Abstract
Chromosomal inversions are a ubiquitous feature of genetic variation. Theoretical models describe several mechanisms by which inversions can drive adaptation and be maintained as polymorphisms. While inversions have been shown previously to be under selection, or contain genetic variation under selection, the specific phenotypic consequences of inversions leading to their maintenance remain unclear. Here we use genomic sequence and expression data from the Drosophila Genetic Reference Panel (DGRP) to explore the effects of two cosmopolitan inversions, In(2L)t and In(3R)Mo, on patterns of transcriptional variation. We demonstrate that each inversion has a significant effect on transcript abundance for hundreds of genes across the genome. Inversion-affected loci (IAL) appear both within inversions as well as on unlinked chromosomes. Importantly, IAL do not appear to be influenced by the previously reported genome-wide expression correlation structure. We found that five genes involved with sterol uptake, four of which are Niemann-Pick Type 2 orthologs, are upregulated in flies with In(3R)Mo but do not have SNPs in linkage disequilibrium (LD) with the inversion. We speculate that this upregulation is driven by genetic variation in mod(mdg4) that is in LD with In(3R)Mo. We find that there is little evidence for a regional or position effect of inversions on gene expression at the chromosomal level, but do find evidence for the distal breakpoint of In(3R)Mo interrupting one gene and possibly disassociating the two flanking genes from regulatory elements.
Keywords: Drosophila, inversion polymorphism, eQTLs, DGRP, DPGP
Chromosomal inversions, in which a portion of linear DNA sequence is flipped in its orientation, are a common member of the menagerie of DNA polymorphisms, and have been found in diverse organismal populations such as humans, plants, and fruit flies (Krimbas and Powell 1992; Kidd et al. 2010; Lowry and Willis 2010) . In many cases, large chromosomal inversions have profound impacts on phenotype and disease (Feuk 2010). For instance, recurrent inversions are responsible for an estimated 43% of hemophilia A cases (Lakich et al. 1993). Inversions can also have beneficial effects. A 900 kb inversion on human chromosome 17 (q21.31) has been shown to be associated with higher female fecundity in the Icelandic population (Stefansson et al. 2005). In populations of the malaria vector Anopheles gambiae, a large chromosomal inversion on chromosome 2L (2La) is associated with desiccation resistance and thus segregates at high frequencies in arid environments (Fouet et al. 2012). These examples are the very tip of the iceberg; inversion polymorphisms have been implicated in numerous phenotypic differences among a host of organisms, however little is known about the mechanisms by which inversions confer their phenotypic effects.
Perhaps the single best studied inversions are those from Drosophila, in part made famous by the pioneering work of Dobzhansky (Dobzhansky and Sturtevant 1938). Dobzhansky focused much attention on spatial and temporal variation in the frequency of large inversions of Drosophila pseudoobscura and showed in broad strokes that clear fitness differences were responsible for the regular patterns of frequency change observed. These findings in turn spurred a large body of population genetics theory to explain the establishment and selective persistence of inversions in natural populations (Levene and Dobzhansky 1958; Fraser et al. 1966; Anderson et al. 1967; Tobari and Kojima 1967). As postulated by Sturtevant (1921), crossover suppression induced in inversion heterozygotes can mean that a single adaptive allele within an inversion may suffice for the selective invasion of that rearrangement (Haldane 1957). Such lowered levels of recombination and attendant increases in linkage disequilibrium (LD) could thus present the opportunity for subsequent coadaptation of multiple genes near inversion breakpoints (Sturtevant and Mather 1938; Dobzhansky 1947). Conversely, locally adapted alleles that predate the rearrangement on the same chromosome might aid the establishment of an inversion simply because of the reduction in recombination rates between such loci (Kirkpatrick and Barton 2006). Further, inversions might have direct fitness effects, for instance by deletion or changes in gene expression near the inversion breakpoints (Kirkpatrick and Kern 2012). At present, we have precious little information as to the variants responsible for differential fitness effects associated with inversions.
In D. melanogaster, paracentric inversions spanning several megabases are common and have been found in populations across the globe (Stalker 1976, 1980; Aulard et al. 2002; Van ’t Land et al. 2000; Knibb et al. 1981; Sezgin et al. 2004; Anderson et al. 2005; Umina et al. 2005). Much of this segregating inversion polymorphism is associated with latitudinal clines in D. melanogaster, a historically tropical species adapting along tropical-to-temperate climatic gradients in Australia and North America (Knibb et al. 1981; Weeks et al. 2002; de Jong and Bochdanovits 2003; Sezgin et al. 2004; Reinhardt et al. 2014; Schrider et al. 2016). Clinically varying phenotypes that are associated with inversions include heat resistance, cold tolerance, and body size (Weeks et al. 2002; Anderson et al. 2005; de Jong and Bochdanovits 2003). Clinal variation of inversion frequency in D. melanogaster has been shown via population genetic approaches to be due to selection independent of demography (Reinhardt et al. 2014; Kapun et al. 2016), though migration has been suggested to generate these patterns along with local adaptation (Bergland et al. 2016). Indeed, inversions have been observed to have a major effect on several phenotypes that vary between temperate and tropical populations across several Drosophila species, and these clines have been stable since their discovery roughly 80 yr ago (Hoffmann et al. 2004; Cogni et al. 2017). Unfortunately, while the associations are known, the molecular mechanisms at work in determining differential phenotypes as a result of inversion status are still unknown.
Recent population genomic projects in D. melanogaster, such as the DGRP and Drosophila Population Genomics Project, have opened up the opportunity to study inversions systematically as these resources have captured segregating inversions from North America and Africa (Langley et al. 2012; Mackay et al. 2012; Pool et al. 2012; Houle and Márquez 2015). Corbett-Detig et al. (2012) bioinformatically mapped previously unknown breakpoints of several inversions, a task that was tedious for even single inversions prior to whole-genome sequencing (Wesley and Eanes 1994; Andolfatto et al. 1999; Matzkin et al. 2005). For instance, Corbett-Detig et al. (2012) discovered the breakpoints associated with numerous inversions, demonstrated the expected increase in LD near inversion breakpoints, and elevated differentiation between inverted and standard arrangement chromosomes at the nucleotide level. In parallel with the exponential increase in population genomic resources, large-scale phenotypic association studies of these same genotypes have been accumulating (Mackay et al. 2012; Telonis-Scott et al. 2016; Vonesch et al. 2016). These include numerous phenotypes previously associated with inversion polymorphism such as body size (Weeks et al. 2002) and desiccation resistance (Hoffmann et al. 2005).
A logical place to look for inversion effects that may influence suites of phenotypes would be transcript level variation. Previous findings strongly suggest that inversions could be important drivers of adaptation with gene expression variation as a potential molecular mechanism (Chambers 1991; López-Maury et al. 2008; Fraser 2013). Indeed, inversions could affect patterns of transcript variation in a number of ways: (1) genes at or near inversion breakpoints may become disabled or separated from their regulatory apparatus, thus inversions may have direct effects on transcription; (2) increased LD in inversions due to crossover suppression may increase linkage with gene expression Quantitative Trait Loci (eQTL), and thus alternative alleles of the inversion may be associated with differential expression of genes within the inversion (i.e., indirect, cis-eQTL associated with the inversion); (3) eQTL in LD with the inversion might themselves regulate genes outside of the inversion (i.e., indirect, trans-eQTL associated with the inversion); or (4) the large-scale nature of Drosophila inversions may create global changes in the organization of chromatin or nuclear localization of the chromosomes such that genes are differentially regulated between inversion and standard karyotypes. Thus, inversions may have a direct effect on global patterns of transcription and act as trans-eQTL themselves. Indeed, earlier studies of transcriptional variation in D. melanogaster have hinted at the influence of inversions on genome-wide patterns of gene expression (Ayroles et al. 2009; Massouras et al. 2012; Huang et al. 2015). Here, we address the effect of two cosmopolitan inversions, In(2L)t and In(3R)Mo, on patterns of transcription by using whole-genome sequence, gene expression, and inversion call data from the DGRP.
