Abstract
Adverse immune reactions prevent clinical translation of numerous implantable devices and materials. Although inflammation is an essential part of tissue regeneration, chronic inflammation ultimately leads to implant failure. In particular, macrophage polarity steers the microenvironment towards inflammation or wound healing via the induction of M1 and M2 macrophages, respectively. Here, we demonstrated that macrophage polarity within biomaterials can be controlled through integrin mediated interactions between human monocytic THP-1 cells and collagen-derived matrix. Surface marker, gene expression, biochemical and cytokine profiling consistently indicated that THP-1 cells within a biomaterial lacking cell attachment motifs yielded pro-inflammatory M1 macrophages, whereas biomaterials with attachment sites in the presence of IL-4 induced an anti-inflammatory M2 like phenotype and propagated the effect of IL-4 in induction of M2 like macrophages. Importantly, integrin α2β1 played a pivotal role as its inhibition blocked the induction of M2 macrophages. The influence of the microenvironment of the biomaterial over macrophage polarity was further confirmed by its ability to modulate the effect of IL-4 and lipopolysaccharide, which are potent inducers of M2 or M1 phenotypes, respectively. Thus, our study represents a novel, versatile and effective strategy to steer macrophage polarity through integrin mediated three-dimensional (3D) microenvironment for biomaterial-based programming.
Keywords: Integrin, Macrophage polarization, Hydrogel, Immune modulation, M1, M2
1. Introduction
Inflammation is an inevitable consequence of implantation and is closely linked to the implant’s clinical outcome. Upon implantation, immune cells migrate to the implantation site and initiate a localized inflammatory response [1]. Although inflammation is an indispensable element in tissue regeneration, an intense or chronic inflammatory response will significantly limit natural healing. Moreover, detrimental inflammatory responses can result in fibrotic capsule formation around the implant and result in ultimately failure of the implants [1b]. Thus, the ability to actively control inflammation in regenerating tissues and implanted medical devices represents a major yet unsolved challenge.
Among the variety of immune cells, monocytes and macrophages play a particularly critical role that determines successful tissue-implant integration or implant failure [2]. Specifically, macrophage polarity strongly influences clinical outcome through the balance between pro-inflammatory M1 and regenerative M2 macrophages. Classically activated M1 macrophages are associated with a pro-inflammatory response [3]. In contrast, alternatively activated M2 macrophages are associated with an anti-inflammatory response, which induces angiogenesis and proliferation [3b, 4]. Therefore, harnessing macrophage polarity presents a unique opportunity to control inflammation, prevent rejection and accelerate integration of biomaterials and medical devices.
Macrophage polarization is most commonly controlled via exposure to biochemical factors. Specifically, the M1 macrophage phenotype is typically induced through interferon gamma (IFNγ) or lipopolysaccharide (LPS) stimulation, while the M2 macrophage phenotype is typically induced through Interleukin-4 (IL-4) or Interleukin-13 (IL-13) stimulation [5]. However, these approaches are not easily translated into in vivo approaches due to the challenge of delivery, temporal nature of the stimulus and risk of adverse effects.
Recent advances in biomaterials science have identified that a biomaterial’s design can be leveraged to instruct the host’s immune system [6]. For example, novel biomaterial surfaces [7], improved immune-instructive biomaterials [8] and incorporated immune modulating cells (e.g. stem cells) [9] could influence the wound healing process. Within the same context, there are several studies in which the phenotype of macrophages inside different 3D biomaterial environment has been studied [10]. In addition, several physical factors have recently been reported to regulate macrophage polarization [11]. These factors include biomaterial pore size [12], mechanical stimulation [13] and extracellular matrix proteins (ECM) [14] amonst others. However, the underlying mechanisms of how biomaterials steer macrophage polarity has remained poorly understood. This has obscured the underlying biomaterial design principles, which has limited our capability to engineer smart biomaterials with the ability to steer macrophage polarity.
Cell adhesion is an essential communication between cells and their microenvironment. This form of interaction is known to regulate numerous aspects of cellular behavior including migration, proliferation, morphogenesis and differentiation [15]. It is well established that surface structure, pore size and ECM influence a cell’s ability to adhere to substrates through e.g. integrin interactions [16]. Indeed, integrins are well-known to influence inflammation [17] and fibrosis [18]. However, their role in monocyte to macrophage differentiation and particularly macrophage polarization is yet to be fully understood. Here, we hypothesized that integrin mediated cell-biomaterial interactions could play a key role in macrophage polarization. Given that in vivo these events take place in the context of ECM and in 3D, obtaining a clear understanding of the role of integrins in macrophage polarization in a 3D microenvironment will be more physilogically relevant than in a 2D environment [19]. Therefore, in this study, we have used two distinct hydrogel systems to probe the effect of cell-biomaterial interactions on macrophage polarization in a 3D environment. Specifically, we have set-out to investigate whether macrophage polarity can be controlled through integrin mediated biomaterial-based programming.
2. Results and Discussion
2.1 Characterization of two distinct monocyte laden 3D hydrogels
The effect of 3D biomaterial environment on monocyte behavior has remained largely unknown. We therefore explored the behavior of human monocytic THP-1 cells encapsulation in two distinct hydrogel systems, namely gelatin methacryloyl (GelMA) and poly (ethylene glycol) diacrylate (PEGDA).
First, we investigated the microenvironment as presented by GelMA by varying the polymer concentrations from 5 to 15% (w/v). Increasing the GelMA concentration from 5 to 15% (w/v) increased the hydrogel’s crosslinking density and compressive modulus due to an inverse relation between the GelMA concentration and its porosity, as previously described by our group [20]. In particular, the compressive modulus increased from 2.96 ± 0.28 kPa to 25.82 ± 1.50 kPa (Fig. S1). Upon encapsulation, THP-1 cells in the softer 5% GelMA hydrogels demonstrated significantly higher levels of cell survival (Fig. S2 A and B) as well as increased metabolic activities compared to those encapsulated in the stiffer 10% and 15% GelMA hydrogels (Fig. S3). In addition, the swelling characteristics of a hydrogel are important in its biocompatibility through affecting various parameters including mass transport and mechanical properties[21]. We performed a swelling test for GelMA hydrogels at various concentrations. The 5% GelMA hydrogels had significantly higher levels of swelling ratio compared to the 10% and 15% GelMA hydrogels. The low cell viability and metabolic activities of THP-1 cells within both 10 and 15% (w/v) GelMA can be explained by increased stiffness values and reduced swelling ratio when using relatively high (w/v) amounts of GelMA. These results underlined THP-1 cells’ sensitivity to their microenvironment’s physical properties (e.g. stiffness and mass transport etc) which could in turn impact the biological processes (Fig. S4). In line with our finding, it has previously been reported that the stiffness of 2D hydrogels correlates with the number of monocytes that are differentiated into macrophages [22]. Based on our results, all subsequent experiments were performed with 5% (w/v) GelMA.
Monocytes can be chemically driven into a regenerative M2 macrophage phenotype through exposure to IL-4 [23]. Analysis of release kinetics demonstrated that GelMA acted as a proper cytokine reservoir with a sustained release of IL-4 over 7 days (Fig. S5). Even though macrophages are highly plastic and can show phenotypic changes within hours; the differentiation or induction in in vitro conditions is typically done for 6–8 days. Future studies could focus on better understanding of the kinetics of the observed phenotypical changes [24]. Thus the IL-4 release profiles from the hydrogel provides an ample time window to present monocytes with a microenvironment that is conducive to macrophage polarization.
Although GelMA appeared highly suitable to study macrophage polarization within a 3D environment, we simultaneously explored a distinct second hydrogel system (10% (w/v) PEGDA) to exclude possible biomaterial-based bias. To maximize the similarities between the two hydrogel systems, we matched the compressive modulus of 10% (w/v) PEGDA to that of 5% GelMA (Fig. S6) and minimized the differences in IL-4 release (Fig. S7). However, the two hydrogel systems remained inherently distinct in that unlike PEGDA, GelMA contains cell attachment sites.
