Abstract
The role of microRNAs in controlling angiogenesis is recognized as a promising therapeutic target in both cancer and cardiovascular disorders. However, understanding a miRNA’s pleiotropic effects on angiogenesis is a limiting factor for these types of therapeutic approaches. Using genome-wide next-generation sequencing, we examined the role of an antiangiogenic miRNA, miR-200b, in primary human endothelial cells. The results indicate that miR-200b has complex effects on hypoxia-induced angiogenesis in human endothelia and importantly, that many of the reported miR-200b effects using miRNA overexpression may not be representative of the physiological role of this miRNA. We also identified the antiangiogenic KLF2 gene as a novel target of miR-200b. Our studies indicate that the physiological changes in miR-200b levels during acute hypoxia may actually have a proangiogenic effect through Klf2 downregulation and subsequent stabilization of HIF-1 signaling. Moreover, we provide a viable approach for differentiating direct from indirect miRNA effects in order to untangle the complexity of individual miRNA networks.
Keywords: micro-RNA 200b, KLF2, HUVEC, HIF-1, HIF-2, hsa-miR-200b-3p
1. Introduction
Angiogenesis promotes new blood vessel development from pre-existing vasculature and is a critical process in wound healing, the menstrual cycle, cancer, and various ischemic and inflammatory diseases. The process of angiogenesis provides cells with a controlled supply of oxygen and requires a complex control system with proangiogenic and antiangiogenic factors. Angiogenesis changes (Bergers and Benjamin, 2003) often accompany cardiovascular disorders, as well as with the development, progression, and metastasis of various human cancers. Hence, the molecular mechanisms that mediate angiogenesis have become promising therapeutic targets and biomarkers for both human cardiovascular diseases and cancer.
Recently, miRNAs that endogenously regulate gene expression via the RNA interference (RNAi) pathway have been shown to play a critical role in angiogenesis (Greco et al., 2014; Greco and Martelli, 2014; Madanecki et al., 2013). However, due to the complexity of the potential miRNA - mRNA interactions, their role in maintaining the angiogenic balance remains unclear. Often conflicting results from different groups have shown that the same miRNAs may have different mRNA targets and thus the effects on angiogenesis may be cell-type specific (Madanecki et al., 2013). Furthermore, for numerous miRNAs, their potential mRNA targets are based on correlative studies in cancer cell lines or by only following the effects of miRNA overexpression, which may be caused through indirect effects by targeting, for example, an upstream regulator or transcription factor.
miR-200b (miRBase id. MIMAT0000318 (Kozomara and Griffiths-Jones, 2014) is a miR-200 family member that is clustered with miR-200a and miR-429 on chromosome 1p36 (Chan et al., 2011). This miRNA is expressed in a variety of endothelial, stem and cancer cells (Brabletz and Brabletz, 2010; Choi et al., 2011), and modulates a wide range of cellular functions including proliferation, motility, apoptosis, and stemness (Brabletz and Brabletz, 2010). Alterations of miR-200b are well described in the context of the progression of epithelial cancers (Zhang et al., 2013b), and have been linked to the acquisition of a migratory, mesenchymal phenotype since miR-200b targets the transcription factors ZEB1 and ZEB2, two master regulators of the epithelial to mesenchymal transition (EMT) (Brabletz and Brabletz, 2010; Zhang et al., 2013b). However, miR-200b function in endothelial cells is less clear. To date, numerous studies have shown that miR-200b overexpression in human endothelial cells has potent antiangiogenic effects and inhibits VEGFA signaling (Chan et al., 2012; Chang et al., 2013; Li et al., 2017; Sinha et al., 2015). Furthermore, a large number of proangiogenic and anti-angiogenic mRNA targets have been proposed for miR-200b in human endothelia (Chan et al., 2011; Chan et al., 2012; Chang et al., 2013; Choi et al., 2011; Li et al., 2017; Sinha et al., 2015). The majority of these studies, however, have focused primarily on one mRNA target for miR-200b and have therefore overlooked the complexity of the angiogenic response. Additionally, many miR-200b overexpression studies often did not consider the physiological alterations of miR-200b levels in human endothelia during hypoxia as well as the wide range of other potential miR-200b target mRNAs that are not directly related to angiogenesis. The complexity of the miRNA networks and angiogenesis suggests that future developments in cancer therapies that are based on miR-200b’s anti-angiogenic properties will require a complete understanding of this miRNA’s physiological role during hypoxic in human endothelium.
To examine miR-200b’s functional role during angiogenesis, we followed its upregulation during hypoxia in primary human umbilical vein endothelial cells (HUVECs). To determine the extent of miR-200b’s regulatory role in these cells, we followed its effects on the transcriptome during miR-200b depletion as well as during overexpression using genome-wide next-generation mRNA sequencing of the transfected HUVECs. Validation of the identified miR-200b network indicated that miR-200b has a pleiotropic effect on hypoxia-induced angiogenesis in human endothelia and that many of the known miR-200b effects using miRNA overexpression may not be representative of the physiological role of this miRNA. Furthermore in primary endothelial cells, we identified antiangiogenic Sp/Kruppel-like factor 2 (KLF2) as a novel miR-200b direct target and provide a viable approach for differentiating direct from indirect miRNA effects in order to untangle the complexity of miRNA networks.
2. Material and methods
2.1. Cell lines and culture conditions
Primary HUVECs (passage 2–6 were used only) pooled from 10 independent donors were obtained from Cellworks (UK, division of Caltag Medsystems Ltd), as well as ATCC (American Type Culture Collection) and maintained until passage five in EGM-2 BulletKit™ medium (Lonza). Cells were split either into 6-well plates or 10 cm dishes and allowed to grow to 70–80% confluence prior to the start of the experiments.
2.2. Induction of hypoxia
Hypoxia was induced in a CO2/O2 incubator for hypoxia research (Tri-gas Binder CB150). Briefly, cells were cultured in 2 cm dishes at 0.9% O2 for the time periods specified. Control cells were maintained in normoxic conditions in the same incubator and harvested at the specified times.
2.3. Isolation of RNA and microRNA
Total RNA containing the microRNA fraction was isolated using miRNeasy kit (Qiagen). RNA concentrations were calculated based on the absorbance at 260 nm. RNA samples were stored at −70°C until use.
2.4. Next generation RNA sequencing analyses
HUVECs (passage 3) were used for the RNA isolation and analyses. Following rRNA (ribosomal RNA) depletion, the remaining RNA fraction was used for library construction and subjected to 100bp paired-end sequencing on an Illumina HiSeq 2000 instrument. Sequencing reads were aligned to the human reference genome assembly (hg19) using TopHat (Trapnell et al., 2009). Transcript assembly and estimation of the relative abundances were carried out with Cufflinks (Trapnell et al., 2010).
