Abstract
Background
The clinical course of Idiopathic Pulmonary Fibrosis (IPF) is unpredictable. Clinical prediction tools are not accurate enough to predict disease outcomes.
Methods
All-comers with Idiopathic Pulmonary Fibrosis diagnosis were enrolled in a six-cohort study. Peripheral blood mononuclear cells or whole blood was collected at baseline from 425 participants and during follow up from 98 patients. The 52-gene signature was measured by the nCounter® analysis system in four cohorts and extracted from microarray data in two others. The Scoring Algorithm for Molecular Subphenotypes (SAMS) was used to classify patients into low or high risk groups based on a 52-gene signature. Mortality and transplant-free survival were studied using Competing risk and Cox proportional-hazard models, respectively. Time course data and response to anti-fibrotic drugs were analyzed using linear mixed-effect models.
Findings
The application of SAMS to the 52-gene signature identified two groups of IPF patients (low and high risk) with significant differences in mortality or transplant-free survival in each of the six cohorts (HR 2·03–4·37). Pooled data revealed similar results for mortality (HR:2·18, 95%CI:1·53–3·09, P<0·0001) or transplant-free survival (HR:2·04, 95%CI: 1·52–2·74, P<0·0001). Adding 52-gene risk profiles to the Gender, Age and Physiology (GAP) index significantly improved its mortality predictive accuracy. Temporal changes in SAMS scores were associated with changes in forced vital capacity (FVC) in two cohorts. Untreated patients did not shift their risk profile over time. A simultaneous increase in up score and decrease in down score was predictive of transplant-free survival (HR:3·18· 95%CI 1·16, 8·76, P=0·025) in the Pittsburgh cohort. A simultaneous decrease in up score and increase in down score after initiation of anti-fibrotic drugs was associated with a significant (P=0·005) improvement in FVC in the Yale cohort.
Interpretation
The peripheral blood 52-gene expression signature is predictive of outcome in patients with IPF. The potential value of the 52-gene signature in predicting response to therapy should be determined in prospective studies.
Introduction
Idiopathic pulmonary fibrosis (IPF) is a progressive and highly lethal interstitial lung disease of unknown etiology. The median survival without transplant is approximately three to four years1. The natural history of the disease is highly variable and unpredictable with some patients demonstrating long term clinical stability and others experiencing a more rapid disease course2. While clinical parameters allow staging of patients, they do not predict outcome accurately3. In recent years, evidence emerged that blood molecular and genetic markers may be indicative of disease outcome and potentially improve the accuracy of clinical predictions4–9. However, the majority of these studies were limited in scope and replication.
We previously identified a 52 gene expression signature in peripheral blood mononuclear cells (PBMC) that predicted transplant-free survival (TFS) in IPF and validated four of these genes (CD28, ICOS, LCK, ITK) by qRT-PCR6. In this study, we hypothesized that genomic risk profiles based on the peripheral blood, 52-gene expression signature, would accurately predict outcome in IPF. Our objectives were to determine the outcome prediction accuracy of 52-gene risk profiles in multiple cohorts, to determine whether adding 52-gene risk profiles to currently accepted clinical staging tools improved their outcome prediction accuracy, and to identify whether genomic risk profiles change with disease progression or in response to anti-fibrotic therapy.
Methods
The following sections summarize our methods. The online supplement includes more details:
Design, settings and participants
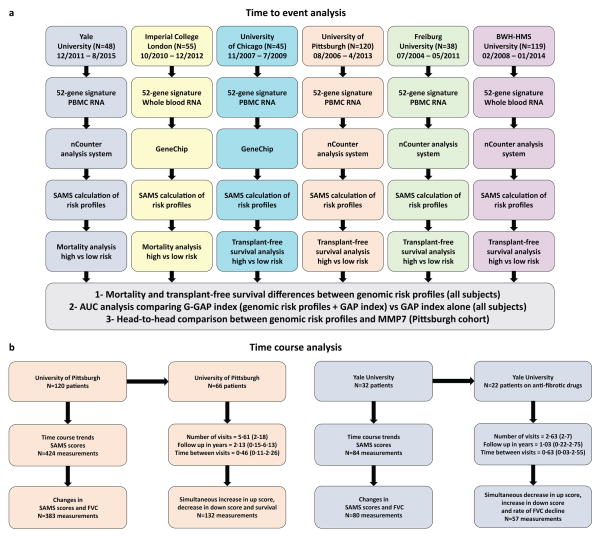
Study design is summarized in figure 1. For time to event analyses, patients were recruited from the Universities of Yale (n=48), Imperial College London (n=55), Chicago (n=45), Pittsburgh (n=120), Freiburg (n=38) and Brigham and Women’s Hospital-Harvard Medical School (BWH-HMS) (n=119) (Table 1). Recruitment started on 07/2004 and ended on 8/2015. For time course analyses, samples were available from Pittsburgh and Yale cohort patients (Figure 1b). IPF diagnosis was established by a multidisciplinary group at each institution following ATS/ERS guidelines10. Studies were approved by Institutional Review Boards at each institution and informed consent was obtained from all patients. Demographic, clinical information, pulmonary function test and diffusion capacity of the lung for carbon monoxide (DLCO) were collected at the time of blood draw. The Gender, Age and Lung Physiology (GAP) index was calculated as reported by Ley and colleagues3.
Figure 1. Study design.
The outline summarizes the (a) time to event and (b) time course analysis design for this study including the cohorts, blood compartments, experiments and statistical methods used in each independent cohort and in the pooled data analysis. PBMC: Peripheral blood mononuclear cells. BWH-HMS: Brigham and Women’s Hospital-Harvard Medical School. Dates of enrollment for each cohort are included in figure 1a. For figure 1b, time is presented in years (average and range, in parenthesis).
Table 1. Clinicopathological characteristics of the IPF patients in the cohorts for time to event analysis.
