Abstract
Intellectual disability (ID) is a prevailing neurodevelopmental condition associated with impaired cognitive and adaptive behaviors. Many chromatin-modifying enzymes and other epigenetic regulators have been genetically associated with ID disorders (IDDs). Here we review how alterations in the function of histone modifiers, chromatin remodelers, and methyl-DNA binding proteins contribute to neurodevelopmental defects and altered brain plasticity. We also discuss how progress in human genetics has led to the generation of mouse models that unveil the molecular etiology of ID, and outline the direction in which this field is moving to identify therapeutic strategies for IDDs. Importantly, because the chromatin regulators linked to IDDs often target common downstream genes and cellular processes, the impact of research in individual syndromes goes well beyond each syndrome and can also contribute to the understanding and therapy of other IDDs. Furthermore, the investigation of these disorders helps us to understand the role of chromatin regulators in brain development, plasticity, and gene expression, thereby answering fundamental questions in neurobiology.
Keywords: α-thalassemia mental retardation syndrome, Claes-Jensen syndrome, DNA methylation, histone posttranslational modification, Kleefstra syndrome, neuroepigenetics, Rett syndrome, Rubinstein-Taybi syndrome, X-linked intellectual disability
Introduction
Intellectual disability (ID) disorders (IDDs) are characterized by impaired cognitive abilities, commonly defined by an IQ < 70, and severe deficits in the capability to adapt to the environment and social milieu. With a prevalence of 2%–3% worldwide, these neurodevelopmental disorders represent one of the biggest medical and social challenges in our society. The causes of IDDs are heterogeneous and include environmental factors, chromosomal aberrations, and single gene mutations. Human genetics and clinical research in the last decade have led to the identification of hundreds of genes responsible for these disorders. Notably, a large number of such genes encode for epigenetic regulators: by proteins that exert their function through genome-wide posttranslational modification of histones, DNA base modifications and other covalent and noncovalent changes of the chromatin (Kramer and van Bokhoven, 2009). Indeed, of the 750 genes currently linked to ID and autism spectrum disorder (ASD) (Kochinke et al., 2016), at least 55 correspond to chromatin regulators (Kleefstra et al., 2014). Several of these ID-linked chromatin factors have been found to directly interact with one another in complexes that regulate chromatin structure at genes important for neurodevelopment and/or neuroplasticity (Kleefstra et al., 2014).
The development of the nervous system is a highly organized process that requires precise spatial and temporal regulation of gene expression programs involved in differentiation, maturation, and survival of neurons, but also the repression of alternative cell fates and restriction of cell type-specific gene expression (Lilja et al., 2013). Such dynamic expression patterns are sustained by extensive changes in the epigenome. It is therefore not surprising that the mutation of genes encoding chromatin modifiers and readers lead to severe neurodevelopmental defects (Kleefstra et al., 2014; Bjornsson, 2015). However, the mechanisms by which these mutations cause IDDs and ASD are still largely unknown. Given the overall prevalence of IDDs and their large medical and social costs, it has become increasingly important to understand their molecular etiology.
Emerging evidence suggests that epigenetic regulation of gene expression plays a crucial role in processing experience-driven synaptic activity required for long-lasting modifications of neural circuits and neuronal properties in the adult brain (Gupta et al., 2010; Baker-Andresen et al., 2013; Sweatt, 2016). Neurons in memory-related networks respond with patterns of activity that relay and encode information in the form of alterations in synaptic strength that modify neuronal connectivity and contribute to cognitive processes, such as learning and memory. Recent findings illustrate the dynamic nature of chromatin marks in mature neurons, demonstrating that DNA methylation and post-translational modifications of histone proteins, such as histone phosphorylation, acetylation, and methylation, actively contribute to the activity-dependent modulation of neuronal networks (Heyward and Sweatt, 2015; Bonnaud et al., 2016). Such changes have also been shown to correlate with memory formation and consolidation (Gupta et al., 2010; Lubin et al., 2011; Mews et al., 2017). As a result, during the last decade, there has been great interest in deciphering the role of chromatin modifications in neuronal plasticity processes in the brain (Zocchi and Sassone-Corsi, 2010; Sweatt, 2013; Lopez-Atalaya and Barco, 2014).
In the next sections, we review recent progress in understanding the molecular etiology of six genetic syndromes associated with ID. These IDDs are caused by mutations in chromatin regulators, ranging from chromatin remodelers to histone-modifying enzymes (Fig. 1). Much of this progress stems from the generation and characterization of appropriate mouse models for these conditions. The study of these genetic models allows us to determine the functions of ID-linked chromatin factors throughout life, dissect the cognitive and neurological defects based on their developmental or adult origin, and identify potentially druggable targets for therapy (Fig. 2). The precise understanding of the chromatin-based etiology of these IDDs will eventually help to cure or ameliorate disease phenotypes.
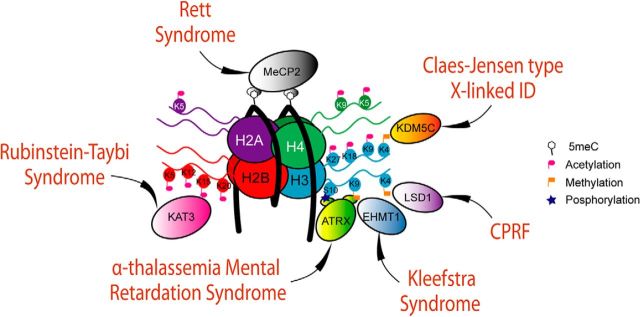
Figure 1.
Six important chromatin-related IDDs. The six IDDs discussed in this review are caused by mutations in different chromatin regulators that converge in related molecular processes. In the scheme, these chromatin factors are depicted in the proximity of their respective chromatin targets. All the acetylation marks presented in the scheme are possible substrates of KAT3 proteins. K, lysine; 5meC, 5-methylcytosine.
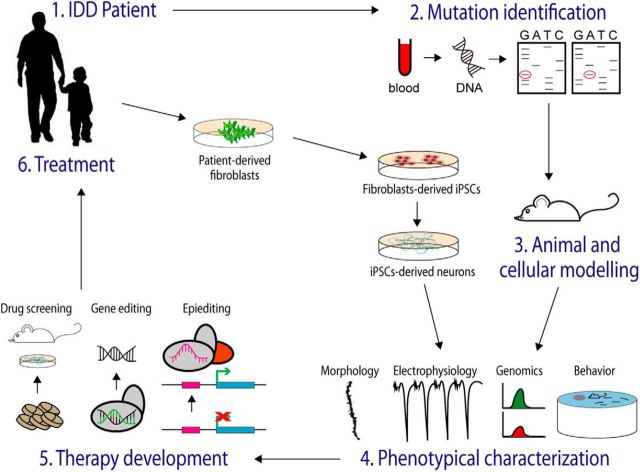
Figure 2.
From identification of ID-related genes to therapy. Schematic representation of the long, and still unaccomplished, path that goes from the identification of the IDD-causing mutation to therapy. After identification of the mutation, the generation and characterization of animal model reproducing the same genetic defects enable the description of the molecular mechanisms underlying the disease and the assessment of therapies. Complementing the studies of animal models, iPSCs derived from the patients can be also used to investigate pathoetiology and assess possible therapies. Therapeutic strategies that provide positive results in the cellular and animal models, such as drug treatment and gene editing or epi-editing, will be eventually evaluated in clinical trials.
ATRX and α-thalassemia mental retardation syndrome
Mutations in the ATRX gene cause α-thalassemia mental retardation syndrome (ATR-X; OMIM #301040). ATR-X syndrome patients present ID, microcephaly, dysmyelination, seizures, autistic-like behavior, microcephaly, α-thalassemia, dysmorphic faces, short stature, skeletal defects, and urogenital abnormalities (Gibbons et al., 2008). The disease is X-linked and confined to males, whereas female carriers display highly skewed X chromosome inactivation toward the mutant allele and are usually phenotypically normal. All inherited ATRX mutations identified to date are hypomorphic, suggesting that null mutations are lethal.
ATRX is a chromatin-remodeling enzyme that uses ATP hydrolysis to move nucleosomes on the DNA template or promote nucleosome exchange (Xue et al., 2003). The protein contains a C-terminal Swi2/Snf2-type ATPase/helicase domain and an N-terminal histone reader domain for the histone post-translational modifications H3K9me3/H3K4me0 and H3K9me3/H3S10P (Picketts et al., 1996; Iwase et al., 2011; Noh et al., 2015). ATRX and the histone chaperone DAXX form a complex that deposits the histone variant H3.3 at pericentric heterochromatin and telomeres (Goldberg et al., 2010; Lewis et al., 2010; Wong et al., 2010). ATRX was also demonstrated to regulate gene expression in the mouse CNS. For a subset of genes, ATRX has a positive effect on transcription, by aiding transcriptional elongation and RNA polymerase II passage through G-rich regions and histone H3.3 incorporation in the gene body. One such gene is Ngln4X, a known autism-related gene (Levy et al., 2008, 2015). Conversely, ATRX has suppressive effects at imprinted genes in the neonatal brain by promoting long-range chromatin interactions mediated by CCCTC-binding factor (CTCF) and cohesin (Kernohan et al., 2010; Kernohan et al., 2014).
