Significance
We sequenced the genome and transcriptomes of the wild olive (oleaster). More than 50,000 genes were predicted, and evidence was found for two relatively recent whole-genome duplication events, dated at approximately 28 and 59 Mya. Whole-genome sequencing, as well as gene expression studies, provide further insights into the evolution of oil biosynthesis, and will aid future studies aimed at further increasing the production of olive oil, which is a key ingredient of the healthy Mediterranean diet and has been granted a qualified health claim by the US Food and Drug Administration.
Keywords: oil crop, whole-genome duplication, siRNA regulation, fatty-acid biosynthesis, polyunsaturated fatty-acid pathway
Abstract
Here we present the genome sequence and annotation of the wild olive tree (Olea europaea var. sylvestris), called oleaster, which is considered an ancestor of cultivated olive trees. More than 50,000 protein-coding genes were predicted, a majority of which could be anchored to 23 pseudochromosomes obtained through a newly constructed genetic map. The oleaster genome contains signatures of two Oleaceae lineage-specific paleopolyploidy events, dated at ∼28 and ∼59 Mya. These events contributed to the expansion and neofunctionalization of genes and gene families that play important roles in oil biosynthesis. The functional divergence of oil biosynthesis pathway genes, such as FAD2, SACPD, EAR, and ACPTE, following duplication, has been responsible for the differential accumulation of oleic and linoleic acids produced in olive compared with sesame, a closely related oil crop. Duplicated oleaster FAD2 genes are regulated by an siRNA derived from a transposable element-rich region, leading to suppressed levels of FAD2 gene expression. Additionally, neofunctionalization of members of the SACPD gene family has led to increased expression of SACPD2, 3, 5, and 7, consequently resulting in an increased desaturation of steric acid. Taken together, decreased FAD2 expression and increased SACPD expression likely explain the accumulation of exceptionally high levels of oleic acid in olive. The oleaster genome thus provides important insights into the evolution of oil biosynthesis and will be a valuable resource for oil crop genomics.
As a symbol of peace, fertility, health, and longevity, the olive tree (Olea europaea L.) is a socioeconomically important oil crop that is widely grown in the Mediterranean Basin. Belonging to the Oleaceae family (order Lamiales), it can biosynthesize essential unsaturated fatty acids and other important secondary metabolites, such as vitamins and phenolic compounds (1). The olive tree is a diploid (2n = 46) allogamous crop that can be vegetatively propagated and live for thousands of years (2). Paleobotanical evidence suggests that olive oil was already produced in the Bronze Age (3). It has been thought that cultivated varieties were derived from the wild olive tree, called oleaster (O. europaea var. sylvestris), in Asia Minor, which then spread to Greece (4). Nevertheless, the exact domestication history of the olive tree is unknown (5). Because of their longevity, oleaster trees might even be related to Neolithic olive tree ancestors (2). Although the natural long generation time of olive trees has traditionally hindered breeding in this species, there are a few breeding programs involving sexual crosses that have generated interesting varieties for novel uses, like “Chiquitita,” specifically selected for high-density hedgerow orchards (6).
The olive is tightly associated with the Mediterranean cuisine. However, its consumption also spread to America (United States, Mexico, Brazil, Argentina, and Peru), Asia (China and India), and Australia. Aside from cultural reasons, this expansion was mainly because of the recognition of the beneficial dietetic properties of olive oil as a source of healthy fatty acids and micronutrients (e.g., antioxidants such as phenolic compounds, including vitamin E). In fact, olive oil has been granted a qualified health claim as reducing the incidence of cardiovascular disease (i.e., coronary heart disease) (7) by the US Food and Drug Administration (FDA; docket no. 2003Q-0559). As such, it represents the third FDA-approved claim for conventional foods, after nuts and omega-3 fatty acids. Moreover, olive tree products and byproducts are also used for pharmaceutical and cosmetic purposes.
Traditionally, olive oil is obtained by pressing olive fruits. Olive fruits consist of 20–30% (wt/wt) oil, 17% cellulose, 4% carbohydrates, 2% protein, and 0.1% micronutrients (1), with the rest (46.9–56.9%) being water. Polyols (mannitol) and oligosaccharides (raffinose and stachyose) are synthesized in olive tree leaves, being further exported with sucrose into the fruits, for general metabolism and as precursors of olive oil biosynthesis (8). Starting from a carbon source such as sucrose, long-chain fatty acids are synthesized, modified, and degraded by the activity of enzymes, including fatty-acid synthases, elongases, desaturases, and carboxylases (9). Fatty acids are the major constituent of triacylglycerols (TAGs). In olive oil, TAGs are mostly composed of monounsaturated oleic acid (C18:1; ∼75% of all TAGs), followed by saturated palmitic acid (C16; ∼13.5%), polyunsaturated linoleic acid (c18:2 ω-6; ∼5.5%), and α-linolenic acid (c18:3ω-3; ∼0.75%) (10).
Results
Assembly of the Oleaster Genome.
The wild olive tree genome was shotgun-sequenced (220× coverage), generating 515.7 Gbp of data (SI Appendix, Table S1). SOAPdenovo (11) was used to assemble the sequence reads, which resulted in a draft genome assembly of 1.48 Gbp, with the scaffold shortest sequence length at 50% of the genome of 228 kbp (SI Appendix, Table S3), which is in agreement with genome size estimations from flow cytometry (SI Appendix, Fig. S1) and k-mer analysis (∼1.46 Gbp; SI Appendix, Fig. S2A and Table S2). By using a newly constructed genetic map, 50% of sequences longer than 1 kbp (∼572 Mbp) could be anchored into 23 linkage groups (Fig. 1 and Tables 1 and 2).
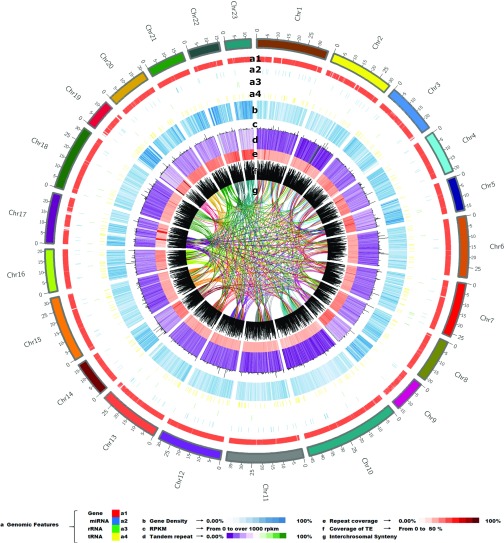
Fig. 1.