FVC%: Forced vital capacity percent predicted, DLCO%: Carbon monoxide diffusing capacity, percent predicted. FEV1% Forced expiratory volume in 1 second, percent predicted. HRCT, high resolution computed tomography. UIP: Usual Interstitial Pneumonia. BWH-HMS: Brigham and Women’s Hospital-Harvard Medical School
| Characteristic | Yale University (N=48) | Imperial College London (n=55) | University of Chicago (n=45) | University of Pittsburgh (n=120) | Freiburg University (n=38) | BWH-HMS (n=119) |
|---|---|---|---|---|---|---|
| Age at enrollment Mean ± SD |
70·8 ± 6·6 | 67·3 ± 8·1 | 67 ± 8·1 | 68·2 ± 8·5 | 66·8 ± 8·8 | 67 ± 8 |
| Gender, n (%) | ||||||
| Males | 39 (81·3) | 36 (65·5) | 40 (88·9) | 87 (72·5) | 34 (89·5) | 82 (68·9) |
| Females | 9 (18·7) | 19 (34·5) | 5 (11·1) | 33 (27·5) | 4 (10·5) | 37 (31·1) |
| Race, n (%) | ||||||
| Caucasian | 47 (97·9) | 52 (94·5) | 37 (82·3) | 118 (98·34) | 38 (100) | 108 (90) |
| Black | 0 (0) | 0 | 3 (6·6) | 1 (0·83) | 0 (0) | 6 (5) |
| Hispanic | 1 (2·1) | 0 | 5 (11·1) | 0 (0) | 0 (0) | 2 (1·5) |
| Other | 0 (0) | 3 (5·5) | 0 (0) | 1 (0·83) | 0 (0) | 3 (2·5) |
| Smoking status, n (%) | ||||||
| Ever smoker | 37 (77·1) | 39 (70·9) | 27 (60) | 80 (66·7) | 27 (71·1) | 82 (68·9) |
| Never smokers | 11 (22·9) | 16 (29·1) | 18 (40) | 40 (33·3) | 11 (28·9) | 37 (31·1) |
| Immunosuppression use, n (%) | ||||||
| No | 45 (93·8) | 46 (83·6) | 43 (95·6) | 106 (88·3) | 25 (65·8) | 90 (75·6) |
| Yes | 3 (6·2) | 9 (16·4) | 2 (4·4) | 14 (11·7) | 13 (34·2) | 29 (24·4) |
| Spirometry, mean ± SD | ||||||
| FVC (%) | 73·6 ± 15·1 | 72·8 ± 20·4 | 61 ± 14·7 | 66·4 ± 18·6 | 65 ± 18 | 65·3 ± 18·5 |
| DLCO (%) | 39·6 ± 12·5 | 39·5 ± 14 | 43·3 ± 17·7 | 50·1 ± 18·9 | 46·8 ± 17·7 | 42·2 ± 16·2 |
| FEV1 (%) | 80·7 ± 19·2 | 73·5 ± 19 | 73·9 ± 17·3 | 78·3 ± 21·2 | 64·6 ± 16·2 | 70·8 ± 18·4 |
| GAP Index | ||||||
| Mean ± SD | 4·3 ± 1·4 | 3·9 ± 1·6 | 4·3 ± 1·6 | 3·8 ± 1·5 | 4·4 ± 1·5 | 3·9 ± 1·3 |
| Diagnosis, n (%) | ||||||
| HRCT + UIP | 16 (33·3) | 0 | 24 (53·3) | 64 (53·3) | 16 (42·1) | 66 (55·5) |
| HRCT Biopsy | 32 (66·7) | 55 (100) | 21 (46·7) | 56 (46·7) | 22 (57·9) | 53 (44·5) |
Sample collection, RNA extraction and quality assessment
Yale, Chicago, Pittsburgh and Freiburg cohorts
PBMC collection, total RNA extraction and quality assessment methods have been previously described6. BWH-HMS and Imperial College Cohorts: Whole blood was collected using PAXgene blood RNA tubes (PreAnalytiX) and total RNA was extracted using the PAXgene Blood RNA Kit, following the manufacturer’s protocol.
52-gene signature measurement
Yale, Pittsburgh, Freiburg and BWH-HMS cohorts
The nCounter® analysis system (Nanostring)11 was used to validate the 52-gene signature. Imperial College London cohort: The 52-gene signature was analyzed from a previously published gene expression dataset of whole blood12 (GEO accession number: GSE93606). University of Chicago cohort: The expression of the 52-gene signature was analyzed from a previously published gene expression dataset of PBMC from IPF patients6 (GEO accession number: GSE27957). Gene expression microarrays were performed in accordance to MIAME guidelines. Gene normalization was performed by cohort (see online supplement for more details). transformed Log2 gene expression values were used for statistical analyses.
MMP7 measurement
Serum samples were obtained from Pittsburgh cohort patients who had PBMC collected simultaneously (N=114) in the time to event analysis. The MMP7 Elisa assay (R&D Systems) has been previously validated by us 13,14
Statistical methods and analysis
Development of the Scoring Algorithm of Molecular Subphenotypes (SAMS)
SAMS is a classification algorithm of gene expression data generated from the calculation of two scores (up and down scores). The following steps summarize the calculation of SAMS Up and Down scores: Step 1: Geometric mean normalization - We subtract the log2 value of the gene from the geometric mean of the same gene in all the samples in the cohort. A gene with a positive value is considered increased, and a gene with a negative value is considered decreased. Step 2: Determination of increased and decreased ratios – This ratio is calculated by dividing the number of genes changed in a certain direction (increased or decreased) in a sample divided by the number of genes expected to change in the same direction. The 52-gene signature contains 7 increased and 45 decreased genes. Thus the increased ratio is calculated by dividing the number of actually increased genes by 7 and the decreased ratio is the number of actually decreased genes divided by 45 (see example in online supplement). Step 3: Sums of the values of increased or decreased genes are calculated per sample. Step 4: Calculation of the scores - the up score is derived by multiplying the sum of the values of the increased genes by the increased ratio and the down score by multiplying the sum of the decreased genes by the decreased ratio. Because the gene expression values are log2 – the up score will be positive and the down score will be negative
To determine 52-gene risk profiles in each independent cohort, patients with up scores above the median value and down scores below the median value in each cohort were classified as “high risk”. Patients without this pattern of expression were classified as “low risk”. Analysis of variance (ANOVA) was used to identify significant differences in SAMS scores between cohorts. The SAMS calculator is publicly available at http://gem.med.yale.edu/SAMSWeb3/index.jsp
Time to event analysis
Patients were followed from study entry until death, loss of follow up, or transplant. Because two cohorts (Yale and Imperial college) did not contain transplants, we used different outcome definitions, transplant-free survival (TFS) in Chicago, Pittsburgh, Freiburg and BWH-HMS and mortality in Yale and Imperial College. The association between genomic risk profiles and outcomes was determined by univariate Cox Proportional-Hazard models. For TFS, both transplants and deaths were considered events. To determine whether genomic risk profiles were predictive of the predetermined clinical outcomes, we pooled the data from all cohorts and adjusted for age, gender, percent predicted forced vital capacity (FVC%) and immunosuppressive therapy, defined as the use of prednisone, azathioprine, or a combination of both at the time of blood draw. Multivariate competing risk15 and Cox proportional-hazard16 models were applied to the pooled data to determine association with mortality or TFS respectively. For mortality analyses in the pooled data, transplants were considered a competing risk (Figure 1a). Differences in mortality and TFS between patients with high and low risk genomic profiles were evaluated using cumulative incidence and Kaplan Meier curves, respectively. To test whether 52-gene risk profiles could improve outcome prediction when used in combination with the GAP index3, we fit competing risk15 and Cox proportional-hazard16 models as follows: GAP only, genomic only, GAP and genomic or the G-GAP index. The G-GAP index was calculated by adding three points (the maximum score in the GAP index) to the GAP index if a patient had a high risk genomic profile and no points if they had a low risk profile. To determine the prediction accuracy of these models and to compare their predictive performance, we used time-dependent Receiver Operating Characteristic (ROC) for censored data17 and Area Under the Curve (AUC) using a 10-fold cross validation procedure. Pooled data analysis results were adjusted by patient’s age, gender, immunosuppression use and percent predicted forced vital capacity (FVC%). MMP7 and 52-gene risk profiles where compared head-to-head using the Concordance index (C-index), an equivalent of the area under the curve (AUC) in a receiver operator curve (ROC), a well-accepted measure of the probability that predicting the outcome is better than chance. 18. GAP index was not included in the comparisons between 52-gene risk profiles and MMP7.