Multiple conditional Atrx knock-out (KO) mouse models have been developed, allowing for temporal and spatial control over gene inactivation. Conditional deletion of Atrx in the developing forebrain results in extensive neuronal apoptosis, microcephaly, and reduced postnatal survival (Bérubé et al., 2005; Seah et al., 2008). Loss of ATRX in this mouse model causes DNA replication stress in neural progenitor cells, resulting in increased DNA damage and p53 activation (Watson et al., 2013). Overall, the identified defects in the ATRX-null developing brain are intimately linked to cell proliferation and might provide an explanation for the microcephaly phenotype observed in a subset of ATR-X syndrome patients. However, whether loss of ATRX strictly in postmitotic cells of the CNS leads to cognitive deficits had not yet been investigated. To specifically interrogate the importance of ATRX in learning and memory, N.G.B. and colleagues mated Atrx floxed mice to mice expressing Cre recombinase in postmitotic forebrain pyramidal neurons under the control of the CaMKII gene promoter (Casanova et al., 2001). These conditional KOs (referred to as Atrx-CaMKIICre) survived to adulthood and did not display microcephaly. Normal activity levels were observed in the open field test and in the Y-maze test for working memory. However, hippocampal-dependent spatial learning and memory were impaired in Atrx-CaMKIICre mice when tested in different memory task (unpublished results). These results would suggest that ATRX in differentiated neurons is required for normal spatial learning and memory. The Atrx-CaMKIICre mice, therefore, represent an ideal model to further investigate the molecular and cellular underpinnings of cognitive defects caused by ATRX mutations and could be used in preclinical trials.
MeCP2 and Rett syndrome
Rett syndrome (RTT; OMIM #312750) is an X-linked neurological disorder characterized by regressive loss of neurodevelopmental milestones and acquired motor and language skills. It represents one of the most common causes of ID among young girls (Chahrour and Zoghbi, 2007). Approximately 95% of RTT cases are caused by mutations in the X-linked gene encoding methyl-CpG binding protein 2 (MECP2) (Amir et al., 1999). MeCP2 is a ubiquitously expressed nuclear protein that binds to methylated DNA and is thought to mediate transcriptional repression or activation, but the identification bona fide transcriptional targets of MeCP2 has been challenging because hundreds of genes have been found to be slightly altered in MeCP2 mutant cells, and the subtlety of these gene expression changes varies among different studies (Lyst and Bird, 2015). In addition, the apparently ubiquitous distribution of MeCP2 across the genome remains perplexing and further complicates the identification of direct targets and, therefore, the understanding of the molecular etiology of RTT (Skene et al., 2010; Guo et al., 2014; Chen et al., 2015; Gabel et al., 2015; Rube et al., 2016).
The complexity in identifying MeCP2 transcriptional targets is further confounded by the heterogeneity of cellular transcriptomes in the mammalian brain. The brain comprises a myriad of intermixed cell types that differ in morphology, function, and electrophysiological properties. Recent studies have reported that cellular identity and diversity are established and maintained by distinctive transcriptional and epigenomic programs, including the cell type-specific genomic distributions of methylated and hydroxymethylated cytosines (Lister et al., 2013; Mo et al., 2015). Thus, MeCP2, as a methyl-cytosine binding protein, may exhibit distinctive binding patterns across different cell types and thus regulate different genes in different types of cells. Overcoming cellular heterogeneity, particularly for neurons in the brain, would be an imperative first step to identify the target genes of MeCP2. Other than cellular heterogeneity, the cells themselves comprise a heterogeneous pool of nuclear and cytoplasmic RNAs at different stages of synthesis, processing, transport, translation, and degradation (Maniatis and Reed, 2002). The bulk of protein-coding mRNAs are enriched in the cytoplasm and subjected to extensive post-transcriptional regulation. In contrast, protein-coding nuclear RNA transcripts lie upstream of most post-transcriptional mechanisms and are thus ideal for studying transcriptional dynamics in the cell (Buxbaum et al., 2015). Therefore, whole-cell mRNA from individual brain regions reflects composite profiles that can obscure cell type-specific expression changes due to the loss of MeCP2 and impede appropriate assessment of MeCP2 function at the transcriptional level. Finally, RTT is an X-linked dominant disorder in heterozygous females, but the majority of RTT research has focused on hemizygous male mouse models because of the limits imposed by the mosaic expression of MeCP2 in females as a consequence of random X-chromosome inactivation (Lyst and Bird, 2015). Thus, overcoming X-linked cellular heterogeneity and distinguishing cell autonomous from non–cell-autonomous effects upon MeCP2 loss in ∼50% of the cells are particularly pertinent for RTT research.
To address the confounding effects of cellular heterogeneity at multiple levels, Z.Z. and colleagues have engineered genetically modified mice whereby MeCP2 is labeled with biotin in a Cre-dependent manner. To understand the molecular impact of RTT-associated mutations on cell type-specific gene expression in vivo, they have also developed tagged knock-in mice bearing one of two frequent and molecularly distinct RTT missense mutations, T158M and R106W. When combined with FACS, this strategy effectively circumvents the cellular heterogeneity of the mouse brain and allows for the isolation of neuronal nuclei from cell types of interest (Johnson et al., 2017). First, they examined nuclear RNAs to assess the primary effects of RTT mutations on transcriptional activity. Upon systematic profiling of nuclear transcriptomes from distinct neuronal cell types in MeCP2 wild-type, T158M, and R106W male and female mice, the laboratory of Z.Z. identified transcriptional features that are specific to each cell type and correlate with the severity of MeCP2 mutation. They found that lowly expressed, cell type-enriched genes are preferentially disrupted by MeCP2 mutations. Upregulated genes demarcate functional categories related to synapse morphology and function, whereas downregulated genes are enriched for functions related to transcription and chromatin regulation. Furthermore, these analyses uncovered that genome-wide transcriptional changes in the nucleus are opposed by post-transcriptional compensation of RNAs in the whole cell in a gene length-dependent manner. This approach effectively circumvented cellular heterogeneity associated with X-chromosome inactivation in heterozygous females through the transcriptional profiling of neighboring wild-type and mutant neurons, thereby discerning cell autonomous from non–cell-autonomous transcriptional effects. The comprehensive analysis across different neuronal settings in an allelic series of RTT mouse models has led to the proposal of a contextualized model by which cell-autonomous and non–cell-autonomous transcriptional changes in different cell types contribute to the molecular severity of neuronal deficits in RTT, providing new directions for therapeutic development (Johnson et al., 2017).
EHMT1 and Kleefstra syndrome (KS)
KS (OMIM#610253) is characterized by moderate to severe ID, autism, microcephaly, and dysmorphic features (Kleefstra et al., 2009). Most individuals have severe expressive speech delay with hardly any speech development. About a decade ago, KS was identified as a neurodevelopmental disorder caused either by a submicroscopic 9q34.3 deletion or an intragenic euchromatin histone methyltransferase 1 (EHMT1) mutations, leading to haploinsufficiency of this gene (Kleefstra et al., 2006). More recently, mutations in EHMT1 have also been associated with isolated idiopathic ASD (Bock et al., 2016) and schizophrenia (Talkowski et al., 2012).
EHMT1 cooperates with its mammalian paralog EHMT2/G9a and exhibits enzymatic activity in histone 3 lysine 9 monomethylation and dimethylation (H3K9me1 and H3K9me2), which is known to promote heterochromatization and gene repression (Tachibana et al., 2008). During development, the EHMT1/EHMT2 repressive complex is important for cell differentiation (Fiszbein et al., 2016). This is exemplified by the fact that both Ehmt1−/− and Ehmt2−/− mice show early embryonic lethality, whereas heterozygous Ehmt1−/+ and Ehmt2−/+ mice are viable and fertile (Tachibana et al., 2008). Loss of EHMT1 function in mice and Drosophila reduces H3K9 methylation and lead to learning and memory impairments (Schaefer et al., 2009; Maze et al., 2010; Kramer et al., 2011). Initial studies reported that Ehmt1−/+ mice show cranial abnormalities (Balemans et al., 2014), hypoactivity, reduced exploration, increased anxiety, and aberrant social behavior compared with their wild-type littermates (Balemans et al., 2010). This recapitulates the autistic-like features and hypoactivity seen in KS patients (Vermeulen et al., 2017). In follow-up studies, the same group showed that Ehmt1−/+ mice were also impaired in fear extinction learning as well as novel and spatial object recognition (Balemans et al., 2013). More recently, they tested whether Ehmt1−/+ and wild-type mice differ in several cognitive tests in a touchscreen-equipped operant chamber. Surprisingly, Ehmt1−/+ mice were mostly unaffected except in the location discrimination test of pattern separation in which they outperformed their wild-type littermates. In line with this, they detected increased cell proliferation in the subgranular zone of the dentate gyrus (Benevento et al., 2017). At the cellular level, a reduction in dendritic branching and spine density, and an increase in paired-pulse ratio indicative of a presynaptic deficit were observed. However, there was no change in long-term plasticity, a Hebbian form of synaptic plasticity (Balemans et al., 2013).
