Significance
We generated genomic data to estimate the population history of grapes, the most economically important horticultural crop in the world. Domesticated grapes experienced a protracted, 22,000-y population decline prior to domestication; we hypothesize that this decline reflects low-intensity cultivation by humans prior to domestication. Domestication altered the mating system of grapes. The sex determination region is detectable as a region of heightened genetic divergence between wild and cultivated accessions. Based on gene expression analyses, we propose candidate genes that alter sex determination. Finally, grapes contain more deleterious mutations in heterozygous states than do their wild ancestors. The accumulation of deleterious mutations is due in part to clonal propagation, which shelters deleterious recessive mutations.
Keywords: demography, sex determination, candidate genes, deleterious variants, clonal propagation
Abstract
We gathered genomic data from grapes (Vitis vinifera ssp. vinifera), a clonally propagated perennial crop, to address three ongoing mysteries about plant domestication. The first is the duration of domestication; archaeological evidence suggests that domestication occurs over millennia, but genetic evidence indicates that it can occur rapidly. We estimated that our wild and cultivated grape samples diverged ∼22,000 years ago and that the cultivated lineage experienced a steady decline in population size (Ne) thereafter. The long decline may reflect low-intensity management by humans before domestication. The second mystery is the identification of genes that contribute to domestication phenotypes. In cultivated grapes, we identified candidate-selected genes that function in sugar metabolism, flower development, and stress responses. In contrast, candidate-selected genes in the wild sample were limited to abiotic and biotic stress responses. A genomic region of high divergence corresponded to the sex determination region and included a candidate male sterility factor and additional genes with sex-specific expression. The third mystery concerns the cost of domestication. Annual crops accumulate putatively deleterious variants, in part due to strong domestication bottlenecks. The domestication of perennial crops differs from that of annuals in several ways, including the intensity of bottlenecks, and it is not yet clear if they accumulate deleterious variants. We found that grape accessions contained 5.2% more deleterious variants than wild individuals, and these were more often in a heterozygous state. Using forward simulations, we confirm that clonal propagation leads to the accumulation of recessive deleterious mutations but without decreasing fitness.
The study of crop domestication has long been used as a proxy for studying evolutionary processes, such as the genetic effects of bottlenecks (1) and the detection of selection to identify agronomically important loci (2–4). Several crops have been studied in this evolutionary context (5), but there are at least two emerging issues. The first is the speed at which domestication occurs. One view, supported primarily by archaeological evidence, is that domestication is a slow process that takes millennia (6–8). Another view, based on genetic evidence and population modeling (9, 10), argues that domestication occurs much more rapidly. The gap between these two views has been bridged, in part, by a recent study of African rice. The study used population genomic data to infer that a bottleneck occurred during domestication ∼3.5 kya and also that the bottleneck was preceded by a long, ∼14,000-y decline in the effective population size (Ne) of the progenitor population (11). The authors hypothesized that the protracted Ne decline reflects a period of low-intensity management and/or cultivation before modern domestication. While an intriguing hypothesis, it is not yet clear whether other crops also have demographic histories marked by protracted Ne declines.
The second emerging issue is the “cost of domestication” (12), which refers to an increased genetic load within cultivars. This cost originates partly from the fact that the decreased Ne during a domestication bottleneck reduces the efficacy of genome-wide selection (13), which may in turn increase the frequency and number of slightly deleterious variants (14, 15). The characterization of deleterious variants is important because they may be fitting targets for crop improvement (16). Consistent with a cost of domestication, annual crops are known to contain an increase in derived, putatively deleterious variants relative to their wild progenitors (17–20). However, it is not yet clear whether these deleterious variants increase genetic load and whether this phenomenon applies to perennial crops.
The distinction between annual and perennial crops is crucial because perennial domestication is expected to differ from annual domestication in at least three aspects (21, 22). The first is clonal propagation; many perennials are propagated clonally but most annuals are not. Clonal propagation maintains genetic diversity in desirous combinations but also limits opportunities for sexual recombination (20, 22). The second aspect is time. Long-lived perennials have extended juvenile stages. As a result, the number of sexual generations is much reduced for perennials relative to annual crops, even for perennials that were domesticated relatively early in human agricultural history. The third aspect is the severity of the domestication bottleneck. A meta-analysis has documented that perennial crops retain 95% of neutral variation from their progenitors, on average, while annuals retain an average of 60% (22). This observation suggests that many (and perhaps most) perennial crops have not experienced severe domestication bottlenecks; as a consequence, their domestication may not come with a cost.
Here we study the domestication history of the grapevine (Vitis vinifera ssp. vinifera), which is the most economically important horticultural crop in the world (23). Grapes (hereafter vinifera) have been a source of food and wine since their hypothesized domestication ∼8.0 kya from their wild progenitor, V. vinifera ssp. sylvestris (hereafter sylvestris) (24). The exact location of domestication remains uncertain, but most lines of evidence point to a primary domestication event in the Near East (23, 24). Domestication caused morphological shifts that include larger berry and bunch sizes, higher sugar content, altered seed morphology, and a shift from dioecy to a hermaphroditic mating system (25). There is interest in identifying the genes that contribute to these morphological shifts. For example, several papers have attempted to identify the gene(s) that are responsible for the shift to hermaphroditism, which were mapped to an ∼150-kb region on chromosome 2 (26, 27).
Historically, genetic diversity among V. vinifera varieties has been studied with simple sequence repeats (28). More recently, a group genotyped 950 vinifera and 59 sylvestris accessions with a chip containing 9,000 SNPs (23). Their data suggest that grape domestication led to a mild reduction of genetic diversity, indicating that grape is a reasonable perennial model for studying the accumulation of deleterious variation in the absence of a pronounced bottleneck. Still more recent studies have used whole-genome sequencing (WGS) to assess structural variation among grape varieties (29–31). Surprisingly, however, WGS data have not been used to investigate the population genomics of grapes. Here we perform WGS on a sample of vinifera cultivars and on putatively wild sylvestris accessions to focus on three sets of questions. First, what do the data reveal about the demographic history of cultivated grapes, specifically, the timing and severity of a domestication bottleneck? Second, what genes bear the signature of selection in vinifera, and do they provide insights into the agronomic shifts associated with domestication? Finally, do domesticated grapes have more derived, putatively deleterious variants relative to sylvestris, or have the unique features of perennial domestication permitted an escape from this potential cost?
Results
Plant Samples and Population Structure.
