Significance
During development, transcription factors are necessary not only to pattern the body plan but also to control growth. However, the link between these two developmental components has been difficult to establish. Hox genes are involved in the emergence of a functional digestive system in metazoans, thus providing a potential impact on growth through nutrition. Also, genetic conditions involving these genes lead to important growth retardation. We analyzed several targeted mutant lines at the HoxD locus and found that stunted phenotypes can all be explained by the lack of function of Hoxd3, whose role seems to be critical in the developing gut of suckling mice, perhaps as an adaptation to the milk-dependent early postnatal period in mammals.
Keywords: lncRNA, digestive system, Hox regulation, growth control, CRISPR-cas9
Abstract
During embryonic development, Hox genes participate in the building of a functional digestive system in metazoans, and genetic conditions involving these genes lead to important, sometimes lethal, growth retardation. Recently, this phenotype was obtained after deletion of Haglr, the Hoxd antisense growth-associated long noncoding RNA (lncRNA) located between Hoxd1 and Hoxd3. In this study, we have analyzed the function of Hoxd genes in delayed growth trajectories by looking at several nested targeted deficiencies of the mouse HoxD cluster. Mutant pups were severely stunted during the suckling period, but many recovered after weaning. After comparing seven distinct HoxD alleles, including CRISPR/Cas9 deletions involving Haglr, we identified Hoxd3 as the critical component for the gut to maintain milk-digestive competence. This essential function could be abrogated by the dominant-negative effect of HOXD10 as shown by a genetic rescue approach, thus further illustrating the importance of posterior prevalence in Hox gene function. A role for the lncRNA Haglr in the control of postnatal growth could not be corroborated.
Slow postnatal weight gain phenotypes can be induced by mutations in a range of genes important for developmental patterning. However, the molecular and physiological bases of such growth retardations are often difficult to pinpoint due to the high pleiotropy associated with these genes. While none of the 39 gene members of the Hox family has been included in this broad phenotypic class thus far, we noticed two cases of postnatal growth retardation during the several-years-long studies of our allelic series at the mouse HoxD locus. First, animals homozygous for a complete HoxD cluster deficiency showed about 30% body-mass deficit by the time of weaning. However, given that they suffered severe limb and innervation defects, growth retardation seemed compatible with reduced feeding (1). Secondly, a shorter deficiency removing Hoxd1–Hoxd9 [HoxDDel(1–9)] was associated with postnatal growth retardation in heterozygous mice in the absence of mobility defects. However, some specimens also displayed facial defects, again suggesting the existence of a physical problem potentially affecting the feeding behavior (2).
Highly severe postnatal growth retardation in the absence of any obvious malformations was recently reported following a targeted deletion of the Hoxd antisense growth-associated long noncoding RNA (lncRNA) Haglr in mice. This midget (Mdgt) phenotype, which was recorded in homozygous-mutant specimens (3), displayed a reduced viability attributed to a function of the lncRNA distinct from those associated with either Hoxd1 (4, 5) or Hoxd3 (6, 7), the two genes flanking Haglr in the HoxD cluster. Therefore, in these cases, both recessive and dominant phenotypes were recorded, and they were always associated with loci located in the anterior (telomeric) part of the HoxD gene cluster. In addition, other HoxD alleles showed midgut patterning defects, but their detailed analyses were made difficult by lethality at birth (8). Finally, distant cis-acting regulatory regions necessary for the transcriptional control of these genes in midgut were reported within the large regulatory landscape flanking this gene cluster on its telomeric side (9), emphasizing a potential link between growth retardation and alterations in gut development.
In this study, we tried to find a common explanatory framework for the various cases described above. We decided to complete the allelic series with a few more mutant chromosomes necessary to unambiguously interpret the previous observations relating phenotypes to particular genes. By using CRISPR/Cas9 mutagenesis in embryo, we demonstrate that the growth-retardation phenotype observed in HoxDDel(1–9) mice is caused by the dominant-negative effect of the HOXD10 protein. We also report comparable growth deficits in mice lacking the entire region from Hoxd1 to Hoxd4 [HoxDDel(1–4)] (10) and that a mutation in the HOXD3 DNA-binding domain produces mice with similar growth deficits, in contrast to mice mutant for either HOXD1 or HOXD4. We document pathological signs of intestinal failure in three independent nested deletions. Together, these results converge toward a central role for Hoxd3 in the growth-retardation phenotypes previously recorded and further explain the strong functional hierarchy at work among HOX proteins.
To address the potential function of Haglr in this context, we further engineered a pair of alleles carrying either a deficiency or an inversion of the CpG island containing the bidirectional promoter for both Hoxd1 and the antisense lncRNA. Neither of these two alleles affected postnatal growth, even in the resulting absence of the reported Haglr RNA, suggesting that the Haglr-associated phenotype may result from an interference with Hoxd3 transcription or mRNA stabilization rather than from a direct effect of the lncRNA as previously reported (11). As many of the severely stunted mutant mice largely recovered after weaning, we hypothesize that the suckling period and its particular physiological requirements may vitally depend on the function of the Hoxd3 gene in the gut.
Results
Gain of Function-Dependent Growth Retardation.
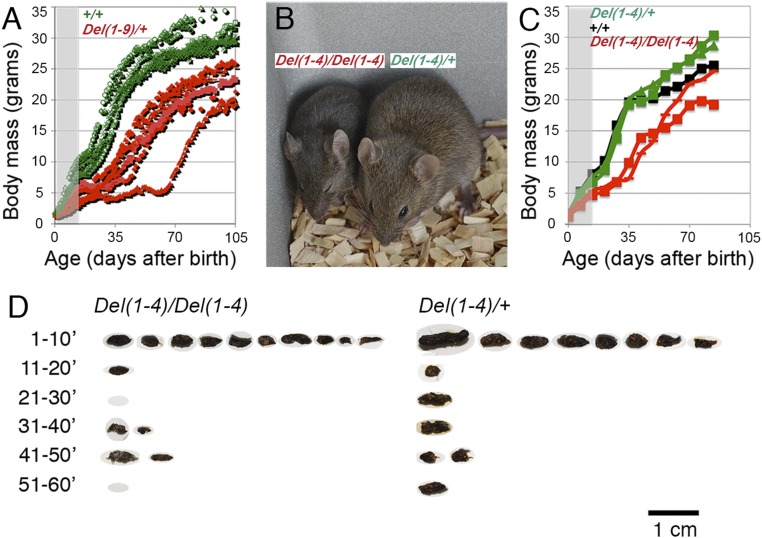
Hoxd deficiencies have been used extensively to study the complex regulation of this gene cluster (12). One of them involving the DNA interval from Hoxd1 to Hoxd9 [HoxDDel(1–9) or Del(1–9), discussed further below] (Fig. S1) resulted in a semilethal slow postnatal weight gain. Heterozygous males crossed with wild-type females produced the predicted 50% proportion of wild-type normal and 50% heterozygous offspring. Soon after birth, growth of half of the progeny lagged. Many growth-retarded individuals dropped below half their normal siblings’ body mass and thus were killed following legal recommendation. However, the majority of growth-retarded specimens survived, and they were genotyped along with their killed littermates. All growth-retarded individuals carried a Del(1–9) chromosome (Fig. 1A; red labels), whereas all normally growing siblings were wild type (Fig. 1A; green labels). Wild-type individuals showed a peak of body growth between postnatal days (P)2 and P14, followed by a few days of slower growth. The growth curve then resumed and continued until the end of the seventh week.
Fig. S1.
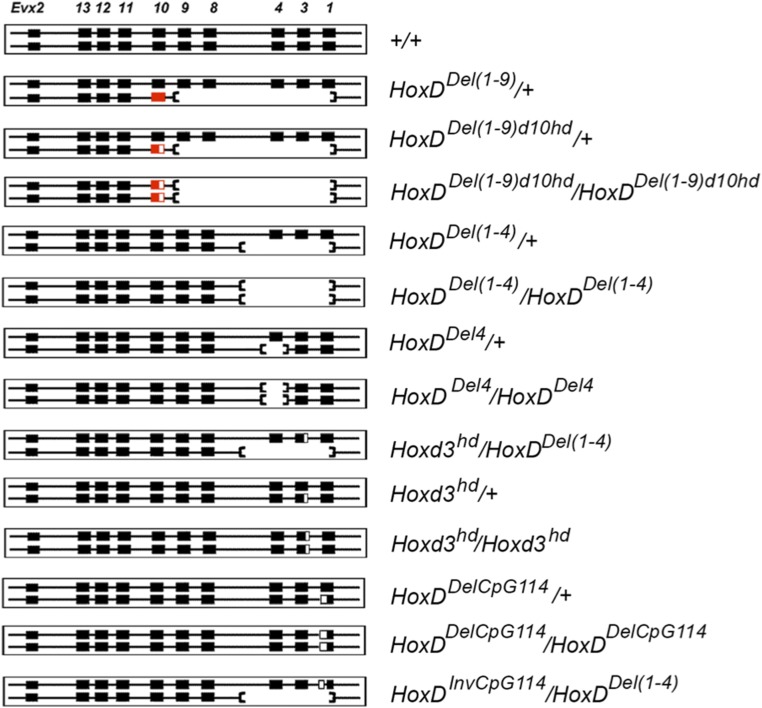
Schematics of the various HoxD-mutant alleles used in this study. On the left are represented the various alleles of the HoxD clusters with the wild-type haplotype at the top. Hoxd1 is on the far right, and Evx2 on the far left, while the other Hoxd genes are indicated by the paralog group. The scheme does not represent relative distances. Coding genes are depicted as single boxes without showing introns. The sequence of the genes reflects the sequence of the homeodomain-containing exons, without reference to alternative promoters. Black boxes indicate the presence of intact Hoxd genes, and red boxes indicate that, in the Del(1–9) allele, Hoxd10 expression was gained outside its normal spatial domain. Half-empty red boxes indicate that the Hoxd10 gene was inactivated by CRISPR/cas9 genome editing. Empty black boxes indicate either that the Hoxd3 homeodomain was deleted or that the CpG114 island was deleted or inverted by CRISPR/cas9 genome editing. Deficiencies are marked by an interruption of the horizontal line and by the absence of gene symbols. The corresponding genotypes are given on the right. Only the superscripts in the names of these alleles were used in Table 1 entries and in the body of the text for brevity.
Fig. 1.
