodies followed by the streptavidin–biotin–peroxidase complex (VectorLaboratories) and were then counter-stained with hematoxylin. Negative controls were incubated in buffered solution without primary antibodies.
Cell Culture.
MCF7, SKBR3, MDA-MB231, MDA-MB468, CV-1, and HEK293T cells were from the American Type Culture Collection (ATCC), E0771 cells were from Tebu-bio and cultured until passage 30. TS/A cells were kindly provided by P. L. Lollini, University of Bologna, Bologna, Italy. Cell lines were tested once a month for mycoplasma contamination using Mycoalert Detection (Lonza). MCF7 cells were grown in RPMI medium 1640 (Lonza) supplemented with 5% FBS (Dutcher), SKBR3 cells were grown in McCoy’s medium (Invitrogen) with 10% FBS, TSA, and MDA-MB468 cells were grown in RPMI with 10% SVF, E0771 were grown in RPMI with 10% FBS and 10 mM Hepes and HEK293T, and MDA-MB231 and CV-1 cells were grown in DMEM (Lonza) with 10% FBS. All cell media contained 2 mM l-glutamine. All cells lines were cultured in a humidified atmosphere with 5% CO2 at 37 °C.
Cell Transfection.
For cell transfection, 5 × 106 MCF7, MDA-MB231, or HEK293T cells were transfected with 5 µg of the indicated plasmid using the NEON Transfection System according to the manufacturer’s recommendations. MCF7 or MDA-MB231 cells were transfected separately with either four different shRNA plasmids targeting the gene encoding the protein of interest (11HSD2 or GR) or with a control scramble shRNA (SureSilencing ShRNA plasmid; Qiagen). Clone selection was performed in the presence of 0.5 mg/mL puromycin (Life Technologies). Several clones were analyzed using immunoblot analysis or real-time qRT-PCR and different shRNAs providing a better extinction of the expression of the protein of interest were selected, as well as control clones, to establish stable clones. MCF7 cells were transfected as described above with a plasmid encoding 11HSD2 (RG207796; OriGene) or the empty plasmid. Stable clones were obtained after growing the cells for 3 wk in the presence of 0.5 mg/mL puromycin and 11HSD2 expression was analyzed by immunoblotting. Two clones overexpressing 11HSD2 and two control clones were selected.
Immunoblotting.
Cells treated as indicated were washed with ice-cold PBS, then scraped and centrifuged at 800 × g for 5 min at 4 °C. Pellets were resuspended in 100 µL of extraction buffer (50 mM Tris pH 7.4, 5 mM NaCl, 1% Triton X-100, 10% glycerol) with 1% protease inhibitor mixture (Sigma-Aldrich), then vortexed and centrifuged at 10,000 × g for 10 min at 4 °C. Whole-cell extracts were fractionated by SDS/PAGE and transferred to a polyvinylidene difluoride membrane using a transfer apparatus according to the manufacturer’s protocols (Life Technologies). After incubation with 5% nonfat milk in TBST (10 mM Tris, pH 8.0, 150 mM NaCl, 1% Tween 20) for 60 min, the membrane was incubated with antibodies against 11HSD2 (1:1,000), 11HSD1 (1:500), H6PD (1:500), D8D7I (EBP) (1: 500), DHCR7 (1: 200), or actin (1/20,000, C4; Merck Millipore) at 4 °C overnight. Membranes were then washed three times for 10 min each and incubated either with a 1:10,000 dilution of horseradish peroxidase-conjugated anti-mouse (W402B; Promega) or anti-rabbit antibodies (W401B; Promega) for 1 h. Blots were then washed three times with TBST and developed with the ECL system (Amersham Biosciences), according to the manufacturer’s protocol.
Chemical Synthesis.
For chemical synthesis, 5,6α-EC, 5,6β-EC, [14C]5,6α-EC, and [14C]5,6β-EC were synthesized as previously reported (5). [25,26,26,26,27,27,27-d7]5,6α-EC, [25,26,26,26,27,27,27-d7]5,6β-EC, were prepared as previously described (2). [25,26,26,26,27,27,27-d7]-CT, [25,26,26,26,27,27,27-d7]OCDO, [14C]CT and [14C]OCDO were prepared as previously described (9). DDA, PBPE, and tesmilifene were synthesized as previously described and were 99% pure (10, 31).
