Abstract
Epigenetic processes have been implicated in the pathophysiology of alcohol dependence, but the specific molecular mechanisms mediating dependence-induced neuroadaptations remain largely unknown. Here, we found that a history of alcohol dependence persistently decreased the expression of Prdm2, a histone methyltransferase that monomethylates histone 3 at the lysine 9 residue (H3K9me1), in the rat dorsomedial prefrontal cortex (dmPFC). Downregulation of Prdm2 was associated with decreased H3K9me1, supporting that changes in Prdm2 mRNA levels affected its activity. Chromatin immunoprecipitation followed by massively parallel DNA sequencing showed that genes involved in synaptic communication are epigenetically regulated by H3K9me1 in dependent rats. In non-dependent rats, viral-vector-mediated knockdown of Prdm2 in the dmPFC resulted in expression changes similar to those observed following a history of alcohol dependence. Prdm2 knockdown resulted in increased alcohol self-administration, increased aversion-resistant alcohol intake and enhanced stress-induced relapse to alcohol seeking, a phenocopy of postdependent rats. Collectively, these results identify a novel epigenetic mechanism that contributes to the development of alcohol-seeking behavior following a history of dependence.
INTRODUCTION
Long-term neuroadaptations induced by chronic alcohol exposure are critical for the development of alcohol addiction (here-after equated with ‘alcoholism’), and can in part be modeled in experimental animals.1,2 Using these models, epigenetic processes have emerged as an important mechanism in the regulation of alcohol seeking and taking, as well as associated changes in gene expression profiles. For instance, inhibition of DNA methyltransferase and histone deacetylase (HDAC) was recently shown to prevent escalation of voluntary alcohol intake, and restored alcohol-induced gene expression changes.3,4 In the amygdala, ethanol exposure was associated with increased histone acetylation, whereas withdrawal was associated with decreased histone acetylation.5 Furthermore, treatment with the HDAC inhibitor trichostatin A prevented the development of alcohol withdrawal-related anxiety in Sprague—Dawley rats.6,7 Similar results were found with the selectively bred alcohol-preferring (P) rats. P rats showed higher HDAC2 expression associated with higher HDAC activity in the central amygdala. Moreover, HDAC2 knockdown in the central amygdala attenuated anxiety-like behaviors and voluntary drinking in P rats, suggesting a role of HDAC2 in anxiety and alcoholism.8
Although the role of DNA methylation and histone acetylation in neuroadaptations underlying alcohol-related behaviors has been well studied, little is known about the role of histone methylation and its related enzymes in this context. Recently, however, two studies demonstrated that the histone methyltransferase G9a may be involved in cocaine-induced plasticity and morphine-associated behaviors,9,10 suggesting that histone methylation may also be relevant for addiction.
In rats, a prolonged history of alcohol dependence results in a persistent increase in alcohol self-administration, increased aversion-resistant alcohol intake and increased propensity to relapse.2,11–15 These traits, collectively labeled ‘postdependent’, are to a large extent driven by processes in the dorsomedial prefrontal cortex (dmPFC).4,16,17 Alcohol vapor inhalation, as opposed to liquid diet administration, more readily leads to physical dependence, as indicated by withdrawal symptoms that peak between 12 and 24 h after cessation of alcohol exposure.18 In addition, prolonged chronic intermittent exposure to alcohol vapor induces neuroadaptations that persist beyond the acute withdrawal symptoms. We have previously shown that alcohol drinking is persistently increased 3 weeks after recovery from vapor exposure-induced dependence.15 At this same time point, we observed changes in DNA methylation and expression of genes related to synaptic plasticity pathways.4 Because the effects of acute alcohol withdrawal did not confound these findings, these studies showed that long-term neuroadaptations underlie enduring increases in alcohol consumption and vulnerability for relapse.
In the present study, we sought to test the hypothesis that histone-modifying enzymes control long-term gene expression changes that contribute to neuroadaptations and enduring alcohol-seeking behaviors in dependent animals. We focused on the dmPFC because of its prominent role in behavioral consequences of alcohol dependence,19 and our recent work indicating persistent dependence-induced reprogramming of gene expression in this region.4,16,20
We found that the histone methyltransferase PR domain containing 2, with ZNF domain (PRDM2, also called RIZ1), was dysregulated in the dmPFC of postdependent animals. PRDM2 is a tumor suppressor protein whose inactivation has an important role in several human cancers.21–25 Unlike some other histone methyltransferases, which can exhibit promiscuity for histone tail modification sites or degree of methylation at a particular site, PRDM2 specifically modifies histone H3 on lysine 9,26 where it preferentially catalyzes the addition of a single methyl group (histone 3 at the lysine 9 residue (H3K9me1)).27 One recent study reported an increase in acetylation specifically at H3K9 in the PFC of alcohol-exposed mice, suggesting that this histone tail residue, in particular, may be important for alcohol-induced gene expression changes.28 It has been shown that a dietary deficiency of the methyl donor S-adenosyl-methionine or treatment with the methyltransferase reaction product S-adenosyl-homocysteine inhibits PRDM2 histone methyltransferase activity, suggesting that PRDM2 activity can be regulated by environmental factors.26 Hypermethylation on the Prdm2 promoter is associated with decreased Prdm2 mRNA expression in cancer.29 Prdm2 downregulation is associated with poor prognosis in gliomas and neuroblastomas, suggesting that it could have a role in the brain.30,31 However, no studies, to date, have identified a role for this enzyme in psychiatric diseases.
Our previous study4 showed that postdependent neuroadaptations involved DNA methylation changes in the mPFC, and previous studies showed that Prdm2 is likely to be regulated by DNA methylation. Here, we hypothesized that downregulation of Prdm2 in the mPFC of postdependent rats may be one of the mechanisms through which DNA hypermethylation induces alcohol-seeking behavior. To address this hypothesis, we examine whether dysregulation of Prdm2 in the mPFC contributes to the long-term behavioral and molecular changes induced by a history of alcohol dependence.
MATERIALS AND METHODS
Animals
Male Wistar rats (200–225 g; Charles River, Wilmington, MA, USA) were housed under a reverse light cycle with food and water ad libitum, and were habituated to the facility and handled before experiments. Testing took place during the dark phase. Procedures were approved by the NIAAA Animal Care and Use Committee.
Dependence induction
Dependence was induced using chronic intermittent alcohol vapor exposure as described.32 Briefly, postdependent rats were exposed to alcohol vapor for 14 h each day (on at 19:30 hours, off at 09:30 hours) for 7 weeks, resulting in blood alcohol concentrations between ~200 and ~300mg dl−1. Controls were kept in identical chambers with normal air flow. Once weekly, blood was collected from the lateral tail vein. Blood alcohol concentrations were assessed using quantitative gas chromatography.16 All molecular and biochemical analyses in these studies were conducted using dorsomedial prefrontal cortex (dmPFC, coordinates relative to bregma: +2.76 to +3.72; Paxinos and Watson33) collected 3 weeks after the end of the last exposure to assess persistent, rather than intoxication- or withdrawal-related, effects of alcohol exposure.2,18
Surgery
For the lentiviral microinjection, rats received two injections bilaterally (1 μl per injection; rate: 0.25 μl min−1) directly into the dmPFC (coordinates relative to bregma: anterior–posterior: +2.5 and +3 mm, medial–lateral: ±0.7 mm, dorsal—ventral −3.5 mm; Paxinos and Watson33) of a lentiviral vector containing a short hairpin RNA (shRNA) targeting PRDM2 (5′-GGAGCCCAAGTCCTTGTAAAT-3′; titer: 9.7 × 109 TU ml−1; Sigma, St Louis, MO, USA; Supplementary Figure 1) and a scrambled control (titer: 2.9 × 109 TU ml−1). Rats were subjected to behavioral studies after a 1-week recovery period.
For the RG-108 experiments, rats received continuous infusion of RG-108 (Sigma; 100 pM dissolved in 5% 2-hydroxypropyl β-cyclodextrin (w v−1)) into the dmPFC (coordinates relative to bregma: anterior—posterior: +2.5 mm, medial-lateral: ± 1.5 mm, dorsal-ventral: −3.5 mm, 10°). The rats underwent surgery 2 weeks after the end of alcohol exposure using the osmotic minipumps 2004 (0.25 μl h−1; 28 days; Alzet, Cupertino, CA, USA) for dmPFC infusion.
RNA extraction
Bilateral samples from the dmPFC were dissected out as described,34 and stored at −80 °C. RNA was extracted from dmPFC using PureLink RNA Mini Kit (Life Technologies, Carlsbad, CA, USA), following the manufacturer’s instructions.
