Abstract
Objective
Although the importance of sleep is increasingly recognized, the lack of robust and efficient algorithms hinders scalable sleep assessment in healthy persons and those with sleep disorders. Polysomnography (PSG) and visual/manual scoring remain the gold standard in sleep evaluation, but more efficient/automated systems are needed. Most previous works have demonstrated algorithms in high agreement with the gold standard in healthy/normal (HN) individuals – not those with sleep disorders.
Methods
This paper presents a statistical framework that automatically estimates whole-night sleep architecture in patients with obstructive sleep apnea (OSA) – the most common sleep disorder. Single-channel frontal electroencephalography was extracted from 65 HN/OSA sleep studies, and decomposed into 11 spectral features in 60,903 30s sleep epochs. The algorithm leveraged kernel density estimation to generate stage-specific likelihoods, and a 5-state hidden Markov model to estimate per-night sleep architecture.
Results
Comparisons to full PSG expert scoring revealed the algorithm was in fair agreement with the gold standard (median Cohen’s kappa = 0.53). Further analysis revealed modest decreases in median scoring agreement as OSA severity increased from HN (kappa = 0.63) to severe (kappa = 0.47). A separate implementation on HN data from the Physionet Sleep-EDF Database resulted in a median kappa = 0.65, further indicating the algorithm’s broad applicability.
Conclusion
Results of this work indicate the proposed single-channel framework can emulate expert-level scoring of sleep architecture in OSA.
Significance
Algorithms constructed to more accurately model physiological variability during sleep may help advance automated sleep assessment, for practical and general use in sleep medicine.
Index Terms: Electroencephalography (EEG), density estimation, hidden Markov model, obstructive sleep apnea (OSA), sleep scoring
I. Introduction
Sleep, like eating and breathing, is an essential part of the daily life cycle. Although the process of sleep is not fully understood, it has been shown to play a vital role in immune, cardiovascular, and neurocognitive function [1]. Despite its great importance, nearly 40% of US adults experience problems with sleep ranging from insufficient total sleep time, trouble initiating or maintaining sleep (insomnia), circadian rhythm disorders, sleep-related movement disorders, and sleep-related breathing disorders such as obstructive sleep apnea (OSA) [2]. All of the above have been shown to take a toll on the affected individual physically, mentally, financially, and/or socially.
Sleep disorders can be diagnosed by an overnight polysomnogram (PSG), which utilizes multiple sensing modalities to measure biophysiological signals, including electroencephalogram (EEG), electrooculogram (EOG), and respiratory rate and flow [2]. Although considered the “gold standard,” there are multiple reasons that hinder more widespread PSG use. First, the cumbersome nature of the equipment interferes with sleep. Second, both the equipment, and the cost/time of a registered polysomnography technician (RPSGT) who performs sleep scoring visually according to standard rules, are expensive. Third, clinical scoring of sleep remains a mundane process with considerable inter-rater variability. To maintain a standard level of clinical sleep scoring, technicians/physicians adhere to rules delineated by Rechtschaffen and Kales (R&K) and the American Academy of Sleep Medicine (AASM), which are designed to visually categorize any epoch of sleep into one of five clinically-recognized sleep stages (Wake, N1, N2, N3, REM) [3], [4], [5]. Despite standardization efforts, the mean inter-rater agreement between experts scoring sleep in OSA is only 71% [6]. For all these reasons, relatively few sleep studies are performed. A robust, yet cost-effective and minimally invasive system to accurately measure sleep would be valuable to better understand sleep in a research and clinical context.
In an attempt to remedy the problems of manual sleep scoring, many in the literature have proposed machine learning and data science techniques for facilitating automated scoring of sleep. Such studies have employed algorithms such as decision trees [7]–[11], support vector machines [12]–[15], Markov models [16]–[21], and neural networks [22], [23], which operate on combinations of the traditional multi-channel PSG biometrics (e.g. EEG, EOG, Respiration) to provide algorithmic and automated assessment of a patient’s underlying sleep architecture. To further simplify the current sleep scoring paradigm, many have presented algorithms which perform on very few or even single-channel recordings from varied modalities during sleep. [10], [12], [13], [16], [22]–[28].
While progress has been made in the multi- and single-channel domain of automated sleep scoring, agreement can be modest – especially when the number of inputs is restricted. Additionally, most of the prior work has focused on sleep in healthy/normal (HN) subjects. These algorithms may not generalize to older individuals with chronic diseases, or those with sleep disorders that cause sleep fragmentation, such as OSA. Given that 25–50% of middle aged men and women may have clinically relevant OSA [29], algorithms will need to be capable of assessing sleep in a wide range of people. Additionally, to be feasible, data will need to be derived from smaller systems and a minimal number of channels.
Presented herein is an algorithmic approach to scoring sleep using only a single frontal channel of EEG that is satisfactory for automated sleep scoring within the context of OSA. The work assesses time-frequency features of sleep EEG generated via the multitaper spectrogram, and leverages a non-parametric likelihood model for each of the five sleep stages via kernel density estimation. Whole-night sleep architecture is estimated using a five-state hidden Markov model and the Viterbi algorithm, designed to operate on the multimodal likelihood structure of different sleep stages. Results are presented for per-night and per-epoch comparisons of algorithm vs. clinical scoring of the sleep data in subjects who are HN as well as those with OSA. The paper concludes with a discussion of the results and insight into algorithm performance as a function of OSA severity.
II. Methodology
The present work includes a retrospective analysis of eighty clinically-scored overnight PSG studies. The analysis was divided into two parts: 1) An analysis of 65 datasets recorded at UC San Diego for 15 HN (HNUCSD) and 50 OSA (OSAUCSD) combined datasets, and 2) an analysis of 15 HN datasets derived from the Physionet Sleep-EDF Database (HNPhysionet) [30], [31].
The first sixty-five datasets were recorded on a 1401-plus interface and Spike 2 software (Cambridge Electronic Design Ltd., Cambridge, UK) at the UCSD Sleep Laboratory in San Diego, California. Ethical approval for these studies was obtained from the Human Research Protections Program at the University of California, San Diego. Manual scoring of sleep was performed by a RPSGT who had access to all modalities included in the full PSG study to create the clinical hypnogram. Fifty of the sixty-five UCSD datasets – comprising a subtotal of 48,819 30s epochs of sleep – contained a mix of OSA severities based on the Apnea Hypopnea Index (AHI); 9 were mild OSA (5 ≤ AHI < 15 events/hour), 9 were moderate OSA (15 ≤ AHI < 30 events/hour), and 32 were severe OSA (AHI ≥ 30 events/hour). Another fifteen of the sixty-five datasets – comprising a subtotal of 12,084 30s epochs of sleep – contained HN data (AHI ≤ 5 events/hour). A total of 60,903 30s epochs were used in the five-fold cross validation scoring analysis described below.
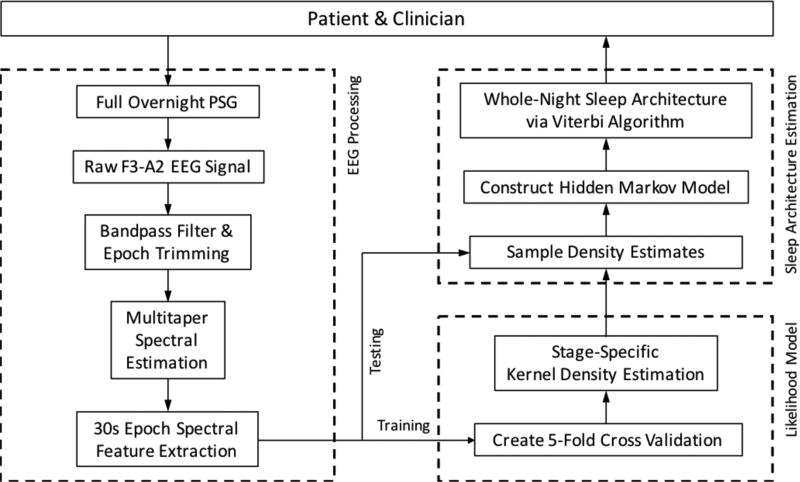
For the purposes of this study, only a single EEG channel (F3-A2) and the clinical hypnogram from the full HNUCSD and OSAUCSD PSG studies were used for training and testing of the automated algorithm. Fig. 1 illustrates the process workflow for automated assessment of sleep via single-channel sleep EEG. The algorithm classifies a continuous sleep EEG signal into a 5-stage sleep paradigm comprised of stages Wake (W), REM (R), N1, N2, and N3. Python 3.4.4 and modified scripts from of the scikit-learn library were used to create the algorithm.
Fig 1.
Process workflow for automated assessment of single-channel sleep EEG.