2.5. Bioinformatic analysis of potential miRNA effects
GeneAnalytics™ (geneanalytics.genecards.org), is a comprehensive gene set analysis tool for rapid contextualization of expression patterns and functional signatures embedded in the postgenomics Big Data domains, such as Next Generation Sequencing (NGS), RNAseq, and microarray experiments (Ben-Ari Fuchs et al., 2016). The webserver was used to place NGS results into physiological context using built in biological pathway algorithm. In GeneAnalytics, matched SuperPaths appear with their matching score and link to the relevant webcard in PathCards, as well as the list of matched genes and total number of genes associated with each SuperPath. The scoring algorithm in the pathways category is based on the algorithm used by the GeneDecks Set Distiller tool (Stelzer et al., 2009). Briefly, all genes in each SuperPath are given a similar weight in the analysis, and the matching score is based on the cumulative binomial distribution, which is used to test the null hypothesis that the queried genes are not over-represented within any SuperPath pathway unification is employed on all of the sources found in GeneCards. The results were independently confirmed with Quiagen Ingenuity® Pathway Analysis (IPA®).
2.6. Measurement of mRNA and miRNA levels using quantitative Real Time PCR (qRT-PCR)
We used TaqManOne-Step RT-PCR Master MixReagents (Applied Biosystems) as described previously (Bartoszewska et al., 2013; Bartoszewski et al., 2011; Bartoszewski et al., 2014) using the manufacturer’s protocol. The relative expressions were calculated using the comparative relative standard curve method (Larionov et al., 2005). We used 18S rRNA as the relative control for our studies. We also validated this relative control against another housekeeping gene, TATA-binding protein (TBP). As relative controls for miRNA quantification, we validated and used RNU44 and RNU48. TaqMan probes ids used were: 18S - Hs99999901_s1; TBP - Hs4332659_m1; HIF1A - Hs00153153_m1; EPAS1 - Hs01026149_m1; KLF2 - Hs00360439_g1; RNU44 - 001094; RNU48 - 001006; miR-200b – 002251. Complete list of TaqMan assays used is provided in Supporting Materials (Supporting Table S3).
2.7. miRNA analogs and target protector transfections
miR-200b mimic (id MC10492) and antagomiR (id MH10492) were purchased from Ambion. HUVECs were transfected using the Lipofectamine RNAiMax according to manufacturer's protocol. miR-200b mimic and antagomiR were used at final concentrations of 10 nM and 20 nM, respectively. The transfected cells were cultured for 2 days prior to further analysis. The degree of miRNA over-expression or knockdown was determined by qRT-PCR. Target protector (TP) were designed with Qiagen and purchased from Qiagen and directed against the KDR (5'-CATTTTGATCTTCTATTTGGTCCGTTACATTCACAAGCTC-3') and KLF2 (5'-GGACCCAGAGAACCGGGCCGGGCACAGCTG-3') mRNAs. TP were used at final concentration of 600 nM. cel-miR-67 was used as a control (Ambion assay id MC22484). As an additional control, Ambion siRNA Negative Control 1 (no. 4390843), Ambion mimic control (no. 4464060) and Ambion antagomiR control (no. 4464076) were used as well.
2.8. Western Blots
Cells were lysed in RIPA buffer (150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris- HCl, pH 8.0) supplemented with protease Inhibitor Complete Mini (Roche) on ice for 15 min. The cell lysates were rotated at 4°C for 30 min and the insoluble material was removed by centrifugation at 15,000 g for 15 min. Protein concentrations were determined by BioRad™ Protein Assay using bovine serum albumin (BSA) as a standard. Following the normalization of protein concentrations, lysates were mixed with an equal volume of 2× Laemmli sample buffer and incubated for 5 min at 95°C prior to separation by SDS PAGE on stain-free TGX gradient gels (BioRad). Following SDS-PAGE, the proteins were transferred to polyvinylidene fluoride membranes (300 mA for 90 min at 4°C). The membranes were then blocked with BSA (Sigma-Aldrich) proteins dissolved in PBS/Tween-20 (3% BSA, 0.5% Tween-20 for 1–2 hours), followed by immunoblotting with the primary antibody specified for each experiment Klf2 (Sigma SAB1403063 diluted at 1:500), Hif-1α (Abcam ab16066, diluted at 1:1000); VEGFA (Abcam ab51745, diluted at 1:250), HIF-2α (Abcam ab199, diluted at 1:800), beta Actin (Abcam ab1801, diluted at 1:1000), VEGFR2 (Abcam ab39256, diluted at 1:500). After the washing steps, the membranes were incubated with goat anti-rabbit IgG (H+L) or with goat anti-mouse IgG (H+L) HRP-conjugated secondary antibodies (BioRad) and detected using ECL (Amresco). Densitometry was performed using Image Lab software v. 4.1 (BioRad).
2.9. Statistical analysis
Results were expressed as means ± standard deviations (SD). Statistical significance among means was determined using the Student’s t-test (two samples, paired and unpaired). The correlation coefficients and p values were calculated according to Spearman Rank-Order Correlation (Spearman, 1904).
3. Results
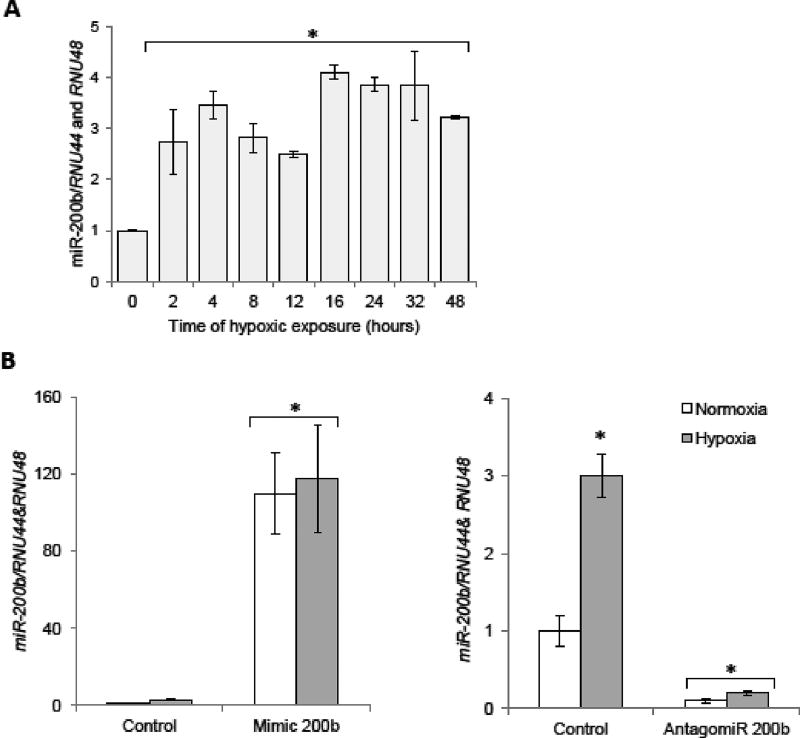
To understand the role of miR-200b in hypoxia-mediated angiogenesis, we first performed a time-course study and monitored miR-200b regulation in primary human endothelial cells. Primary HUVECs (pooled from 10 independent donors) were exposed to hypoxia (0.9% O2) for up to 48 h and miR-200b levels were measured at the specified time points. As shown on Figure 1A, miR-200b expression was induced at 2 h and stayed elevated during the entire 48 h time course. Our initial in silico analysis identified hundreds of potential human genes that could be controlled by miR-200b. The MiRanda algorithm predicts over 7000 mRNAs as having potential miR-200b target sites (Betel et al., 2010), whereas miRDB predicts 757 potential mRNA targets (Wong and Wang, 2015). Given that miR-200b was significantly (from ~2.5 to 4 fold) and continuously upregulated during hypoxia (Figure 1A), a large number of angiogenesis-related mRNA targets could be regulated either directly or indirectly through miR-200b’s actions. To distinguish between those two possibilities, we analyzed how miR-200b mimic overexpression and antagomiR depletion had on the mRNA profiles using genome-wide transcriptome sequencing (RNA-seq) analysis on primary HUVECs under normoxic conditions. Our goal was to determine the effects of this particular miRNA without the complicating effects of hypoxia and hypoxia inducible factor (HIF) expression.
