Abstract
Infant colic is a distressing condition of unknown etiology. An aberrant gastrointestinal microbiota has been associated, and Lactobacillus reuteri supplementation has been shown to reduce crying and/or fussing time (‘crying time’) in some infants with colic. The relationship between L. reuteri gut colonization and crying time has not been examined. We investigated the relationship between L. reuteri colonization and fecal microbiota (microbial diversity and Escherichia coli), intestinal inflammation, and crying time in infants with colic, using a subset of 65 infants from the Baby Biotics trial, which randomized healthy term infants aged <13 weeks with infant colic to receive probiotic L. reuteri DSM 17938 (1 × 108 colony forming units) or placebo daily for 28 days. We observed an overall reduction in median crying time, regardless of L. reuteri colonization status (n = 14 colonized). There were no differences in E. coli colonization rates or densities, microbial diversity or intestinal inflammation by L. reuteri colonization status. We found that L. reuteri density positively correlated with crying time, and E. coli density negatively correlated with microbial diversity. As density of L. reuteri was associated with increased crying time, L. reuteri supplementation may not be an appropriate treatment for all infants with colic.
Introduction
Infant colic is a distressing condition characterized by crying and/or fussing of unknown cause that affects up to 28% of infants under three months of age1. The etiology of infant colic is unclear and there is no widely accepted or effective treatment2,3. Several studies have found differences in gastrointestinal (GI) microbial diversity between infants with colic and healthy infants, suggesting that an aberrant microbial intestinal profile may cause or contribute to infant colic4–6. Infants with colic have higher rates and densities of Escherichia coli and other gas-producing coliforms and lower levels of Lactobacillus spp. compared with healthy infants4–7, with some aerobic bacterial genera not detected in infants with colic8. One study found that infants with colic had lower microbial diversity and elevated fecal calprotectin (a marker of gut inflammation) compared with healthy infants9. However, another study found no difference in fecal calprotectin levels between healthy infants and infants with colic10.
The finding that infants with colic have an altered intestinal microbiota has led to the investigation of probiotic supplementation for the treatment of this condition, with the aim of promoting a healthy intestinal microbiota and reducing intestinal inflammation. Several clinical trials suggest Lactobacillus reuteri may reduce crying and/or fussing time (referred to as ‘crying time’) in some infants with colic11–15. In contrast, we recently reported results of a double blind, randomized, placebo-controlled trial evaluating L. reuteri supplementation for the treatment of infant colic in Melbourne, Australia (Baby Biotics trial, Current Controlled Trials ISRCTN95287767 (25/10/2010)) that found no effect on crying time when compared to placebo16. However, analysis of fecal samples indicated that less than half of the infants in the probiotic treatment group were colonized with L. reuteri at day 28 of the trial, and this low colonization rate may have contributed to the negative trial findings. As probiotics are thought to exert their beneficial effects on the host through transiently colonizing the GI tract, we sought to examine the relationship between L. reuteri colonization and key microbiological, immunological, and clinical characteristics of infant colic. We also investigated the role of E. coli, which has been implicated in infant colic4,7,17. The use of quantitative real-time PCR (qPCR) for detection of L. reuteri and E. coli enabled density-based analysis of bacterial loads. This study examined the relationships between L. reuteri and E. coli colonization, microbial diversity, intestinal inflammation, and crying time in a subset of infants from the Baby Biotics trial cohort. We hypothesized that infants colonized with L. reuteri would have lower E. coli colonization, greater microbial diversity, lower intestinal inflammation (calprotectin levels), and lower crying time, when compared to infants not colonized with L. reuteri.
Results
Sixty-five infants (31 probiotic, 34 placebo) were included in this study. Fecal samples were collected on day 28 of treatment and examined for L. reuteri and E. coli colonization, microbial diversity, and calprotectin. Some infants were excluded from analysis of diversity, calprotectin, and/or crying time due to no detection of a 16S rRNA gene PCR product or >10% of peak profile consisting of large fragments (AluI, n = 20; Sau96I, n = 10), insufficient sample volume (n = 7), or insufficient crying time data (n = 9), respectively.
The two groups in this study, infants colonized by L. reuteri (n = 14) and infants not colonized by L. reuteri (n = 51), had similar clinical characteristics (Table 1). The clinical characteristics were also representative of the original Baby Biotics trial cohort, except for higher rates of family history of allergic disease in this study.
Table 1.
Clinical characteristics of the study population.
| Characteristic | Original trial16 | This study | |||
|---|---|---|---|---|---|
| Probiotic (n = 85) | Placebo (n = 82) | Colonized by L. reuteri (n = 14) | Not colonized by L. reuteri (n = 51) | P-valuea | |
| Gender Male, n (%) | 37 (44) | 48 (59) | 6 (43) | 30 (59) | 0.29b |
| Mode of birth Caesarean section, n (%) | 35 (41) | 32 (39) | 4 (29) | 20 (40) | 0.46b |
| Method of feeding Exclusively breastfed, n (%) | 33 (39) | 35 (43) | 6 (43) | 17 (33) | 0.51b |
| Family history of allergic disease Yes, n (%) | 51 (60) | 50 (61) | 11 (79) | 41 (80) | 0.88b |
| Birth weight (g) Mean ± SD | 3272 ± 406 | 3426 ± 421 | 3425 ± 482 | 3312 ± 398 | 0.57c |
| Infant age at enrolment (weeks) Mean ± SD | 7.5 ± 2.9 | 6.9 ± 2.5 | 8.2 ± 2.8 | 7.4 ± 2.7 | 0.29c |
| Infant crying time at day 0 (min/day) d Mean ± SD | 327.6 ± 151.9 | 329.3 ± 126.4 | 337.2 ± 186.3 | 344.1 ± 124.1 | 0.94c |
aDifferences assessed for infants in the current study (colonized versus not colonized by L. reuteri). bChi-Squared test; c Mann-Whitney U test; dCalculated using probiotic (n = 75), placebo (n = 65), colonized by L. reuteri (n = 13), and not colonized by L. reuteri (n = 47) due to missing data.