The genomic landscape of oleaster. The outer layer represents the karyotype ideogram (colored blocks), with minor and major tick marks labeling each 5 Mbp and 25 Mbp, respectively. Genome features across the 23 chromosomes (distinct characters shown as different colors, as indicated in the legend). Gene density per megabase pair. Gene expression patterns in average RPKM (range of RPKM values plotted from 0 to >1,000). Tandem duplication density per megabase pair. Percentage heat map of repeat coverage per megabase pair. Percentage of TEs per megabase pair (ranges of values plotted from 0 to >50). Inner circular representation shows interchromosomal synteny.
Table 1.
Statistics of the wild olive tree genome and assembly
| Features | Statistics |
| Genome | |
| Size (n, Gbp) | 1.48 |
| Karyotype (chromosomes, 2n) | 46 = 2n |
| GC content, % (with/without Ns) | 36.8/38.8 |
| High-copy repeat no. | |
| No. LTR/Gypsy and Copia | 1,182,454 |
| No. LINE | 43,834 |
| No. DNA TE | 219,901 |
| No. unknown | 42,630 |
| Gene | 50,684 |
| Assembly | |
| No. scaffold >100 bp/>1 kbp | 2,356,597/42,843 |
| N50 > 100 bp/>1 kbp | 228.62/364.6 |
N50, shortest sequence length at 50% of the genome assembly.
Table 2.
Statistics of wild olive tree genome annotation
| Annotation | No. | Total size, Kbp | Size, bp | ||
| Average | Maximum | Minimum | |||
| mRNA | 50,684 | 65,933.6 | 1,300.9 | 48,863 | 99 |
| CDS | 50,684 | 52,756.9 | 1,040.9 | 16,602 | 99 |
| Exon | 235,149 | 65,933.6 | 223.4 | 7,913 | 1 |
| Intron | 184,465 | 87,396.5 | 473.8 | 42,191 | 10 |
| miRNA | 411 | 49.979 | 113.33 | 24 | 21 |
| tRNA | 798 | 59.716 | 74.83 | 95 | 63 |
| rRNA | 773 | 121.906 | 121 | 1,804 | 29 |
| snRNA | 422 | 47.737 | 113 | 217 | 62 |
| Tandem repeat | 454,960 | 372,874.8 | 819.57 | 500,000 | 25 |
| TE protein | 428,172 | 23,958.1 | 559.54 | 5,505 | 24 |
| Transposon | 320,201 | 150,867.9 | 471.16 | 5,928 | 11 |
| 5′-UTR | 15,172 | 8,002.1 | 527.42 | 38,088 | 5 |
| 3′-UTR | 15,075 | 7,337 | 486.7 | 47,263 | 5 |
Genome Annotation.
The annotation of the oleaster genome was carried out by combining three different approaches, namely ab initio prediction, homology-based prediction, and transcriptome mapping (Fig. 1 and Tables 1 and 2). Approximately 51% of the genome assembly was found to be composed of repetitive DNA (Fig. 1), which is less than what was found for the draft genome of a recently published cultivated olive tree (63%) (12). Genome comparisons between oleaster and nine other plant species showed differences in gene numbers, transcript lengths, and proportions of transposable elements (TEs; SI Appendix, Table S5B). TEs and interspersed repeats occupied ∼43% of the genome (Tables 1 and 2 and SI Appendix, Table S7). LTRs were the most abundant type of TE (40.3% of genome), which is in agreement with a previous analysis of a cultivated olive tree (38.8% of genome) (13), followed by DNA-type TEs (4.6%; SI Appendix, Table S7). A total of 50,684 protein-coding genes were predicted on the current assembly, of which 47,124 genes (93%) were confirmed by RNA sequencing (RNA-seq) data. Further, 31,245 genes were located on the anchored pseudochromosomes (Fig. 1 and SI Appendix, Fig. S6 and Tables S8 and S9).
Approximately 90 million small RNA (sRNA) reads from six different tissues were used for noncoding RNA (ncRNA) annotation (SI Appendix, Figs. S8 and S9 and Tables S10 and S11). A total of 498 conserved miRNA families and 125 novel miRNAs were identified. Considering highly conserved miRNAs and their function, 29,842 miRNA–target pairs, including 7,849 unique target genes, were predicted. Totals of 4,606, 1,937, and 630 miRNA targets were associated with transcription factors, stress-response genes, and metabolism genes, respectively (SI Appendix, Table S12).
Oleaster protein-coding genes were functionally characterized through Gene Ontology and Kyoto Encyclopedia of Genes and Genomes (KEGG), which allowed annotation of 72.42% and 50.14% of all genes, respectively (SI Appendix, Table S13). KEGG metabolic pathway annotations of oleaster and 11 other plant species, including other oil crops such as Sesamum indicum (sesame) and Glycine max (soybean), as well as Populus trichocarpa (poplar) as a reference tree genome, Utricularia gibba (bladderwort) and Mimulus guttatus (monkey flower) as close relatives within the Lamiales, and Fraxinus excelsior (European ash tree) as a member of the Oleaceae family, showed a majority of oleaster genes to be involved in folding, sorting, and degradation (n = 4,263); biosynthesis of secondary metabolites (n = 2,236); carbohydrate metabolism (n = 1,905); and lipid metabolism (n = 811). Protein clustering of predicted oleaster genes with genes of other sequenced plant species resulted in 17,208 gene families, 1,070 of which were oleaster-specific and 7,522 were shared with the Lamiales F. excelsior, S. indicum, M. guttatus, and U. gibba. Although the number of gene families is largely consistent across the different species, the oleaster genome contains a large number (n = 8,986) of unique genes (SI Appendix, Fig. S11 and Table S14).
Genome Evolution.