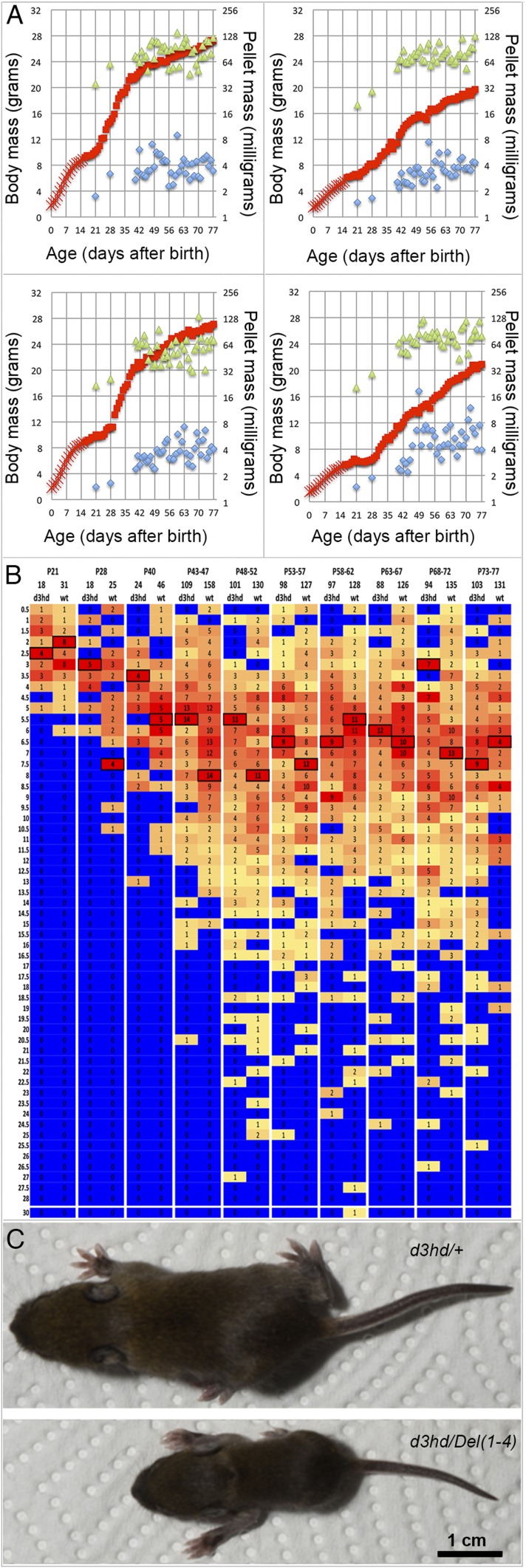
Stunted growth, survival, and vital signs in mouse stocks carrying HoxD cluster deficiencies. (A) Growth trajectories of Del(1–9)-heterozygous (red) and sibling wild-type controls (green) expressed as a scatter diagram of individual daily body mass values. The gray zone on the left indicates that genotypes (determined at P14–P16) are still unknown at these time points, and colors are thus extrapolated from subsequent values. (B) A representative Del(1–4)-homozygous specimen (Left) and a heterozygous sibling on P21. (C) Growth trajectories of Del(1–4)-homozygous (red), -heterozygous (green), and wild-type control (black) siblings expressed as scatter diagrams of individual body mass values obtained on their day of birth and weekly thereafter. The gray zone is as in A. (D) Photographs of droppings released during six consecutive 10-min periods after isolation on P35 by a homozygous and a heterozygous Del(1–4) mutant. Individual dry pellet masses collected during daily 30-min periods of such defecation-reflex episodes were taken as ejected dropping mass, a noninvasive measure of gut function (SI Materials and Methods).
After birth, however, growth of heterozygous Del(1–9) mutants was continuously retarded until the end of the fifth week. For survivors, growth did return, even though it remained depressed in some individuals as late as the end of the tenth week. The analysis of average body mass at 4 wk revealed that heterozygous Del(1–9) individuals had about half the body mass of their wild-type siblings. The comparison between a group of seven wild-type controls and 10 Del(1–9)-heterozygous siblings gave a statistically valid difference (Table 1). It is noteworthy that, once growth of mutant animals resumed, it reached slopes similar to those of their wild-type siblings weeks before. Therefore, while strong growth retardation occurred during milk feeding, growth could catch up after weaning in the surviving specimens.
Table 1.
Body weights of eight cohorts of various HoxD cluster mutants at age 4 wk
| Genotypes | Average weight, g | SD | Fold change | Count | P value |
| +/+ | 14.5 | 1.71 | 1.00 | 7 | |
| Del(1–9)/+ | 7.6 | 2.21 | 0.52 | 10 | 4.55E-06 |
| +/+ | 12.8 | 1.29 | 1.00 | 5 | |
| Del(1–9)d10hd/+ | 10.5 | 1.71 | 0.82 | 11 | 1.33E-02 |
| Del(1–9)d10hd/Del(1–9)d10hd | |||||
| +/+ | 13.8 | 2.2 | 1.00 | 13 | |
| Del(1–4)/+ | 12.5 | 2.55 | 0.91 | 28 | 2.17E-01 |
| Del(1–4)/Del(1–4) | 5.9 | 1.63 | 0.43 | 10 | 9.09E-08 |
| +/+ | 11.3 | 1.82 | 1.00 | 3 | |
| Del (4)/+ | 13.7 | 2.21 | 1.21 | 9 | 1.24E-01 |
| Del (4)/Del (4) | 12.4 | 2.92 | 1.10 | 3 | 5.98E-01 |
| d3hd/+ | 11.0 | 1.96 | 1.00 | 3 | |
| d3hd/Del(1–4) | 7.4 | 0.57 | 0.67 | 4 | 1.46E-02 |
| +/+ | 12.7 | 1.58 | 1.00 | 14 | |
| d3hd/+ | 11.8 | 1.67 | 0.93 | 24 | 1.17E-01 |
| d3hd/d3hd | 8.3 | 2.15 | 0.66 | 20 | 3.30E-07 |
| +/+ | 12.5 | 1.52 | 1.00 | 7 | |
| Del(CpG114)/+ | 12.8 | 1.44 | 1.03 | 13 | 6.00E-01 |
| Del(CpG114)/Del(CpG114) | 11.8 | 0.87 | 0.95 | 8 | 3.04E-01 |
| Del(1–4)/+ | 13.3 | 1.87 | 1.00 | 7 | |
| InvCpG114/Del(1–4) | 12.6 | 1.97 | 0.94 | 8 | 4.69E-01 |
Various HoxD cluster mutant genotypes and body weight statistics are grouped by crosses either between heterozygous and wild-type or between heterozygous animals. Statistical significance was calculated comparing mutants’ body weight values with that of controls from the same cross. Four of fourteen heterozygous individuals in the Del1–9/+ class, all four homozygous individuals in the Del(1–9)d10hd/Del(1–9)d10hd class, and 8 of 23 homozygous individuals in the extended Del(1–4)/Del(1–4) class were killed during the second to fourth weeks due to severe stunting. Several of the Del(1–4)/Del(1–4) escaper individuals of both sexes bred successfully. In the extended Hoxd3hd class, 2 of 24 homozygous individuals were killed during the second to fourth weeks after birth due to severe stunting. One escaper homozygous female bred successfully. P values in bold indicate a significant growth delay.
Previously, we had observed in various alleles carrying partial deletions in the HoxD cluster (13) that the remaining gene located next to the breakpoint after the deletion had occurred was likely to adopt some of the expression specificities of the deleted genes. This observation, along with the dominant inheritance pattern of this Del(1–9) stock, strongly suggested a gain-of-function mechanism, because even a full deficiency of the HoxD cluster did not induce such severe growth retardation in heterozygous-mutant animals (1). Therefore, we suspected Hoxd10 expression was gained in space and time and thus interfered with more anterior Hox gene function following the property of posterior prevalence.
Posterior prevalence was defined as the capacity of some posterior HOX proteins to override the function of more anterior ones when coexpressed in some contexts (14). As this property may rely upon the binding of these proteins on their DNA target sites, we induced a small deletion within the Hoxd10 homeobox sequence in cis with the Del(1–9) deficiency by using the CRISPR/Cas9 technology in fertilized mouse zygotes. In this HoxDDel(1–9)d10hd allele, while the same Del(1–9) deficiency was present, the adjacent Hoxd10 gene produced a protein that lacks its DNA-binding domain. The Del(1–9)d10hd founder animal had growth comparable to its wild-type littermates and was used to establish a stable mutant line.
Early postnatal growth of the Del(1–9)d10hd-heterozygous animals appeared normal, and females no longer showed atypical courtship, as previously reported for the Del(1–9) allele (2). Likewise, facial malformations were no longer recorded. Transheterozygous crosses generated wild-type, heterozygous, and homozygous offspring, as genotyped at 2 wk of age. By the end of the fourth week, the mass of all homozygous animals dropped below the 40% limit in comparison with wild-type siblings, and hence animals were killed before the end of the experiment (Table 1). The comparisons of body mass averages between wild-type and heterozygous mice from two combined litters showed that the mass of heterozygous animals was only about 20% below the average mass of the wild-type controls. This was closer to normal than the nearly 50% deficit seen in presence of the intact Hoxd10 locus in cis with the deficiency. This result demonstrated that a major factor in the dominant effects of the Del(1–9) allele was indeed due to a gain of function of Hoxd10 after its relocation near the deletion breakpoint. The nearly 20% difference recorded between wild-type and Del(1–9)d10hd-heterozygous mice was statistically significant, suggesting that beyond the gain of function of Hoxd10, a combined loss of function of several genes in cis also had an impact upon the growth rate.
Loss of Function-Dependent Growth Retardation.
Next we investigated a smaller deficiency including Hoxd1 to Hoxd4 [HoxDDel(1–4) or Del(1–4)], as homozygous specimens were severely retarded, and many did not survive to adulthood, thus further supporting a loss-of-function mechanism being causative of growth retardation. This was unexpected, since previous targeted loss of function of Hoxd1 (4, 5), Hoxd3 (6, 7), or Hoxd4 (15) did not reveal such growth retardation. This phenotype was highly reproducible and was stable over generations, and homozygous escapers were visibly smaller than normal (Fig. 1B). The growth curves of homozygous, heterozygous, and wild-type controls from the same litter and fed by the same dam showed that homozygous specimens lagged in much the same way as Del(1–9)-heterozygous escapers (Fig. 1C). In both conditions, the retardation was similarly recorded by the end of the first week, and animals continued to grow at a reduced rate compared with heterozygous or wild-type littermates until the end of the seventh week. One homozygous animal continued to grow at a relatively steady rate until it caught up with the wild type by the end of the 11th week, whereas the other reached about 80% of wild-type body weight.