L. reuteri and E. coli colonization
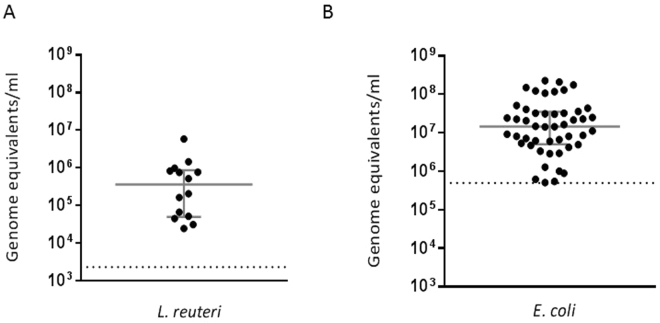
Of the 31 infants that received probiotic, 14 (45%) were colonized with L. reuteri at day 28, whilst none of the 34 infants in the placebo group became colonized (Chi-squared test, P < 0.0001). Of the infants colonized by L. reuteri, 93% (13/14) were also colonized by E. coli, whereas 69% (35/51) of infants not colonized by L. reuteri, were colonized by E. coli, however this difference was not statistically significant (Chi-squared test, P = 0.07). Infants colonized with L. reuteri had a median colonization density of 3.6 × 105 genome equivalents/ml (Interquartile range (IQR): 5.0 × 104, 8.7 × 105 genome equivalents/ml), while infants colonized with E. coli had a median colonization density of 1.5 × 107 genome equivalents/ml (IQR: 5.0 × 106, 3.5 × 107 genome equivalents/ml) (Fig. 1). There was no difference in E. coli density in infants colonized or not colonized by L. reuteri (Table 2). In infants colonized by both species, there was no association between L. reuteri and E. coli densities (Spearman’s rank correlation coefficient, r = 0.16; P = 0.60).
Figure 1.
(A) L. reuteri (n = 14) and (B) E. coli (n = 48) colonization levels in fecal samples collected at day 28 from infants with colic. Bars represent median ± interquartile range; dotted lines represent assay limit of detection.
Table 2.
Outcomes by Lactobacillus reuteri colonization status, at day 28.
| Outcomes | Median (IQR) | P-valuea | |||
|---|---|---|---|---|---|
| n | Colonized by L. reuteri | n | Not colonized by L. reuteri | ||
| Fecal E. coli colonization (genome equivalents/ml) | 13 | 1.4 × 107 (3.6 × 106, 6.9 × 107) | 35 | 1.6 × 107 (5.4 × 106, 3.6 × 107) | 0.71 |
| Fecal microbial diversity score | |||||
| - AluI peaks | 10 | 28.5 (20.5, 35.8) | 35 | 24.0 (21.0, 27.0) | 0.14 |
| - Sau96I peaks | 14 | 32.0 (28.0, 35.3) | 41 | 31.0 (27.0, 36.0) | 0.62 |
| Fecal calprotectin (µg/g) | 13 | 135.4 (89.5, 568.5) | 45 | 114.8 (67.1, 237.9) | 0.37 |
| Infant crying time (min/day) | 13 | 165.0 (100.3, 268.8) | 45 | 180.0 (136.3, 274.1) | 0.72 |
aMann-Whitney U test.
Microbial diversity
Terminal restriction fragment length polymorphism (T-RFLP) was conducted, with peak profiles generated from FAM-labelled PCR product targeting total 16S rRNA genes using two restriction enzymes (AluI and Sau96I) and analyzed separately. Peaks were quantified to provide an estimate of intestinal microbial diversity, with a higher number of peaks representing greater diversity. There was no difference between the number of peaks in infants colonized with L. reuteri when compared to those not colonized using AluI or Sau96I (Table 2). In infants colonized by L. reuteri, there was no association between density of L. reuteri colonization and number of peaks in profiles generated by either AluI (Spearman’s rank correlation coefficient, r = −0.52; P = 0.13) or Sau96I digestion (Spearman’s rank correlation coefficient, r = 0.26; P = 0.36).
Intestinal inflammation
Fecal calprotectin levels were measured to investigate inflammation within the GI tract. All 58 infants assessed had detectable levels of calprotectin (median 116.2 µg of calprotectin per g wet feces (µg/g), IQR: 72.3, 255.8). There was no difference in calprotectin levels between infants colonized with L. reuteri compared to those not colonized (Table 2). In infants colonized by L. reuteri, there was no association between density of L. reuteri colonization and calprotectin (Spearman’s rank correlation coefficient, r = 0.16; P = 0.60).
