Abstract
Organisms have developed behavioral strategies to defend themselves from starvation stress. Despite of their importance in nature, the underlying mechanisms have been poorly understood. Here, we show that Drosophila G9a (dG9a), one of the histone H3 Lys 9-specific histone methyltransferases, functions as a key regulator for the starvation-induced behaviors. RNA-sequencing analyses utilizing dG9a null mutant flies revealed that the expression of some genes relating to gustatory perception are regulated by dG9a under starvation conditions. Reverse transcription quantitative-PCR analyses showed that the expression of gustatory receptor genes for sensing sugar are up-regulated in starved dG9a null mutant. Consistent with this, proboscis extension reflex tests indicated that dG9a depletion increased the sensitivity to sucrose under starvation conditions. Furthermore, the locomotion activity was promoted in starved dG9a null mutant. We also found that dG9a depletion down-regulates the expression of insulin-like peptide genes that are required for the suppression of starvation-induced hyperactivity. Furthermore, refeeding of wild type flies after starvation conditions restores the hyperactivity and increased sensitivity to sucrose as well as dG9a expression level. These data suggest that dG9a functions as a key regulator for the decision of behavioral strategies under starvation conditions.
Introduction
Behavioral epigenetics has attracted attentions for the last decade, because epigenetic regulation can induce rapid and long-lasting effects on gene expression in response to environmental changes. Although a numerous number of studies have revealed the biological roles of epigenetic factors over the last 40 years, how epigenetic regulation affects behaviors of organisms and how behaviors affect epigenetic regulation have just started to be investigated1. Previous studies reported that several epigenetic factors are relevant to behaviors included in learning/memory, neurodevelopmental disorders, drug addiction, parenting and stress responses in mammals1. Some of these relationships have also been found in Drosophila melanogaster, the model organism extensively used for genetic studies because of its short life span and the high homology to human genes2. For example, some Drosophila epigenetic factors like Ash1 and Suppressor of variegation 4–20 homolog 1 are associated with autism spectrum disorders, one of the neurodevelopmental disorders characterized by the impaired communication, restricted interests and hyperactivity3. Especially it is revealed that the dG9a, a Drosophila homolog of mammalian G9a 4 functions as the key regulator of learning and memory through alteration of histone modification5.
G9a has been identified in mammals as one of the histone H3 Lysine 9 (H3K9) specific methyltransferases (HMTases) that catalyzes both H3K9 mono-methylation and H3K9 di-methylation6,7. G9a has various biological roles including DNA replication8, genome imprinting9,10, developmental reprogramming11 and substance addiction12. Especially, G9a plays a critical role in embryogenesis, since G9a knockout mice show embryonic lethality in early stages due to severe growth defects7,13. Generally, G9a functions to suppress expression of its target genes through H3K9 methylation, although from some reports it may act as a co-activator to positively regulate some genes such as targets of the hormone-activated glucocorticoid receptor14,15. However, studies on in vivo functions of G9a utilizing the mouse model have not advanced efficiently because of the embryonic lethality of the G9a knockout mice. On the other hand, dG9a depletion exerted no effect on fly viability at all16. Therefore, Drosophila melanogaster is suitable for the functional analyses of G9a in adults. dG9a can catalyze the methylation of H3K9 in euchromatin regions and the methylated H3K9 contributes to heterochromatin formation and transcriptional repression of specific genes in vivo 5,17. Although the dG9a depleted flies show no viability defect16, previous reports utilizing dG9a knockdown flies or knockout flies revealed that dG9a has a regulatory role in the developmental process of germ cell line like spermatogenesis and oocyte specification18,19 as well as in learning and memory5. In contrast to the mammalian G9a, dG9a was not essential for Drosophila viability. Because of this unexpected finding, one can consider that the epigenetic regulation through H3K9 methylation is not developed to be critical for Drosophila viability and therefore dG9a plays no important role for Drosophila viability under laboratory conditions. However, it was important to note that while there are various environmental stresses in the wild, Drosophila was always maintained under optimal conditions in the laboratory. Therefore, we focused on the analyses under stressed conditions and recently revealed that dG9a has a critical role in acquisition of tolerance under starvation stress in adult stage through regulating the activity of autophagy20.
In terms of behavioral changes under starvation stress, Drosophila developed two behavioral strategies to increase the possibility that they can find new food sources21,22. Firstly, they increase responsiveness for a sugar taste by means of up-regulating the expression of Gustatory receptor (Gr) 64a, a well-known gustatory receptor for sensing sugar23. Secondary, starvation stress induces hyperactivity through activating octopaminergic neurons whose functions can be regulated by glucagon and insulin signals24. However, it remains totally unknown whether there is a key regulator for the decision of these starvation-induced behaviors in spite of its importance in nature.
In this study, RNA-sequencing (RNA-seq) analyses followed by gene ontology (GO) analyses revealed that the expression of genes encoding gustatory receptors and odorant binding proteins are altered in dG9a mutants under starvation conditions. Further genetic analyses revealed that dG9a depletion increases the expression levels of gustatory receptor genes for sensing sucrose. Behavioral analyses revealed that dG9a depletion up-regulates the sucrose sensitivity in response to starvation stress. Our data suggest that dG9a regulates the starvation-induced shift of locomotion activity through controlling the expression of insulin-like peptide (Ilp) genes that are required for the suppression of starvation-induced hyperactivity. Refeeding of wild type flies after starvation conditions restored the hyperactivity and increased sensitivity to sucrose as well as dG9a expression level. Our data suggest that dG9a functions as a key regulator for the decision of behavioral strategies under starvation conditions.
Results
RNA-seq analyses of dG9a-depleted Drosophila under starvation conditions
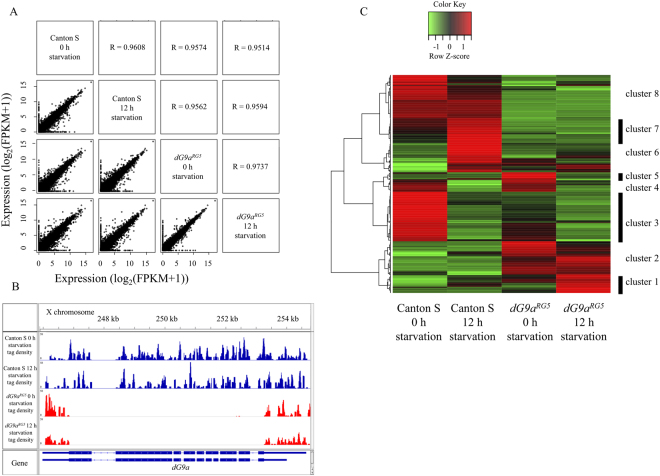
Previous study utilizing dG9a null mutant (dG9a RG5) showed that dG9a plays an important role for the survival of adult fly under starvation stress conditions20. We confirmed that there is no difference in the life span of dG9a RG5 mutant compared to that of Canton S under fed conditions (Fig S1). These results demonstrate that dG9a depletion affects fly viability under starvation conditions, but not under fed conditions. To determine which genes are regulated by dG9a under starvation stress, we performed RNA-seq analyses using adult flies of the dG9a null mutant and the wild type after 0 h and 12 h starvation. Thus, we made 4 groups; 0 h starved Canton S, 12 h starved Canton S, 0 h starved dG9a RG5 and 12 h starved dG9a RG5. Isolated RNAs were sequenced with a deep sequencer (Illumina HiSeq; Illumina, San Diego, CA, USA), which yielded an average of 40,508,694 1 × 51 bp single end reads per sample. These reads were aligned to the Drosophila melanogaster reference genome (UCSC dm6). The expression of in total 404 genes were differentially expressed between Canton S and dG9a null mutant across the analyses (q-value < 0.05). The expression levels of all detected genes were shown as fragments per kilobase of transcript per million mapped fragments (FPKM) in all combination of the groups (Fig. 1A). We calculated Pearson’s correlation coefficient between these groups (Fig. 1A). High correlation was detected in any combination of the groups (R > 0.95). The dG9a null mutant lacks the whole dG9a open reading frame (ORF)16. dG9a expression almost completely disappeared with almost no detectable reads mapped on the dG9a ORF in the dG9a null mutant (Fig. 1B). Among the 404 differentially expressed genes, we generated 8 clusters according to the type of expression changes and created a heatmap with two-dimensional hierarchical cluster analysis (Fig. 1C).