Several such caught-up individuals otherwise proved healthy and fertile. However, more than half of them dropped below 40% of control sibling body mass before the end of the seventh week. Growth trajectories diverged during the period of milk feeding. From these results, a genetically determined period of sensitivity seems to occur between birth and the end of the fourth week. This period was characterized by a reduced capacity to thrive despite the ability to feed. While wild-type and heterozygous body mass values were not appreciably different (Table 1), the average body mass of homozygous individuals was well below half the average of the wild-type individuals. This was comparable to the body mass deficit we observed with Del(1–9)-heterozygous mice, and the recovery of the homozygous Del(1–4) individuals was also similarly shifted (Fig. 1, compare A and C). We also observed that Del(1–9)d10hd-heterozygous displayed twice as great a body mass deficit as Del(1–4)-heterozygous (Discussion).
Coprometric Investigation of Slow Postnatal Weight Gain.
In our daily observations of litters including either Del(1–9)-heterozygous or Del(1–4)-homozygous individuals, we noticed that growth-retarded animals were weak and lethargic. In addition, they dropped few or no fecal pellets. We used the latter observation as a noninvasive measure to characterize and follow the phase of recovery. Analysis of droppings reveals species-specific features of excretory activity (16) and thus reflects the morphology and function of the intestinal tract. Such analysis can start after the end of the third week, once animals switch to solid food. In rodents, the gut content is liquid in the small intestine and paste-like in the cecum. The segmentation into pellets and dehydration occur along more distal colon segments. When we collected the droppings in fractions, we found the majority was released in the first 10 min. Subsequently, their number dropped, indicating that this prolific early release was a defecation reflex (Fig. 1D).
In nearly all cases, the reflex gave comparable numbers of solid pellets per individual released by a similar dynamic. In addition, although the sizes of individual pellets varied, quite a few fell into in a rather narrow range. Finally, animals that suffered growth retardation produced smaller but not necessarily fewer pellets of similar consistency (Fig. 1D). The comparable solidity of up to 16 pellets per single collection suggested that sudden total pellet release represented the content of the terminal gut. On the other hand, the average or most frequent pellet sizes reflected a fairly regular size-setting mechanism that presumably operated at a more proximal segment. Because growth-retarded mutants released smaller pellets, we concluded that the gut was of a different anatomical and/or motile character compared with normal controls, while the mechanism of pellet segmentation and reflexive release were intact. As a consequence, we could evaluate gut performance individually in extended time series and in a reproducible noninvasive manner by measuring both the dried pellet mass collected in 30-min defecation-reflex periods and the body mass (Figs. 2–4 and Fig. S2).
Fig. 2.
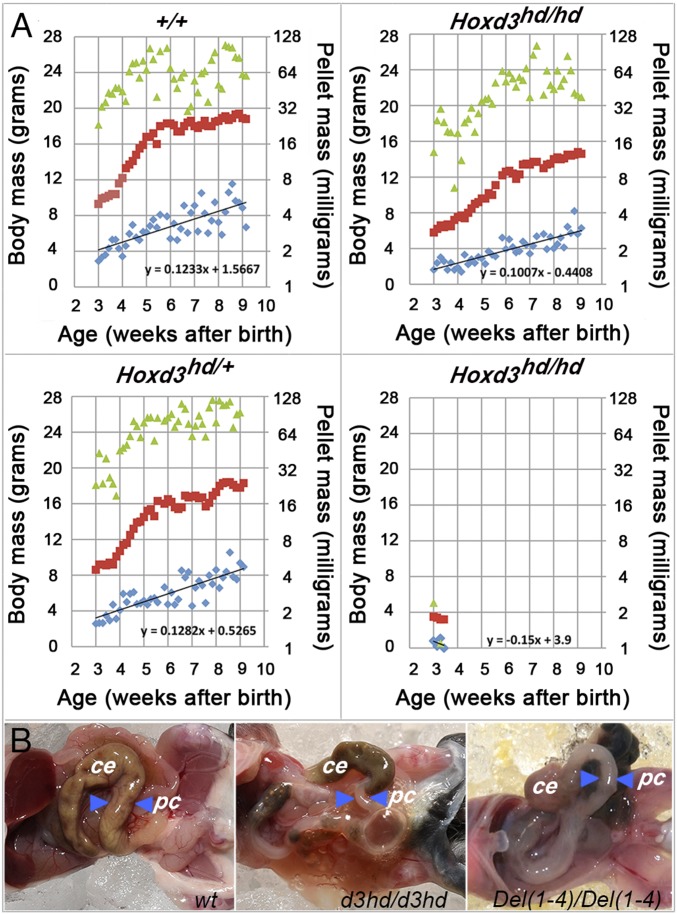
Stunted growth, survival, vital signs, and gut malformation in Hoxd3hd-homozygous mice. (A) Daily body mass (red), total dropping mass (green), and average dropping mass (blue) are plotted for wild-type (Upper Left) and heterozygous (Lower Left) mice and two homozygous siblings (Right). All four were females. The animal on the Bottom Right was killed at P23 due to severe stunting and wasting after weaning. Linear fitted lines show the dampened slope of average pellet mass for both homozygous animals. (B) Dissections of one representative individual of the three indicated genotypes showing the posterior midgut. Blue arrowheads point to the same segment of the proximal colon in each panel. The specimen on the Middle and on the Right were representative of severely wasted animals, usually killed during the fourth week after birth. ce, cecum; pc, proximal colon.
Fig. 4.
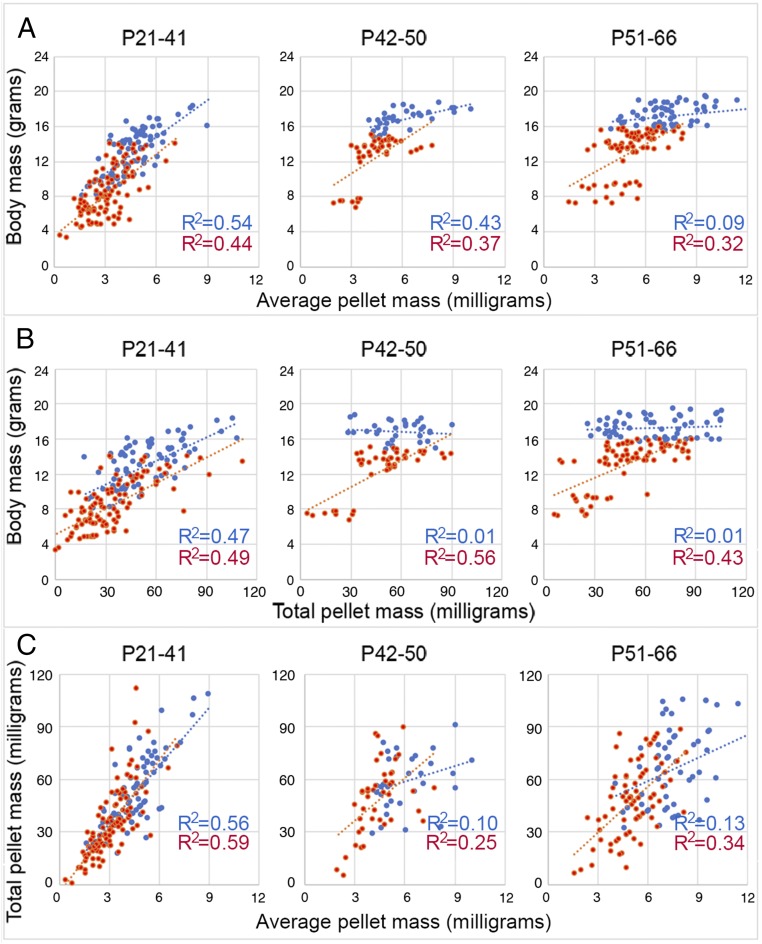
Correlation analysis of body mass and dropping mass measures. Values from five wild-type controls (blue) and four homozygous Hoxd3hd mutants (red) at three characteristic periods of wild-type controls’ postnatal growth were pooled. Values were included from three different litters in which both controls and homozygous siblings occurred as members of the same litter. (A) Plots of body weight as a function of average pellet weight. (B) Plots of body weight as a function of total ejected pellet weight. (C) Plots of total ejected pellet weight as function of average pellet weight. Pellets released during single daily 30-min defecation-reflex episodes were included. In each plot, the R2 values are depicted for wild-type (blue) and homozygous (red) animals to estimate the percentage of the variance due to variation in the respective parameter. R2 values of 0.010 or below were not statistically significant; R2 at 0.13 was statistically significant (P < 0.005); all other higher R2 values were statistically significant (P < 0.0005 or far below).
Fig. S2.
Growth retardation in F0 mice after inactivation of Hoxd3 by CRISPR/Cas9. (A) Daily body weight (red), total weight of dropping (green), and average weight (blue) are plotted for two normal siblings (Left) and two Hoxd3hd mutants (Right). All four were males issued from pronuclear injections of the Hoxd3 homeodomain-targeting plasmid vectors. (B) Frequency distributions of individual dropping masses pooled from the pairs of wild-type control and mutant individuals illustrated in A. Age at the time of collections is indicated above the heat map, total counts by wild-type and mutant genotypes are indicated as pairs below the heat map, the size bins set to 0.5-mg increments are indicated on the left. Heat maps reflect the relative frequency of pellet counts in a given lot; the most frequent values are boxed for better visibility. (C) Photograph of two P12 siblings from a litter born to a Hoxd3hd founder animal and a Del(1–4)-heterozygous male. Genotypes indicated for each specimen. A single picture was cut into two pieces for the ease of figure presentation (the scale is the same for both mice).
Hoxd3-Dependent Growth Retardation.
Prior analysis of the combined deletion of Hoxd1, Hoxd3, and Hoxd4 [Del(1–4)] revealed a drastic growth retardation in homozygous specimens (see above), which was not observed in either Hoxd1 (4, 5) or Hoxd4 (15) single-mutant mice. This could derive either from a combined effect of several gene products and the consequent compensatory mechanisms or from a prominent effect of Hoxd3. To discriminate between these possibilities, we produced a deficiency including the entire Hoxd4 gene, HoxDDel4 [Del (4)], as well as a Hoxd3-mutant allele. In the latter case, we used CRISPR-Cas9 to induce an out-of-frame deletion within the HOXD3 protein homeodomain, which presumably prevented HOXD3 from binding to DNA target sites.