Cell Proliferation Assays.
Cells (5 × 103) cells were seeded in 96-well plates in the appropriate complete media. One day after seeding, the medium was changed for complete medium without Phenol red supplemented with dextran-coated charcoal-stripped FBS. Cells were treated for 24 h with the indicated concentration of OCDO, cortisol, or cortisone. At the end of this time, cells were incubated with BrDU for an additional 8 h and then evaluated for proliferation using an ELISA kit (Roche Diagnostics), according to the manufacturer’s instructions.
Clonogenic Assays.
Cells (5 × 103) were seeded in duplicate in 35-cm2-diameter dishes. Twenty-four hours later, cells were treated with either 1 µM OCDO or solvent vehicle and this treatment was repeated every 3 d. At day 10, colonies were fixed with 3.7% PFA, stained with a 0.05% aqueous Crystal violet solution, and the number of colonies was counted.
RNA Isolation and qPCR Analysis.
Total RNA from cultured cells treated as indicated or not was isolated using TRIzol Reagent (Invitrogen). RNA was quantified (nanodrop; Thermofisher). Total RNA (1 μg) was reverse-transcribed using an iScript cDNA synthesis kit (Bio-Rad), according to the manufacturer’s instructions. qRT-PCR was performed with an iCycler iQreal-time PCR detection system (Bio-Rad) using iQ SYBR Green Supermix (Bio-Rad) and the indicated primers. For 11βHSD1 and 11βHSD2 expression, the threshold cycle (Ct) values of the gene of interest were normalized with the Ct values of Cyclophiline A1. Primers used: cyclophiline A1: F: GCA-TAC-GGG-TCC-TGG-CAT-CTT-GTC-C; R: ATG-GTG-ATC-TTC-TTG-CTG-GTC-TTG-C. 11βHSD1: F: GA-CAG-CGA-GGT-CAA-AAG-AAA; R: GTC-CTC-CCA-TGA-GCT-TTC-CTG. 11βHSD2: F: CCA-CCG-TAT-TGG-AGT-TGA-ACA; R: CGC-GGC-TAA-TGT-CTC-CTG-G. For SGK1, MKP1, and MMP1 expression study, MDA-MB231 cells seeded at a density of 750,000 cells per 100-mm dishes were cultured for 24 h in the appropriate complete medium. After 24 h, the cell medium was changed for serum-free medium for MMP1 expression study or complete medium supplemented with dextran-coated charcoal-stripped FBS for SGK1 or MKP1 expression study. After 24 h, cells were treated for 3 h (MMP1) or 24 h (SGK1 and MKP1) with the indicated compounds. RNA expression of the different genes of interest were determined using the ΔΔCT method, where the difference in the threshold cycle (ΔCT) values of the target gene and the housekeeping genes (36B4 or YWAZ) for each sample was normalized to the ΔCT value of the untreated sample. Primers used: SGK1: F: GGACTACATTAATGGTGGAGAGC; R: AGAATCGAGCCCGTGGTT; MKP1/DUSP1: F: CCTGACAGCGCGGAATCT; R: GATTTCCACCGGGCCAC; YWHAZ: F: ACTTTTGGTACATTGTGGCTTCAA; R: CCGCCAGGACAAACCAGTAT; 36B4: F: GATTGGCTACCCAACTGTTG; R: CAGGGGCAGCAGCCACAAA.
IHC.
