Abstract

Direct analysis by mass spectrometry (imaging) has become increasingly deployed in preclinical and clinical research due to its rapid and accurate readouts. However, when it comes to biomarker discovery or histopathological diagnostics, more sensitive and in-depth profiling from localized areas is required. We developed a comprehensive, fully automated online platform for high-resolution liquid extraction surface analysis (HR-LESA) followed by micro–liquid chromatography (LC) separation and a data-independent acquisition strategy for untargeted and low abundant analyte identification directly from tissue sections. Applied to tissue sections of rat pituitary, the platform demonstrated improved spatial resolution, allowing sample areas as small as 400 μm to be studied, a major advantage over conventional LESA. The platform integrates an online buffer exchange and washing step for removal of salts and other endogenous contamination that originates from local tissue extraction. Our carry over–free platform showed high reproducibility, with an interextraction variability below 30%. Another strength of the platform is the additional selectivity provided by a postsampling gas-phase ion mobility separation. This allowed distinguishing coeluted isobaric compounds without requiring additional separation time. Furthermore, we identified untargeted and low-abundance analytes, including neuropeptides deriving from the pro-opiomelanocortin precursor protein and localized a specific area of the pituitary gland (i.e., adenohypophysis) known to secrete neuropeptides and other small metabolites related to development, growth, and metabolism. This platform can thus be applied for the in-depth study of small samples of complex tissues with histologic features of ∼400 μm or more, including potential neuropeptide markers involved in many diseases such as neurodegenerative diseases, obesity, bulimia, and anorexia nervosa.
Mass spectrometry imaging (MSI) is being used more often in preclinical and clinical research due to its many advantages over conventional imaging techniques.1−3 MSI offers the possibility to correlate distribution maps of multiple molecular species simultaneously with histological and clinical features without labeling. This methodology enables the discovery of potential diagnostic and prognostic markers of diseases in a single experiment. With these unique features, MSI opens new doors for molecular-driven pathology, in various fields of histopathological diagnostics, such as identification and grading of tumors.4,5 MSI has also proven to be a powerful tool in drug discovery as it gives insights not only on drug and metabolite distribution and site(s) of action but also about the biology present at the sites of drug localization relating to treatment efficacy.6
Matrix-assisted laser desorption/ionization (MALDI) is by far the most popular ionization technique used to map the tissue microenvironment. In this way, molecular classification can be accurately established with different tissue types, including tumors, to improve the accuracy of diagnosis and characterize tumor heterogeneity.7 MALDI-MSI allows in situ tissue characterization at different molecular “omics” levels—metabolomics, lipidomics, peptidomics, and proteomics8—and has found numerous clinical applications, including oncology,9 psychiatric and neurodegenerative disorders,10−12 cardiovascular diseases,13,14 ophthalmology,15 and joint and cartilage-related disorders.16 Extensive efforts focused on instrument development to improve spatial resolution, throughput, and sensitivity have placed MALDI-MSI in a competitive position for clinical studies where knowledge about the tumor microenvironment is critical and hundreds of samples require analysis.17−19 One of the drawbacks of MALDI-MSI is the need for the application of homogeneous layers of MALDI matrix on the tissue surface and the ion suppression resulting from the extraction process. This is in part due to the lack of separation technology in an imaging experiment and can degrade the sensitivity and quality of data generated.
Recently, ambient ionization techniques have proven their suitability to extract relevant information from complex biological matrices.20 The main benefit of these techniques is the possibility to analyze directly surfaces with relatively less complex and time-consuming sample preparation compared to MALDI. This considerably reduces sample handling and speeds up the whole analytical workflow. Desorption electrospray ionization (DESI)21,22 is increasing its application in clinical research as it provides relatively fast, sensitive, and complementary analysis to MALDI-MSI.23−29 DESI allows rapid classification of human tumors based on tissue-specific lipid molecular profiling.30 One of the most promising applications would be to use the information at the time of surgery on resected specimens to guide surgical resections that could improve management of patients.31 Liquid extraction surface analysis (LESA), based on a liquid microjunction (LMJ) surface sampling, was introduced in 2008 and has been employed for profiling biological matrices from localized tissue area in various studies, including drug distribution and metabolism,32−36 microbiology,37 small molecule antibody–drug conjugate catabolites,38 lipidomics,32,39 proteomics,40−42 and native mass spectrometry for nonconvalent complex studies.43,44 Although often criticized for the poor spatial resolution achievable (i.e., 1.2–2.0 mm with 1 μL of solvent deposited on the top of the surface), LESA is an excellent tool for conducting profiling experiments from a selected spot, such as quick metabolite screening or parent drug localization to an organ. Complementary to quantitative whole-body autoradiography that provides distribution information on radiolabeled material, LESA followed by MS detection enables differentiation of a parent drug from its metabolites.
Chromatography-free approaches create ionization suppression effects, which enables the identification of analytes of high abundance45 but limits protein/peptide identification because of the high degree of cell or tissue complexity. Frequently, compounds are codesorbed; to deal with structural isomers and/or isobaric compounds (e.g., interfering isotopic clusters), chromatography-free approaches often suffer from the absence of separation before the mass analysis. Indeed, high resolution is not adequate to distinguish isomers and in some cases tandem mass spectrometry (MS/MS) does not provide enough selectivity. Therefore, additional information and improved selectivity are required to provide adequate identification of biologically relevant analytes. The addition of a postdesorption and postionization gas-phase ion mobility separation (IMS) after MALDI, DESI, and LESA has demonstrated to resolve isobaric species and reduce chemical noise.29,34,46 However, this method does not account for tissue-specific ionization suppression.45 Because the surface sampling and the ionization processes are resolved both in space and time dimensions, LESA allows for the manipulation of the extracted material in the liquid phase prior to ionization of molecular content.47 As a consequence, LESA can easily incorporate a liquid-based separation after the surface sampling process, which is not possible with MALDI or DESI ionization techniques. Kertesz et al. developed a LMJ-SS approach followed by HPLC-ESI-MS for the analysis of drugs and metabolites in whole-body thin tissue sections, which helps to distinguish isomeric phase II metabolites of propanolol.48 Continuous-flow LMJ-SS coupled online with HPLC/MS also enables the extraction, separation, and detection of proteins and low-molecular-weight compounds (e.g., drugs of abuse) from tissue sections and dried blood spots. Usually, the surface of dried blood spots is sufficiently hydrophobic for maintaining a stable liquid junction, even with high-aqueous-content solvents.49 This innovative LMJ-SS-HPLC-MS/MS approach was also used to investigate the distribution of specific markers within normal human pituitary gland and pituitary adenoma tissue sections, to discriminate between tumor and nontumor tissues.50 However, such a targeted approach requires sample cleanup prior to analysis and will not allow broad screening for potential biomarkers.
