Significance
Cells communicate with each other via signaling molecules that bind to surface receptors of responding cells. One example is Hedgehog (Hh) signaling, which is known for its regulatory role of tissue development and maintenance. Here we identify a Hh function, which is to regulate the sense of smell. Hh signaling modulates odorant responsiveness by regulating the accumulation of odorant receptors (ORs) in the olfactory ciliary compartment, which is where odorant detection is initiated. Since the mechanism appears evolutionarily conserved, even though the structures of ORs are not, the results open for the interesting possibility that Hh signaling may regulate protein transport and stimuli responsiveness also in other types of neurons.
Keywords: odorant receptors, Hedgehog pathway, Smoothened, vismodegib, cilia transport
Abstract
The ciliary localization of odorant receptors (ORs) is evolutionary conserved and essential for olfactory transduction. However, how the transport of ORs is regulated in mammalian olfactory sensory neurons is poorly understood. Here we demonstrate that odorant responsiveness and OR transport is regulated by the Hedgehog pathway. OR transport is inhibited by conditional gene inactivation of the Hedgehog signal mediator Smoothened (Smo) as well as by systemic administration of the Smo inhibitor vismodegib, a clinically used anticancer drug reported to distort smell perception in patients. The ciliary phenotype of Smo inhibition is haploinsufficient, cell autonomous, and correlates with the accumulation of OR-containing putative transport vesicles in the cytosol. The Smo-dependent OR transport route works in parallel with a low basal transport of vesicle containing both ORs and other olfactory transduction components. These findings both define a physiological function of Hedgehog signaling in olfaction and provide an important evolutionary link between olfaction and the requirement of a ciliary compartment for Hedgehog signaling.
Hedgehog (Hh) signaling is an evolutionarily conserved pathway that controls embryonic patterning and organ development in both vertebrates and insects (1). Hh family members are secreted morphogens, i.e., molecules that elicit distinct cellular responses in a concentration-dependent manner. Hh family members bind to Patched receptors. Binding results in the activation of a transmembrane signal transducer Smoothened (Smo), which induces a cellular response via a canonical pathway that includes glioma-associated transcription factors and/or via noncanonical pathways that can be mediated by l-type Ca2+ channels or Src kinase members (2, 3). Hh signaling continues to play a role after embryogenesis, regulating organ homeostasis and repair. Hh signaling has been shown to regulate proliferation and differentiation in the adult nervous system (4), but little is known about the function of Hh signaling in mature neurons that are part of a circuitry.
It was recently shown that Drosophila melanogaster possesses vertebrate-like cilia-dependent Hh signaling in addition to the canonical cilia-independent Hh signaling, which regulates Drosophila development (5). The identified cilia-dependent signaling in Drosophila is noncanonical and influences odorant-induced behavior and odorant responsiveness by regulating the transport of odorant receptors (ORs) to, and within, the sensory cilia compartment of olfactory sensory neurons (OSNs) (6). While insect and vertebrate ORs display differences in protein sequence, topology, and even transduction mechanisms, their localization to sensory cilia is conserved between the two phyla. Insects and vertebrates also share the general organization of the primary olfactory pathway. In brief, odorant detection is mediated by a mosaic of numerous classes of OSNs. Neurons of each class express one defined OR that determines the odorant response property of the class. Despite a likely impact on odorant detection threshold and odorant discrimination, the molecular mechanisms regulating ciliary localization of ORs in mammals are still unknown (7).
In light of the OR transport regulatory mechanism identified in flies, it is interesting to note that the most extensively studied mammalian Hh family member, Sonic hedgehog (Shh), is present in human nasal mucus as well as in dendrites of OSN target neurons in the rat olfactory bulb of the brain (8–10). The localization in target neurons is in line with the findings that Shh influences OSN axon growth and branching (11). Intriguingly, a low Shh level in human nasal mucus has been associated with reduced sense of smell (hyposmia) (8, 9). In addition, distortion of smell perception (dysosmia) is a side effect of the anticancer drug and Smo inhibitor vismodegib (GDC0449, Erivedge) (9). Taken together, these observations lead to the intriguing question of whether Hh-dependent regulation of OR availability and odorant responsiveness is evolutionary conserved to mammals.
Here we analyze mice and show that systemic i.p. administration of vismodegib as well as inactivation of a single Smo allele selectively in mature OSNs reduce OR levels in olfactory cilia. This reduction correlates with decreased expression of the odorant-inducible gene S100A5 and accumulation of putative OR-specific transport vesicles in the cytosol outside of cilia. Furthermore we identify low basal levels of Smo-independent puncta with ORs that colocalize with the olfactory transduction proteins adenylate cyclase 3 (AC3) and cyclic nucleotide-gated ion channel subunit alpha 2 (CNGA2). Collectively, the results show that Hh signaling regulates OR transport and odorant responsiveness in mammals. Moreover, the overall conclusion that this Smo-regulatory mechanism does not represent an insect specialization provides an important evolutionary link between olfactory transduction and ciliary-dependent Hh signaling that was apparently established in an ancestral organism before the evolutionary split of arthropods and chordates.
Results
Smo Signaling Regulates Ciliary OR Localization.
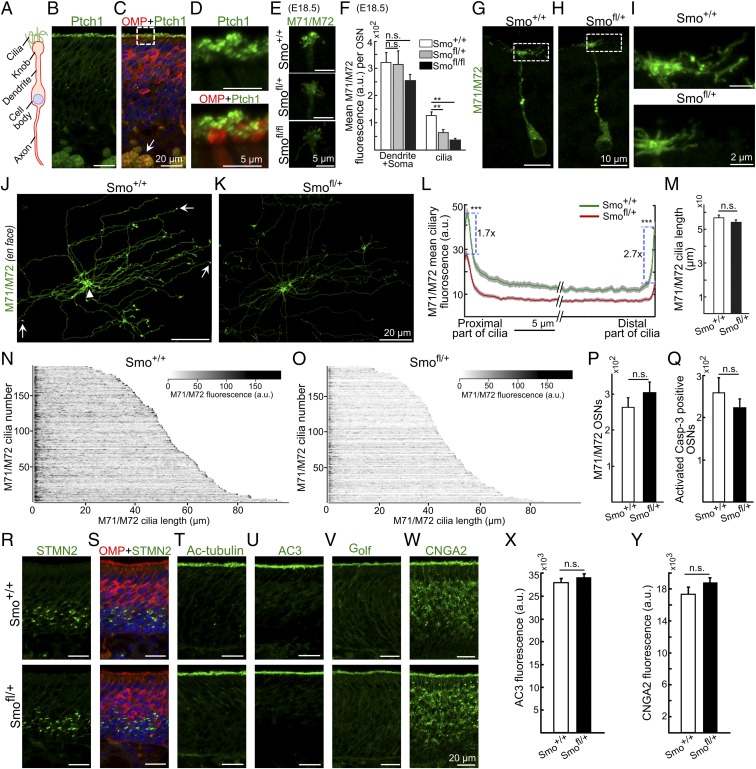
The Hh receptor Patch1 (Ptch1) and the signal transducer Smo have been shown to be expressed in the olfactory epithelium and in OSNs (10, 12). A double olfactory marker protein (OMP)/Ptch1 immunohistochemical analysis showed that Ptch1 was enriched in the dendritic knob and proximal cilia as well as in axons of OSNs (Fig. 1 A–D). To address the function of Hh signaling in OSNs we generated a transgenic mouse line (OMP-CRE), in which the OMP gene promoter drives expression of CRE recombinase in OSNs, but not in basal progenitor cells or other cell types of the primary olfactory system (13, 14). An analysis with a reporter mouse line showed that CRE was functional in OSNs, but not in other cell types of the olfactory epithelium (Fig. S1). To inactivate Smo selectively in OSNs we used mice with a floxed Smo gene (15). Breeding produced offspring with Smo+/+; CRE, Smofl/+; CRE, and Smofl/fl; CRE genotypes that deviated from the expected Mendelian ratio of 1:2:1. Of 73 weaned offspring, the ratios were ∼1:2:0 (27:45:1). An analysis of newborn mice (n = 84) from Smofl/+; CRE × Smofl/+ breeding pairs showed that the mortality of Smofl/fl with a CRE allele (n = 7) was 100% while no Smofl/fl mice without a CRE allele (n = 10) were found dead. It is known that a functional olfactory system somehow is important but not totally required for newborn mice to be successful in getting parental nursing (16, 17). However, some dead newborn Smofl/fl pups had milk in the stomach, indicating that the mortality was not necessarily due to inability to suckle. An alternative possibility was that the neonatal mortality was dependent on a lack of Smo signaling in OMP-positive cells other than OSNs, e.g., vomeronasal sensory neurons or OMP-positive nonolfactory cells (18, 19). Because of the neonatal mortality, we analyzed mice at embryonic day 18.5 (E18.5). To address whether Smo gene inactivation in OSNs influenced ciliary OR localization we used an antibody that recognizes two closely related ORs, M71 and M72 (20). Interestingly, immunohistochemical analysis of coronal tissue sections showed that M71/M72 immunofluorescence was reduced in cilia but not in dendrites or soma in both Smofl/+ (approximately twofold) and Smofl/fl mice (approximately threefold) compared with Smo+/+ control mice (Fig. 1 E and F). This result indicated that Smo signaling regulates accumulation of OR in cilia. That the OR signal was only half in Smofl/+ mice was in line with haplotype insufficiency, i.e., that one functional Smo allele was not enough to bring about wild-type ciliary OR levels. That M71/M72 immunofluorescence in cilia of Smofl/fl mice was further reduced, but not totally abolished, indicated that a low level of M71/M72 OR reached the ciliary compartment by a Smo-independent mechanism(s).