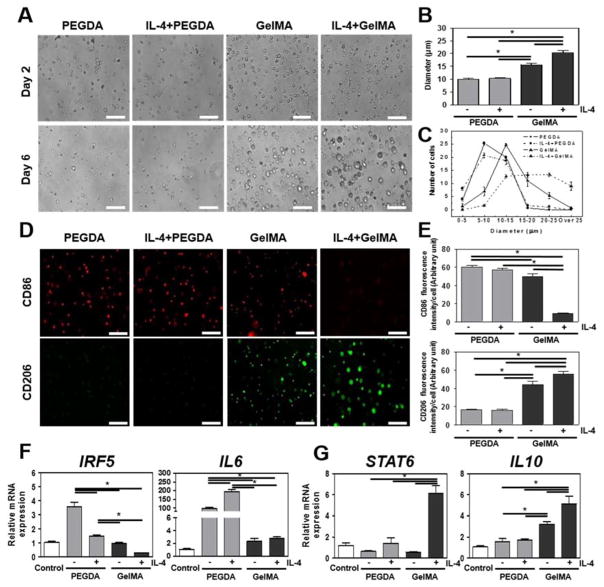
THP-1 cells were used as a surrogate for human monocytes to ensure reproducibility of our findings by eliminating potential donor-to-donor variation. Monocytic cells were encapsulated in PEGDA and GelMA hydrogels with or without IL-4 incorporation to examine their morphological phenotype over 6 days. Intriguingly, the size of the THP-1 cellsin GelMA hydrogels became progressively larger (Fig. 1A). Supplementation of IL-4 to GelMA hydrogels further exacerbated this cell enlargement. In contrast, the size of the THP-1 cellsin PEGDA hydrogels remained the same under all conditions and time points. Quantitative analysis of the cell diameters confirmed that cells within GelMA (15.58 ± 0.63 μm) and IL-4 incorporated GelMA (20.31 ± 0.89 μm) hydrogels became significantly larger than cells within PEGDA (9.99 ± 0.34 μm) and IL-4 incorporated PEGDA (10.41 ± 0.35 μm)) after 6 days of culture (Fig. 1B). Cell size distribution analysis revealed that while GelMA hydrogels created a shift in the general cell size population, IL-4 yielded an additional enrichment of the largest cell fraction (Fig. 1C). Taken together, GelMA hydrogel’s bioactive microenvironment influenced the THP-1 cells’ shape, whereas the bio-inert microenvironment of PEGDA did not. Thus, hydrogel composition has been found to play a significant role on the size of the encapsulated THP-1 cells. Even though no previous study has definitively demonstrated the effect of monocyte size on macrophage polarity, it is well documented that cell size and cell shape [25] are important determinants of cellular events including differentiation, function, proliferation, and apoptosis [26].
Figure 1. Characterization of 3D encapsulated human monocytic THP-1 cells behavior in IL-4 incorporated PEGDA and GelMA hydrogels for 6 days.
(A) THP-1 cells morphology in IL-4 cytokine incorporated PEGDA and GelMA hydrogels. (B) Quantitation of cellsdiameter in IL-4 cytokine incorporated PEGDA or GelMA hydrogel. (C) Distribution of cell diameter in IL-4 cytokine incorporated PEGDA and GelMA hydrogels. (D) Micrographs of cellsin hydrogel constructs stained for M1 surface marker CD86 (red) and M2 surface marker CD206 (green). Scale bar represents 100 μm. (E) Quantitative analysis of micrographs. (F) Real-time PCR of M1 related IRF5 and IL6 and (G) M2 related STAT6 and IL10. Scale bar represents 100 μm. The data are shown as the mean ± SEM (*, p < 0.05).
2.2 Hydrogel composition strongly affects expression of M1-M2 macrophage surface markers
Next, we set out to determine whether the observed differences in cell size between hydrogel systems correlated with a change in macrophage polarity. To this end, the expression of M1-M2 macrophage surface markers on human monocytic THP-1 cell cultured in PEGDA or GelMA hydrogels were visualized in the presence or absence of IL-4, the M2 macrophage inducing agent (Fig. 1D). The well-established surface biomarkers CD86 and CD206 were chosen to identify M1 and M2 macrophages, respectively [27]. Semi-quantitative image analysis demonstrated that after 6 days THP-1 cells in GelMA expressed a notably high level of CD206 as well as less intense yet detectable levels of CD86 (Fig. 1E). As expected, incorporation of IL-4 in GelMA hydrogels drove the induction of M2 macrophages as evidenced by increasing CD206 and decreasing CD86 expression. In sharp contrast, THP-1 cellsin PEGDA hydrogels expressed high levels of CD86 whereas the CD206 levels were undetectable. Surprisingly, incorporation of IL-4 in PEGDA did not alter the expression of either CD86 or CD206.
To further confirm the observed effects of PEGDA and GelMA hydrogels on macrophage polarity, we fingerprinted the THP-1 cells monocytes by profiling their M1-M2 gene expression. Indeed, encapsulation of THP-1 cells in PEGDA resulted in increased expression of the genes encoding M1 inducing transcription factor IRF5 and M1 related cytokine IL6, whereas mRNA levels of IRF5 and IL6 remained unaffected when cells were encapsulated in GelMA hydrogels (Fig. 1F). Furthermore, human monocytic THP-1 cells encapsulated in GelMA hydrogels demonstrated increased gene expression of M2 related cytokine IL10 expression (Fig. 1G) [28]. Similar to our CD86 observations, the incorporation of IL-4 in PEGDA hydrogels was unable to significantly increase IL10 and M2 inducing transcription factor STAT6 expression levels, whereas IL-4 incorporated GelMA hydrogels strongly increased their gene expression. Collectively, these results supported the stipulation that a biomaterial’s composition can prime monocytes towards either a M1 or M2 phenotype.
By extension, it might therefore be possible to program the immune system to either pro or anti-inflammatory responses purely based on the design of a biomaterial. In particular, IL-4 incorporated GelMA hydrogels induce regenerative M2 macrophages, whereas IL-4 incorporated PEGDA hydrogels induced pro-inflammatory M1 macrophages. PEGDA hydrogel’s M1 phenotype inducing effect is further underlined by its ability to block IL-4’s capacity to induce M2 macrophages. This is in line with recent studies showing that bio-inert PEGDA hydrogels can elicit strong immune response [29]. This observation is of interest as PEG is typically used for its immune-shielding properties, which is based on its mesh size and bio-inert nature [30]. Regardless, recent studies are in line with our observation that the bio-inert PEGDA can elicit a strong immune response [29]. Consequently, by understanding how biomaterials affect macrophage polarity, we might improve our capability to design biomaterials with improved immunomodulatory properties.
2.3 Hydrogel composition controls macrophage’s functional properties
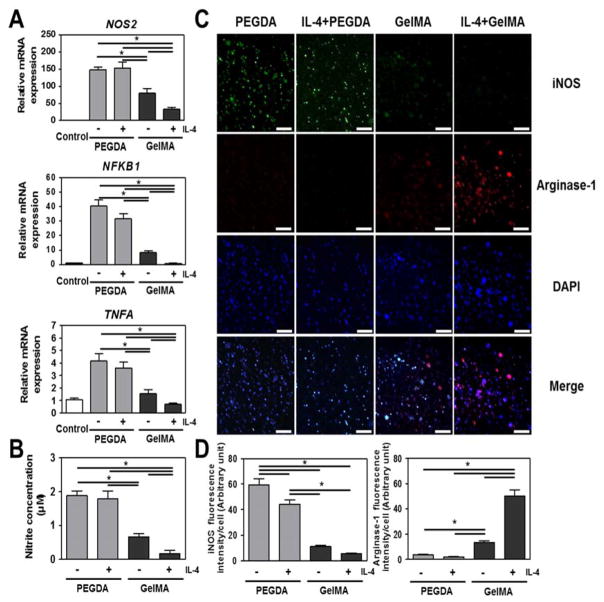
We then set out to confirm that the PEGDA hydrogel indeed robustly drives THP-1 cells into a functional M1 macrophage phenotype. To this end, we determined the expression of NOS2, NFKB and TNFA (Fig. 2A) as well as nitrite production (Fig. 2B), which are all indicative of M1 functionality. Human monocytic THP-1 cells encapsulated in PEGDA hydrogels demonstrated higher expression levels for all four pro-inflammatory markers compared to cellss encapsulated in GelMA hydrogels. Unexpectedly, GelMA in the absence of IL-4 also guided pro-inflammatory responses, as noted by increased NOS2 and NFKB expressions compared to the control group. Nevertheless, IL-4 stimulation decreased the levels of all markers even further in GelMA hydrogels. However, the expression levels of NOS2, NFKB, TNFA, and nitrite from THP-1 cellsin PEGDA hydrogels remained largely unaffected after exposure of THP-1 cells to IL-4. In line with these observations, immunohistochemistry revealed that cells encapsulated in PEGDA hydrogels demonstrated intense positive staining for M1 marker iNOS [28d, 31] and negative for M2 marker Arginase-1 [28d, 32], whereas THP-1 cells in GelMA demonstrated the exact opposite staining pattern (Fig. 2C and 2D). In addition, both PEGDA and GelMA hydrogels appeared to generate pro-inflammatory response even in the absence of a polarizing stimuli, with PEGDA having a stronger effect. Although the expression and role of iNOS and Arginase-1 are better defined in mice models; recent work have also reported their expression and function in human macrophages [31, 33].