Materials and Methods
Materials
Processed expression data previously reported in Ayroles et al. (2009) was downloaded from ArrayExpress (Kolesnikov et al. 2015). We accepted inversion state calls for each of the DGRP lines where cytological and bioinformatic inversion calls for In(2L)t and In(3R)Mo agree and removed lines from analyses where there was any disagreement (Corbett-Detig et al. 2012; Huang et al. 2014; Houle and Márquez 2015). Using the same databases, we removed individuals from lines likely heterozygous for In(2L)t or In(3R)Mo. Expression analyses were performed with 34 lines (136 individuals), with two lines homozygous for In(2L)t and seven lines homozygous for In(3R)Mo. There were 26 lines homozygous for the standard arrangement at both In(2L)t and In(3R)Mo, as one line was homozygous for both inversions. We calculated LD between In(2L)t and SNPs using 181 lines, including 19 inversion-bearing lines. We calculated LD between In(3R)Mo and SNPs using 197 lines, including 17 inversion-bearing lines. To maintain consistency with Affymetrix library files, dm3/BDGP release 5 genomic coordinates and annotations corresponding to BDGP version 5.49 were used in conjunction with the Affymetrix Drosophila 2 Release 35 library file update.
Methods
Statistical analyses were performed in R (R Core Team 2013) using the functions (lm), (anova), (quantile), (qvalue), (phyper), and (ggplot2).
3′ UTR array analysis:
Correlation structure:
Pairwise gene expression correlation coefficients were calculated by linear regression on all unique pairwise combinations of probe sets, excepting self-comparisons. Correlation coefficients are reported here as adjusted r2 from the R function (lm). Gene expression modules used here were reported in Ayroles et al. (2009). Null distributions of uniquely occupied clusters for each inversion were generated by permuting the occupied cluster for each gene 100,000 times and calculating the total unique clusters occupied by IAL for each permutation.
Inversion effect on expression:
Linear regressions of sex, inversion, and Line effects with expression as the response were performed for each probe set:
for individual expression value, Yijkl, in response to ith Sex, Ai, jth In(2L)t state, Bj, kth In(3R)Mo state, Ck, and the lth Line in the jth In(2L)t state, Dkl, with εijkl as the error term in Lines. Model testing was performed using (add1) and (drop1) in R to add interaction terms or remove main effect terms from the above model, respectively. Interaction terms were added one at a time to the main effects and tested for each probe set. An AIC is reported for each model with a lower absolute value being preferred when comparing two models. The effect of adding an interaction term between Sex and In(2L)t or Sex and In(3R)Mo varied by probe set, but the above model was the best fit for 10,082 of the probe sets. Models with and without an interaction term between inversions performed the same for all loci, thus, we chose the less complex model. All of our analyses compare results across all loci; therefore, for meaningful conclusions, we must use the same model for all loci. We chose the simpler model by simple majority rule. It is worth noting that this means that most loci are tested by the best fit model and the rest of the loci are tested by a likely overly conservative model with respect to detecting inversion effects. Similarly, the above model was the best fit for 12,994 probe sets when compared to dropping any of the main effect terms. We calculated P-values of the observed F values as percentiles of the F distributions generated by 10,000 permutations, sampling each inversion independently without replacement. Multiple testing correction was performed by calculating q-values using the R package (q value) with FDR = 0.05 (Storey and Tibshirani 2003) on the permutation-derived P-values. Proportion of variance explained by each effect was calculated as η2. Magnitude and direction of inversion effect was calculated as Cohen’s d (Cohen 1988). Cohen’s d is the difference of the means of two groups scaled to SD by means of the root mean square of the SD of the two groups. Thus, effect sizes as Cohen’s d can be compared across multiple tests, as we present here, even though SD, in addition to means, may differ both within the pairwise comparison of one locus and between loci. For example, for two distributions, each with a SD = 1, Cohen’s d = 1 represents a difference of one SD between the means and a ∼62% expected overlap given a false positive rate of 5%.
Functional annotation enrichment:
Functional annotation profiling was performed using the g:Profiler online portal of g:GOSt using default settings (Reimand et al. 2016) (version r1622_e84_eg31). Ambiguous 3′ UTR probe sets were resolved manually if possible, or ignored if they overlapped transcripts for > 1 gene.
SNP-inversion LD:
LD was calculated as r2 = D’/pS(1-pS)pI(1-pI) for each diallelic SNP S and inversion I, with major allele frequencies pS and pI, using a custom bash script. For each SNP, significance was calculated as a chi-squared (χ2) transformation with one d.f. of r2 as χ2 = Nr2, where N is the sample size. Significant LD was defined as a SNP with sample size of at least 60, minor allele frequency of 10% for both the SNP and inversion state, and χ2 > critical value (P = 0.05, d.f. = 1) with Bonferroni correction for all SNPs on that chromosome arm (n = 967774, χ2 > 29.65329 and 947970, χ2 > 29.61321 for chr2L and chr3R, respectively). The sample size and minor allele frequency cutoffs ensured that there were at least six representative lines bearing minor alleles. Genes were considered in significant LD with inversion state if at least one significant SNP was found within the annotated gene region (FlyBase v5.49).
IAL physical clustering:
To see if IAL were physically clustered within the genome, we examined physical clustering by measuring the coefficient of variance (CV) of distances between genes by chromosome arm. Location and length of each gene was used from FlyBase v5.49. Distance between neighboring genes was calculated as the distance between ends of gene-annotated regions of neighboring genes. Distance from the most distal or proximal genes to the distal or proximal endpoint, respectively, was not included. CV was calculated as the SD divided by the mean of the distribution of intergenic distances for each chromosomal arm (Sokal and Rohlf 1995). We defined the intergenic distance between overlapping gene regions as zero. Null distributions of intergenic distances for each chromosome and each inversion were generated by 100,000 random samples, without replacement, of the same number of genes from a chromosome arm as the number of IAL for that chromosome arm and inversion, then calculating the distances between those genes. C.I.s were calculated as 2.5–97.5% quantiles from the corresponding random sample distribution.
Transcription factor (TF)–target gene (TG) interactions:
TFs and TGs were defined by Drosophila Interaction Database modMine (Contrino et al. 2012) (v2015_12). Genes in this analysis were those that are present in Affymetrix Drosophila2 genome array annotation (Release 35), the DroID TF-TG database v2015_12, and FlyBase v5.49 annotation. Over/underrepresentation of genes with significant inversion effect on expression as targets of TFs with SNPs in LD with inversion state was calculated as the probability of the observation given the hypergeometric distribution.
Data availability
Custom scripts, data, and analysis results can be found online at https://github.com/kern-lab/lavingtonKern, including file descriptions in the AnalysisFiles.readme document.
