Abstract
Hydrogen sulfide is common in the environment, and is also endogenously produced by animal cells. Although hydrogen sulfide is often toxic, exposure to low levels of hydrogen sulfide improves outcomes in a variety of mammalian models of ischemia-reperfusion injury. In Caenorhabditis elegans, the initial transcriptional response to hydrogen sulfide depends on the hif-1 transcription factor, and hif-1 mutant animals die when exposed to hydrogen sulfide. In this study, we use rescue experiments to identify tissues in which hif-1 is required to survive exposure to hydrogen sulfide. We find that expression of hif-1 from the unc-14 promoter is sufficient to survive hydrogen sulfide. Although unc-14 is generally considered to be a pan-neuronal promoter, we show that it is active in many nonneuronal cells as well. Using other promoters, we show that pan-neuronal expression of hif-1 is not sufficient to survive exposure to hydrogen sulfide. Our data suggest that hif-1 is required in many different tissues to direct the essential response to hydrogen sulfide.
Keywords: hydrogen sulfide, hypoxia, tissue-specific expression
Hydrogen sulfide (H2S) in the environment is produced by industrial sources and natural sources, including volcanic deposits and anaerobic bacteria (Beauchamp et al. 1984). In fact, the gut microbiota produces H2S, which can influence the activity of host colonocytes (Beaumont et al. 2016). H2S is also endogenously produced by animals as a product of cysteine biosynthesis through the transsulfuration pathway, and endogenous H2S has important roles in cellular signaling (Li et al. 2011; Vandiver and Snyder 2012; Wang 2012). Chronic exposure to relatively low concentrations of environmental H2S in humans has been associated with neurological, respiratory, and cardiovascular dysfunction (Kilburn and Warshaw 1995; Richardson 1995; Bates et al. 2002). However, transient exposure to low H2S has also been shown to improve outcome in many mammalian models of ischemia-reperfusion injury (Bos et al. 2015; Wu et al. 2015). Although the mechanistic basis of the physiological effects of H2S are poorly understood, it is possible that the biological effects of exogenous H2S exposure, both beneficial and detrimental, result from activation of pathways that are normally regulated by endogenous H2S.
Caenorhabditis elegans is an excellent system to define physiological responses to exogenous H2S. In addition to powerful genetics, all cells are directly exposed to the gaseous environment (Shen and Powell-Coffman 2003). This feature allows for control of cellular H2S exposure without confounding factors from physiological regulation of gas delivery. C. elegans grown in 50 ppm H2S are long-lived, thermotolerant, and resistant to the hypoxia-induced disruption of proteostasis (Miller and Roth 2007; Fawcett et al. 2015). HIF-1 directs the transcriptional response to H2S in C. elegans (Budde and Roth 2010; Miller et al. 2011). HIF-1 is a highly conserved transcription factor best known for regulating the transcriptional response to low oxygen (hypoxia) in metazoans (Semenza 2000, 2001). C. elegans hif-1 mutant animals are viable and fertile in room air but die if exposed to hypoxia during embryogenesis (Jiang et al. 2001; Nystul and Roth 2004). By contrast, exposure to low H2S is lethal for hif-1 mutant animals at all developmental stages (Budde and Roth 2010), and mutations in hif-1 suppress protective effects of some mutations that confer tolerance to H2S (Budde and Roth 2010; Livshits et al. 2017). Moreover, increasing the activity of HIF-1, by mutations in negative regulators VHL-1, EGL-9, or RHY-1, increases the tolerance of C. elegans to otherwise lethally high concentrations of H2S (Budde and Roth 2010; Livshits et al. 2017). These observations indicate that HIF-1 has a central role in the organismal response to H2S.
Several studies have argued for neuronal-specific functions of HIF-1, although the hif-1 promoter is active in most, if not all, cells, and HIF-1 protein is stabilized ubiquitously in C. elegans exposed to either hypoxia or H2S (Jiang et al. 2001; Budde and Roth 2010). Neuronal expression of hif-1 in hypoxia is reported to be sufficient to prevent hypoxia-induced diapause and to increase lifespan through induction of intestinal expression of the flavin monooxygenase FMO-2 (Miller and Roth 2009; Leiser et al. 2015). Furthermore, neuronal expression of the cysteine synthase-like protein CYSL-1 regulates the activity of HIF-1 to modulate behavioral responses to changes in oxygen availability (Ma et al. 2012). These data motivated us to determine whether neuronal HIF-1 activity is sufficient for C. elegans to survive exposure to H2S.