Fig. 1.
Conditional inactivation of the Smo gene in OSNs inhibits OR cilia localization. (A) Schematic illustration of an OSN. (B and C) Double immunohistochemical analysis of postnatal day 15 (PD15) olfactory epithelium showing Ptch1 (green) and OMP-positive (red) immunofluorescence in the ciliary layer (boxed) and axon bundles (arrow). (D) Close-up of boxed area in C. (E) M71/M72 immunofluorescence in olfactory cilia of embryonic day 18.5 (E18.5) Smo+/+, Smofl/+, and Smofl/fl OMP-CRE conditional knockout mice. (F) Mean intensities of M71/M72 immunofluorescence in OSN dendrite/soma and ciliary layer in E18.5 Smo+/+, Smofl/+, and Smofl/fl mice. (G and H) Immunofluorescence in the dendrite/soma and ciliary regions of OSNs in PD15 Smo+/+ and Smofl/+ mice. (I) Close-ups of boxed areas in G and H. (J and K) En face preparations of olfactory epithelium showing M71/M72 immunofluorescence in OSNs of Smo+/+ and Smofl/+ mice. Arrows in J indicate ciliary distal tips which are spoon-like enlargements (45). Arrowhead in J indicates a dendritic knob. (L) Mean intensities of M71/M72 immunofluorescence in the proximal and distal parts of cilia. Mean intensities in the proximal end are 45.9 ± 2.1 and 27.4 ± 1.8 in Smo+/+ and Smofl/+ mice, respectively. In the distal tip, the corresponding intensity values are 39.5 ± 2.7 and 14.6 ± 1.4. (M) Mean cilia lengths in Smo+/+ and Smofl/+ mice. (N and O) Heat maps of M71/M72 immunofluorescence intensity in individual cilia of Smo+/+ and Smofl/+ mice. X axis, length of cilia from proximal part to distal end; y axis, number of individual cilia. (P and Q) Relative quantifications of M71/M72-positive (P) and activated caspase-3/OMP double-positive neurons (Q) in Smo+/+ and Smofl/+ mice. (R and S) Double immunohistochemical analysis Smo+/+ and Smofl/+ mice showing STMN2 (green) and OMP (red) immunofluorescence in immature and mature OSNs, respectively. (T–W) Immunofluorescence corresponding to acetylated-tubulin (Ac-tubulin), adenylyl cyclase-3 (AC3), olfactory type G protein α-subunit (Golf), and cyclic nucleotide-gated ion channel subunit alpha 2 (CNGA2) in Smo+/+ and Smofl/+ mice. (X and Y) Quantifications of AC3 (X) and CNGA2 immunofluorescence (Y) in Smo+/+ and Smofl/+ mice. Values represent mean ± SEM, numbers of animals = 4, n = 34 for Smo+/+, 19 for Smofl/+, and 35 for Smofl/fl in F, 173 Smo+/+ and 157 Smofl/+ in L and M, 4 in P, and 3 in Q. **P < 0.01, ***P < 0.0001. a.u., arbitrary unit; n.s., not significant.
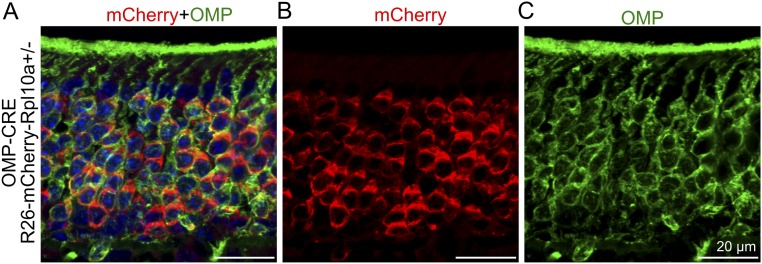
Fig. S1.
CRE-mediated recombination in OSNs of transgenic OMP-CRE mice. OMP-CRE mice were crossed to a reporter mouse line (R26-mCherry-Rpl10a) that carry a mCherry reporter gene under the transcriptional control of a floxed (ROSA)26Sor locus (38). (A–C) A double mCherry and OMP immunohistochemical analysis showing that mCherry expression, activated by CRE-mediated recombination, is specific to OMP-positive OSNs.
The finding that the Smo inhibition gave a haplotype-insufficient phenotype allowed us to focus our continued analyzes on heterozygous postnatal Smofl/+ mice. The postnatal growth of wild-type Smo+/+ and conditional knockout Smofl/+ mice was similar (Fig. S2A). This together with a predicted Mendelian Smo+/+/Smofl/+ ratio indicated that inactivation of one Smo allele did not influence viability. Analysis of coronal epithelial sections showed that ciliary M71/M72 immunofluorescence was reduced in PD15 Smofl/+ mice compared with Smo+/+ controls (Fig. 1 G–I). To analyze cilia along their entire lengths we performed whole mount en face preparations, which allowed for direct visualization of individual OSN cilia at the surface of the olfactory epithelium (Fig. 1 J and K). Quantification of immunofluorescence signals showed that M71/M72 accumulated to relatively high levels at the proximal end as well as at the distal tip of the cilium (Fig. 1L). The results further showed that ciliary M71/M72 immunofluorescence was reduced ∼2-fold in PD15 Smofl/+ compared with Smo+/+ controls (Fig. 1 J–L, N, and O). Median immunofluorescence intensities were 13.8 (11.5–45.9) and 7.5 (6.8–27.4) in Smo+/+ and Smofl/+ mice, respectively (Fig. 1L). The reduction of M71/M72 immunofluorescence was most pronounced in the distal tip (2.7-fold) compared with the proximal end (1.7-fold) of cilia (Fig. 1L). The same investigation revealed no alteration in the length distribution of cilia in Smofl/+ mice compared with Smo+/+ controls (Fig. 1M).
Fig. S2.
Genetic and pharmacological inhibition of Smo does not affect body weight. (A) Body weight change of PD0–PD15 Smo+/+ and Smo+/− mice. Values represent mean ± SEM, n = 7, n.s., not significant. (B) Body weight of vismodegib-treated and vehicle (DMSO)-treated control mice during the treatment period (PD12–PD15). Values represent mean ± SEM, n = 12. n.s., not significant.
The Smo haploinsufficient regulation of ciliary OR accumulation in postnatal mice was in line with the reported Ptc gene-dose sensitivity of Hh-regulated OR transport in adult D. melanogaster (6). One important difference between flies and mice is the continuous postnatal generation of OSNs in the mouse. To control for a possible Smo impact on OSN survival and adult neurogenesis, we quantified the number of M71/M72-positive cells and performed immunohistochemical analyses with antibody markers for apoptotic cells (activated caspase-3), mature OSNs (OMP) and immature OSNs [Stathmin-2 (STMN2)]. The result revealed no differences in the number of M71/M72-positive cells and caspase-3+ cells (Fig. 1 P and Q) or differences in mature and immature OSN cell layers of Smofl/+ mice and littermate Smo+/+ controls (Fig. 1 R and S). Further analyses showed that Smofl/+ and Smo+/+ mice were indistinguishable with respect to the ciliary enrichment of acetylated tubulin, which marks stable microtubules in OSN cilia (Fig. 1T). Immunofluorescence analyses for the olfactory transduction proteins Gαolf, AC3, and CNGA2 were also similar between Smofl/+ and Smo+/+ mice (Fig. 1 U–Y). Taken together these results indicated that inactivation of one Smo allele inhibited ciliary OR transport without influencing ciliary length, OSN differentiation, cell survival, or ciliary accumulation of other olfactory transduction proteins.
Vismodegib, a Smo Antagonist, Mimics the Effect of Smo Gene Inactivation in OSNs.