Contrary to Hebbian synapse-specific mechanisms, homeostatic synaptic plasticity acts to maintain a fine-tuning of overall neuronal excitability by monitoring and scaling globally all synapses (Turrigiano, 2012). Intriguingly, EHMT1 is required for homeostatic synaptic scaling (Benevento et al., 2016). H3K9me2 levels are bidirectionally altered in response to enduring stimulatory or inhibitory neuronal network activity. Specifically, EHMT1 is critical for the repression of Bdnf, which encodes for a neurotrophin critically involved in homeostatic plasticity (Desai et al., 1999), during synaptic scaling up. As a consequence, loss of EHMT1 prevented synaptic scaling up in vitro and in vivo. EHMT1, through the regulation of H3K9me2 levels, functions as a permissive tag to recruit the machinery that adds more stable repressive marks, such as H3K9me3, H3K27me3, and DNA methylation (Mozzetta et al., 2014; Rothbart and Strahl, 2014; Yearim et al., 2015). According to this view, H3K9me2 levels correlate with the activity state of a neuronal network and could serve as a general mechanism for poising a state in the genome in a “ready to repress” mode. This hypothesis is in line with data showing that neurons lacking EHMT1 (and having reduced H3K9me2) show few and modest changes in gene expression under basal conditions but were unable to respond to changes in activity through the repression of genes involved in synaptic scaling (including Bdnf). H3K9me2 can thus be viewed as a dynamic regulatory histone mark in euchromatic regions, and not merely a mark for heterochromatin.
In conclusion, the role of EHMT1 in homeostatic plasticity is in line with recent reports supporting the dynamic nature of histone methylation in activity-dependent gene transcription and neuronal plasticity (Heller et al., 2014; Rusconi et al., 2016; Webb et al., 2017). Impaired chromatin regulation due to mutations in EHMT1 may thus lead to a pathophysiological change in neuronal excitability, resulting in aberrant network activity and seizures, which are present in KS patients. The laboratory data of N.N.K. measuring neuronal network development in vitro do indeed show that loss of Ehmt1 in rat cortical neurons during development leads to neuronal network miswiring (Martens et al., 2016).
LSD1, a novel chromatin regulator associated with ID
Very recently, a novel and still unnamed form of neurodevelopmental disorder featuring distinctive traits of facial dysmorphisms and associated to ID has been linked to point mutations in the Lysine Specific Demethylase 1 (LSD1) gene (Rauch et al., 2012; Tunovic et al., 2014; Chong et al., 2016) (the disorder is referred as cleft palate, psychomotor retardation and distinctive facial features, or CPRF, in the OMIM database; OMIM #616728). This new autosomal dominant pathology, displaying partial phenotypic overlap with Kabuki syndrome, but representing a distinct condition, still needs to be fully characterized at the clinical point of view. Three missense mutations in the LSD1 gene have been described so far, all entailing single amino acid substitution in the catalytic domain of this chromatin modifier (Pilotto et al., 2016).
LSD1 is a flavin-dependent enzyme that erases monomethyl and dimethyl groups from histone H3 Lysine 4 (H3K4me1/2) (Shi et al., 2004). H3K4me1/2 represent “permissive” histone marks, which is why their removal can contribute to transcription repression. Indeed, LSD1 is an epigenetic transcriptional corepressor that participates in a macromolecular complex, including CoREST and histone deacetylases HDAC1/2 (Shi et al., 2005). The three de novo LSD1 point mutations associated with this new form of ID (E403K; D580G; Y785H) partially affect the ability of LSD1 to demethylate H3K4 but do not modify LSD1 binding to other molecular partners, including transcription factors and cofactors, such as CoREST and HDAC1/2 (Pilotto et al., 2016). Relevantly, while LSD1 KOs are embryonic lethal, heterozygous deletion of LSD1 does not trigger pathologic traits in rodent models (Wang et al., 2007), suggesting a possible dominant negative pathogenic function of the annotated human mutations, competing with wild-type allele-derived LSD1 for complex assembly.
The importance of LSD1 as a regulator of neuronal physiology was first unveiled with the discovery of a neurospecific splicing isoform, referred to as neuroLSD1, involved in neuronal maturation (Zibetti et al., 2010). NeuroLSD1 differs from LSD1 by the inclusion of an additional exon (the microexon E8a) at the mRNA level, encoding 4 amino acids that form a protein loop located in the vicinity of the substrate entry site (Zibetti et al., 2010). The neuroLSD1 isoform does not substitute LSD1 in neurons. Rather, both isoforms take part in a neuronal-restricted mechanism of fine transcriptional modulation (Toffolo et al., 2014; Rusconi et al., 2016). Interestingly, whereas LSD1 can exert a repressive action toward transcription, neuroLSD1, which shares the same gene targets, is not able to accomplish this molecular task in vivo (Laurent et al., 2015; Wang et al., 2015). In this frame, it has been proposed that neuroLSD1 represents a dominant negative isoform, competing with LSD1 and modulating LSD1-related repressive strength in accordance to their relative abundance in neurons (Rusconi et al., 2016). Notably, the three LSD1 mutations associated with ID can modify the function of both LSD1 and neuroLSD1 because they map within exons that are shared by the two isoforms (Pilotto et al., 2016).
The study of the physiological role of LSD1 in controlling neuronal plasticity began a few years ago, with two papers describing LSD1/neuroLSD1-mediated modulation of activity-dependent transcription and behavior in mouse models. Because LSD1 KOs are embryonic lethal, the neuroLSD1KO mouse model represents a very important tool to investigate the relevance of LSD1 and neuroLSD1 in neurons and their specific role in ID. Several independent observations provide a perspective on how LSD1 mutations can affect cognition. Mice with ablation of the neuroLSD1-specific microexon E8a, but preserved LSD1 expression, show learning-related disabilities and deficits in stress-related plasticity (Wang et al., 2015; Rusconi et al., 2016). In particular, Rosenfeld's group reported that neuroLSD1KO display significant memory impairment (Wang et al., 2015), whereas Battaglioli's group showed that neuroLSD1 haploinsufficiency translates in an aberrant acquisition of stress-related plasticity, leading to decreased anxiety-like behavior in neuroLSD1HET (Rusconi et al., 2016). Remarkably, convergent evidence in both models indicates that activity-dependent transcription of plasticity-related immediate early genes (IEGs), such as Fos, Egr1, Npas4, Nr4a1, and Arc, is impaired upon neuroLSD1 deletion, which indicates that either neuroLSD1 acts as positive transcriptional regulator for these genes (Wang et al., 2015) or that its ablation results in increased level of LSD1 (Rusconi et al., 2016). Both the formation of new memories and the acquisition of a correct emotional phenotype in terms of anxiety (i.e., the ability to turn stressful experiences into protective warnings) require an efficient experience-driven transactivation of plasticity genes (Rusconi et al., 2016) modulating structural plasticity in the hippocampus, prefrontal cortex, and amygdala in a highly coordinated fashion (Felix-Ortiz et al., 2013; Calhoon and Tye, 2015). Although the origin of IEG transcriptional impairment caused by loss of neuroLSD1 still needs to be fully understood, the dual LSD1/neuroLSD1 system is clearly implicated in the process of activity-dependent transcription in the brain. A formal demonstration that mutations in the LSD1 gene associated to ID alter IEG transcription in neurons is still lacking, but the implication of LSD1 in cognition-related processes via direct control of neuronal plasticity transcriptional programs further envisages aberrant IEG transcriptional modulation in LSD1-related new form of ID.
KDM5C and Claes-Jensen Type X-Linked ID
Mutations into the KDM5C gene account for at least 2% of X-linked IDs, which is more frequent than most of the X-linked ID genes. The KDM5C-associated cases are referred to as mental retardation, X-linked, syndromic, Claes-Jensen type (MRXSCJ; OMIM #300534). Patients with these mutations, in addition to ID, often show epilepsy and aggressive behavior.
KDM5C encodes for the lysine (K)-specific demethylase 5C, an eraser enzyme for dimethylated and trimethylated histone H3 lysine 4 (H3K4me2/3) (Iwase et al., 2007; Tahiliani et al., 2007). ID-associated missense mutations decrease demethylase activity, suggesting that mutations lead to loss of function. Therefore, this disorder, like the one discussed in the previous section, seems to be also caused by altered H3K4 methylation (although the specific forms affected vary since LSD1 removes monomethylations and dimethylations instead of dimethylations and trimethylations). Iwase et al. (2016) have recently shown that Kdm5c-KO mice closely recapitulate the behavioral abnormalities of human patients, including impaired learning and augmented aggression. While gross morphological abnormalities were not noted in the Kdm5c-KO brains, a decreased density of dendritic spines was found in pyramidal neurons of the amygdala. RNA-seq analysis in these animals revealed brain region-specific expression changes in hundreds of genes. In addition, the levels of H3K4me2 and H3K4me3 were increased at the promoters of genes, some of which are relevant to neuronal maturation, in cultured Kdm5c-deficient neurons (Iwase et al., 2016). Kdm5c-KO mice represent the first mouse model of ID caused by defective erasure of histone methylation, thereby providing a link between the dynamic regulation of histone methylation and cognitive development.