In this study, we used tissue-specific rescue of hif-1 to define the site of essential HIF-1 activity in low H2S. We found that expression of hif-1 from the unc-14 promoter was sufficient for survival in H2S. Although it is considered a pan-neuronal promoter (Ogura et al. 1997; Pocock and Hobert 2008), our data indicate that the unc-14 promoter is also broadly expressed in nonneuronal cells. We show that hif-1 expressed from the pan-neuronal rab-3 promoter is not sufficient for viability in H2S. We further demonstrate that expression of hif-1 in muscle, hypodermis, and intestine is not sufficient for viability in low H2S. Together, our data indicate that the activity of HIF-1 may be required in multiple tissues to coordinate the organismal response to H2S.
Materials and Methods
Strains
Strains were grown at room temperature on nematode growth media plates (NGM) seeded with the OP50 strain of Escherichia coli (Brenner 1974). All strains were derived from N2 (Bristol). Full genotypes of strains used in this study are shown in Table 1. To sequence the Punc-14::hif-1 junction of otIs197, the region was amplified with forward primer oET479 (5′-GTTGTCCACCATCACAGTAATACG-3′) and reverse primer oET480 (5′-ACGACGGCGTTCCATG-3′). The oET479 primer was used for sequencing.
Table 1. Strains used in this study.
| ZG31: hif-1(ia4) V |
| DLM25: hif-1(ia4) V; otIs197[Punc-14::hif-1P621A, Pttx-3::RFP] |
| DLM26: hif-1(ia4) V; otEx3165[Punc-120::hif-1P621A, Pttx-3::RFP] |
| XZ2056: hif-1(ia4) V; yakEx126[Punc-17::hif-1cDNA, Pmyo-2::mCherry] |
| XZ2065: hif-1(ia4) V; yakEx131[eef-1A.1::hif-1cDNA, Pmyo-2::mCherry] |
| XZ2073: hif-1(ia4) V; yakEx137[Punc-14::hif-1P621A::YFP, Pmyo-2::mCherry] |
| XZ2074: hif-1(ia4) V; yakEx136[Pvha-6::hif-1cDNA, Pmyo-2::mCherry] |
| XZ2080: yakEx142[Punc-14::GFP, Pmyo-2::mCherry] |
| XZ2081: hif-1(ia4) V; yakEx143[Pdpy-7::hif-1cDNA, Pmyo-2::mCherry] |
| XZ2082: hif-1(ia4) V; yakEx144[Punc-14::hif-1cDNA, Pmyo-2::mCherry] |
| XZ2083: hif-1(ia4) V; yakEx145[Punc-47::hif-1cDNA, Pmyo-2::mCherry] |
| XZ2084: hif-1(ia4) V ; yakEx125[Prab-3::hif-1cDNA, Pmyo-2::mCherry] |
| XZ2085: hif-1(ia4) V ; yakEx146[Pvha-6::hif-1cDNA, Pdpy-7::hif-1cDNA, Prab-3::hif-1cDNA, Pmyo-2::mCherry] |
Constructs and transgenes
All constructs were made using the multisite Gateway system (Invitrogen), where a promoter region, a gene region (hif-1 cDNA or GFP), and a C-terminal 3′ untranslated region (UTR) were cloned into the destination vector pCFJ150 (Frøkjær-Jensen et al. 2008). The hif-1 A isoform was amplified from cDNA using forward primer oET467 (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCAATGGAAGACAATCGGAAAAGAAAC-3′) and reverse primer oET469 (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTGTCAAGAGAGCATTGGAAATGGG-3′). For the tissue-specific rescuing experiments, an operon GFP::H2B was included in the expression constructs downstream of the 3′UTR (Frøkjær-Jensen et al. 2012). This resulted in expression of untagged HIF-1 protein and histone H2B fused to GFP, which allowed for confirmation of promoter expression by monitoring GFP expression. The unc-14 promoter (1425 bp upstream of the start codon) was amplified from genomic DNA using forward primer oET520 (5′-GGGGACAACTTTGTATAGAAAAGTTGGAGAGCAGCAGCATCTCGAG-3′) and reverse primer oET507 (5′-GGGGACTGCTTTTTTGTACAAACTTGTTTTGGTGGAAGAATTGAGGG-3′). All plasmids constructed were verified by sequencing. Constructs used in this study are shown in Table 2. Extrachromosomal arrays were made by standard injection methods (Mello et al. 1991) with 10–15 ng/μl of the expression vector. At least two independent lines were isolated for each construct.