Results
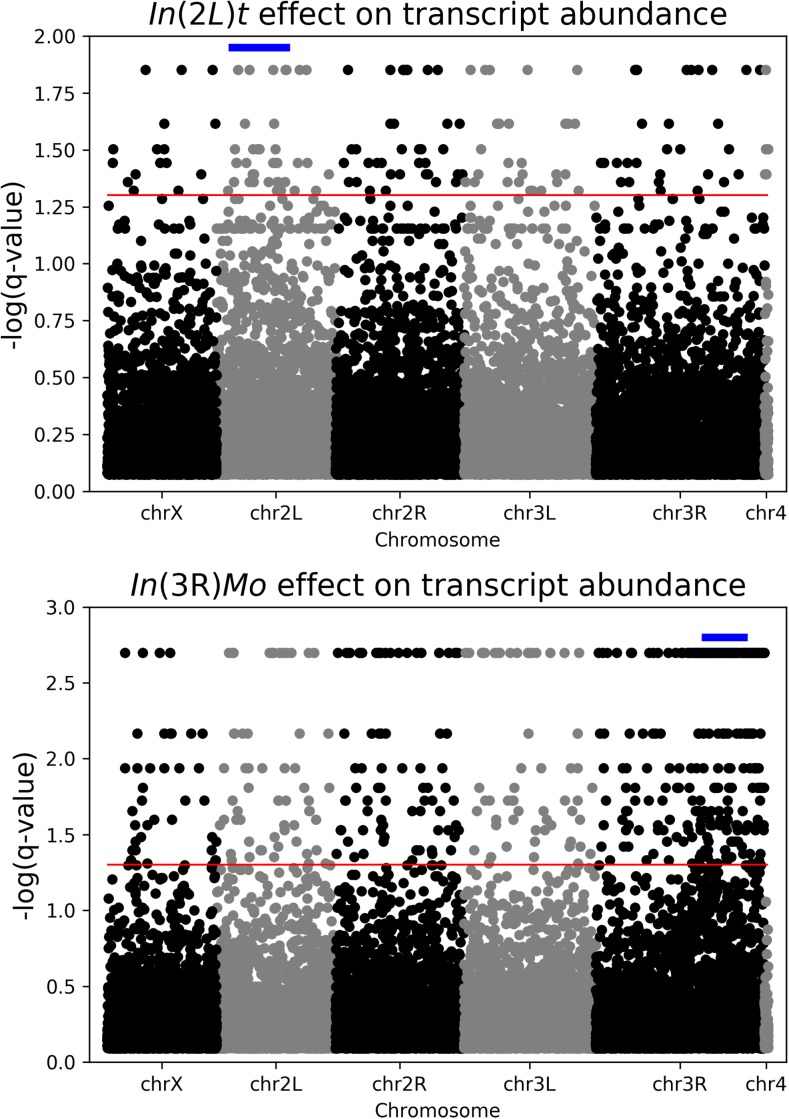
To examine what influence, if any, common inversion polymorphisms have on patterns of transcription in the Drosophila genome, we combined publicly available genome sequences (Mackay et al. 2012), their associated karyotype calls (Corbett-Detig et al. 2012; Huang et al. 2014; Houle and Márquez 2015), and previously published microarray-based expression data (Ayroles et al. 2009). We validated the use of a model with only main effects of Sex, Line, and two cosmopolitan inversions, In(2L)t and In(3R)Mo, using the R functions (add1) and (drop1). This main effects model performed better than any model having any of the main effect terms removed or with the addition of any interaction term (see Materials and Methods). After correction for multiple testing (see Materials and Methods), we found 229 and 498 total probe sets with significant inversion effects for In(2L)t and In(3R)Mo, respectively, hereafter referred to as IAL (Figure 1). These IAL occur both within the inversions themselves (40 In(2L)t and 111 In(3R)Mo), outside the inversions but near the breakpoints (3 In(2L)t and 38 In(3R)Mo within 1 Mb of the breakpoint), and scattered throughout the genome (134 In(2L)t and 181 In(3R)Mo; see Table 1). There are a large number of loci with transcript abundance variation correlating with inversion state; however, we note that the inversion effect contribution to variance is relatively small for the vast majority of loci (Supplemental Material, File S1, Table S1, and Table S2).
Figure 1.
Manhattan plot of -log transformed q-values for In(2L)t (top) and In(3R)Mo (bottom) inversion effect on transcription for each probe set across the genome. The blue bar above the points indicates the genomic location of the inversion. The red horizontal line represents the genome-wide significance threshold of q=0.05.
Table 1. Location of IAL.
| Within Inversion | Outside Inversion, on Same Chr | <1 Mb Outside Inversion | Chr 2L | Chr 2R | Chr 3L | Chr 3R | Chr X | Chr 4 | |
|---|---|---|---|---|---|---|---|---|---|
| In(2L)t | 40 | 18 | 3 | 61 | 42 | 36 | 35 | 17 | 4 |
| In(3R)Mo | 111 | 133 | 38 | 44 | 58 | 49 | 244 | 30 | 0 |
Counts of IAL between breakpoints, on the same chromosome as the inversion but outside of the breakpoints, and those on the same chromosome as the inversion but outside and within 1 Mb of the breakpoints, as well as total IAL on each chromosome arm for each inversion. Chr, chromosome; IAL, inversion-affected loci.
One explanation for the large number of IAL found across the genome is that few loci are directly affected by the inversion and the remaining loci are affected indirectly by an expression variation correlation structure, previously described by Ayroles et al. (2009). We addressed this correlation structure by the numbers of unique expression modules occupied by, and the distribution of correlation coefficients of, IAL as compared to all genes. If a significant portion of the IAL we observe are due to expression variation correlation, then we would expect that IAL occupy fewer expression modules than the same number of genes drawn at random. We would also expect the mean correlation of IAL between IAL to be higher than the genome-wide average. We observe that IAL occupy more modules than expected at random for both In(2L)t (71 obs; 38–56 95% C.I.) and In(3R)Mo (108 obs ; 65–87 95% C.I.). For both inversions, we did not observe higher mean correlation between IAL and non-IAL or the genome-wide mean (Table 2).
Table 2. Pairwise correlation of gene expression.
| Mean r2 | ||
|---|---|---|
| In(2L)t | IAL × IAL | 0.0965 |
| IAL × other | 0.0962 | |
| In(3R)Mo | IAL × IAL | 0.1025 |
| IAL × other | 0.1042 | |
| Genome-wide | 0.1181 |
Mean r2 was calculated from all pairwise correlation for the set description. IAL × IAL is the set of all pairwise correlation coefficients of expression between IAL for that inversion. IAL × other is the set of correlation coefficients of expression between IAL and all other (non-IAL). IAL, inversion-affected loci.
We then examined the inversion effect on gene expression variation for four distinct categories of effect: (1) cis- or (2) trans-inversion effects of SNPs in LD with the inversion, (3) direct effects of the inversion by interrupting genes, and (4) regional effects of chromosomal rearrangement.
Cis-inversion effect of SNPs in LD with the inversion
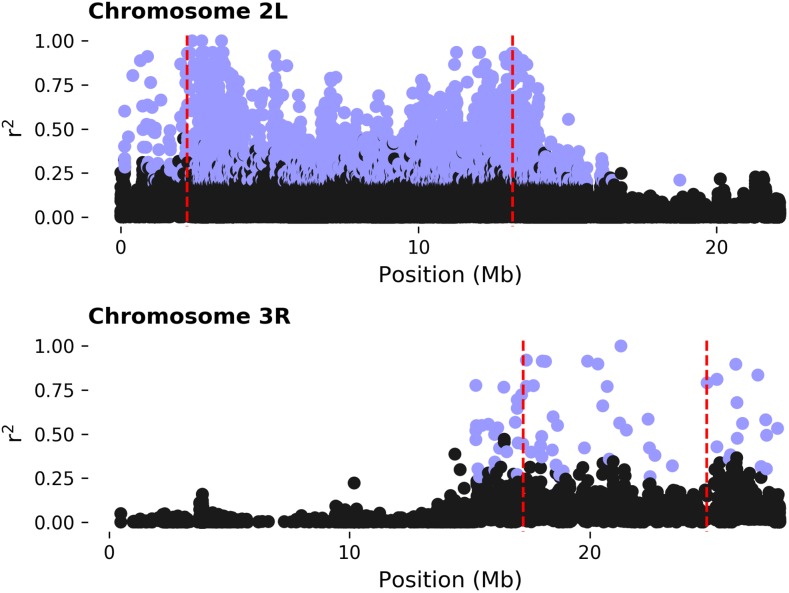
Our model explicitly tests the effect of chromosomal arrangement on expression variation, and here we focus on SNP variation as the main driver of the inversion effect by taking advantage of LD between the inversion state and SNP variants. LD with inversions is highest at the breakpoints and decays in both directions from each breakpoint as expected (Figure 2) (Wesley and Eanes 1994; Andolfatto et al. 2001; Langley et al. 2012; Corbett-Detig and Hartl 2012). To determine cis-effects of the inversions, we tested for overrepresentation of IAL among loci in LD with inversion state. This ignores the location of the loci and focuses on the correlation of SNP alleles with the inversion state. As expected, few loci in LD with the inversion were located on a different chromosomal arm [three with In(2L)t, and two with In(3R)Mo)] and IAL in LD are located only on the same chromosomal arm (Table 3). We observed a significant overrepresentation of IAL with SNPs in LD with inversion state (Table 4).