Figure 2. In vitro immune response of human monocytic THP-1 cells encapsulated in PEGDA and GelMA hydrogels in the absence or presence of IL-4 for 6 days.
(A) Real-time PCR of inflammatory genes. (B) Biochemical quantification of nitrite production. (C) Micrographs of THP-1 cells fluorescently stained for M1 related iNOS, M2 related Arginase-1 and DAPI. (D) Quantitative analysis of micrographs. Scale bar represents 100 μm. The data are shown as the mean ± SEM (*, p < 0.05).
Our results demonstrate the potency of cell-biomaterial interactions to program monocytes into a specific polarized macrophage phenotype. Paradoxically, the data also suggests that bio-inert materials such as PEGDA hydrogels can elicit a pro-inflammatory immune response [29]. However, these pro-inflammatory responses induced in GelMA in the absence of IL-4 stimulation would not be able to explain the mechanism, clearly. Moreover, biomaterial composition can even prevent monocytes from responding to cytokines such as IL-4 that would drive implant integration and tissue regeneration. The mechanism by which biomaterials can drive monocytes into such distinct macrophage phenotypes has remained largely unknown. However, we reasoned that our approach of simultaneously using two comparable (e.g. in terms of their mechanical properties and IL-4 release profiles) yet distinct hydrogel systems might yield valuable information on how biomaterials composition can control macrophage polarization.
2.4 Hydrogel composition determines macrophage cytokine releaseprofile
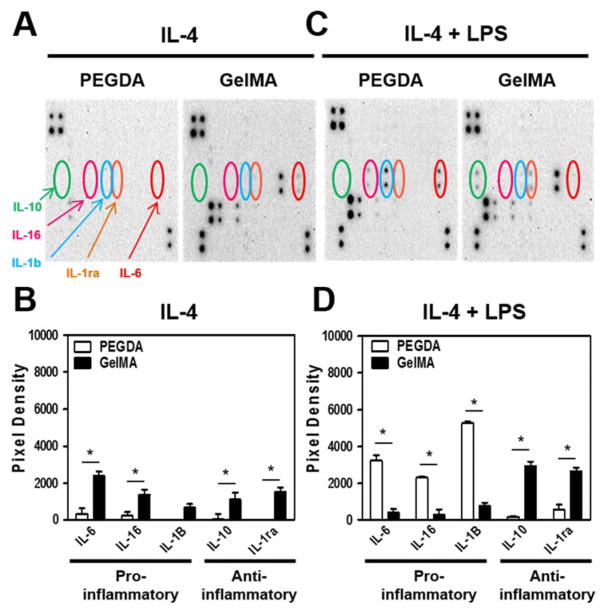
To confirm the M1 and M2 fingerprints, we additionally visualized and semi-quantified the M1-M2 macrophage specific cytokine release profiles from human monocytic THP-1 cells encapsulated in either PEGDA or GelMA hydrogels in the presence of IL-4. In particular, the secretion of pro-inflammatory cytokines IL-1B, IL-6, and IL-16 as well as the anti-inflammatory cytokines IL-1RA and IL-10 were investigated (Fig. 3A and 3B). THP-1 cells encapsulated in GelMA demonstrated significantly higher levels of all tested anti-inflammatory cytokines as compared to those encapsulated in PEGDA. Interestingly, no LPS stimulated THP-1 cells encapsulated in GelMA also demonstrated a modest trend of higher expression levels of pro-inflammatory cytokines as compared to the M1 inducing PEGDA.
Figure 3. Pro- and anti-inflammatory cytokine release of human monocytic THP-1 cells in IL-4 incorporated PEGDA and GelMA hydrogels for 6 days.
(A and C) Cytokine release assay and its (B and D) Quantitative analysis of THP-1 cellsexposed to both M1 inducing lipopolysaccharides (LPS) and M2 macrophage inducing IL-4 in PEGDA and GelMA hydrogels. The data are shown as the mean ± SEM (*, p < 0.05).
It is of note that these experiments were performed in the presence of M2 inducing IL-4, but in the absence of an M1 inducing factor. However, wound and implant sites naturally contain a combination of stimuli that favor both M1 and M2 polarization at different stages of healing. We therefore encapsulated THP-1 cells in PEGDA or GelMA hydrogels and exposed them to both LPS and IL-4, which drove M1 and M2 polarization, respectively. Remarkably, under these more biologically complex conditions, we observed that THP-1 cells in PEGDA hydrogels expressed significantly higher levels of all M1 related cytokines, whereas those in GelMA hydrogels expressed significantly higher levels of all M2 related cytokines (Fig. 3C and 3D). In accordance with previous studies, although in 2D culture, the secretion of the pro-inflammatory molecules IL-1β and TNF-α increased dramatically following human primary monocyte interactions with PEG-only hydrogel films as compared with tissue culture polystyrene (TCPS) [34]. In fact, supplementation of the M1 inducing LPS to THP-1 cells in GelMA hydrogels further increased the expression levels of M2 related cytokines while decreasing those of M1 related cytokines, as compared to IL-4 alone. In essence, these results demonstrated that LPS simulation did not revert the M2 polarization of macrophages in GelMA in the presence of IL-4 and unexpectedly enhanced the polarization.
2.5 Hydrogel composition affects on the THP-1 cells’ integrin expression and cytoskeletal organization
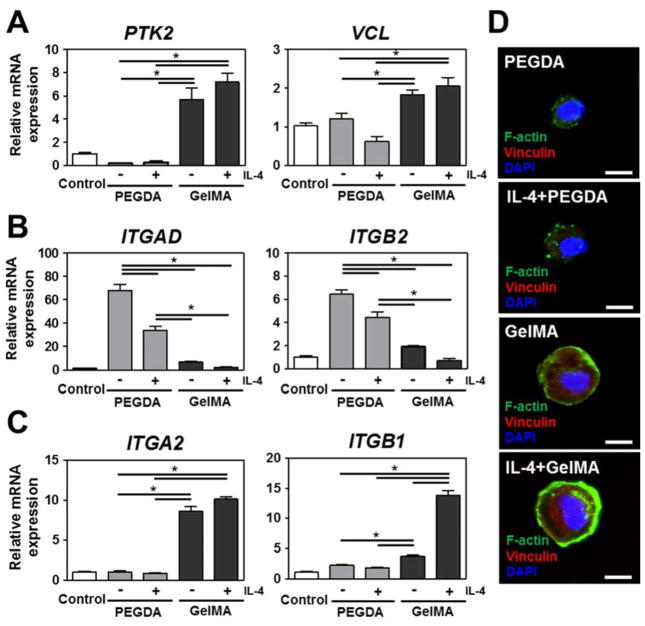
A key difference between the hydrogels used in this study is the presence or absence of cell-adhesive motifs. Specifically, GelMA hydrogels contain cell adhesive sequences while PEGDA hydrogels do not. We therefore hypothesized that the difference in macrophage phenotype between the two hydrogel systems might be regulated through attachment and downstream signaling, which is mediated via integrin subunits and focal adhesions. To this end, we determined the relative gene expression levels of Focal Adhesion Kinase (PTK2), Vinculin (VCL), monocyte-related integrin receptors Integrin αD (ITGAD) and β2 (ITGB2), and collagen matrix-related integrin receptors Integrin α2 (ITGA2) and β1 (ITGB1) in cells encapsulated in PEGDA and GelMA hydrogels with or without IL-4. PEGDA hydrogel’s inability to provide binding sites was mirrored by the strong decrease in PTK2 expression levels compared to GelMA hydrogels and tissue culture plastic grown control samples. In contrast, GelMA hydrogels induced an increase in monocytes’ expression of PTK2 and VCL (Fig. 4A). Low levels of integrin αD and β2 – alternatively known as CD11d and CD18 – are correlated with monocyte migration, whereas high levels of these integrins are associated with pro-inflammatory macrophages [35]. In line with this, monocytes encapsulated in PEGDA hydrogels showed significantly high levels of ITGAD and ITGB2 expressions, whereas the expressions of these integrin proteins were found to be low when encapsulated in GelMA hydrogels (Fig. 4B). Such an increase in upregulation of integrin αDβ2 on monocytes/macrophages might be due to the stimulation of pro-inflammatory responses resulting from the IL-4 presence with lesser effects arising from the absence of available binding sites in PEGDA hydrogels. Migration and collagen attachment is mediated via integrins such as α2 and β1, which are also known as CD49b and CD29 [36]. The expression of ITGA2 and ITGB1 were drastically upregulated in GelMA hydrogels but remained unchanged in PEGDA hydrogels (Fig. 4C). These results demonstrated that the composition of the biomaterial effectively determines human monocytic THP-1 cells’ expression of adhesion molecules. In addition to facilitating cell attachment, integrins also play an important and versatile role in multiple signaling pathways. Therefore, biomaterials can program the cellular response to a given microenvironment, which includes their response to e.g. macrophage polarizing cytokines.