The final fifteen of eighty datasets were derived from the public Sleep-EDF Database [30], [31]. Specifically, EEG channel Fpz-Cz and clinical hypnograms were extracted from Sleep Telemetry subjects 01–02, 04–14, and 16–17. All recordings were obtained from subjects who had mild difficulty falling asleep, but who were otherwise healthy. Processing of these datasets followed suit with the HNUCSD and OSAUCSD data. Sleep architecture estimation of the HNPhysionet sleep EEG datasets was performed: 1) as a training-testing analysis entirely separate from the UCSD-trained algorithm, and 2) by treating the HNPhysionet data as test data against the UCSD-trained algorithm. The former assessed the generalizability of the raw algorithm, while the latter assessed generalizability of the F3-A2 training for classification of data derived from other EEG montages.
A. EEG Pre-Processing
Raw single channel F3-A2 EEG data were derived from full PSG recordings in each of the 65 UCSD datasets. Single-channel EEG was originally sampled at 125Hz. Time series EEG data was bandpass filtered between 0.1Hz and 50Hz using a zero-phase forward-backward filter (Python, SciPy module). After filtering, 30s epochs of sleep deemed as “NO STAGE” in the clinical hypnogram were trimmed from both the hypnogram and at corresponding points in the time series EEG data. “NO STAGE” epochs only appeared at the beginning or end of clinical hypnograms (accounting for subject wiring and disconnection during the overnight PSG), so EEG signal continuity during epoch trimming was preserved.
Similarly, raw single channel Fpz-Cz EEG data were derived from each of the 15 Physionet datasets. Single-channel EEG was originally sampled at 100Hz. The first 6-hours of each HNPhysionet dataset was used, to ensure alignment between the EEG data and corresponding hypnograms. In these hypnograms, epochs “Stage 3” and “Stage 4” were replaced by “N3”, to be consistent with the analysis of UCSD data.
B. Multitaper Spectral Estimation
Filtered EEG signals were spectrally decomposed using multitaper (MT) spectral estimation. Like the conventional fast Fourier transform (FFT), MT spectral estimation is an approach for constructing a time-frequency representation of non-stationary time series signal. The advantage in using the MT approach is in its use of orthonormal bases to serve as different, uncorrelated “tapers” (hence multitaper), resulting in a modulation of spectral estimation variance and bias [32], [33], [34]. MT spectral estimation also boasts better frequency resolution than some overlapping segment average approaches, such as Welch’s method, for the same spectral leakage and variance estimators; specifically, the resolution bandwidth for Welch’s method is 20–60% wider than the MT approach [35].
In essence, these multiple tapers are auxiliary to the standard FFT – each taper augments the FFT separately; the outputs of which are averaged across the total number of tapers used to assemble the MT spectral estimate (1, 2). If x(n) is the time series acquisition of sleep EEG with discrete samples n = 0, 1, …, N, Δ represents the time interval between recorded samples, and denotes the set of orthonormal tapers i = 1, 2, …, L at each time sample n, then the MT power spectral density (PSD) estimate of the sleep EEG signal, S, was given by
| (1) |
where
| (2) |
For a mathematical narrative on MT spectral estimation, Babadi and Brown provide a brief derivation of the MT method and a comparison to other non-parametric spectral estimation techniques [33]. In the proposed algorithm, EEG MT spectral estimation was implemented in Python via the Spectrum module available in the Python Package Index. A 30s non-overlapping window and a suggested time half-bandwidth parameter NW = 3 were used, which resulted in L = (NW) (2) − 1 = 5 tapers used for EEG MT spectral estimation. Discrete prolate spheroidal sequences – or Slepian sequences – were used as the orthonormal set of tapers. Finally, S was converted to a log-PSD:
| (3) |
C. EEG Spectral Feature Extraction
Eleven spectral features were extracted on an epoch-by-epoch basis from the log-MT spectral estimate of sleep EEG (Table I). Frequency bands were chosen based on the previous literature and guidance from the AASM Sleep Scoring Manual [4], [5], [14], [20], [21], [23].
Table I.
Spectral Features used for automated classification of sleep EEG.
| EEG Frequency Bands/Features |
Spectral Edges (Hz) | Characteristic Sleep Stage |
|---|---|---|
| Broadband (broad) | (0.1, 50) | W (Motion Artifact) |
| Gamma (γ) | (30, 50) | W |
| Beta (β) | (20, 30) | W |
| Sigma (σ) | (11, 14) | N2 (Sleep Spindles) |
| Alpha (α1, α2, α3, α4) | (7, 8), (8, 9), (9, 10), (10, 11) | W, N1, R |
| Theta (θ) | (4, 7) | N1, R (Sawtooth Waves) |
| Delta (δ) | (1, 4) | N3 (Slow Waves) |
| Very-Low Frequency (vlf) | (0.1, 1) | W (Eye Blinks), R (Rapid Eye Movements), N2 (K-Complexes) |
Of interest here was the decision to split the 7–11 Hz alpha band into four equally spaced bands of 1 Hz bandwidth. This was done to implement insight from the AASM Scoring Manual, which states, “The alpha frequency in stage R often is 1–2 Hz slower than during wakefulness.” [4] Moreover, stages R and N1 often resemble each other in low-amplitude, mixed frequency activity. Therefore, a segmentation of the alpha band was performed in an attempt to better discern these three often misclassified stages.
In this work, the spectral feature represented a mean PSD value for frequency band k during epoch t of an overnight sleep EEG dataset. Denote the set of frequencies in frequency band k as ℱ(k) and the size of ℱ(k) as |ℱ(k)|. For instance, for k = 0, the broad feature, we have that ℱ(0) = {0.1, 0.13, 0.16, …, 49.96, 49.99} and |ℱ(0)| = 1663. For k = 1, 2,…,10, the frequency bands pertaining to ℱ(k) are given in Table I. For k = 0, 1,…, 10, each spectral feature was calculated as follows:
| (4) |
One feature (k = 0, broadband EEG activity) was simply calculated as the mean PSD value between spectral edges (0.1Hz, 50Hz). The remaining features were calculated as relative spectral values – the difference between activity in frequency band k (for k ≠ 0) and broadband activity . The result is a feature vector yt ∈ ℝ11 for each 30s clinically-scored epoch of sleep. Over the entire UCSD dataset of 65 overnight studies, a total of 60,903 feature vectors were extracted. For Physionet data, a total of 10,800 feature vectors were extracted.
D. Kernel Density Estimation
Following epoch-by-epoch spectral feature extraction, the 65 nights of UCSD EEG feature vectors were equally segmented into five separate folds (13 nights per fold: 3 HNUCSD and 10 OSAUCSD), defining the 5-fold cross validation paradigm for algorithm training and testing. Separately, the 15 nights of Physionet EEG feature vectors were equally segmented into five separate folds (3 HNPhysionet datasets per fold). To construct likelihood models for the feature vector yt, kernel density estimation (KDE) was used to estimate the conditional probability density function of observing EEG spectral features during a specific stage of sleep.
KDE is a non-parametric method for estimating the probability density function of a continuous random variable. In this formulation, we treat as sample vectors of dimension d = 11, drawn from the ith class of an unknown density function . Generally speaking, it is difficult to determine the true distribution , so the following kernel density estimate is used for approximation:
| (6) |
where K is the kernel function – a d-dimensional, non-negative, zero-mean function that integrates to one – and bi is a non-negative, non-zero bandwidth parameter corresponding to the ith class.
KDE is an attractive means to approximate the true topology of a density. Its formulation is similar to that of a histogram of the data, except that it performs a weighted average of many kernel functions centered about each data point in the sample space. In this way, Ri(y) leverages properties of the chosen kernel K to enforce smoothness and continuity on the likelihood surface.
Moreover, unlike the multivariate Gaussian distribution, Ri(y) can exhibit multimodal behavior, which is necessary for encoding the variations in sleep architecture within and across different patients, pathologies, and nights of sleep. For example, the same stage of sleep could display variants of sleep EEG activity based on age, sex, mental state, and overall health [36]. Inter-individual variability in sleep and frequency of sleep arousals increases as a function of age [37], [38], and is prominent in diseases such as Parkinson’s Disease [39] and Rheumatoid Arthritis [40]. Moreover, such variations in regard to sleep arousals and sleep continuity are mitigated by non-anatomical features such as the arousal threshold, which is considered an important contributor to the pathogenesis of sleep breathing disorders such as OSA [41]. By utilizing a density estimation approach, such as KDE, the goal is to model appropriately the heterogeneity of sleep EEG activity within each stage of sleep for varied classes of subjects and sleep physiology.
To construct the trained likelihood models for each sleep stage class, KDE was implemented using the SciPy stats.gaussian_kde package. An 11-dimensional 𝒩 (0,1) Gaussian was used as the kernel function, and the optimal bandwidth parameter bi was automatically determined for each sleep stage i ∈ {W, R, N1, N2, N3} via Scott’s Rule [42]. For an arbitrary fold of test data, Ri(y) was constructed with the remaining 4 folds of data, so to train the likelihood models with data separate from the testing set. The result per-fold is a set of stage-specific conditional probability density functions fY|X (y|x), where the sleep stage x probabilistically exhibits EEG spectral activity y.