Table 2. Plasmids and constructs used in this study.
| Gateway entry clones | |
|---|---|
| pCFJ326 | tbb-2 3′UTR::OPERON::GFP [2-3] |
| pCFJ386 | eef-1A.1 [4-1] 625 bp upstream of and including the ATG |
| pCR110 | GFP [1-2] |
| pEGB05 | Prab-3 [4-1] 1232 bp upstream of the ATG |
| pET168 | hif-1 cDNA A isoform [1-2] |
| pET210 | Punc-14 [4-1] 1425 bp upstream of the ATG |
| pGH1 | Punc-17 [4-1] 3229 bp upstream of and including the ATG |
| pMH522 | Punc-47 [4-1] 1254 bp upstream of and including the ATG |
| pET187 | Pdpy-7 [4-1] 350 bp upstream of and including the ATG |
| pET188 | Pvha-6 [4-1] 881 bp upstream of and including the ATG |
| Gateway expression constructs | |
| pET171 | Punc-47::hif-1 cDNA::tbb-2 3′UTR::OPERON::GFP_pCFJ150 |
| pET172 | Punc-17::hif-1 cDNA::tbb-2 3′UTR::OPERON::GFP_pCFJ150 |
| pET182 | Prab-3::hif-1 cDNA:tbb-2 3′UTR::OPERON::GFP_pCFJ150 |
| pET187 | Pdpy-7::hif-1 cDNA::tbb-2 3′UTR::OPERON::GFP_pCFJ150 |
| pET188 | Pvha-6::hif-1 cDNA::tbb-2 3′UTR::OPERON::GFP_pCFJ150 |
| pET212 | Punc-14::GFP::let-858 3′UTR_pCFJ150 |
| pET213 | Punc-14::hif-1 cDNA::tbb-2 3′UTR::OPERON::GFP_pCFJ150 |
| pET216 | eef-1A.1::hif-1 cDNA::tbb-2 3′UTR::OPERON::GFP_pCFJ150 |
H2S atmospheres
Construction of atmospheric chambers was as previously described (Miller and Roth 2007; Fawcett et al. 2012). In short, H2S (5000 ppm with balance N2) was diluted continuously with room air to a final concentration of 50 ppm. Final H2S concentration was monitored using a custom-built H2S detector containing a three-electrode electrochemical SureCell H2S detector (Sixth Sense) as described (Miller and Roth 2007), calibrated with 100 ppm H2S with balance N2. Compressed gas mixtures were obtained from Airgas (Radnor, PA) and certified as standard to within 2% of the indicated concentration. H2S atmospheres were maintained at 20°.
Survival assays
20 to 40 L4 animals were picked to plates seeded with OP50. Plates were exposed to 50 ppm H2S for 20–24 hr in a 20° incubator, and then returned to room air to score viability. Death was defined as failure to move when probed with a platinum wire on the head or tail. Animals were scored 30 min after removal from H2S, and plates with dead animals were reexamined after several hours to ensure animals had not reanimated.
Imaging
For imaging expression of GFP, larval stage 1 (L1) or first-day adult animals were mounted on 2% agarose pads and anesthetized with 50 mM sodium azide for 10 min before placing the cover slip. The images were obtained using a Nikon 80i wide-field compound microscope.
Data availability
Strains are available upon request and have been deposited at the Caenorhabditis Genetics Center (cgc.umn.edu). Plasmid constructs are available upon request.
Results and Discussion
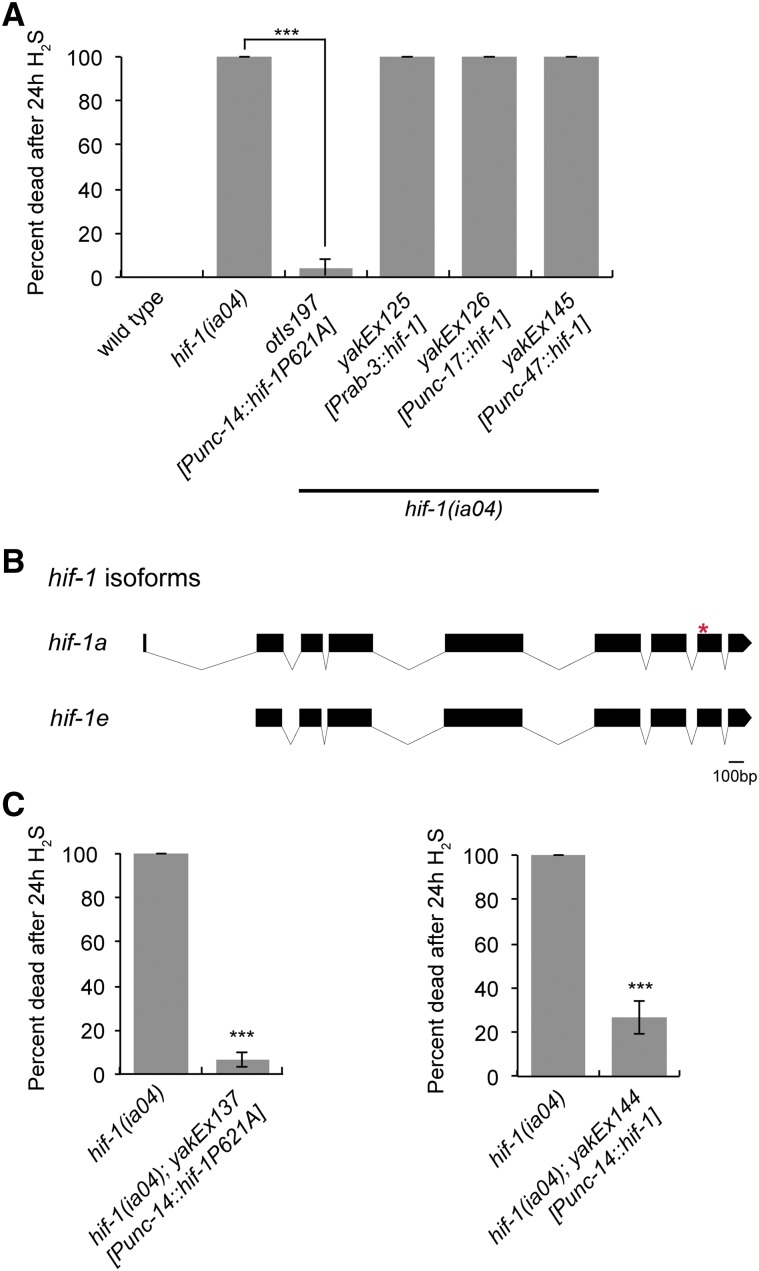