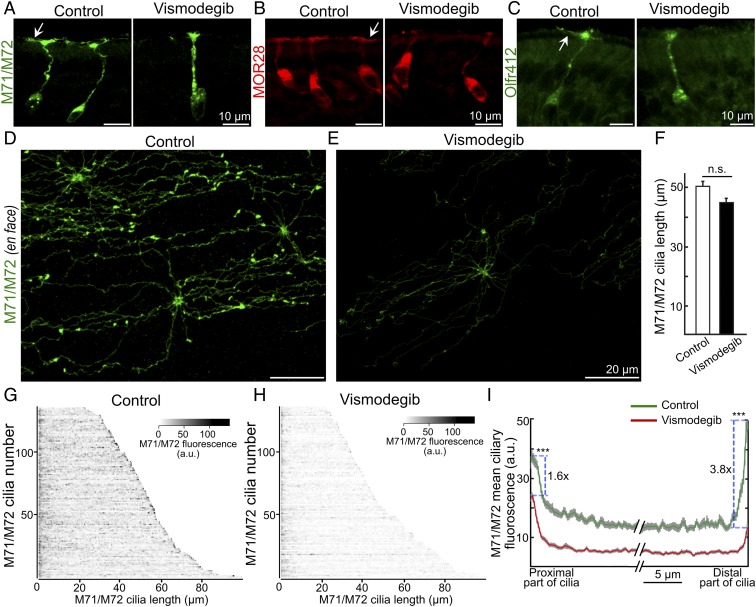
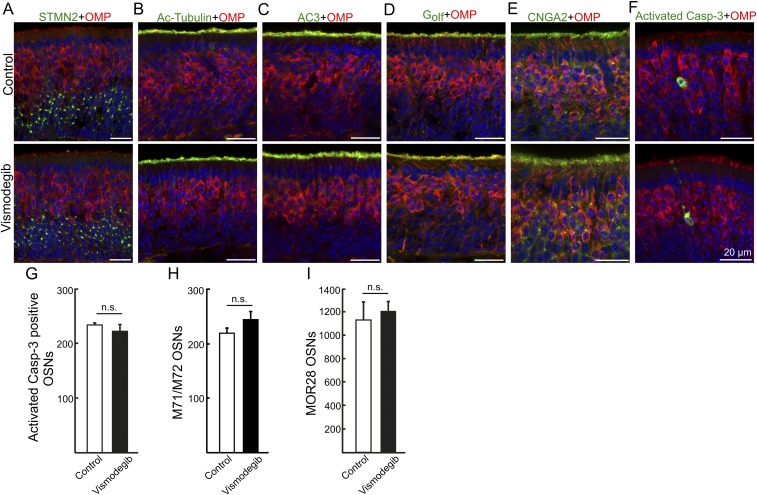
We next addressed whether Smo exerted a direct effect on ciliary OR transport in mature OSNs postnatally using a pharmacological approach. The ciliary OR accumulation following systemic administration of the Smo inhibitor vismodegib was investigated. Two-week-old mice received 40 mg/kg vismodegib i.p., twice a day for 3 consecutive days, which corresponded to a dose and administration scheme that have been shown to efficiently inhibit Smo signaling in mice (21). Upon killing, vismodegib-treated mice and vehicle-treated (DMSO) littermate controls showed no signs of apparent weight loss (Fig. S2B) or adverse health effects. Immunohistochemical analyses of coronal epithelial sections showed that vismodegib led to decreased ciliary immunofluorescence for M71/M72 ORs as well as decreased immunofluorescence using two other anti-OR antibodies recognizing ORs MOR28 and Olfr412, respectively (Fig. 2 A–C). Quantification of immunofluorescence and ciliary length in whole-mount en face preparations of vismodegib-treated mice showed, in similarity to the observations in Smofl/+ mice, that ciliary M71/M72 OR was reduced, while cilia lengths were not changed (Fig. 2 D–I). Immunofluorescence was reduced ∼3-fold and the median intensities were 14.0 (11.3–50.6) and 5.0 (3.8–23.9) in DMSO- and vismodegib-treated mice, respectively (Fig. 2I). As for Smofl/+ mice, the vismodegib-dependent reduction of M71/M72 immunoreactivity was most pronounced in the distal tip (3.8-fold) compared with the proximal end (1.6-fold) of cilia (Fig. 2I). Moreover, the epithelial integrity was unchanged in vismodegib-treated mice compared with control mice with regard to the expression of OMP, STMN2, acetylated tubulin, AC3, Gαolf, and CNGA2 as well as numbers of OSNs expressing activated caspase-3, M71/M72, and MOR28 (Fig. S3). Collectively, these results showed that systemic administration, in a postdevelopmental context, of a nontoxic dose of a Smo inhibitor phenocopied the effect of Smo gene inactivation.
Fig. 2.
Vismodegib mimics the effect of Smo gene inactivation. (A–C) Immunofluorescence corresponding to the ORs M71/M72, MOR28, and Olfr412 in coronal olfactory epithelial sections of vehicle (DMSO)-treated control and vismodegib-treated mice. Arrows indicate immunopositive olfactory cilia. (D and E) En face olfactory epithelium preparations showing M71/M72 immunofluorescence in the cilia layer of control and vismodegib-treated mice. (F) Mean ciliary length in control and vismodegib-treated mice. (G and H) Heat maps of M71/M72 immunofluorescence intensities in cilia of control mice and vismodegib-treated mice. (I) Mean intensities of M71/M72 immunofluorescence in proximal and distal parts of cilia. Mean intensities in the proximal ciliary end are 37.4 ± 1.9 and 23.9 ± 1.1 in DMSO-treated and vismodegib-treated mice, respectively. In the distal tip the corresponding intensity values are 50.6 ± 3.5 and 13.2 ± 1.3. Values represent mean ± SEM, numbers of animals = 4, n = 136 (control mice) and 132 (vismodegib-treated mice). ***P < 0.0001. a.u., arbitrary unit; n.s., not significant.
Fig. S3.
OSN differentiation, survival, and ciliary expression of Ac-Tubulin, AC3, Gαolf, and CNGA2 are not affected by vismodegib. Double immunohistochemical analyses showing similar expression in DMSO (vehicle)-treated control mice and vismodegib-treated mice of (A) STMN2 and OMP in immature and mature OSNs, respectively. (B) Ac-Tubulin in cilia, (C) AC3, (D) Gαolf, (E) CNGA2, and (F) activated caspase-3. (G and H) Quantification of OSNs immunopositive for activated caspase-3, M71/M72, and MOR28. All values represent mean ± SEM, n = 3. n.s., not significant.
Smo Inhibition Results in Decreased Expression of the Odorant-Inducible Gene S100A5.
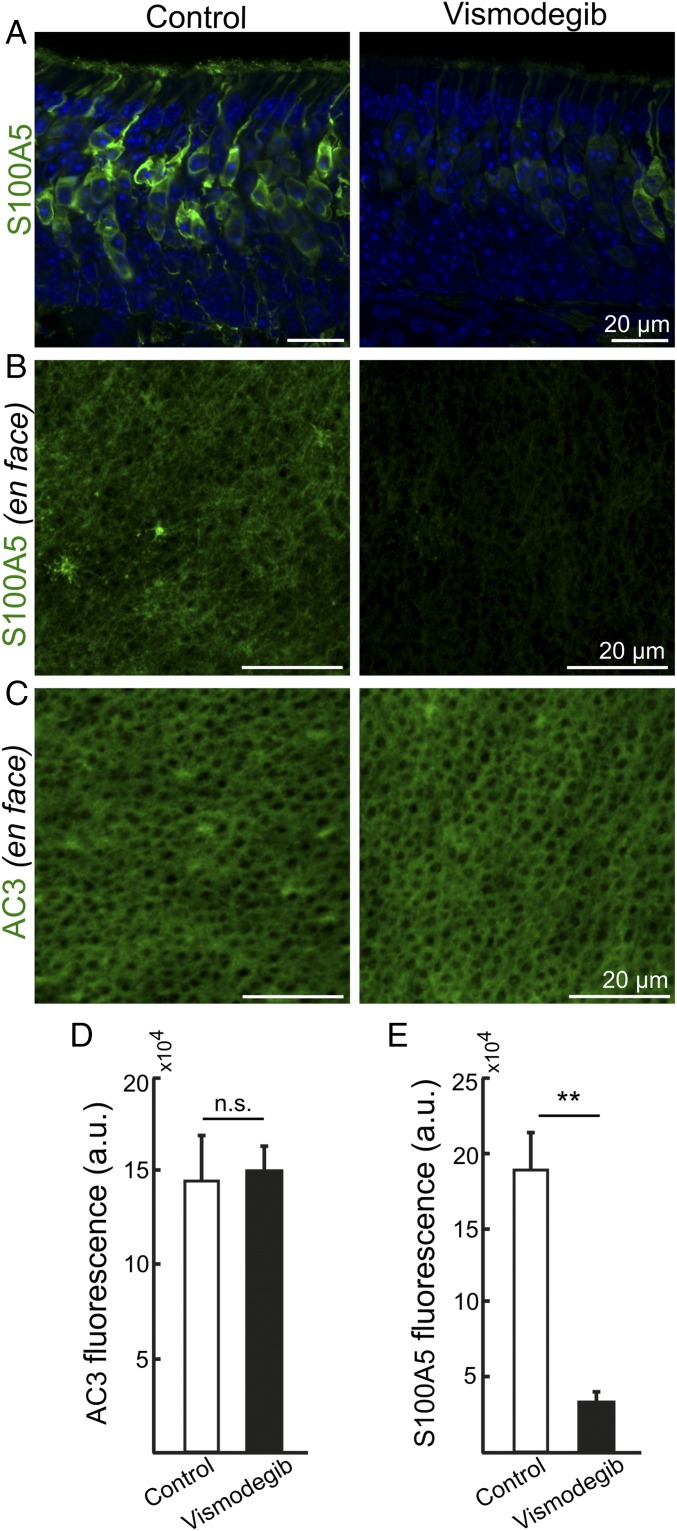
To address whether the reduction in ciliary OR accumulation affected the function of OSNs, we analyzed the S100 calcium binding protein A5 (S100A5) that is transiently expressed to high levels in odorant-stimulated OSNs and that requires intact olfactory cilia to be expressed (22, 23). In concordance with reduced olfactory transduction in vismodegib-treated mice we found that the number of S100A5-positive OSNs was reduced in vismodegib-treated mice compared with controls (Fig. 3A). Quantification of immunofluorescence in whole-mount en face preparations of olfactory epithelia confirmed that S100A5 was down-regulated by vismodegib, while AC3 was unaltered (Fig. 3 B–E). Thus, the Smo-dependent OR down-regulation correlated with reduced odorant responsiveness. Moreover, the reduction of S100A5 across the whole epithelium indicated that multiple OSN classes depend on Smo signaling for S100A5 expression.
