Significance
During malaria infections, Plasmodium falciparum parasites invade RBCs. Identification of host factors for parasite invasion guides the development of vaccines and host-targeted therapeutics. Here we describe the development of an in vitro culture system for the functional analysis of RBC determinants using the immortal erythroleukemia cell line JK-1. JK-1 cells can be induced to differentiate synchronously, support parasite invasion, and are amenable to genetic manipulation. Using this system, we validated two host factors, basigin and CD44, as strain-transcendent host factors for parasite invasion, and we demonstrated a functional interaction between these two proteins. The ability to perform gene editing to produce RBC mutants will augment our ability to study malaria infection.
Keywords: BSG, CD44, CRISPR/Cas9, Plasmodium falciparum, parasite invasion
Abstract
During malaria blood-stage infections, Plasmodium parasites interact with the RBC surface to enable invasion followed by intracellular proliferation. Critical factors involved in invasion have been identified using biochemical and genetic approaches including specific knockdowns of genes of interest from primary CD34+ hematopoietic stem cells (cRBCs). Here we report the development of a robust in vitro culture system to produce RBCs that allow the generation of gene knockouts via CRISPR/Cas9 using the immortal JK-1 erythroleukemia line. JK-1 cells spontaneously differentiate, generating cells at different stages of erythropoiesis, including terminally differentiated nucleated RBCs that we term “jkRBCs.” A screen of small-molecule epigenetic regulators identified several bromodomain-specific inhibitors that promote differentiation and enable production of synchronous populations of jkRBCs. Global surface proteomic profiling revealed that jkRBCs express all known P. falciparum host receptors in a similar fashion to cRBCs and that multiple P. falciparum strains invade jkRBCs at comparable levels to cRBCs and RBCs. Using CRISPR/Cas9, we deleted two host factors, basigin (BSG) and CD44, for which no natural nulls exist. BSG interacts with the parasite ligand Rh5, a prominent vaccine candidate. A BSG knockout was completely refractory to parasite invasion in a strain-transcendent manner, confirming the essential role for BSG during invasion. CD44 was recently identified in an RNAi screen of blood group genes as a host factor for invasion, and we show that CD44 knockout results in strain-transcendent reduction in invasion. Furthermore, we demonstrate a functional interaction between these two determinants in mediating P. falciparum erythrocyte invasion.
Malaria is an infectious disease caused by Plasmodium parasites and is a major public health burden with upwards of 200 million cases and over 400,000 deaths annually (1). Upon infection of a new host, the parasite replicates in a liver cell, following which it establishes a cyclical infection of RBCs, leading to all the clinical symptoms of disease (2). Invasion of new RBCs occurs rapidly after the release of daughter merozoites from mature schizonts (3), and during the invasion process parasites use multiple invasion ligands to bind to the host RBC by interacting with specific host receptors (4–6). Blocking these interactions can lead to a reduction in parasite invasion (7), a strategy underlying blood-stage vaccine design (8, 9).
Fundamental insights into host–parasite interactions during invasion have come from the analysis of rare, naturally occurring RBC polymorphisms (10) or through biochemical interaction studies using recombinant invasion ligands and recombinant host receptor panels (7, 11). We have focused on a genetic approach, which requires using CD34+ hematopoietic stem cells (HSCs) (12, 13) that allows systematic generation of RBC genetic mutants. Using this system, we have functionally characterized the effects of knockdown of the host receptor GypA on the invasion of the sialic acid-dependent Plasmodium falciparum strain W2mef (14).
There are several challenges in using primary CD34+ HSCs: (i) the short time frame for introducing gene knockdowns during erythroid differentiation may limit the extent of knockdown and precludes obtaining clonal cell populations; (ii) primary cell differentiation is terminal, leading to the need to repeatedly generate gene knockdowns for each assay; and (iii) invasion screening and functional characterization of gene knockdowns require large numbers of cells, which can be costly to generate. The broad range of genetic techniques facilitated by the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 system (15–19) is highly desirable for modifying RBC host factors to investigate Plasmodium invasion, but the use of these techniques remains challenging in primary CD34+ cells (20, 21).
Here we have developed an in vitro culture system using the immortal JK-1 erythroleukemia cell line (22) that permits the rapid and efficient generation of RBC genetic mutants and overcomes the challenges of using primary CD34+ HSCs. JK-1 cells spontaneously differentiate at low rates to form cells that resemble young, nucleated RBCs. We have developed methods for enriching differentiated cells, and to reduce heterogeneity we screened a library of epigenetic regulators for compounds that induce differentiation. Importantly, the differentiated JK-1 cells support invasion by P. falciparum, and, combined with the ability to genetically modify the cells, provides a platform for the functional characterization of host factors important for parasite invasion. Using this system, we have generated a knockout of the essential host receptor basigin (BSG), for which no natural nulls exist and which binds the parasite invasion ligand Rh5 (23, 24), now a leading vaccine candidate (9). We show that the ΔBSG-knockout line is completely refractory for parasite invasion, thus validating BSG as an essential receptor for P. falciparum invasion (9, 25).
In a recent shRNA-based forward genetic screen of 42 blood group genes, we identified two host factors important for parasite invasion, CD55 and CD44 (26). CD55 was functionally characterized as an essential host factor for invasion through use of natural CD55 RBC-null cells; however, similar natural nulls were not available for CD44. Using the JK-1 cell system, we have generated a CD44 knockout, and we show that this knockout line displays a pronounced reduction in invasion across multiple parasite strains, confirming the importance of CD44 for P. falciparum invasion. As CD44 has been reported to interact with BSG (27–29), we investigated the functional significance of this interaction by using an α-BSG antibody to inhibit invasion. We find that the CD44 knockout enhances the α-BSG–dependent inhibition of invasion, indicating a functional interaction between BSG and CD44 during parasite invasion.
Results
JK-1 Erythroleukemia Cells Models Erythropoiesis in Vitro.
While P. falciparum preferentially invades mature RBCs, it is also capable of invading nucleated RBCs, primarily orthochromatic erythroblasts (14, 26, 30). As such, we were interested in testing the ability of immortal erythroleukemia cell lines to differentiate, form RBCs, and support parasite invasion. A search of the literature identified 10 different erythroleukemia cell lines that we were able to obtain and culture in the laboratory: Ery-1 (31), K562 (32), KH88/C2F8 (33), B4D6 (33), LAMA-84 (34), TF-1A (35), HEL92.1.7 (36), OCIM (37), OCIM-2 (37), and JK-1 (22) (Fig. S1A). During routine culture, we observed spontaneous differentiation into predominantly polychromatic-like nucleated RBCs (38) only in the JK-1 cell line. The JK-1 erythroleukemia cell line was isolated from an individual with Philadelphia chromosome-positive chronic myelogenous leukemia and is reported to express HbF (22). A typical JK-1 culture produced a stochastically fluctuating mixture of erythroid-like cells of different sizes and at different stages of differentiation (Fig. 1). A majority (>80%) of the actively dividing cells was composed of less differentiated proerythroblasts and basophilic erythroblasts. The differentiated nucleated RBCs in the JK-1 cell-line consisted of primarily early- and late-stage polychromatic erythroblasts, with a very small fraction (<1%) of orthochromatic erythroblasts [characterized by fully condensed nuclei (38)] and occasional (<0.5%) enucleated cells (resembling reticulocytes).
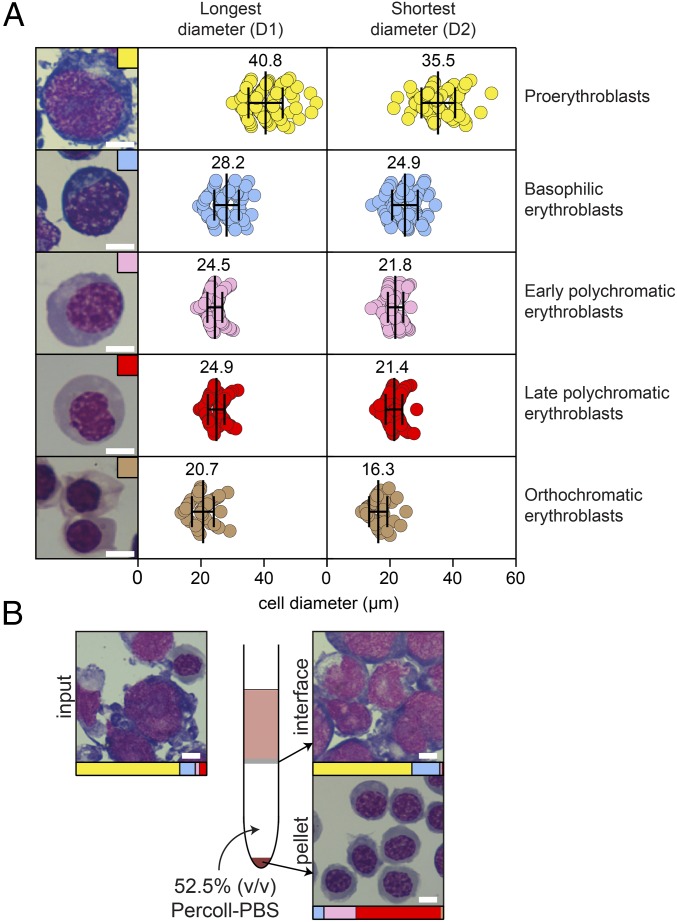
Fig. 1.
The JK-1 erythroleukemia cell line models erythropoiesis, and homogenous populations of differentiated cells can be obtained by density sedimentation and FACS. (A) Cells at different stages of differentiation, including proerythroblasts, basophilic erythroblasts, polychromatic erythroblasts (early and late), and orthochromatic erythroblasts (38, 94), were observed in a typical JK-1 cell culture, and representative images of each stage are shown in the left panel. The dimensions of 50–100 cells of each stage were measured from stained images using Photoshop version 13.0. For each cell, a measurement was made of both the longest (D1) and shortest (D2) diameter, and the average (indicated numerically and by the vertical line in the diagram) and SD (indicated by whiskers) for each range are shown. (B) Layering of a mixed population of JK-1 cells on a 52.5% (vol/vol) Percoll–PBS gradient leads to the enrichment of early- and late-stage polychromatic cells in the pellet fraction and retention of undifferentiated proerythroblast and basophilic cells at the interface. For all images, cells were stained with May–Grünwald Giemsa. (Scale bars: 10 μm.)
