Significance
DNA polymerases frequently incorporate ribonucleotides in place of deoxyribonucleotides during genome replication. RNase HII is responsible for initiating the removal of ribonucleotide errors across all three domains of life. Ribonucleotides that persist in genomic DNA due to defects in RNase HII result in strand breaks, mutagenesis, and neurodevelopmental disease in humans. Here, we define the proteins important for ribonucleotide excision repair in Bacillus subtilis and use genome-wide mutational profiling to determine the mutagenic cost of ribonucleotides in RNase HII-deficient cells. We show that the absence of RNase HII yields error-prone ribonucleotide correction via a pathway that relies on an essential DNA polymerase. We further demonstrate that error-prone ribonucleotide removal causes sequence context-dependent GC → AT transitions on the lagging strand.
Keywords: ribonucleotide excision repair, DNA polymerase, mutagenesis, RNase HII
Abstract
Replicative DNA polymerases misincorporate ribonucleoside triphosphates (rNTPs) into DNA approximately once every 2,000 base pairs synthesized. Ribonucleotide excision repair (RER) removes ribonucleoside monophosphates (rNMPs) from genomic DNA, replacing the error with the appropriate deoxyribonucleoside triphosphate (dNTP). Ribonucleotides represent a major threat to genome integrity with the potential to cause strand breaks. Furthermore, it has been shown in the bacterium Bacillus subtilis that loss of RER increases spontaneous mutagenesis. Despite the high rNTP error rate and the effect on genome integrity, the mechanism underlying mutagenesis in RER-deficient bacterial cells remains unknown. We performed mutation accumulation lines and genome-wide mutational profiling of B. subtilis lacking RNase HII, the enzyme that incises at single rNMP residues initiating RER. We show that loss of RER in B. subtilis causes strand- and sequence-context–dependent GC → AT transitions. Using purified proteins, we show that the replicative polymerase DnaE is mutagenic within the sequence context identified in RER-deficient cells. We also found that DnaE does not perform strand displacement synthesis. Given the use of nucleotide excision repair (NER) as a backup pathway for RER in RNase HII-deficient cells and the known mutagenic profile of DnaE, we propose that misincorporated ribonucleotides are removed by NER followed by error-prone resynthesis with DnaE.
Replicative DNA polymerases duplicate genomes with high fidelity (1). In bacteria, it is estimated that base-pairing errors between deoxyribonucleoside triphosphates (dNTP) occur approximately once every 60 million properly paired bases (2). Such a high degree of accuracy is due to the intrinsic fidelity of the DNA polymerases from factors including induced fit in the active site and 3′ to 5′ exonuclease “proofreading” activity (1). Interestingly, replicative DNA polymerases are far more likely to incorporate sugar errors as opposed to dNTP base-pairing errors (3, 4). Sugar errors represent the insertion of a ribonucleoside triphosphate (rNTP) in place of its corresponding dNTP (5). With the exception of uridine triphosphate (rUTP), the difference between each dNTP and its corresponding rNTP is the presence of a single oxygen atom at the 2′ position of the ribose sugar. Many DNA polymerases have a steric gate residue that limits the use of rNTPs as a substrate (6). The steric gate is often a bulky amino acid side chain that clashes with the 2′-OH on the ribose sugar of rNTPs, limiting their incorporation into DNA (5, 6). The intracellular abundance of rNTPs presents a challenge for sugar specificity during DNA replication as polymerases needed to select the proper dNTP are out numbered 10- to 100-fold by rNTPs (3, 7). The imbalance in nucleotide concentration causes rNTPs to be incorporated into genomic DNA in eukaryotes and bacteria (4, 8).
In Saccharomyces cerevisiae, the DNA polymerases required for genome replication combine for an error rate of ∼10,000 sugar errors per round of replication (3). Specifically, the DNA polymerases δ and ε are responsible for incorporation of 1 ribonucleoside monophosphate (rNMP) every 5,000 and 1,250 base pairs replicated, respectively (3). In S. cerevisiae, ribonuclease (RNase) H2 is responsible for initiating removal of single rNMPs from DNA as part of the ribonucleotide excision repair (RER) pathway (9, 10). During RER in S. cerevisiae, RNase H2 incises the sugar-phosphate backbone 5′ to a single rNMP followed by strand displacement synthesis with Pol δ or Pol ε. The resulting 5′ flap is removed by FEN1 or ExoI with the nick sealed by DNA ligase (10). Loss of RER is mutagenic in S. cerevisiae, resulting in topoisomerase I-dependent 2- to 5-bp deletions (8).
Much less is known about rNTP incorporation, RER, or the consequences of rNMPs nested in bacterial DNA. In Escherichia coli, DNA polymerase III holoenzyme incorporates sugar errors at a rate of one misinserted rNMP every 2.3 kb replicated (4). Based on the selectivity of Pol III for dNTPs relative to rNTPs, it is expected that ∼2,000 rNMPs would be incorporated per round of replication (4). Furthermore, Y-family translesion DNA polymerases have been shown to readily incorporate rNMPs in place of deoxyribonucleoside monophosphates (dNMPs) (11, 12). Therefore, rNMPs likely represent the most frequent nucleotide targeted for removal from bacterial DNA.
Although it is clear that rNMPs are frequently incorporated into DNA, the consequences of embedded rNMPs for genome integrity in bacteria remain unclear. Interestingly, RNase HII is broadly conserved in eubacteria (13), yet loss of RER in E. coli does not increase mutagenesis; however, B. subtilis cells lacking RNase HII (ΔrnhB) show an increase in mutation rate (4). In E. coli nucleotide excision repair (NER) removes ribonucleotides from DNA in cells lacking RNase HII (ΔrnhB) (14). Therefore, in E. coli it seems that RNase HII-dependent RER provides the primary pathway for rNMP removal and that NER serves as a backup (14). Furthermore, genetic evidence taking advantage of a Pol V variant adept at rNMP incorporation showed that DNA polymerase I is important for RER in E. coli with other gene products providing redundant functions (15). E. coli and B. subtilis differ in that loss of RNase HII is mutagenic in B. subtilis, but not in E. coli, suggesting a fundamental difference in RER or the backup pathways used (4). The source of mutagenesis due to rNMPs in genomic DNA is unknown, and the overall RER pathway has not been defined genetically or reconstituted in vitro for B. subtilis.