Fig. 3.
Smo inhibition results in diminished expression of the odorant-inducible gene S100A5. (A) Coronal olfactory epithelial sections showing reduced S100A5 immunofluorescence in OSNs of vismodegib-treated mouse compared with DMSO (vehicle)-treated control mouse. (B) En face olfactory epithelial preparations showing reduced S100A5 immunofluorescence in the cilia layer of vismodegib-treated mouse compared with control mouse. (C) En face olfactory epithelial preparations showing similar AC3 immunofluorescence in control mouse and vismodegib-treated mouse. (D and E) Quantification of AC3 and S100A5 immunofluorescence in en face epithelial preparations of control mice and vismodegib-treated mice. Values represent mean ± SEM, n = 3. **P < 0.01. a.u., arbitrary unit; n.s., not significant.
Smo Inhibition Results in Accumulation of OR-Positive Puncta in the Cytosol.
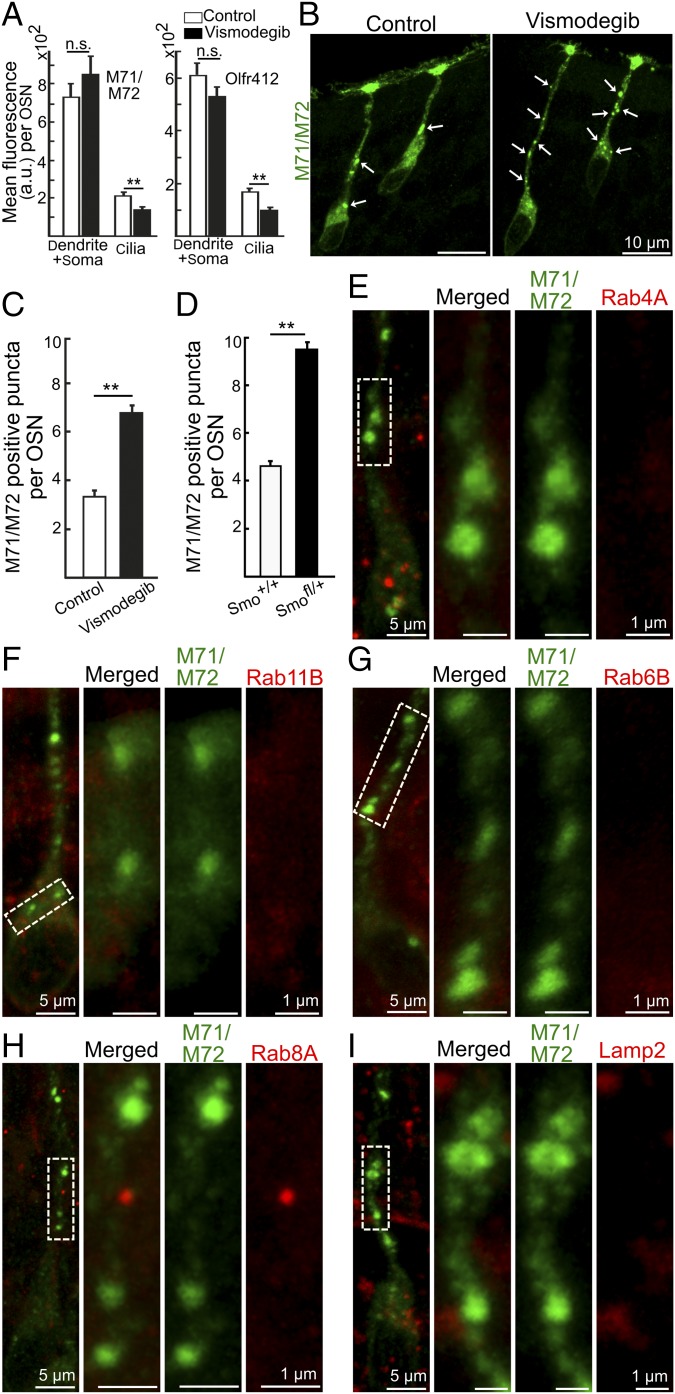
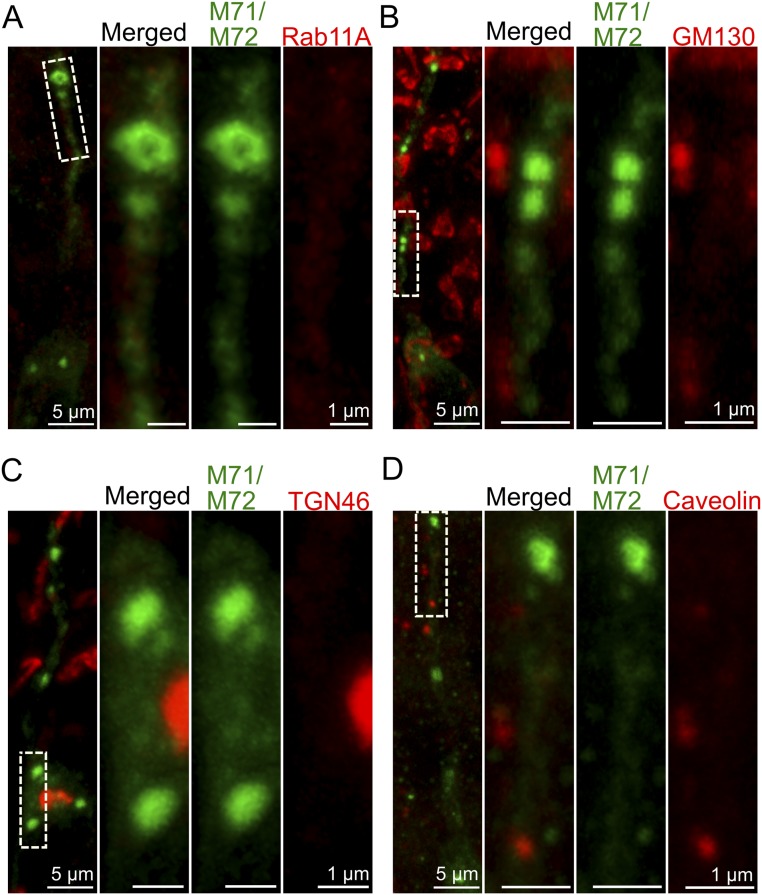
Analyses of coronal sections with anti-M71/M72 and anti-Olfr412 antibodies showed that the immunofluorescence in the ciliary layer was reduced in vismodegib-treated mice while the combined levels of immunofluorescence in the dendrite and cell body were similar in treated and control mice (Fig. 4A). Besides finding M71/M72 and Olfr412 in the regions of the endoplasmic reticulum, the Golgi apparatus, and cell and ciliary membranes, we detected OR immunofluorescence in a punctate fashion in the apical soma and along the entire length of the dendrites (Fig. 4B and Fig. S5). These OR-positive puncta have been suggested to represent OR transport vesicles en route between Golgi apparatus and cilia (24). Interestingly, our quantifications showed that the number of OR-positive puncta increased in vismodegib-treated mice as well as in Smofl/+ mice relative to controls (Fig. 4 C and D). To investigate the origin of the OR-positive puncta, we performed double immunohistochemical analyses with markers for different types of organelles and transport vesicles. Surprisingly, 3D rendering and rotation of a stack of high-resolution images revealed no overlap in immunofluorescence between OR-positive puncta and the following small G protein Rabs: RAB4A involved in the sorting and recycling of early endosomes (25) (Fig. 4E), Rab11A/B associated primarily with recycling endosomes (25) (Fig. 4F and Fig. S4), or Rab6B and Rab8A required for multiple apical transport pathways, including rhodopsin transport to the cilium in Drosophila photoreceptors (25, 26) (Fig. 4 G and H). The OR-positive puncta were also negative for the lysosomal marker Lamp2, Caveolin, and the Golgi markers GM130 and TGN46 (Fig. 4I and Figs. S4 and S5). Altogether these results showed that inhibition of Smo resulted in an accumulation OR-positive puncta in the cytosol, which was analogous to the reported effect of Smo inhibition in Drosophila OSNs (6). Moreover, the results indicated that OR transport is mediated by a pathway(s) with molecular properties that are distinct from many known protein transport pathways.