We collected WGS data from nine putatively wild sylvestris individuals from the Near East that represent a single genetic group (23), 18 vinifera individuals representing 14 cultivars, and one outgroup (Vitis rotundifolia) (SI Appendix, Table S1). Our sylvestris accessions are a subset of the wild sample from ref. 23, which was filtered for provenance and authenticity. We nonetheless label the sylvestris sample as “putatively wild,” because it can be difficult to identify truly wild individuals. Reads were mapped to the Pinot Noir reference genome PN40024 (32), resulting in the identification of 3,963,172 and 3,732,107 SNPs across the sylvestris and vinifera samples (Materials and Methods).
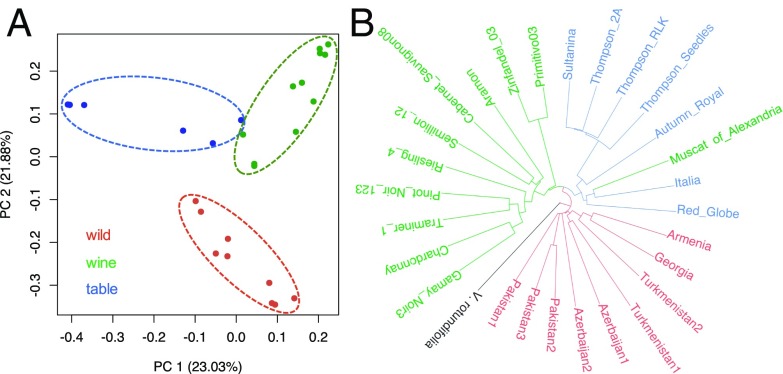
To investigate population structure, we applied principal component analysis (PCA) to genotype likelihoods (33). Only the first two principal components (PCs) were significant (P < 0.001); they explained 23.03% and 21.88% of the total genetic variance, respectively (Fig. 1A). PC1 separated samples of wine and table grapes, except for two accessions (Italia and Muscat of Alexandria) positioned between the two groups. PC2 divided wild and cultivated samples. Wine, table, and wild grapes clustered separately in a neighbor-joining tree, except for Muscat of Alexandria, which has been used historically for both wine and table grapes (Fig. 1B). Finally, STRUCTURE analyses revealed an optimal grouping of K = 4, which separated sylvestris accessions, table grapes, wine grapes, and the Zinfandel/Primitivo subgroup of wine grapes while also identifying admixed individuals (SI Appendix, Fig. S1).
Fig. 1.
Population structure of cultivated and wild samples. (A) PCA plot based on genetic covariance among all individuals of wild samples (red) and cultivars (green for wine grapes and blue for table grapes). (B) Neighbor-joining tree across all samples, rooted by V. rotundifolia.
Nucleotide Diversity and Demographic History.
We estimated population genetic parameters based on the sylvestris accessions (n = 9) and on a cultivated sample of n = 14 that included only one Thompson clone and one Zinfandel/Primitivo clone (SI Appendix, Table S1). Both samples harbored substantial levels of nucleotide diversity across all sites (sylvestris: πw = 0.0147 ± 0.0011; vinifera: πc = 0.0139 ± 0.0014; SI Appendix, Fig. S2). Although π was higher in sylvestris (πc/πw = 0.94 ± 0.14), vinifera had higher levels of heterozygosity and Tajima’s D values (vinifera, D = 0.5421 ± 0.0932; sylvestris, D = −0.4651 ± 0.1577; SI Appendix, Fig. S2). Linkage disequilibrium (LD) decayed to r2 < 0.2 within 20 kb in both samples, but it declined more slowly for vinifera after ∼20 kb (SI Appendix, Fig. S2).
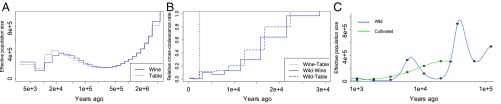
We inferred the demographic history of the vinifera sample using MSMC, a method that infers both population size and gene flow using phased SNPs (34). Assuming a generation time of 3 y (24) and a mutation rate of 2.5 × 10−9 mutations per nucleotide per year (35), we converted scaled population parameters into years and individuals (Ne). Based on these analyses, vinifera experienced a continual reduction of Ne starting ∼22.0 kya until its nadir from ∼7.0 kya to 11.0 kya (Fig. 2A), which corresponds to the time of domestication and implies a mild domestication bottleneck. Notably, there was no evidence for a dramatic expansion of Ne since domestication. MSMC results were similar across two separate analyses (Fig. 2A), based on n = 4 samples of either table or wine grapes (SI Appendix, Table S1), suggesting that analyses captured shared aspects of the samples’ histories. We also used MSMC to compute divergence times. The divergence between sylvestris and vinifera was estimated to be ∼22 kya (Fig. 2B), which corresponds to the onset of the decline of vinifera Ne. Divergence between wine and table grapes was estimated to be ∼2.5 kya, which is well within the hypothesized period of vinifera domestication (Fig. 2B).
Fig. 2.
Intra- and intertaxon analyses of demographic history and divergence. (A) MSMC estimates of the effective population size (Ne) of vinifera based on two separate runs of four individuals. The solid line represents wine grapes, and the dashed line is table grapes. (B) MSMC analysis of cross-coalescence (y axis) based on comparisons between sylvestris and either wine or table grapes. (C) Divergence time and past Ne changes inferred by the SMC++ analyses, based on unphased genotypes.
We repeated demographic analyses with SMC++, which estimates population histories and divergence without phasing (36) (Fig. 2C). This method yielded no evidence for a discrete bottleneck from ∼7.0 kya to 11.0 kya, but SMC++ and MSMC analyses had four similarities: (i) an estimated divergence time (∼30 kya) that greatly predates domestication; (ii) a slow decline in vinifera Ne since divergence; (iii) no evidence for a rapid expansion in Ne after domestication; and (iv) an ∼2.6-kya divergence of wine and table grapes (Fig. 2C and SI Appendix, Fig. S3). We also used SMC++ to infer the demographic history of our sylvestris sample, revealing a complex Ne pattern that corresponds to features of climatic history (Discussion).
Sweep Mapping.
We investigated patterns of selection and interspecific differentiation across the grape genome. All sweep analyses focused on sliding 20-kb windows, reflecting the genome-wide pattern of LD decline (SI Appendix, Fig. S2). Windows that scored in the top 0.5% were considered candidate sweep regions.