Crying time
There was a reduction in infant crying over time, regardless of L. reuteri colonization status. In infants colonized by L. reuteri, median crying reduced from 330.0 min/day (IQR: 211.3, 445.3 min/day) on day 0 to 172.5 min/day (IQR: 92.7, 278.1 min/day) on day 28 (Wilcoxon signed-rank test, P = 0.03) (Fig. 2A). Similarly, median crying reduced from 328.4 min/day (IQR: 237.7, 443.8 min/day) on day 0 to 180.0 min/day (IQR: 135.6, 265.5 min/day) on day 28 in infants not colonized by L. reuteri (Wilcoxon signed-rank test, P < 0.0001). At day 28 there was no difference between median infant crying time between infants colonized and infants not colonized by L. reuteri (Table 2). For the 13 infants colonized by L. reuteri, a positive association was found between L. reuteri colonization density and median infant crying time (Spearman’s rank correlation coefficient, r = 0.68; P = 0.01) (Fig. 2B).
Figure 2.
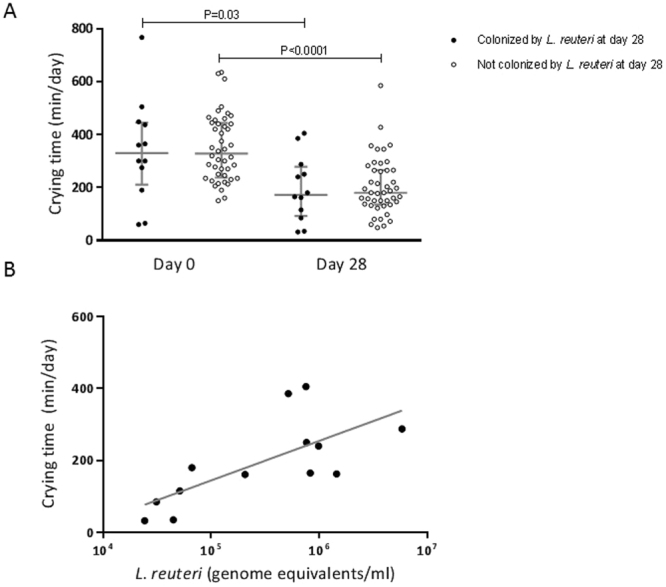
The association between L. reuteri colonization and infant crying time. (A) Crying time in infants colonized (n = 12) and not colonized (n = 44) by L. reuteri. Infants were categorized as colonized or not colonized by L. reuteri at both time points by L. reuteri colonization status at day 28. Bars represent median ± interquartile range. Differences within groups (day 0 to day 28) were assessed using Wilcoxon signed-rank test and differences between groups (colonized and not colonized) using Mann-Whitney U test. (B) The relationship between crying time and L. reuteri colonization density (n = 13); Spearman’s rank correlation coefficient, r = 0.68; P = 0.01.
Secondary analyses
We additionally assessed the relationships between E. coli colonization and microbial diversity, intestinal inflammation and crying time (Table 3). We found no differences in microbial diversity in infants colonized or not colonized by E. coli, and no association between density of E. coli colonization and number of peaks by AluI digestion (Spearman’s rank correlation coefficient, r = −0.12; P = 0.49). However, in infants colonized by E. coli, there was a negative association between E. coli colonization density and number of peaks in profiles generated by Sau96I digestion (Spearman’s rank correlation coefficient, r = −0.33; P = 0.03). We found no association between density of E. coli colonization and calprotectin (Spearman’s rank correlation coefficient, r = −0.06; P = 0.69). Median crying reduced from 307.5 min/day (IQR: 226.3, 439.2 min/day) on day 0 to 180.0 min/day (IQR: 134.0, 282.0 min/day) on day 28 in infants colonized by E. coli (Wilcoxon signed-rank test, P < 0.0001). Similarly, median crying reduced from 352.5 min/day (IQR: 288.8, 460.8 min/day) on day 0 to 168.8 min/day (IQR: 133.1, 220.3 min/day) on day 28 in infants not colonized by E. coli (Wilcoxon signed-rank test, P = 0.0003) (Fig. 3A). There was no statistically significant difference between median change in crying time from day 0 to day 28 between infants colonized (81.3 min/day, IQR: 15.5, 263.6) or not colonized (193.8 min/day, IQR: 125.7, 295.6) by E. coli (Mann-Whitney U test, P = 0.07). For the 42 infants colonized by E. coli, no relationship was found between E. coli colonization density and median infant crying time (Spearman’s rank correlation coefficient, r = 0.01; P = 0.94) (Fig. 3B).
Table 3.
Outcomes by Escherichia coli colonization status, at day 28.
| Outcomes | Median (IQR) | P-valuea | |||
|---|---|---|---|---|---|
| n | Colonized by E. coli | n | Not colonized by E. coli | ||
| Fecal microbial diversity score | |||||
| - AluI peaks | 36 | 25.0 (21.3, 29.0) | 9 | 22.0 (16.5, 26.5) | 0.17 |
| - Sau96I peaks | 44 | 31.0 (27.0, 36.0) | 11 | 30.0 (28.0, 33.0) | 0.87 |
| Fecal calprotectin (µg/g) | 46 | 118.0 (70.2, 267.3) | 12 | 94.2 (74.4, 246.5) | 0.58 |
| Infant crying time (min/day) | 42 | 180.0 (142.0, 288.5) | 16 | 168.8 (133.1, 220.3) | 0.44 |
aMann-Whitney U test.
Figure 3.