To understand how unrepaired rNMPs impact genome stability, we completed mutation accumulation lines in RER-deficient (ΔrnhB) B. subtilis cells. We found that persistent rNMPs in genomic DNA result in a mutagenic signature that is caused by error-prone gap-filling. Futhermore, to understand how rNMPs in genomic DNA are replaced, we reconstituted the minimal set of proteins to replace an rNMP with a dNMP in a primer extension reaction in vitro.
Results and Discussion
RNase HII Incises at Single rNMPs in Duplex DNA.
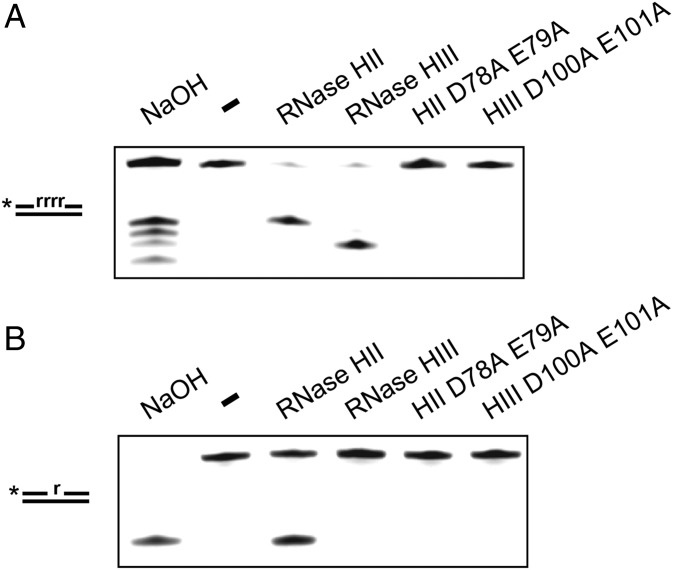
B. subtilis is known to have the RNase H enzymes RNase HII (rnhB) and RNase HIII (rnhC) (16, 17). Based on prior studies, we expected that B. subtilis RNase HII would be able to incise DNA 5′ to a single ribonucleotide in dsDNA (16, 18). To be certain, we purified B. subtilis RNases HII and HIII alongside catalytically inactive variants (DE-AA) to serve as controls (SI Appendix, Fig. S1). Each protein was purified with an N-terminal His6-SUMO tag that was removed during the purification process, yielding recombinant proteins without any additional amino acids (Materials and Methods). We incubated each protein with a dsDNA substrate containing four contiguous rNMPs or a single rNMP with the rNMP-containing strand labeled on the 5′ end. Following incubation, each reaction was resolved in a 20% urea–polyacrylamide gel. We show that RNases HII and HIII cleave the four-rNMP substrate while each catalytically inactive variant failed to show activity (Fig. 1A). Under the conditions tested, RNase HII cleaved the substrate containing a single rNMP, while RNase HIII did not (Fig. 1B). This work supports prior results showing that RNase HII initiates removal of single ribonucleotides from DNA, whereas RNase HIII may be more important for initiating repair of longer stretches of rNMPs embedded in DNA (16, 18).
Fig. 1.
RNase HII cleaves single rNMP in hybrid substrates. (A and B) A 5′-labeled substrate with four rNMPs (A) or a single rNMP (B) in DNA was incubated under the indicated conditions for 5 min followed by electrophoresis in a 20% denaturing urea–PAGE.
RER-Deficient B. subtilis Cells Accumulate GC → AT Transitions.
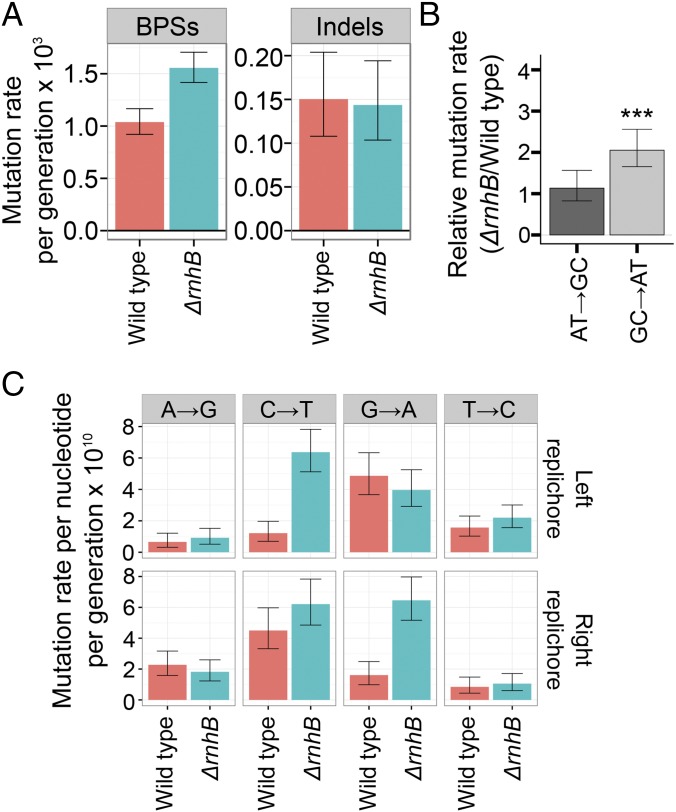
We completed mutation accumulation (MA) lines followed by whole-genome sequencing in cells without RNase HII (ΔrnhB) to determine the consequences to the genome-wide mutation rate and mutation spectrum. Genome-wide MA was compared with a compendium of wild-type B. subtilis MA line data that we had previously compiled (2). Eighty-one individual ΔrnhB (RNase HII) lines were completed for this work, each of which underwent 3,610 generations (Materials and Methods). An overall summary of the MA line results are presented in SI Appendix, Table S1, with all variants presented in Dataset S1. A total of 462 mutations were detected, 420 of which were base-pair substitutions (BPSs). We also detected 42 insertions/deletions (indels) in ΔrnhB lines (SI Appendix, Table S1). In agreement with prior results using a mutational reporter (4), loss of rnhB yielded an increase in the overall genome-wide mutation rate of ∼1.5-fold compared with wild type (SI Appendix, Table S1). BPSs were increased in ΔrnhB (Fig. 2A), with GC → AT transitions occurring approximately twofold more frequently in ΔrnhB than in wild type (Fig. 2B). From these data, we conclude that ribonucleotides persistent in genomic DNA result in a specific genome-wide mutagenic signature of GC → AT transitions. The mutation spectrum reported here is distinct from the 2- to 5-bp deletion spectrum observed for an RNase H2 deficiency in S. cerevisiae (8).
