Deep-sea fishes evolved an unconventional visual system to optimize vision in twilight conditions.
Abstract
Most vertebrates have a duplex retina comprising two photoreceptor types, rods for dim-light (scotopic) vision and cones for bright-light (photopic) and color vision. However, deep-sea fishes are only active in dim-light conditions; hence, most species have lost their cones in favor of a simplex retina composed exclusively of rods. Although the pearlsides, Maurolicus spp., have such a pure rod retina, their behavior is at odds with this simplex visual system. Contrary to other deep-sea fishes, pearlsides are mostly active during dusk and dawn close to the surface, where light levels are intermediate (twilight or mesopic) and require the use of both rod and cone photoreceptors. This study elucidates this paradox by demonstrating that the pearlside retina does not have rod photoreceptors only; instead, it is composed almost exclusively of transmuted cone photoreceptors. These transmuted cells combine the morphological characteristics of a rod photoreceptor with a cone opsin and a cone phototransduction cascade to form a unique photoreceptor type, a rod-like cone, specifically tuned to the light conditions of the pearlsides’ habitat (blue-shifted light at mesopic intensities). Combining properties of both rods and cones into a single cell type, instead of using two photoreceptor types that do not function at their full potential under mesopic conditions, is likely to be the most efficient and economical solution to optimize visual performance. These results challenge the standing paradigm of the function and evolution of the vertebrate duplex retina and emphasize the need for a more comprehensive evaluation of visual systems in general.
INTRODUCTION
The retina of most vertebrates is categorized as being duplex, comprising both rod and cone photoreceptors. Each photoreceptor type differs in morphology and function, allowing animals to switch between the two systems and thus maintain vision during most of the daily (24-hour) period. Rods express the highly sensitive rod opsin gene (rh1), which mediates vision in dim-light (scotopic) conditions, whereas cones express up to four classes of visual pigment genes [short-wavelength sensitive (sws1 and sws2), medium-wavelength sensitive (rh2), and long-wavelength sensitive (lws)], which mediates vision and color discrimination in bright-light (photopic) environments (1). In mesopic conditions, such as during twilight, both rods and cones are active and contribute to vision, but neither work at optimal levels (2, 3). Depending on whether a species is diurnal, nocturnal, or crepuscular, the proportion of each photoreceptor type may vary to maximize visual performance (2). In a few extreme cases, an entire photoreceptor type and function is lost, resulting in a simplex retina, such as the pure cone or pure rod retinas of diurnal lizards and deep-sea fishes, respectively (4).
The classification of photoreceptors into rods and cones is, however, not always straightforward, and some intermediate forms of photoreceptors exist and exhibit morphological, electrophysiological, and/or molecular characteristics of both cell types. This is most notable in squamate reptiles (5–7), amphibians (8), lampreys (9), and skates (10) (Table 1). These intermediate forms were first observed in snakes and geckos by Walls (4), which led to the “transmutation” theory that suggested that rods and cones could evolve or “transmute” from one type to another, via a series of intermediate states, as a result of a major ecological shift in activity pattern. For instance, the gecko ancestor was diurnal and accordingly had a pure cone retina, but cones in modern nocturnal geckos have transmuted into rod-like receptors to regain vision under scotopic light conditions (6). To our knowledge, the study presented here is the first case of photoreceptor transmutation in teleost fishes, specifically the pearlsides, Maurolicus spp. The results suggest that photoreceptor transmutation is not always an evolutionary transition to regain a lost function but also an adaptation to optimize vision in a specific ecological niche, in this case, the twilight environment.
Table 1. Characteristics summary (morphology, opsin, phototransduction cascade, and electrophysiology) of the transmuted photoreceptors of different species compared to true rods and true cones.
Lizard data are for the genus Anolis. Lamprey is the sea lamprey Petromyzon marinus. Salamander is the tiger salamander Ambystoma tigrinum. Pearlside is the Mueller’s pearlside M. muelleri. The snake with the cone-like rods is the diurnal garter snake Thamnophis proximus. The snake with the rod-like cones data are for the nocturnal genus Hypsiglena. The gecko is the nocturnal Tokay gecko Gekko gekko. The skate is the genus Raja. R, true rod; C, true cone; n.a., not available; poly, polysynaptic.
|
Photoreceptor characteristics |
True rod (1, 19) |
True cone (1, 19) |
Cone-like rod | Rod-like cone | ||||||
|
Snake (7, 78) |
Skate (10, 45, 79) |
Lizard (5, 80, 81) |
Lamprey (9, 82, 83) |
Gecko (6, 84–86) |
Snake (4, 35) |
Salamander (8, 87) |
Pearlside (this study) |
|||
| Outer segment shape |
Long, rod- shaped (cylindrical) |
Short, cone- shaped (distally tapering) |
C | R | C | R | R | R | R | R |
| Outer segment discs |
Individual sealed disc, separated from the plasma membrane |
Discs continuous with the plasma membrane (open) |
R | R | n.a. | R C | R C | n.a. | n.a. | R |
| Incisure | Present | Absent | n.a. | n.a. | C | C | R | n.a. | R | R |
| Paraboloid | Absent | Present | C | R | R | R | ||||
| Oil droplet | Absent | Sometimes present | R | n.a. | C | n.a. | R | R | R | R |
| Synaptic ending | Small, spherical, oligosynaptic |
Large, conical, flat-end base, polysynaptic |
n.a. | C | n.a. | R C Small poly |
C | n.a. | R C Small poly |
R C Small poly |
| Opsin | rh1 | sws1, sws2, lws, rh2 | R | R | R | R | C rh2 |
C sws1 lws |
C sws2 |
C rh2-1 rh2-2 |
| Spectral sensitivity (nm) |
480–510 |
rh2, 450–530 sws1, 360–440 sws2, 400–450 m/lws, 510–560 |
R 482 |
R 500 |
R 491 |
n.a. | C 521 |
C 358, 536 |
C 432 |
C 441 (both pigments) |
| Phototransduction cascade |
Rod-like | Cone-like | R | n.a. | n.a. | n.a. | C(R) | n.a. | R | C |
| Cell physiology | Rod properties (high sensitivity) |
Cone properties (fast, never saturate) |
n.a. | R C | n.a. | R | R | n.a. | n.a. | n.a. |
Pearlsides are deep-sea teleost fishes that live in the upper part of the mesopelagic zone (~200 m) and vertically migrate to feed on zooplankton present in the upper layers of the ocean (0 to 100 m). Contrary to other vertically migrating fishes that ascend to feed throughout the night, pearlsides mostly migrate to the surface at dusk and dawn (twilight) and spend the night at greater depths without foraging (11). This behavior is best explained by the “antipredation window” theory, which suggests that it is advantageous to spend short periods of time foraging at twilight when light levels are sufficient for prey detection but low enough to hide from predators (12). Whereas animals that are active during this time period usually use a combination of rods and cones for vision (2), pearlsides, like most deep-sea teleost fishes, have a morphologically pure rod retina implying an rh1-based scotopic visual system (13), which is at odds with their daily behavior. To explore this paradox, the visual system of two species of pearlsides, Maurolicus muelleri from the Norwegian fjords and Maurolicus mucronatus from the Red Sea, was investigated.
RESULTS AND DISCUSSION
Pearlside retinal transcriptomes
Retinal transcriptome sequencing of both species revealed the expression of three distinct opsin genes: rh1 (usually restricted to rods) and, surprisingly, two cone opsins (rh2-1 and rh2-2; Fig. 1A and fig. S1). The rh2 genes were expressed at similar levels and comprised ~99% of the total opsin expression in the retina (Fig. 1B and table S1), suggesting that pearlsides rely almost exclusively on their cone photopigments for vision, a strategy expected in animals devoid of rods. This is further supported at the amino acid level. In general, amino acid sites 122 and 189 confer photoreceptor specificity in vertebrates, with Gln122 and Pro189 sites being cone-specific and Glu122 (except Gln122 in deep-sea fish rods) and Ile189 sites being rod-specific (1). Whereas the pearlside rh1 showed typical deep-sea rod specificity (that is, Gln122 and Ile189), pearlside rh2s showed the cone typical Gln122 and Pro189 combination (fig. S2).
Fig. 1. Vertebrate opsin gene phylogeny and pearlside opsin gene expression.
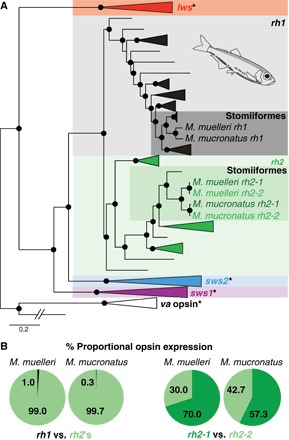
(A) The pearlside retinal transcriptomes (n = 5 per species) contained three opsin genes: one rod opsin (rh1) and two rhodopsin-like 2 (rh2) cone opsins. Black spheres indicate Bayesian posterior probabilities >0.8. Asterisk indicates that this opsin gene class was not present in the pearlside transcriptome. lws, long-wavelength sensitive; sws1 and sws2, short-wavelength sensitive 1 and 2; va, vertebrate ancient opsin (outgroup). A detailed phylogeny and GenBank accession numbers are shown in fig. S1. Pearlside-specific accession numbers are given in table S4. (B) The per-species mean of the proportional opsin gene expression shows the almost exclusive use of cone opsins in pearlside vision.
At the level of the phototransduction cascade, the Gα subunit of transducin (critical for signal transduction; Fig. 2A and fig. S3) and arrestin (involved in pigment recovery; Fig. 2B and fig. S4) were also found to be mainly cone-like (1) and were expressed at a ratio of around 99% cone to 1% rod (tables S2 and S3), strongly supporting the use of a predominantly cone-based visual system, a first in adult teleost fishes. A detailed analysis of the transcriptomes revealed that pearlsides express three Gα transducins: one rod transducin (gnat1) and two cone transducins (gnat2-1 and gnat2-2; Fig. 2A). Although with low support values, the pearlside cone transducins form their own clade, indicating a lineage-specific duplication that might have facilitated photoreceptor transmutation (fig. S3). Alternatively, the gnat-2 duplication might have a much deeper phylogenetic origin that has so far been missed (1, 14) and, as for the opsins (15), would imply that the evolutionary history of these genes is more complex than previously thought. The transcriptomes also contained four arrestin genes: two rod arrestins (saga and sagb) and two cone arrestins (arr3a and arr3b; Fig. 2B). In the medaka (Oryzias latipes), rod arrestin orthologs, saga and sagb (fig. S4), are expressed in different parts of the photoreceptor. Saga is expressed in the outer segments and mediates phototransduction, whereas sagb is expressed in the synapses (function unknown) (16). It is therefore possible that only saga is involved in phototransduction in pearlsides, explaining the presence of two rod arrestin genes but only one rod opsin.