Given the heterogeneity of normal JK-1 cell cultures, we tested a number of different techniques to specifically enrich for different cell populations. We observed that cell size varied based on the stage of differentiation, with undifferentiated proerythroblasts having almost twofold larger diameters than differentiated polychromatic and orthochromatic erythroblasts (Fig. 1A). We first tested whether we could use FACS to separate cells based on size. Using forward-scatter (FSC) and side-scatter (SSC) parameters, we found a gate that resulted in the enrichment of basophilic early- and late-stage polychromatic cells (small cell gate) and a gate that enriched for proerythroblasts (large cell gate) (Fig. S1B). As FACS is time and resource intensive for sorting large numbers of cells, we next tested whether we could enrich cells using a bulk method. As Percoll density gradients have been used to enrich for HSCs from bone marrow extracts (39), we tested whether this method would be feasible for JK-1 cells. Centrifuging a mixed population of JK-1 cells through a 52.5% (vol/vol) Percoll–PBS gradient resulted in an ∼15-fold enrichment of differentiated early- and late-stage polychromatic cells in the cell pellet, while proerythroblasts and basophilic erythroblasts were retained at the interface between the Percoll gradient and the culture medium (Fig. 1B). We found that the Percoll–PBS method was faster than FACS and was simple to scale up for large numbers (>108) of cells.
Bromodomain Inhibition Induces Differentiation of JK-1 Cells.
While FACS and Percoll–PBS allowed us to enrich for terminally differentiated nucleated RBCs (herein, “jkRBCs”), only a relatively small proportion (10–15%) of a typical JK-1 culture contained differentiated cells. Therefore, we were interested in finding ways of increasing the proportion of differentiated nucleated RBCs in a synchronous manner. We hypothesized that, as JK-1 cells display spontaneous differentiation, this process might be under epigenetic control, and indeed epigenetic regulators have been reported to induce cellular differentiation (40–42). We screened an epigenetic library for small-molecule inducers of differentiation. To begin, we required a method of quantitatively monitoring JK-1 differentiation. We observed that expression of glycophorin A (GypA), a cell-surface marker of erythrocyte maturation (43, 44), was correlated with JK-1 differentiation (Fig. S2A). Using a FITC-labeled α-GypA antibody, we observed two distinct populations: the GypA− fraction (gated based on unstained cells) contained predominantly proerythroblasts and basophilic erythroblasts, while the GypA-high fraction was enriched for early- and late-stage polychromatic cells.
Using this method, we sorted for a population of GypA− JK-1 cells and screened these cells with a library of 96 epigenetic modifiers (Cayman Chemicals). This library includes small molecules that target a wide variety of epigenetic regulatory proteins. Two rounds of screening were performed with cells harvested after 5 d in screen 1 and after both 7 and 14 d in screen 2. Upon harvest, the levels of GypA were measured by flow cytometry, and the GypA-high:GypA− ratio was calculated (Fig. S2B). The data from the screen were ranked by hierarchical clustering (Fig. 2A and Dataset S1). Six compounds were identified that displayed substantial induction of JK-1 cell differentiation. Significantly, four of the six top compounds included inhibitors of bromodomain-containing proteins: two compounds, (+)-JQ1 (45) and PFI-1 (46), target mammalian bromodomain and extra terminal domain (BET) proteins; bromosporine is a general bromodomain inhibitor (47); and I-CBP112 targets the bromodomain of cAMP-responsive element-binding protein binding protein (CREBBP)/E1A-associated protein p300 (EP300) (48, 49). The only other bromodomain-specific inhibitor in the library, PFI-3, targets a different category of bromodomain-containing proteins (50) and was not found to be an inducer of JK-1 differentiation. The two other top inducers, GSK343 (51) and UNC1999 (52), both target the catalytic core of the polycomb repressive complex 2 (PRC2) enhancer of zeste homolog 2 (EZH2) histone methyltransferase (53).
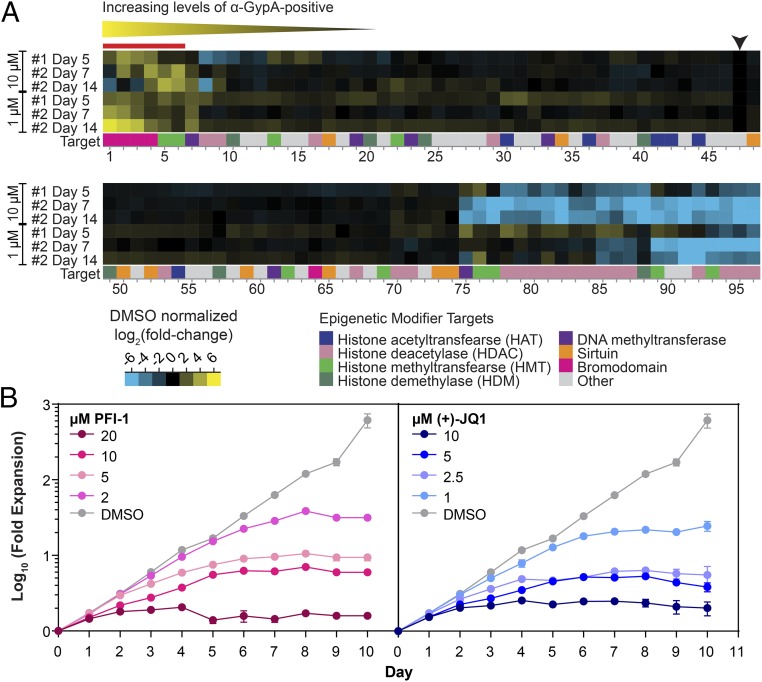
Fig. 2.
Screen of an epigenetic library to identify inducers of JK-1 differentiation. (A) Heat-map showing the results of an epigenetic screen for inducers of JK-1 differentiation. The signals have been normalized to the DMSO control (black arrow). Of the top six inducers (red bar), four of the compounds target bromodomain-containing proteins, indicating a possible conserved mechanism of differentiation induction. The full list of the 96 epigenetic modifiers is provided in Dataset S1. (B) Validation of two of the top hits from the epigenetic screen [PFI-1 and (+)-JQ1] showing the fold expansion of undifferentiated JK-1 cells at different concentrations of compounds. Data shown are averages and SDs from three technical replicates from a representative biological assay.
We validated two top hits, PFI-1 and (+)-JQ1, by testing differentiation with a range of concentrations of compound (Fig. 2B). In both cases, at high concentrations of compound, cell expansion was inhibited, while at the lowest concentration of compound cell growth was similar to that of the DMSO-treated control cells for the first 6 d, after which the cell expansion plateaued. We next monitored cells treated with the optimal concentrations of PFI-1 (2 µM) and (+)-JQ1 (1 µM) during differentiation by staining for GypA and CD34, which is a marker for early hematopoietic stem cells (Fig. 3A) (43). Both PFI-1 and (+)-JQ1 treatment led to a rapid increase of GypA surface levels resulting in a homogenous population of GypA+ cells by 8 d postinduction, while the uninduced control had a broad mixture of cells with different levels of GypA. CD34 levels also decreased rapidly and were almost undetectable by day 4 in the induced conditions but remained at a low level in the control cells. Analysis of cell morphology (Fig. 3B) demonstrated that both the PFI-1 and the (+)-JQ1 treatment resulted in the formation of differentiated cells, whereas the DMSO-treated control cells remained a mixed population. When PFI-1 and (+)-JQ1 were tested in combination (Fig. S2 C and D), differentiation was still observed but with reduced cell expansion.
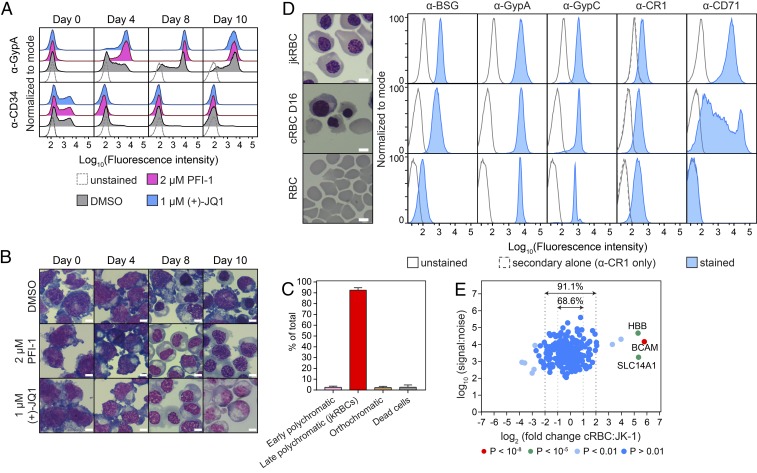
Fig. 3.
Generation of synchronous jkRBCs and comparison with cRBCs and peripheral RBCs. (A) Flow cytometry plots showing changes in expression of GypA and CD34 during differentiation for DMSO-treated control cells and cells treated with 2.0 µM PFI-1 or 1.0 µM (+)-JQ1. (B) Representative microscopy images of differentiating cells stained with May–Grünwald Giemsa. (Scale bars: 10 μm.) (C) Demonstration of the homogeneity of induced JK-1 cells at 12–14 d postinduction with 2 µM PFI-1 and after passage through 52.5% (vol/vol) PBS–Percoll gradients. The average and SD from 10 independent inductions are shown, with counts from at least 1,000 cells per experiment. (D) Comparison of jkRBCs, day 16 cRBCs, and peripheral RBCs. Representative microscopy images are shown alongside flow cytometry plots measuring the expression of the known host receptors GypA, GypC, and CR1 as well as the immature erythroid cell marker CD71. (E) The relative abundance of the 237 surface-membrane proteins identified by quantitative surface proteomics was compared in jkRBCs and day 16 cRBCs. P values were estimated using Benjamini–Hochberg-corrected significance A values as previously described for this approach (56, 87, 95), and proteins with a highly significant fold change are indicated.