Figure 4. Changes in cytoskeletal organization and focal adhesion molecules in IL-4 cytokine incorporated PEGDA and GelMA hydrogels for 6 days.
(A) Real-time PCR of protein tyrosine kinase 2 (PTK2) and Vinculin (VCL). (B) Real-time PCR of monocyte related integrin receptors (Integrin αD (ITGAD) and Integrin β2(ITGB2)). (C) Real-time PCR of collagen matrix related integrin receptors (Integrin α2 (ITGA2) and Integrin β1 (ITGB1)). (D) Micrographs of F-Actin and Vinculin of THP-1 cells encapsulated in IL-4 cytokine incorporated PEGDA and GelMA hydrogels. Scale bar represents 10 μm. The data are shown as the mean ± SEM (*, p < 0.05).
To investigate whether the changes in integrin expression translated to cytoskeletal changes, we measured the expression and distribution of vinculin and filamentous actin (F-Actin) using immunocytochemistry. Vinculin is involved in the linkage of integrin adhesion molecules to the actin cytoskeleton. Therefore, Vinculin and F-Actin are both sensitive and responsive to biological and mechanical stimuli mediated via integrin-based cell attachment [37]. THP-1 cellsencapsulated in GelMA hydrogels showed intense staining for vinculin, whereas the staining of those encapsulated in PEGDA hydrogels were consistently below the detection limit (Fig. 4D). The latter result might be explained by the diminished biomechanical stimulation due to the lack of cell binding sites in PEGDA hydrogels. Furthermore, THP-1 cellsin PEGDA hydrogels demonstrated clump-like cytoplasmic aggregates of F-Actin, which has been reported to correlate with monocyte-to-macrophage transition as well as the migration and function of various immune cells [38]. F-Actin staining of THP-1 cells in GelMA hydrogels revealed the presence of a prominent cortical shell, which has been reported to be indicative of M2 commitment [39]. Together, these results demonstrated that biomaterial induced changes in integrin expression effectively translated to – or tightly correlated with – marked changes in cell behavior and cytoskeletal organization.
2.6 Integrin α2β1 mediated binding of THP-1 cells to biomaterials is required for M2 macrophage polarization
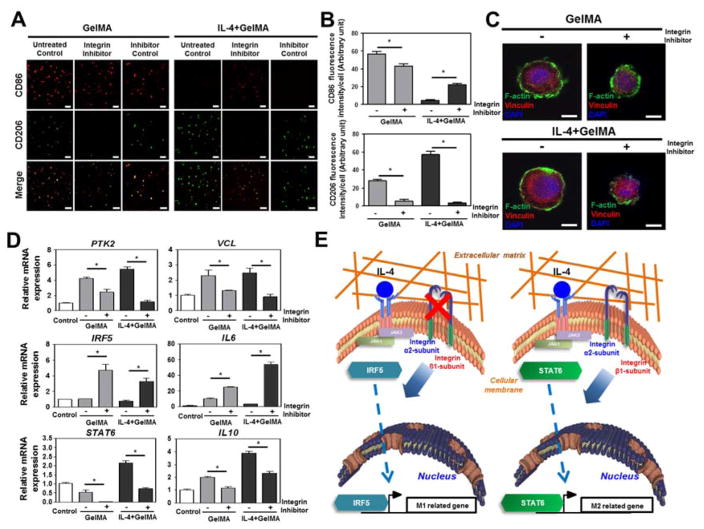
Based on the significant changes in integrin expression and subsequent cytoskeletal reorganization, we hypothesized that integrins might potently affect downstream signaling pathways and thereby control macrophage polarity. To this end, THP-1 cellsencapsulated in GelMA hydrogels were exposed to a neutralizing antibody for integrin α2β1 in the presence or absence of the M2 inducing cytokine IL-4. Similar to our previous experiments, THP-1 cells in GelMA hydrogels expressed high levels of M1-related CD86 and low levels of M2-related CD206, and IL-4 supplementation further decreased CD86 to undetectable levels and significantly increased CD206 expression. Remarkably, blocking integrin α2β1 strongly increased CD86 expression and reduced CD206 expression below the detection limit, even in the presence of IL-4 (Fig. 5A and 5B). THP-1 cells encapsulated in GelMA hydrogels that were exposed to integrin α2β1 blocker therefore mirrored the behavior of human monocytic THP-1 cells encapsulated in PEGDA. Addition of non-inhibitory isotype control antibody was undistinguishable from the untreated control group. This suggested that the integrin α2β1 induced polarization through its integrin α2β1 binding domain rather than its conserved antibody domain. Furthermore, immunohistochemical staining revealed that blocking integrin α2β1 lowered the expression of F-actin and Vinculin while reducing the cell size therefore more closely resembling THP-1 cells encapsulated in PEGDA hydrogels. Moreover, these results suggested the effective blocking of integrin α2β1 mediated binding and thus limiting cell-biomaterial interactions (Fig. 5C). Moreover, this blockage mitigated the high levels of cytoskeletal related PTK2 and VCL expression in GelMA hydrogels, strongly increased expression of M1-related IRF5 and IL6, and decreased expression of M2-related STAT6 and IL10 (Fig. 5D). In summary, integrin α2β1 appeared to play a pivotal role in macrophage polarization (Fig. 5E). In particular, IL-4 incorporated GelMA hydrogel biomaterials were observed to drive monocytes into the M2 macrophage phenotype through integrin α2β1 attachment, most likely via STAT6 activation. The inability of monocytes to attach to biomaterials via integrin α2β1 – either through pharmacological blockage or absence of available binding sites – results in the generation of M1 macrophages, most likely through IRF5 activation. This might also suggest that M1 is potentially a default phenotype.
Figure 5. The effect of integrin α2β1 inhibition on macrophage polarity in IL-4 cytokine incorporated GelMA hydrogel.
(A) Confocal images of M1 and M2 macrophage surface markers CD86 and CD206 with or without integrin α2β1 receptor inhibitor or non-inhibitory isotype control antibody. Scale bar represents 100 μm. (B) Quantitative analysis of micrographs. (C) Confocal images of focal adhesion molecule expression with or without integrin α2β1 receptor inhibitor. Scale bar represents 10 μm. (D) mRNA expression levels of M1 and M2 macrophage markers and focal adhesion molecules in IL-4 incorporated GelMA hydrogel with or without integrin α2β1 receptor inhibitor. (E) Proposed mechanism via which biomaterials are able to prime human monocytic THP-1 cells into either a M1 or M2 macrophage phenotype. The data are shown as the mean ± SEM (*, p < 0.05).
To confirm that the observation of integrin α2β1-mediated macrophage polarization was not restricted to the THP-1 monocytic cell line, we also seeded human primary monocytes on plates coated with integrin α2β1 peptide (the type I collagen α1(I)-CB3 fragment Asp-Gly-Glu-Ala or DGEA) and investigated their polarization status after 6 days (Fig. S8 A and B). The cells cultured on DGEA-coated plates expressed higher levels of the M2 marker CD206, with no noticeable changes in the level of M1 marker expressions (in this case Calprotectin [5]), compared to BSA control group (Fig. S7 A) indicating a shift towards M2 phenotype in cells cultured on DGEA coated surfaces. Notably, cells cultured on DGEA-coated plates also demonstrated significantly higher levels of the anti-inflammatory cytokine, IL-10, compared to the BSA control groups (Fig. S7 B) which again is an indication of a shift towards an M2 phenotype[40]. This data also showed an increase in IL-6 production by cells on the DGEA-coated plates. While IL-6 is typically considered as a pro-inflammatry cytokine, recent in vivo data has clearly shown that it also enhances the polarization of altenatively activated macrophages (i.e. M2 phenotype) [41]. Collectively, this data indicates that simple DGEA motives (which is recognized by integrin α2β1) induced a partial shift of human primary monocytes towards the M2 macrophage phenotype and created a cytokine environment that promoted M2 macrophages polarization, even in a 2D culture environment [41].
