The melibiose symporter MelB couples melibiose transport to that of cations such as Na+. Hariharan and Guan show that the binding of Na+ and melibiose is thermodynamically cooperative and that Na+ coupling is based on ion concentrations and competitive binding, but not ion selectivity.
Abstract
The Na+-coupled melibiose symporter MelB, which can also be coupled to H+ or Li+ transport, is a prototype for the glycoside-pentoside-hexuronide:cation symporter family. Although the 3-D x-ray crystal structure of Salmonella typhimurium MelB (MelBSt) has been determined, the symport mechanisms for the obligatory coupled transport are not well understood. Here, we apply isothermal titration calorimetry to determine the energetics of Na+ and melibiose binding to MelBSt, as well as protonation of this transporter. Studies of the thermodynamic cycle for the formation of the Na+–MelBSt–melibiose ternary complex at pH 7.45 reveal that the binding of Na+ and melibiose is cooperative. The binding affinity for one substrate (Na+ or melibiose) is increased by the presence of the other by about eightfold. The coupling free energies (ΔΔG) of either substrate binding are ∼5 kJ/mol, and binding of both substrates releases a free energy of ∼35 kJ/mol. Measurements of the Na+-binding enthalpy at three different pH values, including the pKa value of MelB, indicate that the binding of one Na+ displaces one H+ per MelBSt molecule. In addition, the absolute dissociation constants for Na+ and H+, determined by competitive binding, show that MelBSt is selective for H+ over Na+ by ∼1,000-fold at a pKa of 6.25. Thus, the Na+ coupling in MelBSt is based not on ion selectivity but on ion concentrations and competitive binding because of a much higher Na+ concentration under physiological conditions. Such a selectivity feature seems to be common for membrane transport proteins that can bind both H+ and Na+ at a common site.
Introduction
Cation-coupled symporters use the energy stored in cation electrochemical gradients across cell membranes to translocate molecules necessary for cellular functions. Most secondary-active transporters in the major facilitator superfamily (MFS), the largest family of transporters containing over 10,000 sequenced members (Saier et al., 1999), use the H+ electrochemical gradient for their functions, but some members are able to couple solute transport to Na+ translocation, such as the bacterial melibiose transporter MelB (Tsuchiya and Wilson, 1978; Wilson and Ding, 2001) and the eukaryotic lysophosphatidylcholine symporter (Nguyen et al., 2014). Both proteins belong to the glycoside-pentoside-hexuronide:cation symporter family (TCDB 2.A.2; Poolman et al., 1996), a subgroup of the MFS family. MelB catalyzes galactoside symport not only with Na+ but also with Li+ or H+ (Tsuchiya and Wilson, 1978; Niiya et al., 1980; Bassilana et al., 1987; Guan et al., 2011); however, this symporter cannot transport sugars with K+, Rb+, or Cs+ (Guan et al., 2011). We have determined the high-resolution x-ray 3-D crystal structure of Salmonella typhimurium MelB (MelBSt) at a resolution of 3.35 Å (Ethayathulla et al., 2014). This is the first high-resolution structure of a member of the MFS family that uses Na+ as a coupling cation. MelBSt was captured in two slightly different conformations (Ethayathulla et al., 2014). Like other MFS-fold transporters (Abramson et al., 2003; Huang et al., 2003; Guan and Kaback, 2006; Guan et al., 2007; Dang et al., 2010), its N- and C-terminal six-helix bundles surround a central aqueous cavity that contains side chains important for the binding of galactoside and Na+, Li+, or H+ and opens to the periplasmic side (Fig. 1, a and b). Previously, using a homology threading approach, we proposed that MelB is an MFS symporter (Yousef and Guan, 2009). The crystal structures confirmed this prediction and cleared up previous controversies about the MelB fold. The structural information is consistent with many previous biochemical and biophysical studies with Escherichia coli MelB (MelBEc; Mus-Veteau et al., 1995; Pourcher et al., 1995; Mus-Veteau and Leblanc, 1996; Maehrel et al., 1998; Ganea et al., 2001; Wilson and Ding, 2001; Meyer-Lipp et al., 2006; Granell et al., 2010). An alternating-access mechanism has been proposed to be involved in the sugar transport process (Meyer-Lipp et al., 2006; Yousef and Guan, 2009; Guan et al., 2012; Ethayathulla et al., 2014), similar to that proposed for other members of this superfamily (Abramson et al., 2003; Huang et al., 2003; Guan and Kaback, 2006; Meyer-Lipp et al., 2006; Kaback, 2015).
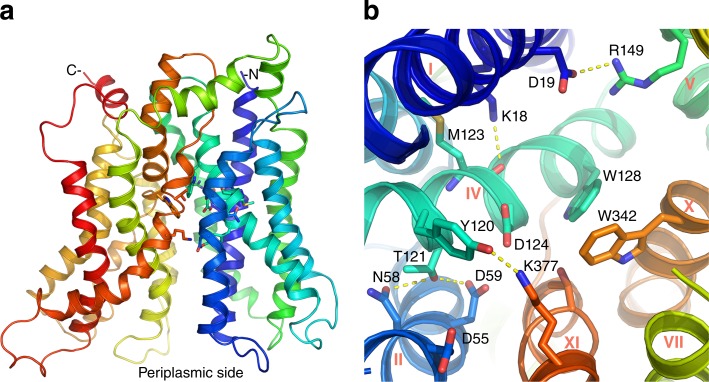
Figure 1.
X-ray crystal structure of MelBSt. See Protein Data Bank accession no. 4M64. (a) The overall fold of MelBSt in a periplasmic-side-open conformation. Helices in rainbow colors from blue (N terminus) to red (C terminus). (b) Cosubstrate-binding sites. The helices are labeled in roman numerals, and side chains potentially involved in the cation binding (residues D55, D59, D124, N58, and T121) or in the galactoside binding (residues D19, R149, Y120, D124, W128, W342, and K377) are highlighted as sticks. Residues Y120, D124, and K373 may be involved in both sites.
The structure shows that Asp residues at positions 55 and 59 (helix II) and 124 (IV) may form a cation-binding site (Fig. 1 b). In both MelBSt and MelBEc, all three Asp residues are required for Na+ stimulation of melibiose transport or galactoside binding (Pourcher et al., 1993; Zani et al., 1994; Granell et al., 2010; Ethayathulla et al., 2014), as well as for the specific Fourier-transform infrared signal change elicited by Na+ (Granell et al., 2010). Functional studies with MelBEc have shown that all three cations (Na+, Li+, and H+) compete for a common binding site (Lopilato et al., 1978; Damiano-Forano et al., 1986; Mus-Veteau et al., 1995). With MelBSt, a common binding site for Na+ or Li+ has been also established (Guan et al., 2011). The crystal structure (Ethayathulla et al., 2014) suggests that the proposed cation-binding pocket could selectively coordinate a Na+ or Li+, but it is not known how many Asp residues among the three are protonated. A single binding site for sugar has been also suggested, and the Na+/galactoside stoichiometric ratio has been determined to be 1:1 (Bassilana et al., 1987; Wilson and Ding, 2001; Guan et al., 2011). This galactoside-binding site, which is in close proximity to the cation site, is surrounded by residues D19 (helix I), R149 (V), Y120, D124, W128 (IV), W342 (X), and K377 (XI; Fig. 1 b). Helix IV physically hosts both cosubstrate sites, and residues Y120, D124, and K373 may contribute to the binding of both substrates, which was proposed to be the structural basis for the observed increase of the galactoside affinity by Na+ or Li+ (Ethayathulla et al., 2014). However, how the two sites cooperate for the binding events, the protonation status of the cation site, and the mechanism of cation selectivity are not well understood.