Inhibition with LSD1 Inhibitors Maintains an Undifferentiated State.
While the primary focus of the epigenetic screen was to identify compounds that induce synchronous differentiation, we were also interested to see if there were compounds that had the opposite effect. In the screen (Fig. 2A) we observed a number of compounds that had low levels of differentiation as measured by α-GypA staining. Examination of the flow cytometry data for these compounds over the different days of the assay indicated that the majority of this effect was due to the toxicity of the compounds, as the initial GypA− population had not expanded. However, we identified one compound, the lysine-specific demethylase 1 (LSD1) inhibitor tranylcypromine, which was able to maintain growth of cells in an undifferentiated state. When a population of GypA− cells was treated with tranylcypromine, the cells grew at the same rate as DMSO-treated control cells, doubling once every ∼30 h (Fig. S2E). Tranylcypromine-treated cells retained similar levels of GypA, CD34, and CD71 (transferrin receptor) over multiple generations, while DMSO-treated controls showed pronounced increases in GypA levels and reductions in CD34 levels over the same period (Fig. S2F).
JkRBCs Functionally Resemble Nucleated RBCs.
Having identified epigenetic factors that could control JK-1 differentiation, we next tested the synchronicity of the differentiated cells. Starting with cells maintained on 10 µM tranylcypromine, we induced differentiation of these cells with 2 µM PFI-1, and at 12–14 d postinduction cells were harvested and passaged through 52.5% (vol/vol) PBS–Percoll. The resulting cells displayed a high degree of homogeneity and consisted of >90% late-stage polychromatic cells (Fig. 3C). Next, we compared these jkRBCs to bone marrow-derived CD34+ HSCs (cRBCs) and peripheral RBCs. The cRBCs were at day 16 post thaw and consisted of a mixture of cells including basophilic erythroblasts, early- and late-stage polychromatic erythroblasts, orthochromatic erythroblasts, reticulocytes, and pyrenocytes (ejected nuclei) (Fig. S3A). Analysis of three independent biological cRBC cultures at 16–17 d post thaw showed that a majority of cells were orthochromatic erythroblasts and reticulocytes (together, >70%), while earlier-stage basophilic and polychromatic erythroblasts were present at much lower frequencies (<10%) (Fig. S3B). A comparison of cell diameter showed that the jkRBCs (Fig. 1A) were ∼1.25-fold larger on average than the dominant cRBCs (orthochromatic erythroblasts and reticulocytes). During the process of P. falciparum invasion, the parasite interacts with numerous host surface-membrane proteins on the surface of peripheral RBCs. To check whether jkRBCs expressed known host receptors, we performed flow cytometry to compare the levels of expression of BSG, GypA, GypC, CR1, and CD71 in jkRBCs, cRBCs, and peripheral RBCs (Fig. 3D). The relative flow cytometry signal for three of the known host receptors (GypA, GypC, and CR1) were tightly correlated in jkRBCs, cRBCs, and peripheral RBCs. The level of BSG was higher in jkRBCs and cRBCs and was about 10-fold lower in peripheral RBCs, suggesting that levels of this protein change substantially during the final stages of erythroid maturation. As a control, we measured the levels of transferrin receptor (CD71), which is abundant on jkRBCs and cRBCs but is absent from peripheral RBCs, as has been observed previously (54, 55).
We next performed a global analysis of the surface-membrane protein composition of jkRBCs by quantitative surface proteomics (26, 56). We identified 237 surface-membrane proteins by two or more peptides from a total of 677 identified proteins (Dataset S2). We compared this dataset with available RBC proteomes (Fig. S3C) and were able to identify 92.2% of the jkRBC proteins in one or more of the published proteomes. The dataset with the greatest overlap (85.9%) included proteomes not only of mature RBCs but also of erythroid progenitors (57). Next, we used quantitative surface proteomics to compare the relative abundance of surface-membrane proteins between jkRBCs and an equal number of day-16 cRBCs. The relative abundance of a large proportion (68.6%) of the cRBC membrane proteins was within a twofold (±) range of the equivalent jkRBC proteins, and 91.1% were within a fourfold (±) range (Fig. 3E). A comparison of the blood group proteins (Fig. S3D) showed a similar pattern. The majority of proteins, including the known P. falciparum host receptors GypA, GypC, CR1, and BSG, were within a twofold range. We are not able to distinguish between GypA and GypB by this method so the signal we observe for GypA is a combination of GypA and GypB. We also identified three proteins with greater than fourfold abundance (BCAM, CD99, and SLC14A1) in cRBCs compared with jkRBCs.
JkRBCs Support Invasion by P. falciparum.
We next tested the ability of jkRBCs to support P. falciparum invasion, as has been observed for other nucleated erythroid precursors (14, 30, 58). Indeed, we observed invasion into jkRBCs by two different strains of P. falciparum: the sialic acid-independent strain 3D7 (59) and the sialic acid-dependent strain Dd2 (Fig. 4A) (60). To compare the invasion efficiency of P. falciparum into jkRBCs, cRBCs, and RBCs, we measured the parasitized erythrocyte multiplication rate (PEMR) (i.e., the percent final ring parasitemia/the percent initial schizontemia) in the different cell types. The invasion rates of P. falciparum strains 3D7 and Dd2 into jkRBCs were comparable to the invasion into cRBCs and RBCs (Fig. 4B), suggesting that jkRBCs express sufficient levels of all relevant host receptors and possess the requisite glycosylation required for parasite binding and invasion (4, 6). Since we often observed multiple parasites invading a single host jkRBC, we quantified the preference for multiple parasite invasion events by determining the selectivity index (Fig. S3E) (61). The selectivity index is a measure of the observed number of multiply infected cells compared with the number that would be expected by chance based on a Poisson distribution and can be used to determine the susceptibility of host cells to invasion by Plasmodium parasites. The jkRBCs showed the highest selectivity index, followed by cRBCs and RBCs. To determine whether P. falciparum parasites grew normally in jkRBCs, we assessed parasite growth during a single cycle (Fig. S4). While parasites were occasionally observed to develop into trophozoites and schizonts, development of these stages was significantly impeded in jkRBCs compared with RBCs.
Fig. 4.
JkRBCs support invasion of multiple strains of P. falciparum at levels comparable to cRBCs and RBCs. (A) Representative images of P. falciparum 3D7 and Dd2 parasites after successful invasion into jkRBCs. (Scale bars: 10 μm.) (B) PEMR values (% final ring parasitemia/% initial schizontemia) for P. falciparum 3D7 and Dd2 strains were similar in jkRBCs, cRBCs, and RBCs. Data shown are the average and SD are from four biological replicates.
Generation of a BSG Knockout via CRISPR/Cas9.
We next tested if it was possible to genetically manipulate the JK-1 cells. We transduced the JK-1 cells with a lentivirus expressing a shRNA targeting GYPA and monitored protein levels by flow cytometry. We were able to detect a substantial decrease in GypA protein expression within about 1 wk posttransduction (Fig. S5A), thus confirming that shRNA gene knockdowns were supported by JK-1 cells. We then attempted to generate gene knockouts using the CRISPR/Cas9 gene-editing system (Fig. S5B) (19). We chose the human BSG gene encoding the basigin receptor [Ok blood group (62)], which is an essential receptor for P. falciparum (7). We first generated lentivirus containing the LentiCas9-Blast plasmid (19) and introduced it by viral transduction into JK-1 cells. Cells were selected by growth on blasticidin until a stable JK-1-Cas9 cell line was obtained. No toxicity or difference in growth rate associated with Cas9 expression was observed. Next, three single-guide RNAs (sgRNAs) targeting BSG were individually cloned into the LentiGuide-Puro vector (19), and these constructs were virally transduced into LentiCas9+ JK-1 cells. After 2–4 wk of selection, single-cell clones were obtained by limiting dilution of the bulk population. The presence of gene knockouts in these clonal cell lines was assessed by loss of α-BSG flow cytometry staining and subsequently was verified by Sanger sequencing and tracking of indels by decomposition (TIDE) analysis (Fig. S5 C–E). Of the three sgRNAs we tested, we only observed one guide that showed a complete loss of α-BSG flow staining in the bulk population (BSG-1 sgRNA), and following cloning two individual clonal lines (ΔBSG-1 and ΔBSG-2) were obtained from this sgRNA, both with different deletions in each gene copy (Fig. 5 C and D). The BSG-1 sgRNA targeted the N terminus of the BSG protein, and the resulting deletions disrupted the initiator methionine ATG codon (Fig. S5E).
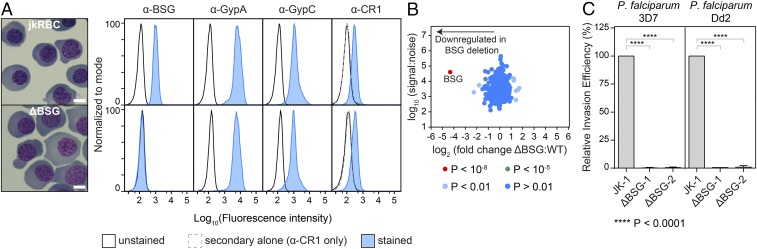
Fig. 5.
Use of CRISPR/Cas9 to generate BSG knockout in JK-1 cells. (A) The ΔBSG clone has no surface expression of BSG but retains expression of known host receptors (GypA, GypC, and CR1) at levels comparable to the levels in jkRBCs. (Scale bars: 10 μm.) (B) Quantitative surface proteomics analysis comparing the abundance of 237 surface-membrane proteins in WT jkRBCs and ΔBSG jkRBCs confirms the specific loss of BSG in the ΔBSG cells. (C) The ΔBSG-knockout JK-1 cell line is refractory to P. falciparum Invasion. Invasion of the sialic acid-independent P. falciparum strain 3D7 and sialic acid-dependent P. falciparum strain Dd2 was completely inhibited in two independent clones of ΔBSG. Data are normalized to the invasion efficiency of WT JK-1 cells and are representative of three biological replicates. Error bars represent SDs from three biological replicates. P values were calculated using a two-tailed t test (GraphPad Prism version 7.01).
