Abstract
Small cell lung cancer (SCLC) accounts for approximately 15% of all lung cancers and demands effective targeted therapeutic strategies. In this meta-analysis study, we aim to identify significantly mutated genes and regulatory pathways to help us better understand the progression of SCLC and to identify potential biomarkers. Besides ranking genes based on their mutation frequencies, we sought to identify statistically significant mutations in SCLC with the MutSigCV software. Our analysis identified several genes with relatively low mutation frequency, including PTEN, as highly significant (p<0.001), suggesting these genes may play an important role in the progression of SCLC. Our results also indicated mutations in genes involved in the axon guidance pathways likely play an important role in SCLC progression. In addition, we observed that the mutation rate was significantly higher in samples with RB1 gene mutated when compared to samples with wild type RB1, suggesting that RB1 status has significant impact on the mutation profile and disease progression in SCLC.
Keywords: Small cell lung cancer, High-throughput sequencing data, Meta-analysis, Significantly mutated genes, PTEN, RB1
1. Introduction
Lung cancer is the leading cause of cancer deaths in men and women in the U.S. and worldwide. Small cell lung cancer (SCLC), a neuroendocrine carcinoma accounts for approximately 15% of all lung cancers. Although SCLC can be initially responsive to standard chemotherapy and radiation therapy, the recurrence rate is very high and there is a lack of effective second-line strategy [1]. Despite the improvements in diagnosis and therapeutic interventions made in the past two decades, the current prognosis for patients with SCLC remains poor. Potential targeted therapies for SCLC include those that target the EGFR pathway, the apoptotic pathway, and VEGF. Immunotherapeutic agents are also under clinical investigation for the treatment of SCLC [2]. However, most of these agents have limited efficacy and there has not yet been a promising candidate target for drug development. Thus, there is an urgent need for novel predictive biomarkers to differentiate subtypes of SCLC for personalized treatments.
Cancer is considered to be a genetic disease with sequential accumulation of somatic mutations and selective sub-clonal evolution of cancer cells. SCLC is known to exhibit very high mutation rates and genomic instability, possibly linked to tobacco carcinogens [3]. TP53 and RB1 are known to be the two most frequently inactivated tumor suppressors in SCLC [4]. Recently, high throughput sequencing (HTS) analysis implicated many other genes for their role in SCLC, including kinases, G-protein-coupled receptors, chromatin-modifying proteins, SOX family members, and genes in the PI3K/AKT/mTOR pathway [5–8]. Many of these analyses were based on mutation frequency of the genes observed in SCLC. However, in the light of recent studies indicating that frequency-based approaches tend to overclassify tumor suppressor genes, it was not clear whether these genes are indeed significantly mutated in SCLC. On the other hand, genes mutated with low frequency, but are nonetheless statistically significant, may be responsible for the progression of SCLC and may serve as good candidates for subtyping.
Mechanism of tumorigenesis and progression may also be revealed by studying patterns of mutation and/or silencing, such as co-occurring and mutually exclusive mutations. For instance, the Kaye group previously observed a mutually exclusive relationship between RB mutation and p16INK4 silencing in lung cancer cell lines [9]. The study found that 10% of small cell lung cancers present silenced p16 and wildtype RB leading to the model that loss of p16 may be functionally equivalent to a null RB phenotype and the inactivation of either gene is sufficient to dysregulate the RB tumor suppressor pathway [10].
In this meta-analysis study, we included data from 4 published SCLC high throughput sequence studies – George et al [8], Rudin et al [5], Umemura et al [7] and Iwakawa et al [11]. Besides ranking genes based on their mutation frequencies, we sought to identify statistically significant mutations with the MutSigCV software package developed by Lawrence et al [12]. In addition, we also sought to identify specific patterns of mutations that might help us understand SCLC progression.
2. Methods
Identifying significantly mutated genes in SCLC
Lists of silent and non-silent mutations were either obtained from published data directly or obtained from database centers for the datasets described in Table 1. FASTQ files obtained from database center (EGAD00001000222) were processed with MuTect 1.1.4 [13] to identify somatic mutations, the functional consequences of the mutations were annotated with Oncotator (v1.8.0.0) with default parameters [14]. Mutation file for Iwakawa et al dataset was obtained from the National Cancer Research Institute, Japan. Finally, combined mutation data was processed with MutSigCV 1.0 [12] with full coverage and covariates files as control. Significantly mutated genes are determined based on p-values calculated by MutSigCV using the 2D projection method as described by Lawrence et al [12].
Table 1.
Datasets and number of samples used for the meta-analysis.
Interdependence analysis
Samples pooled from all 4 datasets were classified into 2 groups – those having mutation in RB1 gene and those without. Mutation rate (average number of genes mutated per sample) was calculated for both categories (λ1 and λ2 respectively).
Poisson likelihood ratio test p-value was obtained using the R package.
Gene ontology and pathway analysis
List of genes with mutation frequencies > 2.5% and identified by MutSigCV as significantly mutated genes (with p<0.05) were used for functional annotation analysis using DAVID version 6.7 [15,16].
3. Results and Discussion
3.1. MutSigCV identified genes with relatively low mutation frequency as highly significant
Not surprisingly, our frequency-based ranking analysis confirmed that TP53 and RB1 are the most frequently mutated genes in SCLC with a mutation frequency of about 85% (231/272) and 57% (156/272) respectively. Overall, 119 genes were found to be mutated in more than 10% of SCLC samples sequenced in the four three studies. Among those, 15 were mutated in more than 20% of the combined samples (Supplementary tables 1,2).