We began with CLR (37), which identifies potentially selected regions by detecting skews in the site frequency spectrum (sfs) within a single taxon, and XP-CLR (38), which detects sfs skews relative to a reference taxon (sylvestris). Within vinifera, CLR identified 117 20-kb windows encompassing 309 candidate-selected genes (SI Appendix, Table S2). Among those detected by CLR, nine functional categories were identified as significantly overrepresented (P ≤ 0.01), including the “alcohol dehydrogenase superfamily,” “monoterpenoid indole alkaloid biosynthesis,” and “flower development” (SI Appendix, Table S3). XP-CLR identified a similar number of genes (367); both tests identified genes involved in berry development and/or quality, including the SWEET1 gene (SI Appendix, Fig. S4), which encodes a bidirectional sugar transporter (39). SWEET1 was overexpressed in full-ripe berries compared with immature berries [adjusted (adj.) P = 9.4E-3; SI Appendix, Fig. S6], suggesting an involvement in sugar accumulation during berry ripening. Additional genes of interest detected by both tests included: (i) a leucoanthocyanidin dioxygenase (LDOX) gene (VIT_08s0105g00380) that peaks in expression at the end of veraison (adj. P = 8.9E-10; SI Appendix, Fig. S6) and may be involved in proanthocyanidin accumulation (40–42); (ii) genes potentially involved in berry softening, such as two pectinesterase-coding genes and a xyloglucan endotransglucosylase/hydrolase gene that exhibited maximal expression in postveraison berry pericarps (SI Appendix, Fig. S6); and (iii) flowering-time genes, including a Phytochrome C homolog.
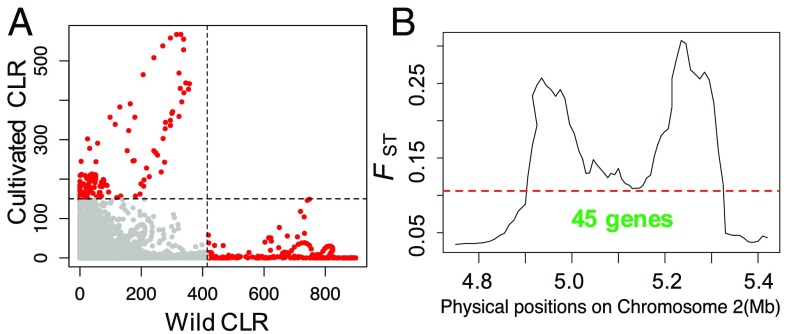
As a comparison, we applied CLR analyses to the sylvestris sample, which were notable for three reasons. First, the top 0.5% of windows yielded far fewer (88 vs. 309) genes (SI Appendix, Table S2). Second, CLR candidate-selected regions within sylvestris were distinct from those in vinifera (Fig. 3A); none of the putatively selected regions overlapped between taxa. Third, candidate-selected genes were enriched primarily for stress resistance (SI Appendix, Table S4), including flavonoid production (P = 6.27E-3), ethylene-mediated signaling pathways (P = 8.76E-6), and the stilbenoid biosynthesis pathway (P = 1.93E-50). Stilbenoids accumulate in response to biotic and abiotic stresses (43).
Fig. 3.
The CLR statistic computed for 20-kbp windows along chromosomes separately for wild samples and cultivars. (A) The scatterplot shows the CLR statistic for corresponding windows for both wild (x axis) and cultivated samples (y axis). The dashed line represents the 99.5% cutoff, and red dots represent outlier regions. (B) FST analyses between vinifera and sylvestris identify two peaks in the sex determination region that encompass 45 annotated genes.
We also detected regions of high divergence between wild and cultivated samples using FST and Dxy, which identified 929 and 546 candidate-selected genes (SI Appendix, Tables S2 and S6). A prominent region of divergence was identified by both methods from ∼4.90 Mb to ∼5.33 Mb on chromosome 2 (Fig. 3B and SI Appendix, Fig. S5), which coincides with the sex determination region (44). With both methods, the region contained two peaks of divergence. In FST analyses, the two peaks contain 13 and 32 genes, respectively. In the first peak, six genes were overexpressed in female (F) compared with both male (M) and hermaphroditic (H) flowers (adj. P ≤ 0.05; SI Appendix, Fig. S7 and Table S6), representing a nonrandom enrichment of F expression under the peak (binomial; P < 10−7). One of these genes had been identified as a candidate male sterility gene (VviFSEX) (45). The second peak included four genes with biased sex expression: one with higher F expression, two with higher H expression, and one with higher M expression (SI Appendix, Table S6).
Deleterious Variants.
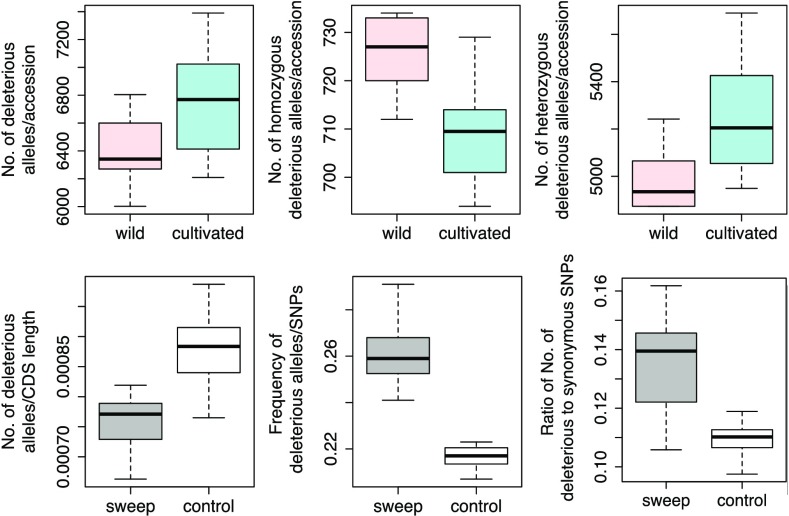
Domesticated annual crops accumulate more deleterious variants than their progenitors (17, 20, 46). To examine the potential increase in the number and frequency of deleterious variants at nonsynonymous sites between vinifera and sylvestris samples, we predicted deleterious SNPs using SIFT (47). A total of 33,653 nonsynonymous mutations were predicted to be deleterious in both samples. The number of derived deleterious variants was 5.2% higher, on average, for vinifera individuals than for sylvestris individuals (Fig. 4), and the ratio of deleterious to synonymous variants was also elevated in vinifera (SI Appendix, Fig. S8). Most (∼77%) deleterious variants were found in a heterozygous state in both samples, but the distribution by state differed between taxa because deleterious variants were more often homozygous in sylvestris (P < 0.001, Fig. 4). Cultivated accessions had a higher proportion of heterozygous deleterious variants (P = 0.002, Fig. 4) and an elevated ratio of deleterious to synonymous variants (P < 0.001, SI Appendix, Fig. S8).