Figure 2.
Linkage disequilibrium (LD) between single nucleotide polymorphisms (SNPs) and inversion state. Disequilibrium is measured as r2. Inversion breakpoints are depicted as red vertical lines. SNPs with at least 10% minor allele frequency, for inversion and SNP, and that are in significant LD with the inversion are shown in light blue (P < 0.05, Bonferroni correction for all SNPs detected by chromosome arm).
Table 3. Location of loci with SNPs in LD with inversions.
| chr2L | chr2R | chr3L | chr3R | chrX | ||
|---|---|---|---|---|---|---|
| In(2L)t | All loci | 370 | 1 | 0 | 1 | 1 |
| IAL | 11 | 0 | 0 | 0 | 0 | |
| In(3R)Mo | All loci | 0 | 2 | 0 | 663 | 0 |
| IAL | 0 | 0 | 0 | 110 | 0 |
Loci are identified as BDGP v5.49 and Affymetrix library v.35 annotated gene regions containing SNPs in significant LD with the inversion (see Materials and Methods). chr, chromosome; IAL, inversion-affected loci; BDGP, Berkeley Drosophila Genome Project; SNP, single nucleotide polymorphism; LD, linkage disequilibrium.
Table 4. IAL in LD with inversion.
| Inversion | Total Genes | Total IAL | IAL in LD with Inversion | Genes in LD with Inversion | E[X] | p(E[X] > x) |
|---|---|---|---|---|---|---|
| In(2L)t | 11969 | 192 | 11 | 372 | 6 | 0.01683 |
| In(3R)Mo | 425 | 102 | 744 | 26 | 6.08 × 10−35 |
Expected counts (E[X]) and P-values are calculated by hypergeometric distribution with uncorrected P-value cutoff of P = 0.025 for the two-tailed test. Genes included in analyses are annotated in both BDGP v5.49 and Affymetrix library version 35. IAL, inversion-affected loci; LD, linkage disequilibrium; BDGP, Berkeley Drosophila Genome Project.
Trans-inversion effect of SNPs in LD with the inversion
Our expectation of a cis-inversion effect is dependent on SNP variation in LD with the inversion. By the same rationale, a trans-inversion effect may be detected as an IAL without SNP variation in LD with the inversion, as we observe with a majority of IAL for each inversion [181 of 192 for In(2L)t and 323 of 425 for In(3R)Mo] (see Table 4). Assuming that SNP variation is the basis of expression variation, one trivial explanation of a trans-inversion effect is SNP variation in TFs in LD with the inversion acting on downstream targets. The TFs in this case need not be IAL as SNPs in protein-coding regions of TFs can give rise to expression variation in downstream targets. However, we found no over- or underrepresentation of IAL that are targets of TFs with SNPs in LD with either inversion (Table 5). A possible example of trans-inversion effect was found by functional analysis of IAL, and is discussed below.
Table 5. IAL as targets of transcription factors with SNPs in LD with inversion.
| Inversion | Total Genes | DroID TFs with Sig SNPs | Total DroID TF Targets | Total DroID TF Targets Also IAL | Targets of Sig SNP DroID TFs Also IAL | E[X] | p(E[X] > x) |
|---|---|---|---|---|---|---|---|
| In(2L)t | 10623 | 3 | 3802 | 181 | 53 | 65 | 0.96260 |
| In(3R)Mo | 8 | 2373 | 385 | 99 | 86 | 0.04801 |
Expected counts E[X] and P-values calculated by hypergeometric distribution with uncorrected P-value cutoff of P = 0.025 for the two-tailed test. Genes included in these analyses are annotated in BDGP v5.49, Affymetrix library v.35, and DroIDb v2015_12. BDGP, Berkeley Drosophila Genome Project; TF, transcription factor; Sig, significant; SNP, single nucleotide polymorphism; IAL, inversion-affected loci.
Direct effect of inversions by disrupting genes at breakpoints
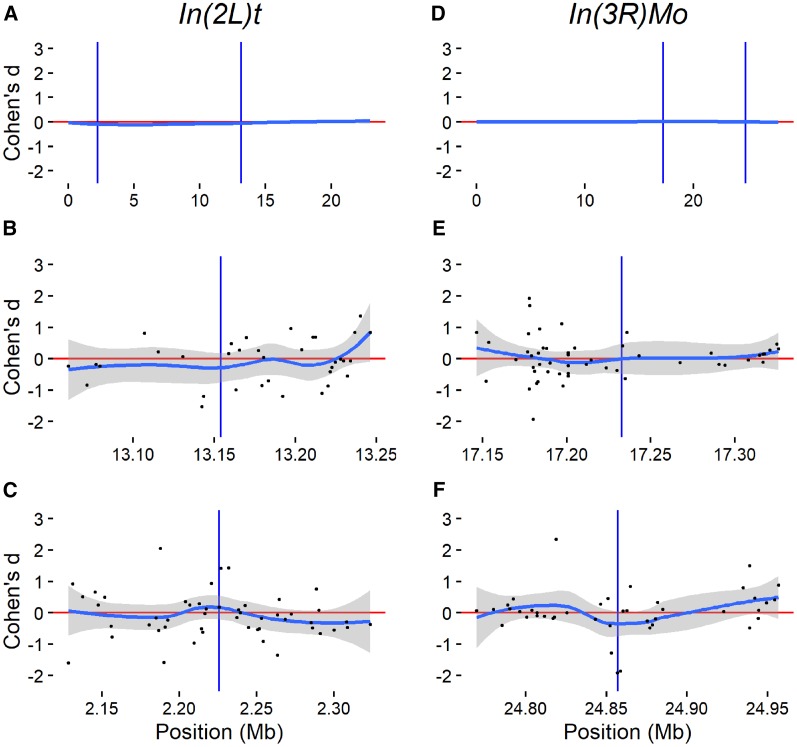
Nucleotide sequence variation at each breakpoint of an inversion can truncate transcribed gene regions and disassociate transcribed regions from TF binding sites and other regulatory elements. Truncation most likely leads to downregulation of genes and rearrangement of regulatory elements, and can lead to up- or downregulation of genes at either breakpoint. The closest IAL to any of the four breakpoints are near the distal breakpoint of In(3R)Mo. CG1951 and βGalNAcTB are within 2.5 kb inside and outside of the inversion region, respectively, and Ssl2 is interrupted by the breakpoint. All three are downregulated in the presence of the inversion. The next closest IAL is 24 kb away. The closest IAL to the proximal breakpoint of In(3R)Mo are 32 kb away. It is also important to note that loci immediately surrounding the proximal breakpoint are not transcriptionally affected by the rearrangement, so the presumed disassociated regulatory elements from the distal end are not altering expression of loci at the proximal end. The closest IAL to the proximal and distal breakpoints of In(2L)t are 34 and 37 kb away, respectively, and thus probably too far to have been affected by direct effects of the breakpoint.
