Figure 3. Analysis of minimal Gle1-binding domain in Nup42.
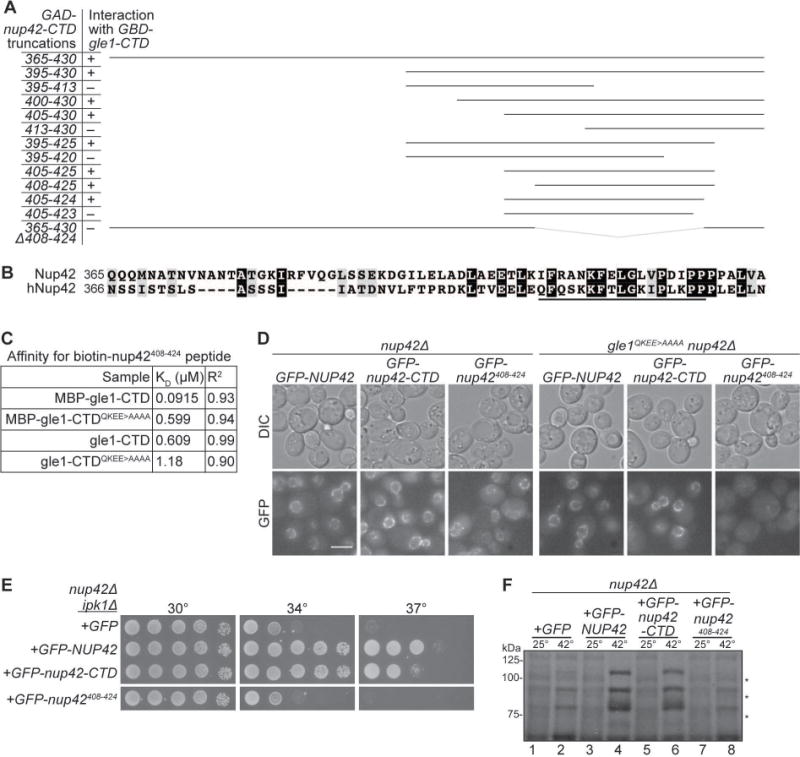
(A) nup42408–424 is required for interaction with Gle1. Indicated plasmids were transformed into the Y2H reporter strain, and struck to –Trp –Leu –His –Ade synthetic media for growth at 23°C. Growth on quadruple drop-out media is indicated as “+”. (B) nup42408–424 lies in a conserved region of the protein. Clustal Omega35 alignment between nup42-CTD and hnup42-CTD. Identical (black) and similar (grey) residues are indicated. The black line above the sequence indicates residues 408–424 of Nup42. (C) Affinity measurements between gle1-CTD and nup42408–424. BioLayer Interferometry was performed with a biotin-nup42408–424 peptide and recombinant gle1-CTD protein. Calculated KD and R2 correlation is indicated. (D) GFP-nup42408–424 localization at the nuclear envelope is disrupted in gle1QKEE>AAAA mutants. Strains containing the indicated plasmids were grown to mid-log phase at 23°C and live cells were imaged by wide-field microscopy. Scale bar = 5μm. (E) nup42408–424 does not rescue the growth defect of nup42Δ ipk1Δ mutants. nup42Δ ipk1Δ was transformed with the indicated GFP-tagged constructs, grown to mid-log phase, and plated on –Leu synthetic media for growth at the indicated temperatures. (F) nup42408–424 is not sufficient for heat shock mRNA export. nup42Δ mutants were transformed with the indicated GFP-tagged constructs, grown at 25°C to early-log phase in synthetic media, kept at 25°C or shifted to 42°C for 15 min, labeled with [35S]methionine for an additional 15 min, and lysed. Lysates were separated by SDS-PAGE, and proteins were visualized by autoradiography. Hsp proteins are indicated by asterisks.