Figure 1.
RNA-seq analyses utilizing starved dG9a depleted adult flies. (A) RNA-seq data using 0 h and 12 h starved adult flies of the dG9a null mutant (dG9a RG5) and the wild type (Canton S). We added 1 to the FPKM value before log2 transformation to avoid a negative value and used this transformed value for the gene expression level. Horizontal axis represents the gene expression level in the group shown above the graph. Vertical axis represents the gene expression level in the group shown in the right of the graph. R value means the Pearson’s correlation coefficient between the group shown in below and in left. (B) RNA-seq analysis of the dG9a gene region. The x-axis represents genomic location on X chromosome. dG9a open reading frame is shown as the blue line in the ‘gene’ column. The y-axis represents tag density in Canton S (Blue) and dG9a RG5 (Red) (C) Heatmap of differentially expressed genes. Eight clusters were generated based on the expression changes between Canton S and dG9a RG5 as well as starvation conditions.
To identify biological features of differentially expressed genes, we carried out a GO analysis. The enriched biological process terms were analyzed with the Database for Annotation, Visualization and Integrated Discovery (DAVID; http://david.abcc.ncifcrf.gov) tool in the context of the GO classification with biological process (BP) terms. We focused on the cluster 1, 2, 6, 7 and 8, because genes included in these clusters are differentially expressed between 12 h starved dG9a null mutant and 12 h starved wild type (Fig. 1C). The all enriched BP terms with differentially expressed genes included in these clusters are shown in Table 1. The most enriched term was ‘innate immune response’ (P = 7.8 × 10−13) and the second was ‘response to bacterium’ (Table 1). These results are consistent with the previous study utilizing mouse model that revealed that G9a is recruited by the transcription factor ATF7 and regulates the expression of a group to genes involved in innate immunity in macrophages25.
Table 1.
GO analysis of differentially expressed genes in starved dG9a RG5.
| Category | Term | P-Value | Benjamini |
|---|---|---|---|
| GOTERM_BP_DIRECT | innate immune response | 7.80E-13 | 1.50E-10 |
| GOTERM_BP_DIRECT | response to bacterium | 6.30E-12 | 6.10E-10 |
| GOTERM_BP_DIRECT | antibacterial humoral response | 1.10E-09 | 6.80E-08 |
| GOTERM_BP_DIRECT | defense response to Gram-positive bacterium | 6.20E-07 | 3.00E-05 |
| GOTERM_BP_DIRECT | cellular response to heat | 2.30E-05 | 8.80E-04 |
| GOTERM_BP_DIRECT | defense response | 2.80E-05 | 9.00E-04 |
| GOTERM_BP_DIRECT | defense response to bacterium | 2.20E-04 | 5.90E-03 |
| GOTERM_BP_DIRECT | response to heat | 4.90E-04 | 1.20E-02 |
| GOTERM_BP_DIRECT | multicellular organism reproduction | 9.50E-04 | 2.00E-02 |
| GOTERM_BP_DIRECT | cellular response to UV | 2.80E-03 | 5.20E-02 |
| GOTERM_BP_DIRECT | humoral immune response | 3.10E-03 | 5.20E-02 |
| GOTERM_BP_DIRECT | oxidation-reduction process | 3.30E-03 | 5.20E-02 |
| GOTERM_BP_DIRECT | response to oxidative stress | 4.60E-03 | 6.60E-02 |
| GOTERM_BP_DIRECT | proteolysis | 5.00E-03 | 6.70E-02 |
| GOTERM_BP_DIRECT | defense response to Gram-negative bacterium | 5.30E-03 | 6.50E-02 |
| GOTERM_BP_DIRECT | carbohydrate metabolic process | 8.70E-03 | 9.90E-02 |
| GOTERM_BP_DIRECT | response to UV | 1.30E-02 | 1.40E-01 |
| GOTERM_BP_DIRECT | nitrogen compound metabolic process | 1.50E-02 | 1.50E-01 |
| GOTERM_BP_DIRECT | defense response to fungus | 2.00E-02 | 1.80E-01 |
| GOTERM_BP_DIRECT | cellular response to unfolded protein | 3.50E-02 | 2.90E-01 |
| GOTERM_BP_DIRECT | defense response to protozoan | 3.50E-02 | 2.90E-01 |
| GOTERM_BP_DIRECT | hexose metabolic process | 5.80E-02 | 4.20E-01 |
| GOTERM_BP_DIRECT | wing disc development | 8.80E-02 | 5.50E-01 |
| GOTERM_BP_DIRECT | sensory perception of chemical stimulus | 9.70E-02 | 5.70E-01 |
dG9a null mutant shows the increased expression of gustatory receptor genes under starvation conditions
Our GO analyses detected the term ‘sensory perception of chemical stimulus’ (P = 9.7 × 10−2). dG9a depletion significantly changed the expression of several Odorant binding proteins (Obps) and Gustatory receptors (Grs) under starved conditions. The RNA-seq results of these genes, Gr93d, Obp46a, Obp56a, Obp56b, Obp56e and Obp56f, as well as other Obps and Grs are summarized in Table S1. We confirmed that the expression of Obp56a, Obp56e, Obp56f and Gr93d is significantly changed by dG9a depletion by the reverse transcription–quantitative PCR (RT-qPCR) analyses (Fig. S2). A number of Drosophila OBPs are expressed both in gustatory sensilla and olfactory sensilla26. Furthermore, some OBPs have been reported to be required for taste sensitivity27–29. Although the detailed functions of most of the differentially expressed Obp and Gr genes have not been clarified yet, we considered that dG9a might be responsible for regulating the expression of genes relating to gustatory senses. To address this point, we performed RT-qPCR analyses to examine the expression levels of the 8 gustatory receptor genes, that are well known to be required for the sense of sweet taste30, in 4 time points after starvation (0, 6, 12, 24 h). Among these genes, Gr64b and Gr64c are translated from the same transcript according to Flybase (http://Flybase.org). Gr64d and Gr64e are also translated from the same transcript.
The RT-qPCR analyses indicated that dG9a depletion significantly up-regulated the expression of Gr64a, Gr64b/Gr64c and Gr64d/Gr64e under starvation conditions as well as fed conditions (P < 0.05), while it showed little effects on the expression of Gr5a, Gr61a and Gr64f (Fig. 2). Gr5a is reported to be necessary for sensing trehalose31–33. Gr61a is functional for sensing trehalose and glucose34. Gr64f is required for sugar detection in combination with other gustatory receptors35. Gr64a-e are also reported to sense sugar taste, especially Gr64a plays a critical role in sensing sucrose, maltose and glucose35. These results indicate that dG9a is responsible for suppressing the expression levels of the genes encoding gustatory receptors for sensing sweet taste. Although dG9a modifies the expression of a wide variety of genes through its epigenetic regulation as shown in RNA-seq analyses, the results of our RT-qPCR analyses led us to further examine the relationship between dG9a and the sense of taste under starvation conditions in the present study.
Figure 2.