Regional effect of chromosomal rearrangement
We tested for regional effects of chromosomal rearrangement by looking for over- or underrepresentation of genes in LD with the inversion as IAL. We did observe more IAL than expected in LD with each inversion (Table 4) and a trend of transcriptional downregulation across the In(2L)t region (Figure 3, A and D), but note that the effect size is relatively small. Near the breakpoints, patterns of inversion effect direction are generally small and not likely significantly different from zero (Figure 3, B, C, E, and F). The pattern of downregulation immediately surrounding the distal breakpoint of In(3R)Mo is moderate and appears to be driven by three loci: CG1951, Ssl2, and β4GalNAcTB. This is likely a direct effect of the inversion on Ssl2, as the breakpoint interrupts this gene proximal to CG1951 (Corbett-Detig et al. 2012), and possibly disassociation of regulatory elements from coding sequence with respect to CG1951 and β4GalNAcTB.
Figure 3.
Cohen’s d of inversion effect for probe sets by location. Cohen’s d for In(2L)t effect across chromosome 2L (A) and for In(3R)Mo effect across chromosome 3R (D). Loess curves (polynomial line) and 95% C.I.s (shaded area) for 200 kb surrounding each breakpoint: In(2L)t (B) proximal and (C) distal, and In(3R)Mo proximal (E) and distal (F). Inversion breakpoints are depicted as vertical lines. Loess curves and C.I.s generated by (geom_smooth) function in R package (ggplot2). Positive values of Cohen’s d represent increased transcript levels associated with the inverted chromosome state and negative values represent decreased transcript levels.
We also examined whether IAL tend to cluster together by physical location along chromosomes, which could arise from more localized regional effects. We measured physical clustering as the CV of distances between genes, for the global CV, and between IAL by chromosome arm. For each arm, IAL for both inversions were less clustered than the distribution of all genes, although In(3R)Mo IAL are more clustered on chromosome 3R than we would expect for the same number of randomly drawn genes, but are still less clustered than the genomic background generally (Table 6).
Table 6. Gene location dispersion as described by the coefficient of variance (CV).
| In(2L)t | Genome | In(3R)Mo | |||
|---|---|---|---|---|---|
| 95% C.I. | Obs | Obs | Obs | 95% C.I. | |
| chr2L | 0.882–1.366 | 1.338 | 2.612 | 1.417 | 0.834–1.305 |
| chr2R | 0.834–1.341 | 1.066 | 3.11 | 1.176 | 0.853–1.287 |
| chr3L | 0.806–1.352 | 0.979 | 2.812 | 1.129 | 0.829–1.278 |
| chr3R | 0.786–1.341 | 1.015 | 2.681 | 1.676 | 1.07–1.312 |
| chrX | 0.724–1.399 | 1.152 | 2.814 | 0.944 | 0.734–1.295 |
Observed (Obs) genome CV’s calculated from loci in both FlyBase ver.5.49 and Affymetrix Drosophila2 r35 annotation databases by chromosome arm. See Materials and Methods for details. chr, chromosome; CV, coefficient of variance.
Functional analysis
Coadapted alleles segregating with an inversion should also be in LD with the inversion breakpoints. We used gProfiler g:GOSt functional profiling to detect overrepresentation of functional groups in sets of IAL. The sets of IAL that we analyzed were those IAL in LD with the inversion (Table 2) or targets of another gene with variation segregating with the inversion (Table 4). Functional analysis of IAL for each inversion yielded significant groups only when considering all In(3R)Mo IAL or only IAL where inversion state explains ≥ 15% of expression variance (Supplemental Material). Sterol transport is significant in both cases (P = 0.022 for all, P = 0.000116 for ≥ 15% variance) and catalytic activity term (GO:0003824) is significant when considering all IAL (P = 0.000146). We found no significant functional groups when considering any similar grouping of In(2L)t IAL.
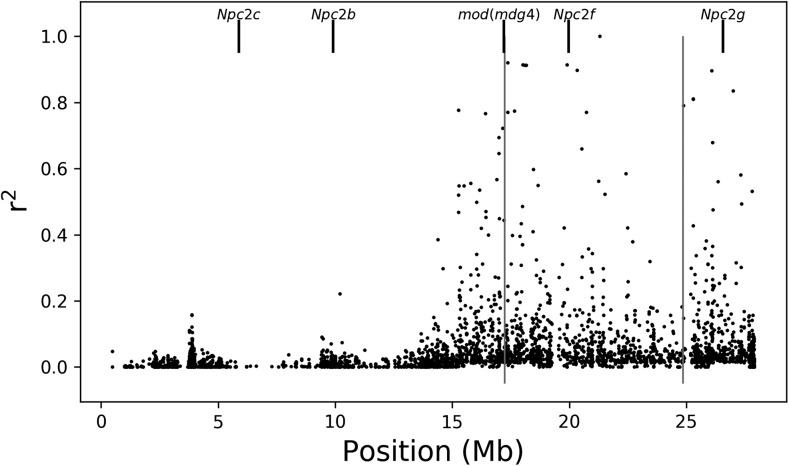
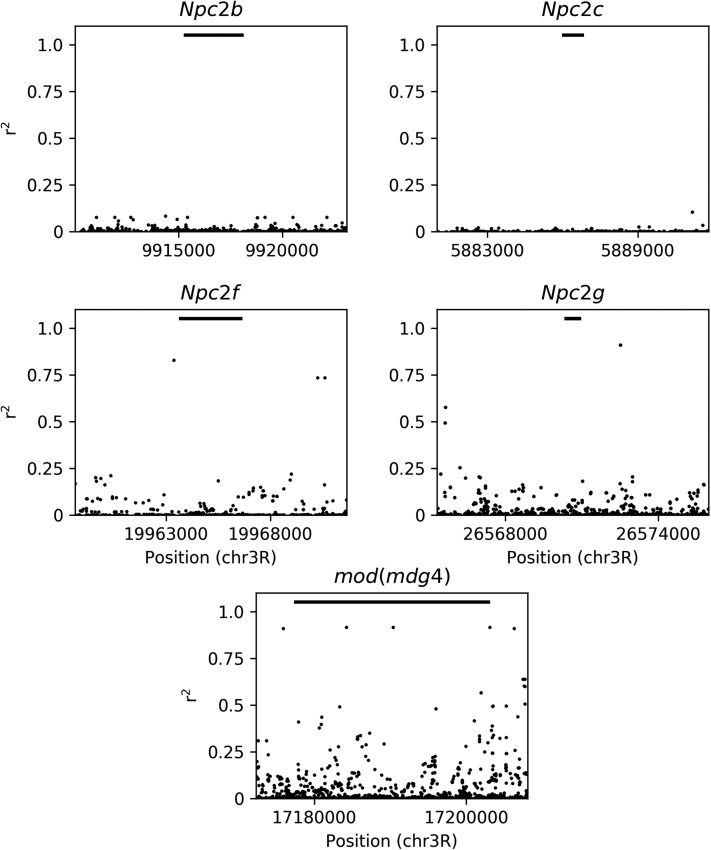
The sterol transport group found to be enriched among In(3R)Mo IAL includes four of the eight Niemann–Pick type 2 orthologs (Npc2b, Npc2c, Npc2f, and Npc2g) (Huang et al. 2007), along with Apoltp. All five genes are upregulated in association with the inverted arrangement, suggesting an increase of sterol uptake in In(3R)Mo-bearing flies. In the context of the expression samples, most of these genes are preferentially expressed in the adult gut (modENCODE, Contrino et al. 2012). While the four NPC2s are on chromosome 3R (Figure 4), Apoltp is on chromosome 2L, and none of these genes contain a SNP in the gene region that is in significant LD with In(3R)Mo, or each other, and only Npc2f is within the inversion region (Figure 5). It is possible that the significant upregulation of the sterol transport group is a downstream effect of mod(mdg4), which is an IAL near the proximal breakpoint (Figure 4) and in LD with In(3R)Mo (Figure 5). mod(mdg4) is a chromatin protein associated with Npc2b as well as other chromatin proteins and TFs associated with all eight Npc2 orthologs (Supplemental Material). We address this exciting finding in further detail in the Discussion.
Figure 4.