In the present work, we describe a platform developed to improve both poor spatial resolution achieved with “conventional” LESA and ionization suppression effect. The platform was modified for high-resolution (HR)-LESA for direct analysis of endogenous peptides from a 400-μm area from preclinical tissue samples.51 The HR-LESA was integrated with an online washing step to remove salts and other contaminants, the key source of ion suppression. Micro-LC (μLC) was used to separate the analytes of interest from endogenous sample matrix compounds, an additional source of ion suppression. This platform allows isomeric separation due to the implementation of μLC further improved by ion mobility. After this μLC separation, the compounds were analyzed by MS, which combines IMS with high-definition (HD) MSE, a data-independent acquisition method. Finally, we automated the entire HR-LESA-μLC-HDMSE platform by implementing new software. This platform is presented here as a complementary addition to the field of mass spectrometry imaging since it opens doors to a more in-depth profiling of spatial extractions of biological tissues.
Experimental Section
Chemicals and Reagents
ULC/MS-grade water, ULC/MS-grade acetonitrile (ACN), and 99% formic acid were purchased from Biosolve (Valkenswaard, NL). Microscopic glass slides were purchased from Thermo Scientific (Braunschweig, DE). Leucine enkephalin standard was provided by a Waters Q-ToF Qualification Standards Kit (Etten-Leur, NL) and prepared at a concentration of 5 ng/μL in ACN/water (50/50; v/v) and used as lock mass.
Murine Tissue Sectioning
Healthy Wistar Han rat pituitary gland tissue samples were provided by the Department of General Surgery of the Maastricht University Medical Center (MUMC+). The fresh-frozen wild type and transgenic APP KM670/671NL/PS1L166P mice were supplied by the Bio-Imaging Lab, University of Antwerp. Tissues were cryo-sectioned (Microtome cryostat Thermo Scientific, Braunschweig, DE) into 12-μm-thick tissue sections and subsequently thaw-mounted on regular microscope glass slides. These tissue sections were stored at −80 °C prior to analysis. Right before the analysis, samples were thawed at room temperature and desiccated for 30 min. Hematoxylin and eosin (H&E) staining was performed on these tissue samples after HR-LESA-μLC-MS analysis.
H&E Staining Protocol
H&E staining was performed on the same sections used for HR-LESA-μLC-HDMSE experiments. After MSI analysis, the residual matrix was gently removed by dipping the glass slides in ethanol for 2 min. Sections were then washed in successive baths (96% EtOH, 70% EtOH, and deionized water, 3 min each). The hematoxylin (Merck, Darmstadt, Germany) staining was then performed for 3 min, and slides were then washed in gently running tap water for 3 min, followed by eosin staining for 30 s, washing under running tap water for 1 min, and finally immersing in 100% EtOH for 2 min. Slides were finally dehydrated in xylene (30 s), covered in Etallen, and coverslipped. The optical images were acquired using a MIRAX Desk scanner (Zeiss, Gottingen, Germany). Images were acquired with a magnification of 40.
Conventional LESA Extraction
The LESA extraction was performed using the automated TriVersa NanoMate Advion robot (Advion, Ithaca, NY). A 0.5-μL volume of the extraction solvent (ACN/water/formic acid, 70/30/0.1 v/v/v) was deposited with a conductive pipet tip onto the tissue section for 5 s repeated 1 times. This extract was directly infused into the mass spectrometer (Waters Synapt G2-Si, USA) using chip-based nano-ESI, by applying a nitrogen gas pressure of 0.3 bar and voltage of 1.40 kV.
HR-LESA Extraction, Online Washing, and μLC Separation
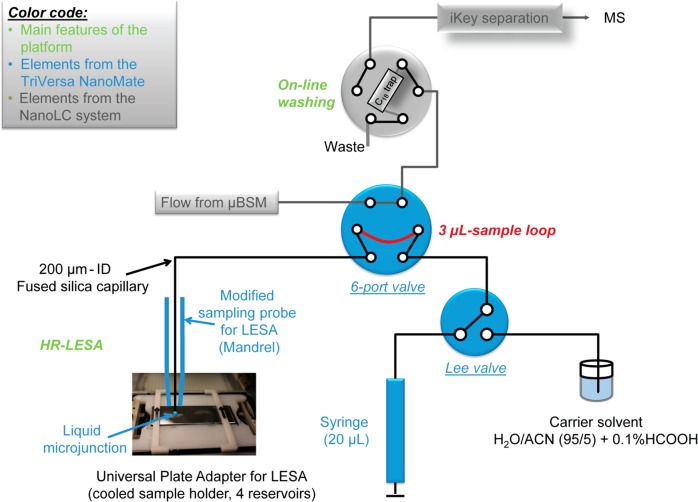
High spatial resolution extraction was performed using the capillary extraction arm usually used for coupling with LC-MS fraction collection. Extraction with this setup was performed with 0.5 μL of extraction solvent (ACN/water/formic acid, 70/30/0.1 v/v/v) after 5 s in contact with the tissue section. An online and automated buffer exchange was then performed, by diluting 10 times the sample with the carrier solvent (ACN/water/formic acid, 5/95/0.1 v/v/v), to ensure compatibility with reversed-phase chromatography, and collected in a 3-μL loop (Figure 1). Under these conditions, the local extraction was achieved at a spatial resolution of 400 μm, and the total workflow from the extraction solvent aspiration to filling in the sample loop takes ∼3 min.