Recent studies have revealed that the genes with high mutation frequency may not necessarily be responsible for cancer initiation and progression. The higher mutation rates associated with these genes are largely due to the varying mutation frequencies associated with gene expression levels and duplication timing [12]. In addition, there is dramatic patient-specific difference in mutation frequency. Advanced methods developed to take into consideration of the mutational heterogeneity, such as MutSigCV [12], have made great advance in filtering out false positive genes and revealed genes associated with disease progression but have relatively low mutation frequency.
Using MutSigCV, we identified significantly mutated genes from the combined data set of SCLC samples previously reported by George et al, Rudin et al and Iwakawa et al (Supplementary table 3). The number of genes with p-values less than 0.001, 0.01, and 0.05 are 15, 56, and 225, respectively. From the list of 8 most significantly mutated genes (p<0.001 and q<1) identified by MutSigCV, 4 were mutated in more than 10% of SCLC samples, which are TP53, RB1, LRP1B, and KIAA1211 (Figure 1a, b). The remaining 4 of the significantly mutated genes have mutation frequencies <10% including PTEN (7.352941%, p=2.55E-07), GABRG1 (7.720588%, p=9.19E-05), TGM3 (5.147059%, p=9.79E-05) and DEFB112(2.941176%, p=1.36E-04). Our analysis with MutsigCV indicated that the majority of genes with high frequency are not statistically significant when mutational heterogeneity was considered. We suggest that these genes, despite of their high mutation rate in SCLC, may not be functionally significant for tumorigenesis.
Figure 1. Significantly mutated genes in SCLC.
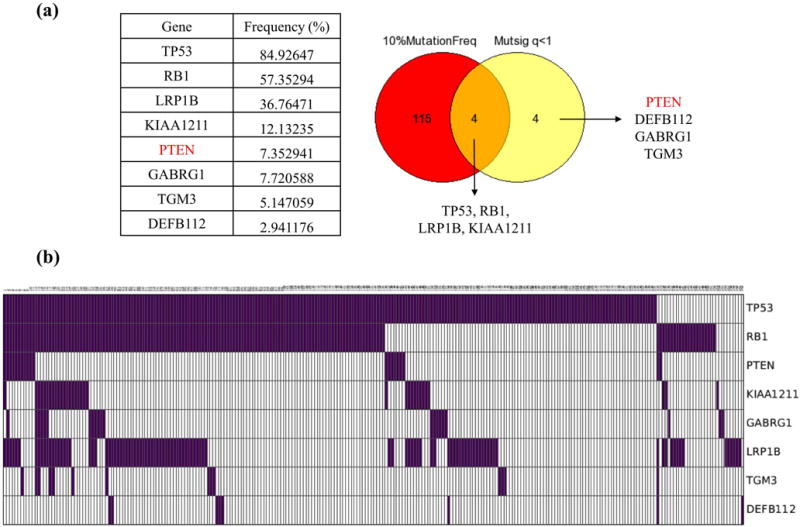
a) Venn diagram and table showing 8 highly significant (p<0.001 and q<1) genes identified by MutSigCV in the combined datasets. 4 of those are mutated with relatively low (<10%) frequency. b) CoMut plot of samples showing mutations for the 8 MutSigCV significant genes.
3.2. Significantly mutated genes with high frequency
TP53 and RB1 are the most frequently inactivated genes in SCLC, and they also have the lowest p-values in our analysis. The importance of these two genes in SCLC tumorigenesis has been documented by numerous functional studies [8,17–19]. For instance, mice with conditional inactivation of P53 and RB1 in lung epithelial cells have shown to develop neuroendocrine lung tumors that morphologically resemble SCLC [20].
LRP1B or Low-density lipoprotein receptor-related protein 1B was observed to be mutated in about 38% of SCLC samples. LRP1B has previously shown to be mutated or epigenetically silenced in Esophageal Squamous Cell Carcinoma [21] and mutated in Lung adenocarcinoma [22]. Downregulation of LRP1B has previously been shown to promote cell migration in renal cell cancer [23]. LRP1B deletion has also been associated with poor survival outcome in glioblastoma patients [24]. However, the functional mechanism of LRP1B as a potential tumor suppressor and conversely the functional significance of LRP1B mutations in SCLC remains to be understood.
KIAA1211 – The was no PubMed record studying this gene and there is a lack of functional annotation for the protein it encodes. However, a single nucleotide polymorphism (SNP) in KIAA1211 was found by large scale GWAS studies to be associated with increased risk of prostate cancer (rs629242) [25].
3.3 Significantly mutated genes with less than 10% frequency
Our analysis identified 4 genes which have mutation frequency between 3% to 8%, but are statistically significant (p<0.001). Mutation of PTEN was found in less than 10% of SCLC samples in the combined dataset we analyzed (~7%). But our analysis identified it as one of the significantly mutated genes in SCLC (p=2.55E−07). The PTEN/PIK3CA pathway has previously been implicated by various studies as a significant player in SCLC progression and metastasis [26,27]. PTEN mutation was found to be specifically associated with SCLC but not with NSCLC (Non-small cell lung cancer) [27]. Loss of function of PTEN was found to accelerate SCLC progression in a AdenoCre-driven mouse model with P53 and RB1 inactivation [26]. Interestingly, PTEN expression level has been suggested as a prognostic biomarker in NSCLC patients since loss of PTEN expression is associated with a more advanced disease and poor survival [28]. It may also worth mentioning that reduced expression of PTEN correlates with a worse prognosis for breast cancer patients [29,30]. Further clinical investigation is needed to document whether PTEN mutation is negatively correlated with prognosis of SCLC.