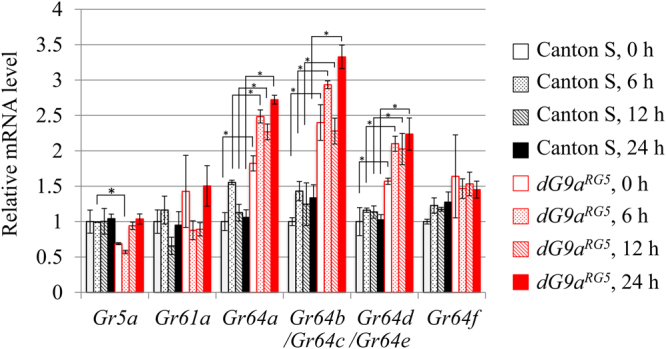
dG9a regulates the expression of gustatory receptor genes. Quantification of mRNA levels by RT-qPCR analyses of Gr5a, Gr61a, Gr64a, Gr64b/c, Gr64d/e and Gr64f in Canton S and dG9a RG5 adult flies after 0 h, 6 h, 12 h and 24 h starvation. According to Flybase (http://Flybase.org), Gr64b and Gr64c are translated from the same transcript shown as Gr64b/Gr64c. Gr64d and Gr64e are also translated from the same transcript shown as Gr64d/Gr64e. Results were normalized to β-tubulin and are displayed as relative values for that of 0 h starved Canton S. n = 3. Error bars represent standard errors. *There is significant difference between Canton S and dG9a RG5 in each time point evaluated by unpaired two-tailed Student t-tests. *P < 0.05. There was also significant difference in Gr64a, Gr64b/c, Gr64d/e between Canton S and dG9a RG5 when a two-way factorial analysis of variance was applied (P < 0.05).
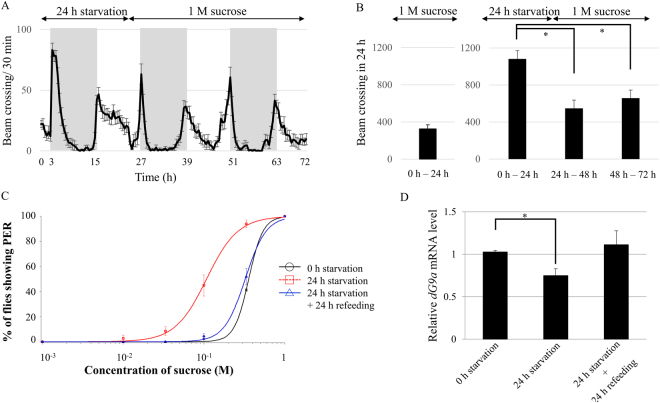
dG9a null mutant shows increased sensitivity to sucrose under starvation conditions
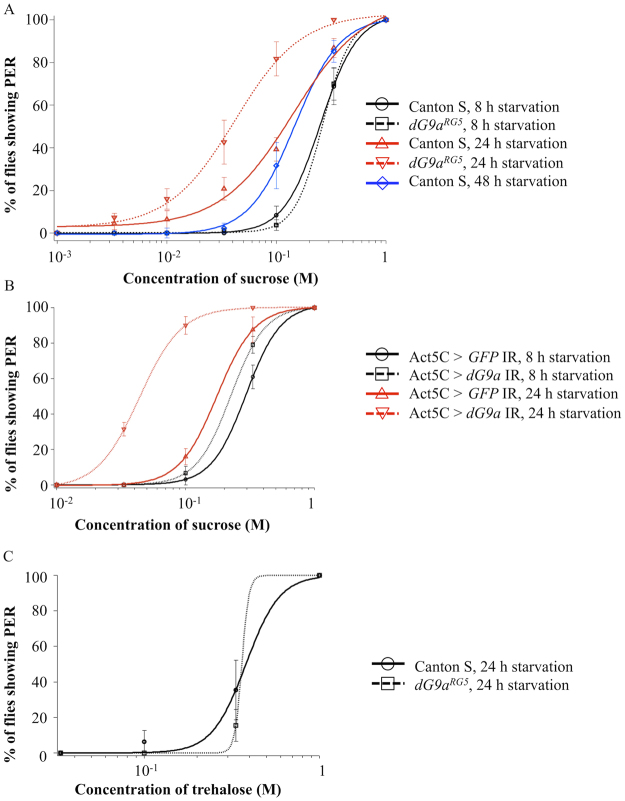
We then examined the sensitivity to sweet tastes in starved wild type and dG9a null mutant by proboscis extension reflex (PER) tests (Fig. 3A), one of the widely used behavioral assays for evaluating gustatory sensitivity in insects including Drosophila 23,36. When an attractive substance like sugar contacts to the chemosensilla, the fly extends its proboscis to feed it. Therefore, the taste sensitivity to specific substances can be evaluated by measuring the threshold changes of PER responses23,36. The Gr64a has been reported to be required for the detection of sugars including sucrose35,37, therefore we first examined PER responses to sucrose. The threshold of PER responses to sucrose of 24 h starved wild type flies (threshold: 0.13 ± 0.023 M (average ± standard error)) was significantly decreased compared to that of 8 h starved wild type flies (threshold: 0.25 ± 0.035 M) (P < 0.05). Such up-regulation of the sensitivity in the wild type (Canton S) under starvation was previously observed38. We also performed PER tests with dG9a null mutant at the same time. dG9a null mutant showed no significant difference in the threshold level of PER responses to sucrose in 8 h starvation conditions (threshold: 0.29 ± 0.023 M) in compared to the wild type (P > 0.05). However, dG9a depletion significantly increased the sensitivity in 24 h starved conditions (threshold: 0.047 ± 0.012 M) (P < 0.05). The sensitivity is not further increased after 24 h starvation in the wild type because the threshold of 48 h starved wild type (threshold: 0.17 ± 0.044 M) is not significantly changed in compared with that of 24 h starved wild type (P > 0.05), indicating that the increase of the sucrose sensitivity in dG9a null mutant is not due to the acceleration of starvation conditions. These results indicate that dG9a depletion increases the sucrose sensitivity in response to starvation.
Figure 3.
dG9a depletion increases the taste sensitivity to sucrose under starvation. (A) Sucrose concentration-PER curves in Canton S and the dG9a RG5 under different starvation conditions (n = 5). (B) Sucrose concentration-PER curves in the dG9a knockdown fly (+; GAL4 Act5C.PI/UAS-dG9a IR;+) and control fly (+; GAL4 Act5C.PI/GFP dsRNA. R. Scer/UAS;+) under different starvation conditions (n = 5). The UAS-dG9a IR strain was created previously (strain number 79)17. (C) Trehalose concentration-PER curves in the wild type and the dG9a null mutant under 24 h starvation conditions (n = 4). These sucrose concentration-PER curves are created by fitting the average of the results in each time point to Hill equation using Igor pro software (WaveMetrics). Error bars represent standard errors.
We further confirmed these results with dG9a knockdown flies of whole bodies (Act5C > dG9a IR) (Fig. 3B). The threshold of PER responses to sucrose in 24 h starved Act5C > dG9a IR flies (threshold: 0.044 ± 0.0036 M) was significantly decreased in compared with that in the control flies (Act5C > GFP IR) (threshold: 0.157 ± 0.029 M) (P < 0.05), while there was no decrease under 8 h starved conditions (Act5C > GFP IR threshold: 0.30 ± 0.031 M, Act5C > dG9a IR: 0.24 ± 0.040 M) (P > 0.05). These data confirmed that the increase of the sucrose sensitivity in dG9a null mutant under 24 h starvation conditions shown in Fig. 3A is not due to the possible background mutation of the dG9a null mutant.
It is noted that disruption of Gr5a decreases the sensitivity to trehalose31–33. dG9a null mutant showed no significant difference in the threshold level of PER responses to trehalose in 24 h starvation conditions (threshold: 0.46 ± 0.068 M) in compared to the wild type (threshold: 0.36 ± 0.084 M) (P > 0.05) (Fig. 3C). This is consistent with the results that dG9a depletion has little effects on regulating the expression of Gr5a (Fig. 2).
dG9a null mutant exhibits the hyperactivity under starvation conditions
The PER tests suggest that dG9a depletion promotes the taste sensitivity to sucrose in response to starvation conditions. Starved flies also promote their locomotion activity to increase the possibility that they can find desirable food21,22. Therefore, we next examined whether dG9a regulates the locomotion activity under starvation conditions or not. The locomotion activity was measured by the frequency that flies cross the infrared beam in the midpoint of the tubes of the Drosophila Activity Monitoring (DAM) system (Trikinetics) as described previously39.