To validate that the BSG-knockout jkRBCs did not have any RBC developmental defects, we compared the expression levels of BSG, GypA, GypC, and CR1 by flow cytometry for JK-1 WT and ΔBSG jkRBCs (Fig. 5A). The ΔBSG cells showed a complete loss of α-BSG signal, confirming a functional loss of BSG protein. The levels of GypA, GpyC, and CR1 were very similar in the WT and ΔBSG lines. To further confirm that deletion of BSG did not result in changes to any other surface-membrane protein, we compared the abundance of surface proteins in ΔBSG jkRBCs and WT jkRBCs using quantitative surface proteomics (Fig. 5B and Dataset S2). Our data demonstrate that the knockout of BSG was specific and did not lead to the significant alteration of other surface-membrane proteins.
BSG Is Essential for P. falciparum Invasion.
Basigin is proposed to be an essential receptor for P. falciparum (7), and we have previously demonstrated that knockdown of BSG in CD34+ HSCs via shRNA leads to a substantial (∼80%) decrease in invasion by multiple strains of P. falciparum (7). While there is strong evidence that BSG is an essential receptor for P. falciparum, the residual invasion observed with the BSG knockdown (7) raised some doubts about whether loss of BSG would completely block P. falciparum invasion. There are natural BSG polymorphisms that occur as part of the Ok blood group (62), but to date no natural BSG nulls have been described. To determine the effect of deleting BSG in jkRBCs, we performed invasion assays with two different strains of P. falciparum, 3D7 and Dd2, using two independent BSG-knockout clones, ΔBSG-1 and ΔBSG-2, both generated using the BSG-1 sgRNA. We observed a complete inhibition of invasion into both ΔBSG clones for both P. falciparum strains (Fig. 5C). This result provides strong evidence that BSG is required for strain-transcendent invasion.
CD44 Is a Strain-Transcendent Invasion Host Factor.
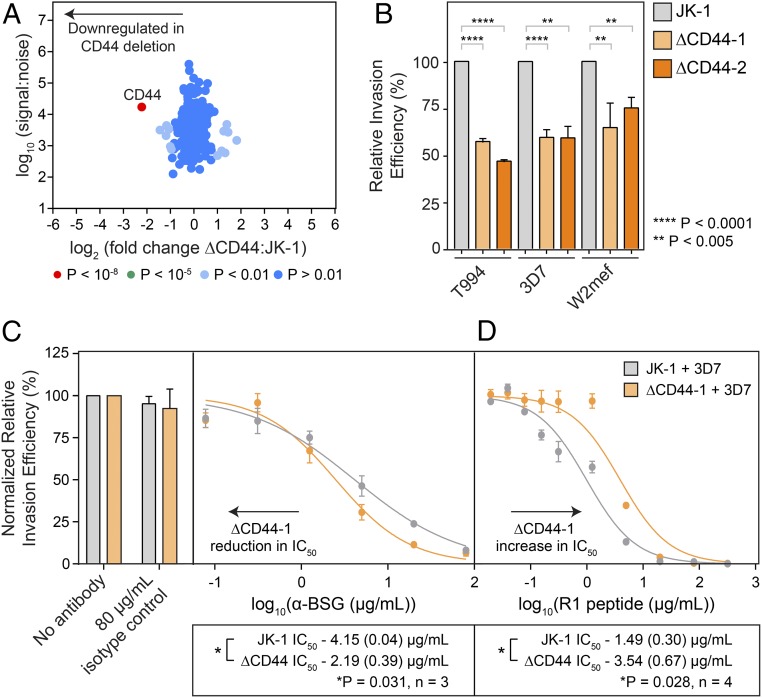
CD44 was identified as a potential host receptor during a screen of blood group gene knockdowns (26). Knockdowns of CD44 in CD34+ HSCs led to a modest reduction in P. falciparum invasion, but this was tested only in the 3D7 strain, and functional characterization of CD44 was limited by the lack of naturally occurring CD44 nulls. Therefore, we generated a knockout of CD44 using CRISPR/Cas9 and obtained mutant cells with an insertion in the exon 2 that leads to the formation of a premature stop codon and truncation of the protein in the N-terminal extracellular domain (Fig. S6 A and B). We confirmed that the knockout of CD44 was specific by flow cytometry (Fig. S6C) and quantitative surface proteomics (Fig. 6A) in which we did not observe a significant change in abundance specifically of any known host receptor (BSG, GypA, GypC, CR1) or other surface-membrane protein. We next tested invasion of multiple P. falciparum strains into two CD44-knockout clonal lines, ΔCD44-1 and ΔCD44-2, and observed a consistent inhibition of invasion (∼30–40%) across multiple parasite strains, including the sialic acid-independent 3D7 and sialic acid-dependent W2mef strains (Fig. 6B), confirming the importance of CD44 in P. falciparum invasion.
Fig. 6.
CD44 knockout results in a strain-transcendent reduction in parasite invasion. (A) Quantitative surface proteomics analysis confirming that the loss of CD44 in the ΔCD44 clone is specific. (B) Comparison of the invasion efficiency in WT and two ΔCD44-knockout clones with multiple P. falciparum strains demonstrates a consistent reduction in invasion efficiency. Data for each parasite strain are normalized to the invasion efficiency of WT JK-1 cells and are representative of two or three biological replicates. Error bars represent SDs; P values were calculated using a two-tailed t test (GraphPad Prism version 7.01). (C) The ΔCD44 knockout has a reduced sensitivity to inhibition of invasion using the MEM6/6 α-BSG antibody, suggesting a functional interaction between BSG and CD44 during parasite invasion. Data were normalized to the invasion efficiency of the isotype controls, and a representative plot is shown in which error bars represent the SD from three technical replicates. (D) Inhibition of invasion via dilution series of R1 peptide shows that the ΔCD44 knockout has a greater resistance to inhibition compared with WT JK-1 cells. A representative plot is shown with error bars representing SD from three technical replicates. For both C and D, IC50 values were calculated using a least-squares fit with log(inhibitor) vs. normalized response and a variable slope using GraphPad Prism version 7.01. Below the plots, the average IC50 values for JK-1 and ΔCD44 are shown for three biological replicates (α-BSG, C) or four biological replicates (R1 peptide, D) with the SEM indicated in brackets. A paired, two-tailed t test was used to calculate the P value between the IC50 values of JK-1 WT and ΔCD44 knockout for each experiment (α-BSG, C; R1 peptide, D), and in both cases this was found to be *P < 0.05.
CD44 Functionally Interacts with BSG.
CD44 has been reported to interact with BSG in multiple cancer cell lines (27–29), prompting us to test whether there was a functional interaction between CD44 and BSG. To do so, we used the monoclonal MEM6/6 α-BSG antibody, which has previously been shown to inhibit parasite invasion (7), to inhibit P. falciparum 3D7 invasion into JK-1 WT and ΔCD44-1 cells (Fig. 6C). We observed an approximately twofold reduction in the IC50 for the ΔCD44-1 knockouts compared with WT JK-1 cells, indicating that the ΔCD44-1–knockout cells were more sensitive to inhibition by the α-BSG antibody. We next checked if this could be explained by differences in levels of BSG on JK-1 WT and ΔCD44-1–knockout cells. However, we did not observe any significant difference in BSG protein levels by either flow cytometry (Fig. S6D) or quantitative surface proteomics (Dataset S2). Next we tested the effect of inhibition of an invasion step downstream of Rh5/BSG by using the R1 peptide that inhibits the interaction between the parasite factors AMA1 and RON2, which are involved in strong attachment of the invading merozoite (Fig. 6D) (63, 64). In this case, we observed an approximately twofold increase in the IC50 in the ΔCD44-1–knockout cells compared with the WT JK-1 cells, indicating an increased utilization of the AMA1/RON2 interaction in the absence of CD44.
Discussion
A major area of interest in Plasmodium biology has been the identification of essential, strain-transcendent host receptors since their cognate invasion ligands may be potent vaccine candidates (9). We have developed an in vitro culture system for functional analysis of the host contribution to blood-stage P. falciparum invasion using the JK-1 erythroleukemia cell line, which displayed unique features. (i) JK-1 cells naturally produce erythroid-lineage cells (proerythroblast-, basophilic-, polychromatic-, and orthochromatic-like cells), and, using small-molecule epigenetic modifiers, we were able either to maintain the cells in an undifferentiated state or to predictably induce synchronous differentiation to produce jkRBCs (nucleated RBCs). (ii) JkRBCs functionally resembled differentiated cRBCs and peripheral RBCs. The surface-membrane protein composition of jkRBCs was comparable to that of cRBCs and peripheral RBCs, and known P. falciparum host receptors were expressed at levels equal to or greater than those in RBCs and cRBCs. Critically, jkRBCs supported robust invasion of multiple P. falciparum strains, implying that all the requisite host factors were present at sufficient levels for parasite invasion. (iii) JK-1 cells were readily amenable to different genetic modifications such as gene knockdowns via RNAi and gene knockouts via CRISPR/Cas9, which have been challenging in primary CD34+ HSCs (20, 21). (iv) As JK-1 cells are immortal, we were able to generate clonal mutant cell lines and were able to freeze down, thaw, and cost-effectively produce large numbers of WT and mutant cells.