Figure 1.
Regulation of miR-200b during hypoxia in primary HUVECs. (A) The miR-200b levels were monitored in qRT-PCR experiments. The results from 3 independent experiments (n=12) are plotted normalized to RNU44 and RNU48 RNA levels and expressed as a fold-change over the normoxic control. (B) HUVECs were transfected with miR-200b mimic or antagomiR, and the miRNA levels were monitored in normoxic conditions and after 4 h of exposure to hypoxia. miRNA levels from 2 independent experiments (n=8) are plotted normalized to RNU44 and RNU48 RNA levels and expressed as a fold change over normoxic control. Error bars represent standard deviations. Significant changes (p<0.05) are marked with an asterisk.
Reduction of miR-200b levels with the antagomiR resulted in the significant (two-fold log2 change) upregulation of 221 mRNAs, and a reduction of 109 mRNAs (Supportive Table 1). Overexpression of miR-200b with mimic caused a downregulation of 230 genes, and upregulation of 252 mRNAs (Supportive Table 1). However, among 221 mRNAs that were upregulated after the antagomiR, 102 were significantly induced with mimic as well. Similarly, among 230 genes downregulated upon miR-200b overexpression, 56 were significantly reduced by antagomiR (Supportive Figure 1). Surprisingly, we did not identify a gene using RNAseq that was either induced or reduced after transfection with antagomiR or mimic, respectively, using our selection criteria. We did, however, identify the control genes (ZEB1 (Chen et al., 2011b) and KDR (Choi et al., 2011)) that showed this reciprocal pattern as expected, but the magnitude of the expression changes was below our selection cut-off criteria. This suggested that modulation of miR-200b levels results in a very wide network of secondary interactions that could mask the miRNA’s direct effects. As an alternative, we lowered our selection criteria to allow for the selection of genes that were either induced by antagomiR and in parallel, reduced or were not affected by mimic, or reduced by mimic and induced or not affected by antagomir. This strategy resulted in the selection of 377 genes that were potentially regulated by miR-200b (Supportive Table 2). Further, using Gene Ontology Software, we narrowed our focus to 16 mRNAs from the RNAseq data that were postulated to be involved in the cellular response to hypoxia and angiogenesis, and the putative angiogenesis-related miR-200b targets.
These mRNAs were further tested with miRNA target sites predicting algorithms for potential miR-200b binding sites (miRanda (Betel et al., 2010), Targetscan7.1 (Agarwal et al., 2015) and RNAhybrid (Kruger and Rehmsmeier, 2006)) and the effects of miR-200b modulation were verified with qPCR both in normoxic and hypoxic conditions (Table 1, Supportive Figure 2). Since mimic transfection caused miRNA overexpression, much higher (> 100-fold) than physiological levels of miR-200b (Figure 1B left panel) and antagomir dramatically inhibited miR-200b expression (Figure 1B right panel), we only considered potential target mRNAs that were reduced on mimic treatment and induced by antagomir treatment, preferably during both normoxia and hypoxia.
Table 1. Genes related to the hypoxia-induced angiogenesis that were altered in HUVECs following miR-200b reduction/induction.
The genes were preselected based on RNASeq experiments and subsequent Gene Ontology analysis. The presence of potential miR-200b target sequences was predicted in bioinformatics approach (Betel et al., 2008; Kruger and Rehmsmeier, 2006; Lewis et al., 2005). The changes in genes expression after miR-200b overexpression and inhibition were accessed by qPCR in normoxia and under hypoxia (Supportive Figure 2). The previously proposed miR-200b targets that are angiogenesis related were included as well and denoted with an asterisk. Opposite mRNA expression patterns after overexpression or inhibition of miR-200b are highlighted in grey.
| Gene | miR- 200b target site |
miR-200b effect in normoxia | miR-200b effect under hypoxia |
Role in angiogenesis |
Ref. |
|---|---|---|---|---|---|
| KLF2 | yes | Downregulated with mimic and upregulated with antagomiR | Downregulated with mimic and upregulated with antagomiR | antiangiogenic | (Bhattacharya et al., 2005; Kawanami et al., 2009) |
| *KDR | yes | Downregulated with mimic and upregulated with antagomiR | Downregulated with mimic and not changed with antagomiR | proangiogenic | (Soldi et al., 1999) |
| *ETS1 | yes | Downregulated with mimic and antagomiR | Downregulated with mimic and antagomiR | proangiogenic | (Hashiya et al., 2004) |
| *VEGFA | yes | Downregulated with mimic and antagomiR | Downregulated with mimic and antagomiR | proangiogenic | (Ferrara et al., 2003) |
| NOS3 | yes | Downregulated with mimic only | Upregulated with antagomiR | proangiogenic | (Amano et al., 2003) |
| THSD7A | yes | Downregulated with mimic | No significant effects | antiangiogenic | (Wang et al., 2010) |
| ITGB3 | yes | Downregulated with mimic | Downregulated with mimic | proangiogenic | (Brooks et al., 1994) |
| MYOF | yes | Downregulated with mimic | No significant effects | proangiogenic | (Yu et al., 2011) |
| NRP2 | yes | Downregulated with mimic | Downregulated with mimic | proangiogenic | (Takashima et al., 2002) |
| FYN | yes | Downregulated with mimic and antagomiR | Downregulated with mimic | proangiogenic | (Sen and Johnson, 2011) |
| FZD4 | yes | Downregulated with mimic | Downregulated with mimic | proangiogenic | (Chen et al., 2011a) |
| GRB10 | yes | Downregulated with mimic | Downregulated with mimic | proangiogenic | (Soriano et al., 2004) |
| EP300 | yes | Downregulated with mimic and antagomiR | Downregulated with mimic and antagomiR | proangiogenic | (Zhang et al., 2013a) |
| *FLT1 | yes | No effect | Downregulated with mimic and antagomiR | proangiogenic | (Birnbaum, 1995) |
| RGS5 | yes | Downregulated with mimic | Downregulated with mimic | proangiogenic | (Mitchell et al., 2008) |
| HMOX1 | yes | Downregulated with mimic and antagomiR | Downregulated with mimic and antagomiR | proangiogenic | (Malaguarnera et al., 2002) |
Although miRNA overexpression (mRNA) negatively affected all of the selected mRNAs, the antagomir only upregulated only 1 of the mRNAs, Krupple-like factor 2 (KLF2), and thus this represented a potential direct target of miR-200b. KDR, which encodes the VEGFR2, and already reported as miR-200b target (Choi et al., 2011), fulfilled our criteria during normoxia, but not during hypoxia (Supportive Figure 2). KLF2 is a novel target that encodes for a transcription factor that is antiangiogenic (Kawanami et al., 2009).