We consider the identification of PTEN as a significantly mutated gene in SCLC, despite its relatively low frequency, to be a validation of the analysis approach we applied. The fact that PTEN has the most significant p-value among genes with less than 10% mutation frequency indicates that it warrants further investigation as a possible biomarker for selecting treatment plans for SCLC patients.
GABRG1 or Gamma-Aminobutyric Acid (GABA) A Receptor Gamma 1 subunit is a membrane protein that plays an important role in inhibition of neurotransmission. It has been found to be mutated in lung adenocarcinoma negative for mutations in EGFR/KRAS/ALK [31].
TGM3 or transglutaminase 3 is downregulated in head and neck cancer (HNC). TGM3 promoter CpG island hypermethylation has been observed in HNC and exogenous expression of TGM3 reduces tumor growth in vivo. Furthermore, low TGM3 expression is associated with poor overall survival [32]. There is also evidence for TGM3 to induce cell proliferation and migration in esophageal cancer by downregulating the NF-κB signaling pathway [33]. Genome-wide association study (GWAS) of 38.5 million SNPs and small indels identified TGM3 variant as a contributor to risk of basal cell carcinoma [34]. The role of TGM3 in small cell lung cancer remains to be established.
DEFB112 or Defensin Beta 112 encodes a peptide that is thought to be involved in antimicrobial response [35]. So far there is no report on its role in cancer.
3.4. MutSigCV identified IL-27 as highly significant in a Japanese SCLC dataset
Using the Iwakawa et al dataset which consisted of 44 SCLC samples from Japanese patients, we performed MutSigCV analysis using default parameters. MutSig identified TP53, RB1, PTEN and IL-27 as the most significantly mutated genes with p<0.001 and q<1. It is interesting to note that IL-27 is mutated with much higher significance (p-value 7.75E-05; 4 out of 44) in the Japanese dataset but was barely significant (p-value ~0.0088, 6 out of 272) with the combined dataset with the other datasets that are predominantly from European/American patients. When the Iwakawa et al cohort was excluded from MutSig analysis, IL-27 was not observed to be among the significantly (p<0.05) mutated genes.
IL-27 is a cytokine produced by macrophages and dendritic cells. It is known to play a critical role in regulating the interaction between innate and adaptive antitumor immunity and has been shown to possess the ability to inhibit tumor growth and invasiveness [36]. There is increasing interest in cancer immunotherapy and modulation of the patients’ immune system to target cancer cells. The role of IL-27 in cancer immunosuppression has been reviewed by Murugaiyan et al [37]. Previous studies have shown that IL-27 signaling is mediated by activation of JAK-STAT pathway resulting in regulation of angiogenesis and EMT in NSCLC [38]. IL-27 performs its functions by suppressing cyclooxygenase-2 and prostaglandin E2 expression, increasing epithelial marker expression and decreasing the levels of mesenchymal markers [38]. Gene transfer of IL-27 to NSCLC lung tumor cells in mice models has been shown to be effective in tumor growth inhibition [39]. IL-27 has been reported to down-regulate stemness- and EMT-related genes and drive myeloid cells towards antitumor activities in NSCLC cells. Airoldi and Tupone et al evaluated IL-27R expression in NSCLC patient samples and suggest that immunocompromised or advanced NSCLC patients may benefit from IL-27 therapy [40]. A recent study has also proved the association of a Single Nucleotide Polymorphism in IL-27 and risk of NSCLC in a Chinese population [41] and might possess potential as a biomarker in SCLC. The fact that our analysis identified it as significantly mutated in the Japanese cohort indicate that it may serve as an important tumor-suppressor for SCLC in specific ethnic group. The various MutSigCV p<0.05 genes clustered by DAVID functional annotation as immune related genes in the datasets including and excluding the Iwakawa et al cohort are mentioned in supplementary table 6.
3.5. Clustering of mutated genes in axon guidance pathways
A caveat to identify individual significantly mutated genes associated with cancer is that the impact of gene mutation on the function of a particular biological system may vary significantly. It is possible that in certain systems, there isn’t a single central rate-limiting factor such as the role played by P53. Rather, it is possible that mutation of a variety of genes on a particular pathway all have impact on the system and thus could be mutated alternatively. To test for pathways or biological processes significantly mutated in SCLC, we analyzed the list of genes with >2.5% mutation frequency and the list of genes with a MutSigCV p-value of less than 0.05 using DAVID (Database for Annotation, Visualization, and Integrated Discovery) functional annotation analysis [15,16].
We found that there is a significant over-representation of genes associated with GO terms ‘GPCR signaling pathway’, ‘Sensory perception of smell’, ‘Cell-cell adhesion’ and ‘Neurological system process’ (Figure 2a) when frequently mutated genes were used. In case of significantly mutated genes (p<0.05), ‘Adaptive immune response’ showed up as significantly enriched pathway in addition to terms ‘GPCR signaling pathway’ and ‘Sensory perception of smell’ (Figure 2c). The role of immune response system players in tumor growth, microenvironment and therapy have been established previously [42,43].
Figure 2. Biological processes and regulatory pathways mutated in SCLC.

a) GO terms and b) Pathways enriched for genes mutated in SCLC (>2.5% mutation frequency). c) GO terms and d) Pathways enriched for genes identified as significantly mutated in SCLC (MutSig p<0.05).