In our screen for epigenetic factors that induce synchronous differentiation of JK-1 cells, the most potent inducers targeted bromodomain-containing proteins. Bromodomain proteins bind to acetylated ε-amino lysine residues on histones and are involved in the regulation of gene expression (65, 66). The two top inducers of JK-1 differentiation, (+)-JQ1 and PFI-1, despite having different chemical scaffolds, target proteins in the BET family, which consists of four members: BRD2, BRD3, BRD4, and BRDT (67, 68). BET family proteins are involved in multiple transcription complexes and help regulate cell growth (69). In the context of erythropoiesis, they promote chromatin occupancy of acetylated forms of the global erythroid transcription factor GATA-1 (70, 71), which itself modulates gene expression during erythropoiesis. With the ability to genetically manipulate JK-1 cells, it may be possible to elicit controlled induction of differentiation independent of epigenetic regulators by directly controlling levels of BET protein expression, e.g., by knockin of destabilization domain tags (72).
The composition of the surface-membrane proteome of jkRBCs shares a high degree of overlap with published RBC proteomes. Our quantitative surface proteomics analysis comparing cRBCs and jkRBCs showed that >90% of proteins were expressed within a fourfold range. Instances of higher protein abundance on jkRBCs compared with cRBCs and RBCs (e.g., BSG) may be explained by (i) the overall larger size of jkRBCs compared with cRBCs and RBCs; (ii) the greater homogeneity and relative immaturity of jkRBCs (late-stage polychromatic cells) compared with cRBCs (orthochromatic cells and reticulocytes); and (iii) the overall decrease in protein abundance per cell during erythropoiesis (57). While we observed variation in the levels of surface-membrane proteins, jkRBCs supported P. falciparum invasion rates equivalent to those in cRBCs, indicating that none of the essential host receptors is limiting. However, it is possible that the variation in surface-membrane protein levels may result in differences in invasion between jkRBCs and cRBCs/peripheral RBCs when host receptor levels are modified (e.g., by enzyme treatment or by knockout of nonessential receptors).
One limitation of jkRBCs is the relative immaturity of these cells compared with cRBCs, as judged by the larger average size, higher levels of CD71, and deficiency in forming reticulocytes. These features likely stem from the cancer-causing mutations (22) that favor continued cell replication instead of terminal differentiation. JK-1 cells have double Philadelphia chromosomes, which is typically linked with the formation of the BCR-ABL kinase oncogene (73). The relative immaturity of jkRBCs and the expression of HbF (22) may explain the delayed growth of P. falciparum postinvasion. A similar effect has been observed with parasite invasion into younger CD34+ basophilic and polychromatic erythroblasts (30). To study parasite growth we could either (i) screen for genetic mutations in JK-1 cells that support parasite growth and/or (ii) adapt parasites to growth in JK-1 cells by long-term propagation, as has been shown for Plasmodium knowlesi adaptation to growth in human reticulocytes (74).
The JK-1 cell-culture system has facilitated functional characterization of two host factors, BSG and CD44, important in P. falciparum invasion. BSG-null cells have not been found naturally, and strain-transcendent inhibition with BSG was demonstrated using anti-BSG or anti-RH5 antibodies, often at high concentrations (7, 25, 75). We have previously generated a BSG knockdown by RNAi in CD34+ HSCs, which showed ∼80% reduction in invasion efficiency (7). We hypothesized that the remaining invasion could be due to residual BSG protein present on the knockdown cells. Using the JK-1 system and CRISPR/Cas9 to generate ΔBSG cell lines has allowed us to confirm that the loss of BSG expression results in complete inhibition of invasion of multiple parasite strains, thus confirming the essential role that BSG plays in parasite invasion.
CD44 was identified as an invasion host factor in a forward genetic RNAi screen of blood group genes (26), but its role in invasion could not be fully characterized due to the absence of natural CD44-null cells. We observed that knockout of CD44 resulted in consistent reduction of invasion efficiency, in a strain-transcendent fashion, confirming the importance of CD44 as a host factor for P. falciparum invasion. Furthermore, we observed a functional interaction between CD44 and BSG, as measured by a reduction in the IC50 of the α-BSG MEM6/6 antibody in ΔCD44-1–knockout cells compared with JK-1 WT cells. This effect is not simply due to decreased levels of BSG in the ΔCD44-1–knockout cells. A possible explanation is CD44 functioning directly as a host receptor at an earlier stage than the Rh5/BSG interaction (76); in this case loss of CD44 would result in a reduced number of parasites successfully reaching the Rh5/BSG step of invasion. Alternatively, based on the reported CD44/BSG interaction (27–29), CD44 might operate directly as a coreceptor with BSG, prompting the parasite to preferentially utilize a subset of BSG bound to CD44 during invasion.
In contrast to the effect of Rh5/BSG inhibition, we observed an increase in the IC50 of the R1 peptide inhibition of AMA1/RON2 in ΔCD44 knockout compared with WT JK-1 cells. AMA1 and RON2 are parasite-derived factors that are host receptor independent and mediate strong attachment of the merozoite (63, 64) at a step downstream of the Rh5/BSG interaction (76). Similar antagonistic effects of inhibition of Rh5/BSG and AMA1/RON2 have been reported previously (77, 78). As the CD44 knockout is synergistic with BSG inhibition, and as AMA1 and RON2 are parasite derived, we suggest that the CD44 function maps with BSG rather than with AMA1/RON2. Therefore, one possible consequence of the loss of CD44 [based on the limited-area model for invasion ligand/host receptor interactions (79)] may be the reduced engagement of an earlier invasion ligand (e.g., Rh5). As such, there would be a subsequent increase in the space available for AMA1/RON2 at the apical end of the merozoite during RBC attachment (77, 79), thus resulting in an increased utilization of the AMA1/RON2 pathway. Of great interest in regard to the function of CD44 during invasion are the identification of any potential parasite invasion ligand, the effect of the previously reported CD44 interaction with cytoskeletal proteins band 4.1 and ankyrin (80), and possible signaling roles of CD44 during invasion, either separately or in parallel with BSG (81).
The versatility of the JK-1 in vitro culture system in supporting both robust parasite invasion and simple genetic manipulation to produce gene knockouts will facilitate the functional analysis of the host contribution to P. falciparum invasion. Indeed, the identification and characterization of essential and strain-transcendent host factors and the parasite molecules with which they interact is a vital aspect of understanding parasite invasion biology and will ultimately aid in the development of vaccines and host-targeted therapeutics.
Materials and Methods
Cell Culture.
The following erythroleukemia cell lines were obtained from the Leibniz Institute Deutsche Sammlung von Mikroorganismen und Zellkulturen collection of microorganisms and cell cultures: JK-1 (catalog no. ACC347), OCIM-1 (catalog no. ACC529), OCIM-2 (catalog no. ACC619), and LAMA84 (catalog no. ACC168). The following erythroleukemia cell lines were obtained from the American Type Culture Collection: HEL 92.1.7 (TIB-180), K562 (CCL-243), and TF-1A (CRL-2451). The C2F8 and B4D6 cell lines were kind gifts of Tatsuo Furukawa, Niigata University School of Medicine, Niigata, Japan (33). The Ery-1 cell line was a kind gift of Michael Arock, Unité CNRS UMR 8147, Paris (31). The erythroleukemia cell lines were propagated in Iscove’s Modified Dulbecco’s Medium (IMDM) with GlutaMAX (Thermo Fisher Scientific) and supplemented with 0.5% (vol/vol) penicillin/streptomycin (Thermo Fisher Scientific) and either 10% AB-positive heat-inactivated serum (Interstate Blood Bank) or 10% AB-positive octaplasLG (OctaPharma) with 2 IU/mL heparin (Affymetrix). Cells were maintained at 1 × 105 to 1 × 106 cells/mL in vented T-flasks (BD Falcon) at 37 °C in a humidified chamber with 5% (vol/vol) CO2. When necessary, cells were frozen in growth medium plus 5% (vol/vol) dimethyl sulfoxide (Sigma-Aldrich). JK-1 clones were obtained by limiting dilution, and all subsequent experiments were performed with the JK-1-7B clone. CD34+ hematopoietic stem cells (Lonza) were cultured as described previously (12, 13, 26). Cytospins were prepared as described previously (26) and stained with May–Grünwald (Sigma-Aldrich) followed by Giemsa (Sigma-Aldrich) according to the manufacturer’s instructions. A double-chamber Neubauer hemocytometer (VWR) was used for live cell counting.
Percoll Density Gradients.
The Percoll (GE Healthcare) density gradients were prepared based on modifications to an existing protocol (39), by mixing stock Percoll (100%) to the indicated final volumetric dilution [e.g., 52.5% (vol/vol)] with one volume of 10× PBS (final concentration 1×) and the remainder with double-distilled H2O. The pH was adjusted to 7.40 with HCl, after which the mixture was filtered through a 0.2-μm sterile filter (Millipore). Typically 4 mL of the gradients were added to a 15-mL Falcon tube, and a suspension of cells in 4 mL IMDM plus 10% AB-positive medium was gently layered on top of the Percoll cushion. The cells were pelleted at 500 × g for 10 min with low acceleration and low braking. Following the centrifugation, the interface and pellet fractions were transferred to separate 15-mL Falcon tubes and washed twice with IMDM.
Flow Cytometry and FACS.
For flow cytometry, 1–5 × 105 cells were washed into flow buffer [PBS plus 0.5% (wt/vol) BSA] and allowed to bind to antibodies for 30 min at room temperature and protected from light. The following antibodies and dilutions were used: Alexa Fluor 647 goat α-mouse, 1:200 (Thermo Fisher Scientific); α-BSG–FITC, 1:100 (Thermo Fisher Scientific); α-CD34–FITC, 1: 20 (Miltenyi Biotech); α-CD71–APC, 1:20 (Miltenyi Biotech); α-CR1, 1:100 (Santa Cruz Biotechnology); α-GypA–FITC, 1:100 (Stem Cell Technologies); α-GypC–FITC, 1:2,000 (Santa Cruz Biotechnology); and α-CD44–APC, 1:20 (Miltenyi Biotech). Samples were washed in flow buffer and analyzed on a Miltenyi MACSQuant instrument equipped with 405-nm, 488-nm, and 638-nm lasers and an autosampler. During flow cytometry measurements, cells were stained with propidium iodide (Miltenyi Biotec) to exclude live/dead cells. Flow cytometry data were analyzed using FlowJo version 10.2. For FACS analysis, cells were sorted on a Bio-Rad S3 cell sorter equipped with both 488-nm and 561-nm lasers.