Location of Npc2s and mod(mdg4) on chromosome 3R. Long vertical lines represent breakpoints of In(3R)Mo. Only mod(mdg4) contains a single nucleotide polymorphism in linkage disequilibrium with In(3R)Mo.
Figure 5.
Correlation of single nucleotide polymorphism (SNPs) with In(3R)Mo by gene. All SNPs within gene regions and within 5 kb up- and downstream of each gene: Npc2b, Npc2c, Npc2f, Npc2g, and mod(mdg4). Horizontal bars on each plot represent the gene region for that gene.
Discussion
Despite decades of research on polymorphic inversions in D. melanogaster, and despite an overwhelming consensus that inversions are maintained due to selection, we have little understanding of the targets of selection in these inversions that lead to their maintenance (cf. Kirkpatrick and Kern 2012). Here, we examine the role of transcriptional variation induced by cosmopolitan inversions to explore what effect, if any, inversions might have on gene expression that itself might be selectively favored. Besides gross rearrangement effects, we examined gene disruption at inversion breakpoints, IAL in LD with the inversion, and IAL not in LD with the inversion. We assumed multiple functionally-linked IAL to be potentially coadapted so long as they contained SNPs in LD with the inverted arrangement. We also assumed that trans-inversion effects, IAL not in LD with the inversion, could be the result of these loci interacting with TFs in LD with the inversion or epistatic interactions with loci in LD. We note that while sample size of In(2L)t-bearing individuals in the expression analysis is small (8 of 136), we were still able to detect a relatively large number of significant loci across the genome. We believe that our methods were conservative and limit the number of false positives at the expense of a likely high number of false negatives. We argue that we are accounting for the small sample sizes in our analyses and interpretations. We also note that we could not address over- or underdominance in this study as we examined only lines known to be homozygous for either arrangement of In(2L)t or In(3R)Mo.
Of particular concern was what role, if any, expression correlation had on our findings. To explore this, we considered pairwise correlation coefficients and previously described expression modules (Ayroles et al. 2009). One could imagine that the true false discovery rate would be much higher than anticipated due to expression correlation of multiple loci from the same expression module. We found that IAL for both inversions occupied more expression modules than we would expect when drawing genes at random. Even when considering just the pairwise expression correlation, we found that IAL for both inversions were slightly less correlated on average than the genome-wide mean. These observations suggest that the inversion effects arose independently of the underlying correlation structure.
We found many loci of significant effect, yet most have a modest contribution to expression variation. Importantly, the only evidence of direct structural influence on patterns of expression was at a single breakpoint that appears to be due to the inversion mutation interrupting gene regions but not moving genes to a different region of the chromosome. That is, there was no appreciable pattern of up- or downregulation of genes along the chromosome with respect to the location of inversion breakpoints. Perhaps unsurprisingly, we did find an overabundance of expression perturbation within the inversion regions themselves. This would suggest that the effect is possibly due to genetic, rather than the structural, variation in LD with the inversion. That is, transcription variation associated with the inversion is due to genetic variation at the gene level and not the rearrangement of loci.
Inversion polymorphism can be maintained by reduced recombination between locally coadapted alleles within inversions (Dobzhansky and Dobzhansky 1970), both with (Nei et al. 1967; Pepper 2003) or without epistasis (Kirkpatrick and Barton 2006). Clinal inversion variation in D. melanogaster within In(3R)Payne in Australian populations and In(3R)Payne, In(2R)NS, and In(2L)t in American populations has been shown to be due to selection rather than demographic history and indicative of coadaptation (Kennington et al. 2006; Kolaczkowski et al. 2011; Kapun et al. 2016). We reasoned that one might be able to detect epistatic coadaptation through functional analysis of IAL. Coadaptation of noninteracting loci would likely result in either no significant functional groups or multiple, unrelated significant groups. We found functional annotation enrichment only when we considered all IALs with respect to In(3R)Mo. This would suggest that either one locus or a few coadapted, but noninteracting, loci segregate with these inversions to maintain polymorphic inversions. However, it is still possible that coadapted loci could be overlooked in our analysis; certainly, the existing annotation is incomplete. Moreover, our statistical power to detect IAL suffers due to the constraints of the number of inversions captured in the DGRP dataset. Nevertheless, our observation that there are multiple IAL within the inversion argues strongly for the role that inversions play as modifiers of recombination, which may hold adaptive haplotypes together.
Our strongest functional finding, that sterol uptake associated with In(3R)Mo, appears to be driven by genetic variation in a single locus as a trans-inversion effect. Four of the five genes in this cluster are located on chromosome 3R; however, none of these genes have a SNP in significant LD with In(3R)Mo, or with each other (Supplemental Material), and only one is found within the inversion region (Npc2f). This would rule out effective coadaptation of these genes and suggest that the location of these genes on the same chromosome as the inversion is a coincidence. Assuming that the upstream effector of the sterol transport is a TF, it could contain a SNP that alters either its protein-coding sequence or expression. Either scenario would require that the genetic variant responsible for the upregulation of the four Npc2s in question would have to be in LD with In(3R)Mo. Only one TF, mod(mdg4), annotated in DroIDb as interacting with any (Npc2b) of the five sterol transport IAL, contains a SNP in LD with In(3R)Mo. Furthermore, mod(mdg4) is itself an IAL and is also located near the proximal breakpoint of In(3R)Mo. We speculate that inversion-associated expression variation detected in this functional group is under the control of mod(mdg4).
Increased sterol uptake fits nicely with the positive correlation of In(3R)Mo frequency with latitude (Kapun et al. 2014). Npc2 genes control sterol homeostasis via uptake of dietary sterols in D. melanogaster (Huang et al. 2007; Carvalho et al. 2010; Niwa et al. 2011). Two species of Drosophila differentially express Npc2 in response to cold acclimation, though the patterns differ between the two (Parker et al. 2015). Increased dietary cholesterol increases cold tolerance in D. melanogaster (Shreve et al. 2007); however, D. melanogaster takes up phytosterols more efficiently than cholesterol (Cooke and Sang 1970). Furthermore, D. melanogaster is a sterol auxotroph (Carvalho et al. 2010; Niwa et al. 2011) that utilizes dietary sterols preferentially to biosynthesizing different sterols from dietary sterols (Carvalho et al. 2012). This would suggest that In(3R)Mo carrying D. melanogaster could be cold-acclimated due to increased uptake of dietary sterols, rather than the upregulation of cholesterol production.
It is difficult to interpret results for In(2L)t without a clear functional annotation group associated with the inverted state. Assuming that In(2L)t is under selection (Kapun et al. 2016), the simplest interpretation is that In(2L)t polymorphism is maintained by only a small number of loci in LD with the inversion. It is possible that one or more loci in LD with In(2L)t contain protein-coding variation under selection and no appreciable transcript abundance variation with respect to chromosomal arrangement. We note that there are only 14 IAL in LD with In(2L)t (Supplemental Material). While a lack of a significant functional group may be dissatisfying, this does provide a manageable candidate list for validation of single targets.
Conclusions
We found that two different cosmopolitan inversions in D. melanogaster have some effect on the expression of hundreds of genes across the genome. While we caution that our sample sizes, particularly for In(2L)t, are very small, the permutation approach that we have taken is conservative. The genetic variation responsible for the observed transcriptional variation is only in small part due to the inversion event itself, with the majority of the variation being the result of either allelic variation in LD with the inversions, trans-effects of regulators that also are in LD with the inversions, or as of yet uncharacterized, indirect effects of the inversions. Our results mirror those of a recent report on transcriptional variation caused by inversions in D. pseudoobscura (Fuller et al. 2016). Fuller et al. (2016) demonstrated quite convincingly that the well-studied polymorphic inversions of D. pseudoobscura are modulating levels of transcription at multiple life history stages and even found hints of trans-effects. However, due to experimental design constraints, they could not examine inversion effects on unlinked chromosomes. Our findings extend this pattern to D. melanogaster and show that inversions are affecting loci genome-wide. While we have begun to parse the possible causes of IAL, our focus on available data limits the statistical power of our study. Thus, it will be important to conduct carefully designed experiments on inversion polymorphism in D. melanogaster to elucidate the true extent of influence of inversions on genome-wide patterns of transcription.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi: 10.1534/g3.117.1133/-/DC1.