By extension, this conclusion stipulates that poly(ethylene glycol) (PEG)-based hydrogels are bio-inert [42] and do not present integrin binding sites and are prone to cause M1 macrophage polarization. Further studies with other bio-inert polymers can elucidate whether this conclusion can be generalized. Several such bio-inert biomaterials are often viewed as immunoprotective or immune-shielding. However, the inability of these materials to interact with immune cells (e.g., macrophages) in a direct manner can potentially drive the immune response in a deleterious direction. Indeed, these materials are more prone to deleterious immune responses and result in relatively intense fibrous capsule formation upon implantation. This foreign body response is at least in part generated via M1 macrophages, which as resported in this work are generated when monocytes are unable to interact with the implant via integrins. Although the concept that the absence of a cell-biomaterial interaction can potently determine cell fates is novel, there is a well-established precedence within the domain of cell-cell interactions. In particular, our observations are reminiscent of the immunological failsafe that safeguards major histocompatibility complex (MHC) function. In this system, the immune system triggers programmed cell death of cells that present unfavorable antigens via their MHC [43]. Intuitively, it might therefore be expected that the absence of MHC expression might provide a degree of immunoprotection [44]. However, the immune system also triggers programmed cell death in cells that do not allow this interaction. Similarly, the immune system might attempt to destroy or isolate (via fibrous capsule formation) implants. In our study, we revealed that biomaterials designed to evade the immune system recognition by not presenting any integrin recognition motifs induce a pro-inflammatory M1 macrophage phenotype. It might therefore be reasoned that our immune system has evolved multiple distinct safeguards, which by default elicit a deleterious immune response when unable to interact with its target, be it a cell or biomaterial. In other words, the presence of encapsulated macrophages, acquiring an M2 phenotype, in vivo can trigger a faster observation of the initial inflammation by facilitating the conversion of M1 macrophages to M2 macrophages. However, the complex nature of the in vivo implant microenvironment and the presence of other immune cells (such as T-cells) must be also taken into account and the means for long term cytokine induction (via controlled delivery systems) must be considered for potent in vivo effects.
As such, biomaterial-based implants can be made truly immune-compatible by including motifs that prime the immune system to drive integration rather than rejection. Incorporating cell attachment sites into the design of biomaterials will thus not only stimulate the encapsulated therapeutic cells, but also steer the host’s immune system towards a healing response. However, the amount of the inflammation must also be controlled; otherwise long-term presence of M2 macrophages can result in fibrosis [45]. Indeed, recent studies have indicated that this approach is both feasible and practical; coating bio-inert biomaterials with decellularized ECM decreased M1 macrophage induction and the subsequent chronic inflammatory response [46]. Although our data indicated that integrin α2β1 played an essential role in M2 macrophage induction, other integrins could potentially affect macrophage proliferation distinctly as they interact with different ligands [47]. Indeed, decorating biomaterials with the integrin binding tripeptide Arg-Gly-Asp (RGD) has been reported to induce fibrosis [17b, 18c]. Systematic analysis of the effects of integrins, and other adhesive moieties, on macrophage polarity could resolve such contradictions and provide design principles on how to create next-generation biomaterials that controllably induce M1 or M2 macrophage polarization.
In summary, incorporating cell binding domains into biomaterials to facilitate integrin interactions in 3D microenvironment, such as those with α2β1, to steer the host’s immune system towards a natural healing response represents an exciting and novel opportunity to control and improve the clinical outcomes of biomaterial-based implants and cell therapies.
3. Conclusion
Biomaterial-mediated immunomodulation by programming macrophage polarity is a promising tool in tissue engineering, regenerative medicine, and implantology to decrease adverse immune reactions, accelerate implant integration, facilitate tissue regeneration, and increase implant lifetime. Here, we present a mechanism and 3D biomaterial-based approach for immunomodulation, which controls the balance between inducing pro-inflammatory M1 macrophages and regenerative M2 macrophages. We have shown the ability of 3D encapsulated human monocytic THP-1 cells to interact with IL-4 stimulation and that the biomaterials through integrin α2β1 has a direct effect on macrophage phenotype by inducing M2 macrophages, whereas blocking this mechanism induces M1 macrophages. We have thereby further uncovered the essential and pivotal role of integrins in the immune response. Overall, biomaterial-based control over macrophages represents a novel technique to obtain a fundamental understanding of macrophage behavior and is a strong therapeutic tool for immunomodulation for implants, drug and cell delivery systems.
4. Experimental section
4.1 Biomaterials
Polyethylene glycol diacrylate (PEGDA), gelatin (Type A, 300 bloom from porcine skin), and methacrylic anhydride (MA) were purchased from Sigma-Aldrich (Wisconsin, USA). Gelatin methacryloyl (GelMA) was synthesized as described previously [48]. Hydrogels were crosslinked using an UV source (Omnicure S2000, Ontario, Canada).
4.2 Fabrication of hydrogel constructs
Freeze dried prepolymer solutions were mixed in DPBS with 2-hydroxy-1-(4-(hydroxyethoxy)phenyl)-2-methyl-1-propanone (Irgacure 2959, CIBA Chemicals) and placed at 80 °C until fully dissolved. Recombinant human IL-4 (R&D Systems) was added when desired at a final concentration of 10 ng/mL. Human monocytic THP-1 cells were encapsulated in the prepolymer at a final concentration of 6 × 106 cells per mL. Prepolymer samples were divided in 20 μL samples and photocrosslinked using 800 mW/cm2 UV light (Omnicure S2000, EXFO Photonic Solutions Inc.) for 10 or 40 seconds for GelMA and PEGDA hydrogels, respectively. Constructs were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Gibco BRL) supplemented with 10% (v/v) fetal bovine serum (Gibco BRL) and 100 units/mL penicillin (Gibco BRL) in an incubator with 5% (v/v) CO2 at 37 °C. Medium was refreshed every two days.
4.3 Compressive modulus
Two hundred microliters were formed and allowed to incubate at 37 °C in DPBS for 24 hours. Hydrogels were tested at a rate of 20% strain/min on an Instron 5542 mechanical tester. The compressive modulus was determined as the slope of the linear region corresponding with 0–5% strain.
4.4 IL-4 release kinetics
Hydrogels were submerged in 200 μL DPBS and incubated at 37 °C without agitation for up to 7 days. At predetermined time points, the supernatant was retrieved and replaced with fresh buffer. The IL-4 concentration in the supernatant was quantified using a human IL-4 ELISA Kit (Quantikine® ELISA, R&D Systems).
4.5 Viability and proliferation of human monocytic THP-1 cells
24 hours post-encapsulation, live and dead cells were visualized using calcein-AM and ethidium homodimer (Invitrogen), photographed using an inverted fluorescence microscope (Nikon TE 2000-U), and quantified using NIH ImageJ software. Cell proliferation was measured up to 7 days of culture using Alamar Blue assay (Invitrogen).
4.6 Hydrogel swelling analysis
Polymerization was performed as described in section 4.2. Immediately following the hydrogel formation, discs (8mm in radius) of each composition was punched from a flat thin sheet and placed in DPBS at 37 °C for 24 hours. Discs were removed from DPBS and blotted with a KimWipe to remove the residual liquid and the swollen weight was recorded. Samples were then lyophilized and weighed once more to determine the dry weight of the polymer. The mass swelling ratio was then calculated as the ratio of swollen hydrogel mass to the mass of dry polymer.
4.7 Immunohistochemical staining
Samples were fixed in 4% (v/v) formalin and permeabilized with 0.1% (v/v) Triton X-100 for 30 minutes. Molecules of interest were labeled using a primary antibody such as Anti-CD86 antibody (Abcam), anti-calprotectin antibody (Thermo Scientific), anti-mannose receptor antibody (CD206, Abcam), anti-vinculin antibody (Sigma), anti-NOS2 antibody (Santa Cruz Biotechnology), and anti-Arginase-1 antibody (Santa Cruz Biotechnology). Target molecules were then visualized using Alexa 488 or Alexa 594 conjugated secondary antibodies (Molecular Probes). F-Actin was visualized using rhodamine-phalloidin (Invitrogen). Samples were counterstained with 4, 6-diamidino-2-phenylindole (DAPI, Vector Laboratories Inc.) and photographed using either a fluorescence microscope (IX71 inverted microscope, Olympus) or an inverted laser scanning confocal microscope (SP5 X MP, Leica Microsystems). Signal intensities of CD86, CD206, iNOS, and arginase-1 of individual cells were quantitatively analyzed using NIH ImageJ software.
4.8 Quantitative real-time PCR
Total RNA was extracted using TRIzol (Invitrogen), quantified using a Nanodrop2000 (Thermo Fisher Scientific), and synthesized into cDNA synthesis using SuperScript™ III First-Strand Synthesis SuperMix (Invitrogen). For quantitative real-time PCR analysis, 20 ng of input RNA was amplified in an IQ5 detection system (Biorad) using SYBR Green Master Mix (Bio-Rad) and 500 nmol/L of gene-specific primers. All mRNA expression levels were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Primer sequences are listed in Supplementary table 1.
4.9 Cytokine expression profiling
Hydrogels embedded THP-1 cellswere cultured for 6 days after which fresh medium was allowed to be conditioned for 24 hours and laden onto a human cytokine antibody array (Human Cytokine Array C6, RayBiotech Inc), processed, and detected according to manufacturer’s protocol. Invariant set normalization was used to normalize the inter-array intensity (Image pro PLUS, Media Cybermetics Inc.).