IHC analysis of patient samples was performed on formalin-fixed, paraffin-embedded sections of the initial tumor biopsies with the indicated antibodies. The same antibodies were used for both the immunoblotting and the IHC assays (Table S5). Their specificities were tested by IHC analysis of different normal human tissues known to express or not the enzyme of interest. In addition, they were tested on HEK293 cells transfected with a plasmid encoding the protein of interest (either DHCR7, H6PD, D8D6I, 11HSD1, or 11HSD2) or the empty vector (negative control) by immunoblotting or by IHC of paraffin-embedded transfected-HEK293 cells. For these assays, HEK293 cells transfected with the enzymes of interest or the empty vector were first pelleted by centrifugation, then washed with PBS and fixed in 4% formalin, embedded in paraffin, and processed in the same manner as patient material. Immunostaining was blindly scored using a multihead microscope by four people (M.P., S.S.-P., M.V. and M.L.-T.), one of whom is an experienced breast pathologist (M.L.-T.). For each marker, the staining intensity was scored as either absent (0), weak (1+), moderate (2+), or intense (3+). The percentage of stained tumor cells, as well as the localization of staining (cytoplasmic, nuclear…), was assessed for each biomarker. The immunoreactive score (ranging from 0 to 12) was calculated by multiplying the staining intensity (0, 1, 2, 3) with the percentage of stained cells scored as follows: <1% stained cells = 0; 1–10% stained cells = 1; 11–50% stained cells = 2; 51–80% stained cells = 3; >80% stained cells = 4.
SPR Assays.
All binding studies based on SPR technology were performed on a BIAcore T200 optical biosensor instrument (GE Healthcare). Immobilization of the GST-tagged LBD of human GR (A15668), human LXRβ (PV4660), LXRα (PV4657), or ERα (15677) (from Thermo Fisher Scientific) was performed by covalent coupling of the indicated ligands to the chip surface using amine coupling (CM5) sensor chips in PBS-P+ buffer (20 mM phosphate buffer, pH 7.4, 2.7 mM KCl, 137 mM NaCl, and 0.05% surfactant P20; GE Healthcare). All immobilization steps were performed at a flow rate of 10 µL/min, with a final concentration of 20 µg/mL. The total amount of immobilization was 11,000–12,000 RU. Channel Fc1 was used as a reference surface for nonspecific binding measurements. A low mass weight–multiple-cycle kinetics analysis to determine affinity constants was carried out by injecting different protein concentrations (6.25 µM–100 µM). Each sensorgram (expressed in RUs as a function of time in seconds) represents a differential response where the response on an empty reference channel (Fc1) was subtracted. Binding parameters were obtained by fitting the overlaid sensorgrams with the 1:1 Langmuir binding model of the Biacore T200 Evaluation software v2.0 or the Steady State affinity model of the BIAevaluation software v3.1.
Gene Reporter Luciferase Assays.
HEK293T cells were transiently cotransfected using the polyethylenimine method with the indicated plasmids. For the human FXR reporter assay, cells were cotransfected with pSG5-FXR, pSG5-RXRγ and the reporter gene for FXR: tk-EcRE-Luc. The day after transfection, cells were seeded in 12-well plates (50,000 cells per well) in complete medium with dextran-coated charcoal-stripped FBS. After 24 h, cells were treated with either solvent-vehicle, 1 or 10 µM OCDO with or without 10 µM chenodeoxycholic acid (CDCA, anFXR agonist) (46). For the human RORα reporter assay, cells were cotransfected with pSG5-RORα and RORE-Luc. The day after transfection, cells were seeded in 12-well plates (50,000 cells per well) in complete medium with dextran-coated charcoal-stripped FBS. For RORα, cells were incubated with or without 10 µM 7-ketocholesterol (7KC, a RORα inverse agonist) (25) or 1 or 10 µM OCDO. For the human RORγ reporter assay, cells were cotransfected with pCMV-RORγ and RORE-Luc. Cells were treated with solvent-vehicle, 1 or 10 µM OCDO with or without 5 µM SR1078 (a RORγ agonist) (47). At the end of each treatment, cells were lysed in 100 μL lysis buffer (Promega). A pCMV-lacZ galactosidase expression plasmid was cotransfected for internal controls. Protein concentrations were measured using the Bradford method to normalize luciferase activities. For each condition, luciferase activity was calculated from the data of three independent wells.
Binding Assay.