Figure 1.
Schematic representation of flow connections between the elements of the μLC system (in gray) and the automated sampler device (blue). Main features of the analytical platform are indicated in green. The route of the sample loop is shown in red. The 6-port valve is in the “sample loading” position. HR: high spatial resolution. LESA: liquid extraction surface analysis. BSM: binary solvent manager.
Following this extraction, the sample was trapped onto a trap column (ACQUITY UPLC M-class Symmetry C18, 100 Å, 5 μm, 300 μm × 50 mm, Waters, City, ST) and washed with the carrier solvent for 2 min to remove salts and other possible interferences/contamination. After the 2 min online washing, the trap column was back-flushed onto the μLC IonKey column (iKey BEH C18 Separation Device, 130 Å, 1.7 μm, 150 μm × 50 mm, Waters, Milford, MA) for chromatographic separation as follow: a 13 min gradient from 1% to 85% solvent B (ACN/formic acid, 100/0.1) was used for elution of peptides. The column was washed for 3 min at 85% solvent B prior to the column equilibration at 1% solvent B for 5 min. The trap column was equilibrated at 1% solvent B for 4 min.
Mass Spectrometry
All MS experiments were conducted on a Waters Synapt G2-Si system operated in positive ionization mode (in sensitivity mode). General operating parameters were as follows: capillary voltage = 4 kV; source temperature = 80 °C; sampling cone voltage = 40 V; and a desolvation temperature = 150 °C. The default collision energy was set at 4 eV in full MS scan mode. IMS was performed using nitrogen as a drift gas at a flow rate of 90 mL/min. The TRAP DC entrance was set to 0 V, and the wave height was set to 40 V. The velocity of the IMS wave was used to separate the ions over the total 200 ms. The start velocity was set at 1200 m/s and the end velocity at 400 m/s. Data-independent HDMSE acquisition was conducted for the analysis of the pituitary gland and further identification of extracted endogenous peptides. These measurements were performed in the TRANSFER T-wave using a collision energy ramp from 20 to 45 eV. The detector voltage was set at 2500 V, and data was acquired within a mass range of m/z 50–2000.
Software
The LESA extraction was controlled by a beta version of the LESA Plus software (Advion, UK), and MassLynx 1.4 (Waters, U.S.A.) was used for controlling the online washing step and the μLC separation. Data were processed and visualized using Mass Lynx 1.4 (Waters, U.S.A.) and DriftScope v2.5 (Waters, U.S.A.). The identification of the neuropeptides and proteins was performed using Progenesis QI for proteomics v2.0.5556.29015 (Non Linear Dynamics, U.S.A.). For this identification, a species-specific FASTA file was created, and a nonspecific digest reagent was selected. The amount of missed cleavages was set at three, and the post-translational modifications (PTMs) allowed in this MSE search were N-acetylation, M-oxidation, and C-carbamidomethylation.
Results and Discussion
Here, we describe the HR-LESA-μLC-MS platform and its performance for spatial analysis of neuropeptides. For this purpose, two animal models were used. The first experiments rely on the investigation of neuropeptides in rat pituitary gland tissues, which consists of two different regions with different biological functionalities: the adenohypophysis (anterior lobe and intermediate lobe) and the neurohypophysis (posterior lobe). Therefore, due to the morphology of the tissue (∼2 × 3 mm), local extraction at high spatial resolution is crucial for molecular characterization of both regions. The second set of experiments is performed on wild type and transgenic mice expressing amyloidosis to assess the potential of the platform to study neuropeptides potentially involved in Alzheimer’s disease progression. In this case, the sampling areas include regions with high expression of amyloidosis, such as the cerebral cortex, hippocampus, and cerebellum, which also require precise sampling.
Analytical Platform
Spatial Resolution
In a conventional LESA setup, the extraction is achieved using a conductive pipet tip–which, depending on the solvent composition, can lead to a large sampled area up to 2 mm.52 This is potentially a limiting factor when analyzing small objects such as the anatomical features of the pituitary gland or any other organs that require precise sampling. In our HR-LESA-μLC-MS platform (Figure 1), we modified the extraction system to improve the spatial resolution and reduce the size of the extracted areas. By using a silica capillary instead, we were able to significantly improve the spatial resolution to 400 μm, as assessed under microscopic evaluation of the tissue section after the liquid extraction took place (Figure 2). With an extracted area of 400 μm, the molecular content from the anterior and posterior lobe of the pituitary gland can be accurately extracted. We illustrate the ability to unambiguously distinguish between the adenohypophysis and neurohypophysis of the pituitary in Figure 2. Equal spot sizes were observed from the mouse brain tissue section in the H&E image after HR-LESA-μLC-HDMSE (Figure S-1).
Figure 2.

H&E stained images of the pituitary gland before (a) and after (b) HR-LESA extraction show an improvement in spatial resolution with HR-LESA (i.e., 400 μm area represented by a green dot in b) compared to conventional LESA (area shows the spot size of a typical LESA extraction (i.e., 1000 μm-diameter, red circle in b)). In (b), the arrow indicates the trace after an extraction with the capillary. Anatomical features of the pituitary gland containing the adenohypophysis and the neurohypophysis. Anatomical features: 1. anterior lobe; 2. posterior lobe; 3. intermediate lobe.