4.10 Integrin receptor blocking
Human monocytic THP-1 cells were pretreated with 10 μg/mL Anti-Integrin alpha 2+beta 1 antibody (Abcam) or non-inhibitory isotype control antibody (Abcam) for 30 minutes at 37 °C, washed, encapsulated in GelMA hydrogels containing IL-4, and cultured for 6 days.
4.11 Nitrite production
After 6 days of culture, 50 μL of supernatant was retrieved and proceed according to manufacturer’s protocols (Griess reagent, Promega). The absorbance of the developed solution was measured at a wavelength of 550 nm using an ELISA Microplate Reader (VersaMax, Molecular Device).
4.12 Fabrication of α2β1 integrin ligand peptide Asp-Gly-Glu-Ala (DGEA) coated substrate
To prepare the DGEA coated coverslips, acid-etched glass coverslips were incubated with 0.1 mg/ml MAPTrix™-C-DGEA peptide (Amsbio) or bovine serum albumin (BSA) (used as a negative control) in 500 mM NaHCO3 solution for 1 hour at room temperature. Coverslips were washed with PBS and used immediately. Buffy coat samples were obtained from healthy volunteers in accordance with the relevant guidelines after obtaining informed written consent and approval of local ethics committee (all approved by the National Blood Service, UK). Human peripheral blood mononuclear cells (PBMCs) were obtained from heparinised blood by Histopaque-1077 density gradient centrifugation. Monocytes were isolated from PBMCs using the Miltenyi Biotec magnetic cell separation system (positive selection with CD14 MicroBeads and LS columns) as per the manufacturer’s instructions. This method yielded 95% pure monocytes as determined by flow cytometric analysis of CD14 expression. Purified monocytes were cultured at 5 × 105 cells/coverslip in RPMI-1640 supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin, 100 μg/mL streptomycin (referred to henceforth as “RPMI complete medium”) and 10 ng/ml Macrophage-Colony Stimulating Factor (M-CSF) (Miltenyi Biotec) in 24-well tissue culture-treated plates. Included controls were: M1 (50 ng/mL granulocyte-macrophage colony-stimulating factor (GM-CSF) (Miltenyi Biotec) + 20 ng/mL IFN-γ (R&D Systems)), M2 (50 ng/mL M-CSF + 20 ng/mL IL-4 (Miltenyi Biotec)), and M-CSF (10 ng/mL M-CSF). Samples were incubated at 37°C, 5% CO2 for 6 days, with fresh complete medium containing cytokines added on Day 3. On Day 6, samples were stained with 1 μg/mL rabbit anti-human mannose receptor antibody (Abcam) and 2 μg/mL mouse anti-human calprotectin antibody (Thermo Scientific) (M2 and M1 markers respectively [5]) diluted in 5% goat serum in PBS. Secondary antibody staining was then carried out using 8 μg/mL each of goat anti-rabbit Alexa Fluor® 488-conjugated antibody and goat anti-mouse Rhodamine Red-X-conjugated antibody (both from Thermo Fisher Scientific). Finally, samples were counterstained with 250 ng/mL DAPI and mounted on microscope slides. Images were captured using an IMSTAR automated fluorescence microscope. Image analysis and quantification was carried out using CellProfiler software v 2.1.1. Furthermore, supernatants were collected and assayed for the cytokines IL-6 and IL-10 using ELISA DuoSet kits (R&D Systems) as per the manufacturer’s instructions.
4.13 Statistical analysis
At least three independent sets of experiments for each condition were performed in triplicate. Data were pooled and statistically expressed as mean ± standard deviation (S.D). Two-way analysis of variance (ANOVA) was used for analysis of quantitative values, and Tukey’s post hoc test was used for all pair-wise comparisons among groups. Differences were considered significant at p < 0.05 and were indicated with an asterisk. The SPSS software package (version 12.0; SPSS Inc.) was used to perform statistical tests.
Supplementary Material
Supplemental table 1. Nucleotide sequences of the primer pairs used for qPCR in this study.
Figure S1. Compressive modulus of 5, 10, and 15% GelMA hydrogel. The data are shown as the mean ± SEM (*, p < 0.05).
Figure S2. (A) Human monocytic THP-1 cell viability in 5, 10, and 15% GelMA hydrogels 24 hours post-encapsulation. (B) Quantification of cell viability demonstrated excellent cell survival in 5% GelMA. Scale bar represents 100 μm. The data are shown as the mean ± SEM (*, p < 0.05).
Figure S3. THP-1 cellsmetabolic activity in 5, 10, and 15% GelMA hydrogels over a period of 7 days. The data are shown as the mean ± SEM (*, p < 0.05; compared to both 10 and 15% GelMA).
Figure S4. (A) Swelling properties of 5, 10, and 15% GelMA hydrogels over a period of 7 days. The swelling ratios of GelMA hydrogels at various GelMA % (w/v) show statistically significant differences. The data are shown as the mean ± SEM (*, p < 0.05; compared to both 10 and 15% GelMA, §, p < 0.05; compared to both 5 and 15% GelMA). (B) Swelling properties of 10% PEGDA and 5% GelMA hydrogels over a period of 5 days. The swelling ratios between PEGDA and GelMA hydrogels show no significant differences.
Figure S5. IL-4 cytokine release from 5, 10, and 15% GelMA hydrogels. The data are shown as the mean ± SEM (*, p < 0.05; compared to both 10 and 15% GelMA).
Figure S6. Compressive modulus of PEGDA and GelMA hydrogels with or without of incorporated IL-4.
Figure S7. IL-4 cytokine release profile when incorporated in PEGDA and GelMA hydrogels.
Figure S8. The effects of α2β1 integrin peptide on human primary monocytes‘ differentiation. (A) Micrographs of macrophages fluorescently stained for M1 marker Calprotectin, M2 marker CD206, and DAPI. Scale bar represents 200 μm. (B) IL-6 and IL-10 cytokine release profile as measured by ELISA. The data are shown as the mean ± SEM (*, p < 0.05).
Acknowledgments
The authors acknowledge funding from the National Science Foundation (EFRI-1240443), UK Engineering and Physical Sciences Research Council (EP/N006615/1), EU FP 7 IMMODGEL (602694), and the National Institutes of Health (EB012597, AR057837, DE021468, HL099073, R56AI105024). Dr. Cha was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (NRF-2016R1D1A1B03934876). Dr. Leijten acknowledges financial support from Innovative Research Incentives Scheme Veni #14328 of the Netherlands Organization for Scientific Research (NWO).
Footnotes
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Dr. Byung-Hyun Cha, Biomaterials Innovation Research Center, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139, USA, Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA. Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, MA 02115, USA.
Dr. Su Ryon Shin, Biomaterials Innovation Research Center, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139, USA, Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA. Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, MA 02115, USA.