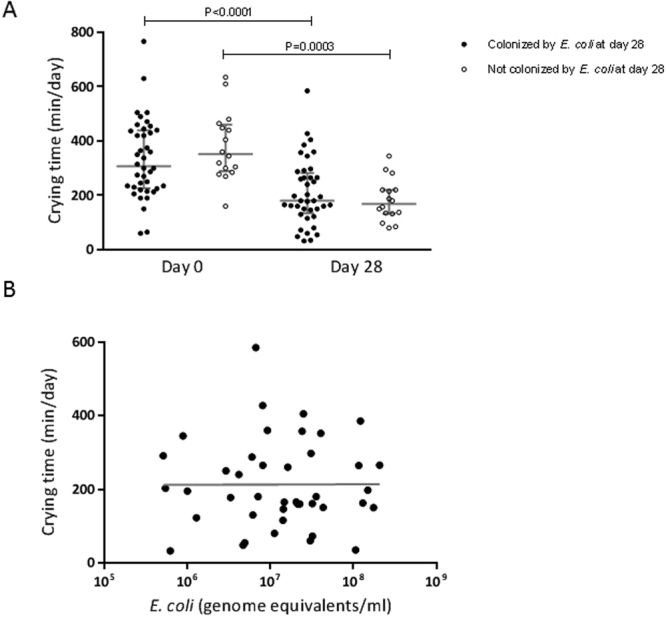
The association between E. coli colonization and crying time. (A) Crying time in infants colonized (n = 40) and not colonized (n = 16) by E. coli. Infants were categorized as colonized or not colonized by E. coli at both time points by E. coli colonization status at day 28. Bars represent median ± interquartile range. Differences within groups (day 0 to day 28) were assessed using Wilcoxon signed-rank test and differences between groups (colonized or not colonized) using Mann-Whitney U test. (B) The relationship between crying time and E. coli colonization density (n = 42); Spearman’s rank correlation coefficient, r = 0.01; P = 0.94.
Discussion
We previously reported that supplementation with L. reuteri DSM 17938 did not reduce crying time in breastfed and formula-fed infants with colic16. However, the L. reuteri colonization rates were lower than expected, potentially affecting the Baby Biotics trial results. In this substudy, we examined the relationship between L. reuteri colonization and microbiological outcomes (E. coli colonization and microbial diversity), intestinal inflammation, and crying time in a subset of infants from the trial. L. reuteri colonization rates (45% at ~ 4 months of age) in the current study were lower than those previously reported in other pediatric studies (92% at ~ 2 months11, and 65% at ~3 months18). The reduction in colonization rates with increasing infant age suggest age at probiotic administration and/or age at sample collection, as well as the use of different microbiological detection methods (culture-dependent versus culture-independent methods in our study), may affect L. reuteri detection11,18.We found median colonization density of L. reuteri to be 3.6 × 105 genome equivalents/ml in infants who were colonized with L. reuteri, which is in line with previous research that found mean L. reuteri colonization densities of 104–105 CFU/g of feces after supplementation11,19. Microbial diversity was similar in infants colonized or not colonized with L. reuteri, consistent with Roos et al. who found that treatment with L. reuteri did not affect the global composition of the bacterial community20. We found no difference in calprotectin levels in infants colonized or not colonized by L. reuteri, and no association between calprotectin levels and crying time at day 28. However, we previously reported that infants who had a reduction in crying time of at least 50% had lower calprotectin levels, suggesting gut inflammation may be implicated in infant colic, although the precise relationship remains unclear16. Crying time decreased in infants from day 0 to day 28 regardless of L. reuteri colonization status, in line with the natural resolution of infant colic that typically occurs when an infant reaches three to four months of age21. There were two infants in the L. reuteri colonized group with lower crying times at day 0 than were observed in the not colonized group (approximately 60 min/day, versus minimum of 150 min/day). This may have decreased the ability to detect a reduction in crying time from day 0 to day 28 in the infants colonized with L. reuteri, however we still found a reduction in median crying time. We found no difference in crying time at day 28 between infants colonized or not colonized by L. reuteri. These findings were consistent with the original Baby Biotics trial results, and in contrast to previous reports that L. reuteri treatment is effective in reducing crying time in infants with colic when crying is assessed by treatment allocation11–15. These divergent findings may reflect differences in geographic location of previous studies (Italy, Poland, China and Canada) and rates of exclusive breastfeeding (majority of infants in previous studies versus 35% in our study). Geography and method of feeding are known to impact on gut microbiota22–25, therefore our study emphasizes that results from other settings may not be generalizable to our study population, or to Australian infants in general. In infants colonized by L. reuteri, we observed an unexpected positive relationship between L. reuteri density and crying time. Although this finding may generate concerns that high density of L. reuteri could exacerbate symptoms of colic, there does not appear to be a link with gut inflammation as there was no association between L. reuteri density and calprotectin. It is plausible that some infants may have an underlying gut environment that is associated with higher crying and also facilitates high-density L. reuteri growth.
In our study, we found no evidence linking E. coli to gut inflammation as measured by calprotectin. We found a negative association between E. coli colonization density and microbial diversity, although this was only detected with one of the enzymes used in the diversity assay. E. coli has been associated with Crohn’s Disease, with decreases in microbial diversity compared with healthy participants26,27, suggesting a possible pathophysiological relationship between GI E. coli colonization and microbial diversity. Savino et al. found that infants who responded to L. reuteri treatment (a reduction of ≥50% crying time) had a reduction in fecal E. coli, so our finding of no difference in E. coli colonization is consistent with the outcome that treatment with L. reuteri did not reduce crying time in this study population. There was moderate evidence that infants colonized with E. coli had a smaller reduction in crying from baseline to day 28, however there was no difference between crying time at day 28 when assessed by E. coli colonization status. It has been previously reported that L. reuteri administration was associated with a decreased presence of E. coli, suggesting a potential negative interaction between these two species11. However, we did not observe a negative relationship between L. reuteri and E. coli, and in fact, 93% of infants colonized with L. reuteri also had E. coli detected. This suggests that the findings of Savino et al. may have been due to an indirect rather than direct relationship between the two species, and/or could be another example of differences attributable to geography and feeding methods.