Reproducibility of the Extraction
To investigate the extraction reproducibility and sample carry-over of the HR-LESA setup, extraction of a leucine enkephalin standard was performed followed by flow injection analysis. A 5-ng leucine enkephalin solution (prepared in ACN/H2O; 50/50; v/v) was spotted onto a hydrophobic plate and air-dried. The extractions of five consecutive spots using 0.9 μL of 50% ACN + 0.1% HCOOH and the modified HR-LESA extraction system resulted in a coefficient of variation of 30% (based on the surface area from the extracted ion chromatogram at m/z 556.28 (protonated species [LeuEnk + H]+) for each extracted spots; n = 5; Figure S-2).
Integration of Online Clean up between Extraction and Chromatographic Separation
The tissue sample was placed in the sample plate holder as shown in the photograph in Figure 1. This tissue extract was then collected in a 384-well plate and diluted with carrier solvent. The diluted extract was “injected” by the same fused silica capillary, collected in the 3-μL loop, and, after switching the 6-port valve, trapped onto the C18 trap column for an online washing with water/ACN (99/1; v/v) to remove all water-soluble matrix compounds. The trap column was back-flushed to separate the remaining sample on an Ionkey separation device using a reversed phase μLC separation.
Reproducibility of the Chromatographic Separation and Sample Carry-Over
HR-LESA combined with the online washing and μLC separation IonKey system was evaluated. The chromatographic peak of leucine enkephalin was found at a retention time of 16 min. This extraction was performed in triplicate to test the retention time reproducibility followed by two blank extractions to study carry-over. The relative standard deviation of the retention time was <0.12% and the carry-over in the first blank extraction was <1.1% using the absolute peak area, which demonstrates a good reproducibility of chromatographic separation (Figure S-3). The minimal occurrence of sample carry-over was due to the online cleanup of the sample loop during the analysis. In addition to the leucine enkephalin standard extraction, reproducibility of the chromatographic separation of neuropeptides from mouse brain and corresponding carry-over has been investigated. Relative standard deviation values vary from 7.1% to 25.4%, and the sample carry-over is <0.8% (Figures S-4, S-5, and S-6). These values were obtained from three tissue extractions followed by a blank extraction in use of the absolute chromatographic peak area. While the implementation of μLC into our platform significantly minimizes the amount of ionization suppression, the reproducibility could be considerably improved by the addition of an isotope-labeled internal standard to the tissue sample.
Postsurface Sampling Chromatographic Separation
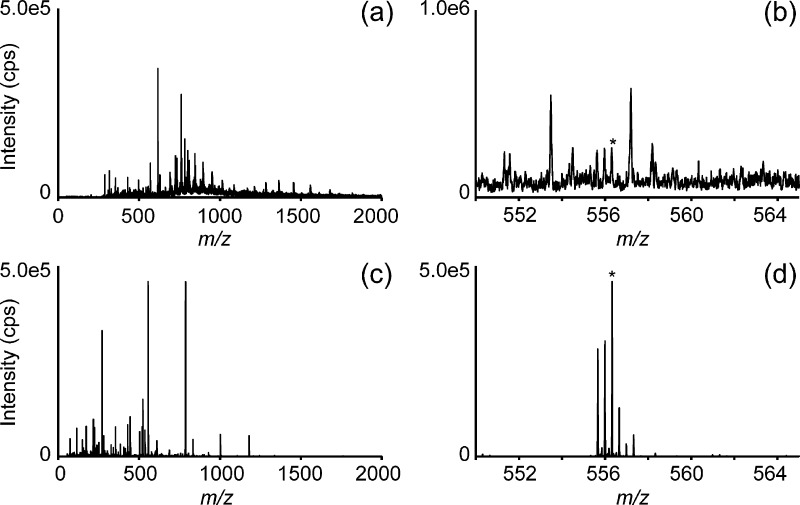
Having demonstrated that the HR-LESA provided higher spatial resolution, good reproducibility, and minimal sample carry-over, we implemented a μLC separation to enable isomeric separation and further sample cleanup. Isomers have the same mass and the same molecular formula, in contrast to isobaric compounds, which have the same nominal mass but differ in molecular formula. MS alone is insufficient to separate and accurately identify isomeric compounds (even with high mass resolution instrumention). Furthermore, ion suppression often occurs and is considered the primary cause of irreproducibility in MS. Because LC is a powerful tool for separation of these isomeric compounds,52 we integrated an online washing step and μLC separation after the surface sampling and prior to electrospray ionization (Figure 1). Compared to conventional LESA, the HR-LESA approach with online washing and μLC separation showed an increase in sensitivity and selectivity for leucine enkephalin in the adenohypophysis region of rat pituitary online (Figure 3). When we performed our HR-LESA-μLC-HDMSE approach for the analysis of pituitary, 67 compounds could be chemically identified, while out of this list (Table S-2) only five and four compounds could be found in the conventional LESA-HDMS and LESA-MS data, respectively. For the mouse brain cerebral cortex analysis, 14 compounds could be identified with HR-LESA-μLC-HDMSE, while out of this list (Table S-4) only three and two compounds were observed in the conventional LESA-HDMS and LESA-MS data, respectively. In view of the even larger spot size of LESA-MS, the minimization of ionization suppression by μLC is crucial for detection of lower abundant compounds. Although the implementation of μLC in our approach requires a longer analysis time (30 min/spot) compared to conventional LESA-MS (1 min/spot), this can be justified due to the enhanced detection limits of the HR-LESA-μLC-HDMSE platform compared to conventional LESA-HDMS and LESA-MS.
Figure 3.