Quantitative real-time PCR
Total RNA was reverse transcribed into cDNA using qScript cDNA SuperMix (Quanta Biosciences, Gaithersburg, MD, USA). The reactions were run on a Veriti 96-well Fast Thermal Cycler (Life Technologies). Inventoried TaqMan Gene expression assay probes (Prdm2: Rn011516793_m1 and Gapdh: Rn4308313; Life Technologies) were used to assess the expression of rat target genes on an ABI 7900HT Fast Real-time PCR System (Life Technologies). Target gene expression was quantified with respect to glyceraldehyde 3-phosphate dehydrogenase using RQ Manager 1.2 (Life Technologies) by calculating an RQ value using 2−ΔΔCT analysis.35
Chromatin immunoprecipitation
Unless specified, reagents were purchased from Sigma. Starting with dmPFC from eight rats per group, two animals were pooled to generate sufficient starting material (~50 mg). This is the only instance of pooling in our studies. Subsequently, four samples per group were processed as individual biological replicates. After fixing the diced tissue in 1% formaldehyde for 15 min, the reaction was quenched with 0.125 M glycine for 5 min and centrifuged at 450 g for 5 min at 4 °C. Pellets were washed 2× in phosphate-buffered saline with Complete, EDTA-free Protease Inhibitor (CPI; Roche, Indianapolis, IN, USA) and suspended in cytoplasmic lysis buffer (10 mM Tris-HCl (pH 7.5), 10 mM NaCl, 0.2% NP-40, 1 × CPI). Cells were lysed with 25 strokes of a Dounce homogenizer (Kimble Chase, small clearance pestle) and centrifuged at 5500 g for 5 min at 4 °C. The nuclear pellet was resuspended in 450 μl of nuclear lysis buffer (50 mM Tris-HCl (pH 8.0), 10 mM EDTA, 1% sodium dodecyl sulfate, 1 × CPI). DNA was sheared to an average length of 400 bp using a Misonix S-4000 microtip probe sonicator (25% amplitude, Farmingdale, NY, USA) for 4 × 20 s pulses with 20 s rest (three total cycles). After centrifugation at 15 700 g for 5 min at 4 °C, the supernatant was diluted 10 times with chromatin immunoprecipitation (ChIP) dilution buffer (0.01% sodium dodecyl sulfate, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl (pH 8.1), 167 mM NaCl, 1 × CPI). Two percent of each sample (genomic DNA input) was saved. Prewashed Protein A Dynabeads (50 μl; Life Technologies) were bound to 4 μg of either normal rabbit immunoglobulin G (Millipore, Billerica, MA, USA) or rabbit anti-H3K9me1 (Abcam, Cambridge, MA, USA) antibodies. The bead—antibody complex was washed and suspended in sample overnight at 4 °C. Each IP complex was washed sequentially with the following solutions: 1 × low salt buffer (0.1% sodium dodecyl sulfate, 0.1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl (pH 8.0), 150 mM NaCl), 1 × high salt buffer (NaCl replaced with 500 mM), 1 × LiCl (Millipore) and 2 × TE buffer. DNA complexes were eluted by adding 150 μl elution buffer (1% sodium dodecyl sulfate, 0.1 M NaHCO3 and 0.2 M NaCl). Samples were heated at 65 °C for 4 h to reverse crosslinks, followed by the addition of 50 μl Proteinase K mix (10 μg Proteinase K, 0.04 M EDTA, 0.2 M Tris-HCl (pH 8.0)) at 42 °C for 2 h. DNA was extracted using the QIAquick PCR Purification Kit (Qiagen, Valencia, CA, USA). Use of the H3K9me1 antibody was validated by quantitative real-time PCR (qPCR) showing enrichment of the ChIP signal compared with immunoglobulin G (Figure 1a).
Figure 1.
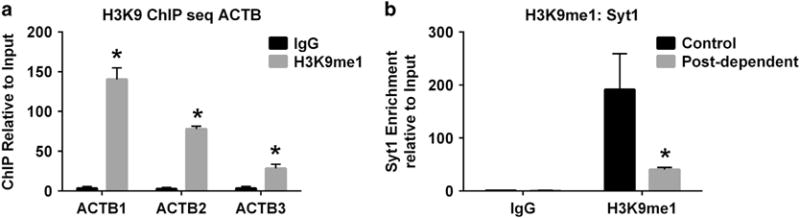
(a) Validation of the histone 3 at the lysine 9 residue (H3K9me1) antibody. (b) Validation of the bioinformatic pipeline using quantitative real-time PCR (qPCR) to verify differential H3K9me1 enrichment at the Syt1 gene in control versus postdependent rats. *P < 0.05.
DNA sequencing
H3K9me1 ChIP samples from control and postdependent rats, as well as each respective genomic DNA input samples (n = 4 per condition for a total of 16 samples), were processed for DNA sequencing by the Center for Genome Technology Sequencing Core (Hussman Institute for Human Genomics, University of Miami Miller School of Medicine, Miami, FL, USA). A High Sensitivity DNA Assay (Agilent Bioanalyzer 2100; Agilent Technologies, Santa Clara, CA, USA) was used to determine DNA concentration and shearing. Thirteen nanograms of input and ChIP-enriched DNA were used for 2 × 101 paired-end multiplexed library preparation using the NEBNext Ultra DNA Kit (New England Biolabs, Ipswich, MA, USA) and 13 cycles of PCR. The libraries were run on an Illumina HiSeq2000 using a PE rapid flow cell (San Diego, CA, USA).
Bioinformatic analysis
Read quality was evaluated using FastQC using default run parameters (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/). Reads were mapped against RGSC6.0 using Bowtie 2 with no mismatches in the seed sequence allowed.36 Average alignment was 94.3 ± 0.003% (mean ± s.e.m.; Supplementary Table 1), which is comparable to previous studies of ChIP-sequencing analyses in the brain.37,38 SICER (Spatial clustering for identification of ChIP-enriched region)39 was used for peak calling to capture the broad nature of H3K9 modifications.40 H3K9me1-enriched regions that exhibited statistically significant differences using a Benjamini-Hochberg false discovery rate correction between ChIP-control compared with ChIP-postdependent samples, but did not deviate between experimental conditions in the input samples were retained for further analysis. BEDTools41 was used to identify genes closest to and under enriched regions. We defined a ‘gene region’ to encompass 3000 bp upstream of the first exon genomic position through 500 bp downstream of the last exon genomic position (http://github.com/Bohdan-Khomtch-ouk/geneXtender). To assess the biological variability, edgeR42 was used to identify H3K9me1-enriched regions that exhibited significant differences in control as compared with postdependent samples (n = 4). The bioinformatic pipeline was initially validated using qPCR to verify differential H3K9me1 enrichment at the Syt1 gene in control versus postdependent rats (Figure 1b). As we previously reported for RNA-sequencing (RNA-seq) analysis,4 we used a nominal significance cutoff (p/q-value < 0.05, fold change > 1.2, n = 4) for our ChIP-seq analysis. WebGestalt gene ontology analysis was conducted using the default settings with the Rattus norvegicus genome (Rn6) as the reference background.43 Selected genes were visualized in the Integrative Genomics Viewer (Cambridge, MA, USA).44,45
Western blotting
Tissue was homogenized in lysis buffer (RIPA buffer, DTT; Cell Signaling, Danvers, MA, USA). Supernatant containing proteins was collected after 10 min centrifugation (10 000 g at 4°C). Protein concentration was assessed using the Pierce BCA Protein Assay Kit (Thermo Scientific, Rochester, NY, USA). Protein samples were denaturated at 70 °C for 10 min and run on a 4–12% Bis-Tris gel (NuPAGE Novex; Life Technologies, Grand Island, NY, USA), and then transferred to a polyvinylidene fluoride membrane (Millipore). The membrane was blocked with 5% nonfat dry milk and incubated overnight with the primary antibody (rabbit anti-H3K9me1 (1/1000; Abcam) and rabbit anti-H3 (1/1000; Cell Signaling). The membrane was washed with TBST and then incubated with secondary antibody anti-rabbit HRP (1:10 000; Cell Signaling) for 1 h. Detection and densitometric evaluations were performed using the ECL western blotting detection reagent (GE Healthcare, Piscataway, NJ, USA) and ImageJ software (Bethesda, MD, USA).
Laser capture microscopy and nanostring nCounter analysis
Arcturus XT (Life Technologies, Carlsbad, CA, USA) was used to perform laser capture microdissection. Cells showing lentiviral vector infection were captured by hand and transferred onto a cap (Capsuret Macro LCM Caps, Life Technologies, catalog number LCM0211) using a near-infrared laser pulse. RNA was extracted from caps using Single-Cell Lysis Kit following the manufacturer’s instruction (Life Technologies, Grand Island, NY, USA). Given the very low amount of RNA collected, we measured mRNA expression using the nCounter Single-Cell Gene Expression assay following the manufacturer’s instruction (Nanostring Technologies, Seattle, WA, USA).
Overview of behavioral testing
Rats underwent a battery of alcohol-associated behavioral tests. First, they were tested for operant alcohol self-administration. This was followed by testing for aversion-resistant alcohol-seeking using quinine adulteration.46 Finally, baseline self-administration was re-established over a week after the last quinine test, extinguished, and rats were tested for stress-induced reinstatement of alcohol seeking.47–49
Alcohol self-administration
Training and testing for operant self-administration of 10% alcohol in water were as described.50 Briefly, rats were trained to self-administer alcohol under a fixed ratio one with a 5 s timeout. Once self-administration rates were stable, rats received a microinjection of PRDM2-targeting or scrambled lentiviral vector directly into the dmPFC, and were allowed to recover for 1 week. Rats were then tested for alcohol self-administration after 1-week recovery.
Quinine adulteration
Rats were assessed for aversion-resistant alcohol consumption, a hallmark of compulsivity-like behavior. Alcohol was mixed with increasing concentration of quinine (0.005, 0.01, 0.025, 0.05 and 0.075 g l−1) during alcohol self-administration (one test session per quinine concentration). Compulsivity-like behavior was assessed by measuring % decreased reward after the addition of quinine.
Stress-induced reinstatement
The experiment was performed as described previously.50 Briefly, extinction sessions started once self-administration rates were stable. During extinction, alcohol was no longer available, and operant responses had no programmed consequences. Stress-induced reinstatement testing started after operant responding was extinguished (< 10 active lever presses/30 min session). Rats were tested four times for four different shock intensities (0.2, 0.4, 0.6 and 0.8 mA) in 15 min sessions. Each test was separated by 3 extinction days.
Statistical analysis
All data results were analyzed using analysis of variance (ANOVA), with factors for the respective analysis indicated in conjunction with its results. When appropriate, post hoc comparisons were performed using Newman—Keuls test. Hypergeometric probabilities were calculated using an online tool: https://www.geneprof.org/GeneProf/tools/hypergeometric.jsp. The accepted level of significance for all tests was P < 0.05.