Fig. 2. Vertebrate phylogenies of phototransduction cascade genes and phototransduction cascade gene expression.
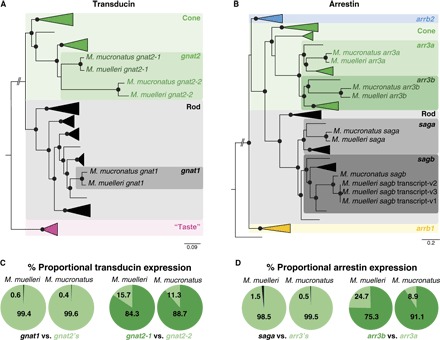
(A and B) Vertebrate Gα transducin (A) and vertebrate arrestin (B) gene phylogenies. Black spheres indicate Bayesian posterior probabilities >0.8. Detailed phylogenies and GenBank accession numbers are shown in figs. S3 and S4. Pearlside-specific accession numbers are given in table S4. gnat2, G protein subunit α transducin 2; gnat1, G protein subunit α transducin 1; “taste,” G protein subunit α transducin 3; arrb2, arrestin β2; arr3, arrestin 3; sag, s-antigen arrestin [saga is present in the outer segment, and sagb is present in the synapses (16)]; arrb1, arrestin β1. (C and D) The per-species mean of the proportional transducin (C) and arrestin (D) gene expression shows the almost exclusive use of cone transducin (gnat2) and cone arrestin (arr3) in pearlside vision.
Pearlside spectral sensitivities
Vertebrates distinguish colors by comparing the visual input between two photoreceptors that are sensitive to different parts of the light spectrum and are active at the same time (17). Accordingly, the expression of two rh2 opsins, presumably with different peak spectral sensitivities (λmax) and expressed in different photoreceptors, could confer color vision to pearlsides. However, the rh2 genes in both species showed very few amino acid differences, with none occurring at key spectral tuning sites (fig. S2) (18–20). Therefore, the two rh2 opsins are likely to have similar λmax values, making color vision an unlikely feature of the pearlside visual system. Instead, as was suggested for rh1-duplicated genes (rh1-1 and rh1-2) in the zebrafish (Danio rerio) (21), the ancestral rh2 function may be partitioned between the two rh2 copies in pearlsides. After comparing the synonymous substitution rate in the coding region [coding DNA sequence (CDS)] of the rh2 paralogs to that in the 3′ untranslated regions (3′UTRs), it seemed plausible that gene conversion might have acted to conserve the rh2 spectral sensitivities (15, 22). The occurrence of gene conversion was confirmed by reconstructing a phylogeny using only the information contained in the 3′UTR and the last 50 base pairs (bp) of exon 5, which revealed that the rh2 duplication occurred in the common ancestor of pearlsides (fig. S5).
In vitro expression was subsequently attempted to determine the spectral sensitivities of the M. muelleri opsins using a vitamin A1 chromophore (that is, 11-cis-retinal chromophore). Although some deep-sea fishes have been found to use both A1- and A2-based chromophores for vision (23), this seems not to be the case for pearlsides based on a lack of cytochrome P450 family 27 subfamily c member 1 (cyp27c1) expression in their transcriptomes. The Cyp27c1 enzyme has been shown to convert vitamin A1 to vitamin A2 in zebrafish, leading to the replacement of 11-cis-retinal with 11-cis-3,4-didehydroretinal: This change results in a long-wavelength shift of the spectral peak for visual photopigments (24). In vitro ultraviolet (UV)–visible spectrophotometry was successfully performed for the rod opsin pigments expressed in M. muelleri (rh1) and human (RH1; control), with peak spectral sensitivity (λmax) values of 494 and 495 nm, respectively (Fig. 3A), closely matching the predicted λmax values of 496 nm for M. muelleri and a previous spectral study of the human RH1 photopigment (25). The predicted value of M. muelleri rh1 (λmax = 496 nm) was calculated on the basis of two other deep-sea teleost fishes from the same family, namely, Argyropelecus aculeatus and Argyropelecus gigas, where their spectral peaks were determined to be 477 nm using microspectrophotometry (26). In comparison to these two fishes, the spectral peak of the M. muelleri rh1 opsin is long-wavelength–shifted by 19 nm, which corresponds to Phe261Tyr (+10 nm) and Ser292Ala (+9 nm) substitutions (20, 26). Unfortunately, both M. muelleri rh2-1 and rh2-2 pigments did not regenerate or reconstitute with 11-cis-retinal in vitro, despite numerous attempts. This is not uncommon with photopigments studied in vitro, where technical artefacts, such as misfolding of overexpressed proteins, may cause experiments to be unsuccessful (27–29). Nonetheless, logical predictions may be made on the basis of published data, as well as those present in this study (Fig. 3B). Although most rh2 pigments yield spectral peaks between 470 and 510 nm (20), those found in deep-sea species are typically short-wavelength–shifted. For example, the deep-sea teleost Scopelarchus analis has an rh2 photopigment with a λmax value of 444 nm (30, 31), and the elephant shark (Callorhinchus milii) has an rh2 photopigment with a λmax value of 442 nm (32); the former was measured by in vivo microspectrophotometry, and the latter was measured by in vitro UV-visible spectrophotometry. When comparing the known tuning sites for rh2 photopigments (18, 20) identified in M. muelleri (this study) and S. analis (30), the only difference found was at site 300 with Ile and Leu in M. muelleri (for both rh2-1 and rh2-2) and S. analis, respectively (fig. S2). From an extensive comparative study of spectral tuning in deep-sea fish visual pigments (26), a Leu300Ile substitution results in a short-wavelength shift of 3 nm. Thus, for M. muelleri, the rh2 pigments are predicted to have spectral peaks at 441 nm (Fig. 3B). When comparing known rh1 tuning sites between pearlsides species, no differences were observed; hence, rh1 in M. mucronatus is predicted to have a λmax value of 494 to 496 nm (that is, measured and predicted). When rh2 pigments were compared between Maurolicus species, only a single difference between known tuning sites was found at site 299, with Ala in M. mucronatus and Ser in M. muelleri. Analyses of spectral tuning in deep-sea fish visual pigments indicate that a Ser299Ala substitution results in a short-wavelength shift of 6 nm (26); therefore, both rh2 pigments of M. mucronatus are predicted to exhibit a spectral peak at 435 nm (Fig. 3B).
Fig. 3. Absorbance spectra of photopigments expressed in two representative Maurolicus spp.
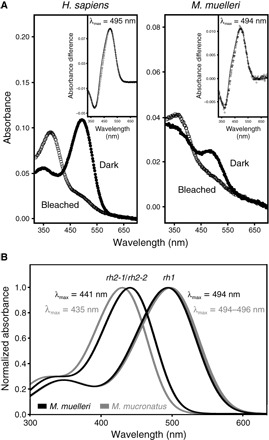
(A) Experimentally determined absorbance spectra of M. muelleri and human (Homo sapiens; control) rod opsin photopigments, reconstituted with 11-cis-retinal. For all pigments, representative dark (filled circles) and light-bleached (open circles) spectra are shown, with difference spectra (open diamonds) that have been fitted with a Govardovskii rhodopsin/vitamin A1 template (63) (black line) in the inset to determine the λmax. (B) Predicted spectral sensitivities of rh1 and the two rh2 opsins found in the two pearlside species, M. muelleri (black) and M. mucronatus (gray).
Distribution and morphology of M. muelleri photoreceptors
Given the high levels of cone-specific pigment expression, it was expected that at least a few cone photoreceptors (based on morphological criteria) would occur in the pearlside retina. However, similar to previous findings (13), no cone-like cells were found, and RNA in situ hybridization (R-ISH) using opsin-specific probes confirmed that both rh1 and rh2s were expressed only in rod-like photoreceptors in M. muelleri (Fig. 4). Moreover, R-ISH and anti-rhodopsin (rod pigment) immunohistochemistry (IHC) corroborated the transcriptomic data, with both rh2 genes being expressed collectively in about 99% of photoreceptor cells and rh1 in the remaining 1% of photoreceptors (Fig. 4).
Fig. 4. Opsin R-ISH and the distribution of the two photoreceptor types in the retina of M. muelleri.
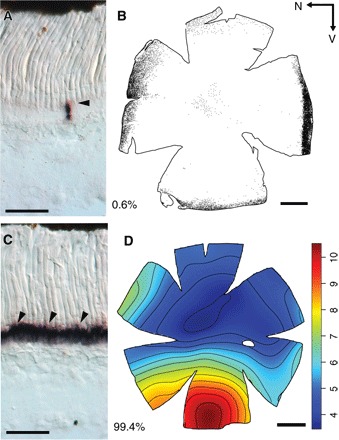
(A and C) Retinal cryosections showing the expression of rh1 (A) and rh2 (C) opsin genes in cryosections. Arrowheads highlight labeled cells. Note that both genes are expressed in rod-like photoreceptor cells. (B) Distribution of rh1 photoreceptors labeled with anti-rhodopsin antibodies. Each dot represents one labeled photoreceptor. Black arrows indicate the orientation of the retina. N, nasal; V, ventral. (D) Topographic map of rh2 photoreceptor densities (cells × 104 mm−2). Percentages indicate the proportion of each cell type. Scale bars, 50 μm (A and C) and 1 mm (B and D).