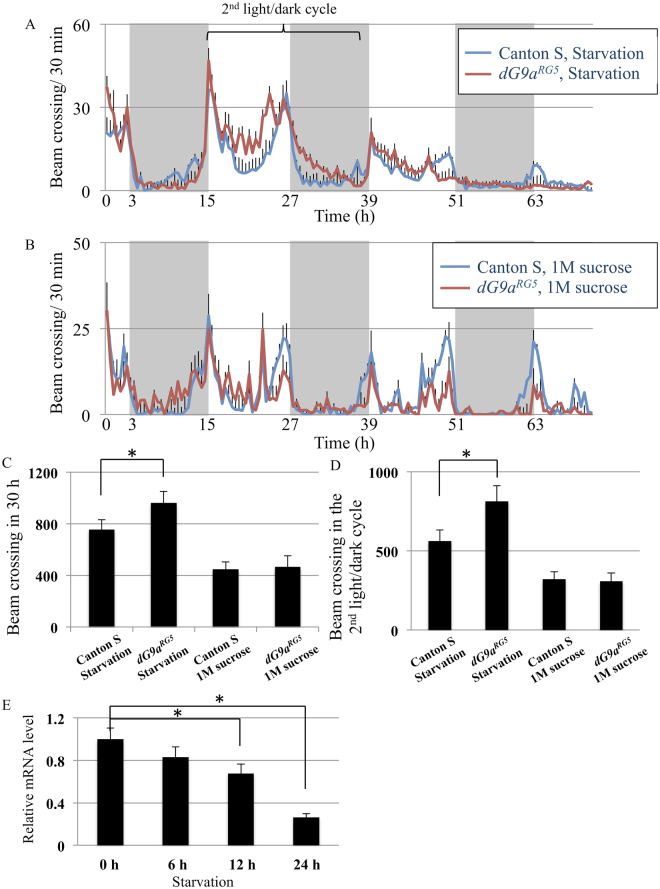
We examined the locomotion activity of the wild type and dG9a null mutant under starvation conditions (Fig. 4A) and that with feeding 1 M sucrose solution as a control experiment (Fig. 4B). Interestingly the hyperactivity was observed with dG9a null mutant remarkably during the 2nd light/dark cycle under starvation conditions, whereas such hyperactivity was not occurred in the case with feeding 1 M sucrose solution (Fig. 4A,B). Since the dG9a null mutant starts to die after 30 h starvation20, we compared the total beam crossing during 30 h after the start point (Fig. 4C). The total beam crossing under starvation conditions in the dG9a null mutant exhibited 27% increase in compared with that in the wild type (Fig. 4C). Remarkably, the total beam crossing under starvation conditions in the dG9a null mutant exhibited 45% increase compared with that in the wild type during the 2nd light/dark cycle (Fig. 4D). These results indicate that dG9a is responsible for suppressing the locomotion activity under starvation and that this suppression is disrupted in the dG9a null mutant. Taken together with the results of PER tests (Fig. 3), we suggest that dG9a is functional for the suppression of both sucrose sensitivity and locomotion activity, which may result in longevity under starvation through saving energy. Consistent with our hypothesis, the expression of dG9a in adult whole body decreases gradually under starvation in wild type flies (Fig. 4D). The mechanisms of the starvation-induced regulation of locomotion activity and sucrose sensitivity may be too complex to be explained solely by dG9a expression in wild type flies, as starvation conditions may change a wide variety of genes. However, our results indicate the possibility that the hyperactivity and higher sucrose sensitivity observed in starved wild type flies occur by the lack of dG9a, as observed in the starved dG9a RG5 mutant.
Figure 4.
dG9a depletion increases the locomotion activity under starvation. (A) The beam crossing activity of Canton S and the dG9a RG5 under starvation conditions. Gray and white background in the graph represents 12 h dark periods and 12 h light periods, respectively. (B) The beam crossing activity of Canton S and dG9a RG5 in the presence of 1 M sucrose. (C,D) Averaged total crossing in 30 h after start point (C) and during 2nd light/dark cycle (D). 0 h starved Canton S: n = 31. 0 h starved dG9a RG5: n = 29. Canton S on 1 M sucrose: n = 24. dG9a RG5 on 1 M sucrose: n = 20. *P < 0.05. (E) Quantification of mRNA levels by RT-qPCR analyses of dG9a in wild type adult flies after 0 h, 6 h, 12 h and 24 h starvation. Results were normalized to β-tubulin and are displayed as relative values for that of 0 h starvation (n = 3). Error bars represent standard errors. *P < 0.05.
dG9a depletion decreases the expression of insulin-like peptides under starvation conditions
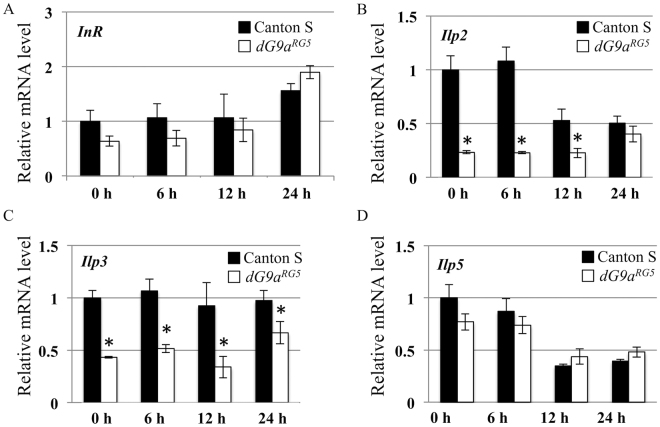
The starvation-induced hyperactivity is caused by activating the octopaminergic neurons in Drosophila melanogaster 24,38. It also has been reported that the starvation-induced hyperactivity can be suppressed by Insulin-like peptides (Ilps) in Drosophila 24. Hence, we examined whether dG9a plays a role in regulating the expression of genes involved in these signals. Among the 7 Ilp genes (Ilp1-7) existing in the Drosophila genome, Ilp2, 3 and 5 are reported to be expressed in insulin producing cells during adult stage and the expression of these Ilps decreases in response to starvation40–42. Interestingly, our RT-qPCR analyses showed that the dG9a depletion significantly reduced the expression levels of Ilp2 and Ilp3 throughout the starvation, whereas it exerted no effect on the expression level of Insulin-like receptor (InR) and Ilp5 (Fig. 5A–D). The hyperactivity was not induced in dG9a null mutant compared to wild type in fed conditions (Fig. 4B), although the expression levels of Ilp2 and Ilp3 were significantly reduced by dG9a depletion (Fig. 5C,D). Consistent with these observations, other studies reported that suppression of Ilps increases the locomotion activity under starvation conditions, but not under fed conditions24. There results suggest that dG9a is responsible for suppressing the locomotion activity under starvation stress by up-regulating the expression levels of Ilps.
Figure 5.
dG9a regulates the expression of Ilp genes. Quantification of mRNA levels by RT-qPCR analyses of InR. (A), Ilp2 (B), Ilp3 (C) and Ilp5 (D) in Canton S and dG9a RG5 adult flies after 0 h, 6 h, 12 h and 24 h starvation. Results were normalized to β-tubulin or Gapdh1 and are displayed as relative values for that of 0 h starved Canton S (n = 3). Error bars represent standard errors. *P < 0.05.
We found that dG9a regulates the expression of Ilp genes as well as Gr64 genes (Figs 2 and 5). To examine whether dG9a regulates the expression of these genes directly or not, we performed Chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR) assay with 0 h and 24 h starved flies. By this assay, we could not detect the dG9a binding to the genomic regions containing the promoters of Ilp2, Ilp3, Ilp5, Gr64a and Gr64b/c (Fig. S3), suggesting that dG9a regulates the expression of these genes rather indirectly at these time points. However, by the ChIP-qPCR assay, we cannot exclude the possibility that dG9a interacts only transiently with the target chromatin loci. Further detailed analyses will be required to reveal the mechanism how dG9a regulates the expression of these genes.