Fig. 4.
The number and frequency of derived deleterious alleles in cultivars and wild samples. (Top) Comparisons between vinifera and sylvestris for the number of deleterious variants per individual overall (Left), as homozygotes (Middle), and as heterozygotes (Right). (Bottom) Comparisons between sweep regions and the rest of the genome (control) for the number (Left), population frequency (Middle), and ratio (Right) of the number deleterious to synonymous variants per vinifera individual.
We also examined the distribution of putatively deleterious variants for vinifera in sweep regions compared with the remainder of the genome (i.e., the “control”). Sweep regions contained a significantly lower number of deleterious mutations when corrected for length (P < 0.001, Fig. 4), but these variants were also found at significantly higher frequencies (P < 0.001, Fig. 4) and in higher numbers relative to synonymous variants (P < 0.001; Fig. 4). All of these trends—including the number of deleterious variants per individual, the distribution by state, and the effects in sweep regions—were qualitatively similar using PROVEAN (48) to identify deleterious variants (SI Appendix, Fig. S9).
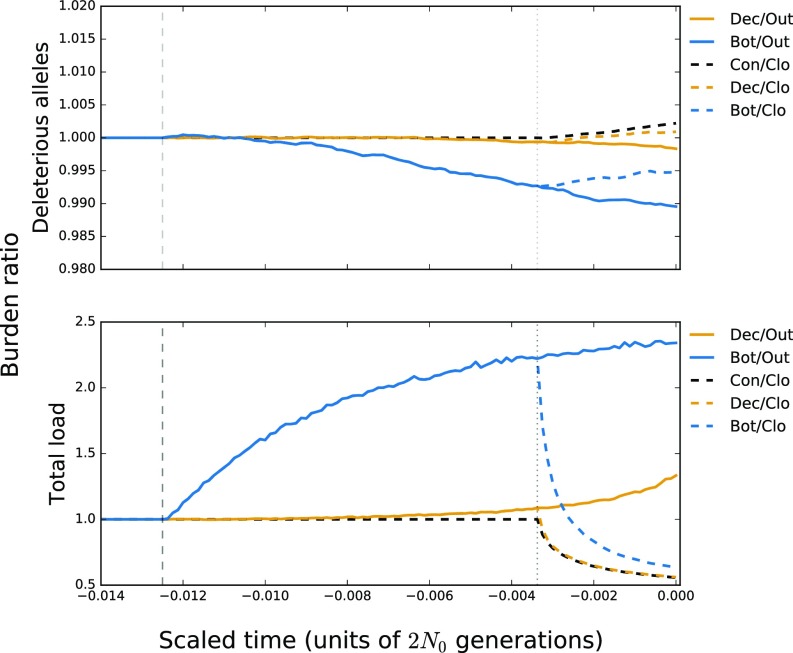
Like grapes, cassava is clonally propagated, and it also has high levels of heterozygous deleterious variants (20). To determine whether clonal propagation can contribute to the accumulation of deleterious variants, we performed forward simulations under two mating systems: outcrossing and clonal propagation that began at the time of domestication (∼8 kya). Each mating system was considered under three demographic models: a constant size population, a long ∼30,000-y population decline similar to that inferred from SMC++ analysis, and a discrete bottleneck (Materials and Methods and SI Appendix, Fig. S10). Under an additive model without back mutation, the discrete bottleneck increased the number of deleterious alleles under both mating systems but with little effect on load (SI Appendix, Fig. S12). Under a recessive model, an outcrossing, bottlenecked population purged deleterious variants (49, 50) (Fig. 5), and clonal propagation increased the number of deleterious variants under all demographic scenarios (Fig. 5). Despite the increase in deleterious variants, clonal propagation decreased load under the recessive model (Fig. 5) because clonality hides deleterious, recessive variants.
Fig. 5.
Forward simulations under a model of recessive selection for three demographic scenarios and two mating systems. (Top) The average number of deleterious alleles per accession relative to an outcrossing population of constant size. (Bottom) The total load relative to an outcrossing population of constant size. The dashed lines represent the time of demographic shift, ∼30 kya, and the onset of clonal propagation during domestication ∼8 kya. Bot, bottleneck; Clo, clonal propagation; Con, constant population size; Dec, declining population size; Out, outcrossing.
Discussion
The Eurasian wild grape (Vitis vinifera subsp. sylvestris) is a dioecious, perennial, forest vine that was widely distributed in the Near East and the northern Mediterranean before its domestication (51). The earliest archaeological evidence of wine production suggests that domestication took place in the Southern Caucasus between the Caspian and Black Seas ∼6.0–8.0 kya (24, 52). After domestication, the cultivars spread south by 5.0 kya to the western side of the Fertile Crescent, the Jordan Valley, and Egypt and finally reached Western Europe by ∼2.8 kya (24, 53). Here, however, we are not concerned with the spread of modern grapes, but rather with demographic history before and during domestication, the identity of genes that may have played a role in domestication, and the potential effects of domestication and breeding on the accumulation of deleterious variants.
A Protracted Predomestication History?
We have gathered genome-wide resequencing data from a sample of table grapes, wine grapes, and putatively wild grapes to investigate population structure and demographic history. These analyses lead to our first conclusion, which is that our sylvestris sample represents bona fide wild grapes, as opposed to feral escapees. This conclusion is evident from the fact that the sylvestris accessions cluster together in population structure analyses (Fig. 1), that they are estimated to have diverged from cultivated grapes ∼22 kya to 30 kya (Fig. 2), and that the set of putatively selected genes differs markedly between the vinifera and sylvestris samples (Fig. 3A). The divergence time between wild and cultivated samples suggests, however, that our sylvestris accessions likely do not represent the progenitor population of domesticated grapes.