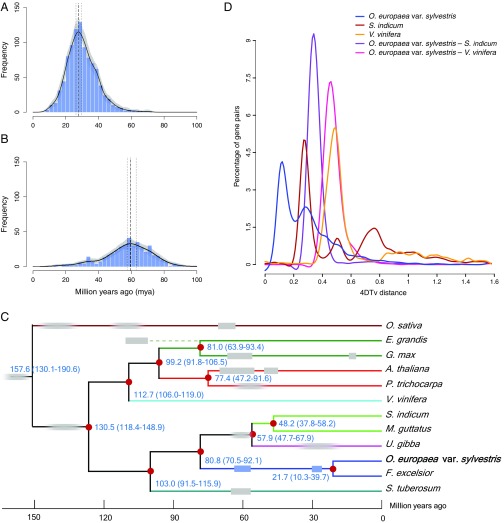
The oleaster genome contains multiple signatures of paleopolyploidy events. Distributions of synonymous substitutions per synonymous site (KS) for the whole paranome (the set of all duplicated genes in the genome; SI Appendix, Fig. S12A) and duplicates retained in colinear regions only (i.e., excluding duplicates from small-scale duplications; SI Appendix, Fig. S12B) consistently showed two clear peaks of duplicates at KS values around 0.25 and 0.75, respectively. Peaks at similar KS values have been reported for duplicated genes in the genome of European ash (F. excelsior, a sister to oleaster in Oleaceae) (14). Most likely, these peaks indicate two rounds of ancient whole-genome duplication (WGD) in the oleaster lineage (15) shared by olive and ash (14). To establish the age of these two WGDs, absolute phylogenomic dating (16) was carried out. Absolute dating suggests that the most recent WGD had occurred approximately 26–30 Mya (Fig. 2A) and the older one approximately 57–63 Mya (Fig. 2B). As with many other WGDs in different plant lineages, the latter event seems to have occurred close to the Cretaceous–Paleogene extinction event, providing additional evidence that WGDs—at least in plants—might be linked with periods of environmental change or upheaval (17).
Fig. 2.
Oleaster genome evolution. (A and B) Phylogenomic dating of O. europaea var. sylvestris paralogs. Absolute age distribution for the most recent WGD event (KS of approximately 0.25; SI Appendix, Fig. S12A), with a consensus WGD age estimate of 28 Mya and 90% CI of 26–30 Mya (A). Absolute age distribution for the older WGD event (KS of approximately 0.75; SI Appendix, Fig. S12B), with a consensus WGD age estimate of 59 Mya and 90% CI of 57–63 Mya (B). The solid black line represents the KDE of dated paralogs, and the vertical dashed black line corresponds to its peak, which was used as the consensus WGD age estimate. Gray lines represent density estimates from 2,500 bootstrap replicates, whereas vertical black dotted lines indicate the corresponding 90% CI for the WGD age estimate. Blue histogram shows the raw distribution of dated paralogs. (C) Estimation of divergence time. Blue numbers on the nodes are divergence time to present (in Mya). The two Oleaceae WGDs are indicated on the tree (blue rectangles), as are other known WGDs described in the literature for the species shown (gray rectangles; faded rectangles indicate that an absolute date has not been estimated). Note discussion of phylogenetic relationships in SI Appendix, S.3.2. (D) Fourfold degenerate (i.e., 4DTv) distributions for S. indicum, V. vinifera, and O. europaea var. sylvestris. Abscissa and ordinate represent 4DTv distance [using the HKY85 (Hasegawa–Kishino–Yano–1985) model] and percentage of homologous gene pairs, respectively.
Paleopolyploidy events of similar age have been reported for other asterids in this period. Within the Solanales, a shared whole-genome triplication has been found in the lineage leading to Solanum tuberosum (potato) and Solanum lycopersicum (tomato), with an estimated age approximately 57–65 Mya, using methods similar to the ones used here (16). Within the Lamiales, multiple WGDs independent from the paleopolyploidy in the Solanales have been described: two or three in the lineage leading to U. gibba (one of which could be shared with M. guttatus) (18) and one in the lineage leading to S. indicum (estimated age similar to tomato) (19). This latter one and the oldest WGD in U. gibba could denote the same event, possibly even shared with the older WGD in the oleaster and ash lineage, or both could be independent ones, partly depending on their phylogenetic relationship (SI Appendix, S.3.2). Mean estimates for the divergence of oleaster from S. indicum are 69–74 Mya (20–22) or even older (23, 24) (Fig. 2C). Duplication and speciation events analyzed using fourfold synonymous third-codon transversion rates (4DTv) also showed that there were probably two WGDs in oleaster and one WGD in S. indicum, and that these likely occurred after their divergence (Fig. 2D). Thus, the aforementioned dates and 4DTv patterns suggest that both WGD events inferred from the oleaster genome (as well as from the ash one) are specific to Oleaceae and occurred independently of the WGD in the lineage leading to S. indicum, M. guttatus, and U. gibba (Fig. 2C; see also ref. 14). This seems further supported by a phylogenomic analysis of duplicates from the older oleaster WGD, in which a majority of trees supported an Oleaceae lineage-specific event (SI Appendix, S.3.4, Fig. S13, and Table S15). High colinearity among oleaster chromosomes forms additional evidence for WGDs. At least 78 duplicated homologous genomic segments, 12 of which are intrachromosomal, were identified in the oleaster genome. Among them, chromosomes 1 and 12 (4,743 genes), 7 and 14 (2,300 genes), and 6 and 21 (1,361 genes) are remarkably colinear (SI Appendix, Fig. S14 and Table S16).
Evolutionary Analysis of Oil Biosynthesis.