In addition, we also searched for KEGG pathways enriched for genes mutated in SCLC based on mutation frequency or MutSigCV significance analysis. Among the pathways enriched with frequently mutated gene were – ‘Olfactory transduction’, ’ECM-receptor interaction’ and ’Axon guidance’ (Figure 2b). In case of the MutSigCV significantly mutated genes, ‘Olfactory transduction’ remained the most common pathway involved followed by immune system related pathways (Figure 2d).
The axon guidance pathway has been previously implicated in lung cancer progression [44]. Figure 3 exhibits the genes mutated in the axon guidance pathway with a mutation frequency > 2.5% in our datasets. Genes mutated in the axon guidance pathway in SCLC include SLITs, ROBOs, Ephrins and Plexins. SLIT/ROBO signaling has been implicated in angiogenesis, cell migration, and metastasis of cancers and may serve as a potential target for anticancer therapy [45–47]. Evidence of somatic inactivation of SLIT2 and its potential role as a tumor suppressor has been found in colorectal cancer [48]. Dallol et al have furthermore identified frequent hypermethylation of the promoter of human SLIT2 in lung, breast, colorectal tumors and gliomas [49]. Understanding the role of axon guidance proteins and their mutations remains an area to be explored for potential biomarkers for SCLC.
Figure 3. Axon guidance pathway genes mutated in SCLC.
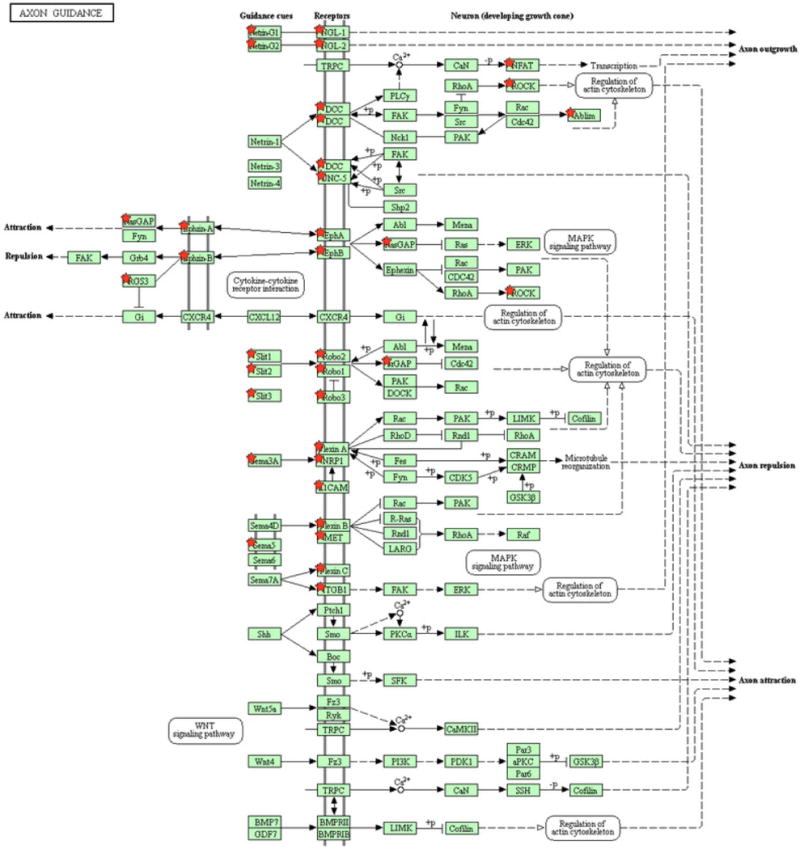
Genes mutated with > 2.5% mutation frequency in the Axon guidance pathway (KEGG).
3.6. Does mutation in RB1 affect mutation profile in SCLC?
Loss of Retinoblastoma 1 (RB1) gene function in the lung has been shown to negatively regulate neuroendocrine cell differentiation, and may explain why RB1 loss is predominately associated with SCLC when compared to other cancer types [17,20]. Somatic inactivation of P53 and RB1 in neuroendocrine and surfactant protein C (SPC)-expressing cells of the lung has been shown to promote tumorigenesis resembling SCLC in murine models [50]. It is possible that loss of RB1 function may predispose the cancer cell to a progression course that is distinct from those that do not have RB1 mutation. To test this hypothesis, we looked at whether RB1(+) and RB1(−) SCLC samples have different rates of mutation.
We found the mutation rate to be significantly higher in RB1(−) samples when compared to samples with wild type RB1 (Poisson likelihood ratio test p-value = 4.630177e−104) (Figure 4a). This difference is still significant when only samples with verified TP53 mutation were considered (Poisson likelihood ratio test p-value = 9.870942e−80) (Figure 4b). This suggests that RB1 mutation significantly alters the subsequent mutation progression of SCLC.
Figure 4. Effect of RB status on mutation profile.

Distribution plot showing number of mutations in samples with or without RB1 mutation in all the samples (a) or in samples that have P53 mutation (b). Statistical analysis (p-value on top of each panel) indicates that RB1 mutation causes increased accumulation of mutations in SCLC.