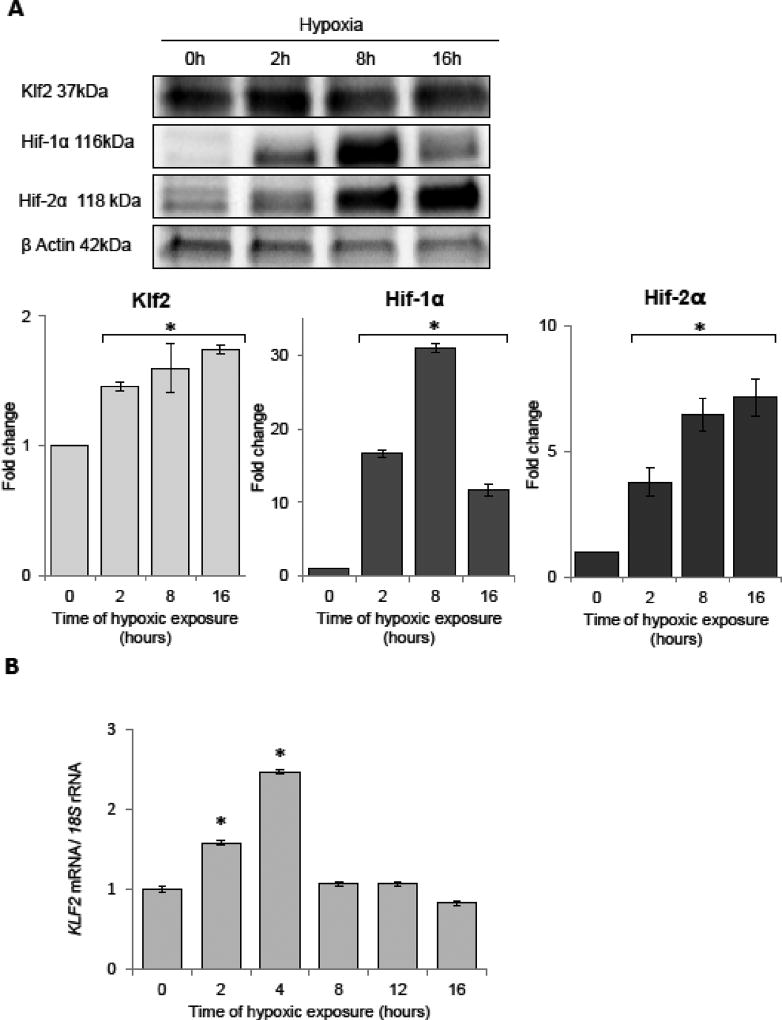
During acute hypoxia, Klf2 protein increases (Figure 2A) as has previously been shown (Bhattacharya et al., 2005; Kawanami et al., 2009) and goes up approximately 2-fold after 16 h. In parallel, Hif-1α and Hif-2α are increased during this same time with Hif-1α maximal at 8 h and Hif-2α continuing to increase at 16 h (Figure 2A). The mRNA changes for KLF2 increase 2.5-fold during hypoxia with significant elevations at 2 and 4 h (Figure 2B), and this agrees with a previous study suggesting that KLF2 is induced during hypoxia (Bhattacharya et al., 2005; Kawanami et al., 2009).
Figure 2.
Acute hypoxia induces dynamic changes in the protein expression profile of the Klf2 in HUVECs. (A) Hypoxia induces dynamic changes of protein levels of Klf2, Hif-1α, and Hif-2α. The protein levels of were detected with SDS-PAGE and Western Blot and related to total protein levels. 2 individual samples (4 µg of total protein per lane) were tested for each time point and the experiments were repeated twice. (B) The KLF2 mRNA levels were monitored in qRT-PCR experiments. The results from 3 independent experiments (n=12) are plotted normalized to 18S rRNA levels and expressed as a fold-change over the normoxic control. Error bars represent standard deviations. Significant changes (p<0.05) are marked with an asterisk.
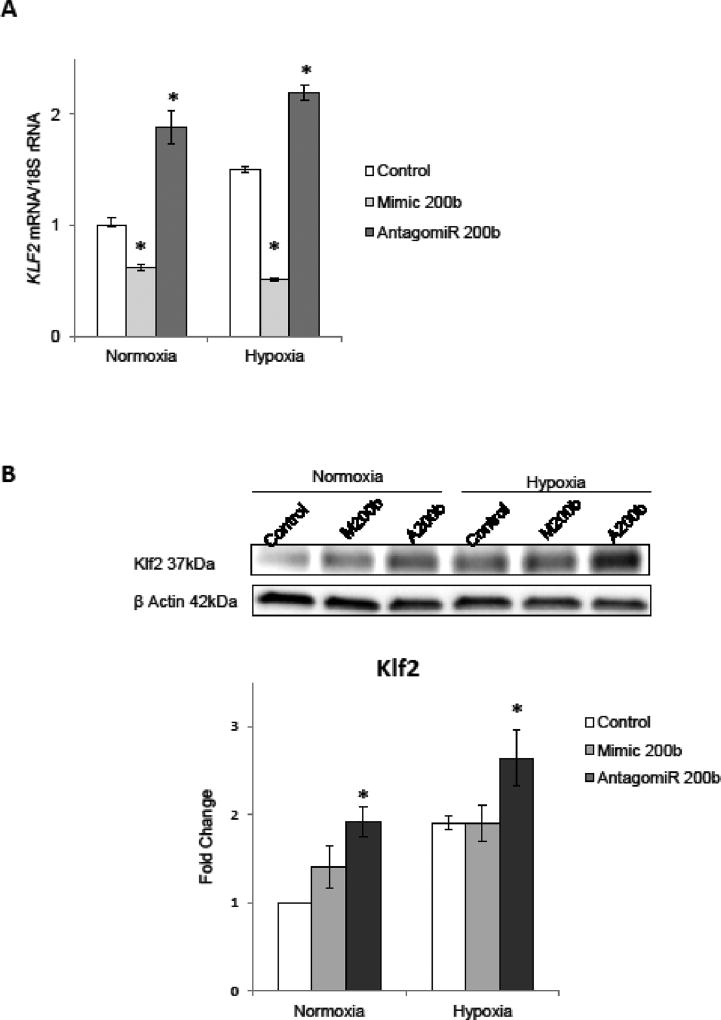
Given that KLF2 mRNA and miR-200b expression profiles during hypoxia were positively correlated (correlation coefficient 0.6 and p value 0.002, Spearman rank order test n=18), and that KLF2 mRNA has a potential miR-200b target site, we next examined effects of miR-200b overexpression (mimic) and depletion (antagomiR) on KLF2 mRNA levels during normoxia and after 4 hours of hypoxia. As shown in Figure 3A, KLF2 mRNA was significantly reduced along with increased miR-200b levels and accumulated upon miR-200b depletion, both during normoxia and hypoxia. These miR-200b-induced changes were preserved on Klf2 protein levels as well. As shown in Figure 3B, reduction of miR-200b expression with antagomiR significantly induced Klf2 protein expression under both conditions. However, miR-200b overexpression didn't decrease the Klf2 protein, suggesting that the proteins levels were stable during the 4-hour time course.