Acknowledgments
We thank Dan Schrider, Steve Schaeffer, and two anonymous reviewers for feedback on the manuscript. A.D.K. and E.L. were supported in part by National Institutes of Health award no. R01GM078204.
Footnotes
Communicating editor: H. Salz
Literature Cited
- Anderson A. R., Hoffmann A. A., McKechnie S. W., Umina P. A., Weeks A. R., 2005. The latitudinal cline in the In(3R)Payne inversion polymorphism has shifted in the last 20 years in Australian Drosophila melanogaster populations. Mol. Ecol. 14: 851–858. [DOI] [PubMed] [Google Scholar]
- Anderson W. W., Dobzhansky T., Kastritsis C. D., 1967. Selection and inversion polymorphism in experimental populations of Drosophila pseudoobscura initiated with the chromosomal constitutions of natural populations. Evolution 21: 664–671. [DOI] [PubMed] [Google Scholar]
- Andolfatto P., Wall J. D., Kreitman M., 1999. Unusual haplotype structure at the proximal breakpoint of In(2L)t in a natural population of Drosophila melanogaster. Genetics 153: 1297–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfatto P., Depaulis F., Navarro A., 2001. Inversion polymorphisms and nucleotide variability in Drosophila. Genet. Res. 77: 1–8. [DOI] [PubMed] [Google Scholar]
- Aulard S., David J. R., Lemeunier F., 2002. Chromosomal inversion polymorphism in Arotropical populations of Drosophila melanogaster. Genet. Res. 79: 49–63. [DOI] [PubMed] [Google Scholar]
- Ayroles J. F., Carbone M. A., Stone E. A., Jordan K. W., Lyman R. F., et al. , 2009. Systems genetics of complex traits in Drosophila melanogaster. Nat. Genet. 41: 299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergland A. O., Tobler R., González J., Schmidt P., Petrov D., 2016. Secondary contact and local adaptation contribute to genome-wide patterns of clinal variation in Drosophila melanogaster. Mol. Ecol. 25: 1157–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho M., Schwudke D., Sampaio J. L., Palm W., Riezman I., et al. , 2010. Survival strategies of a sterol auxotroph. Development 137: 3675–3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho M., Sampaio J. L., Palm W., Brankatschk M., Eaton S., et al. , 2012. Effects of diet and development on the Drosophila lipidome. Mol. Syst. Biol. 8: 600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers G. K., 1991. Gene expression, adaptation and evolution in higher organisms. Evidence from studies of Drosophila alcohol dehydrogenases. Comp. Biochem. Physiol. B. 99: 723–730. [DOI] [PubMed] [Google Scholar]
- Cogni R., Kuczynski K., Koury S., Lavington E., Behrman E. L., et al. , 2017. On the long-term stability of clines in some metabolic genes in Drosophila melanogaster. Sci. Rep. 7: 42766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J., 1988. Statistical Power Analysis for the Behavioral Sciences, Ed. 2. Lawrence Erlbaum Associates, Hillsdale, NJ. [Google Scholar]
- Contrino S., Smith R. N., Butano D., Carr A., Hu F., et al. , 2012. modMine: flexible access to modENCODE data. Nucleic Acids Res. 40: D1082–D1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke J., Sang J. H., 1970. Utilization of sterols by larvae of Drosophila melanogaster. J. Insect Physiol. 16: 801–812. [DOI] [PubMed] [Google Scholar]
- Corbett-Detig R. B., Hartl D. L., 2012. Population genomics of inversion polymorphisms in Drosophila melanogaster. PLoS Genet. 8: e1003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett-Detig R. B., Cardeno C., Langley C. H., 2012. Sequence-based detection and breakpoint assembly of polymorphic inversions. Genetics 192: 131–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong G., Bochdanovits Z., 2003. Latitudinal clines in Drosophila melanogaster: body size, allozyme frequencies, inversion frequencies, and the insulin-signalling pathway. J. Genet. 82: 207–223. [DOI] [PubMed] [Google Scholar]
- Dobzhansky T., 1947. Genetics of natural populations. Xiv. A response of certain gene arrangements in the third chromosome of Drosophila pseudoobscura to natural selection. Genetics 32: 142–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T., Dobzhansky T. G., 1970. Genetics of the Evolutionary Process. Columbia University Press, New York. [Google Scholar]
- Dobzhansky T., Sturtevant H., 1938. Inversions in the chromosomes of Drosophila pseudoobscura. Genetics 23: 28–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuk L., 2010. Inversion variants in the human genome: role in disease and genome architecture. Genome Med. 2: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouet C., Gray E., Besansky N. J., Costantini C., 2012. Adaptation to aridity in the malaria mosquito Anopheles gambiae: chromosomal inversion polymorphism and body size influence resistance to desiccation. PLoS One 7: e34841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser A., Burnell D., Miller D., 1966. Simulation of genetic systems X. Inversion polymorphism. J. Theor. Biol. 13: 1–14. [Google Scholar]
- Fraser H. B., 2013. Gene expression drives local adaptation in humans. Genome Res. 23: 1089–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller Z. L., Haynes G. D., Richards S., Schaeffer S. W., 2016. Genomics of natural populations: how differentially expressed genes shape the evolution of chromosomal inversions in Drosophila pseudoobscura. Genetics 204: 287–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane J. B. S., 1957. The conditions for coadaptation in polymorphism for inversions. J. Genet. 55: 218–225. [Google Scholar]
- Hoffmann A. A., Sgrò C. M., Weeks A. R., 2004. Chromosomal inversion polymorphisms and adaptation. Trends Ecol. Evol. 19: 482–488. [DOI] [PubMed] [Google Scholar]
- Hoffmann A. A., Shirriffs J., Scott M., 2005. Relative importance of plastic vs. genetic factors in adaptive differentiation: geographical variation for stress resistance in Drosophila melanogaster from eastern Australia. Funct. Ecol. 19: 222–227. [Google Scholar]
- Houle D., Márquez E. J., 2015. Linkage disequilibrium and inversion-typing of the Drosophila melanogaster genome reference panel. G3 (Bethesda) 5: 1695–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Massouras A., Inoue Y., Peiffer J., Ràmia M., et al. , 2014. Natural variation in genome architecture among 205 Drosophila melanogaster Genetic Reference Panel lines. Genome Res. 24: 1193–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Carbone M. A., Magwire M. M., Peiffer J. A., Lyman R. F., et al. , 2015. Genetic basis of transcriptome diversity in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 112: E6010–E6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Warren J. T., Buchanan J., Gilbert L. I., Scott M. P., 2007. Drosophila Niemann-Pick type C-2 genes control sterol homeostasis and steroid biosynthesis: a model of human neurodegenerative disease. Development 134: 3733–3742. [DOI] [PubMed] [Google Scholar]
- Kapun M., van Schalkwyk H., McAllister B., Flatt T., Schlötterer C., 2014. Inference of chromosomal inversion dynamics from Pool-Seq data in natural and laboratory populations of Drosophila melanogaster. Mol. Ecol. 23: 1813–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapun M., Fabian D. K., Goudet J., Flatt T., 2016. Genomic evidence for adaptive inversion clines in Drosophila melanogaster. Mol. Biol. Evol. 33: 1317–1336. [DOI] [PubMed] [Google Scholar]