The density of the rh2-expressing photoreceptors varied across the retina with an increase in cell density in the ventral region (that is, elongated area ventralis; Fig. 4D and fig. S6). This pattern matched the retinal ganglion cell topography (fig. S7), and assuming a horizontal position of the fish in the water column, this high photoreceptor density zone is likely to facilitate the detection of silhouettes situated above the fish against the lighter background of the upper mesopelagic zone (33). The small population of rh1-expressing photoreceptors was confined to the peripheral margins of the retina (Fig. 4B and figs. S8 and S9) and might, therefore, be used to detect motion, although their extremely low level of expression questions their ecological significance.
Subsequent immunofluorescence, transmission electron microscopy (TEM) and 3View-based three-dimensional (3D) reconstruction revealed two types of rod photoreceptors at the morphological level: a rod-like cone (the main cell type expressing rh2) and a “true” rod (labeled with anti-rhodopsin antibodies and expressing rh1) (Fig. 5 and movie S1). Both cell types showed the classic anatomical characteristics of a rod (Table 1): a long cylindrical outer segment, individual sealed discs surrounded by a plasma membrane, incisures (Fig. 5F), and the absence of both a paraboloid and an oil droplet. Structurally, the main differences between the two photoreceptor types were restricted to the vitreated region of the receptors (the nucleus and synaptic terminal). In the true rod, the nucleus was displaced toward the inner retina, whereas its synaptic process was located slightly above the processes of rod-like cones (Fig. 5, B and C). Furthermore, although both cell types had a relatively small synaptic terminal (~3 μm) and a single invagination (a characteristic of rod cells), the rod-like cones had a polysynaptic ending with three synaptic ribbons and multiple processes constituting the only cone feature observed in the pearlside retina (Fig. 5, D and E, Table 1, and movie S2).
Fig. 5. Morphology of the two photoreceptor types in M. muelleri.
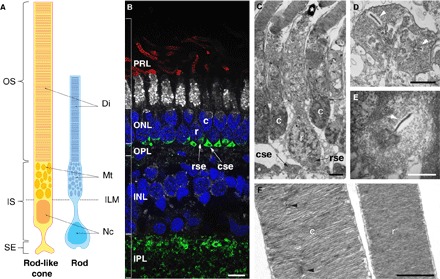
(A) Schematic of the rod-like cone (yellow; left) and rod (blue; right) drawn from the 3D reconstruction using 3View. OS, outer segment; IS, inner segment; SE, synaptic ending; Di, discs; Mt, mitochondria; ILM, inner limiting membrane; Nc, nucleus. Note the displaced nucleus and synaptic ending in the rod. (B) Immunofluorescence labeling of transverse retinal cryosections. Rod outer segments were labeled with anti-rhodopsin antibodies (red), inner segments with NeuN (white), cell nuclei with 4′,6-diamidino-2-phenylindole (DAPI) (blue), and synaptic connections with synapsin (green). Note that NeuN does not usually stain photoreceptor inner segments, but in M. muelleri, the inner segments of the rods were strongly labeled compared to the rod-like cones. PRL, photoreceptor layer; ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer; c, rod-like cone; r, rod; cse, rod-like cone synaptic ending; rce, rod synaptic ending. Scale bar, 5 μm. (C to F) TEM of transverse retinal sections showing the two photoreceptor types (C), the polysynaptic ending of the rod-like cone (D), the oligosynaptic ending of the rod (E), and the sealed discs and incisures of the outer segments (F). The white arrowheads in (D) and (E) show the synaptic ribbons, and the black arrowheads in (F) show the incisures present in the rod-like cone. Scale bars, 2 μm (C) and 1 μm (D to F).
Likely evolutionary scenario leading to photoreceptor transmutation in pearlsides
Two possible evolutionary scenarios could lead to cone opsins being expressed in rod-like photoreceptor cells: (i) Pearlside cone photoreceptors transmuted to morphologically resemble rod cells or (ii) cone opsin(s) and visual phototransduction genes co-adapted and replaced the conventional rod-based machinery in 99% of all rod cells. The occurrence of about 1% of the true rod photoreceptors expressing a rod opsin, the cone-like synaptic ending of the rod-like cones, and cone-specific amino acid residues of rh2 opsins strongly support the first scenario. However, to demonstrate either scenario, a complete ontogenetic analysis of the pearlside retina, at both molecular and morphological levels, is required (34).
Although photoreceptor transmutation normally occurs following the loss of a photoreceptor type or a specific opsin gene (4, 35, 36), this is unlikely to be the case for pearlside fishes. First, they express both rod and cone opsins and associated phototransduction genes. Second, teleosts, including deep-sea species with pure rod retinas as adults, have both cones and rods at some stage during their development (37). Finally, adult individuals from species within the same family (Sternoptychidae) have both photoreceptor types (38). Hence, the transmuted rod-like cones of the pearlside are likely to be an ecological adaptation rather than an evolutionary regain of function.
Ecological significance
The likely explanation for the peculiar visual system of the pearlside is an adaptation to the surrounding light environment. Pearlsides inhabit the upper mesopelagic zone of the ocean where two main sources of light are found: the downwelling light produced by the sun and stars, and bioluminescence produced and emitted by the organisms themselves. However, adaptation to bioluminescent light seems unlikely for the following reasons: (i) Pearlsides are only active in light intensities between 0.2 cd m−2 (full moon) and 127 cd m−2 (sunset/sunrise) (39), that is, in mesopic conditions and/or the lowest intensities of photopic illumination (Fig. 6A), which would severely limit visualization of bioluminescent signals (40). (ii) The predicted peak spectral sensitivity for both rh2 opsins is between λmax = 435 and 441 nm in both species (Fig. 3B), which is offset from most bioluminescent emissions at 475 nm (41) but instead matches the blue-shifted twilight spectrum of downwelling light at 450 nm (Fig. 6B) (42). (iii) Although pearlsides are bioluminescent, the unique ventral placement of their photophores (43) suggests that they are used for camouflage via countershading rather than for communication, implying a dire need to visualize downwelling light (44). (iv) Increased photoreceptor and ganglion cell densities in the ventral part of the retina (figs. S6 and S7) indicate that their visual system is optimized to detect shadows of objects situated above the eye rather than perceive bioluminescent signals directly (33). Therefore, the visual system of pearlsides appears to be specifically adapted to the ambient blue-shifted mesopic light conditions in which they live (Fig. 6).
Fig. 6. Ambient light environment and pearlside visual capabilities.
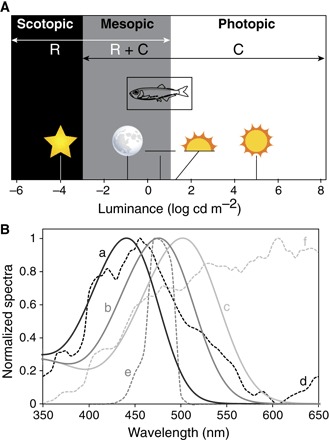
(A) Light levels associated with different photoreceptor functionalities. M. muelleri is only active during mesopic and low-level photopic light intensities (39). R, rod; C, cone. Scotopic vision is defined by the use of rods. Mesopic vision is defined by the use of both rods and cones limited by cone threshold and rod saturation. Photopic vision is defined by the use of cones and ends when light intensities start to be damaging (75). Environmental light sources (from left to right) are as follows: starlight, full moon, civil twilight, sunset/sunrise, and sunlight (76). Figure partially redrawn from Hood and Finkelstein (75). (B) Spectral sensitivity curves of the pearlside M. muelleri rh2 (a, black line; predicted λmax = 441 nm; fig. S7), the deep-sea myctophid Symbolophorus evermanni rh1 [b, dark gray line; λmax = 476 (23)], and the nocturnal squirrelfish Neoniphon sammara rh1 [c, light gray line; λmax = 502 nm (77)] along with the relative downwelling vector irradiance spectra (courtesy of S. Johnsen) of their respective light environments: twilight (−6.5° solar elevation) at the surface (d, black dashed line), downwelling light at 500 m (e, dark gray dashed line), and moonlight (full moon at 70° elevation above horizon) at the surface (f, light gray dashed line). Note how the spectral sensitivity of each species is tuned to the light spectra of their respective habitat.
Instead of having two photoreceptor types that do not function at their full potential under mesopic conditions, pearlsides have evolved a more efficient and economical solution by combining properties of both rods and cones in a single cell type to optimize visual performance. Under this premise, transmuting cones to morphologically resemble rods, while retaining the cone pigment and the cone phototransduction cascade rather than the reverse situation, would result in the greatest gain of sensitivity. Specifically, whereas cone photopigments have a higher tolerance to light intensities before reaching saturation and can be very stable, a rod morphology with a larger outer segment, which allows a greater packing of photopigment molecules, would result in a higher photon catch and hence greater sensitivity (1). Furthermore, whereas rh1 peak spectral sensitivities across vertebrates seem to have a lower limit of around 470 nm (26), rh2 peak spectral sensitivities appear more variable, where a comparable short-wavelength–shifted pigment with a λmax value of 442 nm has previously been described in the deep-sea elephant shark (32). Further studies on the pearlside rod-like cones are, however, needed to verify their predicted spectral sensitivities (that is, using microspectrophotometry) and to assess their physiological properties to determine whether they function as a rod, a cone, or a combination of both, as has been reported for the cone-like rods of the skate Raja spp. (10, 45). Unfortunately, deep-sea fishes are difficult to source and catch alive and are not yet viable in normal aquarium settings, rendering most experimental and behavioral approaches extremely difficult.
Conclusions
Teleost fishes are remarkably adaptive, as revealed by their diverse forms and varied visual specializations. This study represents the first description of a teleost fish visual system that combines the characteristics of both rods and cones into a single photoreceptor type to presumably optimize vision in twilight conditions. This exceptional visual solution challenges the current paradigm of the evolution of the vertebrate duplex retina and the limits of visual adaptation. It also highlights the need for more comprehensive evaluations of visual systems in general and a more cautious approach in classifying photoreceptors into rods and cones.