CV-1 cells were transfected with 15 µg pSG5-hGR plasmid using the lipofectamine methodology according to the manufacturer’s instructions (Life Technologies). Briefly, transfected cells were grown to 80% confluency in complete medium, scraped, and washed twice with PBS. After centrifugation for 10 min at 120 × g, the cells were resuspended in TM buffer (20 mM Tris⋅HCl, pH 7.4; 20 mM sodium molybdate). Cells were broken by freeze–thaw lysis of the cell pellets in an equal volume of TM buffer. Cytosols were prepared by a 105,000 × g centrifugation at 4 °C for 60 min. Glycerol was added to the cytosol to a final concentration of 10% (vol/vol), the extract frozen immediately in liquid nitrogen, and stored at −80 °C until use. For competition assays, 50-µL aliquots (adjusted to 2.5 mg proteins per milliliter) were incubated with 10 nM of [3H]-CRT (specific activity 82 Ci/mmol; PerkinElmer) and various concentrations of unlabeled CRT or OCDO, ranging from 0.1 nM to 10 µM for 16 h at 4 °C. Specific binding was defined as the difference between binding of [3H]-CRT in the absence or in the presence of 1 µM unlabeled cortisol. For saturation assays, cytosols were incubated in the presence of increasing concentrations of [3H]-CRT, ranging from 0.1 to 100 nM in the absence or in the presence of 1 µM unlabeled CRT (nonspecific binding) or 1 µM OCDO for competitivity studies. Unbound [3H]-CRT was removed with dextran-coated charcoal. The quantification of the radioactivity was completed using a Tri-Carb liquid scintillation counter (Perkin-Elmer). Data were plotted as a standard competition curve by GraphPad prism 7. Experiments were repeated three times with duplicate at each time. IC50 value was converted into the apparent Ki using the Cheng–Prusoff equation and the Kd value of CRT (48).
GR Nuclear Localization Assay.
MDA-MB231 cells were plated in the adequate complete medium into six-well culture plates (250,000 cells per well). After 24 h, the medium was changed for complete medium with dextran-coated charcoal-stripped FBS for 24 h and cells were stimulated with the indicated compounds for 24 h. Cells were fixed in 3.7% paraformaldehyde for 15 min, permeabilized with 0.05% Triton X-100, then washed with 10% BSA in PBS. Cells were incubated with anti-GR (1:100) primary rabbit monoclonal antibody (12041 clone (D6H2L); Cell Signaling) for 1 h and then with anti-rabbit Alexa fluor-488 secondary goat polyclonal antibody (A-11008; Thermo Fisher) for 1 h. Finally, cells were stained with DAPI. Fluorescence images of cells were acquired with LSM 780 (Zeiss) confocal microscope with a 63× plan-apochromat objectif (1.4 oil). The microscope was equipped with a diode at 405 nm and argon laser at 488 nm. One-hundred cells per slide were evaluated to calculate Manders coefficient with the Jacop plugin in ImageJ v1.51. The experiments were performed in triplicate.
Cell Cycle Analysis.
Cell cycle analysis was performed by flow cytometry using MACSQuant VYB analyzer (Miltenyi Biotec). shC and shGR6 MDA-MB231 cells were plated in the adequate complete medium at a density of 750,000 cells per plate. After 24 h, the medium was changed for FBS-free medium. After 48 h, cells were incubated with 10 µM BrdU and treated with the solvent vehicle (control) or OCDO for 7 h. Cells were washed with PBS, fixed and permeabilized in 70% ethanol, and stored at −20 °C for subsequent cell cycle analysis. Cells were incubated with 2N HCl for 30 min and then with 0.1 M Na2B4O7, pH 8.5, for 5 min. Cells were then stained with FITC-conjugated anti-BrdU antibody (1:40) in PBS/1% BSA for 30 min in the dark at 25 °C. Finally, cells were incubated with 100 μg/mL RNaseA and 50 μg/mL propidium iodide in PBS/1% BSA (Sigma-Aldrich) for 30 min at 37 °C in the dark. Approximately 10,000 events were analyzed from each samples. The percentages of cells within the G0/G1, S, and G2/M phases of the cell cycle were calculated using FlowJo software.
Statistical Analyses.