Analyses of vinifera data suggest that its historical Ne has experienced a long decline starting from ∼22.0 kya to ∼30.0 kya. MSMC analyses indicate that this decline culminated in a weak bottleneck around the estimated time of domestication (Fig. 2A). The potential bottleneck corresponds to the estimated time of grape domestication and the shift from hunter-gatherer to agrarian societies (6). We note, however, that SMC++ analysis found no evidence for a distinct bottleneck, but instead inferred a consistent Ne decline (Fig. 2C). The question becomes, then, whether the domestication of vinifera included a discrete bottleneck. The evidence is mixed. The positive Tajima’s D for vinifera superficially suggests a population bottleneck, but forward simulations show that positive D values also result from a long population decline (SI Appendix, Fig. S11). If there was a discrete bottleneck for grapes, we join previous studies in concluding that it was weak (23, 54, 55), based on two lines of evidence. First, the diversity level in our vinifera sample is 94% that of sylvestris, representing a far higher cultivated-to-wild ratio than that of maize (83%) (4), indica rice (64%) (17), soybean (83%) (56), cassava (71%) (20), and tomato (54%) (57). Second, MSMC analyses suggest an approximately two- to threefold reduction in Ne at the time of domestication (Fig. 2A). This implies that 33–50% of the progenitor population was retained during domestication, a percentage that contrasts markedly with the <10% estimated for maize (3, 58) and ∼2% for rice (59).
The protracted decline in Ne for vinifera prompts a question about its cause(s). One possibility is that it reflects natural processes that acted on vinifera progenitor populations. For example, climatic shifts may have contributed to the long Ne decline because the Last Glacial Maximum (LGM) occurred between 33.0 and 26.5 kya (60). If the LGM caused vinifera’s population decline, one might expect to see population recovery during glacial retraction from 19.0 kya to 20.0 kya. We detect evidence of recovery in sylvestris but not in vinifera (Fig. 2C). A second possibility is that the domesticated germplasm is derived from a single deme of a larger metapopulation because population structure can produce a signal of apparent Ne decline (61). Finally, it is possible that proto-vinifera populations experienced a long period of human-mediated management, as suggested in the study of African rice (11). It is difficult to prove this proposition, but three factors are consistent with this possibility: (i) the contrasting historical pattern of the wild sample, (ii) the fact that some sites in the Southern Caucasus mountains have evidence of human habitation for >20,000 y (62), and (iii) a growing consensus that humans altered ecosystems long before the onset of agriculture (63).
A surprising feature of demographic inference is the lack of evidence for a postdomestication expansion of vinifera (Fig. 2). This observation contrasts sharply with studies of maize (58) and African rice (11), both of which had greater than fivefold Ne increases following domestication. We hypothesize that the lack of expansion in grapes relates to the dynamics of perennial domestication, specifically clonal propagation and the short time frame (in generations). Data from peach are consistent with our hypothesis, but peach also has extremely low historical levels of Ne (64). Almond, which is another clonally propagated perennial, exhibits an approximately twofold Ne expansion after domestication (64), but it also may have been propagated sexually before the discovery of grafting (65). Clearly more work needs to be done to compare demographic histories across crops with varied demographic and life histories.
Our demographic inferences have caveats. First, our study—along with all previous studies—has likely not measured genetic diversity from the precise progenitor population to vinifera. Indeed, such a population may be extinct or at least substantially modified since domestication. Second, our sample size is modest, but it is sufficient to infer broad historical patterns (34). Consistent with this supposition, the two runs of MSMC with two different samples of n = 4 yielded qualitatively identical inferences about the demographic history of vinifera. Larger samples will be necessary for investigating more recent population history and may provide further insights into the potential for population expansion after domestication. Finally, demographic calculations assume a mutation rate and a generation time that may be incorrect, and they also treat all sites equivalently. Note, however, that masking selected regions provides similar inferences (SI Appendix, Fig. S3) and also that our observations are consistent with independent estimates about domestication times and glacial events.
Selective Sweeps and Agronomically Important Genes.
Selective sweep analyses identified genes and regions that have been previously suspected to mediate agronomic change. One example is that of the SWEET1 gene, which is within a potential vinifera sweep region. The same gene is also within a region of differentiation between nonadmixed table and wine grapes (SI Appendix, Fig. S4). Based on haplotype structures, we hypothesize that at least one difference between wine and table grapes is attributable to the SWEET1 sugar transporter.
A major change during grape domestication was the switch from dioecy to hermaphroditism (66). The sex-determining region resides on chromosome 2, based on quantitative trait locus analyses that fine-mapped the sex locus between ∼4.90 and 5.05 Mbp (26, 27). The region corresponds to a larger chromosomal segment from 4.75 Mb to 5.39 Mb based on Genotype-by-Sequencing data and on segregation patterns from multiple families (44). With WGS data, we have identified a similar region that contains two discrete divergence peaks, from ∼4.90 to 5.05 Mb and from ∼5.2 Mb to 5.3 Mb (Fig. 3B and SI Appendix, Fig. S5). We posit that the two peaks are meaningful because the evolution of dioecy requires two closely linked loci: one that causes loss of M function and another that houses a dominant F sterility mutation (67, 68). The first peak contains six genes overexpressed in F flowers, including VviFSEX, which may abort stamen development (45). We predict that the second peak houses a dominant F sterility factor. The leading candidates are four genes that are differentially expressed among sexes (SI Appendix, Table S2), but none of the four are annotated with an obvious function in sex determination (69) (SI Appendix, Table S6).
Putatively Deleterious Mutations in a Clonally Propagated Perennial.
Like grapes, most perennials have experienced moderate bottlenecks (22), raising the question as to whether they typically have an increased burden of slightly deleterious mutations (21). We find that each vinifera accession contains 5.2% more putatively deleterious SNPs, on average, than the wild individuals in our sample. This difference exceeds that observed for dogs (2.6%) (46) and rice (∼3–4%) (17) but pales in comparison with cassava (26%), a clonally propagated annual (20). Our simulations show that clonal propagation can lead to the accumulation of deleterious recessive mutations and a reduction of load under a recessive model (Fig. 5). We do not know the dominance of variants in grapes, but we predict that most heterozygous, putatively deleterious mutations are recessive and hence do not contribute to increased load or to a cost associated with domestication. These same mutations, however, do provide a genomic explanation of a well-known feature of grape breeding: severe inbreeding depression (70).
Materials and Methods
For full materials and methods, see SI Appendix, Supplementary Text. We collected leaf tissue for 13 individuals from 11 vinifera cultivars, 9 sylvestris accessions, and 1 accession of V. rotundifolia (SI Appendix, Table S1). DNA was extracted from leaf samples, Illumina paired-end sequencing libraries were constructed (TrueSeq), and libraries were sequenced as 150-bp paired reads. Illumina raw reads for five other cultivars were gathered from the Short Read Archive (SRA) at the National Center for Biotechnology Information (SI Appendix, Table S1).