More recently, A.B. and colleagues developed and analyzed inducible and forebrain-restricted cKOs for Kdm5c (referred to as Kdm5c-ifKOs). In these mice, KDM5C was specifically deleted in excitatory forebrain neurons of adult mice. In contrast to the severe phenotype and broad behavioral alterations of KOs, Kdm5c-ifKOs displayed highly specific spatial memory defects (Scandaglia et al., 2017). These results suggest that the most severe impairments observed in Kdm5c-KOs originate during development. It will be important to test in future experiments whether this is also the case after deleting Kdm5c in other cell types, including inhibitory neurons and astrocytes, as KDM5C is also expressed in those cells (Iwase et al., 2016). Parallel genomic screens in Kdm5c-KOs and ifKOs enabled a fine dissection of the genomic actions of Kdm5c during neuronal differentiation, maturation, and maintenance. These analyses indicated that Kdm5c functions as an epigenetic repressor of germline genes during early development, as a fine-tuner of activity-regulated enhancers during neuronal maturation, and it prevents illegitimate activation of non-neuronal and cryptic promoters in mature neurons (Scandaglia et al., 2017).
Before their removal by KDM5C, the H3K4me marks are placed by a group of lysine methyltransferases (KMTs) with specificity for this residue (H3K4me writer enzymes). The human genome encodes seven H3K4me writer genes that are ubiquitously expressed; however, the functional relationship between KDM5C and the H3K4me writer enzymes is not known (Vallianatos and Iwase, 2015; Garay et al., 2016). How does the balance between writers and erasers of histone methylation influence cognitive development? Do one or more specific H3K4me writer enzymes mediate the abnormal brain development caused by the loss of KDM5C? Answering these questions will open the possibility that inhibition of those enzymes compensates for the loss of KDM5C. Identifying H3K4me writer enzymes that counteracts with KDM5C may thereby provide a specific drug target for MRXSCJ.
Moreover, it is possible that the same drug could be also used to ameliorate the form of ID associated with LSD1 deficiency. As aforementioned, KDM5C and LSD1 act on the same target (H3K4), and both suppress transcriptional enhancers of actively transcribed genes and can be found in the same protein complex (Whyte et al., 2012; Shen et al., 2016; Agarwal et al., 2017). Furthermore, similar to the neuroLSD1-deficient neurons, Kdm5c-KO neurons display aberrant expression of some neuronal activity-dependent genes (Iwase et al., 2016; Scandaglia et al., 2017). Further research is warrant to test whether the cleft palate, psychomotor retardation, and distinctive facial features (LSD1 deficiency) and MRXSCJ (KDM5C deficiency) can be grouped as a clinical condition associated with impaired erasure of H3K4me, and similar amelioration strategies may apply to this group of IDD in the future.
KAT3 proteins and the Rubinstein-Taybi syndrome (RSTS)
RSTS is a sporadic autosomal dominant disorder caused by mutation in the genes CREBBP (RSTS1; OMIM #180849) or EP300 (RSTS2; OMIM #613684), which account for ∼70% and 5% of the cases, respectively (Negri et al., 2015; Rusconi et al., 2015; Spena et al., 2015). The syndrome is characterized by mental impairment of variable severity and a wide spectrum of congenital anomalies, being usually diagnosed based on the concurrence of ID and broad thumbs and toes (Rubinstein and Taybi, 1963).
The genes involved in RSTS encode for two very large (>250 kDa) paralog proteins belonging to the lysine acetyltransferase (KAT) 3 family: CBP (aka KAT3a) and p300 (aka KAT3b). Both proteins are known to act as a molecular bridge between different transcription factors and the RNA polII complex, and as molecular scaffolds that bring a variety of enzymatic activities to the promoter. In addition, their KAT domains catalyze the transfer of acetyl groups to lysine residues in the histones tails (likely affecting chromatin accessibility) and in numerous nonhistone substrates, including transcription factors, components of the RNA pol II complex and other chromatin regulators (Valor et al., 2013; Lopez-Atalaya et al., 2014). Both proteins display widespread occupancy of transcriptional regulatory regions, including many putative enhancers (Wang et al., 2009). Although the function of KAT3 proteins has been extensively studied, downstream events of CBP and p300 deficiencies responsible for neurodevelopmental and cognitive deficits in RSTS patients remain obscure. Outstanding questions still under investigation are the unique and redundant roles of these two proteins regulating neuronal chromatin acetylation and their distinct genomic targets during the development of the nervous system and in neuronal plasticity processes in the adult brain.
The use of animal models has demonstrated that CBP and p300 are expressed in almost identical patterns in the mouse embryo, but rather than compensate for each other, both factors are required for embryonic development and viability (Valor et al., 2013). As a result, both the CBP and p300 KOs display defects in neural tube closure and cell proliferation and are early embryonically lethal (Yao et al., 1998; Tanaka et al., 2000), which makes it difficult to ascribe the developmental abnormalities to deregulation of specific genes. This limitation has been circumvented by the investigation of conditional and inducible KOs, focal gene ablation using viral vectors, and tissue-restricted expression of dominant negative transgenes. Despite significant differences across KAT3-deficient strains and laboratories, these approaches have shown that some cognitive impairments associated with CBP deficiency are not due to developmental defects but result from the continuous requirement of CBP and p300 activities throughout life (Korzus et al., 2004; Chen et al., 2010; Barrett et al., 2011; Valor et al., 2011; compare Lopez-Atalaya et al., 2014).
The A.B. laboratory and others have found that CBP deficiency causes severe deacetylation of specific lysine residues at the histone tails in the brain of mouse models (Alarcón et al., 2004; Chen et al., 2010; Barrett et al., 2011; Valor et al., 2011) and in cell lines derived from patients (Lopez-Atalaya et al., 2012). The use of next-generation sequencing-based techniques (Telese et al., 2013; Maze et al., 2014) now provides the ideal conditions for establishing a detailed and multilayered map of these alterations at a genomic scale. These acetylation deficits can have a major role in the etiology of ID because a number of studies have shown that histone acetylation correlates with memory formation in diverse paradigms, pinpointing a role for this process in cognition (Gräff and Tsai, 2013). Supporting this view, inhibitors of Classes I and II histone deacetylases (HDACi), a heterogeneous group of compounds that increase histone acetylation levels, have been found to ameliorate cognitive deficits in RSTS mouse models (Alarcón et al., 2004; Korzus et al., 2004; Stefanko et al., 2009), although there are also some inconsistencies between studies that still need to be clarified (Chen et al., 2010; Barrett et al., 2011). These studies identified HDACi as promising drugs to treat RSTS, a hypothesis that is currently been tested in clinical trials (https://clinicaltrials.gov/ct2/show/NCT01619644). In addition, to pharmacological approaches, recent experiments using the Clustered Regularly Interspaced Short Palindromic Repeats system have opened a new avenue for therapy, although its application in clinical settings seems to be still far away. By directing the KAT activity of p300 to specific loci, Hilton et al. (2015) demonstrated that it is possible to manipulate the acetylation status of a given loci with unprecedented precision. Therefore, upon identifying hypoacetylated loci downstream of CBP/p300 loss involved in neuropathology, the novel tools for epi-editing (Konermann et al., 2013) could be used for correcting these lysine acetylation defects.
Conclusion
Eradicating rare congenital IDDs caused by de novo mutations will require detailed knowledge of their etiology and pathogenesis. The progress brought by studies in animal models, such as those outlined above, together with the availability and characterization of neurons differentiated from induced pluripotent stem cells (iPSC)-derived from patients and the precise description of abnormal chromatin profiles in the different models, should lead to the identification and evaluation of novel drugs, and the implementation of innovative therapeutic strategies based on genetic or epigenetic editing (Fig. 2). However, before reaching that goal, many challenges toward the treatment of IDDs will need to be overcome. Therapeutic time window is one such challenge. As described earlier, adult-neuron-specific KO resulted in cognitive deficits in the model of ATR-X syndrome, RSTS, and MRXSCJ (although deficits were clearly milder than in conventional KOs). The pioneering study by Guy et al. (2007) revealed that reexpression MeCP2 after onset of the symptoms in Mecp2-null mice alleviated motor and cognitive regression, indicated that ID-related phenotypes can be reversed in adult state. These observations point to importance of determining developmental time windows, in which a given chromatin regulator is required and/or sufficient for normal brain development and function in gene-by-gene basis. As discussed in the MeCP2 section, another forthcoming challenge is the ubiquitous presence of chromatin regulators and heterogeneous impacts of their malfunction in different cell types. The presence of both cell autonomous and non–cell-autonomous impacts of deficient chromatin regulation represents perhaps greater hurdle for rationalized treatments. Harnessing state-of-the-art genetics/genomics and cell biology tools combined with mouse models will likely help us to understand the complexity of IDD and ASD pathophysiology.
As clearly illustrated in the examples described here, the development of treatment strategy for IDDs will greatly benefit from discoveries in the molecular biology/biochemistry of individual syndromes. For example, the activity-dependent transcriptional response appears to be a key target for future drug discovery given that different chromatin regulators, such as CBP/p300 (Visel et al., 2009), KDM5C (Shen et al., 2016; Scandaglia et al., 2017), and LSD1 (Whyte et al., 2012), all regulate transcriptional enhancers positively or negatively. The ATR-X and Rett syndromes may also share phenotypic commonality and mechanistic root because ATRX fails to locate at heterochromatin in MeCP-null cells (Nan et al., 2007), indicating the roles of MeCP2 in ATRX localization within heterochromatin. Therefore, they may be treated with the same strategy. These discoveries may also shed light on the etiology of other neuropsychiatric conditions, such as autism and schizophrenia, because the proteins under study play important roles in the regulation of gene expression programs that govern many aspects of brain development and plasticity. Therefore, in addition to clinical insights, the investigation of these genetic disorders can unveil fundamental mechanisms by which chromatin sculpts the complex neuronal networks underlying cognition.