RESULTS
Expression of several epigenetic enzymes is persistently altered following a history of alcohol dependence
Using whole transcriptome sequencing analysis, we previously identified 784 genes with nominally significant persistent expression changes within the dmPFC after a history of alcohol dependence.4 Within this list, we found 22 genes that encode enzymes involved in histone methylation and acetylation, DNA methylation, bromodomain, chromodomain and PHD domain-containing reader proteins, as well as enzymes involved in chromatin remodeling (Table 1). These findings suggest that repeated cycles of alcohol intoxication followed by 3 weeks of protracted abstinence disrupt epigenetic processes.
Table 1.
Genes with known chromatin remodeling functions that were dysregulated according to RNA-seq data from the dmPFC of control versus postdependent rats
| Gene symbol | Epigenetic class | Target site or gene function | Fold change | P-value |
|---|---|---|---|---|
| Smarce1 | Chromatin remodeling | SWI/SNF complex | −3.02 | 0.0003 |
| Brd4 | Bromodomain reader | H4K5/8/12/16 | −1.95 | 0.006 |
| Tet1 | DNA demethylase | 5 hmc | −2.06 | 0.007 |
| Phf2 | Histone demethylase | H3K9me2 | −1.95 | 0.012 |
| Trim33 | Bromodomain reader | E3 Ub ligase, H3K9me3, H3K18ac | 1.35 | 0.016 |
| Brpf1 | Histone acetyltransferase | H2AK5, H4K12, H3K14 | −1.55 | 0.017 |
| Chd8 | Chromodomain reader | DNA helicase | −1.36 | 0.017 |
| Srcap | Chromatin remodeling | SWI/SNF complex, H2AZ/H1AZ exchange | −1.69 | 0.020 |
| Hdac7 | Histone deacetylase | Class IIa HDAC | −2.44 | 0.023 |
| Brd3 | Bromodomain reader | H4K5/8/12/16 | −1.48 | 0.024 |
| Mll5 | Histone methyltransferase | H3K4 | −1.39 | 0.026 |
| Dot1l | Histone methyltransferase | H3K79 | −2.09 | 0.026 |
| Tet3 | DNA demethylase | 5 hmc | −1.73 | 0.027 |
| Dpf2 | PhD domain reader | Acetylated histones | −1.53 | 0.027 |
| Kdm6b | Histone demethylase | H3K27 | −2.33 | 0.029 |
| Chmp1b | Chromatin remodeling | ESCRT-II complex | 1.41 | 0.030 |
| Prdm2 | Histone methyltransferase | H3K9me1 | −1.51 | 0.032 |
| Chd4 | Chromodomain reader | NuRD chromatin remodeling complex | −1.49 | 0.039 |
| Ino80 | Chromatin remodeling | SNF2/SWI2 helicase family | −1.35 | 0.040 |
| Myst3 | Histone acetyltransferase | H3 acetylation | −1.77 | 0.041 |
| Ep300 | Histone acetyltransferase | All core histones, non-histone targets | −1.33 | 0.042 |
| Prmt5 | Histone methyltransferase | H3R8, H4R3, H2A, non-histone targets | 1.31 | 0.050 |
Abbreviation: dmPFC, dorsomedial prefrontal cortex; RNA-seq, RNA-sequencing.
A history of alcohol dependence decreases the expression and function of the histone methyltransferase PRDM2
Although no studies have implicated a function for PRDM2 in the brain outside of cancer, microarray analysis of various human tissues suggested that Prdm2 is strongly enriched in the PFC and other brain regions compared with peripheral tissues (http://biogps.org;Figure 2a). Prdm2 enrichment in the brain was confirmed using qPCR analysis of various tissues isolated from adult mouse (Figures 2b and c). Given its enrichment in the PFC, we selected PRDM2 as a candidate for confirmation and functional validation.
Figure 2.
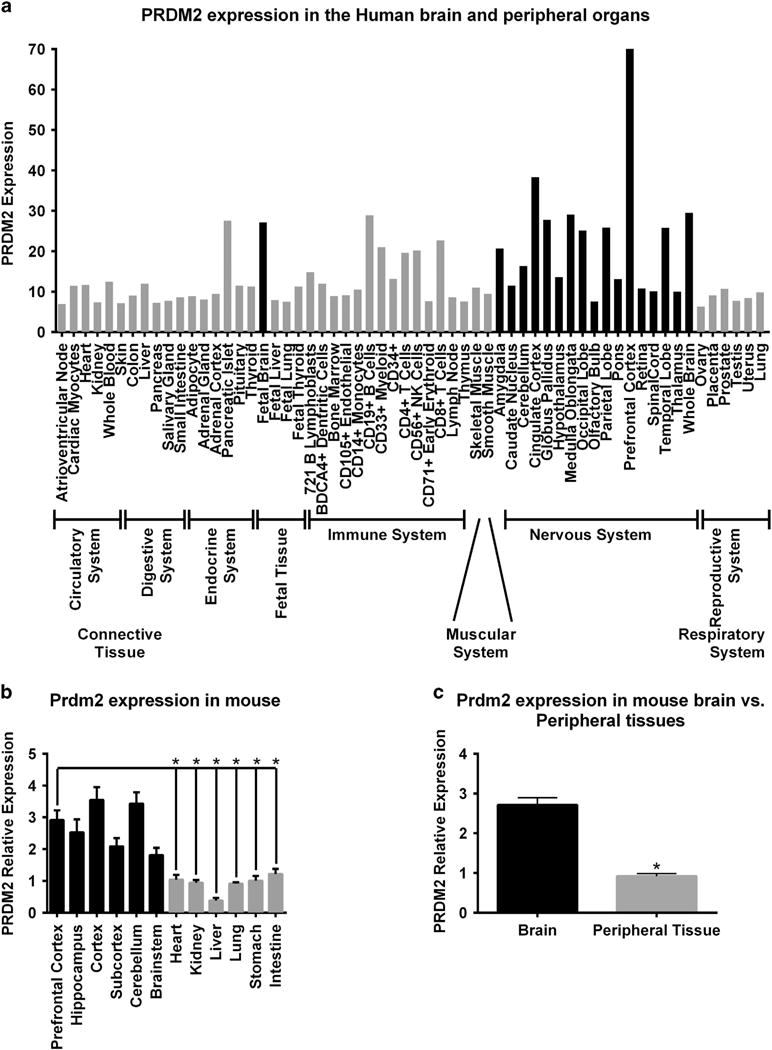
Prdm2 expression in the brain and peripheral organs. (a) Human Affymetrix microarray data (http://www.biogps.org). (b and c) Quantitative real-time PCR (qPCR) of adult mouse tissues. The central nervous system tissue is shown in black bars and the peripheral tissues are shown in gray bars. *P < 0.05.
qPCR analysis confirmed the downregulation of Prdm2 expression in the dmPFC of postdependent rats (Table 1 and Figure 3a). Prdm2 expression in the nucleus accumbens was not affected (scrambled: 100 ± 9.4% of control;Prdm2: 97.6 ± 8.6% of control; P > 0.05), suggesting a region-specific effect. The decreased expression of Prdm2 in postdependent dmPFC was restored by the DNA methyltransferase inhibitor RG-108 (Figure 3a;two-way ANOVA showed a significant effect of alcohol: F(1–29) = 6.326, P = 0.02 and a significant interaction between alcohol and treatment: F(1–29) = 8.177, P = 0.008; n = 6–10 per group). Although the baseline levels of Prdm2 expression in control vehicle and RG-108-treated groups appeared to be slightly different, this was not statistically different (P = 0.2). As RG-108 was previously found to rescue both molecular and behavioral consequences of alcohol dependence in postdependent rats,4 this suggests that alcohol-induced Prdm2 downregulation in the dmPFC may be driven by DNA hypermethylation. MeDIP analysis revealed no difference in DNA methylation level in the promoter region of Prdm2 in the dmPFC of postdependent compared with control rats (Supplementary Figure 2). Taken together, these results suggest that alcohol-induced DNA hypermethylation downregulates Prdm2 expression through intermediate molecular mechanisms.
Figure 3.
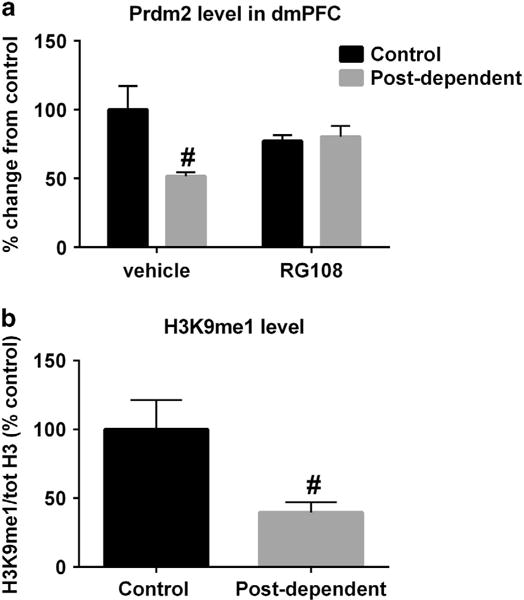
(a) Repeated cycles of alcohol intoxication decreased mRNA levels of Prdm2. The DNA methyltransferase (DNMT) inhibitor RG-108 significantly restored levels of Prdm2 (n = 6–10;two-way analysis of variance (ANOVA): main effect of group;control versus postdependent;F(1–29) = 6.3; P = 0.02; post hoc analysis control vehicle versus postdependent vehicle; P = 0.008; No significant difference between postdependent RG-108 versus control RG-108 or postdependent RG-108 versus control vehicle). The mean values (± s.e.m.) of control rats are shown in black bars. The mean values (± s.e.m.) of postdependent rats are shown in gray bars. mRNA expression is presented as % change from control/vehicle values. (b) Western blot analysis shows that monomethylation at H3K9 (H3K9me1) is significantly reduced in the dorsomedial prefrontal cortex (dmPFC) of postdependent rats (n = 7 per group;one-way ANOVA;main effect of group;control versus postdependent; F(1–12) = 7.1; P < 0.05). #P < 0.05, control versus postdependent rats.