There are several methods to determine the affinity of MelB for Na+ binding; most depend on binding of a sugar (e.g., Na+ stimulation of [3H]α-nitrophenyl galactoside [[3H]α-NPG] binding; Damiano-Forano et al., 1986) or fluorescence resonance energy transfer (FRET) from Trp residues to the dansyl moiety of a fluorescent sugar 2′-(N-dansyl)aminoalkyl-1-thio-β-d-galactopyranoside (Trp→D2G FRET; Maehrel et al., 1998; Ganea et al., 2011; Guan et al., 2011; Jakkula and Guan, 2012; Amin et al., 2014). Isothermal titration calorimetry (ITC) is a label-free technique to measure heat changes (either release or absorption) derived from molecular interactions. It can directly reveal the binding enthalpy and allow for determination of the binding association constant (Cooper, 1999; Leavitt and Freire, 2001). Therefore, the binding free energy (ΔG) can be calculated. Here, we used Trp→D2G FRET and ITC to study Na+ binding, determine the free energy for Na+ and melibiose binding to MelBSt, test the protonation status of MelBSt, and study the competition between Na+ and H+ in the absence or presence of melibiose. The results show that the binding of Na+ and melibiose is thermodynamically cooperative, providing insights into the coupling mechanism of this symporter. Furthermore, the binding stoichiometry of melibiose and Na+ or H+ was also determined, confirming the previous conclusion that MelB catalyzes stoichiometric translocation of a melibiose with a cation (Na+, Li+, or H+). Moreover, by determining the absolute dissociation constants for Na+ and H+, we conclude that the use of Na+ as coupling ion for sugar transport is based not on ion selectivity but on competitive binding under physiological conditions (i.e., with a Na+ concentration five orders of magnitude higher than the H+ concentration).
Materials and methods
Reagents
The detergent undecyl-β-d-maltopyranoside (UDM) was from Anatrace. 2′-(N-dansyl)aminoalkyl-1-thio-β-d-galactopyranoside (D2G, dansyl-galactoside) was obtained from G. Leblanc (Institut de Biologie et Technologies-Saclay, CEA Saclay, France) and H.R. Kaback (University of California, Los Angeles, Los Angeles, CA). Tetramethylammonium hydroxide (TMAOH) and N-(2-acetamido)-2-aminoethanesulfonic acid (ACES) were from Sigma-Aldrich. 2-(N-morpholino)ethanesulfonic acid (MES) was obtained from Research Products International.
MelBSt expression and purification
The plasmid pK95 ΔAH/MelBSt/CHis10 (Pourcher et al., 1995; Guan et al., 2011) was used for the constitutive expression of the WT MelBSt or MelBSt mutants D55C or D59C containing a Cys in the position Asp55 or Asp59 (Ethayathulla et al., 2014), respectively. E. coli DW2 cells (melA+, melB, and lacZY) were used for protein overexpression (Pourcher et al., 1995). The cells were grown in Luria–Bertani broth supplemented with 50 mM KPi (pH 7.0), 45 mM (NH4)SO4, 0.5% glycerol, and 100 mg/L ampicillin. The protocols for membrane preparation and MelBSt purification by cobalt-affinity chromatography after extracted in a detergent UDM have been described previously (Ethayathulla et al., 2014). MelB protein in 20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.035% UDM, and 10% glycerol was concentrated and stored at −80°C. Protein samples were dialyzed against a specific assay buffer before a test.
Protein assay
The Micro BCA Protein Assay (Pierce Biotechnology, Inc.) was used for the protein concentration assay.
Na+ stimulation constant (K0.5(Na+)) on Trp→D2G FRET
Steady-state fluorescence measurements were performed with an AMINCO-Bowman Series 2 Spectrometer with purified MelBSt or MelBSt mutants D55C or D59C in a Na+-free buffer consisting of 20 mM Tris-HCl, pH 7.5, 0.035% UDM, 50 mM choline chloride (ChCl), and 10% glycerol at a protein concentration of 1 µM. The emission intensity was recorded at 490 nm using an excitation wavelength of 290 nm. After the addition of 10 µM D2G, NaCl was consecutively added until no change in fluorescence intensity occurred. The Na+ concentration at the end of titration was ∼50 mM for the WT and 200 mM for the mutants. Melibiose at oversaturating concentration was added after the end of the Na+ titration to displace the bound D2G. On a separate test, an identical volume of water instead of NaCl was used for the correction of dilution effect. For each addition, the intensities were recorded for 60 s, integrated, and averaged. Increases in fluorescence intensity are expressed as differential FRET (diffFRET; the difference before and after the addition of NaCl) and corrected by the dilution effect. The diffFRET values were plotted against the Na+ concentration, and the Na+ activation constant (the Na+ concentration for the half-maximal diffFRET [K0.5(Na+)]) was determined by fitting a hyperbolic function to the data (OriginPro).
Na+ binding and melibiose binding by ITC
ITC measurements were performed in a Nano Isothermal Titration Calorimeter (TA Instruments), and all data were collected at 25°C. MelBSt (40–100 µM) was placed in the sample cell with a reaction volume of 163 µl. The titrant and titrand were prepared in the same dialysis buffers, degassed for 15 min using a TA Instruments Degassing Station model 6326. 2-µl aliquots were injected incrementally into the sample cell at an interval of 300 s with constant stirring at 250 rpm.
To determine Na+ binding to MelBSt, the protein samples were dialyzed against Na+-free buffer (20 mM Tris-HCl unless defined otherwise, 50 mM ChCl, 10% glycerol, and 0.035% UDM) at a given pH in the absence or presence of ∼20–50 mM melibiose. The Na+ contamination is calculated to be lower than 5 µM. NaCl samples at concentrations of 1 to 20 mM were dissolved in a buffer matching the MelBSt buffer and injected incrementally into the sample cell containing MelBSt. To determine melibiose binding in the absence or presence of Na+, the melibiose solutions (10 mM or 80 mM) were buffer matched with the MelBSt sample buffer in the absence or presence of 100 mM NaCl and placed in the syringe.
ITC data processing was performed using the one-site independent binding model in the NanoAnalyze version 3.6.0 software provided by the ITC equipment. The total heat changes were subtracted from the heat of dilution elicited by last few injections, where no further binding occurred, and the corrected heat change was normalized and plotted against the molar ratio of titrant versus titrand, as previously described (Hariharan and Guan, 2014; Hariharan et al., 2015, 2016). The association constant (Ka) and the change in enthalpy (ΔH) were determined by fitting the data with a one-site independent-binding model. The binding stoichiometry (N) was fixed to 1 because it is a known parameter, which can restrain the data fitting and achieve more accurate results (Turnbull and Daranas, 2003). Kd = 1/Ka; ΔG = −RT ln Ka, where R is the gas constant (8.315 J/mol·K) and T is the absolute temperature.
Determination of absolute dissociation constants for Na+ (KD(Na+)) and H+ (KD(H+))
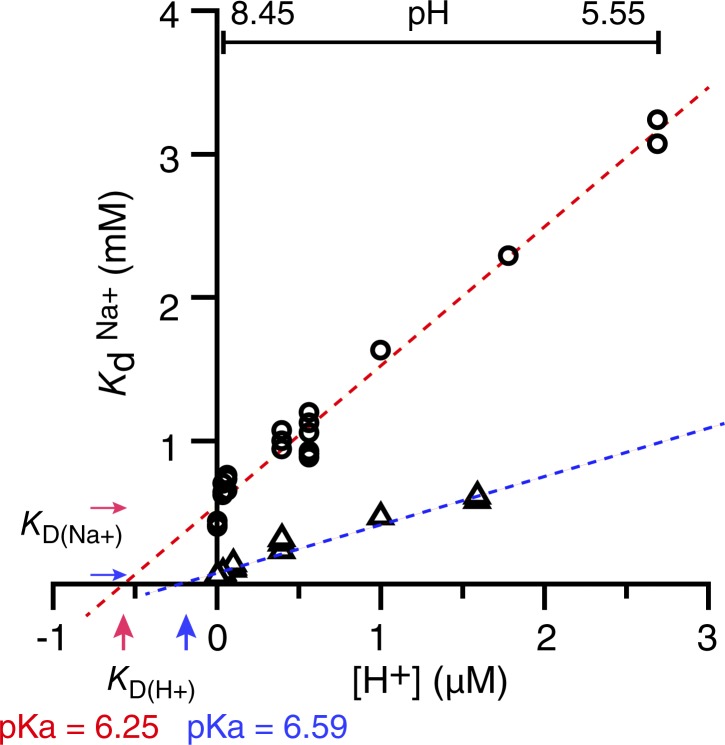
The apparent Kd for Na+ binding (Kd(Na+)) in the absence or presence of 20 mM melibiose was determined in a pH range of 5.55 to 8.45 in one of the following buffers: 20 mM Tris-HCl, MES-TMAOH, potassium phosphate (KPi), Bis-Tris-HCl, or ACES-TMAOH. The apparent Kd(Na+) versus H+ concentration fit linearly, suggesting competition between Na+ and H+ for a common cation-binding site. Thus, values for the absolute KD(Na+) and absolute KD(H+) can be derived from a linear regression (Leone et al., 2015) based on the equation Kd(Na+) = KD(Na+){1+ [H+]/KD(H+)}. On this Kd(Na+) versus H+ concentrations plot, the y intercept (i.e., H+ concentration = 0) corresponds the absolute KD(Na+), and the x intercept (i.e., Na+ concentration = 0 and then Kd(Na+) = 0) corresponds to the absolute KD(H+). pKa = −log KD(H+).