4. Results and Conclusions
Mutational heterogeneity between individual cancers can lead to false discovery of cancer-associated genes and mask the detection of functionally significant genes with relatively low mutation frequency. In this study, we performed a meta-analysis of combined SCLC samples using MutSigCV that takes into account of mutational heterogeneity. This allowed us to detect several genes mutated with relatively low frequency, including PTEN, as significant for SCLC. These genes might serve as potential biomarkers for subtyping of SCLC. Our analysis also provided evidence indicating that RB1 status plays an important role in determining tumor progression and subsequent mutation profile of SCLC.
Supplementary Material
Highlights.
Genes with low mutation frequencies identified as highly significant in SCLC.
Axon guidance pathway genes are likely to play an important role in SCLC.
Mutation rate is higher in samples with RB1 mutation compared to those with wildtype RB1.
Acknowledgments
We would like to acknowledge Adrian R. Acuna Higaki and Andre Revell for their contributions in writing scripts for data analysis at the early stage of the project. We would also like to thank Guanhong Miao and Suwa Xu for their valuable input and assistance in statistical analysis. This work was supported in part by UF Informatics Institute Graduate Student Fellowship (VS) and a grant from NIH GM106174 (LZ).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
All the authors indicate none financial or another interest that is relevant to the subject matter under consideration in this article.
References
- 1.Arcaro A. Targeted therapies for small cell lung cancer: Where do we stand? Crit Rev Oncol Hematol. 2015;95:154–164. doi: 10.1016/j.critrevonc.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 2.Koinis F, Kotsakis A, Georgoulias V. Small cell lung cancer (SCLC): no treatment advances in recent years. Transl Lung Cancer Res. 2016;5:39–50. doi: 10.3978/j.issn.2218-6751.2016.01.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karachaliou N, Pilotto S, Lazzari C, Bria E, de Marinis F, Rosell R. Cellular and molecular biology of small cell lung cancer: an overview. Transl Lung Cancer Res. 2016;5:2–15. doi: 10.3978/j.issn.2218-6751.2016.01.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Byers LA, Rudin CM. Small cell lung cancer: Where do we go from here? Cancer. 2015;121:664–672. doi: 10.1002/cncr.29098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rudin CM, Durinck S, Stawiski EW, Poirier JT, Modrusan Z, Shames DS, Bergbower Ea, Guan Y, Shin J, Guillory J, Rivers CS, Foo CK, Bhatt D, Stinson J, Gnad F, Haverty PM, Gentleman R, Chaudhuri S, Janakiraman V, Jaiswal BS, Parikh C, Yuan W, Zhang Z, Koeppen H, Wu TD, Stern HM, Yauch RL, Huffman KE, Paskulin DD, Illei PB, Varella-Garcia M, Gazdar AF, Sauvage FJ de, Bourgon R, Minna JD, Brock MV, Seshagiri S. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet. 2012;44:1111–1116. doi: 10.1038/ng.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peifer M, Fernández-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, Plenker D, Leenders F, Sun R, Zander T, Menon R, Koker M, Dahmen I, Müller C, Cerbo V Di, Schildhaus H-U, Altmüller J, Baessmann I, Becker C, Wilde B de, Vandesompele J, Böhm D, Ansén S, Gabler F, Wilkening I, Heynck S, Heuckmann JM, Lu X, Carter SL, Cibulskis K, Banerji S, Getz G, Park K-S, Rauh D, Grütter C, Fischer M, Pasqualucci L, Wright G, Wainer Z, Russell P, Petersen I, Chen Y, Stoelben E, Ludwig C, Schnabel P, Hoffmann H, Muley T, Brockmann M, Engel-Riedel W, Muscarella La, Fazio VM, Groen H, Timens W, Sietsma H, Thunnissen E, Smit E, Heideman DaM, Snijders PJF, Cappuzzo F, Ligorio C, Damiani S, Field J, Solberg S, Brustugun OT, Lund-Iversen M, Sänger J, Clement JH, Soltermann A, Moch H, Weder W, Solomon B, Soria J-C, Validire P, Besse B, Brambilla E, Brambilla C, Lantuejoul S, Lorimier P, Schneider PM, Hallek M, Pao W, Meyerson M, Sage J, Shendure J, Schneider R, Büttner R, Wolf J, Nürnberg P, Perner S, Heukamp LC, Brindle PK, Haas S, Thomas RK. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet. 2012;44:1104–1110. doi: 10.1038/ng.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Umemura S, Mimaki S, Makinoshima H, Tada S, Ishii G, Ohmatsu H, Niho S, Yoh K, Matsumoto S, Takahashi A, Morise M, Nakamura Y, Ochiai A, Nagai K, Iwakawa R, Kohno T, Yokota J, Ohe Y, Esumi H, Tsuchihara K, Goto K. Therapeutic priority of the PI3K/AKT/mTOR pathway in small cell lung cancers as revealed by a comprehensive genomic analysis. J Thorac Oncol. 