We then examined whether Prdm2 downregulation in the dmPFC of postdependent rats has functional consequences. Western blot analysis confirmed decreased monomethylation at H3K9 (H3K9me1) relative to total histone 3 levels in the dmPFC of postdependent compared with control rats (Figure 3b; one-way ANOVA showed a significant effect of alcohol, F(1–12) = 7.1; P < 0.05; n = 7 per group). Total levels of histone 3 did not differ between the groups (P = 0.5). This supports the notion that the downregulation of Prdm2 expression was also associated with decreased function.
PRDM2 is selectively expressed in neurons in dmPFC
The expression of Prdm2 in the normal brain has not previously been investigated. In situ hybridization using RNAscope technology revealed that Prdm2 expression was colocalized with the neuronal marker NeuN/RbFox3 (Figures 4a–c). Prdm2 was not expressed in cells that did not express NeuN/RbFox3, suggesting that Prdm2 is specifically expressed in neurons.
Figure 4.
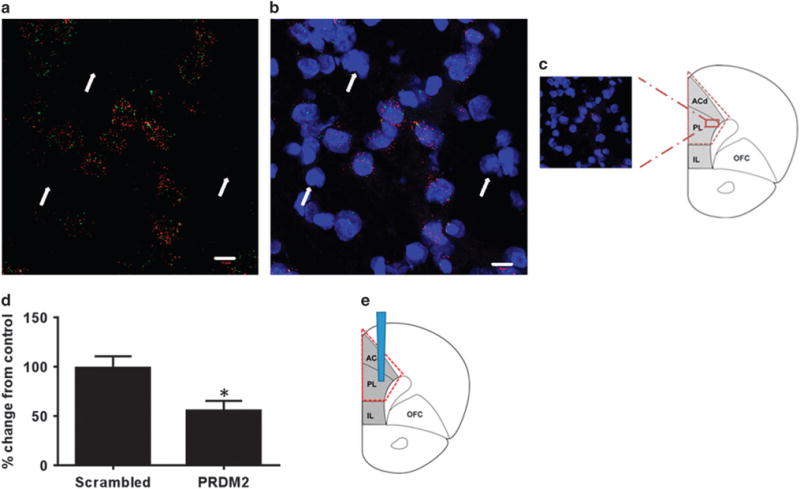
(a–c) Expression of PRDM2 in rat dorsomedial prefrontal cortex (dmPFC) measured by RNAscope. Red dots show the expression of the neuronal marker RbFox3, and the green dots show the expression of Prdm2 and blue labeling represents DAPI (4′,6-diamidino-2-phenylindole) staining. (d) Prdm2 mRNA expression is decreased after Prdm2 knockdown. (e) Site of virus injection. *P < 0.05; Scrambled versus PRDM2.
ChIP-seq identifies genomic loci regulated by H3K9me1 in alcohol dependence
To understand the persistent consequences of Prdm2 downregulation that may confer susceptibility to alcohol dependence, we used ChIP followed by massively parallel DNA sequencing (ChIP-seq). Rather than isolating DNA directly associated with the PRDM2, whose dynamic interaction with target genes is subject to enzyme kinetics, we chose to examine more stable histone methylation changes that are functionally altered as a consequence of the known enzymatic activity of PRDM2. Because PRDM2 specifically monomethylates H3K9,26,27 we used ChIP-seq to identify genomic loci subject to differential enrichment of H3K9me1 in the dmPFC of postdependent rats.
ChIP-seq analysis identified 3449 regions that exhibited decreased enrichment of H3K9me1 in postdependent rats. Only 1350 regions were more enriched in postdependent rats compared with controls, confirming an alcohol-induced global decrease of H3K9me1 consistent with PRDM2 downregulation (Supplementary Table 2 and Figure 5a). To minimize the potential for false positives and narrow our focus on H3K9me1 target genes that are functionally dysregulated in alcohol dependence, we overlaid our ChIP-seq findings with RNA-seq analysis of control versus postdependent dmPFC4 (Figure 5b). We found 156 regions within 119 genes that were less enriched for H3K9me1 in postdependent rats and were also dysregulated by RNA-seq analysis (Supplementary Table 3 and Figure 5b). The majority of these genes (79%) were downregulated in postdependent rats (Supplementary Table 3). The degree of overlap between these two approaches was significantly more than that which would be expected by chance (hypergeometric distribution test P = 4.9 × 10−23), suggesting that H3K9me1 is an essential component of alcohol-induced transcriptional dysregulation.
Figure 5.
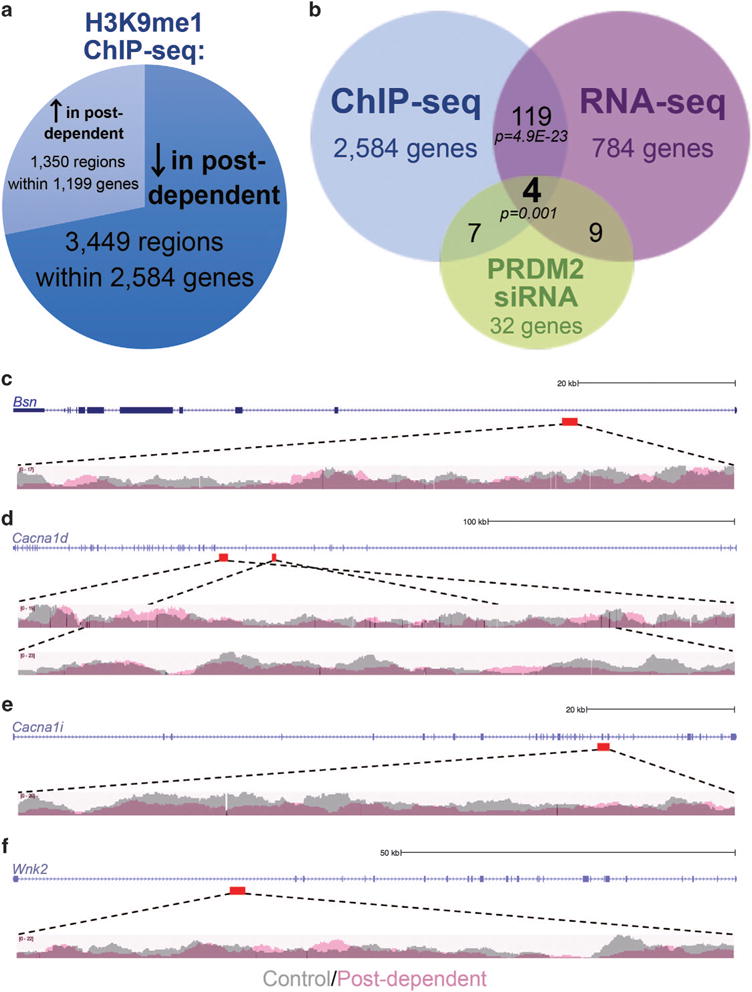
Tripartite approach implicates high confidence targets of PRDM2 in alcohol dependence. (a) Chromatin immunoprecipitation-sequencing (ChIP-seq) analysis identified genomic regions subjected to histone 3 at the lysine 9 residue (H3K9me1) epigenetic regulation in control versus postdependent dorsomedial prefrontal cortex (dmPFC). (b) Overlay of ChIP-seq and RNA-sequencing (RNA-seq) analysis from control versus postdependent dmPFC identified 119 common genes. These analyses were further overlaid with NanoString gene expression analysis from rat mPFC subjected to PRDM2 short hairpin RNA (shRNA) compared with a scramble control (hypergeometric distribution test P = 0.001). (c–f) ChIP-seq data from the four common genes implicated in all three approaches. Top tracks show the UCSC gene relative to the scale shown at the right. The location of the region of differential H3K9me1 enrichment is denoted by the red box below the gene track. The bottom tracks show the Spatial clustering for identification of ChIP-enriched region (SICER)-filtered read enrichment of H3K9me1 ChIP data for the control rats (gray) and postdependent rats (light red) overlaid.
Consistent with our results showing neuronal-specific Prdm2 expression, cellular component analysis of these 119 overlapping genes revealed a significant enrichment of gene ontologies related to neuronal localization (coated vesicle, dendrite, neuron projection, excitatory synapse; Table 2). This analysis further implicated H3K9me1-mediated regulation of genes within the synaptic compartment (coated vesicle, cell junction, voltage-gated calcium channel complex, excitatory synapse, etc.). The molecular function analysis implicated regulation of calcium channel activity (calcium ion transmembrane transporter activity, calcium: cation antiporter activity, divalent inorganic cation transmembrane transporter activity, calcium channel activity, voltage-gated channel activity, etc.).
Table 2.