Time course analysis
Time course analyses were performed in the Pittsburgh and Yale, time course cohorts (Figures 1b and S1). Trends in SAMS scores and forced vital capacity volumes (FVC) were plotted to identify shifts in genomic risk profiles over time. To identify statistically significant differences in up and down scores, and FVC across time between high and low risk patients, we used a linear mixed-effect (LME) model19 with random intercepts. A linear mixed-effect model with random intercepts was also used to study the associations between changes in up and down scores and changes in FVC in patients with simultaneous measurements. LME models were adjusted by patient’s age, gender and therapy (immunosuppression therapy in the Pittsburgh cohort and anti-fibrotic therapy in Yale). Anti-fibrotic therapy was initiated after baseline sample was collected and defined as the use of Pirfenidone or Nintedanib. To determine the association between changes in SAMS scores and survival, we calculated the relative changes in up and down scores for each IPF patient based on their first two visits, adjusting to the duration of time intervals. Thus, for each one of the Pittsburgh cohort patients with at least two visits (N=66), we calculated the relative changes in both scores per month, from one to six months (see supplementary methods). Patients were classified as high risk if relative changes in up and down scores between two subsequent visits occurred simultaneously and were ≥10% (bidirectional changes). A Cox proportional-hazards model was used to determine the association between bidirectional changes in SAMS scores and TFS. Results were adjusted by patient’s age, gender, FVC and immunosuppressive therapy. Finally, we used a LME model to compare the rate of FVC decline per year in IPF patients from the Yale cohort who had a simultaneous decrease in up score and increase in up score (N=6) from those with other time course changes in SAMS scores (N=16), after initiation of anti-fibrotic therapy. Statistical significance was defined as two-sided P<0·05. Analyses were performed using R. Details on the R packages are provided in the online supplement.
Results
52-gene risk profiles are predictive of outcome in IPF and non-inferior to serum MMP7 levels
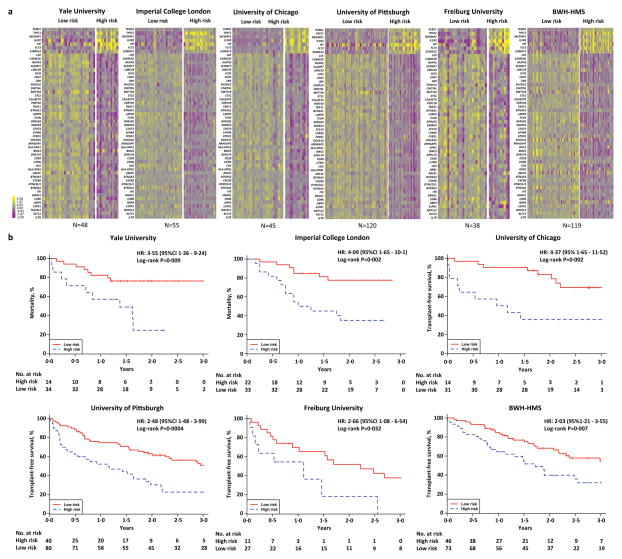
We measured the expression of a peripheral blood 52-gene signature6 in IPF patients from six independent academic centers (Table 2). Gene expression levels, clinical and demographic data were collected at baseline in all patients and across time in patients from the Pittsburgh and Yale cohorts to perform time to event (Figure 1a) and time course analysis (Figure 1b), respectively. To classify patients as high or low risk we calculated up and down scores using SAMS. Up or down scores were not significantly different between cohorts suggesting a similar distribution of patients with 52-gene, high risk profiles in each cohort (Figure S2). SAMS scores separated patients into high and low risk groups with impressive similarity in gene expression patterns within risk groups across the various cohorts (Figure 2a). Univariate Cox Proportional-hazard models demonstrated that patients in the high risk group had significantly (P<0·05) higher mortality (Yale and Imperial College London cohort) or lower TFS (Chicago, Pittsburgh, Freiburg and BWH-HMS cohorts), respectively, when compared to patients in the low risk group (Figure 2b). The hazard ratios (HR) for mortality and TFS ranged from 2·03 to 4·37 indicating that patients with a 52-gene, high risk profile had at least a two-fold increased risk of dying or having a lung transplant during follow-up in each independent cohort.
Table 2. Clinicopathological characteristics of the IPF patients in the two risk groups (pooled data) for time to event analysis.
P-values were calculated using the Fisher’s exact test except for age, pulmonary function tests and GAP index where an unpaired, two tailed, t-test was used. FVC%, forced vital capacity, percent predicted, DLCO%, carbon monoxide diffusing capacity, percent predicted. FEV1%, forced expiratory volume in 1 second, percent predicted. HRCT, high-resolution computed tomography. UIP, usual interstitial pneumonia.
| Characteristics | Low risk (n=278) | High risk (n=147) | P-value† |
|---|---|---|---|
| Age (yr) | |||
| Mean ± SD | 67·4 ± 7·9 | 68·4 ± 8·7 | 0·24 |
| Gender, n (%) | |||
| Males | 198 (71·2) | 120 (81·6) | 0·019 |
| Females | 80 (28·8) | 27 (18·4) | |
| Race, n (%) | 0·077 | ||
| Caucasian | 257 (92·4) | 143 (97·7) | |
| Black | 10 (3·6) | 0 (0) | |
| Hispanic | 5 (1·8) | 3 (2) | |
| Other | 6 (2·2) | 1 (0·7) | |
| Smoking status, n (%) | 0·27 | ||
| Ever smoker | 185 (66·5) | 106 (72·1) | |
| Never smoker | 93 (33·5) | 41 (27·9) | |
| Immunosuppression use, n (%) | |||
| No | 252 (90·6) | 103 (70·1) | <0·0001 |
| Yes | 26 (9·4) | 44 (29·9) | |
| Spirometry (mean ± SD) | |||
| FVC% | 69·3 ± 18·4 | 62·7 ± 17·3 | 0·0004 |
| DLCO% | 46 ± 17·3 | 40·9 ± 16·2 | 0·005 |
| FEV1% | 76 ± 19·8 | 70·6 ± 18·4 | 0·007 |
| GAP Index | 0·002 | ||
| Mean ± SD | 3·9 ± 1·4 | 4·3 ± 1·5 | |
| Diagnosis, n (%) | 0·41 | ||
| HRCT+ UIP biopsy | 126 (45·3) | 60 (40·8) | |
| HRCT | 152 (54·7) | 87 (59·2) |
Figure 2. 52-gene risk profiles are predictive of outcome in IPF.
(a) Clustering of IPF patients based on52-gene risk profiles (high vs low) derived using SAMS in each one of the six cohorts studied. Every row represents a gene and every column a patient. Color scale is shown adjacent to heat maps in log-based two scale; yellow denotes increase over the geometric mean of samples and purple, decrease. (b) Mortality and Transplant-free survival (TFS) differs between high vs low risk profiles based on the 52-gene signature in each independent cohort.