C. elegans requires hif-1 to survive exposure to low H2S (Budde and Roth 2010). To determine whether neuronal expression of hif-1 was sufficient for survival in H2S, we used transgenic hif-1(ia4) mutant animals that expressed hif-1 from heterologous promoters. We first used the available otIs197 transgene, which expresses hif-1 from the putative pan-neuronal unc-14 promoter (Pocock and Hobert 2008). We found that hif-1(ia4); otIs197 animals survived exposure to 50 ppm H2S (Figure 1A). This result suggests that neuronal expression of hif-1, from the unc-14 promoter, is sufficient to survive exposure to H2S.
Figure 1.
HIF-1 expression from the unc-14 promoter rescues the H2S lethality of hif-1(ia4) mutant animals. (A) Survival of animals exposed to H2S. All animals have the null hif-1(ia4) mutation. The otIs197 integrated array expresses a nondegradable HIF-1 variant. Other constructs were extrachromosomal arrays that express wild-type HIF-1. The unc-14 promoter is expressed pan-neuronally (Ogura et al. 1997), the rab-3 promoter is expressed in most, if not all, neurons (Nonet et al. 1997), unc-17 is expressed in cholinergic neurons (Rand et al. 2000), and unc-47 is expressed in GABAergic neurons (Eastman et al. 1999). Animals were exposed to 50 ppm H2S starting at L4. (B) HIF-1 gene structure and predicted A and E isoforms (Wormbase 2017). The P621A mutation that prevents degradation of hif-1 included in otIs197 is marked with *. (C) Survival of animals expressing HIF-1 from unc-14 promoter exposed to H2S. All animals have the null hif-1(ia4) mutation. Expression of HIF-1 was from extrachromosomal arrays. The yakEx137 array expresses nondegradable HIF-1(P621A) and the yakEx144 array expresses wild-type hif-1. For all panels, animals were exposed to 50 ppm H2S starting at L4. Average of three independent experiments is shown, each with n = 20–40 animals. Error bars are SEM. In all panels, statistical comparisons were to hif-1(ia4) controls. Statistically significant differences are indicated with *** P < 0.001 (Fisher’s exact test).
To further dissect in which neuronal cell type(s) HIF-1 activity was required to survive exposure to H2S, we generated transgenic animals that expressed hif-1 cDNA under the control of promoters active in specific neuronal subtypes. We found that expression in neither cholinergic neurons (Punc-17) nor GABAergic neurons (Punc-47) was sufficient to rescue the lethality of hif-1(ia4) mutant animals exposed to H2S (Figure 1A). Curiously, we also observed that expression of hif-1 cDNA from the pan-neuronal rab-3 promoter did not rescue survival of the hif-1(ia4) mutant animals (Figure 1A). This was unexpected, as expression of HIF-1 from the unc-14 promoter (the otIs197 transgene) was sufficient for survival in H2S. We therefore pursued the source of this discrepancy.