Sensitivity of LESA-MS versus HR-LESA-μLC-MS: The total mass spectrum (a and c) and a zoomed-in mass spectrum (b and d) of the adenohypophysis region of rat pituitary gland using LESA-MS (a and b) compared to HR-LESA-μLC-MS (c and d). The zoomed mass range displays a low-intensity peak corresponding to the protonated molecule of leucine enkephalin (marked with *), which is known to be present in this region. This is extracted from the ion chromatogram at a retention time at 16 min. The zoomed mass range from HR-LESA-μLC-MS shows the presence of leucine enkephalin coeluting with a triply charged species, which was not observed with LESA-MS.
Interestingly, in the pituitary gland data, we observed the presence of a triply charged species (Figure 3d) coeluting with the leucine enkephalin, which was not detected with the conventional LESA extraction (Figure 3b). This difference can be explained by the occurrence of ion suppression, which was minimized by HR-LESA-μLC-MS. The extracted ion chromatogram (XIC) based on the protonated molecule signal of leucine enkephalin (m/z 556.28 ± 0.05) extracted from the adenohypophysis region showed a retention time of 16 min, similar to that of the monoisotopic peak of the triply charged species (m/z 555.65, Figure 4a).
Figure 4.

Extraction of two coeluting compounds: the XIC (a) and corresponding mass spectra (b). The green XIC represents the chromatogram extracted from the monoisotopic peak of the triply charged species at m/z 555.65. The red XIC represents the chromatogram extracted from leucine enkephalin at m/z 556.28.
However, the corresponding mass spectrum shows a different isotope pattern belonging to a triply charged species (Figure 4b, green asterisk). This clearly exposes the limitations of LC separation and the need for additional separation power to increase selectivity and accurate identification. For this reason, an additional gas-phase IMS was added to the analytical workflow to increase the analytical content of the data.
Ion Mobility Separation (IMS) of Isobaric Species
We employed triwave ion mobility to achieve additional separation: the TRAP T-Wave region was used for trapping and accumulating ions, IMS T-Wave region for subsequent separation, and the TRANSFER T-Wave region to focus the ions and also as fragmentation cell when performing data-independent HDMSE measurements.
IMS was performed from the anterior lobe HR-LESA extraction (m/z 556.28 eluting at tR = 16 min; Figure 5a). The extracted ion mobility drift time spectrum (Figure 5b) depicts two drift time peaks, which can be labeled using the corresponding mass spectra as leucine enkephalin (Figure 5d) and its coeluting compound (Figure 5c). The mass spectra extracted at this specific retention time displayed clean signals with overall improved sensitivity. The observed isotope pattern of the triply charged species has a mass of 1664 Da after deconvolution. The advantage of the integration of IMS after chromatographic separation is the additional separation power that is gained to separate coeluting isomeric species, without compromising the overall analysis time. For the identification of this coeluting species and other extracted proteins (Figure S-7), HDMSE analysis was performed. HDMSE is a data-independent acquisition (DIA) mode that includes both high and low collision energy measurements alternated in one parallel analysis using the retention time to match precursor and product ions. Species can be identified in one analysis without requiring additional data-dependent MS/MS experiments by performing HDMSE analysis.
Figure 5.

Ion mobilogram (a) of the extract from the anterior lobe of pituitary gland is depicted. The extracted ion mobility drift time spectrum (b) shows the separation of two coeluting compounds. The extracted mass spectra at drift time 3.8 (c) and 5.6 ms (d) show the separation of both compounds, indicated in red (leucine encephalin) and green (N-acetylated alpha-melanocyte stimulating hormone).
Identification of Neuropeptides and Proteins Present in the Pituitary Gland
We sought to identify neuropeptides and proteins in the rat pituitary samples using our platform by integrating data-independent acquisition (DIA). We chose the DIA approach because data-dependent acquisition (DDA) often neglects low-abundance precursor ions, which limits the discovery of untargeted analytes and markers.53
One DIA approach, MSE, collects TOF mass spectra with and without fragmentation by alternating the energy of the collision cell between low and high values.53 Another approach, sequential window acquisition of all theoretical fragment-ion spectra (SWATH), was developed on hybrid QqTOF mass analyzers that offer resolving power of 20 000–40 000.54 Both approaches were demonstrated to be particularly powerful in detecting low-abundance analytes for further metabolite and peptide identifications. We integrated high-definition MSE (HDMSE) acquisition, including ion mobility separation, into our platform.
HDMSE detects both precursor ions and fragments of the precursor ions fragmented independently on their abundance.55 Progenesis QI for proteomics software uses an algorithm that also performs a database search considering retention time, mass accuracy, and PTMs.53 However, due to the DIA nature of these analyses, this algorithm is based on physicochemical properties of the peptides and proteins. These characteristics are used to calculate the correlation to models with regards to hydrophobicity and gas-phase separation and are, therefore, applicable to reversed-phase LC and IMS. In similar strategy to DDA to calculate false-positive identification rates, we applied decoy and species-specific databases. Sensitivity and selectivity are significantly increased because of the repetitive approach of this algorithm. After the identification of the most abundant protein, this data is removed from the database, and another search is executed to identify the second most abundant protein. After removing the data from the second most abundant protein, a third search is performed. This process is continued until all proteins are identified.
In the extracts from the rat pituitary gland, we identified vasopressin and POMC (Figure S-8), based on 17 and 25 peptides, respectively (Tables S-1; Table S-2 lists the peptides found).53 These peptides, which are deriving from the pro-opiomelanocortin (POMC) precursor protein, are important signaling molecules with regard to feeding behavior and, therefore, involved in diseases like obesity, bulimia, and anorexia nervosa.56,57 In the mouse brain cerebral cortex extracts, we identified 14 peptides (Table S-3; Table S-4 lists the peptides found).