Fig. 2.
Persistent ribonucleotides cause strand-dependent transitions. (A) Base-pair substitution rate increases in the absence of RNase HII. (B) GC → AT transition rates are increased approximately twofold in ΔrnhB relative to wild type. (C) Barplots show the mutation rates of the indicated transitions from the perspective of the reference sequence. Persistent ribonucleotides cause GC → AT transitions in a strand-dependent manner.
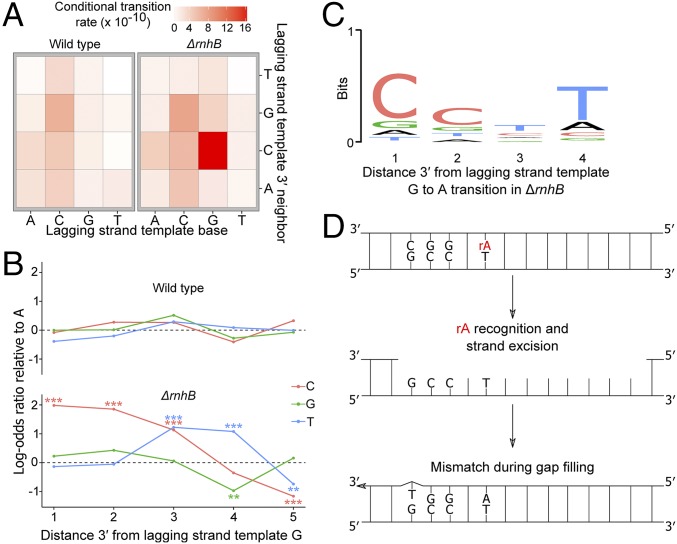
After observing the accumulation of transitions along the genome as cumulative distributions, it became clear that the increase in GC → AT transitions in ΔrnhB cells showed a strand dependence (Fig. 2C). The reference sequence of the right replichore represents the lagging-strand template and the reference sequence of the left replichore represents the leading-strand template. Therefore, the effect of persistent ribonucleotides on the GC → AT transition rate must be strand-dependent. Specifically, genomic loci were at an increased risk of undergoing a transition when RER was inactivated and guanosine was in either the leading strand or the lagging-strand template. We tested whether transitions due to misinserted rNMPs were more likely to occur in certain contexts by calculating conditional mutation rates for transitions in the 16 possible dinucleotide sequence contexts (Fig. 3A). All sequence contexts were considered from the perspective of the lagging-strand template, and the analysis was performed separately for 5′ (SI Appendix, Fig. S2) and 3′ contexts (Fig. 3A). The effects of 5′ neighboring nucleotides on the rNMP-induced transition rate were subtle; however, the 3′ neighboring context had a strong effect (Fig. 3A). Specifically, if guanosine is present in the lagging-strand template (or possibly the leading strand) followed 3′ by cytidine, the guanosine will undergo a transition ∼4 times more frequently in ΔrnhB than in wild type (Fig. 3A and SI Appendix, Fig. S2).
Fig. 3.
Persistent ribonucleotides cause context-dependent transitions. (A) Heatmap showing the transition rates for the 16 possible dinucleotides normalized to the abundance of each dinucleotide in the lagging-strand template. If guanosine is followed 3′ by cytidine in the lagging-strand template, the guanosine is more likely to undergo transition. (B) Logistic regression to determine the effect of sequence context 3′ to guanosine on transition occurrence at a guanosine in the lagging-strand template. All lagging-strand template guanosine positions were included in the regression analysis, and the log-odds of guanosine undergoing transition to adenosine were regressed against nucleotide identities up to five positions 3′ to the guanosine. The log-odds ratios are presented with adenosine as the baseline. (C) A motif logo was generated using the four positions 3′ to all guanosine-to-adenosine transitions in the lagging-strand template in ΔrnhB. The motif that is enriched 3′ to guanosine-to-adenosine transitions in ΔrnhB is 5′-CC(T/C)T-3′. (D) A model for ribonucleotide-mediated mutagenesis in the lagging-strand template 5′-GCC(T/C)T-3′.
Upon finding that GC → AT transitions are strand- and sequence-context–dependent, we further investigated the effect of the 3′ local sequence context on G → A transitions in the lagging-strand template using logistic regression. Logistic regression was performed genome-wide to determine if sequence context in the lagging-strand template at distances up to five nucleotides 3′ to a guanosine could influence mutation occurrence at the position of the guanosine (SI Appendix, SI Materials and Methods, Eq. S1). Impressively, nucleotide identity up to five nucleotides 3′ to a guanosine in the lagging-strand template was associated with G → A transition ocurrence in ΔrnhB (Fig. 3B). Bioniformatic analysis (19) determined a sequence motif to be present at G → A transitions in the lagging-strand template of ΔrnhB as 5′-GCC(T/C)T-3′. The underlined guanosine indicates the position that underwent a transition to adenosine (Fig. 3C). Therefore, genome-wide MA lines show a strong sequence-context–dependent increase in GC → AT transitions in RNase HII (ΔrnhB)-deficient cells (Figs. 2 and 3 and SI Appendix, Fig. S2).