GO functional enrichment analysis of 119 overlapping genes implicated in H3K9me1 ChIP-seq and RNA-seq analysis of post-dependent rat dmPFC.
| Category | GO | GO ID | Number of hit genes versus expected number | Ratio of enrichment | P-value |
|---|---|---|---|---|---|
| Cellular component | Coated vesicle | GO:0030135 | 6/1.33 | 4.51 | 0.00005 |
| Dendrite | GO:0030425 | 8/2.77 | 2.89 | 0.0001 | |
| Cell junction | GO:0030054 | 11/4.10 | 2.69 | 0.0002 | |
| Golgi apparatus part | GO:0044431 | 7/2.26 | 3.10 | 0.0002 | |
| Voltage-gated calcium channel complex | GO:0005891 | 2/0.15 | 13.67 | 0.0002 | |
| Golgi stack | GO:0005795 | 4/0.56 | 7.15 | 0.0003 | |
| Neuron projection | GO:0043005 | 14/5.21 | 2.69 | 0.0004 | |
| Organelle subcompartment | GO:0031984 | 3/0.41 | 7.37 | 0.0004 | |
| Excitatory synapse | GO:0060076 | 2/0.15 | 13.10 | 0.0004 | |
| Trans-Golgi network transport vesicle | GO:0030140 | 2/0.13 | 14.98 | 0.0004 | |
| Molecular function | Methylcytosine dioxygenase activity | GO:0070579 | 2/0.03 | 75.32 | 0.0003 |
| SH3 domain binding | GO:0017124 | 5/0.64 | 7.85 | 0.0004 | |
| Calcium ion transmembrane transporter activity | GO:0015085 | 5/0.64 | 7.76 | 0.0005 | |
| Calcium:cation antiporter activity | GO:0015368 | 2/0.05 | 43.04 | 0.0009 | |
| Divalent inorganic cation transmembrane transporter activity | GO:0072509 | 5/0.82 | 6.12 | 0.0014 | |
| Protein domain-specific binding | GO:0019904 | 11/3.79 | 2.90 | 0.0014 | |
| Calcium channel activity | GO:0005262 | 4/0.54 | 7.44 | 0.0021 | |
| Inositol phosphate phosphatase activity | GO:0052745 | 2/0.09 | 23.17 | 0.0032 | |
| Phosphatase activity | GO:0016791 | 6/1.53 | 3.91 | 0.0044 | |
| Voltage-gated channel activity | GO:0022832 | 5/1.10 | 4.54 | 0.0050 | |
| Biological process | Glomerulus vasculature morphogenesis | GO:0072103 | 2/0.03 | 60.85 | 0.0007 |
| Positive regulation of smooth muscle cell proliferation | GO:0048661 | 5/0.39 | 12.68 | 0.0022 | |
| Artery development | GO:0060840 | 4/0.34 | 11.93 | 0.0024 | |
| Aorta morphogenesis | GO:0035909 | 3/0.11 | 26.84 | 0.0026 | |
| Regulation of smooth muscle cell migration | GO:0014910 | 4/0.26 | 15.60 | 0.0065 | |
| Muscle cell migration | GO:0014812 | 4/0.30 | 13.23 | 0.0075 | |
| Comma-shaped body morphogenesis | GO:0072049 | 2/0.03 | 60.85 | 0.0078 | |
| Regulation of smooth muscle cell proliferation | GO:0048660 | 5/0.63 | 7.92 | 0.0078 | |
| Artery morphogenesis | GO:0048844 | 4/0.28 | 14.15 | 0.0093 | |
| Negative regulation of chromatin silencing | GO:0031936 | 2/0.03 | 60.85 | 0.0101 |
Abbreviations: dmPFC, dorsomedial prefrontal cortex; GO, gene ontology; H3K9me1, histone 3 at the lysine 9 residue; RNA-seq, RNA-sequencing.
Prdm2 knockdown mimics dependence-induced gene expression changes in dmPFC
To identify the signaling pathways functionally affected by Prdm2 dysregulation, we used laser capture microdissection, and specifically isolated cells transduced with Prdm2 shRNA or scrambled control lentivirus in the dmPFC of non-dependent rats. Nanostring nCounter gene expression analysis51 of 340 genes previously found to be dysregulated in the dmPFC of postdependent rats4 or that have been strongly implicated in alcohol dependence in the literature confirmed knockdown of Prdm2 in the dmPFC of ‘PRDM2’ compared with scrambled rats (Figures 4d and e). There were 32 genes that were significantly affected by Prdm2 knockdown (Supplementary Table 4; n = 8–9); nine of these overlapped with RNA-seq analysis from alcohol-dependent rats (n = 8–9; Table 3).
Table 3.
Overlapping dmPFC gene expression changes caused by a history of alcohol dependence (RNA-seq data from PD versus CTL), as well as knockdown of PRDM2 (n = 8–9)
| Gene name | RNA-seq: PD versus CTL
|
PRDM2 shRNA versus scramble
|
H3K9me1 ChIP: PD versus CTL
|
||||
|---|---|---|---|---|---|---|---|
| Fold change | P-value | Fold change | P-value | Fold change | P-value | Region of enrichment | |
| Abcd3 | 1.27 | 0.03 | −17.5 | 0.02 | |||
| Bsn | −1.52 | 0.03 | −143.50 | 0.04 | −1.32 | 0.008 | Intron 1/11 |
| −2.21 | 0.02 | Intron 1/11 | |||||
| Cacna1d | −1.66 | 0.02 | −5.56 | 0.01 | −1.27 | 0.005 | Intron 8/50 |
| −1.51 | 0.006 | Intron 9/50 | |||||
| Cacna1i | −1.71 | 0.01 | −6.75 | 0.03 | −1.27 | 0.0006 | Introns 25/28–27/38 |
| Helz | −1.53 | 0.01 | −4.82 | 0.04 | |||
| Ncor2 | −1.78 | 0.01 | −3.74 | 0.02 | |||
| Nop14 | 1.42 | 0.02 | −8.27 | 0.01 | |||
| Rab3c | 1.41 | 0.02 | 62.73 | 0.01 | |||
| Wnk2 | −1.99 | 0.008 | −11.55 | 0.004 | −2.06 | 0.01 | Intron 1/26 |
Abbreviations: ChIP, chromatin immunoprecipitation; CTL, control; dmPFC, dorsomedial prefrontal cortex; H3K9me1, histone 3 at the lysine 9 residue; PD, postdependent; shRNA, short hairpin RNA. Several of these genes may be direct targets of PRDM2, since EdgeR significance analysis of ChIP-seq data identified differential enrichment of H3K9me1 at the indicated genomic loci in the context of alcohol dependence (n = 4).
Of the overlapping genes affected by Prdm2 knockdown and dysregulated in alcohol-dependent rats; four also exhibited regions of differential H3K9me1 enrichment between control and postdependent rats in the ChIP-seq analysis (Table 3 and Figures 5b–f), suggesting that these genes may be directly regulated by PRDM2 through methylation at H3K9. Again, this overlap was significantly more than would be expected by chance (hypergeometric distribution test P = 0.001), suggesting a true biological enrichment of high confidence PRDM2 target genes in alcohol dependence. Many of the regions of differential H3K9me1 enrichment lie within intron/exon boundaries or intronic regions of the target genes (Table 3 and Figures 5c–f).
Lentiviral knockdown of Prdm2 expression in dmPFC of non-dependent rats mimics the behavioral consequences of alcohol dependence
To investigate a possible causal role of PRDM2 in alcohol-related behaviors, we examined non-dependent animals injected with the shRNA lentiviral vector targeting Prdm2 in the dmPFC, and assessed them for alcohol self-administration, aversion-resistant alcohol consumption and stress-induced reinstatement of alcohol seeking, a model of relapse47,48 (Figure 6). Prdm2 knockdown induced a range of behaviors characteristic of protracted abstinence following a prolonged history of alcohol dependence. First, we observed a modest but characteristic increase in operant alcohol self-administration (one-way ANOVA, main effect of group; scrambled versus PRDM2 rats; Figures 6a and b; n = 22–20 per group; F(1,40) = 5,9; P < 0.02). Second, Prdm2 knockdown increased resistance to quinine adulteration, suggesting a role of Prdm2 in compulsive alcohol drinking (repeated-measures ANOVA: main effect of group; scrambled versus PRDM2 rats, Figure 6b; n = 9–10 per group; F(1,48) = 7; P = 0.02). Third, rats with suppressed Prdm2 expression showed increased sensitivity and response to stress-induced reinstatement (repeated-measures ANOVA: main effect of group, scrambled versus PRDM2 rats; Figure 6c; n = 9–10, F(1,84) = 9.3; P = 0.006). Prdm2 knockdown did not alter locomotor activity or sensitivity to shock intensity, showing that increased stress-induced reinstatement after inhibition of PRDM2 was not caused by altered locomotion or pain sensitivity. Finally, Prdm2 knockdown did not affect sucrose self-administration (Figure 6d–h), suggesting that PRDM2 specifically mediates alcohol-seeking behavior.
Figure 6.
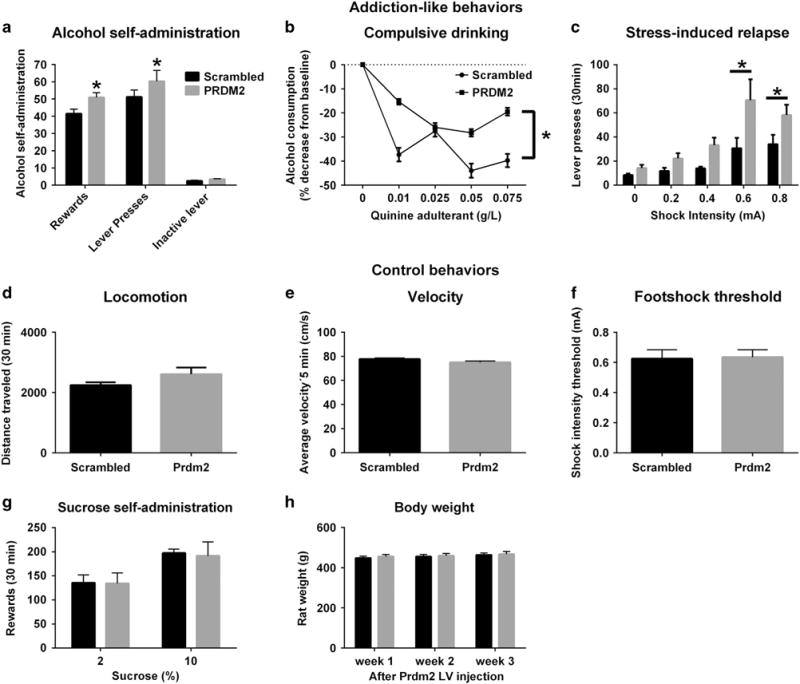
Lentiviral-mediated knockdown of Prdm2 in the dmPFC alters addiction-like behaviors (a–c): Voluntary alcohol-self-administration (a) (One way ANOVA: main effect of group;scrambled versus PRDM2 rats;F(1,40) = 5.9; P < 0.02), compulsive drinking in a quinine adulteration task (b) (Repeated-measure ANOVA: main effect of group;Scrambled versus PRDM2 rats;F(1,48) = 7; P = 0.02), and stress-induced relapse (c) (Repeated-measure ANOVA;main effect of group;scrambled versus PRDM2 rats F(1,84) = 9.3; P = 0.006). Lentiviral knockdown of Prdm2 did not affect control behaviors (d–h) such as locomotor activity (d), average velocity (e), sensitivity to shock intensity (f), sucrose self-administration (g), nor rat weight (h). The mean values (± s.e.m.) of scrambled rats are shown in black bars. The mean values (± s.e.m.) of PRDM2 knockdown rats are shown in gray bars (n = 9–10). *p < 0.05, scrambled versus PRDM2 rats.