Reads were trimmed, filtered, and mapped to the PN40024 reference (12X) (32). Local realignment was performed around indels, reads were filtered for PCR duplicates, and sites with extremely low or high coverage were removed. For population structure analyses, we used ANGSD (33) to generate a BEAGLE file for the variable subset of the genome and then applied NGSadmix (71). To measure genome-wide genetic diversity and other population parameters, we estimated a genome-wide sfs from genotype likelihoods (33).
Functional regions were based on the V. vinifera genome annotation in Ensembl (v34). Nonsynonymous SNPs were predicted to be deleterious based on a SIFT score of ≤0.05 (72). The V. rotundifolia outgroup allele was submitted to prediction programs to avoid reference bias (17, 18). The number of deleterious or synonymous alleles per individual or region was calculated as 2 × the number of homozygous variants + heterozygous variants (73).
We employed MSMC 2.0 to estimate Ne over time (34, 74), based on SNPs called in GATK v3.5 (75) (SI Appendix, Supplementary Text). Segregating sites within each sample were phased and imputed using Shapeit (76) based on a genetic map (44). Demographic history was also inferred with SMC++, which analyzes multiple genotypes without phasing (36). SweeD (37) and XP-CLR (38) were used to detect selective sweeps. FST and Dxy values were averaged within 20 kbp nonoverlapping windows using ANGSD (33).
Functional categories were assigned to genes using VitisNet functional annotations (77). We tested functional category enrichment using Fisher’s exact test with P ≤ 0.01 as significant. Gene expression data used SRA data for berry (SRP049306) and flower (SRP041212) samples. Reads were trimmed for quality and mapped onto the PN40024 transcriptome (v.V1 from genomes.cribi.unipd.it/grape/) using Bowtie2 (78). DESeq2 (79) was used to normalize read counts and to test for differential expression.
Forward-in-time simulations were carried out using fwdpy11 (80). Five hundred replicate simulations were run for each demographic and mating scheme model. The population decline model was based on SMC++ results and rescaled for computational performance. Three demographic models were simulated: constant population size, a linear population decline, and a discrete bottleneck. Two mating schemes were simulated: strict outcrossing for the whole simulation and outcrossing at the onset of domestication. Additional details are available in SI Appendix, Supplementary Text.
Supplementary Material
Acknowledgments
We thank R. Gaut and R. Figueroa-Balderas for generating the data and sampling; two anonymous reviewers, D. Seymour, Q. Liu, K. Roessler, and E. Solares provided comments. Y.Z. is supported by the International Postdoctoral Exchange Fellowship Program, J.S. is supported by the National Science Foundation Graduate Research Fellowships Program, and B.S.G. is supported by the Borchard Foundation. D.C. is supported by J. Lohr Vineyards and Wines, E. & J. Gallo Winery, and the Louis P. Martini Endowment.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the National Center for Biotechnology Information Sequence Read Archive (NCBI SRA) database (accession no. PRJNA388292).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1709257114/-/DCSupplemental.
References
- 1.Eyre-Walker A, Gaut RL, Hilton H, Feldman DL, Gaut BS. Investigation of the bottleneck leading to the domestication of maize. Proc Natl Acad Sci USA. 1998;95:4441–4446. doi: 10.1073/pnas.95.8.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tenaillon MI, U’Ren J, Tenaillon O, Gaut BS. Selection versus demography: A multilocus investigation of the domestication process in maize. Mol Biol Evol. 2004;21:1214–1225. doi: 10.1093/molbev/msh102. [DOI] [PubMed] [Google Scholar]
- 3.Wright SI, et al. The effects of artificial selection on the maize genome. Science. 2005;308:1310–1314. doi: 10.1126/science.1107891. [DOI] [PubMed] [Google Scholar]
- 4.Hufford MB, et al. Comparative population genomics of maize domestication and improvement. Nat Genet. 2012;44:808–811. doi: 10.1038/ng.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meyer RS, Purugganan MD. Evolution of crop species: Genetics of domestication and diversification. Nat Rev Genet. 2013;14:840–852. doi: 10.1038/nrg3605. [DOI] [PubMed] [Google Scholar]
- 6.Purugganan MD, Fuller DQ. The nature of selection during plant domestication. Nature. 2009;457:843–848. doi: 10.1038/nature07895. [DOI] [PubMed] [Google Scholar]
- 7.Purugganan MD, Fuller DQ. Archaeological data reveal slow rates of evolution during plant domestication. Evolution. 2011;65:171–183. doi: 10.1111/j.1558-5646.2010.01093.x. [DOI] [PubMed] [Google Scholar]
- 8.Fuller DQ, et al. Convergent evolution and parallelism in plant domestication revealed by an expanding archaeological record. Proc Natl Acad Sci USA. 2014;111:6147–6152. doi: 10.1073/pnas.1308937110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaut BS. Evolution is an experiment: Assessing parallelism in crop domestication and experimental evolution: (Nei Lecture, SMBE 2014, Puerto Rico) Mol Biol Evol. 2015;32:1661–1671. doi: 10.1093/molbev/msv105. [DOI] [PubMed] [Google Scholar]
- 10.Zhang LB, et al. Selection on grain shattering genes and rates of rice domestication. New Phytol. 2009;184:708–720. doi: 10.1111/j.1469-8137.2009.02984.x. [DOI] [PubMed] [Google Scholar]
- 11.Meyer RS, et al. Domestication history and geographical adaptation inferred from a SNP map of African rice. Nat Genet. 2016;48:1083–1088. doi: 10.1038/ng.3633. [DOI] [PubMed] [Google Scholar]
- 12.Lu J, et al. The accumulation of deleterious mutations in rice genomes: A hypothesis on the cost of domestication. Trends Genet. 2006;22:126–131. doi: 10.1016/j.tig.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 13.Charlesworth D, Willis JH. The genetics of inbreeding depression. Nat Rev Genet. 2009;10:783–796. doi: 10.1038/nrg2664. [DOI] [PubMed] [Google Scholar]