Refeeding after fasting restores the hyperactivity and increased sensitivity to sucrose as well as dG9a expression
We observed that starvation conditions induce hyperactivity (Fig. 4C) and increased sensitivity to sucrose (Fig. 3A) as well as decreased dG9a mRNA level (Fig. 4D). Next, we examined whether refeeding after starvation conditions restores these changes or not. Firstly, we examined the locomotion activity of the wild type under 24 h starvation followed by refeeding of 1 M sucrose (Fig. 6A). The total beam crossing during 24 h starvation conditions exhibited 70% increase in compared with that in 1 M sucrose fed conditions (Fig. 6B). The total beam crossing during 24 h refeeding after 24 h starvation conditions exhibited 49% decrease in compared with that in 24 h starvation conditions (Fig. 6B). Secondly, we examined the sensitivity to sucrose by PER tests. The threshold of PER responses of wild type flies under 24 h starvation conditions followed by 24 h refeeding of 1 M sucrose (threshold: 0.31 ± 0.018 M) was significantly increased compared to that of 24 h starved wild type flies (threshold: 0.11 ± 0.014 M) (P < 0.05) (Fig. 6C). There was no significant difference between PER thresholds of refed flies and that of 0 h starved flies (threshold: 0.34 ± 0.010 M) (P = 0.13) (Fig. 6C). Finally, we examined the mRNA level of dG9a by RT-qPCR. Remarkably, 24 h refeeding rescued the reduced mRNA level of 24 h starved flies to the similar level to that of 0 h starved flies (P = 0.31 between 0 h starved flies and refed flies) (Fig. 6D). These results indicate that refeeding after fasting restores the hyperactivity and increased sensitivity to sucrose as well as dG9a expression. Moreover, these results support the idea that the hyperactivity and higher sucrose sensitivity observed in starved wild type flies are dependent on the expression level of dG9a and that the regulation of behaviors by dG9a is highly responsive to the surrounding nutrient conditions.
Figure 6.
Refeeding after fasting restores the hyperactivity and increased sensitivity to sucrose as well as dG9a expression. (A) The beam crossing activity of Canton S under 24 h starvation conditions followed by 1 M sucrose refeeding. Gray and white background in the graph represents 12 h dark periods and 12 h light periods, respectively. (B) (left) Averaged total crossing in 24 h under 1 M sucrose feeding conditions. n = 24. (right) Averaged total crossing in 24 h under starvation conditions and refed conditions calculated from the data shown in Fig. 6A. n = 17. *P < 0.05. (C) Sucrose concentration-PER curves in Canton S under different starvation conditions and refed conditions (n = 3). (D) Quantification of mRNA levels by RT-qPCR analyses of dG9a in wild type adult flies after 0 h, 24 h starvation and after 24 h starvation followed by 24 h 1 M sucrose refeeding. Results were normalized to β-tubulin and are displayed as relative values for that of 0 h starvation (n = 3). Error bars represent standard errors. *P < 0.05.
Discussion
In the present study, we found that dG9a depletion increases the sucrose sensitivity in response to starvation conditions. We also found that dG9a regulates the locomotion activity under starvation conditions by controlling the expression of Ilp genes.
Prior to this study, we found that dG9a null mutant flies are sensitive to starvation and explored the underlying mechanism. dG9a is functional for saving energy through recycling cellular components by regulating the expression of genes required for autophagy20. In addition to this process, we here further found that dG9a functions as a suppressor of starvation-induced hyperactivity. This is also preferable for saving energy under starvation conditions. In nature, animals are exposed to starvation frequently, however, foraging require the costs of food-seeking energy as well as the threats from predation and environmental changes along with their migration43–45. Therefore, this foraging strategy requires assumption that the nutrient-poor conditions do not last long. Moreover, the strategy appears to be not effective under the conditions that there is no food available near them. Another way to survive under starvation is saving energy without moving like the hibernation associated with seasonal fluctuations of food availability, which can be observed in a wide range of animals including Drosophila 46,47. Our data indicated that dG9a suppresses the starvation-induced hyperactivity and that wild type flies exhibit the hyperactivity along with reduction of dG9a expression (Fig. 4). These data suggest that the wild type flies save energy without moving at the early phase of starvation and they become active to seek foods with risks along with the reduction of dG9a expression at the late phase of starvation. Therefore, our data suggest that dG9a functions as a key regulator for flies to decide these strategies depending on the time course of starvation and has an adaptive advantage to survive starvation conditions.
We performed RNA-seq analyses to identify which genes are regulated by dG9a under starvation stress (Fig. 1). Our GO analyses indicated that the most enriched term was ‘innate immune response’ and the second was ‘response to bacterium’ (Table 1), which suggests that there is strong relationship between dG9a and innate immune responses. In Drosophila, starved conditions and the following disruption of insulin signaling induce the expression of 4 antimicrobial peptides (AMPs), Metchnikowin (Mtk), Drosocin (Dro), Drosomycin (Drs) and Attacin-A (AttA)48. Our RNA-seq analyses detected the significant increase in the expression of all of these 4 antimicrobial genes under 12 h starved dG9a null mutant (Table S1). We examined the expression of these representative antibacterial genes by RT-qPCR using Canton S and dG9a RG5 kept under same conditions. The expression of 3 representative genes, Mtk, Dro and Drs, showed similar expression pattern to that observed in the results of RNA-seq analyses (Fig. S4). These results indicate that the expression pattern of antibacterial genes was not due to accidental infection of one set of the flies, but truly due to dG9a mutation. These observations suggest a link between activation of innate immune system and starvation. Previous reports suggested that the induction of AMPs may help maintaining and enhancing the defense activity in particular when organisms are exposed to poor conditions like starvation48. Our data indicates the possibility that dG9a null mutant acquires the excess defense activity to bacterial infection under starvation conditions.
We revealed that the dG9a depletion increases taste sensitivity to sucrose under 24 h starved conditions (Fig. 3). Our RT-qPCR results indicated that dG9a depletion significantly increased the expression levels of genes encoding gustatory receptors for sensing sugar taste under 24 h starved conditions (Fig. 2). These results suggest that the sucrose sensitivity under 24 h starved conditions is dependent on the expression levels of gustatory receptor genes regulated by dG9a. However, the dG9a depletion does not increase sucrose sensitivity under earlier (8 h) starved conditions (Fig. 3), although we detected the significant increases in gustatory receptor genes under 0 h and 6 h starved dG9a null mutant (Fig. 2). Moreover, none of these genes in wild type flies are up-regulated under 12 h and 24 h starvation conditions in compared to non-starved conditions (Fig. 2). These results suggest that the starvation-induced increase of the sucrose sensitivity in wild type flies as well as increase of the sucrose sensitivity under non-starved conditions are dependent on underlying mechanisms other than expression changes of gustatory receptor genes, including the alterations of translation levels of Gr mRNAs, localization of Gr mRNA/proteins and the responses of higher order gustatory circuits in the brain like the feeding behavior control by neuropeptides like dRYamides49–51. Further analyses are required to clarify these mechanisms.
Materials and Methods
Fly stocks
Fly stocks were cultured at 25 °C on standard food. Canton S was used as the wild-type strain. The dG9a RG5 flies were kindly provided by Dr. P. Spierer and used as the dG9a null mutant52. The dG9a RG5 flies show no defects in viability compared to Canton S in 1st instar larvae, pupae and adult stages52. The dG9a RG5 flies were back-crossed 10 times with Canton S to adjust the genetic background to Canton S. These backcrossed flies showed decreased viability under starvation conditions with similar extent of those with heterozygous mutant (dG9a RG5 /dG9a del34) and Fat-body specific knockdown of dG9a by RNAi20. The UAS-FLAG-IR dG9a (strain #79) fly stock was produced previously17 and used as the dG9a IR flies in this paper. The y1 w *; GAL4 Act5C.PI /CyO flies were used as the Act5C-GAL4 flies. The w 111; GFP dsRNA. R. Scer/UAS flies were used as the GFP IR flies. All other stocks used in this study were obtained from the Drosophila Genomics and Genetic Resources, Bloomington Drosophila Stock Center and Vienna Drosophila RNAi Center.