2014;9:1324–31. doi: 10.1097/JTO.0000000000000250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.George J, Lim JS, Jang SJ, Cun Y, Ozretic L, Kong G, Leenders F, Lu X, Fernandez-Cuesta L, Bosco G, Muller C, Dahmen I, Jahchan NS, Park KS, Yang D, Karnezis AN, Vaka D, Torres A, Wang MS, Korbel JO, Menon R, Chun SM, Kim D, Wilkerson M, Hayes N, Engelmann D, Putzer B, Bos M, Michels S, Vlasic I, Seidel D, Pinther B, Schaub P, Becker C, Altmuller J, Yokota J, Kohno T, Iwakawa R, Tsuta K, Noguchi M, Muley T, Hoffmann H, Schnabel PA, Petersen I, Chen Y, Soltermann A, Tischler V, Choi C, Kim YH, Massion PP, Zou Y, Jovanovic D, Kontic M, Wright GM, Russell PA, Solomon B, Koch I, Lindner M, Muscarella LA, la Torre A, Field JK, Jakopovic M, Knezevic J, Castanos-Velez E, Roz L, Pastorino U, Brustugun OT, Lund-Iversen M, Thunnissen E, Kohler J, Schuler M, Botling J, Sandelin M, Sanchez-Cespedes M, Salvesen HB, Achter V, Lang U, Bogus M, Schneider PM, Zander T, Ansen S, Hallek M, Wolf J, Vingron M, Yatabe Y, Travis WD, Nurnberg P, Reinhardt C, Perner S, Heukamp L, Buttner R, Haas SA, Brambilla E, Peifer M, Sage J, Thomas RK. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524:47–53. doi: 10.1038/nature14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Otterson GA, Kratzke RA, Coxon A, Kim YW, Kaye FJ. Absence of p16INK4 protein is restricted to the subset of lung cancer lines that retains wildtype RB. Oncogene. 1994;9:3375–8. http://www.ncbi.nlm.nih.gov/pubmed/7936665 (accessed August 6, 2016) [PubMed] [Google Scholar]
- 10.Kaye FJ. RB and cyclin dependent kinase pathways: defining a distinction between RB and p16 loss in lung cancer. Oncogene. 2002;21:6908–6914. doi: 10.1038/sj.onc.1205834. [DOI] [PubMed] [Google Scholar]
- 11.Iwakawa R, Kohno T, Totoki Y, Shibata T, Tsuchihara K, Mimaki S, Tsuta K, Narita Y, Nishikawa R, Noguchi M, Harris CC, Robles AI, Yamaguchi R, Imoto S, Miyano S, Totsuka H, Yoshida T, Yokota J. Expression and clinical significance of genes frequently mutated in small cell lung cancers defined by whole exome/RNA sequencing. Carcinogenesis. 2015;36:616–621. doi: 10.1093/carcin/bgv026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts Sa, Kiezun A, Hammerman PS, McKenna A, Drier Y, Zou L, Ramos AH, Pugh TJ, Stransky N, Helman E, Kim J, Sougnez C, Ambrogio L, Nickerson E, Shefler E, Cortés ML, Auclair D, Saksena G, Voet D, Noble M, DiCara D, Lin P, Lichtenstein L, Heiman DI, Fennell T, Imielinski M, Hernandez B, Hodis E, Baca S, Dulak AM, Lohr J, Landau D-A, Wu CJ, Melendez-Zajgla J, Hidalgo-Miranda A, Koren A, McCarroll Sa, Mora J, Lee RS, Crompton B, Onofrio R, Parkin M, Winckler W, Ardlie K, Gabriel SB, Roberts CWM, Biegel Ja, Stegmaier K, Bass AJ, Garraway La, Meyerson M, Golub TR, Gordenin Da, Sunyaev S, Lander ES, Getz G. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–8. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramos AH, Lichtenstein L, Gupta M, Lawrence MS, Pugh TJ, Saksena G, Meyerson M, Getz G. Oncotator: Cancer variant annotation tool. Hum Mutat. 2015;36:E2423–E2429. doi: 10.1002/humu.22771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 16.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Semenova EA, Nagel R, Berns A. Origins, genetic landscape, and emerging therapies of small cell lung cancer. Genes Dev. 2015;29:1447–1462. doi: 10.1101/gad.263145.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harbour JW, Lai SL, Whang-Peng J, Gazdar AF, Minna JD, Kaye FJ. Abnormalities in structure and expression of the human retinoblastoma gene in SCLC. Science. 1988;241:353–7. doi: 10.1126/science.2838909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takahashi T, Nau MM, Chiba I, Birrer MJ, Rosenberg RK, Vinocour M, Levitt M, Pass H, Gazdar AF, Minna JD. p53: a frequent target for genetic abnormalities in lung cancer. Science. 1989;246:491–4. doi: 10.1126/science.2554494. [DOI] [PubMed] [Google Scholar]
- 20.Meuwissen R, Linn SC, Linnoila RI, Zevenhoven J, Mooi WJ, Berns A. Induction of small cell lung cancer by somatic inactivation of both Trp53 and Rb1 in a conditional mouse model. Cancer Cell. 2003;4:181–189. doi: 10.1016/S1535-6108(03)00220-4. [DOI] [PubMed] [Google Scholar]
- 21.Sonoda I, Imoto I, Inoue J, Shibata T, Shimada Y, Chin K, Imamura M, Amagasa T, Gray JW, Hirohashi S, Inazawa J. Frequent silencing of low density lipoprotein receptor-related protein 1B (LRP1B) expression by genetic and epigenetic mechanisms in esophageal squamous cell carcinoma. Cancer Res. 2004;64:3741–3747. doi: 10.1158/0008-5472.CAN-04-0172. [DOI] [PubMed] [Google Scholar]