- 14.Lohmueller KE. The distribution of deleterious genetic variation in human populations. Curr Opin Genet Dev. 2014;29:139–146. doi: 10.1016/j.gde.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 15.Henn BM, Botigué LR, Bustamante CD, Clark AG, Gravel S. Estimating the mutation load in human genomes. Nat Rev Genet. 2015;16:333–343. doi: 10.1038/nrg3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morrell PL, Buckler ES, Ross-Ibarra J. Crop genomics: Advances and applications. Nat Rev Genet. 2011;13:85–96. doi: 10.1038/nrg3097. [DOI] [PubMed] [Google Scholar]
- 17.Liu Q, Zhou Y, Morrell PL, Gaut BS. Deleterious variants in Asian rice and the potential cost of domestication. Mol Biol Evol. 2017;34:908–924. doi: 10.1093/molbev/msw296. [DOI] [PubMed] [Google Scholar]
- 18.Kono TJ, et al. The role of deleterious substitutions in crop genomes. Mol Biol Evol. 2016;33:2307–2317. doi: 10.1093/molbev/msw102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Renaut S, Rieseberg LH. The accumulation of deleterious mutations as a consequence of domestication and improvement in sunflowers and other compositae crops. Mol Biol Evol. 2015;32:2273–2283. doi: 10.1093/molbev/msv106. [DOI] [PubMed] [Google Scholar]
- 20.Ramu P, et al. Cassava haplotype map highlights fixation of deleterious mutations during clonal propagation. Nat Genet. 2017;49:959–963. doi: 10.1038/ng.3845. [DOI] [PubMed] [Google Scholar]
- 21.Gaut BS, Díez CM, Morrell PL. Genomics and the contrasting dynamics of annual and perennial domestication. Trends Genet. 2015;31:709–719. doi: 10.1016/j.tig.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 22.Miller AJ, Gross BL. From forest to field: Perennial fruit crop domestication. Am J Bot. 2011;98:1389–1414. doi: 10.3732/ajb.1000522. [DOI] [PubMed] [Google Scholar]
- 23.Myles S, et al. Genetic structure and domestication history of the grape. Proc Natl Acad Sci USA. 2011;108:3530–3535. doi: 10.1073/pnas.1009363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGovern PE, Fleming SJ, Katz SH. The Origins and Ancient History of Wine: Food and Nutrition in History and Anthropology. Routledge; Amsterdam: 2003. [Google Scholar]
- 25.This P, Lacombe T, Cadle-Davidson M, Owens CL. Wine grape (Vitis vinifera L.) color associates with allelic variation in the domestication gene VvmybA1. Theor Appl Genet. 2007;114:723–730. doi: 10.1007/s00122-006-0472-2. [DOI] [PubMed] [Google Scholar]
- 26.Fechter I, et al. Candidate genes within a 143 kb region of the flower sex locus in Vitis. Mol Genet Genomics. 2012;287:247–259. doi: 10.1007/s00438-012-0674-z. [DOI] [PubMed] [Google Scholar]
- 27.Picq S, et al. A small XY chromosomal region explains sex determination in wild dioecious V. vinifera and the reversal to hermaphroditism in domesticated grapevines. BMC Plant Biol. 2014;14:229. doi: 10.1186/s12870-014-0229-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bowers J, et al. Historical genetics: The parentage of Chardonnay, Gamay, and other wine grapes of Northeastern France. Science. 1999;285:1562–1565. doi: 10.1126/science.285.5433.1562. [DOI] [PubMed] [Google Scholar]
- 29.Xu Y, et al. Genome-wide detection of SNP and SV variations to reveal early ripening-related genes in grape. PLoS One. 2016;11:e0147749. doi: 10.1371/journal.pone.0147749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cardone MF, et al. Inter-varietal structural variation in grapevine genomes. Plant J. 2016;88:648–661. doi: 10.1111/tpj.13274. [DOI] [PubMed] [Google Scholar]
- 31.Di Genova A, et al. Whole genome comparison between table and wine grapes reveals a comprehensive catalog of structural variants. BMC Plant Biol. 2014;14:7. doi: 10.1186/1471-2229-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jaillon O, et al. French-Italian Public Consortium for Grapevine Genome Characterization The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- 33.Korneliussen TS, Albrechtsen A, Nielsen R. ANGSD: Analysis of next generation sequencing data. BMC Bioinformatics. 2014;15:356. doi: 10.1186/s12859-014-0356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schiffels S, Durbin R. Inferring human population size and separation history from multiple genome sequences. Nat Genet. 2014;46:919–925. doi: 10.1038/ng.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koch MA, Haubold B, Mitchell-Olds T. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae) Mol Biol Evol. 2000;17:1483–1498. doi: 10.1093/oxfordjournals.molbev.a026248. [DOI] [PubMed] [Google Scholar]
- 36.Terhorst J, Kamm JA, Song YS. Robust and scalable inference of population history from hundreds of unphased whole genomes. Nat Genet. 2017;49:303–309. doi: 10.1038/ng.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pavlidis P, Živkovic D, Stamatakis A, Alachiotis N. SweeD: Likelihood-based detection of selective sweeps in thousands of genomes. Mol Biol Evol. 2013;30:2224–2234. doi: 10.1093/molbev/mst112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen H, Patterson N, Reich D. Population differentiation as a test for selective sweeps. Genome Res. 2010;20:393–402. doi: 10.1101/gr.100545.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chong J, et al. The SWEET family of sugar transporters in grapevine: VvSWEET4 is involved in the interaction with Botrytis cinerea. J Exp Bot. 2014;65:6589–6601. doi: 10.1093/jxb/eru375. [DOI] [PubMed] [Google Scholar]
- 40.Bogs J, et al. Proanthocyanidin synthesis and expression of genes encoding leucoanthocyanidin reductase and anthocyanidin reductase in developing grape berries and grapevine leaves. Plant Physiol. 2005;139:652–663. doi: 10.1104/pp.105.064238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Savoi S, et al. Transcriptome and metabolite profiling reveals that prolonged drought modulates the phenylpropanoid and terpenoid pathway in white grapes (Vitis vinifera L.) BMC Plant Biol. 2016;16:67. doi: 10.1186/s12870-016-0760-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blanco-Ulate B, et al. Developmental and metabolic plasticity of white-skinned grape berries in response to Botrytis cinerea during noble rot. Plant Physiol. 2015;169:2422–2443. doi: 10.1104/pp.15.00852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chong J, Poutaraud A, Huegeney P. Metabolism and roles of stilbenes in plants. Plant Sci. 2009;3:143–155. [Google Scholar]