To our knowledge, this is the first study to investigate microbiological changes in infant colic by L. reuteri colonization status. The participants in our study are representative of help-seeking parents and carers from the general population. Because Australian pediatricians do not deliver primary care, families often present to the emergency department for common childhood concerns, especially for conditions such as colic where the burden typically occurs outside business hours. Additionally, this study included both breast and formula-fed infants and did not restrict the mother’s diet to make the probiotic intervention more generalizable. This is in contrast to the majority of infant colic studies that use populations of exclusively (or predominately) breastfed infants11–15, and have mothers or infants on a cow’s milk-free diet11,12,14. However, this study had several limitations. Only a subset of samples from the original trial were available for the analyses conducted in this pilot project. We performed laboratory analyses at a single time point at the end of treatment when crying time had already reduced, which may have blunted the ability to detect potential differences linked to crying time. The infant microbiome is highly variable, and it is plausible that transient changes due to probiotic colonization may have occurred earlier. Additionally, while we used robust culture-independent molecular methods to identify L. reuteri and E. coli, the qPCR primers and probes may detect other closely related species, such as the closely related Shigella spp. for the E. coli assay28–30. Nevertheless, only infants in the probiotic group had detectable L. reuteri, suggesting our assay was specific for the probiotic strain. Lastly, parents were instructed to deliver the supplement using a spoon or dropper; it is not known whether the route of administration impacted L. reuteri colonization. Advances in molecular methods and sequencing technology have enabled more detailed characterization of the microbiome including identification of more subtle differences in the bacterial communities present. Employing one of these methods may provide further information on differences in the GI microbiota and greater insight into potential impacts of L. reuteri treatment on the gut microbiome.
We found no reduction in crying time in infants colonized with L. reuteri and identified an unexpected positive association between L. reuteri density and crying time. Our results support the finding of the original Baby Biotics trial that L. reuteri supplementation did not reduce crying time in infants with colic in Melbourne, Australia. We also found a negative association between E. coli colonization density and microbial diversity, suggesting a possible pathophysiological relationship between E. coli colonization and microbial diversity. Our findings suggest L. reuteri supplementation may not be an appropriate treatment for infants with colic in some geographic settings, and that the impact of such an intervention on intestinal microbiota composition remains unclear. As our study included a small subset of infants from the Baby Biotics randomized trial, additional studies with larger sample size are required to further investigate the role of the gut microbiome in infant colic and the utility of probiotic treatment in different populations.
Methods
Study participants and design
The 65 infants included in this study were part of a previously described placebo-controlled trial in Melbourne, Australia that assessed L. reuteri supplementation as a treatment for infant colic16,31. In brief, 167 healthy term infants aged <13 weeks with infant colic (modified Wessel’s criteria, crying and/or fussing for ≥3 hours a day, for ≥3 days over seven days) were randomized to receive five drops of probiotic (1 × 108 colony forming units (CFU)/day L. reuteri DSM 17938; BioGaia AB, Stockholm, Sweden) or placebo daily for 28 days. A random sample of eight returned probiotic bottles were tested to confirm the probiotic suspension was above the required dose; mean L. reuteri recovery was 1.8 × 108 CFU/5 drops. The trial was approved by the Human Research Ethics Committee of The Royal Children’s Hospital (HREC 30111), and informed consent was obtained for all participants. 127 infants completed the original trial (67 probiotic, 60 placebo). For the current study, fecal samples were analyzed from the first 65 infants enrolled in the original trial who produced adequate fecal sample volume (31 probiotic, 34 placebo; Table 1). All methods were carried out in accordance with relevant guidelines and regulations. All laboratory analyses were performed blinded.
Infant crying and/or fussing time
Infant crying and/or fussing time (referred to as ‘crying time’) was measured using the Barr diary and analyzed as described elsewhere32,33. Infant crying and fussing times were reported by the primary caregiver and collected over a 24 hour period at baseline, and as an average of 48 hours for day 28 of the study (measured over days 28 and 29). The diaries were adjusted for missing information, with diaries that had ≥30% missing data excluded from analyses at the relevant time point. For diaries with <30% missing data, mean values of crying time were generated using the equation: [episodes of daily cry/fuss bouts]*[1440/(1440 – minutes of missing data)].
Fecal sample collection and storage
Fecal samples were collected on day 28 of the trial. Samples were collected from the infant’s diaper, transferred to a sterile container, and placed immediately into the caregiver’s freezer. Alternatively, the whole diaper was placed into the freezer. Frozen samples were transported on ice in an insulated container from the caregiver’s freezer to the laboratory within a median of 3 days (range 0–20 days). Once received at the laboratory, diaper samples were transferred into a container. All samples were stored at −80 °C until use (maximum storage of 9 months from collection to DNA extraction).