E. Hidden Markov Model
During each 30s epoch of sleep, a hidden stage of sleep emits observable multivariate EEG spectral activity, giving an indication of the underlying sleep state. The observed EEG signal varies for different stages of sleep, as well as for different nights of sleep and sleep pathologies. As sleep evolves over the course of the night, discrete sleep stage transitions occur between neighboring epochs, constrained by time-varying physiological phenomena governing the sleep process. These transitions are traditionally scored such that only the previous epoch can influence the transition to another sleep stage in the current epoch [4]. To encompass these properties of sleep and sleep scoring, a state space model was utilized to represent per-night sleep architecture as a 5-state, transition-constrained, Markov chain. The likelihood model from section D and the Markov model jointly comprise a hidden Markov model (HMM) [43].
To construct sleep architecture as a HMM, the following variables and parameters are defined for epochs t = 0, 1, …, T and sleep states i, j ∈ {W, R, N1, N2, N3}:
| yt: | Multivariate observation vector of single-channel EEG spectral feature at time t. |
| xt: | Hidden sleep state i at epoch t. |
| y0:T: | (y0, y1, …, yT−1, yT). Sequence of observed multivariate EEG spectral activity. |
| x0:T: | (x0, x1, …, xT−1, xT). Sequence of hidden sleep states composing whole-night sleep architecture. |
| πi: | P(x0 = i). Initial probability of sleep state i at time t = 0. |
| Qi,j: | P(Xt = j|Xt−1 = i). Probability of transitioning to state j at time t from state i at time t − 1. |
| Ri(y): | P(Yt = yt|Xt = i). Probability of observing EEG features yt in sleep state i. |
The goal is to generate a model for which x0:T can be estimated through a corresponding sequence of observed EEG activity and prior knowledge of sleep stage transitioning constraints.
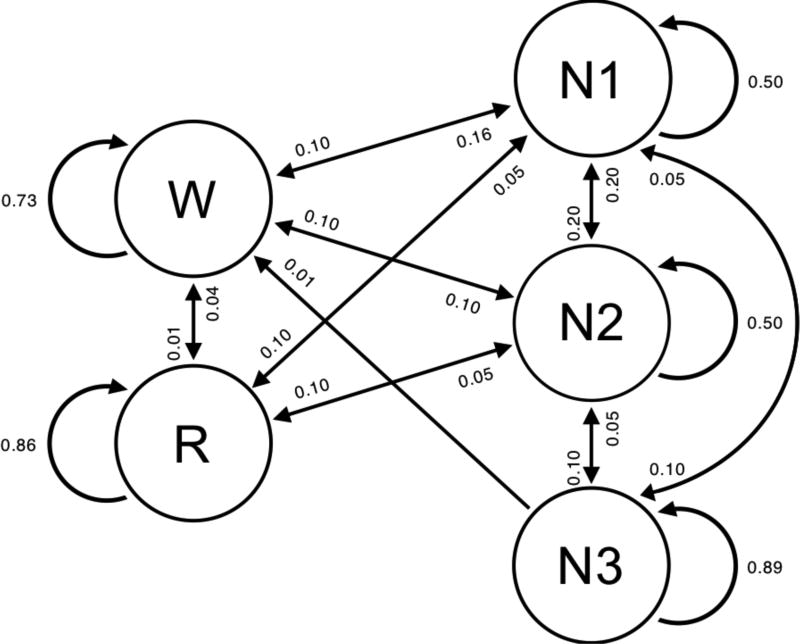
The HMM algorithm presented here was formulated using a modified version of the framework available in the hmmlearn python module. The modifications allowed for the use of alternative likelihood models, which are framed as the set of stage-specific KDE likelihoods Ri (y) generated during the training phase. Since all PSG studies begin before the onset of sleep, the only non-zero initial probability corresponds to the sleep state i = W, such that the initial probability vector πi = [1,0,0,0,0]. Values from the work of [44] provide insight on the transition properties of sleep in clinical populations of healthy subjects and OSA subjects. As the work was performed for a 4-stage sleep model, OSA-specific values were extrapolated to create a 5-state transition probability matrix for nights of sleep in OSA subjects; transition probabilities are graphically illustrated in Fig 2. To demonstrate the generalizability of the presented algorithm, these values were fixed for all nights of sleep in the analysis and were not adjusted when testing on untrained or new data. With that said, these transition probabilities can be determined on a per-night and/or per-subject basis for analyzing sleep architecture that departs from the typical HN or OSA sleep structure shown here.
Fig 2.
Graphical model of HMM sleep state transitions with corresponding probabilities. Absence of directed arrow indicates a transition probability Qi,j = 0.00.
Following the HMM formulation, the Viterbi algorithm (VA) was used to generate an algorithmic representation of the 5-state clinical hypnogram. The VA is a recursive decoding method for determining the sequence of latent (hidden) variables most likely associated with a corresponding sequence of observations [43]. In the case of sleep staging, the VA uses the HMM to identify an optimal sequence of hidden sleep stages x0:T that best fit the observed set of EEG signals y0:T during a whole night of sleep via maximum a posteriori sequence estimation. The final output is the Viterbi path – a sequence of values x0:T = x0, x1, …, xT that represents the automated sleep staging for a single night of sleep. This process is performed on a per-night basis.
F. Comparison to Clinical Hypnogram
To assess the accuracy of the described algorithm, agreement between automated sleep scoring and clinical sleep scoring was determined via Cohen’s kappa. Cohen’s kappa (κ) measures the inter-rater agreement between two scorers that classify items into a number of mutually exclusive categories [45]:
| (7) |
Here, po is the observed probability of agreement between scorers and pc is the probability of agreement due to chance. In this case, κ is thought to be a more robust measure than raw accuracy. A κ value of 0–0.2 is considered essentially no agreement, 0.2–0.4 slight agreement, 0.4–0.6 fair agreement, 0.6–0.8 high agreement and 0.8–1.0 nearly perfect agreement [46].
III. Results
A. Whole-Night EEG Multitaper Decomposition
To perform automated classification of whole-night sleep architecture, F3-A2 single-channel sleep EEG was spectrally decomposed via the conventional FFT spectrogram and MT spectral estimate.
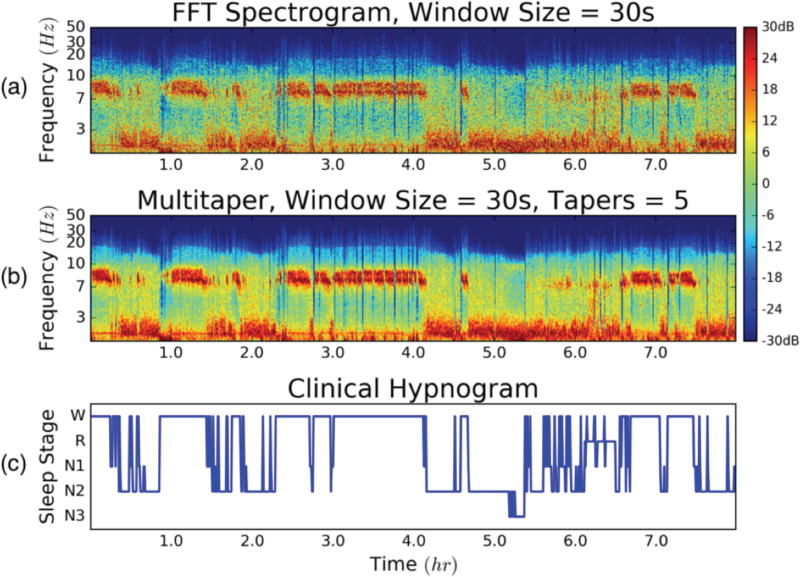
Fig. 3 illustrates an example of time-frequency outputs of both methods for 30s, non-overlapping windows over a whole night of sleep. The corresponding manually-scored hypnogram is aligned with both representations of the single-channel EEG data, revealing the connection between EEG spectral features and full PSG-based sleep scored architecture. Conventional spectral decomposition of the sleep EEG signal visually exhibited noisier outputs, as compared to the MT approach. Specifically, spectral bleeding was prevalent in frequency bands between 3–7 Hz (i.e. θ and δ waves) and higher frequency components (β and γ) when using the FFT. This is significant since, β, and γ waves are essential in distinguishing between sleep stages W, R, and N1, as previously noted. Though the MT approach resolved this problem and provided a more de-noised time-frequency image of sleep EEG, both methods provided clear association between spectral EEG features and manually-scored sleep architecture.
Fig 3.
(a) Conventional FFT spectrogram of channel F3 EEG data for a full night of sleep; 30s, 0s-overlapping windows. (b) Multitaper spectral estimate of the same data set; 30s windows and 5 tapers. (c) Ground Truth – full PSG, manually-scored clinical hypnogram; 30s epochs.