Determination of the binding stoichiometry of Na+, H+, and MelBSt
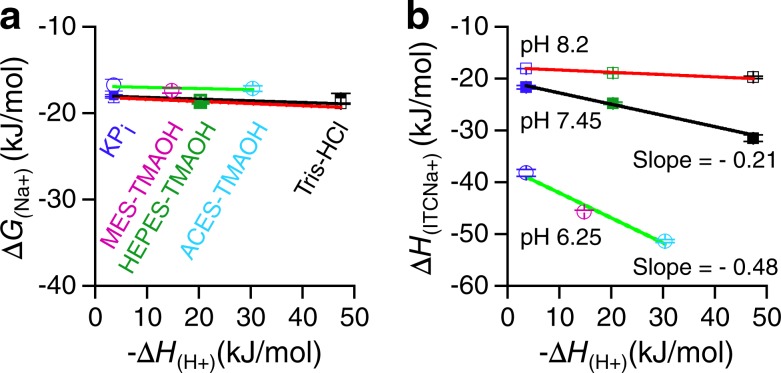
ITC was used to determine the binding enthalpies for Na+ (ΔHITC(Na+)) at three pHs (6.25, 7.45, and 8.2) in five buffer systems. KPi, HEPES-TMAOH, and Tris-HCl buffers were used to test the buffer effect on ΔHITC(Na+) at pH 7.45 or 8.2. The KPi, MES-TMAOH, and ACES-TMAOH buffers were used for the test at pH 6.25, which is the pKa value for MelBSt. The selections for these buffers are mainly based on their protonation enthalpy (ΔH(H+)) values and lesser Na+ contamination. The −ΔH(H+) values for phosphate buffer, MES-TMAOH, HEPES-TMAOH, ACES-TMAOH, and Tris-HCl are 3.6, 14.8, 20.4, 30.43, and 47.4 kJ/mol, respectively (Goldberg et al., 2002; Bianconi, 2003). Purified MelBSt samples were dialyzed against a specific Na+-free buffer system containing 50 mM ChCl, 0.035% UDM and 10% glycerol before ITC measurements. The ITC-determined ΔHITC(Na+) values were plotted against the corresponding standards −ΔH(H+) from each buffer. By fitting a linear function to the data, the negative sign of the slope indicates the release of H+ by Na+ binding, and the slope reflects the number of H+ replaced by Na+.
Statistics
An unpaired t test was used for data analysis. P-values <0.05 were considered statistically significant.
Results
Determination of Na+ binding by Trp→D2G FRET
Trp emission wavelength of MelB overlaps with the excitation wavelength of a dansyl group. Using an excitation wavelength of 290 nm, we detect the FRET from Trp→dansyl moiety on a fluorescent sugar analogue D2G bound to the galactoside-binding site of MelB (Maehrel et al., 1998; Guan et al., 2011, 2012). Na+ stimulates Trp→D2G FRET, and the differential intensity (diffFRET) induced by Na+ is concentration-dependent and saturable. The Kd for D2G binding to MelBSt in membrane vesicles is ∼10 µM in the presence of Na+ and was not determined in the absence of Na+ because of poor D2G affinity (Guan et al., 2011). The increase in the FRET signal induced by Na+ is due mainly to the increase in the number of D2G molecules bound to MelBSt. This method has been widely used to determine Na+ binding in the WT MelBSt and MelBEc and their mutants (Cordat et al., 1998; Maehrel et al., 1998; Meyer-Lipp et al., 2006; Ganea et al., 2011; Guan et al., 2011, 2012; Jakkula and Guan, 2012; Amin et al., 2014). Most of these reported tests were performed with membrane preparations, either right-side-out or inside-out membrane vesicles or reconstituted proteoliposomes (Maehrel et al., 1998; Guan et al., 2011; Jakkula and Guan, 2012). Notably, MelBEc purified with conventional detergents, such as DDM, does not bind either D2G or melibiose, but sugar binding has been detected using these new detergents with strong stabilizing capabilities (Amin et al., 2015; Sadaf et al., 2016; Das et al., 2017; Hussain et al., 2017). Because of greater stability, MelBSt in most detergent solutions binds sugar substrates, so the Trp→D2G FRET assay has been used to determine the melibiose binding to purified MelBSt in the presence of Na+ or Li+ (Ethayathulla et al., 2014; Amin et al., 2015). However, there is no study showing whether the purified MelBSt also binds Na+.
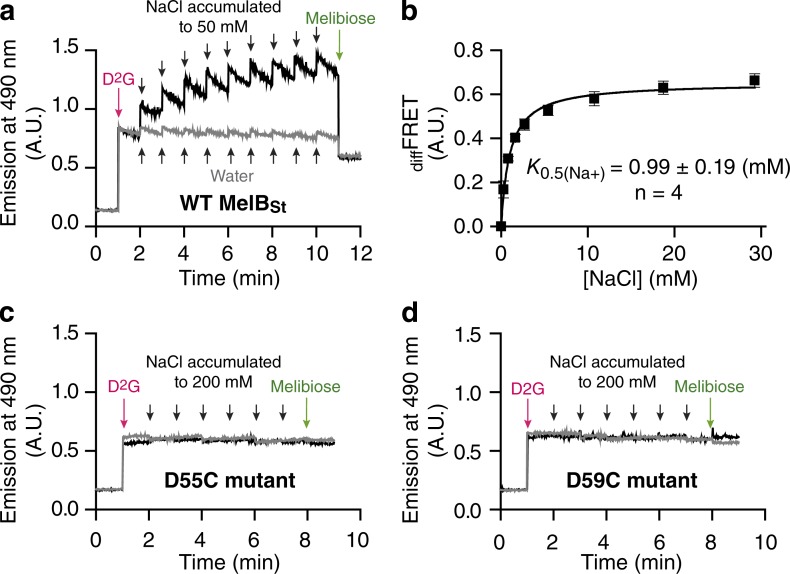
A “Na+-free” MelBSt sample in 20 mM Tris-HCl, pH 7.45, 50 mM ChCl, 10% glycerol, and 0.035% UDM is stable and can be used for measuring the diffFRET stimulated by Na+. We recorded emission at 490 nm during stepwise additions of sugar and Na+ (Fig. 2 a). 10 µM D2G (near the Kd value in the presence of Na+) was added at the 1-min time point (Fig. 2, red arrows), the fluorescence intensity was recorded for 1 min, and then NaCl was incrementally added up to a final concentration of ~50 mM (Fig. 2 a, black arrows). Finally, melibiose at oversaturating concentration was added into the same solution to displace the bound D2G, and the decrease in fluorescence intensity after the addition of melibiose reflects the magnitude of Trp→D2G FRET (Fig. 2, green arrows). Na+ stimulation of the Trp→D2G FRET was observed with purified MelBSt (Fig. 2 a). The Na+ activation constant (K0.5(Na+)) was determined by fitting a hyperbolic function to the data. The resulting value of 0.99 mM for K0.5(Na+) (Fig. 2 b) is similar to the value obtained with membrane vesicles (Guan et al., 2011; Jakkula and Guan, 2012). The Na+-binding affinity in the absence of sugar cannot be obtained by this method, so development of a direct Na+ binding assay was necessary.
Figure 2.