We first sought to verify the molecular nature of the otIs197 integrated transgene. We used PCR to amplify a region from the unc-14 promoter and the hif-1 coding region from the otIs197 transgenic animals. As expected, this reaction generated a single band of approximately 500 bp. However, when we sequenced the resulting PCR product, we discovered an insertion of an extra G immediately following the ATG of the hif-1 cDNA. This insertion causes a frame-shift and results in a stop codon after 13 amino acids. However, the otIs197 transgene must express some HIF-1 protein, as it can rescue many phenotypes of hif-1 mutant animals (Pocock and Hobert 2008; Miller and Roth 2009; Ma et al. 2012; Leiser et al. 2015). The otIs197 transgene was constructed to express isoform A of hif-1, though there are six predicted isoforms (WormBase 2017). We noted that the ATG for isoform E is 21 bp downstream of the original ATG in the hif-1 cDNA. Thus, it could be that expression of the hif-1e isoform is the basis of the activity of the otIs197 transgene. Because our Prab-3::hif-1 transgene expressed the hif-1a isoform, it was possible that the differences we observed from otIs197 were due to the expression of different hif-1 isoforms. To test this possibility we created transgenic strains expressing hif-1a under control of the unc-14 promoter using a Punc-14::hif-1a(P621A)::YFP plasmid (Pocock and Hobert 2008), which we verified had had the expected hif-1a(P621A) sequence. We injected this plasmid into hif-1(ia4) mutant animals to generate the yakEx137 transgene. If the rescue we observed in otIs197 was due to expression of hif1e rather than hif1a, then the animals expressing Punc-14::hif-1a(P621A)::YFP would die in H2S. However, these animals survived exposure to H2S (Figure 1C), indicating that potential expression of different isoforms did not underlie differences in survival of exposure to H2S.
The HIF-1 protein expressed by the otIs197 transgene has a P621A mutation that prevents it from being hydroxylated and degraded by the proteasome (Pocock and Hobert 2008). By contrast, the constructs we generated produced wild-type HIF-1 protein. We did not expect this feature to be salient for our experiments, since HIF-1 protein is stabilized in H2S due to inhibition of the hydroxylation reaction (Budde and Roth 2010; Ma et al. 2012). However, it is possible that constitutive stabilization of HIF-1 protein in neurons promotes survival in H2S. To evaluate this possibility, we cloned wild-type hif-1 cDNA under control of the unc-14 promoter, including 1.4 kb upstream of the transcription start site (Ogura et al. 1997). We found that hif-1(ia4); Punc-14::hif-1 (yakEx144) animals survived exposure to H2S, similar to hif-1(ia4); otIs197 animals (Figure 1C). We conclude that the P621A mutation in otIs197 does not underlie the difference in survival in H2S that we observed for animals expressing hif-1 from rab-3 and unc-14 promoters.
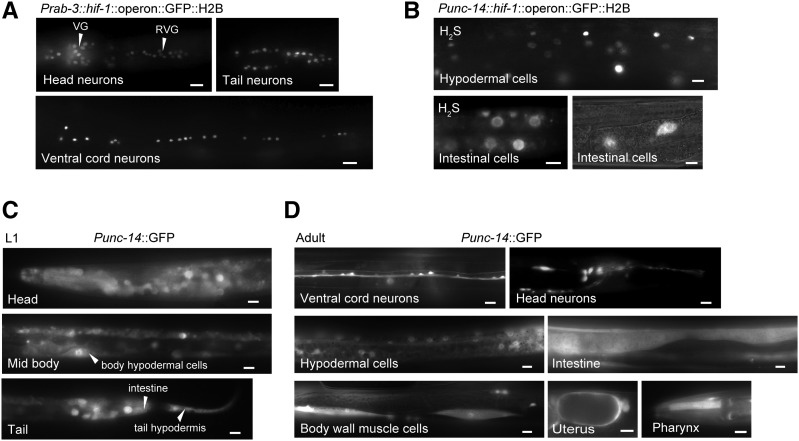
Given that the only other notable difference between the Prab-3::hif-1 and Punc-14::hif-1 constructs is the promoter elements, we hypothesized that differences between either the levels of expression from these promoters or the identity of the cells where these promoters are expressed should account for their different behavior. The transgenic constructs we generated all included an operon GFP::H2B downstream of the 3′UTR (Frøkjær-Jensen et al. 2012). This resulted in expression of untagged HIF-1 protein as well as GFP::H2B. We therefore visualized GFP expression to evaluate the expression levels and cellular patterns of promoter activity. As expected, GFP expression from adult hif-1(ia4); Prab-3::hif-1::operon::GFP::H2B was exclusively in neurons (Figure 2A). However, when we imaged adult hif-1(ia4); Punc-14::hif-1::operon::GFP::H2B animals that had survived exposure to H2S, we observed GFP expression in neurons, as expected, but also in intestinal and hypodermal cells (Figure 2B). We saw similar expression in animals that had not been exposed to H2S. To corroborate this observation, we cloned the unc-14 promoter upstream of GFP and injected it into wild-type animals. We then imaged larvae (Figure 2C) and adult animals (Figure 2D) from three separate lines. We observed expression of GFP in numerous cells other than neurons including intestine, hypodermis, muscle, and the uterus. Every animal that we imaged had expression in at least one cell type other than neurons (n = 50).