Figure 3.
miR-200b alters the expression of KLF2. (A) HUVECs were transfected with miR-200b mimic or antagomiR, and the mRNA levels were monitored in normoxic conditions and after 4 h exposure to hypoxia. KLF2 mRNA levels from 3 independent experiments (n=12) are plotted normalized to 18S rRNA or TBP mRNA levels and expressed as a fold change over the normoxic control. Significant changes (p<0.05) are marked with an asterisk. (B) The corresponding changes of Klf2 protein levels were detected with SDS-PAGE and Western Blot and normalized to the β-Actin and 2 individual samples (4 µg of total protein per lane) were tested for each treatment and the experiments were repeated twice.
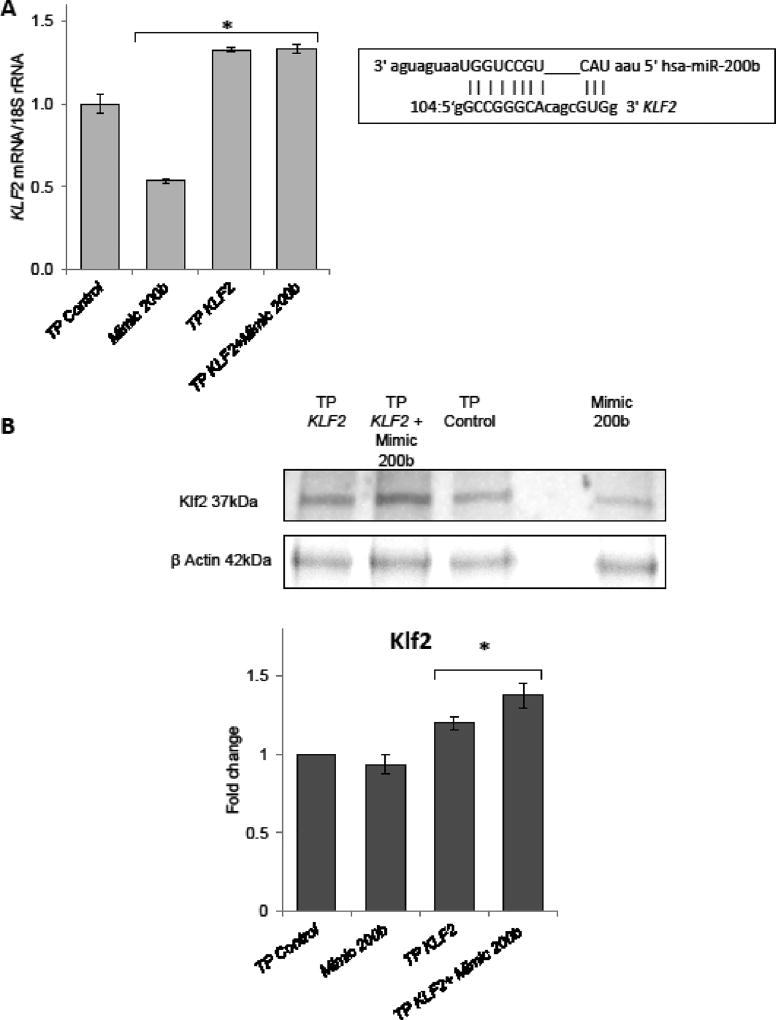
We further verified the miR-200b direct interaction with KLF2 mRNA target sequence with use of specific Target Protector (TP) (Bartoszewska et al., 2015; Janaszak-Jasiecka et al., 2016; Summerton, 2007). TPs bind to specific complementary RNA sequences and block miRNA binding; however, they do not trigger the RNAi (Love et al., 2008). Target protectors increase mRNA target expression by a modest amount and only in cells where the mRNA is already expressed (Staton and Giraldez, 2011). As shown in Figure 4, both KLF2 mRNA and protein were resistant to miR-200b overexpression in the presence of specific TP morpholino, confirming the direct interaction between miR-200b and this mRNA. Importantly, the KLF2 TP increased the physiological mRNA and protein levels of KLF2. This observation is consistent with our previous experiments where miR-200b overexpression had little effect on Klf2 protein. Thus physiological levels of miR-200b efficiently control the Klf2 protein levels both in normoxic and hypoxic conditions. KDR mRNA, being a previously validated target of miR-200b was used as a control for both mimic and anatgomiR as well as target protector experiments (Supportive Figure 3AB).
Figure 4.
miR-200b binds to predicted target sequence in the KLF2 3’UTR (A).The RNAhybrid predicted miR-200b target site at 3' UTR of KLF2 mRNA is presented at the top right. HUVECs were transfected with KLF2 target sequence-specific target protector and/or the miR-200b analog (Mimic 200b) and exposed to hypoxia for 4 h. The KLF2 mRNA levels were monitored in qRT-PCR experiments. The KLF2 levels results from 2 independent experiments (n=8) are plotted normalized to 18S rRNA levels and expressed as a fold change over the transfection control. Significant changes (p<0.05) are marked with an asterisk. (B). The corresponding changes of Klf2 protein levels were detected with SDSPAGE and Western Blot and normalized to the β-actin levels 2 µg of total protein per lane was loaded for each sample and the experiments were repeated twice.
4. Discussion
The results of recent reports suggest that miR- 200b negatively regulates angiogenesis in human endothelial cells. Chan and coworkers showed in HMECs (human microvascular endothelial cells) that miR-200b is downregulated by hypoxia and thus, the levels of its target, the proangiogenic ETS1 gene, are induced (Chan et al., 2011). Another study reported that in A549 cells, miR-200b targeted the predicted binding sites in the 3'UTRs of VEGF, FLT1, and KDR using miRNA overexpression-based luciferase reporter assays (Chan et al., 2011; Choi et al., 2011; Roybal et al., 2011).
In our present and previous studies (Bartoszewska et al., 2015) in primary HUVECs demonstrated that miR-200b levels are dynamically induced during a hypoxia time course and illustrated that HIF-1 and HIF-2 expression was biphasic with HIF-2 going up as HIF-1 went down. Here we demonstrate that the miR-200b expression profile during hypoxia in primary HUVECs strongly resembles both HIFs and proangiogenic signaling. However, these data do not support miR-200b’s antiangiogenic function, at least in HUVECs. In HMECs, miR-200b was shown to be downregulated after 24h of hypoxia only, whereas its levels remained constant during 6 and 12 hours of hypoxia. Although HMECs and HUVECs are both primary human endothelial cells, they are from different vascular beds and therefore have differential miR-200b expression profiles during hypoxia.