Na+ stimulation constant of the Trp→D2G FRET (K0.5(Na+)) with MelBSt WT and mutants D55C and D59C. The FRET signals from Trp residues of MelBSt to the dansyl moiety of a fluorescent sugar (D2G) in response to increasing Na+ concentration were measured as described in Materials and methods. (a, c, and d) The purified WT MelBSt (a) or MelBSt single-site mutants D55C (c) or D59C (d) in a Na+-free buffer containing 20 mM Tris-HCl, pH 7.5, 50 mM ChCl, 0.035% UDM, and 10% glycerol were adjusted to a protein concentration of 1 µM. D2G in 20% DMSO was added at 1 min (red arrows) at a concentration of 10 µM (Kd value for D2G binding to MelBSt in the presence of Na+). NaCl solutions of increasing concentrations were consecutively added, up to a final concentration of ~50 mM for the WT MelBSt and ~200 mM for the mutants (black arrows). Finally, melibiose was added at an oversaturating concertation (green arrows). In control experiments, identical water volumes, instead of NaCl solution, was used (gray arrows) to control for sample dilution. Data collection and correction were as described in Materials and methods. (b) An increase in FRET intensity is expressed as diffFRET (the difference before and after the addition of NaCl), and the value for K0.5(Na+) was determined by fitting a hyperbolic function to the diffFRET versus Na+ concentration. Error bars, standard error (SEM); the number of tests = 4.
Structural and functional studies indicate that Asp55 and Asp59 on helix II may coordinate Na+ (Fig. 1; Pourcher et al., 1993; Zani et al., 1994; Granell et al., 2010; Ethayathulla et al., 2014). Two MelBSt single-site mutants (D55C and D59C), in which a Cys residue replaces an Asp residue in positions 55 or 59, respectively, were purified, and the Na+-binding assay was tested by Trp→D2G FRET. Na+ stimulation was absent in both mutants, even with high Na+ concentrations (Fig. 2, c and d) or D2G concentration (not depicted). However, the data also show that there was no melibiose displacement in both cases, which underscores the need for a direct Na+-binding assay.
ITC measurements of Na+ binding in the absence or presence of melibiose
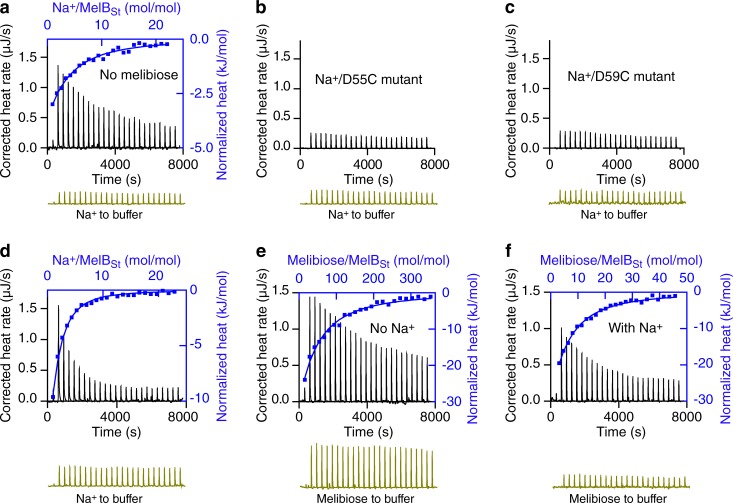
A Nano ITC device was used for data collection. Positive curves denote heat release (exothermic reactions). Both NaCl solutions and purified MelBSt samples were prepared in a Na+-free buffer containing 20 mM Tris-HCl, pH 7.45, 50 mM ChCl, 10% glycerol, and 0.035% UDM. NaCl solutions were injected into the ITC sample cell containing MelBSt at 25°C, and heat release and the exothermic titration thermogram were obtained (Fig. 3 a, black). As a control, parallel experiments with buffer present in the sample cell without protein yielded small exothermic peaks (Fig. 3 a, dark yellow), supporting that the thermogram is generated by Na+ binding to MelBSt. Measurements with the two MelBSt mutants (D55C and D59C) reveal nearly flat thermograms, and the heat releases are much smaller and indistinguishable from the buffer controls (Fig. 3, b and c), indicating that Na+ binding in both mutants is dramatically decreased. Collectively, these experiments strongly indicate that the Na+ titration curve shown in Fig. 3 a originates from Na+ binding to the cation site on MelBSt.
Figure 3.
Cooperative binding of Na+ and melibiose to MelBSt. (a–f) ITC was used to determine the binding of Na+ (a–d) or melibiose (e and f) to MelBSt at 25°C. Data collection was performed with a Nano ITC instrument at 25°C as described in Materials and methods. Titrant and titrand samples were subjected to buffer matching and degassing before each test. The samples containing MelBSt (a and d–f), or MelBSt mutants D55C (b) or D59C (c), at a protein concentration of 80 µM were placed in the sample cell. NaCl samples (5 mM) in the absence or presence of 50 mM melibiose placed in the syringe were injected into the MelBSt samples in the absence or presence of 50 mM melibiose or corresponding buffers without protein (controls, bottom of each panel, dark yellow). The melibiose binding to MelBSt was measured in the absence or presence of 100 mM NaCl by placing melibiose solutions (10–80 mM) in the syringe. The normalized heat changes (kJ/mol) were plotted against the Na+/MelBSt (a and d; top/right axis, blue curves) or melibiose/MelBSt molar ratio (e and f; top/right axis, blue curves), and fitted with a one-site independent-binding model with fixed stoichiometry (n = 1).
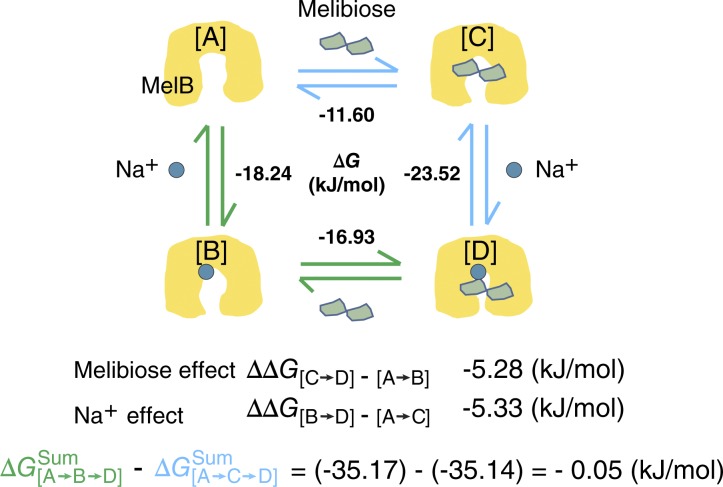
The normalized heat change was plotted against the molar ratio of the titrant Na+ to the titrand MelBSt (Fig. 3 a, top/right, blue). The binding isotherm is not sigmoidal, even after exhaustive tests. To increase the fitting accuracy, the stoichiometric number of 1 for Na+ binding to MelB was used to fit the data with a single-site independent binding model. The results are similar to those obtained without fixing the n value. The apparent Kd for Na+ binding to MelBSt (Kd(Na+)) in the absence of galactoside at pH 7.45 and 25°C is ∼0.64 mM (Fig. 3 a and Table 1), and ΔG is −18.24 kJ/mol. The result is close to the K0.5(Na+) value for Na+ stimulation on the Trp→D2G FRET (Fig. 2). When the Na+ binding measurements were performed in the presence of 50 mM melibiose (Fig. 3 d), the apparent Kd(Na+) value decreased by eightfold, from 0.64 mM to 0.08 mM, and ΔG changed from −18.24 to −23.52 kJ/mol, yielding the thermodynamic coupling free energy (ΔΔG) of −5.28 kJ/mol (Table 1). This result indicates that the Na+ binding affinity is increased by approximately eightfold by melibiose binding to MelBSt.