Tumor growth curves in animals were analyzed for significance by two-way ANOVA followed by a Bonferroni posttest. In other experiments, as indicated, significant differences in the quantitative data were analyzed either using the Student’s t test for unpaired variables or by one-way ANOVA followed by a Tukey’s posttest. Immunostaining data were summarized by the frequency and percentage for categorical variables. Differences between the expression of the enzymes in tumor and normal tissues were analyzed using the McNemar test for paired samples. Comparisons between the immunoreactive score (multiplication between the percentage of positive cells and the staining intensity) dichotomized in two groups [(0–6) vs. (7–12)] of tumor samples and their pathological characteristics were analyzed using the χ2 test or Fisher’s exact test. For these statistical tests, differences were considered significant at the 5% level. Statistical analysis of the immunostaining data were performed using Stata 13.0 software and Prism software was used for the other analyses. A nonparametric exact paired Wilcoxon test was used to compare the levels of oxysterols between patient tumors and normal matched tissues. In all figures, *P < 0.05, **P < 0.01, and ***P < 0.001, compared with controls (solvent vehicle), unless otherwise specified.
Breast Cancer Gene-Expression Miner mining module.
The Breast Cancer Gene-Expression Miner v3 statistical mining module (bcgenex.centregauducheau.fr/BC-GEM/GEM-Accueil.php?js=1) was used to determine gene-expression correlations or correlations with survival outcomes (49). For gene correlation-targeted analysis, the Pearson’s correlation coefficient was computed with the associated P value for each pair of genes and Pearson’s pairwise correlation plots were generated to illustrate each pairwise correlation. For targeted prognostic analyses, data from all selected studies were converted to a common scale with suitable normalization to generate a set denoted “pool.” The prognostic impact of each gene was evaluated by means of a univariate Cox proportional hazards model. Kaplan–Meier curves were performed on the pool with the gene values dichotomized according to the gene median (calculated from the pool). Cox results corresponding to dichotomized values are displayed on the curve. To minimize unreliability at the end of the curve, the 15% of patients with the longest follow-up were not plotted.
Kaplan–Meier plotter.
The Kaplan–Meier plotter (kmplot.com) allows meta-analysis–based biomarker assessment using a background database, which is manually curated (50). Gene-expression data and relapse-free and overall survival information was downloaded from the Gene Expression Omnibus (Affymetrix microarrays only), European Genome-phenome Archive, and The Cancer Genome Atlas. To analyze the prognostic value of a particular gene, patient samples were split into two groups according to median expression. The two patient cohorts were compared using a Kaplan–Meier survival plot, and the HR with 95% confidence intervals and logrank P values were calculated.
BreastMark.
BreastMark (glados.ucd.ie/BreastMark) is an algorithm that allows the identification of genes that are associated with disease progression in breast cancer (16). It integrates gene-expression microarrays and detailed clinical data to correlate clinical outcome with differential gene-expression levels. The search was performed at the median threshold with disease-free survival as the survival endpoint. Survival curves were based on Kaplan–Meier estimates and the log-rank P values are shown for differences in survival. Cox regression analysis was used to calculate HRs.
Acknowledgments
We thank R. M. Evans, H. Richard-Foy, V. Laudet, B. Staels, and M. Oulad-Abdelghani for the generous gifts of the indicated plasmids; Dr. N. Elarouci from the Carte d'Identité des Tumeurs (CIT) for performing the analyses according to tumor molecular subtype; and Dr. Stoney Simons Jr. (NIH) for critical reading of our manuscript. This work was funded by the Institut National de la Santé et de la Recherche Médicale, the Université de Toulouse; the Ministère Français de la Recherche et de l’Enseignement Supérieur through a predoctoral fellowship for 3 y (to M.V.); the fondation Association pour la Recherche sur le Cancer (ARC) Projet PJA 20131200342, plus the allocation of an ARC young researcher grant for 1 y (to M.V.); and International Neuroendocrine Cancer Alliance Translational Grant PRTK-K15-118 and the Initiatives d'Excellence (IDEX), Actions Thématiques Stratégiques (ATS) InnoVinBC, including a predoctoral fellowship for 3 y (to A. Mallinger).
Footnotes
Conflict of interest statement: P.d.M., E.N., and L.M. are employees of the company Affichem, of which S.S.-P. and M.P. are founders.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1707965114/-/DCSupplemental.