After applying de novo peptide sequencing to the fragmentation spectrum of the coeluting species at m/z 1664 at the retention time of 16 min, this compound was identified as N-acetylated alpha-melanocyte stimulating hormone based on nine fragment ions: y1, b1, y2, y7+2, y5, y10+2, y12+2, y7, and y8, deriving from P01194 (124–136). The MS/MS spectrum can be found in the Supporting Information (Figure S-9). Based on the MSE data, the singly charged species was identified as leucine enkephalin (Figure S-10).
Conclusion
This work reports the development of an automated and integrated platform combining the advantages of both spatial sampling with LESA and chromatographic separation (μLC) for the direct analysis of tissue sections with potential clinical, preclinical relevance (Figure 1). In addition, the platform was strengthened by the integration of ion mobility spectrometry (IMS) and data-independent acquisition (DIA) for both the separation and the identification of neuropeptides from a complex tissue extract from a 400-μm area. Improvement over the “conventional LESA” of the spatial resolution capabilities of the surface-sampling process was achieved, down to a spatial resolution of 400 μm, by modifying the sampling probe (Figure 2). The platform demonstrated strong reproducibility, minimal carry-over, increased sensitivity, technical reproducibility, and identification of isobaric compounds (Figures 3–5).
The advantages of this integrated platform hold strong potential for preclinical and clinical applications. Currently, the extraction efficacy is limited to soluble proteins with the number of protein and peptide identifications at fewer than 500. In particular, low-abundance analytes can be identified in complex and small tissue samples, as demonstrated with rat pituitary and mouse brain here. Information about protein isoforms, important in many neurodegenerative diseases, is predicted to be elucidated with the application of our platform with classical in-gel proteomics.42 This approach paves the way for imaging researchers to increase the total number of proteins and peptides identified and enable regional quantification-based on-tissue analysis.
Acknowledgments
This research was made possible with the support of the LINK program of the Dutch Province of Limburg. N.O.P. acknowledges support from FP7 European Union Marie Curie IAPP Program, BRAINPATH. T.P. acknowledges support from the Swiss National Science Foundation (P2GEP2_148527) and from Janssen R&D (Beerse, Belgium). We are grateful to Audrey Jongen (General Surgery, MUMC+) for providing us with the rat pituitary gland samples. We would like to thank Dr. Jelle Praet and Prof. Annemie van Der Linden (Bio-Imaging Lab, University of Antwerp, Belgium) for supplying the wildtype and transgenic mice samples.We are indebted to Daniel Eikel for his help in developing the LESA plus software (Advion). Eric van Beelen, Perry Derwig, Jasper Vanderfeesten, Aurélien Boland, Marco Kools, and Laurence Van Oudenhove (Waters) are acknowledged for their support in setting up the IonKey-Synapt G2Si platform at M4I. We are grateful to Lee Gethings (Waters) for his help with the Progenesis software. We are grateful to Hang Nguyen (M4I) for proof-reading our manuscript.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.analchem.7b03512.
Additional materials including Figures S-1 to S-10 and Tables S1 to S4 as discussed in the text. (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Schwamborn K. J. Proteomics 2012, 75, 4990–4998. 10.1016/j.jprot.2012.06.015. [DOI] [PubMed] [Google Scholar]
- Ye H.; Gemperline E.; Li L. Clin. Chim. Acta 2013, 420, 11–22. 10.1016/j.cca.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addie R. D.; Balluff B.; Bovee J. V.; Morreau H.; McDonnell L. A. Anal. Chem. 2015, 87, 6426–6433. 10.1021/acs.analchem.5b00416. [DOI] [PubMed] [Google Scholar]
- Kriegsmann J.; Kriegsmann M.; Casadonte R. Int. J. Oncol. 2015, 46, 893–906. 10.3892/ijo.2014.2788. [DOI] [PubMed] [Google Scholar]
- Longuespee R.; Casadonte R.; Kriegsmann M.; Pottier C.; Picard de Muller G.; Delvenne P.; Kriegsmann J.; De Pauw E. Proteomics: Clin. Appl. 2016, 10, 701–719. 10.1002/prca.201500140. [DOI] [PubMed] [Google Scholar]
- Kwon H. J.; Kim Y.; Sugihara Y.; Baldetorp B.; Welinder C.; Watanabe K.; Nishimura T.; Malm J.; Torok S.; Dome B.; Vegvari A.; Gustavsson L.; Fehniger T. E.; Marko-Varga G. Arch. Pharmacal Res. 2015, 38, 1718–1727. 10.1007/s12272-015-0627-2. [DOI] [PubMed] [Google Scholar]