- Kennington W. J., Partridge L., Hoffmann A. A., 2006. Patterns of diversity and linkage disequilibrium within the cosmopolitan inversion In(3R)Payne in Drosophila melanogaster are indicative of coadaptation. Genetics 172: 1655–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd J. M., Graves T., Newman T. L., Fulton R., Hayden H. S., et al. , 2010. A human genome structural variation sequencing resource reveals insights into mutational mechanisms. Cell 143: 837–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M., Barton N., 2006. Chromosome inversions, local adaptation and speciation. Genetics 173: 419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M., Kern A., 2012. Where’s the money? Inversions, genes, and the hunt for genomic targets of selection. Genetics 190: 1153–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knibb W. R., Oakeshott J. G., Gibson J. B., 1981. Chromosome inversion polymorphism in Drosophila melanogaster. I. Latitudinal clines and associations between inversions in Australian populations. Genetics 98: 833–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolaczkowski B., Kern A. D., Holloway A. K., Begun D. J., 2011. Genomic differentiation between temperate and tropical Australian populations of Drosophila melanogaster. Genetics 187: 245–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesnikov N., Hastings E., Keays M., Melnichuk O., Tang Y. A., et al. , 2015. ArrayExpress update–simplifying data submissions. Nucleic Acids Res. 43: D1113–D1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimbas C. B., Powell J. R., 1992. Drosophila Inversion Polymorphism. CRC Press, Boca Raton, FL. [Google Scholar]
- Lakich D., Kazazian H. H., Antonarakis S. E., Gitschier J., 1993. Inversions disrupting the factor VIII gene are a common cause of severe haemophilia A. Nat. Genet. 5: 236–241. [DOI] [PubMed] [Google Scholar]
- Langley C. H., Stevens K., Cardeno C., Lee Y. C. G., Schrider D. R., et al. , 2012. Genomic variation in natural populations of Drosophila melanogaster. Genetics 192: 533–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levene H., Dobzhansky T., 1958. New evidence of heterosis in naturally occurring inversion heterozygotes in Drosophila pseudoobscura. Heredity (Edinb) 12: 37–49. [Google Scholar]
- López-Maury L., Marguerat S., Bähler J., 2008. Tuning gene expression to changing environments: from rapid responses to evolutionary adaptation. Nat. Rev. Genet. 9: 583–593. [DOI] [PubMed] [Google Scholar]
- Lowry D. B., Willis J. H., 2010. A widespread chromosomal inversion polymorphism contributes to a major life-history transition, local adaptation, and reproductive isolation. PLoS Biol. 8: e1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T. F. C., Richards S., Stone E. A., Barbadilla A., Ayroles J. F., et al. , 2012. The Drosophila melanogaster Genetic Reference Panel. Nature 482: 173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massouras A., Waszak S. M., Albarca-Aguilera M., Hens K., Holcombe W., et al. , 2012. Genomic variation and its impact on gene expression in Drosophila melanogaster. PLoS Genet. 8: e1003055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzkin L. M., Merritt T. J. S., Zhu C.-T., Eanes W. F., 2005. The structure and population genetics of the breakpoints associated with the cosmopolitan chromosomal inversion In(3R)Payne in Drosophila melanogaster. Genetics 170: 1143–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Kojima K. I., Schaffer H. E., 1967. Frequency changes of new inversions in populations under mutation-selection equilibria. Genetics 57: 741–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa R., Niwa Y. S., Niwa R., Niwa Y. S., 2011. The fruit fly Drosophila melanogaster as a model system to study cholesterol metabolism and homeostasis. Cholesterol 2011: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker D. J., Vesala L., Ritchie M. G., Laiho A., Hoikkala A., et al. , 2015. How consistent are the transcriptome changes associated with cold acclimation in two species of the Drosophila virilis group? Heredity (Edinb) 115: 13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper J. W., 2003. The evolution of evolvability in genetic linkage patterns. Biosystems 69: 115–126. [DOI] [PubMed] [Google Scholar]
- Pool J. E., Corbett-Detig R. B., Sugino R. P., Stevens K. A., Cardeno C. M., et al. , 2012. Population genomics of sub-Saharan Drosophila melanogaster: African diversity and non-African admixture. PLoS Genet. 8: e1003080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team , 2013. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Reimand J., Arak T., Adler P., Kolberg L., Reisberg S., et al. , 2016. g:Profiler-a web server for functional interpretation of gene lists (2016 update). Nucleic Acids Res. 44: W83–W89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt J. A., Kolaczkowski B., Jones C. D., Begun D. J., Kern A. D., 2014. Parallel geographic variation in Drosophila melanogaster. Genetics 197: 361–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrider D. R., Hahn M. W., Begun D. J., 2016. Parallel evolution of copy-number variation across continents in Drosophila melanogaster. Mol. Biol. Evol. 33: 1308–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sezgin E., Duvernell D. D., Matzkin L. M., Duan Y., Zhu C.-T., et al. , 2004. Single-locus latitudinal clines and their relationship to temperate adaptation in metabolic genes and derived alleles in Drosophila melanogaster. Genetics 168: 923–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shreve S. M., Yi S. X., Lee R. E., 2007. Increased dietary cholesterol enhances cold tolerance in Drosophila melanogaster. Cryo Letters. 28: 33–37. [PubMed] [Google Scholar]
- Sokal R. R., Rohlf F. J., 1995. Biometry. W. H. Freeman, San Francisco. [Google Scholar]
- Stalker H. D., 1976 Chromosome studies in wild populations of D. melanogaster. Genetics 82: 323–47. [DOI] [PMC free article] [PubMed]
- Stalker H. D., 1980 Chromosome studies in wild populations of Drosophila melanogaster. II. Relationship of inversion frequencies to latitude, season, wing-loading and flight activity. Genetics 95: 211–223. [DOI] [PMC free article] [PubMed]
- Stefansson H., Helgason A., Thorleifsson G., Steinthorsdottir V., Masson G., et al. , 2005. A common inversion under selection in Europeans. Nat. Genet. 37: 129–137. [DOI] [PubMed] [Google Scholar]
- Storey J. D., Tibshirani R., 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100: 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevart A. H., 1921. A case of rearrangement of genes in Drosophila. Proc. Natl. Acad. Sci. USA 7: 235–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant A. H., Mather K., 1938. The interrelations of unversions, heterosis and recombination. Am. Nat. 72: 447–452. [Google Scholar]
- Telonis-Scott M., Sgrò C. M., Hoffmann A. A., Griffin P. C., 2016. Cross-study comparison reveals common genomic, network, and functional signatures of desiccation resistance in Drosophila melanogaster. Mol. Biol. Evol. 33: 1053–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobari Y. N., Kojima K. I., 1967. Selective modes associated with inversion karyotypes in Drosophila ananassae. I. Frequency-dependant selection. Genetics 57: 179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umina P. A., Weeks A. R., Kearney M. R., McKechnie S. W., Hoffmann A. A., 2005. A rapid shift in a classic clinal pattern in Drosophila reflecting climate change. Science 308: 691–693. [DOI] [PubMed] [Google Scholar]
- Van ’t Land J., Van Putten W. F., Villarroel H., Kamping A., Van Delden W., 2000. Latitudinal variation for two enzyme loci and an inversion polymorphism in Drosophila melanogaster from Central and South America. Evolution 54: 201–209. [PubMed] [Google Scholar]
- Vonesch S. C., Lamparter D., Mackay T. F. C., Bergmann S., Hafen E., et al. , 2016. Genome-wide analysis reveals novel regulators of growth in Drosophila melanogaster. PLoS Genet. 12: e1005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks A. R., McKechnie S. W., Hoffmann A. A., 2002. Dissecting adaptive clinal variation: markers, inversions and size/stress associations in Drosophila melanogaster from a central field population. Ecol. Lett. 5: 756–763. [Google Scholar]
- Wesley C. S., Eanes W. F., 1994. Isolation and analysis of the breakpoint sequences of chromosome inversion In(3L)Payne in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 91: 3132–3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Custom scripts, data, and analysis results can be found online at https://github.com/kern-lab/lavingtonKern, including file descriptions in the AnalysisFiles.readme document.