References
- 1.Poirot M, Silvente-Poirot S. Cholesterol-5,6-epoxides: Chemistry, biochemistry, metabolic fate and cancer. Biochimie. 2013;95:622–631. doi: 10.1016/j.biochi.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 2.Segala G, et al. 5,6-Epoxy-cholesterols contribute to the anticancer pharmacology of tamoxifen in breast cancer cells. Biochem Pharmacol. 2013;86:175–189. doi: 10.1016/j.bcp.2013.02.031. [DOI] [PubMed] [Google Scholar]
- 3.de Medina P, et al. Dendrogenin A arises from cholesterol and histamine metabolism and shows cell differentiation and anti-tumour properties. Nat Commun. 2013;4:1840. doi: 10.1038/ncomms2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silvente-Poirot S, Poirot M. Cancer. Cholesterol and cancer, in the balance. Science. 2014;343:1445–1446. doi: 10.1126/science.1252787. [DOI] [PubMed] [Google Scholar]
- 5.de Medina P, Paillasse MR, Segala G, Poirot M, Silvente-Poirot S. Identification and pharmacological characterization of cholesterol-5,6-epoxide hydrolase as a target for tamoxifen and AEBS ligands. Proc Natl Acad Sci USA. 2010;107:13520–13525. doi: 10.1073/pnas.1002922107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silvente-Poirot S, Poirot M. Cholesterol metabolism and cancer: The good, the bad and the ugly. Curr Opin Pharmacol. 2012;12:673–676. doi: 10.1016/j.coph.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 7.Leignadier J, Dalenc F, Poirot M, Silvente-Poirot S. Improving the efficacy of hormone therapy in breast cancer: The role of cholesterol metabolism in SERM-mediated autophagy, cell differentiation and death. Biochem Pharmacol. June 19, 2017;144c:18–28. doi: 10.1016/j.bcp.2017.06.120. [DOI] [PubMed] [Google Scholar]
- 8.Sevanian A, McLeod LL. Catalytic properties and inhibition of hepatic cholesterol-epoxide hydrolase. J Biol Chem. 1986;261:54–59. [PubMed] [Google Scholar]
- 9.Voisin M, Silvente-Poirot S, Poirot M. One step synthesis of 6-oxo-cholestan-3β,5α-diol. Biochem Biophys Res Commun. 2014;446:782–785. doi: 10.1016/j.bbrc.2014.01.138. [DOI] [PubMed] [Google Scholar]
- 10.Poirot M, et al. Synthesis, binding and structure-affinity studies of new ligands for the microsomal anti-estrogen binding site (AEBS) Bioorg Med Chem. 2000;8:2007–2016. doi: 10.1016/s0968-0896(00)00119-x. [DOI] [PubMed] [Google Scholar]
- 11.Lathe R, Kotelevtsev Y. Steroid signaling: Ligand-binding promiscuity, molecular symmetry, and the need for gating. Steroids. 2014;82:14–22. doi: 10.1016/j.steroids.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 12.Chapman K, Holmes M, Seckl J. 11β-hydroxysteroid dehydrogenases: Intracellular gate-keepers of tissue glucocorticoid action. Physiol Rev. 2013;93:1139–1206. doi: 10.1152/physrev.00020.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Odermatt A, Arnold P, Stauffer A, Frey BM, Frey FJ. The N-terminal anchor sequences of 11beta-hydroxysteroid dehydrogenases determine their orientation in the endoplasmic reticulum membrane. J Biol Chem. 1999;274:28762–28770. doi: 10.1074/jbc.274.40.28762. [DOI] [PubMed] [Google Scholar]
- 14.Atanasov AG, Nashev LG, Schweizer RA, Frick C, Odermatt A. Hexose-6-phosphate dehydrogenase determines the reaction direction of 11beta-hydroxysteroid dehydrogenase type 1 as an oxoreductase. FEBS Lett. 2004;571:129–133. doi: 10.1016/j.febslet.2004.06.065. [DOI] [PubMed] [Google Scholar]