Table 1. Binding cooperativity of melibiose and Na+ to MelBSt.
| Parameter | Na+ binding (NaCl in syringe) | Melibiose effect on Na+ binding | Melibiose binding (melibiose in syringe) | Na+ effect on melibiose binding | ||
|---|---|---|---|---|---|---|
| No sugar (n = 4) | With melibiose (n = 2) | No NaCl (n = 3) | With NaCl (n = 5) | |||
| Apparent Kd (mM) or affinity increase | 0.64 ± 0.02a | 0.08 ± 0.01 | 8-fold | 9.28 ± 0.23 | 1.09 ± 0.06 | 8.5-fold |
| ΔG or ΔΔG (kJ/mol) | −18.24 ± 0.08 | −23.52 ± 0.13 | −5.28 (P < 0.05)b | −11.60 ± 0.06 | −16.93 ± 0.14 | −5.33 (P < 0.05) |
ITC data were collected at pH 7.45 and 25°C with MelBSt in sample cell as described in Materials and methods. ΔG, binding free energy; ΔΔG, thermodynamics coupling free energy. n = number of test from a total of 5 different batches of MelBSt purification.
Standard error (SEM).
Unpaired t test.
Melibiose binding in the absence or presence of Na+
Using ITC measurements, the Kd for melibiose binding to MelBSt in the Na+-free buffer at pH 7.45 and 25°C is 9.28 mM and ΔG is −11.60 kJ/mol (Fig. 3 e and Table 1). In the presence of Na+ (Fig. 3 f), the melibiose-binding affinity was substantially increased; the Kd value was decreased by 8.51-fold from 9.28 mM to 1.09 mM, yielding the ΔΔG value of −5.33 kJ/mol, which is virtually equal to the coupling free energy determined for Na+ binding in the presence of melibiose (Table 1 and see Fig. 6). This result indicates that melibiose binding to MelBSt is increased by eightfold by Na+ binding. Thus, the coupling binding free energy had similar values for both processes.
Figure 6.
Thermodynamic cycle for the formation of the melibiose–Na+–MelBSt ternary complex at pH 7.5. Symbols for MelBSt, melibiose or Na+ are indicated. State [A], cartoon for MelBSt from the crystal structure (Protein Data Bank accession no. 4M64) represents a periplasmic side-open apo state. States [B] and [C] represent a Na+-bound or melibiose-bound binary complex, respectively. State [D] represents a Na+–melibiose–MelBSt ternary complex. ΔG values are listed for each binding step. The thermodynamic coupling free energy (ΔΔG) reflects the effect of one substrate on the binding of the other. ΔGsum, the sums of free energies for the path [A→B→D] or [A→C→D]. The difference in the ΔG from paths [A→B→D] and [A→C→D] is close to zero.
Determination of the absolute dissociation constants for Na+ (KD(Na+)) and H+ (KD(H+))
It has been shown that Na+ and Li+ compete for a common binding site on MelBSt (Guan et al., 2011). To study the competition between Na+ and H+, Na+ binding at pH values ranging from 5.5 to 8.45 was measured by ITC, in the absence of melibiose, at 25°C. The data show that the apparent Kd(Na+) value increases (i.e., the affinity for Na+ decreases) linearly with increasing H+ concentration (Fig. 4, open circles), supporting the idea that Na+ and H+ compete for a common binding site. Thus, it is important to determine the absolute dissociation constants for both cations KD(Na+) and KD(H+). As discussed in Materials and methods, the absolute KD(Na+) value corresponds to the y-intercept gained from the extrapolation of the linear fit where the H+ concentration is zero, and the absolute KD(H+) corresponds to the x-intercept (Fig. 4; Leone et al., 2015). The results show that the absolute KD(Na+) value for MelBSt under the experimental condition is 0.54 ± 0.03 mM and the absolute KD(H+) value is 0.56 ± 0.03 µM (Table 2). Calculated from the KD(H+) value by the equation pKa = −log KD(H+), the pKa of the cation-binding site in MelBSt is 6.25 in the absence of melibiose. Therefore, the cation selectivity ratio (KD(Na+)/KD(H+)) = slope = 540/0.56 = 964 (Fig. 4 and Table 2), indicating that MelBSt’s intrinsic selectivity for H+ over Na+ is almost 1,000-fold.
Figure 4.
Determination of the absolute dissociation constants for Na+ (KD(Na+)) and H+ (KD(H+)). The apparent Na+-binding dissociation constants (Kd(Na+)) in the absence or presence of melibiose in the pH range from 5.55 to 8.45 were determined by ITC at 25°C. 1–20 mM NaCl was placed in the syringe. All apparent Kd(Na+) data were plotted against the H+ concentration without averaging and fitted by a linear function; the absolute KD(Na+) value corresponds to the y-axis intercept, and the absolute KD(H+) value corresponds to the x-axis intercept (see Materials and methods). pKa = −log KD(H+).
Table 2. Absolute dissociation constants for Na+ or H+ binding to MelBSt.
| Parameter | No sugar | With melibiose |
|---|---|---|
| Absolute KD(Na+) (mM)a | 0.54 ± 0.03b | 0.09 ± 0.02 |
| Absolute KD(H+) (μM) | 0.56 ± 0.03 | 0.26 ± 0.02 |
| Cation selectivity ratio (slope = KD(Na+)/KD(H+)) | 964 | 346 |
| pKa | 6.25 | 6.59 |
Data were extracted from Fig. 4.
Standard error from curve fitting.
When similar measurements were performed in the presence of melibiose, the absolute KD(Na+) value was decreased from 0.54 ± 0.03 mM to 0.09 ± 0.02 mM (Fig. 4 and Table 2). Interestingly, the absolute KD(H+) decreased from 0.56 µM to 0.26 µM, only 2.1-fold increase for the H+ affinity, and the pKa changed from pH 6.25 to 6.59, an ∼0.3-unit increase (Table 2). Accordingly, the cation selectivity ratio was reduced from 964 to 346. These data clearly show that melibiose has a stronger cooperativity with Na+ than with H+. This is also implied from the nonparallel shift induced by melibiose on the Kd(Na+) versus H+ concentration curve (Fig. 4).
Determination of stoichiometric ratios between Na+ and H+ and between H+ and MelBSt
The heat changes caused by Na+ binding may have several origins; one of them could be protonation of the reaction buffer if MelBSt undergoes deprotonation during Na+ binding. If so, the ITC-measured Na+-binding enthalpy (ΔHITC(Na+)) should be buffer dependent with an unchanged ΔG(Na+). This is because the intrinsic protonation enthalpies (ΔH(H+)) of various buffer systems differ (i.e., the ΔH(H+) values for phosphate and Tris-HCl buffer are −3.6 kJ/mol and −47.4 kJ/mol, respectively; Table 3; Goldberg et al., 2002; Bianconi, 2003). To determine how many H+ were displaced by the binding of one Na+, Na+ binding in five buffer systems was performed at three pH including a pKa for MelBSt via ITC measurements. Overall, the results show that, at each pH, the apparent Kd(Na+) or ΔG values are similar in different buffers, but the binding enthalpy ΔHITC(Na+) values vary (Fig. 5, a and b; and Table 3). The determined ΔHITC(Na+) values plotted against the standard protonation enthalpies of each buffer (−ΔH(H+)) yield linear relationships (Fig. 5 b).
Table 3. Buffer effects of Na+-binding enthalpy.
| Buffer | Buffer pH | −ΔH(H+)a | Kd(Na+) (mM) | ΔG(Na+) | ΔHITC(Na+) | Slope |
|---|---|---|---|---|---|---|
| kJ/mol | mM | kJ/mol | kJ/mol | |||
| KPi | 6.45 (nb = 2) | 3.6 | 1.16 ± 0.04c | −16.76 ± 0.08 | −38.22 ± 0.67 | |
| MES-TMAOH | 6.25 (n = 3) | 14.8 | 0.90 ± 0.01 | −17.38 ± 0.02 | −45.70 ± 0.28 | −0.48 ± 0.09 |
| ACES-TMAOH | 6.25 (n = 2) | 30.43 | 0.99 ± 0.06 | −17.16 ± 0.16 | −51.37 ± 0.32 | |
| KPi | 7.45 (n = 3) | 3.6 | 0.65 ± 0.02 | −18.19 ± 0.09 | −21.67 ± 0.34 | |
| HEPES-TMAOH | 7.45 (n = 3) | 20.3 | 0.52 ± 0.03 | −18.75 ± 0.12 | −24.79 ± 0.23 | −0.21 ± 0.03 |
| Tris-HCl | 7.45 (n = 4) | 47.4 | 0.64 ± 0.02 | −18.24 ± 0.08 | −31.50 ± 0.63 | |
| KPi | 8.2 (n = 1) | 3.6 | 0.72 | −17.94 | -18.06 | |
| HEPES-TMAOH | 8.2 (n = 2) | 20.3 | 0.56 ± 0.01 | −18.55 ± 0.11 | −18.93 ± 0.12 | −0.04 ± 0.01 |
| Tris-HCl | 8.2 (n = 2) | 47.4 | 0.50 ± 0.01 | −18.90 ± 0.05 | −19.76 ± 0.26 |
Standards for the buffer protonation enthalpy (Goldberg et al., 2002).
n = number of tests.