Olive oil is mainly composed of TAG formed by fatty acids (10). Here, genes involved in oil biosynthesis were annotated and grouped according to their sequence identity, pathway, and enzyme codes. KEGG pathway analysis of genes related to oil biosynthesis in oleaster and 11 other species showed that the oleaster genome has the highest fraction of pathways related to lipid metabolism and secondary metabolite biosynthesis. Among 308 described pathway annotations, some of them, such as Ca2+-transporting ATPase (K01537), acyl-CoA oxidase (K00232), and phosphatidylserine decarboxylase (K01613), are highly represented in the oleaster genome compared with others. To further compare the evolution of oil biosynthesis between oleaster and another major oil-bearing crop, oleaster and sesame genes were subjected to InParanoid ortholog analysis (25). Among 2,327 oil biosynthesis genes in oleaster, 2,025 seem to have homologs in sesame. After excluding outparalogs, 911 groups of orthologs could be built, with 1,232 inparalogs for olive tree and 1,171 inparalogs from sesame. Interestingly, 563 oil biosynthesis genes showed a strict one-to-one orthology between oleaster and sesame (despite independent WGD in oleaster and sesame), whereas the rest of the inparalogs (669 in oleaster and 608 in sesame) were the result of independent and lineage-specific duplication events (see Fig. 2 C and D). Furthermore, 94 of 267 genes (35%) were found to be unique to oleaster, in comparison with sesame, in terms of oil biosynthesis metabolic pathway annotation. Comparing orthologous genes between oleaster and sesame showed that a large proportion of genes required for oil biosynthesis have been maintained as duplicated genes in the oleaster genome (1,962 genes in 221 families). In contrast, only a small number of gene families (54 genes in 27 families) showed contraction in oleaster.
Fatty-acid biosynthesis is one of the major steps of complex oil biosynthesis (26). It includes elongation, degradation, and biosynthesis of unsaturated fatty acids and is carried out through the activity of a large number of genes encoding fatty acid synthases, elongases, desaturases, and carboxylases. Although the polyunsaturated fatty acid (PUFA) pathway is common in plants, and a considerable number of orthologous gene families (n = 911, as detailed above) are shared between oleaster and sesame, many important gene families involved in the oil biosynthesis pathway were found to be expanded in the oleaster genome compared with sesame (Fig. 3 and SI Appendix, Fig. S17). Besides the expansion of some oil biosynthesis gene families in the oleaster genome, the contraction of gene families encoding degrading/catabolic enzymes (such as dehydrogenases and hydrolases) may also be responsible for the differential fatty-acid accumulation in oleaster and sesame. For instance, the number of linoleic acid metabolism genes was found to be significantly smaller for oleaster (n = 20) than for sesame (n = 164).
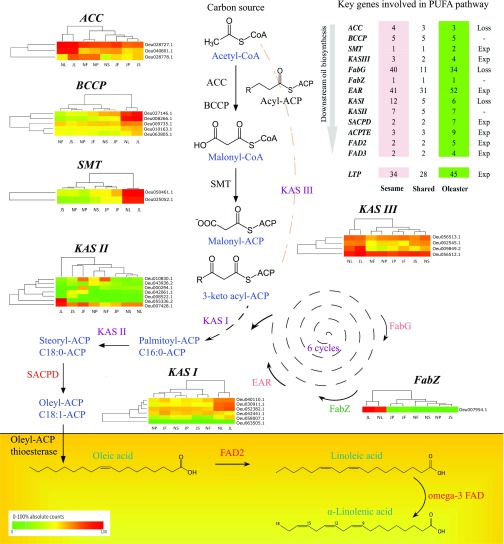
Fig. 3.
Oleic-acid biosynthesis pathway in oleaster. Genes involved in oleic-acid biosynthesis with their differential expression patterns in stem (marked as “S”), leaf (“L”), pedicel (“P”), and fruit ( “F”) tissues are shown. Heat-map data correspond to start (July; “J”) and end (November; “N”) time points for olive oil biosynthesis. The first step of such biosynthesis is catalyzed by Acetyl-CoA carboxylase (ACC), carboxylating Acetyl-CoA to form malonyl-CoA, which is converted to malonyl-acyl carrier protein (ACP) by S-malonyltransferase (SMT). Malonyl-ACP first reacts with 3-keto acyl-ACP, which is elongated by six reaction cycles in which chain-extender units are added. Then, fatty-acid synthases (FASs) act on that substrate to produce saturated fatty-acid 16-carbon palmitate, which will be desaturated to form unsaturated fatty acids, such as oleic acid in oleaster. ACPTE, ACP-hydrolase/thioesterase; BCCP, biotin carboxyl carrier protein; EAR, enoyl-ACP reductase; Exp, expanded; FabG, β-ketoacyl-ACP reductase; FabZ, β-hydroxyacyl-ACP dehydrase; FAD, fatty-acid desaturase; KAS, β-ketoacyl-ACP synthase; SACPD, stearoyl-ACP desaturase; SMT, S-malonyltransferase. Sesame expression data were retrieved from the Sesame Functional Genomics Database (SesameFG; www.sesame-bioinfo.org/SesameFG/).
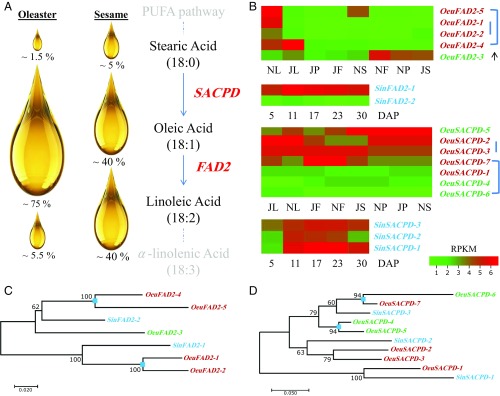
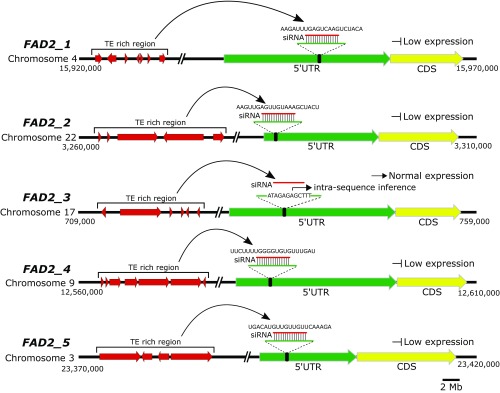
To explore functional divergence following duplication, expression analyses were performed in different tissues collected from ripe and unripe fruits. Interestingly, it was observed that the expression of duplicated oleaster fatty-acid desaturase (FAD2) genes (FAD2-1, FAD2-2, FAD2-4, and FAD2-5) was down-regulated in fruit tissues, especially during the lipid-accumulation ripening stage. Suppression of the expression of these genes causes reduced desaturation of oleic acid into linoleic acid (Fig. 4). FAD2 genes underwent at least two rounds of WGD events in oleaster, but only one duplication event in sesame (19) (Fig. 4 B–D). By mapping sRNA reads to 10-kbp regions encompassing the oleaster FAD2 genes (SI Appendix, Fig. S26), we discovered that an siRNA, which originated from a TE-rich region (27), may bind specifically to the 5′-UTR region of duplicated copies of the FAD2 gene transcripts, repressing expression in the fruit tissues. Because of the presence of an additional 12 nt at the siRNA-binding site, the FAD2-3 transcript, unlike the other FAD2 transcripts, may not be regulated by the activity of the siRNA in ripe fruit (Fig. 5 and SI Appendix, Fig. S27). The FAD2-3 gene is actively expressed in fruits and is responsible for the conversion of only a relatively low amount of oleic acid into linoleic acid (Fig. 4B). Sesame seeds also showed a differential expression pattern for FAD2 genes (FAD2-1 and FAD2-2), but with low diversity (FAD2, π = 0.0016), as reported previously (19). Consequently, silencing effects caused by siRNA on FAD2 olive gene transcripts (FAD2-1, -2, -4, and -5; Fig. 5), and the low diversity in FAD2 genes of sesame (19), are likely responsible for the higher accumulation of oleic acid in oleaster with respect to sesame.