Footnotes
S.I. was supported by National Institutes of Health Grant NS089896, University of Michigan Medical School, Cooley's Anemia Foundation Fellowship, March of Dimes Foundation, and the Farrehi Research Fund. N.G.B. was supported by Canadian Institutes of Health Research MOP 142268. Z.Z. was supported by National Institutes of Health Grants R01MH091850 and R01NS081054. N.N.K. was supported by The Netherlands Organization for Scientific Research Grants ALW2PJ/13082 and 012.200.001. E.B. was supported by Telethon Grant GGP14074, ONLUS Insieme per la Ricerca Grant PCDH19, and MIUR (Progetto Bandiera Epigenomica-EPIGEN). A.B. was supported by Spanish Ministry of Economy and Competitivity Grants SAF2014-56197-R, PCIN-2015-192-C02-01 (part of the coordinated project ERA-Net NEURON8-2015), and SEV-2013-0317, cofinanced by the European Regional Development Fund, Generalitat Valenciana Grant PROMETEO/2016/006, and the Alicia Koplowitz Foundation. M.S. received Formación de Personal Investigador fellowship given by Spanish Ministry of Economy and Competitivity. The Instituto de Neurociencias is a Centre of Excellence Severo Ochoa.
The authors declare no competing financial interests.
References
- Agarwal S, Garay PM, Porter RS, Brookes E, Murata-Nakamura Y, Mcfarlan TS, Bing R, Iwase S (2017) LSD1/KDM1A maintains genome-wide homeostasis of transcriptional enhancers. bioRxiv 146357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcón JM, Malleret G, Touzani K, Vronskaya S, Ishii S, Kandel ER, Barco A (2004) Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron 42:947–959. 10.1016/j.neuron.2004.05.021 [DOI] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY (1999) Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet 23:185–188. 10.1038/13810 [DOI] [PubMed] [Google Scholar]
- Baker-Andresen D, Ratnu VS, Bredy TW (2013) Dynamic DNA methylation: a prime candidate for genomic metaplasticity and behavioral adaptation. Trends Neurosci 36:3–13. 10.1016/j.tins.2012.09.003 [DOI] [PubMed] [Google Scholar]
- Balemans MC, Huibers MM, Eikelenboom NW, Kuipers AJ, van Summeren RC, Pijpers MM, Tachibana M, Shinkai Y, van Bokhoven H, Van der Zee CE (2010) Reduced exploration, increased anxiety, and altered social behavior: autistic-like features of euchromatin histone methyltransferase 1 heterozygous knockout mice. Behav Brain Res 208:47–55. 10.1016/j.bbr.2009.11.008 [DOI] [PubMed] [Google Scholar]
- Balemans MC, Kasri NN, Kopanitsa MV, Afinowi NO, Ramakers G, Peters TA, Beynon AJ, Janssen SM, van Summeren RC, Eeftens JM, Eikelenboom N, Benevento M, Tachibana M, Shinkai Y, Kleefstra T, van Bokhoven H, Van der Zee CE (2013) Hippocampal dysfunction in the Euchromatin histone methyltransferase 1 heterozygous knockout mouse model for Kleefstra syndrome. Hum Mol Genet 22:852–866. 10.1093/hmg/dds490 [DOI] [PubMed] [Google Scholar]
- Balemans MC, Ansar M, Oudakker AR, van Caam AP, Bakker B, Vitters EL, van der Kraan PM, de Bruijn DR, Janssen SM, Kuipers AJ, Huibers MM, Maliepaard EM, Walboomers XF, Benevento M, Nadif Kasri N, Kleefstra T, Zhou H, Van der Zee CE, van Bokhoven H (2014) Reduced Euchromatin histone methyltransferase 1 causes developmental delay, hypotonia, and cranial abnormalities associated with increased bone gene expression in Kleefstra syndrome mice. Dev Biol 386:395–407. 10.1016/j.ydbio.2013.12.016 [DOI] [PubMed] [Google Scholar]
- Barrett RM, Malvaez M, Kramar E, Matheos DP, Arrizon A, Cabrera SM, Lynch G, Greene RW, Wood MA (2011) Hippocampal focal knockout of CBP affects specific histone modifications, long-term potentiation, and long-term memory. Neuropsychopharmacology 36:1545–1556. 10.1038/npp.2011.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benevento M, Iacono G, Selten M, Ba W, Oudakker A, Frega M, Keller J, Mancini R, Lewerissa E, Kleefstra T, Stunnenberg HG, Zhou H, van Bokhoven H, Nadif Kasri N (2016) Histone methylation by the Kleefstra syndrome protein EHMT1 mediates homeostatic synaptic scaling. Neuron 91:341–355. 10.1016/j.neuron.2016.06.003 [DOI] [PubMed] [Google Scholar]
- Benevento M, Oomen CA, Horner AE, Amiri H, Jacobs T, Pauwels C, Frega M, Kleefstra T, Kopanitsa MV, Grant SG, Bussey TJ, Saksida LM, Van der Zee CE, van Bokhoven H, Glennon JC, Kasri NN (2017) Haploinsufficiency of EHMT1 improves pattern separation and increases hippocampal cell proliferation. Sci Rep 7:40284. 10.1038/srep40284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bérubé NG, Mangelsdorf M, Jagla M, Vanderluit J, Garrick D, Gibbons RJ, Higgs DR, Slack RS, Picketts DJ (2005) The chromatin-remodeling protein ATRX is critical for neuronal survival during corticogenesis. J Clin Invest 115:258–267. 10.1172/JCI200522329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornsson HT. (2015) The Mendelian disorders of the epigenetic machinery. Genome Res 25:1473–1481. 10.1101/gr.190629.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock I, Németh K, Pentelényi K, Balicza P, Balázs A, Molnár MJ, Román V, Nagy J, Lévay G, Kobolák J, Dinnyés A (2016) Targeted next generation sequencing of a panel of autism-related genes identifies an EHMT1 mutation in a Kleefstra syndrome patient with autism and normal intellectual performance. Gene 595:131–141. 10.1016/j.gene.2016.09.027 [DOI] [PubMed] [Google Scholar]
- Bonnaud EM, Suberbielle E, Malnou CE (2016) Histone acetylation in neuronal (dys)function. Biomol Concepts 7:103–116. 10.1515/bmc-2016-0002 [DOI] [PubMed] [Google Scholar]
- Buxbaum AR, Yoon YJ, Singer RH, Park HY (2015) Single-molecule insights into mRNA dynamics in neurons. Trends Cell Biol 25:468–475. 10.1016/j.tcb.2015.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoon GG, Tye KM (2015) Resolving the neural circuits of anxiety. Nat Neurosci 18:1394–1404. 10.1038/nn.4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova E, Fehsenfeld S, Mantamadiotis T, Lemberger T, Greiner E, Stewart AF, Schütz G (2001) A CamKIIalpha iCre BAC allows brain-specific gene inactivation. Genesis 31:37–42. 10.1002/gene.1078 [DOI] [PubMed] [Google Scholar]
- Chahrour M, Zoghbi HY (2007) The story of Rett syndrome: from clinic to neurobiology. Neuron 56:422–437. 10.1016/j.neuron.2007.10.001 [DOI] [PubMed] [Google Scholar]
- Chen G, Zou X, Watanabe H, van Deursen JM, Shen J (2010) CREB binding protein is required for both short-term and long-term memory formation. J Neurosci 30:13066–13077. 10.1523/JNEUROSCI.2378-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Chen K, Lavery LA, Baker SA, Shaw CA, Li W, Zoghbi HY (2015) MeCP2 binds to non-CG methylated DNA as neurons mature, influencing transcription and the timing of onset for Rett syndrome. Proc Natl Acad Sci U S A 112:5509–5514. 10.1073/pnas.1505909112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong JX, Yu JH, Lorentzen P, Park KM, Jamal SM, Tabor HK, Rauch A, Saenz MS, Boltshauser E, Patterson KE, Nickerson DA, Bamshad MJ (2016) Gene discovery for Mendelian conditions via social networking: de novo variants in KDM1A cause developmental delay and distinctive facial features. Genet Med 18:788–795. 10.1038/gim.2015.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai NS, Rutherford LC, Turrigiano GG (1999) BDNF regulates the intrinsic excitability of cortical neurons. Learn Mem 6:284–291. [PMC free article] [PubMed] [Google Scholar]
- Felix-Ortiz AC, Beyeler A, Seo C, Leppla CA, Wildes CP, Tye KM (2013) BLA to vHPC inputs modulate anxiety-related behaviors. Neuron 79:658–664. 10.1016/j.neuron.2013.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiszbein A, Giono LE, Quaglino A, Berardino BG, Sigaut L, von Bilderling C, Schor IE, Steinberg JH, Rossi M, Pietrasanta LI, Caramelo JJ, Srebrow A, Kornblihtt AR (2016) Alternative splicing of G9a regulates neuronal differentiation. Cell Rep 14:2797–2808. 10.1016/j.celrep.2016.02.063 [DOI] [PubMed] [Google Scholar]