To determine how outcome prediction using 52-gene risk profiles compared to serum MMP7, we measured MMP7 concentrations by ELISA in Pittsburgh cohort patients with simultaneous PBMC and serum collections (N=114) and compared their TFS prediction performance using the C-index. Our analysis demonstrated that the C-index for TFS prediction in the Pittsburgh cohort was significantly higher (P=0·011) when using 52-gene, genomic risk profiles (C-index=0·72 95%CI 0.659, 0.779) versus MMP7 levels in serum (C-Index=0·61, 95%CI 0.535, 0.683).
52-gene, genomic risk profiles are predictive of outcome independent of demographic and clinical variables
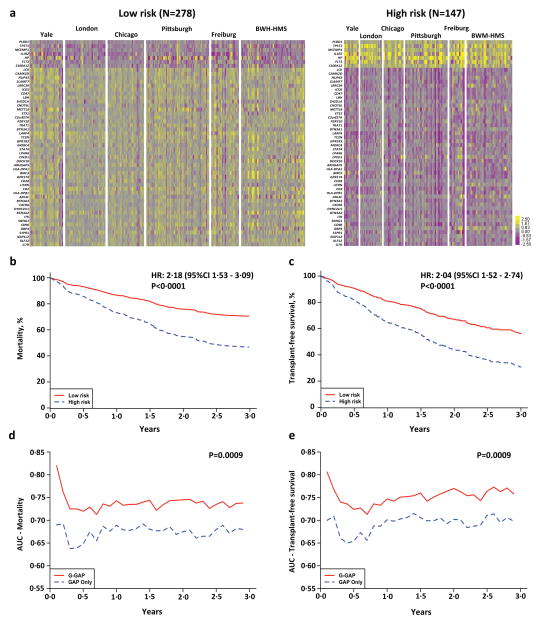
To identify demographic and clinical characteristic differences between 52-gene risk profiles, a pooled data analysis was performed using data from all 425 IPF patients (Figure 3a). High risk patients were predominantly Caucasian males with lower FVC% and DLCO% at presentation. There were more high-risk patients under immunosuppressants (Table 3). A high risk, 52-gene profile was independently predictive of mortality (HR 2·18, 95% CI 1·53, 3·09, P<0·0001) or TFS (HR 2·04, 95%CI 1·52, 2·74, P<0·0001) (Figure 3b and c) after adjusting for age, gender FVC% and immunosuppressive therapy in the pooled dataset. To account for possible cohort heterogeneity, we also performed multivariate competing risk and Cox PH models stratified by cohort in the pooled data and the results did not differ significantly (HR 2·36, 95% CI 1·67, 3·35, P= 1.3e-6 for mortality and HR 2·08, 95% CI 1·54, 2·80, P= 1.6e-6 for TFS). Because of the known adverse effects of immunosuppressive therapy on survival of patients with IPF20, we repeated the analysis only on patients that did not receive immunosuppression. A 52-gene, high risk genomic profile was also independently predictive of mortality (HR 2·27, 95% CI 1·54, 3·35, P<0·0001) or TFS (HR 2·13, 95%CI 1·54, 2·96 P<0·0001) in this dataset, after excluding patients under immunosuppressants (Figure S3). A prediction model based on the calculated G-GAP index outperformed all other prediction models studied (Supplementary Tables 1 and 2) and significantly improved accuracy prediction of mortality or TFS (Figure 3d and e). The maximal Area Under the Curve changed by 13% (69% to 82%) or 10·6% (70% to 80·6%) for a 30-day mortality and TFS prediction, respectively.
Figure 3. 52-gene risk profiles are predictive of outcome independent of demographic and clinical variables.
(a) Pooled data analysis comparing high vs low risk profile patients from all cohorts. Color scale is shown adjacent to heat maps in log-based two scale. (b) Mortality and (c) Transplant-free survival (TFS) differs between high vs low risk patients from all cohorts after adjusting for age, gender, FVC% and immunosuppressive therapy. (d) Area Under the Curve (AUC) of time-dependent ROC analysis for (d) mortality and (e)TFS based on the GAP index alone or the G-GAP index in all patients.
Association of 52-gene expression trends over time with disease progression and survival
For time course analyses, we measured the expression of the 52-gene signature in RNA isolated from PBMC using the nCounter system, calculated up and down scores at each time point and collected FVC values over time in two cohorts (Pittsburgh and Yale, Figure 1b). Details about number of visits and follow up duration can be seen in Figure 1.
To determine the association between changes in up and down scores over time with FVC, we performed a LME model adjusted for age and gender in Pittsburgh and Yale cohorts. In both cohorts, up scores were negatively associated with FVC and down scores were positively associated with FVC. The association of up scores with FVC was −0·025 (95%CI −0·039, −0·011, P=0·0004) in the Pittsburgh cohort and −0·010 (95%CI −0·017, −0·004, P=0·004) in the Yale cohort. Similarly, the association of down scores with FVC was 0·008 (95%CI 0·005, 0·011, P<0·0001) in the Pittsburgh cohort, and 0·027 (95%CI 0·004, 0·051, P=0·029) in the Yale cohort.
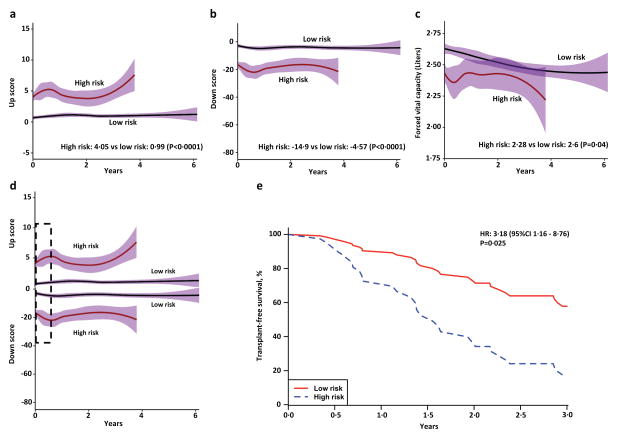
To determine whether 52-gene, high or low risk patients, not on anti-fibrotic drugs (Pittsburgh cohort) shifted their risk profile, we plotted up and down scores and FVC trends and compared their values across time in high versus low risk groups using a LME model. Our results indicate no shift in risk profiles or FVC trends (Figure 4a, b and c), results confirmed by the LME model. This model demonstrated a significant difference for up scores (high risk: 4·05 vs low risk: 0·99, P<0·0001), down scores (high risk: −14·9 versus low risk: −4·57, P<0·0001) and FVC (high risk: 2·28 liters versus low risk: 2·60 liters, P=0·04) between high and low risk groups across time in this cohort.
Figure 4. 52-gene signature trends over time demonstrate association with disease progression and survival.