NER is able to remove ribonucletoides from DNA when RER is deficient by excising a tract of DNA extending from eight phosphodiester bonds 5′ to the rNMP through four or five phosphodiester bonds 3′ to the rNMP (14). Adenosine is by far the most frequently misinserted ribonucleotide by E. coli Pol III (4). The motif that we identified to be associated with G → A transitions in the lagging-strand template includes thymidine three to four nucleotides (a distance of four to five phosphodiester bonds) 3′ to the G → A transition. In the absence of RNase HII, if adenosine monophosphate (rAMP) were misinserted across from a thymidine in 5′-GCC(T/C)T-3′ during lagging-strand synthesis, NER could excise the rAMP-containing strand (Fig. 3D). NER-dependent removal would generate a gap that must be filled. Because high-GC content near a given position contributes to a higher rate of mispairing by DNA polymerases (20), we hypothesized that gap filling after rNMP removal is more error-prone in part due to the high-GC content at the motif near the G → A transitions in the lagging-strand template. Below, we test the hypothesis that error-prone resynthesis results in mutagenesis.
DNA Polymerase I Participates in RNase HII-Dependent RER in B. subtilis.
In E. coli, DNA polymerase I (Pol I) has been shown to participate in RER in vivo (15), while in B. subtilis the DNA polymerase participating in RER is unknown. B. subtilis has three enzymes that we considered candidates for resynthesis of the DNA at an RNase HII incision. These polymerases are the replicative enzymes PolC, DnaE, and Pol I. B. subtilis also has two translesion DNA polymerases, PolY1 and PolY2 (21). Due to the specific nature of the GC → AT transition in ΔrnhB cells, the observed genome-wide spectrum is inconsistent with activity of PolY1 and PolY2, which would include transversion mutations (21).
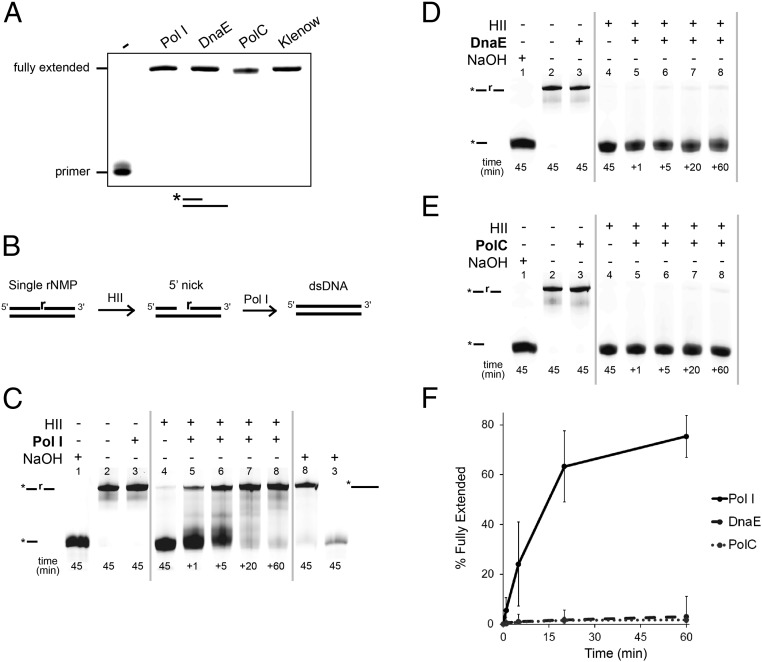
We began by testing which DNA polymerase(s) could function in the canonical RNase HII-dependent RER pathway. We purified Pol I and received DnaE and PolC from Charles McHenry, University of Colorado, Boulder, for extension reactions (Materials and Methods). The purity of Pol I, DnaE, and PolC was verified (SI Appendix, Fig. S3). All three B. subtilis Pols were active in a positive control assay for primer extension (Fig. 4A). We show a schematic to determine the Pol capable of extending a 3′-OH end generated after RNase HII incision at a single rNMP (Fig. 4B). We incubated a 5′ end-labeled RNase HII-incised dsDNA substrate with each DNA polymerase. The RNase HII reactions were quenched after 45 min and Pol I, DnaE, or PolC were added to the RNase HII reaction followed by quenching at 1, 5, 20, and 60 min (Fig. 4 C–F). Pol I extended the substrate after RNase HII incision at the single rNMP (Fig. 4C). Furthermore, the Pol I product (Fig. 4C, lane 8) was refractory to alkaline hydrolysis, demonstrating that rNMP was removed and replaced. As a control, we show that the substrate with the embedded rNMP from lane 3 (Fig. 4C) is alkali-sensitive.
Fig. 4.
Pol I removes and replaces a single rNMP after processing by RNase HII. (A) Primed DNA substrate was incubated with Pol I, DnaE, PolC, or Klenow fragment. Replication products were resolved using 17% urea–PAGE. (B) Schematic representation of substrate, RNase HII treatment, and subsequent DNA synthesis used in C–E. (C–E) A 5′-labeled RNA/DNA hybrid substrate was incubated with 3 M NaOH, no protein, or the indicated DNA polymerase: (C) Pol I, (D) DnaE, or (E) PolC. (C) Pol I, (D) DnaE, or (E) PolC was then added to the RNase HII reaction and stopped at 1, 5, 20, and 60 min. (C) Samples from lanes 8 and 3 were then incubated with 3 M NaOH for 45 min followed by resolution using a denaturing urea–PAGE. (F) Mean percentage of substrate extended is presented for triplicate samples for all polymerases at the indicated times. Error bars represent 95% CI.
Neither DnaE nor PolC were able to catalyze DNA synthesis from a nick generated by RNase HII (Fig. 4 D and E). Pol I was able to extend 78% of an RNase HII-nicked substrate after 60 min whereas DnaE and PolC were able to extend only 3% and 2% of the substrate, respectively (Fig. 4F). We conclude that RNase HII and Pol I work in conjunction to remove and replace a single misincorporated rNMP in DNA and that PolC and DnaE are not effective at strand-displacement synthesis from an RNase HII-incised nick.
DnaE Catalyzes Error-Prone Resynthesis of a Gapped Substrate Resulting in a G → A Transition.