Figure 2.
The unc-14 promoter is active in many nonneuronal cells. (A) Visualization of GFP expressed from Prab-3::hif-1::operon::GFP::H2B (transgene yakEx125). Tail, head, and ventral cord neurons are shown from the ventral aspect of the same animal. VG, ventral ganglia; RVG, retrovesicular ganglia. In all images: bar, 10 μm. (B) Representative images of adult hif-1(ia4); Punc-14::hif-1::operon::GFP::H2B (transgene yakEx144) animals. GFP expression in hypodermal and intestinal cells is shown. Bar, 10 μm. (C and D) Representative images of (C) L1 and (D) adult transgenic animals expressing Punc-14::GFP (transgene yakEx142). Representative animals are shown with GFP expression in hypodermis, intestine, muscle, uterus, pharynx, and neurons. Bar, 5 μm in (C) and 10 μm in (D).
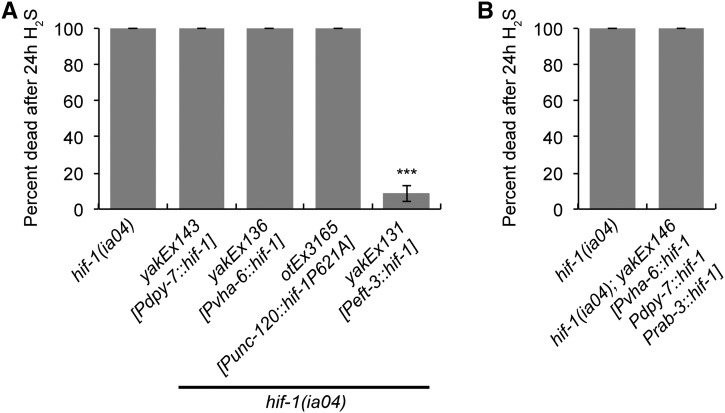
Based on our understanding of Punc-14 expression and the fact that hif-1(ia4); Prab-3::hif-1 animals die when exposed to H2S (Figure 1A), we inferred that neuronal HIF-1 activity is not sufficient for survival in H2S. We therefore explored whether expression of hif-1 exclusively in nonneuronal tissues was sufficient for survival in H2S. For these experiments, we generated transgenes with hif-1 expressed under control of the unc-120 promoter, which is active in body-wall and vulval muscle; the dpy-7 promoter, which is active in hypodermis; the vha-6 promoter, which is active in intestine; and the ubiquitous eef-1A.1 promoter. We chose these promoters because they included many of the tissues that had unc-14-driven expression of GFP (Figure 2B). As shown in Figure 3, only the ubiquitously expressed eef-1A.1::hif-1 rescued the lethality of hif-1(ia4) mutants exposed to H2S. Although we did not test all possible cell and tissue types, these data suggest that HIF-1 activity in a single tissue cannot support survival in H2S.
Figure 3.
Survival in H2S requires broad expression of hif-1. Survival of animals exposed to H2S. All animals have the null hif-1(ia4) mutation. (A) Lethality of animals that express hif-1 only in hypodermis (Pdpy-7::hif-1; yakEx143), intestine (Pvha-6::hif-1; yakEx136), or muscle (Punc-120::hif-1(P621A); otEx3165). As a control, hif-1 was expressed from a ubiquitous promoter (eef-1A.1::hif-1; yakEx131). Expression was from extrachromosomal arrays. Wild-type hif-1 was used for all constructs except the Punc-120::hif-1(P621A), which expresses the nondegradable variant. (B) Survival of hif-1(ia4); yakEx146 animals exposed to H2S that express hif-1 simultaneously in intestine (Pvha-6::hif-1), hypodermis (Pdpy-7::hif-1), and neurons (Prab-3::hif-1). Average of three independent experiments is shown, each with n = 20–35 animals. Error bars are SEM. In all panels, statistical comparisons were to hif-1(ia4) controls. Statistically significant differences are indicated with *** P < 0.001 (Fisher’s exact test).
The fact that Punc-14::hif-1 was sufficient for survival in H2S (Figure 1A) suggests that activity of HIF-1 may not be required in all cells. Since we did not observe rescue when hif-1 was expressed in a single tissue, we made transgenic animals with expression of hif-1 in >1 tissue to determine whether we could find a minimal expression that was sufficient for survival in H2S. We found that even animals with hif-1 expression in neurons, hypodermis, and intestine—hif-1(ia4); yakEx146[Prab-3::hif-1, Pvha-6::hif-1, Pdpy-7::hif-1]—did not survive exposure to H2S (Figure 3B). Together, our data suggest that that HIF-1 activity is required in many tissues to coordinate the essential response to H2S. This could indicate that HIF-1 acts cell-autonomously to direct expression of many tissue-specific transcripts that are required to survive exposure to H2S.