In the present studies, we have validated miR-200b expression profile during hypoxia in at least 6 independent experiments using primary cells from different batches and sources, and we have also observed that primary HUVECs exhibit the significant miR-200b induction in response to hypoxia until passage 6, whereas higher passages show no effect on miR-200b levels. Finally we have followed changes in ETS1 levels not only during miR-200b overexpression, but also up on depletion of physiological levels of miR-200b during both in normoxia and under hypoxia. Our results indicated that miR-200b inhibition did not result in ETS1 mRNA stabilization in HUVECs. Similar results were obtained for FLT1 and VEGFA, which demonstrated that only miR-200b overexpression affected their mRNA levels during hypoxia (Supp. Figure 2).
To re-evaluate miR-200b role in regulation of hypoxia induced angiogenesis, we accessed the genome wide consequences of miR-200b modulation in primary HUVECs. Following the miR-200b inhibition and overexpression consequences on HUVECs transcriptome, we observed and although miR-200b levels modulation altered expression of large number of genes, none of these genes showed a pattern of opposite effects when transfected with antagomiR and mimics. After changing our selection criteria, we analyzed an extended list of miR-200b affected genes this resulted in the identification of 12 angiogenesis-related transcripts that could be potentially miR-200b targets, some of which had already been identified as miR-200b targets.
We have independently verified these targets with qPCR during hypoxia and normoxia in primary HUVECs. Importantly, although miR-200b overexpression resulted in downregulation of majority of the targets, the inhibition of physiological miR-200b levels resulted in induction of only 2 of them, both under normoxic and hypoxic conditions. One was KDR, while the other novel miR-200b target was KLF2. Finally using specific target protectors, we have confirmed the direct binding of miR-200b to these 2 mRNAs.
KDR mRNA expression was elevated on miR-200b inhibition suggesting that this gene is miR-200b target indeed. Interestingly, although miR-200b was specifically modulating KDR mRNA levels (Supportive Figure 3AB), the impact of this miRNA on VEGFR2 protein was less direct (Supportive Figure 3CDE). Despite the KDR mRNA levels being downregulated during the hypoxia time course (that could correspond to miR-200b induction), the VEGFR2 protein shown was transiently changed. Finally, despite miR-200b antagomiR rescuing VEGFR2 expression during hypoxia, the mimic treatment resulted in even more significant VEGFR2 accumulation, whereas no significant change in VEGFR2 expression was observed up on miR-200b modulation in normoxia. Although these observations require further studies, in HUVECs the miR-200b impact on VEGR2 protein levels is probably rather limited.
KLF2 is strongly expressed in endothelial cells and is necessary for normal vessel formation (Anderson et al., 1995; Kuo et al., 1997; SenBanerjee et al., 2004). Overexpression of this transcription factor inhibits Hif-1α and its target genes (Kawanami et al., 2009). Importantly, Klf2 was shown to selectively promote Hif-1α degradation during hypoxia in a von Hippel-Lindau-independent but proteasome-dependent manner through disruption of the interaction between Hif-1α and its chaperone Hsp90 (Bhattacharya et al., 2005; Kawanami et al., 2009). Interestingly, Klf2 has no effect on Hif-2α protein stability (Kawanami et al., 2009). Finally, Klf2 was shown to reduce VEGF receptor 2 (VEGFR2) expression, which directly affected VEGFA signaling (Bhattacharya et al., 2005; Kawanami et al., 2009). Thus Klf2, as a selective natural HIF-1 activity inhibitor, was proposed as a "molecular switch" modulating endothelial angiogenic balance during hypoxia (Feinberg et al., 2004; Kawanami et al., 2009; Lin et al., 2005; SenBanerjee et al., 2004). Here, we demonstrate that acute hypoxia-induced miR-200b expression modulates Klf2 mRNA levels directly (Figures 3 and 4), and thus contributes to the HIF-1/HIF-2 switch in human endothelia during hypoxia.
5. Conclusions
In summary, our studies suggest that the physiological changes in miR-200b levels during hypoxia have a proangiogenic effect through Klf2 downregulation and stabilization of HIF-1 signaling. Furthermore, we show that majority of known antiangiogenic effects of miR-200b could result from the artificial overexpression models used for both target selection and assessment of angiogenesis. Whereas, our studies indicate that they are not affected by miR-200b at physiological levels, and thus some of these gene targets are not functional targets as has been suggested in other studies. Furthermore, our data clearly point out that understanding the complexity of the potential single miRNA - mRNA interactions and related pleiotropic effects on cell signaling is a limiting factor in the development of novel miRNAbased therapies in a number of human diseases.
Supplementary Material
Acknowledgments
This work has been supported by National Science Center "SONATA BIS" Program under contract UMO-2015/18/E/NZ3/00687 (to R.B.) and NIH P30 DK072482 (to J.F.C.). We would also like to thank Dawid Lejnowski for his kind assistance.
Abbreviations used
- KLF2
Kruppel Like Factor 2
- HUVEC
human umbilical vascular endothelial cells
- miRNA
microRNA
- TP
target protectors
- VEGFA
vascular endothelial growth factor A
- VEGFR2
vascular endothelial growth factor A receptor 2
- KDR
Kinase insert domain receptor
- HIF
hypoxia inducible factor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions: Conceived and designed the experiments: R.B. Performed the experiments: M.S. A.J.J. S.B. R.B. K.K. All authors analyzed the data. Contributed reagents/materials/analysis tools: R.B. Wrote the paper: R.B. and J.F.C. All authors read and revised the final version of the manuscript.
Competing financial interests: The authors declare no competing financial interests.