Epigenetic Library Screening.
A focused library of 96 epigenetic modifiers (Cayman Chemicals) was screened for the ability to induce differentiation of JK-1 cells. Undifferentiated JK-1 cells were obtained by FACS by gating for an α-GypA–FITC–negative population. In the first experimental run, cells were diluted to 4.0 × 104 cells per well in 200 μL JK-1 growth medium in 96-well flat-bottomed plates (Falcon), and epigenetic modifiers were added to 10 μM or 1 μM final concentration using the robotics facility at the Institute of Chemistry and Cell Biology at Harvard Medical School. Cells were grown for 5 d under standard growth conditions before harvesting. In the second experimental run, cells were diluted to 8.0 × 103 cells per well with the same two concentrations (10 μM and 1 μM) of epigenetic modifiers. Half of the cells were harvested at day 7 post setup, and the medium was refreshed for the remainder of the cells, which were allowed to grow until day 14 post setup. The harvested cells were stained with α-GypA–FITC, and the level of GypA was measured by flow cytometry on a Miltenyi MACSQuant flow cytometer (Miltenyi). The GypA-high:GypA− ratio was calculated from plots of SSC vs. α-GypA–FITC (Fig. S2 A and B) for each compound, and the values were normalized to the highest ratio for each concentration of each experimental run. The data were clustered using Gene Cluster version 3.0 (82) by hierarchical clustering with a Euclidean distance similarity metric and complete linkage. The data were visualized using TreeView version 1.1.6r4 (83).
Cloning and Lentivirus Generation.
GuideRNA target sequences were identified bioinformatically using the Broad Institute Genetic Perturbation Platform sgRNA designer tool (https://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design) (84). Primers for the top three hits for BSG and CD44 were synthesized (Integrated DNA Technologies): BSG-1-F 5ʹ–CACCGGCGAGGAATAGGAATCATGG; BSG-1-RC 5ʹ–AAACCCATGATTCCTATTCCTCGCC; BSG-2-F 5ʹ–CACCGTCTTCATCTACGAGAAGCGC; BSG-2-RC 5ʹ–AAACGCGCTTCTCGTAGATGAAGAC; BSG-3-F 5ʹ–CACCGCGTTGCACCGGTACTGGCCG; BSG-3-RC 5ʹ–AAACCGGCCAGTACCGGTGCAACGC; CD44-1-F 5′–CACCGCGTGGAATACACCTGCAAAG; CD44-1-RC 5′–AAACCTTTGCAGGTGTATTCCACGC; CD44-2-F 5′–CACCGACTGATGATGACGTGAGCAG; CD44-2-RC 5′–AAACCTGCTCACGTCATCATCAGTC; CD44-3-F 5′–CACCGCTGTGCAGCAAACAACACAG; and CD44-3-RC 5′–AAACCTGTGTTGTTTGCTGCACAGC. Primer pairs were phosphorylated using T4 polynucleotide kinase (New England Biolabs) and ligated using Quick Ligase (New England Biolabs) into the lentiGuide-Puro vector (19), which had previously been digested with BsmBI (Thermo Fisher Scientific) and dephosphorylated with FastAP alkaline phosphatase (Thermo Fisher Scientific). Ligated plasmids were transformed into Stbl3 bacteria (Thermo Fisher Scientific) and were selected with 100 μg/mL carbenicillin on LB agar plates. Correctly integrated sgRNAs were confirmed by Sanger sequencing using the U6 promoter 5ʹ–GACTATCATATGCTTACCGT (19). Plasmid DNA was purified using MaxiPreps (Qiagen) and was used to generate lentivirus using established protocols (85).
CRISPR/Cas9 Knockouts.
CRISPR/Cas9 knockouts were generated following existing protocols (17, 19). First JK-1 cells were transduced with the LentiCRISPR-Blast vector and were selected with 6 μg/mL blasticidin (Sigma-Aldrich). The blasticidin-resistant cells were next transduced with the LentiGuide-Puro vectors containing each of the three BSG sgRNAs, and cells were selected for by growing with both 6 μg/mL blasticidin and 2 μg/mL puromycin (Sigma-Aldrich). Knockout generation was monitored by flow cytometry, and once cultures had >50% knockout cells, the population was cloned by limiting dilution. Of the three sgRNAs tested only BSG-1 resulted in the generation of BSG knockouts. Individual clones were screened by Sanger sequencing using the primer set BSG-1-seq-F 5ʹ–AAGCAGGAAGGAAGAAATG; BSG-1-seq-RC 5ʹ–TTCACGCCCACACACAGAG followed by TIDE analysis (86) to find biallelic knockouts. Of the three sgRNAs tested for CD44, only CD44-1 resulted in the generation of CD44 knockouts. The DNA region around the target site was PCR amplified using the primers CD44-1-seq-F 5′–AGCGAATTCTGGGATTGTAGGCATGAG and CD44-1-seq-RC 5′–TGTCTAGAGGTGCTGGTCTCTTACCTG, digested with EcoRI (New England Biolabs) and XbaI (New England Biolabs), ligated into a carrier plasmid, and transformed into XL-10 Gold cells (Agilent) to obtain bacterial clones. The DNA sequence around the sgRNA cut site was subsequently obtained by Sanger sequencing.
Quantitative Surface Proteomics.
Plasma membrane profiling was performed as described (56, 87) using 2 × 107 of two batches of WT jkRBCs, one batch each of the two different ΔBSG clones, one batch each of the two different CD44 clones, and one batch of day 16 CD34+ cRBCs. Surface-membrane proteins were identified following labeling of sialic acid residues with aminooxy-biotin and after processing and generation of tryptic peptides were labeled with isobaric tandem mass tag (TMT) reagents (Thermo Fisher) (56) in a 1:1:1:1:1:1:1 ratio. These labeled peptides were enriched and subjected to mass spectrometry as described in SI Materials and Methods.
Invasion Assays.
All parasite assays were performed with either P. falciparum 3D7 attB::TdTomato or P. falciparum Dd2 attB::TdTomato strains unless otherwise indicated (SI Materials and Methods for a description of these lines). Parasites were cultured following established protocols (88, 89) at 2% hematocrit in O-positive blood (Interstate Blood Bank) in complete RPMI medium with 0.5% (wt/vol) albumax and 0.2% (wt/vol) sodium bicarbonate at 37 °C with 5% (vol/vol) CO2 and 1% (vol/vol) O2. Invasion assays were performed as described (14, 26, 77). Typically invasion assays were prepared with 0.5–1.5 × 106 cells in 50 μL complete IMDM medium in a half-area 96-well plate (Corning) with 0.5–2.0% schizonts [enriched by magnetic LD columns (Miltenyi Biotec) (90, 91)]. Cytospins were prepared immediately upon mixing the schizonts and target cells as well as 18–24 h post invasion. Slides were stained with May–Grünwald Giemsa as described, and parasitemia was evaluated by reticle counting (92, 93). For invasion inhibition assays, the MEM6/6 clone of the α-BSG antibody (preservative free) was used (Invitrogen) along with a matched isotype control antibody (preservative free) (Invitrogen). R1 peptide (63, 64) was prepared in complete RPMI medium with 0.5% (wt/vol) albumax and 0.2% (wt/vol) sodium bicarbonate.
Supplementary Material
Acknowledgments
The P. falciparum 3D7attB and Dd2attB parasite lines were a kind gift from Prof. David Fidock (Columbia University). The TdTomato plasmid was a kind gift from Prof. Matthias Marti (University of Glasgow). The LentiCas9-Blast and LentiGuide-Puro vectors were gifts from John Doench and David Root (Broad Institute). We thank James Williamson (Cambridge Institute for Medical Research) for assistance with mass spectrometry and Stewart Rudnicki and Katrina Rudnicki (Institute of Chemistry and Cell Biology at Harvard Medical School) for assistance with preparing the chemical library for screening. U.K. was supported by a Canadian Institutes of Health Research Postdoctoral Fellowship. C.G. was supported by a Swiss National Science Foundation Postdoctoral Fellowship. M.P.W. was supported by Wellcome Trust Senior Clinical Research Fellowship 108070/Z/15/Z. This work was supported by NIH Grants R01AI091787 and R01HL139337 (to M.T.D.) and by Bill and Melinda Gates Foundation Grant OPP1023594 (to M.T.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1711310114/-/DCSupplemental.