Fig. 4.
Smo inhibition results in accumulation of OR-positive puncta in the cytosol. (A) Quantifications of M71/M72 and Olfr412 immunofluorescence in the dendrite/soma and ciliary regions of OSNs in coronal tissue sections from DMSO (vehicle)-treated control and vismodegib-treated mice. (B) M71/M72 immunopositive puncta (arrows). (C and D) Quantification of M71/M72-positive puncta in dendrites and apical soma of OSNs in DMSO-treated control, vismodegib-treated Smo+/+ and Smofl/+ mice. All values in A, C, and D represent mean ± SEM, n = 62 (control M71/M72), 71 (vismodegib-treated M71/M72), 43 (control Olfr412), 46 (vismodegib-treated Olfr412), 144 (Smo+/+) and 122 (Smofl/+). Five pairs of mice for both the control/vismodegib comparison and the Smo+/+/Smofl/+ comparison were analyzed. a.u., arbitrary unit; n.s., not significant, **P < 0.01. (E–I) Double immunohistochemical analyses of OSNs, in vismodegib-treated mice, with anti-M71/M72 antibody and antibodies for Rab4A (E), Rab11B (F), Rab6B (G), Rab8A (H), and Lamp2 (I). In each panel, the three images to the Right are a close-up of the area that is boxed in the image to the Left.
Fig. S5.
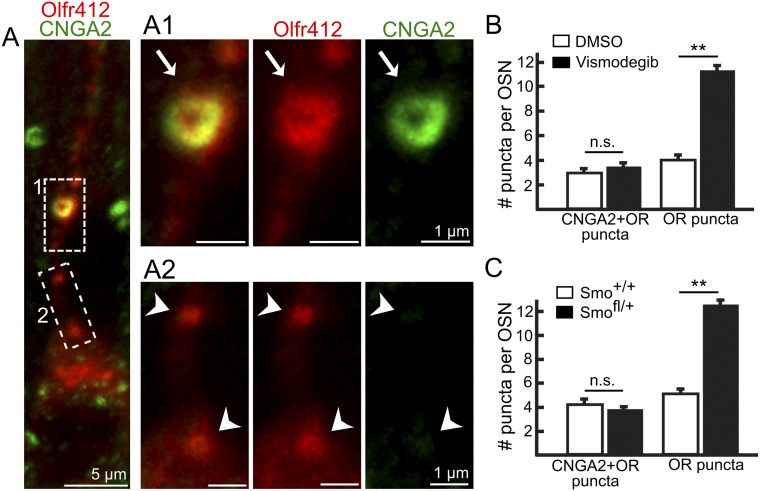
Olfr412-positive puncta that colocalize with CNGA2 do not increase in number. (A) Identification of Olfr412-positive puncta in vismodegib-treated mice that colocalize with CNGA2 (arrow in A1) as well as Olfr412-positive puncta that do not colocalize with CNGA2 (arrowheads in A2). In each panel, the three images to the Right are a close-up of the area that is boxed in the image to the Left. (B and C) Quantification of Olfr412-positive puncta that are negative and double-positive CNGA2 in vehicle (DMSO)-treated, visomdegib-treated, Smo+/+, and Smofl/+ mice. All values represent mean ± SEM. The n values in B and C are 43 (DMSO treated), 46 (vismodegib treated), 52 (Smo+/+), and 63 (Smofl/+). Three pairs of mice for both the DMSO/vismodegib comparison and the Smo+/+/Smofl/+ comparison were analyzed. **P < 0.01. n.s., not significant.
Fig. S4.
OR-positive puncta do not colocalize with Rab11A, Golgi markers, and Caveolin. Double immunohistochemical analyses of OSNs, in vismodegib-treated mice, with anti-M71/M72 antibody and antibodies for (A) Rab11A, (B) GM130 (cis-Golgi marker), (C) TGN46 (trans-Golgi marker), and (D) Caveolin. In each panel, the three images to the Right are a close-up of the area that is boxed in the image to the Left.
Smo Regulates One of at Least Two Different Types of OR Containing Putative Transport Vesicles.
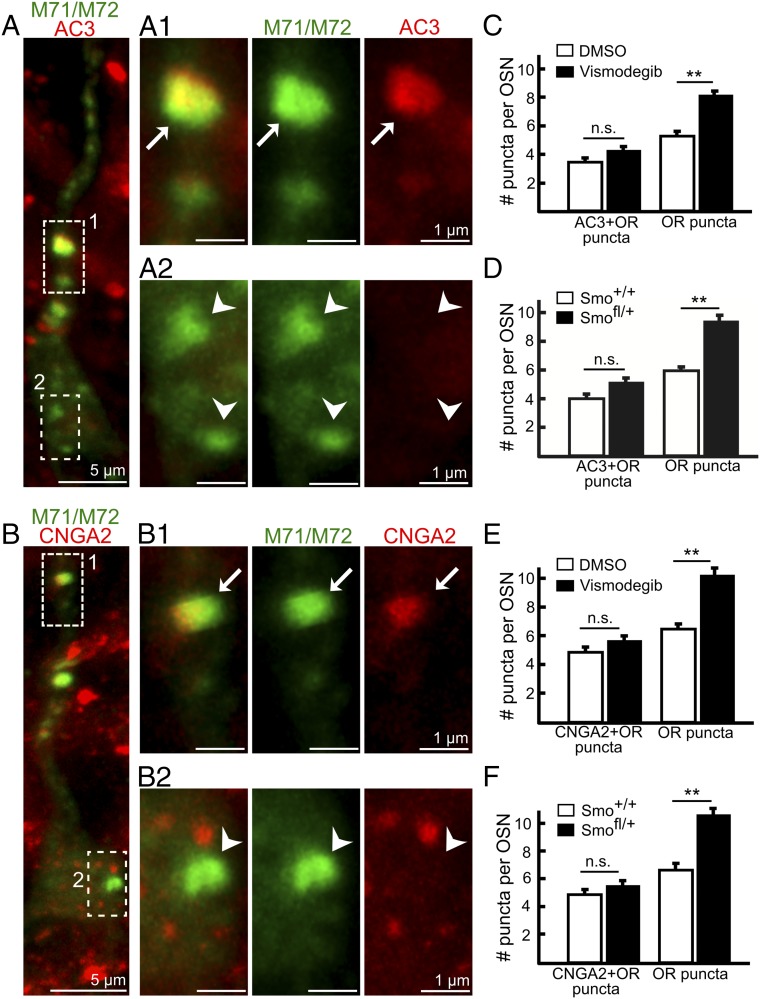
OR-positive puncta in Drosophila OSNs can colocalize with Orco (6), which is a coreceptor in insect OSNs that regulates OR ciliary localization and forms homomers as well as OR–Orco heteromers that can function as ion channels (27). The OR-positive puncta that increase in number after Smo knockdown in Drosophila OSNs belong to a fraction of putative transport vesicles that do not contain the olfactory transduction protein Orco (6). We likewise identified a fraction of the M71/M72-positive puncta that colocalized with the olfactory transduction proteins AC3 and CNGA2 while another fraction of puncta did not (Fig. 5 A and B). Strikingly, and analogous to Drosophila, a quantification showed that it was the OR-positive puncta without AC3 and CNGA2 that selectively increased in number in Smofl/+ conditional knockout mice as well as in vismodegib-treated mice (Fig. 5 C–F). Similar results were obtained using the OR Olfr412 antibody (Fig. S5). Overall these results showed that the decrease of ORs in cilia in response to Smo inhibition correlated with an increased accumulation of “OR-specific” puncta in the cytoplasm. The unaltered number of OR-positive puncta that colocalized with CNGA2 and AC3 indicated the existence of at least one additional trafficking route that provides basal and/or Smo-independent dendritic OR transport.
Fig. 5.
OR-positive puncta that colocalizes with AC3 and CNGA2 do not increase in number. (A and B) Identification of M71/M72-positive puncta in vismodegib-treated mice that colocalize with AC3 (arrow in A1) or CNGA2 (arrow in B1) as well as M71/M72-positive puncta that do not colocalize with AC3 or CNGA2 (arrowheads in A2 and B2). In each panel, the three images to the Right are a close-up of the area that is boxed in the image to the Left. (C–F) Quantification of M71/M72-positive puncta that are negative and double positive for AC3 and CNGA2 in vehicle (DMSO)-treated, visomdegib-treated, Smo+/+, and Smofl/+ mice. All values represent mean ± SEM; the n values in C and D are 65 (DMSO treated), 63 (vismodegib treated), 64 (Smo+/+), and 68 (Smofl/+). The n values in E and F are 57 (DMSO treated), 54 (vismodegib treated), 61 (Smo+/+), and 84 (Smofl/+). Three pairs of mice for both the DMSO/vismodegib comparison and the Smo+/+/Smofl/+ comparison were analyzed. **P < 0.01. n.s., not significant.