Extraction of fecal genomic DNA
On the day of DNA extraction, samples were thawed and homogenized before use. Genomic DNA was extracted using the UltraClean® Fecal DNA Isolation Kit (MO BIO Laboratories, Inc., Carlsbad, USA). Briefly, 0.15 g of homogenized fecal sample was added to a bead/ball mixture containing 0.5 g of 0.1 mm Zirconia/Silica beads (BioSpec Products Inc., Bartlesville, USA) and four 3 mm, undrilled, glass balls (Ajax Finechem Pty Ltd., Taren Point, Australia). DNA isolation was completed following the manufacturer’s protocol from the addition of Solution S1 of the MO BIO kit. If pellets were not formed, the sample was spun in a centrifuge for an additional 3 min at 11,300 x g before transferring the supernatant. DNA was eluted in a total volume of 100 µl.
Quantification of L. reuteri and E. coli
L. reuteri and E. coli were quantified by qPCR using previously published primers and probes targeting the tuf and uidA genes, respectively28–30. Each 25 µl reaction consisted of 2 µl of genomic DNA, 1X Brilliant III Ultra-Fast QPCR Master Mix (Agilent Technologies, Santa Clara, USA), 100 µM probe and either 300 µM of L. reuteri primers or 100 µM of E. coli primers. Assays were performed using Stratagene MX3005P qPCR instrument with an initial activation of 95 °C for 3 min followed by 40 cycles of 95 °C for 20 s and 60 °C for 20 s. Reactions were performed in duplicate and the mean cycle threshold (Ct) value calculated. Genomic DNA from L. reuteri DSM 17938 and E. coli ATCC 25922 were used for standard curves; a mean Ct of <35 and <34, respectively, were considered positive. The assay detection limits were determined by validation against a panel of related bacteria, and no template controls. Samples with a higher Ct than an extraction control were considered negative. Bacterial density data are reported as genome equivalents/ml (assuming one copy of the target gene per genome, one genome per CFU, and a genome size of 2.0 Mb for L. reuteri and 5.24 Mb for E. coli).
Microbial diversity
Microbial diversity was examined using T-RFLP, using a modified version of a method previously described34,35. Briefly, the 16S rRNA gene was amplified using universal primers FAM27f and 519r (Sigma-Aldrich, Castle Hill, Australia) and the HotStarTaq® kit (Qiagen). Each 25 µl reaction contained 5 µl genomic DNA, 1.25 U DNA Polymerase, 1 X PCR Buffer, 2.5 mM MgCl2, 200 µM dNTPs, 500 nM fluorescently labelled forward primer and 500 nM reverse primer. PCR cycling conditions were denaturation at 95 °C for 5 min, followed by 25 cycles of 94 °C for 15 s, 50 °C for 15 s, 72 °C for 1 min, and an extension of 50 °C for 1 min 30 s and 72 °C for 6 min. Triplicate reactions were pooled. Samples with a PCR product of 480–540 base pairs were used for further processing. PCR products were purified using a Wizard SV Gel and PCR Clean-Up System column (Promega, Annandale, Australia). T-RFLP fragments were generated using separate reactions for enzymes AluI and Sau96I (New England Biolabs, Ipswich, USA). In 25 µl reactions, approximately 50–100 ng of purified PCR product was digested with 5 U of enzyme and 1 X NEBuffer 4 (New England Biolabs) in a water bath (37 °C overnight). PCR products were precipitated and analysed by an AB3730 DNA analyser using AB GeneMapper software (Applied Biosystems, Carlsbad, USA) at the Australian Genome Research Facility, Parkville, Australia. Separate peak profiles were generated for each enzyme. Analysis of T-RFLP peak profiles was performed as described previously34.
Calprotectin
100 mg of fecal sample and 4.9 ml of Extraction Buffer were mixed for 3 min in the Fecal Extraction Device (CALPRO AS, Lysaker, Norway) on a Ratek MPS1 plate shaker. Fecal extracts were stored at 4 °C and used within 5 days. Calprotectin levels were measured by enzyme linked immunosorbent assay (CALPROLAB™, CALPRO AS) according to manufacturer’s protocol. The optical density was measured at 405 nm using an ELx808™ Absorbance Microplate Reader (BioTek) and the KCjunior™ (BioTek) package to determine calprotectin levels.
Statistical analyses
Analyses were completed using GraphPad Prism version 7.01 for Windows (GraphPad Software, San Diego California USA). The measures of E. coli colonization, microbial diversity, intestinal inflammation, and crying time were stratified by L. reuteri colonization status and tested for normality using the D’Agostino-Pearson K-squared test. As the majority of data sets were not normally distributed, non-parametric tests were used throughout. Differences between categorical variables were assessed using the Chi-Squared test. Differences within groups (colonized or not colonized by L. reuteri; colonized or not colonized by E. coli) were assessed using the Wilcoxon signed-rank test, and between groups using the Mann-Whitney U test. Correlations were assessed using Spearman’s rank correlation coefficient. All tests were two-tailed and P < 0.05 was considered statistically significant.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Acknowledgements
We thank all study participants and families who took part in the Baby Biotics trial, Melissa Wake and Harriet Hiscock for their contribution into the design of the Baby Biotics trial, Frances Oppedisano, Susie Germano and Anne Balloch for assistance with laboratory testing, Elissa York for patient recruitment, and CALPRO for providing Fecal Extraction Devices and Calprotectin ELISAs.
Author Contributions
M.N. developed and performed laboratory assays, completed statistical analysis and drafted the manuscript. M.T. designed and oversaw the study, advised on laboratory outcome measures and interpretation, and oversaw manuscript development. C.S. and E.D. oversaw laboratory assays and their interpretation, and assisted with manuscript development. FM provided statistical advice. S.J. completed the analysis of terminal restriction fragment polymorphism peak profiles. V.S. designed the original Baby Biotics trial with M.T. and F.M. All authors read and approved the final manuscript.