Life span assay
In the life span assay, newly merged adult flies were placed into vials on standard food at 25 °C. Every 3 days, they were transferred to new vials containing fresh standard food and the number of living flies was monitored until all had died. All assays were performed under non-crowded conditions (<20 flies per vial) and the results from 5 independent assays were combined. Graphs were generated with the Kaplan-Meier method by GraphPad Prism 6 software. Significance was calculated with Log-rank tests using GraphPad Prism 6 software.
RNA-seq analysis
Five days old adult flies were placed into vials including a piece of paper soaked with 1.0 mL PBS under non-crowded conditions (20 flies per vial) and were collected after 0 h and 12 h with each fly line. Thus, we made 4 groups; 0 h starved Canton S, 12 h starved Canton S, 0 h starved dG9a RG5 and 12 h starved dG9a RG5. RNAs were isolated using Trizol® reagent (Invitrogen) from 10 flies. The extraction procedures were repeated twice and samples were combined in each group for the analysis. Isolated RNAs were used for comparison with Illumina mRNA-seq libraries using a TruSeq Stranded mRNA Sample Prep Kits (Illumina) according to the manufacturer’s instructions. The obtained libraries were subjected to sequencing on a HiSeq. 1500 system (Illumina) for 51 cycles. We aligned the mRNA-seq reads to the reference Drosophila melanogaster genome sequence (UCSC dm6) using the TopHat53 program version 2.1.0 with the option ‘–library-type fr-secondstrand’. Cufflinks54 (version 2.2.1) was then employed with the option ‘-u–library-type fr-secondstrand’ to assemble transcripts and to calculate FPKM. Differentially expressed genes were also identified using Cuffdiff in the Cufflinks program package. The datasets generated during our RNA-seq analyses are available in the NCBIs Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession number GSE93144.
Cluster analysis
Two-dimensional hierarchical cluster analysis of differentially expressed genes was performed to generate a heatmap. The distance between every pair of genes was calculated as 1.0 - (Pearson’s correlation coefficient in FPKM between the 2 genes). The Ward’s linkage algorithm was used for cluster analysis. The heatmap was created using the heatmap.2 function of the ‘gplots’ packages in R language (https://www.r-project.org).
Gene ontology analysis of differentially expressed genes
To gain biological insights into differentially expressed genes, we analyzed genes that were significantly altered with q-values of <0.05. For this the Gene Ontology (GO) classification system was applied, employing the database for annotation, visualization and integrated discovery, DAVID version 6.855.
Reverse transcription–quantitative PCR (RT-qPCR)
Three replicates of 20 starved adult male flies were collected at each starvation periods. The starvation conditions were the same to those written in the RNA-seq analysis. Total RNA was isolated using Trizol® reagent (Invitrogen) from whole body and cDNA was synthesized using PrimeScript RT reagent kit (TaKaRa) according to the manufacturer’s instructions. Samples were run in duplicates with SYBR® Premix Ex TaqTM II (TaKaRa) using CFX96 touchTM (Biorad) and the data were analyzed with standard curve-based method calculated with CFX ManagerTM software. Specificity of primers was tested with melt curves created by CFX ManagerTM software and agarose gel electrophoresis of amplified fragments. β-tubulin and Gapdh1 were used as internal controls. Primer sequences of examined genes are listed below:
β-tubulin forward 5′-GAGACGTACTGCATCGACAAC-3′
β-tubulin reverse 5′-CAGGGAGACAAGATGGTTCAG-3′
Gapdh1 forward 5′-GGAGCCACCTATGACGAAATC-3′
Gapdh1 reverse 5′-TCGAACACAGACGAATGGG-3′
Gr5a forward 5′-CGCACTCTTTATCGCTCCATAG-3′
Gr5a reverse 5′-GCAGGCAGATGAAGTACAGATT-3′
Gr61a forward 5′-GAGGGTCTGAATGCCAAGAATA-3′
Gr61a reverse 5′-GAACTCCAGGGCAGCATTAT-3′
Gr64a forward 5′-ATGTTCACCCACCTGCTAAA-3′
Gr64a reverse 5′-AGGCGAATGAAGACCACATAG-3′
Gr64b/c forward 5′-GCAGCAAATGCCGAATGAA-3′
Gr64b/c reverse 5′-ATAGCACGATCAAACCGGATAA-3′
Gr64d/e forward 5′-CGCCTGGATGGTGTTCTTTA-3′
Gr64d/e reverse 5′-CAAGCCTCGACACATGAGAA-3′
Gr64f forward 5′-CTGGTCTTGATAGTGGCTCTTAAT-3′
Gr64f reverse 5′-TGGTCAACTGTGTCTTGTACTG-3′
dG9a forward 5′-TCAGATGGCCTATCTCCTTC-3′
dG9a reverse 5′-CAGTCCGCAGTTCATAATCC-3′
Akh forward 5′-CGTCCAGTGTCAATTGACCTTCTC-3′
Akh reverse 5′-AGCAGCATTTCGTTGGAGGTCTTG-3′
InR forward 5′-GGTACCAGAAGTCAGAGAACAAG-3′
InR reverse 5′-CGCTCCAAACTGTCGATAAGA-3′
Ilp2 forward 5′-ACGAGGTGCTGAGTATGGTGTGCG-3′
Ilp2 reverse 5′-CACTTCGCAGCGGTTCCGATATCG-3′
Ilp3 forward 5′-GTCCAGGCCACCATGAAGTTGTGC-3′
Ilp3 reverse 5′-CTTTCCAGCAGGGAACGGTCTTCG-3′
Ilp5 forward 5′-ATGGACATGCTGAGGGTTG-3′
Ilp5 reverse 5′-CGCCAAGTGGTCCTCATAAT-3′
Mtk forward 5′-GCAACTTAATCTTGGAGCGATTT-3′
Mtk reverse 5′-GGTCTTGGTTGGTTAGGATTGA-3′
Dro forward 5′-ATTCTGCCCGCCTAAAGATG-3′
Dro reverse 5′-TGCTGTCTTTCGTGTGTTTATTG-3′
Drs forward 5′-AAGTACTTGTTCGCCCTCTTC-3′
Drs reverse 5′-CACAGGGACCCTTGTATCTTC-3′
Proboscis extension reflex (PER) test
The PER tests23,36 were performed using flies after 8 h or 24 h starvation for Fig. 3. We used 8 h starved flies for evaluating the PER responses during the earlier stage of starvation to reduce the variations in the data. Five days old flies were used for PER tests. To induce dG9a knockdown in whole body, Act5C-GAL4 flies were crossed with dG9a IR flies and F1 progenies were used for tests. Act5C-GAL4 flies were crossed with GFP IR flies and F1 progenies were used as a control. The starvation conditions were the same to those written in the RNA-seq analysis. Seven concentration steps of sucrose or trehalose solutions (1, 1/3, 1/10, 1/30, 1/100, 1/300 and 1/1000 M) were prepared with distilled water for stimulation. Each fly was fixed at the tip of the 200 μL yellow tip. They were satiated with distilled water prior to the PER tests. Labellar gustatory sensilla were stimulated with a droplet of sugar solution with a pipette tip under a binocular microscope. Ingestion of the sucrose solutions may affect the starvation conditions and PER. Therefore, we stimulated the sensilla by touching the tip of them with a droplet of sugar solution in a moment not to feed flies. 15–20 flies were tested with the concentration series of sucrose starting with the lowest concentration with >5-min interval to eliminate the adaptation effect of the last stimulation. PER thresholds were calculated by fitting the data to the Hill equation using Igor pro software (WaveMetrics). The PER thresholds were defined as the concentration of sucrose which induced PER in the half of the population. We repeated this PER tests 3–5 times and calculated the average threshold.