DISCUSSION
We report that a history of alcohol dependence results in persistently decreased expression of the H3K9 histone methyltransferase PRDM2 in the dmPFC of rats. Decreased expression of Prdm2 was associated with decreased levels of the H3K9me1 mark, supporting the notion that downregulated expression of Prdm2 results in decreased enzymatic activity. Our molecular analysis suggested that some of the gene expression changes observed after history of alcohol dependence are likely driven by Prdm2 downregulation. Most importantly, behavioral analysis demonstrated that a viral-vector-mediated knockdown of Prdm2 in the dmPFC of rats without a history of dependence resulted in several phenotypic traits that mimicked the consequences of a dependence history, and that are thought to be characteristic of alcohol addiction. Collectively, these findings support a role of Prdm2 downregulation as a causal factor in alcohol-induced neuroadaptations.
Previous data from whole transcriptome analysis suggest that a history of alcohol dependence disrupts epigenetic processes.4 Epigenetic mRNAs dysregulated in alcohol-dependent rats oversee diverse epigenetic functions, including histone acetylation, histone methylation and non-enzymatic ‘reader’ function, suggesting a possible interplay between several epigenetic processes that takes place to regulate concerted and persistent gene expression changes observed after a history of alcohol dependence. A majority of the epigenetic enzymes were downregulated after chronic intermittent alcohol exposure. This is consistent with prior work, which has shown that a majority of dysregulated transcripts in the mPFC are downregulated after a history of alcohol dependence in human52 and postdependent rats.4 This observation could reflect a coordinated epigenetic network that represses global gene transcription through DNA methylation. Accordingly, we previously found that DNA methylation in postdependent rats is persistently increased in the mPFC.4
The wide-ranging dysregulation observed poses a considerable challenge for demonstrating a causal contribution by any individual enzyme to the behavioral phenotype induced by a history of dependence. Here, we focused on PRDM2, and examined its possible mechanistic contribution to several key behaviors induced by alcohol dependence. Previous studies suggest that Prdm2 expression and its histone methyltransferase activity are dependent on the level of promoter DNA methylation.26,29 Because alcohol-induced DNA hypermethylation has been shown to be involved in increased alcohol drinking,3,4 we hypothesized that Prdm2 might contribute to gene expression and behavioral changes underlying alcohol addiction. Accordingly, we found that the DNA methyltransferase inhibitor RG-108 directly infused into the dmPFC restored the levels of Prdm2 in the postdependent rats, demonstrating a regulatory effect of DNA methylation on Prdm2 levels. However, we did not observe changes in DNA methylation level in the promoter region of Prdm2 itself following a history of dependence. This indicates an indirect regulation of Prdm2 by alcohol-induced hypermethylation. Further investigation will be needed to identify the intermediate steps involved in this regulatory mechanism.
It has been shown that PRDM2 specifically monomethylates histone H3 on its lysine 9 residue.27 Both western blot and ChIP-seq analysis showed that H3K9me1 is decreased following a history of alcohol dependence. Our initial expression analysis only identified two H3K9-targeted epigenetic enzymes, Prdm2 and Phf2, as being altered in control versus postdependent rats. Because Phf2 is a histone demethylase, the observed downregulation of Phf2 in postdependent rats would predict increased H3K9 methylation, which was not consistent with our observations. Our findings do not preclude the potential for involvement of other H3K9-targeted enzymes. However, the fact that Prdm2 knockdown is sufficient to mimic several key behavioral and transcriptional characteristics of postdependent animals suggests that changes in activity of this enzyme may be a primary mechanism driving a transition into alcohol dependence.
Viral knockdown of Prdm2 expression in the present study resulted in behaviors characteristic of rats with a history of dependence. For instance, rats with decreased expression of Prdm2 in the dmPFC showed increased alcohol self-administration and increased aversion-resistant alcohol intake, as shown by increased resistance to quinine adulteration. Continued drug seeking despite aversive consequences, frequently referred to as ‘compulsive drug seeking’, is increasingly thought to be a hallmark of addiction and has been interpreted as a model for compulsive behavior.46,53–55 Identifying molecular mechanisms that underlie aversion-resistant alcohol seeking is therefore critical for developing novel effective alcoholism medications. Our data suggest that Prdm2 is a key player in modulating aversion-resistant alcohol intake. However, Prdm2 knockdown did not fully prevent the suppression of alcohol intake observed as a result of quinine adulteration, suggesting that multiple genes work in concert to modulate aversion-resistant alcohol intake. In accordance with this hypothesis, we previously found that inhibition of synaptotagmin 2 expression was also able to increase aversion-resistant alcohol intake.4
Prdm2 appears to regulate a broader range of alcohol-addiction-related behaviors, suggesting that it has a key role in the neuroadaptive processes within the dmPFC that are involved in the emergence of addiction. In addition to elevated self-administration and aversion-resistant intake, we also observed a role of PRDM2 in stress-induced reinstatement of alcohol seeking, an established model of relapse;48 Prdm2 knockdown increased sensitivity to stress-induced reinstatement. In control experiments, this manipulation did not alter locomotor activity, sensitivity to shock intensity or sucrose seeking, making nonspecific behavioral effects of Prdm2 unlikely as a cause of the behavioral findings. Collectively, these observations suggest that long-term repression of Prdm2 is a key epigenetic mechanism contributing to a cluster of behaviors thought to be at the core of alcohol addiction.
To identify PRDM2 target genes that may underlie alcohol-seeking phenotypes, we overlaid H3K9me1 ChIP-seq and RNA-seq data from control versus postdependent dmPFC. In accordance with previous findings that H3K9me1 is associated with gene activation,56 the majority of genes that showed decreased H3K9me1 in postdependent rats were downregulated in post-dependent rats.4 The regions of differential H3K9me1 enrichment predominantly lie within gene bodies rather than promoter regions. While methylation at H3K9 in a gene promoter is commonly associated with transcriptional repression, methylation within the gene body promotes transcriptional elongation.57 Epigenetic regulation in regions outside of gene promoters has previously been shown to be affected by drug exposure. For example, differentially methylated DNA regions within introns, exons and intergenic intervals rather than within gene promoters was associated with parental THC exposure.58 Future studies will have to determine the functional implications of the regions subject to H3K9me1 regulation in the context of alcohol dependence. Another implication of our findings to be explored in future studies is whether these intronic regions of enrichment could mediate alternative promoter usage, cotranscriptional splicing of pre-mRNA or expression of noncoding transcripts that often arise from intronic regions.59
Pathway analysis of converging results from our epigenomics and transcriptomic molecular approaches suggested that PRDM2-mediated histone methylation may contribute to gene expression changes that alter synaptic functioning, particularly by affecting calcium channel activity. When our ChIP-seq and RNA-seq analysis of control versus postdependent rats were further overlaid with genes that were functionally dysregulated as a result of Prdm2 knockdown in vivo, we identified four common genes, suggesting that long-term intermittent exposure to alcohol regulates these genes in part through Prdm2 downregulation. Collectively, these observations support the hypothesis that Prdm2 contributes to a reprogramming of the genome within the dmPFC that contributes to long-term neuroadaptations following a history of alcohol dependence.
Among the four overlapping genes, we found that the voltage-gated L- and T-type calcium channel subunits Cacna1i (Cav3.3) and Cacna1d (Cav1.3), respectively, were downregulated in Prdm2 knockdown rats, paralleling alcohol dependence-induced downregulation of these genes. Cacna1i, Cacna1d and another PRDM2 target gene Wnk2 were part of a DNA methylation-regulated transcriptional network that is disrupted in alcohol dependence.4 These genes are part of a gene network that may contribute to altered calcium release and neurotransmitter exocytosis in the mPFC of alcohol postdependent rats.4 Calcium channels have previously been implicated in altered patterns of cortical excitability that may underlie excessive alcohol intake and increase the propensity for relapse. For instance, non-human primates that chronically self-administer alcohol display plasticity in T-type calcium channel currents that is sensitive to periods of intoxication and withdrawal.60,61 In addition to the calcium channel subunits Cacna1i and Cacna1d, Wnk2 is also an essential regulator of neuronal excitability through regulation of cationchloride cotransporters.62 The fourth overlapping gene in our tripartite approach was Bsn, the Bassoon scaffolding protein that modulates calcium channel activity at the presynaptic active zone.63,64 Our findings are in accordance with prior post-mortem studies, which have shown changes in the expression pattern of genes involved in synaptic neurotransmission.65,66 Taken together, these studies and our results indicate impaired neurotransmitter release in the mPFC as a potential key mechanism that contributes to the emergence of alcohol addiction.