Fig. 4.
Oleic-acid biosynthesis pathway in oleaster. (A) Oil content of oleaster and sesame with major genes involved in oil biosynthesis. (B) Heat-map analyses of oleaster and sesame FAD2 and SACPD genes. Blue lines indicate paralogs, which share orthologs with sesame. The arrow represents up-regulation of FAD2-3 gene, compared with other paralogs, in July unripe and November ripe fruits. Genes with green font color indicate unique genes in the wild olive tree, which have no orthologous counterpart in sesame, whereas red font color represents orthologous genes. Sesame genes are labeled with turquoise color. (C and D) Phylogenetic trees showing the duplication history of sesame and oleaster FAD2 and SACPD genes. Blue squares show duplicated genes after WGD and tandem duplications (SI Appendix, Fig. S28A). DAP, days after pollination.
Fig. 5.
Regulation of FAD2 gene expression by siRNA. Possible siRNA-binding sites are marked on 5′-UTRs. Interestingly, siRNA can bind to FAD2-1, FAD2-2, FAD2-4, and FAD2-5 transcripts but cannot bind to FAD2-3 transcripts because of the presence of 12 additional nucleotides in the binding site (SI Appendix, Fig. S27). Red lines show siRNA molecules. CDS, coding sequence.
Oleic acid as a major component of olive oil is formed by dehydrogenation from stearic acid by stearoyl-ACP desaturase (SACPD), after which it is desaturated into linoleic acid by FAD2 (7). Expression measurement of oleaster SACPD genes showed that SACPD1 and 2 have up-regulated expression in leaf tissues, whereas SACPD7 is highly expressed in fruit tissues. On the contrary, SACPD5 was found to be overexpressed in stem and pedicel tissues. Additionally, expression patterns of SACPD1, 5, and 6 were found at relatively low levels in other tissues (Fig. 4B).
It appears that the oleaster key genes involved in the PUFA pathway such as enoyl-ACP reductase (EAR), β-ketoacyl-ACP synthase II (KASII), β-ketoacyl-ACP reductase (FabG), acyl carrier protein (ACP)-hydrolase/thioesterase (ACPTE), SACPD, and FAD2 have been expanded by WGD and/or segmental duplications (SI Appendix, Figs. S28 and S29 and Table S17). Synteny analysis suggests that oleaster FAD2-1/-2 and SACPD6/7 paralogs have been duplicated through WGD (SI Appendix, Fig. S29A). Furthermore, EAR (52 genes), ACPTE (9 genes), FabG (34 genes), and KASII (7 genes) were shown to be expanded by WGD (SI Appendix, Figs. S28 and S29 B–E) and tandem and segmental duplications and now have different expression patterns (Figs. 3 and 4).
Discussion
To date, besides the wild olive tree, the sequencing and assembly of two cultivated olive tree genomes have been reported, namely O. europaea cv. Leccino (13) and O. europaea cv. Farga (12), at ∼4× and ∼150× coverage, respectively. The latter, with a size of 1.31 Gbp, was preliminary annotated solely by using RNA-seq data, which resulted in more than 56,000 protein-coding genes (12). Compared with the oleaster genome presented here, the cultivated olive tree has a smaller genome size, albeit with a higher number of genes. Unlike some previous reports on olive tree genome data, which lacked chromosome anchoring and genome-wide functional annotation (12, 13), our study includes a near-complete representation and localization of genes, repeat elements, and sRNA, as well as functional and metabolic annotations and an evolutionary analysis of oil biosynthesis genes.
Absolute dating of the two identified WGD events in oleaster and 4DTv patterns suggest that both WGDs, which seem to be shared with the ash tree, are specific to Oleaceae and independent from WGDs reported in other non-Oleaceaen Lamiales, including S. indicum (sesame; Fig. 2C). This is also consistent with synteny results from the ash tree genome (14). The age of the older WGD is close to the Cretaceous–Paleogene boundary. Additional Oleaceaen genomes will be required to determine which of the other lineages within Oleaceae, apart from ash, share either of the two WGDs, and whether one or both are related to patterns of diversification within the family (28).
The expansion of gene families and the functional divergence of genes playing important roles in oil biosynthesis may explain the higher accumulation of oleic acid (∼75% of olive oil) rather than linoleic acid (∼5.5% of olive oil) in oleaster (10). In sesame seed oil, both types of fatty acids are more evenly present (∼40%) with lower variation (±5%; Figs. 3 and 4A) (19, 29). As a result of gene expansion and loss events in oleaster with respect to the PUFA pathway genes responsible for the accumulation of oleic and linoleic acids, the fatty-acid content of olive oil greatly differs from that of sesame seed oil (10, 19) (Fig. 4A).