References
- 1.WHO . World Malaria Report 2015. WHO; Geneva: 2015. [Google Scholar]
- 2.White NJ, et al. Malaria. Lancet. 2014;383:723–735. doi: 10.1016/S0140-6736(13)60024-0. [DOI] [PubMed] [Google Scholar]
- 3.Dvorak JA, Miller LH, Whitehouse WC, Shiroishi T. Invasion of erythrocytes by malaria merozoites. Science. 1975;187:748–750. doi: 10.1126/science.803712. [DOI] [PubMed] [Google Scholar]
- 4.Cowman AF, Crabb BS. Invasion of red blood cells by malaria parasites. Cell. 2006;124:755–766. doi: 10.1016/j.cell.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Weiss GE, Crabb BS, Gilson PR. Overlaying molecular and temporal aspects of malaria parasite invasion. Trends Parasitol. 2016;32:284–295. doi: 10.1016/j.pt.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 6.Tham WH, Healer J, Cowman AF. Erythrocyte and reticulocyte binding-like proteins of Plasmodium falciparum. Trends Parasitol. 2012;28:23–30. doi: 10.1016/j.pt.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 7.Crosnier C, et al. Basigin is a receptor essential for erythrocyte invasion by Plasmodium falciparum. Nature. 2011;480:534–537. doi: 10.1038/nature10606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richards JS, Beeson JG. The future for blood-stage vaccines against malaria. Immunol Cell Biol. 2009;87:377–390. doi: 10.1038/icb.2009.27. [DOI] [PubMed] [Google Scholar]
- 9.Ord RL, Rodriguez M, Lobo CA. Malaria invasion ligand RH5 and its prime candidacy in blood-stage malaria vaccine design. Hum Vaccin Immunother. 2015;11:1465–1473. doi: 10.1080/21645515.2015.1026496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sim BK, Chitnis CE, Wasniowska K, Hadley TJ, Miller LH. Receptor and ligand domains for invasion of erythrocytes by Plasmodium falciparum. Science. 1994;264:1941–1944. doi: 10.1126/science.8009226. [DOI] [PubMed] [Google Scholar]
- 11.Crosnier C, et al. A library of functional recombinant cell-surface and secreted P. falciparum merozoite proteins. Mol Cell Proteomics. 2013;12:3976–3986. doi: 10.1074/mcp.O113.028357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giarratana MC, et al. Ex vivo generation of fully mature human red blood cells from hematopoietic stem cells. Nat Biotechnol. 2005;23:69–74. doi: 10.1038/nbt1047. [DOI] [PubMed] [Google Scholar]
- 13.Giarratana MC, et al. Proof of principle for transfusion of in vitro-generated red blood cells. Blood. 2011;118:5071–5079. doi: 10.1182/blood-2011-06-362038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bei AK, Brugnara C, Duraisingh MT. In vitro genetic analysis of an erythrocyte determinant of malaria infection. J Infect Dis. 2010;202:1722–1727. doi: 10.1086/657157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cong L, Zhang F. Genome engineering using CRISPR-Cas9 system. Methods Mol Biol. 2015;1239:197–217. doi: 10.1007/978-1-4939-1862-1_10. [DOI] [PubMed] [Google Scholar]
- 19.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mandal PK, et al. Efficient ablation of genes in human hematopoietic stem and effector cells using CRISPR/Cas9. Cell Stem Cell. 2014;15:643–652. doi: 10.1016/j.stem.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hendel A, et al. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat Biotechnol. 2015;33:985–989. doi: 10.1038/nbt.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okuno Y, et al. Establishment of an erythroid cell line (JK-1) that spontaneously differentiates to red cells. Cancer. 1990;66:1544–1551. doi: 10.1002/1097-0142(19901001)66:7<1544::aid-cncr2820660719>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 23.Chen L, et al. Crystal structure of PfRh5, an essential P. falciparum ligand for invasion of human erythrocytes. Elife. 2014;3 doi: 10.7554/eLife.04187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wright KE, et al. Structure of malaria invasion protein RH5 with erythrocyte basigin and blocking antibodies. Nature. 2014;515:427–430. doi: 10.1038/nature13715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Douglas AD, et al. Neutralization of Plasmodium falciparum merozoites by antibodies against PfRH5. J Immunol. 2014;192:245–258. doi: 10.4049/jimmunol.1302045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Egan ES, et al. Malaria. A forward genetic screen identifies erythrocyte CD55 as essential for Plasmodium falciparum invasion. Science. 2015;348:711–714. doi: 10.1126/science.aaa3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slomiany MG, et al. Hyaluronan, CD44, and emmprin regulate lactate efflux and membrane localization of monocarboxylate transporters in human breast carcinoma cells. Cancer Res. 2009;69:1293–1301. doi: 10.1158/0008-5472.CAN-08-2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hao J, et al. Co-expression of CD147 (EMMPRIN), CD44v3-10, MDR1 and monocarboxylate transporters is associated with prostate cancer drug resistance and progression. Br J Cancer. 2010;103:1008–1018. doi: 10.1038/sj.bjc.6605839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grass GD, Tolliver LB, Bratoeva M, Toole BP. CD147, CD44, and the epidermal growth factor receptor (EGFR) signaling pathway cooperate to regulate breast epithelial cell invasiveness. J Biol Chem. 2013;288:26089–26104. doi: 10.1074/jbc.M113.497685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamez PA, Liu H, Fernandez-Pol S, Haldar K, Wickrema A. Stage-specific susceptibility of human erythroblasts to Plasmodium falciparum malaria infection. Blood. 2009;114:3652–3655. doi: 10.1182/blood-2009-07-231894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ribadeau Dumas A, et al. Establishment and characterization of a new human erythroleukemic cell line, ERY-1. Leuk Res. 2004;28:1329–1339. doi: 10.1016/j.leukres.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 32.Lozzio CB, Lozzio BB. Human chronic myelogenous leukemia cell-line with positive Philadelphia chromosome. Blood. 1975;45:321–334. [PubMed] [Google Scholar]
- 33.Furukawa T, et al. Establishment of a new cell line with the characteristics of a multipotential progenitor from a patient with chronic myelogenous leukemia in early erythroblastic crisis. Leukemia. 1994;8:171–180. [PubMed] [Google Scholar]
- 34.Seigneurin D, et al. Human chronic myeloid leukemic cell line with positive Philadelphia chromosome exhibits megakaryocytic and erythroid characteristics. Exp Hematol. 1987;15:822–832. [PubMed] [Google Scholar]
- 35.Hu X, et al. Characterization of a unique factor-independent variant derived from human factor-dependent TF-1 cells: A transformed event. Leuk Res. 1998;22:817–826. doi: 10.1016/s0145-2126(98)00073-3. [DOI] [PubMed] [Google Scholar]
- 36.Martin P, Papayannopoulou T. HEL cells: A new human erythroleukemia cell line with spontaneous and induced globin expression. Science. 1982;216:1233–1235. doi: 10.1126/science.6177045. [DOI] [PubMed] [Google Scholar]
- 37.Papayannopoulou T, Nakamoto B, Kurachi S, Tweeddale M, Messner H. Surface antigenic profile and globin phenotype of two new human erythroleukemia lines: Characterization and interpretations. Blood. 1988;72:1029–1038. [PubMed] [Google Scholar]
- 38.Kaushansky K, et al. Williams Hematology. 9th Ed McGraw-Hill; New York: 2015. [Google Scholar]
- 39.Harrison FL, Beswick TM, Chesterton CJ. Separation of haemopoietic cells for biochemical investigation. Preparation of erythroid and myeloid cells from human and laboratory-animal bone marrow and the separation of erythroblasts according to their state of maturation. Biochem J. 1981;194:789–796. doi: 10.1042/bj1940789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lotem J, Sachs L. Epigenetics and the plasticity of differentiation in normal and cancer stem cells. Oncogene. 2006;25:7663–7672. doi: 10.1038/sj.onc.1209816. [DOI] [PubMed] [Google Scholar]
- 41.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lotem J, Sachs L. Epigenetics wins over genetics: Induction of differentiation in tumor cells. Semin Cancer Biol. 2002;12:339–346. doi: 10.1016/s1044-579x(02)00054-8. [DOI] [PubMed] [Google Scholar]
- 43.Hu J, et al. Isolation and functional characterization of human erythroblasts at distinct stages: Implications for understanding of normal and disordered erythropoiesis in vivo. Blood. 2013;121:3246–3253. doi: 10.1182/blood-2013-01-476390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li J, et al. Isolation and transcriptome analyses of human erythroid progenitors: BFU-E and CFU-E. Blood. 2014;124:3636–3645. doi: 10.1182/blood-2014-07-588806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Picaud S, et al. PFI-1, a highly selective protein interaction inhibitor, targeting BET Bromodomains. Cancer Res. 2013;73:3336–3346. doi: 10.1158/0008-5472.CAN-12-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gallenkamp D, Gelato KA, Haendler B, Weinmann H. Bromodomains and their pharmacological inhibitors. ChemMedChem. 2014;9:438–464. doi: 10.1002/cmdc.201300434. [DOI] [PubMed] [Google Scholar]
- 48.Picaud S, et al. Generation of a selective small molecule inhibitor of the CBP/p300 bromodomain for leukemia therapy. Cancer Res. 2015;75:5106–5119. doi: 10.1158/0008-5472.CAN-15-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zucconi BE, et al. Modulation of p300/CBP acetylation of nucleosomes by bromodomain ligand I-CBP112. Biochemistry. 2016;55:3727–3734. doi: 10.1021/acs.biochem.6b00480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vangamudi B, et al. The SMARCA2/4 ATPase domain surpasses the bromodomain as a drug target in SWI/SNF-mutant cancers: Insights from cDNA rescue and PFI-3 inhibitor studies. Cancer Res. 2015;75:3865–3878. doi: 10.1158/0008-5472.CAN-14-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verma SK, et al. Identification of potent, selective, cell-active inhibitors of the histone lysine methyltransferase EZH2. ACS Med Chem Lett. 2012;3:1091–1096. doi: 10.1021/ml3003346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Konze KD, et al. An orally bioavailable chemical probe of the lysine methyltransferases EZH2 and EZH1. ACS Chem Biol. 2013;8:1324–1334. doi: 10.1021/cb400133j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim KH, Roberts CW. Targeting EZH2 in cancer. Nat Med. 2016;22:128–134. doi: 10.1038/nm.4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pan BT, Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: Selective externalization of the receptor. Cell. 1983;33:967–978. doi: 10.1016/0092-8674(83)90040-5. [DOI] [PubMed] [Google Scholar]