References
- Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4 doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano K, Matsubara H, Iba O, Okigaki M, Fujiyama S, Imada T, Kojima H, Nozawa Y, Kawashima S, Yokoyama M, Iwasaka T. Enhancement of ischemia-induced angiogenesis by eNOS overexpression. Hypertension. 2003;41:156–162. doi: 10.1161/01.hyp.0000053552.86367.12. [DOI] [PubMed] [Google Scholar]
- Anderson KP, Kern CB, Crable SC, Lingrel JB. Isolation of a gene encoding a functional zinc finger protein homologous to erythroid Kruppel-like factor: identification of a new multigene family. Mol Cell Biol. 1995;15:5957–5965. doi: 10.1128/mcb.15.11.5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartoszewska S, Kochan K, Madanecki P, Piotrowski A, Ochocka R, Collawn JF, Bartoszewski R. Regulation of the unfolded protein response by microRNAs. Cellular & molecular biology letters. 2013;18:555–578. doi: 10.2478/s11658-013-0106-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartoszewska S, Kochan K, Piotrowski A, Kamysz W, Ochocka RJ, Collawn JF, Bartoszewski R. The hypoxia-inducible miR-429 regulates hypoxia-inducible factor-1alpha expression in human endothelial cells through a negative feedback loop. Faseb J. 2015;29:1467–1479. doi: 10.1096/fj.14-267054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartoszewski R, Brewer JW, Rab A, Crossman DK, Bartoszewska S, Kapoor N, Fuller C, Collawn JF, Bebok Z. The unfolded protein response (UPR)-activated transcription factor X-box-binding protein 1 (XBP1) induces microRNA-346 expression that targets the human antigen peptide transporter 1 (TAP1) mRNA and governs immune regulatory genes. J Biol Chem. 2011;286:41862–41870. doi: 10.1074/jbc.M111.304956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartoszewski R, Hering A, Marszall M, Stefanowicz Hajduk J, Bartoszewska S, Kapoor N, Kochan K, Ochocka R. Mangiferin has an additive effect on the apoptotic properties of hesperidin in Cyclopia sp. tea extracts. PLoS ONE. 2014;9:e92128. doi: 10.1371/journal.pone.0092128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Ari Fuchs S, Lieder I, Stelzer G, Mazor Y, Buzhor E, Kaplan S, Bogoch Y, Plaschkes I, Shitrit A, Rappaport N, Kohn A, Edgar R, Shenhav L, Safran M, Lancet D, Guan-Golan Y, Warshawsky D, Shtrichman R. GeneAnalytics: An Integrative Gene Set Analysis Tool for Next Generation Sequencing, RNAseq and Microarray Data. Omics. 2016;20:139–151. doi: 10.1089/omi.2015.0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergers G, Benjamin LE. Tumorigenesis and the angiogenic switch. Nat Rev Cancer. 2003;3:401–410. doi: 10.1038/nrc1093. [DOI] [PubMed] [Google Scholar]
- Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome biology. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic acids research. 2008;36:D149–153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya R, Senbanerjee S, Lin Z, Mir S, Hamik A, Wang P, Mukherjee P, Mukhopadhyay D, Jain MK. Inhibition of vascular permeability factor/vascular endothelial growth factor-mediated angiogenesis by the Kruppel-like factor KLF2. J Biol Chem. 2005;280:28848–28851. doi: 10.1074/jbc.C500200200. [DOI] [PubMed] [Google Scholar]
- Birnbaum D. VEGF-FLT1 receptor system: a new ligand-receptor system involved in normal and tumor angiogenesis. Japanese journal of cancer research : Gann. 1995;86 inside cover. [PubMed] [Google Scholar]
- Brabletz S, Brabletz T. The ZEB/miR-200 feedback loop-a motor of cellular plasticity in development and cancer? Embo Rep. 2010;11:670–677. doi: 10.1038/embor.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks PC, Clark RA, Cheresh DA. Requirement of vascular integrin alpha v beta 3 for angiogenesis. Science. 1994;264:569–571. doi: 10.1126/science.7512751. [DOI] [PubMed] [Google Scholar]
- Chan YC, Khanna S, Roy S, Sen CK. miR-200b Targets Ets-1 and Is Downregulated by Hypoxia to Induce Angiogenic Response of Endothelial Cells. J Biol Chem. 2011;286:2047–2056. doi: 10.1074/jbc.M110.158790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YC, Roy S, Khanna S, Sen CK. Downregulation of Endothelial MicroRNA-200b Supports Cutaneous Wound Angiogenesis By Desilencing GATA Binding Protein 2 and Vascular Endothelial Growth Factor Receptor 2. Arteriosclerosis, Thrombosis, and Vascular Biology. 2012;32:1372–1382. doi: 10.1161/ATVBAHA.112.248583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S-H, Lu Y-C, Li X, Hsieh W-Y, Xiong Y, Ghosh M, Evans T, Elemento O, Hla T. Antagonistic Function of the RNA-binding Protein HuR and miR-200b in Post-transcriptional Regulation of Vascular Endothelial Growth Factor-A Expression and Angiogenesis. J Biol Chem. 2013;288:4908–4921. doi: 10.1074/jbc.M112.423871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Stahl A, Krah NM, Seaward MR, Dennison RJ, Sapieha P, Hua J, Hatton CJ, Juan AM, Aderman CM, Willett KL, Guerin KI, Mammoto A, Campbell M, Smith LE. Wnt signaling mediates pathological vascular growth in proliferative retinopathy. Circulation. 2011a;124:1871–1881. doi: 10.1161/CIRCULATIONAHA.111.040337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Wang L, Matyunina LV, Hill CG, McDonald JF. Overexpression of miR-429 induces mesenchymal-to-epithelial transition (MET) in metastatic ovarian cancer cells. Gynecologic oncology. 2011b;121:200–205. doi: 10.1016/j.ygyno.2010.12.339. [DOI] [PubMed] [Google Scholar]
- Choi YC, Yoon S, Jeong Y, Yoon J, Baek K. Regulation of vascular endothelial growth factor signaling by miR-200b. Mol Cells. 2011;32:77–82. doi: 10.1007/s10059-011-1042-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg MW, Lin Z, Fisch S, Jain MK. An emerging role for Kruppel-like factors in vascular biology. Trends in cardiovascular medicine. 2004;14:241–246. doi: 10.1016/j.tcm.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Ferrara N, Gerber HP, LeCouter J. The biology of VEGF and its receptors. Nat Med. 2003;9:669–676. doi: 10.1038/nm0603-669. [DOI] [PubMed] [Google Scholar]
- Greco S, Gaetano C, Martelli F. HypoxamiR regulation and function in ischemic cardiovascular diseases. Antioxidants & redox signaling. 2014;21:1202–1219. doi: 10.1089/ars.2013.5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco S, Martelli F. MicroRNAs in Hypoxia Response. Antioxidants & redox signaling. 2014;21:1164–1166. doi: 10.1089/ars.2014.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashiya N, Jo N, Aoki M, Matsumoto K, Nakamura T, Sato Y, Ogata N, Ogihara T, Kaneda Y, Morishita R. In vivo evidence of angiogenesis induced by transcription factor Ets-1: Ets-1 is located upstream of angiogenesis cascade. Circulation. 2004;109:3035–3041. doi: 10.1161/01.CIR.0000130643.41587.DB. [DOI] [PubMed] [Google Scholar]