Discussion
We have analyzed conditional knockout mice with OSNs deficient in the Shh signaling transducer Smo as well as mice treated with the Smo inhibitor vismodegib. Both approaches resulted in lower OR levels in the ciliary compartment. The reduction of ciliary OR is not associated with changes in OSN differentiation, ciliary length, cell death, or ciliary reduction of other olfactory transduction proteins. Moreover, the reduction of OR correlates with a reduced odorant responsiveness, as indicated by diminished expression of the odorant-induced activity marker S100A5. We find that the Hh receptor Ptch1 is enriched in the dendritc knob and cilia of mature OSNs, which is a suitable localization with respect to the control of protein transport to cilia. Olfactory cilia are immersed in mucus, which in humans has been shown to contain Shh (28). Interestingly, data that Shh indeed may regulate olfactory perception comes from clinical studies which have found a correlation between hyposmia and low Shh levels in nasal mucosa (8, 28). Further evidence that the Hh pathway regulates olfactory perception in humans comes from the finding that dysosmia is a side effect of the anticancer drug, and Smo inhibitor, vismodegib (9). These observations also imply that Hh regulation of ciliary OR transport is evolutionarily conserved between the mouse and humans. The recent discovery of cilia-mediated Hh signaling in Drosophila (5), which regulates ciliary OR transport, indicates that the evolutionary conservation is extended to include insects (6).
The Hh family members are morphogens that mediate graded, concentration-dependent cellular responses where unbound Ptch1 represses Smo signaling. Analysis of Drosophila OSNs has demonstrated that increasing Smo signaling by decreasing Ptc gene dosage, increases ciliary OR transport and odorant responsiveness in heterozygous Ptc+/− flies compared with wild type (6). In mouse, we find that inactivation of one Smo allele results in ∼50% reduction of ciliary OR levels. Vismodegib reduced the ciliary OR levels more efficiently (by ∼75%) but not completely. Due to postnatal mortality, our analysis of OSNs lacking both Smo alleles is restricted to prenatal Smofl/fl knockout mice. In these OSNs, we find ciliary OR immunofluorescence to be reduced (∼75%) but not absolutely abolished. Taken together, these results show that more than one mechanism provide OR to cilia. The other(s) mechanism is constitutive and/or regulated in a Smo-independent manner and perhaps may be sufficient to allow for detection of odorant at relatively high concentrations. Compared with the situation when prolonged odorant stimuli allow for induction of rapid endocytic retrieval of ORs (29), it is likely that activation of Smo signaling by Hh ligands mediates more “long-term” tonic changes in odorant responsiveness. A speculative function of the Smo-dependent signaling may be to enhance responsiveness by increasing OR accumulation in cilia above basal ciliary OR levels. This model is in line with the known graded response properties of Hh signaling, the gene-dosage sensitivity in mice and flies, as well as olfactory transduction, which is characterized by linearity of the response at low odorant concentrations (30, 31).
Smo regulates OR transport to the cilium as well as within the ciliary compartment of Drosophila OSNs (6). OR transport within the cilia depends on the intraflagellar transport system as well as Cos2, which is a known kinesin-like Smo pathway effector. Cos2 regulates OR distribution in the cilium, as there is a shift of ORs from the distal part of the cilium as in control flies, to even distribution of ORs in the entire cilium in Smo knockdown flies (6). We found that the OR distribution in olfactory cilia was not uniform and that the reduction of OR immunofluorescence in response to Smo inhibition was more pronounced in the ciliary tip than at the proximal end of the cilium. It remains to be determined whether the regulation of the distribution in the cilia is due to an effect on the intraflagellar transport system and/or the mouse orthologs of Cos2 (Kif7/Kif27).
We find that the reduced accumulation of OR specifically in cilia after Smo inhibition correlates with an increased number of OR-positive puncta located outside of the cilia, i.e., in the cytosol in the apical part of the soma and in dendrites. Unexpectedly, we found that OR-positive puncta did not colocalize with markers for many known transport paths. While the molecular nature of the OR-positive puncta is elusive, our results show that there are at least two different types of OR-positive puncta. One type is “heterogeneous puncta,” in which OR colocalizes with CNGA2 and AC3, while another type of puncta is OR-specific, at least with respect to the absence of CNGA2 and AC3. Similar to observations of Orco and OR transport in Drosophila (6), we find that it is the OR-specific puncta that accumulate in dendrites in a Smo-dependent manner. Our result indicate that Smo-dependent transport of OR-specific vesicles take place in a background of a Smo-independent transport route(s) of heterogeneous OR transport vesicles.
ORs in insects differ from mammalian ORs in that they have an inverted membrane topology and lack sequence homologies with G protein-coupled receptors (32). These differences make it less likely that common transport sequence motifs play a role in Smo-dependent regulation of OR transport. Speculatively, the knowledge that the transport mechanism appears to be evolutionarily conserved while the structure of the cargo is not, may indicate that the Hh signaling pathway has evolved independently in insects and mammals to adopt an OR transport regulatory function. However, the evolution of cilia for environmental sensing is likely to have occurred before evolution of Hh-mediated cell–cell signaling. Instead of a parallel evolutionary scenario, it is therefore possible that ciliary Hh signaling has evolved in an ancestral species as a consequence of an adaptive advantage of Hh-regulated chemosensation. Support for divergent evolution is that ORs as well as Smo-mediated Hh signaling are first found in the phylum Cnidaria, in species that have ciliated chemoreceptor cells (33–35). There is a striking evolutionary convergence toward a conserved organization of the peripheral olfactory pathways of insects and vertebrates. For example, a single mature OSN in Drosophila and mammals expresses only one specific OR and synapse with precision to topographically defined neurons in the central target area. By contrast, the nematode Caenorhabditis elegans, which lacks the Hh signaling pathway, has chemosensory neurons that express ∼1,000 different chemoreceptors in only 11 pairs of chemosensory neurons. Cell-specific control of ciliary OR transport by Hh signaling may thus have evolved as a mechanism that improves odorant identification and discrimination.
That the regulatory mechanism appears not to be dependent on the structure of the cargo may have contributed to an adaptive advantage, as it allows for rapid evolution of OR gene families to meet niche- and species-specific demands on odorant detection. OR gene families have evolved independently many times (36). Even within the same species, odorant detection is mediated by several distinct gene families, e.g., trace amine-associated receptors and guanylyl cyclase type D receptors mediate odorant recognition in the cilia of mouse OSNs in addition to the canonical G protein-coupled ORs. In insect OSNs, odorant detection can also be mediated by ionotropic receptors that are not related to canonical insect ORs, but rather have evolved from ionotropic glutamate receptors (37). In light of the finding that Smo regulates ciliary accumulation of two very disparate OR families in insects and mammals, respectively, an interesting area for future research is to address whether the mechanism has a more general function, regulating dendritic transport and/or ciliary targeting of not only other OR families but also proteins different from ORs and in cell types other than OSNs.
Materials and Methods
Animals.
All animal experiments were approved by the local ethics committee for animal research at the court of appeal for the upper northern area of Norrland (Umeå, Sweden). Mice were kept in open cages with 12 h light/dark cycle housed at the Umeå Center for Comparative Biology, Umeå. The generation of transgenic mice expressing CRE under the OMP gene promoter was done essentially as previously described (13). Briefly, the coding sequence of CRE was ligated 3′ to a 6-kb fragment of the mouse OMP promoter and 5′ to a simian virus 40 polyadenylation site (SV40polyA+) in the cloning vector. After DNA sequencing, a DNA fragment corresponding to the OMP-CRE-SV40polyA+ transgene was gel purified and microinjected into pronuclei of C57BL/6/CBA one-cell embryos at the transgene core facility at the Umeå Center of Comparative Biology. A founder line (OMP-CRE) was backcrossed to C57BL/6J mice (Taconic M&B), for more than five generations. The progeny of OMP-CRE and a mouse line that carried a Gt(ROSA)26Sor-mCherry allele (38), was analyzed to verify selective CRE-dependent recombination in OSNs (Fig. S1). Conditionally, Smo knockout mice were generated with the Smotm2Amc/J mouse line (15). Mice of either sex were analyzed.
Drug Administration.
Vismodegib (GDC-0449) was purchased from Selleckchem (catalog no. S1082, lot nos. 05 and 06) and dissolved in DMSO just before injecting in mice. Mice were injected i.p. with five doses of vismodegib (40 mg/kg body weight) at intervals of 12 h from postnatal day 12. Mice were killed 12 h after last dose. For each control group, the same number of mice were injected with equivalent volume of DMSO. There were at least five mice per group.
Immunohistochemistry.