- 22.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, Fulton L, Fulton RS, Zhang Q, Wendl MC, Lawrence MS, Larson DE, Chen K, Dooling DJ, Sabo A, Hawes AC, Shen H, Jhangiani SN, Lewis LR, Hall O, Zhu Y, Mathew T, Ren Y, Yao J, Scherer SE, Clerc K, Metcalf GA, Ng B, Milosavljevic A, Gonzalez-Garay ML, Osborne JR, Meyer R, Shi X, Tang Y, Koboldt DC, Lin L, Abbott R, Miner TL, Pohl C, Fewell G, Haipek C, Schmidt H, Dunford-Shore BH, Kraja A, Crosby SD, Sawyer CS, Vickery T, Sander S, Robinson J, Winckler W, Baldwin J, Chirieac LR, Dutt A, Fennell T, Hanna M, Johnson BE, Onofrio RC, Thomas RK, Tonon G, Weir BA, Zhao X, Ziaugra L, Zody MC, Giordano T, Orringer MB, Roth JA, Spitz MR, Wistuba II, Ozenberger B, Good PJ, Chang AC, Beer DG, Watson MA, Ladanyi M, Broderick S, Yoshizawa A, Travis WD, Pao W, Province MA, Weinstock GM, Varmus HE, Gabriel SB, Lander ES, Gibbs RA, Meyerson M, Wilson RK. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ni S, Hu J, Duan Y, Shi S, Li R, Wu H, Qu Y, Li Y. Down expression of LRP1B promotes cell migration via RhoA/Cdc42 pathway and actin cytoskeleton remodeling in renal cell cancer. Cancer Sci. 2013;104:817–825. doi: 10.1111/cas.12157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tabouret E, Labussi??re M, Alentorn A, Schmitt Y, Marie Y, Sanson M. LRP1B deletion is associated with poor outcome for glioblastoma patients. J Neurol Sci. 2015 doi: 10.1016/j.jns.2015.09.345. [DOI] [PubMed] [Google Scholar]
- 25.Thomas G, Jacobs KB, Yeager M, Kraft P, Wacholder S, Orr N, Yu K, Chatterjee N, Welch R, Hutchinson A, Crenshaw A, Cancel-Tassin G, Staats BJ, Wang Z, Gonzalez-Bosquet J, Fang J, Deng X, Berndt SI, Calle EE, Feigelson HS, Thun MJ, Rodriguez C, Albanes D, Virtamo J, Weinstein S, Schumacher FR, Giovannucci E, Willett WC, Cussenot O, Valeri A, Andriole GL, Crawford ED, Tucker M, Gerhard DS, Fraumeni JF, Hoover R, Hayes RB, Hunter DJ, Chanock SJ. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40:310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 26.Cui M, Augert A, Rongione M, Conkrite K, Parazzoli S, Nikitin AY, Ingolia N, MacPherson D. PTEN Is a Potent Suppressor of Small Cell Lung Cancer. Mol Cancer Res. 2014;12:654–659. doi: 10.1158/1541-7786.MCR-13-0554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yokomizo A, Tindall DJ, Drabkin H, Gemmill R, Franklin W, Yang P, Sugio K, Smith DI, Liu W. PTEN/MMAC1 mutations identified in small cell, but not in non-small cell lung cancers. Oncogene. 1998;17:475–9. doi: 10.1038/sj.onc.1201956. [DOI] [PubMed] [Google Scholar]
- 28.Ji Y, Zheng M, Ye S, Chen J, Chen Y. PTEN and Ki67 expression is associated with clinicopathologic features of non-small cell lung cancer. J Biomed Res. 2014;28:462–7. doi: 10.7555/JBR.27.20130084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Depowski PL, Rosenthal SI, Ross JS. Loss of Expression of the PTEN Gene Protein Product Is Associated with Poor Outcome in Breast Cancer. Mod Pathol. 2001;14:672–676. doi: 10.1038/modpathol.3880371. [DOI] [PubMed] [Google Scholar]
- 30.Tsutsui S, Inoue H, Yasuda K, Suzuki K, Higashi H, Era S, Mori M. Reduced Expression of PTEN Protein and Its Prognostic Implications in Invasive Ductal Carcinoma of the Breast. Oncology. 2005;68:398–404. doi: 10.1159/000086981. [DOI] [PubMed] [Google Scholar]
- 31.Ahn JW, Kim HS, Yoon JK, Jang H, Han SM, Eun S, Shim HS, Kim HJ, Kim DJ, Lee JG, Lee CY, Bae MK, Chung KY, Jung JY, Kim EY, Kim SK, Chang J, Kim HR, Kim JH, Lee MG, Cho BC, Lee JH, Bang D. Identification of somatic mutations in EGFR/KRAS/ALK-negative lung adenocarcinoma in never-smokers. Genome Med. 2014;6:18. doi: 10.1186/gm535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu X, Cao W, Wang X, Zhang J, Lv Z, Qin X, Wu Y, Chen W. TGM3, a candidate tumor suppressor gene, contributes to human head and neck cancer. Mol Cancer. 2013;12:151. doi: 10.1186/1476-4598-12-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li W, Zhang Z, Zhao W, Han N. Transglutaminase 3 protein modulates human esophageal cancer cell growth by targeting the NF-κB signaling pathway. Oncol Rep. 2016 doi: 10.3892/or.2016.4921. [DOI] [PubMed] [Google Scholar]