Extensive analyzes of Del (4) mice revealed no significant growth defect in homozygous mice (Table 1). In contrast, F0 animals derived from the CRISPR/Cas9 mutation in Hoxd3 (SI Materials and Methods) showed slow postnatal weight gain. Five apparently homozygous F0 animals indeed displayed this growth-retardation problem, even though this was not reported in previous evaluations of mutant Hoxd3-null alleles (6, 7). We crossed these F0 animals and analyzed progenies to confirm the link to Hoxd3. We first mated a Hoxd3hd-mutant F0 female with a Del(1–4)-heterozygous male to obtain transheterozygous individuals and heterozygous controls, as well as to eliminate potential recessive off-target mutations since the Del(1–4) stock was generated without CRISPR/Cas9 exposure. However, Del(1–4)/+ animals were not recovered in the F1 progeny, showing that the F0 Hoxd3hd female was homozygous in the germ line. All Hoxd3hd/Del(1–4) compound-mutant individuals were severely stunted (Table 1), and by P28, they reached only two-thirds of the body mass of their Hoxd3hd/+-heterozygous siblings. This difference matched that observed between heterozygous and homozygous Del(1–4) specimens and was statistically significant (Figs. S1 and S2C).
Litters of F2 pups including all three possible Hoxd3hd genotypes were monitored, and wild-type and heterozygous females showed similar characteristics as we observed with F0 control males. However, their body weight at the end of the fast growth period at P42 was around 16 g, compared with the average of 20 g recorded with males (Fig. 2A and Fig. S2A). The body weight of wild-type and heterozygous individuals at the end of the third week was typically above 8 g Fig. 2A, Left), whereas most homozygous-mutant animals weighed less than 7 g (Fig. 2A, Right), and the most severely affected weighed less than 4 g (a more than 60% deficit compared with siblings of that age) (Fig. 2A). The body weight of the least-affected surviving homozygous individual (“escaper”) rarely exceeded 12 g at the end of the seventh week (a deficit of ca. 25%) (Fig. 2A, Upper). To uncover a potential association with a gut defect, we recorded pellet masses from the beginning of the fourth week.
Like their wild-type siblings, heterozygous-mutant female mice regularly released more than 32 mg of total pellet weight on most days of the fourth week. During the fifth week and later, the total ejected weight was often in excess of 64 mg. In contrast, homozygous surviving females consistently released less than 32 mg of pellets during both the fourth and the fifth week and rarely reached 64 mg output by the end of the seventh week (Fig. 2A, Upper). Similarly, the average pellet weight of heterozygous and wild-type animals was about 4 mg by the end of the seventh week, while pellets of homozygous animals hardly ever reached this value even after the ninth week of age. It is noteworthy that most affected homozygous individuals released far fewer and smaller pellets and, during the terminal periods, released none at all (Fig. 2A, Lower Right).
Examination of the growth deficit including all F2 individuals at P28 also revealed a slight body mass deficit (7%) in the subpopulation of Hoxd3hd-heterozygous individuals compared with wild-type mice, even though this was of poor statistical significance (P value = 1.17−01). In the homozygous subpopulation, however, the average body mass deficit was one-third of the average control body mass, a difference that was statistically significant. These observations supported the conclusion that Hoxd3 is required for postnatal growth during the suckling period. Again, the distinctly reduced total ejected pellet mass during the fourth week suggested a decrease in the volume of the terminal segments of the gut. While the total ejected pellet mass slowly increased to reach nearly normal levels, the average size of pellets remained significantly smaller even at late stages (Fig. 3). This suggested a permanent loss of isometry in the initial formation of droppings and indicated that postnatal large bowel growth was likely delayed.
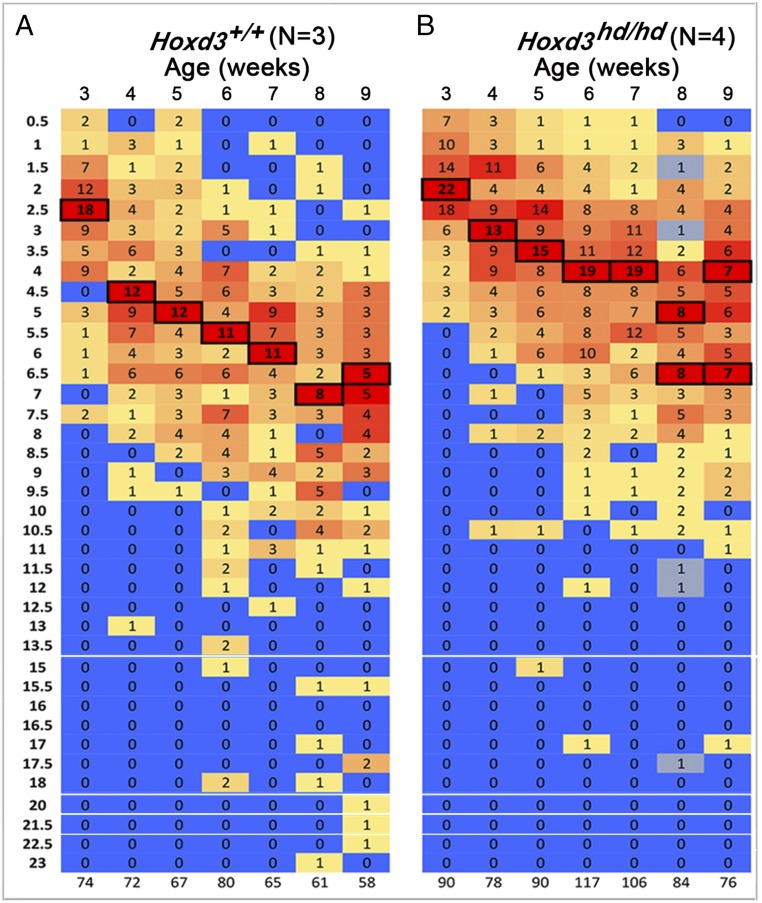
Fig. 3.
Frequency distributions of dropping weights pooled from three wild-type controls (A) and four homozygous mutants (B). Age in weeks at the time of collection is indicated above, and the total counts of pellets collected during the last two consecutive days are shown below. The size bins set to 0.5-mg increments are indicated on the left. The heat maps reflect the relative frequency of pellet counts in a given lot; the most frequent values are boxed for better visibility.
Morphological Alterations.
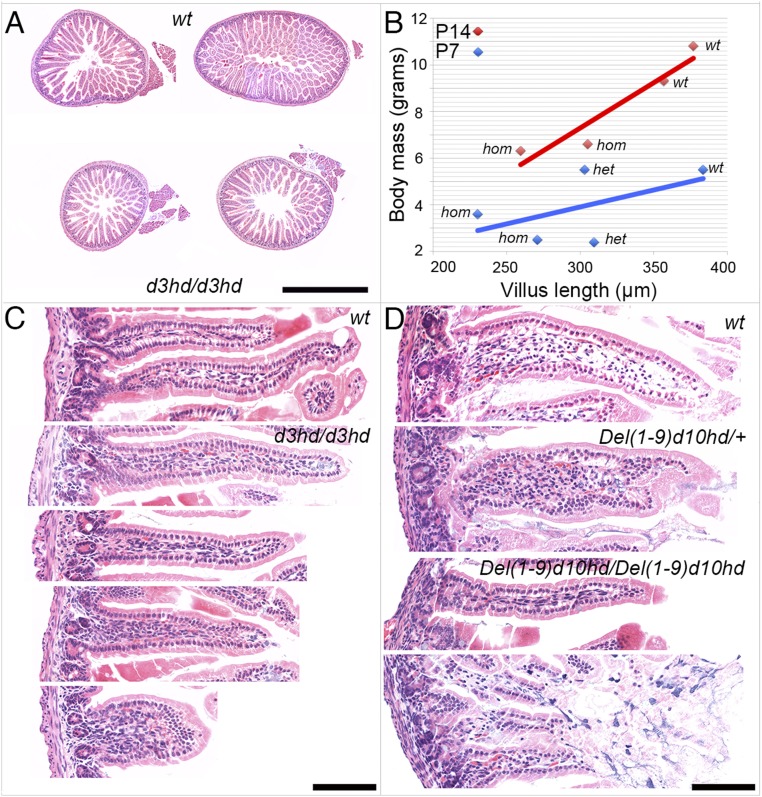
The anatomical analysis of specimens from various genotypes revealed that droppings were already distinct in the more anterior part of the large bowel in the proximal colon. Consequently, we suspected that the proximal colon was morphologically or functionally insufficient in Hoxd3hd-homozygous mutants. Also, in one particularly stunted mutant animal, while all abdominal organs were expectedly reduced in size (the total body mass was decreased by 60%), a clear retention of gut content in the terminal small intestine and in the cecum was recorded (Fig. 2B, Right). This distended region continued in a narrow empty proximal colon segment. Such a severe narrowing was also observed in Del(1–4)-homozygous stunted specimens (Fig. 2B, Middle). In agreement with the coprometric analyses, we concluded that this limitation in proximal colon diameter impeded the passage of luminal content and led to both reduced dropping size and reduced dropping deposition during an induced defecation reflex, leading over time to chronic constipation, malnutrition, and pronounced stunting.
Such a severe condition was further studied, in particular regarding the relative contribution of proximal and terminal colon defects to this slow postnatal weight gain. To this aim, we performed a correlation analysis of Hoxd3hd-homozygous and wild-type control siblings. We plotted the body mass against average pellet mass, body mass against total ejected pellet mass, and the total ejected pellet mass against the average pellet mass. The R2 values of wild-type controls were higher in the body mass versus average pellet mass comparison in all three phases than in the body mass versus total pellet mass comparisons (Fig. 4 A and B, blue). In the average pellet mass versus total pellet mass comparison, the R2 value reached 0.56 during the growth period (P < 1.17332−15) but remained nonsignificant at later stages (Fig. 4C, blue). Average pellet mass was a better predictor of body mass than total ejected pellet mass.