(a) up and down (b) scores from SAMS, and (c) FVC volumes do not shift their trends over time in high (continuous red line) vs low (continuous black line) risk groups (Pittsburgh cohort). Pointwise confidence intervals are represented in purple. (d) Bidirectional changes in SAMS scores (Simultaneous increase in up score and decrease in down score) can be observed during disease course in IPF and are more prominent in high risk individuals (example shown in dotted black line box). (e) Bidirectional changes in SAMS scores are predictive of Transplant-free survival (TFS). Dotted blue line (high risk) –Pittsburgh cohort patients with 30-day bidirectional changes in SAMS scores ≥10%. Continuous red line (low risk) – Pittsburgh cohort patients with 30-day bidirectional changes in SAMS scores <10%. Results adjusted by age, gender, FVC and immunosuppressive therapy.
We also assessed whether substantial changes in SAMS scores over time were predictive of IPF survival in patients not on anti-fibrotic drugs (Pittsburgh cohort). Since relative changes in FVC ≥10% have been associated with decreased IPF survival 21,22, we hypothesized that a relative increase in up score and a simultaneous decline in down score ≥10%, was also predictive of IPF survival. Univariate and multivariate Cox models (Supplementary Table 3) demonstrated that a simultaneous ≥10% increase in up score and decrease in down score (Bidirectional changes), between two measurements obtained 30-days apart (Figure 4d), was significantly predictive of future transplant-free survival (HR: 3·18· 95%CI 1·16, 8·76, P=0·025) (Figure 4e). Only three out of 32 patients in the Yale time course cohort had ≥10% bidirectional changes across time thus we could not assess the relationship between bidirectional changes and survival in this cohort.
Changes in 52-gene expression trends over time are associated with clinical response to anti-fibrotic agents
To determine the effect of anti-fibrotic drugs on 52-gene risk profiles, we first plotted up and down score trends over time in the Yale time course cohort. Low risk profile patients exhibited the same patterns as observed in the Pittsburgh cohort, but high risk profile patients exhibited shifts in up scores (Figure S4a) and down scores (Figure S4b). Because a higher proportion of high risk patients were initiated on anti-fibrotic therapy (90%) compared to low risk patients (59%) (Supplementary table 4), we analyzed the interaction between changes in scores and response to therapy. Impressively, in patients who exhibited a simultaneous decrease in up score and increase in down score, we observed an average increase in FVC (0·06 liters per year), while in patients that did not exhibit these changes in scores, we observed an average decrease in FVC (−0.21 liters per year). The difference was statistically significant (P=0·005) (Figure S4c).
Discussion
We have previously identified a 52-gene signature predictive of TFS in two IPF patient cohorts by using microarray analysis of PBMC6. Here, we analyzed the 52-gene signature in the peripheral blood from 425 IPF patients from six independent cohorts. Using the novel Scoring Algorithm of Molecular Subphenotypes (SAMS), we derived risk profiles from the 52-gene signature that identified two classes of IPF patients with significant differences in outcome in all six cohorts. The prediction accuracy of 52-gene risk profiles was better than serum concentrations of MMP7 and adding 52-gene risk profile information to the clinical GAP index significantly increased its prediction accuracy. Temporal analysis revealed that untreated patients generally did not change their risk profiles; however, simultaneous increase in up score and decrease in down scores was predictive of subsequent transplant-free survival. In patients initiated on anti-fibrotic therapy, a simultaneous decrease in up score and increase in down score was associated with stabilization of FVC.
The recognition of the variable clinical course in IPF led to a substantial effort to identify clinical tools and reliable peripheral blood biomarkers for risk stratification. Changes in peripheral blood proteins such as MMP74,13, ICAM and IL84, SP-A and SP-D5, KL-623, CCL-1824, YKL4025, CXCL1326, POSTN27, anti-hsp70 IgG antibodies28 and protease degradation products29, have been found to be predictive of poor IPF outcomes. Changes in circulating cells (CD4+CD28+ T cells6, fibrocytes30 and Semaphorin 7a+ regulatory T cells31), gene polymorphisms (TOLLIP32, TLR333 and MUC5B7) and aging biomarkers (Telomere length8, free mitochondrial DNA34) have also been associated with mortality in IPF. While these studies strongly suggested the value of peripheral blood biomarkers for risk stratification in IPF, no marker is currently used in clinical practice. This is, in part, because the majority of the studies did not have truly independent replication cohorts, nor did they demonstrate added value over clinical staging tools. In contrast to previous studies, our study provides validation of our 52-gene expression signature in six independent IPF cohorts and demonstrates a substantial improved accuracy when incorporated with currently used clinical tools. This is important, because accurate outcome prediction has very practical implications for IPF patients. Based on the current lung allocation score, and on their clinical characteristics, nearly all of the patients in our study would be referred for transplant evaluation, and many would be eligible for lung transplantation. However, our data suggests that only patients with a high risk genomic profile could require this evaluation urgently, and many may not require lung transplantation even three to five years after diagnosis. Thus, incorporating 52-gene risk profiles in the evaluation of IPF patients, may enhance the precision of lung transplantation referral – avoiding delays in transplants to those who need it early, and delaying those who may not need it. Similarly, when lung transplantation is not an option, this test could also help physicians deciding when to refer IPF patients to palliative care, a currently significant unmet need35 or distinguish patients who respond to drug therapy from those who do not. Similarly, the majority of previous studies did not assess the change of markers over time. This is important, as it is unknown whether IPF patients shift their risk profiles. We demonstrate that a patient’s 52-gene, genomic risk profile rarely changes in the absence of anti-fibrotic therapy. However, when the profile does change it is important. In untreated patients, a simultaneous increase in up score and decrease in down score reflects subsequent increased mortality.
In patients treated with anti-fibrotic agents, a simultaneous decrease in up score and increase in down score, reflects stabilization or even increase in FVC. Thus, our study demonstrates that 52-gene risk profiles at presentation are predictive of outcome and changes in a patient’s genomic risk profile are informative of clinical deterioration as well as potential response to anti-fibrotic therapies.
While our study focuses on the biomarker applications of the 52-gene signature for risk stratification in IPF, it could also serve to generate hypotheses for follow up studies. We have previously shown that four genes of this signature (CD28, ICOS, LCK and ITK), that belong to the T-cell co-stimulatory signaling pathway, were correlated with the percentage of CD4+CD28+ T cells in the circulation of these patients6. Similarly, previous reports have demonstrated that changes in circulating CD4+ T cells with CD28 down-regulation36 of IPF patients are also associated with poor disease outcomes. These reports suggest a potential link between changes in the expression of genes in the 52-gene signature with phenotypic shifts in circulating immune cells. Similarly, a recent report suggested that down-regulation of T cell co-stimulation markers is associated with T cell exhaustion and poor outcomes in inflammatory and autoimmune diseases37. While IPF is not generally considered an autoimmune disease, T cell exhaustion is a mechanism that should be explored as a potential explanation of our findings. Additionally, other members of the 52-gene expression signature may have also some clues about the role of immune aberrations in IPF. As an example, MCEMP1 (mast cell-expressed membrane protein 1) one of the outcome predictive genes when overexpressed, encodes a transmembrane protein isolated from human mast cells38, known to work in concert with fibroblasts to aggravate pulmonary fibrosis39 or FLT3 (Fms-related tyrosine kinase 3) a strong Nintedanib-responsive tyrosine kinase with unknown roles in pulmonary fibrosis. While such studies were beyond the scope of this paper, they could potentially shed light on the role of immune aberrations in IPF.