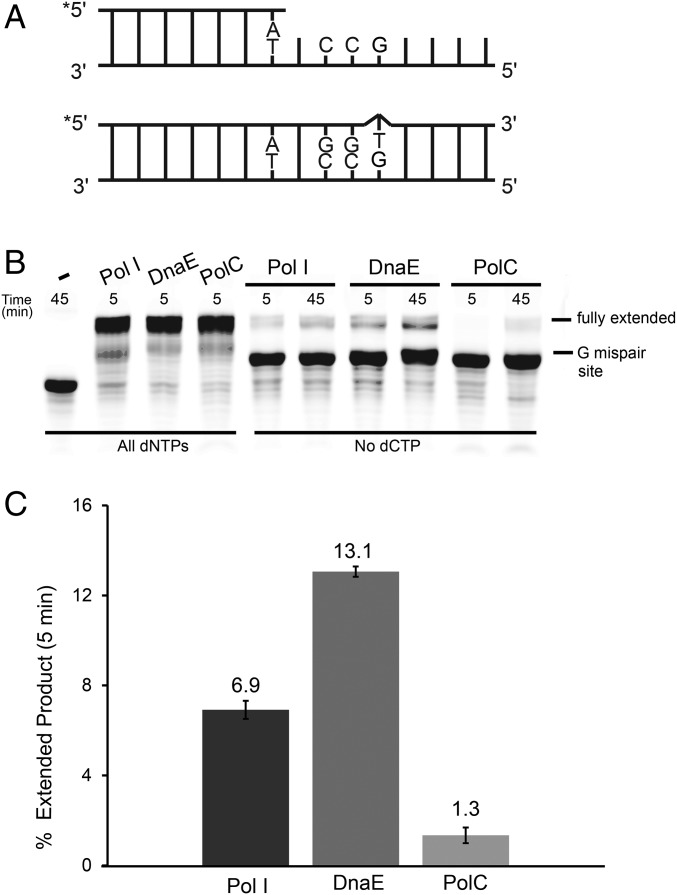
After establishing that Pol I participated in RNase HII-dependent RER and that PolC and DnaE did not, we asked if Pol I, DnaE, or PolC catalyzed mutagenic resynthesis in vitro. Because we identified the sequence context [5′-GCC(T/C)T-3′] in vivo (Fig. 3), we assembled a 5′ end-labeled substrate to more closely model the resynthesis step following NER-mediated excision (Fig. 5A). We provided all four dNTPs to demonstrate that each polymerase could extend the primer during a 5-min reaction (Fig. 5B). With activity established, we tested the ability of Pol I, DnaE, and PolC to generate a mismatch and then fully extend the mismatched product in the absence of deoxycytidine triphosphate (dCTP) (Fig. 5B). Pol I and DnaE were able to misincorporate and extend from a mismatch in the absence of dCTP. B. subtilis PolC has intrinsic 3′ to 5′ exonuclease proofreading activity (22) while B. subtilis DnaE lacks a known proofreading-associated protein (23). Furthermore, although E. coli Pol I possesses proofreading (24), B. subtilis Pol I lacks proofreading activity (25). Quantification of extension from a mismatch showed that DnaE caused an approximately twofold increase in error relative to Pol I (Fig. 5 B and C). We found that DnaE extended 13% of the substrate, whereas Pol I extended ∼7% of the substrate when reactions lacked dCTP (Fig. 5C). PolC was ineffective at mismatch formation and extension, with only 1.3% of the primer extended after 5 min (Fig. 5C). Therefore, DnaE was approximately twice as likely to form a mispair across from guanosine in the 5′-GCC(T/C)T-3′ sequence context than Pol I and 10 times more likely than PolC (Fig. 5C).
Fig. 5.
DnaE and Pol I mispair T across from G in a 5′-GCCTT-3′ sequence context. (A, Top) A schematic of the 5′ end-labeled substrate designed to mimic NER-mediated excision of the rA-containing strand across from 5′-GCC(C/T)T-3′. (A, Bottom) The fully extended primer in the absence of dCTP. (B) The subtrate shown in A was treated with no protein, Pol I, DnaE, or PolC with all four dNTPs or with dCTP omitted from the reactions for the indicated times followed by electrophoresis through a 17% urea–PAGE. (C) A barplot showing the mean of two independent experiments with the proportion of primer fully extended after 5 min in the absence of dCTP relative to reactions in which all nucleotides were present. Bar height and error bars represent the mean and range, respectively.
Interestingly, in the absence of RNase HII (ΔrnhB), we observed a twofold increase in GC → AT transitions, a result supportive of the error rate that we observed in vitro following DnaE extension. We propose that, following excision of rNMPs in cells that lack RNase HII activity (ΔrnhB), the gap is filled by DnaE, resulting in an increased genome-wide BPS rate.
Loss of NER Alters the Mutation Spectrum in ΔrnhB.
To test if rnhB and uvrA interact genetically, we used rifampin resistance as an indicator for mutation rate (SI Appendix, Table S2). We found that cells with ΔrnhB or ΔuvrA showed a mutation rate 1.7- and 1.6-fold, respectively, above that of wild type (SI Appendix, Table S2). The mutation rate of the double mutant (ΔrnhB, ΔuvrA) was increased 3.9-fold relative to wild type, suggesting an additive or synergistic affect. Because the mutation rate suggests genetic interaction between rnhB and NER, we tested the mutation spectrum of a reporter gene to determine if ΔuvrA altered the mutation spectrum of ΔrnhB. Loss-of-function mutations in genes encoding thymidylate synthase yield resistance to the drug trimethoprim (26). Because B. subtilis encodes two thymidylate synthases (27), we disrupted thyB and determined the mutation spectrum of thyA mutants in trimethoprim-resistant colonies. We tested strains that were ΔrnhB, ΔuvrA, ΔrnhB ΔuvrA, or otherwise wild type. Dataset S2 includes information on all thyA variants detected. Comparison of the thyA mutation spectrum from each strain showed that the only significant difference in mutation spectra was between ΔrnhB and ΔrnhB ΔuvrA strains (P = 0.04) (SI Appendix, Fig. S4). These data support the conclusion that the mutation spectrum in ΔrnhB cells is altered when NER is inactivated (ΔuvrA), suggesting that NER may provide a backup role for rNMP removal as shown for E. coli (14).
Model for RER in B. subtilis.