B. Sleep Stage Spectral Density Estimation
Following EEG spectral feature extraction, the 5-fold cross validation for 65 nights of HNUCSD and OSAUCSD sleep was constructed. Density estimation was implemented in the training phase of the proposed algorithm to construct the stage-specific EEG likelihood models Ri(y). All 11 features were used for density estimation, culminating in 5 probability density functions specific to W, R, N1, N2, and N3 for each fold of data.
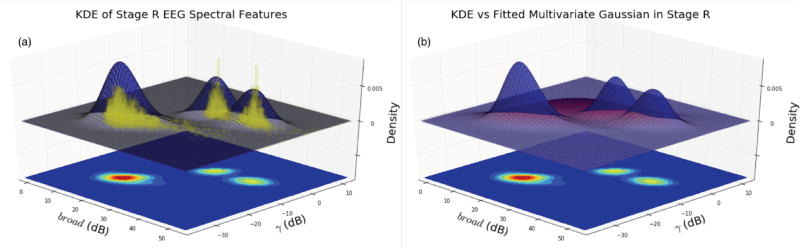
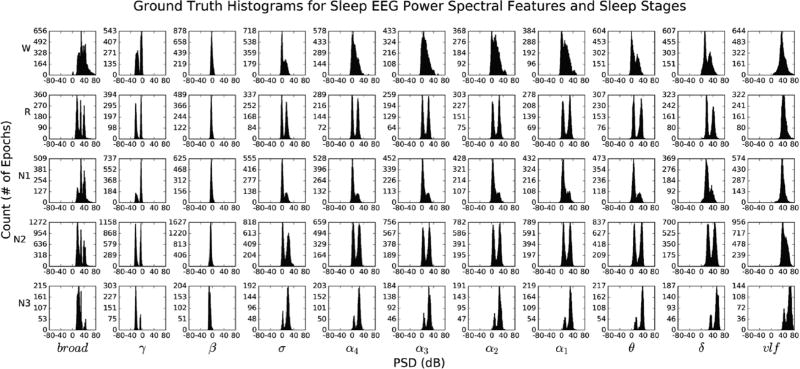
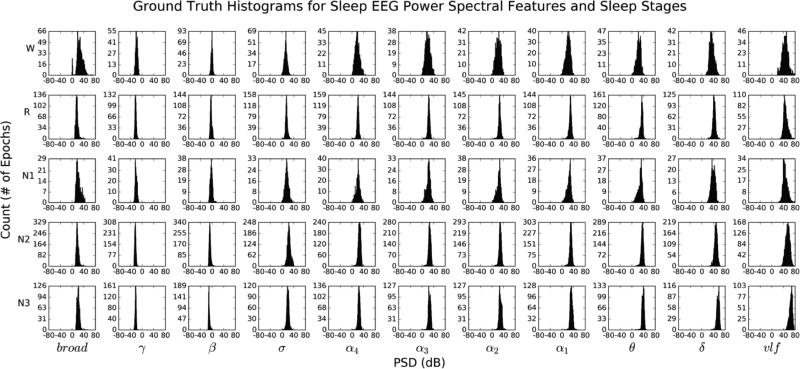
Fig. 4 (given in the Appendix) illustrates “ground truth” univariate histograms of all 60,903 extracted spectral features per each sleep stage (55 histograms total). Many EEG features exhibit bimodal structure within the same sleep stage, supporting the need to go beyond multivariate Gaussian modeling of intra-stage sleep EEG activity. Conversely, Fig. 5 (given in the Appendix) illustrates histograms for 12,084 epochs of HNUCSD data only, revealing unimodal Gaussian-like structure across all sleep stage-EEG feature combinations. Fig. 6 illustrates an example of the 3D likelihood surfaces of both density estimation and fitted multivariate Gaussian approaches for the domain of γ and broad EEG spectral features. EEG spectral data were first used to construct a “ground truth” histogram of the likelihood surface during stage R (yellow bars). Similarly, the data were used in density estimation to generate a likelihood surface (blue), which closely followed the histogram’s intricate trimodal structure. Conversely, the fitted Gaussian likelihood surface (red) failed to depict accurately the underlying distribution of γ and broad features, instead modeling it as a single wide peak between the three true modes.
Fig 6.
(a) Yellow = Ground truth histogram of broad and γ spectral data in stage R. Blue = bivariate, multimodal distribution of the log-power data generated via density estimation. (b) Blue = bivariate, multimodal distribution of the log-power data generated via density estimation. Red = bivariate, unimodal distribution of the same data generated via fitted Gaussian. Floor projections depict the blue estimated surface topography.
C. Whole-Night Sleep Architecture Estimation
Results from stage-specific density estimation were implemented into the 5-state HMM, along with initial probabilities πi, transition probabilities Qi,j, and testing feature vectors yt. In concert with the VA, the result was an estimation of whole-night sleep architecture derived from single-channel F3-A2 and Fpz-Cz EEG.
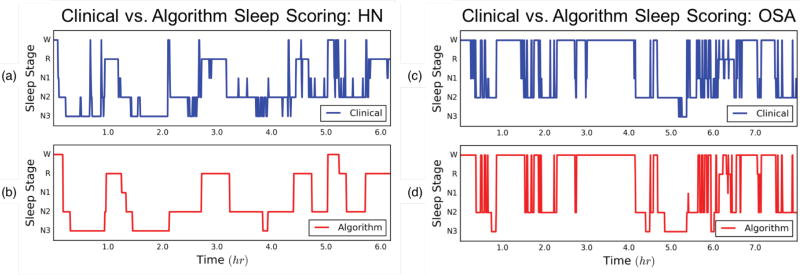
An example of the final algorithmic output is shown in Fig. 7. Panels (a) and (b) illustrate the expert-scored clinical hypnogram and corresponding automated algorithm score for HN sleep, respectively. Additionally, the same hypnogram from Fig. 3 is shown again here in panel (c), with the corresponding automated score shown in panel (d). As is evident, the algorithm was able to follow closely the macrostructure of expert-scored sleep architecture despite using only a single channel of EEG. The algorithm was also able to capture many nuances in sleep microstructure such as the many arousals from stage N2 to stage W, and reversions back to sleep evident in the OSA hypnogram. An exception of this was the algorithm under-scoring of stage N2 epochs, which were scored instead as N3 at moments throughout the night of sleep. For the night of sleep in panels (c) and (d), the subject had an AHI = 63.1 events/hr, i.e. severe OSA. In spite of this finding, the algorithm was able to score accurately whole-night sleep architecture with a κ = 0.70. To put this into perspective, the mean inter-rater κ between two experts scoring OSA sleep using full PSG is 0.59 [6].
Fig 7.
(a) HN clinical sleep hypnogram from full PSG and technician manual scoring. (b) HMM-based Algorithm using single-lead forehead EEG. Subject AHI = 2.1/hour; Cohen’s Kappa = 0.69. (c) OSA clinical sleep hypnogram from full PSG and technician manual scoring. (d) HMM-based Algorithm using single-lead forehead EEG. Subject AHI = 63.1/hour; Cohen’s Kappa = 0.70.
D. Per-Night & Per-Epoch Sleep Staging Comparison
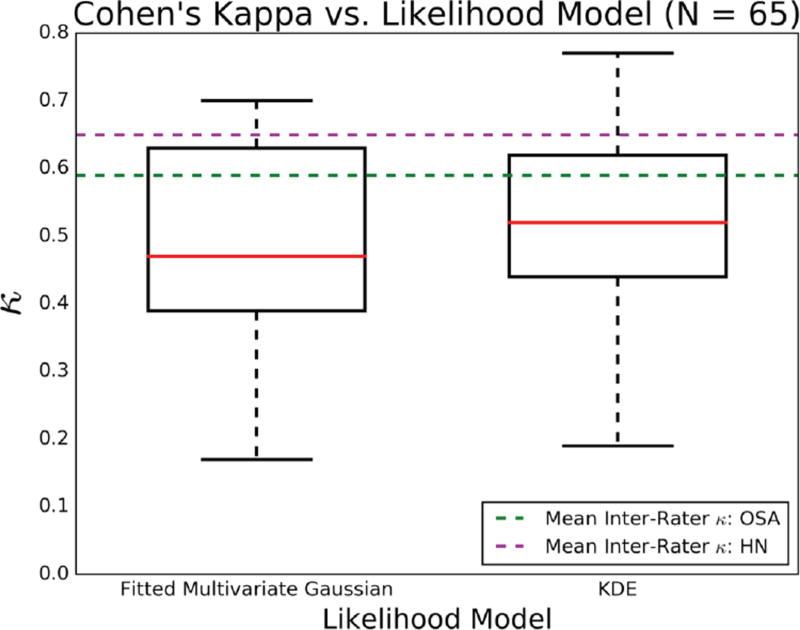
Cohen’s kappa was used to investigate the algorithm’s per-night classification performance against corresponding experts-cored hypnograms. Furthermore, two instantiations of the proposed algorithm – one using a fitted multivariate Gaussian likelihood model and another using KDE – were employed to investigate the utility of density estimation in modeling the expected multimodal structure of sleep EEG. Fig. 8 shows a box plot of the per-night κ values generated for each of the two likelihood models. Each model made use of all 65 whole-night UCSD datasets (HNUCSD and OSAUCSD) originally separated in the 5-fold cross validation.