Despite the impressive reproducibility of our findings, we need to recognize some of the limitations of our study. First, SAMS scores were calculated for each individual after normalization within each cohort. The normalization within cohort was required because the data was obtained by different technologies using RNA extracted from whole blood or PBMC (Figure 1). This of course limits the clinical applicability of our results because the expressions of the 52 genes of an entire IPF cohort need to be available for the calculation of the genomic risk profile of an individual patient. For our results to be implemented in the clinic, we would need to generate a set of reference values for the 52 genes in IPF patients. Such reference values could be used to calculate SAMS up and down scores for every new sample and determine the 52-gene risk profile of patients, independently of a specific cohort. The significant reproducible performance of the 52-gene signature, should encourage the development of this reference set and the standardization as an essay for clinical use. Second, we did not determine the specificity of the 52-gene signature to IPF. To assess the effect of aging, we analyzed the 52-gene signature in control individuals older than 90 years of age40,41 (Figure S5), and found that it was not predictive of mortality in the aged, but we did not study other chronic lung disease. Third, treatment guidelines have changed in IPF in some cohorts, patients were at least initially on immunosuppressive therapy, which it is well known, affect outcome. However, the 52-gene signature was originally discovered in a cohort (Chicago cohort) where only two out of 45 patients were on immunosuppressive therapy at study entry. Such small number of patients under immunosuppression should not account for the transplant-free survival and mortality prediction accuracy of the signature. To further address this, we performed a separate analysis in which we excluded all patients on immunosuppressive therapy at the time of blood draw. The 52-gene signature was predictive of outcome in this population indicating that immunosuppression did not confound our results. Fourth, our initial predictive model was not adjusted to DLCO because we had missing data especially among high risk patients who did not have DLCO measurements performed at the time of blood draw. However, we did address the effect of DLCO indirectly, through the comparison to the GAP index. DLCO is a component of the GAP index, and adding the 52-gene risk profile to the GAP index significantly improved its outcome predictive accuracy. Finally, our longitudinal analysis was limited by the size of cohorts and the difference between them, however we have demonstrated significant reproducibility on two observations, that untreated IPF patients do not generally shift their genomic risk profile and that 52-gene SAMS scores are significantly associated with FVC. The observation that in treated high risk patients, a simultaneous decrease in up score and increase in down score is associated with a significant stabilization of FVC is intriguing, but will require replication, as it is based on a very small number of patients.
In conclusion, our study demonstrates that the 52-gene risk profiles are reproducible predictors of outcome in IPF patients. The enhanced outcome prediction accuracy when 52-gene risk profiles are added to the GAP index (G-GAP index) and the association of changes in genomic risk profiles with changes in FVC, survival and potential response to anti-fibrotic therapy, indicate the potential value of the 52-gene signature as a blood test to risk stratify and monitor disease in IPF. To develop this blood test, we would need prospective studies that specifically address some of the limitations of our study including, the establishment of universal reference values for the 52 genes, a prospective comparison to other molecular markers, and determination whether the 52 gene signature is predictive or associates with acute exacerbations.
Supplementary Material
Evidence before this study
We searched the scientific literature using PubMed to identify studies that use gene expression in the peripheral blood to identify outcome prediction markers in IPF. We used the search terms “Pulmonary Fibrosis”, “biomarkers”, “outcome prediction”, and “blood” and did not use language or date restrictions. We identified multiple studies that assessed the value of proteins carried in the blood stream to predict outcome in IPF. When we added the term gene expression we identified two relevant studies, one that assessed the correlation of the peripheral blood transcriptome with extent of fibrosis, and our own previous study that discovered the 52 gene signature, but did not include a complete validation of the signature or assessment of its change over time and in response to novel therapies.
Added value of this study
In this study, we developed a genomic risk scoring system (SAMS) based on the 52-gene signature, and tested it on 425 patients from six independent cohorts from Academic Centers in the United States, United Kingdom and Germany. We identified two groups of IPF patients (low and high risk) with significant differences in mortality or transplant-free survival in each of the six cohorts (HR 2·03–4·37). Pooled data revealed similar results for mortality (HR 2·18, 95%CI:1·53–3·09, P<0·0001) or transplant-free survival (HR: 2·04, 95%CI: 1·52–2·74, P<0·0001). Adding 52-gene risk profiles to the Gender, Age and Physiology (GAP) index significantly improved its outcome predictive accuracy. Temporal changes in SAMS were associated with changes in forced vital capacity in two cohorts. Untreated patients tended not to change their risk profiles, but some high risk patients started on antifibrotic therapy experienced a reversal of their high risk profile. Change in 52-gene risk profiles after initiation of anti-fibrotic therapy was associated with a significant (P=0·005) improvement in Forced Vital Capacity.
Implications of all the available evidence
The 52-gene signature is a reproducible predictor of mortality and transplant-free survival in patients with IPF that can improve the performance of the GAP index. The signature correlates with Forced Vital Capacity (FVC) and without therapy, patients do not shift their risk profile. Limited data suggest that a reversal of a high-risk genomic profile is associated with stabilization of FVC. Prospective studies are required to establish the value of the 52-gene signature as a marker for response to antifibrotic therapy in IPF.
Acknowledgments
Funding:
Jose D. Herazo-Maya: The Pulmonary Fibrosis Foundation and the Robert Wood Johnson Foundation under the Harold Amos Medical Faculty Development Program. Philip L. Molyneaux: Asmarley trust grant. Antje Prasse: E-RARE project, JRC 2011 IPF-AE (DLR 01GM1210A). Erica L. Herzog: RO1 HL109233 and RO1 HL125850. Ivan O. Rosas: P01 HL11450 and RO1 HL15024. Toby M. Maher: NIHR Clinician Scientist Fellowship (NIHR Ref: CS-2013-13-017). Naftali Kaminski: U01 HL112707, R01 HL127349, U01 HL108642 and UH3 HL123886. The funding bodies had no role on the conception of this manuscript, they did not participate in any way in the design or performance of the experiments, analysis of the results or writing or revision of the manuscript. The corresponding authors had full access to all of the data and the final responsibility to submit for publication.