The data that we present shows that RNase HII is responsible for incision at a single embedded rNMP and that Pol I is efficient at resynthesis. Based on these results, we propose that the minimal set of proteins for RER in B. subtilis includes RNase HII, Pol I, and DNA ligase to seal the remaining nick. Our results indicate that PolC and DnaE are largely incapable of replication from an RNase HII-incised nick. NER has been shown to function as a backup pathway for removal of rNMPs in E. coli lacking RNase HII (14), and we provide evidence suggesting that B. subtilis NER functions in a similar capacity. Our findings indicate that, in the absence of RNase HII, the backup pathway for removal of rNMPs in the B. subtilis genome comes at a mutagenic cost (4). Here, we show that DnaE is capable of causing the increase in genome-wide BPS rate in B. subtilis cells lacking RNase HII. Therefore, we propose that in wild-type cells RNase HII incises 5′ to a single rNMP followed by removal and resynthesis by Pol I. In the absence of RNase HII, NER likely removes the rNMP, leaving a gap allowing access and mutagenic resynthesis by DnaE. DnaE is inactive from a nick (Fig. 4), but should be active on a gapped substrate (Fig. 5). Considering our data and prior studies (14), we propose that NER provides the five to eight nucleotide gap needed for resynthesis by DnaE resulting in mutagenesis in vivo. Because the mutation rate increased in cells deficient for NER and RER, it remains possible that, in ΔrnhB uvrA+ cells, mutations are generated by DnaE, but in ΔrnhB ΔuvrA strains, the mutations that are observed are generated by alternate mechanisms.
Our prior work and results herein measured spontaneous mutagenesis for ΔrnhB B. subtilis and E. coli cells (SI Appendix, Table S2) (4). These results show that ΔrnhB cells have a 1.7-twofold higher mutation rate than that of wild type. In contrast, the mutation rate of ΔrnhB E. coli cells was indistinguishable from that of the wild-type control (4). We propose that the difference in mutagenic cost of ΔrnhB B. subtilis cells relative to E. coli is due to differences in proofreading activity associated with DnaE. E. coli uses DnaE for replication of the leading and lagging strands with ε providing proofreading (28). In contrast, B. subtilis uses proofreading-proficient PolC for replication of the leading and lagging strands after DnaE extends the RNA primer (29). We propose that, because B. subtilis DnaE lacks proofreading, it causes mutagenic resynthesis of gapped repair intermediates. During RNase HII-dependent RER, DnaE does not cause mutagenesis because DnaE is unable to extend the 3′ end of a nicked substrate. In contrast, E. coli ΔrnhB cells do not show an increase in mutagenesis because, if Pol III gains access to the repair intermediate, it has an associated proofreading domain that limits errors during the resynthesis step. In further support of this model, it has been shown that Pol III-dependent resynthesis during NER is mutagenic when proofreading is inactivated in E. coli (15).
Embedded Ribonucleotides Are Unlikely to Provide Significant Participation in Nascent Strand Recognition During Mismatch Repair.
E. coli uses DNA methylation as a marker for strand recognition during mismatch repair (MMR) (30). Most bacteria and all eukaryotes lack a methylation-directed pathway (31). Biochemical results show that a nick will direct excision to the incised strand for mismatch removal (32). It is reasonable to hypothesize that 5′ and 3′ termini of Okazaki fragments serve as nascent-strand signals for correction of errors in the lagging strand. The signal that directs mismatch excision to the leading strand is unclear. It has recently been proposed that removal of rNMPs from DNA in eukaryotes could provide a strand discrimination signal for mismatch correction (33, 34). We considered that bacteria like B. subtilis, which lack a methylation-directed MMR pathway, could also use RNase HII incision at rNMP errors as a mechanism to direct mismatch repair to the leading strand (4). If rNMPs provided such a signal, spontaneous mutagenesis of the ΔrnhB and ΔrnhB ΔuvrA strains should be higher than 1.5- to 3.9-fold. Furthermore, an MMR deficiency should cause an increase in transition rates for A·T → G·C and G·C → A·T in a variety of sequence contexts. To the contrary, we observed genome-wide that G·C → A·T transitions are increased within a specific sequence context in the absence of RNase HII (ΔrnhB).
With these results we argue against the model that excision at embedded rNMPs provides a substantial strand discrimination signal for MMR in bacteria that lack a methylation-directed pathway. For B. subtilis, it has been shown that MutL-dependent incision is stimulated by interaction with the replication sliding clamp (β-clamp) (35). Furthermore, MutL variants unable to interact with β-clamp show defects in MMR in vivo and fail to nick a linear substrate in vitro (35). Studies of the eukaryotic proteins showed that MutLα incision was also stimulated by the replication sliding clamp, proliferating cell nuclear antigen (PCNA) and variants of MutLα impaired for interaction with PCNA prevented MMR in vitro and in vivo (36). It seems that the strand discrimination mechanism in organisms lacking a methylation signal may instead rely on orientation imparted to the MMR machinery by replication sliding clamps, allowing for stimulation of strand-dependent incision (37).
Materials and Methods
Plasmid Cloning and Proteins.
All primers used in this study are listed in SI Appendix, Table S3, all plasmids in SI Appendix, Table S4 and strains in SI Appendix, Table S5. Briefly, pJR17, pJR19, and pJR22 were cloned by first amplifying the pE-SUMO linear vector via PCR using primers oJR46 and oJR47. Each insert was amplified via PCR from PY79 genomic DNA with 15- to 20-bp overlaps complementary to the pE-SUMO linear fragment (rnhB: oJR88, oJR89; rnhC: oJR90, oJR91; polA: oJR102, oJR103). Plasmids pJR18 and pJR20 were generated via overlap PCR using oJR94 and oJR95 on pJR17 (pJR18) and oJR96 and oJR97 on pJR19 (pJR20). All PCR-generated plasmid DNA was Sanger-sequenced.
Plasmids used for overexpression of recombinant proteins RnhB, RnhC, and Pol I and catalytically inactive variants of RnhB and RnhC were used to transform BL21(DE3+) cells. Proteins were isolated as described in SI Appendix, SI Materials and Methods.