MATERIALS AND METHODS
Sample collection and ocular tissue preservation
Adult individuals of similar sizes of the Mueller’s pearlside (M. muelleri) were collected in Masfjorden, Norway, aboard the research vessel G.O. Sars in October 2014. For each individual, the standard length and rostro-caudal eye diameter were measured with digital calipers to an accuracy of 0.1 mm before dissection. Eyes were enucleated, the cornea and lens were dissected free of the posterior chamber, and the lens diameter was measured as above. The eyecups were fixed either in 4% paraformaldehyde (PFA) in 0.1 M phosphate buffer (PB; pH 7.4) or in RNAlater (Thermo Fisher). RNAlater samples were stored at −80°C until further processing. The whole heads of three specimens of M. muelleri were also fixed in 4% PFA at 4°C for 48 hours.
Adult individuals of the pearlside, M. mucronatus, were collected in the Red Sea, Saudi Arabia, aboard the research vessel Thuwal in November 2014. Each individual was measured, and the eyes were enucleated in a similar fashion as for M. muelleri and preserved in RNAlater only.
The research undertaken in Saudi Arabia was in accordance with the policies and procedures of the King Abdullah University of Science and Technology. Permissions relevant for the research have been obtained from the applicable governmental agencies in the Kingdom of Saudi Arabia. The research undertaken in Norway followed the local animal care guidelines approval by the Norwegian Veterinary Authorities.
Transcriptome sequencing, quality filtering, and de novo assembly
Total RNA of five individuals of M. muelleri and five individuals of M. mucronatus were processed from RNAlater-fixed eyes and sequenced separately. Briefly, each eyecup was homogenized in TRIzol reagent, and total RNA was extracted using the PureLink RNA Mini Kit (Thermo Fisher) following the manufacturer’s protocol. Total RNA quality was checked using a Eukaryotic Total RNA chip on the Bioanalyzer 2100 (Agilent Technologies). mRNA was subsequently extracted from 2 μg of total RNA using a Dynabeads mRNA Purification kit (Thermo Fisher), and complementary DNA (cDNA) libraries were prepared from 50 ng of mRNA following a NEBNext Ultra Directional RNA Library Prep Kit protocol (item: E7420, New England Biolabs). The quality of libraries was checked using a Bioanalyzer DNA 1000 chip. Library concentrations were measured using a Qubit dsDNA BR Assay kit (Thermo Fisher). Samples were pooled in equimolar ratios, and 100-bp paired-end sequence reads were obtained using Illumina HiSeq 2000.
Transcriptomes were uploaded to the Genomics Virtual Laboratory (GVL 4.0.0) (46) on the Queensland Galaxy Server (http://galaxy-qld.genome.edu.au/galaxy/), and data were converted using FASTQ Groomer (Galaxy v.1.0.4) (47). Sequences were quality-checked with FastQC (Galaxy v.0.53) and trimmed with Trimmomatic (Galaxy v.0.32.2) (48) using customized settings [ILLUMINACLIP (TrueSeq3); HEADCROP (10 bp); LEADING (Q20), TRAILING (Q20); SLIDINGWINDOW (average of 4 bp with Q20); and MINLEN (80 bp)]. Trimming and quality filtering removed overrepresented sequences, retaining only those sequences with a minimum length of 80 bp and a quality score of ≥20. Two transcriptomes per species were subsequently assembled de novo using Trinity (Galaxy v.0.0.2) (49) with default settings, a k-mer coverage of at least four for initial contig construction, and a minimum contig length of 200 bp. Transcriptomes are available on the short-read archive database (www.ncbi.nlm.nih.gov/sra/) (tables S1 to S3) and assemblies on Dryad (http://datadryad.org/).
Gene extraction, phylogenetic reconstruction, and expression analyses
Putative opsin sequences of the two Maurolicus spp. were identified by initially mapping assembled contigs to publicly available opsin coding sequences (CDS) of the dusky dottyback, Pseudochromis fuscus (50), in Geneious v.9.1.5 (www.geneious.com) using customized sensitivity settings (fine tuning, none; maximum gap per read, 30%; word length, 14; maximum mismatches per read, 40%; maximum gap size, 1000 bp; and index word length, 12). Similarly, publicly available zebrafish (D. rerio) CDS was used to identify rod- and cone-specific signal transduction genes (gnat1 and gnat2) and pigment recovery genes (sag and arr3) (1). Mapped contigs were extracted and scored for similarity to publicly available gene sequences using BLASTN (https://blast.ncbi.nlm.nih.gov/Blast.cgi). However, de novo assembly of highly similar genes, in this case, opsin gene paralogs, using short-read libraries is prone to errors, often causing misassemblies (chimeric genes) or missing lowly expressed “rare” paralogs/isoforms/alleles (51, 52). Therefore, a second approach was used to corroborate the initial results by manually extracting pearlside opsin genes from back-mapped reads [similar to the study of Cortesi et al. (15)].
In this manual approach, unassembled reads were first mapped against the dusky dottyback opsin CDS using medium-sensitivity settings (70% identity threshold for mapping) in Geneious v.9.1.5. Matching reads were manually split and extracted by copy-moving from single polynucleotide polymorphism (SNP) to SNP and taking advantage of paired-end matching to cover gaps between SNPs (fig. S10). Extracted reads were assembled de novo, and their consensus was used as a species-specific reference for low-sensitivity (highly accurate) mapping using customized identity parameters (100% identity threshold for mapping). During this cyclic mapping, unassembled reads were mapped and assembled repeatedly against an extending reference until the assembled gene included >300 bp of the 3′UTR. Alignments were continuously inspected visually to ensure unambiguous mapping of genes. Both approaches were also used to search for the expression of cyp27c1.
Pearlside opsin and phototransduction gene sequences were confirmed and assigned to a particular class based on phylogenetic relationships with gene sequences from a number of species obtained from GenBank (www.ncbi.nlm.nih.gov/genbank/) or Ensembl (www.ensembl.org/) (Figs. 1A and 2, A and B, and figs. S1 and S3 to S5). The CDS of gene-specific data sets (that is, opsin, transducin, and arrestin) was aligned using the L-INS-I algorithm in MAFFT v.7.222 (53), and the most appropriate model of sequence evolution was estimated in jModeltest v.2.1.6 (54), using the Akaike information criterion for model selection. Bayesian inference phylogenetic hypothesis was calculated on the CIPRES platform (55), using GTR+I+γ models and Markov chain Monte Carlo searches with two independent runs and four chains each in MrBayes v.3.2.6 (56). Each run was set to 10 million generations, with trees sampled every 1000 generations (that is, 10,000 trees per run) and a burn-in of 25%. A similar approach was used to test for gene conversion between rh2 paralogs, only in this case the phylogeny was reconstructed using the information contained in the 3′UTR and the last 50 bp of exon 5 alone (fig. S5). To corroborate the phylogenetic hypotheses, we reran our data sets using Bayesian inference with a GTR+γ model (using parameters as above) as wells as maximum likelihood using RAxML v.8.2.9 (57) and 1000 iterations for bootstrapping. However, no substantial differences in tree structure or node support could be found, and the resulting trees and data alignments have been deposited in Dryad. GenBank accession numbers for genes used in different phylogenies are depicted after the species names in figs. S1 and S3 to S5.
To analyze differences in gene expression of pearlside opsins and phototransduction genes, unassembled reads were mapped against pearlside genes (CDS and 3′UTRs for duplicated rh2 genes and only CDS for all other genes) using customized low-sensitivity settings in Geneious v.9.1.5 (a minimum overlap between reads of 80 bp and 98% and 95% identity thresholds for opsin and phototransduction gene mapping, respectively). A higher identity threshold was used for opsin genes to assure the unambiguous mapping of reads against highly similar rh2 copies, but allowing for 2% differences to enable the mapping of reads containing heterozygous positions. Proportional gene expression (opsin, tranducin, or arrestin) was then calculated according to , where Ti/Tall is the gene expression ratio for a given gene Ti normalized by the total genes expressed Tall, and Ni is the number of mapped reads for a given gene divided by its length. First, the proportional expression of rod-specific genes was compared to the combined proportional expression of cone-specific genes (that is, rh1 versus rh2, gnat1 versus gnat2, and saga versus arr3), and second, the proportional expression of cone-specific genes was compared among themselves (Figs. 1B and 2, C and D). However, visual inspection of reads showed that, despite using high identity thresholds, read mapping against the CDS of rh2 paralogs remained inaccurate. Therefore, to resolve ambiguous mapping between cone opsin paralogs, all rh2-specific reads were extracted and submapped only against the 3′UTR of the genes. Proportional gene expression of rh2 paralogs was then recalculated using normalized read counts from this submapping approach (Fig. 1B and table S1).
Molecular cloning of visual opsins and R-ISH
Using one eyecup of M. muelleri fixed in RNAlater, total RNA was extracted using TRI reagent (Sigma), and DNase treatment (TURBO DNA-free, Ambion) was performed. Subsequently, cDNA was synthesized using SuperScript III (Invitrogen) reverse transcriptase with oligo(dT) primers following the manufacturer’s instructions. Gene-specific primers for M. muelleri visual opsin genes [rh1: forward primer MMRH1F1, GAGCCCGTATGAGTACCCTCAG; reverse primer MMRH1R1, CCACAGATGACGTGGAGGAG (gives a 1004-bp product); rh2: forward primer MMRH2x1F3, AACGCATCTGGGCTTGTGAG; reverse primer MMRH2x1R3, CTGGGGACACGGAAGAGAC (gives a 1000-bp product)] were used to amplify opsin sequences from cDNA derived from retinal tissue using polymerase chain reaction (PCR). Bands of the correct size were excised from the agarose gel, purified (MinElute Gel Extraction Kit, Qiagen), and cloned into StrataClone vector (Stratagene). To extract plasmids, a Miniprep (QIAprep Spin Kit, Qiagen) was used with cultures grown overnight from positive colonies (PCR-screened with M13 TOPO primers) following the manufacturer’s instructions. All positive plasmids were sequenced using a 3730XL analyzer (Applied Biosystems) at the University of Bergen Sequencing Facility (Norway). Sequenced opsins were confirmed by comparing the sequences to annotated M. muelleri opsin genes (table S4) and orthologs in other species using BLASTN and TBLASTX algorithms.