Although it was reported that otIs197 expresses hif-1 selectively in neurons (Pocock and Hobert 2008), our data show that the unc-14 promoter is more broadly expressed. In fact, others have reported nonneuronal expression of transgenes expressed under the control of the unc-14 promoter (Ogura et al. 1997; Wolkow et al. 2000; da Graca et al. 2004). However, the nonneuronal expression we have demonstrated is much more penetrant than has been previously acknowledged. This is an important consideration when interpreting the results of experiments using transgenes driven by unc-14, including hif-1 from otIs197. Our data show that nonneuronal expression from the unc-14 promoter is significant, and that rescue by unc-14-driven transgenes is not sufficient to infer neuronal function of HIF-1 and, presumably, other proteins.
Acknowledgments
We thank Michael Ailion (University of Washington) for sharing plasmids, discussing ideas, helping identify the different C. elegans tissues, and providing useful feedback on drafts of this manuscript. We are also grateful to Roger Pocock (Monash University) and Oliver Hobert (Columbia University) for sharing plasmids and strains. We thank Suzanne Hoppins (University of Washington) and Andrea Wills (University of Washington) for critical reading of the manuscript. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health (NIH) Office of Research Infrastructure Programs (P40 OD010440). This work was supported by NIH grant R01 ES024958 to D.L.M. D.L.M. is a New Scholar in Aging of the Ellison Medical Foundation.
Footnotes
Communicating editor: S. Lee
Literature Cited
- Bates M. N., Garrett N., Shoemack P., 2002. Investigation of health effects of hydrogen sulfide from a geothermal source. Arch. Environ. Health 57: 405–411. [DOI] [PubMed] [Google Scholar]
- Beauchamp R. O., Bus J. S., Popp J. A., Boreiko C. J., Andjelkovich D. A., 1984. A critical review of the literature on hydrogen sulfide toxicity. Crit. Rev. Toxicol. 13: 25–97. [DOI] [PubMed] [Google Scholar]
- Beaumont M., Andriamihaja M., Lan A., Khodorova N., Audebert M., et al. , 2016. Detrimental effects for colonocytes of an increased exposure to luminal hydrogen sulfide: the adaptive response. Free Radic. Biol. Med. 93: 155–164. [DOI] [PubMed] [Google Scholar]
- Bos E. M., Van Goor H., Joles J. A., Whiteman M., Leuvenink H. G., 2015. Hydrogen sulfide: physiological properties and therapeutic potential in ischaemia. Br. J. Pharmacol. 172: 1479–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budde M. W., Roth M. B., 2010. Hydrogen sulfide increases hypoxia-inducible factor-1 activity independently of von Hippel-Lindau tumor suppressor-1 in C. elegans. Mol. Biol. Cell 21: 212–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Graca L. S., Zimmerman K. K., Mitchell M. C., Kozhan-Gorodetska M., Sekiewicz K., et al. , 2004. DAF-5 is a Ski oncoprotein homolog that functions in a neuronal TGF beta pathway to regulate C. elegans dauer development. Development 131: 435–446. [DOI] [PubMed] [Google Scholar]
- Eastman C., Horvitz H. R., Jin Y., 1999. Coordinated transcriptional regulation of the unc-25 glutamic acid decarboxylase and the unc-47 GABA vesicular transporter by the Caenorhabditis elegans UNC-30 homeodomain protein. J. Neurosci. 19: 6225–6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett E. M., Horsman J. W., Miller D. L., 2012. Creating defined gaseous environments to study the effects of hypoxia on C. elegans. J. Vis. Exp. 65: e4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett E. M., Hoyt J. M., Johnson J. K., Miller D. L., 2015. Hypoxia disrupts proteostasis in Caenorhabditis elegans. Aging Cell 14: 92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjær-Jensen C., Davis M. W., Hopkins C. E., Newman B. J., Thummel J. M., et al. , 2008. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 40: 1375–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjær-Jensen C., Davis M. W., Ailion M., Jorgensen E. M., 2012. Improved Mos1-mediated transgenesis in C. elegans. Nat. Methods 9: 117–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Guo R., Powell-Coffman J. A., 2001. The Caenorhabditis elegans hif-1 gene encodes a bHLH-PAS protein that is required for adaptation to hypoxia. Proc. Natl. Acad. Sci. USA 98: 7916–7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilburn K. H., Warshaw R. H., 1995. Hydrogen sulfide and reduced-sulfur gases adversely affect neurophysiological functions. Toxicol. Ind. Health 11: 185–197. [DOI] [PubMed] [Google Scholar]