Mice were killed by cervical dislocation followed by decapitation. Nasal tissue was dissected, fixed for 4 h in 4% PFA (4% paraformaldehyde, wt/vol in PBS), incubated in 30% sucrose (wt/vol in PBS) overnight, and frozen in OCT (Histolab). Air dried 14-µm thick cryosections were incubated with 0.5% Triton X-100 (vol/vol) in PBS for 20 min followed by 2-h incubation in blocking solution [2% FCS (vol/vol) + 0.2% Triton X-100 (vol/vol) in PBS]. For Olfr412 immunohistochemistry, sections were treated with citrate buffer (10 Mm, pH 6.0) for 5 min in 100 °C before incubation in blocking solution. Sections were washed in PBS, incubated overnight with primary antibodies in blocking solution, washed, incubated with secondary antibodies diluted in T-PBS [0.1% Tween-20 (vol/vol) in PBS]. Sections were then stained in Hoechst (0.1 µg/mL) and mounted with fluorescence mounting media (Dako). Primary antibodies used were anti-MOR28 (20) (1:10,000), anti-M71/72 (20) (1:2,000), anti-Olfr412 (1:10,000, catalog no. OSR00211W; Thermo Fisher Scientific), Gαolf/s (1:200, catalog no. SC55545; Santa Cruz Biotechnology), anti-OMP (1:2,000, catalog no. 544–10001; Wako), anti-CNGA2 (1:100, catalog no. APC-045; Alomone Labs and 1:100, catalog no. SC13700; Santa Cruz Biotechnology for the experiment in Fig. S5), anti-Rab4A (1:150, catalog no. sc-312; Santa Cruz), anti-Rab8A (1:200, clone 23, 610844; BD Biosciences), anti-Rab11A (1:200, 71-5300; Invitrogen), anti-Rab11B (1:100, MBS830790; MyBioSource) anti-Caveolin (1:200, catalog no. 610406; BD Biosciences), anti-Lamp2 (1:100, catalog no. 1921-01; Southern Biotech), anti-GM130 (1:150, catalog no. 610823; BD Biosciences), anti-TGN46 (1:200, catalog no. ab16059; Abcam), anti-STMN2 (1:1,000, catalog no. PA5-23049; Thermo Fisher Scientific), anti-acTub (1:100, catalog no. T7451; Sigma-Aldrich), anti-activated caspase-3 (1:500, catalog no. 559565; BD Biosciences), anti-AC3 (1:100, catalog no. sc-588; Santa Cruz Biotechnology), anti-S100A5 (39) (62 FAC1, 1:2,000), and anti-Ptch1 (1:200, catalog no. MAB41051; R&D Systems). Supportive validation data for the anti-Ptch1 antibody have been published by Katic et al. (40). Secondary antibodies used were donkey anti-guinea pig Alexa 488 (catalog no. 706–545-148, Jackson ImmunoResearch Europe), donkey anti-rabbit Alexa 488 (catalog no. A-21206, Life Technologies), donkey anti-rabbit Alexa 546 (catalog no. A-10040, Life Technologies), donkey anti-goat Alexa 546 (catalog no. A-11056, Life Technologies), donkey anti-mouse Alexa 488 (catalog no. AS101201, Agrisera AB Sweden), and donkey anti-rat Alexa 546 (catalog no. A-21208, Life Technologies).
Tissue Preparations.
En face preparation of olfactory epithelium was done essentially as described by Challis et al. (41). Snouts were fixed in 4% PFA (wt/vol) + 20% sucrose (wt/vol) in PBS for 3 h. Nasal septum was dissected, washed in PBS, immersed for 2 h in blocking solution [3% FCS (vol/vol) + 0.2% Triton X-100 (vol/vol) in PBS], incubated overnight in blocking solution with primary antibodies, washes three times in TPBS, incubated for 2 h with secondary antibodies, and washed three times in TPBS. Epithelia were then dissected and mounted in mounting media (Dako).
Image Analysis and Quantifications.
Imaging was done with Nikon D eclipse C1 confocal system equipped with Nikon eclipse 90i microscope using plan APO VC 60× 1.40 N.A. objective and with a Leica TCS SPE confocal system equipped with a Leica DM5500Q microscope using PL APO 63× 1.4 N.A. objective. Leica TCS SPE confocal images were acquired with 0.50-µm voxel size either with 0.10 × 0.10 or 0.07 × 0.07 µm2 pixel size. Nikon EZ-C1 confocal images were acquired with 0.75-µm voxel and 0.14 × 0.14 µm2 pixel size. For coronal sections, confocal images were acquired of ∼7-µm z axis thickness using Nikon EZ-C1 software. Approximately 3-µm z axis thick confocal images were acquired for en face preparations using Leica LAS AF software. All images for comparative analysis were acquired with the same laser, gain, offset, pinhole, and zoom settings of confocal acquisitions. Confocal images were processed using the ImagJ image processing package Fiji (42). Z stacks were collapsed using the “z-project” tool with “average intensity” option. Counting of OR-positive puncta was done using the “cell counter” tool after “maximum intensity” z-stack projection. Colocalization was determined for OR-positive puncta >0.75 µm. X-Y-Z images and 3D rotation images were generated using Vaa3D software version 3.20 (43). Cilia length and fluorescence intensity measurements from en face preparations were done using the “plot profile” tool within Fiji. Fluorescence intensity was measured from maximum intensity projection of z axis according to Dummer et al. (44). Heat maps were prepared using R version 3.2.3 with package “gplots” function “heatmap.2.” For the analysis of average intensity profiles of ciliary ends from cilia with different lengths, all cilia were aligned and mean intensity fluorescence and SE was measured up to 15 µm of the proximal part of the cilia. Cilia were subsequently aligned from distal end and average intensity and SE was measured up to 15 µm of the distal end. The measurements of immunofluorescence corresponding to S100A5, AC3, and CNGA2 expression were done with Leica 63× objective with ∼1.7 zoom. The x and y axes dimensions of acquired images were ∼105 µm. Fluorescence intensity was measured after maximum projection of z stack with Fiji. The olfactory epithelium from right and left nasal cavities of the same individual were analyzed with anti-S100A5 and anti-AC3 antibodies, respectively. Images were assembled using Inkscape version 0.91.
Numbers of OR-positive cells and anti-activated caspase-3/OMP double immunopositive OSNs were determined from six coronal tissue sections at five positions with intervals of 140 μm throughout the rostrocaudal extent of the nasal cavity. Three pairs of mice for both the DMSO/vismodegib comparison and the Smo+/+/Smofl/+ comparison were analyzed.
Statistics.
Data were expressed as mean ± SEM or median (range). Student’s unpaired t test was used. If P values were >0.01, differences were noted nonsignificant (n.s.). Microsoft Office Excel 2013 was used for statistical analysis. In each experimental group, three or more mice were analyzed. For OSNs and cilia comparison, sample size was >50. No statistical method was used to determine the sample size. Sample sizes were in accordance to standard in field. No randomization was used to collect data. None of the data points were excluded from the study. For microscopy acquisition and analysis most of the time the user was blind.
Data Availability.
All data generated or analyzed during this study are included in this published article (and its Supporting Information).
Supplementary Material
Acknowledgments
We thank Dr. Gilad Barnea (Brown University) for anti-M71/M72 and anti-Mor28 antibodies; Dr. Claus W. Heizmann (University of Zürich) for the anti-S100A5 antibody; Drs. Katarina Kågedal, Karin Öllinger, and Richard Lundmark for reagents; and Drs. Sara Wilson and Anna Berghard for critical reading of the manuscript. This work was supported by the Swedish Natural Science Research Council (project no. 2016-05208). D.K.M. was supported by the Carl Tryggers Foundation and the Kempe Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1708321114/-/DCSupplemental.