Antisense digoxigenin-labeled riboprobes for rh1 and rh2 opsins were prepared following the manufacturer’s instructions (Roche Diagnostics). For the synthesis of the riboprobes, opsin-specific PCR products using forward and reverse primers with T3 and T7 RNA polymerase sites (underlined) (rh1: MMRH1F1T3, CATTAACCCTCACTAAAGGGAAGAGCCCGTATGAGTACCCTCAG and MMRH1R1T7, TAATACGACTCACTATAGGGCCACAGATGACGTGGAGGAG; rh2: MMRH2x1F3T3, CATTAACCCTCACTAAAGGGAAAACGCATCTGGGCTTGTGAG and MMRH2x1R3T7, TAATACGACTCACTATAGGGCTGGGGACACGGAAGAGAC) were used as template, as described by Thisse and Thisse (58), and the synthesized probes were precipitated by 0.1× volume of 4 M LiCl and 3× volume of 100% ethanol together with transfer RNA (Roche Diagnostics).
The head of three M. muelleri fixed in 4% PFA was briefly washed in phosphate-buffered saline (PBS), and the eyes were dissected and incubated in a solution of 25% sucrose and 25% Tissue-Tek (Sakura Finetek) overnight at 4°C. Each eye was then mounted in a mold of Tissue-Tek and rapidly frozen on an iron block precooled in liquid nitrogen. Parallel sectioning (10 μm) series of the eyes was performed with a Leica CM3050 S cryostat. Before storage at −22°C, the tissue was air-dried for 1 hour at room temperature and for 10 min at 65°C. One parallel series of the sectioned eye was stained for the presence of rh1 expression, and the other parallel series was stained for rh2 transcripts by R-ISH, as described by Sandbakken et al. (59). Photographs were taken with a Leica 6000B microscope mounted with a Leica DFC digital camera.
In vitro expression and spectral sensitivity
Total RNA was extracted from a single M. muelleri eye previously fixed in RNAlater (Thermo Fisher), using the PureLink RNA Mini Kit (Thermo Fisher) following the manufacturer’s protocol. Oligo(dT)-primed retinal mRNA (2 μg) was converted to cDNA using the Transcriptor First Strand cDNA Synthesis Kit (Roche), according to the manufacturer’s instructions. Both rh1 and rh2 coding sequences were amplified using the following primer sets (where rh2 forward and reverse oligonucleotides were designed to detect both rh2-1 and rh2-2 gene paralogs): rh1, 5′-GCGCGAATTCCACCATGAACGGCACGGAGGGAC-3′ (PE_MMuelleri_RH1_F) and 5′-CGGCGTCGACGCTGCAGGGCCCACAGATGAC-3′ (PE_MMuelleri_RH1_R); rh2, 5′-GCGCGAATTCCACCATGAACGGAACGGAGGGAG-3′ (PE_MMuelleri_RH2_F) and 5′-CGGCGTCGACGCTGCTGGGGACACGGAAGAG-3′ (PE_MMuelleri_RH2_R).
PCR reagents and conditions were used as previously described (27, 60, 61). Opsin amplicons comprising the full-length coding sequences were excised from a 1.2% agarose gel and purified using a QIAquick Gel Extraction kit (Qiagen) before digestion with 1 U each of Eco RI and Sal I restriction enzymes and repurification with a QIAquick PCR Purification kit (Qiagen). These digested fragments were cloned into a pMT4 mammalian expression vector via the same restriction enzyme sites mentioned above, as previously described (60), and sequenced by the Australian Genome Research Facility to confirm the presence and fidelity of all three M. muelleri visual opsin sequences.
Each pearlside opsin plasmid (210 μg of DNA in total) was separately mixed with GeneJuice Transfection Reagent at a DNA/GeneJuice ratio of 3:1, according to the manufacturer’s instructions (Novagen), and used to transfect 12 large tissue culture plates (140 mm) that were preseeded with a nearly confluent monolayer of human embryonic kidney (HEK) 293T cells. The transfected cells were harvested after 48 hours, pelleted, and washed four times in PBS to remove all dead cells and any remaining traces of cell culture medium. Visual photopigments were repeatedly regenerated with excess 11-cis-retinal (>20 μM) under dark conditions. Cells were lysed with dodecyl maltoside detergent (1%), in the presence of the protease inhibitor phenylmethylsulfonyl fluoride (20 mg/ml), to release membrane fractions containing the opsin proteins. Lysate was then subjected to affinity chromatography over a CNBr-activated Sepharose-binding column coupled to an anti-1D4 monoclonal antibody (62), as previously described (60). Triplicate absorbance spectra were recorded in complete darkness using a Shimadzu UV-visible spectrophotometer (UV-2700) before bleaching with white light for 1 hour. A difference spectrum was calculated by subtracting the bleached spectrum from that measured in the dark, which was then fitted to a modified Govardovskii rhodopsin A1 template (63) using Microsoft Excel to determine the λmax, as previously shown (18, 32, 60).
Spectral sensitivities were predicted by analyzing amino acid substitution at 13 known key spectral tuning sites [83, 122, 124, 132, 207, 208, 211, 261, 265, 292, 295, 299, and 300 (18, 20, 26)] (fig. S2). Because several studies thus far suggest that similar tuning sites are used for both rh1 and rh2 pigments (18–20), all 13 sites were analyzed to predict spectral sensitivities for each gene.
Preparation of retinal whole mounts
Retinal whole mounts were prepared according to standard protocols (64–66). Radial cuts were performed to flatten the eye and subsequently the retina in toto onto a glass slide, where the orientation was confirmed by making a small additional cut in the nasal or dorsal part of the eye. The sclera was gently removed, and the retina was bleached overnight in a solution of 3% hydrogen peroxide in 0.1 M PB.
Immunohistochemistry
Anti-rhodopsin (rod opsin) antibody was used to selectively label the true rod photoreceptor population from whole mounts of three individuals of M. muelleri. Eyes fixed in 4% PFA were dissected and bleached as described previously. To optimize immunolabeling, two pretreatments were performed after bleaching: antigen retrieval [incubation in sodium citrate buffer (pH 6) at 60°C for 30 min] followed by endogenous peroxidase inactivation (15 min in a solution of 10% methanol and 3% hydrogen peroxide in 0.1 M PB) (67). Retinas were then rinsed three times (15 min each) in 0.1 M PBS and incubated for 30 min at room temperature in a blocking solution (5% goat serum and 0.3% Triton X-100 in 0.1 M PBS) with 50 μM glycine. Retinal whole mounts were incubated under gentle rocking at room temperature for 48 hours in a blocking solution containing anti-rhodopsin (mouse monoclonal, 1:500; MAB5316, Millipore). Thereafter, the retinas were washed three times in 0.1 M PB (15 min each) and further incubated for 24 hours in a mixture of horseradish peroxidase–conjugated anti-mouse secondary antibody (1:500, Jackson ImmunoResearch), 5% goat serum, and 0.05% thiomersal in 0.1 M PB. Finally, after being washed three times (15 min) in 0.1 M PB azide and twice (5 min) in 0.1 M sodium acetate buffer, retinal whole mounts were incubated in a solution containing 5% of 3,3′-diaminobenzidine solution, 2% nickel sulfate, and NH2Cl (100 mg/ml) in 0.1 M sodium acetate sodium for 5 min before adding 30% hydrogen peroxide and incubating for another 6 min. Finally, retinas were rinsed three times in 0.1 M sodium acetate buffer (15 min each) and mounted in 100% glycerol on a glass slide for analysis.
Immunofluorescence was performed on three retinas cut transversely using a vibratome (Leica 1000S). Thick sections (50 μm) were collected into a multiwall chamber and incubated in a blocking solution (0.5% bovine serum albumin, 0.05% saponin, 0.1% Triton X-100, and 0.05% sodium azide in 0.1 M PBS) for 30 min at room temperature. Sections were incubated with the following primary antibodies for 48 hours at room temperature: the synaptic vesicle marker anti-SV2 (mouse monoclonal at 1:300, Developmental Studies Hybridoma Bank) and the neuronal marker NeuN (rabbit polyclonal used at 1:1000, Merck Millipore). Subsequently, sections were washed three times in 0.1 M PBS (15 min) and incubated for 5 hours at room temperature with species-specific immunoglobulin G (IgG) secondary antibodies: specifically, these comprised either anti-mouse Alexa Fluor 488 or anti-rabbit Alexa Fluor 555 (1:1000, Thermo Fisher) diluted in a blocking solution. Sections were incubated in donkey anti-mouse IgG (1:500, Jackson ImmunoResearch) overnight to absorb any unbound mouse IgG and cross-linked with 4% PFA for 15 min. Sections were washed three times in PBS (15 min each) and incubated with the mouse monoclonal anti-rhodopsin antibody as described above at 1:500 for 48 hours at room temperature. Sections were washed three times in PBS (15 min each) and incubated with an anti-mouse IgG Alexa Fluor 647 secondary antibody (1:1000, Thermo Fisher) made in a blocking solution. Sections were washed in PBS and incubated with DAPI to stain the nuclei. Sections were mounted in vector shield and imaged using a Yokogawa CSU-W1 spinning disc confocal microscope.
Photoreceptor morphology
TEM and serial block-face scanning electron microscopy (3View) were used to assess the photoreceptor morphology of M. muelleri. Each analysis was performed on retinas fixed in 4% PFA and subsequently cross-linked with 2.5% glutaraldehyde in 0.1 M PB (pH 7.4). Because both rods and cones are only present in the periphery of the retina, pieces of tissue from the retinal margins were processed and analyzed. For TEM analyses, pieces of retina were postfixed in 1% osmium tetroxide in 0.15 M PB, dehydrated through an alcohol and acetone series, and infiltrated with Epon resin (ProSciTech) using a BioWave tissue processor. Resin samples were then polymerized at 60°C for 48 hours. Ultrathin sections (90 nm) were cut using a diamond knife, mounted on a 100-mesh square copper grid, and stained with uranyl acetate and Reynold’s lead citrate. Examination of the sections was conducted using a JEOL 1010 transmission electron microscope operating at 80 kV, and images were taken using an Olympus Soft Imaging Solutions Veleta 2K × 2K wide-angle digital camera. To be able to identify morphological differences between distinct photoreceptor types, one of the retinas previously labeled with anti-rhodopsin was first analyzed. This allowed the true rods (expressing rh1) to be confidently distinguished from rod-like cones (expressing either rh2-1 or rh2-2) and to identify morphological characteristics specific to each photoreceptor type. Further TEM analyses on unlabeled retinas were performed to acquire high-resolution and magnification images of the different parts and components of each photoreceptor type.