Here, consistent with a previous report (27), we also describe an siRNA sequence that originated from a TE-rich genomic region. To inhibit expression of duplicated copies of FAD2 gene transcripts, this regulatory siRNA may specifically bind to the 5′-UTR region of the transcripts in fruit tissues during the oil production period. In a previous study (30), it was reported that mutations associated with a duplication of the Oleate Desaturase (OD) gene caused its silencing by binding of an siRNA, further promoting accumulation of high levels of oleic acid in sunflower seeds. Similarly, suppression of FAD2 gene expression as a result of gene expansion probably leads to the high oleic acid content in oleaster.
Based on expression analysis, SACPD6/7 may have evolved through subfunctionalization or neofunctionalization events following their duplication (Fig. 4B). On the contrary, FAD2-1/-2 have probably retained similar functions, as their expression patterns have not changed (Fig. 4B). Compared with sesame, expansion of SACPD genes (SACPD1–7) in oleaster has likely led to increased desaturation activity and increased expression through neofunctionalization of SACPD2, 3, 5, and 7 in fruit and stem tissues (Fig. 4B). Thus, neofunctionalized SACPD gene copies in oleaster are likely also responsible for the differences in oleic and linoleic acid contents of olive and sesame (19, 30). Recently, it was observed that mutations in the soybean SACPD-C gene promote higher accumulation of leaf stearic acid content, as well as changes in leaf structure and morphology (31). Therefore, SACPD1 and 2, which are highly expressed in leaves, might be related to leaf morphology as well as oleic acid accumulation in fruit with overexpressed levels of SACPD7 (Fig. 4B).
Methods
A full description of the study methods is provided in the SI Appendix.
Plant Material.
A wild olive tree (2n = 46) was selected for whole-genome shotgun and transcriptome sequencing. Genomic DNA was isolated from leaf tissue (32).
Genome and Transcriptome Sequencing.
Sequencing libraries were prepared and sequenced on the Illumina HiSEq 2000 platform, followed by assembly with SOAPdenovo (11). Transcriptome libraries of four tissues including leaf, stem, pedicel, and fruit (ripe and unripe), collected from two different seasons, were also sequenced.
Genome Assembly.
All sequence reads were assembled with the SOAPdenovo software (11, 33) producing a reference sequence of the oleaster genome. A total of 319.39 Gbp of clean data were assembled into contigs and scaffolds by using the de Bruijn graph-based assembler of SOAPdenovo with the following four steps: (i) building contigs and scaffolds, (ii) filling gaps, (iii) removing redundancy, and (iv) reconstructing scaffolds.
Genetic Map Construction and Chromosome Anchoring.
DNA samples of each F1 individual and parents were digested with PstI-MseI restriction enzymes and then ligated with enzyme-compatible adapters. To increase the number of PstI-MseI fragments, PCR amplifications were performed as described (34). The DArTsEq (35) genotyping-by-sequencing (GBS) approach was used to identify SNPs. GBS data were analyzed by using a regression-mapping algorithm of JoinMap 4.0 software (Kyazma) to enable linkage-map construction. MapChart 2.0 (36) was used for the graphical presentation of linkage maps. Genetic linkage maps were constructed to develop the integrated genome map for anchoring the scaffolds by using 94 individuals from a cross-pollinated population of a cross between cultivars Memecik and Uslu. For chromosome-scale pseudomolecule construction, two maps were established from two progenies: both F1 progenies of 92 individuals (Memecik × Uslu). An integrated map including 1,307 markers was established (37) based on double heterozygous loci (38, 39). Genetic markers were mapped onto the scaffolds by using the Burrows–Wheeler Aligner software module for alignment (40) with default parameters. Afterward, anchoring of assembled scaffolds to genetic maps was achieved by applying the ALLMAPS software (41).
Repeat Element Analyses.
Homology-based and de novo approaches were used to find TEs in the oleaster genome. The homology-based approach involved applying commonly used databases of known repetitive sequences, along with programs such as RepeatProteinMask and RepeatMasker (42). RepeatModeler (www.repeatmasker.org/RepeatModeler.html) was used with two ab initio repeat-prediction programs (RECON and RepeatScout) to identify repeat-element boundaries and family relationships among sequences. Tandem repeats were also searched for in the genome by using Tandem Repeats Finder (43).
Gene Prediction.
Homology-based and de novo methods, as well as RNA-seq data, were used to predict genes in the O. europaea var. sylvestris genome. GLEAN (44) was used to consolidate results. Protein sequences of Arabidopsis thaliana, S. indicum, S. tuberosum, and Vitis vinifera were aligned with TBLASTN and genBLASTA (45) against the matching genomic sequence by using GeneWise (46) for accurate spliced alignments. Next, the de novo gene-prediction methods GlimmerHMM (47) (https://ccb.jhu.edu/software/glimmerhmm) and Augustus (48) were used to predict protein-coding genes, with parameters trained for O. europaea var. sylvestris, A. thaliana, S. indicum, S. tuberosum, and V. vinifera.
Genome Annotation.
Functional annotation was achieved by comparing predicted proteins against public databases, including UniProtor the UniProt Knowledgebase, KEGG, and InterPro. Results are available online at the Olive Genome Browser (olivegenome.org) and Online Resource for Community Annotation of Eukaryotes (ORCAE; bioinformatics.psb.ugent.be/orcae). Gene-family clustering was performed by OrthoMCL (49).
Evolutionary Analyses.
The GTR+gamma evolutionary model was applied to reconstruct a phylogenetic tree by using 231 single-copy orthologous genes from 12 different plant genomes. KS-based age distributions of oleaster were also constructed to unveil WGD events in oleaster (15). Absolute dating of two identified WGD events in the oleaster genome was performed as previously described (16) (SI Appendix, S.3). SyMAP (50) was used to identify synteny with other species (i.e., S. indicum, V. vinifera, P. trichocarpa, and S. tuberosum). Circos (51) was applied to generate a circular visualization of the oleaster genome features. InParanoid was used to identify orthologs and paralogs with sesame involved in the oil biosynthesis pathways. Additional information is provided in SI Appendix, S.3.
Availability of Data.
The oleaster genome assembly has been deposited in the National Center for Biotechnology Information (NCBI) GenBank database (https://www.ncbi.nlm.nih.gov/genbank; accession no. MSRW00000000; BioProject record ID PRJNA350614). Transcriptome datasets were deposited in the NCBI Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra; accession nos. SRR4473639, SRR4473641, SRR44742, SRR4473643, SRR4473644, SRR4473645, SRR4473646, and SRR4473647). The genome and annotation files were also uploaded into ORCAE (bioinformatics.psb.ugent.be/orcae), Phytozome (https://phytozome.jgi.doe.gov), and the olive genome consortium Web site (olivegenome.org).