Of note, the same comparisons applied to the Hoxd3hd-homozygous samples diverged from this overall picture. The R2 values were as high or indeed slightly higher in the body mass versus total pellet mass comparison than in the body mass versus average pellet mass comparison (Fig. 4 A and B, red). This was also confirmed in the average pellet mass versus total pellet mass analysis, and a positive correlation with delayed growth was established during all three periods. Average pellet mass and total pellet mass were positively correlated, and the correlation was of a higher degree at all stages compared with wild-type control (Fig. 4C, red). Average pellet mass as well as total ejected pellet mass were similarly good predictors of body mass in Hoxd3hd-homozygous individuals. This correlation analysis therefore supported the inference that Hoxd3hd-homozygous guts performed differently from those of wild-type control animals. Absolute values were nevertheless lower compared with controls, which was consistent with the idea that the reduced dimension of both proximal colon, as reflected in average pellet mass, and of the terminal colon, as reflected in total ejected pellet mass, contributed simultaneously to a strong constraint on organismal growth.
Epithelial Defects in Hoxd3hd- and HoxDDel(1–9)d10hd-Mutant Small Intestines.
Following the abrogation of Hoxd3 function, only about half of the variance in body mass could be accounted for by noninvasive coprometry. As body mass deficit first occurred before excrement could be collected, we investigated juvenile gut histology of stunted animals and normal littermates on P14 and P7. The duodenum of Hoxd3hd-homozygous mutants and wild-type control siblings showed a reduction in diameter and in epithelium thickness (Fig. 5 A and B). When the body mass was plotted as a function of the average length of villi, it showed a positive correlation with the length of duodenum villi (Fig. 5B). Up to 90% of the variance in body mass could be accounted for by variation in duodenum epithelial structure.
Fig. 5.
Reduced small intestine diameter and epithelial dysgenesis in Hoxd3hd and HoxDDel(1–9)d10hd milk-fed pups. (A) Two wild-type controls (Upper) and two Hoxd3hd-homozygous siblings (Lower) are shown. (B) The longest villi from at least six available sections from each individual were selected and photographed, and their longest axis was measured. In the diagram the body weight was plotted as a function of the average longest villus length. The Hoxd3hd stock at P14 is shown as red diamonds and a red line. The two diamonds on the right represent wild-type (wt) mice, and the two on the left represent homozygous (hom) siblings. A similar analysis was carried out with Del(1–9)d10hd at 7 d after birth and is depicted as blue diamonds and a blue line. The two values at the left represent homozygous animals (hom), the two in the middle represent heterozygous animals (het), and the one on the right is a wild-type control (wt). (C) H&E-stained paraffin sections of gut villi from wild-type control and two pairs of sections from two Hoxd3hd-homozygous specimens showing representative morphologies. (D) High-power photomicrographs of H&E sections of wild-type and heterozygous specimens and from two Del(1–9)d10hd-homozygous animals showing representative morphologies. (Scale bar, 1 mm in A and 100 µm in C and D.)
Next we extended this analysis to Del(1–9)d10hd specimens and investigated wild-type control, heterozygous, and homozygous siblings 1 wk after birth. Examination of villus length also revealed a positive correlation between body mass and villus length (Fig. 5B). Body mass values were lower than at P14, consistent with the animals’ younger age. Although a positive correlation was obtained between body mass and villus length, the slope of the fit line was clearly less steep than in the case of Hoxd3hd mice at P14, and the R2 value was only 0.28. Since body mass reduction and lethality was more severe in Del(1–9)d10hd animals, we surmised that changes beyond villus length contributed to the severity of this allele. More detailed observations using high-power images revealed reduced maximal villus length in both cases (Fig. 5 C and D). Del(1–9)d10hd-homozygous individuals also showed an extended region of villus disintegration (Fig. 5D, Bottom). The tips of villi were ill-formed, and more basal surfaces were devoid of the enterocyte layer, indicative of epithelial necrosis. From these histological observations, we concluded that early during milk feeding, epithelial dysgenesis occurred in Hoxd3hd-homozygous mutants, which hindered proper alimentation of pups. This condition was further aggravated in the absence of an extended genomic locus, which in addition to Hoxd3 removed Hoxd10, Hoxd9, Hoxd8, Hoxd4, and Hoxd1 functions. These results further supported the hypothesis that gut defects were a cause rather than a consequence of small body size.
Contribution of the LncRNA Haglr to Growth Retardation.
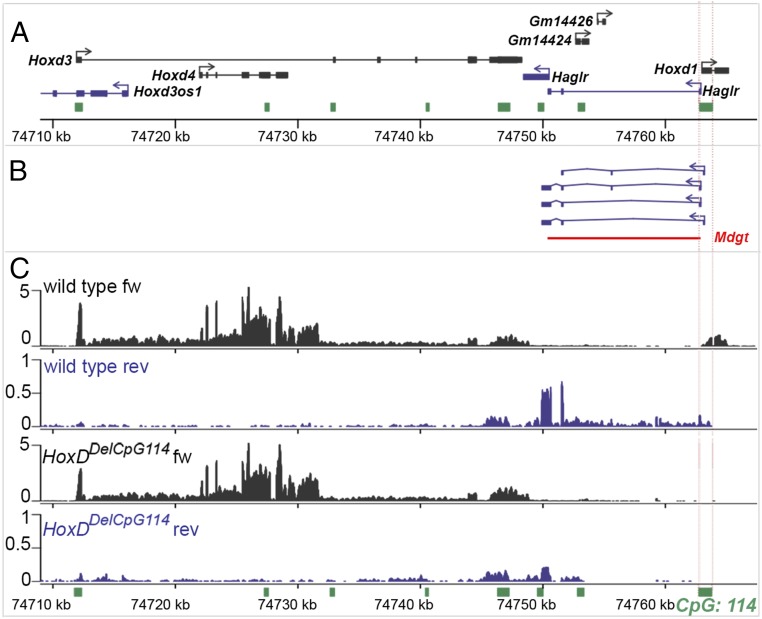
In a recent report, a similar semilethal recessive slow postnatal weight gain was observed upon disruption of the Haglr lncRNA, leading to the midget (Mdgt) phenotype (3). The HoxD antisense Haglr RNA starts from the Hoxd1 bidirectional promoter and extends to the Hoxd3 transcription unit. To further evaluate a potential function of this lncRNA on the growth-retardation phenotype, we produced two mutant alleles in which Haglr was presumably affected without removing the entire Hoxd1–Hoxd3 intergenic region, which includes two other noncoding Gm14426 and Gm14424 RNAs (Fig. 6A). Transcriptome analysis of E8.5 embryos revealed that Hoxd1 and Haglr share the same bidirectional start site located within the CpG island 114. On the reverse Hox strand, two Haglr start sites were mapped, as well as alternatively spliced transcripts including a short Haglr exon (Fig. 6B). We used CRISPR/Cas9 to produce two lines of mice, the first carrying a deletion of the CpG island 114 [HoxDDel(CpG114) or Del(CpG114)] and the second carrying an inversion of the same DNA fragment [HoxDInv(CpG114) or Inv(CpG114)]. Nine Del(CpG114) F0 founders were obtained that were homozygous for the deficiency, and none of them suffered from growth retardation.
Fig. 6.
Targeted deletion and inversion of the CpG114 island that contains the bidirectional Hoxd1/Haglr promoter. (A) Map of the Hoxd3-Hoxd1 genomic locus showing annotated transcripts in the Ensembl database and eight CpG islands (green). The CpG114 is shown between two vertical dotted red lines and includes the first exons of both Hoxd1 and the Haglr lncRNA. Nucleotide positions below are on the mm10 genome assembly. (B) Splice variants of the Haglr transcripts, which were added to the Ensembl transcript annotation list in quantifying FPKM values in RNA-seq analyses. (C) BED graphs of uniquely mapped reads to the Hoxd3–Hoxd1 genomic region on chromosome 2 from E9.5 mouse ribo-depleted RNA samples derived from two wild-type controls and two Del(CpG114)-homozygous embryos. Forward (black) and reverse (purple) strands from two biological replicates are combined.
Clean stocks for both the deficiency and the inversion were further established, and RNAs were isolated from pairs of homozygous and wild-type control siblings at E9 for RNA-sequencing (RNA-seq). Browser Extensible Data (BED) graphs of mapped reads on both the forward and the reverse strands of the HoxD cluster indicated that in wild-type mice Hoxd1, Hoxd3, and Hoxd4 were all transcribed as expected from the Hox forward strand (Fig. 6C). On the reverse strand, reads mapping to the Haglr locus were clearly recovered, even though the fragments per kilobase of transcript per million mapped reads (FPKM) value was five- to 10-fold reduced compared with Hox genes transcripts. In Del(CpG114)-homozygous animals, Hoxd1 transcripts were missing, as were the long isoforms of Haglr on the opposite strand. Nonetheless, a signal was present over the third and fourth exons of the Haglr locus. In terms of FPKM values, this was around one order of magnitude below that the wild-type Haglr signal. This observation showed that, besides the two Haglr promoters in the CpG island 114, at least one other start site contributed to the expression of the last two exons of Haglr. When interrogating the transcriptome genome-wide, only two loci were found with a statistically significant difference after adjusting the P values for multiple testing. One was Hoxd1 and the other Haglr, as predicted by the genomic deficiency. Both Del and Inv(CpG114) alleles were bred to homozygosity, but these animals failed to show any slow postnatal weight gain, lethality, or loss of fertility (Table 1). We concluded that neither the deficiency nor the inversion alleles of the CpG1 island 114 had any effect on organismal growth.
Discussion
HoxD Cluster-Dependent Gut Maturation and Organismal Growth.
Even though mammalian Hox gene functions have been scrutinized in great detail, a potential involvement in postnatal growth has remained undocumented. Here we report that four distinct mutant alleles associated with the HoxD cluster induced an early postnatal growth deficit. Growth retardation set in early, and a proportion of animals became fatally wasted. It is therefore likely that an important function for this gene cluster, probably linked to the proper morphogenesis of the gut, was impaired in these various alleles. We followed surviving individuals and determined that growth was systematically resumed once milk feeding was terminated. During this recovery period, the initially reduced excrement size and mass grew, showing an improvement of gut function along with an increase in body mass. Escapers formed small pellets reflecting a retarded expansion of their proximal colon. In addition, the initially meager total dropping mass improved progressively, indicating a delayed expansion of their distal colon.
Likewise, we also noticed that fatally stunted individuals suffered from partial atresia of the proximal colon. In addition, epithelial dysgenesis in the small bowel was noticed in both homozygous Hoxd3hd- and HoxDDel(1–9)d10hd-mutant specimens, in the latter case with severe enterocyte necrosis at the end of the first week of age. This extensive epithelial cell erosion in HoxDDel(1–9)d10hd-homozygous mice is somewhat reminiscent of necrotizing enterocolitis mouse models, which present similar pathological features (17). The repeated occurrence of such pathological deviations in small and large intestine in HoxD mutants provides evidence for a broad dysfunction along the intestine at the time the animals fed on milk. Interestingly, previous work involving Hox genes reported problems either in producing milk or in the proper regulation of these genes’ expression during mammary gland development (18, 19), showing the diverse panel of functions that these genes likely acquired to accompany the emergence of lactation (20).