- 15.Guedj M, et al. A refined molecular taxonomy of breast cancer. Oncogene. 2012;31:1196–1206. doi: 10.1038/onc.2011.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Madden SF, et al. BreastMark: An integrated approach to mining publicly available transcriptomic datasets relating to breast cancer outcome. Breast Cancer Res. 2013;15:R52. doi: 10.1186/bcr3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nelson ER, et al. 27-Hydroxycholesterol links hypercholesterolemia and breast cancer pathophysiology. Science. 2013;342:1094–1098. doi: 10.1126/science.1241908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu Q, et al. 27-Hydroxycholesterol promotes cell-autonomous, ER-positive breast cancer growth. Cell Rep. 2013;5:637–645. doi: 10.1016/j.celrep.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holbeck S, et al. Expression profiling of nuclear receptors in the NCI60 cancer cell panel reveals receptor-drug and receptor-gene interactions. Mol Endocrinol. 2010;24:1287–1296. doi: 10.1210/me.2010-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ratman D, et al. How glucocorticoid receptors modulate the activity of other transcription factors: A scope beyond tethering. Mol Cell Endocrinol. 2013;380:41–54. doi: 10.1016/j.mce.2012.12.014. [DOI] [PubMed] [Google Scholar]
- 21.Weikum ER, Knuesel MT, Ortlund EA, Yamamoto KR. Glucocorticoid receptor control of transcription: Precision and plasticity via allostery. Nat Rev Mol Cell Biol. 2017;18:159–174. doi: 10.1038/nrm.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kassel O, et al. Glucocorticoids inhibit MAP kinase via increased expression and decreased degradation of MKP-1. EMBO J. 2001;20:7108–7116. doi: 10.1093/emboj/20.24.7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Webster MK, Goya L, Ge Y, Maiyar AC, Firestone GL. Characterization of sgk, a novel member of the serine/threonine protein kinase gene family which is transcriptionally induced by glucocorticoids and serum. Mol Cell Biol. 1993;13:2031–2040. doi: 10.1128/mcb.13.4.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jonat C, et al. Antitumor promotion and antiinflammation: Down-modulation of AP-1 (Fos/Jun) activity by glucocorticoid hormone. Cell. 1990;62:1189–1204. doi: 10.1016/0092-8674(90)90395-u. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, et al. Modulation of retinoic acid receptor-related orphan receptor alpha and gamma activity by 7-oxygenated sterol ligands. J Biol Chem. 2010;285:5013–5025. doi: 10.1074/jbc.M109.080614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soroosh P, et al. Oxysterols are agonist ligands of RORγt and drive Th17 cell differentiation. Proc Natl Acad Sci USA. 2014;111:12163–12168. doi: 10.1073/pnas.1322807111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deng R, Yang D, Yang J, Yan B. Oxysterol 22(R)-hydroxycholesterol induces the expression of the bile salt export pump through nuclear receptor farsenoid X receptor but not liver X receptor. J Pharmacol Exp Ther. 2006;317:317–325. doi: 10.1124/jpet.105.097758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vilasco M, et al. Glucocorticoid receptor and breast cancer. Breast Cancer Res Treat. 2011;130:1–10. doi: 10.1007/s10549-011-1689-6. [DOI] [PubMed] [Google Scholar]
- 29.Keenan CR, Lew MJ, Stewart AG. Biased signalling from the glucocorticoid receptor: Renewed opportunity for tailoring glucocorticoid activity. Biochem Pharmacol. 2016;112:6–12. doi: 10.1016/j.bcp.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 30.Joshi T, Johnson M, Newton R, Giembycz M. An analysis of glucocorticoid receptor-mediated gene expression in BEAS-2B human airway epithelial cells identifies distinct, ligand-directed, transcription profiles with implications for asthma therapeutics. Br J Pharmacol. 2015;172:1360–1378. doi: 10.1111/bph.13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Medina P, Paillasse MR, Payré B, Silvente-Poirot S, Poirot M. Synthesis of new alkylaminooxysterols with potent cell differentiating activities: Identification of leads for the treatment of cancer and neurodegenerative diseases. J Med Chem. 2009;52:7765–7777. doi: 10.1021/jm901063e. [DOI] [PubMed] [Google Scholar]
- 32.Gundimeda U, Chen ZH, Gopalakrishna R. Tamoxifen modulates protein kinase C via oxidative stress in estrogen receptor-negative breast cancer cells. J Biol Chem. 1996;271:13504–13514. doi: 10.1074/jbc.271.23.13504. [DOI] [PubMed] [Google Scholar]