Supplementary Material
Acknowledgments
This project was initiated in Cankiri Karatekin University and finalized in Dokuz Eylul University. The authors acknowledge funding from the Cankiri Karatekin University, Bilimsel Arastirma Projeleri Birimi (BAP) (Grant 2012-10, FF12035L19); Ankara University, BAP (Project 14B0447004); Mustafa Kemal University, BAP (Project 12022); Gaziosman Pasa University, BAP (Grant 2013/27); Turkish Academy of Sciences (Outstanding Young Scientists Award); Ministry of Food, Agriculture and Livestock of Turkey (Grant TAGEM/BBAD/12/A08/P06/3); Consejería de Agricultura y Pesci (Grants 041/C/2007, 75/C/2009, and 56/C/2010); Grupo del Plan Andaluz de Investigación (PAI) (Grant AGR-248) of Junta de Andalucía and Universidad de Córdoba (Ayuda a Grupos of Spain), Spain; the Multidisciplinary Research Partnership “Bioinformatics: From Nucleotides to Networks” (Project 01MR0310W) of Ghent University; and European Union Seventh Framework Program Grant FP7/2007-2013 under European Research Council Advanced Grant Agreement 322739–DOUBLEUP.
Footnotes
The authors declare no conflict of interest.
Data deposition: The oleaster genome assembly has been deposited in the GenBank database, https://www.ncbi.nlm.nih.gov/genbank (accession no. MSRW00000000; BioProject record ID PRJNA350614). Transcriptome datasets were deposited in the at National Center for Biotechnology Information Sequence Read Archive, https://www.ncbi.nlm.nih.gov/sra (accession nos. SRR4473639, SRR4473641, SRR44742, SRR4473643, SRR4473644, SRR4473645, SRR4473646, and SRR4473647). The genome and annotation files were uploaded to Online Resource for Community Annotation of Eukaryotes (ORCAE), bioinformatics.psb.ugent.be/orcae; Phytozome, https://phytozome.jgi.doe.gov; and the olive genome consortium Web site, olivegenome.org. .
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1708621114/-/DCSupplemental.
References
- 1.Tripoli E, et al. The phenolic compounds of olive oil: Structure, biological activity and beneficial effects on human health. Nutr Res Rev. 2005;18:98–112. doi: 10.1079/NRR200495. [DOI] [PubMed] [Google Scholar]
- 2.Lumaret R, Ouazzani N. Plant genetics. Ancient wild olives in Mediterranean forests. Nature. 2001;413:700. doi: 10.1038/35099680. [DOI] [PubMed] [Google Scholar]
- 3.Riley FR. Olive oil production on bronze age Crete: nutritional properties, processing methods and storage life of Minoan olive oil. Oxf J Archaeol. 2002;21:63–75. [Google Scholar]
- 4.de Candolle A. Origine des Plantes Cultivées. Librairie Germer Baillière et Cie; Paris: 1883. [Google Scholar]
- 5.Diez CM, et al. Olive domestication and diversification in the Mediterranean Basin. New Phytol. 2015;206:436–447. doi: 10.1111/nph.13181. [DOI] [PubMed] [Google Scholar]
- 6.Rallo L, Barranco D, de la Rosa R, León L. ‘Chiquitita’olive. HortScience. 2008;43:529–531. [Google Scholar]
- 7.Estruch R, et al. PREDIMED Study Investigators Primary prevention of cardiovascular disease with a Mediterranean diet. N Engl J Med. 2013;368:1279–1290. doi: 10.1056/NEJMoa1200303. [DOI] [PubMed] [Google Scholar]
- 8.Conde C, Delrot S, Gerós H. Physiological, biochemical and molecular changes occurring during olive development and ripening. J Plant Physiol. 2008;165:1545–1562. doi: 10.1016/j.jplph.2008.04.018. [DOI] [PubMed] [Google Scholar]
- 9.Bates PD, Stymne S, Ohlrogge J. Biochemical pathways in seed oil synthesis. Curr Opin Plant Biol. 2013;16:358–364. doi: 10.1016/j.pbi.2013.02.015. [DOI] [PubMed] [Google Scholar]
- 10.Rueda A, et al. Characterization of fatty acid profile of argan oil and other edible vegetable oils by gas chromatography and discriminant analysis. J Chem. 2014;2014:843908. [Google Scholar]
- 11.Li R, et al. The sequence and de novo assembly of the giant panda genome. Nature. 2010;463:311–317. doi: 10.1038/nature08696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cruz F, et al. Genome sequence of the olive tree, Olea europaea. Gigascience. 2016;5:29. doi: 10.1186/s13742-016-0134-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barghini E, et al. The peculiar landscape of repetitive sequences in the olive (Olea europaea L.) genome. Genome Biol Evol. 2014;6:776–791. doi: 10.1093/gbe/evu058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sollars ES, et al. Genome sequence and genetic diversity of European ash trees. Nature. 2017;541:212–216. doi: 10.1038/nature20786. [DOI] [PubMed] [Google Scholar]
- 15.Vanneste K, Van de Peer Y, Maere S. Inference of genome duplications from age distributions revisited. Mol Biol Evol. 2013;30:177–190. doi: 10.1093/molbev/mss214. [DOI] [PubMed] [Google Scholar]
- 16.Vanneste K, Baele G, Maere S, Van de Peer Y. Analysis of 41 plant genomes supports a wave of successful genome duplications in association with the Cretaceous-Paleogene boundary. Genome Res. 2014;24:1334–1347. doi: 10.1101/gr.168997.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van de Peer Y, Mizrachi E, Marchal K. The evolutionary significance of polyploidy. Nat Rev Genet. 2017;18:411–424. doi: 10.1038/nrg.2017.26. [DOI] [PubMed] [Google Scholar]
- 18.Ibarra-Laclette E, et al. Architecture and evolution of a minute plant genome. Nature. 2013;498:94–98. doi: 10.1038/nature12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L, et al. Genome sequencing of the high oil crop sesame provides insight into oil biosynthesis. Genome Biol. 2014;15:R39. doi: 10.1186/gb-2014-15-2-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bell CD, Soltis DE, Soltis PS. The age and diversification of the angiosperms re-revisited. Am J Bot. 2010;97:1296–1303. doi: 10.3732/ajb.0900346. [DOI] [PubMed] [Google Scholar]