Locomotion activity test
Fly strains were kept on 12 h light/dark cycles for at least 5 generations before performing locomotion activity tests. Individual 3–5 day-old adult male fly was placed in glass tube and its locomotion activity was continuously recorded by DAM2 Drosophila Activity Monitor (TriKinetics). One end of glass tubes was filled with cotton wool soaked with 0 M or 1 M sucrose solution and the other was sealed with PARAFILM® (Bemis). These instruments were put in the incubator at 25 °C and kept on 12 h light/dark cycles.
Chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR)
Fourty adult flies (3–5 days old) were collected after 0 h and 24 h starvation. The starvation conditions were the same to those written in the RNA-seq analysis. The flies were homogenized in a mortar after freezing with liquid N2 and DNA was cross-linked with 16% formaldehyde for 5 min. After cross-linking, we performed ChIP using ChIP Assay Kit (Millipore) according to the manufacturer’s instructions. In brief, cross-linked DNA was cut into fragments by sonication (Bioruptor, COSMO BIO) and immunoprecipitated with 0.2 μg of anti-dG9a antibody17 for 16 h. After washing and reverse cross-linking, DNA was isolated and performed qPCR with SYBR® Premix Ex TaqTM II (TaKaRa) using CFX96 touchTM (Biorad). The data were analyzed with standard curve-based method. The primers were designed in the site of −600 to −100 bp from the transcriptional start site, because dG9a is reported to bind to these upstream regions in the target genes by ChIP-sequencing analyses performed utilizing larvae5. Primer sequences used in this study are listed below:
Ilp2 forward 5′-GGCGCGGATTGGTAAGTATAG-3′
Ilp2 reverse 5′-CCGTACTAAGCCCTGCATTTAT-3′
Ilp3 forward 5′-CTCCTTGGGCGAGAAAGTAAA-3′
Ilp3 reverse 5′-CTGGCATGGGATGTCTGAAA-3′
Ilp5 forward 5′-TACGCTAATCCATGGGCATAAA-3′
Ilp5 reverse 5′-GCAGGAGCGTACTTGCTAAT-3′
Gr64a forward 5′-ACATGTATCTCGGCAGCTAATC-3′
Gr64a reverse 5′-GACACTTCTGTGGGAATTGGA-3′
Gr64b/c forward 5′-GACTGCTCTTTGGAGTGAGTAA-3′
Gr64b/c reverse 5′-GACAATGGTTCCAGCCATCTA-3′
MAGE forward 5′-CAGACGAAGAACATGGCTACTT-3′
MAGE reverse 5′-GCGTTCCCGCTTCAGATATT-3′
Refeeding assay
Five days old adult Canton S were placed into vials including a piece of paper soaked with 1.0 mL PBS under non-crowded conditions (20 flies per vial) for 24 h and subsequently into the vials including a piece of paper soaked with 1.0 mL 1 M sucrose for 24 h. We performed PER tests and RT-qPCR analyses utilizing flies collected in 24 h and 48 h after the start point. For locomotion activity tests, the wild type flies were put under starvation conditions for the first 24 h and then put under 1 M sucrose feeding conditions.
Statistical analysis
Unpaired two-tailed Student t-tests were performed to evaluate the difference of the average thresholds in PER assay, and to analyze the results of locomotion analyses and qPCR analyses. Compared results were considered statistically significant when the P-value < 0.05.
Electronic supplementary material
Acknowledgements
We thank Drs P. Spierer and C. Seum for kindly providing the dG9a RG5 fly. We acknowledge all members of the Chromosome Engineering Laboratory for helpful discussion and advice. We also thank Medical English Service for the English language review. This study was partially supported by the JSPS Core-to-Core Program, Asia-Africa Science Platforms and JSPS KAKENHI Grant Number 15J0948 and 16K07346.
Author Contributions
K.S., R.T., T.M. and T.S. performed the experiments. K.S., R.T., T.M., M.O., H.Y., Y.O., T.S., M.S. and M.Y. designed and analyzed the experiments. K.S. and M.Y. wrote the paper. K.S., T.M., M.O., H.Y., Y.O., T.S., M.S. and M.Y. revised the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-15344-2.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lester BM, et al. Behavioral epigenetics. Ann. N. Y. Acad. Sci. 2011;1226:14–33. doi: 10.1111/j.1749-6632.2011.06037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pandey UB, Nichols CD. Human disease models in Drosophila melanogaster and the role of the fly in therapeutic drug discovery. Pharmacol Rev. 2011;63:411–436. doi: 10.1124/pr.110.003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loke YJ, Hannan AJ, Craig JM. The role of epigenetic change in autism spectrum disorders. Front. Neurol. 2015;6:107. doi: 10.3389/fneur.2015.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mis J, Ner SS, Grigliatti TA. Identification of three histone methyltransferases in Drosophila: dG9a is a suppressor of PEV and is required for gene silencing. Mol. Genet. Genomics. 2006;275:513–526. doi: 10.1007/s00438-006-0116-x. [DOI] [PubMed] [Google Scholar]
- 5.Kramer JM, et al. Epigenetic regulation of learning and memory by Drosophila EHMT/G9a. PLoS Biol. 2011;9:e1000569. doi: 10.1371/journal.pbio.1000569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tachibana M, Sugimoto K, Fukushima T, Shinkai Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J. Biol. Chem. 2001;276:25309–25317. doi: 10.1074/jbc.M101914200. [DOI] [PubMed] [Google Scholar]
- 7.Tachibana M, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Estève PO, et al. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev. 2006;20:3089–3103. doi: 10.1101/gad.1463706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagano T, et al. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–1720. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- 10.Wagschal A, et al. G9a histone methyltransferase contributes to imprinting in the mouse placenta. Mol. Cell Biol. 2008;28:1104–1113. doi: 10.1128/MCB.01111-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Myant K, et al. LSH and G9a/GLP complex are required for developmentally programmed DNA methylation. Genome Res. 2011;21:83–94. doi: 10.1101/gr.108498.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maze I, et al. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science. 2010;327:213–216. doi: 10.1126/science.1179438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tachibana M, et al. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev. 2005;19:815–826. doi: 10.1101/gad.1284005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee DY, Northrop JP, Kuo MH, Stallcup MR. Histone H3 lysine 9 methyltransferase G9a is a transcriptional coactivator for nuclear receptors. J. Biol. Chem. 2006;281:8476–8485. doi: 10.1074/jbc.M511093200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bittencourt D, et al. G9a functions as a molecular scaffold for assembly of transcriptional coactivators on a subset of glucocorticoid receptor target genes. Proc. Natl. Acad. Sci. USA. 2012;109:19673–19678. doi: 10.1073/pnas.1211803109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seum C, Bontron S, Reo E, Delattre M, Spierer P. Drosophila G9a is a nonessential gene. Genetics. 2007;177:1955–1957. doi: 10.1534/genetics.107.078220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kato Y, Kato M, Tachibana M, Shinkai Y, Yamaguchi M. Characterization of Drosophila G9a in vivo and identification of genetic interactants. Genes Cells. 2008;13:703–722. doi: 10.1111/j.1365-2443.2008.01199.x. [DOI] [PubMed] [Google Scholar]
- 18.Lee KS, Yoon J, Park JS, Kang YK. Drosophila G9a is implicated in germ cell development. Insect Mol. Biol. 2010;19:131–139. doi: 10.1111/j.1365-2583.2009.00955.x. [DOI] [PubMed] [Google Scholar]
- 19.Ushijima Y, et al. Roles of histone H3K9 methyltransferases during Drosophila spermatogenesis. Chromosome Res. 2012;20:319–331. doi: 10.1007/s10577-012-9276-1. [DOI] [PubMed] [Google Scholar]