3View analysis was performed to reconstruct and view the two photoreceptor types in 3D. Briefly, retinal tissue was postfixed in 2% osmium tetroxide/1.5% potassium ferricyanide solution, followed by 1% thiocarbohydrazide and 2% osmium tetroxide, then stained overnight with 1% uranyl acetate, and incubated in lead aspartate for 1 hour at 60°C. Each step was followed by 3 × 5-min rinse in ultrapure water. The tissue was dehydrated with increasing concentrations of ethanol before infiltration and embedding in Durcupan resin [protocol adapted from Nguyen et al. (68)]. The resin-embedded tissue was trimmed, mounted on aluminum pins, and coated with conductive silver. Retinal tissue was imaged on a Zeiss Sigma SEM fitted with a 3View (Gatan). Imaging was conducted at 2 kV with section thickness of 50 or 100 nm. From the images, the surfaces of each photoreceptor type were manually segmented and rendered for 3D visualization using the image analysis software IMOD (69).
Distribution of the different neural cell types across the retina
For topographic analysis of photoreceptors, retinas were whole-mounted (photoreceptor layer up) in 100% glycerol on a microscopic slide. For similar analysis of ganglion cells, retinas were flat-mounted on a gelatinized slide (ganglion cell layer facing up) and stained for 5 min in 0.1% cresyl violet, following the protocol of Coimbra et al. (65). Shrinkage using this technique was deemed negligible because the retinal whole mounts were attached to the slides during all steps (65). Different types of analyses were performed for high-density cell types (that is, rod-like cone photoreceptors and ganglion cells) and low-density cell types (rod photoreceptors).
Following the protocols described by de Busserolles et al. (33, 70), topographic distribution of rod-like cone photoreceptor and ganglion cell populations was assessed using the optical fractionator technique (71) modified by Coimbra et al. (67, 72) for its use in whole-mounted retinas. Briefly, the outline of the retinal whole mounts was digitized using a 4× objective (numerical aperture, 0.13) mounted on a compound microscope (Olympus BX50) equipped with a motorized stage (MAC5000, Ludl Electronic Products), a digital video camera (MicroFire, Optronics), and a computer running Stereo Investigator software (MicroBrightField). Using a 60× oil immersion objective (numerical aperture, 1.35) for ganglion cell counts and 100× oil objective (numerical aperture, 1.40) for photoreceptor counts, cells were randomly and systematically counted using the parameters listed in table S5. The counting frame and grid size were chosen carefully to maintain the highest level of sampling (~200 sampling sites) and achieve an acceptable Schaeffer coefficient of error (CE). The CE is a measure of the accuracy of the total number of cell estimates and is considered acceptable below 0.1. Topographic maps were constructed using the statistical program R v.2.15.0 (R Foundation for Statistical Computing, 2012) with the results exported from the Stereo Investigator Software according to Garza-Gisholt et al. (73). The Gaussian kernel smoother from the Spatstat package (74) was used and adjusted the sigma value to the distance between points (that is, grid size).
The distribution of the rod photoreceptors labeled with anti-rhodopsin was mapped from one retina using the Neurolucida software (MicroBrightField). Although IHC was performed on three retinas and gave similar results visually (that is, labeling at the peripheral margins), only one whole mount preparation was deemed good enough for quantitative analysis due to the challenge of flattening the retinal margins in this type of preparation. The outline of the retinal whole mounts was digitized using a 5× objective (numerical aperture, 0.13). The entire retina was then scanned in contiguous steps using a 20× objective (numerical aperture, 0.8), and each labeled cell was marked. Results were exported from the Neurolucida software, and a dot map representing the location of each labeled cell was constructed using the statistical program R v.2.15.0 (R Foundation for Statistical Computing) and a customized version of the Garza-Gisholt et al. (73) script.
Supplementary Material
Acknowledgments
We thank S. Johnsen for providing downwelling vector irradiance spectra. We gratefully acknowledge A. G. Salvanes, the captain and crew of the G.O. SARS and R. V. Thuwal for sea time opportunities, and G. Ben-Ari for providing access to a Stereo Investigator system at the University of Western Australia. We also thank R. Karlsen for technical support with cloning and ISH, R. Chapman for technical assistance with the 3View analysis, and R. Crouch and the National Eye Institute, NIH, for the provision of 11-cis-retinal. We also acknowledge the facilities and the scientific and technical assistance of the Australian Microscopy and Microanalysis Research Facility at the Centre for Microscopy and Microanalysis, and the Queensland Brain Institute’s Advanced Microscopy Facility at the University of Queensland. Funding: This work was supported by King Abdullah University of Science and Technology, the Air Force Office of Scientific Research, and the Australian Research Council via grants awarded to J.M.; a Discovery Project grant (DP140102117) and Future Fellowship (FT110100176) awarded to W.I.L.D.; and a Linkage Infrastructure, Equipment and Facilities (LIEF) grant (LE100100074) awarded to the Queensland Brain Institute (Neurolucida software). F.C. was supported by a Swiss National Science Foundation Early Postdoctoral Mobility Fellowship (165364) and a University of Queensland Development Fellowship. J.V.H. was supported by a start-up grant from the University of Bergen. Author contributions: F.d.B., F.C., and S.K. designed the study. F.d.B., F.C., J.V.H., W.I.L.D., R.M.T., R.K.P.S., C.T.M., and J.K.M. performed the experiments. F.d.B., F.C., J.V.H., and W.I.L.D. analyzed the data. S.P.C., X.I., S.K., and J.M. supervised the project. All authors wrote the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper, Supplementary Materials, Dryad repository, and GenBank (single gene accession no. MF805819-MF805840 and transcriptome accession no. for the project PRJNA401503 and for the samples SRX3161870, SRX3161890, SRX3161913-SRX3161915, and SRX3162896-SRX3162900). Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/11/eaao4709/DC1
fig. S1. Gene coding region (CDS) inferred vertebrate opsin gene phylogeny.
fig. S2. Reconstruction of amino acid changes at known key spectral tuning sites of pearlside opsins.
fig. S3. Vertebrate Gα transducin gene phylogeny.
fig. S4. Vertebrate arrestin gene phylogeny.
fig. S5. rh2 opsin gene conversion phylogeny.
fig. S6. Topographic distribution of rod-like cone photoreceptors in three individuals of M. muelleri.
fig. S7. Topographic distribution of ganglion cells (excluding amacrine cells and glial cells) in three individuals of M. muelleri.
fig. S8. Expression of rh2 and rh1 opsin genes from sectional RNA in situ hybridization analyses of the eye of M. muelleri.
fig. S9. Location of the ”true“ rod photoreceptors and their distribution across the retina in M. muelleri.
fig. S10. Manual approach to extract genes from back-mapped reads.
table S1. Summary of transcriptomes, GenBank accession numbers, opsin mapping (including base pair coverage), and proportional opsin gene expression.
table S2. Summary of transcriptomes, GenBank accession numbers, transducin mapping (including base pair coverage), and proportional transducin gene expression.
table S3. Summary of transcriptomes, GenBank accession numbers, arrestin mapping (including base pair coverage), and proportional arrestin gene expression.
table S4. Single-gene GenBank accession numbers of gene sequences produced during this study.
table S5. Summary of the stereology parameters used for the analysis of the rod-like cone photoreceptors and ganglion cell distribution along with the quantitative results obtained using the optical fractionator methods in six retinas of M. muelleri.
movie S1. 3D reconstruction of the two photoreceptor types in M. muelleri.
movie S2. Close-up 3D reconstruction of the nucleus and synaptic terminal of the two photoreceptor types in M. muelleri.