Dr. Jeroen Leijten, Biomaterials Innovation Research Center, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139, USA, Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA. Department of Developmental BioEngineering, MIRA Institute for Biomedical Technology and Technical Medicine, University of Twente, Enschede, The Netherlands
Dr. Yi-Chen Li, Biomaterials Innovation Research Center, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139, USA, Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA. Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, MA 02115, USA
Dr. Sonali Singh, Biomaterials Innovation Research Center, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139, USA, Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA. Division of Immunology, School of Life Sciences, Faculty of Medicine and Health Sciences, University of Nottingham, Nottingham, UK
Dr. Julie C. Liu, Biomaterials Innovation Research Center, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139, USA, Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA. School of Chemical Engineering and Weldon School of Biomedical Engineering, Purdue University, West Lafayette, IN 47907, USA
Dr. Nasim Annabi, Biomaterials Innovation Research Center, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139, USA, Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA. Department of Chemical Engineering, Northeastern University, Boston, MA, 02115, USA
Dr. Reza Abdi, Biomaterials Innovation Research Center, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139, USA, Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA. Transplant Research Center, Renal Division, Brigham and Women’s Hospital and Children’s Hospital, Boston, MA 02115, USA
Dr. Mehmet R. Dokmeci, Biomaterials Innovation Research Center, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139, USA, Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA. Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, MA 02115, USA
Dr. Nihal Engin Vrana, Biomaterials Innovation Research Center, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139, USA, Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA. Protip Medical, 8 Place de l’Hôpital, 67000 Strasbourg, France, Institut National de la Sant’e et de la RechercheM’edicale (INSERM), UMR-S 1121, “Biomatériaux et Bioingénierie”, 11 rue Humann, 67085 Strasbourg Cedex, France
Dr. Amir M. Ghaemmaghami, Biomaterials Innovation Research Center, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139, USA, Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA. Division of Immunology, School of Life Sciences, Faculty of Medicine and Health Sciences, University of Nottingham, Nottingham, UK
Dr. Prof. Ali Khademhosseini, Biomaterials Innovation Research Center, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139, USA, Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA. Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, MA 02115, USA. Department of Bioindustrial Technologies, College of Animal Bioscience and Technology, Konkuk University, Seoul 143-701, Republic of Korea, Department of Physics, King Abdulaziz University, Jeddah 21569, Saudi Arabia
References
- 1.a) Gretzer C, Emanuelsson L, Liljensten E, Thomsen P. J Biomater Sci Polym Ed. 2006;17:669. doi: 10.1163/156856206777346340. [DOI] [PubMed] [Google Scholar]; b) Luttikhuizen DT, Harmsen MC, Van Luyn MJ. Tissue Eng. 2006;12:1955. doi: 10.1089/ten.2006.12.1955. [DOI] [PubMed] [Google Scholar]
- 2.a) Anderson JM, Rodriguez A, Chang DT. Semin Immunol. 2008;20:86. doi: 10.1016/j.smim.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Broughton G, 2nd, Janis JE, Attinger CE. Plast Reconstr Surg. 2006;117:12S. doi: 10.1097/01.prs.0000225430.42531.c2. [DOI] [PubMed] [Google Scholar]
- 3.a) Porcheray F, Viaud S, Rimaniol AC, Leone C, Samah B, Dereuddre-Bosquet N, Dormont D, Gras G. Clin Exp Immunol. 2005;142:481. doi: 10.1111/j.1365-2249.2005.02934.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. Trends Immunol. 2004;25:677. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 4.Yakubenko VP, Bhattacharjee A, Pluskota E, Cathcart MK. Circ Res. 2011;108:544. doi: 10.1161/CIRCRESAHA.110.231803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rostam HM, Singh S, Salazar F, Magennis P, Hook A, Singh T, Vrana NE, Alexander MR, Ghaemmaghami AM. Immunobiology. 2016;221:1237. doi: 10.1016/j.imbio.2016.06.010. [DOI] [PubMed] [Google Scholar]
- 6.Anderson JM, Rodriguez A, Chang DT. Seminars in Immunology. 2008;20:86. doi: 10.1016/j.smim.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brodbeck WG, Patel J, Voskerician G, Christenson E, Shive MS, Nakayama Y, Matsuda T, Ziats NP, Anderson JM. Proceedings of the National Academy of Sciences. 2002;99:10287. doi: 10.1073/pnas.162124199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown BN, Badylak SF. Acta biomaterialia. 2013;9:4948. doi: 10.1016/j.actbio.2012.10.025. [DOI] [PubMed] [Google Scholar]
- 9.Moshaverinia A, Chen C, Xu X, Ansari S, Zadeh HH, Schricker SR, Paine ML, Moradian-Oldak J, Khademhosseini A, Snead ML, Shi S. Advanced functional materials. 2015;25:2296. doi: 10.1002/adfm.201500055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.a) Pinto ML, Rios E, Silva AC, Neves SC, Caires HR, Pinto AT, Duraes C, Carvalho FA, Cardoso AP, Santos NC, Barrias CC, Nascimento DS, Pinto-do OP, Barbosa MA, Carneiro F, Oliveira MJ. Biomaterials. 2017;124:211. doi: 10.1016/j.biomaterials.2017.02.004. [DOI] [PubMed] [Google Scholar]; b) Jiang J, Li Z, Wang H, Wang Y, Carlson MA, Teusink MJ, MacEwan MR, Gu L, Xie J. Advanced healthcare materials. 2016;5:2993. doi: 10.1002/adhm.201600808. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Minardi S, Corradetti B, Taraballi F, Byun JH, Cabrera F, Liu X, Ferrari M, Weiner BK, Tasciotti E. Annals of biomedical engineering. 2016;44:2008. doi: 10.1007/s10439-016-1580-z. [DOI] [PubMed] [Google Scholar]
- 11.Rostam HM, Singh S, Vrana NE, Alexander MR, Ghaemmaghami AM. Biomater Sci. 2015;3:424. doi: 10.1039/c4bm00375f. [DOI] [PubMed] [Google Scholar]
- 12.Madden LR, Mortisen DJ, Sussman EM, Dupras SK, Fugate JA, Cuy JL, Hauch KD, Laflamme MA, Murry CE, Ratner BD. Proc Natl Acad Sci U S A. 2010;107:15211. doi: 10.1073/pnas.1006442107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong VW, Rustad KC, Akaishi S, Sorkin M, Glotzbach JP, Januszyk M, Nelson ER, Levi K, Paterno J, Vial IN, Kuang AA, Longaker MT, Gurtner GC. Nat Med. 2012;18:148. doi: 10.1038/nm.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sicari BM, Dziki JL, Siu BF, Medberry CJ, Dearth CL, Badylak SF. Biomaterials. 2014;35:8605. doi: 10.1016/j.biomaterials.2014.06.060. [DOI] [PubMed] [Google Scholar]
- 15.a) Nuttelman CR, Tripodi MC, Anseth KS. Matrix Biol. 2005;24:208. doi: 10.1016/j.matbio.2005.03.004. [DOI] [PubMed] [Google Scholar]; b) VandeVondele S, Voros J, Hubbell JA. Biotechnol Bioeng. 2003;82:784. doi: 10.1002/bit.10625. [DOI] [PubMed] [Google Scholar]; c) Wang N, Butler JP, Ingber DE. Science. 1993;260:1124. doi: 10.1126/science.7684161. [DOI] [PubMed] [Google Scholar]
- 16.Giancotti FG, Ruoslahti E. Science. 1999;285:1028. doi: 10.1126/science.285.5430.1028. [DOI] [PubMed] [Google Scholar]
- 17.a) Luzina IG, Todd NW, Nacu N, Lockatell V, Choi J, Hummers LK, Atamas SP. Arthritis Rheum. 2009;60:1530. doi: 10.1002/art.24435. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zaveri TD, Lewis JS, Dolgova NV, Clare-Salzler MJ, Keselowsky BG. Biomaterials. 2014;35:3504. doi: 10.1016/j.biomaterials.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.a) O’Toole EA. Clin Exp Dermatol. 2001;26:525. doi: 10.1046/j.1365-2230.2001.00891.x. [DOI] [PubMed] [Google Scholar]; b) Nakayama Y, Kon S, Kurotaki D, Morimoto J, Matsui Y, Uede T. Lab Invest. 2010;90:881. doi: 10.1038/labinvest.2010.69. [DOI] [PubMed] [Google Scholar]; c) Lee TT, Garcia JR, Paez JI, Singh A, Phelps EA, Weis S, Shafiq Z, Shekaran A, Del Campo A, Garcia AJ. Nature materials. 2015;14:352. doi: 10.1038/nmat4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.a) Gordon S, Taylor PR. Nature reviews Immunology. 2005;5:953. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]; b) Harrington H, Cato P, Salazar F, Wilkinson M, Knox A, Haycock JW, Rose F, Aylott JW, Ghaemmaghami AM. Molecular pharmaceutics. 2014;11:2082. doi: 10.1021/mp5000295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shin SR, Bae H, Cha JM, Mun JY, Chen YC, Tekin H, Shin H, Farshchi S, Dokmeci MR, Tang S, Khademhosseini A. ACS nano. 2012;6:362. doi: 10.1021/nn203711s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peppas NA, Huang Y, Torres-Lugo M, Ward JH, Zhang J. Annu Rev Biomed Eng. 2000;2:9. doi: 10.1146/annurev.bioeng.2.1.9. [DOI] [PubMed] [Google Scholar]