Competing Interests
The Baby Biotics clinical trial was supported by the Georgina Menzies Maconachie Charitable Trust administered by Equity Trustees. VS (Postgraduate Scholarship 607447) and FM (ECF: 1037449, CDF: 1111160) have support from the Australian National Health and Medical Research Council (NHMRC) for the submitted work. VS is supported by a NHMRC ECF (1125687), a Royal Australasian College of Physicians Cottrell Research Establishment Fellowship, and has been supported by a Melbourne Children’s Clinician Scientist Fellowship. CS holds an Australian NHMRC CDF: 1087957. BioGaia Sweden supplied the investigational product and placebo at no cost. Calpro AS supplied CalproLab ELISA kits for the analysis of calprotectin levels at no cost. BioGaia, Calpro AS, and Equitee Trustees were independent of the trial and its researchers and played no role in the trial design; in the collection, analysis, and interpretation of data; in the writing of the report; and in the decision to submit the article for publication. MT is an inventor on a patent owned by the Murdoch Children’s Research Institute “A method of inducing tolerance”, and is an employee of and holds share interest in ProTA. MT is a member of medical advisory board for Nestle Nutrition Institute, past member of Global Scientific Advisory Board for Danone Nutricia, received honoraria for presentations at symposia sponsored by Danone Nutricia, received probiotic and placebo research products from Nestlé Research Centre Switzerland and Dicofarm Italy for studies unrelated to this trial. CS and ED received probiotic strains from Blis Technologies for studies unrelated to this trial. Murdoch Children’s Research Institute is supported by the Victorian Government’s Operational Infrastructure Support Program. The other authors have indicated they have no financial relationships relevant to this article to disclose.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lucassen PL, et al. Systematic review of the occurrence of infantile colic in the community. Arch. Dis. Childh., Lond. 2001;84:398–403. doi: 10.1136/adc.84.5.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sung V, et al. Probiotics to prevent or treat excessive infant crying: systematic review and meta-analysis. JAMA Pediatr. 2013;167:1150–1157. doi: 10.1001/jamapediatrics.2013.2572. [DOI] [PubMed] [Google Scholar]
- 3.Garrison MM, Christakis DA. A systematic review of treatments for infant colic. Pediatrics. 2000;106:184–190. [PubMed] [Google Scholar]
- 4.Savino F, et al. Molecular identification of coliform bacteria from colicky breastfed infants. Acta Paediatr. 2009;98:1582–1588. doi: 10.1111/j.1651-2227.2009.01419.x. [DOI] [PubMed] [Google Scholar]
- 5.Savino F, et al. Intestinal microflora in breastfed colicky and non-colicky infants. Acta Paediatr. 2004;93:825–829. doi: 10.1111/j.1651-2227.2004.tb03025.x. [DOI] [PubMed] [Google Scholar]
- 6.Savino F, et al. Antagonistic effect of Lactobacillus strains against gas-producing coliforms isolated from colicky infants. BMC Microbiol. 2011;11:157. doi: 10.1186/1471-2180-11-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Savino F, et al. Comparison of formula-fed infants with and without colic revealed significant differences in total bacteria, Enterobacteriaceae and faecal ammonia. Acta Paediatr. 2017;106:573–578. doi: 10.1111/apa.13642. [DOI] [PubMed] [Google Scholar]
- 8.Mentula S, Tuure T, Koskenala R, Korpela R, Kononen E. Microbial composition and fecal fermentation end products from colicky infants- a probiotic supplementation pilot. Micro. Ecol. Health Dis. 2008;20:37–47. doi: 10.1080/08910600801933846. [DOI] [Google Scholar]
- 9.Rhoads JM, et al. Altered fecal microflora and increased fecal calprotectin in infants with colic. J. Pediatr. 2009;155:823–828. doi: 10.1016/j.jpeds.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 10.Olafsdottir E, Aksnes L, Fluge G, Berstad A. Faecal calprotectin levels in infants with infantile colic, healthy infants, children with inflammatory bowel disease, children with recurrent abdominal pain and healthy children. Acta Paediatr. 2002;91:45–50. doi: 10.1111/j.1651-2227.2002.tb01638.x. [DOI] [PubMed] [Google Scholar]
- 11.Savino F, et al. Lactobacillus reuteri DSM 17938 in infantile colic: a randomized, double-blind, placebo-controlled trial. Pediatrics. 2010;126:526–533. doi: 10.1542/peds.2010-0433. [DOI] [PubMed] [Google Scholar]
- 12.Savino F, Pelle E, Palumeri E, Oggero R, Miniero R. Lactobacillus reuteri (American Type Culture Collection Strain 55730) versus simethicone in the treatment of infantile colic: a prospective randomized study. Pediatrics. 2007;119:124–130. doi: 10.1542/peds.2006-1222. [DOI] [PubMed] [Google Scholar]
- 13.Szajewska H, Gyrczuk E, Horvath A. Lactobacillus reuteri DSM 17938 for the Management of Infantile Colic in Breastfed Infants: A Randomized, Double-Blind, Placebo-Controlled Trial. J. Pediatr. 2013;162:257–262. doi: 10.1016/j.jpeds.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 14.Mi GL, et al. Effectiveness of Lactobacillus reuteri in infantile colic and colicky induced maternal depression: a prospective single blind randomized trial. Antonie van Leeuwenhoek. 2015;107:1547–1553. doi: 10.1007/s10482-015-0448-9. [DOI] [PubMed] [Google Scholar]