Notably, this Hox-dependent inability to thrive on a milk diet followed by a more-or-less efficient recovery after weaning adds a further piece to the genetic study of milk-adapted gut function. Previous work has shown that a distinct genetic program was in place in mice to ensure that milk digestion remained active for the necessary period of time. In the absence of Blimp1, for example, early enterocytes are lost prematurely, precluding growth of the organism due to the incompetence of the adult-type enterocytes to digest milk (21, 22). Our observations suggest that, similar to Blimp1, Hoxd3 may promote the maintenance of milk-digesting enterocytes, raising the possibility that the two genes belong to the same genetic pathway, even if Blimp1 function is required in the epithelium whereas Hoxd3 gene expression is restricted to mesenchymal cells (22–24). The importance of Hoxd genes for the patterning of gut mesenchyme derivatives, such as sphincters (25, 26), has been well documented. However, with exception of the ano-rectal transitory epithelium (27), direct function of the HoxD cluster in gut epithelium seems unlikely, and hence any functional mechanism linking mesenchymal Hoxd3 expression to enterocyte survival may imply epithelial–mesenchymal interactions.
Essential Function of Hoxd3 in Gut Development.
Clearly, Hoxd3 is the major Hoxd gene involved in this particular aspect of gut development, as determined by our genetic approaches. Indeed, the loss of function of this gene alone was sufficient to elicit severe growth retardation in homozygous animals. While this phenotype was not described in previously reported studies in which Hoxd3 was inactivated (6, 7), it was likely observed but not characterized in detail. Hoxd3 is nevertheless not the only Hoxd gene to contribute to proper gut formation, since, although the single loss of function of Hoxd1, Hoxd4, Hoxd8, or Hoxd9 did not generate any discernable phenotype, combined loss of function in cis in addition to Hoxd3 strengthened the severity of the alterations. For instance, homozygous HoxDDel(1–4) animals in which Hoxd1, Hoxd3, and Hoxd4 were all missing displayed a phenotype stronger than that in the simple Hoxd3-mutant specimens. However, growth retardation in the latter condition was not as strong as in homozygous HoxDDel(1–9)d10hd animals in which Hoxd8 and Hoxd9 had also been removed. It is possible that Hoxd genes other than Hoxd3 can compensate for the lack of function of the latter, even though they may not functionally contribute in the wild-type context. Alternatively, they may have a genuine function that is perhaps too subtle to be revealed in the presence of the HOXD3 protein.
Remarkably, the strongest allele in the series was the HoxDDel(1–9) chromosome, as even heterozygous animals displayed a strong growth deficit. This heterozygous condition was largely rescued when the Hoxd10 gene was inactivated on the same chromosome, demonstrating that most of the dominant phenotype was induced by a gain of function of this gene, likely following its relocation as the now-leading Hoxd gene on the telomeric side of the gene cluster. While such gain of expression of Hoxd genes flanking the breakpoints of internal cluster deletions have been documented in the past in a variety of developmental contexts (28–30), the mechanism underlying this dominant-negative effect remains to be fully clarified. The HOXD10 protein ectopically expressed with the Hoxd3 tissue specificity may trigger an inappropriate genetic program leading to the misspecification of gut morphology.
Nevertheless, we favor an alternative explanation whereby HOXD10 exerts its dominant-negative effect upon other, more anterior Hox genes such as Hoxd3. This dominant-negative effect of some posterior HOX proteins over more anterior members of the same family [“posterior prevalence” (31–33)] has been documented in a variety of situations (8, 30, 34, 35). In this view, the gain of Hoxd10 expression following its new genomic position within the gene cluster would affect the function of all anterior HOX proteins, somewhat equivalent to a global homozygous loss of function of Hox genes. The fact that many heterozygous HoxDDel(1–9) animals were as strongly affected as homozygous HoxDDel(1–9)10hd animals and displayed a very related phenotype involving gut morphology supports this explanation. As a consequence, the HoxDDel(1–9)-heterozygous condition would be merely yet another loss-of-function HoxD allele, one produced by the dominant-negative interference of the HOXD10 protein. Since rescuing the mutation of Hoxd10 in the HoxDDel(1–9) chromosome involves a microdeletion into its DNA-binding domain, it is possible that this dominant effect requires the binding of HOXD10 to DNA target sites normally occupied by other HOX proteins. In this particular case, we consider other proposed mechanisms to trigger this negative effect, such as disturbed interactions with cofactors of the TALE homeodomain protein family (36, 37) or the involvement of microRNAs (35), as less plausible.
Is the LncRNA Haglr Involved in Postnatal Growth?
According to the description of Mdgt mice, i.e., animals lacking the lncRNA Haglr (3), HoxDDel(1–9)-heterozygous animals and Hoxd3hd-homozygous specimens suffered from a comparable slow postnatal weight gain. In the HoxDDel(1–9) animals, however, a full wild-type haplotype of the HoxD cluster was present, including an intact copy of Haglr, whereas in Hoxd3hd mice both Haglr loci were intact. The Hoxd3hd phenotype was particularly intriguing, since the Haglr RNA extends up to the 3′ end of the Hoxd3 UTR, thus raising the possibility that the deletion of Haglr would, in fact, affect the function of Hoxd3 (11). To verify this hypothesis, a transcriptome analysis of Midget mice would be necessary. On our side, we investigated this alternative by trying to knock down Haglr transcription while preserving the activity of Hoxd3. The targeted deletion of CpG114 indeed abrogated all long isoforms of the Haglr transcript in E9.5 embryos, corresponding to the extent of the deletion reported in ref. 3. In these embryos, the Hoxd3 transcripts were normal in all respects. Despite the absence of Haglr transcripts, growth deficit was not observed in homozygous mice, indicating that variation of Haglr RNA level had no appreciable consequence in organismal growth.
Shorter transcripts were nevertheless still initiated from the anti-Hox strand in the Del(CpG114) chromosome (although at a much lower level) and terminated exactly at the position of the termination site of the Hoxd3 UTR (Fig. 6). These transcripts, which were also named “Haglr” in the Ensembl database even though they have an independent start site, initiated within the last exon of Haglr. It is not known whether such transcripts are still present in Midget mice after the large targeted deletion and lacZ reporter replacement (3). If these additional RNAs were not maintained in Mdgt embryos, it is possible that their absence would perturb the transcription of Hoxd3, thus leading to growth retardation. In such a scenario, however, the function of these RNAs would be exerted in cis rather than in trans (38), as a result of transcriptional interference. Taking these results together, we conclude that the effect of the Mdgt deletion is more likely due to the extent of the genomic deficiency or the insertion of foreign DNA such as the LacZ transcription unit over the Hoxd3 gene rather than to the absence of the specific Haglr transcript. This lncRNA may result from a bidirectional transcription at the Hoxd1 start site (39) and may not have any particular function in controlling postnatal growth.
Materials and Methods
Mouse Strains.
Mice were handled according to the Swiss law on animal protection (LPA) with the authorizations GE/81/14 (to D.D.) and GE/29/26 (to B.M.). Animal experiments were agreed upon by the ethical committee of the Canton of Geneva. Genetically modified mice were maintained and crossed in heterozygosis. All CRISPR/Cas9 alleles were produced by pronuclear injection (40) of the pX330:hSpCas9 (Addgene ID 42230) vector with the appropriate guide-sequence oligos cloned as recommended (41). When deletions or inversions between two guide sequences were intended, the appropriate plasmids were injected in equimolar solution.
Zygotes were derived from (B6CBA)F1 females crossed to (B6CBA)F1 males. Founder animals were crossed with males or females of the (B6CBA)F1 wild-type genotype, and routine strain maintenance was continued by crossing heterozygous males of the allele in question to (B6CBA)F1 wild-type females. Target sequences were identified using the web tool at crispr.mit.edu during the year 2013. Nucleotide positions are based on GRCm38/mm10.
The production of the various mutant alleles used in this study is described in the SI Materials and Methods section.
RNA Extraction and RNA-Seq.
RNA extraction and RNA-seq were as described in ref. 18. Haglr transcript isoforms were cloned and sequenced starting from an E8 mouse tail-bud total RNA preparation as a source for first-strand cDNA synthesis using SuperScript II reverse transcriptase from Invitrogen, following the manufacturer’s protocol. cDNA was amplified using Phusion High Fidelity DNA polymerase from Thermo Scientific. Amplicons were cloned into the pGEM-T Easy plasmid vector (A1360; Promega) and transformed into JM109 cells; cloned and plasmid DNA samples were isolated by the Qiagen Midi protocol. RNA-seq–processed and raw datasets were deposited in the Gene Expression Omnibus (GEO) database with accession number GSE103086.
Histology and Coprometric Analyses.
For histology and coprometric analyses, see SI Materials and Methods.
SI Results
At the day of birth, no significant differences in body mass were recorded between F0 mutant pups and their control littermates. Normal control sibling mice showed steady postnatal body mass growth. They showed fast growth both between P0 and P14, surpassing 8 g body weight by P14, and between P22 and P42, surpassing 20 g by P42 (Fig. S2A). The transition between the early and later fast growth periods corresponded to the progressive reduction of milk feeding, when pups started adapting to solid food. Slow body mass increase, even stagnation for a few days, was observed during this transition period. After P42, approaching adulthood, a reduced but steady growth rate ensued, and body weight surpassed 25 g by P70.
Two apparently Hoxd3hd-homozygous mutant F0 specimens were analyzed using the same protocol, and slow weight gain was clearly documented (Fig. S2A, Right). Growth retardation occurred both during the P0–P14 milk-feeding period, with body weight staying below 7 g at P14, and during the second fast-growth period, P22–P42, when body weight hardly reached 12 g by P42 and stayed well below 20 g up until P70. The total pellet weight showed a rather similar distribution between all four animals from P42 onwards, from 32 to 128 mg total ejected pellet mass, and average pellet masses did not indicate significant differences either, due to the high variance of individual pellet sizes. At P28, we nevertheless observed a statistically significant reduction of average pellet size in mutants, taking all five animals into account (3.2 mg versus 4.7 mg, P < 0.03). This amounted to a 32% reduction of average pellet weight at P28, which was nearly normalized by P40.