- 21.Magallón S, Gómez-Acevedo S, Sánchez-Reyes LL, Hernández-Hernández T. A metacalibrated time-tree documents the early rise of flowering plant phylogenetic diversity. New Phytol. 2015;207:437–453. doi: 10.1111/nph.13264. [DOI] [PubMed] [Google Scholar]
- 22.Yi D-K, Kim K-J. Complete chloroplast genome sequences of important oilseed crop Sesamum indicum L. PLoS ONE. 2012;7:e35872. doi: 10.1371/journal.pone.0035872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bremer K, Friis EM, Bremer B. Molecular phylogenetic dating of asterid flowering plants shows early Cretaceous diversification. Syst Biol. 2004;53:496–505. doi: 10.1080/10635150490445913. [DOI] [PubMed] [Google Scholar]
- 24.Wikström N, Kainulainen K, Razafimandimbison SG, Smedmark JE, Bremer B. A revised time tree of the asterids: Establishing a temporal framework for evolutionary studies of the coffee family (Rubiaceae) PLoS One. 2015;10:e0126690, and erratum (2015) 11:e0157206. doi: 10.1371/journal.pone.0126690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Remm M, Storm CE, Sonnhammer EL. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J Mol Biol. 2001;314:1041–1052. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- 26.Harwood JL, Guschina IA. Regulation of lipid synthesis in oil crops. FEBS Lett. 2013;587:2079–2081. doi: 10.1016/j.febslet.2013.05.018. [DOI] [PubMed] [Google Scholar]
- 27.Kuang H, et al. Identification of miniature inverted-repeat transposable elements (MITEs) and biogenesis of their siRNAs in the Solanaceae: New functional implications for MITEs. Genome Res. 2009;19:42–56. doi: 10.1101/gr.078196.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Besnard G, Rubio de Casas R, Christin PA, Vargas P. Phylogenetics of Olea (Oleaceae) based on plastid and nuclear ribosomal DNA sequences: tertiary climatic shifts and lineage differentiation times. Ann Bot (Lond) 2009;104:143–160. doi: 10.1093/aob/mcp105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei WL, et al. Association analysis for quality traits in a diverse panel of Chinese sesame (Sesamum indicum L.) germplasm. J Integr Plant Biol. 2013;55:745–758. doi: 10.1111/jipb.12049. [DOI] [PubMed] [Google Scholar]
- 30.Lacombe S, Souyris I, Bervillé AJ. An insertion of oleate desaturase homologous sequence silences via siRNA the functional gene leading to high oleic acid content in sunflower seed oil. Mol Genet Genomics. 2009;281:43–54. doi: 10.1007/s00438-008-0391-9. [DOI] [PubMed] [Google Scholar]
- 31.Lakhssassi N, et al. Stearoyl-acyl carrier protein desaturase mutations uncover an impact of stearic acid in leaf and nodule structure. Plant Physiol. 2017;174:1531–1543. doi: 10.1104/pp.16.01929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sahu SK, Thangaraj M, Kathiresan K. DNA extraction protocol for plants with high levels of secondary metabolites and polysaccharides without using liquid nitrogen and phenol. ISRN Mol Biol. 2012;2012:205049. doi: 10.5402/2012/205049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li R, et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010;20:265–272. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raman H, et al. Genome-wide delineation of natural variation for pod shatter resistance in Brassica napus. PLoS One. 2014;9:e101673. doi: 10.1371/journal.pone.0101673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elshire RJ, et al. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One. 2011;6:e19379. doi: 10.1371/journal.pone.0019379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Voorrips RE. MapChart: Software for the graphical presentation of linkage maps and QTLs. J Hered. 2002;93:77–78. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- 37.Risterucci A, et al. A high-density linkage map of Theobroma cacao L. Theor Appl Genet. 2000;101:948–955. doi: 10.1007/BF00223910. [DOI] [PubMed] [Google Scholar]
- 38.Pugh T, et al. A new cacao linkage map based on codominant markers: Development and integration of 201 new microsatellite markers. Theor Appl Genet. 2004;108:1151–1161. doi: 10.1007/s00122-003-1533-4. [DOI] [PubMed] [Google Scholar]
- 39.Fouet O, et al. Structural characterization and mapping of functional EST-SSR markers in Theobroma cacao. Tree Genet Genomes. 2011;7:799–817. [Google Scholar]
- 40.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang H, et al. ALLMAPS: Robust scaffold ordering based on multiple maps. Genome Biol. 2015;16:3. doi: 10.1186/s13059-014-0573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tarailo‐Graovac M, Chen N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinformatics. 2009;25:4.10.1–4.10.14. doi: 10.1002/0471250953.bi0410s25. [DOI] [PubMed] [Google Scholar]
- 43.Benson G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elsik CG, et al. Creating a honey bee consensus gene set. Genome Biol. 2007;8:R13. doi: 10.1186/gb-2007-8-1-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.She R, Chu JS, Wang K, Pei J, Chen N. GenBlastA: Enabling BLAST to identify homologous gene sequences. Genome Res. 2009;19:143–149. doi: 10.1101/gr.082081.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Birney E, Clamp M, Durbin R. GeneWise and Genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Majoros WH, Pertea M, Salzberg SL. TigrScan and GlimmerHMM: Two open source ab initio eukaryotic gene-finders. Bioinformatics. 2004;20:2878–2879. doi: 10.1093/bioinformatics/bth315. [DOI] [PubMed] [Google Scholar]
- 48.Stanke M, et al. AUGUSTUS: Ab initio prediction of alternative transcripts. Nucleic Acids Res. 2006;34:W435–W439. doi: 10.1093/nar/gkl200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soderlund C, Bomhoff M, Nelson WM. SyMAP v3.4: A turnkey synteny system with application to plant genomes. Nucleic Acids Res. 2011;39:e68. doi: 10.1093/nar/gkr123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krzywinski M, et al. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.