Importantly, H3K9me1 alone might, in some cases, be insufficient to regulate gene expression. For instance, Sims et al.67 showed that H3K9me1 cooperated with H4 lysine 20 monomethylation to silence gene expression. In another study, the number of neurons positive for H3 phosphorylation at serine 10 as well as acetylation at lysine 14 was low under basal conditions. However, both of these marks increased considerably after psychological stress, and this was associated with increased expression of c-fos and egr-1.68 A similar situation might be at play in our study. Under baseline condition, the H3K9me1 mark may not influence gene expression, but could render genes more susceptible to expression changes after alcohol or stress exposure. Furthermore, other genes identified in the Prdm2 knockdown study that did not overlap with the H3K9me1 ChIP-seq analysis could be indirectly regulated by PRDM2 or could be regulated by an alternate function of PRDM2. For example, PRDM2 has been shown to act upstream of the repressive PRC2 complex in proliferating cells.69 More experiments are needed to fully understand how PRDM2 regulates gene expression in alcohol dependence.
In conclusion, we demonstrate a novel role of PRDM2 in behaviors thought to be critical for alcohol addiction. Our findings indicate that PRDM2-mediated reprogramming of the genome in neurons of the dmPFC is involved in multiple aspects of alcohol dependence, including increase in alcohol self-administration, compulsive-like drinking and stress-induced relapse. This broad spectrum of actions provides a strong rationale to explore PRMD2 itself or some of its downstream targets as candidate targets for novel alcoholism medications. Of note, although some of these therapies may be able to compensate for the neuroadaptive consequences of alcohol dependence, strategies that restore PRDM2 function might have a unique potential to reverse those changes, and promote a transition back to a preaddicted state.
Supplementary Material
Acknowledgments
Work in the Heilig laboratory was supported by the NIAAA division of Intramural Research and the Swedish Research Council. Work in the Wahlestedt laboratory was supported by NIAAA R01 no. 1R01AA023781-01A1 and by the United States Department of Defense (DoD), through the National Defense Science and Engineering Graduate Fellowship (NDSEG) Program: this research was conducted with Government support under and awarded by DoD, Army Research Office (ARO), National Defense Science and Engineering Graduate (NDSEG) Fellowship, 32 CFR 168a. Related epigenomics and RNA work in the Wahlestedt laboratory has received funding by US National Institute of Health awards DA035592, MH084880 and NS071674.
Footnotes
CONFLICT OF INTEREST
The authors declare that, except for income received from our primary employer, no financial support or compensation has been received from any individual or corporate entity over the past 3 years for research or professional service, and there are no personal financial holdings that could be perceived as constituting a potential conflict of interest.
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
References
- 1.Heilig M, Egli M, Crabbe JC, Becker HC. Acute withdrawal, protracted abstinence and negative affect in alcoholism: are they linked? Addict Biol. 2010;15:169–184. doi: 10.1111/j.1369-1600.2009.00194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meinhardt MW, Sommer WH. Postdependent state in rats as a model for medication development in alcoholism. Addict Biol. 2015;20:1–21. doi: 10.1111/adb.12187. [DOI] [PubMed] [Google Scholar]
- 3.Warnault V, Darcq E, Levine A, Barak S, Ron D. Chromatin remodeling—a novel strategy to control excessive alcohol drinking. Transl Psychiatry. 2013;3:e231. doi: 10.1038/tp.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbier E, Tapocik JD, Juergens N, Pitcairn C, Borich A, Schank JR, et al. DNA methylation in the medial prefrontal cortex regulates alcohol-induced behavior and plasticity. J Neurosci. 2015;35:6153–6164. doi: 10.1523/JNEUROSCI.4571-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pandey SC, Ugale R, Zhang H, Tang L, Prakash A. Brain chromatin remodeling: a novel mechanism of alcoholism. J Neurosci. 2008;28:3729–3737. doi: 10.1523/JNEUROSCI.5731-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sakharkar AJ, Zhang H, Tang L, Pandey SC. A role for histone acetylation in intermittent ethanol exposure-induced changes in BDNF expression and anxietylike behaviors in adolescent rats. Alcohol Clin Exp Res. 2014;38:79a–79a. [Google Scholar]
- 7.Sakharkar AJ, Zhang HB, Tang L, Baxstrom K, Shi GB, Moonat S, et al. Effects of histone deacetylase inhibitors on amygdaloid histone acetylation and neuropeptide Y expression: a role in anxiety-like and alcohol-drinking behaviours. Int J Neuropsychopharmacol. 2014;17:1207–1220. doi: 10.1017/S1461145714000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moonat S, Sakharkar AJ, Zhang H, Tang L, Pandey SC. Aberrant histone deacetylase2-mediated histone modifications and synaptic plasticity in the amygdala predisposes to anxiety and alcoholism. Biol Psychiatry. 2013;73:763–773. doi: 10.1016/j.biopsych.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun H, Maze I, Dietz DM, Scobie KN, Kennedy PJ, Damez-Werno D, et al. Morphine epigenomically regulates behavior through alterations in histone H3 lysine 9 dimethylation in the nucleus accumbens. J Neurosci. 2012;32:17454–17464. doi: 10.1523/JNEUROSCI.1357-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maze I, Covington HE, III, Dietz DM, LaPlant Q, Renthal W, Russo SJ, et al. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science. 2010;327:213–216. doi: 10.1126/science.1179438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roberts AJ, Heyser CJ, Cole M, Griffin P, Koob GF. Excessive ethanol drinking following a history of dependence: animal model of allostasis. Neuropsychopharmacology. 2000;22:581–594. doi: 10.1016/S0893-133X(99)00167-0. [DOI] [PubMed] [Google Scholar]
- 12.Rimondini R, Arlinde C, Sommer W, Heilig M. Long-lasting increase in voluntary ethanol consumption and transcriptional regulation in the rat brain after intermittent exposure to alcohol. FASEB J. 2002;16:27–35. doi: 10.1096/fj.01-0593com. [DOI] [PubMed] [Google Scholar]
- 13.Heilig M, Koob GF. A key role for corticotropin-releasing factor in alcohol dependence. Trends Neurosci. 2007;30:399–406. doi: 10.1016/j.tins.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vendruscolo LF, Barbier E, Schlosburg JE, Misra KK, Whitfield TW, Jr, Logrip ML, et al. Corticosteroid-dependent plasticity mediates compulsive alcohol drinking in rats. J Neurosci. 2012;32:7563–7571. doi: 10.1523/JNEUROSCI.0069-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sommer WH, Rimondini R, Hansson AC, Hipskind PA, Gehlert DR, Barr CS, et al. Upregulation of voluntary alcohol intake, behavioral sensitivity to stress, and amygdala crhr1 expression following a history of dependence. Biol Psychiatry. 2008;63:139–145. doi: 10.1016/j.biopsych.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 16.Tapocik JD, Solomon M, Flanigan M, Meinhardt M, Barbier E, Schank JR, et al. Coordinated dysregulation of mRNAs and microRNAs in the rat medial prefrontal cortex following a history of alcohol dependence. Pharmacogenom J. 2013;13:286–296. doi: 10.1038/tpj.2012.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meinhardt MW, Hansson AC, Perreau-Lenz S, Bauder-Wenz C, Stahlin O, Heilig M, et al. Rescue of infralimbic mGluR2 deficit restores control over drug-seeking behavior in alcohol dependence. J Neurosci. 2013;33:2794–2806. doi: 10.1523/JNEUROSCI.4062-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Macey DJ, Schulteis G, Heinrichs SC, Koob GF. Time-dependent quantifiable withdrawal from ethanol in the rat: effect of method of dependence induction. Alcohol. 1996;13:163–170. doi: 10.1016/0741-8329(95)02030-6. [DOI] [PubMed] [Google Scholar]
- 19.George O, Sanders C, Freiling J, Grigoryan E, Vu S, Allen CD, et al. Recruitment of medial prefrontal cortex neurons during alcohol withdrawal predicts cognitive impairment and excessive alcohol drinking. Proc Natl Acad Sci USA. 2012;109:18156–18161. doi: 10.1073/pnas.1116523109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tapocik JD, Barbier E, Flanigan M, Solomon M, Pincus A, Pilling A, et al. microRNA-206 in rat medial prefrontal cortex regulates BDNF expression and alcohol drinking. J Neurosci. 2014;34:4581–4588. doi: 10.1523/JNEUROSCI.0445-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poetsch M, Dittberner T, Woenckhaus C. Frameshift mutations of RIZ, but no point mutations in RIZ1 exons in malignant melanomas with deletions in 1p36. Oncogene. 2002;21:3038–3042. doi: 10.1038/sj.onc.1205457. [DOI] [PubMed] [Google Scholar]
- 22.Sasaki O, Meguro K, Tohmiya Y, Funato T, Shibahara S, Sasaki T. Altered expression of retinoblastoma protein-interacting zinc finger gene, RIZ, in human leukaemia. Br J Haematol. 2002;119:940–948. doi: 10.1046/j.1365-2141.2002.03972.x. [DOI] [PubMed] [Google Scholar]
- 23.Chadwick RB, Jiang GL, Bennington GA, Yuan B, Johnson CK, Stevens MW, et al. Candidate tumor suppressor RIZ is frequently involved in colorectal carcinogenesis. Proc Natl Acad Sci USA. 2000;97:2662–2667. doi: 10.1073/pnas.040579497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang G, Liu L, Buyse IM, Simon D, Huang S. Decreased RIZ1 expression but not RIZ2 in hepatoma and suppression of hepatoma tumorigenicity by RIZ1. Int J Cancer. 1999;83:541–546. doi: 10.1002/(sici)1097-0215(19991112)83:4<541::aid-ijc17>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 25.He L, Yu JX, Liu L, Buyse IM, Wang MS, Yang QC, et al. RIZ1, but not the alternative RIZ2 product of the same gene, is underexpressed in breast cancer, and forced RIZ1 expression causes G2-M cell cycle arrest and/or apoptosis. Cancer Res. 1998;58:4238–4244. [PubMed] [Google Scholar]