Standard error (SEM).
Figure 5.
Buffer effects on Na+ binding enthalpy (ΔHITC(Na+)). ITC was used to determine Na+ binding ΔHITC(Na+) to MelBSt in a total of five buffer systems in the absence of melibiose at pH 6.25, 7.45, and 8.2 at 25°C. 5–15 mM NaCl was placed in the syringe. (a) ΔG versus standard buffer −ΔH(H+) plot. (b) ΔHITC(Na+) determined by ITC versus buffer standard −ΔH(H+) plot, with linear fits to the data.
At pH 7.45 in KPi, HEPES-TMAOH, or Tris-HCl buffer, a negative slope of ∼0.21 is obtained (Fig. 5 b). The negative sign of the slope suggests the release of H+ from MelBSt, and the value of the slope correlates with the number of H+ replaced by Na+. Because there is only one Na+ binding to the MelB cation site, this result suggests that only a portion of MelBSt molecules are protonated at pH 7.45.
At pH 8.2, the majority of MelBSt proteins should be deprotonated because the pKa is 6.25 and no further change in Na+ affinity was observed (Fig. 4); i.e., the ΔHITC(Na+) values measured with the three buffers at this pH should be similar, and this is in fact the case (Table 3), generating a nearly flat curve (Fig. 5 b). These data support the conclusion that the cation-binding site on MelBSt is unprotonated at a pH greater than 8.2.
There is no suitable Na+-free buffer system at an acidic pH where all MelB molecules are protonated. Taking advantage of the determined pKa value of 6.25, the stoichiometry between H+ and Na+, as well as between H+ and MelBSt, can be established by determining the slope at this specific pH. Using KPi, MES-TMAOH, and ACES-TMAOH buffers adjusted to pH 6.25, where the protonated and unprotonated MelBSt levels are equal, the obtained slope is 0.48, which is very close to 0.5 (Fig. 5 b and Table 3). This result confirms that binding of one Na+ displaces one proton, and the binding stoichiometry among H+, Na+, and MelBSt is unity.
Melibiose binding to unprotonated MelBSt
The results of Na+ binding at pH 8.2 (Fig. 5 b) indicate that at pH 8.2 or above, MelBSt is nearly completely unprotonated. To analyze the melibiose binding to unprotonated MelBSt, the Kd values for melibiose in the absence or presence of NaCl at pH 8.45 were determined by ITC to be 9.42 ± 0.28 mM (SEM, n = 3) and 0.95 ± 0.06 mM (SEM, n = 4), respectively. Accordingly, the ΔG values are −11.57 and −17.26 kJ/mol, respectively.
Discussion
It has been well documented for both MelBEc and MelBSt that Na+ stimulates galactoside binding and transport (Bassilana et al., 1985; Wilson and Ding, 2001; Guan et al., 2011); however, how the Na+-binding affinity is affected by melibiose is not clear, because previous quantitative measurements of Na+ binding were based on sugar binding (e.g., the Na+ stimulation of D2G FRET; Fig. 2; Maehrel et al., 1998; Guan et al., 2011) or [3H]α-NPG binding (Damiano-Forano et al., 1986). Here, we present detailed data from direct measurements of Na+ and melibiose binding to MelBSt, which allow us to construct thermodynamic cycles to analyze the formation of the Na+–MelB–melibiose ternary complex (Fig. 6). The thermodynamic coupling free energy (ΔΔG) values (i.e., the differences in binding free energies [ΔG]) obtained from binding of one component in the absence or presence of the other are quite similar (approximately −5 kJ/mol or eightfold increases in affinity); therefore, the binding of Na+ and melibiose to MelBSt is thermodynamically cooperative (Table 1 and Fig. 6). Helix IV in the N-terminal domain contains residues critical for the binding of both Na+ and galactoside, and both binding sites are physically connected (Fig. 1 b; Ethayathulla et al., 2014), which may be the structural basis for the positive cooperativity of galactoside and cation binding in MelB.
Free-energy changes in a thermodynamic cycle are state functions independent of their paths because of thermodynamic equilibrium. This is the case for the binding of the two cotransporting substrates, Na+ and galactoside, to the symporter MelBSt. The sum of ΔG at −35 kJ/mol from the path [A→B→D] (i.e., from the empty state [A] to the binary Na+-bound state [B] and then from this to the Na+- and melibiose-bound ternary complex [D]) is nearly equal to that from the path [A→C→D] (i.e., binding of melibiose before Na+ binding; Fig. 6). Thus, binding of both substrates releases free energy of ∼35 kJ/mol, and it is likely that the released energy fuels the conformational changes required for transport.
MelB also catalyzes H+-coupled melibiose transport, so it is important to determine its affinity to H+ and protonation status. Analysis of the competitive binding between H+ and Na+ by measuring the apparent Kd(Na+) at a range of pH values indicates that the pKa for MelBSt cation site is 6.25, and the absolute KD(H+) is 0.56 µM. This result is in agreement with data previously obtained from MelBEc by an indirect assay (i.e., Na+ stimulation of [3H]α-NPG binding; Damiano-Forano et al., 1986). Melibiose and H+ are also cooperative; however, the galactoside effect on MelB affinity for H+ is smaller (i.e., the pKa increases by only ∼0.3 units, which is equivalent to a twofold decrease in KD(H+); Table 2). Compared with the much greater effect on Na+ binding, it is clear that melibiose cooperates with Na+ stronger than with H+, although the protein is intrinsically selective for H+.
ITC is also a useful tool for the study of protein protonation and stoichiometry between H+ and its competitive cation. The observed negative linear relationships between the Na+ binding enthalpy and the buffer intrinsic protonation enthalpy at pH 6.25 and pH 7.45 suggest that buffer protonation occurs upon Na+ binding (i.e., MelBSt releases H+ when it binds Na+). The demonstration of competitive binding between Na+ and H+ (Fig. 4), as well as between Na+ and Li+ (Guan et al., 2011), indicates that all three cations (Na+, H+, and Li+) compete for a common binding site in MelBSt. A similar conclusion was previously drawn in MelBEc (Bassilana et al., 1987). Furthermore, the slope of −0.48 is obtained from the linear fit when the tests were done at a buffer pH equal to the pKa value, suggesting that the binding of one Na+ displaces one H+ from the cation binding site per MelBSt, because at this pH, only half of the MelB molecules are protonated. At pH 7.45, the slope value of −0.21 is obtained, which suggests that only ∼20% MelBSt proteins are protonated at this pH. These data may explain the results on lower transport rates and the lower melibiose binding at pH 7.5 in the absence of Na+ or Li+. The initial rate and the steady-state level of H+-coupled melibiose transport were ∼30% of Na+-coupled melibiose transport (Guan et al., 2011). With an empty cation site, MelBSt still binds melibiose, but with very low affinity. Thus, at pH 7.5, only a small portion of MelBSt molecules are able to perform the symport function, which is consistent with the notion that concurrent binding of both substrates is required for the symport process (Yousef and Guan, 2009).