- 44.Hyma KE, et al. Heterozygous mapping strategy (HetMappS) for high resolution genotyping-by-sequencing markers: A case study in grapevine. PLoS One. 2015;10:e0134880. doi: 10.1371/journal.pone.0134880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coito JL, et al. VviAPRT3 and VviFSEX: Two genes involved in sex specification able to distinguish different flower types in Vitis. Front Plant Sci. 2017;8:98. doi: 10.3389/fpls.2017.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marsden CD, et al. Bottlenecks and selective sweeps during domestication have increased deleterious genetic variation in dogs. Proc Natl Acad Sci USA. 2016;113:152–157. doi: 10.1073/pnas.1512501113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simons YB, Turchin MC, Pritchard JK, Sella G. The deleterious mutation load is insensitive to recent population history. Nat Genet. 2014;46:220–224. doi: 10.1038/ng.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kirkpatrick M, Jarne P. The effects of a bottleneck on inbreeding depression and the genetic load. Am Nat. 2000;155:154–167. doi: 10.1086/303312. [DOI] [PubMed] [Google Scholar]
- 51.Zohary D, Spiegel-Roy P. Beginnings of fruit growing in the old world. Science. 1975;187:319–327. doi: 10.1126/science.187.4174.319. [DOI] [PubMed] [Google Scholar]
- 52.McGovern PE, Glusker DL, Exner LJ, Voigt MM. Neolithic resinated wine. Nature. 1996;381:480–481. [Google Scholar]
- 53.Olmo H. Evolution of Crop Plants: Grapes. Longman; New York: 1995. [Google Scholar]
- 54.Barnaud A, Laucou V, This P, Lacombe T, Doligez A. Linkage disequilibrium in wild French grapevine, Vitis vinifera L. subsp. silvestris. Heredity (Edinb) 2010;104:431–437. doi: 10.1038/hdy.2009.143. [DOI] [PubMed] [Google Scholar]
- 55.Grassi F, et al. Evidence of a secondary grapevine domestication centre detected by SSR analysis. Theor Appl Genet. 2003;107:1315–1320. doi: 10.1007/s00122-003-1321-1. [DOI] [PubMed] [Google Scholar]
- 56.Lam HM, et al. Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat Genet. 2010;42:1053–1059. doi: 10.1038/ng.715. [DOI] [PubMed] [Google Scholar]
- 57.Lin T, et al. Genomic analyses provide insights into the history of tomato breeding. Nat Genet. 2014;46:1220–1226. doi: 10.1038/ng.3117. [DOI] [PubMed] [Google Scholar]
- 58.Beissinger TM, et al. Recent demography drives changes in linked selection across the maize genome. Nat Plants. 2016;2:16084. doi: 10.1038/nplants.2016.84. [DOI] [PubMed] [Google Scholar]
- 59.Zhu Q, Zheng X, Luo J, Gaut BS, Ge S. Multilocus analysis of nucleotide variation of Oryza sativa and its wild relatives: Severe bottleneck during domestication of rice. Mol Biol Evol. 2007;24:875–888. doi: 10.1093/molbev/msm005. [DOI] [PubMed] [Google Scholar]
- 60.Clark PU, et al. The last glacial maximum. Science. 2009;325:710–714. doi: 10.1126/science.1172873. [DOI] [PubMed] [Google Scholar]
- 61.Nielsen R, Beaumont MA. Statistical inferences in phylogeography. Mol Ecol. 2009;18:1034–1047. doi: 10.1111/j.1365-294X.2008.04059.x. [DOI] [PubMed] [Google Scholar]
- 62.Adler DS, Tushabramishvili N. Middle Palaeolithic patterns of settlement and subsistence in the southern Caucasus. Middle Palaeolithic Settlement Dynamics. 2004:91–132. [Google Scholar]
- 63.Roberts P, Hunt C, Arroyo-Kalin M, Evans D, Boivin N. The deep human prehistory of global tropical forests and its relevance for modern conservation. Nat Plants. 2017;3:17093. doi: 10.1038/nplants.2017.93. [DOI] [PubMed] [Google Scholar]
- 64.Velasco D, Hough J, Aradhya M, Ross-Ibarra J. Evolutionary genomics of peach and almond domestication. G3 (Bethesda) 2016;6:3985–3993. doi: 10.1534/g3.116.032672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zohary D, Hopf M. Domestication of Plants in the Old World. Oxford Univ Press; Oxford: 2000. [Google Scholar]
- 66.This P, Lacombe T, Thomas MR. Historical origins and genetic diversity of wine grapes. Trends Genet. 2006;22:511–519. doi: 10.1016/j.tig.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 67.Charlesworth D, Charlesworth B, Marais G. Steps in the evolution of heteromorphic sex chromosomes. Heredity (Edinb) 2005;95:118–128. doi: 10.1038/sj.hdy.6800697. [DOI] [PubMed] [Google Scholar]
- 68.Charlesworth D. Plant sex chromosome evolution. J Exp Bot. 2013;64:405–420. doi: 10.1093/jxb/ers322. [DOI] [PubMed] [Google Scholar]
- 69.Ramos MJ, et al. Deep analysis of wild Vitis flower transcriptome reveals unexplored genome regions associated with sex specification. Plant Mol Biol. 2017;93:151–170. doi: 10.1007/s11103-016-0553-9. [DOI] [PubMed] [Google Scholar]
- 70.Kole C. Genetics, Genomics, and Breeding of Grapes. Science Publishers; Enfield, NH: 2011. pp. 160–185. [Google Scholar]
- 71.Skotte L, Korneliussen TS, Albrechtsen A. Estimating individual admixture proportions from next generation sequencing data. Genetics. 2013;195:693–702. doi: 10.1534/genetics.113.154138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 73.Henn BM, et al. Distance from sub-Saharan Africa predicts mutational load in diverse human genomes. Proc Natl Acad Sci USA. 2016;113:E440–E449. doi: 10.1073/pnas.1510805112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li H, Durbin R. Inference of human population history from individual whole-genome sequences. Nature. 2011;475:493–496. doi: 10.1038/nature10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Delaneau O, Zagury JF, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods. 2013;10:5–6. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- 77.Grimplet J, et al. VitisNet: “Omics” integration through grapevine molecular networks. PLoS One. 2009;4:e8365. doi: 10.1371/journal.pone.0008365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thornton KR. A C++ template library for efficient forward-time population genetic simulation of large populations. Genetics. 2014;198:157–166. doi: 10.1534/genetics.114.165019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.