Despite a rather large variance in pellet size in both homozygous Hoxd3hd-mutant and control phenotypes, the increase in size over time until P48–P52, reaching 8 mg in control animals, was slower in mutant animals from P28 until P53–P57 (Fig. S2A), even though the total ejected mass was not remarkably different at these stages. After P48–52 the pellet mass became reduced in controls, coinciding with the subadult and adult slower body mass growth phase, while they essentially stagnated in mutants. This differential dynamic of pellet size was most likely a consequence of an altered proximal colon molding function in Hoxd3hd mutants. The similar total ejected pellet mass, on the other hand, likely reflected similar volumes of distal colon, sigma, and rectum in mutant and control specimen. A similar ejected volume can hardly be indicative of daily total output in mice, as the total transit time from mouth to anus is of about 6 h (42). This allows for variable daily transit rates, even if defecation-reflex–prompted releases were of the same quantity.
SI Materials and Methods
Mutant Alleles.
The Hoxd10hd allele was produced in cis with the HoxDDel(1–9) deficiency [Del(2Hoxd1-Hoxd9)63Ddu; location: Chr2:74,697,727–74,765,142] (2). We cloned and Sanger sequenced the mutant HoxDDel(1–9)d10hd allele from homozygous genomic DNA. A small deletion occurred involving the guide sequence AGAGCGTTAACCTCACCGAC, as induced by Cas9 activity. This indel removed 10 nucleotides (CCGACAGGCA) corresponding to positions chr2: +74,695,262–74,695,271. Based on translation of the consensus CCDS 16141.1, the last 34 residues of the HOXD10 protein were thus replaced by 10 other residues as a consequence of a frame shift, thus resulting in a truncated protein product with a disrupted homeodomain after position 40. This deletion eliminated the third α-helix of the homeodomain. In the mutant protein, the resulting shorter C-terminal end of the sequence is underlined:
MSFPNSSPAANTFLVDSLISACRSDSFYSSSASMYMPPPSADMGTYGMQTCGLLPSLAKREVNHQNMGMNVHPYIPQVDSWTDPNRSCRIEQPVTQQVPTCSFTANIKEESNCCMYSDKRNKLISAEVPSYQRLVPESCPVENPEVPVPGYFRLSQTYATGKTQEYNNSPEGSSTVMLQLNPRGAAKPQLSAAQLQMEKKMNESASGQEPTKVSQVESPEAKGGLPEDRSCLAEVSVSSPEVQEKESKEEIKSDTPTSNWLTAKSGRKKRCPYTKHQTLELEKEFLFNMYLTRERRLEISKSVNLRSRFGFKTAE.
For the Hoxd3hd mutation, we cloned and Sanger sequenced the mutant allele from homozygous genomic DNA. Induced Cas9 activity resulted in a short deletion comprising the intervening region between the two guide sequences, GTACGCCGTGCGCACCCTCT (chr2: +74,746,364–74,746,383) and GGCGCGGCCGGCACAGATAG (chr2: −74,746,431–74,746,450). As a consequence, a 67-bp deletion occurred. This deficiency truncated the second exon of the Hoxd3 locus by the sequence GGGTGCGCACGGCGTACACGAGTGCTCAGCTGGTGGAGCTGGAGAAGGAGTTCCACTTCAACCGCTA (chr2: +74,746,367–74,746,433), thus inducing a truncation from the third amino acid residue position of the homeodomain onwards, replacing the rest of the 187-aa consensus sequence by the ICAGRAAWRWPTC frameshifted sequence. In the mutant protein, the resulting shorter C-terminal end of the sequence is underlined:
MLFEQGQQALELPECTMQKAAYYENPGLFGGYGYSKTTDTYGYSTPHQPYPPPAAASSLDTDYPGSACSIQSSAPLRAPAHKGAELNGSCMRPGTGNSQGGGGGSQPPGLNSEQQPPQPPPPPPTLPPSSPTNPGGGVPAKKPKGGPNASSSSATISKQIFPWMKESRQNSKQKNSCATAGESCEDKSPPGPASKICAGRAAWRWPTC.
PCR genotyping was carried out using primers ACCCCACCCTGCTCTAGGTAGG (chr2: +74,746,192–74,746,213) and CGGCTGGAGAATGCAGGATGC (chr2: −74,746,550–74,746,570). Thus, the wild-type allele was 379 bp, and the deficiency was 312-bp long in a multiplex amplification.
For the HoxDDel(CpG114) and HoxDInv(CpG114) alleles, we cloned and Sanger sequenced the mutant alleles from homozygous genomic DNA derived from descendants of seven different primary events. Six of these involved small deletions between the target sequences GAGATTCTACTCCTAAGCCC (chr2: + 74,763,853–74,763,872) and ATTCCCCGGGAAATAGCCCA (chr2: + 74,762,690–74,762,709) due to recombination as a consequence of Cas9 activity. Seven distinct end-joining reactions that removed the CpG114 island were identified; one of these reactions represented a precise predicted deletion of 1,163 bp. This Del(CpG114) founder was bred to homozygosity and gave stocks with no appreciable growth or fertility anomalies in homozygous animals. It was genotyped by using primers TGGTGCTGAAATTTGTGCGG (chr2: − 74,764,098–74,764,117) and AGGTGGTCCCAGGAGTCATT (chr2: + 74,762,465–74,762,484). The HoxDInv(CpG114) allele was obtained by screening for inversion of the genomic fragment between the target sequences GAGATTCTACTCCTAAGCCC (chr2: + 74,763,853–74,763,872) and ATTCCCCGGGAAATAGCCCA (chr2: + 74,762,690–74,762,709) due to recombination as a consequence of Cas9 activity. Sanger sequencing of this allele did not reveal any obvious DNA loss at either breakpoint. This allele together with the wild-type locus was routinely genotyped in a multiplex PCR using four primers: a wild-type–specific pair AGGTGGTCCCAGGAGTCATT (chr2: + 74,762,465–74,762,484) and CGCGCCTGCCTTGCTGCCAACTTAG (chr2: − 74,762,872–74,762,896) as well as a Inv(CpG114)-specific pair CGCGCCTGCCTTGCTGCCAACTTAG (chr2: −74,762,872–74,762,896) and ACCTCCAGCCCAAACTAAGC (chr2: −74,764,465–74,764,484). The Inv(CPG114) allele was bred to homozygosity with no obvious growth or fertility anomalies.
The HoxDDel (4) allele was generated by targeted meiotic recombination (43) between appropriate LoxP sites located immediately upstream and downstream of Hoxd4 and using the HPRT-Cre deleter strain. Genotyping primers were TTTATGGTTCTGCAGAGC (chr2: + 74,721,933–74,721,952) and CACCCTCACATCCTCATCCT (chr2: − 74,733,314–74,733,333).
The HoxDDel(1–4) allele corresponds to Del(2Hoxd1–Hoxd4)55Ddu, a deficiency of chr2: 74,711,929–74,765,142 previously reported (4).
The HoxDDel(1–9) allele corresponds to Del(2Hoxd1–Hoxd9)63Ddu, a deficiency of chr2: 74,697,727–74,765,142 previously reported (2).
Coprometric Analyses.
As a test arena we used a covered mouse holding cage with a 215 × 150 mm base surface area with a blotting paper sheet (3M paper) placed on the bottom. A high-resolution Nikon D810 digital camera was used to document freshly collected droppings minutes after collection. The camera was set on a stand, such that the 215 × 150 mm blotting paper filled the picture frame allowing about an extra centimeter on both sides to incorporate the object with scale arms. After photography, the cages with the droppings on the blotting paper were placed in a chemical hood for drying overnight. A Mettler XSE104 electronic balance with a 0–100 mg scale range (precision 0.1 mg) was imported into Excel for numerical analysis. Test groups were formed at the time genotyping results were recorded. On the day of sample collection, between 3 and 5 PM, we set up the test arena as well as a separate cage for each individual to be tested during that session. We placed individuals one by one into each cage, as quickly as possible, to reduce the chances of losing the first release. We set a timer for the intended time period (10–30 min). During the time-period run, we removed each individual and measured its body mass. We collected any droppings released during these manipulations and deposited them onto the blotting sheet. We placed the test individual back into the test arena and continued recording individual data of the remaining subjects until all datasheets were consistently filled. At the end of the pellet-collection period we removed animals from their respective arenas and placed all members belonging to the initial group housing cages back together. We stacked all cages containing the datasheets and droppings and moved them to the photography setup. We took pictures of each data sheet with well-controlled focus. After the photograph was taken we placed the datasheets back into the cages, and at the end of the photograph session we restacked cages and placed them into a chemical hood running at low speed for 16–48 h. Approximately 16 h later the samples were ready for pellet mass measurements. After the drying, we collected all pellets from each cage and datasheet onto 40 × 40 mm plastic weighing boats and measured the weight of each individual pellet on the milligram scales to 0.1-mg precision. We recorded each individual measurement and then moved all pellets from a single cage into plastic sample tubes and placed those tubes with the datasheet and sealed them into airtight soft polyethylene terephthalate PET plastic bags. We imported each record into an Excel sheet marking all entries with the appropriate set of ID, body mass, and date parameters. We calculated and displayed in Pivot Table the pellet count and weight, individual pellet weight, maximum, mean, and constructed frequency distribution in 0.5- or 1-mg bins (coproplot). All statistical analysis and plotting were done in Microsoft Excel, Office 365, version 15.34(170515). In correlation analyses, the growth trajectory was broken down into three phases, which reflect the fast and early postnatal growth, the later slow juvenile growth, and the essentially stationary adult phases observed with wild-type normal laboratory mice (44, 45). The average pellet mass and most frequent pellet mass variables reported here for laboratory mice are reminiscent of the species-specific measures taken of field-collected droppings to identify nests of different species of rodents in ethology studies (16).
Histological analyses were carried out following standard procedures of fixation, paraffin embedding, and sectioning. Sections (4 µm) were processed for standard H&E staining.
Acknowledgments
We thank members of the D.D. laboratories for sharing material and discussions, the histology platform at Ecole Polytechnique Fédérale (Lausanne) (EPFL), the genomics platform in Geneva, Olivier Schaad for advice on statistical analyses, and Thomas Alexander McKee for discussions. This work was supported by funds from the EPFL, the University of Geneva, Swiss National Research Fund Grant 310030B_138662, European Research Council Grants SystemHox 232790 and RegulHox 588029, and the Claraz Foundation (D.D.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE103086).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1712511114/-/DCSupplemental.