- Gabel HW, Kinde B, Stroud H, Gilbert CS, Harmin DA, Kastan NR, Hemberg M, Ebert DH, Greenberg ME (2015) Disruption of DNA-methylation-dependent long gene repression in Rett syndrome. Nature 522:89–93. 10.1038/nature14319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garay PM, Wallner MA, Iwase S (2016) Yin-yang actions of histone methylation regulatory complexes in the brain. Epigenomics 8:1689–1708. 10.2217/epi-2016-0090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons RJ, Wada T, Fisher CA, Malik N, Mitson MJ, Steensma DP, Fryer A, Goudie DR, Krantz ID, Traeger-Synodinos J (2008) Mutations in the chromatin-associated protein ATRX. Hum Mutat 29:796–802. 10.1002/humu.20734 [DOI] [PubMed] [Google Scholar]
- Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, Wen D, Chapgier A, DeKelver RC, Miller JC, Lee YL, Boydston EA, Holmes MC, Gregory PD, Greally JM, Rafii S, et al. (2010) Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140:678–691. 10.1016/j.cell.2010.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gräff J, Tsai LH (2013) Histone acetylation: molecular mnemonics on the chromatin. Nat Rev Neurosci 14:97–111. 10.1038/nrn3427 [DOI] [PubMed] [Google Scholar]
- Guo JU, Su Y, Shin JH, Shin J, Li H, Xie B, Zhong C, Hu S, Le T, Fan G, Zhu H, Chang Q, Gao Y, Ming GL, Song H (2014) Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat Neurosci 17:215–222. 10.1038/nn.3607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Kim SY, Artis S, Molfese DL, Schumacher A, Sweatt JD, Paylor RE, Lubin FD (2010) Histone methylation regulates memory formation. J Neurosci 30:3589–3599. 10.1523/JNEUROSCI.3732-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy J, Gan J, Selfridge J, Cobb S, Bird A (2007) Reversal of neurological defects in a mouse model of Rett syndrome. Science 315:1143–1147. 10.1126/science.1138389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller EA, Cates HM, Peña CJ, Sun H, Shao N, Feng J, Golden SA, Herman JP, Walsh JJ, Mazei-Robison M, Ferguson D, Knight S, Gerber MA, Nievera C, Han MH, Russo SJ, Tamminga CS, Neve RL, Shen L, Zhang HS, et al. (2014) Locus-specific epigenetic remodeling controls addiction- and depression-related behaviors. Nat Neurosci 17:1720–1727. 10.1038/nn.3871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyward FD, Sweatt JD (2015) DNA methylation in memory formation: emerging insights. Neuroscientist 21:475–489. 10.1177/1073858415579635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton IB, D'Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, Gersbach CA (2015) Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol 33:510–517. 10.1038/nbt.3199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase S, Lan F, Bayliss P, de la Torre-Ubieta L, Huarte M, Qi HH, Whetstine JR, Bonni A, Roberts TM, Shi Y (2007) The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell 128:1077–1088. 10.1016/j.cell.2007.02.017 [DOI] [PubMed] [Google Scholar]
- Iwase S, Xiang B, Ghosh S, Ren T, Lewis PW, Cochrane JC, Allis CD, Picketts DJ, Patel DJ, Li H, Shi Y (2011) ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome. Nat Struct Mol Biol 18:769–776. 10.1038/nsmb.2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase S, Brookes E, Agarwal S, Badeaux AI, Ito H, Vallianatos CN, Tomassy GS, Kasza T, Lin G, Thompson A, Gu L, Kwan KY, Chen C, Sartor MA, Egan B, Xu J, Shi Y (2016) A mouse model of X-linked intellectual disability associated with impaired removal of histone methylation. Cell Rep 14:1000–1009. 10.1016/j.celrep.2015.12.091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BS, Zhao YT, Fasolino M, Lamonica JM, Kim YJ, Georgakilas G, Wood KH, Bu D, Cui Y, Goffin D, Vahedi G, Kim TH, Zhou Z (2017) Biotin tagging of MeCP2 in mice reveals contextual insights into the Rett syndrome transcriptome. Nat Med. Advance online publication. Retrieved Sept. 18, 2017. doi: 10.1038/nm.4406. 10.1038/nm.4406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kernohan KD, Jiang Y, Tremblay DC, Bonvissuto AC, Eubanks JH, Mann MR, Bérubé NG (2010) ATRX partners with cohesin and MeCP2 and contributes to developmental silencing of imprinted genes in the brain. Dev Cell 18:191–202. 10.1016/j.devcel.2009.12.017 [DOI] [PubMed] [Google Scholar]
- Kernohan KD, Vernimmen D, Gloor GB, Bérubé NG (2014) Analysis of neonatal brain lacking ATRX or MeCP2 reveals changes in nucleosome density, CTCF binding and chromatin looping. Nucleic Acids Res 42:8356–8368. 10.1093/nar/gku564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleefstra T, van Zelst-Stams WA, Nillesen WM, Cormier-Daire V, Houge G, Foulds N, van Dooren M, Willemsen MH, Pfundt R, Turner A, Wilson M, McGaughran J, Rauch A, Zenker M, Adam MP, Innes M, Davies C, López AG, Casalone R, Weber A, et al. (2009) Further clinical and molecular delineation of the 9q subtelomeric deletion syndrome supports a major contribution of EHMT1 haploinsufficiency to the core phenotype. J Med Genet 46:598–606. 10.1136/jmg.2008.062950 [DOI] [PubMed] [Google Scholar]
- Kleefstra T, Brunner HG, Amiel J, Oudakker AR, Nillesen WM, Magee A, Geneviève D, Cormier-Daire V, van Esch H, Fryns JP, Hamel BC, Sistermans EA, de Vries BB, van Bokhoven H (2006) Loss-of-function mutations in euchromatin histone methyl transferase 1 (EHMT1) cause the 9q34 subtelomeric deletion syndrome. Am J Hum Genet 79:370–377. 10.1086/505693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleefstra T, Schenck A, Kramer JM, van Bokhoven H (2014) The genetics of cognitive epigenetics. Neuropharmacology 80:83–94. 10.1016/j.neuropharm.2013.12.025 [DOI] [PubMed] [Google Scholar]
- Kochinke K, Zweier C, Nijhof B, Fenckova M, Cizek P, Honti F, Keerthikumar S, Oortveld MA, Kleefstra T, Kramer JM, Webber C, Huynen MA, Schenck A (2016) Systematic phenomics analysis deconvolutes genes mutated in intellectual disability into biologically coherent modules. Am J Hum Genet 98:149–164. 10.1016/j.ajhg.2015.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino A, Hsu PD, Heidenreich M, Cong L, Platt RJ, Scott DA, Church GM, Zhang F (2013) Optical control of mammalian endogenous transcription and epigenetic states. Nature 500:472–476. 10.1038/nature12466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzus E, Rosenfeld MG, Mayford M (2004) CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron 42:961–972. 10.1016/j.neuron.2004.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer JM, van Bokhoven H (2009) Genetic and epigenetic defects in mental retardation. Int J Biochem Cell Biol 41:96–107. 10.1016/j.biocel.2008.08.009 [DOI] [PubMed] [Google Scholar]
- Kramer JM, Kochinke K, Oortveld MA, Marks H, Kramer D, de Jong EK, Asztalos Z, Westwood JT, Stunnenberg HG, Sokolowski MB, Keleman K, Zhou H, van Bokhoven H, Schenck A (2011) Epigenetic regulation of learning and memory by Drosophila EHMT/G9a. PLoS Biol 9:e1000569. 10.1371/journal.pbio.1000569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent B, Ruitu L, Murn J, Hempel K, Ferrao R, Xiang Y, Liu S, Garcia BA, Wu H, Wu F, Steen H, Shi Y (2015) A specific LSD1/KDM1A isoform regulates neuronal differentiation through H3K9 demethylation. Mol Cell 57:957–970. 10.1016/j.molcel.2015.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy MA, Fernandes AD, Tremblay DC, Seah C, Bérubé NG (2008) The SWI/SNF protein ATRX co-regulates pseudoautosomal genes that have translocated to autosomes in the mouse genome. BMC Genomics 9:468. 10.1186/1471-2164-9-468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy MA, Kernohan KD, Jiang Y, Bérubé NG (2015) ATRX promotes gene expression by facilitating transcriptional elongation through guanine-rich coding regions. Hum Mol Genet 24:1824–1835. 10.1093/hmg/ddu596 [DOI] [PubMed] [Google Scholar]
- Lewis PW, Elsaesser SJ, Noh KM, Stadler SC, Allis CD (2010) Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc Natl Acad Sci U S A 107:14075–14080. 10.1073/pnas.1008850107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilja T, Heldring N, Hermanson O (2013) Like a rolling histone: epigenetic regulation of neural stem cells and brain development by factors controlling histone acetylation and methylation. Biochim Biophys Acta 1830:2354–2360. 10.1016/j.bbagen.2012.08.011 [DOI] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y, Dwork AJ, Schultz MD, Yu M, Tonti-Filippini J, Heyn H, Hu S, Wu JC, Rao A, Esteller M, He C, Haghighi FG, Sejnowski TJ, et al. (2013) Global epigenomic reconfiguration during mammalian brain development. Science 341:1237905. 10.1126/science.1237905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Atalaya JP, Barco A (2014) Can changes in histone acetylation contribute to memory formation? Trends Genet 30:529–539. 10.1016/j.tig.2014.09.003 [DOI] [PubMed] [Google Scholar]