- 55.Liu J, Guo X, Mohandas N, Chasis JA, An X. Membrane remodeling during reticulocyte maturation. Blood. 2010;115:2021–2027. doi: 10.1182/blood-2009-08-241182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weekes MP, et al. Quantitative temporal viromics: An approach to investigate host-pathogen interaction. Cell. 2014;157:1460–1472. doi: 10.1016/j.cell.2014.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gautier EF, et al. Comprehensive proteomic analysis of human erythropoiesis. Cell Rep. 2016;16:1470–1484. doi: 10.1016/j.celrep.2016.06.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fernandez-Becerra C, et al. Red blood cells derived from peripheral blood and bone marrow CD34+ human haematopoietic stem cells are permissive to Plasmodium parasites infection. Mem Inst Oswaldo Cruz. 2013;108:801–803. doi: 10.1590/0074-0276108062013019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walliker D, et al. Genetic analysis of the human malaria parasite Plasmodium falciparum. Science. 1987;236:1661–1666. doi: 10.1126/science.3299700. [DOI] [PubMed] [Google Scholar]
- 60.Guinet F, et al. A developmental defect in Plasmodium falciparum male gametogenesis. J Cell Biol. 1996;135:269–278. doi: 10.1083/jcb.135.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Simpson JA, Silamut K, Chotivanich K, Pukrittayakamee S, White NJ. Red cell selectivity in malaria: A study of multiple-infected erythrocytes. Trans R Soc Trop Med Hyg. 1999;93:165–168. doi: 10.1016/s0035-9203(99)90295-x. [DOI] [PubMed] [Google Scholar]
- 62.Spring FA, et al. The Oka blood group antigen is a marker for the M6 leukocyte activation antigen, the human homolog of OX-47 antigen, basigin and neurothelin, an immunoglobulin superfamily molecule that is widely expressed in human cells and tissues. Eur J Immunol. 1997;27:891–897. doi: 10.1002/eji.1830270414. [DOI] [PubMed] [Google Scholar]
- 63.Lamarque M, et al. The RON2-AMA1 interaction is a critical step in moving junction-dependent invasion by apicomplexan parasites. PLoS Pathog. 2011;7:e1001276. doi: 10.1371/journal.ppat.1001276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Paul AS, Egan ES, Duraisingh MT. Host-parasite interactions that guide red blood cell invasion by malaria parasites. Curr Opin Hematol. 2015;22:220–226. doi: 10.1097/MOH.0000000000000135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dhalluin C, et al. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 66.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 67.Florence B, Faller DV. You bet-cha: A novel family of transcriptional regulators. Front Biosci. 2001;6:D1008–1018. doi: 10.2741/florence. [DOI] [PubMed] [Google Scholar]
- 68.Filippakopoulos P, Knapp S. The bromodomain interaction module. FEBS Lett. 2012;586:2692–2704. doi: 10.1016/j.febslet.2012.04.045. [DOI] [PubMed] [Google Scholar]
- 69.Dawson MA, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lamonica JM, et al. Bromodomain protein Brd3 associates with acetylated GATA1 to promote its chromatin occupancy at erythroid target genes. Proc Natl Acad Sci USA. 2011;108:E159–E168. doi: 10.1073/pnas.1102140108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stonestrom AJ, et al. Functions of BET proteins in erythroid gene expression. Blood. 2015;125:2825–2834. doi: 10.1182/blood-2014-10-607309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Park A, Won ST, Pentecost M, Bartkowski W, Lee B. CRISPR/Cas9 allows efficient and complete knock-in of a destabilization domain-tagged essential protein in a human cell line, allowing rapid knockdown of protein function. PLoS One. 2014;9:e95101. doi: 10.1371/journal.pone.0095101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Salesse S, Verfaillie CM. BCR/ABL: From molecular mechanisms of leukemia induction to treatment of chronic myelogenous leukemia. Oncogene. 2002;21:8547–8559. doi: 10.1038/sj.onc.1206082. [DOI] [PubMed] [Google Scholar]
- 74.Lim C, et al. Expansion of host cellular niche can drive adaptation of a zoonotic malaria parasite to humans. Nat Commun. 2013;4:1638. doi: 10.1038/ncomms2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bustamante LY, et al. A full-length recombinant Plasmodium falciparum PfRH5 protein induces inhibitory antibodies that are effective across common PfRH5 genetic variants. Vaccine. 2013;31:373–379. doi: 10.1016/j.vaccine.2012.10.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Weiss GE, et al. Revealing the sequence and resulting cellular morphology of receptor-ligand interactions during Plasmodium falciparum invasion of erythrocytes. PLoS Pathog. 2015;11:e1004670. doi: 10.1371/journal.ppat.1004670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Paul AS, et al. Parasite calcineurin regulates host cell recognition and attachment by apicomplexans. Cell Host Microbe. 2015;18:49–60. doi: 10.1016/j.chom.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Williams AR, et al. Enhancing blockade of Plasmodium falciparum erythrocyte invasion: Assessing combinations of antibodies against PfRH5 and other merozoite antigens. PLoS Pathog. 2012;8:e1002991. doi: 10.1371/journal.ppat.1002991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Duraisingh MT, Maier AG, Triglia T, Cowman AF. Erythrocyte-binding antigen 175 mediates invasion in Plasmodium falciparum utilizing sialic acid-dependent and -independent pathways. Proc Natl Acad Sci USA. 2003;100:4796–4801. doi: 10.1073/pnas.0730883100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nunomura W, et al. Regulation of CD44-protein 4.1 interaction by Ca2+ and calmodulin. Implications for modulation of CD44-ankyrin interaction. J Biol Chem. 1997;272:30322–30328. doi: 10.1074/jbc.272.48.30322. [DOI] [PubMed] [Google Scholar]
- 81.Aniweh Y, et al. P. falciparum RH5-Basigin interaction induces changes in the cytoskeleton of the host RBC. Cell Microbiol. 2017;19:e12747. doi: 10.1111/cmi.12747. [DOI] [PubMed] [Google Scholar]
- 82.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 83.Saldanha AJ. Java Treeview–Extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 84.Doench JG, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Moffat J, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 86.Brinkman EK, Chen T, Amendola M, van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42:e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Weekes MP, et al. Latency-associated degradation of the MRP1 drug transporter during latent human cytomegalovirus infection. Science. 2013;340:199–202. doi: 10.1126/science.1235047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 89.Cranmer SL, Magowan C, Liang J, Coppel RL, Cooke BM. An alternative to serum for cultivation of Plasmodium falciparum in vitro. Trans R Soc Trop Med Hyg. 1997;91:363–365. doi: 10.1016/s0035-9203(97)90110-3. [DOI] [PubMed] [Google Scholar]
- 90.Trang DT, Huy NT, Kariu T, Tajima K, Kamei K. One-step concentration of malarial parasite-infected red blood cells and removal of contaminating white blood cells. Malar J. 2004;3:7. doi: 10.1186/1475-2875-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ribaut C, et al. Concentration and purification by magnetic separation of the erythrocytic stages of all human Plasmodium species. Malar J. 2008;7:45. doi: 10.1186/1475-2875-7-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lim C, et al. Improved light microscopy counting method for accurately counting Plasmodium parasitemia and reticulocytemia. Am J Hematol. 2016;91:852–855. doi: 10.1002/ajh.24383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Brecher G, Schneiderman M. A time-saving device for the counting of reticulocytes. Am J Clin Pathol. 1950;20:1079–1083. doi: 10.1093/ajcp/20.11_ts.1079. [DOI] [PubMed] [Google Scholar]
- 94.Rozenberg G. Microscopic Haematology: A Practical Guide for the Laboratory. 3rd Ed Elsevier Health Sciences; Chatswood, Australia: 2011. [Google Scholar]
- 95.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 96.Aingaran M, et al. Host cell deformability is linked to transmission in the human malaria parasite. Plasmodium falciparum. Cell Microbiol. 2012;14:983–993. doi: 10.1111/j.1462-5822.2012.01786.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Coleman BI, et al. Nuclear repositioning precedes promoter accessibility and is linked to the switching frequency of a Plasmodium falciparum invasion gene. Cell Host Microbe. 2012;12:739–750. doi: 10.1016/j.chom.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nkrumah LJ, et al. Efficient site-specific integration in Plasmodium falciparum chromosomes mediated by mycobacteriophage Bxb1 integrase. Nat Methods. 2006;3:615–621. doi: 10.1038/nmeth904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pasini EM, et al. In-depth analysis of the membrane and cytosolic proteome of red blood cells. Blood. 2006;108:791–801. doi: 10.1182/blood-2005-11-007799. [DOI] [PubMed] [Google Scholar]
- 100.Bosman GJ, et al. The proteome of red cell membranes and vesicles during storage in blood bank conditions. Transfusion. 2008;48:827–835. doi: 10.1111/j.1537-2995.2007.01630.x. [DOI] [PubMed] [Google Scholar]
- 101.Lange PF, Huesgen PF, Nguyen K, Overall CM. Annotating N termini for the human proteome project: N termini and Nalpha-acetylation status differentiate stable cleaved protein species from degradation remnants in the human erythrocyte proteome. J Proteome Res. 2014;13:2028–2044. doi: 10.1021/pr401191w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pesciotta EN, et al. A label-free proteome analysis strategy for identifying quantitative changes in erythrocyte membranes induced by red cell disorders. J Proteomics. 2012;76:194–202. doi: 10.1016/j.jprot.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.van Gestel RA, et al. Quantitative erythrocyte membrane proteome analysis with Blue-native/SDS PAGE. J Proteomics. 2010;73:456–465. doi: 10.1016/j.jprot.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 104.Ringrose JH, et al. Highly efficient depletion strategy for the two most abundant erythrocyte soluble proteins improves proteome coverage dramatically. J Proteome Res. 2008;7:3060–3063. doi: 10.1021/pr8001029. [DOI] [PubMed] [Google Scholar]
- 105.Roux-Dalvai F, et al. Extensive analysis of the cytoplasmic proteome of human erythrocytes using the peptide ligand library technology and advanced mass spectrometry. Mol Cell Proteomics. 2008;7:2254–2269. doi: 10.1074/mcp.M800037-MCP200. [DOI] [PubMed] [Google Scholar]
- 106.Wilson MC, et al. Comparison of the proteome of adult and cord erythroid cells, and changes in the proteome following reticulocyte maturation. Mol Cell Proteomics. 2016;15:1938–1946. doi: 10.1074/mcp.M115.057315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Weekes MP, et al. Proteomic plasma membrane profiling reveals an essential role for gp96 in the cell surface expression of LDLR family members, including the LDL receptor and LRP6. J Proteome Res. 2012;11:1475–1484. doi: 10.1021/pr201135e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Boyle MJ, Richards JS, Gilson PR, Chai W, Beeson JG. Interactions with heparin-like molecules during erythrocyte invasion by Plasmodium falciparum merozoites. Blood. 2010;115:4559–4568. doi: 10.1182/blood-2009-09-243725. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.