REFERENCES AND NOTES
- 1.Lamb T. D., Evolution of phototransduction, vertebrate photoreceptors and retina. Prog. Retin. Eye Res. 36, 52–119 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Munz F. W., McFarland W. N., The significance of spectral position in the rhodopsins of tropical marine fishes. Vision Res. 13, 1829–1874 (1973). [DOI] [PubMed] [Google Scholar]
- 3.Stockman A., Sharpe L. T., Into the twilight zone: The complexities of mesopic vision and luminous efficiency. Ophthalmic Physiol. Opt. 26, 225–239 (2006). [DOI] [PubMed] [Google Scholar]
- 4.G. L. Walls, The Vertebrate Eye and Its Adaptive Radiation (The Cranbrook Institute of Science, 1942), p. 785. [Google Scholar]
- 5.McDevitt D. S., Brahma S. K., Jeanny J.-C., Hicks D., Presence and foveal enrichment of rod opsin in the “all cone” retina of the American chameleon. Anat. Rec. 237, 299–307 (1993). [DOI] [PubMed] [Google Scholar]
- 6.Röll B., Gecko vision—Visual cells, evolution, and ecological constraints. J. Neurocytol. 29, 471–484 (2000). [DOI] [PubMed] [Google Scholar]
- 7.Schott R. K., Müller J., Yang C. G. Y., Bhattacharyya N., Chan N., Xu M., Morrow J. M., Ghenu A.-H., Loew E. R., Tropepe V., Chang B. S. W., Evolutionary transformation of rod photoreceptors in the all-cone retina of a diurnal garter snake. Proc. Natl. Acad. Sci. U.S.A. 113, 356–361 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma J.-x., Znoiko S., Othersen K. L., Ryan J. C., Das J., Isayama T., Kono M., Oprian D. D., Corson D. W., Cornwall M. C., Cameron D. A., Harosi F. I., Makino C. L., Crouch R. K., A visual pigment expressed in both rod and cone photoreceptors. Neuron 32, 451–461 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Dickson D. H., Graves D. A., Fine structure of the lamprey photoreceptors and retinal pigment epithelium (Petromyzon marinus L.). Exp. Eye Res. 29, 45–60 (1979). [DOI] [PubMed] [Google Scholar]
- 10.Dowling J. E., Ripps H., On the duplex nature of the skate retina. J. Exp. Zool. 256, 55–65 (1990). [DOI] [PubMed] [Google Scholar]
- 11.Giske J., Aksnes D. L., Baliño B. M., Kaartvedt S., Lie U., Nordeide J. T., Salvanes A. G. V., Wakili S. M., Aadnesen A., Vertical distribution and trophic interactions of zooplankton and fish in Masfjorden, Norway. Sarsia 75, 65–81 (1990). [Google Scholar]
- 12.Clark C. W., Levy D. A., Diel vertical migrations by juvenile sockeye salmon and the antipredation window. Am. Nat. 131, 271–290 (1988). [Google Scholar]
- 13.M. A. Ali, M. Anctil, Retinas of Fishes: An Atlas (Springer Science & Business Media, 1976). [Google Scholar]
- 14.Lamb T. D., Patel H., Chuah A., Natoli R. C., Davies W. I. L., Hart N. S., Collin S. P., Hunt D. M., Evolution of vertebrate phototransduction: Cascade activation. Mol. Biol. Evol. 33, 2064–2087 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cortesi F., Musilová Z., Stieb S. M., Hart N. S., Siebeck U. E., Malmstrøm M., Tørresen O. K., Jentoft S., Cheney K. L., Marshall N. J., Carleton K. L., Salzburger W., Ancestral duplications and highly dynamic opsin gene evolution in percomorph fishes. Proc. Natl. Acad. Sci. U.S.A. 112, 1493–1498 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imanishi Y., Hisatomi O., Tokunaga F., Two types of arrestins expressed in medaka rod photoreceptors. FEBS Lett. 462, 31–36 (1999). [DOI] [PubMed] [Google Scholar]
- 17.Kelber A., Vorobyev M., Osorio D., Animal colour vision—Behavioural tests and physiological concepts. Biol. Rev. 78, 81–118 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Davies W. L., Cowing J. A., Carvalho L. S., Potter I. C., Trezise A. E. O., Hunt D. M., Collin S. P., Functional characterization, tuning, and regulation of visual pigment gene expression in an anadromous lamprey. FASEB J. 21, 2713–2724 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Yokoyama S., Evolution of dim-light and color vision pigments. Annu. Rev. Genomics Hum. Genet. 9, 259–282 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Yokoyama S., Molecular evolution of vertebrate visual pigments. Prog. Retin. Eye Res. 19, 385–419 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Morrow J. M., Lazic S., Dixon Fox M., Kuo C., Schott R. K., de A. Gutierrez E., Santini F., Tropepe V., Chang B. S. W., A second visual rhodopsin gene, rh1-2, is expressed in zebrafish photoreceptors and found in other ray-finned fishes. J. Exp. Biol. 220, 294–303 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Escobar-Camacho D., Ramos E., Martins C., Carleton K. L., The opsin genes of Amazonian cichlids. Mol. Ecol. 26, 1343–1356 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Busserolles F., Hart N. S., Hunt D. M., Davies W. I., Marshall N. J., Clarke M. W., Hahne D., Collin S. P., Spectral tuning in the eyes of deep-sea lanternfishes (Myctophidae): A novel sexually dimorphic intra-ocular filter. Brain Behav. Evol. 85, 77–93 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Enright J. M., Toomey M. B., Sato S.-y., Temple S. E., Allen J. R., Fujiwara R., Kramlinger V. M., Nagy L. D., Johnson K. M., Xiao Y., How M. J., Johnson S. L., Roberts N. W., Kefalov V. J., Guengerich F. P., Corbo J. C., Cyp27c1 red-shifts the spectral sensitivity of photoreceptors by converting vitamin A1 into A2. Curr. Biol. 25, 3048–3057 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davies W. I. L., Downes S. M., Fu J. K., Shanks M. E., Copley R. R., Lise S., Ramsden S. C., Black G. C. M., Gibson K., Foster R. G., Hankins M. W., Németh A. H., Next-generation sequencing in health-care delivery: Lessons from the functional analysis of rhodopsin. Genet. Med. 14, 891–899 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Hunt D. M., Dulai K. S., Partridge J. C., Cottrill P., Bowmaker J. K., The molecular basis for spectral tuning of rod visual pigments in deep-sea fish. J. Exp. Biol. 204, 3333–3344 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Davies W. I. L., Tamai T. K., Zheng L., Fu J. K., Rihel J., Foster R. G., Whitmore D., Hankins M. W., An extended family of novel vertebrate photopigments is widely expressed and displays a diversity of function. Genome Res. 25, 1666–1679 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davies W. I. L., Turton M., Peirson S. N., Follett B. K., Halford S., Garcia-Fernandez J. M., Sharp P. J., Hankins M. W., Foster R. G., Vertebrate ancient opsin photopigment spectra and the avian photoperiodic response. Biol. Lett. 8, 291–294 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davies W. L., Hankins M. W., Foster R. G., Vertebrate ancient opsin and melanopsin: Divergent irradiance detectors. Photochem. Photobiol. Sci. 9, 1444–1457 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Pointer M. A., Carvalho L. S., Cowing J. A., Bowmaker J. K., Hunt D. M., The visual pigments of a deep-sea teleost, the pearl eye Scopelarchus analis. J. Exp. Biol. 210, 2829–2835 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Partridge J. C., Archer S. N., Vanoostrum J., Single and multiple visual pigments in deep-sea fishes. J. Mar. Biol. Assoc. U.K. 72, 113–130 (1992). [Google Scholar]
- 32.Davies W. L., Carvalho L. S., Tay B.-H., Brenner S., Hunt D. M., Venkatesh B., Into the blue: Gene duplication and loss underlie color vision adaptations in a deep-sea chimaera, the elephant shark Callorhinchus milii. Genome Res. 19, 415–426 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Busserolles F., Marshall N. J., Collin S. P., Retinal ganglion cell distribution and spatial resolving power in deep-sea lanternfishes (Myctophidae). Brain Behav. Evol. 84, 262–276 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Kim J.-W., Yang H.-J., Oel A. P., Brooks M. J., Jia L., Plachetzki D. C., Li W., Allison W. T., Swaroop A., Recruitment of rod photoreceptors from short-wavelength-sensitive cones during the evolution of nocturnal vision in mammals. Dev. Cell 37, 520–532 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simões B. F., Sampaio F. L., Loew E. R., Sanders K. L., Fisher R. N., Hart N. S., Hunt D. M., Partridge J. C., Gower D. J., Multiple rod–cone and cone–rod photoreceptor transmutations in snakes: Evidence from visual opsin gene expression. Proc. Biol. Sci. 283, 20152624 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhattacharyya N., Darren B., Schott R. K., Tropepe V., Chang B. S. W., Cone-like rhodopsin expressed in the all-cone retina of the colubrid pine snake as a potential adaptation to diurnality. J. Exp. Biol. 220, 2418–2425 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Bozzano A., Pankhurst P. M., Sabatés A., Early development of eye and retina in lanternfish larvae. Vis. Neurosci. 24, 423–436 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Biagioni L. M., Hunt D. M., Collin S. P., Morphological characterization and topographic analysis of multiple photoreceptor types in the retinae of mesopelagic hatchetfishes with tubular eyes. Front. Ecol. Evol. 4, 25 (2016). [Google Scholar]
- 39.Staby A., Aksnes D. L., Follow the light—Diurnal and seasonal variations in vertical distribution of the mesopelagic fish Maurolicus muelleri. Mar. Ecol. Prog. Ser. 422, 265–273 (2011). [Google Scholar]
- 40.Warrant E. J., Locket N. A., Vision in the deep sea. Biol. Rev. 79, 671–712 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Widder E. A., Bioluminescence in the ocean: Origins of biological, chemical, and ecological diversity. Science 328, 704–708 (2010). [DOI] [PubMed] [Google Scholar]
- 42.Sweeney A. M., Boch C. A., Johnsen S., Morse D. E., Twilight spectral dynamics and the coral reef invertebrate spawning response. J. Exp. Biol. 214, 770–777 (2011). [DOI] [PubMed] [Google Scholar]
- 43.Parin N. V., Kobyliansky S. G., Diagnoses and distribution of fifteen species recognized in genus Maurolicus Cocco (Sternoptychidae, Stomiiformes) with a key to their identification. Cybium 20, 185–195 (1996). [Google Scholar]
- 44.P. J. Herring, How to survive in the dark: Bioluminescence in the deep sea, in Physiological Adaptations in Marine Animals, M. S. Laverack, Ed. (Society for Experimental Biology, 1985), vol. 39, pp. 323–350. [PubMed] [Google Scholar]
- 45.Ripps H., Dowling J. E., Structural features and adaptive properties of photoreceptors in the skate retina. J. Exp. Zool. 256, 46–54 (1990). [DOI] [PubMed] [Google Scholar]