References
- 1.Ingham PW, Nakano Y, Seger C. Mechanisms and functions of hedgehog signalling across the metazoa. Nat Rev Genet. 2011;12:393–406. doi: 10.1038/nrg2984. [DOI] [PubMed] [Google Scholar]
- 2.Teperino R, et al. Hedgehog partial agonism drives Warburg-like metabolism in muscle and brown fat. Cell. 2012;151:414–426. doi: 10.1016/j.cell.2012.09.021. [DOI] [PubMed] [Google Scholar]
- 3.Yam PT, Langlois SD, Morin S, Charron F. Sonic hedgehog guides axons through a noncanonical, Src-family-kinase-dependent signaling pathway. Neuron. 2009;62:349–362. doi: 10.1016/j.neuron.2009.03.022. [DOI] [PubMed] [Google Scholar]
- 4.Yao PJ, Petralia RS, Mattson MP. Sonic hedgehog signaling and hippocampal neuroplasticity. Trends Neurosci. 2016;39:840–850. doi: 10.1016/j.tins.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuzhandaivel A, Schultz SW, Alkhori L, Alenius M. Cilia-mediated hedgehog signaling in Drosophila. Cell Reports. 2014;7:672–680. doi: 10.1016/j.celrep.2014.03.052. [DOI] [PubMed] [Google Scholar]
- 6.Sanchez GM, et al. Hedgehog signaling regulates the ciliary transport of odorant receptors in Drosophila. Cell Rep. 2016;14:464–470. doi: 10.1016/j.celrep.2015.12.059. [DOI] [PubMed] [Google Scholar]
- 7.McIntyre JC, Hege MM, Berbari NF. Trafficking of ciliary G protein-coupled receptors. Methods Cell Biol. 2016;132:35–54. doi: 10.1016/bs.mcb.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 8.Henkin RI, Abdelmeguid M, Knöppel AB. On the mechanism of smell loss in patients with Type II congenital hyposmia. Am J Otolaryngol. 2016;37:436–441. doi: 10.1016/j.amjoto.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 9.Demirci H, Worden F, Nelson CC, Elner VM, Kahana A. Efficacy of vismodegib (Erivedge) for basal cell carcinoma involving the orbit and periocular area. Ophthal Plast Reconstr Surg. 2015;31:463–466. doi: 10.1097/IOP.0000000000000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gong Q, Chen H, Farbman AI. Olfactory sensory axon growth and branching is influenced by sonic hedgehog. Dev Dyn. 2009;238:1768–1776. doi: 10.1002/dvdy.22005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Persson L, et al. Shh-proteoglycan interactions regulate maturation of olfactory glomerular circuitry. Dev Neurobiol. 2014;74:1255–1267. doi: 10.1002/dneu.22202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saraiva LR, et al. Hierarchical deconstruction of mouse olfactory sensory neurons: From whole mucosa to single-cell RNA-seq. Sci Rep. 2015;5:18178. doi: 10.1038/srep18178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alenius M, Bohm S. Differential function of RNCAM isoforms in precise target selection of olfactory sensory neurons. Development. 2003;130:917–927. doi: 10.1242/dev.00317. [DOI] [PubMed] [Google Scholar]
- 14.Login H, Håglin S, Berghard A, Bohm S. The stimulus-dependent gradient of Cyp26B1+ olfactory sensory neurons is necessary for the functional integrity of the olfactory sensory map. J Neurosci. 2015;35:13807–13818. doi: 10.1523/JNEUROSCI.2247-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Long F, Zhang XM, Karp S, Yang Y, McMahon AP. Genetic manipulation of hedgehog signaling in the endochondral skeleton reveals a direct role in the regulation of chondrocyte proliferation. Development. 2001;128:5099–5108. doi: 10.1242/dev.128.24.5099. [DOI] [PubMed] [Google Scholar]
- 16.Belluscio L, Gold GH, Nemes A, Axel R. Mice deficient in G(olf) are anosmic. Neuron. 1998;20:69–81. doi: 10.1016/s0896-6273(00)80435-3. [DOI] [PubMed] [Google Scholar]
- 17.Brunet LJ, Gold GH, Ngai J. General anosmia caused by a targeted disruption of the mouse olfactory cyclic nucleotide-gated cation channel. Neuron. 1996;17:681–693. doi: 10.1016/s0896-6273(00)80200-7. [DOI] [PubMed] [Google Scholar]
- 18.Baker H, Grillo M, Margolis FL. Biochemical and immunocytochemical characterization of olfactory marker protein in the rodent central nervous system. J Comp Neurol. 1989;285:246–261. doi: 10.1002/cne.902850207. [DOI] [PubMed] [Google Scholar]
- 19.Kang N, et al. Olfactory marker protein expression is an indicator of olfactory receptor-associated events in non-olfactory tissues. PLoS One. 2015;10:e0116097. doi: 10.1371/journal.pone.0116097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lomvardas S, et al. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 21.Wong H, et al. Pharmacokinetic-pharmacodynamic analysis of vismodegib in preclinical models of mutational and ligand-dependent hedgehog pathway activation. Clin Cancer Res. 2011;17:4682–4692. doi: 10.1158/1078-0432.CCR-11-0975. [DOI] [PubMed] [Google Scholar]
- 22.McClintock TS, et al. In vivo identification of eugenol-responsive and muscone-responsive mouse odorant receptors. J Neurosci. 2014;34:15669–15678. doi: 10.1523/JNEUROSCI.3625-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McIntyre JC, et al. NISC Comparative Sequencing Program Gene therapy rescues cilia defects and restores olfactory function in a mammalian ciliopathy model. Nat Med. 2012;18:1423–1428. doi: 10.1038/nm.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strotmann J, Levai O, Fleischer J, Schwarzenbacher K, Breer H. Olfactory receptor proteins in axonal processes of chemosensory neurons. J Neurosci. 2004;24:7754–7761. doi: 10.1523/JNEUROSCI.2588-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li G, Marlin MC. Rab family of GTPases. Methods Mol Biol. 2015;1298:1–15. doi: 10.1007/978-1-4939-2569-8_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iwanami N, Nakamura Y, Satoh T, Liu Z, Satoh AK. Rab6 is required for multiple apical transport pathways but not the basolateral transport pathway in Drosophila photoreceptors. PLoS Genet. 2016;12:e1005828. doi: 10.1371/journal.pgen.1005828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vosshall LB, Hansson BS. A unified nomenclature system for the insect olfactory coreceptor. Chem Senses. 2011;36:497–498. doi: 10.1093/chemse/bjr022. [DOI] [PubMed] [Google Scholar]
- 28.Henkin RI, Hosein S, Stateman WA, Knöppel AB, Abdelmeguid M. Improved smell function with increased nasal mucus sonic hedgehog in hyposmic patients after treatment with oral theophylline. Am J Otolaryngol. 2017;38:143–147. doi: 10.1016/j.amjoto.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 29.Mashukova A, Spehr M, Hatt H, Neuhaus EM. Beta-arrestin2-mediated internalization of mammalian odorant receptors. J Neurosci. 2006;26:9902–9912. doi: 10.1523/JNEUROSCI.2897-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhandawat V, Reisert J, Yau KW. Elementary response of olfactory receptor neurons to odorants. Science. 2005;308:1931–1934. doi: 10.1126/science.1109886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Castillo K, Restrepo D, Bacigalupo J. Cellular and molecular Ca2+ microdomains in olfactory cilia support low signaling amplification of odor transduction. Eur J Neurosci. 2010;32:932–938. doi: 10.1111/j.1460-9568.2010.07393.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benton R, Sachse S, Michnick SW, Vosshall LB. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 2006;4:e20. doi: 10.1371/journal.pbio.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watson GM, Hessinger DA. Cnidocyte mechanoreceptors are tuned to the movements of swimming prey by chemoreceptors. Science. 1989;243:1589–1591. doi: 10.1126/science.2564698. [DOI] [PubMed] [Google Scholar]
- 34.Churcher AM, Taylor JS. The antiquity of chordate odorant receptors is revealed by the discovery of orthologs in the cnidarian Nematostella vectensis. Genome Biol Evol. 2011;3:36–43. doi: 10.1093/gbe/evq079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matus DQ, Magie CR, Pang K, Martindale MQ, Thomsen GH. The hedgehog gene family of the cnidarian, Nematostella vectensis, and implications for understanding metazoan hedgehog pathway evolution. Dev Biol. 2008;313:501–518. doi: 10.1016/j.ydbio.2007.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fleischer J, Breer H, Strotmann J. Mammalian olfactory receptors. Front Cell Neurosci. 2009;3:9. doi: 10.3389/neuro.03.009.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell. 2009;136:149–162. doi: 10.1016/j.cell.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hupe M, Li MX, Gertow Gillner K, Adams RH, Stenman JM. Evaluation of TRAP-sequencing technology with a versatile conditional mouse model. Nucleic Acids Res. 2014;42:e14. doi: 10.1093/nar/gkt995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schäfer BW, et al. Brain S100A5 is a novel calcium-, zinc-, and copper ion-binding protein of the EF-hand superfamily. J Biol Chem. 2000;275:30623–30630. doi: 10.1074/jbc.M002260200. [DOI] [PubMed] [Google Scholar]
- 40.Katic J, Loers G, Tosic J, Schachner M, Kleene R. The cell adhesion molecule CHL1 interacts with patched-1 to regulate apoptosis during postnatal cerebellar development. J Cell Sci. 2017;130:2606–2619. doi: 10.1242/jcs.194563. [DOI] [PubMed] [Google Scholar]
- 41.Challis RC, et al. An olfactory cilia pattern in the mammalian nose ensures high sensitivity to odors. Curr Biol. 2015;25:2503–2512. doi: 10.1016/j.cub.2015.07.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schindelin J, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peng H, Bria A, Zhou Z, Iannello G, Long F. Extensible visualization and analysis for multidimensional images using Vaa3D. Nat Protoc. 2014;9:193–208. doi: 10.1038/nprot.2014.011. [DOI] [PubMed] [Google Scholar]
- 44.Dummer A, Poelma C, DeRuiter MC, Goumans MJ, Hierck BP. Measuring the primary cilium length: Improved method for unbiased high-throughput analysis. Cilia. 2016;5:7. doi: 10.1186/s13630-016-0028-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Noda M, Harada Y. Development of olfactory epithelium in the mouse: Scanning electron-microscopy. Biomed Res. 1981;2:449–454. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its Supporting Information).