- 34.Stacey SN, Sulem P, Gudbjartsson DF, Jonasdottir A, Thorleifsson G, Gudjonsson SA, Masson G, Gudmundsson J, Sigurgeirsson B, Benediktsdottir KR, Thorisdottir K, Ragnarsson R, Fuentelsaz V, Corredera C, Grasa M, Planelles D, Sanmartin O, Rudnai P, Gurzau E, Koppova K, Hemminki K, Nexo BA, Tjonneland A, Overvad K, Johannsdottir H, Helgadottir HT, Thorsteinsdottir U, Kong A, Vogel U, Kumar R, Nagore E, Mayordomo JI, Rafnar T, Olafsson JH, Stefansson K. Germline sequence variants in TGM3 and RGS22 confer risk of basal cell carcinoma. Hum Mol Genet. 2014;23:3045–3053. doi: 10.1093/hmg/ddt671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patil AA, Cai Y, Sang Y, Blecha F, Zhang G. Cross-species analysis of the mammalian β-defensin gene family: presence of syntenic gene clusters and preferential expression in the male reproductive tract. Physiol Genomics. 2005;23:5–17. doi: 10.1152/physiolgenomics.00104.2005. [DOI] [PubMed] [Google Scholar]
- 36.Carbotti G, Barisione G, Airoldi I, Mezzanzanica D, Bagnoli M, Ferrero S, Petretto A, Fabbi M, Ferrini S, Carbotti G, Barisione G, Airoldi I, Mezzanzanica D, Bagnoli M, Ferrero S, Petretto A, Fabbi M, Ferrini S. IL-27 induces the expression of IDO and PD-L1 in human cancer cells. Oncotarget. 2014;6:43267–43280. doi: 10.18632/oncotarget.6530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murugaiyan G, Saha B. IL-27 in tumor immunity and immunotherapy. Trends Mol Med. 2013;19:108–116. doi: 10.1016/j.molmed.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 38.Kachroo P, Lee MH, Zhang L, Baratelli F, Lee G, Srivastava MK, Wang G, Walser TC, Krysan K, Sharma S, Dubinett SM, Lee JM. IL-27 inhibits epithelial-mesenchymal transition and angiogenic factor production in a STAT1-dominant pathway in human non-small cell lung cancer. J Exp Clin Cancer Res. 2013;32:97. doi: 10.1186/1756-9966-32-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ho MY, Leu SJJ, Sun GH, Tao MH, Tang SJ, Sun KH. IL-27 Directly Restrains Lung Tumorigenicity by Suppressing Cyclooxygenase-2-Mediated Activities. J Immunol. 2009;183:6217–6226. doi: 10.4049/jimmunol.0901272. [DOI] [PubMed] [Google Scholar]
- 40.Airoldi I, Tupone MG, Esposito S, Russo MV, Barbarito G, Cipollone G, Di Carlo E, Airoldi I, Tupone MG, Esposito S, Russo MV, Barbarito G, Cipollone G, Di Carlo E. Interleukin-27 re-educates intratumoral myeloid cells and down-regulates stemness genes in non-small cell lung cancer. Oncotarget. 2015;6:3694–3708. doi: 10.18632/oncotarget.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ge P, Xiao G. Interleukin-27 rs153109 polymorphism and the risk of non-small-cell lung cancer in a Chinese population. Onco Targets Ther. 2016;9:895–8. doi: 10.2147/OTT.S93226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spurrell EL, Lockley M. Adaptive immunity in cancer immunology and therapeutics. 2014 doi: 10.3332/ecancer.2014.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu M Michelle, Pu Y, Zhang Y, Fu Y-X. The Role of Adaptive Immunity in the Efficacy of Targeted Cancer Therapies. 2016 doi: 10.1016/j.it.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dallol A, Da Silva NF, Viacava P, Minna JD, Bieche I, Maher ER, Latif F. SLIT2, a human homologue of the Drosophila Slit2 gene, has tumor suppressor activity and is frequently inactivated in lung and breast cancers. Cancer Res. 2002;62:5874–5880. http://www.ncbi.nlm.nih.gov/pubmed/12384551 (accessed April 1, 2016) [PubMed] [Google Scholar]
- 45.Fujiwara M, Ghazizadeh M, Kawanami O. Potential role of the Slit/Robo signal pathway in angiogenesis. Vasc Med. 2006;11:115–121. doi: 10.1191/1358863x06vm658ra. [DOI] [PubMed] [Google Scholar]
- 46.Legg JA, Herbert JMJ, Clissold P, Bicknell R. Slits and Roundabouts in cancer, tumour angiogenesis and endothelial cell migration. Angiogenesis. 2008;11:13–21. doi: 10.1007/s10456-008-9100-x. [DOI] [PubMed] [Google Scholar]
- 47.Gara RK, Kumari S, Ganju A, Yallapu MM, Jaggi M, Chauhan SC. Slit/Robo pathway: A promising therapeutic target for cancer. Drug Discov Today. 2015;20:156–164. doi: 10.1016/j.drudis.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dallol A, Morton D, Maher ER, Latif F. SLIT2 Axon Guidance Molecule Is Frequently Inactivated in Colorectal Cancer and Suppresses Growth of Colorectal Carcinoma Cells. Cancer Res. 2003;63:1054–1058. http://cancerres.aacrjournals.org/content/63/5/1054.long (accessed May 29, 2016) [PubMed] [Google Scholar]
- 49.Dallol A, Krex D, Hesson L, Eng C, Maher ER, Latif F. Frequent epigenetic inactivation of the SLIT2 gene in gliomas. Oncogene. 2003;22:4611–6. doi: 10.1038/sj.onc.1206687. [DOI] [PubMed] [Google Scholar]
- 50.Sutherland KD, Proost N, Brouns I, Adriaensen D, Song JY, Berns A. Cell of origin of small cell lung cancer: Inactivation of Trp53 and Rb1 in distinct cell types of adult mouse lung. Cancer Cell. 2011;19:754–764. doi: 10.1016/j.ccr.2011.04.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