- 46.Afgan E., Sloggett C., Goonasekera N., Makunin I., Benson D., Crowe M., Gladman S., Kowsar Y., Pheasant M., Horst R., Lonie A., Genomics virtual laboratory: A practical bioinformatics workbench for the cloud. PLOS ONE 10, e0140829 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blankenberg D., Gordon A., Von Kuster G., Coraor N., Taylor J., Nekrutenko A. Galaxy Team , Manipulation of FASTQ data with Galaxy. Bioinformatics 26, 1783–1785 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haas B. J., Papanicolaou A., Yassour M., Grabherr M., Blood P. D., Bowden J., Couger M. B., Eccles D., Li B., Lieber M., MacManes M. D., Ott M., Orvis J., Pochet N., Strozzi F., Weeks N., Westerman R., William T., Dewey C. N., Henschel R., LeDuc R. D., Friedman N., Regev A., De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cortesi F., Musilová Z., Stieb S. M., Hart N. S., Siebeck U. E., Cheney K. L., Salzburger W., Marshall N. J., From crypsis to mimicry: Changes in colour and the configuration of the visual system during ontogenetic habitat transitions in a coral reef fish. J. Exp. Biol. 219, 2545–2558 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Conesa A., Madrigal P., Tarazona S., Gomez-Cabrero D., Cervera A., McPherson A., Szcześniak M. W., Gaffney D. J., Elo L. L., Zhang X., Mortazavi A., A survey of best practices for RNA-seq data analysis. Genome Biol. 17, 13 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vijay N., Poelstra J. W., Künstner A., Wolf J. B. W., Challenges and strategies in transcriptome assembly and differential gene expression quantification. A comprehensive in silico assessment of RNA-seq experiments. Mol. Ecol. 22, 620–634 (2013). [DOI] [PubMed] [Google Scholar]
- 53.Katoh K., Misawa K., Kuma K.-i., Miyata T., MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Darriba D., Taboada G. L., Doallo R., Posada D., jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 9, 772 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.M. A. Miller, W. Pfeiffer, T. Schwartz, Creating the CIPRES Science Gateway for inference of large phylogenetic trees, in 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, 14 November 2010 (IEEE, 2010), pp. 1–8. [Google Scholar]
- 56.Ronquist F., Teslenko M., van der Mark P., Ayres D. L., Darling A., Höhna S., Larget B., Liu L., Suchard M. A., Huelsenbeck J. P., MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stamatakis A., RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thisse C., Thisse B., High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59–69 (2008). [DOI] [PubMed] [Google Scholar]
- 59.Sandbakken M., Ebbesson L., Stefansson S., Helvik J. V., Isolation and characterization of melanopsin photoreceptors of Atlantic salmon (Salmo salar). J. Comp. Neurol. 520, 3727–3744 (2012). [DOI] [PubMed] [Google Scholar]
- 60.Davies W. I. L., Zheng L., Hughes S., Tamai T. K., Turton M., Halford S., Foster R. G., Whitmore D., Hankins M. W., Functional diversity of melanopsins and their global expression in the teleost retina. Cell. Mol. Life Sci. 68, 4115–4132 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Davies W. L., Cowing J. A., Bowmaker J. K., Carvalho L. S., Gower D. J., Hunt D. M., Shedding light on serpent sight: The visual pigments of henophidian snakes. J. Neurosci. 29, 7519–7525 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Molday R. S., MacKenzie D., Monoclonal antibodies to rhodopsin: Characterization, cross-reactivity, and application as structural probes. Biochemistry 22, 653–660 (1983). [DOI] [PubMed] [Google Scholar]
- 63.Govardovskii V. I., Fyhrquist N., Reuter T., Kuzmin D. G., Donner K., In search of the visual pigment template. Vis. Neurosci. 17, 509–528 (2000). [DOI] [PubMed] [Google Scholar]
- 64.J. Stone, The Whole Mount Handbook: A Guide to the Preparation and Analysis of Retinal Whole Mounts (Maitland Publications, 1981), p. 128. [Google Scholar]
- 65.Coimbra J. P., Marceliano M. L. V., Andrade-da-Costa B. L. S., Yamada E. S., The retina of tyrant flycatchers: Topographic organization of neuronal density and size in the ganglion cell layer of the great kiskadee Pitangus sulphuratus and the rusty margined flycatcher Myiozetetes cayanensis (Aves: Tyrannidae). Brain Behav. Evol. 68, 15–25 (2006). [DOI] [PubMed] [Google Scholar]
- 66.Ullmann J. F. P., Moore B. A., Temple S. E., Fernández-Juricic E., Collin S. P., The retinal wholemount technique: A window to understanding the brain and behaviour. Brain Behav. Evol. 79, 26–44 (2012). [DOI] [PubMed] [Google Scholar]
- 67.Coimbra J. P., Nolan P. M., Collin S. P., Hart N. S., Retinal ganglion cell topography and spatial resolving power in penguins. Brain Behav. Evol. 80, 254–268 (2012). [DOI] [PubMed] [Google Scholar]
- 68.Nguyen J. V., Soto I., Kim K.-Y., Bushong E. A., Oglesby E., Valiente-Soriano F. J., Yang Z., Davis C.-h. O., Bedont J. L., Son J. L., Wei J. O., Buchman V. L., Zack D. J., Vidal-Sanz M., Ellisman M. H., Marsh-Armstrong N., Myelination transition zone astrocytes are constitutively phagocytic and have synuclein dependent reactivity in glaucoma. Proc. Natl. Acad. Sci. U.S.A. 108, 1176–1181 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kremer J. R., Mastronarde D. N., McIntosh J. R., Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76 (1996). [DOI] [PubMed] [Google Scholar]
- 70.de Busserolles F., Fitzpatrick J. L., Marshall N. J., Collin S. P., The influence of photoreceptor size and distribution on optical sensitivity in the eyes of lanternfishes (Myctophidae). PLOS ONE 9, e99957 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.West M. J., Slomianka L., Gundersen H. J. G., Unbiased stereological estimation of the total number of neurons in the subdivisions of the rat hippocampus using the optical fractionator. Anat. Rec. 231, 482–497 (1991). [DOI] [PubMed] [Google Scholar]
- 72.Coimbra J. P., Trévia N., Marceliano M. L. V., da Silveira Andrade-Da-Costa B. L., Picanço-Diniz C. W., Yamada E. S., Number and distribution of neurons in the retinal ganglion cell layer in relation to foraging behaviors of tyrant flycatchers. J. Comp. Neurol. 514, 66–73 (2009). [DOI] [PubMed] [Google Scholar]
- 73.Garza-Gisholt E., Hemmi J. M., Hart N. S., Collin S. P., A comparison of spatial analysis methods for the construction of topographic maps of retinal cell density. PLOS ONE 9, e93485 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Baddeley A., Turner R., Spatstat: An R package for analyzing spatial point patterns. J. Stat. Softw. 12, 1–42 (2005). [Google Scholar]
- 75.D. C. Hood, M. A. Finkelstein, Sensitivity to light, in Handbook of Perception and Human Performance, K. Boff, L. Kaufman, J. Thomas, Eds. (Wiley, 1986), vol. 1. [Google Scholar]
- 76.H. W. Leibowitz, Ambient illuminance during twilight and from the moon, in Night Vision: Current Research and Future Directions. Symposium Proceedings (National Academies Press, 1987), pp. 19–22. [Google Scholar]
- 77.N. J. Marshall, M. Vorobyev, U. Siebeck, What does a reef fish see when it sees a reef fish? Eating “Nemo,” in Communication in Fishes, F. Ladich, S. P. Collin, P. Moller, B. G. Kapoor, Eds. (Science Publisher, 2006). [Google Scholar]
- 78.C. G. Y. Yang, thesis, University of Toronto (2010). [Google Scholar]
- 79.Brin K. P., Ripps H., Rhodopsin photoproducts and rod sensitivity in the skate retina. J. Gen. Physiol. 69, 97–120 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kawamura S., Yokoyama S., Functional characterization of visual and nonvisual pigments of American chameleon (Anolis carolinensis). Vision Res. 38, 37–44 (1998). [DOI] [PubMed] [Google Scholar]
- 81.F. Crescitelli, The visual cells and visual pigments of the vertebrate eye, in Photochemistry of Vision, H. J. A. Dartnall, Ed. (Springer, 1972), pp. 245–363. [Google Scholar]
- 82.Morshedian A., Fain G. L., Single-photon sensitivity of lamprey rods with cone-like outer segments. Curr. Biol. 25, 484–487 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang H., Yokoyama S., Molecular evolution of the rhodopsin gene of marine lamprey, Petromyzon marinus. Gene 191, 1–6 (1997). [DOI] [PubMed] [Google Scholar]
- 84.Kojima D., Okano T., Fukada Y., Shichida Y., Yoshizawa T., Ebrey T. G., Cone visual pigments are present in gecko rod cells. Proc. Natl. Acad. Sci. U.S.A. 89, 6841–6845 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang X., Wensel T. G., Yuan C., Tokay gecko photoreceptors achieve rod-like physiology with cone-like proteins. Photochem. Photobiol. 82, 1452–1460 (2006). [DOI] [PubMed] [Google Scholar]
- 86.Kleinschmidt J., Dowling J. E., Intracellular recordings from gecko photoreceptors during light and dark adaptation. J. Gen. Physiol. 66, 617–648 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mariani A. P., Photoreceptors of the larval tiger salamander retina. Proc. R. Soc. Lond. B Biol. Sci. 227, 483–492 (1986). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/11/eaao4709/DC1
fig. S1. Gene coding region (CDS) inferred vertebrate opsin gene phylogeny.
fig. S2. Reconstruction of amino acid changes at known key spectral tuning sites of pearlside opsins.
fig. S3. Vertebrate Gα transducin gene phylogeny.
fig. S4. Vertebrate arrestin gene phylogeny.
fig. S5. rh2 opsin gene conversion phylogeny.
fig. S6. Topographic distribution of rod-like cone photoreceptors in three individuals of M. muelleri.
fig. S7. Topographic distribution of ganglion cells (excluding amacrine cells and glial cells) in three individuals of M. muelleri.
fig. S8. Expression of rh2 and rh1 opsin genes from sectional RNA in situ hybridization analyses of the eye of M. muelleri.
fig. S9. Location of the ”true“ rod photoreceptors and their distribution across the retina in M. muelleri.
fig. S10. Manual approach to extract genes from back-mapped reads.
table S1. Summary of transcriptomes, GenBank accession numbers, opsin mapping (including base pair coverage), and proportional opsin gene expression.
table S2. Summary of transcriptomes, GenBank accession numbers, transducin mapping (including base pair coverage), and proportional transducin gene expression.
table S3. Summary of transcriptomes, GenBank accession numbers, arrestin mapping (including base pair coverage), and proportional arrestin gene expression.
table S4. Single-gene GenBank accession numbers of gene sequences produced during this study.
table S5. Summary of the stereology parameters used for the analysis of the rod-like cone photoreceptors and ganglion cell distribution along with the quantitative results obtained using the optical fractionator methods in six retinas of M. muelleri.
movie S1. 3D reconstruction of the two photoreceptor types in M. muelleri.
movie S2. Close-up 3D reconstruction of the nucleus and synaptic terminal of the two photoreceptor types in M. muelleri.