References
- 1.Spitz F, et al. Large scale transgenic and cluster deletion analysis of the HoxD complex separate an ancestral regulatory module from evolutionary innovations. Genes Dev. 2001;15:2209–2214. doi: 10.1101/gad.205701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zakany J, Duboule D. A genetic basis for altered sexual behavior in mutant female mice. Curr Biol. 2012;22:1676–1680. doi: 10.1016/j.cub.2012.06.067. [DOI] [PubMed] [Google Scholar]
- 3.Sauvageau M, et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife. 2013;2:e01749. doi: 10.7554/eLife.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo T, et al. An evolving NGF-Hoxd1 signaling pathway mediates development of divergent neural circuits in vertebrates. Nat Neurosci. 2011;14:31–36. doi: 10.1038/nn.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zákány J, Kmita M, Alarcon P, de la Pompa JL, Duboule D. Localized and transient transcription of Hox genes suggests a link between patterning and the segmentation clock. Cell. 2001;106:207–217. doi: 10.1016/s0092-8674(01)00436-6. [DOI] [PubMed] [Google Scholar]
- 6.Condie BG, Capecchi MR. Mice homozygous for a targeted disruption of Hoxd-3 (Hox-4.1) exhibit anterior transformations of the first and second cervical vertebrae, the atlas and the axis. Development. 1993;119:579–595. doi: 10.1242/dev.119.3.579. [DOI] [PubMed] [Google Scholar]
- 7.Greer JM, Puetz J, Thomas KR, Capecchi MR. Maintenance of functional equivalence during paralogous Hox gene evolution. Nature. 2000;403:661–665. doi: 10.1038/35001077. [DOI] [PubMed] [Google Scholar]
- 8.Zacchetti G, Duboule D, Zakany J. Hox gene function in vertebrate gut morphogenesis: The case of the caecum. Development. 2007;134:3967–3973. doi: 10.1242/dev.010991. [DOI] [PubMed] [Google Scholar]
- 9.Delpretti S, et al. Multiple enhancers regulate Hoxd genes and the Hotdog LncRNA during cecum budding. Cell Rep. 2013;5:137–150. doi: 10.1016/j.celrep.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 10.Tschopp P, Christen AJ, Duboule D. Bimodal control of Hoxd gene transcription in the spinal cord defines two regulatory subclusters. Development. 2012;139:929–939. doi: 10.1242/dev.076794. [DOI] [PubMed] [Google Scholar]
- 11.Bassett AR, et al. Considerations when investigating lncRNA function in vivo. Elife. 2014;3:e03058. doi: 10.7554/eLife.03058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tschopp P, Duboule D. A genetic approach to the transcriptional regulation of Hox gene clusters. Annu Rev Genet. 2011;45:145–166. doi: 10.1146/annurev-genet-102209-163429. [DOI] [PubMed] [Google Scholar]
- 13.Tarchini B, Duboule D. Control of Hoxd genes’ collinearity during early limb development. Dev Cell. 2006;10:93–103. doi: 10.1016/j.devcel.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 14.Duboule D, Morata G. Colinearity and functional hierarchy among genes of the homeotic complexes. Trends Genet. 1994;10:358–364. doi: 10.1016/0168-9525(94)90132-5. [DOI] [PubMed] [Google Scholar]
- 15.Horan GS, Kovàcs EN, Behringer RR, Featherstone MS. Mutations in paralogous Hox genes result in overlapping homeotic transformations of the axial skeleton: Evidence for unique and redundant function. Dev Biol. 1995;169:359–372. doi: 10.1006/dbio.1995.1150. [DOI] [PubMed] [Google Scholar]
- 16.Gouat P. Faecal pellet size difference as a field criterion to distinguish between two Ctenodactylus species (Mammalia, Rodentia) Z Saugetierkd. 1992;57:183–185. [Google Scholar]
- 17.Neal MD, et al. A critical role for TLR4 induction of autophagy in the regulation of enterocyte migration and the pathogenesis of necrotizing enterocolitis. J Immunol. 2013;190:3541–3551. doi: 10.4049/jimmunol.1202264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen F, Capecchi MR. Paralogous mouse Hox genes, Hoxa9, Hoxb9, and Hoxd9, function together to control development of the mammary gland in response to pregnancy. Proc Natl Acad Sci USA. 1999;96:541–546. doi: 10.1073/pnas.96.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schep R, et al. Control of Hoxd gene transcription in the mammary bud by hijacking a preexisting regulatory landscape. Proc Natl Acad Sci USA. 2016;113:E7720–E7729. doi: 10.1073/pnas.1617141113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duboule D. No milk today (my Hox have gone away) Proc Natl Acad Sci USA. 1999;96:322–323. doi: 10.1073/pnas.96.2.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harper J, Mould A, Andrews RM, Bikoff EK, Robertson EJ. The transcriptional repressor Blimp1/Prdm1 regulates postnatal reprogramming of intestinal enterocytes. Proc Natl Acad Sci USA. 2011;108:10585–10590. doi: 10.1073/pnas.1105852108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muncan V, et al. Blimp1 regulates the transition of neonatal to adult intestinal epithelium. Nat Commun. 2011;2:452. doi: 10.1038/ncomms1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gurdziel K, Vogt KR, Walton KD, Schneider GK, Gumucio DL. Transcriptome of the inner circular smooth muscle of the developing mouse intestine: Evidence for regulation of visceral smooth muscle genes by the hedgehog target gene, cJun. Dev Dyn. 2016;245:614–626. doi: 10.1002/dvdy.24399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan DP, Shao X, Pu L, Guo V, Nirenberg M. Sequence and expression of the murine Hoxd-3 homeobox gene. Proc Natl Acad Sci USA. 1996;93:8247–8252. doi: 10.1073/pnas.93.16.8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kondo T, Dollé P, Zákány J, Duboule D. Function of posterior HoxD genes in the morphogenesis of the anal sphincter. Development. 1996;122:2651–2659. doi: 10.1242/dev.122.9.2651. [DOI] [PubMed] [Google Scholar]
- 26.Zákány J, Duboule D. Hox genes and the making of sphincters. Nature. 1999;401:761–762. doi: 10.1038/44511. [DOI] [PubMed] [Google Scholar]
- 27.Dollé P, Izpisúa-Belmonte JC, Boncinelli E, Duboule D. The Hox-4.8 gene is localized at the 5′ extremity of the Hox-4 complex and is expressed in the most posterior parts of the body during development. Mech Dev. 1991;36:3–13. doi: 10.1016/0925-4773(91)90067-g. [DOI] [PubMed] [Google Scholar]
- 28.Kmita M, Tarchini B, Duboule D, Hérault Y. Evolutionary conserved sequences are required for the insulation of the vertebrate Hoxd complex in neural cells. Development. 2002;129:5521–5528. doi: 10.1242/dev.00151. [DOI] [PubMed] [Google Scholar]
- 29.Zákány J, Kmita M, Duboule D. A dual role for Hox genes in limb anterior-posterior asymmetry. Science. 2004;304:1669–1672. doi: 10.1126/science.1096049. [DOI] [PubMed] [Google Scholar]
- 30.Kmita M, van Der Hoeven F, Zákány J, Krumlauf R, Duboule D. Mechanisms of Hox gene colinearity: Transposition of the anterior Hoxb1 gene into the posterior HoxD complex. Genes Dev. 2000;14:198–211. [PMC free article] [PubMed] [Google Scholar]
- 31.Struhl G. Role of the esc+ gene product in ensuring the selective expression of segment-specific homeotic genes in Drosophila. J Embryol Exp Morphol. 1983;76:297–331. [PubMed] [Google Scholar]
- 32.González-Reyes A, Urquia N, Gehring WJ, Struhl G, Morata G. Are cross-regulatory interactions between homoeotic genes functionally significant? Nature. 1990;344:78–80. doi: 10.1038/344078a0. [DOI] [PubMed] [Google Scholar]
- 33.Bachiller D, Macías A, Duboule D, Morata G. Conservation of a functional hierarchy between mammalian and insect Hox/HOM genes. EMBO J. 1994;13:1930–1941. doi: 10.1002/j.1460-2075.1994.tb06462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Hoeven F, Zákány J, Duboule D. Gene transpositions in the HoxD complex reveal a hierarchy of regulatory controls. Cell. 1996;85:1025–1035. doi: 10.1016/s0092-8674(00)81303-3. [DOI] [PubMed] [Google Scholar]
- 35.Yekta S, Tabin CJ, Bartel DP. MicroRNAs in the Hox network: An apparent link to posterior prevalence. Nat Rev Genet. 2008;9:789–796. doi: 10.1038/nrg2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rivas ML, et al. Antagonism versus cooperativity with TALE cofactors at the base of the functional diversification of Hox protein function. PLoS Genet. 2013;9:e1003252. doi: 10.1371/journal.pgen.1003252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bürglin TR, Affolter M. Homeodomain proteins: An update. Chromosoma. 2016;125:497–521. doi: 10.1007/s00412-015-0543-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gummalla M, et al. abd-A regulation by the iab-8 noncoding RNA. PLoS Genet. 2012;8:e1002720. doi: 10.1371/journal.pgen.1002720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sigova AA, et al. Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proc Natl Acad Sci USA. 2013;110:2876–2881. doi: 10.1073/pnas.1221904110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mashiko D, et al. Generation of mutant mice by pronuclear injection of circular plasmid expressing Cas9 and single guided RNA. Sci Rep. 2013;3:3355. doi: 10.1038/srep03355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Padmanabhan P, Grosse J, Asad AB, Radda GK, Golay X. Gastrointestinal transit measurements in mice with 99mTc-DTPA-labeled activated charcoal using NanoSPECT-CT. EJNMMI Res. 2013;3:60. doi: 10.1186/2191-219X-3-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hérault Y, Rassoulzadegan M, Cuzin F, Duboule D. Engineering chromosomes in mice through targeted meiotic recombination (TAMERE) Nat Genet. 1998;20:381–384. doi: 10.1038/3861. [DOI] [PubMed] [Google Scholar]
- 44.Theiler K. The House Mouse. Atlas of Embryonic Development. Springer; New York: 1989. p. 150. [Google Scholar]
- 45.Walthall K, Cappon GD, Hurtt ME, Zoetis T. Postnatal development of the gastrointestinal system: A species comparison. Birth Defects Res B Dev Reprod Toxicol. 2005;74:132–156. doi: 10.1002/bdrb.20040. [DOI] [PubMed] [Google Scholar]