- 26.Kim KC, Geng L, Huang S. Inactivation of a histone methyltransferase by mutations in human cancers. Cancer Res. 2003;63:7619–7623. [PubMed] [Google Scholar]
- 27.Congdon LM, Sims JK, Tuzon CT, Rice JC. The PR-Set7 binding domain of Riz1 is required for the H4K20me1-H3K9me1 trans-tail ‘histone code’ and Riz1 tumor suppressor function. Nucleic Acids Res. 2014;42:3580–3589. doi: 10.1093/nar/gkt1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Finegersh A, Ferguson C, Maxwell S, Mazariegos D, Farrell D, Homanics GE. Repeated vapor ethanol exposure induces transient histone modifications in the brain that are modified by genotype and brain region. Front Mol Neurosci. 2015;8:39. doi: 10.3389/fnmol.2015.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Du Y, Carling T, Fang W, Piao Z, Sheu JC, Huang S. Hypermethylation in human cancers of the RIZ1 tumor suppressor gene, a member of a histone/protein methyltransferase superfamily. Cancer Res. 2001;61:8094–8099. [PubMed] [Google Scholar]
- 30.Zhang C, Zhu Q, He H, Jiang L, Qiang Q, Hu L, et al. RIZ1: a potential tumor suppressor in glioma. BMC Cancer. 2015;15:990. doi: 10.1186/s12885-015-2023-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geli J, Kiss N, Kogner P, Larsson C. Suppression of RIZ in biologically unfavourable neuroblastomas. Int J Oncol. 2010;37:1323–1330. doi: 10.3892/ijo_00000784. [DOI] [PubMed] [Google Scholar]
- 32.Rimondini R, Arlinde C, Sommer W, Heilig M. Long-lasting increase in voluntary ethanol consumption and transcriptional regulation in the rat brain after intermittent exposure to alcohol. FASEB J. 2002;16:27–35. doi: 10.1096/fj.01-0593com. [DOI] [PubMed] [Google Scholar]
- 33.Paxinos G, Watson C. The Rat Brain in Stereotaxic Coordinates: Hard Cover Edition. Academic Press; New York, NY, USA: 2007. [Google Scholar]
- 34.Bjork K, Saarikoski ST, Arlinde C, Kovanen L, Osei-Hyiaman D, Ubaldi M, et al. Glutathione-S-transferase expression in the brain: possible role in ethanol preference and longevity. FASEB J. 2006;20:1826–1835. doi: 10.1096/fj.06-5896com. [DOI] [PubMed] [Google Scholar]
- 35.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(—Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 36.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Connor CM, Dincer A, Straubhaar J, Galler JR, Houston IB, Akbarian S. Maternal immune activation alters behavior in adult offspring, with subtle changes in the cortical transcriptome and epigenome. Schizophr Res. 2012;140:175–184. doi: 10.1016/j.schres.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen EY, Ahern TH, Cheung I, Straubhaar J, Dincer A, Houston I, et al. Epigenetics and sex differences in the brain: a genome-wide comparison of histone-3 lysine-4 trimethylation (H3K4me3) in male and female mice. Exp Neurol. 2015;268:21–29. doi: 10.1016/j.expneurol.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zang C, Schones DE, Zeng C, Cui K, Zhao K, Peng W. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu S, Grullon S, Ge K, Peng W. Spatial clustering for identification ofChIP-enriched regions (SICER) to map regions of histone methylation patterns in embryonic stem cells. Methods Mol Biol. 2014;1150:97–111. doi: 10.1007/978-1-4939-0512-6_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang J, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 2013;41:W77–W83. doi: 10.1093/nar/gkt439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seif T, Chang SJ, Simms JA, Gibb SL, Dadgar J, Chen BT, et al. Cortical activation of accumbens hyperpolarization-active NMDARs mediates aversion-resistant alcohol intake. Nat Neurosci. 2013;16:1094–1100. doi: 10.1038/nn.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Epstein DH, Preston KL, Stewart J, Shaham Y. Toward a model of drug relapse: an assessment of the validity of the reinstatement procedure. Psychopharmacology (Berl) 2006;189:1–16. doi: 10.1007/s00213-006-0529-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Le A, Shaham Y. Neurobiology of relapse to alcohol in rats. Pharmacol Ther. 2002;94:137–156. doi: 10.1016/s0163-7258(02)00200-0. [DOI] [PubMed] [Google Scholar]
- 49.Le AD, Quan B, Juzytch W, Fletcher PJ, Joharchi N, Shaham Y. Reinstatement of alcohol-seeking by priming injections of alcohol and exposure to stress in rats. Psychopharmacology (Berl) 1998;135:169–174. doi: 10.1007/s002130050498. [DOI] [PubMed] [Google Scholar]
- 50.Cippitelli A, Karlsson C, Shaw JL, Thorsell A, Gehlert DR, Heilig M. Suppression of alcohol self-administration and reinstatement of alcohol seeking by melanin-concentrating hormone receptor 1 (MCH1-R) antagonism in Wistar rats. Psychopharmacology. 2010;211:367–375. doi: 10.1007/s00213-010-1891-y. [DOI] [PubMed] [Google Scholar]
- 51.Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 52.Farris SP, Arasappan D, Hunicke-Smith S, Harris RA, Mayfield RD. Transcriptome organization for chronic alcohol abuse in human brain. Mol Psychiatry. 2015;20:1438–1447. doi: 10.1038/mp.2014.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deroche-Gamonet V, Belin D, Piazza PV. Evidence for addiction-like behavior in the rat. Science. 2004;305:1014–1017. doi: 10.1126/science.1099020. [DOI] [PubMed] [Google Scholar]
- 54.Vengeliene V, Celerier E, Chaskiel L, Penzo F, Spanagel R. Compulsive alcohol drinking in rodents. Addict Biol. 2009;14:384–396. doi: 10.1111/j.1369-1600.2009.00177.x. [DOI] [PubMed] [Google Scholar]
- 55.Wolffgramm J, Galli G, Thimm F, Heyne A. Animal models of addiction: models for therapeutic strategies? J Neural Transm. 2000;107:649–668. doi: 10.1007/s007020070067. [DOI] [PubMed] [Google Scholar]
- 56.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 57.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 58.Watson CT, Szutorisz H, Garg P, Martin Q, Landry JA, Sharp AJ, et al. Genome-wide DNA methylation profiling reveals epigenetic changes in the rat nucleus accumbens associated with cross-generational effects of adolescent THC exposure. Neuropsychopharmacology. 2015;40:2993–3005. doi: 10.1038/npp.2015.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brown SJ, Stoilov P, Xing Y. Chromatin and epigenetic regulation of pre-mRNA processing. Hum Mol Genet. 2012;21:R90–R96. doi: 10.1093/hmg/dds353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carden WB, Alexander GM, Friedman DP, Daunais JB, Grant KA, Mu J, et al. Chronic ethanol drinking reduces native T-type calcium current in the thalamus of non-human primates. Brain Res. 2006;1089:92–100. doi: 10.1016/j.brainres.2006.02.135. [DOI] [PubMed] [Google Scholar]
- 61.Welsh JP, Han VZ, Rossi DJ, Mohr C, Odagiri M, Daunais JB, et al. Bidirectional plasticity in the primate inferior olive induced by chronic ethanol intoxication and sustained abstinence. Proc Natl Acad Sci USA. 2011;108:10314–10319. doi: 10.1073/pnas.1017079108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rinehart J, Vazquez N, Kahle KT, Hodson CA, Ring AM, Gulcicek EE, et al. WNK2 kinase is a novel regulator of essential neuronal cation-chloride cotransporters. J Biol Chem. 2011;286:30171–30180. doi: 10.1074/jbc.M111.222893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nishimune H, Numata T, Chen J, Aoki Y, Wang Y, Starr MP, et al. Active zone protein Bassoon co-localizes with presynaptic calcium channel, modifies channel function, and recovers from aging related loss by exercise. PLoS One. 2012;7:e38029. doi: 10.1371/journal.pone.0038029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Frank T, Rutherford MA, Strenzke N, Neef A, Pangrsic T, Khimich D, et al. Bassoon and the synaptic ribbon organize Ca(2)+ channels and vesicles to add release sites and promote refilling. Neuron. 2010;68:724–738. doi: 10.1016/j.neuron.2010.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu J, Lewohl JM, Harris RA, Iyer VR, Dodd PR, Randall PK, et al. Patterns of gene expression in the frontal cortex discriminate alcoholic from nonalcoholic individuals. Neuropsychopharmacology. 2006;31:1574–1582. doi: 10.1038/sj.npp.1300947. [DOI] [PubMed] [Google Scholar]
- 66.Liu J, Lewohl JM, Harris RA, Dodd PR, Mayfield RD. Altered gene expression profiles in the frontal cortex of cirrhotic alcoholics. Alcohol Clin Exp Res. 2007;31:1460–1466. doi: 10.1111/j.1530-0277.2007.00444.x. [DOI] [PubMed] [Google Scholar]
- 67.Sims JK, Houston SI, Magazinnik T, Rice JC. A trans-tail histone code defined by monomethylated H4 Lys-20 and H3 Lys-9 demarcates distinct regions of silent chromatin. J Biol Chem. 2006;281:12760–12766. doi: 10.1074/jbc.M513462200. [DOI] [PubMed] [Google Scholar]
- 68.Bilang-Bleuel A, Ulbricht S, Chandramohan Y, De Carli S, Droste SK, Reul JM. Psychological stress increases histone H3 phosphorylation in adult dentate gyrus granule neurons: involvement in a glucocorticoid receptor-dependent behavioural response. Eur J Neurosci. 2005;22:1691–1700. doi: 10.1111/j.1460-9568.2005.04358.x. [DOI] [PubMed] [Google Scholar]
- 69.Cheedipudi S, Puri D, Saleh A, Gala HP, Rumman M, Pillai MS, et al. A fine balance: epigenetic control of cellular quiescence by the tumor suppressor PRDM2/RIZ at a bivalent domain in the cyclin a gene. Nucleic Acids Res. 2015;43:6236–6256. doi: 10.1093/nar/gkv567. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