- Lopez-Atalaya JP, Gervasini C, Mottadelli F, Spena S, Piccione M, Scarano G, Selicorni A, Barco A, Larizza L (2012) Histone acetylation deficits in lymphoblastoid cell lines from patients with Rubinstein-Taybi syndrome. J Med Genet 49:66–74. 10.1136/jmedgenet-2011-100354 [DOI] [PubMed] [Google Scholar]
- Lopez-Atalaya JP, Valor LM, Barco A (2014) Epigenetic factors in intellectual disability: the Rubinstein-Taybi syndrome as a paradigm of neurodevelopmental disorder with epigenetic origin. Prog Mol Biol Transl Sci 128:139–176. 10.1016/B978-0-12-800977-2.00006-1 [DOI] [PubMed] [Google Scholar]
- Lubin FD, Gupta S, Parrish RR, Grissom NM, Davis RL (2011) Epigenetic mechanisms: critical contributors to long-term memory formation. Neuroscientist 17:616–632. 10.1177/1073858410386967 [DOI] [PubMed] [Google Scholar]
- Lyst MJ, Bird A (2015) Rett syndrome: a complex disorder with simple roots. Nat Rev Genet 16:261–275. 10.1038/nrg3897 [DOI] [PubMed] [Google Scholar]
- Maniatis T, Reed R (2002) An extensive network of coupling among gene expression machines. Nature 416:499–506. 10.1038/416499a [DOI] [PubMed] [Google Scholar]
- Martens MB, Frega M, Classen J, Epping L, Bijvank E, Benevento M, van Bokhoven H, Tiesinga P, Schubert D, Nadif Kasri N (2016) Euchromatin histone methyltransferase 1 regulates cortical neuronal network development. Sci Rep 6:35756. 10.1038/srep35756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maze I, Covington HE 3rd, Dietz DM, LaPlant Q, Renthal W, Russo SJ, Mechanic M, Mouzon E, Neve RL, Haggarty SJ, Ren Y, Sampath SC, Hurd YL, Greengard P, Tarakhovsky A, Schaefer A, Nestler EJ (2010) Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science 327:213–216. 10.1126/science.1179438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maze I, Shen L, Zhang B, Garcia BA, Shao N, Mitchell A, Sun H, Akbarian S, Allis CD, Nestler EJ (2014) Analytical tools and current challenges in the modern era of neuroepigenomics. Nat Neurosci 17:1476–1490. 10.1038/nn.3816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mews P, Donahue G, Drake AM, Luczak V, Abel T, Berger SL (2017) Acetyl-CoA synthetase regulates histone acetylation and hippocampal memory. Nature 546:381–386. 10.1038/nature22405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo A, Mukamel EA, Davis FP, Luo C, Henry GL, Picard S, Urich MA, Nery JR, Sejnowski TJ, Lister R, Eddy SR, Ecker JR, Nathans J (2015) Epigenomic signatures of neuronal diversity in the mammalian brain. Neuron 86:1369–1384. 10.1016/j.neuron.2015.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton AJ, Skillings E, Bussey TJ, Saksida LM (2006) Measuring cognitive deficits in disabled mice using an automated interactive touchscreen system. Nat Methods 3:767. 10.1038/nmeth1006-767 [DOI] [PubMed] [Google Scholar]
- Mozzetta C, Pontis J, Fritsch L, Robin P, Portoso M, Proux C, Margueron R, Ait-Si-Ali S (2014) The histone H3 lysine 9 methyltransferases G9a and GLP regulate polycomb repressive complex 2-mediated gene silencing. Mol Cell 53:277–289. 10.1016/j.molcel.2013.12.005 [DOI] [PubMed] [Google Scholar]
- Nan X, Hou J, Maclean A, Nasir J, Lafuente MJ, Shu X, Kriaucionis S, Bird A (2007) Interaction between chromatin proteins MECP2 and ATRX is disrupted by mutations that cause inherited mental retardation. Proc Natl Acad Sci U S A 104:2709–2714. 10.1073/pnas.0608056104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negri G, Milani D, Colapietro P, Forzano F, Della Monica M, Rusconi D, Consonni L, Caffi LG, Finelli P, Scarano G, Magnani C, Selicorni A, Spena S, Larizza L, Gervasini C (2015) Clinical and molecular characterization of Rubinstein-Taybi syndrome patients carrying distinct novel mutations of the EP300 gene. Clin Genet 87:148–154. 10.1111/cge.12348 [DOI] [PubMed] [Google Scholar]
- Noh KM, Maze I, Zhao D, Xiang B, Wenderski W, Lewis PW, Shen L, Li H, Allis CD (2015) ATRX tolerates activity-dependent histone H3 methyl/phos switching to maintain repetitive element silencing in neurons. Proc Natl Acad Sci U S A 112:6820–6827. 10.1073/pnas.1411258112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picketts DJ, Higgs DR, Bachoo S, Blake DJ, Quarrell OW, Gibbons RJ (1996) ATRX encodes a novel member of the SNF2 family of proteins: mutations point to a common mechanism underlying the ATR-X syndrome. Hum Mol Genet 5:1899–1907. 10.1093/hmg/5.12.1899 [DOI] [PubMed] [Google Scholar]
- Pilotto S, Speranzini V, Marabelli C, Rusconi F, Toffolo E, Grillo B, Battaglioli E, Mattevi A (2016) LSD1/KDM1A mutations associated to a newly described form of intellectual disability impair demethylase activity and binding to transcription factors. Hum Mol Genet 25:2578–2587. 10.1093/hmg/ddw120 [DOI] [PubMed] [Google Scholar]
- Rauch A, Wieczorek D, Graf E, Wieland T, Endele S, Schwarzmayr T, Albrecht B, Bartholdi D, Beygo J, Di Donato N, Dufke A, Cremer K, Hempel M, Horn D, Hoyer J, Joset P, Röpke A, Moog U, Riess A, Thiel CT, et al. (2012) Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet 380:1674–1682. 10.1016/S0140-6736(12)61480-9 [DOI] [PubMed] [Google Scholar]
- Rothbart SB, Strahl BD (2014) Interpreting the language of histone and DNA modifications. Biochim Biophys Acta 1839:627–643. 10.1016/j.bbagrm.2014.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rube HT, Lee W, Hejna M, Chen H, Yasui DH, Hess JF, LaSalle JM, Song JS, Gong Q (2016) Sequence features accurately predict genome-wide MeCP2 binding in vivo. Nat Commun 7:11025. 10.1038/ncomms11025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein JH, Taybi H (1963) Broad thumbs and toes and facial abnormalities: a possible mental retardation syndrome. Am J Dis Child 105:588–608. 10.1001/archpedi.1963.02080040590010 [DOI] [PubMed] [Google Scholar]
- Rusconi D, Negri G, Colapietro P, Picinelli C, Milani D, Spena S, Magnani C, Silengo MC, Sorasio L, Curtisova V, Cavaliere ML, Prontera P, Stangoni G, Ferrero GB, Biamino E, Fischetto R, Piccione M, Gasparini P, Salviati L, Selicorni A, et al. (2015) Characterization of 14 novel deletions underlying Rubinstein-Taybi syndrome: an update of the CREBBP deletion repertoire. Hum Genet 134:613–626. 10.1007/s00439-015-1542-9 [DOI] [PubMed] [Google Scholar]
- Rusconi F, Grillo B, Ponzoni L, Bassani S, Toffolo E, Paganini L, Mallei A, Braida D, Passafaro M, Popoli M, Sala M, Battaglioli E (2016) LSD1 modulates stress-evoked transcription of immediate early genes and emotional behavior. Proc Natl Acad Sci U S A 113:3651–3656. 10.1073/pnas.1511974113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scandaglia M, Lopez-Atalaya JP, Medrano-Fernandez A, Lopez-Cascales MT, del Blanco B, Lipinski M, Benito E, Olivares R, Iwase S, Shi Y, Barco A (2017) Loss of Kdm5c causes spurious transcription and prevents the fine-tuning of activity-regulated enhancers in neurons. Cell Rep 21:47–59. 10.1016/j.celrep.2017.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer A, Sampath SC, Intrator A, Min A, Gertler TS, Surmeier DJ, Tarakhovsky A, Greengard P (2009) Control of cognition and adaptive behavior by the GLP/G9a epigenetic suppressor complex. Neuron 64:678–691. 10.1016/j.neuron.2009.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seah C, Levy MA, Jiang Y, Mokhtarzada S, Higgs DR, Gibbons RJ, Bérubé NG (2008) Neuronal death resulting from targeted disruption of the Snf2 protein ATRX is mediated by p53. J Neurosci 28:12570–12580. 10.1523/JNEUROSCI.4048-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Xu W, Guo R, Rong B, Gu L, Wang Z, He C, Zheng L, Hu X, Hu Z, Shao ZM, Yang P, Wu F, Shi YG, Shi Y, Lan F (2016) Suppression of enhancer overactivation by a RACK7-histone demethylase complex. Cell 165:331–342. 10.1016/j.cell.2016.02.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y (2005) Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell 19:857–864. 10.1016/j.molcel.2005.08.027 [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y (2004) Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119:941–953. 10.1016/j.cell.2004.12.012 [DOI] [PubMed] [Google Scholar]