The study of competitive binding between Na+ and H+ yields an absolute KD(Na+) of 0.54 mM and KD(H+) of 0.56 µM; thus, the Na+ affinity is ∼1,000-fold lower than that for H+ in the absence of melibiose, and the MelBSt cation site is intrinsically selective for H+ over Na+. Even in the presence of melibiose, the H+ selectivity persists, with a greater than 300-fold higher affinity. In MelBEc, previous studies yielded an KD(Na) of 0.3 mM and a pKa of 6.3, also suggesting that the cation site in MelBEc is intrinsically selective for H+ (Damiano-Forano et al., 1986). Such a selectivity feature has been recognized in several ATP synthases (Krah et al., 2010; Schlegel et al., 2012; Leone et al., 2015), such as in the F-type ATP synthase of Ilyobacter tartaricus, where the ion-driven membrane rotor exhibits very similar value of KD(Na+) (0.29 mM) and pKa (6.5; Leone et al., 2015). Collectively, these studies from radically different membrane transporters reveal a common principle of cation selectivity in membrane proteins with a common cation site used by both Na+ and H+. Although intrinsically selective for H+, the availability of H+ in physiological environments (pH 7.5, [H+] = 32 nM) is very low, and the availability of Na+ is often high enough to ensure that Na+ can effectively compete for the cation site. Such a cation site thus appears to have evolved for the effective use of the metal cation Na+ under physiological conditions. Because the living environments for bacteria are not always Na+ rich, an elevated pKa value for Asp residues in the cation site allows the bacteria to use H+ as the coupling cation for melibiose transport, albeit with less efficiency. This elegant mechanism secures MelB’s important biological function.
Acknowledgments
The authors thank Drs. Gerard Leblanc and H. Ronald Kaback for providing D2G, Dr. José Faraldo-Gómez for his suggestions on the determination of absolute dissociation constants, and Dr. Luis Reuss for his critical reading of the manuscript.
This work was partially supported by the National Science Foundation (grant MCB-1158085 to L. Guan).
The authors declare no competing financial interests.
Author contributions: L. Guan conceived and directed this research. P. Hariharan performed all experiments and data processing. L. Guan and P. Hariharan analyzed the results, and L. Guan wrote the manuscript.
Merritt Maduke served as editor.
Footnotes
Abbreviations used:
- ACES
- N-(2-acetamido)-2-aminoethanesulfonic acid
- α-NPG
- α-nitrophenyl galactoside
- D2G
- 2′-(N-dansyl)aminoalkyl-1-thio-β-d-galactopyranoside
- diffFRET
- differential FRET
- FRET
- fluorescence resonance energy transfer
- ITC
- isothermal titration calorimetry
- MelBEc
- Escherichia coli melibiose permease
- MelBSt
- Salmonella typhimurium melibiose permease
- MFS
- major facilitator superfamily
- TMAOH
- tetramethylammonium hydroxide
- UDM
- undecyl-β-d-maltopyranoside
References
- Abramson J., Smirnova I., Kasho V., Verner G., Kaback H.R., and Iwata S.. 2003. Structure and mechanism of the lactose permease of Escherichia coli. Science. 301:610–615. 10.1126/science.1088196 [DOI] [PubMed] [Google Scholar]
- Amin A., Ethayathulla A.S., and Guan L.. 2014. Suppression of conformation-compromised mutants of Salmonella enterica serovar Typhimurium MelB. J. Bacteriol. 196:3134–3139. 10.1128/JB.01868-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin A., Hariharan P., Chae P.S., and Guan L.. 2015. Effect of Detergents on Galactoside Binding by Melibiose Permeases. Biochemistry. 54:5849–5855. 10.1021/acs.biochem.5b00660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassilana M., Damiano-Forano E., and Leblanc G.. 1985. Effect of membrane potential on the kinetic parameters of the Na+ or H+ melibiose symport in Escherichia coli membrane vesicles. Biochem. Biophys. Res. Commun. 129:626–631. 10.1016/0006-291X(85)91937-0 [DOI] [PubMed] [Google Scholar]
- Bassilana M., Pourcher T., and Leblanc G.. 1987. Facilitated diffusion properties of melibiose permease in Escherichia coli membrane vesicles. Release of co-substrates is rate limiting for permease cycling. J. Biol. Chem. 262:16865–16870. [PubMed] [Google Scholar]
- Bianconi M.L. 2003. Calorimetric determination of thermodynamic parameters of reaction reveals different enthalpic compensations of the yeast hexokinase isozymes. J. Biol. Chem. 278:18709–18713. 10.1074/jbc.M211103200 [DOI] [PubMed] [Google Scholar]
- Cooper A. 1999. Thermodynamic analysis of biomolecular interactions. Curr. Opin. Chem. Biol. 3:557–563. 10.1016/S1367-5931(99)00008-3 [DOI] [PubMed] [Google Scholar]
- Cordat E., Mus-Veteau I., and Leblanc G.. 1998. Structural studies of the melibiose permease of Escherichia coli by fluorescence resonance energy transfer. II. Identification of the tryptophan residues acting as energy donors. J. Biol. Chem. 273:33198–33202. 10.1074/jbc.273.50.33198 [DOI] [PubMed] [Google Scholar]
- Damiano-Forano E., Bassilana M., and Leblanc G.. 1986. Sugar binding properties of the melibiose permease in Escherichia coli membrane vesicles. Effects of Na+ and H+ concentrations. J. Biol. Chem. 261:6893–6899. [PubMed] [Google Scholar]
- Dang S., Sun L., Huang Y., Lu F., Liu Y., Gong H., Wang J., and Yan N.. 2010. Structure of a fucose transporter in an outward-open conformation. Nature. 467:734–738. 10.1038/nature09406 [DOI] [PubMed] [Google Scholar]
- Das M., Du Y., Ribeiro O., Hariharan P., Mortensen J.S., Patra D., Skiniotis G., Loland C.J., Guan L., Kobilka B.K., et al. . 2017. Conformationally Preorganized Diastereomeric Norbornane-Based Maltosides for Membrane Protein Study: Implications of Detergent Kink for Micellar Properties. J. Am. Chem. Soc. 139:3072–3081. 10.1021/jacs.6b11997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ethayathulla A.S., Yousef M.S., Amin A., Leblanc G., Kaback H.R., and Guan L.. 2014. Structure-based mechanism for Na(+)/melibiose symport by MelB. Nat. Commun. 5:3009 10.1038/ncomms4009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganea C., Pourcher T., Leblanc G., and Fendler K.. 2001. Evidence for intraprotein charge transfer during the transport activity of the melibiose permease from Escherichia coli. Biochemistry. 40:13744–13752. 10.1021/bi011223k [DOI] [PubMed] [Google Scholar]
- Ganea C., Meyer-Lipp K., Lemonnier R., Krah A., Leblanc G., and Fendler K.. 2011. G117C MelB, a mutant melibiose permease with a changed conformational equilibrium. Biochim. Biophys. Acta. 1808:2508–2516. 10.1016/j.bbamem.2011.07.017 [DOI] [PubMed] [Google Scholar]
- Goldberg R.N., Kishore N., and Lennen R.M.. 2002. Thermodynamic Quantities for the Ionization Reactions of Buffers. J. Phys. Chem. Ref. Data. 31:231–370. 10.1063/1.1416902 [DOI] [Google Scholar]