- Balluff B.; Frese C. K.; Maier S. K.; Schone C.; Kuster B.; Schmitt M.; Aubele M.; Hofler H.; Deelder A. M.; Heck A. Jr.; Hogendoorn P. C.; Morreau J.; Maarten Altelaar A. F.; Walch A.; McDonnell L. A. J. Pathol 2015, 235, 3–13. 10.1002/path.4436. [DOI] [PubMed] [Google Scholar]
- Crecelius A. C.; Schubert U. S.; von Eggeling F. Analyst 2015, 140, 5806–5820. 10.1039/C5AN00990A. [DOI] [PubMed] [Google Scholar]
- Cole L. M.; Clench M. R. Biomarkers Med. 2015, 9, 863–868. 10.2217/bmm.15.61. [DOI] [PubMed] [Google Scholar]
- Meriaux C.; Franck J.; Park D. B.; Quanico J.; Kim Y. H.; Chung C. K.; Park Y. M.; Steinbusch H.; Salzet M.; Fournier I. Hippocampus 2014, 24, 628–642. 10.1002/hipo.22246. [DOI] [PubMed] [Google Scholar]
- Hanrieder J.; Malmberg P.; Ewing A. G. Biochim. Biophys. Acta, Proteins Proteomics 2015, 1854, 718–731. 10.1016/j.bbapap.2014.12.026. [DOI] [PubMed] [Google Scholar]
- Schubert K. O.; Weiland F.; Baune B. T.; Hoffmann P. Proteomics 2016, 16, 1747–1758. 10.1002/pmic.201500460. [DOI] [PubMed] [Google Scholar]
- Martin-Lorenzo M.; Alvarez-Llamas G.; McDonnell L. A.; Vivanco F. Expert Rev. Proteomics 2016, 13, 69–81. 10.1586/14789450.2016.1116944. [DOI] [PubMed] [Google Scholar]
- Dona A. C.; Coffey S.; Figtree G. Eur. J. Prev. Cardiol. 2016, 23 (15), 1578–1589. 10.1177/2047487316645469. [DOI] [PubMed] [Google Scholar]
- Bowrey H. E.; Anderson D. M.; Pallitto P.; Gutierrez D. B.; Fan J.; Crouch R. K.; Schey K. L.; Ablonczy Z. Proteomics: Clin. Appl. 2016, 10, 391–402. 10.1002/prca.201500103. [DOI] [PubMed] [Google Scholar]
- Cillero-Pastor B.; Eijkel G. B.; Blanco F. J.; Heeren R. M. Anal. Bioanal. Chem. 2015, 407, 2213–2222. 10.1007/s00216-014-8342-2. [DOI] [PubMed] [Google Scholar]
- Spraggins J. M.; Rizzo D. G.; Moore J. L.; Noto M. J.; Skaar E. P.; Caprioli R. M. Proteomics 2016, 16, 1678–1689. 10.1002/pmic.201600003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis S. R.; Cappell J.; Potocnik N. O.; Balluff B.; Hamaide J.; Van der Linden A.; Heeren R. M. Analyst 2016, 141, 3832–3841. 10.1039/C6AN00169F. [DOI] [PubMed] [Google Scholar]
- Ogrinc Potocnik N.; Porta T.; Becker M.; Heeren R. M.; Ellis S. R. Rapid Commun. Mass Spectrom. 2015, 29, 2195–2203. 10.1002/rcm.7379. [DOI] [PubMed] [Google Scholar]
- Ifa D. R.; Eberlin L. S. Clin. Chem. 2016, 62, 111–123. 10.1373/clinchem.2014.237172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takats Z.; Wiseman J. M.; Cooks R. G. J. Mass Spectrom. 2005, 40, 1261–1275. 10.1002/jms.922. [DOI] [PubMed] [Google Scholar]
- Cabral E. C.; Ifa D. R. Methods Mol. Biol. 2015, 1203, 63–77. 10.1007/978-1-4939-1357-2_7. [DOI] [PubMed] [Google Scholar]
- Gerbig S.; Golf O.; Balog J.; Denes J.; Baranyai Z.; Zarand A.; Raso E.; Timar J.; Takats Z. Anal. Bioanal. Chem. 2012, 403, 2315–2325. 10.1007/s00216-012-5841-x. [DOI] [PubMed] [Google Scholar]
- Eberlin L. S.; Norton I.; Orringer D.; Dunn I. F.; Liu X.; Ide J. L.; Jarmusch A. K.; Ligon K. L.; Jolesz F. A.; Golby A. J.; Santagata S.; Agar N. Y.; Cooks R. G. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 1611–1616. 10.1073/pnas.1215687110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbassi-Ghadi N.; Golf O.; Kumar S.; Antonowicz S.; McKenzie J. S.; Huang J.; Strittmatter N.; Kudo H.; Jones E. A.; Veselkov K.; Goldin R.; Takats Z.; Hanna G. B. Cancer Res. 2016, 76 (19), 5647–5656. 10.1158/0008-5472.CAN-16-0699. [DOI] [PubMed] [Google Scholar]
- Alfaro C. M.; Jarmusch A. K.; Pirro V.; Kerian K. S.; Masterson T. A.; Cheng L.; Cooks R. G. Anal. Bioanal. Chem. 2016, 408, 5407–5414. 10.1007/s00216-016-9627-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmusch A. K.; Pirro V.; Baird Z.; Hattab E. M.; Cohen-Gadol A. A.; Cooks R. G. Proc. Natl. Acad. Sci. U. S. A. 2016, 113, 1486–1491. 10.1073/pnas.1523306113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shariatgorji M.; Strittmatter N.; Nilsson A.; Kallback P.; Alvarsson A.; Zhang X.; Vallianatou T.; Svenningsson P.; Goodwin R. J.; Andren P. E. NeuroImage 2016, 136, 129–138. 10.1016/j.neuroimage.2016.05.004. [DOI] [PubMed] [Google Scholar]
- Skraskova K.; Claude E.; Jones E. A.; Towers M.; Ellis S. R.; Heeren R. M. Methods 2016, 104, 69–78. 10.1016/j.ymeth.2016.02.014. [DOI] [PubMed] [Google Scholar]
- Zhang J.; Feider C. L.; Nagi C.; Yu W.; Carter S. A.; Suliburk J.; Cao H. S.; Eberlin L. S. J. Am. Soc. Mass Spectrom. 2017, 28 (6), 1166–1174. 10.1007/s13361-016-1570-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberlin L. S.; Norton I.; Dill A. L.; Golby A. J.; Ligon K. L.; Santagata S.; Cooks R. G.; Agar N. Y. Cancer Res. 2012, 72, 645–654. 10.1158/0008-5472.CAN-11-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt S.; Kallbach S.; Almeida R.; Sandel J. Drug Metab. Dispos. 2012, 40, 419–425. 10.1124/dmd.111.043562. [DOI] [PubMed] [Google Scholar]