- 33.Huang B, Warner M, Gustafsson JA. Estrogen receptors in breast carcinogenesis and endocrine therapy. Mol Cell Endocrinol. 2015;418:240–244. doi: 10.1016/j.mce.2014.11.015. [DOI] [PubMed] [Google Scholar]
- 34.Payré B, et al. Microsomal antiestrogen-binding site ligands induce growth control and differentiation of human breast cancer cells through the modulation of cholesterol metabolism. Mol Cancer Ther. 2008;7:3707–3718. doi: 10.1158/1535-7163.MCT-08-0507. [DOI] [PubMed] [Google Scholar]
- 35.Pulfer MK, Taube C, Gelfand E, Murphy RC. Ozone exposure in vivo and formation of biologically active oxysterols in the lung. J Pharmacol Exp Ther. 2005;312:256–264. doi: 10.1124/jpet.104.073437. [DOI] [PubMed] [Google Scholar]
- 36.Chen Z, et al. Ligand-dependent genomic function of glucocorticoid receptor in triple-negative breast cancer. Nat Commun. 2015;6:8323. doi: 10.1038/ncomms9323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pan D, Kocherginsky M, Conzen SD. Activation of the glucocorticoid receptor is associated with poor prognosis in estrogen receptor-negative breast cancer. Cancer Res. 2011;71:6360–6370. doi: 10.1158/0008-5472.CAN-11-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skor MN, et al. Glucocorticoid receptor antagonism as a novel therapy for triple-negative breast cancer. Clin Cancer Res. 2013;19:6163–6172. doi: 10.1158/1078-0432.CCR-12-3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sorrentino G, et al. Glucocorticoid receptor signalling activates YAP in breast cancer. Nat Commun. 2017;8:14073. doi: 10.1038/ncomms14073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang C, et al. Clinical and mechanistic aspects of glucocorticoid-induced chemotherapy resistance in the majority of solid tumors. Cancer Biol Ther. 2007;6:278–287. doi: 10.4161/cbt.6.2.3652. [DOI] [PubMed] [Google Scholar]
- 41.Karmakar S, Jin Y, Nagaich AK. Interaction of glucocorticoid receptor (GR) with estrogen receptor (ER) α and activator protein 1 (AP1) in dexamethasone-mediated interference of ERα activity. J Biol Chem. 2013;288:24020–24034. doi: 10.1074/jbc.M113.473819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gong H, et al. Glucocorticoids antagonize estrogens by glucocorticoid receptor-mediated activation of estrogen sulfotransferase. Cancer Res. 2008;68:7386–7393. doi: 10.1158/0008-5472.CAN-08-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang F, et al. Glucocorticoid receptor:MegaTrans switching mediates the repression of an ERalpha-regulated transcriptional program. Mol Cell. 2017;66:321–331.e6. doi: 10.1016/j.molcel.2017.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alikhani-Koupaei R, et al. Identification of polymorphisms in the human 11beta-hydroxysteroid dehydrogenase type 2 gene promoter: Functional characterization and relevance for salt sensitivity. FASEB J. 2007;21:3618–3628. doi: 10.1096/fj.07-8140com. [DOI] [PubMed] [Google Scholar]
- 45.Iuliano L, et al. Cholesterol metabolites exported from human brain. Steroids. 2015;99:189–193. doi: 10.1016/j.steroids.2015.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parks DJ, et al. Bile acids: Natural ligands for an orphan nuclear receptor. Science. 1999;284:1365–1368. doi: 10.1126/science.284.5418.1365. [DOI] [PubMed] [Google Scholar]
- 47.Wang Y, et al. Identification of SR1078, a synthetic agonist for the orphan nuclear receptors RORα and RORγ. ACS Chem Biol. 2010;5:1029–1034. doi: 10.1021/cb100223d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 49.Jézéquel P, et al. bc-GenExMiner 3.0: New mining module computes breast cancer gene expression correlation analyses. Database (Oxford) 2013;2013:bas060. doi: 10.1093/database/bas060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Györffy B, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]