- Janaszak-Jasiecka A, Bartoszewska S, Kochan K, Piotrowski A, Kalinowski L, Kamysz W, Ochocka RJ, Bartoszewski R, Collawn JF. miR-429 regulates the transition between Hypoxia-Inducible Factor (HIF)1A and HIF3A expression in human endothelial cells. Sci Rep-Uk. 2016;6 doi: 10.1038/srep22775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawanami D, Mahabeleshwar GH, Lin Z, Atkins GB, Hamik A, Haldar SM, Maemura K, Lamanna JC, Jain MK. Kruppel-like factor 2 inhibits hypoxia-inducible factor 1alpha expression and function in the endothelium. J Biol Chem. 2009;284:20522–20530. doi: 10.1074/jbc.M109.025346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic acids research. 2014;42:D68–73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic acids research. 2006;34:W451–454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CT, Veselits ML, Barton KP, Lu MM, Clendenin C, Leiden JM. The LKLF transcription factor is required for normal tunica media formation and blood vessel stabilization during murine embryogenesis. Genes & development. 1997;11:2996–3006. doi: 10.1101/gad.11.22.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larionov A, Krause A, Miller W. A standard curve based method for relative real time PCR data processing. BMC bioinformatics. 2005;6:62. doi: 10.1186/1471-2105-6-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li EH, Huang QZ, Li GC, Xiang ZY, Zhang X. Effects of miRNA-200b on the development of diabetic retinopathy by targeting VEGFA gene. Bioscience reports. 2017;37 doi: 10.1042/BSR20160572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Kumar A, SenBanerjee S, Staniszewski K, Parmar K, Vaughan DE, Gimbrone MA, Jr, Balasubramanian V, Garcia-Cardena G, Jain MK. Kruppel-like factor 2 (KLF2) regulates endothelial thrombotic function. Circulation research. 2005;96:e48–57. doi: 10.1161/01.RES.0000159707.05637.a1. [DOI] [PubMed] [Google Scholar]
- Love TM, Moffett HF, Novina CD. Not miR-ly small RNAs: big potential for microRNAs in therapy. The Journal of allergy and clinical immunology. 2008;121:309–319. doi: 10.1016/j.jaci.2007.12.1167. [DOI] [PubMed] [Google Scholar]
- Madanecki P, Kapoor N, Bebok Z, Ochocka R, Collawn JF, Bartoszewski R. Regulation of angiogenesis by hypoxia: the role of microRNA. Cellular & molecular biology letters. 2013;18:47–57. doi: 10.2478/s11658-012-0037-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malaguarnera L, Pilastro MR, Quan S, Ghattas MH, Yang L, Mezentsev AV, Kushida T, Abraham NG, Kappas A. Significance of heme oxygenase in prolactin-mediated cell proliferation and angiogenesis in human endothelial cells. Int J Mol Med. 2002;10:433–440. [PubMed] [Google Scholar]
- Mitchell TS, Bradley J, Robinson GS, Shima DT, Ng YS. RGS5 expression is a quantitative measure of pericyte coverage of blood vessels. Angiogenesis. 2008;11:141–151. doi: 10.1007/s10456-007-9085-x. [DOI] [PubMed] [Google Scholar]
- Roybal JD, Zang Y, Ahn YH, Yang YN, Gibbons DL, Baird BN, Alvarez C, Thilaganathan N, Liu DD, Saintigny P, Heymach JV, Creighton CJ, Kurie JM. miR-200 Inhibits Lung Adenocarcinoma Cell Invasion and Metastasis by Targeting Flt1/VEGFR1. Mol Cancer Res. 2011;9:25–35. doi: 10.1158/1541-7786.MCR-10-0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen B, Johnson FM. Regulation of SRC family kinases in human cancers. Journal of signal transduction. 2011;2011:865819. doi: 10.1155/2011/865819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SenBanerjee S, Lin Z, Atkins GB, Greif DM, Rao RM, Kumar A, Feinberg MW, Chen Z, Simon DI, Luscinskas FW, Michel TM, Gimbrone MA, Jr, Garcia-Cardena G, Jain MK. KLF2 Is a novel transcriptional regulator of endothelial proinflammatory activation. The Journal of experimental medicine. 2004;199:1305–1315. doi: 10.1084/jem.20031132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha M, Ghatak S, Roy S, Sen CK. microRNA–200b as a Switch for Inducible Adult Angiogenesis. Antioxidants & redox signaling. 2015;22:1257–1272. doi: 10.1089/ars.2014.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldi R, Mitola S, Strasly M, Defilippi P, Tarone G, Bussolino F. Role of alphavbeta3 integrin in the activation of vascular endothelial growth factor receptor-2. EMBO J. 1999;18:882–892. doi: 10.1093/emboj/18.4.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano JV, Liu N, Gao Y, Yao ZJ, Ishibashi T, Underhill C, Burke TR, Jr, Bottaro DP. Inhibition of angiogenesis by growth factor receptor bound protein 2-Src homology 2 domain bound antagonists. Molecular cancer therapeutics. 2004;3:1289–1299. [PubMed] [Google Scholar]
- Spearman C. The Proof and Measurement of Association between Two Things. The American Journal of Psychology. 1904;15:72–101. [PubMed] [Google Scholar]
- Staton AA, Giraldez AJ. Use of target protector morpholinos to analyze the physiological roles of specific miRNA-mRNA pairs in vivo. Nat Protoc. 2011;6:2035–2049. doi: 10.1038/nprot.2011.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzer G, Inger A, Olender T, Iny-Stein T, Dalah I, Harel A, Safran M, Lancet D. GeneDecks: Paralog Hunting and Gene-Set Distillation with GeneCards Annotation. Omics. 2009;13:477–487. doi: 10.1089/omi.2009.0069. [DOI] [PubMed] [Google Scholar]
- Summerton JE. Morpholino, siRNA, and S-DNA compared: impact of structure and mechanism of action on off-target effects and sequence specificity. Current topics in medicinal chemistry. 2007;7:651–660. doi: 10.2174/156802607780487740. [DOI] [PubMed] [Google Scholar]
- Takashima S, Kitakaze M, Asakura M, Asanuma H, Sanada S, Tashiro F, Niwa H, Miyazaki Ji J, Hirota S, Kitamura Y, Kitsukawa T, Fujisawa H, Klagsbrun M, Hori M. Targeting of both mouse neuropilin-1 and neuropilin-2 genes severely impairs developmental yolk sac and embryonic angiogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:3657–3662. doi: 10.1073/pnas.022017899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotech. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CH, Su PT, Du XY, Kuo MW, Lin CY, Yang CC, Chan HS, Chang SJ, Kuo C, Seo K, Leung LL, Chuang YJ. Thrombospondin type I domain containing 7A (THSD7A) mediates endothelial cell migration and tube formation. Journal of cellular physiology. 2010;222:685–694. doi: 10.1002/jcp.21990. [DOI] [PubMed] [Google Scholar]
- Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic acids research. 2015;43:D146–152. doi: 10.1093/nar/gku1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Sharma A, Trane A, Utokaparch S, Leung C, Bernatchez P. Myoferlin gene silencing decreases Tie-2 expression in vitro and angiogenesis in vivo. Vascular pharmacology. 2011;55:26–33. doi: 10.1016/j.vph.2011.04.001. [DOI] [PubMed] [Google Scholar]
- Zhang B, Day DS, Ho JW, Song L, Cao J, Christodoulou D, Seidman JG, Crawford GE, Park PJ, Pu WT. A dynamic H3K27ac signature identifies VEGFA-stimulated endothelial enhancers and requires EP300 activity. Genome research. 2013a;23:917–927. doi: 10.1101/gr.149674.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang BT, Zhang ZQ, Xia SY, Xing CS, Ci XP, Li X, Zhao RR, Tian S, Ma G, Zhu ZM, Fu LY, Dong JT. KLF5 Activates MicroRNA 200 Transcription To Maintain Epithelial Characteristics and Prevent Induced Epithelial-Mesenchymal Transition in Epithelial Cells. Mol Cell Biol. 2013b;33:4919–4935. doi: 10.1128/MCB.00787-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.