- 15.Chau K, et al. Probiotics for infantile colic: a randomized, double-blind, placebo-controlled trial investigating Lactobacillus reuteri DSM 17938. J. Pediatr. 2015;166:74–78. doi: 10.1016/j.jpeds.2014.09.020. [DOI] [PubMed] [Google Scholar]
- 16.Sung V, et al. Treating infant colic with the probiotic Lactobacillus reuteri: double blind, placebo controlled randomised trial. BMJ. 2014;348:g2107. doi: 10.1136/bmj.g2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Weerth C, Fuentes S, Puylaert P, de Vos WM. Intestinal microbiota of infants with colic: development and specific signatures. Pediatrics. 2013;131:e550–558. doi: 10.1542/peds.2012-1449. [DOI] [PubMed] [Google Scholar]
- 18.Abrahamsson TR, Sinkiewicz G, Jakobsson T, Fredrikson M, Bjorksten B. Probiotic lactobacilli in breast milk and infant stool in relation to oral intake during the first year of life. J. Pediatr. Gastroenterol. Nutr. 2009;49:349–354. doi: 10.1097/MPG.0b013e31818f091b. [DOI] [PubMed] [Google Scholar]
- 19.Shornikova AV, Casas IA, Isolauri E, Mykkanen H, Vesikari T. Lactobacillus reuteri as a therapeutic agent in acute diarrhea in young children. J. Pediatr. Gastroenterol. Nutr. 1997;24:399–404. doi: 10.1097/00005176-199704000-00008. [DOI] [PubMed] [Google Scholar]
- 20.Roos S, et al. 454 pyrosequencing analysis on faecal samples from a randomized DBPC trial of colicky infants treated with Lactobacillus reuteri DSM 17938. PloS one. 2013;8:e56710. doi: 10.1371/journal.pone.0056710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Illingworth RS. Three-months’ colic. Arch. Dis. Childh., Lond. 1954;29:165–174. doi: 10.1136/adc.29.145.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fallani M, et al. Intestinal microbiota of 6-week-old infants across Europe: geographic influence beyond delivery mode, breast-feeding, and antibiotics. J. Pediatr. Gastroenterol. Nutr. 2010;51:77–84. doi: 10.1097/MPG.0b013e3181d1b11e. [DOI] [PubMed] [Google Scholar]
- 23.Prideaux L, et al. Impact of ethnicity, geography, and disease on the microbiota in health and inflammatory bowel disease. Inflamm. Bowel Dis. 2013;19:2906–2918. doi: 10.1097/01.MIB.0000435759.05577.12. [DOI] [PubMed] [Google Scholar]
- 24.Suzuki TA, Worobey M. Geographical variation of human gut microbial composition. Biol. Lett. 2014;10:20131037. doi: 10.1098/rsbl.2013.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yatsunenko T, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Willing B, et al. Twin studies reveal specific imbalances in the mucosa-associated microbiota of patients with ileal Crohn’s disease. Inflamm. Bowel Dis. 2009;15:653–660. doi: 10.1002/ibd.20783. [DOI] [PubMed] [Google Scholar]
- 27.Willing BP, et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010;139:1844–1854e1. doi: 10.1053/j.gastro.2010.08.049. [DOI] [PubMed] [Google Scholar]
- 28.Million M, et al. Obesity-associated gut microbiota is enriched in Lactobacillus reuteri and depleted in Bifidobacterium animalis and Methanobrevibacter smithii. Int. J. Obes. (Lond.) 2012;36:817–825. doi: 10.1038/ijo.2011.153. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Chern EC, Siefring S, Paar J, Doolittle M, Haugland RA. Comparison of quantitative PCR assays for Escherichia coli targeting ribosomal RNA and single copy genes. Lett. Appl. Microbiol. 2011;52:298–306. doi: 10.1111/j.1472-765X.2010.03001.x. [DOI] [PubMed] [Google Scholar]
- 30.Chern EC, Brenner KP, Wymer L, Haugland RA. Comparison of Fecal Indicator Bacteria Densities in Marine Recreational Waters by QPCR. Water Qual. Expo. Health. 2009;1:203–214. doi: 10.1007/s12403-009-0019-2. [DOI] [Google Scholar]
- 31.Sung V, et al. Probiotics to improve outcomes of colic in the community: Protocol for the Baby Biotics randomised controlled trial. BMC Pediatr. 2012;12:135. doi: 10.1186/1471-2431-12-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barr RG, Kramer MS, Pless IB, Boisjoly C, Leduc D. Feeding and temperament as determinants of early infant crying/fussing behavior. Pediatrics. 1989;84:514–521. [PubMed] [Google Scholar]
- 33.Barr RG, Kramer MS, Boisjoly C, McVey-White L, Pless IB. Parental diary of infant cry and fuss behaviour. Arch. Dis. Childh., Lond. 1988;63:380–387. doi: 10.1136/adc.63.4.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ismail IH, et al. Reduced gut microbial diversity in early life is associated with later development of eczema but not atopy in high-risk infants. Pediatr. Allergy Immunol. 2012;23:674–681. doi: 10.1111/j.1399-3038.2012.01328.x. [DOI] [PubMed] [Google Scholar]
- 35.Osborne CA, Rees GN, Bernstein Y, Janssen PH. New threshold and confidence estimates for terminal restriction fragment length polymorphism analysis of complex bacterial communities. Appl. Environ. Microbiol. 2006;72:1270–1278. doi: 10.1128/AEM.72.2.1270-1278.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.