RNase H Cleavage Assays.
The 5′ IR dye-labeled substrates with either one or four consecutive rNMPs were prepared by incubating complementary strands (either oJR209 or oJR210 with oJR145) in a 98 °C water bath for 1 min in a buffer containing 20 mM Tris⋅HCl, pH 8, 100 mM NaCl, 10 mM MgCl2, and 1 mM DTT. Strands were then annealed by slowly cooling the solution to room temperature. Reactions were performed in the same buffer with 1 µM substrate and 200 nM protein for 10 min at 30 °C in 10 µL total volume. For NaOH samples, 1 µM of substrate was placed into 300 mM NaOH and incubated at 55 °C for 30 min, followed by neutralization with 2 M Tris⋅HCl, pH 7.5. Reactions were stopped by addition of 10 µL formamide loading dye (95% formamide, 20 mM EDTA, 1% SDS, 0.01% bromophenol blue), followed by denaturation at 100 °C for 2 min and immediate snap cooling in an ice-water bath. Following cooling, 2 µL of each reaction was electrophoresed in a denaturing 20% urea–polyacrylamide gel followed by visualization with the LI-COR Odyssey imager.
DNA Polymerase Activity Assay.
A 5′ IR dye-labeled primer was annealed to the template strand by incubating oJS895 (1 μM) and oJR283 (1.25 μM) together in a 98 °C water bath for 1 min in a buffer containing 40 mM Tris–acetate, pH 7.8, 12 mM magnesium acetate, 200 µM dNTPs, 300 mM potassium glutamate, 3 µM ZnSO4, 2% wt/vol PEG, and 1 mM DTT and then allowing the mixture to cool slowly to room temperature. Reactions were carried out in a 10-µL volume in the same buffer with 100 nM polymerase (Pol I, DnaE, PolC, or Klenow fragment) and 100 nM substrate at 25 °C for 5 min. Reactions were quenched with 10 µL of formamide dye (see above), denatured at 100 °C for 2 min, and immediately snap cooled in an ice-water bath. Products were resolved by electrophoresis through a 17% urea–polyacrylamide gel, followed by visualization with the LI-COR Odyssey imager.
Mutation Accumulation Line Protocol.
MA lines on ΔrnhB, sequence alignments, and variant detection and conditional mutation rate were performed as previously described (2). Wild-type (B. subtilis PY79) MA line data were previously published (2). Detailed description of the MA line procedure is described in SI Appendix, SI Materials and Methods.
Data-Sharing Plan.
High-throughput sequencing data used in this study have been deposited in the Sequence Read Archive under accession no. SRP117359. Equations are described in the SI Appendix, SI Materials and Methods; all code used is available upon request.
Statistical Analysis.
Statistical analysis and plotting were performed using the statistical computing software R. Throughout this work, *** denotes P ≤ 0.001, ** denotes P ≤ 0.01, and * denotes P ≤ 0.05.
Supplementary Material
Acknowledgments
We thank Dr. Charles McHenry for his generous gifts of DnaE and PolC, Heather Schroeder for help with statistical analyses, and Peter Burby for constructing the ΔuvrA strain. J.W.S. and W.G.H. were each supported in part by NIH National Research Service Award T32 GM007544. J.R.R. was supported in part by NIH Cellular Biotechnology Training Grant (T32 GM008353) and a predoctoral fellowship from Rackham Graduate School, University of Michigan. Additionally, this work was supported by NIH Grant R01 GM107312 (to L.A.S.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the Sequence Read Archive database (accession no. SRP117359).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1710995114/-/DCSupplemental.
References
- 1.Kunkel TA, Bebenek K. DNA replication fidelity. Annu Rev Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 2.Schroeder JW, Hirst WG, Szewczyk GA, Simmons LA. The effect of local sequence context on mutational bias of genes encoded on the leading and lagging strands. Curr Biol. 2016;26:692–697. doi: 10.1016/j.cub.2016.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nick McElhinny SA, et al. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc Natl Acad Sci USA. 2010;107:4949–4954. doi: 10.1073/pnas.0914857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yao NY, Schroeder JW, Yurieva O, Simmons LA, O’Donnell ME. Cost of rNTP/dNTP pool imbalance at the replication fork. Proc Natl Acad Sci USA. 2013;110:12942–12947. doi: 10.1073/pnas.1309506110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown JA, Suo Z. Unlocking the sugar “steric gate” of DNA polymerases. Biochemistry. 2011;50:1135–1142. doi: 10.1021/bi101915z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeLucia AM, Grindley ND, Joyce CM. An error-prone family Y DNA polymerase (DinB homolog from Sulfolobus solfataricus) uses a ‘steric gate’ residue for discrimination against ribonucleotides. Nucleic Acids Res. 2003;31:4129–4137. doi: 10.1093/nar/gkg417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Traut TW. Physiological concentrations of purines and pyrimidines. Mol Cell Biochem. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 8.Kim N, et al. Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science. 2011;332:1561–1564. doi: 10.1126/science.1205016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chon H, et al. RNase H2 roles in genome integrity revealed by unlinking its activities. Nucleic Acids Res. 2013;41:3130–3143. doi: 10.1093/nar/gkt027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sparks JL, et al. RNase H2-initiated ribonucleotide excision repair. Mol Cell. 2012;47:980–986. doi: 10.1016/j.molcel.2012.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McDonald JP, Vaisman A, Kuban W, Goodman MF, Woodgate R. Mechanisms employed by Escherichia coli to prevent ribonucleotide incorporation into genomic DNA by Pol V. PLoS Genet. 