- 22.Blakney AK, Swartzlander MD, Bryant SJ. Journal of Biomedical Materials Research. Part a. 2012;100:1375. doi: 10.1002/jbm.a.34104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mantovani A, Sica A, Locati M. Immunity. 2005;23:344. doi: 10.1016/j.immuni.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 24.a) Cassol E, Cassetta L, Rizzi C, Alfano M, Poli G. Journal of immunology. 2009;182:6237. doi: 10.4049/jimmunol.0803447. [DOI] [PubMed] [Google Scholar]; b) Daigneault M, Preston JA, Marriott HM, Whyte MK, Dockrell DH. PloS one. 2010;5:e8668. doi: 10.1371/journal.pone.0008668. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Armstead AL, Li B. International journal of nanomedicine. 2016;11:6195. doi: 10.2147/IJN.S121141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.a) McWhorter FY, Wang T, Nguyen P, Chung T, Liu WF. Proc Natl Acad Sci U S A. 2013;110:17253. doi: 10.1073/pnas.1308887110. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Bartneck M, Schulte VA, Paul NE, Diez M, Lensen MC, Zwadlo-Klarwasser G. Acta biomaterialia. 2010;6:3864. doi: 10.1016/j.actbio.2010.04.025. [DOI] [PubMed] [Google Scholar]
- 26.a) Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Science. 1997;276:1425. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]; b) Engler AJ, Sen S, Sweeney HL, Discher DE. Cell. 2006;126:677. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 27.Beljaars L, Schippers M, Reker-Smit C, Martinez FO, Helming L, Poelstra K, Melgert BN. Front Immunol. 2014;5:430. doi: 10.3389/fimmu.2014.00430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.a) Genin M, Clement F, Fattaccioli A, Raes M, Michiels C. BMC cancer. 2015;15:577. doi: 10.1186/s12885-015-1546-9. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Biswas SK, Lopez-Collazo E. Trends Immunol. 2009;30:475. doi: 10.1016/j.it.2009.07.009. [DOI] [PubMed] [Google Scholar]; c) Porta C, Rimoldi M, Raes G, Brys L, Ghezzi P, Di Liberto D, Dieli F, Ghisletti S, Natoli G, De Baetselier P, Mantovani A, Sica A. Proc Natl Acad Sci U S A. 2009;106:14978. doi: 10.1073/pnas.0809784106. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Bi J, Zeng X, Zhao L, Wei Q, Yu L, Wang X, Yu Z, Cao Y, Shan F, Wei M. Molecular therapy Nucleic acids. 2016;5:e368. doi: 10.1038/mtna.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.a) Lynn AD, Kyriakides TR, Bryant SJ. J Biomed Mater Res A. 2010;93:941. doi: 10.1002/jbm.a.32595. [DOI] [PubMed] [Google Scholar]; b) Lynn AD, Bryant SJ. Acta biomaterialia. 2011;7:123. doi: 10.1016/j.actbio.2010.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tomei AA, Villa C, Ricordi C. Expert opinion on biological therapy. 2015 doi: 10.1517/14712598.2015.10552421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bertholet S, Tzeng E, Felley-Bosco E, Mauel J. Journal of leukocyte biology. 1999;65:50. doi: 10.1002/jlb.65.1.50. [DOI] [PubMed] [Google Scholar]
- 32.Pesce JT, Ramalingam TR, Mentink-Kane MM, Wilson MS, El Kasmi KC, Smith AM, Thompson RW, Cheever AW, Murray PJ, Wynn TA. PLoS pathogens. 2009;5:e1000371. doi: 10.1371/journal.ppat.1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.a) Zhang Q, Wang Y, Zhai N, Song H, Li H, Yang Y, Li T, Guo X, Chi B, Niu J, Crispe IN, Su L, Tu Z. Scientific reports. 2016;6:36160. doi: 10.1038/srep36160. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Thomas AC, Mattila JT. Frontiers in immunology. 2014:5. [Google Scholar]
- 34.Schmidt DR, Kao WJ. J Biomed Mater Res A. 2007;83:617. doi: 10.1002/jbm.a.31270. [DOI] [PubMed] [Google Scholar]
- 35.Yakubenko VP, Belevych N, Mishchuk D, Schurin A, Lam SC, Ugarova TP. Exp Cell Res. 2008;314:2569. doi: 10.1016/j.yexcr.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eckes B, Zweers MC, Zhang ZG, Hallinger R, Mauch C, Aumailley M, Krieg T. The journal of investigative dermatology. Symposium proceedings / the Society for Investigative Dermatology, Inc. [and] European Society for Dermatological Research. 2006;11:66. doi: 10.1038/sj.jidsymp.5650003. [DOI] [PubMed] [Google Scholar]
- 37.Humphrey JD, Dufresne ER, Schwartz MA. Nature reviews. Molecular cell biology. 2014;15:802. doi: 10.1038/nrm3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.a) Lehto VP, Hovi T, Vartio T, Badley RA, Virtanen I. Lab Invest. 1982;47:391. [PubMed] [Google Scholar]; b) Taylor A, Tang W, Bruscia EM, Zhang PX, Lin A, Gaines P, Wu D, Halene S. Blood. 2014;123:3027. doi: 10.1182/blood-2013-06-507582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vogel DY, Heijnen PD, Breur M, de Vries HE, Tool AT, Amor S, Dijkstra CD. Journal of neuroinflammation. 2014;11:23. doi: 10.1186/1742-2094-11-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rostam HM, Reynolds PM, Alexander MR, Gadegaard N, Ghaemmaghami AM. Sci Rep. 2017;7:3521. doi: 10.1038/s41598-017-03780-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fernando MR, Reyes JL, Iannuzzi J, Leung G, McKay DM. PLoS One. 2014;9:e94188. doi: 10.1371/journal.pone.0094188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heath DE, Sharif AR, Ng CP, Rhoads MG, Griffith LG, Hammond PT, Chan-Park MB. Lab on a chip. 2015;15:2073. doi: 10.1039/c4lc01416b. [DOI] [PubMed] [Google Scholar]
- 43.Albert ML, Pearce SF, Francisco LM, Sauter B, Roy P, Silverstein RL, Bhardwaj N. The Journal of experimental medicine. 1998;188:1359. doi: 10.1084/jem.188.7.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.a) Ferguson TA, Herndon J, Elzey B, Griffith TS, Schoenberger S, Green DR. Journal of immunology. 2002;168:5589. doi: 10.4049/jimmunol.168.11.5589. [DOI] [PubMed] [Google Scholar]; b) Heath WR, Carbone FR. Annual review of immunology. 2001;19:47. doi: 10.1146/annurev.immunol.19.1.47. [DOI] [PubMed] [Google Scholar]
- 45.a) Anders HJ, Ryu M. Kidney international. 2011;80:915. doi: 10.1038/ki.2011.217. [DOI] [PubMed] [Google Scholar]; b) Huen SC, Cantley LG. Pediatr Nephrol. 2015;30:199. doi: 10.1007/s00467-013-2726-y. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Ricardo SD, van Goor H, Eddy AA. The Journal of clinical investigation. 2008;118:3522. doi: 10.1172/JCI36150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.a) Faulk DM, Londono R, Wolf MT, Ranallo CA, Carruthers CA, Wildemann JD, Dearth CL, Badylak SF. Biomaterials. 2014;35:8585. doi: 10.1016/j.biomaterials.2014.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wolf MT, Dearth CL, Ranallo CA, LoPresti ST, Carey LE, Daly KA, Brown BN, Badylak SF. Biomaterials. 2014;35:6838. doi: 10.1016/j.biomaterials.2014.04.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Plow EF, Haas TA, Zhang L, Loftus J, Smith JW. The Journal of biological chemistry. 2000;275:21785. doi: 10.1074/jbc.R000003200. [DOI] [PubMed] [Google Scholar]
- 48.Van Den Bulcke AI, Bogdanov B, De Rooze N, Schacht EH, Cornelissen M, Berghmans H. Biomacromolecules. 2000;1:31. doi: 10.1021/bm990017d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental table 1. Nucleotide sequences of the primer pairs used for qPCR in this study.
Figure S1. Compressive modulus of 5, 10, and 15% GelMA hydrogel. The data are shown as the mean ± SEM (*, p < 0.05).
Figure S2. (A) Human monocytic THP-1 cell viability in 5, 10, and 15% GelMA hydrogels 24 hours post-encapsulation. (B) Quantification of cell viability demonstrated excellent cell survival in 5% GelMA. Scale bar represents 100 μm. The data are shown as the mean ± SEM (*, p < 0.05).
Figure S3. THP-1 cellsmetabolic activity in 5, 10, and 15% GelMA hydrogels over a period of 7 days. The data are shown as the mean ± SEM (*, p < 0.05; compared to both 10 and 15% GelMA).
Figure S4. (A) Swelling properties of 5, 10, and 15% GelMA hydrogels over a period of 7 days. The swelling ratios of GelMA hydrogels at various GelMA % (w/v) show statistically significant differences. The data are shown as the mean ± SEM (*, p < 0.05; compared to both 10 and 15% GelMA, §, p < 0.05; compared to both 5 and 15% GelMA). (B) Swelling properties of 10% PEGDA and 5% GelMA hydrogels over a period of 5 days. The swelling ratios between PEGDA and GelMA hydrogels show no significant differences.
Figure S5. IL-4 cytokine release from 5, 10, and 15% GelMA hydrogels. The data are shown as the mean ± SEM (*, p < 0.05; compared to both 10 and 15% GelMA).
Figure S6. Compressive modulus of PEGDA and GelMA hydrogels with or without of incorporated IL-4.
Figure S7. IL-4 cytokine release profile when incorporated in PEGDA and GelMA hydrogels.
Figure S8. The effects of α2β1 integrin peptide on human primary monocytes‘ differentiation. (A) Micrographs of macrophages fluorescently stained for M1 marker Calprotectin, M2 marker CD206, and DAPI. Scale bar represents 200 μm. (B) IL-6 and IL-10 cytokine release profile as measured by ELISA. The data are shown as the mean ± SEM (*, p < 0.05).