- Leiser S. F., Miller H., Rossner R., Fletcher M., Leonard A., et al. , 2015. Cell nonautonomous activation of flavin-containing monooxygenase promotes longevity and health span. Science 350: 1375–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Rose P., Moore P. K., 2011. Hydrogen sulfide and cell signaling. Annu. Rev. Pharmacol. Toxicol. 51: 169–187. [DOI] [PubMed] [Google Scholar]
- Livshits L., Chatterjee A. K., Karbian N., Abergel R., Abergel Z., et al. , 2017. Mechanisms of defense against products of cysteine catabolism in the nematode Caenorhabditis elegans. Free Radic. Biol. Med. 104: 346–359. [DOI] [PubMed] [Google Scholar]
- Ma D. K., Vozdek R., Bhatla N., Horvitz H. R., 2012. CYSL-1 interacts with the O2-sensing hydroxylase EGL-9 to promote H2S-modulated hypoxia-induced behavioral plasticity in C. elegans. Neuron 73: 925–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C. C., Kramer J. M., Stinchcomb D., Ambros V., 1991. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10: 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D. L., Roth M. B., 2007. Hydrogen sulfide increases thermotolerance and lifespan in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 104: 20618–20622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D. L., Roth M. B., 2009. C. elegans are protected from lethal hypoxia by an embryonic diapause. Curr. Biol. 19: 1233–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D. L., Budde M. W., Roth M. B., 2011. HIF-1 and SKN-1 coordinate the transcriptional response to hydrogen sulfide in Caenorhabditis elegans. PLoS One 6: e25476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet M. L., Staunton J. E., Kilgard M. P., Fergestad T., Hartwieg E., et al. , 1997. Caenorhabditis elegans rab-3 mutant synapses exhibit impaired function and are partially depleted of vesicles. J. Neurosci. 17: 8061–8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystul T. G., Roth M. B., 2004. Carbon monoxide-induced suspended animation protects against hypoxic damage in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 101: 9133–9136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura K., Shirakawa M., Barnes T. M., Hekimi S., Ohshima Y., 1997. The UNC-14 protein required for axonal elongation and guidance in Caenorhabditis elegans interacts with the serine/threonine kinase UNC-51. Genes Dev. 11: 1801–1811. [DOI] [PubMed] [Google Scholar]
- Pocock R., Hobert O., 2008. Oxygen levels affect axon guidance and neuronal migration in Caenorhabditis elegans. Nat. Neurosci. 11: 894–900. [DOI] [PubMed] [Google Scholar]
- Rand J. B., Duerr J. S., Frisby D. L., 2000. Neurogenetics of vesicular transporters in C. elegans. FASEB J. 14: 2414–2422. [DOI] [PubMed] [Google Scholar]
- Richardson D. B., 1995. Respiratory effects of chronic hydrogen sulfide exposure. Am. J. Ind. Med. 28: 99–108. [DOI] [PubMed] [Google Scholar]
- Semenza G. L., 2000. HIF-1: mediator of physiological and pathophysiological responses to hypoxia. J. Appl. Physiol. 88: 1474–1480. [DOI] [PubMed] [Google Scholar]
- Semenza G. L., 2001. HIF-1, O2, and the 3 PHDs: how animal cells signal hypoxia to the nucleus. Cell 107: 1–3. [DOI] [PubMed] [Google Scholar]
- Shen C., Powell-Coffman J. A., 2003. Genetic analysis of hypoxia signaling and response in C. elegans. Ann. N. Y. Acad. Sci. 995: 191–199. [DOI] [PubMed] [Google Scholar]
- Vandiver M. S., Snyder S. H., 2012. Hydrogen sulfide: a gasotransmitter of clinical relevance. J. Mol. Med. (Berl.) 90: 255–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., 2012. Physiological implications of hydrogen sulfide: a whiff exploration that blossomed. Physiol. Rev. 92: 791–896. [DOI] [PubMed] [Google Scholar]
- Wolkow C. A., Kimura K. D., Lee M. S., Ruvkun G., 2000. Regulation of C. elegans life-span by insulinlike signaling in the nervous system. Science 290: 147–150. [DOI] [PubMed] [Google Scholar]
- WormBase, 2017 Release WS259. Available at: http://www.wormbase.org. Accessed: July 20, 2017.
- Wu D., Wang J., Li H., Xue M., Ji A., et al. , 2015. Role of hydrogen sulfide in ischemia-reperfusion injury. Oxid. Med. Cell. Longev. 2015: 186908. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains are available upon request and have been deposited at the Caenorhabditis Genetics Center (cgc.umn.edu). Plasmid constructs are available upon request.