Footnotes
Authors contributions: JHD* conceived and designed the project, performed experiments, developed algorithms, analyzed data and wrote the manuscript; JS* performed analyzed data, generated figures and revised manuscript, PLM recruited subjects, procured biospecimens, and analyzed data, QL performed experiments, and collected data, JVN recruited subjects, procured biospecimens, and collected data, AT performed experiments, HL extracted RNA, performed experiments and analyzed data, BMJD performed experiments, collected data and participated in design, CR recruited subjects, and collected data, JCO procured biospecimens, and analyzed data, XY supervised statistical analysis, GM performed experiments, and generated data, NA handled samples, generated data, KOL designed study, collected data, MJK procured biospecimens, collected data, MFM designed experiments, collected and analyzed data, WOC designed experiments, collected and analyzed data, YZ characterized patients, analyzed data, JGNG participated in in project conception, analysis, IN contributed data, participated in design of project, AP participated in design of project, data analysis and collection, ZBJ developed analytical approaches, KFG participated in project conception, data collection and analysis, HZ supervised and critiqued analytical approaches, developed algorithms and data analysis, ELH supervised sample procurement, data collection and analysis, manuscript preparation, IOR supervised sample procurement, data collection and analysis, participated in manuscript preparation, TMM participated in project design, data collection and analysis, and manuscript preparation, *NK conceived and designed the project, supervised data collection, participated in analysis and figure generation and revised the manuscript. All authors participated in manuscript preparation and provided final approval of the submitted work. *JHD, *JS, equally contributed to this work.
Conflict of interest statements
JHD has a patent on marker panels for Idiopathic Pulmonary Fibrosis diagnosis and evaluation pending. WOC reports grants from Wellcome Trust, during the conduct of the study. MFM reports grants from Wellcome Trust, during the conduct of the study. AP reports personal fees from Boehringer Ingelheim, personal fees from Roche Pharma, personal fees from Sanofi Aventis, personal fees from Bayer, personal fees from AstraZeneca, outside the submitted work. ION reports grants and personal fees from Veracyte, grants and personal fees from Boehringer, grants and personal fees from Genentech, personal fees from Immuneworks, personal fees from Global blood therapeutics, personal fees from Sanofi, outside the submitted work; In addition, Dr. Noth has a patent TOLLIP in IPF pending, and a patent Plasma proteins in IPF MMP7 issued. ELH reports grants from NIH/NHLBI, grants from Greenfield Foundation, during the conduct of the study; personal fees from Boehringer lngelheim, grants from Sanofi, grants from Biogen ldec, grants from Bristol Myers, grants from Navitor, grants from Promedior, outside the submitted work. AP reports personal fees from Boehringer Ingelheim, personal fees from Roche Pharma, personal fees from Sanofi Aventis, personal fees from Bayer, personal fees from AstraZeneca, outside the submitted work. TMM has, via his institution, received industry-academic funding from GlaxoSmithKline R&D and UCB and has received consultancy or speakers fees from Apellis, Astra Zeneca, Bayer, Biogen Idec, Boehringer Ingelheim, Cipla, GlaxoSmithKline R&D, InterMune, ProMetic, Roche, Sanofi-Aventis, Samumed and UCB. NK reports grants and personal fees from Biogen Idec, personal fees from Boehringer Ingelheim, stock options from Moereae Matrix, personal fees and stock options from Pliant, no funds from Samumed, non-financial support from Actelion and Miragen, past personal fees from Third Rock, all outside the submitted work; In addition, NK has patents on new therapies in pulmonary fibrosis issued, and biomarker panels in pulmonary fibrosis. NK is a member of the Scientific Advisory Committee, the Research Advisory Forum and the Board of the Pulmonary Fibrosis Foundation. Serves as Deputy Editor of Thorax, BMJ. The rest of the authors report no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Raghu G, Chen SY, Yeh WS, et al. Idiopathic pulmonary fibrosis in US Medicare beneficiaries aged 65 years and older: incidence, prevalence, and survival, 2001–11. Lancet Respir Med. 2014;2(7):566–72. doi: 10.1016/S2213-2600(14)70101-8. [DOI] [PubMed] [Google Scholar]
- 2.Martinez FJ, Safrin S, Weycker D, et al. The clinical course of patients with idiopathic pulmonary fibrosis. Annals of internal medicine. 2005;142(12 Pt 1):963–7. doi: 10.7326/0003-4819-142-12_part_1-200506210-00005. [DOI] [PubMed] [Google Scholar]
- 3.Ley B, Ryerson CJ, Vittinghoff E, et al. A multidimensional index and staging system for idiopathic pulmonary fibrosis. Annals of internal medicine. 2012;156(10):684–91. doi: 10.7326/0003-4819-156-10-201205150-00004. [DOI] [PubMed] [Google Scholar]
- 4.Richards TJ, Kaminski N, Baribaud F, et al. Peripheral blood proteins predict mortality in idiopathic pulmonary fibrosis. American journal of respiratory and critical care medicine. 2012;185(1):67–76. doi: 10.1164/rccm.201101-0058OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greene KE, King TE, Jr, Kuroki Y, et al. Serum surfactant proteins-A and -D as biomarkers in idiopathic pulmonary fibrosis. The European respiratory journal. 2002;19(3):439–46. doi: 10.1183/09031936.02.00081102. [DOI] [PubMed] [Google Scholar]
- 6.Herazo-Maya JD, Noth I, Duncan SR, et al. Peripheral blood mononuclear cell gene expression profiles predict poor outcome in idiopathic pulmonary fibrosis. Science translational medicine. 2013;5(205):205ra136. doi: 10.1126/scitranslmed.3005964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peljto AL, Zhang Y, Fingerlin TE, et al. Association between the MUC5B promoter polymorphism and survival in patients with idiopathic pulmonary fibrosis. Jama. 2013;309(21):2232–9. doi: 10.1001/jama.2013.5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stuart BD, Lee JS, Kozlitina J, et al. Effect of telomere length on survival in patients with idiopathic pulmonary fibrosis: an observational cohort study with independent validation. Lancet Respir Med. 2014;2(7):557–65. doi: 10.1016/S2213-2600(14)70124-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ley B, Brown KK, Collard HR. Molecular biomarkers in idiopathic pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol. 2014;307(9):L681–91. doi: 10.1152/ajplung.00014.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raghu G, Collard HR, Egan JJ, et al. An official ATS/ERS/JRS/ALAT statement: idiopathic pulmonary fibrosis: evidence-based guidelines for diagnosis and management. American journal of respiratory and critical care medicine. 2011;183(6):788–824. doi: 10.1164/rccm.2009-040GL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geiss GK, Bumgarner RE, Birditt B, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nature biotechnology. 2008;26(3):317–25. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 12.Molyneaux PL, Willis-Owen SAG, Cox MJ, et al. Host-Microbial Interactions in Idiopathic Pulmonary Fibrosis. American journal of respiratory and critical care medicine. 2017;195(12):1640–50. doi: 10.1164/rccm.201607-1408OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tzouvelekis A, Herazo-Maya JD, Slade M, et al. Validation of the prognostic value of MMP-7 in idiopathic pulmonary fibrosis. Respirology. 2017;22(3):486–93. doi: 10.1111/resp.12920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosas IO, Richards TJ, Konishi K, et al. MMP1 and MMP7 as potential peripheral blood biomarkers in idiopathic pulmonary fibrosis. PLoS Med. 2008;5(4):e93. doi: 10.1371/journal.pmed.0050093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fine JPGR. A proportional hazards model for the subdistribution of a competing risk. J Am Stat Assoc. 1999;94:496–509. [Google Scholar]