Fig 8.
Box plots of per-night Cohen’s Kappa values, for two likelihood models: fitted multivariate Gaussian and density estimation. Red line = Median. Box edges = 1st and 3rd Quartiles. Whiskers = (1.5 × IQR). Dashed green and magenta lines = mean inter-rater Cohen’s Kappa between two experts using full PSG in HN and OSA subjects, respectively [6].
The framework utilizing density estimation exhibited a slightly higher median (κ = 0.52) than the alternative using a fitted Gaussian (κ = 0.47). Median values for both frameworks would classify as “fair agreement”, and were in the same agreement domain as the mean inter-rater agreement between two experts scoring sleep in OSA (κ = 0.59). The inter-quartile range (IQR) of the density estimation model was also narrower, suggesting less variability in the model’s ability to classify accurately whole-night sleep architecture. Moreover, whisker edges of the density estimation-based model were both higher than the fitted Gaussian approach, with the 4th quartile of κ values entirely higher than the mean inter-rater κ for OSA. When only inspecting the OSAUCSD results, the median κ and IQR for the fitted Gaussian were 0.43 and 0.15, respectively, while for the density estimation approach were 0.48 and 0.15, respectively. These results suggest that density estimation procedures have the potential to better statistically encode the structure of sleep, and thus are appropriate for use in single-channel automated sleep scoring. In addition to per-night assessment of the density estimation-based algorithm, sensitivity and specificity values were calculated on a per-epoch basis. Table II displays a 5-stage confusion matrix between Clinical and Algorithm scores for each of the 60,903, 30s epochs of sleep. The following is the true per-stage breakdown of the 30s epochs: Wc = 14,582, Rc = 7,105, N1c = 11,398, N2c = 23,021, N3c = 3,797.
Table II.
Confusion Matrix for epoch-by-epoch comparison of Clinical PSG-based sleep scoring vs. Algorithm scoring.
| Algorithm | |||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| WA | RA | N1A | N2A | N3A | Sensitivity | ||
|
| |||||||
| Clinical | Wc | 12,367 | 783 | 1,132 | 261 | 39 | 85% |
|
|
|
||||||
| Rc | 466 | 5,651 | 646 | 308 | 34 | 80% | |
|
|
|
||||||
| N1c | 2,059 | 2,556 | 4,823 | 1,894 | 66 | 42% | |
|
|
|
||||||
| N2c | 913 | 2,586 | 3,913 | 12,815 | 2,794 | 56% | |
|
|
|
||||||
| N3c | 35 | 22 | 12 | 426 | 3,302 | 87% | |
|
| |||||||
| Specificity | 78% | 49% | 46% | 82% | 53% | ||
|
|
|||||||
Using a single-channel of EEG, the proposed algorithm performed exceptionally well in per-epoch recall of stages W, R, and N3 (85%, 80%, and 80%, respectively). Recall that the mean inter-rater agreement for OSA data is just above 70%; by this metric, the sensitivities for W, R, and N3 were on-par with full PSG expert scoring. Stages N1 and N2 reported lower sensitivities values (42% and 56%, respectively). This is expected for stage N1, as it often resembles stages W and R; this misclassification is evident in the spread of W-R-N1 values in the confusion matrix.
Regarding specificity, the algorithm performed best in stages W and N2 (78% and 82%, respectively), with the remaining three stages reporting values between 45–55%. For stage R, the lower specificity is accounted for by misclassifications of stages N1 and N2, while stage N1 was misclassified most as N2 and R. For stage N3, specificity was low due to misclassification with stage N2, though algorithm sensitivity for stage N3 was high.
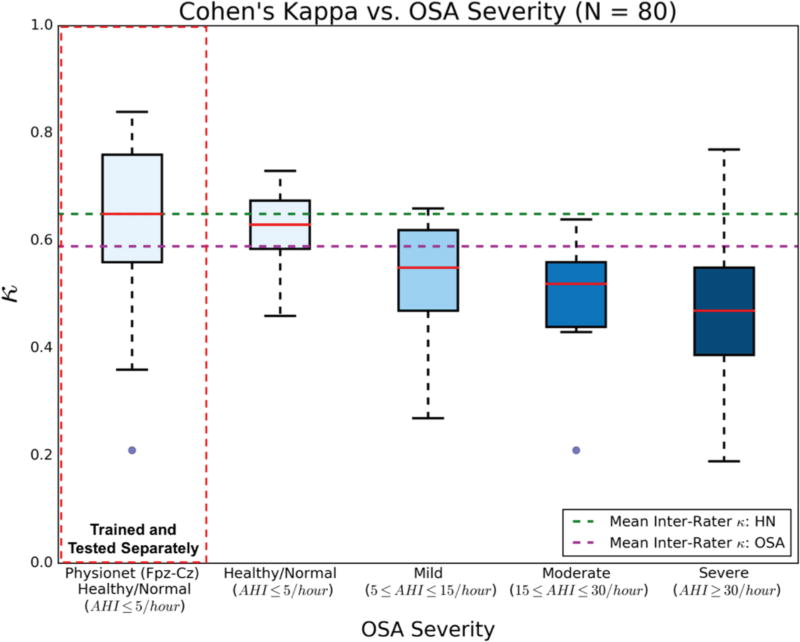
E. Algorithm Performance vs. OSA Severity
To determine the effect of OSA on algorithm performance, the per-night κ values generated via the density estimation-based algorithm were compared across healthy/normal (N = 15), mild (N = 9), moderate (N = 9), and severe OSA (N = 32) categories (Fig. 9). Overall, the downward trend of κ as a function of OSA severity was modest, which indicates a robustness in the algorithm’s ability to score appropriately degrees of fragmented sleep architecture.
Fig 9.
Box plots of per-night Cohen’s Kappa values, for four categories of OSA severity. All values were generated via the density estimation-based algorithm. HNPhysionet Fpz-Cz data were trained and tested separately.
Data extracted from Physionet were separately used for algorithm training and whole-night sleep architecture classification, using the five-fold cross validation method described above. The HNPhysionet data (N = 15) were trained and tested separately due to the difference in sensing montage (Fpz-Cz) used to acquire the public EEG data. The results of the HNPhysionet analysis are also illustrated in Fig. 9, juxtaposed with HNUCSD results to indicate the algorithm performance based on differing healthy/normal EEG acquisition. Using the Fpz-Cz single-channel data, the algorithm produced a median κ exactly equal to the mean inter-rater agreement between two experts scoring sleep in HN subjects (κ = 0.65). Alternatively, to test the generalizability of the trained algorithm, HNPhysionet data was used as test data in the F3-A2-trained algorithm. As expected, algorithm performance on the HNPhysionet data dropped to a median κ = 0.47, with an IQR = 0.31, similar to the results for severe OSA F3-A2 data. Still, more than half of the HNPhysionet classification were considered to be in at least fair agreement, which suggests the algorithm is able to reconcile similar sleep EEG features in data from different sensing montages.
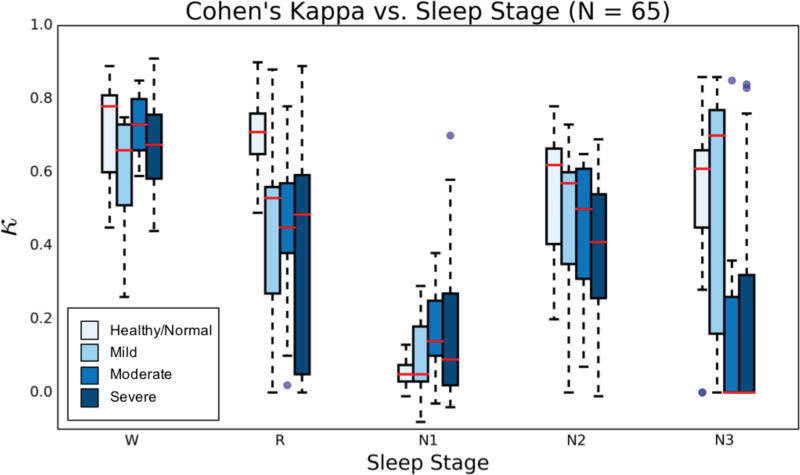
In addition to stratifying performance across OSA severity, κ values were further partitioned based on sleep stages, from whole-night sleep architecture results. Fig. 10 illustrates the stage-specific algorithm performance with increasing OSA severity. Only HNUCSD and OSAUCSD data was included (N = 65). As expected, per-stage κ performance trends downward as OSA increases from HN to severe OSA. An exception here is stage N1, which exhibits a modest increase in κ spread, the largest occurring for severe OSA classification. Stages W, R, and N2 maintain median κ values in at least fair agreement across OSA severity, with a large increase in IQR for stage R in the severe case. Stage N3 appears as the most variable in performance, with an abrupt drop in median κ upon transition from mild to moderate OSA. Despite this, about one quarter (≈ 8 nights) of all N3 κ values for severe OSA lie within the fair/high agreement range. This observation, combined with sustained κ values in the other stages of sleep, further suggests the single-channel algorithm demonstrates classification robustness across OSA severities.