2012;8:e1003030. doi: 10.1371/journal.pgen.1003030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ordonez H, Uson ML, Shuman S. Characterization of three mycobacterial DinB (DNA polymerase IV) paralogs highlights DinB2 as naturally adept at ribonucleotide incorporation. Nucleic Acids Res. 2014;42:11056–11070. doi: 10.1093/nar/gku752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kochiwa H, Tomita M, Kanai A. Evolution of ribonuclease H genes in prokaryotes to avoid inheritance of redundant genes. BMC Evol Biol. 2007;7:128. doi: 10.1186/1471-2148-7-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vaisman A, et al. Removal of misincorporated ribonucleotides from prokaryotic genomes: An unexpected role for nucleotide excision repair. PLoS Genet. 2013;9:e1003878. doi: 10.1371/journal.pgen.1003878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vaisman A, et al. Investigating the mechanisms of ribonucleotide excision repair in Escherichia coli. Mutat Res. 2014;761:21–33. doi: 10.1016/j.mrfmmm.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohtani N, et al. Identification of the genes encoding Mn2+-dependent RNase HII and Mg2+-dependent RNase HIII from Bacillus subtilis: Classification of RNases H into three families. Biochemistry. 1999;38:605–618. doi: 10.1021/bi982207z. [DOI] [PubMed] [Google Scholar]
- 17.Itaya M, et al. Isolation of RNase H genes that are essential for growth of Bacillus subtilis 168. J Bacteriol. 1999;181:2118–2123. doi: 10.1128/jb.181.7.2118-2123.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haruki M, Tsunaka Y, Morikawa M, Kanaya S. Cleavage of a DNA-RNA-DNA/DNA chimeric substrate containing a single ribonucleotide at the DNA-RNA junction with prokaryotic RNases HII. FEBS Lett. 2002;531:204–208. doi: 10.1016/s0014-5793(02)03503-2. [DOI] [PubMed] [Google Scholar]
- 19.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 20.Petruska J, Goodman MF. Influence of neighboring bases on DNA polymerase insertion and proofreading fidelity. J Biol Chem. 1985;260:7533–7539. [PubMed] [Google Scholar]
- 21.Sung HM, Yeamans G, Ross CA, Yasbin RE. Roles of YqjH and YqjW, homologs of the Escherichia coli UmuC/DinB or Y superfamily of DNA polymerases, in stationary-phase mutagenesis and UV-induced mutagenesis of Bacillus subtilis. J Bacteriol. 2003;185:2153–2160. doi: 10.1128/JB.185.7.2153-2160.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanjanwala B, Ganesan AT. Genetic structure and domains of DNA polymerase III of Bacillus subtilis. Mol Gen Genet. 1991;226:467–472. doi: 10.1007/BF00260660. [DOI] [PubMed] [Google Scholar]
- 23.Le Chatelier E, et al. Involvement of DnaE, the second replicative DNA polymerase from Bacillus subtilis, in DNA mutagenesis. J Biol Chem. 2004;279:1757–1767. doi: 10.1074/jbc.M310719200. [DOI] [PubMed] [Google Scholar]
- 24.Setlow P, Brutlag D, Kornberg A. Deoxyribonucleic acid polymerase: Two distinct enzymes in one polypeptide. I. A proteolytic fragment containing the polymerase and 3′ leads to 5′ exonuclease functions. J Biol Chem. 1972;247:224–231. [PubMed] [Google Scholar]
- 25.Duigou S, Ehrlich SD, Noirot P, Noirot-Gros MF. DNA polymerase I acts in translesion synthesis mediated by the Y-polymerases in Bacillus subtilis. Mol Microbiol. 2005;57:678–690. doi: 10.1111/j.1365-2958.2005.04725.x. [DOI] [PubMed] [Google Scholar]
- 26.Dutra BE, Lovett ST. Cis and trans-acting effects on a mutational hotspot involving a replication template switch. J Mol Biol. 2006;356:300–311. doi: 10.1016/j.jmb.2005.11.071. [DOI] [PubMed] [Google Scholar]
- 27.Neuhard J, Price AR, Schack L, Thomassen E. Two thymidylate synthetases in Bacillus subtilis. Proc Natl Acad Sci USA. 1978;75:1194–1198. doi: 10.1073/pnas.75.3.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson A, O’Donnell M. Cellular DNA replicases: Components and dynamics at the replication fork. Annu Rev Biochem. 2005;74:283–315. doi: 10.1146/annurev.biochem.73.011303.073859. [DOI] [PubMed] [Google Scholar]
- 29.Sanders GM, Dallmann HG, McHenry CS. Reconstitution of the B. subtilis replisome with 13 proteins including two distinct replicases. Mol Cell. 2010;37:273–281. doi: 10.1016/j.molcel.2009.12.025. [DOI] [PubMed] [Google Scholar]
- 30.Lahue RS, Au KG, Modrich P. DNA mismatch correction in a defined system. Science. 1989;245:160–164. doi: 10.1126/science.2665076. [DOI] [PubMed] [Google Scholar]
- 31.Lenhart JS, Pillon MC, Guarné A, Biteen JS, Simmons LA. Mismatch repair in Gram-positive bacteria. Res Microbiol. 2016;167:4–12. doi: 10.1016/j.resmic.2015.08.006. [DOI] [PubMed] [Google Scholar]
- 32.Genschel J, Modrich P. Mechanism of 5′-directed excision in human mismatch repair. Mol Cell. 2003;12:1077–1086. doi: 10.1016/s1097-2765(03)00428-3. [DOI] [PubMed] [Google Scholar]
- 33.Lujan SA, Williams JS, Clausen AR, Clark AB, Kunkel TA. Ribonucleotides are signals for mismatch repair of leading-strand replication errors. Mol Cell. 2013;50:437–443. doi: 10.1016/j.molcel.2013.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghodgaonkar MM, et al. Ribonucleotides misincorporated into DNA act as strand-discrimination signals in eukaryotic mismatch repair. Mol Cell. 2013;50:323–332. doi: 10.1016/j.molcel.2013.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pillon MC, et al. The sliding clamp tethers the endonuclease domain of MutL to DNA. Nucleic Acids Res. 2015;43:10746–10759. doi: 10.1093/nar/gkv918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Genschel J, et al. Interaction of proliferating cell nuclear antigen with PMS2 is required for MutLα activation and function in mismatch repair. Proc Natl Acad Sci USA. 2017;114:4930–4935. doi: 10.1073/pnas.1702561114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pluciennik A, et al. PCNA function in the activation and strand direction of MutLα endonuclease in mismatch repair. Proc Natl Acad Sci USA. 2010;107:16066–16071. doi: 10.1073/pnas.1010662107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.