- 16.Therneau TMGP. Modeling survival data: extending the Cox model. New York: Springer; 2000. [Google Scholar]
- 17.Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56(2):337–44. doi: 10.1111/j.0006-341x.2000.00337.x. [DOI] [PubMed] [Google Scholar]
- 18.Harrell FE, Jr, Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996;15(4):361–87. doi: 10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 19.Bates D, Maechler M, Bolker B, et al. lme4: Linear Mixed-Effects Models using ‘Eigen’ and S4. R package. 2015 [Google Scholar]
- 20.Idiopathic Pulmonary Fibrosis Clinical Research N. Raghu G, Anstrom KJ, King TE, Jr, Lasky JA, Martinez FJ. Prednisone, azathioprine, and N-acetylcysteine for pulmonary fibrosis. N Engl J Med. 2012;366(21):1968–77. doi: 10.1056/NEJMoa1113354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richeldi L, Ryerson CJ, Lee JS, et al. Relative versus absolute change in forced vital capacity in idiopathic pulmonary fibrosis. Thorax. 2012;67(5):407–11. doi: 10.1136/thoraxjnl-2011-201184. [DOI] [PubMed] [Google Scholar]
- 22.du Bois RM, Weycker D, Albera C, et al. Forced vital capacity in patients with idiopathic pulmonary fibrosis: test properties and minimal clinically important difference. American journal of respiratory and critical care medicine. 2011;184(12):1382–9. doi: 10.1164/rccm.201105-0840OC. [DOI] [PubMed] [Google Scholar]
- 23.Yokoyama A, Kondo K, Nakajima M, et al. Prognostic value of circulating KL-6 in idiopathic pulmonary fibrosis. Respirology. 2006;11(2):164–8. doi: 10.1111/j.1440-1843.2006.00834.x. [DOI] [PubMed] [Google Scholar]
- 24.Prasse A, Probst C, Bargagli E, et al. Serum CC-chemokine ligand 18 concentration predicts outcome in idiopathic pulmonary fibrosis. American journal of respiratory and critical care medicine. 2009;179(8):717–23. doi: 10.1164/rccm.200808-1201OC. [DOI] [PubMed] [Google Scholar]
- 25.Korthagen NM, van Moorsel CH, Barlo NP, et al. Serum and BALF YKL-40 levels are predictors of survival in idiopathic pulmonary fibrosis. Respiratory medicine. 2011;105(1):106–13. doi: 10.1016/j.rmed.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 26.Vuga LJ, Tedrow JR, Pandit KV, et al. C-X-C motif chemokine 13 (CXCL13) is a prognostic biomarker of idiopathic pulmonary fibrosis. American journal of respiratory and critical care medicine. 2014;189(8):966–74. doi: 10.1164/rccm.201309-1592OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tajiri M, Okamoto M, Fujimoto K, et al. Serum level of periostin can predict long-term outcome of idiopathic pulmonary fibrosis. Respiratory investigation. 2015;53(2):73–81. doi: 10.1016/j.resinv.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 28.Kahloon RA, Xue J, Bhargava A, et al. Patients with idiopathic pulmonary fibrosis with antibodies to heat shock protein 70 have poor prognoses. American journal of respiratory and critical care medicine. 2013;187(7):768–75. doi: 10.1164/rccm.201203-0506OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jenkins RG, Simpson JK, Saini G, et al. Longitudinal change in collagen degradation biomarkers in idiopathic pulmonary fibrosis: an analysis from the prospective, multicentre PROFILE study. Lancet Resp Med. 2015;3(6):462–72. doi: 10.1016/S2213-2600(15)00048-X. [DOI] [PubMed] [Google Scholar]
- 30.Moeller A, Gilpin SE, Ask K, et al. Circulating fibrocytes are an indicator of poor prognosis in idiopathic pulmonary fibrosis. American journal of respiratory and critical care medicine. 2009;179(7):588–94. doi: 10.1164/rccm.200810-1534OC. [DOI] [PubMed] [Google Scholar]
- 31.Reilkoff RA, Peng H, Murray LA, et al. Semaphorin 7a+ regulatory T cells are associated with progressive idiopathic pulmonary fibrosis and are implicated in transforming growth factor-beta1-induced pulmonary fibrosis. American journal of respiratory and critical care medicine. 2013;187(2):180–8. doi: 10.1164/rccm.201206-1109OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noth I, Zhang Y, Ma SF, et al. Genetic variants associated with idiopathic pulmonary fibrosis susceptibility and mortality: a genome-wide association study. Lancet Respir Med. 2013;1(4):309–17. doi: 10.1016/S2213-2600(13)70045-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Dwyer DN, Armstrong ME, Trujillo G, et al. The Toll-like receptor 3 L412F polymorphism and disease progression in idiopathic pulmonary fibrosis. American journal of respiratory and critical care medicine. 2013;188(12):1442–50. doi: 10.1164/rccm.201304-0760OC. [DOI] [PubMed] [Google Scholar]
- 34.Ryu C, Sun H, Gulati M, et al. Extracellular Mitochondrial DNA is Generated by Fibroblasts and Predicts Death in Idiopathic Pulmonary Fibrosis. American journal of respiratory and critical care medicine. 2017 doi: 10.1164/rccm.201612-2480OC. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lindell KO, Liang Z, Hoffman LA, et al. Palliative Care and Location of Death in Decedents With Idiopathic Pulmonary Fibrosis. Chest. 2015;147(2):423–9. doi: 10.1378/chest.14-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gilani SR, Vuga LJ, Lindell KO, et al. CD28 down-regulation on circulating CD4 T-cells is associated with poor prognoses of patients with idiopathic pulmonary fibrosis. PloS one. 2010;5(1):e8959. doi: 10.1371/journal.pone.0008959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKinney EF, Lee JC, Jayne DR, Lyons PA, Smith KG. T-cell exhaustion, co-stimulation and clinical outcome in autoimmunity and infection. Nature. 2015;523(7562):612–6. doi: 10.1038/nature14468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li K, Wang SW, Li Y, et al. Identification and expression of a new type II transmembrane protein in human mast cells. Genomics. 2005;86(1):68–75. doi: 10.1016/j.ygeno.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 39.Wygrecka M, Dahal BK, Kosanovic D, et al. Mast cells and fibroblasts work in concert to aggravate pulmonary fibrosis: role of transmembrane SCF and the PAR-2/PKC-alpha/Raf-1/p44/42 signaling pathway. Am J Pathol. 2013;182(6):2094–108. doi: 10.1016/j.ajpath.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 40.Jylha M, Paavilainen P, Lehtimaki T, et al. Interleukin-1 receptor antagonist, interleukin-6, and C-reactive protein as predictors of mortality in nonagenarians: the vitality 90+ study. J Gerontol A Biol Sci Med Sci. 2007;62(9):1016–21. doi: 10.1093/gerona/62.9.1016. [DOI] [PubMed] [Google Scholar]
- 41.Jylhava J, Raitanen J, Marttila S, Hervonen A, Jylha M, Hurme M. Identification of a prognostic signature for old-age mortality by integrating genome-wide transcriptomic data with the conventional predictors: the Vitality 90+ Study. BMC Med Genomics. 2014;7:54. doi: 10.1186/1755-8794-7-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.