Fig 10.
Box plots of per-night Cohen’s Kappa values, for each stage of sleep and across OSA severity. HNPhysionet data not included because of the different sensing montage.
IV. Discussion
To improve the current state of automated sleep scoring and provide a means for assessing pathological sleep, an algorithm is presented that utilizes a limited physiologic dataset (i.e. single-channel EEG) to estimate whole-night sleep architecture in OSA and HN subjects. The algorithm makes use of KDE to generate statistical models based on single-channel sleep EEG spectral features, and a HMM to formulate whole-night sleep architecture as a state space, transition-constrained process. Few studies have focused automated sleep scoring efforts on OSA subjects [10], [12], [24], [27], and none have implemented a multimodal statistical framework such as that presented here for investigating OSA sleep architecture. The results of this study indicate that this statistical approach to scoring sleep in OSA subjects with single-channel sleep EEG is effective and promising as a means to emulate expert-level scoring in an automated fashion.
Spectral EEG features were generated via the MT spectrogram, which has been shown to produce more accurate spectral estimates of EEG, as compared to the standard FFT, wavelet transform, and other spectral decomposition techniques [33, 34]. Each feature was extracted to embody the rules used for visual scoring of sleep EEG, specifically extracting the activity in EEG frequency bands that possess information about each or a combination of the five stages of sleep. For example, a 0.1–50 Hz broad power feature was used to quantify motion artifact as sharp deflections spanning most frequency bands in the recorded EEG, which typically appear during stage W and at the onset of arousal. Other well-known EEG rhythms (e.g. α, θ, and δ) were used to quantify activity in corresponding characteristic sleep stages, as per Table I. Unique to this work is the separation of the α band into four 1 Hz bands (α1–α4) to capture the nuanced α activity in stage R, which is expected as a 1–2 Hz slower α rhythm compared to stage W [4].
To develop the distribution of spectral features within each stage of sleep, density estimation was used over standard fitted Gaussian approaches. Specifically, KDE was implemented to generate likelihood estimates capturing multimodal structure of the joint density surface in regards to spectral variability within a stage of sleep. For example, approximately 20% of the adult U.S. population generates little or no α activity during wakefulness [4], [47]. In this context, a standard Gaussian model might not accurately represent both the “presence” and “absence” of α activity within stage W, incorrectly approximating a bimodal distribution as a single over-smoothed mode in the domain of α activity. In addition, multimodal statistical modeling of sleep stages can begin to quantify the errors/variability in human sleep scoring within sleep stages. Since visual, per-epoch sleep scoring is not an exact science, small variations in intra-stage human sleep scoring can manifest as large discrepancies in standard Gaussian modeling, resulting in poor algorithmic sleep scoring performance.
Fig. 4 and 5 (given in the Appendix) illustrate univariate histograms of all eleven spectral features (Table I) for each sleep stage, depicting the unimodality of HN sleep EEG and the sometimes subtle changes in EEG spectra as OSA subjects go into deeper modes of sleep. In some instances, a standard Gaussian would represent the data distribution accurately (e.g. most HN sleep and the unimodal distributions of β and vlf features in most stages for OSA). Conversely, other features – in particular, γ, δ, σ, and θ – exhibited distinct multimodal structure across all stages of OSA sleep, which cannot be correctly captured by the standard Gaussian (Fig. 6).
In OSA, each of the four α band features extracted display a transformation from unimodal, skewed Gaussian-like structure in stage W, to bimodal structure in the positive PSD domain during stage N3. This is interesting since α-type rhythms are not typically considered key indicators of deeper sleep, yet distinct peaks centered around 0 dB and 30 dB are present in N2 and N3 sleep, the latter value of which is similar to that in stage W and stage R. This suggests that the histograms (and as a consequence, the KDE likelihoods) capture two different populations of spectral EEG – one centered about 0 dB and another centered about 30 dB. The former might portray the “typical” suppressed EEG signature of α activity in N2–N3 sleep, while the latter might reflect an increase in α activity related to respiratory-based arousals and increased sympathetic activation during these epochs of sleep in OSA subjects [2].
Whole-night sleep architecture was modeled as a HMM transition-constrained process, with conditional likelihoods dictating physiologic transitions during sleep. Previous work in the literature has used HMM to model and score sleep [16]–[21], though none has focused on multimodal class conditional densities, nor have they exclusively focused on OSA subject data as used in the presented HMM framework. The 5-stage transition values used in the HMM were extrapolated from previous work on a 4-stage transition model – Wake, REM, Light (N1/N2), Deep (N3) – of sleep in OSA subjects [44]. Specifically, the proposed algorithm implemented transition likelihoods for N1 and N2, each stage with identical transition probabilities to ensure parity when expanding the “light” stage values to the new 5-stage model. Transitions between N1 and N2 were made more probable (e.g. Qi,j = 0.20), compared to transitions to other stages (e.g. Qi,j = 0.10), to reflect the increased fragmentation and wake-sleep characteristics of sleep in OSA subjects.
Cohen’s kappa values for per-night, KDE-based classification of sleep architecture are shown in Fig. 8. Values range from κ = 0.20 to κ = 0.77, with a median κ = 0.52 and more than three quartiles of the values demonstrating at least “fair agreement” in classification accuracy when compared to expert-scored hypnograms. For perspective, the mean inter-rater Cohen’s kappa between experts scoring with full PSG in HN and OSA patients is κ = 0.65 and κ = 0.59, respectively [6]. While the fitted Gaussian model also produced κ values in fair agreement, this is suspected to be due largely in part to the inclusion of HN EEG datasets, which stand to benefit less from a density estimation-based approach when a simpler unimodal Gaussian will suffice, as depicted in Fig. 5. It seems that density estimation plays a smaller role in improving sleep scoring in HN subjects, instead excelling when implemented on sleep that is heavily fragmented (such as in OSA). These results suggest that the proposed algorithm performs quite well in scoring sleep architecture in a mix of HN and OSA sleep, despite only using a single channel of EEG data.
Further investigation of algorithm performance revealed a modest inverse relationship between per-night κ agreement and OSA severity (Fig. 9). Increased sleep fragmentation equates to more wake-sleep transitioning and a general difficulty in sleep architecture classification. While agreement between the algorithm and clinical scoring decreases as OSA severity increases, Fig. 9 illustrates that the algorithm achieved fair agreement values above κ = 0.50 for almost half of the 32 total nights of sleep with severe OSA. An example of the algorithm’s ability to accurately classify sleep architecture in a subject with severe OSA (AHI = 63.1 events/hr) is shown in Fig. 7. Further speaking to performance on HN data, results from a separate 5-fold cross validation on HNPhysionet data (Fig. 9) show that the algorithm works equally well on data derived from another EEG montage (i.e. Fpz-Cz), emphasizing the generalizability of the described methods. This suggests that the algorithm may not only be robust to certain degrees of OSA severity, but can also be improved to appropriately score sleep in a manner agnostic to sleep fragmentation and EEG acquisition.
A stage-specific analysis of the combined HNUCSD and OSAUCSD results further revealed modest deterioration of the algorithmic single-channel scoring for increasing OSA severity (Fig. 10). As expected, results for stages W, R, and N2 are primarily in good agreement, with little deterioration across OSA severity. Interestingly, stage N1 agreement increased slightly as AHI increased, running counter to other sleep stages. As OSA worsens, an increase in sleep fragmentation generally leads to an increased frequency of stage N1, as patients arouse from sleep more often throughout the night. As a consequence, EEG spectral features related to stage N1 may become more prominent, which may accommodate increased classification accuracy of stage N1 in this analysis.
An increase in N1 scoring during OSA would elicit an infrequency of other sleep stages for the same total sleep time, such as stage N3 and stage R (whose specific discrimination from N1 is already difficult in HN patients). For stage N3, many agreement values in moderate and severe OSA dropped dramatically to κ = 0.00, though some values extend past fair agreement and well into high agreement. The same occurs in stage R for mild and severe OSA. Based on the large degree of κ spread, it appears low Cohen’s κ values not only arise from sheer misclassification between two classes, but also from an uneven distribution of samples between two classes (e.g. in a whole night of sleep, Rc = 25 epochs, Non-Rc = 600 epochs). The result is a trade-off in stage-specific κ performance due to OSA severity, specifically with infrequent sleep stages demonstrating a high agreement due to chance, which by virtue of the numerator of equation (7), results in a low κ score. This happens to be an example of low κ values resulting from imbalance/low prevalence of an observation, a limitation of Cohen’s κ which has been discussed extensively in the literature [48].