- Skene PJ, Illingworth RS, Webb S, Kerr AR, James KD, Turner DJ, Andrews R, Bird AP (2010) Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol Cell 37:457–468. 10.1016/j.molcel.2010.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spena S, Milani D, Rusconi D, Negri G, Colapietro P, Elcioglu N, Bedeschi F, Pilotta A, Spaccini L, Ficcadenti A, Magnani C, Scarano G, Selicorni A, Larizza L, Gervasini C (2015) Insights into genotype-phenotype correlations from CREBBP point mutation screening in a cohort of 46 Rubinstein-Taybi syndrome patients. Clin Genet 88:431–440. 10.1111/cge.12537 [DOI] [PubMed] [Google Scholar]
- Stefanko DP, Barrett RM, Ly AR, Reolon GK, Wood MA (2009) Modulation of long-term memory for object recognition via HDAC inhibition. Proc Natl Acad Sci U S A 106:9447–9452. 10.1073/pnas.0903964106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweatt JD. (2013) The emerging field of neuroepigenetics. Neuron 80:624–632. 10.1016/j.neuron.2013.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweatt JD. (2016) Dynamic DNA methylation controls glutamate receptor trafficking and synaptic scaling. J Neurochem 137:312–330. 10.1111/jnc.13564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Matsumura Y, Fukuda M, Kimura H, Shinkai Y (2008) G9a/GLP complexes independently mediate H3K9 and DNA methylation to silence transcription. EMBO J 27:2681–2690. 10.1038/emboj.2008.192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Mei P, Fang R, Leonor T, Rutenberg M, Shimizu F, Li J, Rao A, Shi Y (2007) The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature 447:601–605. 10.1038/nature05823 [DOI] [PubMed] [Google Scholar]
- Talkowski ME, Rosenfeld JA, Blumenthal I, Pillalamarri V, Chiang C, Heilbut A, Ernst C, Hanscom C, Rossin E, Lindgren AM, Pereira S, Ruderfer D, Kirby A, Ripke S, Harris DJ, Lee JH, Ha K, Kim HG, Solomon BD, Gropman AL, et al. (2012) Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell 149:525–537. 10.1016/j.cell.2012.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Naruse I, Hongo T, Xu M, Nakahata T, Maekawa T, Ishii S (2000) Extensive brain hemorrhage and embryonic lethality in a mouse null mutant of CREB-binding protein. Mech Dev 95:133–145. 10.1016/S0925-4773(00)00360-9 [DOI] [PubMed] [Google Scholar]
- Telese F, Gamliel A, Skowronska-Krawczyk D, Garcia-Bassets I, Rosenfeld MG (2013) “Seq-ing” insights into the epigenetics of neuronal gene regulation. Neuron 77:606–623. 10.1016/j.neuron.2013.01.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toffolo E, Rusconi F, Paganini L, Tortorici M, Pilotto S, Heise C, Verpelli C, Tedeschi G, Maffioli E, Sala C, Mattevi A, Battaglioli E (2014) Phosphorylation of neuronal Lysine-Specific Demethylase 1LSD1/KDM1A impairs transcriptional repression by regulating interaction with CoREST and histone deacetylases HDAC1/2. J Neurochem 128:603–616. 10.1111/jnc.12457 [DOI] [PubMed] [Google Scholar]
- Tunovic S, Barkovich J, Sherr EH, Slavotinek AM (2014) De novo ANKRD11 and KDM1A gene mutations in a male with features of KBG syndrome and Kabuki syndrome. Am J Med Genet A 164A:1744–1749. 10.1002/ajmg.a.36450 [DOI] [PubMed] [Google Scholar]
- Turrigiano G. (2012) Homeostatic synaptic plasticity: local and global mechanisms for stabilizing neuronal function. Cold Spring Harb Perspect Biol 4:a005736. 10.1101/cshperspect.a005736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallianatos CN, Iwase S (2015) Disrupted intricacy of histone H3K4 methylation in neurodevelopmental disorders. Epigenomics 7:503–519. 10.2217/epi.15.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valor LM, Pulopulos MM, Jimenez-Minchan M, Olivares R, Lutz B, Barco A (2011) Ablation of CBP in forebrain principal neurons causes modest memory and transcriptional defects and a dramatic reduction of histone acetylation, but does not affect cell viability. J Neurosci 31:1652–1663. 10.1523/JNEUROSCI.4737-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valor LM, Viosca J, Lopez-Atalaya JP, Barco A (2013) Lysine acetyltransferases CBP and p300 as therapeutic targets in cognitive and neurodegenerative disorders. Curr Pharm Des 19:5051–5064. 10.2174/13816128113199990382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen K, Egger JI, Janzing JG, van Dongen L, van Bokhoven H, Kleefstra T, Staal WG (2017) The context of symptom measures: interpretation and clinical diagnosis of autism spectrum disorders in intellectual disabilities. J Am Acad Child Adolesc Psychiatry 56:618–619. 10.1016/j.jaac.2017.05.009 [DOI] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Ren B, Rubin EM, Pennacchio LA (2009) ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457:854–858. 10.1038/nature07730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Scully K, Zhu X, Cai L, Zhang J, Prefontaine GG, Krones A, Ohgi KA, Zhu P, Garcia-Bassets I, Liu F, Taylor H, Lozach J, Jayes FL, Korach KS, Glass CK, Fu XD, Rosenfeld MG (2007) Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature 446:882–887. 10.1038/nature05671 [DOI] [PubMed] [Google Scholar]
- Wang J, Telese F, Tan Y, Li W, Jin C, He X, Basnet H, Ma Q, Merkurjev D, Zhu X, Liu Z, Zhang J, Ohgi K, Taylor H, White RR, Tazearslan C, Suh Y, Macfarlan TS, Pfaff SL, Rosenfeld MG (2015) LSD1n is an H4K20 demethylase regulating memory formation via transcriptional elongation control. Nat Neurosci 18:1256–1264. 10.1038/nn.4069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K (2009) Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138:1019–1031. 10.1016/j.cell.2009.06.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson LA, Solomon LA, Li JR, Jiang Y, Edwards M, Shin-ya K, Beier F, Bérubé NG (2013) Atrx deficiency induces telomere dysfunction, endocrine defects, and reduced life span. J Clin Invest 123:2049–2063. 10.1172/JCI65634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb WM, Sanchez RG, Perez G, Butler AA, Hauser RM, Rich MC, O'Bierne AL, Jarome TJ, Lubin FD (2017) Dynamic association of epigenetic H3K4me3 and DNA 5hmC marks in the dorsal hippocampus and anterior cingulate cortex following reactivation of a fear memory. Neurobiol Learn Mem 142:66–78. 10.1016/j.nlm.2017.02.010 [DOI] [PubMed] [Google Scholar]
- Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, Cowley SM, Young RA (2012) Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature 482:221–225. 10.1038/nature10805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong LH, McGhie JD, Sim M, Anderson MA, Ahn S, Hannan RD, George AJ, Morgan KA, Mann JR, Choo KH (2010) ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res 20:351–360. 10.1101/gr.101477.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Gibbons R, Yan Z, Yang D, McDowell TL, Sechi S, Qin J, Zhou S, Higgs D, Wang W (2003) The ATRX syndrome protein forms a chromatin-remodeling complex with Daxx and localizes in promyelocytic leukemia nuclear bodies. Proc Natl Acad Sci U S A 100:10635–10640. 10.1073/pnas.1937626100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao TP, Oh SP, Fuchs M, Zhou ND, Ch'ng LE, Newsome D, Bronson RT, Li E, Livingston DM, Eckner R (1998) Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell 93:361–372. 10.1016/S0092-8674(00)81165-4 [DOI] [PubMed] [Google Scholar]
- Yearim A, Gelfman S, Shayevitch R, Melcer S, Glaich O, Mallm JP, Nissim-Rafinia M, Cohen AH, Rippe K, Meshorer E, Ast G (2015) HP1 is involved in regulating the global impact of DNA methylation on alternative splicing. Cell Rep 10:1122–1134. 10.1016/j.celrep.2015.01.038 [DOI] [PubMed] [Google Scholar]
- Zibetti C, Adamo A, Binda C, Forneris F, Toffolo E, Verpelli C, Ginelli E, Mattevi A, Sala C, Battaglioli E (2010) Alternative splicing of the histone demethylase LSD1/KDM1 contributes to the modulation of neurite morphogenesis in the mammalian nervous system. J Neurosci 30:2521–2532. 10.1523/JNEUROSCI.5500-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zocchi L, Sassone-Corsi P (2010) Joining the dots: from chromatin remodeling to neuronal plasticity. Curr Opin Neurobiol 20:432–440. 10.1016/j.conb.2010.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]