- Granell M., León X., Leblanc G., Padrós E., and Lórenz-Fonfría V.A.. 2010. Structural insights into the activation mechanism of melibiose permease by sodium binding. Proc. Natl. Acad. Sci. USA. 107:22078–22083. 10.1073/pnas.1008649107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan L., and Kaback H.R.. 2006. Lessons from lactose permease. Annu. Rev. Biophys. Biomol. Struct. 35:67–91. 10.1146/annurev.biophys.35.040405.102005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan L., Mirza O., Verner G., Iwata S., and Kaback H.R.. 2007. Structural determination of wild-type lactose permease. Proc. Natl. Acad. Sci. USA. 104:15294–15298. 10.1073/pnas.0707688104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan L., Nurva S., and Ankeshwarapu S.P.. 2011. Mechanism of melibiose/cation symport of the melibiose permease of Salmonella typhimurium. J. Biol. Chem. 286:6367–6374. 10.1074/jbc.M110.206227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan L., Jakkula S.V., Hodkoff A.A., and Su Y.. 2012. Role of Gly117 in the cation/melibiose symport of MelB of Salmonella typhimurium. Biochemistry. 51:2950–2957. 10.1021/bi300230h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariharan P., and Guan L.. 2014. Insights into the inhibitory mechanisms of the regulatory protein IIA(Glc) on melibiose permease activity. J. Biol. Chem. 289:33012–33019. 10.1074/jbc.M114.609255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariharan P., Balasubramaniam D., Peterkofsky A., Kaback H.R., and Guan L.. 2015. Thermodynamic mechanism for inhibition of lactose permease by the phosphotransferase protein IIAGlc. Proc. Natl. Acad. Sci. USA. 112:2407–2412. 10.1073/pnas.1500891112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariharan P., Andersson M., Jiang X., Pardon E., Steyaert J., Kaback H.R., and Guan L.. 2016. Thermodynamics of Nanobody Binding to Lactose Permease. Biochemistry. 55:5917–5926. 10.1021/acs.biochem.6b00826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Lemieux M.J., Song J., Auer M., and Wang D.N.. 2003. Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli. Science. 301:616–620. 10.1126/science.1087619 [DOI] [PubMed] [Google Scholar]
- Hussain H., Mortensen J.S., Du Y., Santillan C., Ribeiro O., Go J., Hariharan P., Loland C.J., Guan L., Kobilka B.K., et al. . 2017. Tandem malonate-based glucosides (TMGs) for membrane protein structural studies. Sci. Rep. 7:3963 10.1038/s41598-017-03809-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakkula S.V., and Guan L.. 2012. Reduced Na+ affinity increases turnover of Salmonella enterica serovar Typhimurium MelB. J. Bacteriol. 194:5538–5544. 10.1128/JB.01206-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaback H.R. 2015. A chemiosmotic mechanism of symport. Proc. Natl. Acad. Sci. USA. 112:1259–1264. 10.1073/pnas.1419325112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krah A., Pogoryelov D., Langer J.D., Bond P.J., Meier T., and Faraldo-Gómez J.D.. 2010. Structural and energetic basis for H+ versus Na+ binding selectivity in ATP synthase Fo rotors. Biochim. Biophys. Acta. 1797:763–772. 10.1016/j.bbabio.2010.04.014 [DOI] [PubMed] [Google Scholar]
- Leavitt S., and Freire E.. 2001. Direct measurement of protein binding energetics by isothermal titration calorimetry. Curr. Opin. Struct. Biol. 11:560–566. 10.1016/S0959-440X(00)00248-7 [DOI] [PubMed] [Google Scholar]
- Leone V., Pogoryelov D., Meier T., and Faraldo-Gómez J.D.. 2015. On the principle of ion selectivity in Na+/H+-coupled membrane proteins: experimental and theoretical studies of an ATP synthase rotor. Proc. Natl. Acad. Sci. USA. 112:E1057–E1066. 10.1073/pnas.1421202112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopilato J., Tsuchiya T., and Wilson T.H.. 1978. Role of Na+ and Li+ in thiomethylgalactoside transport by the melibiose transport system of Escherichia coli. J. Bacteriol. 134:147–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maehrel C., Cordat E., Mus-Veteau I., and Leblanc G.. 1998. Structural studies of the melibiose permease of Escherichia coli by fluorescence resonance energy transfer. I. Evidence for ion-induced conformational change. J. Biol. Chem. 273:33192–33197. 10.1074/jbc.273.50.33192 [DOI] [PubMed] [Google Scholar]
- Meyer-Lipp K., Séry N., Ganea C., Basquin C., Fendler K., and Leblanc G.. 2006. The inner interhelix loop 4-5 of the melibiose permease from Escherichia coli takes part in conformational changes after sugar binding. J. Biol. Chem. 281:25882–25892. 10.1074/jbc.M601259200 [DOI] [PubMed] [Google Scholar]
- Mus-Veteau I., and Leblanc G.. 1996. Melibiose permease of Escherichia coli: structural organization of cosubstrate binding sites as deduced from tryptophan fluorescence analyses. Biochemistry. 35:12053–12060. 10.1021/bi961372g [DOI] [PubMed] [Google Scholar]
- Mus-Veteau I., Pourcher T., and Leblanc G.. 1995. Melibiose permease of Escherichia coli: substrate-induced conformational changes monitored by tryptophan fluorescence spectroscopy. Biochemistry. 34:6775–6783. 10.1021/bi00020a024 [DOI] [PubMed] [Google Scholar]
- Nguyen L.N., Ma D., Shui G., Wong P., Cazenave-Gassiot A., Zhang X., Wenk M.R., Goh E.L., and Silver D.L.. 2014. Mfsd2a is a transporter for the essential omega-3 fatty acid docosahexaenoic acid. Nature. 509:503–506. 10.1038/nature13241 [DOI] [PubMed] [Google Scholar]
- Niiya S., Moriyama Y., Futai M., and Tsuchiya T.. 1980. Cation coupling to melibiose transport in Salmonella typhimurium. J. Bacteriol. 144:192–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poolman B., Knol J., van der Does C., Henderson P.J., Liang W.J., Leblanc G., Pourcher T., and Mus-Veteau I.. 1996. Cation and sugar selectivity determinants in a novel family of transport proteins. Mol. Microbiol. 19:911–922. 10.1046/j.1365-2958.1996.397949.x [DOI] [PubMed] [Google Scholar]
- Pourcher T., Zani M.L., and Leblanc G.. 1993. Mutagenesis of acidic residues in putative membrane-spanning segments of the melibiose permease of Escherichia coli. I. Effect on Na(+)-dependent transport and binding properties. J. Biol. Chem. 268:3209–3215. [PubMed] [Google Scholar]
- Pourcher T., Leclercq S., Brandolin G., and Leblanc G.. 1995. Melibiose permease of Escherichia coli: large scale purification and evidence that H+, Na+, and Li+ sugar symport is catalyzed by a single polypeptide. Biochemistry. 34:4412–4420. 10.1021/bi00013a033 [DOI] [PubMed] [Google Scholar]
- Sadaf A., Mortensen J.S., Capaldi S., Tikhonova E., Hariharan P., de Castro Ribeiro O., Loland C.J., Guan L., Byrne B., and Chae P.S.. 2016. A Class of Rigid Linker-bearing Glucosides for Membrane Protein Structural Study. Chem. Sci. (Camb.). 7:1933–1939. 10.1039/C5SC02900G [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier M.H. Jr., Beatty J.T., Goffeau A., Harley K.T., Heijne W.H., Huang S.C., Jack D.L., Jähn P.S., Lew K., Liu J., et al. . 1999. The major facilitator superfamily. J. Mol. Microbiol. Biotechnol. 1:257–279. [PubMed] [Google Scholar]
- Schlegel K., Leone V., Faraldo-Gómez J. D., and Müller V.. 2012. Promiscuous archaeal ATP synthase concurrently coupled to Na+ and H+ translocation. Proc. Natl. Acad. Sci. USA. 109:947–952. 10.1073/pnas.1115796109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya T., and Wilson T.H.. 1978. Cation-sugar cotransport in the melibiose transport system of Escherichia coli. Membr. Biochem. 2:63–79. 10.3109/09687687809063858 [DOI] [PubMed] [Google Scholar]
- Turnbull W.B., and Daranas A.H.. 2003. On the value of c: can low affinity systems be studied by isothermal titration calorimetry? J. Am. Chem. Soc. 125:14859–14866. 10.1021/ja036166s [DOI] [PubMed] [Google Scholar]
- Wilson T.H., and Ding P.Z.. 2001. Sodium-substrate cotransport in bacteria. Biochim. Biophys. Acta. 1505:121–130. 10.1016/S0005-2728(00)00282-6 [DOI] [PubMed] [Google Scholar]
- Yousef M.S., and Guan L.. 2009. A 3D structure model of the melibiose permease of Escherichia coli represents a distinctive fold for Na+ symporters. Proc. Natl. Acad. Sci. USA. 106:15291–15296. 10.1073/pnas.0905516106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zani M.L., Pourcher T., and Leblanc G.. 1994. Mutation of polar and charged residues in the hydrophobic NH2-terminal domains of the melibiose permease of Escherichia coli. J. Biol. Chem. 269:24883–24889. [PubMed] [Google Scholar]