Whole-night classification results demonstrate improved N1 scoring over the literature, while maintaining high degrees of classification for other sleep stages. This suggests that the algorithm has the potential to accurately and automatically generate desirable sleep metrics such as “Total Sleep Time”, “Wake After Sleep Onset”, and “Sleep Efficiency”. Even so, improvement is necessary, in particular to address the difficulty in classification for datasets with increased N1–N2 transitioning. As discussed, this is a general problem of automated sleep scoring, even for scoring in HN subjects, demonstrated by low-sensitivity results for N1 staging in the single-channel algorithm literature [10], [13], [16], [23], [24], [25], [27], [28].
Another area to be addressed is the algorithm sensitivity and specificity between stages N2 and N3. Differences in the accuracy of N2–N3 scoring have been observed before, in particular the over-scoring of N3 compared to N2 for data derived from frontal sensors, as compared to central derivations [49]. More generally, a marked difference in N2–N3 scoring has been observed between automated and manual scoring of sleep [11]. It is difficult to ascertain if automated algorithms such as the proposed are incorrectly scoring N2–N3 epochs of sleep, if the discrepancy is due solely to bias in the manual scoring performed by the expert, or a combination of the two. Because an automated algorithm can quantify minute differences in EEG (e.g. presence and strength of delta waves) more easily and efficiently than a visual scorer, it has been suggested that automated scoring is possibly more precise in N2–N3 classification [11].
The presented work utilized the MT spectral estimate to generate and extract frontal EEG spectral features. Implementation of novel spectrotemporal decomposition techniques [50] might serve to improve algorithm performance through integrated knowledge of the sparse macrostructure of sleep EEG when rendering spectral estimates. Regarding EEG-based features, frontal EEG-derived eye movement and K-complex information can be extracted via cross-correlation approaches to improve the specificity in scoring stages R and N2, respectively [51], [52]. Moreover, a natural extension of the proposed work is the automated detection of arousals and apneas/hypopneas based on single-channel/limited physiologic data streams. A method for detection of relevant sleep phenomena, and subsequent generation of clinical criterion for OSA screening, could be formulated by closer inspection and characterization of the multimodal distributions of EEG spectra described here. By using appropriate statistical methods that accommodate multimodal distributions, it may be possible to categorize an arbitrary epoch of sleep EEG as HN or OSA-like. Moreover, it may be possible to use such per-epoch categorizations to estimate whole-night OSA severity. As such, the resulting paradigm, using only single-channel EEG, has the potential to serve as a surrogate for clinical assessment of arousal indices or AHI. In addition, it may even help characterize different phenotypes of OSA and other sleep disorders.
Finally, to facilitate frontal-based sensing of physiologic signals during sleep, novel technologies in the field of wearable sensors and systems [53], [54], [55] can be leveraged as tools for unobtrusive, peel-and-stick sleep monitoring. Combined with low-resource algorithms – such as the proposed statistical methods – wearable systems can begin to monitor sleep objectively, thus allowing clinical metrics that go beyond the current standard of subjective recall.
V. Conclusion
New technologies have the potential to disrupt the clinic, and the field of sleep medicine may be able to move beyond the limitations of the “gold standard” PSG through smaller and more efficient devices for recording and generating clinical sleep metrics. While the recent surge of minimalistic, at-home sleep monitoring devices aims to improve sleep medicine practices, these endeavors lack analytic techniques that efficiently estimate sleep architecture from reduced data streams. This work outlines a statistical framework for classifying whole-night sleep architecture from single-channel EEG spectral features. The algorithm formulated sleep architecture and the five clinical stages of sleep as a transition-constrained, state space process with intra-stage multimodality in the domain of EEG spectra. Results of the study show the algorithm is able to utilize single-channel EEG to automatically discriminate and score whole-night sleep architecture in both HN and OSA sleep, in many cases with high Cohen’s kappa agreement, when compared to clinical scoring from experts using full PSG. Moreover, the algorithmic approach sustains fair scoring agreement for increased OSA severity, demonstrating potential for generalizability and objectivity in the evaluation of the many intricacies of sleep and sleep disorders. This is one of just a few studies that have implemented state space modeling for single-channel sleep scoring, and the first known study to implement such statistical methods for automated sleep architecture performance in OSA subjects. The continued development of such low-resource algorithms – guided by clinical expertise and emphasizing clinical practicality – will help realize automated tools for assessing sleep and sleep disorders in inpatient and outpatient populations.
Appendix
Fig 4.
Univariate, multimodal histograms of the eleven extracted EEG spectral features listed in Table I, for each true stage of sleep (55 histograms total). Distributions were generated using all 60,903 30s epochs from 65 total HN/ OSA datasets and their true corresponding labels from expert scoring. Per-stage breakdown of all labeled 30s epochs: W = 14,582, R = 7,105, N1 = 11,398, N2 = 23,021, N3 = 3,797.
Fig 5.
Univariate, multimodal histograms of the eleven extracted EEG spectral features listed in Table I, for each true stage of sleep (55 histograms total). Distributions were generated using all 12,084 30s epochs from the 15 HN datasets and their true corresponding labels from expert scoring. Per-stage breakdown of all labeled 30s epochs: W = 1,247, R = 2,351, N1 = 721, N2 = 5,609, N3 = 2,156.
References
- 1.Kryger MH, Roth T, Dement WC. Principles and Practice of Sleep Medicine. 5. Philadelphia, PA: W.B. Saunders; 2010. [Google Scholar]
- 2.Roebuck A, et al. A review of signals used in sleep analysis. Physiological Measurement. 2014;35(1):R1–R57. doi: 10.1088/0967-3334/35/1/R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rechtschaffen A, Kales A. A Manual Of Standardized Terminology, Techniques and Scoring Systems for Sleep Stages of Human Subjects. U. G. P. Office, Washington DC: Public Health Service, U.S. Government Printing Service; 1968. [Google Scholar]
- 4.Berry RB, Brooks R, Gamaldo CE, Harding SM, Marcus CL, Vaughn BV for the American Academy of Sleep Medicine. The AASM Manual for the Scoring of Sleep and Associated Events: Rules, Terminology and Technical Specification, Version 2.0. Darien, IL: American Academy of Sleep Medicine; 2007. [Google Scholar]
- 5.Iber C, et al., editors. The AASM Manual for the Scoring of Sleep and Associated Events: Rules, Terminology, and Technical Specification. American Academy of Sleep Medicine; Westchester: 2007. [Google Scholar]
- 6.Norman RG, et al. Interobserver agreement among sleep scorers from different centers in a large dataset. Sleep. 2000;23(1):901–908. [PubMed] [Google Scholar]
- 7.Agarwal R, et al. Computer-Assisted Sleep Staging. IEEE Transactions on Biomedical Engineering. 2001;48(12):1421–1423. doi: 10.1109/10.966600. [DOI] [PubMed] [Google Scholar]
- 8.Virkkala J, et al. Automatic sleep stage classification using two-channel electro-oculography. Journal of Neuroscience Methods. 2007;166(1):109–155. doi: 10.1016/j.jneumeth.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 9.Liang S-F, et al. A rule-based automatic sleep staging method. Journal of Neuroscience Methods. 2012;205(1):169–176. doi: 10.1016/j.jneumeth.2011.12.022. [DOI] [PubMed] [Google Scholar]
- 10.Stepnowsky C, et al. Scoring accuracy of automated sleep staging from a bipolar electroocular recording compared to manual scoring by multiple raters. Sleep Medicine. 2013;14(11):1199–1207. doi: 10.1016/j.sleep.2013.04.022. [DOI] [PubMed] [Google Scholar]
- 11.Malhotra A, et al. Performance of an Automated Polysomnography Scoring System Versus Computer-Assisted Manual Scoring. Sleep. 2013;36(4):573–582. doi: 10.5665/sleep.2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koley B, et al. An ensemble system for automatic sleep stage classification using single channel EEG signal. Computers in Biology and Medicine. 2012;42(12):1186–1195. doi: 10.1016/j.compbiomed.2012.09.012. [DOI] [PubMed] [Google Scholar]