- Parson W. B.; Koeniger S. L.; Johnson R. W.; Erickson J.; Tian Y.; Stedman C.; Schwartz A.; Tarcsa E.; Cole R.; Van Berkel G. J. J. Mass Spectrom. 2012, 47, 1420–1428. 10.1002/jms.3068. [DOI] [PubMed] [Google Scholar]
- Porta T.; Varesio E.; Hopfgartner G. Anal. Chem. 2013, 85, 11771–11779. 10.1021/ac4020353. [DOI] [PubMed] [Google Scholar]
- Kertesz V.; Paranthaman N.; Moench P.; Catoire A.; Flarakos J.; Van Berkel G. J. Bioanalysis 2014, 6, 2599–2606. 10.4155/bio.14.130. [DOI] [PubMed] [Google Scholar]
- Swales J. G.; Tucker J. W.; Spreadborough M. J.; Iverson S. L.; Clench M. R.; Webborn P. J.; Goodwin R. J. Anal. Chem. 2015, 87, 10146–10152. 10.1021/acs.analchem.5b02998. [DOI] [PubMed] [Google Scholar]
- Randall E. C.; Bunch J.; Cooper H. J. Anal. Chem. 2014, 86, 10504–10510. 10.1021/ac503349d. [DOI] [PubMed] [Google Scholar]
- Lanshoeft C.; Stutz G.; Elbast W.; Wolf T.; Walles M.; Stoeckli M.; Picard F.; Kretz O. Rapid Commun. Mass Spectrom. 2016, 30, 823–832. 10.1002/rcm.7511. [DOI] [PubMed] [Google Scholar]
- Almeida R.; Berzina Z.; Arnspang E. C.; Baumgart J.; Vogt J.; Nitsch R.; Ejsing C. S. Anal. Chem. 2015, 87, 1749–1756. 10.1021/ac503627z. [DOI] [PubMed] [Google Scholar]
- Quanico J.; Franck J.; Dauly C.; Strupat K.; Dupuy J.; Day R.; Salzet M.; Fournier I.; Wisztorski M. J. Proteomics 2013, 79, 200–218. 10.1016/j.jprot.2012.11.025. [DOI] [PubMed] [Google Scholar]
- Wisztorski M.; Fatou B.; Franck J.; Desmons A.; Farre I.; Leblanc E.; Fournier I.; Salzet M. Proteomics: Clin. Appl. 2013, 7, 234–240. 10.1002/prca.201200070. [DOI] [PubMed] [Google Scholar]
- Wisztorski M.; Desmons A.; Quanico J.; Fatou B.; Gimeno J. P.; Franck J.; Salzet M.; Fournier I. Proteomics 2016, 16, 1622–1632. 10.1002/pmic.201500508. [DOI] [PubMed] [Google Scholar]
- Sarsby J.; Griffiths R. L.; Race A. M.; Bunch J.; Randall E. C.; Creese A. J.; Cooper H. J. Anal. Chem. 2015, 87, 6794–6800. 10.1021/acs.analchem.5b01151. [DOI] [PubMed] [Google Scholar]
- Martin N. J.; Griffiths R. L.; Edwards R. L.; Cooper H. J. J. Am. Soc. Mass Spectrom. 2015, 26, 1320–1327. 10.1007/s13361-015-1152-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson L.; Fuchser J.; Futterer A.; Baumert M.; Hassall D. G.; West A.; Marshall P. S. Rapid Commun. Mass Spectrom. 2014, 28, 995–1003. 10.1002/rcm.6869. [DOI] [PubMed] [Google Scholar]
- Feider C. L.; Elizondo N.; Eberlin L. S. Anal. Chem. 2016, 88, 11533–11541. 10.1021/acs.analchem.6b02798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz V.; Weiskittel T. M.; Van Berkel G. J. Anal. Bioanal. Chem. 2015, 407, 2117–2125. 10.1007/s00216-014-8287-5. [DOI] [PubMed] [Google Scholar]
- Kertesz V.; Van Berkel G. J. Anal. Chem. 2010, 82, 5917–5921. 10.1021/ac100954p. [DOI] [PubMed] [Google Scholar]
- Kertesz V.; Van Berkel G. J. J. Mass Spectrom. 2010, 45, 252–260. 10.1002/jms.1709. [DOI] [PubMed] [Google Scholar]
- Kertesz V.; Calligaris D.; Feldman D. R.; Changelian A.; Laws E. R.; Santagata S.; Agar N. Y.; Van Berkel G. J. Anal. Bioanal. Chem. 2015, 407, 5989–5998. 10.1007/s00216-015-8803-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida R.; Vogt J.; Hannibal-Bach K. H.; Baumgart J.; Nitsch R.; Ejsing C. In 59th Conference on Mass Spectrometry and Allied Topics; Denver, CO, 2011.
- Laskin J.; Lanekoff I. Anal. Chem. 2016, 88, 52–73. 10.1021/acs.analchem.5b04188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G. Z.; Vissers J. P.; Silva J. C.; Golick D.; Gorenstein M. V.; Geromanos S. J. Proteomics 2009, 9, 1696–1719. 10.1002/pmic.200800564. [DOI] [PubMed] [Google Scholar]
- Wu Y.; Williams E. G.; Aebersold R. Current protocols in mouse biology 2017, 7, 130–143. 10.1002/cpmo.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distler U.; Kuharev J.; Navarro P.; Levin Y.; Schild H.; Tenzer S. Nat. Methods 2013, 11, 167–170. 10.1038/nmeth.2767. [DOI] [PubMed] [Google Scholar]
- Altelaar A. F.; Luxembourg S. L.; McDonnell L. A.; Piersma S. R.; Heeren R. M. Nat. Protoc. 2007, 2, 1185–1196. 10.1038/nprot.2007.117. [DOI] [PubMed] [Google Scholar]
- Taban I. M.; Altelaar A. F.; Burgt Y. E. M.; McDonnell L. A.; Heeren R. M.; Fuchser J.; Baykut G. J. Am. Soc. Mass Spectrom. 2007, 18, 145–151. 10.1016/j.jasms.2006.09.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.