- 20.An PNT, et al. Epigenetic regulation of starvation-induced autophagy in Drosophila by histone methyltransferase G9a. Sci. Rep. 2017;7:7343. doi: 10.1038/s41598-017-07566-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee G, Park JH. Hemolymph sugar homeostasis and starvation-induced hyperactivity affected by genetic manipulations of the adipokinetic hormone-encoding gene in Drosophila melanogaster. Genetics. 2004;167:311–323. doi: 10.1534/genetics.167.1.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Isabel G, Martin JR, Chidami S, Veenstra JA, Rosay P. AKH-producing neuroendocrine cell ablation decreases trehalose and induces behavioral changes in Drosophila. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2005;288:R531–R538. doi: 10.1152/ajpregu.00158.2004. [DOI] [PubMed] [Google Scholar]
- 23.Nishimura A, et al. Starvation-induced elevation of taste responsiveness and expression of a sugar taste receptor gene in Drosophila melanogaster. J. Neurogenet. 2012;26:206–215. doi: 10.3109/01677063.2012.694931. [DOI] [PubMed] [Google Scholar]
- 24.Yu Y, et al. Regulation of starvation-induced hyperactivity by insulin and glucagon signaling in adult. Drosophila. Elife. 2016;5:e15693. doi: 10.7554/eLife.15693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshida K, et al. The transcription factor ATF7 mediates lipopolysaccharide-induced epigenetic changes in macrophages involved in innate immunological memory. Nat. Immunol. 2015;16:1034–1043. doi: 10.1038/ni.3257. [DOI] [PubMed] [Google Scholar]
- 26.Galindo K, Smith DP. A large family of divergent Drosophila odorant-binding proteins expressed in gustatory and olfactory sensilla. Genetics. 2001;159:1059–1072. doi: 10.1093/genetics/159.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsuo T, Sugaya S, Yasukawa J, Aigaki T, Fuyama Y. Odorant-binding proteins OBP57d and OBP57e affect taste perception and host-plant preference in Drosophila sechellia. PLoS Biol. 2007;5:e118. doi: 10.1371/journal.pbio.0050118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harada E, Haba D, Aigaki T, Matsuo T. Behavioral analyses of mutants for two odorant-binding protein genes, Obp57d and Obp57e. Drosophila melanogaster. Genes Genet. Syst. 2008;83:257–264. doi: 10.1266/ggs.83.257. [DOI] [PubMed] [Google Scholar]
- 29.Jeong YT, et al. An odorant-binding protein required for suppression of sweet taste by bitter chemicals. Neuron. 2013;79:725–737. doi: 10.1016/j.neuron.2013.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fujii S, et al. Drosophila sugar receptors in sweet taste perception, olfaction, and internal nutrient sensing. Curr Biol. 2015;25:621–627. doi: 10.1016/j.cub.2014.12.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dahanukar A, Foster K, van der Goes van Naters WM, Carlson JR. A Gr receptor is required for response to the sugar trehalose in taste neurons of Drosophila. Nat. Neurosci. 2001;4:1182–1186. doi: 10.1038/nn765. [DOI] [PubMed] [Google Scholar]
- 32.Ueno K, et al. Trehalose sensitivity in Drosophila correlates with mutations in and expression of the gustatory receptor gene Gr5a. Curr. Biol. 2001;11:1451–1455. doi: 10.1016/S0960-9822(01)00450-X. [DOI] [PubMed] [Google Scholar]
- 33.Chyb S, Dahanukar A, Wickens A, Carlson JR. Drosophila Gr5a encodes a taste receptor tuned to trehalose. Proc. Natl. Acad. Sci. USA. 2003;100:14526–14530. doi: 10.1073/pnas.2135339100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Freeman EG, Wisotsky Z, Dahanukar A. Detection of sweet tastants by a conserved group of insect gustatory receptors. Proc. Natl. Acad. Sci. USA. 2014;111:1598–1603. doi: 10.1073/pnas.1311724111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiao Y, Moon SJ, Montell CA. Drosophila gustatory receptor required for the responses to sucrose, glucose, and maltose identified by mRNA tagging. Proc. Natl. Acad. Sci. USA. 2007;104:14110–14115. doi: 10.1073/pnas.0702421104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shiraiwa T, Carlson JR. Proboscis extension response (PER) assay in Drosophila. J. Vis. Exp. 2007;3:193. doi: 10.3791/193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dahanukar A, Lei YT, Kwon JY, Carlson JR. Two Gr genes underlie sugar reception in Drosophila. Neuron. 2007;56:503–516. doi: 10.1016/j.neuron.2007.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Z, et al. Octopamine mediates starvation-induced hyperactivity in adult Drosophila. Proc. Natl. Acad. Sci. USA. 2015;112:5219–5224. doi: 10.1073/pnas.1417838112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pfeiffenberger C, Lear BC, Keegan KP, Allada R. Locomotor activity level monitoring using the Drosophila Activity Monitoring (DAM) System. Cold Spring Harb. Protoc. 2010;2010:pdb.prot5518. doi: 10.1101/pdb.prot5518. [DOI] [PubMed] [Google Scholar]
- 40.Ikeya T, Galic M, Belawat P, Nairz K, Hafen E. Nutrient-dependent expression of insulin-like peptides from neuroendocrine cells in the CNS contributes to growth regulation in Drosophila. Curr. Biol. 2002;12:1293–1300. doi: 10.1016/S0960-9822(02)01043-6. [DOI] [PubMed] [Google Scholar]
- 41.Broughton SJ, et al. Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc Natl. Acad. Sci. USA. 2005;102:3105–3110. doi: 10.1073/pnas.0405775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okamoto N, Nishimura T. Signaling from glia and cholinergic neurons controls nutrient-dependent production of an insulin-like peptide for Drosophila body growth. Dev. Cell. 2015;35:295–310. doi: 10.1016/j.devcel.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 43.Anderson PK. Foraging range in mice and voles: the role of risk. Can. J. Zoology. 1986;64:2645–2653. doi: 10.1139/z86-384. [DOI] [Google Scholar]
- 44.Kohler SL, Mcpeek MA. Predation risk and the foraging behavior of competing stream insects. Ecology. 1989;70:1811–1825. doi: 10.2307/1938114. [DOI] [Google Scholar]
- 45.Padilla SL, et al. Agouti-related peptide neural circuits mediate adaptive behaviors in the starved state. Nat. Neurosci. 2016;19:734–741. doi: 10.1038/nn.4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCue MD. Starvation physiology: reviewing the different strategies animals use to survive a common challenge. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2010;156:1–18. doi: 10.1016/j.cbpa.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 47.Dumlupinara R, et al. Trace element changes during hibernation of Drosophila melanogaster by WDXRF analyses at chilling temperature. J. Quantitative Spectroscopy and Radiative Transfer. 2006;102:492–498. doi: 10.1016/j.jqsrt.2006.02.030. [DOI] [Google Scholar]
- 48.Becker T, et al. FOXO-dependent regulation of innate immune homeostasis. Nature. 2010;463:369–373. doi: 10.1038/nature08698. [DOI] [PubMed] [Google Scholar]
- 49.Ida T, et al. Identification of the novel bioactive peptides dRYamide-1 and dRYamide-2, ligands for a neuropeptide Y-like receptor in Drosophila. Biochem. Biophys. Res. Commun. 2011;410:872–877. doi: 10.1016/j.bbrc.2011.06.081. [DOI] [PubMed] [Google Scholar]
- 50.Kain P, Dahanukar A. Secondary taste neurons that convey sweet taste and starvation in the Drosophila brain. Neuron. 2015;85:819–832. doi: 10.1016/j.neuron.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 51.Maeda T, et al. Suppressive effects of dRYamides on feeding behavior of the blowfly. Phormia. regina. Zoological Lett. 2015;1:35. doi: 10.1186/s40851-015-0034-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shimaji K, et al. Genomewide identification of target genes of histone methyltransferase dG9a during Drosophila embryogenesis. Genes Cells. 2015;20:902–914. doi: 10.1111/gtc.12281. [DOI] [PubMed] [Google Scholar]
- 53.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trapnell C, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dennis G, Jr., et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.