- 13.Zhu G, et al. Analysis and Classification of Sleep Stages Based on Difference Visibility Graphs From a Single-Channel EEG Signal. IEEE Transaction on Biomedical Engineering. 2014;18(6):1813–1821. doi: 10.1109/JBHI.2014.2303991. [DOI] [PubMed] [Google Scholar]
- 14.Radha M, et al. Comparison of Feature and Classifier Algorithms for Online Automatic Sleep Staging Based on a Single EEG Signal. Presented at The 36th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC 2014) 2014:1876–1880. doi: 10.1109/EMBC.2014.6943976. [DOI] [PubMed] [Google Scholar]
- 15.Bajaj V, et al. Automatic classification of sleep stages based on the time-frequency image of EEG signals. Computer Methods and Programs in Biomedicine. 2013;112(3):320–328. doi: 10.1016/j.cmpb.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 16.Flexer A, et al. A reliable probabilistic sleep stager based on a single EEG signal. Artificial Intelligence in Medicine. 2005;33(3):199–207. doi: 10.1016/j.artmed.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 17.Doroshenkov LG, et al. Classification of Human Sleep Stages Based on EEG Processing Using Hidden Markov Models. Biomedical Engineering. 2007;41(1):25–28. [PubMed] [Google Scholar]
- 18.Bianchi MT, et al. Probabilistic sleep architecture models in patients with and without sleep apnea. J. Sleep Res. 2012;21(3):330–341. doi: 10.1111/j.1365-2869.2011.00937.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan S-T, et al. A transition-constrained discrete hidden Markov model for automatic sleep staging. Biomedical Engineering Online. 2012;52(1):52. doi: 10.1186/1475-925X-11-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yaghouby F, et al. Quasi-supervised scoring of human sleep in polysomnograms using augmented input variables. Computers in Biology and Medicine. 2015;59(1):54–63. doi: 10.1016/j.compbiomed.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Onton J, et al. Visualization of Whole-Night Sleep EEG From 2-Channel Mobile Recording Device Reveals Distinct Deep Sleep Stages with Differential Electrodermal Activity. Frontiers in Human Neuroscience. 2016;10(605):1–12. doi: 10.3389/fnhum.2016.00605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shambroom, et al. Validation of an automated wireless system to monitor sleep in healthy adults. Journal of Sleep Research. 2012;21(2):221–230. doi: 10.1111/j.1365-2869.2011.00944.x. [DOI] [PubMed] [Google Scholar]
- 23.Hsu Y-L, et al. Automatic sleep stage recurrent neural classifier using energy features of EEG signals. Neurocomputing. 2013;104(1):105–114. [Google Scholar]
- 24.Fraiwan, et al. Automated sleep stage identification system based on time–frequency analysis of a single EEG channel and random forest classifier. Computer Methods and Programs in Biomedicine. 2012;108(1):10–19. doi: 10.1016/j.cmpb.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 25.Liang S-F, et al. Automatic Stage Scoring of Single-Channel Sleep EEG by Using Multiscale Entropy and Autoregressive Models. IEEE Transactions on Instrumentation and Measurement. 2012;61(6):1649–1657. [Google Scholar]
- 26.Garcia-Molina G, et al. Online Single EEG Channel Based Automatic Sleep Staging. Presented at International Conference on Engineering Psychology and Cognitive Ergonomics (EPCE 2013) 2013:333–342. [Google Scholar]
- 27.Lainscsek C, et al. Applied Non-Linear Dynamical Systems. Cham, Switzerland: Springer International Publishing; 2014. Automatic Sleep Scoring from a Single Electrode Using Delay Differential Equations; pp. 371–382. [Google Scholar]
- 28.Hassan AR, et al. On the Classification of Sleep States By Means of Statistical and Spectral Features from Single Channel Electroencephalogram. Presented at International Conference on Advances in Computing, Communications, and Informatics (ICACCI 2015) 2015:2238–2243. [Google Scholar]
- 29.Heinzer R, et al. Prevalence of sleep-disordered breathing in the general population: the HypnoLaus study. Lancet Respiratory Medicine. 2015;3(4):310–318. doi: 10.1016/S2213-2600(15)00043-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kemp B, et al. Analysis of a sleep-dependent neuronal feedback loop: the slow-wave microcontinuity of the EEG. IEEE Transactions on Biomedical Engineering. 2000;47(9):1185–1194. doi: 10.1109/10.867928. [DOI] [PubMed] [Google Scholar]
- 31.Goldberger AL, et al. PhysioBank, PhysioToolkit, and PhysioNet: Components of a New Research Resource for Complex Physiologic Signals. Circulation. 2000;101(23):e215–e220. doi: 10.1161/01.cir.101.23.e215. [DOI] [PubMed] [Google Scholar]
- 32.Thomson DJ. Spectrum estimation and harmonic analysis. Proceedings of the IEEE. 1982;70(9):1055–1096. [Google Scholar]
- 33.Babadi B, et al. A review of multitaper spectral analysis. IEEE Transactions on Biomedical Engineering. 2014;61(5):1555–64. doi: 10.1109/TBME.2014.2311996. [DOI] [PubMed] [Google Scholar]
- 34.Prerau MJ, et al. Sleep Neurophysiology Dynamics Through the Lens of Multitaper Spectral Analysis. Physiology. 2016;32(1):60–92. doi: 10.1152/physiol.00062.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bronez T. On the Performance Advantage of Multitaper Spectral Analysis. IEEE Transactions on Signal Processing. 1992;40(12):2941–2946. [Google Scholar]
- 36.Cantero JL, et al. Human alpha oscillations in wakefulness, drowsiness period, and REM sleep: different electroencephalographic phenomena within the alpha band. Neurophysiologie Clinique. 2002;32(1):54–71. doi: 10.1016/s0987-7053(01)00289-1. [DOI] [PubMed] [Google Scholar]
- 37.Williams RL, Karacan I, Hursch CJ. EEG of human sleep: clinical applications. New York: John Wiley & Sons; 1974. [Google Scholar]
- 38.Carskasdon MA, et al. Sleep fragmentation in the elderly: Relationship to daytime sleep tendency. Neurobiology of Aging. 1982;4(4):321–327. doi: 10.1016/0197-4580(82)90020-3. [DOI] [PubMed] [Google Scholar]
- 39.Stocchi F, et al. Sleep disorder in Parkinson’s disease. Journal of Neurology. 1998;245(1):S15–S18. doi: 10.1007/pl00007731. [DOI] [PubMed] [Google Scholar]
- 40.Zamir G, et al. Sleep fragmentation in children with juvenile rheumatoid arthritis. Journal of Rheumatology. 1998;25(6):1191–1197. [PubMed] [Google Scholar]
- 41.Eckert DP, et al. Trazodone increases the respiratory arousal threshold in patients with obstructive sleep apnea and a low arousal threshold. Sleep. 2014;37(4):811–819. doi: 10.5665/sleep.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scott DW, et al. On optimal and data-based histograms. Biometrika. 1979;66(1):605–610. [Google Scholar]
- 43.Rabiner LR, et al. An Introduction to Hidden Markov Models. IEEE ASSP Magazine. 1986;3(1):4–16. [Google Scholar]
- 44.Lo C-C, et al. Asymmetry and basic pathways in sleep-stage transitions. Europhysics Letters. 2013;102:10008–1. doi: 10.1209/0295-5075/102/10008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cohen J, et al. A Coefficient of Agreement for Nominal Scales. Educational and Psychological Measurement. 1960;20(1):37–46. [Google Scholar]
- 46.Landis JR, et al. The Measurement of Observer Agreement for Categorical Data. Biometrics. 1977;33(1):159–174. [PubMed] [Google Scholar]
- 47.Silber MH, et al. The visual scoring of sleep in adults. Journal of Clinical Sleep Medicine. 2007;3(2):121–131. [PubMed] [Google Scholar]
- 48.Feinstein AR, Cicchetti DM. High Agreement But Low Kappa: I. The Problems Of Two Paradoxes. Journal of Clinical Epidemiology. 1990;43(6):543–549. doi: 10.1016/0895-4356(90)90158-l. [DOI] [PubMed] [Google Scholar]
- 49.Younes M, et al. Accuracy of Automatic Polysomnography Scoring Using Frontal Electrodes. Journal of Clinical Sleep Medicine. 2016;12(5):735–746. doi: 10.5664/jcsm.5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ba D, et al. Robust spectrotemporal decomposition by iteratively reweighted least squares. Proceedings of the National Academy of Sciences. 2014;111(50):E5336–E5345. doi: 10.1073/pnas.1320637111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jobert M, et al. Pattern Recognition by Matched Filtering: An Analysis of Sleep Spindle and K-Complex Density under the Influence of Lormetazepam and Zopiclone. Neuropsychobiology. 1992;26(1–2):100–107. doi: 10.1159/000118902. [DOI] [PubMed] [Google Scholar]
- 52.Hatzilabrou GM, et al. A Comparison of Conventional and Matched Filtering Techniques for Rapid Eye Movement Detection of the Newborn. IEEE Transactions on Biomedical Engineering. 1994;41(10):990–995. doi: 10.1109/10.324532. [DOI] [PubMed] [Google Scholar]
- 53.Kim D-H, et al. Epidermal Electronics. Science. 2011;333(6044):838–843. doi: 10.1126/science.1206157. [DOI] [PubMed] [Google Scholar]
- 54.Kang DY, et al. Scalable Microfabrication Procedures for Adhesive-Integrated Flexible and Stretchable Electronic Sensors. Sensors. 2015;15(9):23459–23476. doi: 10.3390/s150923459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kao H-L, et al. DuoSkin: rapidly prototyping on-skin user interfaces using skin-friendly materials. Presented at Proceedings of the 2016 ACM International Symposium on Wearable Computers (ISWC 2016) 2016:16–23. [Google Scholar]